Abstract
Spindle formation in mammalian cells requires precise spatial and temporal regulation of the kinesin-5, Eg5, which generates outward force to establish spindle bipolarity. Our results demonstrate that Eg5 is phosphorylated in cultured cells by Src family kinases (SFKs) at three sites in the motor head: Y125, Y211, and Y231. Mutation of these sites diminishes motor activity in vitro, and replacement of endogenous Eg5 with phosphomimetic Y211 in LLC-Pk1 cells results in monopolar spindles, consistent with loss of Eg5 activity. Cells treated with SFK inhibitors show defects in spindle formation, similar to those in cells expressing the non-phosphorylatable Y211 mutant, and distinct from inhibition of other mitotic kinases. We propose that this phosphoregulatory mechanism tunes Eg5 enzymatic activity for optimal spindle morphology.
Introduction
Chromosome segregation during mitosis requires the mitotic spindle, a dynamic structure composed of microtubules (MTs), motor proteins, and non-motor MT-associated proteins. All spindles are bipolar and in most cell types, spindle bipolarity relies on the activity of kinesin-5 motor proteins (Blangy et al. 1995; Hoyt et al. 1992; Goshima and Vale 2003; Enos and Morris 1990; Hagan and Yanagida 1990; Scholey et al. 2014; Kapitein et al. 2005). The bipolar arrangement of tetrameric kinesin-5 family members allows them to crosslink and slide MTs originating from each of the two centrosomes, thus establishing the bipolar spindle (Kapitein et al. 2005). Inhibition of the mammalian kinesin-5, Eg5, early in mitosis induces the formation of monopolar spindles that are incapable of proper chromosome segregation (Goshima and Vale 2003; Maliga et al. 2002).
During spindle formation, outward forces generated by kinesin-5 and other motors are opposed by motor-dependent inward forces. How these forces are balanced and regulated remains incompletely understood (Tanenbaum et al. 2008; Brust-Mascher et al. 2009; Saunders et al. 1997; Ferenz et al. 2010). Past studies have shown that phosphorylation of the kinesin-5 tail domain and interaction with binding partners such as TPX2 are important for motor localization to the spindle (Blangy et al. 1995; Ma et al. 2011; Ma et al. 2010; Blangy et al. 1997). Phosphatase and tensin homolog (PTEN) activity has also been shown to contribute to Eg5 localization (He et al. 2016). Other work demonstrated that phosphorylation of the motor domain contributes to kinesin-5 regulation in yeast and Drosophila (Avunie-Masala et al. 2011; Shapira and Gheber 2016; Garcia et al. 2009; Shapira et al. 2017); whether similar modifications affect Eg5 is not yet established.
Although Eg5 is mostly degraded as cells exit mitosis (Uzbekov et al. 1999; Venere et al. 2015), some studies show an interphase function for the motor in neurons, where it contributes to neuronal migration and growth cone behavior (Myers and Baas 2007; Nadar et al. 2008; Falnikar et al. 2011; Venere et al. 2015). Eg5 fails to undergo cell-cycle regulated degradation in patient-derived glioblastoma cells and contributes to the invasive behavior of these cells (Venere et al. 2015). These data suggest that Eg5 function is precisely regulated by a variety of mechanisms, and that dysregulation of Eg5 function can contribute to human disease.
Here we present evidence that Eg5 is phosphorylated at three sites in its motor domain by Src family kinases (SFKs) in mammalian cells. This phosphorylation modulates Eg5 activity in vitro and spindle morphology in vivo. Several SFKs, particularly those that are activated and upregulated in mitosis (c-Src, Fyn, c-Yes, and Lyn; Kuga et al. 2007) overlap in substrate and inhibitor specificity (Thomas and Brugge 1997). Therefore, in this work we refer to the SFKs as kinases collectively acting on Eg5, except when discussing experiments that specifically use c-Src. SFKs are best known for activating cell proliferation, migration, and cytoskeletal reorganization (Sen and Johnson 2011). Their dysregulation also contributes to oncogenesis (Kim et al. 2009) and recent data points to a new role for SFKs in regulating spindle establishment and orientation (Nakayama et al. 2012). Other recent work suggests that phosphotyrosine (pTyr) modification is more prevalent than previously appreciated, particularly in the kinetochore/spindle region and particularly by SFKs (Caron et al. 2016). To date, however, few mitotic SFK targets have been identified and none of them are known to regulate the MT cytoskeleton (Fumagalli et al. 1994; Bhatt et al. 2005; Wang et al. 2008). SFK phosphorylation of the Eg5 motor domain is potentially a novel regulatory mode that links SFK activity to the MT cytoskeleton during spindle establishment and may provide insight into how Eg5 becomes dysregulated in the context of cancer.
Results and Discussion
Endogenous Eg5 is phosphorylated on motor domain tyrosines in mammalian cells
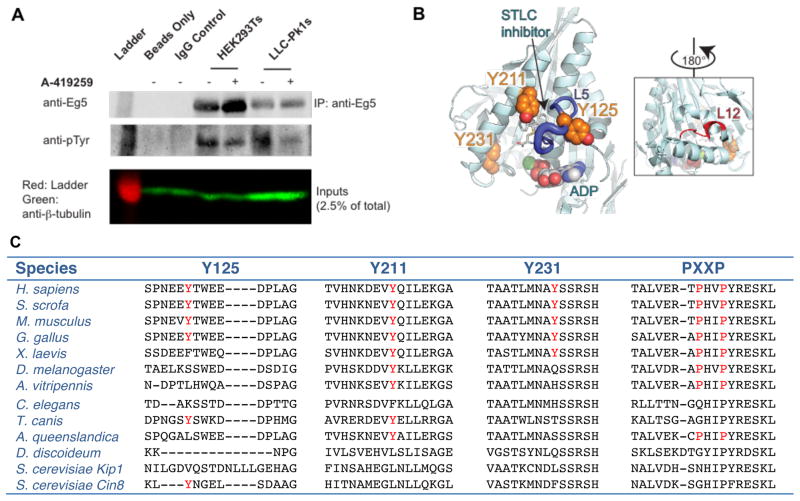
To test whether Eg5 is phosphorylated on tyrosine residues, we immunoprecipitated Eg5 from HEK293T cells and used a two-color Western blot to probe for tyrosine phosphorylation (Fig. 1A; signals from red (Eg5) and green (pTyr) channels are displayed separately). We observed co-localization of pTyr and Eg5 signals, suggesting that human Eg5 is phosphorylated on tyrosines. Additionally, we observed tyrosine phosphorylation of Eg5 immunoprecipitated from pig-derived LLC-Pk1 cells (Fig. 1A). Treatment of immunoprecipitated Eg5 with lambda phosphatase diminished pTyr signal (Fig. S1 A), confirming that the anti-pTyr antibody binds specifically to phosphorylated protein. These data and previous work showing that Eg5 is phosphorylated at multiple tyrosines in the motor domain (Fig. 1B, S1 B; Hornbeck et al. 2015; Li et al. 2009; Kim et al. 2010; Iliuk et al. 2010; Luo et al. 2008; Han et al. 2010) establish that mammalian Eg5 is phosphorylated on tyrosines.
Figure 1. Human Eg5 is phosphorylated on tyrosine residues.
A. Two-color Western blot showing endogenous Eg5 immunoprecipitated from HEK293T or LLC-Pk1 cell lysates, with each channel displayed separately in black and white. A-419259 was added as indicated; the lower panel shows a β-tubulin loading control (green). B. The structure of Eg5 bound to S-trityl-L-cysteine (STLC) is marked with tyrosines Y125, Y211, and Y231 (orange space fill, PDB: 3KEN). A predicted SH3 binding site in the MT-binding site of the Eg5 motor domain is shown in the inset. L5 is shown in dark blue; Loop 12 within the MT binding domain is shown in red. C. Sequence alignment comparing the putative phosphorylation sites and the –PXXP– SH3 targeting domain. Putative phosphorylated tyrosines and the –PXXP– SH3 targeting motifs are shown in red. The accession numbers for each protein are listed in Fig. S1 C.
Src kinase phosphorylates Eg5 more efficiently than Wee1 in vitro
Previous data reported that the mitotic kinase Wee1 phosphorylates the Drosophila kinesin-5, Klp61F, at three tyrosines in the motor head, including the tyrosine homologous to mammalian Y211 (Garcia et al. 2009). However, querying the complete Eg5 peptide sequence in the Scansite 3 kinase predictor site (Methods, (Obenauer et al. 2003) suggested SFKs as potential kinases targeting Y211. In addition, a Src homology domain 3 (SH3) targeting sequence (–PXXP–) is located in the MT binding face of several kinesin-5s, including Eg5 (Fig. 1B, inset, Fig. 1C and Fig. S1 C–D; (Kim et al. 2010). Furthermore, post-translational modification databases recorded Y125, Y211, and Y231 as phosphorylation sites in the motor domain (Fig. S1 B; Hornbeck et al. 2015; Li et al. 2009). We performed in vitro kinase assays to test whether Eg5 motor heads could be phosphorylated on these residues and to compare the ability of c-Src and Wee1 to phosphorylate Eg5 motor heads in vitro (Fig. 2A, B).
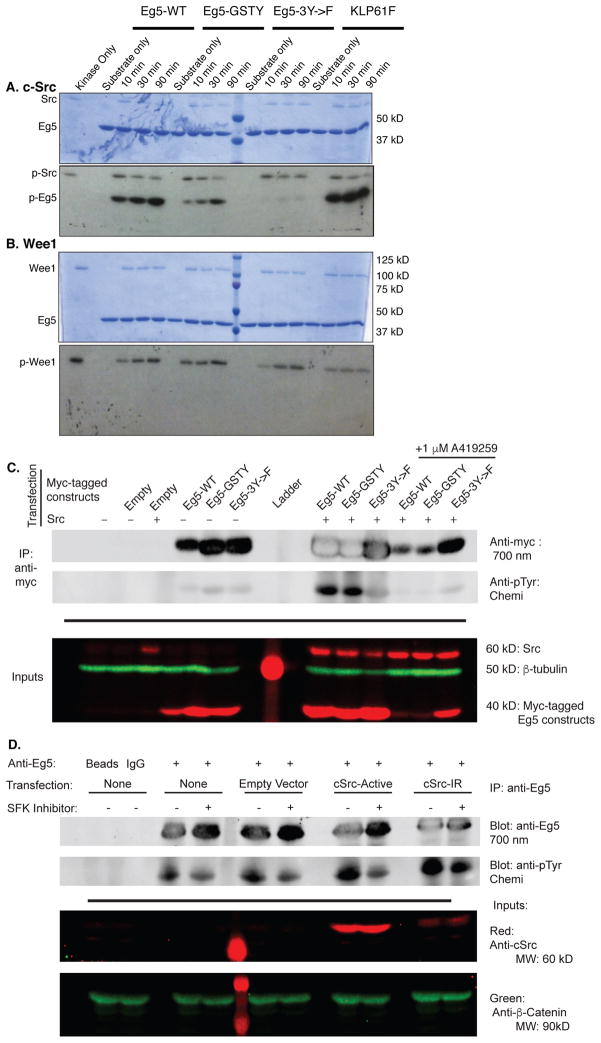
Figure 2. SFK dependent phosphorylation of Eg5 in vitro and in mammalian cells.
Kinesin-5 constructs (denoted across the top) were incubated with human c-Src (A) or human Wee1 kinase (B) and radiolabeled ATP for the times indicated. Reactions were quenched, run on an SDS-PAGE (top) that was then dried and exposed to film (bottom). Position of c-Src, Eg5, and Wee1 marked on the left side. C. Immunoprecipitation of Eg5 from HEK293T cells co-transfected with myc-tagged Eg5 motor head and c-Src constructs; A-419259 added as indicated. Western blot stained using anti-myc and anti-pTyr antibodies which here are displayed separately in black and white. The lower panel shows inputs and β-tubulin level as a loading control. D. Endogenous Eg5 was immunoprecipitated from HEK293T cells transfected with the indicated constructs; A-419259 was added as indicated. pTyr was detected using a two-color Western blot. Each channel is displayed separately in black and white. Transfection efficacy was verified by detection of c-Src in the lysates from transfected cells. β-catenin level is shown as a loading control.
