Abstract
A novel cationic NHC-Au(I) complex was synthesized and studied for its antitumor activity. For all the cell lines tested, cationic NHC-Au(I) complex 2 shows much higher cytotoxicity than its neutral analogue 1. To achieve selective cancer cell targeting, complex 2 was covalently conjugated to aptamer AS1411, a DNA aptamer with strong binding affinity for nucleolin. The successful conjugation was confirmed by HPLC, gel electrophoresis, fluorescence spectroscopy and UV-Vis absorption. Conjugate AS1411-2 was then examined for its specific targeting and binding ability towards cancer cells over human normal cells using flow cytometry analysis and confocal microscopy. The cytotoxicity of AS1411-2 was then estimated by MTS assay. It was found that AS1411-2 exhibits higher activity than complex 2 towards targeted cells. Importantly, AS1411-2 exhibits much lower cytotoxicity towards healthy normal cell lines. Concurrently, the control groups without the AS1411 aptamer or without the NHC-Au(I) complex do have significant impact on cancer cell viability.
Introduction
Targeted drug delivery remains a challenging task. Peptides,1 antibodies,2 and other nanocarriers3 are strategies used in drug delivery. Exploiting antibody recognition of cell surface targets present obstacles for realizing their full potential including the challenging synthesis of antibody-drug conjugates, inconsistent binding affinities,4, 5 and immunogenicity.6 DNA aptamers are a group of excellent escorting agents for targeted drug delivery, providing great alternatives to antibodies. Aptamers are short single-strand oligonucleotides capable of specifically recognizing and effectively binding to their cell-surface targets, ranging from small organic molecules to proteins.7-11 As oligonucleotides, aptamers are biocompatible and generally have a longer shelf life than protein-based targeting ligands.9, 12 Because of these advantages, a wide range of biological studies employ aptamers,12-17 including cancer detection and therapy,18-21 and tumor imaging.22-25
Metal-based cancer drugs, such as cisplatin,26 highlight the importance and continued need for selective delivery vehicles. Despite their notable success, platinum based anticancer drugs suffer from adverse side effects such as heavy metal accumulation and drug resistance.27, 28 Cytotoxic inorganic and organometallic complexes designed for anti-cancer applications continue to receive intense interest.29-31 N-heterocyclic carbene (NHC) metal complexes, especially NHC-Au(I) complexes, are gaining considerable attention, in part due to their stability and cytotoxicity towards tumor cells both in vivo and in vitro.32-36 The NHC ligand is a strong σ-donor and forms stable M-ligand bonds with a large variety of metal ions.36, 37 The straightforward synthesis and ease of modification of NHC ligands make them an excellent choice for drug candidates. High affinity for selenol groups in the TrXR enzyme is the proposed reason for elevated Au(I) cytotoxicity towards cancer cells.38 Although NHC-Au(I) complexes exhibit promise as anticancer drug candidates, their major limitation, is a lack of selectivity towards specific cancer cells over normal cells.27, 37, 39 In previous work, we bioconjugated a neutral NHC-Au(I) complex to the leukemia cell CCRF-CEM specific aptamer sgc8c, and the aptamer-NHC-A(I) conjugate exhibited higher binding affinity and cytotoxicity towards its target cell than off-target cells in vitro.40 This current work employs a cationic NHC-Au(I) complex conjugated to the aptamer AS1411 to improve upon and broaden the scope of this targeted drug delivering strategy.
Aptamer AS1411 is the first aptamer to enter clinical oncology trials.41, 42 This 26-base guanine-rich aptamer binds selectively to nucleolin,43 a protein overexpressed on the cell membrane of many types of cancers, but importantly, not human normal cells.43-45 AS1411 is remarkably stable in serum and blood,43, 45, 46 a prerequisite for practical application. Previous work indicates AS1411 works as a great delivery agent to selectively deliver nanoparticles,47-50 quantum dots25, 51-53 and liposomes4, 54, 55 into cancer cells. The toxic agents in the aforementioned studies are typically cisplatin or doxorubicin; two agents that have severe side effects and exhibit resistance.56-59
Previous reports on NHC-Au(I) complexes indicate that changing the substitution groups on the imidazolium nitrogen minimally improves the cytotoxicity of the NHC-Au(I) complexes,60 cationic NHC complexes; however, show much higher cytotoxicity towards cancer cells compared to their neutral analogue.61, 62 Delocalized Lipophilic Cations (DLCs) are responsible for the increased cytotoxicity in cationic NHC complexes.38, 61-63 Cationic species concentrate in cells and into mitochondria in response to large transmembrane potentials. Higher cellular uptake causes higher systematic cytotoxicity towards the cell as a result. In addition, elevated mitochondria membrane potentials are characteristic for many tumors cells compared to normal cells and can lead to selective accumulation of DLCs in cancer cell mitochondria.38, 63
Herein, we report the synthesis of a highly cytotoxic cationic NHC-Au(I) complex and its subsequent bioconjugation to aptamer AS1411. Evidence from flow cytometry and confocal laser scanning microscopy indicates bioconjugate AS1411-NHC-Au(I) is highly selective and readily enters cancer cells. MTS assays confirm the bioconjugate exhibits selective toxicity towards cancer cells versus human normal cells.
