Abstract
LCK is a tyrosine kinase essential for initiating T-cell antigen receptor (TCR) signaling. A complete understanding of LCK function is constrained by a paucity of methods to quantitatively study its function within live cells. To address this limitation, we generated LCK*, in which a key active site lysine is replaced by a photo-caged equivalent, using genetic code expansion. This enabled fine temporal and spatial control over kinase activity, allowing us to quantify phosphorylation kinetics in situ using biochemical and imaging approaches. We find that auto-phosphorylation of the LCK active site loop is indispensable for its catalytic activity and that LCK can stimulate its own activation by adopting a more open conformation, which can be modulated by point mutations. We then show that CD4 and CD8, the T cell coreceptors, can enhance LCK activity, helping to explain their effect in physiological TCR signaling. Our approach also provides general insights into SRC-family kinase dynamics.
Introduction
Biological systems rely on enzymes such as kinases to transmit information between the nodes of cell signaling networks, often to transduce extracellular ligand binding events into intracellular information. An important example of this is found in T cells, an essential cell-type of our adaptive immune system that can discriminate between healthy cells and those that are infected by pathogens. Expression of the T cell antigen receptor complex (TCR) at the cell surface allows the T cell to probe potentially infected host cells by scrutinizing their surface for expression of peptide fragments of pathogens presented within the MHC protein (pMHC). On binding cognate pMHC, a cascade of intracellular signaling is initiated from the TCR that either leads to the T cell directly killing the infected cells, or instructing other cell-types to do so1.
The most proximal event following pMHC binding is the phosphorylation of the immunoreceptor tyrosine-based activation motifs (ITAMs) in the intracellular tails of the TCR by LCK, a prototypic member of the SRC-family tyrosine kinases (SFK) that is almost exclusively expressed in T cells2. The phosphorylated ITAMs then recruit proteins with SRC-homology 2 (SH2) domains such as ZAP70, a cytoplasmic tyrosine kinase. Bound ZAP70 is phosphorylated by LCK, primarily at tyrosine-319 (Y319) that leads to its activation and subsequent phosphorylation of downstream effector molecules that drive multiple signaling pathways. LCK kinase activity is therefore crucial in translating the TCR–pMHC interaction into downstream signals in T cells. Understanding how the kinase activity of LCK is controlled within T cells at the molecular level is important not just for our fundamental understanding of TCR signal transduction but for suggesting new means by which its activity could be modulated therapeutically, given the deleterious effect of T cell mediated auto-immunity3 and its aberrant regulation in certain leukemias4,5.
Previous studies have shown that the SH2 domain of LCK can bind intramolecularly to a phosphorylated residue (Y505) at the C-terminus to adopt a closed auto-inhibitory conformation, which is a general feature of SFK regulatory mechanism6,7. Phosphorylation of Y505 is catalyzed by C terminal SRC kinase (CSK)8,9 and antagonized primarily by the membrane-bound tyrosine phosphatase CD4510. This modification can regulate the conformations that LCK can adopt, affecting its activity11–13. Full activation of LCK also requires phosphorylation at Y394 in the activation loop of the kinase domain14,15. In addition, LCK can be bound by the T-cell coreceptors CD4 and CD8, transmembrane proteins that can both bind to the MHC protein16 and engage with LCK17,18 through a Zn2+ ‘clasp’19. The functional effect of the coreceptors on T-cell signaling has been extensively studied during thymocyte development16 but it remains unclear whether they have a direct influence on LCK kinase activity.
Current in vitro methods to investigate how LCK, or indeed any SFK, functions at the molecular level invariably depend on assaying its kinase activity after removal from the cellular environment. Experiments are invariably performed in solution on non-physiological substrates that are unlikely to faithfully replicate kinase function when normally constrained to the plasma membrane. A recent study did address this latter issue, by tethering LCK to lipid vesicles14 but this accomplishment required altering the N terminal structure of the kinase to anchor it to the bilayer.
Conversely, most in vivo studies of LCK function have been limited by the inability to initiate kinase activity directly and so normally rely on steady-state measures of catalytic activity that do not provide the quantitative detail required for a mechanistic understanding. Recent methods have been designed to address this, principally by inserting chemically- or optically-controlled domains into kinases to allosterically modulate its activity20–22. This has found some success, although not with LCK, but all require extensive alterations to the native kinase structure that could interfere sterically with potential protein interactions, and may not represent the ‘true’ kinase in situ.
Encoding new functionality at specific sites of a protein when expressed in cells, through the use of genetically encoded unnatural amino acid incorporation23,24 has enabled the in vivo control of protein function with a precision more commonly associated with in vitro approaches25. By combining this approach with a cellular reconstitution of proximal TCR triggering with defined components26, we have developed a direct and quantitative method to assay the kinase activity of native LCK within the cellular environment. Engineering LCK so that its catalytic activity can be spatio-temporally controlled within live cells by brief light stimulation provides a new platform for ‘in vivo biochemistry’, which we use to show that LCK is critically dependent on Y394 phosphorylation in its active site loop for its kinase activity, and that the T cell coreceptors can enhance LCK function at limiting kinase levels.
Results
Engineering a photo-caged LCK kinase using unnatural amino acid incorporation
To quantitatively investigate LCK kinase function when localized in the plasma membrane of live cells, we needed precise temporal control over the initiation of its enzymatic activity. We noted that lysine at position 273 (K273) in the active site of human LCK is essential for its enzymatic activity, where mutation of this residue disrupts ATP binding and substrate phosphate transfer27. We anticipated that engineering control of substrate binding at this position would provide a mechanism to modulate LCK kinase activity. We used unnatural amino acid (UAA) mutagenesis to genetically incorporate a photo-caged variant of lysine (pc-Lys) at K273, which can be rapidly uncaged under ultraviolet (~350-400 nm) illumination, leaving a scar-less lysine residue25,28 (Fig. 1a). The pc-Lys residue can be easily accommodated in the active site of LCK without alteration to the kinase structure and is expected to sterically occlude ATP binding (Supplementary Fig. 1a).
Figure 1. Engineering of a photo-caged LCK kinase using unnatural amino acid incorporation.
(a) Diagram showing photo-caged lysine (pc-Lys) and the removal of its caging group (red) by ultraviolet light.
(b) Flow cytometry of HEK cells transfected with ZAP70-mRuby2 and the components required for pc-Lys incorporation into LCK-eGFP (LCK*), grown in the absence or presence of pc-Lys.
(c) Histogram showing percentage of transfected cells (ZAP70+) that expressed LCK*.
(d) Fluorescence micrographs showing localization of LCK* and wildtype LCK-eGFP in HEK cells. Scale bar, 5 μm.
(e) Western blot of LCK* using LCK-specific antibody. Band at ~82 kDa is LCK* and ~31 kDa is truncated LCK, terminated at the internal amber stop codon.
(f) Representative blot showing ZAP70 phosphorylation at Y319 using a phospho-specific antibody (green), and LCK* expression (red). HEK-TCR cells co-expressing LCK* and ZAP70 were illuminated at different intensities for indicated period. Final lane shows phosphorylation by wildtype LCK-eGFP at steady-state.
(g) Quantification of blots in f. Intensity of pY319 band was normalized to the corresponding intensity of LCK* band in each lane, and scaled relative to the normalized value of Y319 phosphorylation by wildtype LCK (final lane in f).
(h) Representative blot of ZAP70 Y319 phosphorylation by LCK* after photo-uncaging at 12 mW/cm2 for 30 s. Time after initiation of uncaging when cells were flash-frozen is shown.
(i) Quantification of blots in h. ZAP70 Y319 phosphorylation by LCK* as a function of time following illumination. Data are scaled relative to the final time point.
White dotted lines in blots indicate sections from the same blot presented at different brightness for visual clarity. Data in g and i were fit using a 3-parameter logistic function, presented as mean ± s.e.m. from independent experiments (n = 3 in g; 4 in i).
Uncropped blots are shown in Supplementary Data Set 1.
Source data for graphs are in Supplementary Data Set 2.
Using an UAA system optimized for efficient incorporation in mammalian cells25,29, we expressed human LCK fused to eGFP with the codon for K273 mutated to the amber stop codon (LCK K273 TAG) in HEK 293T cells. Cells were transfected with all components required for UAA incorporation, as well as ZAP70 kinase fused to the mRuby2 fluorophore. Cells transfected in normal medium showed expression of ZAP70 alone, but the addition of pc-Lys led to the co-expression of photo-caged LCK (LCK*) (Fig. 1b). This confirmed that in the absence of pc-Lys, translation of LCK K273 TAG was efficiently terminated at the internal stop codon. We found that ~40% of ZAP70-expressing cells also incorporated pc-Lys into LCK* (Fig. 1c), which localized correctly to the plasma membrane (Fig. 1d). Western analysis using an anti-LCK antibody shows that LCK* was expressed at the expected molecular weight (Fig. 1e).