For all in vitro kinase assays we used a previously well-characterized 367-amino acid monomeric Eg5 motor head construct (Eg5-367; Maliga et al. 2002; Cochran et al. 2004; Cochran and Gilbert 2005) that additionally harbored an E270A mutation in the active site (termed Eg5-367 E270A; in Klp61F-364, E266A was mutated, Fig. S1 E, F). This mutation served to abolish the basal ATPase activity of Eg5 and thus to prevent motor heads from depleting the kinase’s supply of ATP during the assay (Methods; Kull et al. 1996). We incubated either human c-Src or human Wee1 (Fig. 2A, B) with the indicated kinesin-5 substrates and radiolabeled ATP. In addition to Eg5-367 E270A and Klp61F E266A, we also tested a non-phosphorylatable Eg5 mutant with phenylalanines at the three putative sites (Eg5-367-3Y->F E270A), and a -PXXP-null mutant (Eg5-367-GSTY E270A) as kinase substrates (Methods; Fig. S1 E). c-Src robustly phosphorylated Eg5-367 E270A and Klp61F E266A motor heads under these conditions (Fig. 2A). c-Src phosphorylated the -PXXP-null construct Eg5-367-GSTY E270A in vitro, albeit markedly less efficiently than wild-type (Fig. 2A), suggesting a role for the –PXXP- targeting motif in Eg5 phosphorylation. In contrast to c-Src, Wee1 showed minimal phosphorylation of all motor head constructs, including Drosophila Klp61F E266A, despite robust autophosphorylation (Fig. 2B).
To identify the residues phosphorylated by c-Src, we performed an in vitro phosphorylation assay with purified Eg5 motor heads and c-Src kinase, and performed liquid chromatography-mass spectrometry (LC-MS) on the trypsinized protein products. The LC-MS data confirmed that c-Src phosphorylated Y211 and Y231 (Fig. S2 A). We generated a construct harboring Y211F and Y231F mutations as well as the E270A mutation and showed that it was still robustly phosphorylated by c-Src. However, an additional Y125F mutation diminished c-Src phosphorylation of Eg5 to near background levels (Fig. S2 B). Notably, c-Src showed no phosphorylation of Eg5-367-3Y->F E270A (Fig. 2A, Fig. S2 B), despite the presence of 7 other tyrosines in the motor head, confirming c-Src phosphorylates Y125. These data show that c-Src phosphorylates Eg5 on Y125, Y211, and Y231, and that this effect is aided by the presence of the SH3-targeting –PXXP- motif in the Eg5 MT binding domain.
SFKs phosphorylate Eg5 in mammalian cells
To examine the ability of c-Src to phosphorylate Eg5 motor heads on Y125, Y211, and Y231 in the cellular environment, we transfected HEK293T cells with either a constitutively active human c-Src construct (c-Src-Active, Fig. S1 F), a C-terminal myc-tagged Eg5 motor head construct (Eg5-367myc), or both. We then immunoprecipitated Eg5-367myc from the cells and analyzed the motor heads for phosphorylation by two-color Western blot (Fig. 2C, Methods). In cells co-transfected with c-Src-Active and either Eg5-367myc-WT or Eg5-367myc-GSTY, robust phosphorylation was observed. However, c-Src-Active did not phosphorylate Eg5-367myc-3Y->F. This result shows that c-Src is capable of phosphorylating Eg5 motor heads in a cellular environment and that it likely phosphorylates Eg5 on the Y125, Y211, and Y231 residues. We did not detect phosphorylation of transfected Eg5-367myc by endogenous SFKs in cells lacking a transfected c-Src-Active construct, as we did for endogenous Eg5. This apparent difference may be because the expressed motor heads lack the tail domain, which is required for localization of Eg5 to spindles (Blangy et al. 1995; Rapley et al. 2008), where a subset of SFKs is known to localize during mitosis (Ley et al. 1994; Levi et al. 2010; David-Pfeuty et al. 1993). Phosphorylation was not observed in cells co-transfected with c-Src-Active and Eg5 motor heads and treated with the SFK-specific inhibitor, A-419259 (Fig. 2C; Calderwood et al. 2002). This confirms that phosphorylation of Eg5-WT and Eg5-GSTY motor heads was due to c-Src-Active.
Finally, we used a chemical genetics approach to determine whether SFKs phosphorylated endogenous Eg5 in cells. We transfected HEK293Ts with either an empty vector control, cSrc-Active, or a constitutively active point mutant of c-Src that is resistant to A-419259 (cSrc-IR, Fig. S1 F), with or without treatment with A-419259, and measured endogenous Eg5 phosphorylation (Fig. 2D). Phosphorylation of endogenous Eg5 was detected even in the absence of transfected c-Src but was significantly enhanced following transfection of cSrc-Active. In both cases treatment with A-419259 abrogated Eg5 phosphorylation. In contrast, Eg5 from cells transfected with the resistant cSrc-IR showed robust phosphorylation regardless of whether they were treated with A-419259. These results strongly suggest that Eg5 tyrosine phosphorylation in cells is dependent on SFK activity.
In summary, these data show Eg5 is phosphorylated in an SFK-dependent manner at the same three residues both in vitro and in cells. Also, since the A-419259 inhibitor that blocked Eg5 phosphorylation in cells is relatively specific for SFKs (Calderwood et al. 2002; Wilson et al. 2002), these results give us confidence that SFKs phosphorylate Eg5.
Tyrosine phosphomimetic mutants alter Eg5 activity in vitro
Since Eg5 motor domains are phosphorylated in an SFK-dependent manner, we tested how phosphomimetic mutations at Y125, Y211, and Y231 affect Eg5 motor activity. For these experiments we generated phosphomimetic (E) and non-phosphorylatable (F) mutants of Eg5-367, as well as the -PXXP-null mutant, Eg5-367-GSTY (Fig. S1 E). We measured both the MT-stimulated ATPase rate and MT-sliding motility velocities (Methods) for each mutant and compared these rates to both wild-type monomeric Eg5 motor heads and an Eg5-367 construct lacking the eight residues 125YTWEEDPL132 from loop L5 (Eg5-367-DL5, Fig. S1 E; Maliga et al. 2002). L5 includes Y125 and lies near Y211. The Eg5-Y211E phosphomimetic mutant exhibited the greatest changes in activity, with an ATPase rate and sliding velocity that were two-fold and three-fold decreased compared to wild-type, respectively (Table I). In fact, Eg5-367-Y211E motor properties were quite similar to those of Eg5-367-DL5 (Table I), consistent with the idea that SFK-dependent phosphorylation may regulate Eg5 by directly altering its motor characteristics, although other mechanisms are possible.
Table I. Effects of phosphomimetic and non-phosphorylatable mutations on Eg5 motor characteristics and STLC binding.
Steady-state ATPase rates and MT sliding velocities of Eg5 phosphomimetic (E) and non-phosphorylatable (F) mutants were measured and compared to those of Eg5-367-WT and Eg5-367-DL5 mutants using standard in vitro assays (Methods). Dissociation constants of the L5 inhibitor STLC to Eg5-367 phosphomimetic and non-phosphorylatable mutants (KD) was calculated from ITC titrations as described in the Methods. Errors in KD were estimated based on the nonlinear least-squares fits to raw ITC data. Stoichiometries (N) show some variability reflecting protein concentration determination, but are generally consistent with single-site binding.
| Construct | Velocity (nm/s) | kcat (s−1) | K0.5(MT) (μM) | KD - STLC (nm) | N (STLC) |
|---|---|---|---|---|---|
| WT | 11.7 ± 3.4 | 7.01 ± 0.15 | 0.073 ± 0.008 | 86±21 | 1.07 |
| DL5 | 4.2 ± 1.3 | 2.09 ± 0.07 | 0.181 ± 0.023 | -- | -- |
| GSTY | 12.3 ± 3.2 | 7.60 ± 0.38 | 0.261 ± 0.047 | -- | -- |
| Y125E | 4.9 ± 1.2 | 6.47 ± 0.25 | 0.128 ± 0.022 | 540±150 | 0.95 |
| Y211E | 4.4 ± 1.2 | 2.95 ± 0.15 | 0.336 ± 0.053 | 1600±250 | 0.74 |
| Y231E | 11.0 ± 3.0 | 8.68 ± 0.28 | 0.231 ± 0.024 | 304±32 | 0.89 |
| Y125F | 12.5 ± 3.5 | 5.42 ± 0.11 | 0.063 ± 0.008 | 42±18 | 1.03 |
| Y211F | 12.5 ± 3.4 | 4.94 ± 0.07 | 0.089 ± 0.009 | 45±6.0 | 0.62 |
| Y231F | 14.9 ± 3.8 | 7.33 ± 0.25 | 0.086 ± 0.017 | 55±19 | 1.09 |
L5 is the binding site for many small molecule inhibitors of Eg5 (Maliga et al. 2002) some of which are in clinical trials for use as cancer therapy (Sarli and Giannis 2008). One could speculate based on our results that Eg5 phosphorylation may affect inhibitor efficacy, and vice-versa (Smith et al. 2015). As a preliminary test of this, we conducted isothermal calorimetry (ITC) experiments (Methods) to measure the binding affinity of the Eg5-367 phosphomimetic and non-phosphorylatable mutants for the inhibitor s-trityl-L-cysteine (STLC), which binds near L5 in human Eg5 (Skoufias et al. 2006; Kim et al. 2010). The ITC data showed that each of the phosphomimetic mutations significantly diminished STLC binding to Eg-367, and the largest effect was observed for the Y211 E mutant (Table I, Fig. S3 A). Binding of STLC to the non-phosphorylatable mutants was similar to wild-type (Table I, Figure S3 A).
Phosphomimetic mutations give only a first approximation of the effects of a uniformly phosphorylated protein sample, but the latter is nearly impossible to generate. Corroborating our results, L5 is a major conformational regulator of the Eg5 mechanochemical cycle, and several mutations and deletions in this region diminish motor activity (Waitzman et al. 2011; Behnke-Parks et al. 2011; Muretta et al. 2013; Maliga et al. 2006; Maliga and Mitchison 2006; Kaan et al. 2009). Furthermore, key structural transitions during the Eg5 mechanochemical cycle require pi-stacking and hydrophobic interactions between Y211 and residues in L5, specifically W127; L5 inhibitors bind through similar interactions (Muretta et al. 2015). By introducing a negatively-charged glutamate residue at position 211 we are most likely abolishing those interactions. It is worth noting that phosphate groups have double the negative charge and increased bulk relative to glutamate (Waksman et al. 1992). In summary, the available structural data suggest that any substantial modification in the L5 region, including phosphomimetic mutation or phosphorylation, is likely to affect Eg5 motor properties, and that phosphorylating Y211 would be at least as disruptive to the mechanochemical cycle as a glutamate phosphomimetic mutation. Based on these results and our ITC data, we would expect that phosphorylation would similarly disrupt L5 inhibitor binding.
