Abstract
Accurate species determination of plant pathogens is a prerequisite for their control and quarantine, and further for assessing their potential threat to crops. The family Peronosporaceae (Straminipila; Oomycota) consists of obligate biotrophic pathogens that cause downy mildew disease on angiosperms, including a large number of cultivated plants. In the largest downy mildew genus Peronospora, a phylogenetically complex clade includes the economically important downy mildew pathogens of spinach and beet, as well as the type species of the genus Peronospora. To resolve this complex clade at the species level and to infer evolutionary relationships among them, we used multi-locus phylogenetic analysis and species tree estimation. Both approaches discriminated all nine currently accepted species and revealed four previously unrecognized lineages, which are specific to a host genus or species. This is in line with a narrow species concept, i.e. that a downy mildew species is associated with only a particular host plant genus or species. Instead of applying the dubious name Peronospora farinosa, which has been proposed for formal rejection, our results provide strong evidence that Peronospora schachtii is an independent species from lineages on Atriplex and apparently occurs exclusively on Beta vulgaris. The members of the clade investigated, the Peronospora rumicis clade, associate with three different host plant families, Amaranthaceae, Caryophyllaceae, and Polygonaceae, suggesting that they may have speciated following at least two recent inter-family host shifts, rather than contemporary cospeciation with the host plants.
Keywords: Cospeciation, Host shift, Multi-locus phylogeny, Oomycetes, Peronospora farinosa, Species tree
1. Introduction
Downy mildew is one of the most important oomycete plant diseases and causes serious damage to a variety of cultivated and ornamental plants. In the cultivation of sugar beet (Beta vulgaris) and spinach (Spinacia oleracea), downy mildew is probably the most widespread and a potentially destructive disease worldwide (Byford, 1981). Although occurrences of this disease have been reported in most countries where these crops are cultivated, a proper nomenclature and the identity of these pathogens has remained controversial (Francis and Byford, 1983; Brandenberger et al., 1991), reflected by the plethora of taxonomic changes that have previously been proposed (Yerkes and Shaw, 1959; Byford, 1967). Fifty-three Peronospora names were validly published for species occurring on particular genera or species of plants included in the family Chenopodiaceae s. str. (Constantinescu, 1991), including the species occurring on Beta and Spinacia. Since Yerkes and Shaw (1959) advocated the synonymization of all of these names under a single species, Peronospora farinosa (Fr.) Fr., the oldest available name thought to be attributable to a Peronospora species on Chenopodiaceae (but see Choi and Thines (2014) for a discussion of its status), P. farinosa has often been regarded as the sole parasitic fungus causing downy mildew disease on the host family. However, this view has been challenged with the advent of phylogenetic approaches, which revealed that Peronospora samples from various chenopodiaceous hosts represent phylogenetically distinct lineages according to the specific host genera or host species and together even do not form a monophyletic group (Voglmayr, 2003; Choi et al., 2007c, 2008a, 2010). This is in line with the narrow species concept of Gäumann (1919, 1923). As a result, Choi et al. (2007c) have reinstated the name P. effusa (Grev.) Rabenh. for Peronospora specimens on S. oleracea, and Choi et al. (2008a) noted the existence of host-specific Peronospora species infecting Chenopodium, that are distinct from lineages occurring on Atriplex, the type host genus for P. farinosa. Importantly, the Latin description provided by Fries (1832) for P. farinosa, for which apparently no type material is extant, has insufficient detail for the name to be associated with any particular species of Peronospora. In addition, it describes several features that are atypical for a Peronospora species (Choi and Thines, 2014). Therefore, the name Botrytis farinosa Fr. (Peronospora farinosa) has recently been proposed to be rejected to enable a detailed taxonomic revision of the more than fifty species of downy mildew pathogens to which the name P. farinosa has often been misapplied (Choi and Thines, 2014).
Beet downy mildew (BDM; Peronospora strains parasitic to sugar beet and other varieties of Beta vulgaris) occurs in almost every country where sugar beet and other varieties of Beta vulgaris are cultivated. However, the nomenclature of BDM has not yet been settled with respect to species designation, being mostly still designated as P. farinosa f. sp. betae Byford. This is again due to the two conflicting views regarding species differentiation in downy mildews as referred to above. The name Peronospora schachtii Fuckel was first introduced to accommodate the pathogen on Beta vulgaris (Fuckel, 1866). Adopting this narrow species concept, most monographs reported the causal agent of BDM as P. schachtii (Gustavsson, 1959; Kochman and Majewski, 1970; Ul’yanishchev et al., 1985; Vanev et al., 1993; Mazelaitis and Staneviciené, 1995). Conversely, Yerkes and Shaw (1959) proposed the synonymization of P. schachtii under P. farinosa, which was widely accepted by plant pathologists, but also some taxonomists. As a consequence, most studies erroneously referred to BDM as P. farinosa (or P. farinosa f. sp. betae) (Byford, 1981; Constantinescu and Negrean, 1983; Francis and Byford, 1983; Francis and Waterhouse, 1988; Yu et al., 1998; García-Blázquez et al., 2006; Kim et al., 2010). Hence, the latter name is widely used when referring to BDM (11,500 hits in Google using the search string “Peronospora farinosa” AND beet, 3510 hits in Google using the search string “Peronospora schachtii” AND beet, at 18 Aug 2014).
In a previous ITS rDNA sequence-based study (Choi et al., 2007c), BDM isolates formed a monophyletic cluster with three other Peronospora species, P. effusa parasitic to spinach (Spinacia oleracea), P. rumicis Corda (the type species of the genus Peronospora) parasitic to Rumex spp., and P. obovata Bonord. parasitic to Spergula arvensis. Additionally, five other Peronospora species were also placed within the same clade in a preliminary survey; P. lepigoni Fuckel parasitic to Spergularia rubra, P. polycarponis Mayor & Vienn.-Bourg. to Polycarpon diphyllum, P. minor (Casp.) Gäum. to Atriplex patula, P. atriplicis-hastatae Săvul. & Rayss to A. prostrata, and P. litoralis Gäum. to A. littoralis. The close phylogenetic relationship among these nine species is not surprising. Morphologically, these species share several similarities: all show monopodial branching pattern in the conidiophores, all have straight to sub-straight branches, and all produce ellipsoidal to broadly ellipsoidal and brownish conidia. The morphological similarity between BDM and P. effusa has been previously described (Berlese and De Toni, 1888; Wilson, 1914). Choi et al. (2007c) also recognized this similarity, although they also found a slight difference in the length to width ratio of the conidia. Nonetheless, a decision regarding the conspecificity or distinctiveness of the species could not be reached, as too few specimens and loci were included. However, it became apparent that at least P. effusa was separate from all of the closely related species (Choi et al., 2007c).
For the usefulness in identification, taxonomy, and phylogeny of oomycetes and fungi, the ITS rDNA region has become the standard nuclear DNA barcode marker (Schoch et al., 2012). However, the lack of ITS variability does not always automatically confirm their conspecifity; there are several cases like the one investigated here (Choi et al., 2007c) where the ITS region exhibits insufficient variability for phylogenetic distinction in closely related species, e.g. in Peronospora (Voglmayr et al., 2014b), Pseudoperonospora (Choi et al., 2005) and Phytophthora (Goodwin et al., 1999; Cooke et al., 2000; Jung and Burgess, 2009). Due to the limitations of the ITS region, further multi-locus phylogenies are required. Therefore, in the current study concatenation-based phylogenetic analyses of two nuclear (ITS, heat shock protein 90 [hsp90]) and five mitochondrial loci (cytochrome c oxidase subunit II and I [cox2 and cox1], a spacer region between cox2 and cox1 genes [coxS], NADH dehydrogenase subunit I [nad1], ribosomal protein S10 and its flanking region [rps10]) were conducted. As there are concerns about the accuracy of species relationships inferred from concatenation method (Edwards et al., 2007; Kubatko and Degnan, 2007; Edwards, 2009; Blair et al., 2012), a coalescent-based approach to estimates gene trees and species tree topologies simultaneously (Heled and Drummond, 2010; Bouckaert et al., 2014) was additionally performed. The aim of the present study was to resolve one of the most complex phylogenetic clades of downy mildews using multi-locus sequence data, and to develop evolutionary hypotheses related to speciation and host shifts in this group.
2. Materials and methods
2.1. Oomycete specimens
In total 90 specimens of Peronospora originating from three host families, Amaranthaceae (including Chenopodiaceae), Caryophyllaceae, and Polygonaceae, which all belong to the order Caryophyllales, were included in the present study. On the Amaranthaceae, 12 beet (Beta vulgaris var. cicla and var. saccharifera) specimens, originating from Asia, Europe, North and South America, and 24 spinach (Spinacia oleracea) specimens from Asia, Europe, North America, and Oceania were analyzed, along with eight samples occurring on Atriplex. In addition, three species parasitic on the Caryophyllaceae, P. polycarponis, P. lepigoni, and P. obovata, and the P. rumicis complex parasitic on the Polygonaceae were included for phylogenetic study. Peronospora boni-henrici Gäum. (parasitic to Chenopodium bonus-henricus) was used as an outgroup. The host plants and geographic samplings of downy mildew specimens were summarized in Table S1, in addition with taxonomic changes of Peronospora names by this study. The samples were either collected by the authors or loaned from herbaria BPI, DM, GLM, HMAS, HMR, KUS-F, UPS, VPRI, and WU. Herbarium abbreviations are according to Thiers (2014). Acronyms of private collections include those of the following: DML, from the USDA ARS laboratory of Steven Klosterman in Salinas California, USA; RD, R. Delhey in the Phytopathology Lab of Bahía Blanca, Argentina; JK-F, Julia Kruse in Biodiversity and Climate Research Centre (BiK-F), Frankfurt am Main, Germany; VK, Volker Kummer in University Potsdam, Germany. Information on the specimens sequenced in this study is shown in Table 1.
Table 1.
