Abstract
Spaceflight is known to affect immune cell populations. In particular, splenic B cell numbers decrease during spaceflight and in ground-based physiological models. Although antibody isotype changes have been assessed during and after space flight, an extensive characterization of the impact of spaceflight on antibody composition has not been conducted in mice. Next Generation Sequencing and bioinformatic tools are now available to assess antibody repertoires. We can now identify immunoglobulin gene- segment usage, junctional regions, and modifications that contribute to specificity and diversity. Due to limitations on the International Space Station, alternate sample collection and storage methods must be employed. Our group compared Illumina MiSeq sequencing data from multiple sample preparation methods in normal C57Bl/6J mice to validate that sample preparation and storage would not bias the outcome of antibody repertoire characterization. In this report, we also compared sequencing techniques and a bioinformatic workflow on the data output when we assessed the IgH and Igκ variable gene usage. This included assessments of our bioinformatic workflow on Illumina HiSeq and MiSeq datasets and is specifically designed to reduce bias, capture the most information from Ig sequences, and produce a data set that provides other data mining options. We validated our workflow by comparing our normal mouse MiSeq data to existing murine antibody repertoire studies validating it for future antibody repertoire studies.
Keywords: Immunoglobulin Gene Use, Next Generation Sequencing, Bioinformatics
INTRODUCTION
For B cell development and specificity, there are a large number of heavy chain (IgH) and kappa light chain (Igκ) gene segments that are used to produce the Ig (antibody) receptor population repertoire (de Bono et al., 2004). This antibody repertoire is quite large and the possible specificities can theoretically exceed the number of actual antibody molecules in the host (Georgiou et al., 2014). In antibodies, the antigen binding region is formed by six complementarity determining regions (CDRs) that loop out from the V region backbone formed by two beta-pleated sheets (Haidar et al., 2014; Saul and Poljak, 1992). The germline V gene segment repertoire is necessary for host responses to pathogens and CDR1 and CDR2 are completely encoded for by variable (V region) gene segments (Lu et al., 2014). Therefore, knowing which V gene segments are utilized is fundamental to understanding B-cell specificity and the development of effective immune responses. The CDR3 of both the heavy and light chains are highly variable due to their unique generation. During the creation of each Ig sequence, partially random splicing between V, D (heavy chain), and J gene segments occurs and random base insertions occur, called “n-nucleotide” additions. One hypothesis is that V gene segments have been maintained in the genome because of their importance in binding specific pathogens and provide essential host defense functions. However, CDR3 may be important because it is highly variable. Its role as a highly diverse and variable region is what provides the essential key to determining antigen specificity (Xu and Davis, 2000).
The spaceflight environment can impact many parameters critical to the host immune response. In multiple species spaceflight affects the total body, thymus and spleen mass (Allebban et al., 1996; Chapes et al., 1999; Congdon et al., 1996; Durnova et al., 1976; Grindeland et al., 1990; Grove et al., 1995; Jahns et al., 1992; Nash and Mastro, 1992; Pecaut et al., 2000; Serova, 1980; Udden et al., 1995; Wronski et al., 1998), circulating corticosterone (Blanc et al., 1998; Chapes et al., 1999; Lesnyak et al., 1993; Meehan et al., 1993; Merrill et al., 1992; Stein and Schluter, 1994; Stowe et al., 2001a; Stowe et al., 2001b; Stowe et al., 1999; Wronski et al., 1998), mitogen-induced proliferation, cytokine production and reactivity (Bikle et al., 1994; Chapes et al., 1999; Cogoli et al., 1990; Fuchs and Medvedev, 1993; Gould et al., 1987; Grigoriev et al., 1993; Hughes-Fulford, 1991; Konstantinova et al., 1973; Lesnyak et al., 1996; Lesnyak et al., 1993; Mandel and Balish, 1977; Miller et al., 1995; Nash et al., 1992; Nash and Mastro, 1992; Sonnenfeld et al., 1998; Sonnenfeld et al., 1992; Sonnenfeld et al., 1990; Stein and Schluter, 1994; Taylor and Dardano, 1983; Taylor et al., 1986), and lymphocyte subpopulation distributions (Allebban et al., 1994; Berry, 1970; Ichiki et al., 1996; Meehan et al., 1992; Pecaut et al., 2000; Sonnenfeld et al., 1998; Sonnenfeld et al., 1992).