SFK phosphorylation of Eg5 regulates spindle morphology
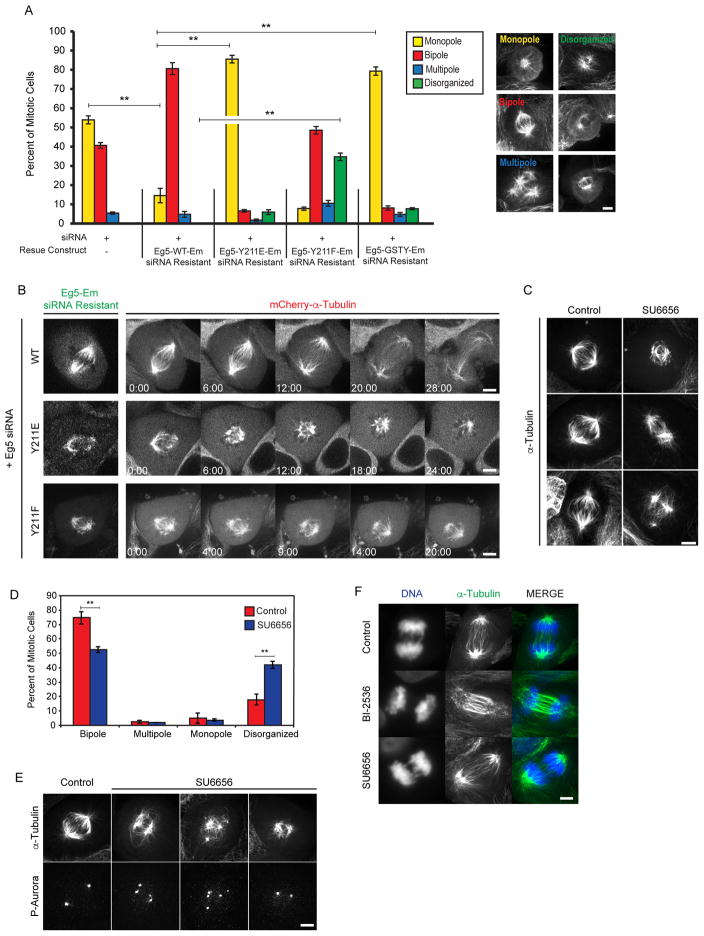
Because Eg5 plays a critical role in mitotic spindle assembly and maintenance, we next assessed the effects of SFK phosphorylation of Eg5 on mitotic spindle morphology. LLC-Pk1 cells were used for these studies because they remain relatively flat during mitosis, facilitating imaging. In initial experiments, we transfected cells with plasmids encoding Emerald-tagged Eg5 with mutations at Y125, Y211, and Y231, to generate stable cell lines for use in experiments. Despite multiple attempts, we were unable to achieve this, suggesting that these mutants have deleterious effects on cell division. Next we adapted and optimized a previously described protein replacement strategy (Zaytsev et al. 2014; Gable et al. 2012), in which we expressed Emerald-tagged Eg5 wild-type, phosphomimetic and non-phosphorylatable mutants while simultaneously inhibiting endogenous Eg5 expression using siRNA (Methods). For these experiments, we targeted Y211, which has been shown to alter mitosis in Drosophila (Garcia et al. 2009) and which resulted in the most pronounced defects in Eg5 motor behavior in vitro (Table I). Using this protocol, endogenous Eg5 protein levels decreased to approximately 50% of wild-type (Fig. S3 B) which caused cells to exhibit a large percentage of monopolar spindles (56% of cells), consistent with previous work (Goshima and Vale 2003; Ma et al. 2010; Skoufias et al. 2006; Mayer et al. 1999). Spindle bipolarity was rescued (81%) when LLC-Pk1 cells were co-transfected with Eg5 siRNA and an siRNA-resistant Eg5-WT-Emerald construct (Eg5-WT-Em, Fig. 3A). In contrast, co-transfection of cells with siRNA and siRNA-resistant phosphomimetic Eg5-Y211E-Em resulted in a significant increase in monopolar spindles as compared to the wild-type rescue construct (p < 0.01), suggesting that modification of this site inhibits Eg5 activity in mitosis (Fig. 3A). When this site was made non-phosphorylatable (Y211F) there was also a significant increase in spindle defects, specifically disorganized spindles (p<0.01). Aberrant spindles that could not be designated as monopoles or multipoles and included spindles with extra foci, fragmented poles, shorter length, and bent morphology were classified as disorganized (Fig. 3A). In addition to these phosphomimetic and non-phosphorylatable mutants, we also tested the -PXXP-null mutant (GSTY) which alters the MT binding site. As expected, since the GSTY mutation weakens MT binding by Eg5 (Table 1), there was a significant increase in monopolar spindles as compared to the wild-type rescue construct (p<0.01).
Figure 3. Mitotic spindle defects in cells expressing phosphomimetic and non-phosphorylatable mutants of Eg5.
A. Percent of mitotic phenotypes in LLC-Pk1 cells transfected with siRNA targeting endogenous Eg5 alone or co-transfected with an siRNA resistant Eg5 Emerald construct (WT, Y211E, Y211F, GSTY). Monopole (yellow), bipole (red), muitipole (blue), disorganized (green). Examples of each phenotype are shown on right. B. Time-lapse imaging of LLC-Pk1 cells expressing mCherry-α-tubulin (right panels) co-transfected with Eg5 siRNA and siRNA resistant Eg5 Emerald constructs (WT, Y211E, and Y211F left panels). C. Immunofluorescence staining for MTs in control (left) and SU6656-treated (right) parental LLC-Pk1 cells. D. Quantification of mitotic spindle phenotypes shown in C. E. Immunofluorescence staining for MTs (top) and phospho-Aurora (bottom) for control and SU6656 treated cells. (F) Immunofluorescence staining of α-tubulin in anaphase LLC-Pk1 cells: control (top), BI-2536 (middle), SU6656 (bottom). ** = p ≤ 0.01. Scale bars in A, B, C, E, F = 5 μm. Time in B (min:sec). Error Bars = St Dev.
To determine whether the monopolar spindle phenotype resulted from spindle collapse or from failure of centrosome separation we performed live cell imaging of mCherry-tubulin-expressing LLC-Pk1 cells. Cells co-transfected with siRNA targeting Eg5 and rescued with Eg5-WT-Em progressed through mitosis (Fig. 3B). In contrast, cells rescued with Eg5-Y211E-Em initially formed a bipolar spindle that eventually collapsed into a monopolar spindle. Residual endogenous Eg5 in the siRNA treated cells, or the presence of Kif15, which functions redundantly with Eg5, could support the initial bipolarization in these cells (Tanenbaum et al. 2009; Vanneste et al. 2009). Distinct from the monopolar spindles observed in cells rescued with Eg5-Y211E-Em, cells rescued with Eg5-Y211F-Em formed disorganized spindles, consistent with the disorganized phenotype observed in fixed cells (Fig. 3B).
While phosphomimetic Eg5 is an imperfect substitution for phosphorylated protein, generating cells with hyper-phosphorylated Eg5 is not trivial. Mitosis involves a complicated and inter-connected network of kinase signaling that is highly regulated (Caron et al. 2016). Simply over-expressing c-Src kinase would not guarantee that Eg5 is hyper-phosphorylated at Y211 and the interpretation of spindle phenotypes would be complicated by the effect of c-Src overactivation on other mitotic targets, potentially including other mitotic kinases. Thus, the use of phosphomimetics allows us to examine the effects in cells of introducing a negative charge at position 211 directly.
In addition to evaluating spindle phenotypes using cells expressing non-phosphorylatable and phosphomimetic mutants of Eg5, we treated non-synchronized LLC-Pk1 cells with the SFK inhibitor SU6656. Similar to our observations in LLC-Pk1 cells expressing the non-phosphorylatable Eg5-Y211F-Em, we observed that LLC-Pk1 cells treated with SU6656 (Methods) displayed high percentages of disorganized mitotic spindles (Fig. 3C, D). Consistent with this, Nakayama et al. observed mis-oriented spindles in HeLa cells treated with the SFK inhibitor PP2 (Nakayama et al. 2012). The SU6656 inhibitor we used has been reported to have some activity on other kinases that contribute to spindle formation, for example, Aurora kinases (Bain et al. 2007). To determine if treatment with SU6656 inhibits Aurora A, we stained LLC-Pk1 cells for phosphorylated Aurora A after treatment with SU6656 (Fig. 3E). Phosphorylated Aurora A was detected at spindle poles/centrosomes, similar to controls. Additionally, cells treated with BI-2536, an inhibitor of the mitotic kinase Plk1, showed a phenotype distinct from cells treated with SU6656, with pronounced bundling of interzonal microtubules in anaphase (Fig. 3F; Brennan et al. 2007). Under these conditions (Methods), we did not observe a statistically significant increase in disorganized spindles as with SU6656 or monopolar spindles as had been previously reported (Lenart et al. 2007). These results suggest that SFK inhibition alters spindle phenotypes in a manner distinct from inhibition of other mitotic kinases indicating that the phenotype of cells treated with SU6656 is not due to off-target effects.
Distinct spindle phenotypes were observed in LLC-Pk1 cells transfected with either Eg5-Y211E-Em or Eg5-Y211F-Em mutants suggesting that the optimal properties of Eg5 are tuned by phosphorylation such that abnormal mitotic phenotypes can occur when Eg5 is either hyperphosphorylated or hypophosphorylated at this site. The simple model is that Eg5 phosphorylation at Y211 alters spindle phenotypes by inhibiting its motor activity, because of the inhibitory effects seen in Table I and the monopolar phenotype in cells expressing Eg5-Y211-E-Em (Goshima and Vale 2003; Mayer et al. 1999; Ma et al. 2010; Skoufias et al. 2006). Furthermore, the largely monopolar spindle phenotype of cells expressing the phosphomimetic Y211E mutant is consistent with the decrease in Eg5 motor activity that is observed when cells are treated with L5 inhibitors, which are thought to act by a similar mechanism (Maliga et al. 2002; Muretta et al. 2015). Finally, it is worth noting that in many systems, Eg5 plays an important role in centrosome separation (Tanenbaum et al. 2008; Whalley et al. 2015; van Ree et al. 2016) and the monopolar phenotype observed in cells expressing Eg5-Y211E-Em could be due to abnormal Eg5 activity during this earlier phase of mitosis.
It is less simple to speculate about how overactive Eg5 would cause the multipolar/disorganized spindle phenotype observed in the Eg5-Y211F-Em transfected LLC-Pk1 cells. One possibility is that excessive force from Eg5 in the spindle midzone could lead to disorganized spindles. A second possibility is that phosphorylation of Eg5 Y211 could also modulate Eg5 localization, protein turnover rates, or its ability to bind MTs, as was observed for the yeast kinesin 5, Cin8p (Shapira and Gheber 2016). Regardless of how the multipolar/disorganized spindle phenotype arises, its physiological relevance is reinforced by its similarity to the spindle phenotype observed in cells in which SFKs are inhibited, which has both been observed by other groups and is distinct from the phenotypes observed when other mitotic kinases are inhibited (Nakayama et al. 2012; Brennan et al. 2007; Bain et al. 2007).
Given that endogenous Eg5 is homotetrameric (Kapitein et al. 2005; van den Wildenberg et al. 2008; Scholey et al. 2014), it is likely that not all of the Eg5 motor heads in a homotetramer are phosphorylated. Eg5 motor heads are highly cooperative when assembled into dimers (Krzysiak and Gilbert 2006; Krzysiak et al. 2008), with dimers having distinct kinetic properties from monomers (Krzysiak and Gilbert 2006; Cochran et al. 2006). Additionally, recent structural studies of the Eg5 coiled-coil domain responsible for the assembly of Eg5 into homotetramers suggests that instead of being a dimer of dimers, each subunit in an Eg5 homotetramer directly contacts every other subunit in a highly intertwined and unique coiled-coil structure (Scholey et al. 2014). These data suggest that phosphorylation of even one motor head within the Eg5 homotetramer could alter the function of the molecule. There is also a substantial body of evidence that mitotic spindle establishment and maintenance involves a balance of forces (Tanenbaum et al. 2008; Saunders et al. 1997; Brust-Mascher et al. 2009), making it feasible that even small changes to Eg5 motor activity could disrupt this balance. In support of this possibility, we observed that endogenous Eg5 was only reduced to 50% of wild-type levels in our experiments using LLC-Pk1 cells, so one can imagine that many Eg5 homotetramers in our experiments had both mutant and wild-type subunits. Despite this, nearly 90% of cells in those experiments had monopolar spindles (Fig. 3A). The severity of this defect supports the view that either not all of the motors in a heterotetrameric motor are simultaneously modified, or that Eg5 undergoes a cycle of phosphorylation and dephosphorylation in vivo.
In summary, these experiments revealed a significant mitotic phenotype in LLC-Pk1 cells expressing Eg5 with phosphomimetic and non-phosphorylatable mutations at Y211, the same site that was shown to impact spindle assembly in Drosophila (Garcia et al. 2009). Y211 is a particularly interesting site because it is conserved in both insects and vertebrates, coinciding almost without exception with the presence of a –PXXP– SH3-targeting domain (Fig. 1B, Fig. S1 D). Conversely, neither Y211 nor the -PXXP– motif is found in worms, which are viable with diminished levels of kinesin-5, suggesting that this organism has evolved alternative pathways for establishing bipolar spindles (Bishop et al. 2005).