Results and Discussion
Synthesis and characterization of cationic NHC-Au(I) complex 2
The synthesis62 of cationic NHC-Au(I) complex 2 is straightforward and involves treating complex 140 with potassium hexafluorophosphate and triphenylphosphine in acetone for 30 min. NMR, HRMS and elemental analysis confirm the identity and purity of complex 2. In the 1H NMR spectrum of 2, two resonances appear at 6.56 and 5.86 ppm as singlets and correspond to the methylene groups attached to the NHC. Compared to compound 1 (6.58 and 5.92 ppm), the singlets are slightly upfield. The 13C{1H} NMR spectrum of 2 exhibits a characteristic carbene resonance (C-Au) at 190.14 ppm. Compared to the carbene of 1 at 180.6 ppm, the shift downfield for 2 is a consequence of the stronger trans influence of the PPh3 ligand compared to Cl−. Further confirmation of the identity of 2 comes from a resonance at 38.34 ppm in its 31P NMR spectrum that matches well with similar cationic NHC-Au(I) complexes.61, 64 Figure 1 depicts the UV-Vis absorption and emission spectra of 2. The spectrum reveals a strong absorption maximum at 256 nm and three weak absorptions centered at 375 nm. The absorptions at 375 nm are characteristic of the anthracene moiety. The emission maximum for 2 appears at 416 nm.
Figure 1.
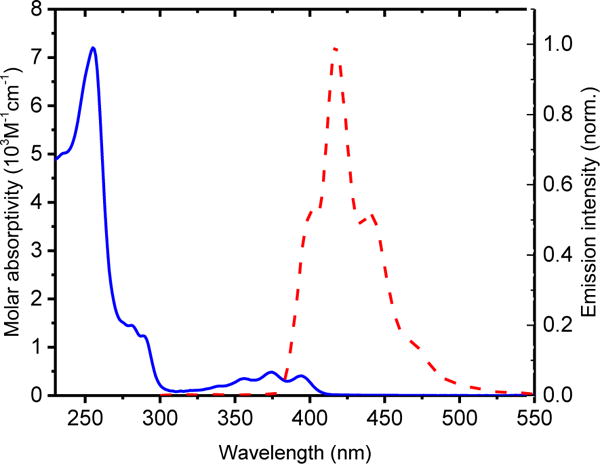
The UV-Vis absorption (blue line) and emission (red dash) of cationic NHC-Au(I) complex 2. (Solvent:VH2O:VAcetonitrile=1:1)
MTS (3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium) assays provide a measure of the cytotoxicity for cationic NHC-Au(I) complex 2. In this experiment, four cancer cell lines: CCRF-CEM (T-leukemia), MDA-MB-231 (breast cancer), DU145 (prostate cancer), and HeLa (cervical cancer), and two normal cell lines: HEK293 (human kidney cell) and HU1545v (human liver cell), were employed. The cells were incubated at 37 °C for 4 h in the presence of 1, 2, ligand, and Auranofin at various concentrations, then washed and allowed to grow in fresh medium for another 48 h. Complex 1 is inefficient for killing the cells lines tested within the incubation timeline, except CCRF-CEM. Under the same conditions, the cationic NHC-Au(I) complex 2 exhibits much higher cytotoxicity (>3 fold) for all the cell lines tested, a result attributable to its lipophilic cationic character and the known “delocalized lipophilic cation” (DLC) effect. DLCs can easily penetrate the hydrophobic barriers of cell membranes and accumulate in mitochondria, causing higher cellular uptake and systematic cytotoxicity.61 Thus, the cationic NHC-Au(I) complex 2 is the more promising candidate for bioconjugation and biological studies.
Preparation and characterization of aptamer NHC-Au(I) conjugate
The AS1411 aptamer sequence was modified with FITC (fluorescein isothiocyanate) on its 3′ end as a fluorescent probe, an amine group on its 5′ end to react with the NHC-Au(I) complex, and six extra T bases to prevent fluorescent quenching by G bases65 to give (5′- TTT TTT GGT GGT GGT GGT TGT GGT GGT GGT GG TTT TTT- FITC 3′). The aptamer-NHC-Au(I) conjugate was synthesized by suspending 0.1 μmol of solid supported aptamer in 1 mL of THF, followed by addition of complex 2 (10 μmol, 100 equiv) and slow addition of 1.4 μL of Et3N (Scheme 2). The mixture was shaken at ambient temperature for 18 h, then the solid support was removed by mixing with AMA solution (Vammonium hydroxide: Vmethylamine = 1:1). Finally, the aptamer-NHC-Au(I) conjugate AS1411-2 was purified via HPLC separation. HPLC, gel electrophoresis, emission, and UV-Vis absorption spectroscopies confirm the successful conjugation. The HPLC retention time of conjugate AS1411-2 (23.1 min) is 7.7 minutes longer than the retention time of the aptamer AS1411 alone (15.4 min). A minor product appears at 20.1 min, but is well separated. Gel electrophoresis of AS1411-2 and the control AS1411 aptamer results in a single band at the predicted size (39 bp), while the component with retention time at 20.1 min migrates much slower (see ESI). The slower gel migration of the minor component suggests it is a larger entity, possibly the result of crosslinking of AS1411.66, 67
Scheme 2.
