Abstract
The early-matured japonica (Geng) rice variety, Suijing18 (SJ18), carries multiple elite traits including durable blast resistance, good grain quality, and high yield. Using PacBio SMRT technology, we produced over 25 Gb of long-read sequencing raw data from SJ18 with a coverage of 62×. Using Illumina paired-end whole-genome shotgun sequencing technology, we generated 59 Gb of short-read sequencing data from SJ18 (23.6 Gb from a 200 bp library with a coverage of 59× and 35.4 Gb from an 800 bp library with a coverage of 88×). With these data, we assembled a single SJ18 genome and then generated a set of annotation data. These data sets can be used to test new programs for variation deep mining, and will provide new insights into the genome structure, function, and evolution of SJ18, and will provide essential support for biological research in general.
Subject terms: Structural variation, Plant breeding
Background & Summary
As the leading staple food resource for humans, rice has been adopted as an important model organism for biological research, especially for monocots. Asian cultivated rice (Oryza sativa L.) comprises two subspecies: O. sativa subsp. japonica (also known as Keng1 with the corresponding Pinyin, Geng) and subsp. indica (also known as Hsien1 with the corresponding Pinyin, Xian). Currently, japonica/Geng, especially the early-matured type, is becoming more and more important in rice production. In 2016, the cultivation area of early-mature japonica/Geng was more than 4 million ha in Northeast China.
It has become clear that one single genome is not enough to represent the huge amount of variation in rice genomes. Recently, in addition to the previously published de novo assemblies, including 93–11 (indica/Xian, two-line hybrid restorer), PA64S (admixture type with roughly 55% indica/Xian, 25% japonica/Geng, and 20% javanica, a two-line hybrid sterile line), IR64 (indica/Xian), DJ123 (aus type indica/Xian)2, HR-12 (indica/Xian)3, and Swarna (indica/Xian)4, three new sets of indica/Xian genomes have been released with the aid of third-generation sequencing technology for variation deep mining, including MH63RS1 (indica/Xian, three-line hybrid restorer), ZS97RS1 (indica/Xian, three-line hybrid maintainer)5, and R498 (indica/Xian, three-line hybrid restorer)6. These data sets have enriched our knowledge of the genomic variations of indica/Xian rice. Nevertheless, the genome of japonica/Geng is quite different from that of indica/Xian. Since the release of the gold standard genome of Nipponbare7,8, a medium-matured japonica/Geng variety with photosensitivity, the public availability of japonica/Geng genomes, especially for the early-mature type, remains largely blank.
According to our breeder’s experiences, early-matured japonica/Geng is a relatively unique type compared with the medium-matured japonica/Geng. In addition, common variations, such as single nucleotide polymorphisms (SNPs) within early-matured japonica/Geng group are relatively sparse. To improve the efficiency of molecular breeding in early-matured japonica/Geng, deep mining of further genome variations is urgently required. Short-read sequencing (SRS) technologies, such as Illumina HiSeq, have offered us an opportunity to access huge amounts of variations, including SNPs and short InDels, instantly from large sets of genomes9; however, to perform deeper mining of complex but critical variations, such as repeat sequence variations, long InDels, and structure variations (SVs), the technical bottleneck of the short sequencing read length remains a challenge. Currently, long-read sequencing (LRS) data are available with the aid of new technology, such as PacBio. However, the cost and error rate still remain relatively high. Thus, a scheme comprising LRS amended by SRS would represent a balanced choice for deep mining of genome variations6,10.
Early-mature japonica/Geng cultivar Suijing18 (SJ18) was newly developed by our joint project and was licensed for release in Northern China in 2014. It is a representative early-matured japonica/Geng cultivar harboring multiple elite traits (such as durable blast resistance, good grain quality, and high yield) and now represents more than 10% of the planting area of early-matured japonica/Geng in China. Therefore, we initiated a collaborative project to generate one high quality genome assembly for SJ18 to be used as a fundamental tool to help us investigate underlying genome variations in early-matured japonica/Geng. In this study, we report the resources and data sets that were generated and used for the deep mining of SJ18 genome variations: (1) raw PacBio LRS data, (2) Illumina whole-genome shotgun (WGS) SRS data, (3) the amended assembly of SJ18, (4) the annotation data based on the amended assembly of SJ18, and (5) the functional analysis results based on this annotation.
