Abstract
Genetically identical individuals growing in the same environment often show substantial phenotypic variation within populations of organisms as diverse as bacteria1, nematodes2, rodents3 and humans4. With some exceptions5, the causes are poorly understood. We show here that isogenic Caenorhabditis elegans nematodes vary in their size at hatching, speed of development, growth rate, starvation resistance, fecundity, and also in the rate of development of their germline relative to that of somatic tissues. Surprisingly, we show that the primary cause of this variation is the age of an individual’s mother, with young mothers producing progeny impaired for many traits. We identify age-dependent changes in maternal provisioning of a lipoprotein complex (vitellogenin) to embryos as the molecular mechanism underlying variation in multiple traits throughout the life of an animal. The production of sub-optimal progeny by young mothers likely reflects a trade-off between the competing fitness traits of a short generation time and progeny survival and fecundity.
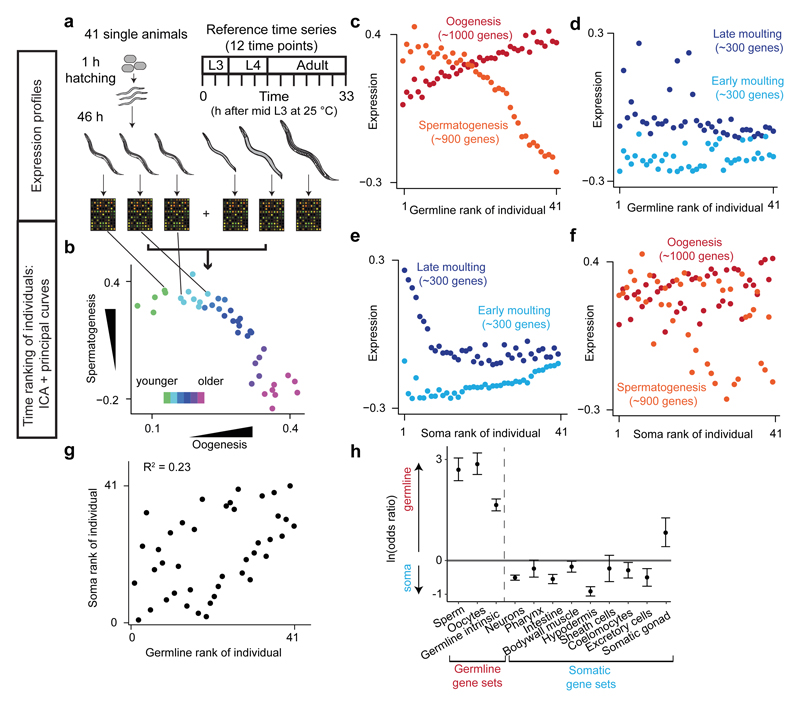
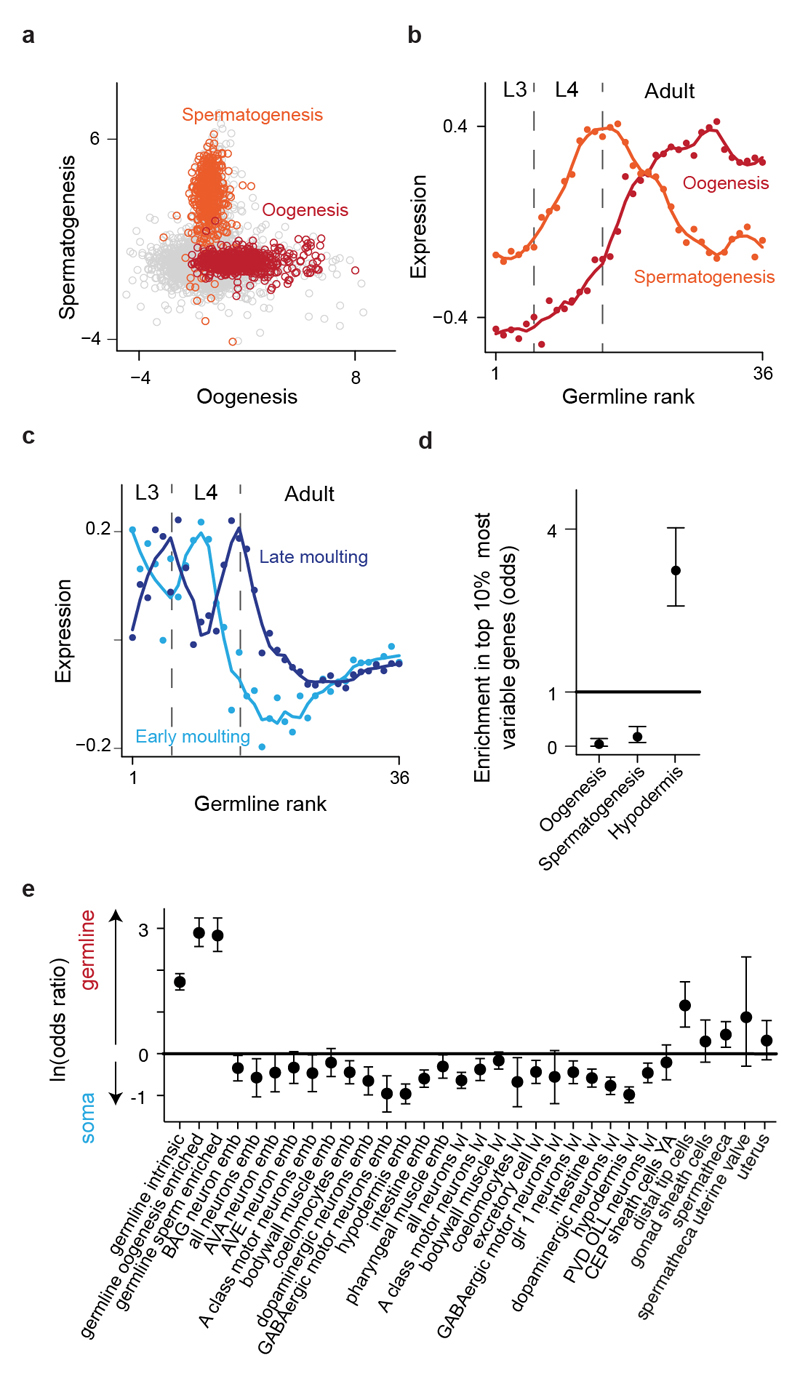
To characterize molecular differences between isogenic individuals, we profiled genome-wide gene expression in single young adult C. elegans worms synchronized at hatching (Fig. 1a). Employing our previously developed strategy6, we used spermatogenesis- and oogenesis- specific expression signatures (Extended Data Fig. 1, a and b) to accurately stage germline development in each animal (Fig. 1b; Supplementary Table 1). However, germline expression only poorly predicted time-dependent expression in the hypodermis (Fig. 1c, d; Extended Data Fig. 1c, d) and vice versa (Fig. 1e, f, g). Expression in other tissues was better predicted by that in the hypodermis than the germline, with the exception of the somatic gonad (Fig. 1h; Extended Data Fig. 1e). Thus soma-germline heterochrony arises not only due to genetic mutation7 and adaptive evolution8 but also varies among individuals in a wildtype population.
Fig 1. Gene expression profiling reveals inter-individual variation in soma-germline phasing.
(a, b) Reference time series and principal curves rank individual worms from younger to older by germline-specific expression.
(c, d, e, f) Individual germline (c) and hypodermal (d) expression ordered by germline expression and vice versa (e, f).
(g) Correlation of individual germline or hypodermal ranks.
(h) Enrichment for genes better explained by germline or hypodermal rank in tissue-specific gene sets. Error bars, 95 % C.I.
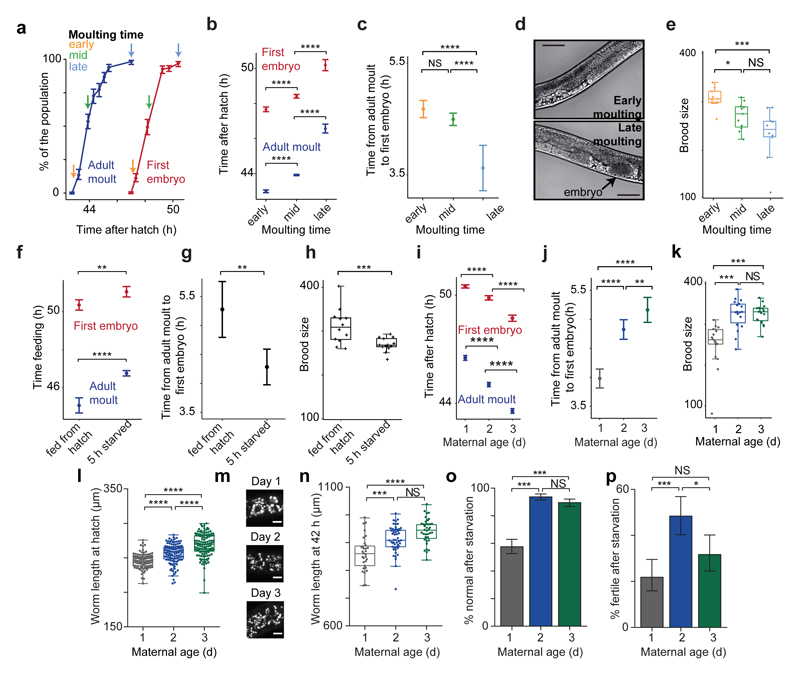
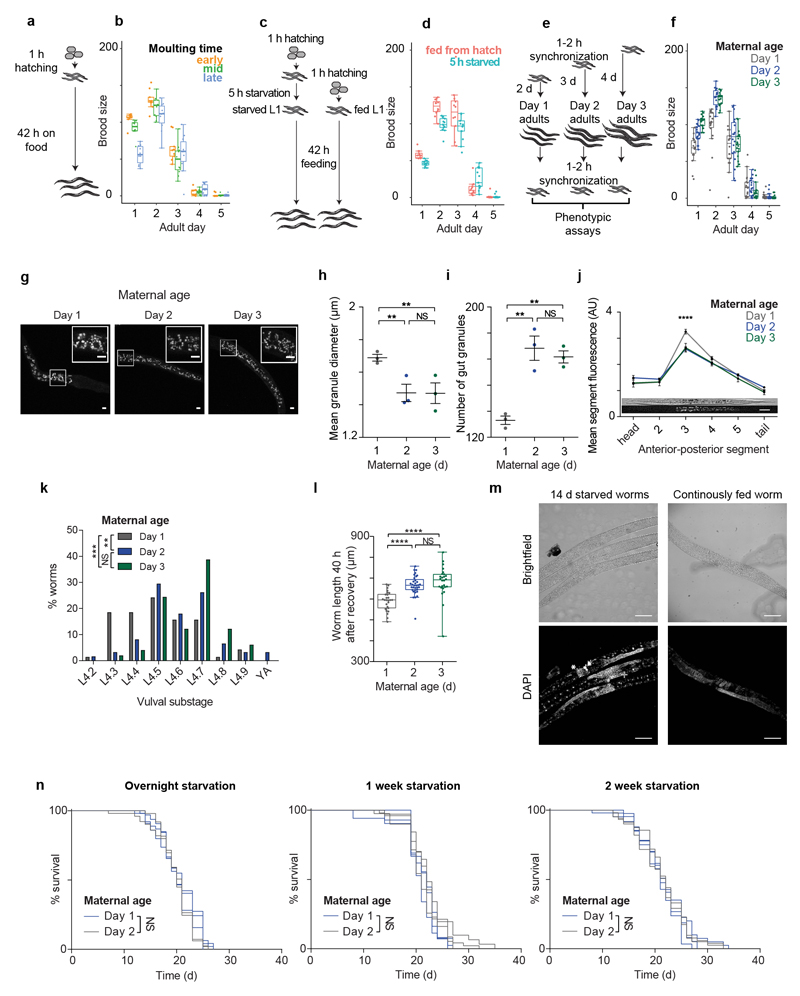
Isogenic animals synchronized at hatching from a proliferating population (Extended Data Fig. 2a) vary in their developmental speed (Fig. 2a, b). Visually following somatic and germline development showed that slower-developing animals have a germline maturation that, although delayed in absolute time, is advanced relative to somatic development (Fig. 2b, c, d). Slower-developing animals also have a lower brood size (Fig. 2e; Extended Data Fig. 2b).
