Abstract
Misfolded endoplasmic reticulum (ER) proteins are retro-translocated through the membrane into the cytosol, where they are poly-ubiquitinated, extracted from the ER membrane, and degraded by the proteasome 1–4, a pathway termed ER-associated protein degradation (ERAD). Proteins with misfolded domains in the ER lumen or membrane are discarded through the ERAD-L and –M pathways, respectively. In S. cerevisiae, both pathways require the ubiquitin ligase Hrd1, a multi-spanning membrane protein with a cytosolic RING finger domain 5,6. Hrd1 is the crucial membrane component for retro-translocation 7,8, but whether it forms a protein-conducting channel is unclear. Here, we report a cryo-electron microscopy (cryo-EM) structure of S. cerevisiae Hrd1 in complex with its ER luminal binding partner Hrd3. Hrd1 forms a dimer within the membrane with one or two Hrd3 molecules associated at its luminal side. Each Hrd1 molecule has eight trans-membrane segments, five of which form an aqueous cavity extending from the cytosol almost to the ER lumen, while a segment of the neighboring Hrd1 molecule forms a lateral seal. The aqueous cavity and lateral gate are reminiscent of features in protein-conducting conduits that facilitate polypeptide movement in the opposite direction, i.e. from the cytosol into or across membranes 9–11. Our results suggest that Hrd1 forms a retro-translocation channel for the movement of misfolded polypeptides through the ER membrane.
The ubiquitin ligase Hrd1 is in a complex with three other membrane proteins (Hrd3, Usa1, and Der1) and a luminal protein (Yos9) 6,12,13. In wild type yeast cells, all these components are required for the retro-translocation of proteins with misfolded luminal domains (ERAD-L substrates). ERAD-M substrates, which contain misfolded domains inside the membrane, also depend on Hrd1 and Hrd3, but not on Der1 6, and only in some cases on Usa114. Among the components of the Hrd1 complex, Hrd3 is of particular importance; it cooperates with Yos9 in substrate binding and regulates the ligase activity of Hrd1 15–17. Both Hrd1 and Hrd3 (called Sel1 in mammals) are conserved in all eukaryotes.
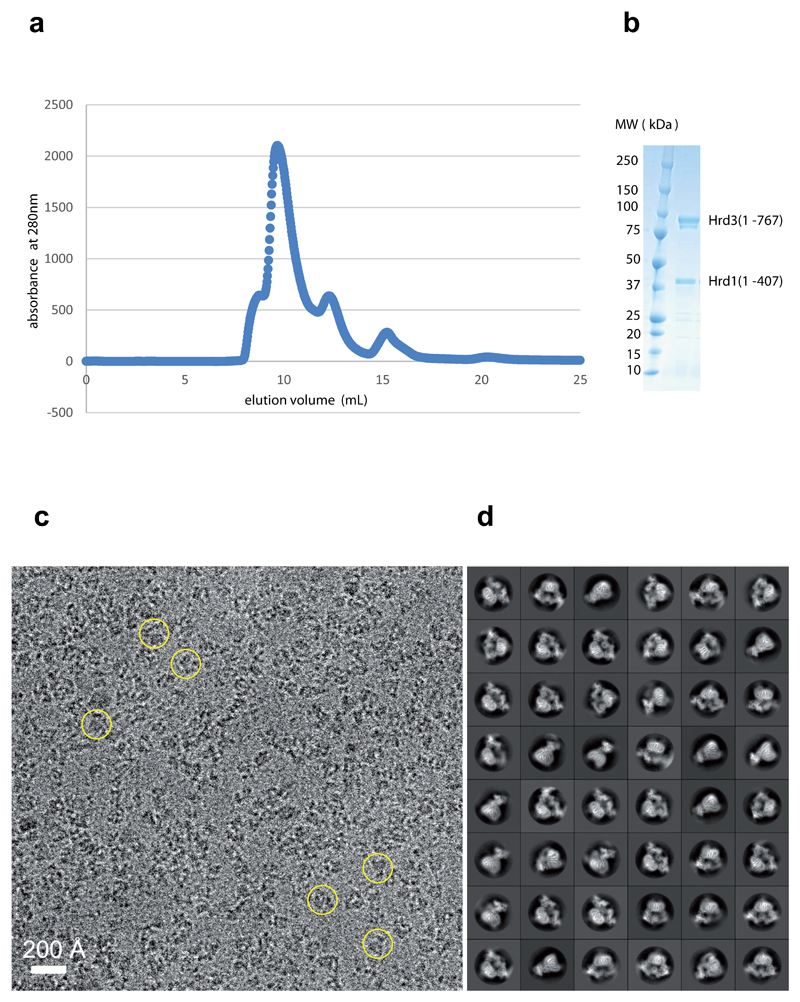
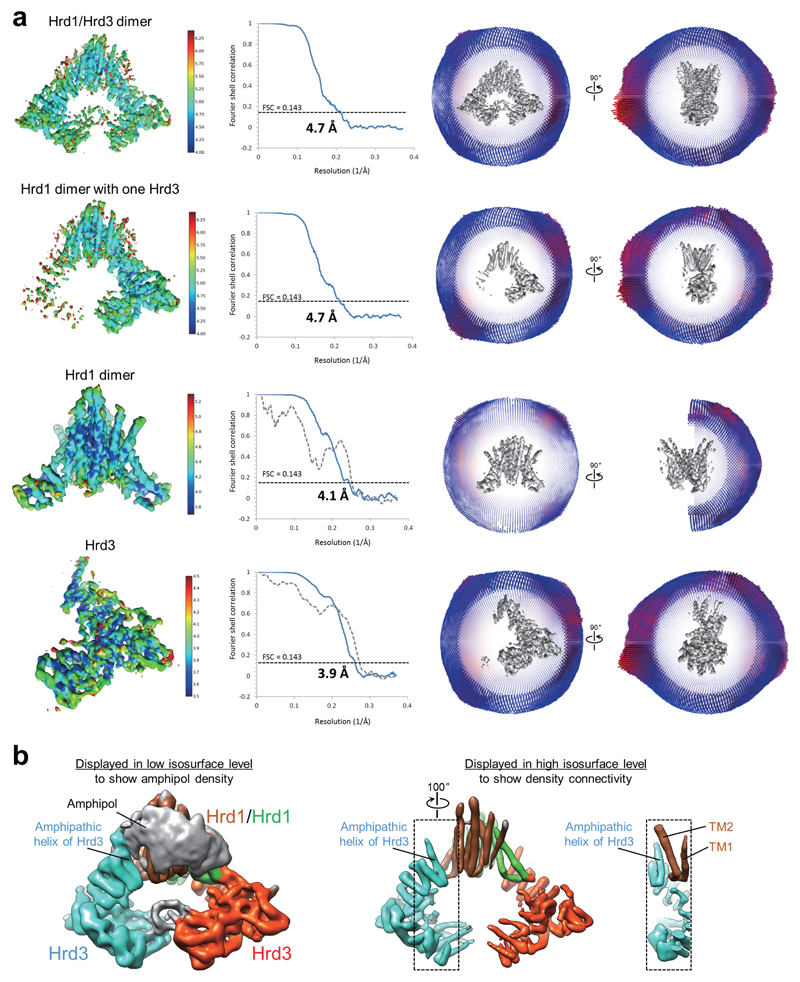
To obtain structural information for Hrd1 and Hrd3, we co-expressed in S. cerevisiae Hrd1, truncated after the RING finger domain (amino acids 1-407), together with a luminal fragment of Hrd3 (amino acids 1-767). The Hrd3 construct lacks the C-terminal trans-membrane (TM) segment, which is not essential for its function in vivo 7. In contrast to Hrd1 alone, which forms heterogeneous oligomers 18, the Hrd1/Hrd3 complex eluted in gel filtration as a single major peak (Extended Data Fig. 1). After transfer from detergent into amphipol, the complex was analyzed by single-particle cryo-EM. The reconstructions showed a Hrd1 dimer associated with either two or one Hrd3 molecules, the latter probably originating from some dissociation during purification. Cryo-EM maps representing these two complexes were refined to 4.7 Å resolution (Extended Data Figs. 2,3; Extended Data Table1). To improve the reconstructions, we performed Hrd1 dimer- and Hrd3 monomer- focused 3D classifications with signal subtraction 19. The resulting homogeneous sets of particle images of Hrd1 dimer and Hrd3 monomer were used to refine the density maps to 4.1Å and 3.9Å resolution, respectively. Models were built into these maps and are based on the agreement between density and the prediction of TMs and helices, the density for some large amino acid side chains and N-linked carbohydrates (Extended Data Fig. 4), evolutionary coupling of amino acids (Extended Data Fig. 5) 20, and energy minimization with the Rosetta program 21.