With the resources and data generated in this study, not only were we able assemble de novo a good quality genome sequence for early-matured japonica/Geng, but also were able to provide the scientific community with data to advance biological research at the genomic level, especially for the deep mining of genetic variations, and provided more information for genome-based molecular breeding of crops.
Methods
Plant material and library construction
The early-matured japonica/Geng cultivar SJ18, which was developed by our own group, was licensed for release in 2014 and is now widely planted (more than 0.8 million hectare) in Heilongjiang province in Northeast China. High-molecular-weight genomic DNA was extracted from 10-day-old leaves of SJ18 (multiple seeds) using the modified CTAB method11, followed by 0.5× bead purification twice. The quality of the DNA sample was assessed using 0.75% agarose gel assays and Nanodrop (Nanodrop Technologies, Wilmington, DE, US), and was quantified using Qubit system (Thermo Fisher Scientific, Waltham, MA). The sample that met the quantity and quality standards was split into two parts, which were used to construct PacBio Sequel and Illumina libraries for LRS and SRS, respectively (Fig. 1).
Figure 1. Outline of the workflow used to generate and analyze the genome data for Suijing18 (SJ18).
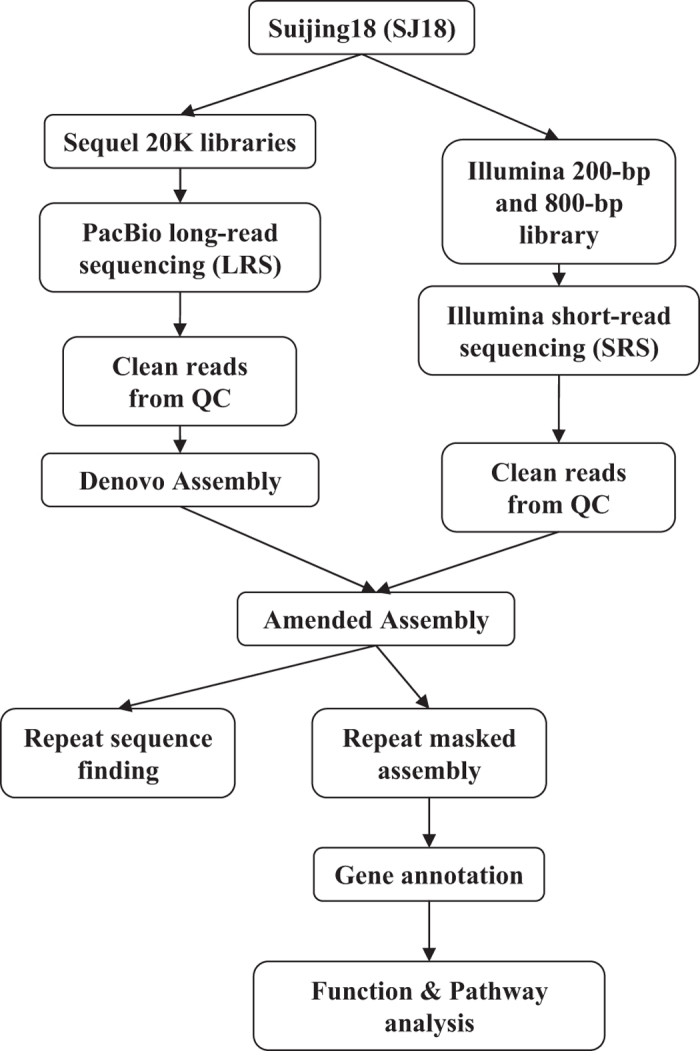
The Sequel 20 K libraries were prepared using the standard protocol from PacBio and sequenced in the wet laboratory department of the Beijing Computing Center (http://www.bcc.ac.cn/) using a PacBio LRS instrument, model Sequel. The 200 bp- and 800 bp-libraries, with peak insert sizes of ~200 bp and ~800 bp, respectively, were prepared using an Illumina Truseq DNA library protocol (Illumina Kit FC-121-4001; Illumina Inc., San Diego, CA, USA). The qualities of libraries were checked using a standard protocol involving an Agilent 2,100 Bioanalyzer High Sensitivity Kit. After library profile analysis, the libraries were sequenced using 150 bp pair-end strategies with the Illumina HiSeq X10 platform (Illumina Inc.).