Fig 2. Slow development, early starvation and young mothers are associated with relative germline acceleration and lower brood size, with early progeny also short and sensitive to starvation.
(a) Variation in timing of L4/adult moult and first embryo. Arrows, worm collection.
(b) Time (T50) by which 50% of worms moulted or contained an embryo.
(c) Time between T50s for (b).
(d) 4 h after moulting, only late-moulting worms have embryos (arrow). Scale, 50 μm.
(e) Brood size (n = 9, 11, 10 worms).
(f) Moulting/embryo T50 for L1-starved worms.
(g) Time between T50s for (f).
(h) Brood size (n = 13, 13 worms).
(i) Moulting/embryo T50 for day 1-3 progeny.
(j) Time between T50s for (i).
(k) Brood size (n = 18, 18, 19 worms).
(l) Hatching length (n = 95, 109, 124 larvae).
(m) Gut granules of L1s (see Extended Data Fig. 2g). Scale, 5 μm.
(n) Length after 42 h (n = 31, 50, 37 larvae).
(o) Normal phenotypes after 10 d L1 starvation (n = 349, 462, 516 worms).
(p) Fertility after 14 d L1 starvation (n = 141, 138, 135 worms).
Error bars, 95 % C.I., Tukey boxplots. Kruskal-Wallis (e, k, l, n), Mann-Whitney (h), ratio tests (b c, f, g, i, j), Fisher’s exact tests (o, p). Data represent typical outcomes, replicated 3 times independently. NS, not significant, * P < 0.05, ** P < 0.01, *** P < 0.001, **** P < 0.0001 for all figures.
We noticed that worms hatching on plates with an exhausted food supply had altered soma-germline synchrony when development was re-initiated. Indeed, starving newly hatched worms for as little as 5 h (Extended Data Fig. 2c) delayed adult moulting more than the time to the appearance of the first embryo (Fig. 2f, g). A brief starvation at the L1 stage also caused a reduced brood size (Fig. 2h) and an altered temporal distribution of progeny production (Extended Data Fig. 2d). Thus, a transient early life environmental perturbation is sufficient to alter multiple phenotypes throughout an individual’s life.
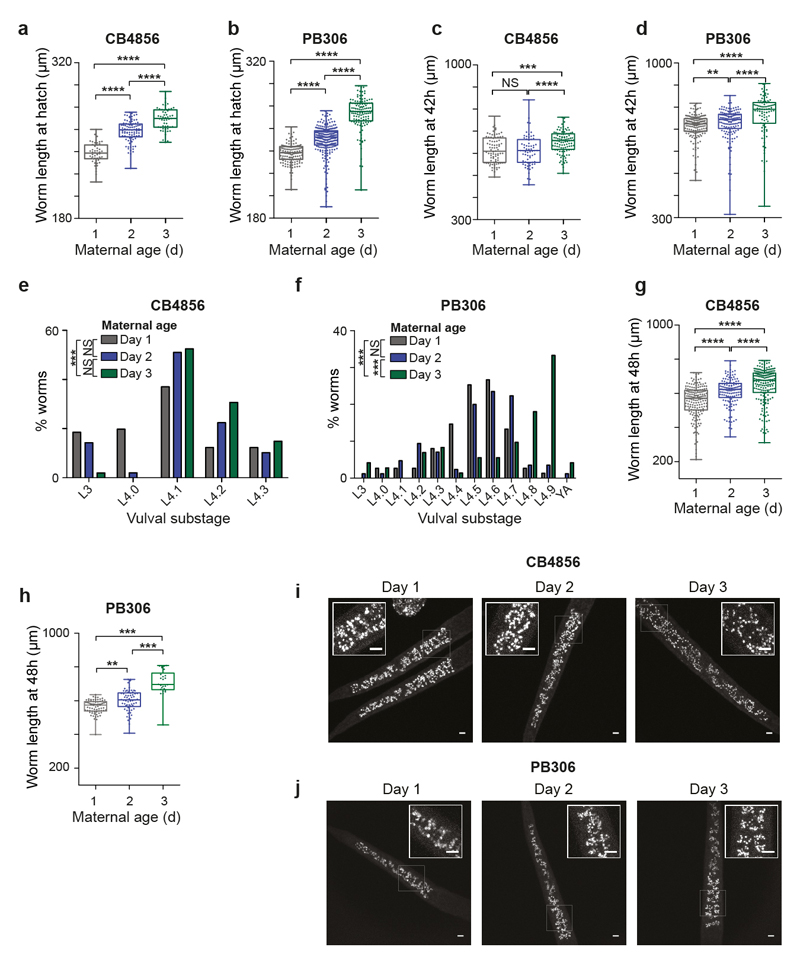
The single worms used for expression profiling and the early- and late-molting worms examined above came from well-fed populations, so early starvation is unlikely to explain the variation in germline-soma phasing. At 20 °C, self-fertilized hermaphrodites produce around 300 progeny over a 4 day period. As parental age has been suggested to influence progeny phenotypes in C. elegans and other organisms9,10,11, we tested whether progeny differ depending upon the day of adult life on which they were produced (Extended Data Fig. 2e). Surprisingly, we found that the progeny of mothers in the first day of adulthood reached adulthood later and had altered soma-germline phasing (Fig. 2i, j) and a lower brood size (Fig. 2k; Extended Data Fig. 2f), phenocopying the effects of early transient starvation. Further, early progeny were shorter at hatching (Fig. 2l), had darker intestines due to larger (though fewer) ‘gut granules’12,13 (Fig. 2m; Extended Data Fig. 2g, h, i, j) and grew (Fig. 2n) and developed (Extended Data Fig. 2k) slower. Moreover, after recovery from extended L1 starvation, day 1 progeny grew slower (Extended Data Fig. 2l) and exhibited more phenotypic abnormalities (Fig. 2o) and sterility (Fig. 2p; Extended Data Fig. 2m). Lifespan was unaffected by maternal age or extended starvation (Extended Data Fig. 2n), in keeping with previous work14,15. We found similar effects of maternal age in two wild isolates of C. elegans, CB4856 and PB306 (Extended Data Fig. 3), indicating that our findings are not restricted to the domesticated16 laboratory strain N2.
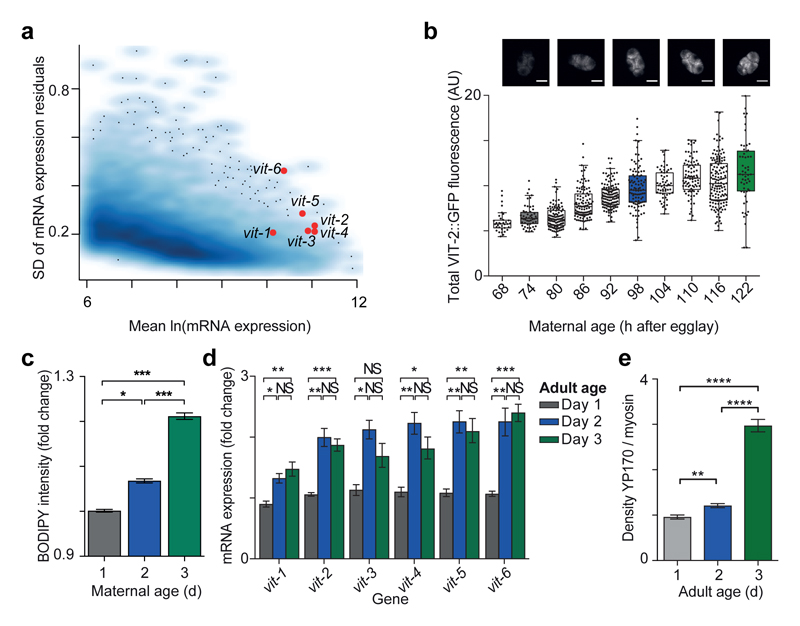
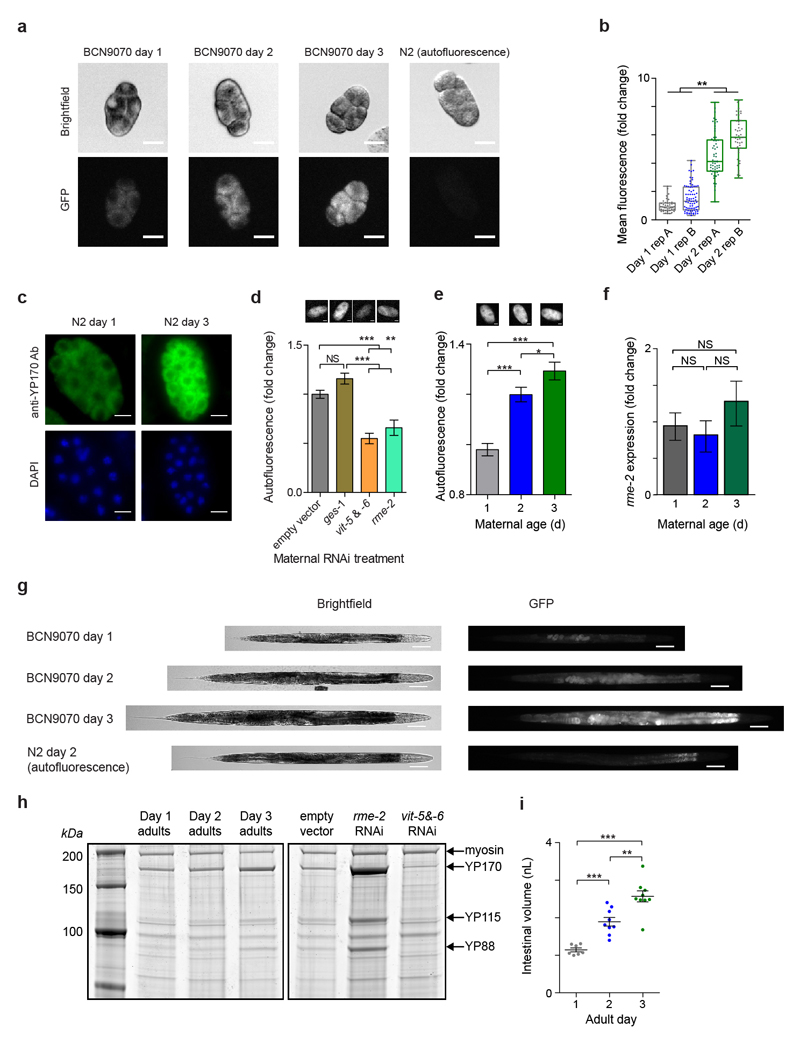
A substantial fraction of progeny (around 10 %) are produced before the end of our day 1 window and so exhibit phenotypic impairments as extensive as those reported above. What could underlie the impairment of so many traits in the progeny of young mothers? One possibility is that early progeny are less well provisioned by young mothers. Supporting this hypothesis, some of the largest changes in gene expression in the transition to adulthood are in the six vitellogenin genes encoding yolk proteins, reflecting their activation during this period17. We reasoned that this trend could continue throughout the reproductive period, leading to greater yolk provisioning to embryos of older mothers. Vitellogenin in C. elegans is synthesized in the intestine and transported into maturing oocytes by endocytosis18. Yolk loading into embryos, whilst not essential for development18, has been implicated in L1 starvation survival19,20. Vitellogenin expression is also extremely variable among single worms, even after correcting for differences in germline and somatic development as appropriate (Fig. 3a).