To demonstrate that LCK* was inactive but could be rendered functional by illumination, we used HEK 293T cells stably expressing the complete TCR complex (HEK TCR; Supplementary Fig. 2a) and measured ZAP70 phosphorylation by LCK* upon illumination of the cells. We have previously shown that LCK shows constitutive activity towards the TCR and other downstream signaling molecules in HEK 293T26 due to the near-complete absence of inhibitory proteins that normally restrain its activity (Supplementary Fig. 2b,c). ZAP70 Y319 is a physiological substrate of LCK and is not associated with any ZAP70 auto-catalytic mechanism30,31, making it a good readout for LCK kinase activity. We first optimized the length and intensity of the illumination to uncage pc-Lys in LCK* by measuring Y319 phosphorylation by phospho-Western analysis (Fig. 1f). Blot quantification showed that prior to illumination no phosphorylation was detectable but 30 s illumination at 12 mW/cm2 could uncage LCK* sufficiently to cause phosphorylation 15 min post-illumination without signal saturation (Fig. 1g). This condition was preferred as it limited any cytotoxic effects of high intensity light and allowed us to capture the dynamic range of the phosphorylation events over the assay time-course. Control experiments confirmed that the observed Y319 phosphorylation was specific to the activity of LCK* uncaged by 350-400 nm light illumination. Phosphorylation was not observed when a catalytically inactive variant of LCK (K273R) was equivalently illuminated, nor when LCK* was illuminated by different wavelengths of light (500-550 nm) (Supplementary Fig. 1b).
Using this optimized system, we measured the kinetics of LCK* activity by quenching the phosphorylation reaction at various time points after pc-Lys uncaging. We could now observe the expected increase in Y319 phosphorylation over time (Fig. 1h) and a progress curve of the wildtype enzyme activity in live cells could be plotted (Fig. 1i). In subsequent experiments, we decreased the density of time points to improve our throughput to analyze several LCK* variants simultaneously, which did not compromise the experimental results (Supplementary Fig. 3a).
Phosphorylation of LCK active site loop is essential for its kinase activity
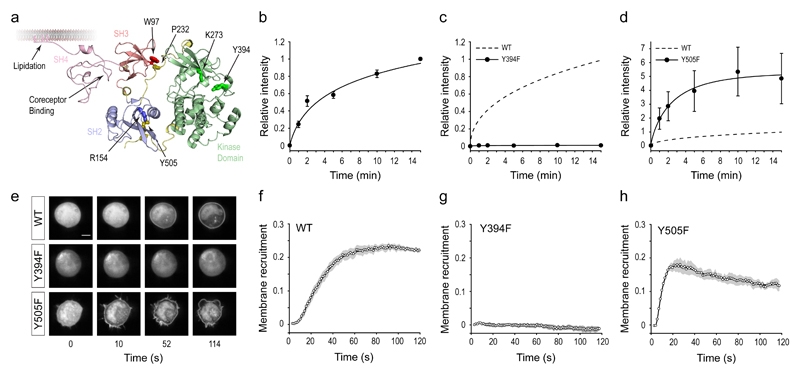
LCK, like many members of the tyrosine kinase family, has an activation loop within the kinase domain (Fig. 2a) that must be phosphorylated at a specific tyrosine residue to ensure full enzymatic activity. However, it remains unresolved whether the unphosphorylated form of LCK retains significant activity in vivo, given a recent in vitro study that showed only a two-fold increase in kinase activity when the active site loop was phosphorylated14. We substituted the tyrosine in the activation loop of LCK to phenylalanine (Y394F), which abolishes its phosphorylation. We found that this substitution rendered the kinase essentially inactive (Fig. 2c), which was not caused by the endogenous expression of the inhibitory kinase CSK by the HEK TCR cells (Supplementary Fig. 3b). This suggested that when not phosphorylated at Y394, LCK activity is almost completely absent when present in its native cellular environment. Conversely, the Y505F mutation in the C-terminal tail of LCK is known to substantially increase its kinase activity32,33 by removing CSK-mediated phosphorylation of Y505 that drives intramolecular inhibition, and this is what we observed (Fig. 2d). To provide a quantitative comparison between the LCK* variants, we derived the maximal reaction rate from the kinetic data, which should be proportional to the catalytic activity of the kinase assuming equivalent enzyme and substrate concentrations. This data pointed to the Y505F mutant having a ~8 fold increased rate compared to wildtype LCK*, whereas the rate for the Y394F mutant was undetectable (Supplementary Fig. 3c).
Figure 2. Phosphorylation of LCK active site loop is essential for its kinase activity.
(a) Cartoon representation of membrane-bound LCK showing key residues in the regulatory SRC-homology (SH) and kinase domains. Structure shown is based on the closely related kinase HCK (PDB: 1QCF) and Phyre2-modeled SH4 domain of LCK. Numbering is based on human LCK sequence. (b-d) Plots of ZAP70 Y319 phosphorylation kinetics by wildtype LCK* (b), Y394F LCK* (c) and Y505F LCK* (d), normalized relative to final time point (15 min) of wildtype LCK* (dashed curve). Data were fit using 3-parameter logistic function, and are presented as mean ± s.e.m. from independent experiments (n = 12 in b; 4 in c and d). (e) Representative microscopy images showing ZAP70 recruitment to the plasma membrane of HEK TCRH cells over time by different LCK* proteins, after photo-uncaging at 5 mW/cm2 for 2 s. Scale bar, 5 μm. (f-h) Quantification of microscopy data to measure ZAP70 recruitment to the plasma membrane over time by wildtype LCK* (n = 4) (f), Y394F LCK* (n = 3) (g) and Y505F LCK* (n = 4) (h). Data were collected using 4-8 cells per experiment, with data points represent mean and filled areas represent s.e.m. from independent experiments at each time point. A smoothed line using a moving-average filter is also shown.
Source data for graphs are in Supplementary Data Set 2.
We used a complementary assay to provide further evidence for the complete inhibition of LCK in the absence of Y394 phosphorylation. Prior to Y319 phosphorylation, ZAP70 is recruited to the phosphorylated ITAMs of the TCR complex at the plasma membrane, an event which also depends on LCK activity. We used live-cell imaging of ZAP70 recruitment to the cell surface as an alternative means to probe LCK kinase activity towards the TCR. We used an equivalent transfection strategy as above, but used HEK-TCR cells expressing higher TCR density (HEK-TCRH) to increase the membrane signal (Supplementary Fig. 2d). As expected, ZAP70 was initially localized to the cytoplasm but rapidly translocated to the plasma membrane on uncaging wildtype LCK* (Fig. 2e and Supplementary Video 1). Quantitative image analysis to measure the relative fluorescence intensity of ZAP70 at the plasma membrane showed that it was efficiently recruited to the plasma membrane (Fig. 2f). The Y394F mutation of LCK* caused undetectable ZAP70 recruitment (Fig. 2g and Supplementary Video 1). Conversely, the Y505F LCK* variant showed even faster dynamics (Fig. 2h and Supplementary Video 1), saturating ZAP70 binding within 20 s that subsequently decreased, presumably due to deleterious effects of the over-active kinase on cell integrity, such as membrane blebbing (Supplementary Video 1). Because the kinetics of ZAP70 recruitment to the membrane did not follow a simple profile, we quantified LCK* activity by determining the time required for half-maximal ZAP70 recruitment, with a shorter time corresponding to increased LCK* function (Supplementary Fig. 3d). The results from these imaging experiments are in strong agreement with the conclusions from the phospho-Western experiments.
Intramolecular SH3 interaction restrains LCK kinase activity
The SH2 domain of LCK is known to not only interact with phosphorylated tyrosine motifs in other proteins but also intramolecularly with phosphorylated Y505, which inhibits LCK activity. The generally agreed explanation for why the Y505F mutation of LCK enhances catalytic activity compared to the wildtype kinase is the disruption of this intramolecular inhibition. We wanted to test whether the converse was true; does disrupting binding by the SH2 domain cause equivalent enhancement of LCK function? The R154K substitution in the SH2 domain of LCK is known to disable binding of this domain to phosphorylated polypeptides34. We anticipated that this mutation would mimic the Y505F mutant by releasing intramolecular inhibition. However, the R154K mutation significantly decreased ZAP70 Y319 phosphorylation to ~25% compared to wildtype (Fig. 3a and Supplementary Fig. 3c), implying that the SH2 domain interaction with binding partners, such as the TCR, is more critical to LCK function than its role in intramolecular inhibition.
Figure 3. Intramolecular SH3 interaction restrains LCK kinase activity.
(a-c) Plots of ZAP70 Y319 phosphorylation kinetics by R154K LCK* (a), W97A LCK* (b) and P232A LCK* (c), normalized relative to final time point (15 min) of wildtype LCK* (dashed curve). (d) ZAP70 Y319 phosphorylation kinetics by wildtype and mutant LCK* kinases are plotted in one graph for direct comparison. Data were fit using 3-parameter logistic function, and are presented as mean ± s.e.m. from independent experiments (n = 12 for wildtype LCK*; 4 for mutant LCK*).
Source data for graphs are in Supplementary Data Set 2.