The amount of raw data from LRS was no less than 25 Gb, with a coverage of 62×. Using SRS, 59 Gb of raw data was generated, including 23.6 and 35.4 Gb of data from the 200 and 800 bp libraries, respectively. The total coverage of SRS was about 147×.
Data analysis
The LRS data was screened and adjusted by the procedures embed in CANU12. Data that met the threshold of Q20 (corresponding to a 1% error rate) were adopted. De novo assembly was carried out for the LRS data using the CANU pipeline with default parameters, except for errorRate=0.045 and genomeSize=350 m. The SRS data were then aligned to the preliminary assembly using BWA13. In addition, the pilon package14 was adopted for the amendment process. The amended assembly represented the submitted version of the SJ18 sequence.
Based on the amended version of the SJ18 assembly, genome annotation was carried out using the following steps with default parameters, except for those indicated:
Tandem repeats were recognized by the TRF package15 with the following parameter settings: Match=2, Mismatch=7, Delta=7, PM=80, PI=10, Minscore=50, MaxPeriod=2,000. Other types of repeat sequences were recognized by RepeatModeler (http://www.repeatmasker.org/RepeatModeler/) with default settings. The database adopted for RepeatModeler analysis was an integrated library comprising Repbase16 (updated in January 2017), Dfam2 (ref. 17), and publicly available libraries containing de novo information for rice.
Annotation for non-coding RNA (ncRNA) was carried out by using cmsearch in Infernal18, searching the Rfam database V12.2 (http://rfam.xfam.org/) with a parameter setting of ‘—cyk -T10’ for microRNAs (miRNAs), small nuclear ribonucleic acid (snRNA), and small RNA (sRNA). The transfer RNA (tRNA) annotation was carried out using tRNAscan-SE19 with default settings.
We masked the repeats using RepeatMask with the parameter setting of ‘-nolow -no_is -norna’ and then annotated the SJ18 genome using multiple tools, including GENEID20 with parameter settings for rice, GeneMark21 with a setting of —ES —cores 24 —min_contig 100, SNAP22 with default setting, and AUGUSTUS23 with -species=rice. We compared the coding sequences (CDSs) and protein sequences from other rice genomes using PASA24 with default settings and GeneWise25 with a setting of ‘splice_gtag -sum -gff -quiet’. All the annotation results were integrated and screened using EVidenceModeler (EVM)26.
The predicted coding genes from SJ18 were translated into protein sequences and aligned to the proteins from plant species in the Uniprot database (http://www.uniprot.org/) and Kyoto Encyclopedia of Genes and Genomes (KEGG) database (http://www.genome.jp/kegg/), respectively, using BLASTP. The threshold was set to e-value<1e-8, and the best hits were submitted for further analysis.
Gene ontology (GO) analysis was carried out based on the above functional annotation results by using topGO27. The biological process (BP), cellular component (CC), and molecular function (MF) matches were listed. The secondary binding point was chosen in the analysis. The annotated proteins from SJ18 were submitted for pathway analysis using KEGG.