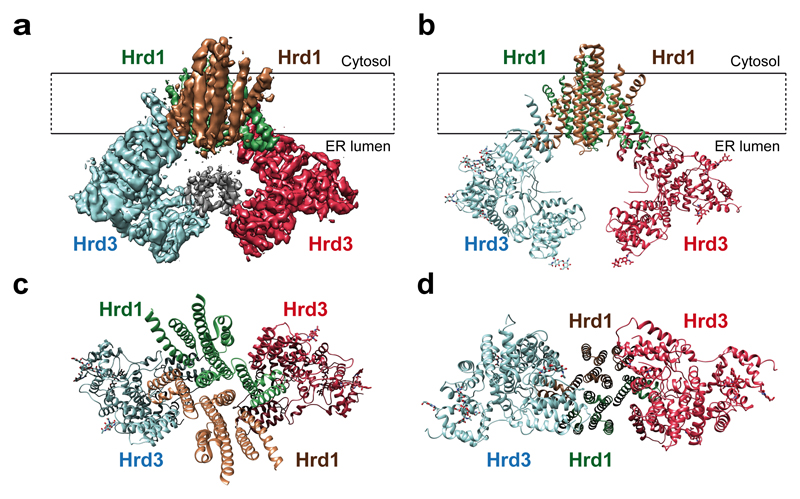
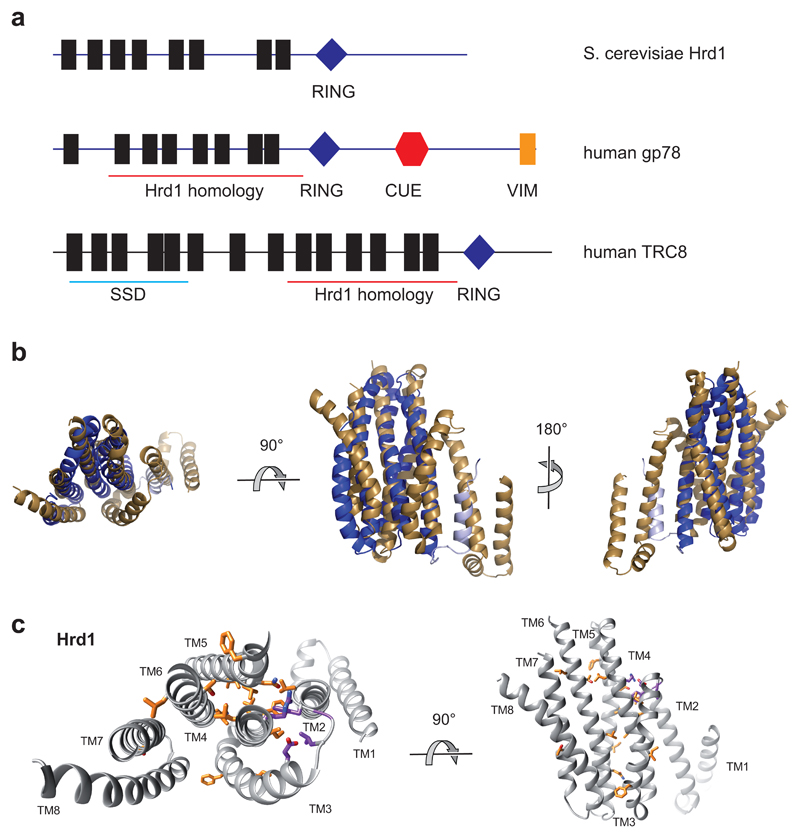
In the complex containing two molecules of both Hrd1 and Hrd3, the Hrd1 molecules interact through their TMs, and the Hrd3 molecules form an arch on the luminal side (Fig. 1a-d). The Hrd1 dimer has essentially the same structure when only one Hrd3 molecule is bound, and Hrd3 is only slightly tilted towards the Hrd1 dimer (not shown). None of the reconstructions showed density for the cytoplasmic RING finger domains of Hrd1 (Fig. 1a), suggesting that they are flexibly attached to the membrane domains.
Figure 1. Architecture of the Hrd1/Hrd3 complex.
a, Side view of the density map of the dimeric Hrd1/Hrd3 complex at 4.7Å resolution. The Hrd1 molecules are shown in brown and green, and the Hrd3 molecules in blue and red. Weak density between the Hrd3 molecules is in grey. The approximate membrane boundaries are indicated. b, Side view of the models for Hrd1 and Hrd3 in ribbon presentation, colored as in a. The grey region in a, corresponding to residues 268-316 of Hrd3, could not be traced. c, As in b, but view from the cytosol. d, As in b, but view from the ER lumen.
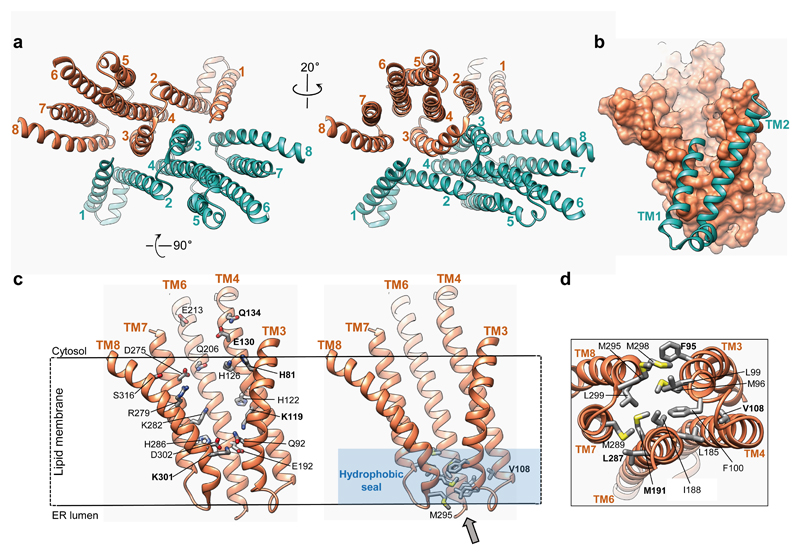
Each Hrd1 molecule has eight helical TMs (Fig. 2a), rather than six, as previously thought 22. Consistent with Hrd1 being a channel, the membrane domains of Hrd1 form a funnel that extends from the cytosol almost to the luminal side of the membrane (Fig. 2a-c). Each of the two symmetry-related funnels is lined by TMs 3, 4, 6, 7, and 8 of one Hrd1 molecule and TM1 of the other; TM1 sits between TMs 3 and 8 and, in an intact membrane, would laterally seal the funnel in the cytosolic leaflet of the bilayer (Fig. 2b). Several TMs extend from the membrane into the cytosol; TM 8 bends away from the funnel center on the cytosolic side, so that the following RING finger domains of the Hrd1 molecules are kept far apart. The funnels are likely filled with water, as they contain several conserved hydrophilic and charged residues, mostly contributed by the multi-TM surface from one Hrd1 molecule (Fig. 2c). These residues show little side chain density by comparison with those involved in interaction between helices (Extended Data Fig. 4), suggesting that they are flexible. The funnels are sealed towards the luminal aqueous phase by two layers of hydrophobic residues (Fig. 2c, d). Dimerization between the two Hrd1 molecules is mediated by interfaces between TMs 1 and 2 of one Hrd1 molecule and TMs 8 and 3 of the other, and between TMs 3 of the two Hrd1 molecules (Fig. 2a).
Figure 2. Structure of Hrd1.
a, Ribbon presentation of the Hrd1 dimer (brown, green) in two different views from the cytosol. Hrd3 was omitted for clarity. The TMs are numbered. b, Side view of a space-filling model of TMs 3-8 of one Hrd1 molecule together with a ribbon presentation of TMs 1 and 2 of the other. c, Left: conserved hydrophilic residues inside the Hrd1 funnel shown as sticks. The position of these residues is deduced from the orientation of TM helices. Right: hydrophobic residues sealing the funnel towards the ER lumen. d, Hydrophobic seal residues viewed along the grey arrow shown in c (right panel).
The structure of Hrd1 is likely conserved among all eukaryotes (Extended Data Fig. 6). Hrd1 contains conserved amino acids in the membrane-embedded domain, particularly in residues involved in the interaction among TMs (Extended Data Fig. 7). This conservation extends to the Hrd1 homologue gp78, another ER-resident ubiquitin ligase that is found in metazoans, plants and other eukaryotes, but seems to have been lost in fungi. Interestingly, the metazoan ubiquitin ligases RNF145 and RNF139 (alternatively called TRC8) also show sequence similarity to TMs 3-8 of Hrd1 and gp78, and are predicted to form similar structures (Extended Data Figs. 6, 7). Thus, all these ligases probably function in a similar way.
