Abstract
The TEAD transcription factor family is best known for transcriptional output of the Hippo signaling pathway and has been implicated in processes such as development, cell growth and proliferation, tissue homeostasis, and regeneration. Our understanding of the functional importance of TEADs has increased dramatically since its initial discovery three decades ago. The majority of our knowledge of TEADs is in the context of Hippo signaling as nuclear DNA binding proteins passively activated by YAP and TAZ, transcription coactivators downstream of the Hippo pathway. However, recent studies suggest that TEAD itself is actively regulated. Here, we highlight evidence demonstrating Hippo-independent regulation of TEADs and the potential impacts these studies may have on new cancer therapeutics.
Keywords: TEAD, YAP/TAZ, Hippo, cancer, cytoplasmic-nuclear shuttling
The TEAD family of transcription factors
Studies of the TEAD (TEA/ATTS domain) transcription factor family began with the identification of TEAD1 which was first discovered in an attempt to identify nuclear proteins that could bind to the SV40 enhancer and activate transcription[1]. Further studies showed that TEAD could not only bind to GT-IIC and Sph motifs on the SV40 enhancer but also human papillomavirus-16 (HPV-16) oncogenes[2–4] and M-CAT motifs[5]. Since their initial discovery, TEADs have been found to be evolutionarily conserved, and have been shown to play important roles in various biological processes and human disease [6, 7].
Mammals express four TEAD genes, TEAD1-4. TEADs are broadly expressed but each TEAD has tissue specific expression, which indicates tissue specific roles for each TEAD[5, 8–10]. In particular, TEADs have been shown to play important roles in development, with activity detected at the two-cell embryo stage and maintained for cardiogenesis[11], neural crest and notochord development[12, 13], and trophectoderm lineage determination[14]. Tead1 null mice are embryonic lethal due to defective maturation during cardiac development[11]. Knockout of Tead2 in mice leads to defects in neural development with an increased risk for defects in neural tube closure[12]. In contrast, another study showed redundant functions for TEAD1 and TEAD2; Tead2 null mice showed no phenotype but knockout of both Tead1 and Tead2 was embryonic lethal with embryos lacking a closed neural tube, notochord, and somites[13]. Tead4 null mice are also embryonic lethal due to failure in embryo implantation, however, disruption of Tead4 after embryo implantation results in normal development[14, 15]. In humans, an inactivating missense mutation of TEAD1 (Y421H) is associated with Sveinsson’s chorioretinal atrophy, a genetic disorder that results in degeneration of the choroid and retina [16, 17].
TEADs seem to have important biological functions, but studies thoroughly characterizing TEAD function and regulation are lacking. Our knowledge of TEADs has developed largely from work that focuses on TEADs in the context of the Hippo pathway signaling [18] (Box 1). TEAD transcriptional activity is broadly believed to be regulated by the presence or the absence of nuclear YAP/TAZ. However, accumulating evidence shows that TEAD itself is regulated through other mechanisms. In particular, this review will focus on recent work that has uncovered Hippo-independent regulation of TEADs and how these studies are helping to shape the development of cancer therapeutics.
Box 1. Hippo Pathway Overview.
The Hippo pathway is a regulator of cell growth, proliferation and homeostasis and has been shown to be essential in development, stem cell function, and tissue regeneration[79, 80]. In recent years, studies have revealed a vast array of regulators upstream of the Hippo pathway[81], however, the main components are comprised of serine/threonine kinases Mammalian STE20-like kinases (MST1/2), mitogen activated protein kinase kinase kinase kinases (MAP4Ks), Large Tumor Suppressor kinases (LATS1/2), and transcription co-activators Yes-associated protein (YAP) and its paralog Transcriptional Activator with PDZ binding domain (TAZ). When complexed with adaptor protein Salvador Homolog (SAV1), MST1/2 phosphorylate and activate LATS1/2 and their adaptor protein Mob1 Homolog (MOB1) [82–85]. LATS1/2 have also been shown to be phosphorylated by the MAP4K4 family[86]. Phosphorylation of YAP/TAZ by activated LATS1/2 results in cytoplasmic sequestration due to binding to 14-3-3 or ubiquitinylation and degradation of YAP/TAZ [29, 87–90]. When the Hippo pathway is turned off, LATS1/2 are inactive, YAP/TAZ are dephosphorylated and accumulate in the nucleus where they bind to TEAD to drive expression of target genes such as CTGF and Cyr61 [27, 28].
Regulation of TEAD by Coactivators
When TEADs were identified, they were found to have little transcriptional activity by themselves and were predicted to require the presence of coactivators to induce target gene transcription [3]. TEAD proteins have an N terminal TEA/ATTS domain which binds to DNA as a homeodomain fold and a C terminal transactivation domain with which coactivators bind in order to transcribe target genes [8, 19–21]. The TEA/ATTS domain of TEAD is highly conserved in all TEAD family members and recognizes the sequence motif 5′-GGAATG-3′ [1, 8, 22]. The C terminal transactivation domain of TEAD is also highly conserved, especially residues that are necessary for coactivator binding [23, 24]. The structure of the TEA domain of TEAD4 bound to DNA has recently been resolved and shows that the α3 helix formed by the TEA domain is the most important interface for DNA binding. While mutations at residues Ser100 and Gln103 completely abolished TEAD4’s DNA binding ability, mutations at other interface residues did not significantly inhibit DNA binding ability. The flexibility of these mutated residues indicates that TEAD-DNA binding sites may be diverse and specificity may be regulated by binding of different coactivators [25]. Several TEAD binding proteins and cofactors have been identified and are discussed in the sections below.
Hippo-dependent coactivators
The Hippo pathway transcriptional coactivators, YAP and its paralog TAZ, were among the cofactors identified [20, 26] and are now the most well-established activators of TEAD. When phosphorylated by LATS 1/2, YAP/TAZ are localized in the cytoplasm and incapable of binding TEAD, thus rendering TEAD transcriptionally inactive. Upon dephosphorylation, YAP/TAZ are translocated to the nucleus to bind TEAD and drive transcription of target genes that are critical for cell growth, proliferation, and survival [27–29]. Structural studies show that the TEAD binding domain of YAP is located in the protein’s N terminus, while the YAP binding region is located in the C terminal, transactivation domain of TEAD. One molecule of YAP and one molecule of TEAD bind to form a heterodimer complex. The N-terminus of YAP wraps around the globular C-terminal structure of TEAD and binds through three major interfaces [23, 24]. Importantly, mutations at Y421 of TEAD1, found in Sveinsson’s chorioretinal atrophy, were discovered to disrupt a hydrogen bond that is essential in mediating TEAD-YAP interaction [16, 17, 23, 24]. Residues critical for the interactions are evolutionarily conserved on both YAP and TEAD [23, 24]. TAZ and TEAD binding has been shown to have two conformations. In one conformation, one molecule of TAZ binds to one molecule of TEAD forming a heterodimer similar to that of YAP-TEAD. In a second conformation, two molecules of TAZ bind two molecules of TEAD. In addition, the two TAZ molecules interact with each other to bridge the two TEAD molecules, forming a heterotetramer complex [30]. Further studies are needed to validate the physiological and functional significance of the difference in YAP-TEAD and TAZ-TEAD complexes. However, human interactome studies show interaction between TEADs, suggesting that homo- and hetero- complexes may differentially regulate TEAD transcriptional activity [31, 32]. As TEAD is the major transcriptional partner of YAP/TAZ [27, 33, 34], Hippo-regulated YAP/TAZ nuclear-cytoplasmic shuttling has served as a proxy for regulation of TEAD activity.