The SH3 domain of LCK can also participate in intramolecular binding with the conserved proline-rich (PxxP) motif in the linker region, although this interaction is significantly less well characterized compared to the SH2 interaction. We expected that SH3-mediated intramolecular binding would also restrain LCK* kinase activity. We first disrupted a key residue (W97A) in the SH3 domain and found that LCK* kinase activity was substantially increased compared to wildtype LCK* (Fig. 3b). This suggested that, at least within the transfected HEK TCR cells, the primary function of the SH3 domain is to inhibit LCK kinase activity rather than bind another target protein. A prediction from this result is that mutating the proline-rich motif that interacts with the SH3 domain within LCK should have an equivalent effect on the activity of LCK. Through structural modeling of full length human LCK based on a related kinase, we identified P232 within the conserved PxxP motif of LCK that should interact with W97 of the SH3 domain (Supplementary Fig. 3e). Mutating this residue (P232A) dramatically increased LCK* kinase activity (Fig. 3c). Both W97A and P232A substitutions led to an equivalent increase in LCK* activity that was quantitatively comparable to the Y505F mutant (Fig. 3d and Supplementary Fig. 3c), suggesting the primary function of the intramolecular SH3-mediated interaction is to restrain LCK catalytic activity.
LCK activation by auto-phosphorylation
It has been shown that Y394 in the activation loop of LCK is auto-phosphorylated in trans12,14,35,36 but the kinetics of this reaction have not been studied in situ. To address this, we first investigated the kinetics of LCK* Y394 auto-phosphorylation following pc-Lys uncaging. We found that the process occurred at a much faster rate compared to that of ZAP70 Y319 phosphorylation (Fig. 4a). To confirm that this rapid auto-phosphorylation was occurring predominantly in trans, we co-expressed wildtype LCK* and a catalytically-inactive variant of LCK (K273R) in cells and measured the phosphorylation of Y394 in both forms by LCK* (Fig. 4b). Since LCK (K273R) is devoid of kinase activity, any phosphorylation observed could only be attributed to the in trans activity of uncaged LCK*, which was indeed the case (Fig. 4c). However, although the relative rates of Y394 auto-phosphorylation on both kinase variants were very similar, the absolute level of LCK (K273R) phosphorylation was much lower than when compared to uncaged LCK* (Fig. 4c), which was not due to differences in their expression (Fig. 4b).
Figure 4. Active LCK conformation increases substrate availability for auto-phosphorylation at Y394.
(a) Plot showing kinetics of wildtype LCK* auto-phosphorylation at Y394 after photo-uncaging.
(b) Representative blots showing auto-phosphorylation at Y394 of both wildtype LCK* and untagged kinase-inactive LCK (K273R) at the indicated time after LCK* uncaging. ZAP70-mRuby2 was not included in these experiments. Dotted lines indicate sections from the same blot but presented at different brightness for visual clarity only.
(c) Quantification of blots in b to show kinetics of LCK* and LCK (K273R) auto-phosphorylation at Y394.
(d) Maximal auto-phosphorylation of Y394 achieved by wildtype LCK* by varying uncaging period of illumination. Separate plots of Y394 auto-phosphorylation were fit to derive the maximum asymptote values, which were plotted against illumination period.
(e) Kinetics of wildtype LCK* and Y505F LCK* auto-phosphorylation at Y394.
For all plots except d, data are normalized relative to wildtype LCK* at 1 min and fit by 3-parameter logistic function, and presented as mean ± s.e.m. from independent experiments (n = 9 in a; 3 in c; 4 in e). For d, error bars represent 95% confidence interval of estimated parameter value from independent experiments (n = 3).
Uncropped blot images are shown in Supplementary Data Set 1.
Source data for graphs are in Supplementary Data Set 2.
The preceding results suggested that the catalytically-inactive LCK (K273R) was less susceptible to auto-phosphorylation at Y394, perhaps by adopting a conformation where Y394 was less accessible, compared to the uncaged LCK*. We tested this hypothesis by varying the illumination period used to uncage LCK*. Cells exposed to shorter illumination would have less uncaged, active LCK*. As predicted, shorter illumination led to a lower maximal pY394 signal compared to the samples that had been illuminated for longer (Fig. 4d). If the effect of different illumination periods was to only vary the level of LCK* as an active enzyme, but not as a ‘substrate’, there would have been differences in the rate of phosphorylation and not the maximal phosphorylation, but this is not what we observed. We hypothesized that because the Y505F LCK* mutant should be in a more ‘open’ conformation, it should have increased accessibility of Y394 and this was exactly what we observed (Fig. 4e).
ZAP70 depends on auto-phosphorylation for activation
Full activation of ZAP70 kinase requires phosphorylation of its activation loop at Y493, in an equivalent manner to LCK37. Although it is accepted that ZAP70 Y493 phosphorylation requires the presence of active LCK, it is less clear whether LCK itself directly phosphorylates Y49338,39 or whether this modification primarily relies on ZAP70 auto-phosphorylation31. We used our LCK* uncaging approach to follow the kinetics of ZAP70 phosphorylation at both Y319 and Y493. We found that LCK*-mediated Y319 phosphorylation had significantly faster kinetics than Y493 phosphorylation, with a delay evident between the initiation of ZAP70 Y319 and Y493 phosphorylation (Fig. 5a). We speculated that this difference was due to Y493 phosphorylation being mediated by ZAP70 auto-phosphorylation rather than LCK*. To investigate this, we repeated the kinetic assay but now included a kinase-dead version of ZAP70 (K369R). Compared to wildtype ZAP70, the catalytically inactive version had a far lower rate of Y493 phosphorylation (Fig. 5b). The data show that the primary kinase responsible for ZAP70 Y493 phosphorylation is ZAP70 itself.
Figure 5. Differential phosphorylation kinetics between Y319 and Y493 of ZAP70.
(a) Plot showing kinetics of ZAP70 phosphorylation at Y319 and Y493 after LCK* photo-uncaging. Data are normalized relative to final time point (15 min) for each phosphorylation site.
(b) Plot showing kinetics of ZAP70 phosphorylation at Y493 after LCK* uncaging in HEK-TCR cells expressing wildtype ZAP70-mRuby2, kinase-inactive ZAP70 (K369R)-mRuby2 or mRuby2 alone as a control. Data are normalized relative to final time point (15 min) of wildtype ZAP70.
All data were fit using 3-parameter logistic function, and are presented as mean ± s.e.m. from independent experiments (n = 7 in a; 3 in b).
Source data for graphs are in Supplementary Data Set 2.
Activity of other SRC-family kinases expressed in T cells
There are nine members of SFK family but only three are expressed at significant levels in T cells: LCK, FYN and to a lesser extent SRC. While both LCK and FYN play roles during early TCR signaling, it is believed that LCK is the major kinase involved in TCR-mediated signaling40,41. To test whether this was because LCK was more efficient at initiating T cell signaling, we constructed the photo-caged versions of FYN and SRC by mutating the codons for K299 (FYN*) and K298 (SRC*) to the amber stop codon, in an equivalent manner as for LCK*. We found that ZAP70 Y319 is phosphorylated by LCK* at a much greater rate when compared with FYN* or SRC* (Supplementary Fig. 4), demonstrating that ZAP70 is a better substrate for LCK.
Spatial control of LCK activation using the photo-caged system
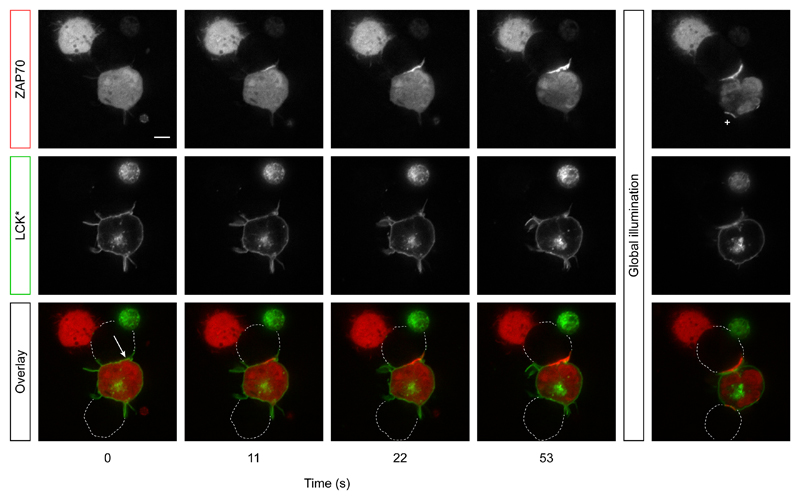
To demonstrate that we could both spatially and temporally control over LCK activation, we took advantage of a cell-cell conjugation system where the TCR on the HEK cells interacted with its specific peptide (derived from NY-ESO) covalently bound into the MHC protein and expressed on the surface of Raji B cells, which act as efficient ligand presenting cells26. The inhibitory proteins CD45 and CSK-CBP are now also expressed in HEK TCRH cells to prevent TCR triggering outside the conjugate interface, where cells adhere through co-expression of ICAM 1, which can interact with the integrin LFA-1 on Raji B cells and the TCR–pMHC interaction itself26. We uncaged LCK* by illuminating a diffraction-limited (<1 μm) region of the conjugate interface with a 405 nm laser beam, pulsed for 1 ms. This spatio-temporally localized activation of LCK* initially led to the recruitment of ZAP70 only to this region, before increasing throughout the cell interface, presumably due to LCK* lateral diffusion (Fig. 6 and Supplementary Video 2). Fortuitously, the presented LCK*-expressing cell was conjugated to two separate B cells, which allowed us to confirm the spatial restriction of LCK* uncaging since no ZAP70 was recruited to the untargeted conjugate interface (Fig. 6 and Supplementary Video 2). To confirm that this conjugate region nonetheless had the potential to be activated, we subsequently used global illumination and observed ZAP70 recruitment to the second conjugate interface (Fig. 6). This demonstrated that we could both spatially and temporally control LCK activation at subcellular and millisecond resolution in live cells.