Fig 3. Vitellogenin expression and provisioning is variable between individuals and increases with age.
(a) SD of residual gene expression vs expression from single-worm microarrays.
(b) Embryo VIT-2::GFP (n = 34, 51, 108, 108, 125, 97, 57, 85, 146, 56 embryos). Representative images scale, 20 μm.
(c) Lipid staining of day 1-3 embryos. (10 biological replicates, 2 trials, n = 1924, 1320, 902 embryos).
(d) vit mRNA (qPCR) in day 1-3 N2 adults (3 biological replicates).
(e) Densitometry of YP170 (6 biological replicates).
Error bars, s.e.m. GLMM analysis (c), one-way ANOVA (d, e). AU, arbitrary units.
Expressing GFP-tagged VIT-2 from the endogenous vit-2 locus revealed that the level of VIT-2 in embryos strongly increased with maternal age across the reproductive period of self-fertilized hermaphrodites (Fig. 3b; Extended Data Fig. 4a). Consistently, YP170, the protein encoded by vit-1 to vit-5, showed a sharp increase in immunostaining intensity from day 1 to day 3 embryos (Extended Data Fig 4b, c), as did green autofluorescence, with maternal RNAi treatments that reduced yolk loading strongly reducing this autofluorescence (Extended Data Fig 4d, e). As vitellogenins transport triglycerides and other lipids into oocytes21, an increase in yolk loading might increase embryonic lipid content. Indeed, we found a progressive increase in lipid content with maternal age in wildtype embryos (Fig. 3c).
Expression of the yolk receptor rme-2 did not change with maternal age (Extended Data Fig. 4f) but all 6 vit transcripts substantially increased (Fig. 3d), suggesting increased intestinal vitellogenin synthesis underlies the increased yolk provisioning. Indeed, VIT-2::GFP fluorescence in hermaphrodites increased substantially from day 1 to day 3 (Extended Data Fig 4g), as did the total yolk content (Fig. 3e; Extended Data Fig 4h) and intestinal volume (Extended Data Fig 4i). We confirmed that provisioning to embryos also increases with maternal age in the C. elegans wild isolates CB4856 and PB306 (Extended Data Fig 5).
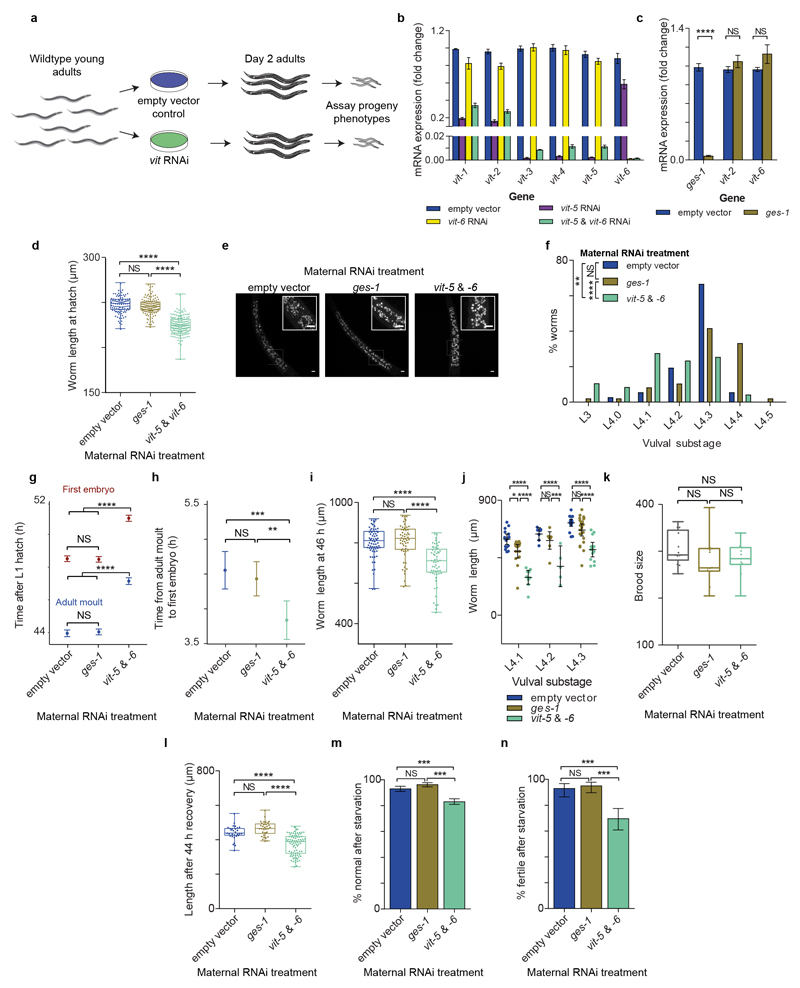
We tested whether reduced yolk provisioning can cause the phenotypic impairments observed in early progeny (Extended Data Fig. 6a, b). Progeny of vit RNAi-treated day 2 mothers hatched smaller with enlarged gut granules (Extended Data Fig. 6d, e), developed slowly with a disproportionate somatic impact (Extended Data Fig. 6f, g, h), grew slower even accounting for developmental stage (Extended Data Fig. 6i, j) but had a normal brood size (Extended Data Fig. 6k). Yolk-depleted progeny were also more sensitive to L1 starvation (Extended Data Fig. 6l, m, n). Yolk depletion therefore phenocopies nearly all of the impairments observed in progeny of young mothers.
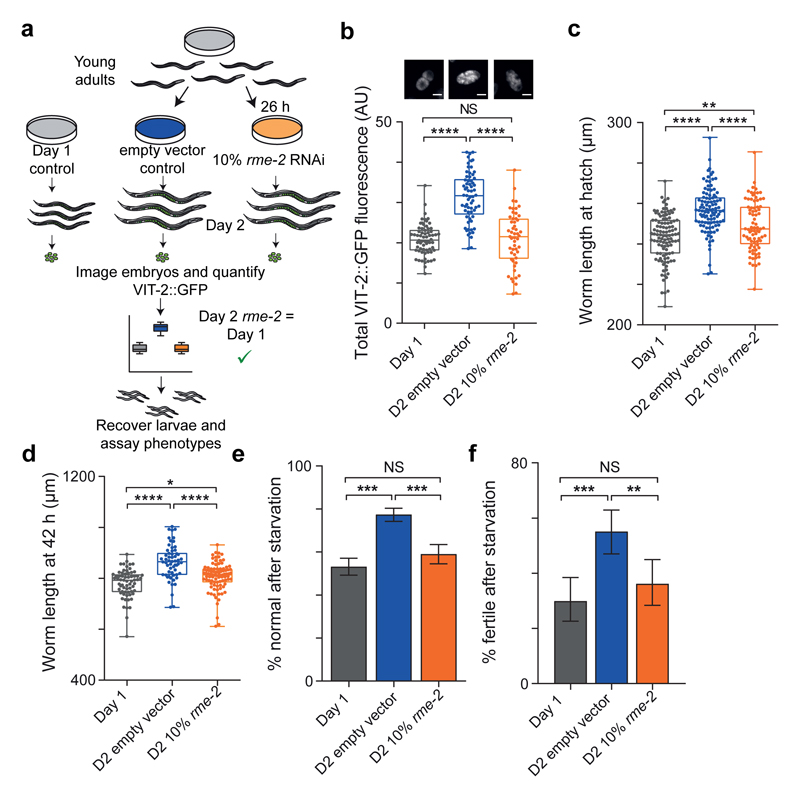
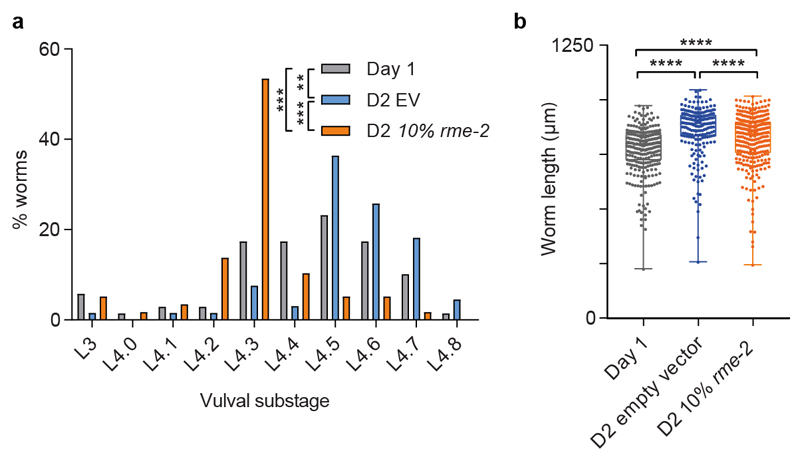
To test whether the differences in yolk levels between day 2 and day 1 progeny are sufficient to confer the impaired phenotypes, we tested a range of timings and dilutions of RNAi inhibition of rme-2, the single yolk receptor in C. elegans18, to produce day 2 progeny with embryonic yolk levels equivalent to day 1 progeny (Fig. 4a, b). This largely abrogated the increased hatching length (Fig. 4c), growth (Fig. 4d), developmental speed (Extended Data Fig. 7a) and starvation resistance (Fig. 4e, f; Extended Data Fig. 7b) of day 2 progeny.
Fig 4. Increased yolk provisioning explains phenotypic differences between day 1 and day 2 progeny.
(a) Equalising day 1/2 vitellogenin using RNAi.
(b) Embryo VIT-2::GFP from day 1/2 control or day 2 mothers treated with diluted rme-2 RNAi (n = 66, 65, 53 embryos). Representative images, scale 20 μm.
(c) Hatching length (n = 109, 103, 80 larvae).
(d) Length after 42 h (n = 65, 58, 93 larvae).
(e) Normal phenotypes after 11 d L1 starvation (n = 609, 717, 459 worms).
(f) Fertility after 14 d L1 starvation (n = 127, 147, 127 worms).
Error bars, 95 % C.I. One-way ANOVA (b, c), Kruskal-Wallis (d), Fisher’s exact test (e, f). Data from single trial, replicated twice independently. AU, arbitrary units.
We conclude that differential provisioning of vitellogenin underlies many of the phenotypic effects of maternal age. However, maternal vit RNAi did not affect progeny brood size and only modestly affected soma-germline phasing. Additionally, despite more yolk and faster development, day 3 progeny are more prone to L1 starvation-induced sterility than day 2 progeny. We speculate that this reflects early signs of reproductive aging22 in day 3 mothers.