Hippo-independent coactivators
Though YAP/TAZ are currently the most well-studied coactivators and regulators of TEAD transcriptional activity, several other cofactors have been identified as TEAD binding partners. The Vestigial-like (VGLL) protein family consists of four members, VGLL1-4. VGLL has been shown to interact with TEAD to regulate gene expression [35–38]. Studies show that VGLL family proteins have binding sites on TEAD that overlap with YAP/TAZ binding sites and thus compete with YAP/TAZ for TEAD binding [21, 39]. Binding of VGLL4 to TEAD inhibits YAP/TAZ-TEAD target gene expression and suppresses tumor growth [21, 40]. In contrast to VGLL4, overexpression of VGLL1 promoted anchorage-independent cell growth and upregulated target genes different from that of YAP/TAZ-TEAD4 [39]. Suppression of canonical YAP/TAZ target genes was not analyzed with overexpression of VGLL1, however as VGLL1 was shown compete with YAP/TAZ for TEAD binding, upregulation of VGLL1 target genes by VGLL1 overexpression likely suppresses YAP/TAZ target genes. Though there are a few studies implicating the functional role of VGLL and TEAD [21, 35–38], further studies are needed to understand the opposing effects on cell growth by VGLL1 and VGLL4. It is not clear if all VGLL proteins broadly compete with YAP/TAZ for TEAD binding or if under different physiological contexts TEAD preferentially binds to different VGLL proteins to carry out YAP/TAZ independent cellular functions. However, binding of VGLL to TEAD occupies the YAP/TAZ-TEAD binding site and prevents TEAD from binding to YAP or TAZ, thereby inhibiting YAP/TAZ-TEAD specific transcriptional activity (Figure 1). As in the case with VGLL1, TEADs may not only regulate YAP/TAZ-driven target gene expression, but under different contexts may mediate transcriptional output of YAP/TAZ independent signaling pathways. In line with this notion, TEAD4 was recently implicated in the regulation of Wnt target genes. TEAD4 directly interacts with transcription factor 4 (TCF4) through its TEA domain to facilitate transactivation of TCF4 and mediate expression of Wnt target genes. VGLL4 binding to TEAD4 inhibits TEAD4-TCF4 driven target gene expression as it does for TEAD-YAP/TAZ target gene expression, but does not compete with TCF4 for TEAD binding. Instead, VGLL4 inhibition of TEAD4-TCF4 transcriptional activity is due to formation of a TEAD4/TCF4/VGLL4 ternary complex [41]. The p160 family of steroid receptor coactivators was also identified to interact with TEAD. In a yeast two-hybrid screen using the bHLH-PAS domain of steroid receptor coactivator 1 (SRC1), TEAD was identified as an interacting partner [42]. Moreover, all members of the p160 family could potentiate TEAD transcriptional activity [42]. In recent studies activator protein-1 (AP-1) was demonstrated to directly interact with TEAD [43, 44]. AP-1 was also shown to co-occupy the same chromatin sites as TEAD and presence of AP-1 was necessary to activate target genes important for tumor growth and progression[43, 44]. Other cofactors identified include poly-ADP ribose polymerase (PARP) [45], serum response factor (SRF) [46], myocyte enhancer factor 2 (MEF2) [47], and myc-associated factor X (MAX) [48]. These cofactors have been shown to aid TEAD transcriptional activity and regulate the transcriptional program necessary for muscle homeostasis and differentiation[45–48]. Although many TEAD interacting proteins have been implicated, it is clear that YAP/TAZ are the most important in stimulating TEAD transcriptional activity as binding of YAP/TAZ potently enhances TEAD reporter activity by several hundred folds. Moreover, inhibition of YAP/TAZ by either knockdown or knockout strongly abolishes endogenous expression of TEAD target genes [27].
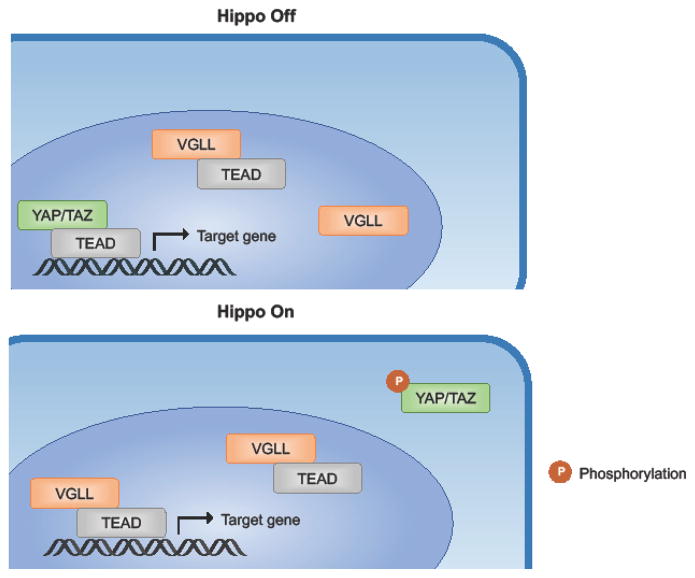
Figure 1. Coactivator binding stimulates TEAD transcriptional activity.
YAP/TAZ and VGLL competitively bind TEAD to regulate its transcriptional activity. When the Hippo pathway is “off” YAP/TAZ are dephosphorylated and translocated to the nucleus to bind TEAD and activate transcription of downstream target genes. Abundance of nuclear YAP/TAZ outcompetes VGLL-TEAD binding. When the Hippo pathway is “on”, YAP/TAZ are phosphorylated and sequestered in the cytoplasm. Absence of YAP/TAZ in the nucleus allows VGLL-TEAD binding.
Regulation of TEAD by post-translational modifications
The regulation of TEAD through binding of coactivators has, until now, been the primary mechanism of modulating TEAD transcriptional activity. However, recent studies suggest that TEAD transcriptional activity is also regulated by post-translational modifications as well as changes in subcellular localization.