Figure 6. Precise spatio-temporal control of LCK kinase activity in live cells.
A single HEK-TCRH cell expressing LCK* and ZAP70-mRuby2 formed two separate conjugates with Raji B cells (outlined in overlay). Initially, a specific region of the upper conjugate (white arrow in overlay) was exposed to a focused 405 nm laser pulse to uncage LCK* in that region only, and ZAP70 recruitment to the region was observed over time. A second, global illumination was applied on the same cells to uncage the remaining LCK*. Subsequently, ZAP70 recruitment to the lower conjugate was now observed (plus sign in ZAP70 image). Twenty cells were analyzed over 3 independent experiments that show similar spatio-temporal control of LCK* kinase activity within individual conjugates.
Effect of CD4 and CD8 coreceptors on LCK function
It is established that the T cell coreceptors have a significant influence on thymocyte development and T cell activation16, and the interaction between the cytosolic tails of the membrane-bound coreceptors and the SH4 domain of LCK (Fig. 2a) has been confirmed19,42. In addition to the intracellular LCK interaction, the coreceptors bind the pMHC protein expressed on antigen-presenting cells. It remains uncertain whether the coreceptor’s effect on LCK activity is mediated solely by influencing the localization, or dwell time, of the kinase at the engaged TCR complex, a question which sought to directly address using the approaches described above. We first used our cell conjugate assay to investigate how CD8 modulates LCK*-mediated ZAP70 membrane recruitment. LCK* uncaging caused the translocation of ZAP70 to the entire conjugate site (Fig. 7a and Supplementary Video 3), the kinetics of which were quantified over many conjugates (Fig. 7b). Repeating the assay using CD8+ HEK-TCRH cells (Supplementary Fig. 2e) now caused an increased rate of ZAP70 translocation following LCK* uncaging (Fig. 7b and Supplementary Fig. 5a). An equivalent CD4 assay was not possible because the TCR used is restricted by class-I MHC.
Figure 7. T-cell coreceptors CD4 and CD8 directly enhance LCK activity.
(a) Representative microscopy images of ZAP70 recruitment to conjugate interface between HEK-TCRH and APCs (Raji B cells) over time after wildtype LCK* photo-uncaging. Colored boxes denote protein representation in the overlay image, green is LCK* (not shown). Outline of APC is shown as dotted line. Scale bar, 5 μm.
(b) Quantification of ZAP70 recruitment to conjugate interface over time, as depicted in a, in HEK-TCRH and CD8+ HEK-TCRH cells after LCK* uncaging.
(c) Representative microscopy images of accumulation of eGFP-tagged proteins, as illustrated within the panel, at the interface between HEK-TCRH or CD8+HEK-TCRH and APC. The ‘Myr’ control uses the first 12 amino acids of LCK sequence fused to eGFP to target the fluorophore to the plasma membrane by myristoylation and palmitoylation. The ‘CaaX’ control uses the C-terminal sequence from K-RAS which is prenylated to target the fused eGFP to the plasma membrane. Outline of APC is shown as dotted line. Scale bar, 5 μm.
(d) Ratio of fluorescence intensity of eGFP-tagged protein inside the conjugate region over that outside the interface in HEK cells. Data are presented as mean ± s.e.m. of 25-45 cells in each condition from 3 independent experiments.
(e-h) Microscopy image quantification showing ZAP70 recruitment to plasma membrane after LCK* uncaging in the absence or presence of CD8 (e, g), CD4 (f, h), CD86ExCD8Int (g), and CD86ExCD4Int (h) in HEK-TCRH cells. Cells were exposed to light at 5 mW/cm2 for 2 s (a, b, e-h). Data in b, e-h are normalized to maximum asymptote values for each dataset. Lines show data smoothed used a moving-average filter, data points represent mean and filled areas represent s.e.m. from independent experiments (n = 4 for all conditions), where 4-8 cells were used in each independent experiment.
Source data for graphs are in Supplementary Data Set 2.
CD8 could enhance LCK activity by simply increasing the kinase density within the conjugate interface. We did indeed observe increased LCK* in this region, compared to the rest of the plasma membrane, but this effect was not CD8-dependent (Fig. 7c,d). Instructively, the lipid-modified N-terminus of LCK alone (Myr eGFP) was sufficient to replicate the increased LCK* density in the conjugate interface (Fig. 7c,d), as was a prenylated eGFP (eGFP-CaaX), which rather suggested an alteration to the membrane lipid environment around the clustered TCR as the primary cause. To test whether coreceptor binding directly enhanced LCK kinase activity independently from pMHC binding, we repeated the ZAP70 membrane recruitment assay in unconjugated CD8-expressing HEK TCRH cells. CD8 expression still enhanced the activity of LCK* (Fig. 7e and Supplementary Fig. 5b), which we also found for CD4 (Fig. 7f and Supplementary Fig. 5c), suggesting both coreceptors use a similar mechanism to enhance LCK function.
The possibility remained that the observed results arose from a direct interaction of the coreceptors with the TCR complex itself, as previously suggested for CD443. To test this hypothesis, we created chimeric coreceptors (CD86ExCD8Int and CD86ExCD4Int) by substituting their extracellular domains with the monomer CD8644, whilst maintaining their interaction with LCK. Repeating the ZAP70 recruitment experiment with these constructs showed that the increased LCK* activity on coreceptor binding could be entirely attributed to the intracellular LCK* interaction (Fig. 7g,h and Supplementary Fig. 5d,e). Furthermore, the coreceptor effect on LCK* function was almost completely attenuated when a larger fraction of LCK* was uncaged (Supplementary Fig. 5f-i), which agrees with previous work that coreceptor enhancement of TCR signaling is only observed at physiologically low levels of ligand binding45.
Discussion
By developing a means to both spatially and temporally activate tyrosine kinases in live cells, we have been able to quantitatively measure the kinetics of LCK catalytic activity in situ, with a level of control over kinase activity not readily achievable by conventional in vivo and in vitro approaches, as previously demonstrated28. In agreement with previous biochemical studies11–13, we found that LCK can adopt a range of conformations that have a strong influence on its intrinsic kinase activity46. These conformations are primarily driven by intramolecular interactions mediated by both the regulatory SH2 and SH3 domains. We showed that the kinase activity of LCK* mutants Y505F, W97A and P232A were quantitatively equivalent (Fig. 3d and Supplementary Fig. 3c), implying they all biased the kinase structure to the same ‘open’ conformation. However, the concordance of reciprocal mutations affecting the SH3 intramolecular interaction (W97A and P232A) was not found for the equivalent SH2 interaction, where the R154K mutation that disrupted this inhibitory interaction actually led to a decrease in activity. This suggested that losing the ability of LCK to bind a phosphorylated substrate, likely the TCR itself in our assay, was more deleterious than decreased inhibition, perhaps because the intermolecular SH2 interaction also stabilizes the active conformation of the kinase domain47,48.
Phosphorylation of the active site loop of LCK leads to rearrangement and stabilization of the catalytic residues into their optimal positions, and drives the full activation of the kinase6,7,49. We found that this phosphorylation was completely indispensable for LCK kinase activity, as the Y394F mutant was essentially inert in both our in situ assays. This is at odds with a recent in vitro study14 that only found a two-fold difference on Y394 phosphorylation, which could be explained by the additional constraints placed on LCK when localized to the plasma membrane. The kinetics of Y394 auto-phosphorylation were rapid, saturating approximately 1 min after LCK* photo-uncaging, which we hypothesize is required to ensure a kinetic advantage for LCK activation over the abundant phosphatases such as CD45 that would antagonize this modification50, as suggested previously12. This result was in striking contrast to the delayed ZAP70 auto-phosphorylation we measured, which could be rate-limiting in TCR triggering and may have a role in ligand discrimination through kinetic proofreading. The decreased Y394 phosphorylation on a kinase-inactive version of LCK (K273R) suggested that LCK in an active open conformation is a better substrate for auto-phosphorylation at Y394, which is supported by structural data of various inactive states of SRC kinase domains that show sequestration of Y394 within the active site, preventing its auto-phosphorylation6. A recent study also suggested a negative-feedback loop may control LCK activity through Y192 phosphorylation by ZAP7051.
Finally, we found that the T-cell coreceptors have a stimulatory effect on LCK kinase activity even in the absence of pMHC binding. This effect could be ascribed solely to the intracellular interaction between the coreceptors and LCK (Fig. 7), a result that cannot easily be explained by current models of coreceptor function45,52. We speculate that the physical interaction between LCK and the coreceptors induces a conformational rearrangement that enhances LCK function and implies that the unique SH4 domain of LCK could also form an intramolecular interaction that restrains kinase activity that is released on coreceptor binding.