In summary, we have shown here that maternal age is a major source of phenotypic variation in isogenic C. elegans populations living in a controlled environment. The progeny of young mothers (~10% of all progeny) are impaired for multiple fitness traits throughout their lives, including developmental rate, starvation resistance and fecundity. Maternal age also controls the rate of development of one organ relative to the others. An age-dependent change in maternal provisioning of a lipoprotein complex is therefore an important source of phenotypic variation in an animal, and maternal age is an important experimental variable to control in this species. This study adds to a growing body of work highlighting the influence of the physiology and environment of previous generations on phenotypic traits23–27.
The advantage of producing sub-optimal early progeny is likely to be simply that they come into existence sooner28. The production of lower-quality early progeny and higher-quality later progeny may therefore reflect a trade-off between the competing fitness traits of a short generation time and progeny quality.
Materials and Methods
Worm culture
All worms were cultured using standard methods at 20 ºC. Worms were fed on nematode growth medium (NGM) plates seeded with Escherichia coli OP50. To produce synchronized cohorts of worms, 20-60 gravid, synchronized hermaphrodites were picked onto a fresh plate and allowed to lay eggs for 2 h before being removed. Alternatively, synchronized gravid hermaphrodites and their previously hatched progeny were washed from a plate, leaving laid embryos on the solid media. After 1 h, larvae that had hatched in this period were collected and transferred to a fresh plate to form a more tightly synchronized cohort. Either protocol was used to synchronize the parental generation, whereas the more accurate hatching synchronization protocol was always used where developmental speeds were to be compared directly. In this paper ‘day 1 adults’ refer to adults 56-62 h after hatching from the egg or 68-74 h after the egg was laid. Day 2 adults and day 3 adults in each trial are 24 h and 48 h older, respectively. The density of worms on plates was matched between parallel cohorts within trials. All worms were self-fertilized hermaphrodites.
Strains
The following C. elegans strains were used in this study: BCN9070 vit-2(crg9070[vit-2::gfp]), CB4856, N2, PB306. The vit-2(crg9070[vit-2::gfp]) C terminus knock-in allele was generated by CRISPR-Cas9 genome editing using the self-excising drug selection cassette approach described in 29. Unless otherwise specified, experiments were performed in an N2 genetic background and N2 is referred to as ‘wildtype’.
Single worm gene expression profiling
Worms were collected as young adults 46 h after a 1 h synchronization by collecting hatching L1 larvae as described above. Single worms were then washed, collected in a 1.5 ml microcentrifuge tube (Eppendorf) and snap-frozen in liquid nitrogen. Total RNA was extracted using an RNAClean XP kit (Agentcourt) following the manufacturer’s instructions and amplified using the TransPlex Complete Whole Transcriptome Amplification Kit (Sigma Aldrich) to generate cDNA. Cy3-labeled cDNA was prepared from 500 ng of double-stranded cDNA using the DNA Enzymatic Labelling Kit (Agilent) according to the manufacturer’s instructions, followed by purification with a 30 kDa column (Amicon). Dye incorporation and cDNA yield were checked with the NanoDrop ND-1000 Spectrophotometer (ThermoScientific). cDNA was mixed with hybridization buffer and blocking agent (Agilent) and incubated at 95 °C for 3 min before cooling on ice. cDNA was then hybridized to a custom C. elegans 4x44K microarray (Agilent) for 40 h at 65 °C. Microarrays were washed 1 min at room temperature with GE wash buffer 1 (Agilent) and 1 min at 37 °C with GE wash buffer 2 (Agilent), then dried immediately by brief centrifugation. Microarrays were scanned on a G2539A scanner (Agilent) at 5 µm resolution and 100 % PMT. Probe intensities were extracted and their quality was assessed with the Feature Extraction software 10.7 (Agilent).
Staging somatic and germline development in vivo
Vulval substages were scored by assessing vulva morphology by brightfield microscopy30. Moulting and appearance of the first fertilized egg were used as developmental transitions to stage somatic and germline development, respectively. These were monitored in animals on agar plates using brightfield microscopy with an MZ9S stereo microscope (Leica). The fraction of the population that had undergone a developmental transition was estimated by counting worms before or after the transition in samples of between 50 and 400 worms. To estimate the time at which half of the population had undergone the developmental transition, each population was scored at least twice, once before half of the population had undergone the transition and once afterwards. Count data was then modelled using a binomial generalized linear model and the time at which half of the population had undergone the developmental transition (T50) was estimated using the function ‘dose.p’ of the library ‘MASS’ in R. 95 % confidence intervals were estimated by multiplying the standard error obtained by the ‘dose.p’ function by 1.96.
The significance of the difference between two calculated T50 values was determined by using the ‘comped’ function in the library ‘drc’ in R, which takes as input the two T50 values, their standard error and a desired probability level (for example 0.95). The output is the confidence interval of the ratio of the two T50 values at the chosen probability level. The difference between two T50 is not significant at a given probability level when the confidence interval of the ratio contains 131.
Microscopy and image analysis
Brightfield and epifluorescence microscopy were conducted with a DMI6000B inverted microscope (Leica) fitted with an ORCA-Flash4.0 V2 Digital CMOS camera (Hamamatsu). N2 embryos for measuring green autofluorescence as a proxy for yolk loading were imaged with a 20 x objective. Embryos imaged for immunofluorescence were imaged with a 40 x objective. In all other experiments, embryos and L1 larvae were imaged with a 10 x objective lens and developed worms were imaged with a 5 x objective lens. Worms and embryos were imaged in M9 buffer in an 8-well μ-slide (Ibidi), with worms anaesthetized with 10 mM NaN3. Embryo preps for imaging were prepared by alkaline hypochlorite treatment of synchronized gravid adult hermaphrodites and were imaged immediately in order to capture early embryos. Autofluorescence from N2 embryos was also measured as a control. For representative images blue autofluorescence from L1 gut granules was imaged using a TCS SPE confocal microscope (Leica; wavelengths: excitation 405 nm, emission 390-450 nm). All confocal images presented of gut granules are maximum projections of Z-stacks. Quantification of gut granule fluorescence intensity in straightened worms was performed by epifluorescence microscopy. For all epifluorescence microscopy experiments, all settings and exposure times were held constant within each experiment.
Quantitative analysis of fluorescence images was conducted with CellProfiler software32. Worm length measurement and digital worm straightening were achieved with the Cell Profiler Worm Toolbox package33. Data were manually curated to ensure accurate thresholding of worms and embryos. For the experiment shown in Extended Data Fig. 6j, worm lengths were measured manually using the segmented line drawing tool in ImageJ software. Intestinal volumes were estimated by measuring the length and width of the intestine on brightfield images of adult worms using the segmented line drawing tool in ImageJ and assuming a cylindrical shape. Presented epifluorescence data from embryos are all from early embryos. This was achieved by post-hoc synchronization by selecting early embryos from corresponding brightfield images of mixed-stage embryo preps obtained by alkaline hypochlorite treatment. Image analysis pipelines can be made available on request. For the experiment showing early embryo green autofluorescence increasing with maternal age, 2 trials were analysed together by generalized linear mixed model analysis with the ‘lme4’ package in R, with the following terms included in the model: maternal age, embryo size as fixed effects and trial as a random effect. For the analysis of gut granule size from confocal microscopy images, data were analysed by generalized linear model analysis with the ‘lme4’ package in R, with the following terms included in the model: maternal age as a fixed effect and individual worm as a random effect.
Nuclear and lipid staining
To perform nuclear staining of worms with 4′,6-diamidino-2-phenylindole (DAPI), worms were fixed in 40 % isopropanol before incubation with shaking for 30 min in 75 ng / ml DAPI solution. Worms were washed twice before epifluorescence microscopy imaging with a standard DAPI filter cube.
Our fixative embryo lipid staining protocol was based on 34. Lipid staining was always performed using non-transgenic worms. Briefly, embryo preps in tubes obtained by alkaline hypochlorite treatment of adults were subjected to 3 freeze-thaw cycles in liquid nitrogen and a 37 ºC water batch in the presence of 4 % formaldehyde in M9 buffer in a PCR tube (Eppendorf). After fixation, embryos were incubated with 1 μg / ml BODIPY 493/503 (ThermoFisher; 1000 x stock solution prepared in dimethyl sulfoxide) in M9 buffer for 1 h and washed 3 times with M9 + 0.01% Triton X-100 (Sigma Aldrich) before imaging by epifluorescence microscopy with a standard GFP filter set. Nuclei were concurrently stained with 15 ng / ml DAPI. Data presented are for intact embryos with between 6 and 20 automatically identified nuclei. As there was substantial batch variability in fixation and staining, volumes of fixative and stain were reproduced as consistently as possible. 5 biological replicates were stained and imaged in parallel for each maternal age group. For data presented in the N2 strain, 2 trials of 5 biological replicates each were analysed together after normalisation to the mean day 1 fluorescence within each trial by generalized linear mixed model analysis with the ‘lme4’ package in R, with the following terms included in the model: maternal age, embryo size, embryo nuclei count as fixed effects and trial and replicate as random effects. For CB4856 and PB306, the same analysis was applied on data from a single trial.
Immunofluorescence
Gravid hermaphrodites were picked into 5 µl of M9 buffer onto a poly-lysine coated slide (Sigma Aldrich). A coverslip was placed on top and worms were gently compressed to allow embryos to extrude. Embryos were immediately freeze-cracked in liquid nitrogen and fixed with methanol for 10 min followed by acetone for an additional 10 min. After 3 washes in PBS containing 0.25 % Triton X-100 (PBS-T), slides were blocked in PBS-T with 0.5 % BSA before overnight incubation with mouse monoclonal primary antibody diluted 1/200 (PIIA3 against YP17021, a kind gift of Susan Strome) at 4 °C. The slides were then washed with PBS-T, and incubated for 2 h with Alexa-488 anti-mouse secondary antibody (Invitrogen) at room temperature. After 3 washes in PBS-T, the samples were mounted in Fluoroshield with DAPI mounting medium (Sigma Aldrich). Stained embryos were imaged by epifluorescence microscopy. Quantification was performed using ImageJ. Data were analysed by generalized linear model analysis with the ‘lme4’ package in R, with the following terms included in the model: maternal age, embryo nuclei count as fixed effects and staining replicate and mother worm as random effects.
Quantification of yolk by SDS-PAGE and densitometry
Protein extraction from hermaphrodites was based on previously published work35. Adults were washed from plates in M9 buffer and separated from progeny by 3 rounds of gravity sedimentation for 3 min. Remaining larvae were washed through a 50 µm CellTrics filter (Sysmex Partec) with abundant M9 buffer before recovering adults. Adults were centrifuged and 80 µl of adults in M9 buffer were added to a 1.5 ml microcentrifuge tube containing 80 µl of 2x Laemmli buffer (Sigma Aldrich). Samples were heated to 70 °C in a ThermoMixer heating block (Eppendorf) with shaking at maximum speed for 15 min. Samples were then heated to 95 ºC for 3 min before centrifugation at 20,000 g for 3 min to remove any insoluble debris. Sample supernatant was further diluted 10-fold in 1x Laemmli buffer before loading 10 to 20 µl of the diluted sample in 10-well NuPAGE 4-12 % Bis-Tris denaturing protein gels (Invitrogen) and running according to manufacturer´s instructions. Gels were stained with SYPRO Ruby fluorescent protein stain (ThermoFisher) according to manufacturer’s instructions and imaged on a Typhoon Trio scanner (GE Healthcare). Densitometry was performed with ImageQuant TL software v8.1 (GE Healthcare) to quantify peaks corresponding to myosin and YP170 (VIT-1 to VIT-5) identified with reference to 35. Additionally, protein samples from hermaphrodites treated with rme-2 RNAi (to induce yolk accumulation) or vit-5 & vit-6 RNAi (to deplete yolk) were run to confirm the identity of the peaks corresponding to yolk proteins.