Phosphorylation
In cardiac myocytes, an overlapping Max binding, E-box motif and a TEAD binding, M-CAT motif were identified on the α-myosin heavy chain (α-MHC) promoter, a promoter that is responsible for cAMP-induced gene expression [49]. This hybrid motif, found in several muscle specific genes, is regulated by a TEAD1-MAX complex [48]. Though interaction with MAX regulates TEAD1 by potentiating target gene expression [48], TEAD1-MAX target gene expression is also regulated by TEAD1 phosphorylation [50]. Protein kinase A (PKA) phosphorylation of TEAD1 at serine 102 inhibited TEAD1 DNA binding ability but did not disrupt TEAD1-MAX interaction [50]. In addition to phosphorylation by PKA, TEAD has also been shown to be a phosphorylation substrate of Protein Kinase C (PKC) [51]. Phosphorylation of TEAD by PKC also resulted in a decrease in its DNA binding ability [51]. Thus, phosphorylation is an alternative mechanism of modulating TEAD activity independent of interaction with coactivators.
Palmitoylation
Recent studies have identified S-palmitoylation as a post-translational modification of the TEAD family and that the palmitoylation of TEADs regulates protein stability [52] and transcriptional activity [53]. Interestingly, TEAD palmitoylation is autocatalytic as the abundance of palmitoylation increased significantly with the addition of palmitoyl-CoA to purified TEAD in vitro, despite the absence of palmitoyltransferases [53]. However, it is possible that palmitoylation of TEAD may require palmitoyltransferase in vivo because bacterially expressed TEAD is not efficiently palmitoylated. Three cysteine residues, conserved among TEADs, were identified as sites of palmitoylation. Mutation of any one cysteine residue on TEAD1 decreased palmitoylation while mutations at all three residues completely ablated TEAD1 palmitoylation [53]. Functionally, S-palmitoylation of TEAD1 is important for YAP/TAZ binding and transcriptional activity. Palmitoylation deficient mutant TEAD1 showed a substantial decrease in YAP binding, diminished transcriptional activity as assessed by a TEAD reporter assay, and inhibited C2C12 myoblast cell differentiation by blocking expression of muscle differentiation genes [53]. Interestingly, despite loss of YAP binding, the TEAD1 palmitoylation mutants retained VGLL4 binding ability [53]. Disruption of TEAD2 palmitoylation decreased protein stability and resulted in a significant loss of TEAD2 protein abundance [52]. Palmitoylation is important for protein trafficking and membrane localization [54], however, palmitoylation of TEAD does not affect TEAD localization or membrane binding [52, 53]. Consistently, the palmitoyl group is buried inside a deep hydrophobic pocket of TEAD as revealed by structure studies. It is still unknown whether TEAD palmitoylation is a dynamic process and whether mechanisms of TEAD depalmitoylation may be manipulated to regulate TEAD coactivator binding and transcriptional activity.
Regulation of TEAD by subcellular localization
Along with post-translational modifications, spatial regulation of transcription factors is also a common mechanism of altering transcriptional activity. Though there are few studies of subcellular changes in TEAD localization, accumulating data suggests that nucleocytoplasmic translocation of TEAD serves as a mechanism of modulating target gene expression.
Trophectoderm differentiation
Mouse knockout studies showed that Tead4 is specifically required for embryo implantation and trophectoderm lineage determination [14, 15]. To elucidate how TEAD4 regulates trophectoderm (TE) and inner cell mass (ICM) lineage in the preimplantation mouse embryo, Home et al. performed ChIP-seq to determine TEAD4 target genes in mouse trophoblast stem cells (mTSCs) and preimplantation mouse embryos. TEAD4 was shown to directly regulate a trophectoderm specific transcriptional program that included genes such as Gata3 and Cdx2. Although TEAD4 was found to be expressed in both the TE and the ICM, Gata3 and Cdx2 were not expressed in the mouse ICM or ICM-derived mouse embryonic stem cells (mESCs). Interestingly, TEAD4 was found to be localized exclusively in the cytoplasm of mESCs compared to mTSCs in which TEAD4 was enriched in the nucleus. Importantly, YAP remained nuclear in both mESCs and mTSCs, indicating that regulation of TEAD by subcellular localization is the primary mechanism of TE and ICM cell lineage determination. In human ESCs that were induced to a trophoblast fate, TEAD4 was found to localize to the nucleus along with an increase of GATA3 expression. Forced expression of nuclear TEAD4 in the inner blastomeres of a developing embryo activated CDX2 and inhibited proper blastocyst formation. At different developmental stages of the embryo, TEAD4 nuclear localization correlated with TE lineage cells expressing CDX2 while TEAD4 cytoplasmic localization correlated with ICM lineage cells. This TEAD4 expression pattern was conserved across various mammalian species, including human. The data suggests that TEAD subcellular localization regulates its transcriptional activity, turning TE-specific transcriptional programs on or off to determine specification of TE vs. ICM lineage differentiation for embryo maturation [55]. However, neither the signal nor the molecular mechanism that regulate the subcellular localization of TEADs in embryos is known.
p38 MAPK regulates the nuclear-cytoplasmic shuttling of TEAD in response to cellular stress
Recently, certain environmental stresses were identified that induce changes in TEAD subcellular localization. Although many extracellular signals, such as serum and glucose, are known to regulate subcellular localization of YAP/TAZ, they have no effect on TEAD localization. However, hyperosmotic stress, high cell density, and cell detachment potently induce TEAD cytoplasmic localization and inhibit its transcriptional activity. Therefore, in addition to regulation by the nuclear-cytoplasmic shuttling of YAP/TAZ, TEAD activity is also regulated by nuclear-cytoplasmic localization, similar to many other transcription factors involved in signal transduction, such as STAT, NF-κB, SMAD, and NFAT. Unlike YAP/TAZ, the cytoplasmic localization of TEAD is independent of the Hippo pathway kinase LATS. Because TEAD is absolutely required for transcriptional induction by YAP/TAZ, signals affecting TEAD localization impact the functional output of the Hippo pathway [56].
The mechanism of TEAD cytoplasmic localization by hyperosmotic stress is mediated by p38, but independent of the Hippo pathway components. TEAD cytoplasmic shuttling is not impaired in LATS1/2 knockout (KO), MST1/2 KO, and MAP4K4 KO cells. Notably, acute osmotic stress activates TEAD transcription via NLK-YAP activation [57, 58] however, during the adaptive response to prolonged osmotic stress [59], p38 directly binds TEAD and subsequently translocates TEAD to the cytoplasm, resulting in inhibition of TEAD transcriptional activity. Upon osmotic stress, the direct interaction between p38 and TEADs via a canonical p38-binding, D-domain on TEAD, is required for cytoplasmic translocation of TEAD. Though TEAD is not a direct phosphorylation substrate of p38, the kinase activity of p38 is required for p38-TEAD interaction and for p38 cytoplasmic translocation post-osmotic stress [59, 60].