There are naturally caveats to the approaches in this study. Although assaying kinase function within the cellular context provides a more physiological representation of the environment in situ, this means the system is less ‘clean’ than true in vitro experiments since endogenous proteins have the potential to affect our results. We used a non-immune cell-line to mitigate as many of these confounding effects as possible. Another limitation is that the absolute concentrations of the active enzyme and substrate cannot easily be determined, forcing us to measure relative rather than absolute enzyme kinetics. The extremely rapid auto-phosphorylation of LCK* at Y394 (Fig. 4) and ZAP70 recruitment to plasma membrane by Y505F LCK* (Fig. 2e,h) upon uncaging indicate that the presence of pc-Lys within the active site of LCK* did not result in a gross conformational change of the enzyme and that the observed kinetics reflect the true substrate phosphorylation kinetics rather than the recovery of kinase conformation upon uncaging.
In summary, our quantitative investigation of LCK kinase activity has provided new insights into how this important SFK functions, with the conclusions likely to translate to other members of the family. Furthermore, the effect of the coreceptors on LCK activity we observed showcases the power of measuring enzyme kinetics in situ, where the modulatory effect of membrane-bound interacting proteins can now be directly addressed. We demonstrated that this approach could be applied to other members of the SFK family (Supplementary Fig. 4) and could easily be extended to many other kinases that contain a critical lysine within their active site, including ZAP70 kinase. Further developments to express the photo-caged proteins in other more physiological cell-types such as primary T cells are ongoing and will enable us to perform ‘in vivo’ biochemistry experiments to modulate protein function with unprecedented spatio-temporal control.
Materials and Methods
Vector constructs
All constructs, unless described otherwise below, were created by amplifying gene sequences using the polymerase chain reaction (PCR), incorporating MluI and [BamHI-STOP-NotI] restriction sites at the 5′ and 3′ ends of the gene, respectively. Following MluI and NotI double digest of the PCR product, it was ligated into pHR-SIN lentiviral vector. Where required, a gene encoding a fluorescent protein was inserted in-frame between BamHI and NotI sites. All photo-caged kinase constructs (wildtype and mutant LCK*, and SRC* and FYN*) were created by mutating the codon encoding for lysine in the active site of the kinase (K273 in LCK, K298 in SRC and K299 in FYN) to the amber (TAG) stop codon using overlap extension PCR. SRC* sequence was gene-synthesized (Thermo Fisher Scientific) to remove restriction sites by synonymous mutations. The modified genes along with gene encoding eGFP were inserted into pHR′ vector as described above.
The components required for unnatural amino acid (UAA) incorporation of photo-caged lysine (pc-Lys) in proteins expressed by mammalian cells have been described previously25,29. Briefly, the orthogonal pair of tRNACUA (U25C) and aminoacyl-tRNA synthetase required to charge the tRNACUA with photo-caged lysine (pc-Lys) are encoded within a PiggyBac vector, (U6 PylT*)4-EF1α-PylRS. It contains 4 copies of the tRNA along with the synthetase, under the control of the U6 and EF1α promoter, respectively. The E55D mutant of eukaryotic release factor (eRF)-1 was additionally used to increase efficiency of pc-Lys incorporation.
pHR-IFP2.0-CaaX: The IFP2.0 fluorescent protein was fused at the C-terminus to the K-Ras prenylation sequence (LEKMSKDGKKKKKKSKTKCVIM). The gene was inserted between the BamHI and NotI of the pHR-SIN vector.
pHCM-CD45RO: The gene for CD45RO (including a N-terminal His tag) was amplified from a published construct53. The SFFV promoter was replaced with the CMV equivalent. To drive high expression, the SV40 introns (16S) were synthesized as a GeneArt String and inserted as BssHII-MluI fragment at the 5’-end of CD45RO gene.
pHR-CBP-CSK: The genes for CBP and CSK were amplified from pHR-CBP and pHR-CSK26 and inserted in-frame into a bicistronic vector with the P2A self-cleaving peptide sequence linking them.
pHR-CD8(βα): The genes for CD8β and CD8α were amplified from gene-synthesized fragments and inserted in-frame into a bicistronic vector with the P2A self-cleaving peptide sequence linking them.
pHR-CD86ExCD4Int and pHR-CD86ExCD8Int: The chimeric sequences were gene-synthesized (Thermo Fisher Scientific) to fuse the extracellular domain of CD86 with the transmembrane domain and cytosolic tail of CD4 or CD8α. The synthesized genes were inserted into pHR vector as described above.
Cell culture
HEK-TCR and HEK-TCRH cell lines were created by lentiviral transduction of HEK-293T cell line (purchased from ATCC to ensure identity) to stably express 1G4 TCRαβ chains and CD3γδεζ chains as previously described26, sorted based on their cell surface expression and made clonal by limiting dilution. All HEK cell variants were grown in DMEM (Sigma) and Raji B cells (purchased from ATCC) were grown in RPMI-1640 (Sigma), both supplemented with 10% FCS (Gibco), and 2 mM L-glutamine, 100 U/ml penicillin and 100 μg/ml Streptomycin (Sigma). All cells were incubated at 37°C, 5% CO2 under full humidity and tested for mycoplasma contamination using MycoAlert kit (Lonza).
Transient transfection
One day prior to transfection, HEK-293T cells were seeded into 6-well plates or 35 mm poly-L-lysine coated glass-bottom imaging dishes (MatTek) so that the cells reach 75% confluency on the day of transfection. For all transfections performed here, a total of 2 μg of DNA per well or dish was transfected using GeneJuice (Merck) except for transfection of cells used in Fig. 4b,c, which was 1 μg of DNA per well. Transfected cells were usually analyzed 40-48 h after transfection.
In general, the main components to be transfected into HEK-TCR cells were pHR-LCK*, (U6 PylT*)4-EF1α-PylRS, pcDNA5-eRF1 (E55D) and pHR-ZAP70-mRuby2 at a DNA ratio of 2:2:2:1. Any additional components were included in the mix with a ratio of 1 while maintaining the total of 2 μg DNA per transfection. For microscopy experiments of unconjugated HEK cells, pHR-IFP2.0-CaaX was transfected in addition to the main components to identify plasma membrane. Biliverdin was added to the medium at 6 h after transfection at a final concentration of 20 μM to enhance IFP2.0 fluorescence. For microscopy experiments of HEK cells conjugated to Raji B cells, additional components of pHCM-CD45RO, pHR-CBP-CSK and pHR-ICAM-1 were transfected in addition to the core LCK* components.
Lentiviral transduction and generation of stable expressing cells
To produce lentivirus, the gene of interest in a lentiviral backbone vector, pCMVΔ8.91 (encoding essential packaging genes) and pMD2.G (encoding VSV-G gene to pseudotype virus) were transfected into HEK-293T cells at a ratio of 2:2:1. A total of 1.5 μg of DNA was transfected per well using GeneJuice (Merck). After 48-72 h, the lentivirus-containing medium was collected, centrifuged at 4,300g for 2 min to remove debris, and incubated with HEK cells to be transduced for 16 h. Fresh medium was then added to the cells and expression of the protein of interest was monitored regularly and cell population was expanded. Once the expression level was stable, the cells were bulk-sorted to isolate cell populations with desired protein expression level.
Uncaging of LCK* by illumination for Western blot analysis
After transfection for 40-48 h, HEK-TCR cells were trypsinized to detach them from the wells for 5 min at 37°C and neutralized by the addition of an equal volume of fresh medium. Cells were pipetted several times to obtain a homogenous single-cell suspension and collected in a FACS tube, after an aliquot was removed for analysis of protein expression by flow cytometry. The rest of the cells were then centrifuged at 800g for 5 min and the supernatant was discarded. Cells were washed once in sterile PBS and resuspended in an appropriate volume of DMEMgfp2 anti-bleaching medium (Evrogen) supplemented with 10% FCS.
Cells were illuminated with light from an AMH-200 light source (Andor) using a ‘DAPI’ (377±25nm) filter at 2.5 or 12 mW/cm2 for the specified illumination period. To facilitate this, a custom-designed illumination chamber was built around a Nikon Ti Epi-FL illuminator so that a tube could be positioned at the focal point of the illuminator while providing a means to sample the cells during illumination. Aliquots of cell suspension (50 μl) were collected at defined time intervals in 1.5 ml tubes pre-frozen on a metal block with dry ice to immediately quench the phosphorylation reaction. An equal volume of 2× lysis buffer [2% Nonidet P40 (AppliChem), 2 mM sodium orthovanadate (Sigma), 2 mM phenylmethanesulfonylfluoride, and 2× Complete, EDTA-free protease inhibitor cocktail (Roche) in 2× Tris-buffered saline (40 mM Tris, 300 mM NaCl, pH7.4)] was overlaid on the frozen cell suspension and the samples were let to thaw slowly on ice. After 1 h incubation on ice with occasional agitation to mix the samples, cell lysates were centrifuged at 20,000g for 10 min at 4°C. The supernatant fraction was then mixed with LDS sample buffer and dithiothreitol, and boiled at 95°C for 10 min before analyzed by SDS-PAGE and Western blotting.