Information on downy mildew specimens used in this study.
| Taxon | Host plant | Geographic origin | Year collected | Herbarium voucher | DNA accession | GenBank accession number |
||||
|---|---|---|---|---|---|---|---|---|---|---|
| ITS | hsp90 | cox2-spacer-cox1a | nad1 | rps10 | ||||||
| Peronospora litoralis | Atriplex littoralis | Sweden, Oland | 1959 | BPI789214 | BP69 | KP330778 | KP330688 | KP330598 | KP330868 | KP330958 |
| P. minor | Atriplex patula | Sweden, Västergötland | 1962 | UPS | S29 | KP330779 | KP330689 | KP330599 | KP330869 | KP330959 |
| P. minor | Atriplex patula | Germany, Sachsen | 1993 | GLM68024 | 39 | KP330780 | KP330690 | KP330600 | KP330870 | KP330960 |
| P. minor | Atriplex patula | Germany, Sachsen | 1999 | GLM77764 | 42 | KP330781 | KP330691 | KP330601 | KP330871 | KP330961 |
| P. minor | Atriplex patula | Germany, Sachsen-Anhalt | 2003 | GLM73462 | 37 | KP330782 | KP330692 | KP330602 | KP330872 | KP330962 |
| P. atriplicis-hastatae | Atriplex prostrata | Germany, Schleswig-Holstein | 2012 | JK-F0359 | 2663 | KP330783 | KP330693 | KP330603 | KP330873 | KP330963 |
| P. atriplicis-hastatae | Atriplex prostrata | Germany, Sachsen-Anhalt | 2003 | GLM73408 | 55 | KP330784 | KP330694 | KP330604 | KP330874 | KP330964 |
| P. atriplicis-hastatae | Atriplex prostrata | Germany, Sachsen-Anhalt | 1990 | GLM67010 | 53 | KP330785 | KP330695 | KP330605 | KP330875 | KP330965 |
| P. schachtii | Beta vulgaris var. cicla | Korea, Namyangju | 2007 | KUS-F23163 | 2737 | KP330786 | KP330696 | KP330606 | KP330876 | KP330966 |
| P. schachtii | Beta vulgaris var. cicla | Korea, Yongin | 2007 | KUS-F23152 | D335 | KP330787 | KP330697 | KP330607 | KP330877 | KP330967 |
| P. schachtii | Beta vulgaris var. cicla | Korea, Yongin | 2007 | KUS-F23153 | D336 | KP330788 | KP330698 | KP330608 | KP330878 | KP330968 |
| P. schachtii | Beta vulgaris var. cicla | USA, California | 2012 | DML3 | CAL3 | KP330789 | KP330699 | KP330609 | KP330879 | KP330969 |
| P. schachtii | Beta vulgaris var. cicla | USA, California | 2012 | DML4 | CAL17 | KP330790 | KP330700 | KP330610 | KP330880 | KP330970 |
| P. schachtii | Beta vulgaris var. cicla | USA, California | 2012 | DML38 | CAL38 | KP330791 | KP330701 | KP330611 | KP330881 | KP330971 |
| P. schachtii | Beta vulgaris var. saccharifera | Argentina, Bahía Blanca | 1991 | RD864 | AG4 | KP330792 | KP330702 | KP330612 | KP330882 | KP330972 |
| P. schachtii | Beta vulgaris var. saccharifera | China, Sichuan | 1979 | HMAS57030 | C3 | KP330793 | KP330703 | KP330613 | KP330883 | KP330973 |
| P. schachtii | Beta vulgaris var. saccharifera | China, Yunnan | 1979 | HMAS57033 | C2 | KP330794 | KP330704 | KP330614 | KP330884 | KP330974 |
| P. schachtii | Beta vulgaris var. saccharifera | Germany, Nossen | 1888 | BPI790777 | BP106 | KP330795 | KP330705 | KP330615 | KP330885 | KP330975 |
| P. schachtii | Beta vulgaris var. saccharifera | USA, California | 2012 | DML17 | CAL4 | KP330796 | KP330706 | KP330616 | KP330886 | KP330976 |
| P. schachtii | Beta vulgaris var. saccharifera | Unknown | 1931 | BPI790781 | BP109 | KP330797 | KP330707 | KP330617 | KP330887 | KP330977 |
| P. sp. (formerly as P. rumicis) | Emex spinosa | Greece, Tsambika | 2011 | VK 0471/1 | 2558 | KP330798 | KP330708 | KP330618 | KP330888 | KP330978 |
| P. sp. (formerly as P. rumicis) | Emex spinosa | Greece, Tsambika | 2012 | VK 0471/2 | 2559 | KP330799 | KP330709 | KP330619 | KP330889 | KP330979 |
| P. polycarponis | Polycarpon diphyllum | France, Montpellier | 2001 | WU33676 | HVF32.2 | KP330800 | KP330710 | KP330620 | KP330890 | KP330980 |
| P. sp. (formerly as P. rumicis) | Rumex acetosa | Austria, Oberösterreich | 2011 | WU34310 | HV2617 | KP330801 | KP330711 | KP330621 | KP330891 | KP330981 |
| P. sp. (formerly as P. rumicis) | Rumex acetosa | Austria, Oberösterreich | 2001 | WU22925 | HV0312 | KP330802 | KP330712 | KP330622 | KP330892 | KP330982 |
| P. sp. (formerly as P. rumicis) | Rumex acetosa | Austria, Wien | 2004 | WU33899 | HV2117 | KP330803 | KP330713 | KP330623 | KP330893 | KP330983 |
| P. sp. (formerly as P. rumicis) | Rumex acetosa | England, South Yorkshire | 2011 | WU34347 | HV2657 | KP330804 | KP330714 | KP330624 | KP330894 | KP330984 |
| P. sp. (formerly as P. rumicis) | Rumex acetosa | Finland, Nylandia | 1960 | UPS | S27 | KP330805 | KP330715 | KP330625 | KP330895 | KP330985 |
| P. sp. (formerly as P. rumicis) | Rumex acetosa | Finland, Nylandia | 1970 | BPI790739 | BP124 | KP330806 | KP330716 | KP330626 | KP330896 | KP330986 |
| P. sp. (formerly as P. rumicis) | Rumex acetosa | Germany,Sachsen-Anhalt | 2004 | GLM62941 | 1029 | KP330807 | KP330717 | KP330627 | KP330897 | KP330987 |
| P. sp. (formerly as P. rumicis) | Rumex acetosa | Germany, Baden-Württemberg | 2001 | WU33730 | HV0852 | KP330808 | KP330718 | KP330628 | KP330898 | KP330988 |
| P. sp. (formerly as P. rumicis) | Rumex acetosa | Germany, Sachsen-Anhalt | 2005 | GLM75948 | 1031 | KP330809 | KP330719 | KP330629 | KP330899 | KP330989 |
| P. sp. (formerly as P. rumicis) | Rumex acetosa | Sweden, Härjedalen | 1975 | UPS | S25 | KP330810 | KP330720 | KP330630 | KP330900 | KP330990 |
| P. rumicis | Rumex acetosella | Denmark, Ulvshale | 1913 | BPI790753 | BP118 | KP330811 | KP330721 | KP330631 | KP330901 | KP330991 |
| P. sp. (formerly as P. rumicis) | Rumex arifolius | Austria, Kärnten | 2000 | WU32786 | HV0507 | KP330812 | KP330722 | KP330632 | KP330902 | KP330992 |
| P. sp. (formerly as P. rumicis) | Rumex arifolius | Italy, Lombardia | 2012 | WU35780 | HV2939 | KP330813 | KP330723 | KP330633 | KP330903 | KP330993 |
| P. sp. (formerly as P. rumicis) | Rumex arifolius | Italy, Valle d’Aosta | 2013 | JK-F0513 | 2709 | KP330814 | KP330724 | KP330634 | KP330904 | KP330994 |
| P. sp. (formerly as P. rumicis) | Rumex arifolius | Romania, Harghita | 1980 | HMR2940 | 2859 | KP330815 | KP330725 | KP330635 | KP330905 | KP330995 |
| P. sp. (formerly as P. rumicis) | Rumex thyrsiflorus | Austria, Niederösterreich | 2012 | WU35760 | HV2908 | KP330816 | KP330726 | KP330636 | KP330906 | KP330996 |
| P. sp. (formerly as P. rumicis) | Rumex thyrsiflorus | Germany, Niedersachsen | 2009 | JK-F0051 | 2710 | KP330817 | KP330727 | KP330637 | KP330907 | KP330997 |
| P. sp. (formerly as P. rumicis) | Rumex thyrsiflorus | Germany, Sachsen | 2008 | GLM90812 | 1023 | KP330818 | KP330728 | KP330638 | KP330908 | KP330998 |
| P. sp. (formerly as P. rumicis) | Rumex thyrsiflorus | Germany, Sachsen | 2008 | GLM90995 | 1025 | KP330819 | KP330729 | KP330639 | KP330909 | KP330999 |
| P. sp. (formerly as P. rumicis) | Rumex thyrsiflorus | Germany, Sachsen | 2003 | GLM74313 | 1035 | KP330820 | KP330730 | KP330640 | KP330910 | KP331000 |
| P. sp. (formerly as P. rumicis) | Rumex thyrsiflorus | Germany, Sachsen-Anhalt | 2004 | GLM64100 | 1042 | KP330821 | KP330731 | KP330641 | KP330911 | KP331001 |
| P. sp. (formerly as P. rumicis) | Rumex thyrsiflorus | Germany, Sachsen-Anhalt | 2003 | GLM73428 | 1046 | KP330822 | KP330732 | KP330642 | KP330912 | KP331002 |
| P. sp. (formerly as P. rumicis) | Rumex thyrsiflorus | Germany, Sachsen-Anhalt | 2003 | GLM73434 | 1041 | KP330823 | KP330733 | KP330643 | KP330913 | KP331003 |
| P. sp. (formerly as P. rumicis) | Rumex thyrsiflorus | Germany, Sachsen-Anhalt | 2003 | GLM74510 | 1044 | KP330824 | KP330734 | KP330644 | KP330914 | KP331004 |
| P. obovata | Spergula arvensis | Austria, Niederösterreich | 2011 | WU34372 | HV2683 | KP330825 | KP330735 | KP330645 | KP330915 | KP331005 |
| P. obovata | Spergula arvensis | Austria, Oberösterreich | 2000 | WU32858 | HV0437 | KP330826 | KP330736 | KP330646 | KP330916 | KP331006 |
| P. obovata | Spergula arvensis | Austria, Oberösterreich | 2000 | WU22914 | HV0457 | KP330827 | KP330737 | KP330647 | KP330917 | KP331007 |