Motor domain phosphorylation has also been described for the yeast kinesin-5, Cin8p (Shapira et al. 2017; Shapira and Gheber 2016; Avunie-Masala et al. 2011). Only one of these sites, S337 (H. sapiens numbering) is in a region of the motor that is conserved in Eg5. Phosphorylation of each of these sites has unique effects on motor behavior including Cin8p microtubule binding, motor directionality, and velocity (Shapira and Gheber 2016; Shapira et al. 2017). Although the precise locations of these modifications are not conserved from yeast to humans, one notable similarity amongst these modifications is that the changes to Cin8p motor behavior are primarily mediated by electrostatic interactions, which we find to be a compelling hypothesis for the effects of Eg5 Y211 phosphorylation given the available structural data. Future experiments examining phosphorylation of kinesin-5 motors could illuminate the extent of their modifications and could reveal a potential mechanism by which kinesin-5s are differentially regulated to play similar, but non-identical, roles in varying cell types and species.
Our results support a growing body of data identifying phosphoregulatory mechanisms governing the activity of several different kinesin motors (see, for example, Garcia et al. 2009; Chee and Haase 2010; DeBerg et al. 2013). Previously identified Eg5 phosphoregulatory mechanisms target serine or threonine residues in the motor stalk and tail, and have been reported to affect Eg5 localization to the spindle or centrosome during mitosis (Blangy et al. 1995; Rapley et al. 2008). Our results showing phosphorylation of Eg5 in its motor domain at Y125, Y211, and Y231 suggest that in addition to altering motor localization, phosphoregulatory mechanisms can tune Eg5 enzymatic activity for optimal spindle morphology (Garcia et al. 2009; Avunie-Masala et al. 2011). Furthermore, our data suggest this post-translational modification could affect the efficacy of small molecule inhibitors that bind to L5, although further study is required to gauge whether this has any practical implications for use of Eg5 inhibitors as cancer therapy.
Materials and Methods
In silico prediction of phosphorylated residues in Eg5 and targeting kinases
There are several databases summarizing the results of large-scale proteomics experiments that provide evidence for the post-translational modification of specific residues in thousands of proteins. We searched PhosphositePlus (Hornbeck et al. 2015) and SysPTM (Li et al. 2009) for modifications entered for human Eg5 and narrowed the list of modifications down to tyrosine phosphorylations. We also did a manual search of PubMed articles for entries presenting phosphoproteomics experiments that included human Eg5 in their results. These searches generated a list of putative tyrosine phosphorylation sites in Eg5 (summarized in Fig. S1 B).
To generate hypotheses regarding possible kinases targeting human Eg5, we entered its full sequence as found in the UniProt database (accession number P52732, UniProt 2015) into the search engine found in the Scansite3 kinase predictor site (Obenauer et al. 2003). We used the “medium stringency” setting, which returns kinases for which the queried protein sequence is in the top percentile of sequences in the vertebrate subset of SWISS-PROT matching the optimal targeting motif (Obenauer et al. 2003). This search revealed three SFKs as possible kinases targeting the Y211 location. It also revealed a –PXXP– SH3 targeting site.
Cloning
Mutagenesis of the Eg5-367 monomer construct has been described previously (Larson et al. 2010). Briefly, Eg5-367 constructs for bacterial expression include the first 367 amino acids of H. sapiens Eg5 immediately followed by a C-terminal 6X-histidine tag in a pRSET plasmid. KLP61F-364 constructs include the first 364 amino acids of the Drosophila kinesin-5, KLP61F similarly followed by a 6X-histidine tag. Phosphomimetic (Y->E) and non-phosphorylatable (Y->F) point mutations were made using Quikchange site-directed mutagenesis kit (Agilent Technologies, Santa Clara, CA), as were enzymatically inactive mutations (E270A in Eg5-367 and E266A in KLP61F). To generate an Eg5-367 mutant lacking the -PXXP- SH3-targeting motif in its MT-binding domain, site-directed mutagenesis was used to replace residues 305RTPH308 with the homologous residues in H. sapiens Kinesin-1 heavy chain (GSTY). This removes the initial proline from the SH3 targeting motif. The resulting motor can still hydrolyze ATP and bind MTs, albeit at reduced affinity (Table I).
For expression in mammalian cells, we replaced the C-terminal 6X-histidine tag of Eg5-367 constructs with a 10-residue Myc tag (EQKLISEEDL). Myc-tagged Eg5-367 constructs were then cloned into the pcDNA3 vector (gift of Dr. Cara Gottardi, Northwestern University) between the Xho1 and HindIII restriction sites using Phusion polymerase (New England Biolabs, Ipswich, MA). All constructs were verified by sequencing. A mammalian c-Src construct in the pCMV-SPORT6 mammalian expression plasmid was a kind gift from Dr. Thomas Smithgall, University of Pittsburgh. This construct was mutated using Quikchange site-directed mutagenesis to generate a constitutively active c-Src construct (Y527F). This constitutively active construct was then further mutated to render it resistant to treatment with A419259 (T338M, Meyn and Smithgall 2009). All c-Src construct numbering refers to the structure of human c-Src (PDB: 1FMK).
The Eg5-Emerald wild-type (Eg5-WT-Em) construct consisted of full-length human kinesin 11 fused to pmEmerald with an 18 amino acid linker; expression is under the control of a pCMV promoter. This construct was used to express fluorescent Eg5 in LLC-Pk1 cells, was a gift from the late Dr. Michael Davidson Florida State University and was made siRNA resistant using PCR site-directed mutagenesis (Forward primer: GTCACAAAAGCAATGTGGAAACCTAACTGAAGATCTCAAGACTATAAAGCAGACCC; reverse primer: CAAAGTTCCTGGGAATGGGTCTGCTTTATAGTCTTGAGATCTTCAGTTAGGTTTCC) and verified by sequencing. Each mutant was then made in this backbone using PCR site-directed mutagenesis and verified by sequencing.
Protein Expression and Purification
Expression and purification of Eg5-367 constructs has been described previously (Larson et al. 2010). Briefly, 6x-His-tagged Eg5-367 and KLP61F-364 constructs were transformed into E. coli BL21-CodonPlus(DE3)-RP cells for expression. Cells were grown in TPM media (2% tryptone, 1.5% yeast extract, 137 mM NaCl, 14 m2HPO4) with in 50 μg/mL carbenicillin and 34 μg/mL chloramphenicol at 37 ° C while shaking at 200 rpm until cells reached an OD600 between 0.6 and 1.0. Plasmid expression was induced through the addition of 0.125 mM IPTG. Cells were allowed to express at 18 ° C overnight. 2 L cultures were then harvested by centrifugation (6,300 rpm for 10 min at 4 ° C) and re-suspended in 20 mL Eg5 lysis buffer (10 mM HEPES, 2 mM MgCl2, 1 mM EGTA, 5% sucrose, 0.02% polyoxyethylenesorbitan monolaurate (TWEEN-20), 10 μM ATP, leupeptin (1 μg/mL), aprotinin (1 μg/mL), pepstatin (1 μg/mL), and 100 μM PMSF, pH 8). Cells were lysed by sonication and the clarified lysate was batch-bound with pre-equlibrated nickel-nitrilotriacetic acid resin (Qiagen, Valencia, CA) for 2 h at 4 ° C. The resin was washed with nickel wash buffer (10 mM HEPES, 2 mM MgCl2, 1 mM EGTA, 5% sucrose, 0.02% TWEEN-20, 10 μM ATP, 300 mM NaCl, and 20 mM imidazole, pH 6) and bound protein eluted in 5 mLs using nickel elution buffer (10 mM HEPES, 2 mM MgCl2, 1 mM EGTA, 5% sucrose, 0.02% TWEEN-20, 10 μM ATP, 300 mM NaCl, and 400 mM imidazole, pH 6). Peak fractions were collected and diluted 20 fold in Buffer A (10 mM HEPES, 2 mM MgCl2, 1 mM EGTA, 5% sucrose, 0.02% TWEEN-20, 10 μM ATP, 1 mM DTT pH 6) to decrease the ionic strength of the buffer. Diluted fractions were then purified further on a 5 mL HiTrap S-Sepharose cation exchange column (GE Healthcare, Little Chalfont, UK). Protein was eluted using a linear 0.05–1 M NaCl gradient. Peak fractions were identified by SDS-PAGE and pooled. After adding an additional 15% sucrose, protein was flash-frozen in liquid nitrogen and stored at −80 ° C until use.
In vitro kinase assay
E. coli-purified Eg5-367 E270A and KLP61F-364 E266A proteins were first dialyzed against Src kinase assay buffer (10 mM HEPES pH 6.8, 2 mM MgCl2, 1 mM EGTA, 2.0 mM DTT, 10% glycerol, 0.02% TWEEN-20). Each reaction contained 3 μM Eg5 protein, [γ32P]-ATP (Perkin-Elmer) to 50 nCi acitivity per reaction, and 200 μM ATP. Src kinase (Invitrogen, Carlsbad, CA) was diluted to a concentration of 2 μM in assay buffer with 0.2 mg/mL BSA (Sigma-Aldrich, St. Louis, MO), and 2 μL of this solution was added to each 20 μL reaction. Reactions were incubated at 30 ° C for the indicated reaction times, quenched with SDS sample buffer and run on an SDS-PAGE gels using the formulation of Laemmli (Laemmli 1970). Gels were dried and exposed to film for 30 min.
Wee1 kinase assays were performed identically except for the buffer used. A Wee1 kinase assay buffer based on conditions cited by Garcia et al. (Garcia et al. 2009) was used instead (50 mM HEPES ph 6.8, 15 mM MgCl2, 1 mM EGTA, 2 mM DTT, 10% glycerol).
Mass Spectrometry
Samples for MS were prepared in an identical manner to those for in vitro kinase assays, but without [γ32P]-ATP. All reactions were incubated for 5 min at 30 ° C before quenching. Samples were trypsinized in-gel and analyzed via LC-MS. Briefly, each sample (6.4 μL) was injected onto a 2.5 cm, 100-μm i.d. C-18 trap column and washed at 5 μL/min with buffer A (95% H2O, 5% acetonitrile, 0.2% formic acid) using a Dionex UltiMate 3000 nanoLC (Thermo Scientific Dionex). A 10 cm, 100 μm-i.d. C-18 column was used for tryptic peptide separation. The gradient was delivered at 300 nL/min starting at 5% B (95% acetonitrile, 5% H2O, and 0.2% formic acid) and rose to 10% at 20 min, 60% B at 90 min, and 90% B at 100 min. The column was returned to 5% B over 5 min and re-equilibrated for 10 min.
Data were collected with a Thermo Fisher Velos Pro Orbitrap using a custom-built nanospray ionization source operating at a spray voltage of 1.9kV. MS1 data were collected over a scan range of m/z 400–2000 using the FT Orbitrap (1 microscan, 120 000 resolving power at m/z 400). MS2 data were collected in the ion trap (3 microscan) and fragmentation was achieved using collision induced dissociation (CID) set at 35. A data-dependent acquisition mode was used to select the top 9 peaks for fragmentation. Phosphopeptides were detected as variable modifications using Mascot (Matrix Science, Inc) and confirmed manually. The data were visualized using Scaffold (Proteome Software, Inc., Portland, OR). These experiments were done with the help of the Northwestern Proteomics Core.
Coupled-enzyme ATPase assay
MTs were purified from porcine brains according to published protocol (Mitchison and Kirschner 1984). A subset were labeled with tetramethylrhodamine (as described in Hyman et al. 1991). For use in coupled-enzyme ATPase assays, MTs were prepared exactly as described in (Woehlke et al. 1997). ATPase assays were performed as described previously, with 10–60 nM Eg5 protein (Woehlke et al. 1997; Huang and Hackney 1994). Briefly, reactions were conducted in ATPase assay buffer (10 mM HEPES, 2 mM MgCl2, 1 mM EGTA, 5% sucrose, 50 mM KCl, 500 nM ATP). Eg5-367 protein and MTs were incubated with a coupled NADH oxidation system (0.3 μM phosphoenolpyruvate, 0.5 μM NADH, pyruvate kinase (11 U/mL), and lactate dehydrogenase (10 U/mL)). We calculated the decrease in absorbance at 340 nm over time to determine the ATPase rate. ATPase rates were determined at MT concentrations from 60 nM - 4 μM, and data were fit to a Michaelis-Menten shown below (R2 > 0.8) with kcat and K0.5, MTs as the only two fit parameters using KaleidaGraph software (Synergy Software, Reading, PA). Errors shown are errors in fit parameters.