Western blotting
SDS-PAGE was performed using NuPAGE Novex 4-12% Bis-Tris Midi protein gels (26 wells) and XCell4 SureLock Midi-Cell Electrophoresis System according to the manufacturer’s instructions (Thermo Fisher Scientific). Gel transfer to nitrocellulose was performed using the iBlot device (Thermo Fisher Scientific). The primary antibodies used in this study were: anti-pY319-ZAP70 (cat#: 2701), anti-pY493-ZAP70 (cat#: 2704), anti-pY416-SRC (reacts with LCK pY394; cat#: 2101), anti-LCK (cat#: 2657), anti-SHP 1 (cat#: 3759), anti-SHP 2 (cat#: 3397) and anti-CSK mAb (cat#: 4980), which were all purchased from Cell Signaling Technology and are all raised in the rabbit except anti-LCK, which is raised in the mouse. Anti-beta actin mAb is raised in mouse (cat#: AB11003; abcam). For the combined detection of LCK*, FYN* and SRC*, a mouse mAb against eGFP (cat#: A11120; Life Technologies) was used to give equivalent sensitivity. All primary antibodies were used at 1:1000 dilution. Secondary antibodies were used at 1:20,000 dilution: anti-rabbit or mouse IgG (H+L) DyLight 800 (cat#: 5151 & 5257; Cell Signaling) and AlexaFluor 680 goat anti-rabbit or mouse IgG (H+L) (cat#: A21109 & A21058; Life Technologies). The transferred blot membrane was first blocked with 2.5% non-fat milk (Sigma) in Tris-buffered saline (MTBS) before incubation with primary antibody in MTBS including 0.1% Tween 20 (MTBS T) at 4°C overnight with agitation. The membrane was then washed 3 times in TBS T, followed by 1 h incubation in the appropriate secondary antibody at room temperature in MTBS T before washing 3 times in TBS. Fluorescent protein bands were visualized using the Odyssey CLx imaging system (LI-COR) that enables two-color detection at 700 nm and 800 nm on the same blot over a >5-decade linear range.
Flow cytometry
An LSRII flow cytometer (Beckton Dickinson) was used to measure expression levels of proteins in transfected or transduced HEK cells. For detection of fluorescently-tagged proteins, the cells were washed once in FACS buffer [5% (v/v) FBS, 0.1% (w/v) NaN3 in PBS, pH 7.4] and fixed in FACS Fix buffer [1.6% (v/v) formaldehyde, 2% (w/v) glucose, 0.1% (w/v) NaN3 in PBS, pH 7.4] prior to flow cytometry experiment. For detection of untagged proteins, the primary antibodies used (all from BioLegend) were AlexaFluor 647-conjugated anti-TCR (cat#: 306714), AlexaFluor 488-conjugated anti-CD4 (cat#: 317419) or anti-CD8 (cat#: 301024) and an AlexaFluor 647-conjugated anti-CD45 (cat#: 304056) antibody that recognizes all isoforms. Antibodies were diluted 1:50 in FACS buffer and incubated with the cells for 30 min on ice. Labeled cells were washed with FACS buffer once, cells were centrifuged at 800g for 5 min and the resuspended pellet fixed in FACS Fix buffer. Routinely, 20,000 live cells gated by scatter were collected for each analysis.
Cell conjugation
HEK-TCR cells for conjugation experiments were grown and transfected in 6-well plates as described above. After 40-48 h, cells were trypsinized to detach them from the wells for 5 min at 37°C before neutralization by the addition of an equal volume of fresh medium. Cells were pipetted several times to obtain a homogenous single-cell suspension and collected in a 1.5 ml Eppendorf tube. Approximately 1×106 Raji B cells expressing the appropriate pMHC were collected in a separate tube. Both tubes were centrifuged at 800g for 3 min to pellet the cells. Resuspended pellets were then washed in fresh complete DMEM and centrifuged again. After this, both pellets were resuspended in residual medium and combined to a total volume of ~200 μl with medium in a 0.2 ml PCR tube. The cells were incubated in a 37°C water bath for 30 min to drive cell conjugation. Following incubation, conjugated cells were washed once in Dulbecco’s PBS with CaCl2 and MgCl2 (DPBS), resuspended in 1 ml of DMEMgfp2 anti-bleaching medium (Evrogen) and transferred to a MatTek imaging dish.
Spinning disc confocal microscopy
For imaging of unconjugated HEK cells to study ZAP70-mRuby2 recruitment to plasma membrane, cells were seeded directly into the imaging dish and transfected as described above. Prior to imaging, cells were washed gently in DPBS once and 1 ml of imaging medium was added to cover the cells in the dish. All live-cell imaging experiments in this study used a Nikon Ti inverted microscope in a thermally-controlled enclosure (OKOLabs), equipped with a CSU-X1 spinning-disc confocal head (Yokogawa). Laser lines used to excite samples were: 405 nm (100 mW), 488 nm (60 mW), 561 nm (50 mW) and 640 nm (100 mW), controlled by an acousto-optic tunable filter (Andor). Fluorescence emission was collected through filters for mTagBFP (460±15 nm), eGFP (525±25 nm), mRuby2 (607±18 nm) and IFP2.0 (708±38 nm). All images were collected using a Plan Apo VC 100× 1.4 NA oil-immersion objective (Nikon) onto an iXon Ultra EM-CCD camera (Andor) with a calculated pixel size of 85 nm. Stage movement was controlled by a Prior motorized stage with a Piezo Z-drive. The entire microscope system was controlled by μManager software that was used to create multi-channel, multi-time point image data-sets and at multiple positions when required. The built-in perfect-focus unit of the microscope was used to correct for axial focus drift due to fluctuations in temperature.
Global photo-uncaging
HEK cells expressing both LCK* and ZAP70-mRuby2 were identified under microscope based on the expression of the tagged fluorescent proteins. In the cell conjugation experiment, conjugated cells were identified based on the presence of pMHC-BFP-expressing Raji B cells forming a cup-shaped contact with LCK* and ZAP70-mRuby2-expressing HEK cells. To follow ZAP70-mRuby2 recruitment to the plasma membrane or cell conjugate interface after LCK* activation, cells were illuminated by light from an AMH-200 light source (Andor) through a DAPI filter cube (excitation at 377±25 nm) in the microscope. A script was written in μManager software to capture 5-10 images before photo-uncaging by illuminating cells through the objective at 5 mW/cm2 or 500 mW/cm2 for a duration of 2 s. Following this, images were acquired continuously every ~1.4 s for approximately 2 min. Images from all relevant fluorescence channels were captured throughout the experiment. Images were analyzed as described below. Acquisition parameters were set up to ensure the best dynamic range to avoid signal saturation in the fluorescence channels. Due to variability in the expression of proteins using transient transfection, individual laser powers were adjusted between experiments while maintaining the exposure times.
Focused photo-uncaging
To activate LCK* spatially with subcellular accuracy in the cell-cell conjugate region, a similar protocol was used as for the global photo-uncaging. To uncage LCK* now though, a defined diffraction-limited spot in the image was excited with a 405 nm laser at full power (1.2 mW at objective) using a dwell-time of 100 μs repeated 10 times. Images were subsequently acquired as above.
Data analysis
Western blot images
High resolution Western blot images from the Odyssey imager were acquired as 32-bit grayscale files for quantification with ImageJ software. Equivalent regions of interest (ROIs) were drawn around the bands of interest to measure the integrated density of each ROI. To express the level of phosphorylation relative to LCK*, to correct for loading and expression, the density value of the phosphorylated band was divided by that of the corresponding LCK* band to obtain the relative phosphorylation of each sample. To allow comparison between experiments, these values were then normalized according to the figure legends to create a plot of phosphorylation kinetics. For every experiment, a wildtype dataset was also collected so that different conditions could be directly compared through this standard. Further processing of the Western blots shown in figures (rotation, cropping and contrasting) was performed uniformly over images solely for better visualization. Where needed, white dotted lines denote sections of the same blot with different contrast settings for visual clarity only. Image quantification, however, always used the values from original image files.
Microscopy images
Processing and quantitative analyses of microscopy images were performed using scripts written in ImageJ. Data from the ZAP70-mRuby2 channel were corrected for photo-bleaching prior to quantification, which followed a single-exponential decay process. ROIs were drawn around the cell edge or the cell conjugate region to measure the signal for ZAP70-mRuby2 recruitment to these regions over time. To define the plasma membrane ROIs, images from the ZAP70-mRuby2, LCK*, and IFP2.0-CaaX stacks were combined, background subtracted and contrast enhanced to clearly define a cell shape that could be thresholded to create a binary mask of the cell. This mask was then outlined to describe the cell edge as a single-pixel width ROI, which was re-mapped to the range [0, 1]. The pixel count in the ROI was used as the mask area. To draw an ROI in the cell conjugate region, the images from pMHC-BFP channel were used. Prior to quantification, where needed, image stacks were cropped to isolate the relevant conjugated cells to remove signal from nearby unconjugated cells. Upon background subtraction, contrast enhancement and Gaussian smoothing, the conjugate region was thresholded to create a binary mask. This mask was skeletonized to create a single-pixel width ROI and mapped over [0,1]. The masked images and corresponding ZAP70-mRuby2 stack were then multiplied to create a stack that only contained the fluorescence intensity values in the ROI only. To obtain the mean value of mRuby2 intensity in the ROI, the summed intensities were divided by the ROI area. A similar workflow was performed to obtain the mean value of mRuby2 over the entire cell by drawing an ROI of the area bounded by the cell mask. The mean intensity at the cell edge or conjugate region was divided by the corresponding mean intensity over the whole cell to obtain the relative fraction for ZAP70 membrane or conjugate recruitment in each image, plotted against time in the main text. To measure the ratio of eGFP signal inside over outside the conjugate, line ROIs were drawn manually to define the conjugate region and the edge of eGFP-expressing HEK cells prior to illumination. Mean intensities of eGFP were then measured at these ROIs.