| P. obovata | Spergula arvensis | Germany, Sachsen-Anhalt | 2003 | GLM73399 | 1135 | KP330828 | KP330738 | KP330648 | KP330918 | KP331008 |
| P. obovata | Spergula arvensis | Germany, Sachsen-Anhalt | 2001 | GLM73791 | 1138 | KP330829 | KP330739 | KP330649 | KP330919 | KP331009 |
| P. obovata | Spergula arvensis | Germany, Sachsen-Anhalt | 2005 | GLM75937 | 1136 | KP330830 | KP330740 | KP330650 | KP330920 | KP331010 |
| P. obovata | Spergula arvensis | Germany, Sachsen-Anhalt | 2002 | GLM76294 | 1137 | KP330831 | KP330741 | KP330651 | KP330921 | KP331011 |
| P. obovata | Spergula arvensis | Germany, Sachsen-Anhalt | 2002 | GLM76717 | 1139 | KP330832 | KP330742 | KP330652 | KP330922 | KP331012 |
| P. obovata | Spergula arvensis | Germany, Sachsen-Anhalt | 2000 | GLM76830 | 1133 | KP330833 | KP330743 | KP330653 | KP330923 | KP331013 |
| P. sp. (formerly as P. lepigoni) | Spergularia marina | Austria, Burgenland | 2001 | WU33808 | HV1019 | KP330834 | KP330744 | KP330654 | KP330924 | KP331014 |
| P. sp. (formerly as P. lepigoni) | Spergularia marina | Germany, Sachsen-Anhalt | 2001 | GLM73809 | 1165 | KP330835 | KP330745 | KP330655 | KP330925 | KP331015 |
| P. sp. (formerly as P. lepigoni) | Spergularia marina | Germany, Sachsen-Anhalt | 2005 | GLM75894 | 1164 | KP330836 | KP330746 | KP330656 | KP330926 | KP331016 |
| P. lepigoni | Spergularia rubra | Germany, Sachsen | 2005 | GLM75718 | 1153 | KP330837 | KP330747 | KP330657 | KP330927 | KP331017 |
| P. lepigoni | Spergularia rubra | Germany, Sachsen | 1998 | GLM77927 | 1154 | KP330838 | KP330748 | KP330658 | KP330928 | KP331018 |
| P. lepigoni | Spergularia rubra | Germany, Sachsen-Anhalt | 2004 | GLM64180 | 1160 | KP330839 | KP330749 | KP330659 | KP330929 | KP331019 |
| P. lepigoni | Spergularia rubra | Germany, Sachsen-Anhalt | 1991 | GLM68688 | 1161 | KP330840 | KP330750 | KP330660 | KP330930 | KP331020 |
| P. lepigoni | Spergularia rubra | Germany, Sachsen-Anhalt | 2001 | GLM73623 | 1157 | KP330841 | KP330751 | KP330661 | KP330931 | KP331021 |
| P. lepigoni | Spergularia rubra | Germany, Sachsen-Anhalt | 2002 | GLM76298 | 1158 | KP330842 | KP330752 | KP330662 | KP330932 | KP331022 |
| P. effusa | Spinacia oleracea | Australia, Leppington | 2002 | VPRI30202 | Au6 | KP330843 | KP330753 | KP330663 | KP330933 | KP331023 |
| P. effusa | Spinacia oleracea | Australia, Turna | 2000 | VPRI22523 | Au5 | KP330844 | KP330754 | KP330664 | KP330934 | KP331024 |
| P. effusa | Spinacia oleracea | Australia, V.I.C. | 2003 | VPRI31625 | Au8 | KP330845 | KP330755 | KP330665 | KP330935 | KP331025 |
| P. effusa | Spinacia oleracea | China, Sichuan | 1958 | HMAS57074 | C6 | KP330846 | KP330756 | KP330666 | KP330936 | KP331026 |
| P. effusa | Spinacia oleracea | Japan, Fukui | 2000 | DM68 | J4 | KP330847 | KP330757 | KP330667 | KP330937 | KP331027 |
| P. effusa | Spinacia oleracea | Japan, Gifu | Unknown | DM22 | J1 | KP330848 | KP330758 | KP330668 | KP330938 | KP331028 |
| P. effusa | Spinacia oleracea | Japan, Hokkaido | 2004 | DM81 | J7 | KP330849 | KP330759 | KP330669 | KP330939 | KP331029 |
| P. effusa | Spinacia oleracea | Japan, Shiga | Unknown | DM34 | J2 | KP330850 | KP330760 | KP330670 | KP330940 | KP331030 |
| P. effusa | Spinacia oleracea | Japan, Shiga | 2000 | DM65 | J3 | KP330851 | KP330761 | KP330671 | KP330941 | KP331031 |
| P. effusa | Spinacia oleracea | Korea, Namyangju | 1999 | KUS-F15680 | 2748 | KP330852 | KP330762 | KP330672 | KP330942 | KP331032 |
| P. effusa | Spinacia oleracea | Korea, Seoul | 2001 | KUS-F18808 | D53 | KP330853 | KP330763 | KP330673 | KP330943 | KP331033 |
| P. effusa | Spinacia oleracea | Korea, Sinan | 2001 | KUS-F18809 | D54 | KP330854 | KP330764 | KP330674 | KP330944 | KP331034 |
| P. effusa | Spinacia oleracea | Mexico, Nogales | 1953 | BPI788308 | BP37 | KP330855 | KP330765 | KP330675 | KP330945 | KP331035 |
| P. effusa | Spinacia oleracea | Mexico, Nogales | 1949 | BPI788314 | BP43 | KP330856 | KP330766 | KP330676 | KP330946 | KP331036 |
| P. effusa | Spinacia oleracea | Sweden, Gästrikland | 1942 | UPS | S19 | KP330857 | KP330767 | KP330677 | KP330947 | KP331037 |
| P. effusa | Spinacia oleracea | USA, California | 2012 | DML2 | CAL2 | KP330858 | KP330768 | KP330678 | KP330948 | KP331038 |
| P. effusa | Spinacia oleracea | USA, California | 2012 | DML39 | CAL39 | KP330859 | KP330769 | KP330679 | KP330949 | KP331039 |
| P. effusa | Spinacia oleracea | USA, California | 2012 | DML40 | CAL40 | KP330860 | KP330770 | KP330680 | KP330950 | KP331040 |
| P. effusa | Spinacia oleracea | USA, California | 2012 | DML5 | CAL5 | KP330861 | KP330771 | KP330681 | KP330951 | KP331041 |
| P. effusa | Spinacia oleracea | USA, Kingston | 1896 | UPS | S20 | KP330862 | KP330772 | KP330682 | KP330952 | KP331042 |
| P. effusa | Spinacia oleracea | USA, Maryland | 1958 | BPI788300 | BP29 | KP330863 | KP330773 | KP330683 | KP330953 | KP331043 |
| P. effusa | Spinacia oleracea | USA, Maryland | 1951 | BPI788309 | BP38 | KP330864 | KP330774 | KP330684 | KP330954 | KP331044 |
| P. effusa | Spinacia oleracea | USA, Oklahoma | 1943 | BPI791055 | BP81 | KP330865 | KP330775 | KP330685 | KP330955 | KP331045 |
| P. effusa | Spinacia oleracea | USA, Virginia | 1932 | BPI788361 | BP57 | KP330866 | KP330776 | KP330686 | KP330956 | KP331046 |
| P. boni-henrici | Chenopodium bonus-henricus | Austria, Tirol | 1991 | GLM51002 | 177 | KP330867 | KP330777 | KP330687 | KP330957 | KP331047 |
Including cox2, cox1, and the spacer region between cox2 and cox1.
2.2. DNA extraction, PCR amplification, and sequencing
In total, 5–20 mg of infected plant tissue from herbarium specimens were disrupted in a mixer mill (MM2, Retsch, Germany), using three iron beads of each 3 mm and 1 mm diameters per sample and shaking at maximum speed for 3 min. Genomic DNA was extracted using the BioSprint 96 DNA Plant Kit (Qiagen) on a KingFisher Flex (Thermo Scientific) robot or using the NucleoSpin® Plant II kit (Machery-Nagel). Five mitochondrial and two nuclear markers were amplified by PCR using combinations of Oomycete-specific primers (Table S2); primers ITS1-O (Bachofer, 2004) and LR-0 (reverse complementary to LR-0R (Moncalvo et al., 1995)) for the complete ITS rDNA (amplicon length of 854 bp), HSP90-F1 (Blair et al., 2008) and HSP90-R1C (designed here: ACMCCCTTGACRAASGABAGRTAC) for hsp90 nDNA (926 bp), cox2-F (Hudspeth et al., 2000) and FMPh-10b (Martin et al., 2004) for cox2 mtDNA and the spacer region between cox2 and cox1 genes (1015–1089 bp), OomCox1-levup and OomCox1-levlo (Robideau et al., 2011) for the cox1 mtDNA (680 bp), Prv9-F and Prv9-R (Blair et al., 2012) for rps10 mtDNA (approx. 601 bp), NADHF3C (designed here: AGGWGCDTTAAGATCAACDGCWCA) and NADHR1 (Kroon et al., 2004) for nad1 mtDNA (548 bp). In some cases, the cox2 gene and the spacer region had to be separately amplified with two primer sets, cox2-F & cox2-R (Hudspeth et al., 2000) and FM79 & FMPh-10b (Martin et al., 2004). Amplification reactions were carried out in 25 μL with genomic DNA from 1 to 3 ng, 1× Mango PCR Buffer, 0.2 mM dNTPs, 2 mM MgCl2, 0.8 mg/mL BSA, 0.4 μM forward and reverse primers, and 0.5 Unit Mango Taq Polymerase (Bioline GmbH, Luckenwalde, Germany). PCR conditions were as follows: an initial denaturation step of 95 °C for 4 min; 36 cycles of 95 °C for 40 s, primer set specific annealing temperature for 40 s (see Table S2), 72 °C extension for 60 s; final extension of 5 min at 72 °C. For hsp90 gene and a spanning region of cox2 and cox2-1 spacer, the extension time was extended to 90 s. For rps10 gene, the extension time was shortened to 40 s, and the cycle numbers were increased to 40 cycles. Amplicons were sequenced at the Biodiversity and Climate Research Centre (BiK-F) laboratory using primers identical to those used for amplifications.
