Abstract
10.1601/nm.2199 CLST is an extremely acidophilic gamma-proteobacteria that was isolated from the Gorbea salt flat, an acidic hypersaline environment in northern Chile. This kind of environment is considered a terrestrial analog of ancient Martian terrains and a source of new material for biotechnological applications. 10.1601/nm.2199 plays a key role in industrial bioleaching; it has the capacity of generating and maintaining acidic conditions by producing sulfuric acid and it can also remove sulfur layers from the surface of minerals, which are detrimental for their dissolution. CLST is a strain of 10.1601/nm.2199 able to tolerate moderate chloride concentrations (up to 15 g L−1 Cl−), a feature that is quite unusual in extreme acidophilic microorganisms. Basic microbiological features and genomic properties of this biotechnologically relevant strain are described in this work. The 3,974,949 bp draft genome is arranged into 40 scaffolds of 389 contigs containing 3866 protein-coding genes and 75 RNAs encoding genes. This is the first draft genome of a halotolerant 10.1601/nm.2199 strain. The release of the genome sequence of this strain improves representation of these extreme acidophilic Gram negative bacteria in public databases and strengthens the framework for further investigation of the physiological diversity and ecological function of 10.1601/nm.2199 populations.
Electronic supplementary material
The online version of this article (10.1186/s40793-017-0305-8) contains supplementary material, which is available to authorized users.
Keywords: Acidithiobacillaceae, Halotolerance, Osmotolerance, Sulfur oxidization, Flexible gene complement, Bioleaching, Mars analog, Salar de Gorbea
Introduction
The genus 10.1601/nm.2198 comprises a group of obligatory acidophilic, Gram negative, rod shaped bacteria that derive energy from the aerobic oxidation of reduced sulfur compounds (RISCs) to support autotrophic growth. In the process of oxidizing RISCs, these bacteria produce sulfuric acid and contribute to the bioleaching of ores. Currently, the genus comprises seven described species, 10.1601/nm.2199 ATCC 19377, 10.1601/nm.2202 ATCC2327, 10.1601/nm.2200 ATCC35403, 10.1601/nm.2201 DSM 8584 [1], 10.1601/nm.17776 [2], Acidtithiobacillus ferridurans [3] and 10.1601/nm.27980 [4]. Despite being the first acidophile ever isolated [5], 10.1601/nm.2199 investigation lags behind other members of the genus, especially when compared to the iron oxidizer 10.1601/nm.2202, for which extensive knowledge on its basic ecophysiology and biotechnological use has been gathered [6].
The draft genomes of ten isolates of 10.1601/nm.2199 are available: the type strain 10.1601/strainfinder?urlappend=%3Fid%3DATCC+19377 obtained from the Kimmeridge clay formation in England [7], the strain DSM 17318 named Licanantay isolated from a copper mine in northern Chile [8], the A01 strain isolated from wastewater of a coal dump in China [9] and seven other isolates obtained from copper mines (BY-02, DXS-W, GD1-3, TYC-17, ZBY) and coal heaps (A02, DMC) in China [10].
The 10.1601/nm.2199 type strain (10.1601/strainfinder?urlappend=%3Fid%3DATCC+19377) is motile, grows on elemental sulfur, thiosulfate or tetrathionate, and has temperature optimum of 30 °C and a pH optimum of 2.0 to 3.0 [1]. Members of the species have been found to occur in a variety of natural-acidic and man-made environments, including sulfidic caves [11], shales [12], fresh water [13], sea water [14], sewer pipes [15], mineral leaching heaps [16], mine dumps [17] and mine wastes [18] from different parts of the world. With the exception of 10.1601/nm.2199 strain SH isolated from sea water, which has a confirmed requirement of NaCl (2%; 0.35 M) for growth in synthetic media [14], all characterized 10.1601/nm.2199 strains are inhibited by even moderate NaCl concentrations [19].
10.1601/nm.2199 CLST is a new NaCl tolerant strain (15 g L−1 Cl−) isolated from the Gorbea salt flat in the Central Andean plateau (Bolivia, Chile and Argentina, between 19° and 27° S latitude). This salt flat is located in an endorheic basin displaying strongly acidic brines (with a pH between 2 and 4 and a salinity ranging between 1.7 - 76.9 g L−1 NaCl) and one of the few acid saline systems known worldwide [20–22]. These uncommon types of natural extreme environments are considered terrestrial analogs to certain ancient Martian terrains and a source of new material for biotechnological applications [23, 24].
This work reports the microbiological properties of this NaCl-tolerant acidophilic sulfur-oxidizing 10.1601/nm.2198 from the saline environment in northern Chile, together with its draft genomic sequence and annotation. The release of the genome of the CLST strain will contribute to a better understanding of the ecophysiology of extreme acidophiles inhabiting saline environments and of sodium-requiring processes (e.g. symport, antiport, flagellar rotation, etc.), in acidophilic chemolithotrophic bacteria. Knowledge derived from the study may also provide new opportunities in biotechnological and astrobiological endeavors.