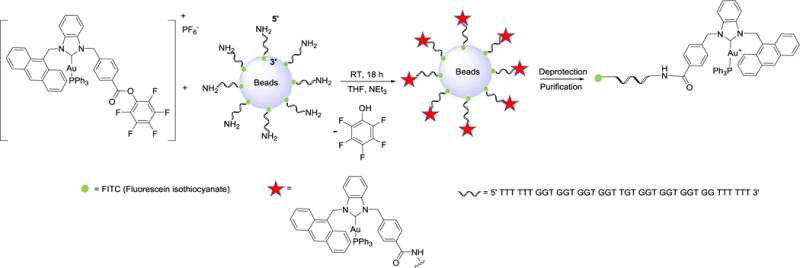
Synthesis strategy for AS1411-2. DNA is synthesized on the supporting beads using an automated ABI3400 DNA/RNA synthesizer.
Evidence for successful bioconjugation comes from an emission at 517 nm for the anthracene unit within AS1411-2 that is absent in the AS1411 aptamer (Figure 2A). Unique as well to the AS1411-2 conjugate is a characteristic weak absorption in the UV-Vis spectrum at 375 nm for the NHC-Au(I) complex (ε375 = 5000 cm−1 M−1: Figure 2B). By applying the Beer–Lambert law, it is possible to calculate the amount of NHC-Au(I) complex contained within the AS1411-2 conjugate. The calculated NHC-Au(I)/aptamer ratio in the conjugate was 1.07(±0.09), indicating that one NHC-Au(I) complex conjugates to one aptamer via the amide bond in the site-specific conjugation reaction (see ESI for calculation details).
Figure 2.
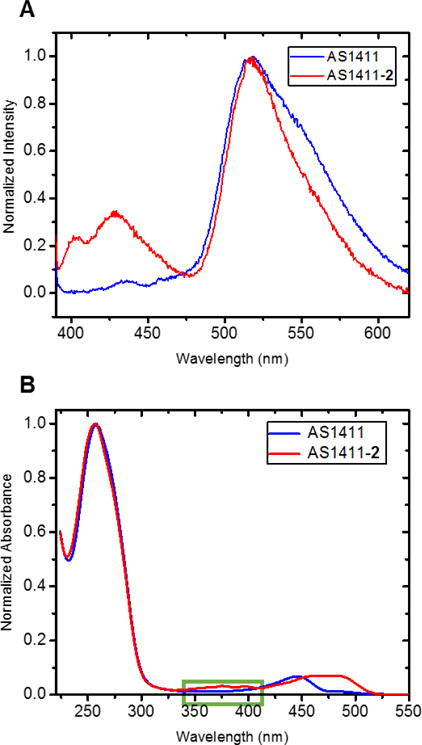
A) Emission spectrum of AS1411-2. (Solvent: VH2O:VAcetonitrile=1:1) B). UV-Vis absorption spectrum of AS1411-2 (H2O: Acetonitrile=1:1)
Targeted cellular uptake and in vitro cytotoxicity of AS1411-2
Flow cytometry and confocal laser scanning microscopy confirms the targeting specificity of the synthetic aptamer-NHC-Au(I) conjugate AS1411-2. In the flow cytometry experiment, FITC labelled AS1411 aptamer is the positive control. A library of random DNA sequences labelled with FITC and conjugated with complex 2 (LIB-2) serves as a non-selective control. Three cancer cell lines with nucleolin overexpression, Hela, DU145, and MDA-MB-231, are the target cancer cells. Normal cells HEK293 and HU1545v, having low nucleolin expression serve as “off-target” control cells. Each cell line was incubated with AS1411, AS1411-2, and LIB-2 individually, for 30 min. For the “off-target” control cells HEK293 and HU1545v, results from the flow cytometry experiments indicate only a marginal increase in fluorescence intensity was detected (Figure 3_(4) and 3_(5)) when treated with AS1411-2. In contrast, MDA-MB-231 cells (Figure 3_(1)), HeLa cells (Figure 3_(2)) and DU145 cells (Figure 3_(3)) exhibit enhanced fluorescence intensity when treated with AS1411-2. Clearly, the conjugate AS1411-2 exhibits higher recognition and binding to the target cancer cells than the “off-target” normal cells. Interestingly, AS1411-2 conjugate exhibits noticeably stronger binding affinity to the cancer cells than AS1411 aptamer itself. This phenomenon may be caused by the cationic character of the NHC-Au(I) complex.61
Figure 3.