Importantly, TEAD subcellular localization was shown to predominate YAP/TAZ-regulating signals and inhibit YAP-driven cancer cell growth. In LATS1/2 KO cells in which YAP is constitutively dephosphorylated, osmotic stress was able to induce YAP cytoplasmic translocation. It is proposed that cytoplasmic translocation of YAP/TAZ under osmotic stress is due to TEAD translocation [56]. Without nuclear localization of TEAD, YAP/TAZ are unable to accumulate in the nucleus. This notion is supported by data showing that TEAD 1/2/4 KO cells fail to accumulate YAP/TAZ in the nucleus, even upon dephosphorylation of YAP/TAZ by stimulating signals, such as LPA or serum. Thus, TEAD subcellular localization is an important factor in YAP/TAZ nuclear retention. Additionally, p38-mediated TEAD cytoplasmic translocation remains intact in both YAP-dependent and independent cancer cell lines. However, YAP-driven cancers were particularly susceptible to TEAD inhibition. Inhibition of TEAD by hyperosmolarity or p38 preferentially decreased growth of YAP-driven cancers both in vitro and in vivo. These growth inhibitory effects of YAP-driven cancers were rescued by expression of constitutively active TEAD. Therefore, though YAP/TAZ can regulate TEAD transcriptional activity, changes in TEAD subcellular localization can also modulate its transcriptional activity and determine Hippo signaling output [56] (Figure 2).
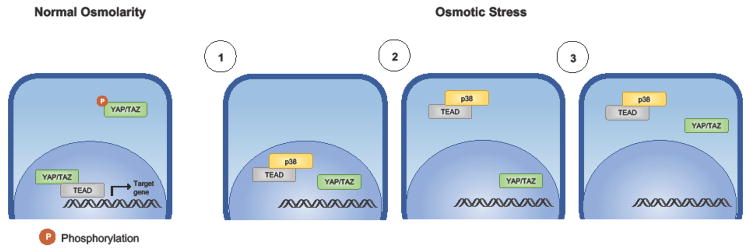
Figure 2. Osmotic stress promotes p38-mediated TEAD cytoplasmic localization.
Under conditions of normal osmolarity, TEAD is localized in the nucleus and its activity is regulated by the nuclear-cytoplasmic shuttling of YAP/TAZ. Under conditions of osmotic stress: 1) p38 is activated and binds to TEAD; 2) the p38-TEAD complex translocates to the cytoplasm; 3) In cancers in which YAP is constitutively dephosphorylated and localized in the nucleus, cytoplasmic translocation of TEAD results in the inability to retain nuclear YAP/TAZ.
While high density is well known to induce YAP/TAZ cytoplasmic localization, we also observed that high density induces cytoplasmic TEAD. However, the cell density signals that regulate YAP/TAZ and TEAD might be different. Moderate cell density is sufficient to induce cytoplasmic YAP/TAZ, however TEADs remain nuclear. TEAD cytoplasmic localization requires a much higher cell density and occurs later than YAP/TAZ cytoplasmic translocation. Importantly, LATS is involved in density-induced cytoplasmic localization of YAP/TAZ, but not TEAD (unpublished). The biochemical mechanism for density-induced TEAD cytoplasmic localization remains to be elucidated.
Regulation of Drosophila Scalloped
Studies of the Hippo pathway in drosophila have characterized Scalloped (Sd), the homolog of TEAD, to bind to Yorkie (Yki), the homolog of YAP, and mediate the growth regulatory effects of the Hippo pathway [61–64]. Interestingly, when analyzing sd;yki double mutant clones, loss of sd rescued yki mutant undergrowth phenotypes in the eye and in ovarian follicle cells [38]. This observation indicates that Sd has a repressor function when not bound with Yki. Thus, Yki may promote normal tissue growth by relieving the default repressor activity of Sd. Furthermore, Tondu-domain-containing growth inhibitor (Tgi), the homolog of VGLL4, was identified as a cofactor mediating Sd default repressor function [38]. Vestigial (Vg), the homolog of VGLL1, did not induce Sd repressor function [38]. Currently, it is unclear whether this mechanism of default repression by Sd is conserved in mammalian TEADs. In addition, Hippo (Hpo), the homolog of MST1/2, promotes cytoplasmic translocation of Sd to suppress Sd-Vg mediated proliferation in the wing, independently of Yki [65].
TEAD Regulation in Cancers
Numerous studies have suggested the importance of Hippo signaling in the development of cancer. These studies have emphasized the role of YAP/TAZ amplification and hyperactivity in various cancers [66], however increased TEAD expression and activity, both dependent and independent of YAP/TAZ, have also been implicated in the progression of several solid tumors [7] (Table 1). High TEAD expression levels are seen in prostate, colorectal, and breast cancers and, concordantly, are an indicator of poor clinical outcome [67–71]. In breast cancer cells, induction of epithelial to mesenchymal transition (EMT) resulted in upregulation of TEAD2 and a marked increase in YAP/TAZ nuclear accumulation despite decreases in overall YAP/TAZ protein levels [70]. The increase in TEAD2 expression resulted in increased YAP/TAZ binding which retained YAP/TAZ in the nucleus and drove TEAD transcriptional activity [70]. TEAD2 and TEAD4 were also found to be overexpressed in colorectal cancer, particularly in metastatic tissues, and knockdown of TEAD4 in vitro and in vivo reduced cell migration and metastasis [71]. Furthermore, the increase in metastatic potential in colorectal cancer was YAP independent as both wild-type TEAD4 and the YAP-binding deficient Y429 TEAD4 mutant rescued the effects of TEAD4 knockdown[71]. TEAD1 has been reported to play a role in conferring resistance to apoptosis in a YAP independent manner [72]. Additionally, TEAD1 was shown to regulate mesothelin, a gene that serves as a cancer biomarker due to its overexpression in many tumors [73]. The importance of TEAD-driven transcriptional programs has further been highlighted in several recent studies. ChIP-seq studies have shown that TEAD binds not only promoters but also distal enhancer elements [43, 44, 74]. Binding of YAP/TAZ-TEAD and AP-1 to enhancers synergistically activates target genes important for oncogenic growth, invasion, and migration in vitro and in vivo [43, 44]. In pre-B cells, YAP-TEAD binds superenhancer networks and contributes to aberrant pre-B cell phenotypes [74]. TEADs have also been reported to drive the transcriptional program responsible for increased invasiveness and resistance to MAPK inhibition in melanomas [75].
Table 1.