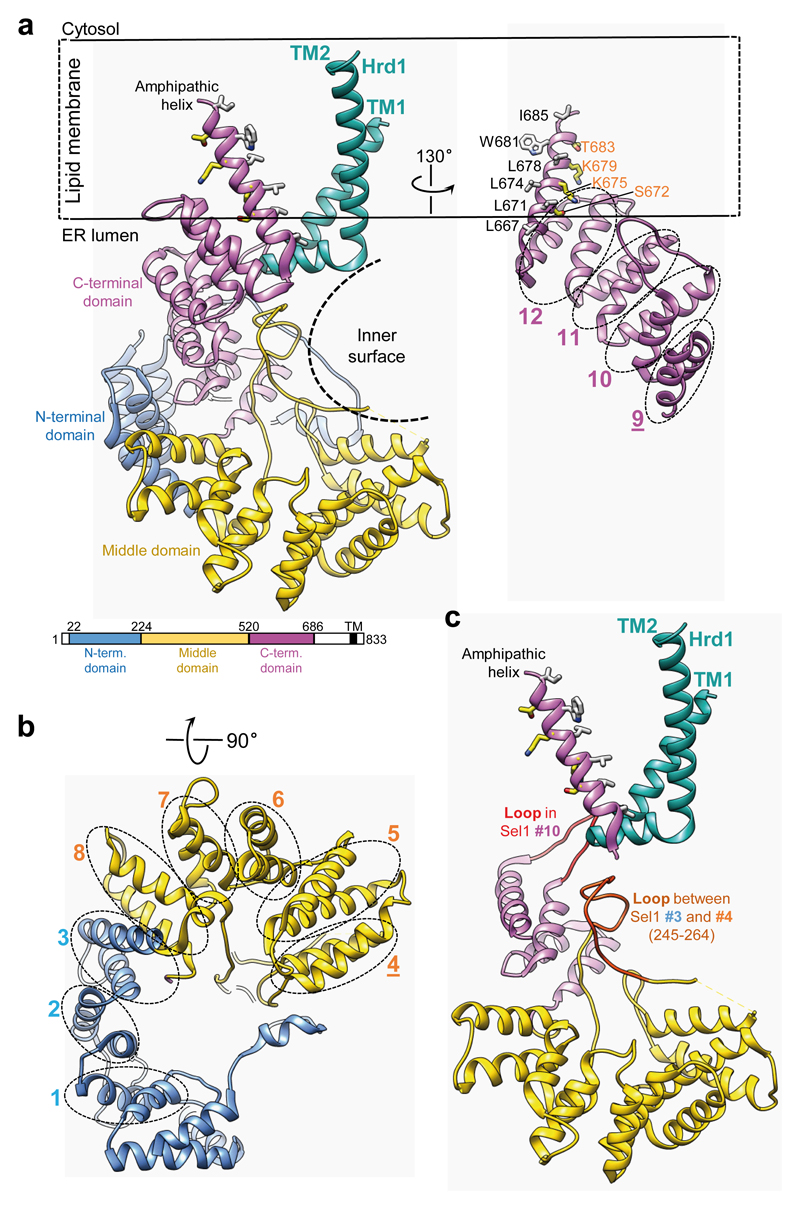
Hrd3 contains 12 Sel1 motifs (Fig. 3a, b), each consisting of a helix, a loop and another helix, which form N-terminal, middle and C-terminal domains that together give Hrd3 an L-shape with inner and outer surfaces (Fig. 3a). The inner surface contains a groove (Extended Data Fig. 8), which might bind substrate. Several patches of conserved residues are also seen on the outer surface of Hrd3 (Extended Data Fig. 8). The patch formed by the last two Sel1 motifs likely interacts with Yos9 17. Hrd3 binds to the loop between TM1 and TM2 of Hrd1, utilizing the concave face of the most C-terminal Sel1 repeats and two loops (Fig. 3c). Our structure is consistent with the reported interaction between the last Sel1 motifs and the TM1/2 loop of Hrd1 23. Surprisingly, the density map shows an additional, amphipathic helix that immediately follows the last Sel1 repeat of Hrd3 and would reach into the hydrophobic interior of an intact membrane, although it is not predicted to be a TM (Fig. 3a). The amphipathic helix makes contact with the C-terminal helix of the last Sel1 motif of Hrd3 and with the loop between TM1 and TM2 of Hrd1 (Fig. 3c). The helix is conserved (Extended Data Fig. 9) and its deletion abolishes Hrd1/Hrd3 interaction 17. Its position in our structure might be stabilized by amphipols (Extended Data Fig. 3), but based on crosslinking data 24, it seems possible that the helix would normally interact with Der1. Residues 687-767 between the amphipathic helix and the TM segment (deleted in our construct) are predicted to be in the ER lumen, but we were unable to find clear density for a segment linking the C-terminal end of the amphipathic helix back to the luminal space.
Figure 3. Structure of Hrd3.
a, Left: ribbon presentation of the Hrd3 model. Domain colors correspond to those in the sequence diagram. TM1-TM2 of Hrd1 are in green. The Hrd3 structure lacks the signal sequence and the segment following residue 686. Black dashed arc: inner surface of Hrd3. Right: Sel1 motifs in the C-terminal domain of Hrd3 (ovals). Underlined motifs were not predicted from primary sequence. Hydrophobic and hydrophilic residues of the amphipathic helix are shown in black and yellow, respectively. b, Sel1 motifs in the N terminal and middle domains of Hrd3. c, Hrd3 segments interacting with Hrd1 (in green). The N-terminal domain and Sel1 motifs #11 and #12 were omitted.
Hrd1 and Hrd3 may be the minimum components required for ERAD-M, although Usa1 might stabilize the complex 14. The Hrd1 channel must allow membrane-spanning segments of ERAD-M substrates to enter sideways from the lipid phase. Such a lateral gate is likely located where TM1 is seen in our structure. TM1 would serve as a space holder until an ERAD-M substrate arrives and TM1 is displaced. TM2 would stay put, associated with TMs 3 and 4 through conserved amino acids on the cytosolic side of the membrane (Extended Data Figs. 6,7). These interactions can explain why mutations in this region affect some ERAD-M substrates 25. Interestingly, the ligases TRC8 and RNF145 show sequence homology to Hrd1 only in the cavity-forming TMs 3-8; these proteins contain an additional multi-spanning sterol-sensing domain (Extended Data Fig. 7), suggesting that their lateral gating is regulated by ligands.
The significance of pairing two Hrd1 channels is currently unknown; only one channel might be active at any given time, or the channels could function independently of each other, as in other oligomeric channels and transporters 26–28. How exactly the Hrd1 channel would operate in ERAD-L also remains unclear, because additional components are required (Usa1, Der1, and Yos9), Hrd1 dimerization in vivo requires Usa1 7,14, and channel opening involves auto-ubiquitination 8. Nevertheless, only a small conformational change at the luminal side of Hrd1 seems to be required to open a pore across the membrane. Channel opening likely requires substrate binding to Hrd3, which in turn would affect Hrd1, as Hrd3 sits on the loop between TMs 1 and 2.
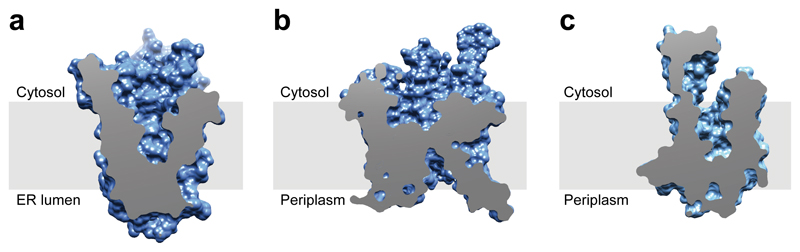
The Hrd1 channel has features reminiscent of the Sec61/SecY channel that transports polypeptides in the opposite direction, i.e., from the cytosol across the eukaryotic ER or prokaryotic plasma membrane 9,29. In both cases, the channels have aqueous interiors (Fig. 4a, b) and lateral gates, and hydrophobic residues provide the membrane barrier, a pore ring in Sec61/SecY and a two-layer seal in Hrd1. Hrd1 also bears intriguing similarity with the bacterial YidC protein and its homologs in plants and mitochondria 10,11, as these also have deep cytosolic invaginations that contain polar residues (Fig. 4c). These proteins allow hydrophobic TM segments to move from the cytosol into the lipid bilayer, whereas Hrd1 facilitates the reverse process during ERAD-M. Thus, the thinning of the membrane barrier might be a general principle employed by protein-conducting conduits to facilitate polypeptide movement in and out of a membrane.
Figure 4. Hydrophilic funnels in protein-conducting channels.
a, Cut-away side view of a space-filling model of Hrd1. The membrane boundaries were determined with the OPM server (http://opm.phar.umich.edu/server.php). b, As in a, but for the SecY channel (PDB code 1RH5). c, As in a, but for YidC (PDB code 3WO6).
Methods and Materials
Yeast Strains and Plasmids
The Hrd1/Hrd3 complex was expressed in the S. cerevisiae strain INVSc1 (Invitrogen) from 2μ plasmids of the pRS42X series under the Gal1 promoter 18. Hrd1 was expressed as a C-terminally truncated version (amino acids 1-407) from a plasmid carrying an Ura marker. The Hrd1 fragment 1-407 corresponds to a stable tryptic fragment. Hrd3 was expressed as a luminal fragment (amino acids 1-767), in which the C-terminal TM segment was replaced with a tobacco etch virus (TEV) protease cleavage site followed by a streptavidin binding peptide (SBP). The plasmid carried a Trp marker.
Protein Purification
Yeast cells were transformed with plasmids encoding Hrd1(1-407) and Hrd3(1-767-TEV-SBP). A starter culture was inoculated and grown for 24 h at 30°C in synthetic dropout medium with amino acid supplements and 2% (w/v) glucose. The culture was diluted 1:40 into fresh medium and grown for additional 24 h. Expression was induced by adding 1/4 of the volume of 5x YEP broth containing 10% (w/v) galactose. The culture was incubated for 14–16 h at 25°C, and the cells were harvested by centrifugation for 10 min at 4000 x g. A 150g cell pellet was resuspended in 150 mL buffer A (50 mM HEPES pH 7.5, 500 mM NaCl, 5mM β-mercaptoethanol) supplemented with 1 mM phenylmethane sulfonyl fluoride (PMSF) and 1.5 μM pepstatin A. Glass beads were added to about 1/2 of the volume, and the cells were lysed in a BioSpec BeadBeater for 30 min with 30 s/60 s on/off cycles in a water/ice bath. After removal of the glass beads, the lysate was centrifuged twice in 250 ml tubes at 4000 x g for 10 min at 4°C. The supernatant was subjected to centrifugation in a Ti45 rotor at 42,000 x g for 45 min at 4°C. The membrane fraction was collected and flash-frozen in liquid nitrogen and stored at −80°C.