Starvation assays
For starvation assays, batches of larvae were prepared by alkaline hypochlorite treatment of synchronized adult worms. Embryo preps were washed thoroughly and passed through a 30 μM CellTrics filter (Sysmex Partec) to exclude any debris or worm corpses. Embryos were suspended in M9 buffer supplemented with streptomycin at 50 μg / ml to prevent contamination. Larvae were starved at 20 ºC with shaking in plastic 15 ml tubes that had been pre-treated with BSA to prevent worm loss by adherence to plastic36. After hatching, larval preps were counted and worm density was adjusted such that it was equal among all treatment groups within each individual trial. Larval preps were thoroughly washed after density adjustment.
Worms resuming development after extended L1 starvation often exhibit phenotypic defects such as extreme developmental delay24, abnormal or absent gonads37 and death due to vulval rupture. To score for penetrance of abnormal phenotypes after extended starvation, starved worms were recovered on food for 96 h before plates were scored blind by a single experimenter. Plates were passed to a third party researcher with no stake in the current study, who obscured the original labelling, randomised the plates and assigned new labels which were only decoded after the assay had been fully scored. Worms were picked off plates to ensure no double counting and tallied in the following categories: normal gravid adult, burst/protruding vulva, internal hatching, multivulva, sick/extremely delayed and gonads disorganized. Where worms fell in multiple categories (e.g. protruding vulva worms were nearly always sterile) they were binned consistently according to the phenotype considered more unusual or extreme (in this case, protruding vulva). The penetrance of abnormal phenotypic outcomes after a defined period of starvation was variable between trials.
After extended L1 starvation, worms often suffer from sterility due to apparent defects in gonad migration and proliferation. To assay sterility, starved worms were recovered on solid NGM plates seeded with E. coli OP50 for around 48 h. At this point, worms were washed from the plates, counted and diluted to an estimated density of 1 live worm per 15 μl in M9 buffer supplemented with 50 μg / ml streptomycin. Streptomycin-resistant E. coli OP50-1 was added to a density of 250 μg / ml. Using a multichannel pipette, 15 μl per well was deposited in a 384-well plate (Corning). The plate was sealed and incubated at 20 ºC without shaking. 5 days later, wells containing a single developed worm were scored as sterile or fertile depending on the presence of hatched larvae. The quantity of food was such that the full development of only 1 worm per well was supported. With such a low concentration of food added, hypoxia did not appear to affect development or embryo hatching. In the absence of extended L1 starvation, worms developed at a normal rate, appeared healthy and no sterility was observed. The penetrance of sterility after a defined period of starvation was variable between trials.
95 % confidence intervals for count data were calculated using ‘binom.confint’ function in the ‘binom’ package in R.
Lifespan assay
Lifespan was scored according to standard methods38. Worms were grown on NGM plates supplemented with streptomycin at 300 μg / ml and seeded with the streptomycin-resistant strain E. coli OP50-1. No fluorodeoxyuridine (FuDR) was used; worms were transferred to new plates every 1-2 days until reproduction ceased.
Brood size assay
Young adult worms were transferred singly to 12-well NGM plates seeded with E. coli OP50. Worms were transferred every 24 h into a new well. Progeny were counted after 2-3 days. For the experiment shown in Fig. 2n and Extended Data Fig. 2c, the first day of egg-laying was counted from the point where the majority of each cohort had at least 1 embryo in utero.
Yolk depletion by vit RNAi
Yolk-depleted progeny were obtained by collecting hatching larvae from plates or alkaline hypochlorite treatment of day 2 adult hermaphrodites shifted 24 h previously to NGM plates supplemented with 1 mM IPTG and 100 μg / ml ampicillin seeded with overnight cultures of the RNAi feeding strain E. coli OP50(xu363)39 transformed with appropriate RNAi plasmids. If hatching larvae were collected, embryos were first subjected to a brief hypochlorite treatment before transfer to OP50 feeding plates to ensure no direct exposure of hatching larvae to dsRNA containing bacteria. All RNAi plasmids used in this study were prepared by miniprep from the Ahringer RNAi library40, with the exception of the vit-6 RNAi plasmid. The vit-6 RNAi plasmid was made by PCR amplification of an 816 bp fragment of the vit-6 gene from C. elegans N2 genomic DNA with primers 5´-TCAGACTAGTACCGCTACCTCTTCAATGGC-3´ and 5´-AGCTCTCGAGGCTCCATGGTGTTCAAGGTG-3´ (underlining indicates complementarity with C. elegans vit-6) before insertion between the SpeI and Xho1 restriction sites of the empty vector plasmid L4440 (a gift from Andrew Fire, Addgene plasmid # 1654). vit-5 & vit-6 combined RNAi plates were spotted with a mix of an equal volume of density-adjusted vit-5 and vit-6 RNAi overnight cultures. All RNAi plasmids were sequenced to ensure that the clones used were correct.
Titration of embryonic yolk by diluted rme-2 RNAi
To titrate down the level of yolk loading into progeny of day 2 mothers, we used plates seeded with a mixture of E. coli OP50(xu363) containing rme-2 RNAi plasmid and L4440 (empty vector), following the RNAi dilution approach of 41. Density-adjusted stationary overnight cultures of rme-2 RNAi were diluted 1/10 by L4440 cultures prior to seeding plates. Appropriate RNAi conditions were variable between trials and plate batches. To control for this, BCN9070 hermaphrodites were shifted to these 10 % rme-2 plates at a range of timings (typically 18-30 h) prior to harvesting embryos when mothers were around 96 h post-lay. The embryos extracted were immediately imaged, alongside day 2 empty vector control and day 1 (around 72 h post-lay) control groups. After quantifying fluorescence levels in the early embryos, the treatment regime which had most accurately titrated day 2 VIT-2::GFP loading to day 1 levels was chosen and the embryos were recovered and used for all further phenotypic assays.
Quantitative real-time PCR
Quantitative real-time PCR (qPCR) was always conducted on non-transgenic worms. qPCR was performed using SYBR Green Master Mix (Roche) in a LightCycler 480 machine (Roche). An absolute quantification workflow was used, utilizing standard curves created with a reference sample that was run on all plates. The mean concentration of technical triplicate reactions was used for all samples and primer pairs for each of 3 biological replicates. Gene expression fold changes were normalized to the geometric mean of 3 housekeeping genes cdc-42, rpl-27 and Y45F10D.442, except for the data shown in Fig. 4d and Extended Data Figure 5f, where only cdc-42 and rpl-27 were used. All statistics, including error bars shown on plots, were calculated using logged fold change values. qPCR primer sequences were obtained from the GETPrime database43, with the exception of primers for rpl-27. Primer sequences are available in Extended Data Table 1.
Microarray data pre-processing
Log-transformed intensities were quantile normalized using the ‘normalizeBetweenArrays’ function in ‘limma’ package of R. Normalized log intensities of different probes for the same genes were averaged.
Reference developmental time courses
The developmental time courses from 44 consist of gene expression profiles of pools of synchronized worms reared at 25 °C and collected every 3 h, for a total of 12 time points in 3 replicates, encompassing development from the middle of the L3 larval stage to the young adult stage. The second time-resolved dataset consists of microarray expression profiles for 208 recombinant inbred advanced intercross lines from 45 for which developmental stages were inferred in 6.
Tissue specificity of gene expression
Sets of genes highly expressed in individual tissues were reported in 46 using cell sorting or polyA-binding protein (PAB-1) immunoprecipitation. In summary, embryonic cells expressing tissue-specific fluorescent reporter constructs were sorted by FACS analysis to define embryo-specific gene expression from the germline precursors, all neurons, BAG neurons, AVA neurons, AVE neurons, A-class motor neurons, dopaminergic and GABAergic neurons, body wall muscle, coelomocytes, hypodermis, intestine and pharyngeal muscle. An mRNA-tagging strategy was used to isolate RNA from specific larval and adult cells: an epitope-tagged PAB-1 was expressed under control of cell-specific promoters and PAB-1–RNA complexes were immunoprecipitated. Larval-specific gene expression profiles were generated for all neurons, A-class motor neurons, dopaminergic and glutamatergic neurons, PVD and OLL neurons, body wall muscle, coelomocytes, hypodermis, intestine, pharyngeal muscle, excretory cells and sheath cells. In all cases, extracted RNA was amplified and hybridized to tiling arrays (Affymetrix). Genes were defined to be tissue-enriched if they showed at least 2-fold enrichment versus a reference sample at FDR 0.05. Somatic gonad gene sets were taken from the gold standard compiled in 47 by parsing tissue localization data limited to single-gene GFP or in situ experiments from WormBase for a total of 2872 genes.
Ranking samples by developmental progression onto tissue-specific developmental trajectories
Independent component analysis (‘fastICA’ function in R library ‘fastICA’) was used on the combined dataset of single worm microarray profiles and the 2 reference time courses to decompose gene expression into mutually independent components. 20 components were extracted. 3 components enriched in germline-specific gene expression, - 1 in oocyte-specific and 2 in spermatocyte-specific gene expression as defined in 44 - were selected to rank samples along a germline developmental trajectory. 3 components enriched in hypodermis-specific expression and oscillating with moulting were selected to rank samples along a somatic-specific developmental trajectory. Samples were ranked onto tissue-specific developmental trajectories using principal curves48 (function ‘prcurve’ in R library ‘analogue’). Principal curves are smooth one-dimensional curves that pass through the middle of multidimensional data providing a non-parametric and non-linear generalization of principal component analysis. The curve is constructed through an iterative algorithm that is initialized with some prior summary (that could be the principal component line) and at each iteration the curve is constructed as a smooth local average of the data where the definition of local is based on the distance in arc length of the projections of the data points onto the curve found in the previous iteration. The complexity of the fitted curve is controlled by a ‘complexity’ parameter. For germline ranking the algorithm curve was initialized using the first principal component (‘prcurve’ function with method = ‘pca’, axis=1, and complexity = 4). For soma ranking, the algorithm was initialized using the germline ranking (‘prcurve’ function with method = ‘user’ and complexity = 4, start=germline ranking).
Accounting for soma or germline developmental stages in gene expression
The effect of either soma- or germline-inferred rank was modelled for each gene as following: a model including somatic rank and a model including germline rank were fitted (‘LmFit’ function in R library ‘limma’) using splines with 3 degrees of freedom (‘bs’ function in R library ‘splines’). The best model was selected using Akaike information criterion (AIC). Residuals of gene expression after correcting for either somatic or germline developmental stage were extracted using ‘resid ‘function in R.