Organism information
Classification and features
10.1601/nm.2199 CLST was isolated at the Biotechnology Center (CBAR-UCN) from a sulfur enrichment culture designed to select acidophilic bacteria that could oxidize RISCs under saline conditions. Briefly, salt-water samples obtained from the Gorbea salt flat were inoculated in a batch reactor containing minimal medium [25] and elemental sulfur as energy source. Phylogenetic analysis of the 16S rRNA sequence indicated that the CLST strain (10.1601/strainfinder?urlappend=%3Fid%3DDSM+103717) is related to 10.1601/nm.2199 (Fig. 1). CLST cells are Gram-negative, rod-shaped (0.4 μm × 1-1.5 μm) and motile (Fig. 2). Optimal growth occurs at 28 °C and pH 1.7. It grows autotrophically using sulfur as electron donor and oxygen as the electron acceptor. It is also a facultative anaerobe capable of using RISCs as electron donors and ferric iron as an electron acceptor. Strain CLST forms small white colonies when grown autotrophically on solid medium containing RISCs. It differs from closely related strains, Licanantay and A01 (JMEB00000000 and FJ154514, respectively), in its capacity to grow in 15 g L−1 of chloride. The microorganism information is presented in Table 1.
Fig. 1.
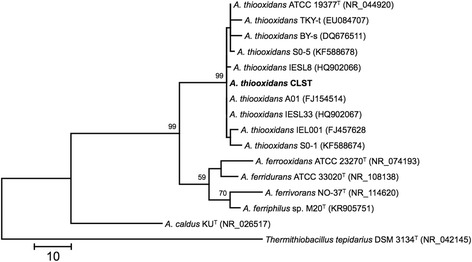
Phylogenetic tree based on the 16S rRNA gene sequences highlighting the position of 10.1601/nm.2199 strain CLST relative to other type and non-type strains of the genus 10.1601/nm.2198. The GenBank database accession codes are indicated between brackets. The evolutionary history was inferred by using the Maximum Parsimony and the Subtree-Pruning-Regrafting (SPR) algorithm with search level 1 [52]. The initial trees were obtained by the random addition of sequences. The analysis involved 16 nucleotide sequences and a total of 1307 non-ambiguous positions in the final dataset. Evolutionary analyses were conducted in MEGA version 6.22 [53]. Tree construction used a bootstrapping process repeated 1000 times to generate a majority consensus tree. A sequence from 10.1601/nm.2205 was used as outgroup. The tree is drawn to scale, with branch lengths calculated using the average pathway method [53]; the scale bar corresponds to the number of changes over the whole sequence
Fig. 2.

Scanning electron microscopy image of 10.1601/nm.2199 strain CLST
Table 1.
Classification and general features of 10.1601/nm.2199 CLST
| MIGS ID | Property | Term | Evidence codea |
|---|---|---|---|
| Classification | Domain Bacteria | TAS [44] | |
| Phylum 10.1601/nm.808 | TAS [45] | ||
| Class 10.1601/nm.24436 | TAS [46] | ||
| Order 10.1601/nm.2196 | TAS [38] | ||
| Family 10.1601/nm.2197 | TAS [39, 47] | ||
| Genus 10.1601/nm.2198 | TAS [1] | ||
| Species 10.1601/nm.2199 | TAS [1, 5] | ||
| Strain: CLST (10.1601/strainfinder?urlappend=%3Fid%3DDSM+103717) | IDA | ||
| Gram stain | Negative | IDA | |
| Cell shape | Rod | IDA | |
| Motility | Motile | IDA | |
| Sporulation | Not reported | IDA | |
| Temperature range | 25 -35 °C | IDA | |
| Optimum temperature | 28 °C | IDA | |
| Optimum pH | 1.7 | IDA | |
| Carbon source | CO2 | TAS [25] | |
| MIGS-6 | Habitat | Brine, acidic hypersaline environment | IDA |
| MIGS-6.3 | Salinity | 10 -15 gL−1 chloride | IDA |
| MIGS-22 | Oxygen requirement | Aerobic and facultative anaerobic | IDA |
| MIGS-15 | Biotic relationship | Free-living | TAS [48] |
| MIGS-14 | Pathogenicity | Non-pathogen | NAS |
| MIGS-4 | Geographic location | Gorbea salt flat, Antofagasta region, Chile | IDA |
| MIGS-5 | Sample collection | 11/20/2007 | IDA |
| MIGS-4.1 | Latitude | 25°25′72.2´´S | IDA |
| MIGS-4.2 | Longitude | 68°41′53.2´´W | IDA |
| MIGS-4.4 | Altitude | 4000 m.a.s.l. | IDA |
aEvidence codes - IDA Inferred from Direct Assay, TAS Traceable Author Statement (i.e., a direct report exists in the literature), NAS Non-traceable Author Statement (i.e., not directly observed for the living, isolated sample, but based on a generally accepted property for the species, or anecdotal evidence). These evidence codes are from the Gene Ontology project [49]. Data is in compliance with MIGS version 2.0 [50] and the NamesforLife database [51]