2.3. Phylogenetic analysis
Sequences were edited using the DNAStar software package (DNAStar, Inc., Madison, Wis., USA), version 5.05. An alignment of each locus was performed using MAFFT 7 (Katoh and Standley, 2013) employing the Q-INS-i algorithm (Katoh and Toh, 2008). SequenceMatrix 1.7.8 (Vaidya et al., 2011) was used for concatenating individual gene sequences and for checking unusually similar or divergent sequences. We made concatenation alignments for each nuclear (ITS and hsp90), mitochondrial (cox2, coxS, cox1, nad1, rps10), and all seven loci. To assess the relative stability of trees with respect to different inference methods, we used four different tree construction methods: Maximum Likelihood (ML), Minimum Evolution (ME), Maximum Parsimony (MP), and Bayesian inference (MCMC). The best-fit substitution models were identified for each locus in MEGA 6.0 (Tamura et al., 2013); these were the general time reversible (GTR) model for mitochondrial loci and the Tamura-Nei (TN) for nuclear loci. Evolutionary rates were estimated at a value of 1.0. For ML analyses, 1000 rounds of random addition of sequences as well as 1000 fast bootstrap replicates were performed using RAxML 7.0.3 (Stamatakis, 2006) as implemented in raxmlGUI 1.3 (Silvestro and Michalak, 2012) using the GTRCAT variant. ME analysis was done using MEGA 6.0, with the default settings of the program, except for using the TN model instead of the maximum composite likelihood model. MP analysis was performed in PAUP 4.0b10 (Swofford, 2002), using 1000 replicates of heuristic search with random addition of sequences and subsequent TBR branch swapping. All sites were treated as unordered and unweighted, with gaps treated as missing data. The reproducibility of the internal nodes of the resulting trees was tested by bootstrap analysis using 1000 replications. MCMC analysis was performed using MrBayes, version 3.2 (Ronquist et al., 2012), with the GTR model and gamma-distributed substitution rates. Four incrementally heated simultaneous Markov chains were run for 10 M generations, with a tree saved every 1000th generation. We assessed convergence and the effective sample size exceeding 200 for each parameter every run, as implemented in Tracer v.1.5 (Rambaut and Drummond, 2009). The AWTY software (Nylander et al., 2008) was also used to confirm the convergence graphically. After checking for convergence and ESS, we discarded the first 10% of trees as a burn-in.
2.4. Species tree estimation
Uncertainty in phylogenetic relationships resulting from the multi-locus concatenation was estimated with *BEAST 2 (Heled and Drummond, 2010). Individual Bayesian trees were inferred for each locus in three datasets (nuclear, mitochondrial, and all seven loci), and relationships based on the combined dataset were estimated using Bayesian analysis. BEAUTi 2 (Bouckaert et al., 2014) was used to create the XML-formatted input files for *BEAST 2. Individual assignments to taxonomic units followed the specific host plant, as well as the clades inferred from the concatenation analysis, under the Taxon sets tab. For the dataset partitioned by locus, the best-fit substitution models and evolutionary rates were identical with ones of the above phylogenetic analysis. The topology and support values were compared under two tree priors (Yule versus birth–death process) and molecular clock models (strict versus relaxed). All datasets were run for 100 M generations, sampling every 5000th generation. Topological convergence and adequate ESS were checked using Tracer and AWTY, as outlined above. After removing the first 25% of trees, a maximum clade credibility tree with the 95% highest probability density was produced by using TreeAnnotator version 2.1.2 (Rambaut and Drummond, 2010), and visualized in FigTree (http://tree.bio.ed.ac.uk/software/figtree/). We used DensiTree (Bouckaert, 2010) to jointly visualize the posterior distribution of the species trees from *BEAST 2. As this program uses transparent lines, thus in areas where many of the trees agree with respect to topology and branch length, there will be many lines drawn and the screen will show a darker color for the respective branch.
3. Results
3.1. ITS rDNA phylogeny
The phylogenetic relationship of ITS sequences was inferred using ML, ME, MP, and MCMC analyses. Since all four analyses did not show support for divergent topologies, only the ML tree is presented in Fig. S1, with the addition of the support values of the other analyses. The Peronospora isolates originating from the genera Beta, Emex, Rumex, Spergula, Spergularia, Polycarpon, and Spinacia grouped together with high support values in all analyses. Within this groups, most sequences were identical, thus relationships in this group could not be resolved. Even though ITS sequence showed a sufficient resolution to distinguish P. effusa isolates ex Spinacia oleracea from other isolates with varying support, discrimination between Peronospora specimens from Polycarpon, Rumex, Spergula, and Spergularia was not possible. Although a group including all BDM specimens was obtained, it also included specimens from Emex spinosa. The two groups representing P. effusa and BDM (including P. sp. ex Emex) revealed a unique single-base substitution in the ITS2 region and the 5.8S gene, respectively. On Atriplex, two distinct lineages representing P. litoralis and P. minor/P. atriplicis-hastatae were found, which were sister to all other groups.
3.2. Multi-locus phylogeny
Trees based on each concatenated alignment for five mitochondrial and two nuclear loci (Fig. S2) showed no strong conflicting support with respect to a phylogeny based on the concatenated alignment of mitochondrial and nuclear loci (fig 1). A concatenated alignment of all mitochondrial and nuclear loci had 4378 total characters, including 311 variable characters, 123 of which were parsimony informative; cox2 (708 characters, 65 variable, 30 parsimony informative), the spacer region between cox2 and cox1 (272, 22, 9), cox1 (701, 58, 28), nad1 (502, 47, 22), rps10 (541, 19, 4), ITS (799, 41, 10), and hsp90 (855, 59, 20). The final matrix was deposited in TreeBASE (http://www.treebase.org) and is available under http://purl.org/phylo/treebase/phylows/study/TB2:S16896.
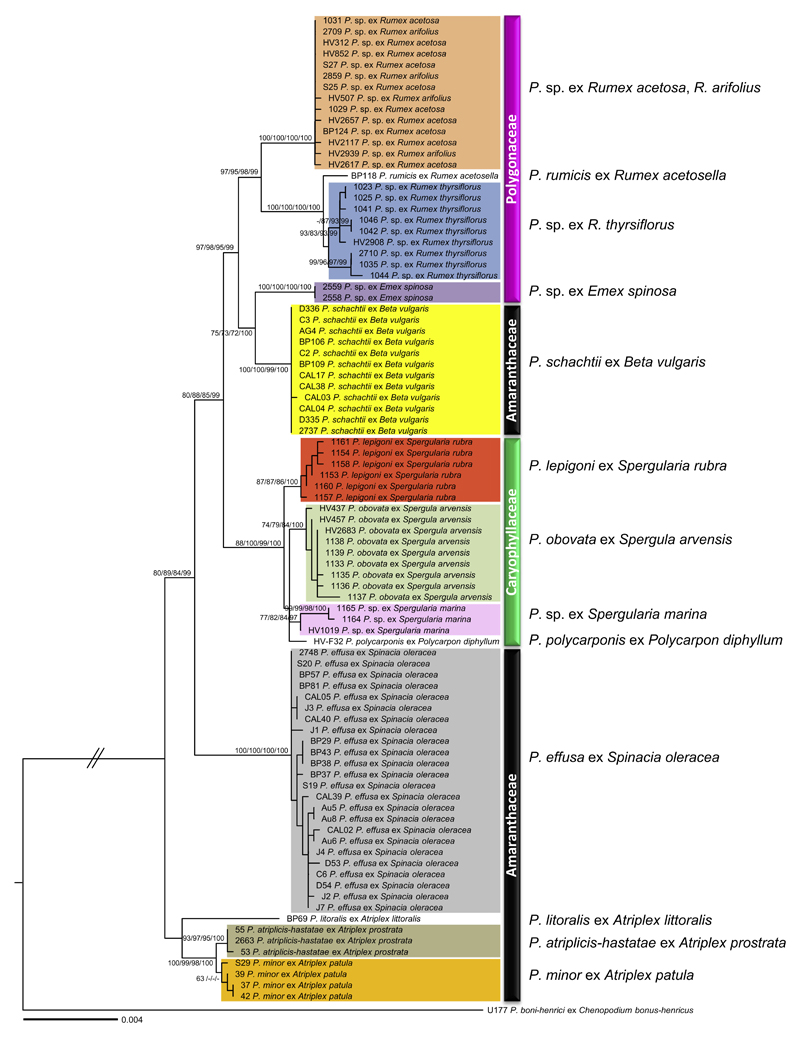
Fig. 1.
Phylogenetic tree inferred from Maximum Likelihood analysis of a concatenated alignment (4378 nucleotides with 123 parsimony informative sites) of five mitochondrial (cox2, coxS, cox1, nad1, rps10) and two nuclear loci (ITS and hsp90). Support values (ML BS/ME BS/ MP BS/MCMC PP) higher than 60% are given above or below the branches. The scale bar equals the number of nucleotide substitutions per site. Specimens originating from different host families are marked with black (Amaranthaceae), pink (Polygonaceae), and green (Caryophyllaceae) bars. ML: Maximum Likelihood, ME: Minimum Evolution, MP: Maximum Parsimony, MCMC: Bayesian inference, BS: Bootstrap support, PP: Posterior Probabilities. (For interpretation of thereferences to color in this figure legend, the reader is referred to the web version of this article.)