The Hrd1/Hrd3 complex was purified as follows. The membrane fraction was resuspended in 1.5 ml of buffer B (25 mM HEPES pH 7.5, 375 mM NaCl, 5 mM β-mercaptoethanol, 2% (w/v) decylmaltoside (DM)) per 1 g of membrane pellet and incubated for 30 min at 4°C. Insoluble material was removed by centrifugation (Ti45, 45min, 42,000 rpm). Six ml of Streptavidin Agarose resin (Goldbio) were added per 100 ml of solubilized membranes and incubated for 3 h on a rolling incubator. Beads were then washed with 5 column volumes (CV) of buffer C (20 mM HEPES pH 7.5, 375 mM NaCl, 5 mM DM, 1 mM tris(2-carboxyethyl)phosphine hydrochloride (TCEP), 0.01 mg/ml yeast polar lipid extract), followed by 10 CV of buffer C supplemented with 0.5 mM ATP and 10 mM MgCl2 and washed again with 35 CV of buffer C. The protein was then eluted with buffer C supplemented with 3 mM biotin. The protein was further purified by size-exclusion chromatography on a Superdex 200 10/300GL Increase column, equilibrated with buffer C without yeast polar lipid extract. Peak fractions were collected and mixed with yeast polar lipid extract (0.1 mg/ml) and Amphipol PMAL C8 (Anatrace) at a 1:3 ratio (w/w) with gentle agitation for 30 min. Detergent was removed by diluting the sample with detergent-free buffer (20 mM HEPES pH 7.5, 375 mM NaCl, 1 mM TCEP) below the CMC (1.8 mM) and subsequent concentration of the sample with an Amicon Ultra Centrifugal Filter (100 kDa cutoff). The protein sample was finally purified by size-exclusion chromatography on a Superdex 200 10/300GL Increase column. The peak fraction was concentrated to 1.4 mg/ml and used for cryo-EM analysis.
EM data acquisition
For cryo-EM, protein samples and freezing conditions were screened on a Tecnai TF20 electron microscope (FEI) operated at 200 kV. Aliquots of 2.5 µl of purified Hrd1/3 complex in PMAL-C8 at a concentration of 0.8 to 1 mg/ml were applied to a glow-discharged Quantifoil holey carbon grid (1.2/1.3, 400 mesh). Grids were blotted for 3 s at ˜90% humidity at room temperature, and plunge-frozen in liquid ethane using a Cryoplunge 3 System (Gatan). Cryo-EM data were recorded on a Titan Krios electron microscope (FEI) at the HHMI Janelia Research Campus, operated at 300 kV and equipped with a K2 Summit direct electron detector (Gatan). A Gatan Imaging filter with a slit width of 15 eV was used to remove inelastically scattered electrons. All cryo-EM movies were recorded in super-resolution counting mode using SerialEM 30. The nominal magnification of 81,000x corresponds to a calibrated pixel size of 1.35 Å on the specimen and 0.675 Å for super-resolution images. The dose rate was set to 5.47 electrons per Å2 and sec. The total exposure time of each movie was 15 s, leading to a total accumulated dose of 82 electrons per Å2, fractionated into 50 frames (300 ms per frame). All movies were recorded in a defocus range between 0.7 and 3.2 µm.
Image processing
Dose-fractionated super-resolution movies were subjected to motion correction using the program MotionCor2 31, and the resulting corrected movies and summed images were binned over 2 x 2 pixels, yielding a pixel size of 1.35 Å. All 50 frames in each movie were summed with or without a dose-weighting scheme 32. The summed images without dose-weighting were used for defocus calculation with the program CTFFIND3 33, while the dose-weighted summed images were used in all other image processing steps. Particle picking and screening, as well as the initial 3D model building were carried out using SamViewer and SAMUEL scripts as previously described 34. 3D classification and refinement were performed in RELION-1.4 35 and GeRelion 36. 3D refinements were completed with the particles summed from all 50 movie frames, and then continued with the particles summed from the movie frames 3-18, which improved the map quality and increased the resolution by ˜0.2 Å. The accumulated dose of the first 18 frames is ˜30 e-/Å2. All reported resolutions are based on gold-standard refinement procedures and the FSC=0.143 criterion. Local resolution was estimated using Resmap 37. The amplitude information of the final maps was corrected by applying a negative B-factor using the program bfactor.exe 38.
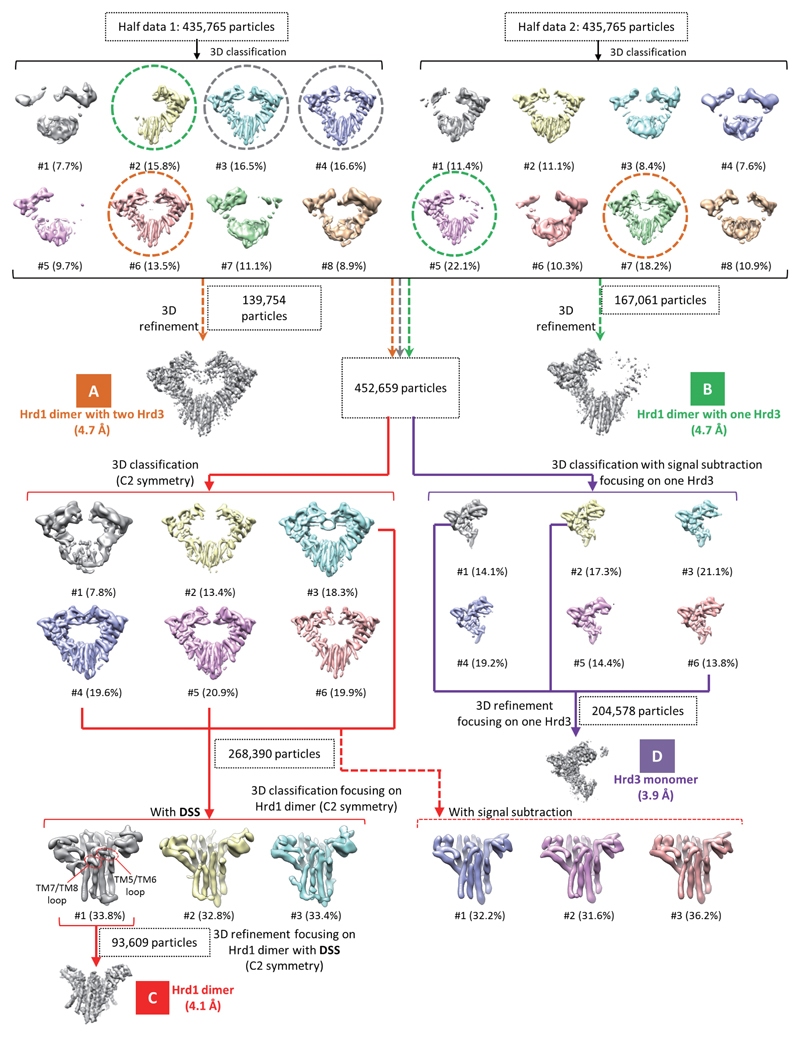
To speed up calculations, the cryo-EM data set (871,530 particles) was divided into two halves for the initial round of 3D classification. No symmetry was applied unless otherwise indicated. 3D reconstructions from these classes display a Hrd1 dimer associated with one or two Hrd3 molecules. We employed different strategies to achieve the best cryo-EM reconstructions for the components of the Hrd1/3 complex (see also Extended Data Fig. 2):
1) Hrd1/Hrd3 complex with two Hrd3 molecules. The 3D classes containing two Hrd3 molecules (class 6 in the first half and class 7 in the second half of the data set; 139,754 particles in total) were combined and refined, generating a reconstruction at 4.7 Å resolution. The major issue in reaching high resolution was the heterogeneity of conformations of the Hrd1/Hrd3 complex. Although in all particles Hrd3 binds to Hrd1 at the same site, alignment of Hrd1/Hrd3 maps from different classes shows that there are small differences in the orientation of Hrd3 relative to Hrd1. For example, classes #3 and #4 of the first half dataset (Extended Data Fig. 2) have a similar overall quality as class #6, but the relative orientation of Hrd3 with respect to Hrd1 is different. We therefore excluded classes #3 and #4 from refinement. Tests showed that including them actually decreased the quality of the map.
2) Hrd1/Hrd3 complex with one Hrd3 molecule. The 3D classes containing only one Hrd3 (class 2 in the first half and class 5 in the second half; 167,061 particles in total) were combined and refined, generating a reconstruction at 4.7 Å resolution.
3) Hrd3 alone. All 3D classes with their reconstructions showing clear densities for Hrd1 and at least one Hrd3 (classes 2, 3, 4, 6 in the first half and classes 5, 7 in the second half; 452,695 particles in total) were combined and refined, followed by Hrd3-focused 3D classification with signal subtraction 19. The resulting 3D classes displaying clear secondary structure features in Hrd3 were combined and refined with a soft mask on the Hrd3 molecule, leading to a density map at 3.9 Å resolution. Class #1 and #2 in the second half dataset were not included because the Hrd1 dimer density in these two classes was not as good as in the other classes, which would compromise signal subtraction and focused classification on Hrd3.
4) Hrd1 dimer. The same set of classes as for Hrd3 alone (classes 2, 3, 4, 6 in the first half and classes 5, 7 in the second half; 452,695 particles in total) were combined, and then subjected to 3D classification without a mask. C2 symmetry was applied in this round of classification and all following steps. Three classes showing clear densities of transmembrane helices were combined and classified based on the Hrd1 dimer, which was done using dynamic signal subtraction (DSS, detailed below). The best 3D class (93,609 particles) was further refined focusing on the Hrd1 dimer with DSS, generating a final reconstruction at 4.1 Å resolution.