TEADs in disease
| Disease | Gene | Alteration (mutation or expression) | Effects | Target gene | Reference |
|---|---|---|---|---|---|
| Sveinsson’s chorioretinal atrophy | TEAD1 | Y421H inactivating mutation | choroidal and retinal degeneration | 14 | |
| Breast cancer, squamous cell carcinoma | TEAD4 | increased expression | increased cell proliferation and tumorigenesis, decreased survival | 42 | |
| Prostate cancer | TEAD1 | increased expression | decreased survival | 62 | |
| Breast cancer | TEAD4 | increased expression | 63 | ||
| Breast cancer | TEAD2 | increased expression | EMT | zyxin | 65 |
| Colorectal cancer | TEAD2, TEAD4 | increased expression | EMT, correlated with decreased survival | vimentin | 66 |
| Cervical cancer, breast cancer | TEAD1 | resistance to apoptosis | livin | 67 | |
| Pancreatic cancer | TEAD1 | increased expression | mesothelin | 68 | |
| Melanoma | TEAD4 | increased invasiveness, resistance to MAPK inhibition | 69 |
Due to the important roles TEAD plays in cancer development and progression, inhibition of TEAD activity in cancers via small molecules and peptides has shown some efficacy in treating cancer in vivo and in vitro. Structural studies of TEAD reveal a central hydrophobic pocket in the transactivation domain [76]. Flufenmate drugs were found to bind in this hydrophobic pocket and inhibit TEAD transcriptional activity, without disrupting YAP-TEAD interaction, leading to decreases in cell migration and proliferation [76]. Palmitoylation of TEAD occurs in this hydrophobic pocket suggesting that flufenmate drugs, despite a low binding affinity, may inhibit TEAD activity by displacing TEAD palmitoylation [52, 53]. In YAP/TAZ driven cancers, studies have explored the effects of disrupting YAP/TAZ-TEAD interaction. Treatment with a VGLL4 mimicking peptide was found to inhibit gastric cancer growth in vitro and in vivo by outcompeting YAP for TEAD binding [21]. Verteporfin, a small molecule found to inhibit YAP-TEAD interaction, also suppressed cancer cell growth [77]. Thus, these attempts at inhibiting TEAD activity show that development of TEAD inhibitors is feasible and is a promising therapeutic strategy for cancer treatment.
Concluding Remarks
TEADs play an important role in development, differentiation, cell growth and proliferation, and tumorigenesis. Though the activity of TEAD is traditionally thought to be regulated through coactivator binding, with the majority of studies placing an emphasis on YAP/TAZ, several studies demonstrate Hippo independent mechanisms of TEAD regulation. Post-translational modifications such as phosphorylation and palmitoylation have been shown to effect TEAD DNA binding ability, protein stability, and coactivator interaction [52, 53, 78]. In addition, changes in TEAD subcellular localization represent an important mechanism to modulate TEAD transcriptional activity in a Hippo-independent manner [55, 56]. Importantly, these changes in TEAD subcellular localization, mediated by upstream regulators such as p38, influence YAP/TAZ localization [55, 56] (Figure 3). These studies show that not only can YAP/TAZ affect TEAD transcriptional activity, but TEAD localization can also regulate YAP/TAZ nuclear retention. Though exciting progress has been made towards understanding TEAD regulation, many key questions remain to be answered (see Outstanding Questions). Future work that elucidates mechanisms of TEAD regulation may be important in developing therapeutic options for cancers, particularly those that rely heavily on TEAD transcriptional activity.
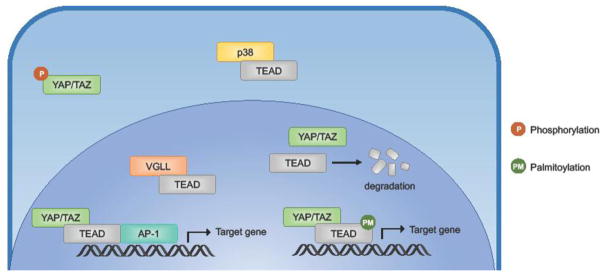
Figure 3. Mechanisms of TEAD regulation.
Several mechanisms have been shown to regulate TEAD transcriptional activity. Coactivator binding is the most important mechanism of altering TEAD transcriptional activity. YAP/TAZ bind TEAD along with AP-1 to activate transcription of downstream target genes. The transcriptional program driven by the YAP/TAZ, TEAD, AP-1 complex has been shown to be important for cancer progression. VGLL has been shown to compete with YAP/TAZ for TEAD binding. Availability of and competition between coactivators drive different TEAD transcriptional programs. Palmitoylation of TEAD in the central hydrophobic pocket is necessary for protein stability and is also suggested to be important for YAP binding. Osmotic stress acts via p38 to induce TEAD cytoplasmic translocation.
Outstanding Questions.
What signals effect TEAD post-translational modifications and subcellular localization? What are the upstream regulators that mediate these changes?
How are TEADs retained in the cytoplasm? Are there TEAD binding partners in the cytoplasm? Do TEADs have non-nuclear function?
How are TEADs regulated in YAP/TAZ independent cancers? What are other modulators that affect TEAD activity?
As individual TEAD expression levels are tissue specific, are all members of the TEAD family regulated by the same signals and mechanisms? Or are there TEAD isoform specific mechanisms?
Trends Box.
The TEAD family of transcription factors (TEAD1-4) is best studied in the context of Hippo signaling. TEADs are the primary transcription factors for the YAP/TAZ transcription co-activators of the Hippo pathway.
TEADs play an important role in development, tissue homeostasis, and tumorigenesis through regulation of processes such as cell growth and proliferation, differentiation, and survival. These processes are largely thought to be regulated by binding of YAP/TAZ.
Recent studies have uncovered new Hippo-independent mechanisms of TEAD regulation including post-translational modifications and changes in subcellular localization.
Acknowledgments
This work was supported by grants from the National Institutes of Health (CA196878, DE15964, and GM51586) to K.-L.G. and by a grant of the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (grant number: HI17C1560), and the National Research Foundation of Korea (NRF) grant funded by the Korea government (MOE) (2017R1D1A1B03034797) and (MSIP) (2017R1A4A1015328) to H.W.P. K.C.L. was supported in part by the University of California, San Diego (UCSD) Graduate Training Program in Cellular and Molecular Pharmacology (T32 GM007752).