Extended feature descriptions
The growth rate of 10.1601/nm.2199 type strain 10.1601/strainfinder?urlappend=%3Fid%3DATCC+19377 undergoes a significant decrease (μ from 0.76 to 0.52 day−1) at NaCl concentration of 325 mM compared with growth on culture medium without the salt (Additional file 1: Figure S1). Meanwhile there is not a significant change in the growth rate of 10.1601/nm.2199 CLST in the same conditions. In addition 10.1601/nm.2199 CLST precipitates CuS when it is grown aerobically in culture medium amended with CuSO4 (Additional file 2: Figure S2). This feature has been already observed in E. coli associated to the heterologous expression of the enzyme cysteine desulfhydrase [26]. We identified the gene for a previously described cysteine desulfhydrase (CdsH) in the genome of 10.1601/nm.2199 CLST strain. CdsH appears to be the major cysteine-degrading and sulfide-producing enzyme aerobically but not anaerobically [27].
Genome sequencing information
Genome project history
The organism was selected for sequencing on the basis of its phylogenetic position and 16S rRNA similarity to members of the genus 10.1601/nm.2198, and for its atypical origin; coming from an extreme acidic and saline biotope. This Whole Genome Shotgun project has been deposited at GenBank under the accession number LGYM00000000. The version described in this paper is the first version, LGYM00000000. The project information is presented in Table 2.
Table 2.
Project information
| MIGS ID | Property | Term |
|---|---|---|
| MIGS 31 | Finishing quality | Draft |
| MIGS-28 | Libraries used | GS FLX Titanium paired end libraries |
| MIGS 29 | Sequencing platforms | Roche 454 GS FLX |
| MIGS 31.2 | Fold coverage | 36 × |
| MIGS 30 | Assemblers | Newbler 2.0.01.14 |
| MIGS 32 | Gene calling method | Glimmer 3.02 |
| Genbank ID | LGYM00000000 | |
| GenBank Date of Release | 2017-04-05 | |
| GOLD ID | Gp0136483 | |
| BIOPROJECT | PRJNA291500 | |
| MIGS 13 | Source Material Identifier | Gorbea-A |
| Project relevance | Territorial biodiversity, Tree of Life, Biomining, Astrobiology |
Growth conditions and genomic DNA preparation
The culture obtained from this reactor grew at 15 g L−1 Cl− and exhibited sulfur oxidizing activity. Strain CLST was isolated by plating the reactors culture medium using Phytagel 1% as gelling agent. Strain CLST was grown in minimal medium (0.4 g L−1, (NH4) 2SO4, 0.4 g L−1, MgSO4 × 7H2O, 0.2 g L−1, K2HPO4 and 3.93 g L−1, CuSO4, pH 1.7) containing NaCl (24.7 g L−1). After successive subculturing (three times), DNA was isolated using High Pure Template Preparation Kit (Roche, Germany) according to the manufacturer instructions.
Genome sequencing and assembly
The genome of 10.1601/nm.2199 strain CLST was sequenced at Beckman Coulter Genomics using 454 sequencing technology and mate pair libraries with insert sizes of ~500 bp [28]. Pyrosequencing reads were assembled de novo using Newbler (v2.0.01.14). The final draft assembly contained 389 contigs in 40 scaffolds ranging in size from 2298 bp to 409,853 bp. The total size of the genome is ~3,9 Mbp and the final assembly is based on 82 Mbp of 454 data, which provides an average 36× coverage of the genome.
Genome annotation
Genes were predicted using Glimmer 3.02 [29] as part of the RAST annotation pipeline [30]. The tRNA and tmRNA identification was achieved using ARAGORN v1.2.36 [31] and the rRNA prediction was carried out with HMMER3 [32]. Additional gene prediction analysis and manual functional annotation was performed at the Center for Bioinformatics and Genome Biology (CBGB-FCV). The predicted CDSs were used to search the NCBI non-redundant database, UniProt, TIGRFam, Pfam, PRIAM, KEGG, COG and InterPro databases. Protein coding genes were analyzed for the presence of signal peptides using SignalP v4.1 [33] and transmembrane helices using TMHMM v2.0 [34].