Dynamic signal subtraction (DSS)
In the previously described method of masked classification with subtraction of residual signal 19, the unwanted signal is subtracted from each particle image based on a predetermined orientation. In this procedure, the orientation angles for signal subtraction are determined using the entire reconstruction as the reference model, and cannot be iteratively optimized based on the region of interest. In order to reduce the bias introduced by using a single fixed orientation for signal subtraction and to achieve better image alignment based on the region of interest, we have extended the signal subtraction algorithm to image alignment in the expectation step of GeRelion. Specifically, during each iteration, the reference model of the Hrd1/Hrd3 complex was subjected to two soft masks, one for Hrd1 and the other for Hrd3 and the amphipol region, generating a Hrd1 map and a non-Hrd1 map, respectively. For image alignment, these two maps generate 2D projections according to all searched orientations. For each search orientation, we subtracted from each original particle image the corresponding 2D projection of the non-Hrd1 map, and then compared it with the corresponding 2D projection of the Hrd1 map. Thus, particle images are dynamically subtracted for more accurate image alignment based on the Hrd1 portion. After alignment, 3D reconstructions were calculated using the original particle images without subtraction or masking.
For 3D classification focusing on the Hrd1 dimer, we obtained the best results by applying the DSS procedure during the local angle search (angular sampling interval: 1.8; local angular search range: 6). Only with DSS were we able to obtain a particle class that resulted in a reconstruction showing clear densities for the TM7/TM8 and TM5/TM6 loops of Hrd1. This class was first refined using the auto-refine procedure without mask or signal subtraction. When the auto-refine procedure reached the local angle search, the DSS procedure was applied to focus the refinement on the Hrd1 dimer region. 3D refinement with DSS improved the map quality, but did not change the nominal resolution.
Model building
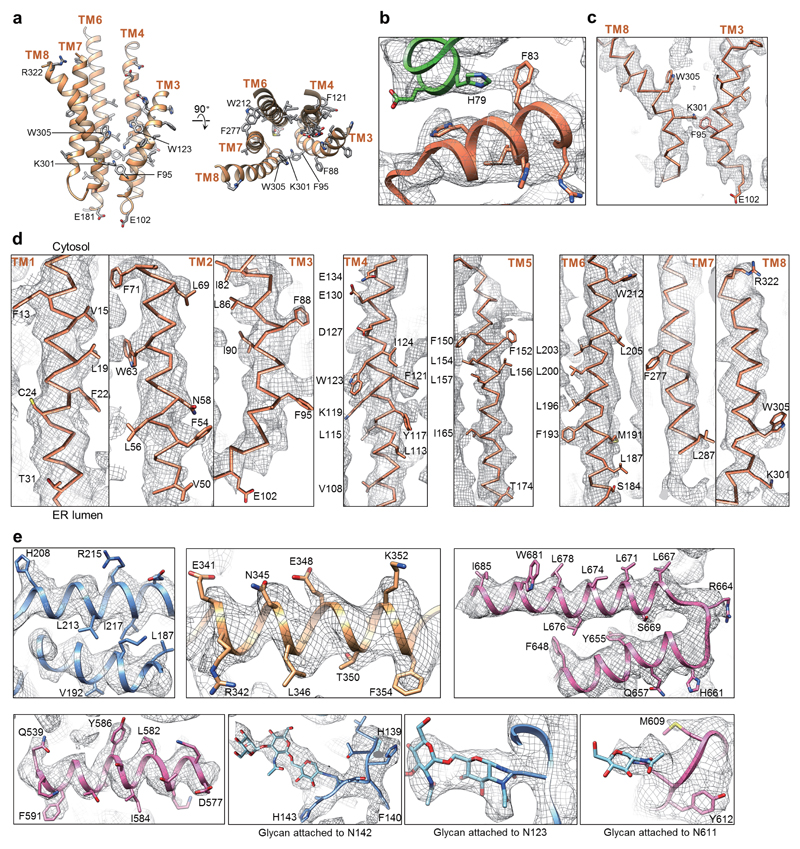
An initial model for Hrd1 was obtained by placing a poly-alanine chain into the density for the TM helices of Hrd1. TMs 1 and 2 could be identified on the basis of the loop between them being involved in the binding to Hrd3 23. The Hrd1 model was further extended manually, using information from TM predictions (Polyphobius, MEMSAT-SVM) and secondary structure predictions (Psipred server). Modeling was facilitated by distance constraints of evolutionarily coupled amino acid pairs (GREMLIN) (Extended Data Fig. 5) 39; these pairs are predicted to have co-evolved based on the analysis of a large dataset of aligned Hrd1 sequences from different species. For the co-evolution analysis by GREMLIN, the alignments were generated using HHblits (from HHsuite version 2.0.15; -n 8 -e 1E-20 -maxfilt ∞ -neffmax 20 -nodiff -realign_max ∞) 40 and run against the clustered UniProt database from 2016 and the fungal database from JGI 41 to generate a multiple sequence alignment. The alignment was then filtered for redundancy and coverage (HHfilter -cov 75 -id 90). In addition, TM helices were oriented in such a way that the exposure of polar residues to the hydrophobic environment of the lipid bilayer was minimized. The identity and registry of the TM helices of Hrd1 were verified on the basis of large amino acid side chains and density for the loops between TMs (Extended Data Fig. 4a, b). The loop between TMs 6 and 7 (residues 222-263) is predicted to be disordered (PSIPRED3v.3) and is invisible in our maps. No density that would fit the RING finger domain of Hrd1 was visible. Overall, a Hrd1 model consisting of residues 5-222 and residues 263-322 was built into the density.
The new topology of Hrd1 is consistent with sequence alignments performed with Hrd1 molecules from many different species, and with the prediction of TMs on the basis of hydrophobicity using a variety of prediction programs (TOPCONS 42, MEMSAT-SVM). For Hrd1 of some species, TMs 3, 7, and 8 are not predicted, as they contain up to 8 polar residues, but it is likely that they all have the same topology. The final model of Hrd1 is a result of refinement into the density (weight on density correlation score term, elec_dens_fast=10) using Rosetta with two-fold symmetry imposed 43.
For Hrd3, we initially built 5-7 helical segments (based on PSIPRED secondary structure prediction) using the AbinitioRelax model building application of Rosetta guided by GREMLIN constraints (weight on distance constraint score term, atom_pair_constraint=3 with a sigmoid function type). These helical segments were then docked into the density map and energy minimized, followed by visual analysis. An initial 7-helix C-terminal segment (residues 536-663) matched a model generated with the PHYRE2 server, providing some confidence of the placement. After extending the initial segment by two helices based on a continuous path in the density, a second 7-helix segment (residues 80-224) was docked into a position that satisfied two predicted long-range GREMLIN contacts (F207&V502 and A218&F509). The overall topology was completed by docking two final overlapping segments into trimmed density: 5 helices from 430-513 and 7 helices from 319-459. The docked segments were then combined together and refined using RosettaCM in an iterative fashion (score term weights: elec_dens_fast=2, atom_pair_constraint=3) 21. After refinement in Rosetta, loop regions in Hrd3 were manually adjusted to better fit the density. The final Hrd3 map at 3.9 Å for Hrd3 allowed the building of a continuous model of Hrd3 with the exception of residues 269-318. Extra density close to N101, N123, N142 and N611 is consistent with predicted N-glycosylation at these sites. A recent crystal structure of a mammalian Hrd3 (Sel1) fragment (PDB code: 5B26) could not be fully docked into the density map, probably because its structure is distorted by artificial dimerization due to crystal packing 23. However, a single chain of this homodimeric Hrd3 structure can be docked into the middle domain of Hrd3 (rmsd of 3.6Å over 144 residues).
To evaluate the fit of the evolutionary coupling data to our models we computed Rc scores (# of contacts made)/(# of expected contact), as described in ref. 44. After additional refinement with density and GREMLIN constraints, the Rc values were 0.710 and 0.757 for Hrd1 and Hrd3, respectively, which is consistent with the values (> 0.7) for the given number of sequences and length.
Generation of Hrd1/gp78/TCR8 sequence alignments
A seed alignment of the transmembrane domain of 20 fungi Hrd1 sequences was used as input for the hmmsearch tool on the Hmmer web server 45. The search was restricted to the rp15 set of representative genomes. This search yielded not only Hrd1 homologs from all branches of the eukaryotic kingdom but also homologs of gp78 (also called AMFR), TRC8 (also called RNF139), and the closely related RNF145. Additional seed alignments of 10 TRC8 sequences from metazoans and 10 gp78 homologs from metazoan and plants were generated and used as inputs for hmmsearch. All hits were combined and aligned with MAFFT using L-INS-I settings 46. The alignments were visually inspected, and sequences with long gaps or insertions were manually removed. Selected sequences of this alignment representing phylogenetically diverse species are shown in Extended Data Fig. 6.
Code availability
GeRelion is an open source and free software, distributed under the GPLv2 licence. It is publicly available for download through https://github.com/gpu-pdl-nudt/GeRelion.
Data availability
The coordinates of the atomic models of the Hrd1 dimer and Hrd3 monomer were deposited in the Protein Data Bank with accession codes 5V6P and 5V7V, respectively. The corresponding cryo-EM maps were deposited in the Electron Microscopy Data Bank with accession codes EMD-8637 and EMD-8642, respectively. The cryo-EM maps of the Hrd1/Hrd3 complexes containing one or two Hrd3 molecules were deposited with accession codes EMD-8639 and EMD-8638, respectively. The raw cryo-EM data were deposited to EMPIAR (accession code EMPIAR-10099). An interactive session of models with co-evolution data can be found at http://gremlin.bakerlab.org/hrd.