Statistical analyses and presentation
Approximate sample sizes were estimated in each experiment according to the assay performed, taking into account technical limitations of the assay. Blinding and randomisation of test samples were performed where assays required a subjective judgment of the researcher (e.g. judging phenotypic outcomes as normal or not) as detailed in the relevant Methods sections. All statistical analyses were performed in R (version 3.4.0) or Graphpad Prism (version 7.03). Shapiro-Wilk tests were performed on data as appropriate to test assumptions of normal distribution; if normality was not supported, alternative nonparametric tests were used. All tests employed were two-sided. In all figures, boxplots represent median values, interquartile ranges and Tukey whiskers with individual data points superimposed. Terms of generalised linear mixed model (GLMM) analysis is described in each relevant experimental section – briefly, P values were obtained by one-way ANOVA comparing a model including maternal age to an otherwise identical model without it. Post-tests for one-way ANOVA were Tukey’s multiple comparisons tests, except for in Fig. 3e where the Games-Howell post-test was used due to heteroscedasticity. Post-tests for Kruskal Wallis analysis were Dunn’s multiple comparisons tests. For count data, P values for pairwise Fisher’s exact tests are reported after Bonferroni correction for multiple comparisons. P values for all figures are as follows: * P < 0.05, ** P < 0.01, *** P < 0.001, **** P < 0.0001.
Data availability
Data for Figure 1 and Extended Data Figure 1 are included as Supplementary Information Tables 1 and 2 or as expression data sets deposited in the National Center for Biotechnology Information Gene Expression Omnibus under accession GSE98747 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE98747). The authors declare that all other data supporting the findings of this study are available within the paper as source data.
Extended Data
Extended data Figure 1. Spermatogenesis and oogenesis gene expression signatures extracted by independent component analysis (ICA).
(a) Scatterplot showing gene loadings on the oogenesis component (x axis) and spermatogenesis component (y axis) extracted by ICA. Spermatogenesis and oogenesis44 genes are highlighted.
(b) The oogenesis and spermatogenesis components in the reference time series44 ranked with principal curves. The 3 replicates per time point in the time series are considered separately.
(c) Early- and late-moulting signatures in the reference time series ranked according to germline rank.
(d) Enrichment (measured by odds) of oogenesis-44, spermatogenesis-44 and hypodermis-specific46 genes in the top 10 % most variable genes among single worms judged by expression residuals after correcting for germline developmental stage.
(e) Enrichment (measured by log odds) for genes whose variance is better explained by germline (positive log odds) or hypodermal (negative log odds) rank in tissue-specific gene sets. Variation in other somatic tissues is best explained by hypodermal gene expression signatures. Error bars represent 95 % confidence intervals. Gene sets are listed in Materials and Methods.
Extended data Figure 2. Slow development, early starvation and young mothers are associated with altered temporal distribution of progeny production, with early progeny displaying enlarged gut granules, slow development and reduced resistance to larval starvation.
(a) Schematic indicating the establishment of a synchronized population by allowing eggs to hatch for 1 h and transferring newly hatched larvae to a new plate.
(b) Progeny production per day for early-, mid- or late-moulting worms (n = 9, 11, 10 worms).
(c) Schematic indicating comparison of worms continuously fed after hatching with worms that underwent a transient 5 h starvation immediately after hatching.
(d) Progeny production per day for worms either continuously fed or transiently starved for 5 h immediately after hatching (n = 12, 13 worms).
(e) Schematic showing establishment of parallel cohorts of progeny from day 1, day 2 or day 3 mothers and subsequent phenotypic assays.
(f) Progeny production per day of offspring of day 1, day 2 or day 3 mothers. The start of the first day of egg laying for each cohort is counted from the point where a majority of the population have at least 1 embryo in utero (n = 18, 18, 19 worms).
(g) Confocal microscopy images of live L1 larvae from day 1, 2 or 3 mothers showing blue autofluorescence from intestinal lysosome-related organelles (‘gut granules’). Insets show expanded section of main image indicated by white box (shown in Fig 2m). All images are maximum projections. Scale bars (main image and inset), 5 μm.
(h, i) Gut granule mean diameter (h) and count (i) in individual newly hatched L1 larvae from day 1, 2 or 3 mothers (n = 3, 3, 3 larvae).
(j) Intensity of blue autofluorescence from gut granules in segments along the anterior-posterior axis of digitally straightened newly hatched L1 larvae from day 1, 2 or 3 mothers (n = 32, 29, 16 larvae). Representative straightened brightfield and fluorescence images are positioned correspondingly along the x axis. Scale bar, 20 μm
(k) Worm developmental stage determined by scoring vulval morphology of L4 larvae from day 1, day 2 or day 3 mothers after 42 h of feeding (n = 70, 61, 49 larvae).
(l) Length of larvae 40 h after recovery from a 7 d L1 starvation from day 1, day 2 or day 3 mothers (n = 26, 33, 26 larvae).
(m) Brightfield and epifluorescence images showing sterile progeny from day 1 mothers recovered for 96 h from a 14 d L1 starvation, or a worm that was continuously fed, with nuclei stained by 4′,6-diamidino-2-phenylindole (DAPI). Gonads of worms recovered from starvation often exhibit severe abnormalities, such as failure to migrate (*), hyper-proliferation (+) and endomitotic oocytes49 (arrowhead). DAPI images are max intensity projection of Z-stacks following rolling ball background subtraction. Scale bars, 100 μm.
(n) Survival curves of progeny of day 1 or day 2 mothers upon recovery from L1 starvation for 12 h (day 1, n= 39, 66 worms, day 2, n = 52, 56, 55 worms), 1 week (day 1, n = 35, 40, 45 worms, day 2, n = 39, 47, 52 worms) or 2 weeks (day 1, n = 56, 65 worms; day 2, n = 51, 61, 68 worms). Each line represents a biological replicate.
Boxplots represent median values, interquartile ranges and Tukey whiskers with individual data points superimposed. Error bars show s.e.m. * P < 0.05, ** P < 0.01, *** P < 0.001, **** P < 0.0001, Kruskal-Wallis test with Dunn’s multiple comparisons tests (k, l), generalised linear mixed model analysis (h, i), two-way ANOVA with Bonferroni multiple comparisons post-tests (j), log-rank test (panel n). Results were replicated at least 2 times independently. NS, not significant. YA, young adult.
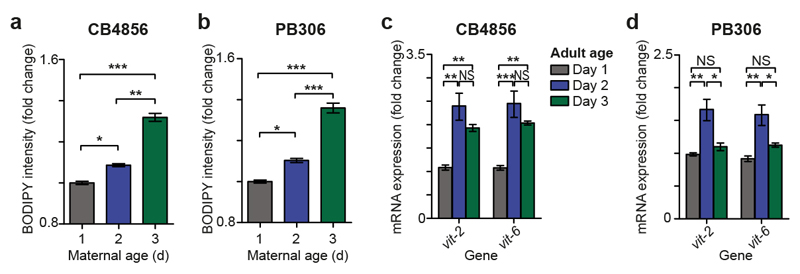
Extended data Figure 3. Maternal age affects progeny phenotypes in C. elegans wild isolates.
(a, b) Boxplot showing length at hatching of L1 larvae from day 1 (grey), day 2 (blue) or day 3 (green) mothers of CB4856 (a, n = 51, 87, 53 larvae) and PB306 (b, n = 134, 237, 120 larvae).
(c, d) Boxplot showing length of larvae after 42 h of feeding from day 1, day 2 or day 3 mothers of CB4856 (c, n = 82, 66, 106 larvae) and PB306 (d, n = 168, 150, 93 larvae).
(e, f) Bar chart indicating worm developmental stage determined by scoring vulval morphology of L4 larvae after 42h of feeding from day 1, day 2 or day 3 mothers of CB4856 (e, n = 81, 49, 101 larvae) and PB306 (f, n = 75, 85, 72 larvae).
(g, h) Boxplot showing length of larvae 48 h after recovery from a 7 day L1 starvation from day 1, day 2 or day 3 mothers of CB4856 (g, n = 174, 132, 185 larvae) and PB306 (h, n = 71, 62, 26 larvae).
(i, j) Confocal microscopy images of live L1 larvae from day 1, 2 or 3 mothers of CB4856 (i) and PB306 (j) showing blue autofluorescence from intestinal lysosome-related organelles (‘gut granules’). Insets show expanded section of main image indicated by white box. All images are maximum projections. Scale bars (main image and inset), 5 μm.
Boxplots represent median values, interquartile ranges and Tukey whiskers with individual data points superimposed. * P < 0.05, ** P < 0.01, *** P < 0.001, Kruskal-Wallis test with Dunn’s multiple comparisons tests (panels a-h). Results were replicated at least twice independently. NS, not significant, YA, young adult.
Extended data Figure 4. Vitellogenin expression and loading into embryos increase with age.
(a) Representative images of early embryos from day 1, day 2 and day 3 BCN9070 (vit-2::gfp) adults and wildtype N2 embryo for autofluorescence. Brightfield and epifluorescence images are shown. Scale bars, 20 μm.
(b) Average fluorescence intensity per embryo from immunostaining with an antibody against YP170, the protein encoded by vit-1 to vit-5, in 2-75 cell wildtype N2 embryos from day 1 or day 3 mothers. Duplicate stainings performed for each age group are shown (n = 41, 76; 58, 48 embryos). Fluorescence values are normalized to the mean of day 1 replicate A.
(c) Representative images of embryos extruded from day 1 or day 3 gravid hermaphrodites and stained with anti-YP170 antibody (green) and DAPI (blue). Scale bars, 20 µm.
(d) Total green autofluorescence of early embryos from wildtype N2 day 2 mothers treated with empty vector, RNAi against the constitutive intestinal gene ges-150 as an additional control, combined vit-5 & vit-6 RNAi and rme-2 RNAi (n = 58, 40, 22, 42 embryos). Fluorescence intensities are normalized to the mean of the empty vector control group. Representative images are shown above the corresponding group.
(e) Total green autofluorescence of early embryos from wildtype N2 day 1, day 2 or day 3 mothers (n = 92, 125, 70 embryos). Data shown are pooled from two trials analysed together. Fluorescence intensities are normalized to the mean of the day 1 group within each trial. Representative images from a single trial are shown above the corresponding group.
(f) mRNA transcripts quantified by qPCR of the yolk receptor rme-2 in day 1, day 2 and day 3 adults. Transcript levels were normalized to the geometric mean of 2 housekeeping genes (cdc-42 and rpl-27). Data shown are from 3 biological replicates.
(g) Representative images of day 1, day 2 and day 3 BCN9070 adults carrying an in-frame gfp knock-in at the C-terminus of vit-2 and wildtype N2 worm for autofluorescence. Brightfield and epifluorescence images are shown. Worms have been digitally straightened. Scale bars, 100 μm.
(h) Representative lysates of day 1, 2 or 3 adult hermaphrodites run on an SDS-PAGE denaturing protein gel and stained with SYPRO Ruby fluorescent protein stain. Additionally, to confirm the identity of peaks corresponding to yolk proteins, samples are shown from hermaphrodites treated with empty vector control RNAi, rme-2 (yolk receptor) RNAi to induce yolk accumulation and vit-5 & vit-6 RNAi to deplete yolk. All samples shown were run on the same gel. Myosin, YP170 (encoded by vit-1 to vit-5) and YP115 and YP88 (both encoded by vit-6) are indicated by arrows. For gel source data, see Supplementary Figure 1.