Since the concatenated dataset for all mitochondrial and nuclear loci revealed no significant conflicts in the tree topologies derived from ML, ME, MP, and MCMC analyses, only the tree from ML inference is shown in Fig. 1. The multi-gene phylogeny showed a high resolution, enabling differentiation between all closely related species of the complex clade, and resolving the uncertainties found in the ITS tree (Fig. S1). The discrimination of the lineages parasitic to specific host genera or species received high to maximum support in all phylogenetic analyses. In the multi-locus tree, all BDM sequences together formed a distinct group, with high support values of 100/100/99/100 in four analyses. All sequences, excluding one specimen ‘CAL03’ (Table 1), were identical to each other, demonstrating the genetic homogeneity of the pathogen causing BDM disease in several regions worldwide. BDM was further distinguishable from Peronospora specimens from Emex spinosa, which had identical ITS sequences. All sequences of P. effusa grouped together with maximum support in all analyses. Also the resolution for Peronospora specimens from caryophyllaceous plants was much improved in the concatenated dataset, compared to the ITS tree. All Peronospora specimens from the Caryophyllaceae, Polycarpon, Spergula, and Spergularia, grouped together with the high support of 88/100/99/100. They further split into three subgroups, corresponding to a particular host plant; P. polycarponis from Polycarpon diphyllum, P. obovata from Spergula arvensis, and P. lepigoni from Spergularia rubra. A fourth subgroup exists with a previously undescribed lineage of Peronospora occurring on Spergularia marina. All specimens from Rumex species formed a monophyletic group with high support values. This group was further subdivided corresponding to particular host species. The type species of Peronospora, P. rumicis, is likely restricted to R. acetosella, as specimens from both R. thyrsiflorus and the other from R. acetosa (including R. arifolius) were highly distinct from P. rumicis from R. acetosella. Peronospora rumicis and Peronospora specimens from R. thyrsiflorus grouped together with maximum support in all analyses. Within specimens identified as R. thyrsiflorus, two distinct groups were observed, but based on the specimens investigated in this study, no morphological differences could be found in preliminary screens. The Peronospora isolates from Emex spinosa and Spergularia marina have respectively been considered to be P. rumicis and P. lepigoni, but are distinct from those species on the basis of the multi-locus phylogeny. Peronospora isolates parasitic to Atriplex were consistently found to be the sister group to all other lineages in the Peronospora clade under investigation. The monophyly of this group received low support values in all analyses, probably due to the slightly different placements of P. litoralis in nuclear (Fig. S2a) and mitochondrial datasets (Fig. S2b). This group was further divided into three clades corresponding to a particular Atriplex species, A. littoralis, A. prostrata, and A. patula, with the latter two clades grouping together with strong support (100/99/98/100).
3.3. Species tree estimation
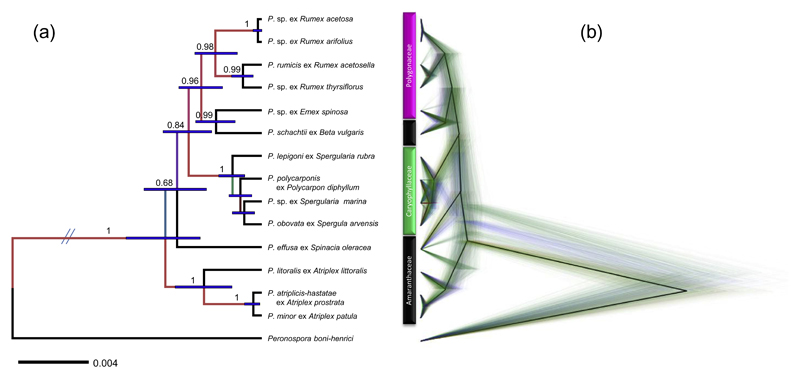
In order to evaluate the results from the concatenated dataset, the species trees of the mitochondrial and nuclear loci were generated by *BEAST, and summarized in a maximum clade credibility tree (fig 2A) and cloudogram (Fig 2B). With an identical tree topology, which was robust to changes in the tree prior (Yule versus birth–death process) and molecular clock model (strict versus relaxed) tested, the species trees showed largely congruent results with the concatenated dataset, except for the grouping of the Peronospora species occurring on members of Atriplex. The species trees strongly supported a grouping of the Peronospora species from Atriplex, with a posterior probability of 1.00, while it was not significantly supported by the concatenated dataset (Fig 1), in which support values were low in all analyses (ML, ME, MP, and MCMC). In both species trees, P. effusa occupied a basal position to specimens from Caryophyllaceae, Polygonaceae and Beta vulgaris, like in the concatenated tree, but the posterior probability is low (0.68) in the maximum clade credibility tree (Fig 2A), due to a slightly different placement of P. effusa and Peronospora species from Atriplex in mitochondrial loci (Fig. S3b). The species tree estimations confirmed that the four Peronospora lineages from Caryophyllaceae form a monophyletic cluster, but along with the concatenation analysis, the evolutionary relationships within this cluster were unclear with regard to none or only low posterior probabilities.
Fig. 2.
Species tree estimation from five mitochondrial (cox2, coxS, cox1, nad1, rps10) and two nuclear loci (ITS and hsp90) using *BEAST. (a) Maximum clade credibility tree visualized by FigTree. Bars correspond to the 95% highest posterior density range, and posterior probabilities higher than 0.60 are shown above branches. (b) Cloudogram of all trees of the MCMC visualized by DensiTree (Bouckaert, 2010). Each possible topology is shown in green with branch lengths averaged among all trees showing that particular topology. Four alternative interpretations are shown in black for the consensus tree, blue for the most frequently occurring topology, red for second, and yellow for third most frequently occurring topology. Higher levels of uncertainty are represented by lower densities of the lines. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
4. Discussion
Despite ITS rDNA being recognized as a potential barcoding marker for identification of Oomycetes (Robideau et al., 2011) and Fungi (Schoch et al., 2012), ITS sequences in some cases may be unsuitable to resolve the phylogenetic relationships among closely related species. In the largest genus of oomycetes, Peronospora, there are a few complex clades containing several distinct species, which were mostly not distinguishable based on ITS sequences, e.g. the Peronospora arborescens complex (Voglmayr et al., 2014b), the P. effusa-rumicis-schachtii complex (Choi et al., 2007c), the P. sparsa complex parasitic to Rosaceae (Cooke et al., 2002; Voglmayr, 2003; Choi et al., 2007a), and the P. viciae complex parasitic to Fabaceae (García-Blázquez et al., 2008). To determine the previously unresolved relationships within the Peronospora effusa-rumicis-schachtii complex (Voglmayr, 2003; Choi et al., 2007c), the present study used multi-locus analyses based on a concatenated dataset and species tree estimations. The nearly identical topologies inferred from the concatenated and coalescent-based analyses illustrate the distinctiveness of all Peronospora species included in this study and correspondence to the specific host genus or species that they infect. Additionally, this study revealed some previously unnoticed, host-specific lineages of Peronospora and that the type species of this genus is likely restricted to R. acetosella. These results support the view that a narrow species concept reflects the evolutionary history of downy mildews much better than a broad species concept, which is in line with recent studies for other species complexes, such as Bremia lactucae (Voglmayr et al., 2004; Choi et al., 2007b, 2011c), Hyaloperonospora parasitica (Choi et al., 2003, 2011b; Göker et al., 2004, 2009; Voglmayr and Göker, 2011; Voglmayr et al., 2014a), Peronospora arborescens (Voglmayr et al., 2014b), Peronospora lamii (Belbahri et al., 2005; Choi et al., 2009c; Thines et al., 2009b), and Plasmopara halstedii (Spring et al., 2003; Komjáti et al., 2007; Choi et al., 2009a).
The taxonomic status of the causal agent for beet downy mildews (BDM) remained obscure due to previous uncertainties on species delimitation of downy mildews. This is due to two conflicting views regarding species differentiation in downy mildews. As a result, BDM is often referred to as either P. schachtii (narrow host range) or P. farinosa (broad host range). The present phylogenetic study has revealed the distinctiveness of BDM from all other Peronospora species. The independence of BDM agrees with previous works about host specialization as several authors were unable to cross-inoculate between BDM and Peronospora isolates from other chenopodiaceous hosts, suggesting a strict host specificity (Leach, 1931; Richards, 1939; Darpoux and Durgéat, 1962; Byford, 1967; Klosterman et al., 2014). Consequently, we suggest that the name P. schachtii should be used for BDM. In addition, P. farinosa is a name that cannot be associated with any type specimen and the description contains insufficient detail to ascertain the pathogen and host species; it is even unclear if the original description refers to a downy mildew. Thus, the name Botrytis farinosa (the basionym of P. farinosa) has been proposed for rejection (Choi and Thines, 2014). None of the species described from Atriplex (the type host of the dubious taxon P. farinosa) is closely related to P. schachtii.
In spite of the economic importance of beet downy mildew, the genetic diversity of P. schachtii has not yet been investigated. In this study, all but one specimens originating from geographically diverse countries (Argentina, China, France, Germany, Korea, and the USA) were identical to each other in ITS, cox2, cox1, nad1, rps10, and hsp90 loci. The one strain (CAL03) that was not identical had only a single base substitution in the spacer region between cox2 and cox1. The Peronospora samples of the two varieties of Beta vulgaris, namely, var. saccharifera (sugar beet) and var. cicla (Swiss chard) did thus not show genetic differentiation from each other in the loci investigated. The beet crop is thought to be native to the Mediterranean (De Bock, 1985; Hanelt, 2001), and has been domesticated over thousands of years from a wild form without swollen roots. Its potential as a source of sugar was not discovered until 1747, and only in the early part of the 19th century when its breeding reached high sugar levels, the beet became a popular vegetable in Europe (Rolph, 1917). In other parts of the world, cultivation has started only recently. This suggests that P. schachtii may have spread together with the crop, possibly starting from an originally very small pathogen population. The resulting comparatively short period of time the crop is grown worldwide might explain the genetic homogeneity of the P. schachtii isolates.
The present study consistently supports a monophyletic grouping of specimens occurring on spinach, but P. effusa isolates appear to be genetically much more heterogeneous compared to P. schachtii. Spinach originated from the Southwest Asian region and has cultivated longer cultivation history outside of its native area. It has become an established vegetable in Europe in the early part of the 16th century, although it has been grown in China since the 7th or 8th century (Sturtevant, 1890). Consequently, the downy mildew pathogens have undergone a recent and major expansion in geographic range throughout the past centuries, possibly also starting from a small area.
The present study also sheds some light on the phylogeny of a taxonomically important species, P. rumicis, the type species of the genus Peronospora. Although Peronospora specimens from Rumex species formed a monophyletic group in all phylogenetic analyses, they are differentiated into several distinct, highly supported lineages. It is noteworthy that the Peronospora specimens on R. acetosa s.l. are genetically distinct from P. rumicis on the type host, R. acetosella. This suggests that the name P. rumicis should not be applied to downy mildew pathogens other than from R. acetosella, even though it has commonly been used to refer to downy mildew pathogens of members of the genus Rumex and closely related genera (Hall, 1994; Constantinescu and Fatehi, 2002).