Motility assay
For use in motility assays, a mixture of tetramethylrhodamine-labeled tubulin (Cytoskeleton, Inc., Denver, CO) and unlabeled tubulin was combined 1:1 with a 2X polymerization mix (80 mM Pipes buffer, 1 mM MgCl2, 1 mM EGTA, 2 mM GTP, 20% DMSO, pH 6.8) and incubated at 37 ° C for 45 min. Paclitaxel (50 μM) was then added to stabilize MTs. For motility assays flow chambers were created using glass coverslips, microscopy slides, and double-sided tape. Anti-His H8 antibody (ab18184, Abcam, Cambridge, MA) in motility buffer (80 mM PIPES, pH 6.8, 2 mM MgCl2, 1 mM EGTA, 0.2 mg/mL BSA, 150 mM sucrose and 1 mM ATP) was incubated in the flow chamber for two minutes. The flow chamber was then washed three times with motility buffer. Next, the flow chamber was incubated with motility buffer containing Eg5-367 proteins for two minutes, before being washed three times with motility buffer. Finally, motility buffer containing an oxygen scavenging system (glucose oxidase (0.432 mg/mL), catalase (0.072 mg/mL), glucose (45 mM), and β-mercaptoethanol (61 mM)), an ATP regenerating system (2 mM creatine phosphate and 810 U/mL creatine phosphate), and tetramethlyrhodamine-labeled polymerized MTs stabilized with GTP (1 mM) and paclitaxel (100 mM) was flowed into the cell, which was then sealed with vacuum grease. MT sliding was visualized on a Nikon TE-2000 E microscope fitted with a X60 objective (N. A. 1.4) using epifluorescence. Images were captured using a Photometrics CoolSnap EZ camera (1392 × 1040 imaging pixels, 6.45 × 6.45 μm/pixel) and Metamorph software. The concentration of MTs and Eg5-367 construct was adjusted to promote sliding populations suitable for tracking and quantification. For Eg5-367, Eg5-367 non-phosphorylatable, and Eg5-367 GSTY mutants, movies were 30 minutes long with a 20 second interval between frames. For Eg5-367 DL5 and Eg5-367 phosphomimetic mutants, movies were one hour long with a 40 second interval between frames to accommodate slower sliding velocities while minimizing photobleaching of MTs. We reported the mean non-zero sliding velocity calculated in the following manner. We tracked the ends of individual fluorescent MTs using the ImageJ plug-in MTrackJ. This plug-in calculates a step velocity based on the difference in location between consecutive frames in a movie. For each movie we tracked 3–7 MTs for a total of ≥ 190 step velocity measurements per slide. The non-zero mean velocity and standard deviation of these measurements for each individual slide using Microsoft Excel. For each Eg5-367 we calculated the weighted average and standard deviation for all step velocity measurements from three different movies to generate the final average velocity and standard deviation reported in Table 1.
Isothermal Titration Calorimetry (ITC)
ITC was performed on an iTC200 instrument (MicroCal, Northampton, MA) at 22°C with both binding partners in buffer containing 10 mM HEPES, pH 6.8, 2 mM MgCl2, 1 mM EGTA, 300 mM NaCl, 150 mM sucrose and 50 μM ADP, with 1.5 % DMSO added (v/v).
In a typical experiment, 150 μM STLC was injected into 10–20 μM Eg5 protein in 2 μL aliquots. For low affinity mutants (Y125E and Y211E), higher STLC and Eg5 concentrations and 1 μL injections were used. The stoichiometry (N), enthalpy (ΔH), entropy (ΔS) and association constant (Ka, presented here as kd=1/Ka) were all obtained by fitting data to a nonlinear, least-squares routine in a single-site binding module in Origin 7.0 software (Microcal, Northampton, MA).
Transient Transfection and Nucleofection of Mammalian Cell Lines
HEK293T cells, the kind gift from Dr. Cara Gottardi, Northwestern University, were cultured in 100 mm dishes containing DMEM medium (Corning Life Sciences, Tewksbury, MA) supplemented with 10% fetal bovine serum (FBS, Atlanta Biologicals, Flowery Branch, GA) and penicillin/streptomycin (200 U/mL, Life Technologies, Carlsbad, CA) in 5% carbon dioxide (CO2) at 37 ° C. LLC-Pk1 cells were cultured in 1:1 Ham’s F-10 medium and Opti-MEM (Life Technologies, Carlsbad, CA) supplemented with 7.5% FBS and 1X antibiotic/antimycotic solution (final concentrations 100 U/mL penicillin, 0.1 mg/mL streptomycin, 0.25 μg/mL amphotericin B; Sigma-Aldrich, St. Louis, MO) at 37 ° C and 5% CO2.
To transfect HEK293T cells, 100 mm plates at 70–80% confluence were transfected with 2 μg of DNA using the Effectene transfection reagent (Qiagen, Redwood City, CA), allowed to express for 24 h.
LLC-Pk1 cells (parental or expressing mCherry- αTubulin) were transfected using an Amaxa nucleofector (Lonza, Portsmouth, NH) using program X-001 and Mirus nucleofection reagent (Mirus Bio LLC, Madison, WI) according to the manufacturers recommendations. siRNA used to target endogenous Eg5 (CUGAAGACCUGAAGACAAU) was obtained from Dharmacon (GE Healthcare Life Sciences, Pittsburgh, PA). Following nucleofection, cells were plated on #1.5 coverslips or Mattek glass bottom dishes (Mattek Corporation, Ashland, MA). Cells were used at 72 hours following nucleofection.
Inhibitors
For treatment of cells with the SFK inhibitor A-419259 (Sigma Aldrich, St. Louis, MO), the protocol developed and verified by the Smithgall lab was followed (Meyn and Smithgall 2009). Specifically, A-419259 was dissolved in water (100 μM stock solution), aliquoted, and stored at −20 ° C. Twenty-four hours prior to harvest, cell culture media was aspirated from plates and carefully replaced with warmed media containing A-419259 (1 μM final concentration). Cells incubated at 37 ° C and 5% CO2 until harvest. In cases where cells were both transiently transfected and treated with A-419259, the SFK inhibitor was added to cell media during the transfection procedure.
Stock solutions of SU6656 and BI-2536 were prepared in DMSO, stored at −20°C and diluted into culture medium before use. SU6656 was used at a final concentration of 500 nM and BI-2536 was used at 2 μM. Each was incubated on cells for 15–30 min prior to fixation.
Immunoprecipitation and 2-color Western Blot
Before harvesting, mammalian cells were first treated with pervanadate to inhibit phosphatase activity. Briefly, hydrogen peroxide (1.7%) was added to PBS containing 5 mM sodium orthovanadate to convert it to pervanadate, after which exposure to light was limited. Next, 0.5 mL of this solution was added to warmed DMEM to generate a final media concentration of 0.25 mM pervanadate. Existing media was then aspirated off each plate of cells and gently replaced with DMEM containing pervanadate. Cells were then incubated at 37 ° C and 5% CO2 for 30 minutes. Next, cells were dislodged from the plate with a cell scraper and pelleted at 1000 x g for 5 minutes. The media was then aspirated off and pellets were re-suspended in PBS containing calcium (0.9 mM) and magnesium (0.49 mM) and washed 3 times. Finally, cells were re-suspended in 1% Triton lysis buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 5 mM EDTA, 5% glycerol, 1% Triton X-100, plus COmplete protease and PhosStop phosphatase inhibitor tablets (Roche, Mannheim, Germany) and incubated on ice for one hour. Total protein concentration of lysate samples was determined using a standard Bradford assay. All samples were normalized to the same protein concentration.
To immunoprecipitate endogenous Eg5, 5 μL of the polyclonal rabbit anti-Eg5 antibody NB500-181 (Novus Biologicals, Littleton, CO) was added to 2.0 mg total protein lysate in the case of HEK293T lysates or 2.5 mg total protein lysate in the case of LLC-Pk1 lysates and incubated while rotating for 2 h at 4 ° C. To create IgG isotype controls, 1 μL of rabbit IgG (EMD Millipore, Billerica, MA) was added to 2.0 mg of total protein lysate and incubated similarly. Then, 60 μL of pre-equilibrated 50% Pierce Protein A agarose resin slurry (ThermoFisher Scientific, Rockford, IL) was added to each sample, which were then incubated while rotating for another 2 h at 4 ° C. To immunoprecipitate transfected myc-tagged Eg5-367 constructs, 25 μL of pre-equilibrated goat anti-myc beads, epitope EQKLISEEDL (Bethyl, Montgomery, TX), were tumbled with 2.0 mg of cell lysate for 3 h at 4 ° C. After 3 washes with 1% Triton lysis buffer and one wash with 0.1% Triton lysis buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 5 mM EDTA, 5% glycerol, 0.1% Triton X-100, plus COmplete protease and PhosStop (Roche, Mannheim, Germany) inhibitor tablets), beads were re-suspended in 30 μL of 2X SDS-PAGE sample buffer (200 mM Tris-Cl, 0. 130 mM SDS, 33 mM DTT, 3 mM bromophenol blue, 20% glycerol, pH 6.8), boiled for 20 minutes, and run on a 6% SDS-PAGE gel overnight. For loading controls and verification of transfection, prior to immunoprecipitation we retained 2.5% of each 2.0 mg total protein lysate sample, to which we added an equal volume of hot 6X SDS-PAGE sample buffer (300 mM Tris-Cl, 0.4 M SDS, 0.1 M DTT, 9 mM bromophenol blue, and 60% glycerol). Each input sample was then boiled for 20 min and run on a 9% SDS-PAGE gel overnight.
After transfer to nitrocellulose (0.45 μm, Thermo Fisher Scientific, Waltham, MA), and blocking with 5% non-fat dehydrated milk in PBS at room temperature for one hour and washing them three times with TBS containing 0.1% TWEEN-20 (TBS-T), we probed with primary antibodies in 5% BSA in TBS while rocking at 4 ° C overnight. For detection of endogenous Eg5 we used 1:5000 polyclonal rabbit anti-Eg5 NB500-181 antibody (Novus Biologicals, Littleton, CO). For detection of myc-tagged Eg5 constructs we used 1:5000 rabbit anti-myc ab9106 antibody (Abcam, Cambridge, MA). To probe for pTyr we used 1:200 mouse anti-pTyr PY20 antibody (sc-508, Santa Cruz Biotechnology, Dallas, TX). To detect β-catenin and β-tubulin as loading controls, we used 1:250 mouse anti-β-catenin BD160154 antibody (BD Biosciences, San Jose, CA) and 1:5000 mouse anti-β-tubulin AA2 antibody (Sigma-Aldrich, St. Louis, MO), respectively. To probe for c-Src, we used 1:200 rabbit anti-c-Src SRC2 antibody (sc-18, Santa Cruz, Dallas, TX). After briefly washing blots three times with TBS containing 0.1% TWEEN-20, we incubated them in secondary antibodies diluted in 5% non-fat dehydrated milk in TBS at room temperature for one hour while rocking. For detection of all rabbit antibodies, we used a 1:5000 dilution of the donkey anti-rabbit IRDye 680RD fluorescent antibody (LI-COR, Lincoln, NE). For detection of all mouse antibodies, except for the anti-pTyr PY20 antibody, we used a 1:5000 dilution of the donkey anti-mouse IRDye 800CW (LI-COR, Lincoln, NE). The pTyr antibody signal was below the minimum detection limit of fluorescent secondary antibodies. Instead, we used a 1:5000 dilution of a goat anti-mouse IgG-HRP conjugate secondary antibody (Bio-Rad, Hercules, CA) for detection by chemiluminescence, which amplifies the signal. After incubation in secondary antibodies, blots were washed three times with TBS-T. Blots exposed only to fluorescent secondary antibodies were then dried for 20 min sandwiched between paper towels in a drawer. Blots exposed to the HRP-conjugate secondary antibody were instead developed for 20 min using the Pierce ECL2 Western Blotting substrate (Thermo Fisher Scientific, Rockford, IL) and imaged while wet. All blots were imaged on a LI-COR Odyssey Fc imaging system (Lincoln, NE). Fluorescent antibody exposure times were 2 min; chemiluminescent detection occurred over 10 min. Entire images were contrast-adjusted using LI-COR Image Studio software without altering gamma settings before being exported to Adobe Illustrator for preparation for publication.