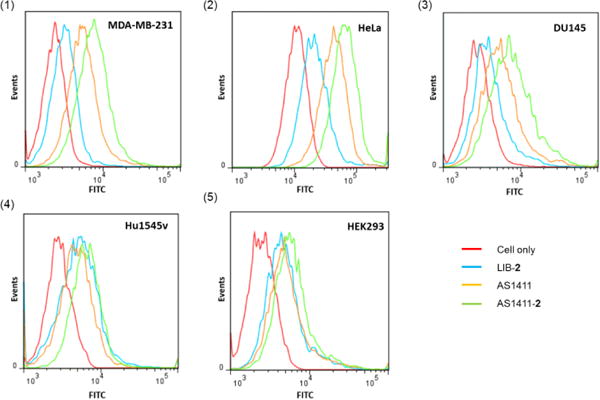
Flow cytometry histogram to monitor the binding of AS1411-2 to (a) MDA-MB-231, (b) HeLa, (c) DU145, (d) HU1545v, and (e) HEK293 cells. LIB-2 and AS1411 were used as controls.
Confocal laser scanning microscopy provides compelling cell-targeting evidence for conjugate AS1411-2. HeLa, MD-MB-231, DU145, HU1545v and HEK293 cells were incubated with 100 nM conjugate AS1411-2 for 30 min. Cancer cells treated with AS1411-2 exhibit strong fluorescence; whereas normal cells provide negligible fluorescence (see ESI). To further study the selective internalization of the conjugate, HeLa cells and HEK293 cells were incubated with AS1411-2 for 2 h at 37 °C, stained with Hoechst 33342 solution to label the nucleus, and then washed with PBS buffer to confirm the selective internalization of the conjugate into the cancer cells. As is evident in Figure 4, HeLa cells treated with conjugate AS1411-2 exhibit strong green fluorescence. Importantly, the cytoplasm and cell membrane are areas that exhibit the most fluorescence; evidence that indicates the conjugate enters the cancer cell, presumably via endocytosis.4 Off target HEK293 cells only display weak fluorescence. These results demonstrate the highly selectivity and effective internalization of conjugate AS1411-2 for targeted cancer cells.
Figure 4.
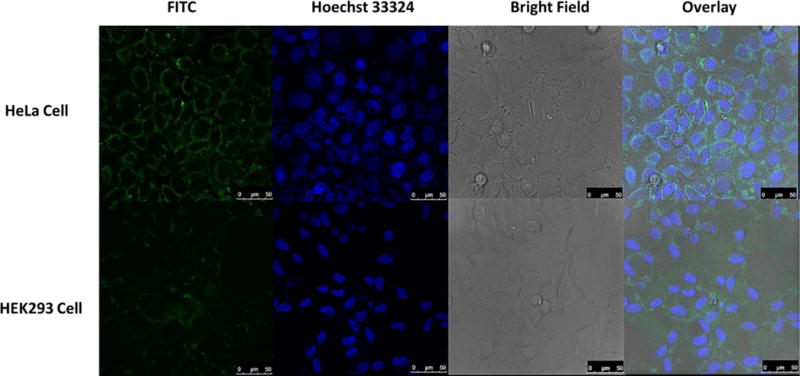
Confocal fluorescence images of living HeLa cells and HEK293 cells incubated with AS1411-2 conjugate. Each series can be sorted as FITC fluorescence, nucleus of cells dyed in blue by Hoechst 33324, optical images of cells in bright field and the overlay of the previous channels, respectively. All scale bars are 50 μm.
More support for the selectivity of AS1411-2 comes from in vitro MTS assays. In the MTS assay, significant inhibition of proliferation was found for all three cancer cell lines by AS1411-2 at 7 μM, while in the case for complex 2 alone, up to 30 μM is required at the incubation time of 4 h. When tested with NHC-Au(I) complex 2 alone, the cell viability percentage of all the five cell lines, including cancer cells and normal cells, decreased synchronously as the concentration increased (Figure 5). In contrast, the nucleolin-targeting aptamer AS1411-2 exhibits cytotoxicities at much lower concentrations, yet leaves normal cells minimally damaged. The cytotoxicity of AS1411-1 conjugate was also tested towards the five cell lines for comparison, yet no significant cytotoxicity was found (Figure S11), which further supports the cationic lipophilic character of complex 2 plays important role in killing cancer cells. Flow cytometry data from figure 4 clearly indicate the aptamer-conjugate AS1411-2 selectively recognizes the target cells; however, it is conceivable that the enhanced cytotoxicity that results is simply a kinetic phenomenon. That is, complex 2 alone, during the timescale of the MTS assay, may not undergo cellular uptake. In an effort to probe the kinetics, HeLa and normal cells (HEK293) were exposed to complex 2 at 4 μmol for 4, 24, and 48 h. The results indicate (Figure S8) that only after prolonged exposure (48 h) does complex 2 achieve the equivalent cytotoxicity of AS1411-2. Thus, the aptamer is important for both cellular recognition and cellular uptake. Finally, to confirm the function of each part of the synthetic conjugate, MTS assays were performed by treating HeLa cells with AS1411 aptamer, AS1411-ligand, LIB-2, and AS1411-2 (Figure 5_bottom). Conjugate AS1411-2 caused the highest cell death, while AS1411 aptamer, AS1411-ligand conjugate, and LIB-2 conjugate did not significantly inhibit cell proliferation. By comparing the red and the black lines in Figure 5_bottom, it is clear the Au(I) ion clearly plays a critical role in the high cytotoxicity of the conjugate. By comparing the black and blue lines, the specific targeting ability of the aptamer is demonstrated since the random sequences do not facilitate targeting.