Data Records
Raw PacBio long-read sequencing (LRS) data are available through the NCBI SRA with the accession number SRR5877285 (Data Citation 1). All Illumina short-read sequencing (SRS) data for SJ18 can be found at the NCBI SRA with accession numbers SRR5880534 (Data Citation 2) and SRR5880533 (Data Citation 3). The assembled SJ18 genome version 1 is available at the NCBI with the accession number PDFQ00000000 (Data Citation 4). All these raw data are also available at figshare (Data Citation 5). The analyzed data are available at figshare (Data Citation 5) or through the URLs offered by figshare and the Rice Functional Genomics and Breeding (RFGB) database28 (Table 1).
Table 1. Analyzed data resources for Suijing18 (SJ18) deposited at figshare or the Rice Functional Genomics and Breeding (RFGB) database.
| Subjects | Title | Public links |
|---|---|---|
| KEGG, Kyoto Encyclopedia of genes and genomes; ncRNA, non-coding RNA. | ||
| De novo assembly | Suijing18 Denovo assembly version 1 | Data Citation 5 or http://www.rmbreeding.cn/downloads/sj18/SJ18_v1.fasta.gz |
| Repeat-masked de novo assembly | Repeat masked data based on Suijing18 Denovo assembly version 1 | Data Citation 5 or http://www.rmbreeding.cn/downloads/sj18/SJ18_v1.masked.fasta.gz |
| Gene annotation results | Annotated genes based on Suijing18 Denovo assembly version 1 | Data Citation 5 or http://www.rmbreeding.cn/downloads/sj18/SJ18.gene.gff3.gz |
| ncRNA annotated | Annotated ncRNAs based on Suijing18 Denovo assembly version 1 | Data Citation 5 or http://www.rmbreeding.cn/downloads/sj18/SJ18.ncRNA.gff3.gz |
| Repeats annotated | Annotated Repeats based on Suijing18 Denovo assembly version 1 | Data Citation 5 or http://www.rmbreeding.cn/downloads/sj18/SJ18.repeat.gff3.gz |
| Functional annotation results based on the alignments from KEGG | Annotated proteins based on Suijing18 Denovo assembly version 1 and KEGG database | Data Citation 5 or http://www.rmbreeding.cn/downloads/sj18/SJ18.kegg.xls.gz |
| Functional annotation results based on the alignments from UniProt | Annotated proteins based on Suijing18 Denovo assembly version 1 and Uniprot database | Data Citation 5 or http://www.rmbreeding.cn/downloads/sj18/SJ18.uniprot.xls.gz |
| Pathway analysis results | Gene ontology analysis results based on Suijing18 Denovo assembly version 1 | Data Citation 5 or http://www.rmbreeding.cn/downloads/sj18/SJ18.pathway.zip |
Technical Validation
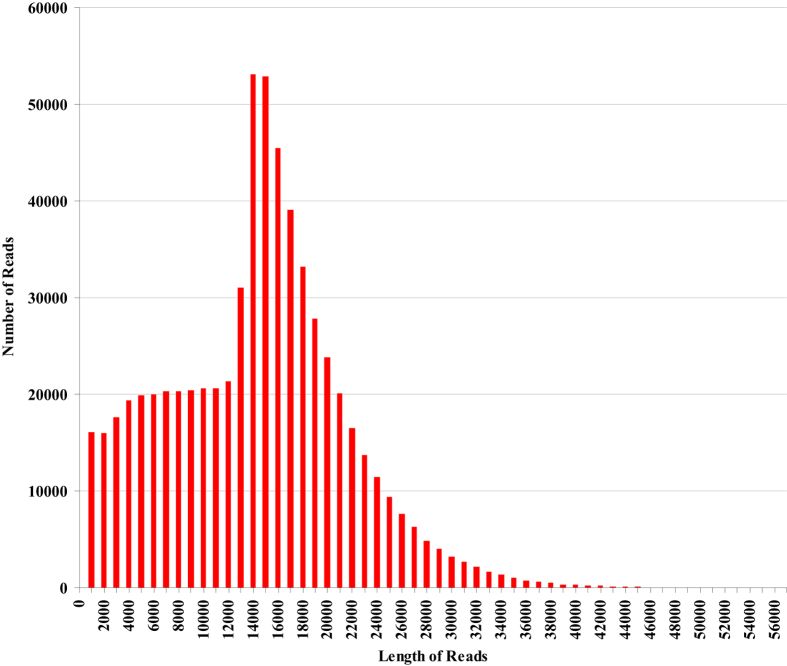
The LRS data were screened and amended with the SRS data using the CANU package with default settings. Possible sequencing errors were further minimized by removing reads that aligned with high scores to the downloaded sequences from bacteria, fungi, or human genomes from GenBank using BWA. Finally, a total of 648,237 high-quality LRS reads that passed this quality check step were submitted for assembly. The distribution of these reads is shown in Fig. 2.