Footnotes
Disclosure Statement: K.L.G. is a co-founder and has equity interest in Vivace Therapeutics, Inc., and OncoImmune, Inc. The terms of this arrangement have been reviewed and approved by the University of California, San Diego in accordance with its conflict of interest policies.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Xiao JH, et al. One cell-specific and three ubiquitous nuclear proteins bind in vitro to overlapping motifs in the domain B1 of the SV40 enhancer. Embo j. 1987;6(10):3005–13. doi: 10.1002/j.1460-2075.1987.tb02606.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davidson I, et al. The HeLa cell protein TEF-1 binds specifically and cooperatively to two SV40 enhancer motifs of unrelated sequence. Cell. 1988;54(7):931–42. doi: 10.1016/0092-8674(88)90108-0. [DOI] [PubMed] [Google Scholar]
- 3.Xiao JH, et al. Cloning, expression, and transcriptional properties of the human enhancer factor TEF-1. Cell. 1991;65(4):551–68. doi: 10.1016/0092-8674(91)90088-g. [DOI] [PubMed] [Google Scholar]
- 4.Ishiji T, et al. Transcriptional enhancer factor (TEF)-1 and its cell-specific co-activator activate human papillomavirus-16 E6 and E7 oncogene transcription in keratinocytes and cervical carcinoma cells. Embo j. 1992;11(6):2271–81. doi: 10.1002/j.1460-2075.1992.tb05286.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Azakie A, et al. DTEF-1, a novel member of the transcription enhancer factor-1 (TEF-1) multigene family. J Biol Chem. 1996;271(14):8260–5. doi: 10.1074/jbc.271.14.8260. [DOI] [PubMed] [Google Scholar]
- 6.Jin Y, et al. The role of transcription enhancer factors in cardiovascular biology. Trends Cardiovasc Med. 2011;21(1):1–5. doi: 10.1016/j.tcm.2011.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pobbati AV, Hong W. Emerging roles of TEAD transcription factors and its coactivators in cancers. Cancer Biol Ther. 2013;14(5):390–8. doi: 10.4161/cbt.23788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anbanandam A, et al. Insights into transcription enhancer factor 1 (TEF-1) activity from the solution structure of the TEA domain. Proc Natl Acad Sci U S A. 2006;103(46):17225–30. doi: 10.1073/pnas.0607171103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jacquemin P, et al. Human TEF-5 is preferentially expressed in placenta and binds to multiple functional elements of the human chorionic somatomammotropin-B gene enhancer. J Biol Chem. 1997;272(20):12928–37. doi: 10.1074/jbc.272.20.12928. [DOI] [PubMed] [Google Scholar]
- 10.Jacquemin P, et al. A novel family of developmentally regulated mammalian transcription factors containing the TEA/ATTS DNA binding domain. J Biol Chem. 1996;271(36):21775–85. doi: 10.1074/jbc.271.36.21775. [DOI] [PubMed] [Google Scholar]
- 11.Chen Z, et al. Transcriptional enhancer factor 1 disruption by a retroviral gene trap leads to heart defects and embryonic lethality in mice. Genes Dev. 1994;8(19):2293–301. doi: 10.1101/gad.8.19.2293. [DOI] [PubMed] [Google Scholar]
- 12.Kaneko KJ, et al. Transcription factor TEAD2 is involved in neural tube closure. Genesis. 2007;45(9):577–87. doi: 10.1002/dvg.20330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sawada A, et al. Redundant roles of Tead1 and Tead2 in notochord development and the regulation of cell proliferation and survival. Mol Cell Biol. 2008;28(10):3177–89. doi: 10.1128/MCB.01759-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yagi R, et al. Transcription factor TEAD4 specifies the trophectoderm lineage at the beginning of mammalian development. Development. 2007;134(21):3827–36. doi: 10.1242/dev.010223. [DOI] [PubMed] [Google Scholar]
- 15.Nishioka N, et al. Tead4 is required for specification of trophectoderm in pre-implantation mouse embryos. Mech Dev. 2008;125(3–4):270–83. doi: 10.1016/j.mod.2007.11.002. [DOI] [PubMed] [Google Scholar]
- 16.Fossdal R, et al. A novel TEAD1 mutation is the causative allele in Sveinsson’s chorioretinal atrophy (helicoid peripapillary chorioretinal degeneration) Hum Mol Genet. 2004;13(9):975–81. doi: 10.1093/hmg/ddh106. [DOI] [PubMed] [Google Scholar]
- 17.Kitagawa M. A Sveinsson’s chorioretinal atrophy-associated missense mutation in mouse Tead1 affects its interaction with the co-factors YAP and TAZ. Biochem Biophys Res Commun. 2007;361(4):1022–6. doi: 10.1016/j.bbrc.2007.07.129. [DOI] [PubMed] [Google Scholar]
- 18.Meng Z, et al. Mechanisms of Hippo pathway regulation. Genes Dev. 2016;30(1):1–17. doi: 10.1101/gad.274027.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burglin TR. The TEA domain: a novel, highly conserved DNA-binding motif. Cell. 1991;66(1):11–2. doi: 10.1016/0092-8674(91)90132-i. [DOI] [PubMed] [Google Scholar]
- 20.Vassilev A, et al. TEAD/TEF transcription factors utilize the activation domain of YAP65, a Src/Yes-associated protein localized in the cytoplasm. Genes Dev. 2001;15(10):1229–41. doi: 10.1101/gad.888601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiao S, et al. A peptide mimicking VGLL4 function acts as a YAP antagonist therapy against gastric cancer. Cancer Cell. 2014;25(2):166–80. doi: 10.1016/j.ccr.2014.01.010. [DOI] [PubMed] [Google Scholar]
- 22.Farrance IK, et al. M-CAT binding factor is related to the SV40 enhancer binding factor, TEF-1. J Biol Chem. 1992;267(24):17234–40. [PubMed] [Google Scholar]
- 23.Chen L, et al. Structural basis of YAP recognition by TEAD4 in the hippo pathway. Genes Dev. 2010;24(3):290–300. doi: 10.1101/gad.1865310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Z, et al. Structural insights into the YAP and TEAD complex. Genes Dev. 2010;24(3):235–40. doi: 10.1101/gad.1865810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shi Z, et al. DNA-binding mechanism of the Hippo pathway transcription factor TEAD4. Oncogene. 2017 doi: 10.1038/onc.2017.24. [DOI] [PubMed] [Google Scholar]
- 26.Mahoney WM, Jr, et al. The transcriptional co-activator TAZ interacts differentially with transcriptional enhancer factor-1 (TEF-1) family members. Biochem J. 2005;388(Pt 1):217–25. doi: 10.1042/BJ20041434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao B, et al. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 2008;22(14):1962–71. doi: 10.1101/gad.1664408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lai D, et al. Taxol resistance in breast cancer cells is mediated by the hippo pathway component TAZ and its downstream transcriptional targets Cyr61 and CTGF. Cancer Res. 2011;71(7):2728–38. doi: 10.1158/0008-5472.CAN-10-2711. [DOI] [PubMed] [Google Scholar]