The multilocus phylogeny and species tree estimations provide additional evidence that distinct species of downy mildews can generally infect only a narrow range of closely related host plants or, sometimes, only a particular host species. However, there is increasing evidence that this is not a curious exception in obligate biotrophic fungi. As these organisms live in intimate and specific associations with their host, their phylogeny often mirrors the phylogeny of their hosts (Fahrenholz, 1913), which has sometimes been referred to as “Fungi as plant taxonomists” (Hijwegen, 1979). Similarly, for the white blister rusts (Albuginaceae), another obligate biotrophic family of oomycetes, several distinct species of Albugo and Pustula were observed to exist on specific host species of the Brassicaceae (Choi et al., 2006, 2007d, 2008b; Thines et al., 2009a; Ploch et al., 2010; Choi et al., 2011a) and Asteridae (Ploch et al., 2011; Rost and Thines, 2012), respectively. Also, in several well-known obligate biotrophic fungi such as rusts (Uredinales), smuts (Ustilaginales), and powdery mildews (Erysiphales), specific associations have been widely recorded, resulting in a high species diversity of about 7000, 1600, and 900 species, respectively (Ainsworth, 2008; Braun and Cook, 2012; Vánky, 2012), which by far surpasses facultative or necrotrophic pathogens. However, there are also a few exceptions from this, such as Pseudoperonospora cubensis in case of the downy mildews (Runge et al., 2011) or Albugo candida in case of the white blister rusts (Choi et al., 2007d, 2008b, 2009b; Thines et al., 2009a; Ploch et al., 2010).
The macro-evolutionary patterns in downy mildews and other biotrophic pathogens are still vastly unexplored. But from the various hosts parasitized by some closely related downy mildew species, e.g. in Peronospora (Voglmayr, 2003) or Hyaloperonospora (Göker et al., 2004), it becomes apparent that downy mildews and their hosts cannot have co-diverged on larger evolutionary scales. In this study, the molecular affinities among Peronospora species roughly mirror that their host plants, but the congruence of downy mildew and host phylogenies is mostly biased to the terminal branches. This phenomenon can probably be explained by the recent radiation and speciation of downy mildews following at least two independent host shifts from Amaranthaceae to two other families, Caryophyllaceae and Polygonaceae, rather than contemporary cospeciation with host plants. This pattern of host shifts and subsequent radiation has also been observed in the downy mildew genus Bremia (Choi & Thines, unpublished). These results suggest that host shifts and radiation are the main evolutionary driving forces for the speciation of downy mildews. Mostly, host shifts occur between closely related species and genera within a host family, which can at first sight be misinterpreted as cospeciation. However, also within the parasites of a specific host family, host and parasite phylogenies usually do not match completely, providing clear evidence for radiation after host shifts in a range of related hosts. Similarly, in white blister rusts (Choi et al., 2007d, 2008b, 2011d; Thines et al., 2009a), powdery mildews (Matsuda and Takamatsu, 2003; Inuma et al., 2007), rusts (Savile, 1979; Van der Merwe et al., 2008) and smuts (Antonovics et al., 2002; Lopez-Villavicencio et al., 2005), it was shown that host shifts may occur frequently throughout the evolutionary history of plant pathogens, although the underlying mechanisms still remain obscure. In summary, it can be assumed that the downy mildews and probably also other biotrophic pathogens evolve by (1) host shifts that expend their host ranges to taxonomically closely related but more rarely to distantly related plants, and (2) subsequent radiation and speciation in a group of closely related hosts that eventually lead to specialization and dependence on a specific host genus or even a single species. This pattern of repeated rare major host jumps followed by radiation might enable biotrophic pathogens to persist in their hosts groups, as this helps to evade effector-triggered immunity. Arguably, this type of immunity becomes more pronounced with increasing time of host-pathogen co-existence, and if the pathogen has a negative effect on plant fitness, it could eventually drive most pathogenic species to extinction. This might explain, why most of the deep-rooted downy mildew genera exhibit broad host ranges, like in Peronospora or Plasmopara, or consist of only a single or few species, like in the case of Benua or Plasmoverna, respectively. Further studies are required to evaluate if this pattern also holds up for other biotrophic pathogens, such as powdery mildew, rust, or smut fungi.
Supplementary Material
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.ympev.2015.03.003.
Acknowledgments
The authors are grateful to curators of BPI (USA), DM (Japan), BRNM (Czech Republic), GLM (Germany), HMAS (China), KUS-F (South Korea), LE (Russia), UPS (Sweden), VPRI (Australia), and WU (Austria) for providing Peronospora specimens. YJC was supported by a fellowship from the Alexander von Humboldt foundation. SJK was supported in part by the California Leafy Greens Research Program. Financial support by the Austrian Science Fund (FWF; project P22739-B20) to HV is gratefully acknowledged. This study was supported by the LOEWE excellence program of the German state of Hessen, in the framework of the Integrative Fungal Research Cluster (IPF) and the Biodiversity and Climate Research Centre (BiK-F). YJC and MT conceived the study with a contribution from SJK. HDS, HV, VK, and YJC provided materials. YJC conducted experiments. YJC analysed and interpreted the data with contributions from MT. YJC and MT wrote the manuscript with contributions from HV, SJK, HDS, VK.
References
- Ainsworth GC. Ainsworth & Bisby’s Dictionary of the Fungi. CAB International; Wallingford: 2008. [Google Scholar]
- Antonovics J, Hood M, Partain J. The ecology and genetics of a host shift: Microbotryum as a model system. Am Nat. 2002;160:S40–S53. doi: 10.1086/342143. [DOI] [PubMed] [Google Scholar]
- Bachofer M. Molekularbiologische Populationsstudien an Plasmopara halstedii, dem Falschen Mehltau der Sonnenblume. University of Hohenheim; Stuttgart: 2004. [Google Scholar]
- Belbahri L, Calmin G, Pawlowski J, Lefort F. Phylogenetic analysis and real time PCR detection of a presumably undescribed Peronospora species on sweet basil and sage. Mycol Res. 2005;109:1276–1287. doi: 10.1017/s0953756205003928. [DOI] [PubMed] [Google Scholar]
- Berlese AN, De Toni JB. In: Peronosporaceae. Saccardo PA, editor. Sylloge Fungorum; Padua: 1888. pp. 233–264. [Google Scholar]
- Blair JE, Coffey MD, Park SY, Geiser DM, Kang S. A multi-locus phylogeny for Phytophthora utilizing markers derived from complete genome sequences. Fungal Genet Biol. 2008;45:266–277. doi: 10.1016/j.fgb.2007.10.010. [DOI] [PubMed] [Google Scholar]
- Blair JE, Coffey MD, Martin FN. Species tree estimation for the late blight pathogen, Phytophthora infestans, and close relatives. PloS One. 2012;7:e37003. doi: 10.1371/journal.pone.0037003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouckaert RR. DensiTree: making sense of sets of phylogenetic trees. Bioinformatics. 2010;26:1372–1373. doi: 10.1093/bioinformatics/btq110. [DOI] [PubMed] [Google Scholar]
- Bouckaert R, Heled J, Kuhnert D, Vaughan T, Wu CH, Xie D, Suchard MA, Rambaut A, Drummond AJ. BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Comp Biol. 2014;10:e1003537. doi: 10.1371/journal.pcbi.1003537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandenberger LP, Correll JC, Morelock TE. Nomenclature of the downy mildew fungus on spinach. Mycotaxon. 1991;41:157–160. [Google Scholar]
- Braun U, Cook RT. Taxonomic Manual of the Erysiphales (powdery mildews) CBS-KNAW Fungal Biodiversity Centre Utrecht; The Netherlands: 2012. [Google Scholar]
- Byford WJ. Host specialization of Peronospora farinosa on Beta, Spinacia and Chenopodium. Trans Brit Mycol Soc. 1967;50:603–607. [Google Scholar]
- Byford WJ. Downy mildews of beet and spinach. In: Spencer DM, editor. The Downy Mildews. Academic Press; London, New York, San Francisco: 1981. pp. 531–543. [Google Scholar]
- Choi YJ, Thines M. (2288) Proposal to reject the name Botrytis farinosa (Peronospora farinosa) (Peronosporaceae: Oomycetes) Taxon. 2014;63:675–676. [Google Scholar]
- Choi YJ, Hong SB, Shin HD. Diversity of the Hyaloperonospora parasitica complex from core brassicaceous hosts based on ITS rDNA sequences. Mycol Res. 2003;107:1314–1322. doi: 10.1017/s0953756203008578. [DOI] [PubMed] [Google Scholar]
- Choi YJ, Hong SB, Shin HD. A reconsideration of Pseudoperonospora cubensis and Ps. humuli based on molecular and morpholgical data. Mycol Res. 2005;109:841–848. doi: 10.1017/s0953756205002534. [DOI] [PubMed] [Google Scholar]
- Choi YJ, Hong SB, Shin HD. Genetic diversity within the Albugo candida complex (Peronosporales, Oomycota) inferred from phylogenetic analysis of ITS rDNA and COX2 mtDNA sequences. Mol Phylogen Evol. 2006;40:400–409. doi: 10.1016/j.ympev.2006.03.023. [DOI] [PubMed] [Google Scholar]