Figure 2. Distribution of high quality reads from PacBio long-read sequencing (LRS) for Suijing18 (SJ18).
The raw SRS data was screened using the Trimmomatic package29, which removed the adaptors and the reads with a quality value lower than 20 (corresponding to a 1% error rate).
We also compared the parameters of SJ18 with other assemblies. The statistics of the assembled contigs are shown in Table 2. The statistics of repeat sequences are shown in Table 3 in comparison with Nipponbare (medium-matured japonica/Geng) and R498 (indica/Xian, the most recently available rice assembly).
Table 2. Comparisons between Suijing18 (SJ18) and the other datasets for representative assembled contigs publicly available and the annotated ncRNAs.
| Subspecies | SJ18 Early-matured japonica/ Geng | IRGSP1.0 Medium-matured japonica/ Geng | R498 Indica/Xian Three-line hybrid restorer | ZS97RS1 Indica/Xian Three-line hybrid maintainer | MH63RS1 Indica/Xian Three-line hybrid restorer | HR-12 Indica/Xian | 9,311 Indica/Xian Two-line hybrid restorer | PA64S Indica/ Xian Two-line hybrid sterile line | IR64 Indica/Xian | DJ123 Aus type of indica/Xian |
|---|---|---|---|---|---|---|---|---|---|---|
| tRNA, transfer RNA; snoRNA, small nucleolar RNA; snRNA, small ribonuclear RNA; rRNA, ribosomal RNA; miRNA, microRNA. | ||||||||||
| Total nucleotides (Mb) | 418.9 | 373.2 | 390.3–423.2 | 346.9 | 359.9 | 389.8 | 374.6–466.0 | 382.0 | 316.3 | 321.2 |
| N50 contig length (bp) | 2,467,626 | 7,711,345 | 1,185,206 | 2,339,070 | 3,097,358 | 28,500 | 6,690 | 17,000 | 22,200 | 25,500 |
| Total genes | 38,456 | 39,045 | 38,714 | 34,610 | 37,324 | 56,284 | 40,745 | 37,162 | 37,768 | 37,812 |
| tRNA | 434 | 244 | NA | 592 | 589 | NA | 734–993 | NA | NA | NA |
| snoRNA | 681 | NA | NA | 449 | 457 | NA | NA | NA | NA | NA |
| snRNA | 108 | NA | NA | 92 | 97 | NA | 3,374 | NA | NA | NA |
| rRNA | 88 | 724 | NA | 40 | 60 | NA | 752 | NA | NA | NA |
| miRNA | 173 | 146 | NA | 341 | 363 | NA | 3,806 | 1,155 | NA | NA |
Table 3. Different types of repeat sequences found in the Suijing18 (SJ18) assembly (version 1).