- 29.Lei QY, et al. TAZ promotes cell proliferation and epithelial-mesenchymal transition and is inhibited by the hippo pathway. Mol Cell Biol. 2008;28(7):2426–36. doi: 10.1128/MCB.01874-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaan HYK, et al. Crystal structure of TAZ-TEAD complex reveals a distinct interaction mode from that of YAP-TEAD complex. Sci Rep. 2017;7(1):2035. doi: 10.1038/s41598-017-02219-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huttlin EL, et al. Architecture of the human interactome defines protein communities and disease networks. Nature. 2017;545(7655):505–509. doi: 10.1038/nature22366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huttlin EL, et al. The BioPlex Network: A Systematic Exploration of the Human Interactome. Cell. 2015;162(2):425–40. doi: 10.1016/j.cell.2015.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chan SW, et al. TEADs mediate nuclear retention of TAZ to promote oncogenic transformation. J Biol Chem. 2009;284(21):14347–58. doi: 10.1074/jbc.M901568200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang H, et al. TEAD transcription factors mediate the function of TAZ in cell growth and epithelial-mesenchymal transition. J Biol Chem. 2009;284(20):13355–62. doi: 10.1074/jbc.M900843200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen HH, et al. Transcription cofactor Vgl-2 is required for skeletal muscle differentiation. Genesis. 2004;39(4):273–9. doi: 10.1002/gene.20055. [DOI] [PubMed] [Google Scholar]
- 36.Chen HH, et al. Vgl-4, a novel member of the vestigial-like family of transcription cofactors, regulates alpha1-adrenergic activation of gene expression in cardiac myocytes. J Biol Chem. 2004;279(29):30800–6. doi: 10.1074/jbc.M400154200. [DOI] [PubMed] [Google Scholar]
- 37.Gunther S, et al. VITO-1 is an essential cofactor of TEF1-dependent muscle-specific gene regulation. Nucleic Acids Res. 2004;32(2):791–802. doi: 10.1093/nar/gkh248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koontz LM, et al. The Hippo effector Yorkie controls normal tissue growth by antagonizing scalloped-mediated default repression. Dev Cell. 2013;25(4):388–401. doi: 10.1016/j.devcel.2013.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pobbati AV, et al. Structural and functional similarity between the Vgll1-TEAD and the YAP-TEAD complexes. Structure. 2012;20(7):1135–40. doi: 10.1016/j.str.2012.04.004. [DOI] [PubMed] [Google Scholar]
- 40.Zhang W, et al. VGLL4 functions as a new tumor suppressor in lung cancer by negatively regulating the YAP-TEAD transcriptional complex. Cell Res. 2014;24(3):331–43. doi: 10.1038/cr.2014.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jiao S, et al. VGLL4 targets a TCF4-TEAD4 complex to coregulate Wnt and Hippo signalling in colorectal cancer. Nat Commun. 2017;8:14058. doi: 10.1038/ncomms14058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Belandia B, Parker MG. Functional interaction between the p160 coactivator proteins and the transcriptional enhancer factor family of transcription factors. J Biol Chem. 2000;275(40):30801–5. doi: 10.1074/jbc.C000484200. [DOI] [PubMed] [Google Scholar]
- 43.Zanconato F, et al. Genome-wide association between YAP/TAZ/TEAD and AP-1 at enhancers drives oncogenic growth. Nat Cell Biol. 2015;17(9):1218–27. doi: 10.1038/ncb3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu X, et al. Tead and AP1 Coordinate Transcription and Motility. Cell Rep. 2016;14(5):1169–80. doi: 10.1016/j.celrep.2015.12.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Butler AJ, Ordahl CP. Poly(ADP-ribose) polymerase binds with transcription enhancer factor 1 to MCAT1 elements to regulate muscle-specific transcription. Mol Cell Biol. 1999;19(1):296–306. doi: 10.1128/mcb.19.1.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.MacLellan WR, et al. Transforming growth factor-beta response elements of the skeletal alpha-actin gene. Combinatorial action of serum response factor, YY1, and the SV40 enhancer-binding protein, TEF-1. J Biol Chem. 1994;269(24):16754–60. [PubMed] [Google Scholar]
- 47.Maeda T, et al. Mammalian vestigial-like 2, a cofactor of TEF-1 and MEF2 transcription factors that promotes skeletal muscle differentiation. J Biol Chem. 2002;277(50):48889–98. doi: 10.1074/jbc.M206858200. [DOI] [PubMed] [Google Scholar]
- 48.Gupta MP, et al. Transcription enhancer factor 1 interacts with a basic helix-loop-helix zipper protein, Max, for positive regulation of cardiac alpha-myosin heavy-chain gene expression. Mol Cell Biol. 1997;17(7):3924–36. doi: 10.1128/mcb.17.7.3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gupta MP, et al. An E-box/M-CAT hybrid motif and cognate binding protein(s) regulate the basal muscle-specific and cAMP-inducible expression of the rat cardiac alpha-myosin heavy chain gene. J Biol Chem. 1994;269(47):29677–87. [PubMed] [Google Scholar]
- 50.Gupta MP, et al. Protein kinase-A dependent phosphorylation of transcription enhancer factor-1 represses its DNA-binding activity but enhances its gene activation ability. Nucleic Acids Res. 2000;28(16):3168–77. doi: 10.1093/nar/28.16.3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jiang SW, et al. DNA binding of TEA/ATTS domain factors is regulated by protein kinase C phosphorylation in human choriocarcinoma cells. J Biol Chem. 2001;276(26):23464–70. doi: 10.1074/jbc.M010934200. [DOI] [PubMed] [Google Scholar]
- 52.Noland CL, et al. Palmitoylation of TEAD Transcription Factors Is Required for Their Stability and Function in Hippo Pathway Signaling. Structure. 2016;24(1):179–86. doi: 10.1016/j.str.2015.11.005. [DOI] [PubMed] [Google Scholar]
- 53.Chan P, et al. Autopalmitoylation of TEAD proteins regulates transcriptional output of the Hippo pathway. Nat Chem Biol. 2016;12(4):282–9. doi: 10.1038/nchembio.2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Resh MD. Trafficking and signaling by fatty-acylated and prenylated proteins. Nat Chem Biol. 2006;2(11):584–90. doi: 10.1038/nchembio834. [DOI] [PubMed] [Google Scholar]
- 55.Home P, et al. Altered subcellular localization of transcription factor TEAD4 regulates first mammalian cell lineage commitment. Proc Natl Acad Sci U S A. 2012;109(19):7362–7. doi: 10.1073/pnas.1201595109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lin KC, et al. Regulation of Hippo pathway transcription factor TEAD by p38 MAPK-induced cytoplasmic translocation. Nat Cell Biol. 2017;19(8):996–1002. doi: 10.1038/ncb3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hong AW, et al. Osmotic stress-induced phosphorylation by NLK at Ser128 activates YAP. EMBO Rep. 2017;18(1):72–86. doi: 10.15252/embr.201642681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Moon S, et al. Phosphorylation by NLK inhibits YAP-14-3-3-interactions and induces its nuclear localization. EMBO Rep. 2017;18(1):61–71. doi: 10.15252/embr.201642683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cargnello M, Roux PP. Activation and function of the MAPKs and their substrates, the MAPK-activated protein kinases. Microbiol Mol Biol Rev. 2011;75(1):50–83. doi: 10.1128/MMBR.00031-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ben-Levy R, et al. Nuclear export of the stress-activated protein kinase p38 mediated by its substrate MAPKAP kinase-2. Curr Biol. 1998;8(19):1049–57. doi: 10.1016/s0960-9822(98)70442-7. [DOI] [PubMed] [Google Scholar]