B cells are among the immune components that are affected by spaceflight. The number of B cells in the spleen was reduced in mice flown on the space shuttle flight, STS-35 (Gridley et al., 2009). The percentage of B cells in the bone marrow and spleen also is reduced in mice subjected to hindlimb unloading (Gaignier et al., 2014; Lescale et al., 2015). When rats were injected with sheep red blood cells 8 days prior to an 18.5-day COSMOS flight there were lower IgG concentrations compared to both immunized and non-immunized ground controls after landing (Lesnyak et al., 1993). IgM production was virtually eliminated in lymphocytes stimulated in vitro with pokeweed mitogen (PWM) on the International Space Station (ISS) when compared to similarly activated ground controls. Furthermore, when cells were activated on Earth, frozen down, and then put back into suspension in space to assess IgM secretion, Fitzgerald et al. found that IgM secretion was significantly slower than similarly treated ground-based controls (Fitzgerald et al., 2009).
Our group is interested in the impact of spaceflight on B-cell immunoglobulin gene usage. Next Generation Sequencing (NGS) now allows us to analyze the repertoire of Ig gene segments that are used in the assembly of immunoglobulins that are transcribed by B cells and that are present in the host. NGS allows the determination of V, D and J gene usage, CDR3 assembly and the assessment of mutations that occur in response to immune challenge. In microgravity, there exist certain limitations associated with tissue collection and storage methods traditionally used on the ground. In preparation for sending mice to the ISS our group needed to validate our procedures and our ability to obtain high-quality RNA that could be used for NGS for the assessment of Ig gene usage. Our group also sought to validate the usage of RNA extracted from whole tissue collected and stored with these limitations in mind. It was also necessary to develop a workflow that would facilitate the analysis of large amounts of data that would be generated during this project. In this manuscript, we describe the development of our workflow and validation of mouse NGS data for use with space flight experiments.
MATERIALS AND METHODS
Sample Preparation and RNA Extraction
Spleens were removed from four 11-week-old, specific pathogen-free, female C57BL/6J mice housed in the Division of Biology vivarium at Kansas State University. One-half of each spleen was homogenized with a 70 µM sieve to generate a single-cell suspension designated, “cells”. Spleen cells were pelleted at 350 × g and were resuspended in 5 mL of ice-cold ACK lysing buffer (155mM NH4Cl, 10mM KHC03, 0.1mM EDTA) to lyse red blood cells. After five minutes, 10 mL of ice-cold isotonic medium was added to the suspension and cells were again pelleted at 350 × g and the supernatant was removed. The pellet was resuspended in Trizol LS (Ambion, Carlsbad, CA, USA) for RNA extraction. The remaining spleen half was immediately placed into Trizol LS for RNA extraction, and designated “tissue”. RNA extraction was performed according to the manufacturer’s instructions. RNAsin (40 units) was added to each RNA aliquot and stored in −80°C. RNA quality was assessed on a 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA).
MiSeq Sequencing
One microgram of high-quality total RNA was used for RNA sequencing (RNA-seq) library construction using the TruSeq RNA Sample Preparation kit v2 (Illumina, San Diego, CA) with the following modification: one minute fragmentation time was applied to allow for longer RNA fragments. The obtained RNA-seq libraries were analyzed with the 2100 Bioanalyzer. The tissue sample described previously was then subjected to size selection and designated “size selected”. Selection of sequences 375–900 nucleotides (nt) in length (275–800 nt sequences plus 50 nt sequencing adaptors on each end of the cDNA) was performed using the Pippin Prep system (Sage Science, Beverly, MA, USA). All libraries were then quantified with qPCR according to Illumina recommendations. The sequencing was performed at the Kansas State University Integrated Genomics Facility on the MiSeq personal sequencing system (Illumina) using the 600 cycles MiSeq reagent v3 kit (Illumina) according to Illumina instructions, resulting in 2 × 300 nt reads.