Phosphatase Assay
To verify that signal from the anti-pTyr antibody was specific to phosphorylated protein and not due to non-specific binding, we submitted our immunoprecipitated endogenous Eg5 to a lambda protein phosphatase assay according to the commercial protocol that came with the lambda protein phosphatase used (New England Biolabs, Ipswich, MA). Briefly, we immunoprecipitated Eg5 from 4.0 mg of HEK293T cell lysate by doubling reagents in the preceding protocol. After incubation with Pierce Protein A agarose resin (Thermo Fisher Scientific, Rockford, IL), we washed the beads 3 times with 1% Triton lysis buffer lacking protease and phosphatase inhibitor tablets, and once with 0.1% Triton lysis buffer lacking protease and phosphatase inhibitors. After the final wash the beads were re-suspended in an equal volume of 0.1% Triton lysis buffer lacking inhibitors, divided into two separate and equal samples. Both samples were spun down briefly in a tabletop microcentrifuge and the supernatants removed by aspiration. The resin in one sample was re-suspended in lambda protein phosphatase assay buffer (50 mM Hepes, 100 nM NaCl, 2 mM DTT, 0.01% Brij 35, 1 mM MnCl2) containing lambda protein phosphatase (8000 U/mL). The resin in the other sample was re-suspended in lambda protein phosphatase buffer without the enzyme as a negative control. Both samples were incubated at 30 ° C for an hour while agitating. Lambda protein phosphatase reactions were quenched by the addition of 50 μL of 2X SDS-PAGE sample buffer, boiled for 20 min, and run on an SDS-PAGE gel overnight. They were blotted for endogenous Eg5 and pTyr as described above.
Mammalian Cell Fixation and Immunofluorescence
LLC-Pk1 cells were rinsed twice with room temperature PBS lacking calcium and magnesium and were then fixed for 10 minutes in 2% paraformaldehyde, 0.25% glutaraldehyde, and 0.5% Triton X 100, made fresh daily in PBS lacking calcium and magnesium. Fixed cells were rinsed in PBS containing 0.02% TWEEN-20 and 0.02% sodium azide (PBS-Tw-Az), treated with sodium borohydride (10mg/10mL H2O) for 10 minutes and then rehydrated in PBS-Tw-Az. The following antibodies were used: Phospho Aurora-A/B/C (Cell Signaling Technology, Danvers, MA); tubulin, DM1α mouse anti-tubulin (Sigma-Aldrich) or YL1/2 rat anti-tubulin (Accurate Chemical and Scientific Corporation, Westbury, NY) and appropriate secondary antibodies as previously described (Ma et al., 2011). Primary antibodies were mixed with 2% BSA in PBS-Tw-Az to block non-specific binding and used at the following final dilutions: Phosoho Aurora-A/B/C 1:1,000, DM1α and YL1/2 1:100; cells were incubated in primary antibodies for 1 hour at 37°C. Stained cells were mounted on glass slides using DAPI Fluomount G (Southern Biotech, Birmingham, AL) to stain DNA.
Immunofluorescence Microscopy/Imaging
To quantify mitotic phenotypes of fixed cells, a Nikon Eclipse Ti with an X-Cite series 120Q excitation light source, and a 100 X, 1.3 N.A., objective lens, was used. Images of fixed cells were acquired using a CSU-10 Yokogawa spinning-disk confocal scan head on a Nikon TE300 as previously described (Tulu et al., 2003). Transfected cells (identified by the Eg5-Emerald signal) were classified by spindle morphology based on microtubule staining as bipole, multipole, monopole, or disorganized. Disorganized spindles included spindles with extra foci, fragmented poles, short spindles, no pole, and bent. For live cell imaging, a Nikon Ti-E microscope with a CSU-X1 Yokogawa spinning-disk confocal scan head (PerkinElmer, Wellesley, MA), an Andor iXon+ electron-multiplying charge-coupled device camera (Andor), and a 100×/1.4 NA objective lens was used. For live-cell imaging, exposures were adjusted without saturating the camera’s pixels; typical exposures were 50–800 ms.
Supplementary Material
Figure S1 Eg5 is phosphorylated on a conserved tyrosine in its motor head A, A single immunoprecipitation reaction from HEK293Ts (3.5 mg total lysate) was divided in two. Half was incubated with l-phosphatase (right); the other was incubated with phosphatase buffer alone (left). B, Table summarizing Eg5 tyrosine phosphorylations as reported in databases collecting results from phosphoproteomics experiments, including PhosphositePlus (Hornbeck et al., 2015). C, Table showing the UniProt Accession numbers used to generate the alignment of Kinesin-5 family members in Figure 1C. D, Alignment of kinesins from families 1–7, highlighting the MT binding region. The –PXXP- SH3 targeting motif, when present, is highlighted in red. Residues that are identical across the alignment are marked in dark grey, while similar residues are marked in light grey. While there is extensive conservation of sequence in this region across kinesin family members, the PXXP motif is only seen in Kinesin-5s of higher order organisms. E List of the sequence mutations made in Eg5 constructs and F, other constructs for experiments using Eg5 mutants presented in the article.
Figure S2 Eg5 motor heads are phosphorylated on Y125, Y211, and Y231 in vitro Purified Eg5 motor heads and purified Src kinase were incubated with ATP (radiolabeled ATP in some cases) for 5 min. Reactions were quenched and then analyzed either A, by LC-MS or B, by radiograph.
Figure S3 Eg5 phosphomimetic mutations decrease L5 inhibitor binding A, ITC plots showing the integrated heats of reaction (main plot) for STLC (around 150 mM) titration into Eg5-367 mutants (10–20 mM, depending on the construct) plotted against the molar ratio (Methods). B, For each experiment, a single-site binding model (insets) was generated using Origin 7.0 that yielded a stoichiometry (N), dissociation constant (KD), enthalpy (H) and entropy (S) displayed in the table. Stoichiometries and dissociations constants are also displayed in Table I. Thermodynamic parameters were obtained from a single-site model of ITC data shown in the plots. Stoichiometries (N) show some variability reflecting protein concentration determination, but are generally consistent with single-site binding. Errors in KD and H were estimated based on the nonlinear least-squares fits to raw ITC data that are shown in the insets.
Figure S4 Eg5-targeting siRNA decreases endogenous Eg5 levels Western blot showing the efficiency of Eg5 knockdown using siRNA in LLC-Pk1 cells. Treatment with siRNA led to an approximately 50% reduction in Eg5 signal as measured by densitometry.
Supplementary Movies. Eg5-367 motility assays were set up as described in Methods. For Eg5-367-WT and Eg5-367-Y211F mutants, movies were 30 minutes long with a 20 second interval between frames. For Eg5-367 DL5 and Eg5-367-Y211E mutants, movies were one hour long with a 40 second interval between frames to accommodate slower sliding velocities while minimizing photobleaching of MTs. In slides with many moving MTs, not all MTs were counted, but for each construct we tracked at least 3 MTs that were visible and moving for at least 50 frames per slide, and our data analysis included 3 slides per construct. MTs were tracked and velocities quantified using the MTrackJ plugin in ImageJ.
Supplementary Movie 5: Eg5-367-WT motility. Total time 30 minutes, scale bar 20 microns. Colored circles indicate MT tracks that were used for velocity analysis.
Supplementary Movie 6: Eg5-367-DL5 mutant motility. Total time 1 hour, scale bar 20 microns. Colored circles indicate MT tracks that were used for velocity analysis. Note that over the long timescale of this movie there was some stage drift; we tracked a stationary particle (dark blue track) to demonstrate that the drift does not significantly impact the analysis of motor velocity.
Supplementary Movie 7: Eg5-367-Y211F mutant motility. Total time 30 minutes, scale bar 20 microns. Colored circles indicate MT tracks that were used for velocity analysis. Due to the high number of moving MTs on this slide, not all tracks were analyzed.
Supplementary Movie 8: Eg5-367-Y211E mutant motility. Total time 1 hour, scale bar 20 microns. Colored circles indicate MT tracks that were used for velocity analysis.
Acknowledgments
We wish to thank Dr. C. Gottardi (Northwestern University) and the members of her lab for the use of facilities for in vitro kinase assays using radiolabeled ATP, and for their intellectual contributions to this work. We also thank Dr. T. E. Smithgall (University of Pittsburgh) for the kind gift of the c-Src constructs used in this work, and Dr. E. Landahl (DePaul University), M. Gonzalez (Northwestern University), and A. DeJesus (Northwestern University) who helped with motility assays. We thank Dr. P. Visconte (University of Mass. Amherst) for the gift of SU6656 (SFK inhibitor), Dr. M. Davidson (Florida State University) for the gift of the mEmerald-labeled Eg5 construct that was used in this work, and Dr. T. Maresca (University of Mass. Amherst) for the Phospho-Aurora antibody. This work was completed with funds from the National Institute of Health (R01 GM107209). J.S.W. was supported by the Myhrvold Family Fellowship from the Fannie and John Hertz Foundation.