Genome properties
The draft genome contains 3,974,949 nucleotides and has an average G + C content of 48.8% (Table 3). From a total of 3941 genes, 3866 are predicted to be protein coding genes and 75 are RNA genes. The RNA genes partitioned into 1 tmRNA, 1 rRNA operon and 71 tRNAs distributed in 17 scaffolds (40% of which map to a single scaffold), suggesting the presence of an additional complete set of tRNAs as in the case of strain Licanantay [8] and 10.1601/nm.2202 type strain (10.1601/strainfinder?urlappend=%3Fid%3DATCC+23270) [35]. Predicted protein functional distributions follow highly similar profiles of other 10.1601/nm.2199 sequenced strains according to COG classification, with 36% of the genes being related to metabolism, 26% to information flux and 15% to cellular structure maintenance. A total of 43.63% of the genes were assigned a putative function while the remaining were annotated as hypotheticals. The distribution of genes in COGs functional categories is presented in Table 4.
Table 3.
Genome statistics
| Attribute | Value | % of Totala |
|---|---|---|
| Genome size (bp) | 3,974,949 | 100.00 |
| DNA coding (bp) | 3,051,435 | 76.76 |
| DNA G + C (bp) | 1,939,775 | 48.80 |
| DNA scaffolds | 40 | 100.00 |
| Total genesb | 3941 | 100.00 |
| Protein coding genes | 3866 | 98.09 |
| RNA genesc | 75 | 1.91 |
| Pseudo genesd | n.d. | n.d. |
| Genes in internal clusters | 2118 | 54.78 |
| Genes with function prediction | 2468 | 63.38 |
| Genes assigned to COGs | 1803 | 46.63 |
| Genes with Pfam domains | 2634 | 68.13 |
| Genes with signal peptides | 292 | 7.55 |
| Genes with transmembrane helices | 880 | 22.33 |
| CRISPR repeats | 0 | 0.00 |
aThe total is based on either the size of the genome in base pairs or the total number of genes in the annotated genome
bIncludes tRNA, tmRNA, rRNA
cIncludes 23S, 16S and 5S rRNA
dn.d.: Not determined
Table 4.
Number of genes associated with general COG functional categories
| Code | Value | %age | Description |
|---|---|---|---|
| J | 165 | 4.27 | Translation, ribosomal structure and biogenesis |
| A | 1 | 0.03 | RNA processing and modification |
| K | 102 | 2.64 | Transcription |
| L | 121 | 3.13 | Replication, recombination and repair |
| B | 0 | 0.00 | Chromatin structure and dynamics |
| D | 31 | 0.80 | Cell cycle control, Cell division, chromosome partitioning |
| V | 59 | 1.53 | Defense mechanisms |
| T | 111 | 2.87 | Signal transduction mechanisms |
| M | 138 | 3.57 | Cell wall/membrane biogenesis |
| N | 69 | 1.78 | Cell motility |
| U | 46 | 1.19 | Intracellular trafficking and secretion |
| O | 88 | 2.28 | Posttranslational modification, protein turnover, chaperones |
| C | 124 | 3.21 | Energy production and conversion |
| G | 75 | 1.94 | Carbohydrate transport and metabolism |
| E | 116 | 3.00 | Amino acid transport and metabolism |
| F | 56 | 1.45 | Nucleotide transport and metabolism |
| H | 103 | 2.66 | Coenzyme transport and metabolism |
| I | 53 | 1.37 | Lipid transport and metabolism |
| P | 93 | 2.41 | Inorganic ion transport and metabolism |
| Q | 22 | 0.57 | Secondary metabolites biosynthesis, transport and catabolism |
| R | 99 | 2.56 | General function prediction only |
| S | 131 | 3.38 | Function unknown |
| – | 2063 | 53.36 | Not in COGs |
The total is based on the total number of protein coding genes in the genome
Insights from the genome sequence
10.1601/nm.2199 CLST predicted gene complement was compared against the genome of the type strain of the species (10.1601/strainfinder?urlappend=%3Fid%3DATCC+19377) and the publically available draft genomes of nine additional strains using the sequence based comparison tools of RAST [36, 37]. CLST shares 86% of its gene complement with the most similar strain in the set (Licanantay) and little over 70% with the type strain of the species (10.1601/strainfinder?urlappend=%3Fid%3DATCC+19377 T). All diagnostic features of 10.1601/nm.2199 strains [1, 38, 39] are encoded in the core genome, and have been described elsewhere [7–10]. The exclusive gene complement of strain CLST encompasses 200 protein-coding genes, 95% of which are hypotheticals. An additional 1234 genes are partially shared with a subset of the strains under comparison (Fig. 3) and thus constitute the flexible gene complement. A number of these exclusive genes can be linked to osmotolerance responses, including active uptake of potassium (kdpFABC), synthesis of the counterion glutamate (glutamate synthase), synthesis of compatible solutes such as the aminoacid Proline (proQ) and possibly also polyamines (carbamoyl-phosphate synthase). Several genes involved in mitigation of other types of stress also formed part of the flexible gene pool of the CLST strain, including the ruberythrin gene cluster and a non-heme chloroperoxidase involved in oxidative stress resistance [40], copper and mercury resistance genes to withstand metal toxicity [41] and genes for the export of protective extracellular polysaccharides (kps system) [42]. Besides, these functions and an extensive number of hypotheticals, the CLST flexible gene complement also includes a variety of functions linked mobile genetic elements of diverse nature [43], suggesting that many of the differentiating features of CLST may have been horizontally transferred from other members of the microbial community.