- Choi YJ, Constantinescu O, Shin HD. A new downy-mildew of the Rosaceae: Peronospora oblatispora sp. nov. (Chromista, Peronosporales) Nova Hedwigia. 2007a;85:93–101. [Google Scholar]
- Choi YJ, Hong SB, Shin HD. Extreme size and sequence variation in the ITS rDNA of Bremia lactucae. Mycopathologia. 2007b;163:91–95. doi: 10.1007/s11046-007-0092-7. [DOI] [PubMed] [Google Scholar]
- Choi YJ, Hong SB, Shin HD. Re-consideration of Peronospora farinosa infecting Spinacia oleracea as distinct species, Peronospora effusa. Mycol Res. 2007c;111:381–391. doi: 10.1016/j.mycres.2007.02.003. [DOI] [PubMed] [Google Scholar]
- Choi YJ, Shin HD, Hong SB, Thines M. Morphological and molecular discrimination among Albugo candida materials infecting Capsella bursa-pastoris world-wide. Fungal Divers. 2007d;27:11–34. [Google Scholar]
- Choi YJ, Denchev CM, Shin HD. Morphological and molecular analyses support the existence of host-specific Peronospora species infecting Chenopodium. Mycopathologia. 2008a;165:155–164. doi: 10.1007/s11046-007-9087-7. [DOI] [PubMed] [Google Scholar]
- Choi YJ, Shin HD, Ploch S, Thines M. Evidence for uncharted biodiversity in Albugo candida complex, with the description of a new species. Mycol Res. 2008b;112:1327–1334. doi: 10.1016/j.mycres.2008.04.015. [DOI] [PubMed] [Google Scholar]
- Choi YJ, Kiss L, Vajna L, Shin HD. Characterization of a Plasmopara species on Ambrosia artemisiifolia, and notes on P. halstedii, based on morphology and multiple gene phylogenies. Mycol Res. 2009a;113:1127–1136. doi: 10.1016/j.mycres.2009.07.010. [DOI] [PubMed] [Google Scholar]
- Choi YJ, Shin HD, Thines M. The host range of Albugo candida extends from Brassicaceae through Cleomaceae to Capparaceae. Mycol Prog. 2009b;8:329–335. [Google Scholar]
- Choi YJ, Shin HD, Thines M. Two novel Peronospora species are associated with recent reports of downy mildew on sages. Mycol Res. 2009c;113:1340–1350. doi: 10.1016/j.mycres.2009.08.010. [DOI] [PubMed] [Google Scholar]
- Choi YJ, Danielsen S, Lübeck M, Hong SB, Delhey R, Shin HD. Morphological and molecular characterization of the causal agent of downy mildew on Quinoa (Chenopodium quinoa) Mycopathologia. 2010;169:403–412. doi: 10.1007/s11046-010-9272-y. [DOI] [PubMed] [Google Scholar]
- Choi YJ, Shin HD, Ploch S, Thines M. Three new phylogenetic lineages are the closest relatives of the widespread species Albugo candida. Fungal Biol. 2011a;115:598–607. doi: 10.1016/j.funbio.2011.02.006. [DOI] [PubMed] [Google Scholar]
- Choi YJ, Shin HD, Voglmayr H. Reclassification of two Peronospora species parasitic on Draba in Hyaloperonospora based on morphological molecular and phylogenetic data. Mycopathologia. 2011b;171:151–159. doi: 10.1007/s11046-010-9340-3. [DOI] [PubMed] [Google Scholar]
- Choi YJ, Thines M, Runge F, Hong SB, Telle S, Shin HD. Evidence for high degrees of specialisation, evolutionary diversity, and morphological distinctiveness in the genus Bremia. Fungal Biol. 2011c;115:102–111. doi: 10.1016/j.funbio.2010.10.010. [DOI] [PubMed] [Google Scholar]
- Choi YJ, Thines M, Shin HD. A new perspective on the evolution of white blister rusts: Albugo s. str. (Albuginales; Oomycota) is not restricted to Brassicales but also present on Fabales. Org Divers Evol. 2011d;11:193–199. [Google Scholar]
- Constantinescu O. An annotated list of Peronospora names. Thunbergia. 1991;15:1–110. [Google Scholar]
- Constantinescu O, Fatehi J. Peronospora-like fungi (Chromista, Peronosporales) parasitic on Brassicaceae and related hosts. Nova Hedwigia. 2002;74:291–338. [Google Scholar]
- Constantinescu O, Negrean G. Check-list of Romanian Peronosporales. Mycotaxon. 1983;16:537–556. [Google Scholar]
- Cooke DEL, Drenth A, Duncan JM, Wagels G, Brasier CM. A molecular phylogeny of Phytophthora and related oomycetes. Fungal Genet Biol. 2000;30:17–32. doi: 10.1006/fgbi.2000.1202. [DOI] [PubMed] [Google Scholar]
- Cooke DEL, Williams NA, Williamson B, Duncan J. An ITS based phylogenetic analysis of the relationships between Peronospora and Phytophthora. In: Spencer-Phillips PTN, Gisi U, Lebeda A, editors. Advances in Downy Mildew Research. Vol. 1. Kluwer Academic; Dordrecht, Boston, London: 2002. pp. 161–165. [Google Scholar]
- Darpoux H, Durgéat L. Studies on the downy mildew of sugar beet; 25th Winter Congress of the Institut International de Recherches Betteraviéres; 1962. [Google Scholar]
- De Bock TS. The genus Beta: domestication, taxonomy and interspecific hybridization for plant breeding; I International Symposium on Taxonomy of Cultivated Plants; 1985. pp. 335–344. [Google Scholar]
- Edwards SV. Is a new and general theory of molecular systematics emerging? Evolution. 2009;63:1–19. doi: 10.1111/j.1558-5646.2008.00549.x. [DOI] [PubMed] [Google Scholar]
- Edwards SV, Liu L, Pearl DK. High-resolution species trees without concatenation. Proc Natl Acad Sci USA. 2007;104:5936–5941. doi: 10.1073/pnas.0607004104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahrenholz H. Ectoparasiten und abstammungslehre. Zool Anz. 1913;41:371–374. [Google Scholar]
- Francis SM, Byford WJ. Peronospora farinosa f. sp. betae. Commonwealth Mycological Institute; Kew: 1983. [Google Scholar]
- Francis SM, Waterhouse GM. List of Peronosporaceae reported from the British Isles. Trans Brit Mycol Soc. 1988;91:1–62. [Google Scholar]
- Fries E. Systema Mycologicum. E. Mauritius; Greifswald: 1832. [Google Scholar]
- Fuckel L. Fungi Rhenani exsiccati. Cent 12–17, No. 1101–1700. Hedwigia. 1866;5:14–16. [Google Scholar]
- García-Blázquez G, Constantinescu O, Tellería MT, Martín MP. Preliminary check list of Albuginales and Peronosporales (Chromista) reported from the Iberian Peninsula and Balearic Islands. Mycotaxon. 2006;98:185–188. [Google Scholar]
- García-Blázquez G, Göker M, Voglmayr H, Martín MP, Tellería MT, Oberwinkler F. Phylogeny of Peronospora, parasitic on Fabaceae, based on ITS sequences. Mycol Res. 2008;112:502–512. doi: 10.1016/j.mycres.2007.10.007. [DOI] [PubMed] [Google Scholar]
- Gäumann E. Zur Kenntnis der Chenopodiaceen bewohnenden Peronospora-Arten. Mittheilungen der naturforschenden Gesellschaft in Bern. 1919:45–66. [Google Scholar]
- Gäumann E. Beiträge zu einer Monographie der Gattung Peronospora Corda. Beiträge zur Kryptogamenflora der Schweiz. 1923;5:1–360. [Google Scholar]
- Göker M, Riethmüller A, Voglmayr H, Weiß M, Oberwinkler F. Phylogeny of Hyaloperonospora based on nuclear ribosomal internal transcribed spacer sequences. Mycol Prog. 2004;3:83–94. [Google Scholar]
- Göker M, Voglmayr H, García-Blázquez G, Oberwinkler F. Species delimitation in downy mildews: the case of Hyaloperonospora in the light of nuclear ribosomal ITS and LSU sequences. Mycol Res. 2009;113:308–325. doi: 10.1016/j.mycres.2008.11.006. [DOI] [PubMed] [Google Scholar]
- Goodwin SB, Legard DE, Smart CD, Levy M, Fry WE. Gene flow analysis of molecular markers confirms that Phytophthora mirabilis and P. infestans are separate species. Mycologia. 1999;91:796–810. [Google Scholar]
- Gustavsson A. Studies on Nordic Peronosporas. I. Taxonomic revision. Opera Bot. 1959;3:1–271. [Google Scholar]
- Hall G. Peronospora rumicis. IMI descriptions of fungi and Bacteria No 1199. Mycopathologia. 1994;126:59–60. [Google Scholar]
- Hanelt P. Mansfeld’s Encyclopedia of Agricultural and Horticultural Crops:(except ornamentals) Springer Press; Berlin, Germany: 2001. [Google Scholar]
- Heled J, Drummond AJ. Bayesian inference of species trees from multilocus data. Mol Biol Evol. 2010;27:570–580. doi: 10.1093/molbev/msp274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hijwegen T. Fungi as plant taxonomists. Symb Bot Upsal. 1979;22:146–165. [Google Scholar]
- Hudspeth DSS, Nadler SA, Hudspeth MES. A COX2 molecular phylogeny of the Peronosporomycetes. Mycologia. 2000;92:674–684. [Google Scholar]
- Inuma T, Khodaparast SA, Takamatsu S. Multilocus phylogenetic analyses within Blumeria graminis, a powdery mildew fungus of cereals. Mol Phylogen Evol. 2007;44:741–751. doi: 10.1016/j.ympev.2007.01.007. [DOI] [PubMed] [Google Scholar]
- Jung T, Burgess TI. Re-evaluation of Phytophthora citricola isolates from multiple woody hosts in Europe and North America reveals a new species, Phytophthora plurivora sp. nov. Persoonia. 2009;22:95–110. doi: 10.3767/003158509X442612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. MAFFT Multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Toh H. Improved accuracy of multiple ncRNA alignment by incorporating structural information into a MAFFT-based framework. BMC Bioinformatics. 2008;9:212. doi: 10.1186/1471-2105-9-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JY, Choi YJ, Shin HD. Downy mildew caused by Peronospora farinosa f. sp. betae newly reported on Swiss chard in Korea. Plant Pathol. 2010;59:405. [Google Scholar]