- 61.Goulev Y, et al. SCALLOPED interacts with YORKIE, the nuclear effector of the hippo tumor-suppressor pathway in Drosophila. Curr Biol. 2008;18(6):435–41. doi: 10.1016/j.cub.2008.02.034. [DOI] [PubMed] [Google Scholar]
- 62.Zhang L, et al. The TEAD/TEF family of transcription factor Scalloped mediates Hippo signaling in organ size control. Dev Cell. 2008;14(3):377–87. doi: 10.1016/j.devcel.2008.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wu S, et al. The TEAD/TEF family protein Scalloped mediates transcriptional output of the Hippo growth-regulatory pathway. Dev Cell. 2008;14(3):388–98. doi: 10.1016/j.devcel.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 64.Huang J, et al. The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila Homolog of YAP. Cell. 2005;122(3):421–34. doi: 10.1016/j.cell.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 65.Cagliero J, et al. The Hippo kinase promotes Scalloped cytoplasmic localization independently of Warts in a CRM1/Exportin1-dependent manner in Drosophila. Faseb j. 2013;27(4):1330–41. doi: 10.1096/fj.12-216424. [DOI] [PubMed] [Google Scholar]
- 66.Moroishi T, et al. The emerging roles of YAP and TAZ in cancer. Nat Rev Cancer. 2015;15(2):73–9. doi: 10.1038/nrc3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Knight JF, et al. TEAD1 and c-Cbl are novel prostate basal cell markers that correlate with poor clinical outcome in prostate cancer. Br J Cancer. 2008;99(11):1849–58. doi: 10.1038/sj.bjc.6604774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Han W, et al. DNA copy number alterations and expression of relevant genes in triple-negative breast cancer. Genes Chromosomes Cancer. 2008;47(6):490–9. doi: 10.1002/gcc.20550. [DOI] [PubMed] [Google Scholar]
- 69.Richardson AL, et al. X chromosomal abnormalities in basal-like human breast cancer. Cancer Cell. 2006;9(2):121–32. doi: 10.1016/j.ccr.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 70.Diepenbruck M, et al. Tead2 expression levels control the subcellular distribution of Yap and Taz, zyxin expression and epithelial-mesenchymal transition. J Cell Sci. 2014;127(Pt 7):1523–36. doi: 10.1242/jcs.139865. [DOI] [PubMed] [Google Scholar]
- 71.Liu Y, et al. Increased TEAD4 expression and nuclear localization in colorectal cancer promote epithelial-mesenchymal transition and metastasis in a YAP-independent manner. Oncogene. 2016;35(21):2789–800. doi: 10.1038/onc.2015.342. [DOI] [PubMed] [Google Scholar]
- 72.Landin Malt A, et al. Alteration of TEAD1 expression levels confers apoptotic resistance through the transcriptional up-regulation of Livin. PLoS One. 2012;7(9):e45498. doi: 10.1371/journal.pone.0045498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hucl T, et al. High cancer-specific expression of mesothelin (MSLN) is attributable to an upstream enhancer containing a transcription enhancer factor dependent MCAT motif. Cancer Res. 2007;67(19):9055–65. doi: 10.1158/0008-5472.CAN-07-0474. [DOI] [PubMed] [Google Scholar]
- 74.Hu Y, et al. Superenhancer reprogramming drives a B-cell-epithelial transition and high-risk leukemia. Genes Dev. 2016;30(17):1971–90. doi: 10.1101/gad.283762.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Verfaillie A, et al. Decoding the regulatory landscape of melanoma reveals TEADS as regulators of the invasive cell state. Nat Commun. 2015;6:6683. doi: 10.1038/ncomms7683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pobbati AV, et al. Targeting the Central Pocket in Human Transcription Factor TEAD as a Potential Cancer Therapeutic Strategy. Structure. 2015;23(11):2076–86. doi: 10.1016/j.str.2015.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Liu-Chittenden Y, et al. Genetic and pharmacological disruption of the TEAD-YAP complex suppresses the oncogenic activity of YAP. Genes Dev. 2012;26(12):1300–5. doi: 10.1101/gad.192856.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yu FX, et al. Protein kinase A activates the Hippo pathway to modulate cell proliferation and differentiation. Genes Dev. 2013;27(11):1223–32. doi: 10.1101/gad.219402.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Johnson R, Halder G. The two faces of Hippo: targeting the Hippo pathway for regenerative medicine and cancer treatment. Nat Rev Drug Discov. 2014;13(1):63–79. doi: 10.1038/nrd4161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mo JS, et al. The Hippo signaling pathway in stem cell biology and cancer. EMBO Rep. 2014;15(6):642–56. doi: 10.15252/embr.201438638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hansen CG, et al. YAP and TAZ: a nexus for Hippo signaling and beyond. Trends Cell Biol. 2015;25(9):499–513. doi: 10.1016/j.tcb.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Callus BA, et al. Association of mammalian sterile twenty kinases, Mst1 and Mst2, with hSalvador via C-terminal coiled-coil domains, leads to its stabilization and phosphorylation. Febs j. 2006;273(18):4264–76. doi: 10.1111/j.1742-4658.2006.05427.x. [DOI] [PubMed] [Google Scholar]
- 83.Praskova M, et al. MOBKL1A/MOBKL1B phosphorylation by MST1 and MST2 inhibits cell proliferation. Curr Biol. 2008;18(5):311–21. doi: 10.1016/j.cub.2008.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tapon N, et al. salvador Promotes both cell cycle exit and apoptosis in Drosophila and is mutated in human cancer cell lines. Cell. 2002;110(4):467–78. doi: 10.1016/s0092-8674(02)00824-3. [DOI] [PubMed] [Google Scholar]
- 85.Wu S, et al. hippo encodes a Ste-20 family protein kinase that restricts cell proliferation and promotes apoptosis in conjunction with salvador and warts. Cell. 2003;114(4):445–56. doi: 10.1016/s0092-8674(03)00549-x. [DOI] [PubMed] [Google Scholar]
- 86.Meng Z, et al. MAP4K family kinases act in parallel to MST1/2 to activate LATS1/2 in the Hippo pathway. Nat Commun. 2015;6:8357. doi: 10.1038/ncomms9357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhao B, et al. A coordinated phosphorylation by Lats and CK1 regulates YAP stability through SCF(beta-TRCP) Genes Dev. 2010;24(1):72–85. doi: 10.1101/gad.1843810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhao B, et al. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 2007;21(21):2747–61. doi: 10.1101/gad.1602907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Liu CY, et al. The hippo tumor pathway promotes TAZ degradation by phosphorylating a phosphodegron and recruiting the SCF{beta}-TrCP E3 ligase. J Biol Chem. 2010;285(48):37159–69. doi: 10.1074/jbc.M110.152942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hao Y, et al. Tumor suppressor LATS1 is a negative regulator of oncogene YAP. J Biol Chem. 2008;283(9):5496–509. doi: 10.1074/jbc.M709037200. [DOI] [PubMed] [Google Scholar]