| Repeat_Type |
SJ18 |
Nipponbare |
R498 |
|||
|---|---|---|---|---|---|---|
| Length (bp) | % | Length (bp) | % | Length (bp) | % | |
| LTR, Long Terminal Repeats; SINE, Short Interspersed Nuclear Element; LINE, Long Interspersed Nuclear Element; EnSpm, Enhancer/Suppressor mutator; hAT, hobo-Ac-Tam3; MuDR, MuDR: A generic notation for a Mu transposon containing a sequence necessary to permit Mu transposition and related behaviors. The ‘DR’ is in honor of Dr Donald S. Robertson, who discovered and characterized the original Mutator lines. | ||||||
| Class I: Retrotransposon | 94,087,436 | 22.3 | 105,098,791 | 28.2 | 117,509,061 | 30.1 |
| LTR-Retrotransposon | 88,392,161 | 21.0 | 98,903,987 | 26.5 | 111,173,484 | 28.4 |
| LTR/Gypsy | 72,477,718 | 17.2 | 65,915,787 | 17.7 | 80,330,011 | 20.6 |
| LTR/Copia | 14,161,220 | 3.4 | 17,931,866 | 4.8 | 15,079,454 | 3.9 |
| LTR/Other | 1,753,223 | 0.4 | 15,056,334 | 4.0 | 15,764,019 | 4.0 |
| Non-LTR Retrotransposon | 5,695,275 | 1.4 | 6,194,804 | 1.7 | 6,335,577 | 1.6 |
| SINE | 362,005 | 0.1 | 796,311 | 0.2 | 848,061 | 0.2 |
| LINE | 5,333,270 | 1.3 | 5,398,493 | 1.5 | 5,487,516 | 1.4 |
| Class II: DNA Transposon | 69,249,868 | 16.4 | 40,716,340 | 10.9 | 40,743,536 | 10.4 |
| EnSpm/CACTA | 10,690,200 | 2.5 | 14,117,095 | 3.8 | 13,264,041 | 3.4 |
| hAT | 5,494,725 | 1.3 | 1,641,580 | 0.4 | 1,897,505 | 0.5 |
| Harbinger | 9,746,844 | 2.3 | 3,729,352 | 1.0 | 3,882,866 | 1.0 |
| Tc1/Mariner | 6,525,750 | 1.6 | 462,697 | 0.1 | 607,903 | 0.2 |
| MuDR | 16,081,157 | 3.8 | 5,872,349 | 1.6 | 5,993,554 | 1.5 |
| Helitron | 12,538,290 | 3.0 | 1,850,232 | 0.5 | 1,702,664 | 0.4 |
| Other | 8,172,902 | 1.9 | 13,043,035 | 3.5 | 13,395,003 | 3.4 |
| Other tandem repeat | 18,811,934 | 4.5 | 3,935,022 | 1.1 | 4,548,484 | 1.2 |
| Low Complexity | 313,050 | 0.1 | 22,478 | 0.0 | 17,672 | 0.0 |
| Unclassified | 13,830,076 | 3.3 | 1,117,819 | 0.3 | 1,607,036 | 0.4 |
| Total | 196,292,364 | 46.5 | 150,890,450 | 40.4 | 164,425,789 | 42.1 |
Additional information
How to cite this article: Nie, S.-J. et al. Assembly of an early-matured japonica (Geng) rice genome, Suijing18, based on PacBio and Illumina sequencing. Sci. Data 4:170195 doi: 10.1038/sdata.2017.195 (2017).
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
We appreciate support from a subprogram of the National Key R&D Program of China (2016YFD0101801) from the Chinese Ministry of Science & Technology to T.-Q.Z. and also the support of the Shenzhen Peacock Plan to Z.-K.L. and J.-L.X.
Footnotes
The authors declare no competing financial interests.