Fig. 3.
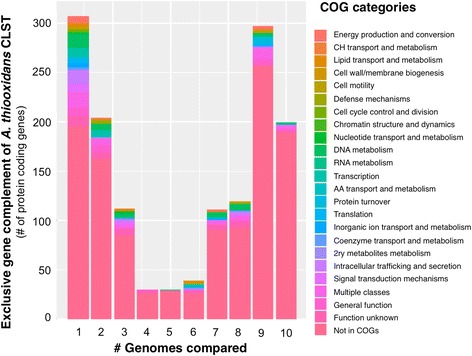
Exclusive gene complement of 10.1601/nm.2199 strain CLST relative to other strains of the species. All possible pairwise comparisons were performed. The number of exclusive genes resulting from the pairwise comparisons between the genomes of CLST and the other 10.1601/nm.2199 strains is plotted against the number of genomes compared. Genes were color coded according to their COG predicted function, as indicated in the lateral bar
Conclusions
This work reports the first draft genome and annotation of a halotolerant acidophilic sulfur-oxidizing 10.1601/nm.2198 (A. thioooxidans strain CLST), together with its basic microbiological properties and fundamental metadata from the saline environment in northern Chile from which it was isolated. The 3.9 Mbp draft genome sequence of strain CLST is arranged in 40 high quality scaffolds, being 24% larger than the genome of the type strain and resembling in size other industrial isolates recently sequenced. It encodes 75 RNAs and 3866 predicted protein-coding genes, 43% of which were assigned putative functions. Over one third of the gene complement is flexible, being represented in few strains other than CLST. Several of the exclusive genes identified in this study can be linked to osmotolerance and other stress responses. Further study of these and other features will likely provide new insights into sodium-requiring processes in acidophilic chemolithotrophic bacteria and further understanding of the mechanisms used by acidophilic bacteria to endure high osmotic stress in natural and industrial saline environments. The release of the genome sequence of this strain improves the representation of these extreme acidophilic Gram negative bacteria in public databases and strengthens the framework for further investigation of the physiological diversity and ecological function of A. thioooxidans.
Additional files
(A, C) A. thiooxidans ATCC 19377 cell growth and growth specific rate with and without NaCl. (B, D) A. thiooxidans CLST cell growth and growth specific rate with and without NaCl. (TIFF 272 kb)
SEM image and EDS spectrum of the precipitate obtained when A. thiooxidans was grown in a medium supplemented with CuSO4. (TIFF 206 kb)
Acknowledgements
We would like to acknowledge Dr. Francisco Remonsellez, Dr. Cristina Dorador and Mónica Gonzalez for technical assistance.
Funding
This work was performed under the auspices of the following projects: Innova CORFO 08CM01-03, Joint BHP Billiton – UCN – FCV Phase I, Fondecyt 1,140,048, Fondecyt 1,130,683, Fondecyt 1,100,795, Basal PFB 16.
Abbreviations
- ATCC
American Type Culture Collection
- CBAR-UNC
Centro de Biotecnología “Profesor Alberto Ruiz”, Universidad Católica del Norte
- DSM
Deutsche Sammlung von Mikroorganismen
- FCV
Fundación Ciencia & Vida
- RISC
Reduced Inorganic Sulfur Compound
Authors’ contributions
LE and CD performed the description of the sampling environment, the sampling and the culture enrichment. PG and MA conducted the isolation, the microbiological characterization of the isolate and purified genomic DNA. CD, RQ and DSH funded the sequencing. AMB and FI did the assembly and annotation. RQ and AMB did the metabolic reconstruction and comparative genomic analysis. JPC, LE and HN did the phylogenetic analysis and typed the strain. RQ, DSH and CD designed the study, and drafted and reviewed the manuscript. All authors read and approved the final manuscript.
Competing interests
The author(s) declare(s) that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s40793-017-0305-8) contains supplementary material, which is available to authorized users.