(i) Estimate of the intestinal volume of adult hermaphrodites (n = 7, 9, 9 worms). Bar shows mean ± s.e.m.
Boxplots represent median values, interquartile ranges and Tukey whiskers with individual data points superimposed. Error bars in barcharts and dot plots show standard error of the mean. * P < 0.05, ** P < 0.01, *** P < 0.001, *** P < 0.0001, one-way ANOVA with Tukey’s multiple comparison tests (f, i), Kruskal Wallis test with Dunn’s multiple comparisons test (d), generalised linear model analysis (b, e). NS, not significant. Ab, antibody.
Extended data Figure 5. Maternal vitellogenin expression and embryonic lipid content increase with maternal age in C. elegans wild isolates.
(a, b) Barchart of total fluorescence of fixed embryos from day 1, 2 or 3 mothers of CB4856 (a, 719, 1315, 249 embryos) and PB306 (b, n = 1090, 839, 330 embryos) stained with BODIPY 493/503, a neutral lipid stain. Data shown are pooled from 4-5 biological replicates.
(c, d) Barchart of mRNA transcripts quantified by qPCR of all vit genes in day 1, day 2 and day 3 adults of CB4856 (c) and PB306 (d). Transcript levels were normalized to the geometric mean of 3 housekeeping genes (cdc-42, rpl-27 and Y45F10D.4). Data shown are from 3 biological replicates.
Error bars in barcharts show standard error of the mean. * P < 0.05, ** P < 0.01, *** P < 0.001, generalised linear mixed model analysis (a, b), one-way ANOVA with Tukey’s multiple comparison tests (panel c, d). NS, not significant.
Extended data Figure 6. Progeny depleted of embryonic yolk by maternal vitellogenin knockdown recapitulate phenotypes observed in progeny of young mothers.
(a) Schematic showing experimental method employed to generate yolk-depleted progeny by maternal vit-5 & vit-6 RNAi treatment.
(b) mRNA transcripts quantified by qPCR of the 6 vit genes in day 2 adults treated with or empty vector control (blue), vit-5 RNAi (magenta), vit-6 RNAi (yellow) or a mixture of vit-5 and vit-6 RNAi (turquoise). Transcript levels were normalized to the geometric mean of 3 housekeeping genes (cdc-42, rpl-27 and Y45F10D.4). Data shown are from 3 biological replicates.
(c) The constitutive intestinal gene ges-150 was employed as an additional control. mRNA transcripts quantified by qPCR for ges-1, vit-2 and vit-6 in day 2 adults treated with empty vector control (blue) or ges-1 RNAi (brown). Transcript levels were normalized to the geometric mean of 3 housekeeping genes (cdc-42, rpl-27 and Y45F10D.4). Data shown are from 3 biological replicates.
(d) Length at hatching of L1 larvae from day 2 mothers treated with empty vector control (blue), ges-1 RNAi (brown) or vit-5 and vit-6 RNAi (turquoise) (n = 103, 130, 159 larvae).
(e) Confocal microscopy images of live L1 larvae from day 2 mothers treated with vit-5 and vit-6 RNAi, ges-1 RNAi or empty vector control showing blue autofluorescence from intestinal lysosome-related organelles (‘gut granules’). Insets show expanded section of main image indicated by white box. All images are maximum projections. Scale bars (main image and inset), 5 μm.
(f) Worm developmental stage determined by scoring vulval morphology of L4 larvae after 46 h of feeding from day 2 mothers treated with empty vector control (blue), ges-1 RNAi (brown) or vit-5 and vit-6 RNAi (turquoise) (n = 48, 36, 47 larvae).
(g) Time after hatching by which half the progeny of mothers treated with empty vector control, ges-1 RNAi or vit-5 and vit-6 RNAi had undergone the L4/adult moult (blue) or carried a fertilised embryo in utero (red). Each time point for each transition is calculated by sampling the population twice. (Sample sizes: empty vector, n = 199 and 198 for L4/adult moult, n = 171 and 168 for first embryo; ges-1, n = 204 and 206 for L4/adult moult, n = 198 and 168 for first embryo; vit-5 and vit-6, n = 201 and 202 for L4/adult moult, n = 179 and 200 for first embryo).
(h) Time between L4/adult moult and first embryo for the data shown in (g).
(i) Length of larvae from day 2 mothers treated with empty vector control (blue), ges-1 RNAi (brown) or vit-5 and vit-6 RNAi (turquoise) after 46 h of feeding (n = 67, 56, 57 larvae).
(j) Length of larvae at early L4 vulval substages in progeny of day 2 mothers treated with empty vector control (blue), ges-1 RNAi (brown) or vit-5 and vit-6 RNAi (turquoise) (L4.1, n = 15, 15, 9 larvae; L4.2, n=7, 8, 5 larvae, L4.3, n= 18, 22, 13 larvae).
(k) Total brood size of progeny of mothers treated with empty vector control (blue), ges-1 RNAi (brown) or vit-5 and vit-6 RNAi (turquoise) (n = 13, 14, 11 worms).
(l) Length 44 h after recovery from a 7 d L1 starvation of larvae from day 2 mothers treated with empty vector control (blue), ges-1 RNAi (brown) or vit-5 and vit-6 RNAi (turquoise) (n = 30, 33, 85 larvae).
(m) Percentage of adult progeny of day 2 mothers treated with empty vector control (blue), ges-1 RNAi (brown) or vit-5 and vit-6 RNAi (turquoise) displaying normal phenotypic outcomes 96 h after recovery from a 10 d L1 starvation (n = 610, 567, 1135 worms).
(n) Percentage of adult progeny of day 2 mothers treated with empty vector control (blue), ges-1 RNAi (brown) or vit-5 and vit-6 RNAi (turquoise) that successfully produced offspring 168 h after recovery from a 11 d L1 starvation (n = 103, 116, 124 worms).
Boxplots represent median values, interquartile ranges and Tukey whiskers with individual data points superimposed. Error bars in panels b and c represent s.e.m.; error bars in panels g, h, j, m and n represent 95 % confidence intervals. * P < 0.05, ** P < 0.01, *** P < 0.001, **** P < 0.0001, two-tailed t-tests (c), Kruskal-Wallis test with Dunn’s multiple comparisons tests (d, f, i, l), ratio tests (g, h), one-way ANOVA with Tukey’s multiple comparisons tests (k), two-way ANOVA with Bonferroni multiple comparisons tests (j), pairwise Fisher’s exact test with Bonferroni multiple corrections (m, n).Data shown are not all from the same trial but represent typical outcomes. All results were replicated at least twice. NS, not significant.
Extended data Figure 7. Increased yolk provisioning accounts for the differences in development and growth between day 1 and day 2 progeny.
(a) Worm developmental stage determined by scoring vulval morphology of L4 larvae after 42 h of feeding from day 1 mothers (grey), day 2 mothers treated with empty vector control (blue) and day 2 mothers treated with 10 % rme-2 RNAi (orange) (n = 69, 66, 52 embryo).
(b) Length 48 h after recovery from a 7 d L1 starvation of larvae from day 1 mothers (grey), day 2 mothers treated with empty vector control (blue) and day 2 mothers treated with 10 % rme-2 RNAi (orange) (n = 295, 206, 293 larvae).
Boxplots represent median values, interquartile ranges and Tukey whiskers with individual data points superimposed. ** P < 0.01, *** P < 0.001, **** P < 0.0001. Kruskal-Wallis test with Dunn’s multiple comparisons tests. Data shown (here and in Fig. 4) are from a single trial. Results were replicated twice independently.
Extended Data Table 1. Sequences of qPCR primers used in this study.
qPCR primers used in this study are listed as forward/reverse primers for each gene, with the corresponding sequences. All primer pairs were obtained from the GETPrime database43, with the exception of primers for rpl-27.
| Primer name | Gene | Sequence (5’ to 3’) |
|---|---|---|
| cdc42_qF1 | cdc-42 | GACAATTACGCCGTCACAG |
| cdc42_qR1 | cdc-42 | CGTAATCTTCCTGTCCAGCA |
| ges-1_qPCR_F | ges-1 | TTGAGATTTGAGAAGCCAGTC |
| ges-1_qPCR_R | ges-1 | GAGGAGTGCAATCGTTTCG |
| rme-2_qPCR_F | rme-2 | CATCATCTGGATCGATTCTTATCAG |
| rme-2_qPCR_R | rme-2 | AGAAGGATTCTGACCTGAGAC |
| rpl-27_F | rpl-27 | AAGCTGAAGCCATTCCTCAA |
| rpl-27_R | rpl-27 | GCTCCTCGAACTTGGACTTG |
| vit-1_qPCR_F | vit-1 | CACATAGAGAAGGTCAATGGA |
| vit-1_qPCR_R | vit-1 | CTTGAAGAAGTCTTCAACGAG |
| vit2_qF1 | vit-2 | CTTCCTTGCTCGTCTTGTC |
| vit2_qR1 | vit-2 | TGATTGGATCTTCTTGTCAGC |
| vit3_qF1 | vit-3 | TGAGACTCGCTCTAAGGTC |
| vit3_qR1 | vit-3 | TCCAGAGACCTTCTTGATCTC |
| vit4_qF1 | vit-4 | TCAAGAAGGTCTCTGGACC |
| vit4_qR1 | vit-4 | GACCTCAGCTTTGTCTCCT |
| vit5_qF1 | vit-5 | AATGAGTTCATGACCGCTG |
| vit5_qR1 | vit-5 | TTCGCATTCCTCATTCTTGAG |
| vit6ac_qF1 | vit-6 | CACCATTCCAAAGTTCGCC |
| vit6ac_qR1 | vit-6 | CGTTGGAAGTTCTCCTCCT |
| Y4510Fd.4_qF1 | Y45F10D.4 | TTCACTGTTCAATGCTCGC |
| Y4510Fd.4_qR1 | Y45F10D.4 | CTTAGGCCTTCTTAGTCTGCT |
Supplementary Material
Acknowledgments
This work was supported by a European Research Council Consolidator grant (616434), the Spanish Ministry of Economy and Competitiveness (BFU2011-26206 and SEV-2012-0208), the AXA Research Fund, the Bettencourt Schueller Foundation, Agència de Gestió d’Ajuts Universitaris i de Recerca (AGAUR), the EMBL-CRG Systems Biology Program, and the CERCA Program/Generalitat de Catalunya. M.F.P. was partially supported by a FPI-Severo Ochoa fellowship. Expression profiling was performed in the CRG Genomics core facility and microscopy in the CRG Advanced Light Microscopy Facility. Some strains were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440).
Footnotes
Author contributions
M.F.P, M.F. and B.L. conceived the model, designed experiments and wrote the manuscript. M.F.P, M.F. and C.H.C. performed experiments and analysed the data.
Author information
Reprints and permissions information is available at www.nature.com/reprints.
The authors declare no competing financial interests.