Figure 5.
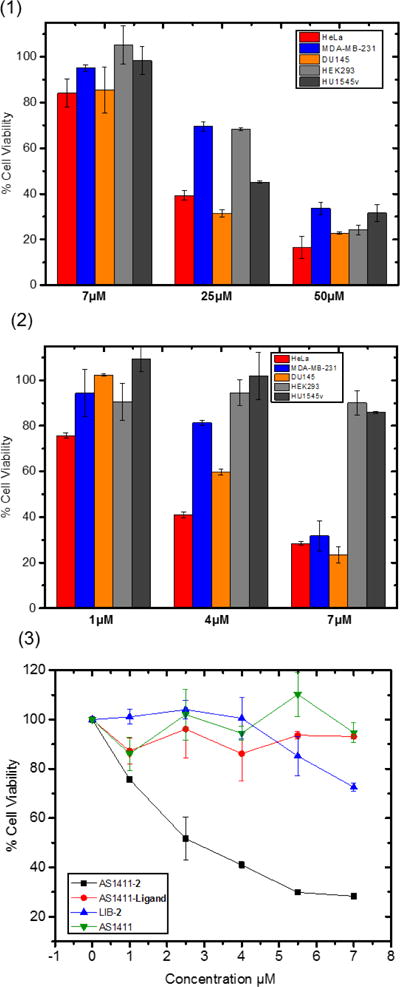
1) In vitro viability of different cell lines after treatment with NHC-Au(I) complex 2. 2) In vitro viability of different cell lines after treatment with AS1411-2 conjugate. 3) In vitro cell viability of HeLa cells after treatment with AS1411-2, LIB-2, AS1411-ligand and AS1411 individually.
In summary, we successfully synthesized the first conjugate of a highly cytotoxic cationic NHC-Au(I) complex with the AS1411 aptamer. Flow cytometry and confocal microscopy experiments demonstrate the specific targeting ability of the AS1411-2 conjugate: AS1411-2 shows much higher binding affinity towards cancer cells over healthy normal cells. Enhanced cytotoxicity was observed when AS1411-2 was applied to cancer cell lines MDA-MB-231, Hela and DU145. AS1411-2 does not significantly reduce proliferation in two healthy normal cell lines HEK293 and HU1545v. In addition, the function of each component in the conjugate was confirmed by comparing the cell viability of HeLa cells after treatment with AS1411, AS1411-2, AS1411-ligand and LIB-2. Considering the vast number of metal complexes already known to be anti-proliferative towards cancer cells, the results presented in this study indicate this method can be exponentially expanded to other metal-ligand constructs, and particular attention should be given to cationic species.
Supplementary Material
Scheme 1.
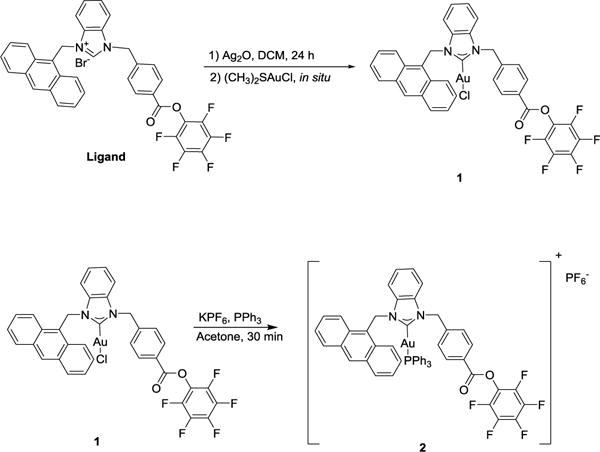
Synthesis of cationic NHC-Au(I) complex 2.
Table 1.