References
- 1.Kelly DP, Wood AP. Reclassification of some species of Thiobacillus to the newly designated genera Acidithiobacillus gen. nov., Halothiobacillus gen. nov., and Thermithiobacillus gen. nov. Int J Syst Evol Microbiol. 2000; doi:10.1099/00207713-50-2-511. [DOI] [PubMed]
- 2.Hallberg KB, González-Toril E, Johnson DB. Acidithiobacillus ferrivorans sp. nov.; facultatively anaerobic, psychrotolerant, iron- and sulfur-oxidizing acidophiles isolated from metal mine-impacted environments. Extremophiles 2010; doi:10.1007/s00792-009-0282-y. [DOI] [PubMed]
- 3.Hedrich S, Johnson DB. Acidithiobacillus ferridurans sp. nov., an acidophilic iron-, sulfur- and hydrogen-metabolizing chemolithotrophic gammaproteobacterium. Int J Syst Evol Microbiol. 2013; doi:10.1099/ijs.0.049759-0. [DOI] [PubMed]
- 4.Falagán C, Johnson DB. Acidithiobacillus ferriphilus sp. nov., a facultatively anaerobic iron- and sulfur-metabolizing extreme acidophile. Int J Syst Evol Microbiol. 2016; doi:10.1099/ijsem.0.000698 [DOI] [PMC free article] [PubMed]
- 5.Waksman SA, Joffe JS. Microorganisms concerned in the oxidation of sulfur in the soil. II. Thiobacillus thioooxidans, a new sulfur-oxidizing organism isolated from the soil. J Bacteriol. 1922;7(2):239–256. doi: 10.1128/jb.7.2.239-256.1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hedrich S, Schlomann M, Johnson DB. The iron-oxidizing proteobacteria. Microbiology. 2011; doi:10.1099/mic.0.045344-0. [DOI] [PubMed]
- 7.Valdés J, Ossandon F, Quatrini R, Dopson M, Holmes DS. Draft genome sequence of the extremely acidophilic biomining bacterium Acidithiobacillus thiooxidans ATCC 19377 provides insights into the evolution of the Acidithiobacillus genus. J Bacteriol. 2011; doi:10.1128/JB.06281-11. [DOI] [PMC free article] [PubMed]
- 8.Travisany D, Cortés MAP, Latorre M, Di Genova A, Budinich M, Bobadilla-Fazzini RA, et al. A new genome of Acidithiobacillus thiooxidans provides insights into adaptation to a bioleaching environment. Res Microbiol. 2014; doi:10.1016/j.resmic.2014.08.004. [DOI] [PubMed]
- 9.Yin H, Zhang X, Liang Y, Xiao Y, Niu J, Liu X. Draft Genome Sequence of the Extremophile Acidithiobacillus thiooxidans A01, Isolated from the Wastewater of a Coal Dump. Genome Announc. 2014; doi:10.1128/genomeA.00222-14. [DOI] [PMC free article] [PubMed]
- 10.Zhang X, Feng X, Tao J, Ma L, Xiao Y, Liang Y, et al. Comparative genomics of the extreme acidophile Acidithiobacillus thiooxidans reveals intraspecific divergence and niche adaptation. Int J Mol Sci. 2016; doi:10.3390/ijms17081355. [DOI] [PMC free article] [PubMed]
- 11.Jones DS, Albrecht HL, Dawson KS, Schaperdoth I, Freeman KH, Pi Y, et al. Community genomic analysis of an extremely acidophilic sulfur-oxidizing biofilm. ISME J. 2012; doi:10.1038/ismej.2011.75. [DOI] [PMC free article] [PubMed]
- 12.Khan S, Haq F, Hasan F, Saeed K, Ullah R. Growth and Biochemical Activities of Acidithiobacillus thiooxidans Collected from Black Shale. J Microbiol Res. 2012; doi:10.5923/j.microbiology.20120204.03.