References and Notes
- 1.Spudich JL, Koshland DE., Jr Non-genetic individuality: chance in the single cell. Nature. 1976;262:467–471. doi: 10.1038/262467a0. [DOI] [PubMed] [Google Scholar]
- 2.Kirkwood TB, et al. What accounts for the wide variation in life span of genetically identical organisms reared in a constant environment? Mechanisms of Ageing and Development. 2005;126:439–443. doi: 10.1016/j.mad.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 3.Gärtner K. A third component causing random variability beside environment and genotype. A reason for the limited success of a 30 year long effort to standardize laboratory animals? Laboratory Animals. 1990;24:71–77. doi: 10.1258/002367790780890347. [DOI] [PubMed] [Google Scholar]
- 4.Wong AH, Gottesman II, Petronis A. Phenotypic differences in genetically identical organisms: the epigenetic perspective. Human Molecular Genetics. 2005;14:R11–R18. doi: 10.1093/hmg/ddi116. [DOI] [PubMed] [Google Scholar]
- 5.Rando OJ. Daddy issues: paternal effects on phenotype. Cell. 2012;151:702–708. doi: 10.1016/j.cell.2012.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Francesconi M, Lehner B. The effects of genetic variation on gene expression dynamics during development. Nature. 2014;505:208–211. doi: 10.1038/nature12772. [DOI] [PubMed] [Google Scholar]
- 7.Ambros V, Horvitz H. Heterochronic mutants of the nematode Caenorhabditis elegans. Science. 1984;226:409–416. doi: 10.1126/science.6494891. [DOI] [PubMed] [Google Scholar]
- 8.Poullet N, et al. Complex heterochrony underlies the evolution of Caenorhabditis elegans hermaphrodite sex allocation. Evolution. 2016;70:2357–2369. doi: 10.1111/evo.13032. [DOI] [PubMed] [Google Scholar]
- 9.Beguet B, Brun J. Influence of parental aging on the reproduction of the F1 generation in a hermaphrodite nematode Caenorhabditis elegans. Experimental Gerontology. 1972;7:195–206. doi: 10.1016/0531-5565(72)90026-5. [DOI] [PubMed] [Google Scholar]
- 10.Seidel HS, et al. A novel sperm-delivered toxin causes late-stage embryo lethality and transmission ratio distortion in C. elegans. PLoS Biology. 2011;9:e1001115. doi: 10.1371/journal.pbio.1001115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Benton T, St Clair J, Plaistow S. Maternal effects mediated by maternal age: from life histories to population dynamics. Journal of Animal Ecology. 2008;77:1038–1046. doi: 10.1111/j.1365-2656.2008.01434.x. [DOI] [PubMed] [Google Scholar]
- 12.Coburn C, Gems D. The mysterious case of the C. elegans gut granule: death fluorescence, anthranilic acid and the kynurenine pathway. Frontiers in Genetics. 2013;4:151. doi: 10.3389/fgene.2013.00151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fouad AD, et al. Quantitative assessment of fat levels in Caenorhabditis elegans using dark field microscopy. G3: Genes, Genomes, Genetics. 2017;7:1811–1818. doi: 10.1534/g3.117.040840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klass MR. Aging in the nematode Caenorhabditis elegans: major biological and environmental factors influencing life span. Mechanisms of Ageing and Development. 1977;6:413–429. doi: 10.1016/0047-6374(77)90043-4. [DOI] [PubMed] [Google Scholar]
- 15.Johnson TE, Mitchell DH, Kline S, Kemal R, Foy J. Arresting development arrests aging in the nematode Caenorhabditis elegans. Mechanisms of Ageing and Development. 1984;28:23–40. doi: 10.1016/0047-6374(84)90150-7. [DOI] [PubMed] [Google Scholar]
- 16.Sterken MG, Snoek LB, Kammenga JE, Andersen EC. The laboratory domestication of Caenorhabditis elegans. Trends in Genetics. 2015;31:224–231. doi: 10.1016/j.tig.2015.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kimble J, Sharrock WJ. Tissue-specific synthesis of yolk proteins in Caenorhabditis elegans. Developmental Biology. 1983;96:189–196. doi: 10.1016/0012-1606(83)90322-6. [DOI] [PubMed] [Google Scholar]
- 18.Grant B, Hirsh D. Receptor-mediated endocytosis in the Caenorhabditis elegans oocyte. Molecular Biology of the Cell. 1999;10:4311–4326. doi: 10.1091/mbc.10.12.4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chotard L, Skorobogata O, Sylvain MA, Shrivastava S, Rocheleau CE. TBC-2 is required for embryonic yolk protein storage and larval survival during L1 diapause in Caenorhabditis elegans. PloS one. 2010;5:e15662. doi: 10.1371/journal.pone.0015662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Van Rompay L, Borghgraef C, Beets I, Caers J, Temmerman L. New genetic regulators question relevance of abundant yolk protein production in C. elegans. Scientific Reports. 2015;5:16381. doi: 10.1038/srep16381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sharrock W, Sutherlin ME, Leske K, Cheng TK, Kim T. Two distinct yolk lipoprotein complexes from Caenorhabditis elegans. Journal of Biological Chemistry. 1990;265:14422–14431. [PubMed] [Google Scholar]
- 22.Luo S, Kleemann GA, Ashraf JM, Shaw WM, Murphy CT. TGF-β and insulin signaling regulate reproductive aging via oocyte and germline quality maintenance. Cell. 2010;143:299–312. doi: 10.1016/j.cell.2010.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rechavi O, et al. Starvation-induced transgenerational inheritance of small RNAs in C. elegans. Cell. 2014;158:277–287. doi: 10.1016/j.cell.2014.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jobson MA, et al. Transgenerational effects of early life starvation on growth, reproduction, and stress resistance in Caenorhabditis elegans. Genetics. 2015;201:201–212. doi: 10.1534/genetics.115.178699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harvey SC, Orbidans HE. All eggs are not equal: the maternal environment affects progeny reproduction and developmental fate in Caenorhabditis elegans. PloS One. 2011;6:e25840. doi: 10.1371/journal.pone.0025840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klosin A, Casas E, Hidalgo-Carcedo C, Vavouri T, Lehner B. Transgenerational transmission of environmental information in C. elegans. Science. 2017;356:320–323. doi: 10.1126/science.aah6412. [DOI] [PubMed] [Google Scholar]
- 27.Klosin A, et al. Impaired DNA replication derepresses chromatin and generates a transgenerationally inherited epigenetic memory. Science Advances. 2017;3:e1701143. doi: 10.1126/sciadv.1701143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hodgkin J, Barnes TM. More is not better: brood size and population growth in a self-fertilizing nematode. Proceedings of the Royal Society of London B: Biological Sciences. 1991;246:19–24. doi: 10.1098/rspb.1991.0119. [DOI] [PubMed] [Google Scholar]
- 29.Dickinson DJ, Pani AM, Heppert JK, Higgins CD, Goldstein B. Streamlined genome engineering with a self-excising drug selection cassette. Genetics. 2015;200:1035–1049. doi: 10.1534/genetics.115.178335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mok DZL, Sternberg PW, Inoue T. Morphologically defined sub-stages of C. elegans vulval development in the fourth larval stage. BMC Developmental Biology. 2015;15:26. doi: 10.1186/s12861-015-0076-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wheeler MW, Park RM, Bailer AJ. Comparing median lethal concentration values using confidence interval overlap or ratio tests. Environmental Toxicology and Chemistry. 2006;25:1441–1444. doi: 10.1897/05-320r.1. [DOI] [PubMed] [Google Scholar]
- 32.Carpenter AE, et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biology. 2006;7:R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wahlby C, et al. An image analysis toolbox for high-throughput C. elegans assays. Nature Methods. 2012;9:714–716. doi: 10.1038/nmeth.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klapper M, et al. Fluorescence-based fixative and vital staining of lipid droplets in Caenorhabditis elegans reveal fat stores using microscopy and flow cytometry approaches. Journal of Lipid Research. 2011;52:1281–1293. doi: 10.1194/jlr.D011940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.DePina AS, et al. Regulation of Caenorhabditis elegans vitellogenesis by DAF-2/IIS through separable transcriptional and posttranscriptional mechanisms. BMC Physiology. 2011;11:11. doi: 10.1186/1472-6793-11-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Artyukhin AB, Schroeder FC, Avery L. Density dependence in Caenorhabditis larval starvation. Scientific Reports. 2013;3:2777. doi: 10.1038/srep02777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee I, Hendrix A, Kim J, Yoshimoto J, You YJ. Metabolic rate regulates L1 longevity in C. elegans. PloS One. 2012;7:e44720. doi: 10.1371/journal.pone.0044720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lionaki E, Tavernarakis N. Assessing aging and senescent decline in Caenorhabditis elegans: cohort survival analysis. Methods in Molecular Biology. 2013;965:473–484. doi: 10.1007/978-1-62703-239-1_31. [DOI] [PubMed] [Google Scholar]
- 39.Xiao R, et al. RNAi interrogation of dietary modulation of development, metabolism, behavior, and aging in C. elegans. Cell Reports. 2015;11:1123–1133. doi: 10.1016/j.celrep.2015.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kamath RS, Ahringer J. Genome-wide RNAi screening in Caenorhabditis elegans. Methods. 2003;30:313–321. doi: 10.1016/s1046-2023(03)00050-1. [DOI] [PubMed] [Google Scholar]
- 41.Rea SL, Ventura N, Johnson TE. Relationship between mitochondrial electron transport chain dysfunction, development, and life extension in Caenorhabditis elegans. PLoS Biology. 2007;5:e259. doi: 10.1371/journal.pbio.0050259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vandesompele J, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biology. 2002;3 doi: 10.1186/gb-2002-3-7-research0034. Research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gubelmann C, et al. GETPrime: a gene- or transcript-specific primer database for quantitative real-time PCR. Database : the Journal of Biological Databases and Curation. 2011;2011:bar040. doi: 10.1093/database/bar040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reinke V, San Gil I, Ward S, Kazmer K. Genome-wide germline-enriched and sex-biased expression profiles in Caenorhabditis elegans. Development. 2004;131:311–323. doi: 10.1242/dev.00914. [DOI] [PubMed] [Google Scholar]
- 45.Rockman MV, Skrovanek SS, Kruglyak L. Selection at linked sites shapes heritable phenotypic variation in C. elegans. Science. 2010;330:372–376. doi: 10.1126/science.1194208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Spencer WC, et al. A spatial and temporal map of C. elegans gene expression. Genome Research. 2011;21:325–341. doi: 10.1101/gr.114595.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chikina MD, Huttenhower C, Murphy CT, Troyanskaya OG. Global prediction of tissue-specific gene expression and context-dependent gene networks in Caenorhabditis elegans. PLoS Computational Biology. 2009;5:e1000417. doi: 10.1371/journal.pcbi.1000417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hastie T, Stuetzle W. Principal curves. Journal of the American Statistical Association. 1989;84:502–516. [Google Scholar]
- 49.Iwasaki K, Mcearter J, Francis R, Schedl T. emo-1, a Caenorhabditis elegans Sec61p gamma homologue, is required for oocyte development and ovulation. The Journal of Cell Biology. 1996;134:699–714. doi: 10.1083/jcb.134.3.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kennedy BP, Aamodt EJ, Allen FL, Chung MA, Heschl MF, McGhee JD. The gut esterase gene (ges-1) from the nematodes Caenorhabditis elegans and Caenorhabditis briggsae. Journal of Molecular Biology. 1993;229:890–908. doi: 10.1006/jmbi.1993.1094. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data for Figure 1 and Extended Data Figure 1 are included as Supplementary Information Tables 1 and 2 or as expression data sets deposited in the National Center for Biotechnology Information Gene Expression Omnibus under accession GSE98747 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE98747). The authors declare that all other data supporting the findings of this study are available within the paper as source data.