- Klosterman SJ, Anchieta AG, McRoberts N, Koike ST, Subbarao KV, VoglMayr H, Choi YJ, Thines M, Martin FN. Coupling spore traps and quantitative PCR assays for detection of the downy mildew pathogens of spinach (Peronospora effusa) and beet (Peronospora schachtii) Phytopathology. 2014;104:1349–1359. doi: 10.1094/PHYTO-02-14-0054-R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochman J, Majewski T. Glonowce (Phycomycetes), Wroslikowe (Peronosporales) Panítwowe Wydawnictwo Naukowe; Warszawa: 1970. [Google Scholar]
- Komjáti H, Walcz I, Virányi F, Zipper R, Thines M, Spring O. Characteristics of a Plasmopara angustiterminalis isolate from Xanthium strumarium. Eur J Plant Pathol. 2007;119:421–428. [Google Scholar]
- Kroon LPNM, Bakker FT, Bosch GBMvd, Bonants PJM, Fliera WG. Phylogenetic analysis of Phytophthora species based on mitochondrial and nuclear DNA sequences. Fungal Genet Biol. 2004;41:766–782. doi: 10.1016/j.fgb.2004.03.007. [DOI] [PubMed] [Google Scholar]
- Kubatko LS, Degnan JH. Inconsistency of phylogenetic estimates from concatenated data under coalescence. Syst Biol. 2007;56:17–24. doi: 10.1080/10635150601146041. [DOI] [PubMed] [Google Scholar]
- Leach LD. Downy mildew of the beet, caused by Peronospora schachtii Fuckel. Hilgardia. 1931;6:203–251. [Google Scholar]
- Lopez-Villavicencio M, Enjalbert J, Hood ME, Shykoff JA, Raquin C, Giraud T. The anther smut disease on Gypsophila repens: a case of parasite sub-optimal performance following a recent host shift? J Evol Biol. 2005;18:1293–1303. doi: 10.1111/j.1420-9101.2005.00924.x. [DOI] [PubMed] [Google Scholar]
- Martin FN, Tooley PW, Blomquist C. Molecular detection of Phytophthora ramorum, the causal agent of sudden oak death in California, and two additional species commonly recovered from diseased plant material. Phytopathology. 2004;94:621–631. doi: 10.1094/PHYTO.2004.94.6.621. [DOI] [PubMed] [Google Scholar]
- Matsuda S, Takamatsu S. Evolution of host-parasite relationships of Golovinomyces (Ascomycete: Erysiphaceae) inferred from nuclear rDNA sequences. Mol Phylogen Evol. 2003;27:314–327. doi: 10.1016/s1055-7903(02)00401-3. [DOI] [PubMed] [Google Scholar]
- Mazelaitis J, Staneviciené S. Gleivunai (Myxomycota), Peronosporieciai (Peronosporales) Mokslo ir Enciklopediju Leidykla; Vilnius: 1995. [Google Scholar]
- Moncalvo JM, Wang HH, Hseu RS. Phylogenetic relationships in Ganoderma Inferred from the internal transcribed spacers and 25s ribosomal DNA sequences. Mycologia. 1995;87:223–238. [Google Scholar]
- Nylander JAA, Wilgenbusch JC, Warren DL, Swofford DL. AWTY (are we there yet?): a system for graphical exploration of MCMC convergence in Bayesian phylogenetics. Bioinformatics. 2008;24:581–583. doi: 10.1093/bioinformatics/btm388. [DOI] [PubMed] [Google Scholar]
- Ploch S, Choi YJ, Rost C, Shin HD, Schilling E, Thines M. Evolution of diversity in Albugo is driven by high host specificity and multiple speciation events on closely related Brassicaceae. Mol Phylogen Evol. 2010;57:812–820. doi: 10.1016/j.ympev.2010.06.026. [DOI] [PubMed] [Google Scholar]
- Ploch S, Telle S, Choi YJ, Cunnington J, Priest M, Rost C, Shin HD, Thines M. The molecular phylogeny of the white blister rust genus Pustula reveals a case of underestimated biodiversity with several undescribed species on ornamentals and crop plants. Fungal Biol. 2011;115:214–219. doi: 10.1016/j.funbio.2010.12.004. [DOI] [PubMed] [Google Scholar]
- Rambaut A, Drummond AJ. Tracer 1.5, 2009. MCMC Trace File Analyser. 2009 < http://tree.bio.ed.ac.uk/software/tracer/>.
- Rambaut A, Drummond AJ. TreeAnnotator, Version 1.7.4. MCMC Output Analysis. 2010 < http:www.beast2.org/wiki/index.php/TreeAnnotator>.
- Richards MC. Downy mildew of spinach and its control. Cornell University Agricultural Experiment Station; Ithaca, New York: 1939. [Google Scholar]
- Robideau GP, de Cock AWAM, Coffey MD, Voglmayr H, Brouwer H, Bala K, Chitty DW, Desaulniers N, Eggertson QA, Gachon CMM, Hu CH, et al. DNA barcoding of oomycetes with cytochrome c oxidase subunit I and internal transcribed spacer. Mol Ecol Resour. 2011;11:1002–1011. doi: 10.1111/j.1755-0998.2011.03041.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolph GM. Something About Sugar: Its History, Growth, Manufacture and Distribution. JJ Newbegin; San Francisco: 1917. [Google Scholar]
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Hohna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rost C, Thines M. A new species of Pustula (Oomycetes, Albuginales) is the causal agent of sunflower white rust. Mycol Prog. 2012;11:351–359. [Google Scholar]
- Runge F, Choi YJ, Thines M. Phylogenetic investigations in the genus Pseudoperonospora reveal overlooked species and cryptic diversity in the P. cubensis species cluster. Eur J Plant Pathol. 2011;129:135–146. [Google Scholar]
- Savile DBO. Fungi as aids in higher plant classification. Bot Rev. 1979;45:377–503. [Google Scholar]
- Schoch CL, Seifert KA, Huhndorf S, Robert V, Spouge JL, Levesque CA, Chen W. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc Natl Acad Sci USA. 2012;109:6241–6246. doi: 10.1073/pnas.1117018109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvestro D, Michalak I. RaxmlGUI: a graphical front-end for RAxML. Org Divers Evol. 2012;12:335–337. [Google Scholar]
- Spring O, Voglmayr H, Riethmüller A, Oberwinkler F. Characterization of a Plasmopara isolate from Helianthus × laetiflorus based on cross infection, morphological, fatty acids and molecular phylogenetic data. Mycol Prog. 2003;2:163–170. [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Sturtevant EL. The history of garden vegetables (continued) Am Nat. 1890:719–744. [Google Scholar]
- Swofford D. PAUP 4.0 b10: Phylogenetic analysis using parsimony. Sinauer Associates; Sunderland, MA, USA: 2002. [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiers B. Index Herbariorum: A global directory of public herbaria and associated staff. New York Botanical Garden’s Virtual Herbarium. 2014 < http://sweetgum.nybg.org/ih/> (Retrieved June 2014) [Google Scholar]
- Thines M, Choi YJ, Kemen E, Ploch S, Holub EB, Shin HD, Jones JDG. A new species of Albugo parasitic to Arabidopsis thaliana reveals new evolutionary patterns in white blister rusts (Albuginaceae) Persoonia. 2009a;22:123–128. doi: 10.3767/003158509X457931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thines M, Telle S, Ploch S, Runge M. Identity of the downy mildew pathogens of basil, coleus, and sage with implications for quarantine measures. Mycol Res. 2009b;113:532–540. doi: 10.1016/j.mycres.2008.12.005. [DOI] [PubMed] [Google Scholar]
- Ul’yanishchev VI, Osipyan LL, Kanchaveli LA, Akhundov TM. In: Peronosporovye Griby. Osipyan LL, editor. Erevan University; Erevan: 1985. [Google Scholar]
- Vaidya G, Lohman DJ, Meier R. SequenceMatrix: concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics. 2011;27:171–180. doi: 10.1111/j.1096-0031.2010.00329.x. [DOI] [PubMed] [Google Scholar]
- Van der Merwe MM, Walker J, Ericson L, Burdon JJ. Coevolution with higher taxonomic host groups within the Puccinia/Uromyces rust lineage obscured by host jumps. Mycol Res. 2008;112:1387–1408. doi: 10.1016/j.mycres.2008.06.027. [DOI] [PubMed] [Google Scholar]
- Vanev SG, Dimitrova EG, Ilieva EI. Razred Peronosporales. In: Fakirova V, editor. Gbite w Bulgarija 2 tom. Bulgarian Academy of Sciences; Sofia: 1993. [Google Scholar]
- Vánky K. Smut fungi of the world. APS Press St. Paul; Minnesota: 2012. [Google Scholar]
- Voglmayr H. Phylogenetic relationships of Peronospora and related genera based on nuclear ribosomal ITS sequences. Mycol Res. 2003;107:1132–1142. doi: 10.1017/s0953756203008438. [DOI] [PubMed] [Google Scholar]
- Voglmayr H, Göker M. Morphology and phylogeny of Hyaloperonospora erophilae and H. praecox sp. nov., two downy mildew species co-occurring on Draba verna sensu lato. Mycol Prog. 2011;10:283–292. [Google Scholar]
- Voglmayr H, Riethmüller A, Göker M, Weiß M, Oberwinkler F. Phylogenetic relationships of Plasmopara, Bremia and other genera of downy mildews with pyriform haustoria based on Bayesian analysis of partial LSU rDNA sequence data. Mycol Res. 2004;108:1011–1024. doi: 10.1017/s0953756204000954. [DOI] [PubMed] [Google Scholar]
- Voglmayr H, Choi YJ, Shin HD. Multigene phylogeny, taxonomy and reclassification of Hyaloperonospora on Cardamine. Mycol Prog. 2014a;13:131–144. doi: 10.1007/s11557-013-0900-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voglmayr H, Montes-Borrego M, Landa BB. Disentangling Peronospora on Papaver: phylogenetics, taxonomy, nomenclature and host range of downy mildew of opium poppy (Papaver somniferum) and related species. PloS One. 2014b;9:e96838. doi: 10.1371/journal.pone.0096838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson GW. Studies in North American Peronosporales. VI. Notes on miscellaneous species. Mycologia. 1914;6:192–210. [Google Scholar]
- Yerkes WD, Shaw CG. Taxonomy of the Peronospora species on Cruciferae and Chenopodiaceae. Phytopathology. 1959;49:499–507. [Google Scholar]
- Yu Y, Zhuang W, Liu X, Ma G, Yang Z, Tao J, Shen Y, Zhang Z, Wang Y, Liu Y. Peronosporales. In: Yu Y, editor. Flora Fungorum Sinicorum. Vol. 6 Science Press; Beijing: 1998. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.