IC50 of 1 and 2 towards different cell lines.
| Cell Lines | Neutral NHC-Au 1 | Cationic NHC-Au 2 | Ligand | Auranofin |
|---|---|---|---|---|
| CCRF-CEM | 31.3±2.1140 | 4.26±0.63 | >100 | 7.03±0.63 |
| MDA-MB-231 | >100 | 29.41±3.44 | >100 | 8.79±3.09 |
| DU145 | >100 | 19.08±2.21 | >100 | 10.71±1.65 |
| Hela | >100 | 15.1±2.27 | >100 | 20.47±3.62 |
| HEK293 | >100 | 32.7±2.37 | >100 | 23.55±0.84 |
| HU1545v | >100 | 19.2±0.87 | >100 | 19.19±1.36 |
IC50 values were calculated as the concentration that causes 50% cell viability compared to untreated control cells, and are given as the means and errors of three independent experiments. The cells were exposed to the gold complexes for 4 h, after which the cells were washed and incubated in fresh medium for another 48 h.
Acknowledgments
ASV and WT acknowledge UF for support. This work was supported in part by the National Institutes of Health (GM079359). The authors thank Dr. Chen Liu’s lab at Rutgers University for providing the Hu1545v cell lines, and Dr. Liqin Zhang for the cultureing of the HU1545v cells.
Footnotes
Electronic Supplementary Information (ESI) available: Full experimental procedures, NMR spectra.
Notes and references
- 1.Boohaker RJ, Lee MW, Vishnubhotla P, Perez JM, Khaled AR. Curr Med Chem. 2012;19:3794–3804. doi: 10.2174/092986712801661004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alley SC, Okeley NM, Senter PD. Curr Opin Chem Biol. 2010;14:529–537. doi: 10.1016/j.cbpa.2010.06.170. [DOI] [PubMed] [Google Scholar]
- 3.Peer D, Karp JM, Hong S, FaroKhzad OC, Margalit R, Langer R. Nat Nanotechnol. 2007;2:751–760. doi: 10.1038/nnano.2007.387. [DOI] [PubMed] [Google Scholar]
- 4.Cao ZH, Tong R, Mishra A, Xu WC, Wong GCL, Cheng JJ, Lu Y. Angew Chem Int ed. 2009;48:6494–6498. doi: 10.1002/anie.200901452. [DOI] [PubMed] [Google Scholar]
- 5.Sapra P, Allen TM. Prog Lipid Res. 2003;42:439–462. doi: 10.1016/s0163-7827(03)00032-8. [DOI] [PubMed] [Google Scholar]
- 6.Peters C, Brown S. Biosci Rep. 2015;35 doi: 10.1042/BSR20150089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iliuk AB, Hu L, Tao WA. Anal Chem. 2011;83:4440–4452. doi: 10.1021/ac201057w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xiao Z, Shangguan D, Cao Z, Fang X, Tan W. Chem Eur J. 2008;14:1769–1775. doi: 10.1002/chem.200701330. [DOI] [PubMed] [Google Scholar]
- 9.Jayasena SD. Clin Chem. 1999;45:1628–1650. [PubMed] [Google Scholar]
- 10.Keefe AD, Pai S, Ellington A. Nat Rev Drug Discov. 2010;9:537–550. doi: 10.1038/nrd3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou JH, Rossi J. Nat Rev Drug Discov. 2017;16:181–202. doi: 10.1038/nrd.2016.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang LQ, Wan S, Jiang Y, Wang YY, Fu T, Liu QL, Cao ZJ, Qiu LP, Tan WH. J Am Chem Soc. 2017;139:2532–2540. doi: 10.1021/jacs.6b10646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bagalkot V, Farokhzad OC, Langer R, Jon S. Angew Chem Int ed. 2006;45:8149–8152. doi: 10.1002/anie.200602251. [DOI] [PubMed] [Google Scholar]
- 14.Oh SS, Lee BF, Leibfarth FA, Eisenstein M, Robb MJ, Lynd NA, Hawker CJ, Soh HT. J Am Chem Soc. 2014;136:15010–15015. doi: 10.1021/ja5079464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu G, Zheng J, Song E, Donovan M, Zhang K, Liu C, Tan W. Proc Natl Acad Sci U S A. 2013;110:7998–8003. doi: 10.1073/pnas.1220817110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang R, Zhu G, Mei L, Xie Y, Ma H, Ye M, Qing FL, Tan W. J Am Chem Soc. 2014;136:2731–2734. doi: 10.1021/ja4117395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dhar S, Gu FX, Langer R, Farokhzad OC, Lippard SJ. Proc Natl Acad Sci U S A. 2008;105:17356–17361. doi: 10.1073/pnas.0809154105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Medley CD, Bamrungsap S, Tan W, Smith JE. Anal Chem. 2011;83:727–734. doi: 10.1021/ac102263v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu CC, Han D, Chen T, Peng L, Zhu GZ, You MX, Qiu LP, Sefah K, Zhang XB, Tan WH. J Am Chem Soc. 2013;135:18644–18650. doi: 10.1021/ja4094617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun HG, Tan WH, Zu YL. Analyst. 