MiSeq Reference Mapping
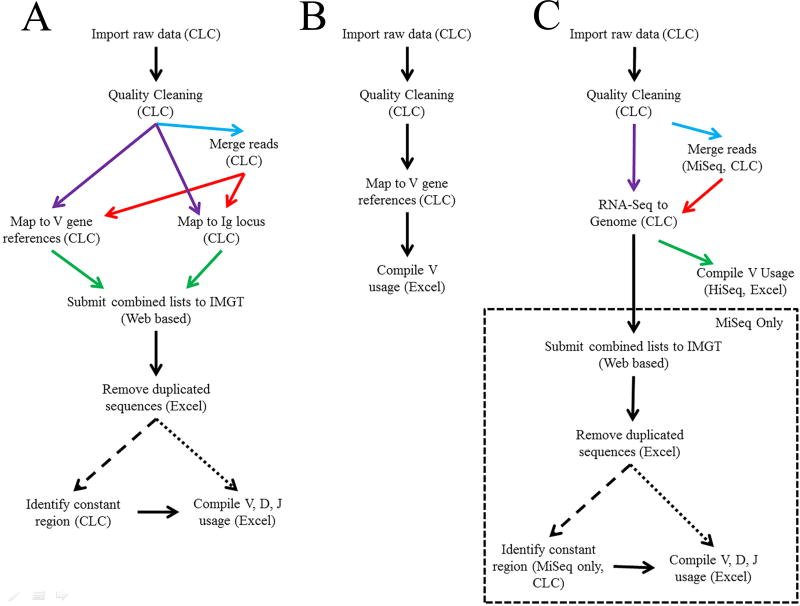
The bioinformatics workflow used in our study is outlined in Figure 1. FASTQ sequencing files were imported into CLC Genomics Workbench v9.5.1 (CLC bio, Aaarhus, Denmark) (https://www.qiagenbioinformatics.com/). Data were cleaned in the CLC program to remove low quality and short sequences. Due to Illumina sequencing artifacts, the first 12 nt were removed from each sequence. Sequences were quality cleaned by retaining the longest region of the sequences with at least 97% of the sequence with Phred scores over 20. Sequences with fewer than 40 nt were removed. Reads remained with paired-end sequences, designated “paired”, and, in cases of overlapping sequence pairs, reads were merged, designated as “merged” (Figure 1a, light blue line). Sequences were merged using a match score of +1, mismatch cost of −2, a gap cost of −3, and a minimum score of 10. Cleaned paired (Figure 1a, purple lines) and merged (Figure 1a, red lines) sequences were mapped to specific C57BL/6 V gene sequences obtained from NCBI for the immunoglobulin heavy (IgH) and light (Igκ) chains. The paired and merged sequences were mapped using a match score of +1 and a mismatch score of −2. Additional putative antibody sequences were obtained by mapping sequences to the IgH and Igκ loci and the whole genome using the same scores. Mapped MiSeq sequences were combined and submitted to ImMunoGeneTics’s (IMGT) High-V Quest (Figure 1a, green lines) (Alamyar et al., 2012). Functionally productive heavy chain sequences were imported into CLC and constant regions were determined using a motif search for the first 20 nt in each constant region that are provided in Table 1 (dashed line). Motifs were reassociated with their original sequences in Microsoft Excel for complete antibody (V[D]JC) identification. While two IgG subclass motifs were used to identify IgGs, they share partial sequence homology, therefore all IgG subclasses were combined resulting in an overarching IgG isotype. Kappa chain sequences were processed directly (Figure 1a, dotted line).
Figure 1. Bioinformatic analysis workflows.
(A) Workflow for MiSeq reference mapping strategy using CLC Genomics Workbench software, the ImMunoGeneTics (IMGT) data base and Excel. (B) Workflow for HiSeq reference mapping strategy. (C) Workflow for MiSeq and HiSeq referenced mapping strategy.
Table 1. Sequences used for heavy chain identification.
Motifs used to determine the constant region of heavy chain Ig sequences.
| Constant Region | Motif Sequence |
|---|---|
| IgA | GAGTCTGCGAGAAATCCCAC |
| IgD | GTAATGAAAAGGGACCTGAC |
| IgE | TCTATCAGGAACCCTCAGCT |
| IgG1/2b/2c | GCCAAAACAACAGCCCCATC |
| IgG3 | AACAACAGCCCCATCGGTCT |
| IgM | TCAGTCCTTCCCAAATGTCT |
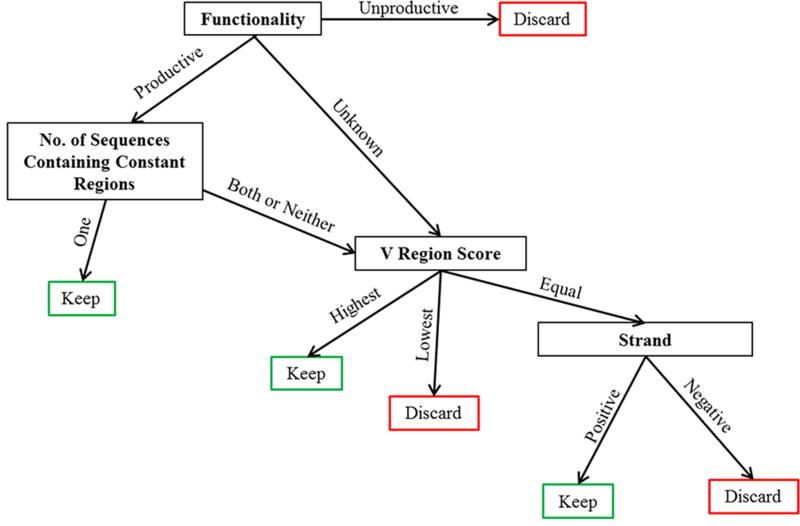
To collect a higher number of putative Ig sequences, our data handling workflow used multiple mapping processes which could result in the same sequence being submitted to IMGT multiple times. Failure to remove these duplicated sequences would lead to incorrect frequency assessments. Figure 2 outlines the procedure for duplicate sequencing read removal. Sequences were identified by their original MiSeq identification numbers for sorting. To retain the sequence with the most information and most accurate mapping, sequences were assessed based on functionality, constant region identification, V region score, and strand. Only one sequence per MiSeq identification number was retained and used for subsequent data compilation. Data from the remaining productive and unknown functionality antibody sequences were compiled for V, D (IgH only), and J gene segment usage, CDR3 length, and CDR3 amino acid (AA) composition.