References
- Avunie-Masala R, Movshovich N, Nissenkorn Y, Gerson-Gurwitz A, Fridman V, Koivomagi M, Loog M, Hoyt MA, Zaritsky A, Gheber L. Phospho-regulation of kinesin-5 during anaphase spindle elongation. J Cell Sci. 2011;124(Pt 6):873–8. doi: 10.1242/jcs.077396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bain J, Plater L, Elliott M, Shpiro N, Hastie CJ, McLauchlan H, Klevernic I, Arthur JS, Alessi DR, Cohen P. The selectivity of protein kinase inhibitors: a further update. Biochem J. 2007;408(3):297–315. doi: 10.1042/BJ20070797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behnke-Parks WM, Vendome J, Honig B, Maliga Z, Moores C, Rosenfeld SS. Loop L5 acts as a conformational latch in the mitotic kinesin Eg5. J Biol Chem. 2011;286(7):5242–53. doi: 10.1074/jbc.M110.192930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatt AS, Erdjument-Bromage H, Tempst P, Craik CS, Moasser MM. Adhesion signaling by a novel mitotic substrate of src kinases. Oncogene. 2005;24(34):5333–43. doi: 10.1038/sj.onc.1208582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop JD, Han Z, Schumacher JM. The Caenorhabditis elegans Aurora B kinase AIR-2 phosphorylates and is required for the localization of a BimC kinesin to meiotic and mitotic spindles. Mol Biol Cell. 2005;16(2):742–56. doi: 10.1091/mbc.E04-08-0682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blangy A, Arnaud L, Nigg EA. Phosphorylation by p34cdc2 protein kinase regulates binding of the kinesin-related motor HsEg5 to the dynactin subunit p150. J Biol Chem. 1997;272(31):19418–24. doi: 10.1074/jbc.272.31.19418. [DOI] [PubMed] [Google Scholar]
- Blangy A, Lane HA, d’Herin P, Harper M, Kress M, Nigg EA. Phosphorylation by p34cdc2 regulates spindle association of human Eg5, a kinesin-related motor essential for bipolar spindle formation in vivo. Cell. 1995;83(7):1159–69. doi: 10.1016/0092-8674(95)90142-6. [DOI] [PubMed] [Google Scholar]
- Brennan IM, Peters U, Kapoor TM, Straight AF. Polo-like kinase controls vertebrate spindle elongation and cytokinesis. PLoS One. 2007;2(5):e409. doi: 10.1371/journal.pone.0000409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brust-Mascher I, Sommi P, Cheerambathur DK, Scholey JM. Kinesin-5-dependent poleward flux and spindle length control in Drosophila embryo mitosis. Mol Biol Cell. 2009;20(6):1749–62. doi: 10.1091/mbc.E08-10-1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderwood DJ, Johnston DN, Munschauer R, Rafferty P. Pyrrolo[2,3-d]pyrimidines containing diverse N-7 substituents as potent inhibitors of Lck. Bioorg Med Chem Lett. 2002;12(12):1683–86. doi: 10.1016/s0960-894x(02)00195-6. [DOI] [PubMed] [Google Scholar]
- Caron D, Byrne DP, Thebault P, Soulet D, Landry CR, Eyers PA, Elowe S. Mitotic phosphotyrosine network analysis reveals that tyrosine phosphorylation regulates Polo-like kinase 1 (PLK1) Science signaling. 2016;9(458):rs14. doi: 10.1126/scisignal.aah3525. [DOI] [PubMed] [Google Scholar]
- Chee MK, Haase SB. B-cyclin/CDKs regulate mitotic spindle assembly by phosphorylating kinesins-5 in budding yeast. PLoS Genet. 2010;6(5):e1000935. doi: 10.1371/journal.pgen.1000935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochran JC, Gilbert SP. ATPase mechanism of Eg5 in the absence of microtubules: insight into microtubule activation and allosteric inhibition by monastrol. Biochemistry. 2005;44(50):16633–48. doi: 10.1021/bi051724w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochran JC, Krzysiak TC, Gilbert SP. Pathway of ATP hydrolysis by monomeric kinesin Eg5. Biochemistry. 2006;45(40):12334–44. doi: 10.1021/bi0608562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochran JC, Sontag CA, Maliga Z, Kapoor TM, Correia JJ, Gilbert SP. Mechanistic analysis of the mitotic kinesin Eg5. J Biol Chem. 2004;279(37):38861–70. doi: 10.1074/jbc.M404203200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David-Pfeuty T, Bagrodia S, Shalloway D. Differential localization patterns of myristoylated and nonmyristoylated c-Src proteins in interphase and mitotic c-Src overexpresser cells. J Cell Sci. 1993;105(Pt 3):613–28. doi: 10.1242/jcs.105.3.613. [DOI] [PubMed] [Google Scholar]
- DeBerg HA, Blehm BH, Sheung J, Thompson AR, Bookwalter CS, Torabi SF, Schroer TA, Berger CL, Lu Y, Trybus KM, et al. Motor domain phosphorylation modulates kinesin-1 transport. J Biol Chem. 2013;288(45):32612–21. doi: 10.1074/jbc.M113.515510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enos AP, Morris NR. Mutation of a gene that encodes a kinesin-like protein blocks nuclear division in A. nidulans. Cell. 1990;60(6):1019–27. doi: 10.1016/0092-8674(90)90350-n. [DOI] [PubMed] [Google Scholar]
- Falnikar A, Tole S, Baas PW. Kinesin-5, a mitotic microtubule-associated motor protein, modulates neuronal migration. Mol Biol Cell. 2011;22(9):1561–74. doi: 10.1091/mbc.E10-11-0905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferenz NP, Gable A, Wadsworth P. Mitotic functions of kinesin-5. Semin Cell Dev Biol. 2010;21(3):255–9. doi: 10.1016/j.semcdb.2010.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fumagalli S, Totty NF, Hsuan JJ, Courtneidge SA. A target for Src in mitosis. Nature. 1994;368(6474):871–4. doi: 10.1038/368871a0. [DOI] [PubMed] [Google Scholar]
- Gable A, Qiu M, Titus J, Balchand S, Ferenz NP, Ma N, Collins ES, Fagerstrom C, Ross JL, Yang G, et al. Dynamic reorganization of Eg5 in the mammalian spindle throughout mitosis requires dynein and TPX2. Mol Biol Cell. 2012;23(7):1254–66. doi: 10.1091/mbc.E11-09-0820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia K, Stumpff J, Duncan T, Su TT. Tyrosines in the kinesin-5 head domain are necessary for phosphorylation by Wee1 and for mitotic spindle integrity. Curr Biol. 2009;19(19):1670–6. doi: 10.1016/j.cub.2009.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G, Vale RD. The roles of microtubule-based motor proteins in mitosis: comprehensive RNAi analysis in the Drosophila S2 cell line. J Cell Biol. 2003;162(6):1003–16. doi: 10.1083/jcb.200303022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan I, Yanagida M. Novel potential mitotic motor protein encoded by the fission yeast cut7+ gene. Nature. 1990;347(6293):563–6. doi: 10.1038/347563a0. [DOI] [PubMed] [Google Scholar]
- Han G, Ye M, Liu H, Song C, Sun D, Wu Y, Jiang X, Chen R, Wang C, Wang L, et al. Phosphoproteome analysis of human liver tissue by long-gradient nanoflow LC coupled with multiple stage MS analysis. Electrophoresis. 2010;31(6):1080–9. doi: 10.1002/elps.200900493. [DOI] [PubMed] [Google Scholar]
- He J, Zhang Z, Ouyang M, Yang F, Hao H, Lamb KL, Yang J, Yin Y, Shen WH. PTEN regulates EG5 to control spindle architecture and chromosome congression during mitosis. Nature communications. 2016;7:12355. doi: 10.1038/ncomms12355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornbeck PV, Zhang B, Murray B, Kornhauser JM, Latham V, Skrzypek E. PhosphoSitePlus, 2014: mutations, PTMs and recalibrations. Nucleic Acids Res. 2015;43(Database issue):D512–20. doi: 10.1093/nar/gku1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyt MA, He L, Loo KK, Saunders WS. Two Saccharomyces cerevisiae kinesin-related gene products required for mitotic spindle assembly. J Cell Biol. 1992;118(1):109–20. doi: 10.1083/jcb.118.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang TG, Hackney DD. Drosophila kinesin minimal motor domain expressed in Escherichia coli. Purification and kinetic characterization. J Biol Chem. 1994;269(23):16493–501. [PubMed] [Google Scholar]
- Hyman A, Drechsel D, Kellogg D, Salser S, Sawin K, Steffen P, Wordeman L, Mitchison T. Methods Enzymol. Vol. 196. Academic Press; 1991. [39] Preparation of modified tubulins; pp. 478–85. [DOI] [PubMed] [Google Scholar]
- Iliuk AB, Martin VA, Alicie BM, Geahlen RL, Tao WA. In-depth analyses of kinase-dependent tyrosine phosphoproteomes based on metal ion-functionalized soluble nanopolymers. Mol Cell Proteomics. 2010;9(10):2162–72. doi: 10.1074/mcp.M110.000091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaan HY, Ulaganathan V, Hackney DD, Kozielski F. An allosteric transition trapped in an intermediate state of a new kinesin-inhibitor complex. Biochem J. 2009;425(1):55–60. doi: 10.1042/BJ20091207. [DOI] [PubMed] [Google Scholar]
- Kapitein LC, Peterman EJ, Kwok BH, Kim JH, Kapoor TM, Schmidt CF. The bipolar mitotic kinesin Eg5 moves on both microtubules that it crosslinks. Nature. 2005;435(7038):114–8. doi: 10.1038/nature03503. [DOI] [PubMed] [Google Scholar]
- Kim ED, Buckley R, Learman S, Richard J, Parke C, Worthylake DK, Wojcik EJ, Walker RA, Kim S. Allosteric drug discrimination is coupled to mechanochemical changes in the kinesin-5 motor core. J Biol Chem. 2010;285(24):18650–61. doi: 10.1074/jbc.M109.092072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim LC, Song LX, Haura EB. Src kinases as therapeutic targets for cancer. Nature Reviews Clinical Oncology. 2009;6(10):587–95. doi: 10.1038/nrclinonc.2009.129. [DOI] [PubMed] [Google Scholar]
- Krzysiak TC, Gilbert SP. Dimeric Eg5 maintains processivity through alternating-site catalysis with rate-limiting ATP hydrolysis. J Biol Chem. 2006;281(51):39444–54. doi: 10.1074/jbc.M608056200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzysiak TC, Grabe M, Gilbert SP. Getting in sync with dimeric Eg5. Initiation and regulation of the processive run. J Biol Chem. 2008;283(4):2078–87. doi: 10.1074/jbc.M708354200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuga T, Nakayama Y, Hoshino M, Higashiyama Y, Obata Y, Matsuda D, Kasahara K, Fukumoto Y, Yamaguchi N. Differential mitotic activation of endogenous c-Src, c-Yes, and Lyn in HeLa cells. Arch Biochem Biophys. 2007;466(1):116–24. doi: 10.1016/j.abb.2007.07.002. [DOI] [PubMed] [Google Scholar]
- Kull FJ, Sablin EP, Lau R, Fletterick RJ, Vale RD. Crystal structure of the kinesin motor domain reveals a structural similarity to myosin. Nature. 1996;380(6574):550–5. doi: 10.1038/380550a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227(5259):680–5. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Larson AG, Naber N, Cooke R, Pate E, Rice SE. The conserved L5 loop establishes the pre-powerstroke conformation of the Kinesin-5 motor, eg5. Biophys J. 2010;98(11):2619–27. doi: 10.1016/j.bpj.2010.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenart P, Petronczki M, Steegmaier M, Di Fiore B, Lipp JJ, Hoffmann M, Rettig WJ, Kraut N, Peters JM. The small-molecule inhibitor BI 2536 reveals novel insights into mitotic roles of polo-like kinase 1. Curr Biol. 2007;17(4):304–15. doi: 10.1016/j.cub.2006.12.046. [DOI] [PubMed] [Google Scholar]
- Levi M, Maro B, Shalgi R. Fyn kinase is involved in cleavage furrow ingression during meiosis and mitosis. Reproduction. 2010;140(6):827–34. doi: 10.1530/REP-10-0312. [DOI] [PubMed] [Google Scholar]
- Ley SC, Marsh M, Bebbington CR, Proudfoot K, Jordan P. Distinct intracellular localization of Lck and Fyn protein tyrosine kinases in human T lymphocytes. J Cell Biol. 1994;125(3):639–49. doi: 10.1083/jcb.125.3.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Xing X, Ding G, Li Q, Wang C, Xie L, Zeng R, Li Y. SysPTM: a systematic resource for proteomic research on post-translational modifications. Mol Cell Proteomics. 2009;8(8):1839–49. doi: 10.1074/mcp.M900030-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W, Slebos RJ, Hill S, Li M, Brabek J, Amanchy R, Chaerkady R, Pandey A, Ham AJ, Hanks SK. Global impact of oncogenic Src on a phosphotyrosine proteome. J Proteome Res. 2008;7(8):3447–60. doi: 10.1021/pr800187n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma N, Titus J, Gable A, Ross JL, Wadsworth P. TPX2 regulates the localization and activity of Eg5 in the mammalian mitotic spindle. J Cell Biol. 2011;195(1):87–98. doi: 10.1083/jcb.201106149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma N, Tulu US, Ferenz NP, Fagerstrom C, Wilde A, Wadsworth P. Poleward transport of TPX2 in the mammalian mitotic spindle requires dynein, Eg5, and microtubule flux. Mol Biol Cell. 2010;21(6):979–88. doi: 10.1091/mbc.E09-07-0601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maliga Z, Mitchison TJ. Small-molecule and mutational analysis of allosteric Eg5 inhibition by monastrol. BMC Chem Biol. 2006;6:2. doi: 10.1186/1472-6769-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maliga Z, Kapoor TM, Mitchison TJ. Evidence that monastrol is an allosteric inhibitor of the mitotic kinesin Eg5. Chem Biol. 2002;9(9):989–96. doi: 10.1016/s1074-5521(02)00212-0. [DOI] [PubMed] [Google Scholar]
- Maliga Z, Xing J, Cheung H, Juszczak LJ, Friedman JM, Rosenfeld SS. A pathway of structural changes produced by monastrol binding to Eg5. J Biol Chem. 2006;281(12):7977–82. doi: 10.1074/jbc.M511955200. [DOI] [PubMed] [Google Scholar]