2016;141:403–415. doi: 10.1039/c5an01995h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shangguan D, Li Y, Tang Z, Cao ZC, Chen HW, Mallikaratchy P, Sefah K, Yang CJ, Tan W. Proc Natl Acad Sci U S A. 2006;103:11838–11843. doi: 10.1073/pnas.0602615103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bagalkot V, Zhang L, Levy-Nissenbaum E, Jon S, Kantoff PW, Langer R, Farokhzad OC. Nano Lett. 2007;7:3065–3070. doi: 10.1021/nl071546n. [DOI] [PubMed] [Google Scholar]
- 23.Chen T, Shukoor MI, Wang R, Zhao Z, Yuan Q, Bamrungsap S, Xiong X, Tan W. ACS Nano. 2011;5:7866–7873. doi: 10.1021/nn202073m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim D, Jeong YY, Jon S. ACS Nano. 2010;4:3689–3696. doi: 10.1021/nn901877h. [DOI] [PubMed] [Google Scholar]
- 25.Wei W, He XW, Ma N. Angew Chem Int ed. 2014;53:5573–5577. doi: 10.1002/anie.201400428. [DOI] [PubMed] [Google Scholar]
- 26.Rosenber B, Vancamp L, Krigas T. Nature. 1965;205:698–&. doi: 10.1038/205698a0. [DOI] [PubMed] [Google Scholar]
- 27.Rabik CA, Dolan ME. Cancer Treat Rev. 2007;33:9–23. doi: 10.1016/j.ctrv.2006.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hartinger CG, Dyson PJ. Chem Soc Rev. 2009;38:391–401. doi: 10.1039/b707077m. [DOI] [PubMed] [Google Scholar]
- 29.Gasser G, Ott I, Metzler-Nolte N. J Med Chem. 2011;54:3–25. doi: 10.1021/jm100020w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu Z, Sadler PJ. Acc Chem Res. 2014;47:1174–1185. doi: 10.1021/ar400266c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hartinger CG, Metzler-Nolte N, Dyson PJ. Organometallics. 2012;31:5677–5685. [Google Scholar]
- 32.Cisnetti F, Gautier A. Angew Chem Int ed. 2013;52:11976–11978. doi: 10.1002/anie.201306682. [DOI] [PubMed] [Google Scholar]
- 33.Hopkinson MN, Richter C, Schedler M, Glorius F. Nature. 2014;510:485–496. doi: 10.1038/nature13384. [DOI] [PubMed] [Google Scholar]
- 34.Liu W, Gust R. Chem Soc Rev. 2013;42:755–773. doi: 10.1039/c2cs35314h. [DOI] [PubMed] [Google Scholar]
- 35.Garner ME, Niu W, Chen X, Ghiviriga I, Abboud KA, Tan W, Veige AS. Dalton Trans. 2015;44:1914–1923. doi: 10.1039/c4dt02850c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Teyssot ML, Jarrousse AS, Manin M, Chevry A, Roche S, Norre F, Beaudoin C, Morel L, Boyer D, Mahiou R, Gautier A. Dalton Trans. 2009;35:6894–6902. doi: 10.1039/b906308k. [DOI] [PubMed] [Google Scholar]
- 37.Liu WK, Gust R. Coord Chem Rev. 2016;329:191–213. [Google Scholar]
- 38.Hickey JL, Ruhayel RA, Barnard PJ, Baker MV, Berners-Price SJ, Filipovska A. J Am Chem Soc. 2008;130:12570–12571. doi: 10.1021/ja804027j. [DOI] [PubMed] [Google Scholar]
- 39.Bruijnincx PCA, Sadler PJ. Curr Opin Chem Biol. 2008;12:197–206. doi: 10.1016/j.cbpa.2007.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Niu WJ, Chen XG, Tan WH, Veige AS. Angew Chem Int ed. 2016;55:8889–8893. doi: 10.1002/anie.201602702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rosenberg JE, Bambury RM, Van Allen EM, Drabkin HA, Lara PN, Harzstark AL, Wagle N, Figlin RA, Smith GW, Garraway LA, Choueiri T, Erlandsson F, Laber DA. Invest New Drugs. 2014;32:178–187. doi: 10.1007/s10637-013-0045-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Laber DA, Sharma VR, Bhupalam L, Taft B, Hendler FJ, Barnhart KM. J Clin Oncol. 2005;23:207S–207S. [Google Scholar]
- 43.Bates PJ, Laber DA, Miller DM, Thomas SD, Trent JO. Exp Mol Pathol. 2009;86:151–164. doi: 10.1016/j.yexmp.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Soundararajan S, Chen WW, Spicer EK, Courtenay-Luck N, Fernandes DJ. Cancer Res. 2008;68:2358–2365. doi: 10.1158/0008-5472.CAN-07-5723. [DOI] [PubMed] [Google Scholar]