- 13.Urbieta MS, González Toril E, Aguilera A, Giaveno MA, Donati E. First prokaryotic biodiversity assessment using molecular techniques of an acidic river in Neuquén, Argentina. Microb Ecol. 2012; doi:10.1007/s00248-011-9997-2. [DOI] [PubMed]
- 14.Kamimura K, Higashino E, Kanao T, Sugio T. Effects of inhibitors and NaCl on the oxidation of reduced inorganic sulfur compounds by a marine acidophilic, sulfur-oxidizing bacterium, Acidithiobacillus thiooxidans strain SH. Extremophiles 2005; doi:10.1007/s00792-004-0420-5. [DOI] [PubMed]
- 15.Okabe S, Odagiri M, Ito T, Satoh H. Succession of sulfur-oxidizing bacteria in the microbial community on corroding concrete in sewer systems. Appl Environ Microbiol. 2007; doi:10.1128/AEM.02054-06. [DOI] [PMC free article] [PubMed]
- 16.Remonsellez F, Galleguillos F, Moreno-Paz M, Parro V, Acosta M, Demergasso C. Dynamic of active microorganisms inhabiting a bioleaching industrial heap of low-grade copper sulfide ore monitored by real-time PCR and oligonucleotide prokaryotic acidophile microarray. Microb Biotechnol. 2009; doi:10.1111/j.1751-7915.2009.00112.x. [DOI] [PMC free article] [PubMed]
- 17.Tan GL, Shu WS, Hallberg KB, Li F, Lan CY, Zhou WH, et al. Culturable and molecular phylogenetic diversity of microorganisms in an open-dumped, extremely acidic Pb/Zn mine tailings. Extremophiles 2008; doi:10.1007/s00792-008-0171-9. [DOI] [PubMed]
- 18.Paulino LC, Bergamo RF, de Mello MP, Garcia Jr O, Manfio GP, Ottoboni LM. Molecular characterization of Acidithiobacillus ferrooxidans and A. thiooxidans strains isolated from mine wastes in Brazil. Ant Van Leeuwenhoek. 2001; doi: 10.1023/A:1012247325537. [DOI] [PubMed]
- 19.Suzuki I, Lee D, Mackay B, Harahuc L, Oh JK. Effect of various ions, pH, and osmotic pressure on oxidation of elemental sulfur by Thiobacillus thiooxidans. Appl Environ Microbiol. 1999;65(11):5163–5168. doi: 10.1128/aem.65.11.5163-5168.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Escudero L, Bijman J, Chong G, Pueyo J, Demergasso C. Geochemistry and microbiology in an acidic, high altitude (4,000 m) salt flat. High Andes, northern Chile. Adv Mat Res. 2013; doi: 10.4028/www.scientific.net/AMR.825.28.
- 21.Benison KC, Bowen BB. Extreme sulfur-cycling in acid brine lake environments of Western Australia. Chem Geol. 2013; doi:10.1016/j.chemgeo.2013.05.018.
- 22.Escudero, L., Bijman, J., Guajardo, M., Pueyo Mur, J., Chong, G., & Demergasso, C. Organotrophic and Mixotrofic Sulfur Oxidation in an Acidic Salt Flat in Northern Chile. Adv Mat Res. 2015; doi: 10.4028/www.scientific.net/AMR.1130.63.
- 23.Mormile MR, Hong BY, Benison KC. Molecular analysis of the microbial communities of Mars analog lakes in western Australia. Astrobiology 2009; doi:10.1089/ast.2008.0293. [DOI] [PubMed]
- 24.West MD, Clarke JDA, Thomas M, Pain CF, Walter MR. The geology of Australian Mars analogue sites. Planet Space Sci. 2010; doi:10.1016/j.pss.2009.06.012.
- 25.Norris PR, Marsh RM, Lindstrôm EB. Growth of mesophilic and thermophilic acidophilic bacteria on sulphur and tetrathionate. Biotechnol Appl Biochem. 1986;8(4):318–329. [Google Scholar]
- 26.Oguri T, Schneider B, Reitzer L. Cysteine catabolism and Cysteine Desulfhydrase (CdsH/STM0458) in Salmonella enterica Serovar Typhimurium. J Bacteriol. 2012;194:4366–4376. doi: 10.1128/JB.00729-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang CL, Lum AM, Ozuna SC, Clark DS, Keasling JD. Aerobic sulfide production and cadmium precipitation by Escherichia coli expressing the Treponema denticola cysteine desulfhydrase gene. Appl Microbiol Biotechnol. 2001;56:425–430. doi: 10.1007/s002530100660. [DOI] [PubMed] [Google Scholar]
- 28.Droege M, Hill B. The Genome Sequencer FLX System-longer reads, more applications, straightforward bioinformatics and more complete data sets. J Biotechnol. 2008; doi:10.1016/j.jbiotec.2008.03.021. [DOI] [PubMed]
- 29.Delcher AL, Bratke KA, Powers EC, Salzberg SL. Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics 2007; doi:10.1093/bioinformatics/btm009. [DOI] [PMC free article] [PubMed]
- 30.Overbeek R, Olson R, Pusch GD, Olsen GJ, Davis JJ, Disz T et al. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res. 2014; doi:10.1093/nar/gkt1226. [DOI] [PMC free article] [PubMed]
- 31.Laslett D, Canback B. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 2004; doi:10.1093/nar/gkh152. [DOI] [PMC free article] [PubMed]
- 32.Huang Y, Gilna P, Li W. Identification of ribosomal RNA genes in metagenomic fragments. Bioinformatics 2009; doi:10.1093/bioinformatics/btp161. [DOI] [PMC free article] [PubMed]
- 33.Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 2011; doi:10.1038/nmeth.1701. [DOI] [PubMed]
- 34.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001; doi:10.1006/jmbi.2000.4315. [DOI] [PubMed]
- 35.Levicán G, Katz A, Valdés J, Quatrini R, Holmes DS, Orellana O. A 300 kpb genome segment, including a complete set of tRNA genes, is dispensable for Acidithiobacillus ferrooxidans. Adv Mat Res. 2009; doi: 10.4028/www.scientific.net/AMR.71-73.187.