Quantification of endogenous protein expression
Expression of endogenous CSK, SHP-1 and SHP-2 in various cell lines were detected by Western blot using monoclonal antibodies specific to the proteins. Samples were prepared as described for the main experiment and the blots were analyzed as described above. Three different dilutions of each soluble cell lysate were measured to obtain an average value. Raw intensity values from the blots were corrected for the dilution factors obtained from the relative expression of an actin control, cell density in sample and the cytoplasmic volumes of the specific cell lines. These corrected values for each cytoplasmic protein are then presented as relative values compared to the protein expression in CD4+ T cells. Cytoplasmic volumes of the different cell lines were 96.1 fL for CD4+ and CD8+ T cells, 664 fL for Jurkat T cells and 790 fL for HEK293T cells54–56.
Data plotting
All data were initially processed in Excel (Microsoft) to compute the mean and SEM values. Subsequent data plotting was performed using MATLAB (MathWorks). Where appropriate, data fitting was performed using a 3-parameter logistic function: where b is the Hill’s coefficient, c is the inflection point, d is the maximum asymptote and t is time (min).The maximum asymptote values calculated were used for normalization of the imaging data in Fig. 7, and the derived 95% confidence interval values were used as the error bar values in Fig. 4d. Otherwise, a moving-average filter was used to smooth the data points. Error bars were used to represent the s.e.m. of discrete data points in phosphorylation analyses. Filled area was used to represent the s.e.m. of the more continuous data points in the ZAP70 recruitment analyses.
Derivation of relative initial rates and time to half-maximal ZAP70 recruitment
Plots from Western and imaging data were analyzed by MATLAB (MathWorks) further to derive a measure of the rate of LCK kinase activity in each experiment. For the Western analyses, the reaction rate was estimated by differentiating the 3-parameter logistic function used to fit the data (see Data Plotting above) with respect to time. Taking this value at t = 0.5 min defined the initial reaction rate (V0), when this parameter is expected to be maximal immediately after end of photo-uncaging period. For the imaging experiments, the ZAP70 recruitment datasets were smoothed and normalized to the corresponding maximum values for datasets that had reached saturation or maximum asymptotes for datasets that had not. The normalized data were then spline-fitted to estimate the time when the recruitment of ZAP70 to the plasma membrane was at half-maximum. In both cases, rate parameters were determined from n number of independent experiments as indicated in the figure legends, and the mean and s.e.m. values were calculated from these values.
A Life Sciences Reporting Summary for this article is available online.
Data Availability Statement
Source data used to generate the graphs are available in Supplementary Data Set 2. Other data that support the findings of this study (e.g. Western blots, microscopy images and pre-normalization raw values) are available from the corresponding author J.R.J. upon reasonable request.
Supplementary Material
Acknowledgements
This work was supported by a Sir Henry Dale Fellowship jointly funded by the Wellcome Trust and the Royal Society (Grant Number: 099966/Z/12/Z to JRJ), and by the Medical Research Council, UK (MC_U105181009 and MC_UP_A024_1008 to JWC). We would like to thank the engineering workshop at the MRC-LMB for manufacturing the illumination device used in this study.
Footnotes
Author Contributions
A.L-J., B.L.M. and J.R.J. designed and performed all the experiments in the study. A.L-J. and J.R.J. analyzed the data. M.M. synthesized the unnatural amino acid pc-Lys. J.W.C. provided scientific input and helped revise the manuscript, which was written by A.L-J. and J.R.J. All authors contributed to the final manuscript. J.R.J. oversaw and supervised the research.
Competing Financial Interests
The authors declare no competing financial interests.
References
- 1.Smith-Garvin JE, Koretzky G, Jordan MS. T cell activation. Annu Rev Immunol. 2009;27:591–619. doi: 10.1146/annurev.immunol.021908.132706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van der Merwe PA, Dushek O. Mechanisms for T cell receptor triggering. Nat Rev Immunol. 2011;11:47–55. doi: 10.1038/nri2887. [DOI] [PubMed] [Google Scholar]
- 3.Ohashi PS. T-cell signalling and autoimmunity: molecular mechanisms of disease. Nat Rev Immunol. 2002;2:427–38. doi: 10.1038/nri822. [DOI] [PubMed] [Google Scholar]
- 4.Talab F, Allen JC, Thompson V, Lin K, Slupsky JR. LCK is an important mediator of B-cell receptor signaling in chronic lymphocytic leukemia cells. Mol Cancer Res. 2013;11:541–54. doi: 10.1158/1541-7786.MCR-12-0415-T. [DOI] [PubMed] [Google Scholar]
- 5.Tycko B, Smith SD, Sklar J. Chromosomal translocations joining LCK and TCRB loci in human T cell leukemia. J Exp Med. 1991;174:867–73. doi: 10.1084/jem.174.4.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu W, Doshi A, Lei M, Eck MJ, Harrison SC. Crystal Structures of c-Src reveal features of its autoinhibitory mechanism. Mol Cell. 1999;3:629–638. doi: 10.1016/s1097-2765(00)80356-1. [DOI] [PubMed] [Google Scholar]
- 7.Boggon TJ, Eck MJ. Structure and regulation of Src family kinases. Oncogene. 2004;23:7918–27. doi: 10.1038/sj.onc.1208081. [DOI] [PubMed] [Google Scholar]
- 8.Bergman M, et al. The human p50csk tyrosine kinase phosphorylates p56lck at Tyr-505 and down regulates its catalytic activity. EMBO J. 1992;11:2919–24. doi: 10.1002/j.1460-2075.1992.tb05361.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chow LM, Fournel M, Davidson D, Veillette A. Negative regulation of T-cell receptor signalling by tyrosine protein kinase p50csk. Nature. 1993;365:156–60. doi: 10.1038/365156a0. [DOI] [PubMed] [Google Scholar]
- 10.Saunders AE, Johnson P. Modulation of immune cell signalling by the leukocyte common tyrosine phosphatase, CD45. Cell Signal. 2010;22:339–48. doi: 10.1016/j.cellsig.2009.10.003. [DOI] [PubMed] [Google Scholar]
- 11.Paster W, et al. Genetically encoded Förster resonance energy transfer sensors for the conformation of the Src family kinase Lck. J Immunol. 2009;182:2160–7. doi: 10.4049/jimmunol.0802639. [DOI] [PubMed] [Google Scholar]
- 12.Nika K, et al. Constitutively active Lck kinase in T cells drives antigen receptor signal transduction. Immunity. 2010;32:766–77. doi: 10.1016/j.immuni.2010.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stirnweiss A, et al. T cell activation results in conformational changes in the Src family kinase Lck to induce its activation. Sci Signal. 2013;6:ra13. doi: 10.1126/scisignal.2003607. [DOI] [PubMed] [Google Scholar]
- 14.Hui E, Vale RD. In vitro membrane reconstitution of the T-cell receptor proximal signaling network. Nat Struct Mol Biol. 2014;21:133–142. doi: 10.1038/nsmb.2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Palacios EH, Weiss A. Function of the Src-family kinases, Lck and Fyn, in T-cell development and activation. Oncogene. 2004;23:7990–8000. doi: 10.1038/sj.onc.1208074. [DOI] [PubMed] [Google Scholar]
- 16.Germain RN. T-cell development and the CD4-CD8 lineage decision. Nat Rev Immunol. 2002;2:309–322. doi: 10.1038/nri798. [DOI] [PubMed] [Google Scholar]
- 17.Veillette A, Bookman MA, Horak EM, Bolen JB. The CD4 and CD8 T cell surface antigens are associated with the internal membrane tyrosine-protein kinase p56lck. Cell. 1988;55:301–8. doi: 10.1016/0092-8674(88)90053-0. [DOI] [PubMed] [Google Scholar]
- 18.Rudd CE, Trevillyan JM, Dasgupta JD, Wong LL, Schlossman SF. The CD4 receptor is complexed in detergent lysates to a protein-tyrosine kinase (pp58) from human T lymphocytes. Proc Natl Acad Sci U S A. 1988;85:5190–4. doi: 10.1073/pnas.85.14.5190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim PW, Sun ZJ, Blacklow SC, Wagner G, Eck MJ. A zinc clasp structure tethers Lck to T cell coreceptors CD4 and CD8. Science. 2003;301:1725–8. doi: 10.1126/science.1085643. [DOI] [PubMed] [Google Scholar]
- 20.Dagliyan O, et al. Engineering extrinsic disorder to control protein activity in living cells. Science. 2016;354:1441–1444. doi: 10.1126/science.aah3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karginov AV, Ding F, Kota P, Dokholyan NV, Hahn KM. Engineered allosteric activation of kinases in living cells. Nat Biotechnol. 2010;28:743–7. doi: 10.1038/nbt.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou XX, Fan LZ, Li P, Shen K, Lin MZ. Optical control of cell signaling by single-chain photoswitchable kinases. Science. 2017;355:836–842. doi: 10.1126/science.aah3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Davis L, Chin JW. Designer proteins: applications of genetic code expansion in cell biology. Nat Rev Mol Cell Biol. 2012;13:168–182. doi: 10.1038/nrm3286. [DOI] [PubMed] [Google Scholar]