- 45.Dapic V, Abdomerovic V, Marrington R, Peberdy J, Rodger A, Trent JO, Bates PJ. Nucleic Acids Res. 2003;31:2097–2107. doi: 10.1093/nar/gkg316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Girvan AC, Teng Y, Casson LK, Thomas SD, Juliger S, Ball MW, Klein JB, Pierce WM, Barve SS, Bates PJ. Mol Cancer Ther. 2006;5:1790–1799. doi: 10.1158/1535-7163.MCT-05-0361. [DOI] [PubMed] [Google Scholar]
- 47.Li LL, Yin Q, Cheng JJ, Lu Y. Adv Heathc Mater. 2012;1:567–572. doi: 10.1002/adhm.201200116. [DOI] [PubMed] [Google Scholar]
- 48.Wu JH, Song CC, Jiang CX, Shen X, Qiao Q, Hu YQ. Mol Pharm. 2013;10:3555–3563. doi: 10.1021/mp300686g. [DOI] [PubMed] [Google Scholar]
- 49.Shieh YA, Yang SJ, Wei MF, Shieh MJ. ACS Nano. 2010;4:1433–1442. doi: 10.1021/nn901374b. [DOI] [PubMed] [Google Scholar]
- 50.Dam DHM, Culver KSB, Odom TW. Mol Pharm. 2014;11:580–587. doi: 10.1021/mp4005657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kang WJ, Chae JR, Cho YL, Lee JD, Kim S. Small. 2009;5:2519–2522. doi: 10.1002/smll.200900848. [DOI] [PubMed] [Google Scholar]
- 52.Hu DH, Zhang PF, Gong P, Lian SH, Lu YY, Gao DY, Cai LT. Nanoscale. 2011;3:4724–4732. doi: 10.1039/c1nr10933b. [DOI] [PubMed] [Google Scholar]
- 53.Zhang PH, Cheng FF, Zhou R, Cao JT, Li JJ, Burda C, Min QH, Zhu JJ. Angew Chem Int ed. 2014;53:2371–2375. doi: 10.1002/anie.201308920. [DOI] [PubMed] [Google Scholar]
- 54.Xing H, Tang L, Yang XJ, Hwang K, Wang WD, Yin Q, Wong NY, Dobrucki LW, Yasui N, Katzenellenbogen JA, Helferich WG, Cheng JJ, Lu Y. J Mater Chem B. 2013;1:5288–5297. doi: 10.1039/C3TB20412J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li LY, Hou JJ, Liu XJ, Guo YJ, Wu Y, Zhang LH, Yang ZJ. Biomaterials. 2014;35:3840–3850. doi: 10.1016/j.biomaterials.2014.01.019. [DOI] [PubMed] [Google Scholar]
- 56.Ahmed LA, El-Maraghy SA. Biochem Pharmacol. 2014;92:517–517. [Google Scholar]
- 57.Pestalozzi BC, Sotos GA, Choyke PL, Fisherman JS, Cowan KH, Oshaughnessy JA. Cancer. 1993;71:1797–1800. doi: 10.1002/1097-0142(19930301)71:5<1797::aid-cncr2820710514>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 58.Smith L, Watson MB, O’Kane SL, Drew PJ, Lind MJ, Cawkwell L. Mol Cancer Ther. 2006;5:2115–2120. doi: 10.1158/1535-7163.MCT-06-0190. [DOI] [PubMed] [Google Scholar]
- 59.Le Trinh T, Zhu GZ, Xiao XL, Puszyk W, Sefah K, Wu QF, Tan WH, Liu C. PLoS One. 2015;10:17. doi: 10.1371/journal.pone.0136673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weaver J, Gaillard S, Toye C, Macpherson S, Nolan SP, Riches A. Chem Eur J. 2011;17:6620–6624. doi: 10.1002/chem.201100321. [DOI] [PubMed] [Google Scholar]
- 61.Rubbiani R, Can S, Kitanovic I, Alborzinia H, Stefanopoulou M, Kokoschka M, Monchgesang S, Sheldrick WS, Wolfl S, Ott I. J Med Chem. 2011;54:8646–8657. doi: 10.1021/jm201220n. [DOI] [PubMed] [Google Scholar]
- 62.Liu WK, Bensdorf K, Proetto M, Hagenbach A, Abram U, Gust R. J Med Chem. 2012;55:3713–3724. doi: 10.1021/jm3000196. [DOI] [PubMed] [Google Scholar]
- 63.Modica-Napolitano JS, Aprille JR. Adv Drug Deliver Rev. 2001;49:63–70. doi: 10.1016/s0169-409x(01)00125-9. [DOI] [PubMed] [Google Scholar]
- 64.Rubbiani R, Salassa L, de Almeida A, Casini A, Ott I. Chemmedchem. 2014;9:1205–1210. doi: 10.1002/cmdc.201400056. [DOI] [PubMed] [Google Scholar]
- 65.Walter NG, Burke JM. RNA-Pub RNA Soc. 1997;3:392–404. [PMC free article] [PubMed] [Google Scholar]
- 66.Dailey MM, Miller MC, Bates PJ, Lane AN, Trent JO. Nucleic Acids Res. 2010;38:4877–4888. doi: 10.1093/nar/gkq166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tseng TY, Wang ZF, Chien CH, Chang TC. Nucleic Acids Res. 2013;41:10605–10618. doi: 10.1093/nar/gkt814. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