Extended Data
Extended Data Figure 1. Purification and cryo-EM of the Hrd1/Hrd3 complex.
a, In the last purification step, the Hrd1/Hrd3 complex was subjected to gel filtration on a Superdex 200 10/300GL Increase column. Shown is the UV elution profile. b, SDS-PAGE gel of the peak fraction, stained with Coomassie blue. For gel source data, see Supplementary Fig. 1. c, Representative cryo-EM image with a few particles marked by circles. A total of 5,361 images were collected. d, 2D class averages of cryo-EM particles.
Extended Data Figure 2. 3D classification and refinement procedure for the Hrd1/Hrd3 complex.
Views parallel to the membrane of 3D reconstructions are shown, and percentages of the particles in each class indicated. Three different classes selected from the first round of 3D classification are encircled with dashed lines in different colors, and were used for further analysis, as indicated by correspondingly colored arrows. The four final maps are labeled A-D, and shown with the resolutions and particle numbers. Maps C and D were used for model building. To obtain the best 3D classification focusing on the Hrd1 dimer, we compared dynamic signal subtraction (DSS) and conventional signal subtraction. Only with DSS was particle class obtained that resulted in a reconstruction showing clear densities for the TM7/TM8 and TM5/TM6 loops of Hrd1.
Extended Data Figure 3. Single particle cryo-EM analysis of Hrd1/Hrd3 complexes.
a, Density maps were generated for the Hrd1/Hrd3 dimer, the Hrd1 dimer with one associated Hrd3 molecule, the Hrd1 dimer, and Hrd3 (see Extended Data Fig. 2). The left panels show the maps in a side view, colored according to local resolution, the middle panels show the gold-standard Fourier shell correlation (FSC) curve (blue) with indicated resolution at FSC = 0.143, and the right panels show the Euler angle distribution in two different views. In the two lower panels, the dashed grey FCS curves were calculated between the atomic model and the corresponding final cryo-EM map. b, The density map for the Hrd1/Hrd3 dimer was filtered to a resolution of 6.8Å without amplitude modification, and is displayed at two different isosurface levels. At a low level (left panel), the weak amphipol density is visible and encloses the density of Hrd1 dimer. The amphipathic helix of Hrd3 only associates with the outer surface of amphipol density. At a high isosurface level (middle and right panels), the density for the amphipathic helix is clearly connected with that of the preceding Sel1 domains and well separated from that of TM1 and TM2 of the nearby Hrd1 molecule. The region in the dashed black box (middle panel) is displayed as a sectional view in the right panel.
Extended Data Figure 4. Examples of the fit of the model and density maps.
a, Amino acids for which side chain density was observed are indicated in side and top views of the Hrd1 model. b, Central interface between the Hrd1 molecules. H79 and F83 from the two Hrd1 molecules (orange and green) probably form cation-pi interactions. c, TMs 3 and 8 of Hrd1. d, Density for the TMs of Hrd1. Amino acids with clear side chain density are indicated. e, Selected areas in Hrd3: N-terminal (blue), central (yellow) and C-terminal domain (purple).
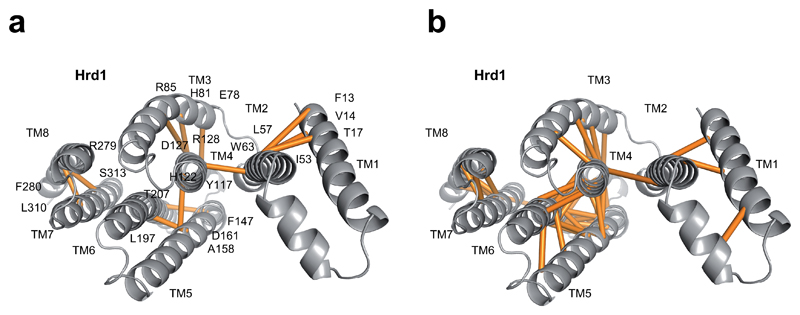
Extended Data Figure 5. Distance constraints between amino acid residues in Hrd1.
a, Evolutionary couplings between amino acids, determined with the program Gremlin 39. Shown is a view from the ER lumen with couplings shown as lines between residues. b, Distance constraints calculated with the program RaptorX-Contact 47,48.
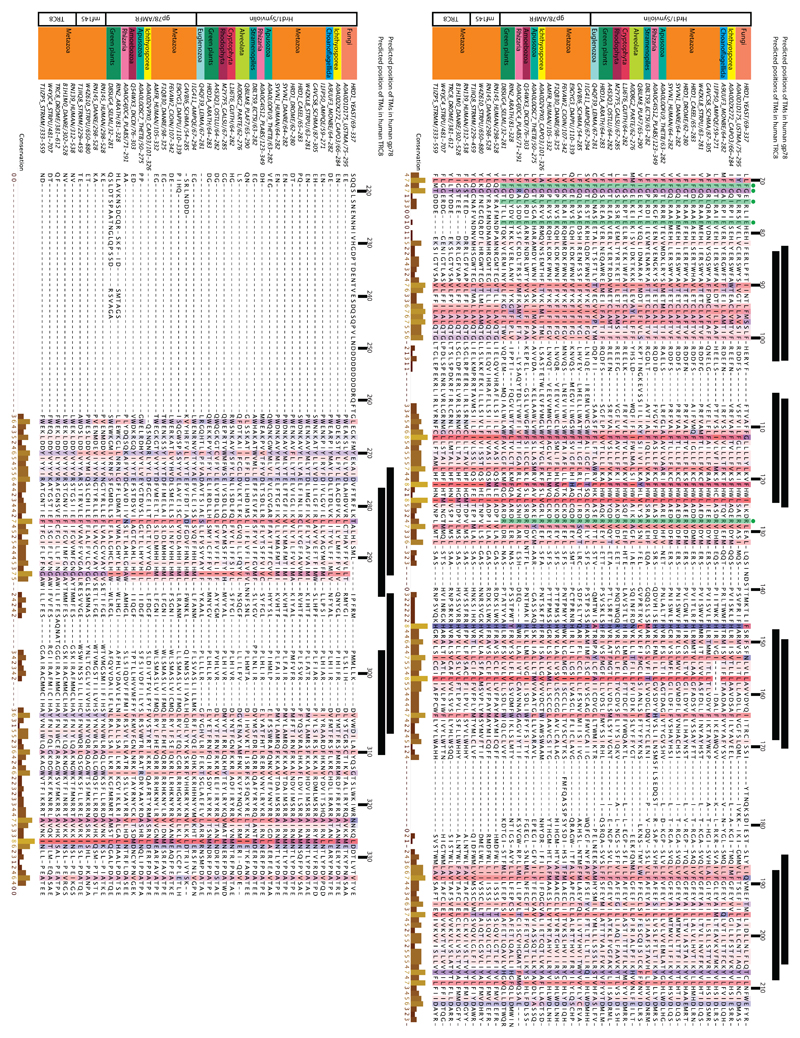
Extended Data Figure 6. Sequence similarities between Hrd1 and other multi-spanning ubiquitin ligases.
Multiple sequence alignment showing amino acid conservation in TMs 3-8 of Hrd1, TMs 3-8 of gp78 (also called AMFR), and TMs 9-14 of TRC8 (also called RNF139) and RNF145. On the left, Uniprot codes for individual sequences are given. Numbers after Uniprot codes indicate the depicted amino acid range. Black bars above the sequences indicate the location of the most C-terminal six transmembrane segments of human gp78 (top), and human TRC8 (bottom) as predicted by TOPCONS. Below that, amino acid numbering for Hrd1p from S. cerevisiae is given. Coloring was edited in JalView according to conservation of hydrophobicity 49. Residues highlighted in green and with green dots are conserved among Hrd1 and gp78 molecules and are involved in the interaction of TMs 2,3, and 4 on the cytosolic side of the membrane (Extended Data Fig. 7c). Species abbreviations in Uniprot codes: YEAST S. cerevisiae, USTMA Ustilago maydis, CAPO3 Capsaspora owczarzaki, MONBE Monosiga brevicollis, AMPQE Amphimedon queenslandica, SCHMA Schistosoma mansoni, STRPU Strongylocentrotus purpuratus, CAEEL Caenorhabditis elegans, DROME Drosophila melanogaster, DANRE Danio rerio, THETB Thecamonas trahens, PLABS Plasmodiophora brassicae, ECTSI Ectocarpus siliculosus, PLAF7 Plasmodium falciparum, PARTE Paramecium tetraurelia, GUITH Guillardia theta, GALSU Galdieria sulphuraria, OSTLU Ostreococcus lucimarinus, ARATH Arabidopsis thaliana, LEIMA Leishmania major, DICDI Dictyostelium discoideum, DAPPU Daphnia pulex, CIOIN Ciona intestinalis, SELML Selaginella moellendorffii, STRMM Strigamia maritima.
Extended Data Figure 7. Structural homology between the ubiquitin ligases Hrd1 and TRC8.
a, Domain organization of human gp78 and TRC8. Black bars indicate the position of TM segments as predicted by TOPCONS. The positions of the VIM (VCP interacting motif), CUE, and RING finger domains were taken from Uniprot entries Q9UKV5 and Q8WU17. The sterol-sensing domain (blue bar) in TRC8 is positioned according to ref.50. Regions with homology to Hrd1 are indicated by red bars. b, Structural model of human TRC 8 (residues 323-516) as generated by RaptorX-Contact (44,45). The structure of S. cerevisiae Hrd1 is shown in brown, the model for TRC in dark blue (residues 345-516). TM9 of TRC8 does not align well with TM3 of Hrd1 and is therefore shown in light blue (residues 323-344). Shown are views from the cytosol (left panel), and views parallel to the membrane either towards the hydrophilic cavity or from the backside (middle and right panels, respectively). The structure of Hrd1 from S. cerevisiae was aligned with the TRC8 model obtained from RaptorX-Contact (44,45) using the command cealign within Pymol 51. c, Views of Hrd1 from the cytosol (left) and the side (right), with residues conserved in the ubiquitin ligases Hrd1, gp78, TRC8, and RNF145 shown in orange, and residues conserved in Hrd1 and gp78 in purple.