- 36.Aziz RK, Devoid S, Disz T, Edwards RA, Henry CS, Olsen GJ, et al. SEED servers: high-performance access to the SEED genomes, annotations, and metabolic models. PLoS One. 2012; doi:10.1371/journal.pone.0048053. [DOI] [PMC free article] [PubMed]
- 37.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, et al. The RAST Server: rapid annotations using subsystems technology. BMC Genomics. 2008; doi:10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed]
- 38.Garrity GM, Bell JA, Lilburn T. Order II. Acidithiobacillales ord. Nov. In: Garrity G, Brenner DJ, Krieg NR, Staley JR, editors. Bergey's manual of systematic bacteriology. New York: Springer; 2005. pp. 60–63. [Google Scholar]
- 39.Garrity GM, Bell JA, Lilburn T. Family I. Acidithiobacillaceae fam. Nov. In: Garrity G, Brenner DJ, Krieg NR, Staley JR, editors. Bergey's manual of systematic bacteriology. New York: Springer; 2005. p. 60. [Google Scholar]
- 40.Cárdenas JP, Quatrini R, Holmes DS. Aerobic lineage of the oxidative stress response protein rubrerythrin emerged in an ancient microaerobic, (hyper)thermophilic environment. Front Microbiol. 2016; doi: 10.3389/fmicb.2016.01822. [DOI] [PMC free article] [PubMed]
- 41.Nies DH, Silver S. Molecular microbiology of heavy metals. 1. NewYork: Springer; 2007. [Google Scholar]
- 42.Silver RP, Prior K, Nsahlai C, Wright LF. ABC transporters and the export of capsular polysaccharides from gram-negative bacteria. Res Microbiol. 2001; 152:357-364. [DOI] [PubMed]
- 43.Quatrini R, Ossandon FJ, Rawlings DE. The flexible genome of acidophilic prokaryotes. In: Quatrini R, Johnson DB, editors. Acidophiles: life in extremely acidic environments. UK: Caister Academic Press; 2016. pp. 199–220. [Google Scholar]
- 44.Garrity GM, Holt JG. The road map to the manual. In: Garrity GM, Boone DR, Castenholz RW, editors. Bergey’s manual of systematic bacteriology. New York: Springer Science+Business Media LLC; 2001. pp. 119–166. [Google Scholar]
- 45.Validation List No. 107: List of new names and new combinations previously effectively, but not validly, published. Int J Syst Evol Microbiol. 2006; doi: 10.1099/ijs.0.64188-0. [DOI] [PubMed]
- 46.Williams KP, Kelly DP. Proposal for a new class (heading leve1 1)within the phylum Proteobacteria, Acidithiobacillia classis nov., with the type order Acidithiobacillales, and emended description of the class Gammaproteobacteria. Int J Syst Evol Microbiol. 2013; doi:10.1099/ijs.0.049270-0. [DOI] [PubMed]
- 47.Kelly DP, Wood AP. The family Acidithiobacillaceae. In: Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F, editors. The prokaryotes. Gammaproteobacteria. 4. New York: Springer; 2014. pp. 15–25. [Google Scholar]
- 48.Bridge TAM, Johnson DB. Reduction of soluble iron and reductive dissolution of ferric iron-containing minerals by moderately thermophilic iron-oxidizing bacteria. Appl Environ Microbiol. 1998;64(6):2181–2186. doi: 10.1128/aem.64.6.2181-2186.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000; doi:10.1038/75556. [DOI] [PMC free article] [PubMed]
- 50.Field D, Garrity G, Gray T, Morrison N, Selengut J, Sterk P, et al. The minimum information about a genome sequence (MIGS) specification. Nat Biotechnol. 2008; doi:10.1038/nbt1360. [DOI] [PMC free article] [PubMed]
- 51.Garrity GM. NamesforLife: browser tool takes expertise out of the database and puts it right in the browser. Microbiol Today. 2010;37(1):9. [Google Scholar]
- 52.Nei M, Kumar S. Molecular evolution and phylogenetics. 1. New York: Oxford University Press; 2000. [Google Scholar]
- 53.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013; doi:10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A, C) A. thiooxidans ATCC 19377 cell growth and growth specific rate with and without NaCl. (B, D) A. thiooxidans CLST cell growth and growth specific rate with and without NaCl. (TIFF 272 kb)
SEM image and EDS spectrum of the precipitate obtained when A. thiooxidans was grown in a medium supplemented with CuSO4. (TIFF 206 kb)