Data Citations
- 2017. NCBI Sequence Read Archive. SRP113746
- 2017. NCBI Sequence Read Archive. SRP113817
- 2017. NCBI Sequence Read Archive. SRP113816
- 2017. NCBI Assembly. GCA_002573525
- Zheng T. Q. 2017. Figshare. https://doi.org/10.6084/m9.figshare.c.3835939
References
- Morishima H. & Oka H.-I. Phylogenetic differentiation of cultivated rice, XXII. Numerical evaluation of the indica-japonica differentiation. Jpn J Breeding 31, 402–413 (1981). [Google Scholar]
- Schatz M. C. et al. Whole genome de novo assemblies of three divergent strains of rice, Oryza sativa, document novel gene space of aus and indica. Genome Biol 15, 506 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahesh H. B. et al. Indica rice genome assembly, annotation and mining of blast disease resistance genes. BMC Genomics 17, 242 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathinasabapathi P., Purushothaman N., Ramprasad V. L. & Parani M. Whole genome sequencing and analysis of Swarna, a widely cultivated indica rice variety with low glycemic index. Sci Rep 5, 11303 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J. et al. Building two indica rice reference genomes with PacBio long-read and Illumina paired-end sequencing data. Scientific Data 3, 160076 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du H. et al. Sequencing and de novo assembly of a near complete indica rice genome. Nat Commun 8, 15324 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara Y. et al. Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice 6, 1–10 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sequencing ProjectInternational Rice, G. The map-based sequence of the rice genome. Nature 436, 793–800 (2005). [DOI] [PubMed] [Google Scholar]
- 3K-RGP. The 3,000 rice genomes project. GigaScience 3, 7 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J. et al. Extensive sequence divergence between the reference genomes of two elite indica rice varieties Zhenshan 97 and Minghui 63. Proc. Natl. Acad. Sci. USA 113, E5163–E5171 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke J. D. Cetyltrimethyl Ammonium Bromide (CTAB) DNA Miniprep for Plant DNA Isolation. CSH Protocols 2009, pdb.prot5177 (2009). [DOI] [PubMed] [Google Scholar]
- Koren S. et al. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res 27, 722–736 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. & Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker B. J. et al. Pilon: An Integrated Tool for Comprehensive Microbial Variant Detection and Genome Assembly Improvement. PLOS ONE 9, e112963 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res 27, 573–580 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurka J. et al. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet Genome Res 110, 462–467 (2005). [DOI] [PubMed] [Google Scholar]
- Wheeler T. J. et al. Dfam: a database of repetitive DNA based on profile hidden Markov models. Nucleic Acids Res 41, D70–D82 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrocki E. P. & Eddy S. R. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics 29, 2933–2935 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe T. M. & Eddy S. R. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25, 955–964 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco E., Parra G. & Guigó R. in Current Protocols in Bioinformatics (John Wiley & Sons, Inc., 2002). [Google Scholar]
- Lomsadze A., Ter-Hovhannisyan V., Chernoff Y. O. & Borodovsky M. Gene identification in novel eukaryotic genomes by self-training algorithm. Nucleic Acids Res 33, 6494–6506 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korf I. Gene finding in novel genomes. BMC Bioinformatics 5, 59 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanke M. & Morgenstern B. AUGUSTUS: a web server for gene prediction in eukaryotes that allows user-defined constraints. Nucleic Acids Res 33, W465–W467 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas B. J. et al. Improving the Arabidopsis genome annotation using maximal transcript alignment assemblies. Nucleic Acids Res 31, 5654–5666 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney E. & Durbin R. Using GeneWise in the Drosophila Annotation Experiment. Genome Res 10, 547–548 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas B. J. et al. Automated eukaryotic gene structure annotation using EVidenceModeler and the Program to Assemble Spliced Alignments. Genome Biol 9, R7 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexa A., Rahnenführer J. & Lengauer T. Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics 22, 1600–1607 (2006). [DOI] [PubMed] [Google Scholar]
- Zheng T. Q. et al. Rice functional genomics and breeding database (RFGB)-3K-rice SNP and InDel sub-database. Chinese Sci Bull 60, 367–371 (2015). [Google Scholar]
- Bolger A. M., Lohse M. & Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- 2017. NCBI Sequence Read Archive. SRP113746
- 2017. NCBI Sequence Read Archive. SRP113817
- 2017. NCBI Sequence Read Archive. SRP113816
- 2017. NCBI Assembly. GCA_002573525
- Zheng T. Q. 2017. Figshare. https://doi.org/10.6084/m9.figshare.c.3835939