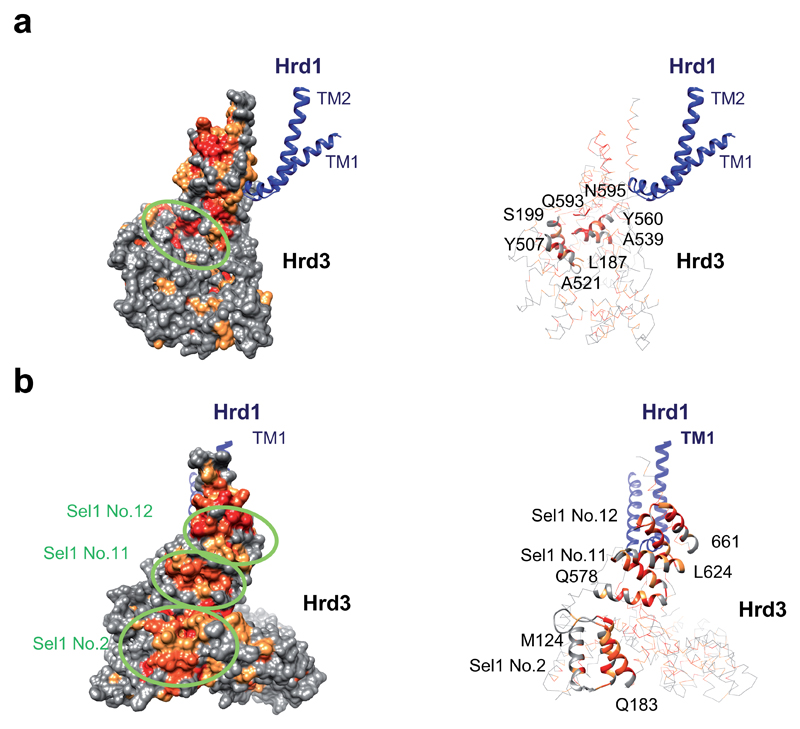
Extended Data Figure 8. Potential Hrd3 binding surfaces for substrate and partners.
a, Groove at the inner surface of Hrd3 (encircled), located at the junction of the N-terminal (residues 187-199), middle (residues 507-521), and C-terminal (residues 539-560, 593-595) domains. The left panel shows a space-filling model with conserved amino acids shown in orange to red (scores 7-9 in the program Consurf). Hrd1 is shown in ribbon presentation. The right panel shows the same view with Hrd3 in cartoon presentation. Helices containing the conserved residues are shown as ribbons. Some residues are labeled for orientation. b, The left panel shows three conserved surface grooves on the outer surface of Hrd3 (encircled), with conserved residues colored as in a. The right panel shows the same view with helices containing conserved residues in ribbon presentation.
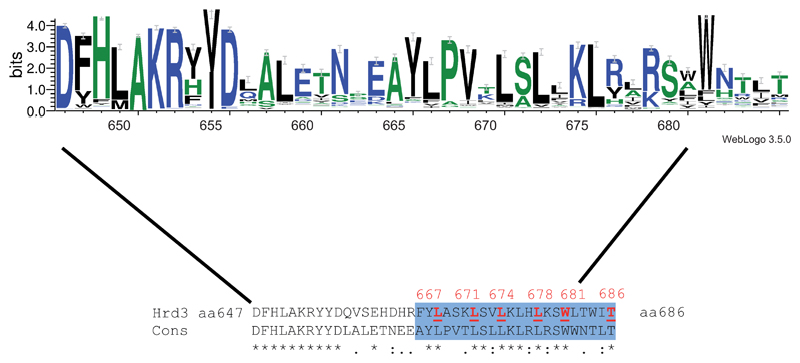
Extended Data Figure 9. Sequence conservation of the amphipathic helix of Hrd3.
Consensus sequence of the amphipathic helix of Hrd3, generated with Weblogo 3.0. The size of the letters correlates with their conservation. Black, blue, and green letters indicate hydrophobic, hydrophilic, and other residues, respectively. The lower panel shows an alignment of the amphipathic helix of S. cerevisiae Hrd3 with the consensus sequence (Cons). The blue area indicates the amphipathic helix, with residues in red forming its hydrophobic face.
Extended Data Table 1. Statistics of the cryo-EM structures presented in this study.
| Cryo-EM data collection and processing | Hrd13 dimer | Hrd1 dimer one Hrd3 | Hrd1 dimer | Hrd3 |
|---|---|---|---|---|
| Voltage (kV) | 300 | |||
| Electron dose (e-/Å2) | 82 | |||
| Number of collected movies | 5,361 | |||
| Particle defocus range (average) (μm) | 0.7-3.2 | |||
| Number of particles for 3D classification | 871,530 | |||
| Number of particles for final map | 139,754 | 167,061 | 93,609 | 204,578 |
| Symmetry for final map | C1 | C1 | C2 | C1 |
| Resolution (Å) | 4.7 | 4.7 | 4.1 | 3.9 |
| Map sharpening B-factor (Å2) | -250 | -250 | -230 | -290 |
| Atomic model refinement | ||||
| Number of protein residues | 542 | 617 | ||
| Number of atoms | 9228 | 10142 | ||
| Geometric parameters (r.m.s.d.) | ||||
| Bond length (Å) | 0.014 | 0.022 | ||
| Bond angle (º) | 1.334 | 1.636 | ||
| Ramachandran statistics | ||||
| Residues in favoured regions (%) | 96.99 | 86.09 | ||
| Residues in allowed regions (%) | 2.63 | 13.75 | ||
| Residues in disallowed regions (%) | 0.38 | 0.16 | ||
| Rotamer outliers (%) | 0 | |||
| MolProbity validation | ||||
| Overall score | 0.94 | 2.21 | ||
| Clash score | 0.87 | 11.34 | ||
Supplementary Material
Acknowledgements
We thank Z. Yu, R. Huang and C. Hong at the HHMI Janelia Cryo-EM Facility for help in microscope operation and data collection, Z. Li for technical support at our in-house EM facility, and S. Andrei Anghel, X. Wu, P. Carvalho and T. Walz for comments on the manuscript. This work was supported by the European Research Council (ERC) under the Horizon 2020 research and innovation program (grant # 677770 to A.S.), by the National Key Research and Development Program of China (grant # 2016YFB1000101 to D.L.) and by NIGMS Award R01GM052586 to T.A.R. T.A.R. is a Howard Hughes Medical Institute Investigator.
Footnotes
Author contributions
S.S. prepared protein and built the models, W.M. and M.L. collected and analyzed EM data, A.S. designed the construct and performed sequence alignments, S.O. and R.P. and their advisors F.D. and D.B. built models based on evolutionary couplings and energy minimization, M.G.C. helped with EM data collection, H.S. and D.L. developed DSS in GeRelion, T.A.R. and M.L. supervised the project. T.A.R. wrote the manuscript.
The authors declare no competing financial interest.