- 24.Chin JW. Expanding and Reprogramming the Genetic Code of Cells and Animals. Annu Rev Biochem. 2014;83:379–408. doi: 10.1146/annurev-biochem-060713-035737. [DOI] [PubMed] [Google Scholar]
- 25.Gautier A, et al. Genetically encoded photocontrol of protein localization in mammalian cells. J Am Chem Soc. 2010;132:4086–8. doi: 10.1021/ja910688s. [DOI] [PubMed] [Google Scholar]
- 26.James JR, Vale RD. Biophysical mechanism of T-cell receptor triggering in a reconstituted system. Nature. 2012;487:64–9. doi: 10.1038/nature11220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carrera AC, Alexandrov K, Roberts TM. The conserved lysine of the catalytic domain of protein kinases is actively involved in the phosphotransfer reaction and not required for anchoring ATP (enzyme mechanism/protein-tyroslne kinase/phosphorylation) Biochemistry. 1993;90:442–446. doi: 10.1073/pnas.90.2.442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gautier A, Deiters A, Chin JW. Light-activated kinases enable temporal dissection of signaling networks in living cells. J Am Chem Soc. 2011;133:2124–2127. doi: 10.1021/ja1109979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schmied WH, Elsässer SJ, Uttamapinant C, Chin JW. Efficient Multisite Unnatural Amino Acid Incorporation in Mammalian Cells via Optimized Pyrrolysyl tRNA Synthetase/tRNA Expression and Engineered eRF1. J Am Chem Soc. 2014;136:15577–15583. doi: 10.1021/ja5069728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Williams BL, et al. Phosphorylation of Tyr319 in ZAP-70 is required for T-cell antigen receptor-dependent phospholipase C-γ1 and Ras activation. EMBO J. 1999;18:1832–44. doi: 10.1093/emboj/18.7.1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brdicka T, Kadlecek TA, Roose JP, Pastuszak AW, Weiss A. Intramolecular Regulatory Switch in ZAP-70: Analogy with Receptor Tyrosine Kinases. Mol Cell Biol. 2005;25:4924–4933. doi: 10.1128/MCB.25.12.4924-4933.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Amrein KE, Sefton BM. Mutation of a site of tyrosine phosphorylation in the lymphocyte- specific tyrosine protein kinase, p56lck, reveals its oncogenic potential in fibroblasts. Proc Natl Acad Sci USA. 1988;85:4247–4251. doi: 10.1073/pnas.85.12.4247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abraham N, Veillette A. Activation of p56lck through mutation of a regulatory carboxy-terminal tyrosine residue requires intact sites of autophosphorylation and myristylation. Mol Cell Biol. 1990;10:5197–206. doi: 10.1128/mcb.10.10.5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rossy J, Owen DM, Williamson DJ, Yang Z, Gaus K. Conformational states of the kinase Lck regulate clustering in early T cell signaling. Nat Immunol. 2013;14:82–9. doi: 10.1038/ni.2488. [DOI] [PubMed] [Google Scholar]
- 35.Caron L, Abraham N, Pawson T, Veillette A. Structural requirements for enhancement of T-cell responsiveness by the lymphocyte-specific tyrosine protein kinase p56lck. Mol Cell Biol. 1992;12:2720–9. doi: 10.1128/mcb.12.6.2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luo KX, Sefton BM. Cross-linking of T-cell surface molecules CD4 and CD8 stimulates phosphorylation of the lck tyrosine protein kinase at the autophosphorylation site. Mol Cell Biol. 1990;10:5305–13. doi: 10.1128/mcb.10.10.5305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chan AC, et al. Activation of ZAP-70 kinase activity by phosphorylation of tyrosine 493 is required for lymphocyte antigen receptor function. EMBO J. 1995;14:2499–2508. doi: 10.1002/j.1460-2075.1995.tb07247.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Watts JD, et al. Identification by electrospray ionization mass spectrometry of the sites of tyrosine phosphorylation induced in activated Jurkat T cells on the protein tyrosine kinase ZAP-70. J Biol Chem. 1994;269:29520–9. [PubMed] [Google Scholar]
- 39.Kong G, et al. Distinct tyrosine phosphorylation sites in ZAP-70 mediate activation and negative regulation of antigen receptor function. Mol Cell Biol. 1996;16:5026–35. doi: 10.1128/mcb.16.9.5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Denny MF, Patai B, Straus DB. Differential T-cell antigen receptor signaling mediated by the Src family kinases Lck and Fyn. Mol Cell Biol. 2000;20:1426–35. doi: 10.1128/mcb.20.4.1426-1435.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lovatt M, et al. Lck Regulates the Threshold of Activation in Primary T Cells, While both Lck and Fyn Contribute to the Magnitude of the Extracellular Signal-Related Kinase Response. Mol Cell Biol. 2006;26:8655–8665. doi: 10.1128/MCB.00168-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huse M, Eck MJ, Harrison SC. A Zn2+ Ion Links the Cytoplasmic Tail of CD4 and the N-terminal Region of Lck. J Biol Chem. 1998;273:18729–18733. doi: 10.1074/jbc.273.30.18729. [DOI] [PubMed] [Google Scholar]
- 43.Vignali DA, Carson RT, Chang B, Mittler RS, Strominger JL. The two membrane proximal domains of CD4 interact with the T cell receptor. J Exp Med. 1996;183:2097–107. doi: 10.1084/jem.183.5.2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.James JR, Oliveira MI, Carmo AM, Iaboni A, Davis SJ. A rigorous experimental framework for detecting protein oligomerization using bioluminescence resonance energy transfer. Nat Methods. 2006;3:1001–6. doi: 10.1038/nmeth978. [DOI] [PubMed] [Google Scholar]
- 45.Artyomov MN, Lis M, Devadas S, Davis MM, Chakraborty AK. CD4 and CD8 binding to MHC molecules primarily acts to enhance Lck delivery. Proc Natl Acad Sci U S A. 2010;107:16916–21. doi: 10.1073/pnas.1010568107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brown MT, Cooper JA. Regulation, substrates and functions of src. Biochim Biophys Acta. 1996;1287:121–49. doi: 10.1016/0304-419x(96)00003-0. [DOI] [PubMed] [Google Scholar]
- 47.Xu B, Miller WT. Src homology domains of v-Src stabilize an active conformation of the tyrosine kinase catalytic domain. Mol Cell Biochem. 1979;158:57–63. doi: 10.1007/BF00225883. [DOI] [PubMed] [Google Scholar]
- 48.Straus DB, Chan AC, Patai B, Weiss A. SH2 domain function is essential for the role of the Lck tyrosine kinase in T cell receptor signal transduction. J Biol Chem. 1996;271:9976–9981. doi: 10.1074/jbc.271.17.9976. [DOI] [PubMed] [Google Scholar]
- 49.Yamaguchi H, Hendrickson Wa. Structural basis for activation of human lymphocyte kinase Lck upon tyrosine phosphorylation. Nature. 1996;384:484–9. doi: 10.1038/384484a0. [DOI] [PubMed] [Google Scholar]
- 50.D’Oro U, Sakaguchi K, Appella E, Ashwell JD. Mutational analysis of Lck in CD45-negative T Cells: dominant role of tyrosine 394 phosphorylation in kinase activity. Mol Cell Biol. 1996;16:4996–5003. doi: 10.1128/mcb.16.9.4996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Courtney AH, et al. A Phosphosite within the SH2 Domain of Lck Regulates Its Activation by CD45. Mol Cell. 2017;67:498–511.e6. doi: 10.1016/j.molcel.2017.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stepanek O, et al. Coreceptor Scanning by the T Cell Receptor Provides a Mechanism for T Cell Tolerance. Cell. 2014;159:333–345. doi: 10.1016/j.cell.2014.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.James JR, et al. The T cell receptor triggering apparatus is composed of monovalent or monomeric proteins. J Biol Chem. 2011;286:31993–2001. doi: 10.1074/jbc.M111.219212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tan CW, et al. Wnt signalling pathway parameters for mammalian cells. PLoS One. 2012;7 doi: 10.1371/journal.pone.0031882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rosenbluth MJ, Lam WA, Fletcher DA. Force microscopy of nonadherent cells: a comparison of leukemia cell deformability. Biophys J. 2006;90:2994–3003. doi: 10.1529/biophysj.105.067496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Loiko VA, et al. Morphometric model of lymphocyte as applied to scanning flow cytometry. J Quant Spectrosc Radiat Transf. 2006;102:73–84. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Source data used to generate the graphs are available in Supplementary Data Set 2. Other data that support the findings of this study (e.g. Western blots, microscopy images and pre-normalization raw values) are available from the corresponding author J.R.J. upon reasonable request.