- Mayer TU, Kapoor TM, Haggarty SJ, King RW, Schreiber SL, Mitchison TJ. Small molecule inhibitor of mitotic spindle bipolarity identified in a phenotype-based screen. Science. 1999;286(5441):971–4. doi: 10.1126/science.286.5441.971. [DOI] [PubMed] [Google Scholar]
- Meyn MA, 3rd, Smithgall TE. Chemical genetics identifies c-Src as an activator of primitive ectoderm formation in murine embryonic stem cells. Science signaling. 2009;2(92):ra64. doi: 10.1126/scisignal.2000311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison T, Kirschner M. Microtubule assembly nucleated by isolated centrosomes. Nature. 1984;312(5991):232–7. doi: 10.1038/312232a0. [DOI] [PubMed] [Google Scholar]
- Muretta JM, Jun Y, Gross SP, Major J, Thomas DD, Rosenfeld SS. The structural kinetics of switch-1 and the neck linker explain the functions of kinesin-1 and Eg5. Proc Natl Acad Sci U S A. 2015;112(48):E6606–13. doi: 10.1073/pnas.1512305112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muretta JM, Behnke-Parks WM, Major J, Petersen KJ, Goulet A, Moores CA, Thomas DD, Rosenfeld SS. Loop L5 assumes three distinct orientations during the ATPase cycle of the mitotic kinesin Eg5: a transient and time-resolved fluorescence study. J Biol Chem. 2013;288(48):34839–49. doi: 10.1074/jbc.M113.518845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers KA, Baas PW. Kinesin-5 regulates the growth of the axon by acting as a brake on its microtubule array. J Cell Biol. 2007;178(6):1081–91. doi: 10.1083/jcb.200702074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadar VC, Ketschek A, Myers KA, Gallo G, Baas PW. Kinesin-5 is essential for growth-cone turning. Curr Biol. 2008;18(24):1972–7. doi: 10.1016/j.cub.2008.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama Y, Matsui Y, Takeda Y, Okamoto M, Abe K, Fukumoto Y, Yamaguchi N. c-Src but not Fyn promotes proper spindle orientation in early prometaphase. J Biol Chem. 2012;287(30):24905–15. doi: 10.1074/jbc.M112.341578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obenauer JC, Cantley LC, Yaffe MB. Scansite 2.0: Proteome-wide prediction of cell signaling interactions using short sequence motifs. Nucleic Acids Res. 2003;31(13):3635–41. doi: 10.1093/nar/gkg584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapley J, Nicolas M, Groen A, Regue L, Bertran MT, Caelles C, Avruch J, Roig J. The NIMA-family kinase Nek6 phosphorylates the kinesin Eg5 at a novel site necessary for mitotic spindle formation. J Cell Sci. 2008;121(Pt 23):3912–21. doi: 10.1242/jcs.035360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarli V, Giannis A. Targeting the kinesin spindle protein: basic principles and clinical implications. Clin Cancer Res. 2008;14(23):7583–7. doi: 10.1158/1078-0432.CCR-08-0120. [DOI] [PubMed] [Google Scholar]
- Saunders W, Lengyel V, Hoyt MA. Mitotic spindle function in Saccharomyces cerevisiae requires a balance between different types of kinesin-related motors. Mol Biol Cell. 1997;8(6):1025–33. doi: 10.1091/mbc.8.6.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholey JE, Nithianantham S, Scholey JM, Al-Bassam J. Structural basis for the assembly of the mitotic motor Kinesin-5 into bipolar tetramers. eLife. 2014;3:e02217. doi: 10.7554/eLife.02217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen B, Johnson FM. Regulation of SRC family kinases in human cancers. Journal of signal transduction. 2011;2011:865819. doi: 10.1155/2011/865819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapira O, Gheber L. Motile properties of the bi-directional kinesin-5 Cin8 are affected by phosphorylation in its motor domain. Sci Rep. 2016;6:25597. doi: 10.1038/srep25597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapira O, Goldstein A, Al-Bassam J, Gheber L. A potential physiological role for bi-directional motility and motor clustering of mitotic kinesin-5 Cin8 in yeast mitosis. J Cell Sci. 2017;130(4):725–34. doi: 10.1242/jcs.195040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoufias DA, DeBonis S, Saoudi Y, Lebeau L, Crevel I, Cross R, Wade RH, Hackney D, Kozielski F. S-trityl-L-cysteine is a reversible, tight binding inhibitor of the human kinesin Eg5 that specifically blocks mitotic progression. J Biol Chem. 2006;281(26):17559–69. doi: 10.1074/jbc.M511735200. [DOI] [PubMed] [Google Scholar]
- Smith KP, Gifford KM, Waitzman JS, Rice SE. Survey of phosphorylation near drug binding sites in the Protein Data Bank (PDB) and their effects. Proteins. 2015;83(1):25–36. doi: 10.1002/prot.24605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanenbaum ME, Macurek L, Galjart N, Medema RH. Dynein, Lis1 and CLIP-170 counteract Eg5-dependent centrosome separation during bipolar spindle assembly. EMBO J. 2008;27(24):3235–45. doi: 10.1038/emboj.2008.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanenbaum ME, Macurek L, Janssen A, Geers EF, Alvarez-Fernandez M, Medema RH. Kif15 cooperates with eg5 to promote bipolar spindle assembly. Curr Biol. 2009;19(20):1703–11. doi: 10.1016/j.cub.2009.08.027. [DOI] [PubMed] [Google Scholar]
- Thomas SM, Brugge JS. Cellular functions regulated by Src family kinases. Annu Rev Cell Dev Biol. 1997;13:513–609. doi: 10.1146/annurev.cellbio.13.1.513. [DOI] [PubMed] [Google Scholar]
- UniProt C. UniProt: a hub for protein information. Nucleic Acids Res. 2015;43(Database issue):D204–12. doi: 10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzbekov R, Prigent C, Arlot-Bonnemains Y. Cell cycle analysis and synchronization of the Xenopus laevis XL2 cell line: study of the kinesin related protein XlEg5. Microsc Res Tech. 1999;45(1):31–42. doi: 10.1002/(SICI)1097-0029(19990401)45:1<31::AID-JEMT3>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- van den Wildenberg SM, Tao L, Kapitein LC, Schmidt CF, Scholey JM, Peterman EJ. The homotetrameric kinesin-5 KLP61F preferentially crosslinks microtubules into antiparallel orientations. Curr Biol. 2008;18(23):1860–4. doi: 10.1016/j.cub.2008.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Ree JH, Nam HJ, Jeganathan KB, Kanakkanthara A, van Deursen JM. Pten regulates spindle pole movement through Dlg1-mediated recruitment of Eg5 to centrosomes. Nat Cell Biol. 2016;18(7):814–21. doi: 10.1038/ncb3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanneste D, Takagi M, Imamoto N, Vernos I. The role of Hklp2 in the stabilization and maintenance of spindle bipolarity. Curr Biol. 2009;19(20):1712–7. doi: 10.1016/j.cub.2009.09.019. [DOI] [PubMed] [Google Scholar]
- Venere M, Horbinski C, Crish JF, Jin X, Vasanji A, Major J, Burrows AC, Chang C, Prokop J, Wu Q, et al. The mitotic kinesin KIF11 is a driver of invasion, proliferation, and self-renewal in glioblastoma. Sci Transl Med. 2015;7(304):304ra143. doi: 10.1126/scitranslmed.aac6762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waitzman JS, Larson AG, Cochran JC, Naber N, Cooke R, Jon Kull F, Pate E, Rice SE. The loop 5 element structurally and kinetically coordinates dimers of the human kinesin-5, Eg5. Biophys J. 2011;101(11):2760–9. doi: 10.1016/j.bpj.2011.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waksman G, Kominos D, Robertson SC, Pant N, Baltimore D, Birge RB, Cowburn D, Hanafusa H, Mayer BJ, Overduin M, et al. Crystal structure of the phosphotyrosine recognition domain SH2 of v-src complexed with tyrosine-phosphorylated peptides. Nature. 1992;358(6388):646–53. doi: 10.1038/358646a0. [DOI] [PubMed] [Google Scholar]
- Wang W, Chen L, Ding Y, Jin J, Liao K. Centrosome separation driven by actin-microfilaments during mitosis is mediated by centrosome-associated tyrosine-phosphorylated cortactin. J Cell Sci. 2008;121(Pt 8):1334–43. doi: 10.1242/jcs.018176. [DOI] [PubMed] [Google Scholar]
- Whalley HJ, Porter AP, Diamantopoulou Z, White GR, Castaneda-Saucedo E, Malliri A. Cdk1 phosphorylates the Rac activator Tiam1 to activate centrosomal Pak and promote mitotic spindle formation. Nature communications. 2015;6:7437. doi: 10.1038/ncomms8437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson MB, Schreiner SJ, Choi HJ, Kamens J, Smithgall TE. Selective pyrrolo-pyrimidine inhibitors reveal a necessary role for Src family kinases in Bcr-Abl signal transduction and oncogenesis. Oncogene. 2002;21(53):8075–88. doi: 10.1038/sj.onc.1206008. [DOI] [PubMed] [Google Scholar]
- Woehlke G, Ruby AK, Hart CL, Ly B, Hom-Booher N, Vale RD. Microtubule interaction site of the kinesin motor. Cell. 1997;90(2):207–16. doi: 10.1016/s0092-8674(00)80329-3. [DOI] [PubMed] [Google Scholar]
- Zaytsev AV, Sundin LJ, DeLuca KF, Grishchuk EL, DeLuca JG. Accurate phosphoregulation of kinetochore-microtubule affinity requires unconstrained molecular interactions. J Cell Biol. 2014;206(1):45–59. doi: 10.1083/jcb.201312107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Eg5 is phosphorylated on a conserved tyrosine in its motor head A, A single immunoprecipitation reaction from HEK293Ts (3.5 mg total lysate) was divided in two. Half was incubated with l-phosphatase (right); the other was incubated with phosphatase buffer alone (left). B, Table summarizing Eg5 tyrosine phosphorylations as reported in databases collecting results from phosphoproteomics experiments, including PhosphositePlus (Hornbeck et al., 2015). C, Table showing the UniProt Accession numbers used to generate the alignment of Kinesin-5 family members in Figure 1C. D, Alignment of kinesins from families 1–7, highlighting the MT binding region. The –PXXP- SH3 targeting motif, when present, is highlighted in red. Residues that are identical across the alignment are marked in dark grey, while similar residues are marked in light grey. While there is extensive conservation of sequence in this region across kinesin family members, the PXXP motif is only seen in Kinesin-5s of higher order organisms. E List of the sequence mutations made in Eg5 constructs and F, other constructs for experiments using Eg5 mutants presented in the article.
Figure S2 Eg5 motor heads are phosphorylated on Y125, Y211, and Y231 in vitro Purified Eg5 motor heads and purified Src kinase were incubated with ATP (radiolabeled ATP in some cases) for 5 min. Reactions were quenched and then analyzed either A, by LC-MS or B, by radiograph.
Figure S3 Eg5 phosphomimetic mutations decrease L5 inhibitor binding A, ITC plots showing the integrated heats of reaction (main plot) for STLC (around 150 mM) titration into Eg5-367 mutants (10–20 mM, depending on the construct) plotted against the molar ratio (Methods). B, For each experiment, a single-site binding model (insets) was generated using Origin 7.0 that yielded a stoichiometry (N), dissociation constant (KD), enthalpy (H) and entropy (S) displayed in the table. Stoichiometries and dissociations constants are also displayed in Table I. Thermodynamic parameters were obtained from a single-site model of ITC data shown in the plots. Stoichiometries (N) show some variability reflecting protein concentration determination, but are generally consistent with single-site binding. Errors in KD and H were estimated based on the nonlinear least-squares fits to raw ITC data that are shown in the insets.
Figure S4 Eg5-targeting siRNA decreases endogenous Eg5 levels Western blot showing the efficiency of Eg5 knockdown using siRNA in LLC-Pk1 cells. Treatment with siRNA led to an approximately 50% reduction in Eg5 signal as measured by densitometry.
Supplementary Movies. Eg5-367 motility assays were set up as described in Methods. For Eg5-367-WT and Eg5-367-Y211F mutants, movies were 30 minutes long with a 20 second interval between frames. For Eg5-367 DL5 and Eg5-367-Y211E mutants, movies were one hour long with a 40 second interval between frames to accommodate slower sliding velocities while minimizing photobleaching of MTs. In slides with many moving MTs, not all MTs were counted, but for each construct we tracked at least 3 MTs that were visible and moving for at least 50 frames per slide, and our data analysis included 3 slides per construct. MTs were tracked and velocities quantified using the MTrackJ plugin in ImageJ.
Supplementary Movie 5: Eg5-367-WT motility. Total time 30 minutes, scale bar 20 microns. Colored circles indicate MT tracks that were used for velocity analysis.
Supplementary Movie 6: Eg5-367-DL5 mutant motility. Total time 1 hour, scale bar 20 microns. Colored circles indicate MT tracks that were used for velocity analysis. Note that over the long timescale of this movie there was some stage drift; we tracked a stationary particle (dark blue track) to demonstrate that the drift does not significantly impact the analysis of motor velocity.
Supplementary Movie 7: Eg5-367-Y211F mutant motility. Total time 30 minutes, scale bar 20 microns. Colored circles indicate MT tracks that were used for velocity analysis. Due to the high number of moving MTs on this slide, not all tracks were analyzed.
Supplementary Movie 8: Eg5-367-Y211E mutant motility. Total time 1 hour, scale bar 20 microns. Colored circles indicate MT tracks that were used for velocity analysis.