References
- 1.Bays NW, Wilhovsky SK, Goradia A, Hodgkiss-Harlow K, Hampton RY. HRD4/NPL4 Is Required for the Proteasomal Processing of Ubiquitinated ER Proteins. Molecular biology of the cell. 2001;12:4114–4128. doi: 10.1091/mbc.12.12.4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ye Y, Meyer HH, Rapoport TA. The AAA ATPase Cdc48/p97 and its partners transport proteins from the ER into the cytosol. Nature. 2001;414:652–656. doi: 10.1038/414652a. [DOI] [PubMed] [Google Scholar]
- 3.Jarosch E, et al. Protein dislocation from the ER requires polyubiquitination and the AAA-ATPase Cdc48. Nature cell biology. 2002;4:134–139. doi: 10.1038/ncb746. [DOI] [PubMed] [Google Scholar]
- 4.Rabinovich E, Kerem A, Frohlich KU, Diamant N, Bar-Nun S. AAA-ATPase p97/Cdc48p, a Cytosolic Chaperone Required for Endoplasmic Reticulum-Associated Protein Degradation. Molecular and cellular biology. 2002;22:626–34. doi: 10.1128/MCB.22.2.626-634.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vashist S, Ng DTW. Misfolded proteins are sorted by a sequential checkpoint mechanism of ER quality control. The Journal of cell biology. 2004;165:41–52. doi: 10.1083/jcb.200309132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carvalho P, Goder V, Rapoport TA. Distinct ubiquitin-ligase complexes define convergent pathways for the degradation of ER proteins. Cell. 2006;126:361–373. doi: 10.1016/j.cell.2006.05.043. [DOI] [PubMed] [Google Scholar]
- 7.Carvalho P, Stanley AM, Rapoport TA. Retrotranslocation of a misfolded luminal ER protein by the ubiquitin-ligase Hrd1p. Cell. 2010;143:579–591. doi: 10.1016/j.cell.2010.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baldridge RD, Rapoport TA. Autoubiquitination of the Hrd1 Ligase Triggers Protein Retrotranslocation in ERAD. Cell. 2016;166:394–407. doi: 10.1016/j.cell.2016.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Park E, Rapoport TA. Mechanisms of Sec61/SecY-mediated protein translocation across membranes. Annual review of biophysics. 2012;41:21–40. doi: 10.1146/annurev-biophys-050511-102312. [DOI] [PubMed] [Google Scholar]
- 10.Kumazaki K, et al. Structural basis of Sec-independent membrane protein insertion by YidC. Nature. 2014;509:516–520. doi: 10.1038/nature13167. [DOI] [PubMed] [Google Scholar]
- 11.Dalbey RE, Kuhn A. Membrane Insertases Are Present in All Three Domains of Life. Structure. 2015;23:1559–1560. doi: 10.1016/j.str.2015.08.002. [DOI] [PubMed] [Google Scholar]
- 12.Bordallo J, Plemper RK, Finger A, Wolf DH. Der3p/Hrd1p is required for endoplasmic reticulum-associated degradation of misfolded lumenal and integral membrane proteins. Molecular biology of the cell. 1998;9:209–222. doi: 10.1091/mbc.9.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bays NW, Gardner RG, Seelig LP, Joazeiro CA, Hampton RY. Hrd1p/Der3p is a membrane-anchored ubiquitin ligase required for ER-associated degradation. Nature cell biology. 2001;3:24–9. doi: 10.1038/35050524. [DOI] [PubMed] [Google Scholar]
- 14.Horn SC, et al. Usa1 functions as a scaffold of the HRD-ubiquitin ligase. Molecular cell. 2009;36:782–793. doi: 10.1016/j.molcel.2009.10.015. [DOI] [PubMed] [Google Scholar]
- 15.Gardner RG, et al. Endoplasmic reticulum degradation requires lumen to cytosol signaling. Transmembrane control of Hrd1p by Hrd3p. The Journal of cell biology. 2000;151:69–82. doi: 10.1083/jcb.151.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Denic V, Quan EM, Weissman JS. A luminal surveillance complex that selects misfolded glycoproteins for ER-associated degradation. Cell. 2006;126:349–359. doi: 10.1016/j.cell.2006.05.045. [DOI] [PubMed] [Google Scholar]
- 17.Gauss R, Jarosch E, Sommer T, Hirsch C. A complex of Yos9p and the HRD ligase integrates endoplasmic reticulum quality control into the degradation machinery. Nature cell biology. 2006;8:849–854. doi: 10.1038/ncb1445. [DOI] [PubMed] [Google Scholar]
- 18.Stein A, Ruggiano A, Carvalho P, Rapoport TA. Key steps in ERAD of luminal ER proteins reconstituted with purified components. Cell. 2014;158:1375–1388. doi: 10.1016/j.cell.2014.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bai X-C, Rajendra E, Yang G, Shi Y, Scheres SHW. Sampling the conformational space of the catalytic subunit of human γ-secretase. eLife. 2015;4:1485. doi: 10.7554/eLife.11182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ovchinnikov S, et al. Protein structure determination using metagenome sequence data. Science. 2017;355:294–298. doi: 10.1126/science.aah4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Song Y, et al. High-resolution comparative modeling with RosettaCM. Structure. 2013;21:1735–1742. doi: 10.1016/j.str.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deak PM, Wolf DH. Membrane topology and function of Der3/Hrd1p as a ubiquitin-protein ligase (E3) involved in endoplasmic reticulum degradation. The Journal of biological chemistry. 2001;276:10663–10669. doi: 10.1074/jbc.M008608200. [DOI] [PubMed] [Google Scholar]
- 23.Jeong H, et al. Crystal structure of SEL1L: Insight into the roles of SLR motifs in ERAD pathway. Sci Rep. 2016;6:20261. doi: 10.1038/srep20261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mehnert M, Sommer T, Jarosch E. Der1 promotes movement of misfolded proteins through the endoplasmic reticulum membrane. Nature cell biology. 2014;16:77–86. doi: 10.1038/ncb2882. [DOI] [PubMed] [Google Scholar]
- 25.Sato BK, Schulz D, Do PH, Hampton RY. Misfolded membrane proteins are specifically recognized by the transmembrane domain of the Hrd1p ubiquitin ligase. Molecular cell. 2009;34:212–222. doi: 10.1016/j.molcel.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gonen T, Walz T. The structure of aquaporins. Quarterly Reviews of Biophysics. 2006;39:361–396. doi: 10.1017/S0033583506004458. [DOI] [PubMed] [Google Scholar]
- 27.Dutzler R, Campbell EB, Cadene M, Chait BT, MacKinnon R. X-ray structure of a ClC chloride channel at 3.0 A reveals the molecular basis of anion selectivity. Nature. 2002;415:287–294. doi: 10.1038/415287a. [DOI] [PubMed] [Google Scholar]
- 28.McCoy JG, Levin EJ, Zhou M. Structural insight into the PTS sugar transporter EIIC. Biochimica et biophysica acta. 2015;1850:577–585. doi: 10.1016/j.bbagen.2014.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van den Berg B, et al. X-ray structure of a protein-conducting channel. Nature. 2004;427:36–44. doi: 10.1038/nature02218. [DOI] [PubMed] [Google Scholar]
- 30.Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. Journal of structural biology. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 31.Zheng S, Palovcak E, Armache J-P, Cheng Y, Agard D. Anisotropic Correction of Beam-induced Motion for Improved Single-particle Electron Cryo-microscopy. 2016 doi: 10.1101/061960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grant T, Grigorieff N. Measuring the optimal exposure for single particle cryo-EM using a 2.6 Å reconstruction of rotavirus VP6. eLife. 2015;4:e06980. doi: 10.7554/eLife.06980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. Journal of structural biology. 2003;142:334–347. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- 34.Ru H, et al. Molecular Mechanism of V(D)J Recombination from Synaptic RAG1-RAG2 Complex Structures. Cell. 2015;163:1138–1152. doi: 10.1016/j.cell.2015.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scheres SHW. RELION: implementation of a Bayesian approach to cryo-EM structure determination. Journal of structural biology. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Su H, et al. GeRelion: GPU-enhanced parallel implementation of single particle cryo-EM image processing. bioRxiv. 2016:075887. doi: 10.1101/075887. [DOI] [Google Scholar]
- 37.Kucukelbir A, Sigworth FJ, Tagare HD. Quantifying the local resolution of cryo-EM density maps. Nat Methods. 2014;11:63–65. doi: 10.1038/nmeth.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lyumkis D, Brilot AF, Theobald DL, Grigorieff N. Likelihood-based classification of cryo-EM images using FREALIGN. Journal of structural biology. 2013;183:377–388. doi: 10.1016/j.jsb.2013.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ovchinnikov S, Kamisetty H, Baker D. Robust and accurate prediction of residue-residue interactions across protein interfaces using evolutionary information. eLife. 2014;3:e02030. doi: 10.7554/eLife.02030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Remmert M, Biegert A, Hauser A, Söding J. HHblits: lightning-fast iterative protein sequence searching by HMM-HMM alignment. Nat Methods. 2011;9:173–175. doi: 10.1038/nmeth.1818. [DOI] [PubMed] [Google Scholar]
- 41.Nordberg H, et al. The genome portal of the Department of Energy Joint Genome Institute: 2014 updates. Nucleic Acids Research. 2014;42:D26–31. doi: 10.1093/nar/gkt1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tsirigos KD, Peters C, Shu N, Käll L, Elofsson A. Nucleic Acids Research. 2015;43(Webserver issue):W401–W407. doi: 10.1093/nar/gkv485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.DiMaio F, Tyka MD, Baker ML, Chiu W, Baker D. Refinement of protein structures into low-resolution density maps using rosetta. Journal of molecular biology. 2009;392:181–190. doi: 10.1016/j.jmb.2009.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ovchinnikov S, et al. Large-scale determination of previously unsolved protein structures using evolutionary information. eLife. 2015;4:e09248. doi: 10.7554/eLife.09248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Finn RD, et al. HMMER web server: 2015 update. Nucleic Acids Research. 2015;43:W30–8. doi: 10.1093/nar/gkv397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7 improvements in performance and usability. Molecular biology and evolution. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang S, Sun S, Li Z, Zhang R, Xu J. Accurate De Novo Prediction of Protein Contact Map by Ultra-Deep Learning Model. PLoS Comput Biol. 2017;13:e1005324. doi: 10.1371/journal.pcbi.1005324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang S, Li W, Zhang R, Liu S, Xu J. CoinFold: a web server for protein contact prediction and contact-assisted protein folding. Nucleic Acids Research. 2016;44:W361–6. doi: 10.1093/nar/gkw307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gemmill RM, et al. The hereditary renal cell carcinoma 3;8 translocation fuses FHIT to a patched-related gene, TRC8. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:9572–9577. doi: 10.1073/pnas.95.16.9572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.The PyMOL Molecular Graphics System. Version 1.8. Schrödinger, LLC; [Accessed: 15 March 2017]. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The coordinates of the atomic models of the Hrd1 dimer and Hrd3 monomer were deposited in the Protein Data Bank with accession codes 5V6P and 5V7V, respectively. The corresponding cryo-EM maps were deposited in the Electron Microscopy Data Bank with accession codes EMD-8637 and EMD-8642, respectively. The cryo-EM maps of the Hrd1/Hrd3 complexes containing one or two Hrd3 molecules were deposited with accession codes EMD-8639 and EMD-8638, respectively. The raw cryo-EM data were deposited to EMPIAR (accession code EMPIAR-10099). An interactive session of models with co-evolution data can be found at http://gremlin.bakerlab.org/hrd.