Figure 2. Decision-making matrix to remove duplicate sequence reads after IMGT processing.
Mapped sequences that were identified using Illumina sequence identification tags and sequences identified multiple times were removed as outlined.
HiSeq Reference Mapping
The MiSeq workflow described above was modified to analyze mouse liver transcriptomic data from the Rodent Research 1 (RR1) NASA validation flight provided by the NASA GeneLab program (https://genelab-data.ndc.nasa.gov/genelab/, Accession Numbers: GLDS-47, GLDS-48). These data include sequences from the livers of ground control and flight mice from two separate cohorts, CASIS (GLDS-47) and NASA (GLDS-48), that were generated using Illumina HiSeq (1 × 50 nt reads). Raw sequencing reads were imported into CLC and quality cleaned as described with the exception of short read removal as quality cleaned reads were below the threshold utilized in the original workflow (Figure 1b). Reads were then mapped to the Vκ references identified above. Total Vκ mapping counts were collected and analyzed in Excel.
MiSeq and HiSeq Genome Mapping
FASTQ files were imported into CLC and quality cleaned as described previously (Figure 1c). MiSeq reads were merged as described previously (Figure 1c, blue arrow). Paired and merged MiSeq and unpaired HiSeq reads (Figure 1c, purple arrow) were mapped using the RNA-Seq tool in CLC to the current mouse reference genome (GRCm38). A match score of +1, a mismatch score of −2, and insertion and deletion scores of −3 were used to map reads to the genome. Due to the short read lengths of the HiSeq data, V gene segment usage was compiled directly after the RNA-Seq analysis (Figure 1c, green arrow). For MiSeq data, reads were collected, submitted to IMGT, duplicates removed and usage statistics compiled as described above (Figure 1c, dashed box).
NASA RNA Preparation and Sequencing
Tissues from two sets of mice were analyzed. The first set of spleens and livers were removed from five 35-week-old female C57BL/6Tac mice aboard the ISS 21–22 days after launch (CASIS Flight, SpaceX-4). Five 35-week old female mice housed in the ISS Environmental Simulator were processed similarly with a four-day delay (CASIS Ground Controls) (Figliozi, 2014). Spleens and livers were placed in RNALater (LifeTechnologies, Carlsbad, CA) for at least 24 hours at 4°C and then stored at −80°C while aboard the ISS, during transport, and upon return to Earth. The second set of tissues were isolated from seven 21-week-old female C57BL/6J were euthanized aboard the ISS 37 days post launch (NASA Flight, SpaceX-4). Carcasses were immediately frozen (−80°C) and after arriving at Earth, were dissected. Livers were preserved in RNALater for at least 24 hours at 4°C and then frozen at −80°C. RNA was extracted from the tissues using Trizol (LifeTechnologies, Carlsbad, CA) according to the manufacturer’s instructions. The resultant RNA was then processed through an RNeasey mini column (QIAGEN, Hilden, Germany) as per manufacturer’s instructions. RNA quality was assessed on a 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA) and stored at −80°C. RNA was sequenced on Illumina HiSeq with single reads of 50nt (1× 50nt).
RESULTS
Size Selection Yields Highest Number of Antibody Sequences
Most studies of Ig gene-segment use frequency have used single-cell suspensions or sorted to isolate specific cell populations (Aoki-Ota et al., 2012; Collins et al., 2015; Kaplinsky et al., 2014; Kramer et al., 2016; Lu et al., 2014). The advantage of these preparations is the exclusion of extraneous tissues and cells, which can enhance recovery of Ig sequences.
Our goal was to assess Ig gene segment usage in mice housed on the ISS. Due to limitations of animal and tissue handling on the ISS, only whole tissues would be available for analysis. To determine if we could obtain a sufficient number of Ig sequences from alternative sample preparations, we examined the total Ig sequence results from three different RNA preparation and sequencing treatment groups. The first two treatment groups comprised of RNA prepared from cells or whole tissue. A third treatment group was the same RNA from the tissue, which was subsequently size selected at 275–800 nt and sequenced independently. We selected this range of lengths because the IgH VDJ recombination sequences are generally require 350–450 nt to gather information on V/D/J/C usage. Size selection also allows us to eliminate the inherent Illumina bias for short reads, while maintaining total transcriptome integrity for later data mining purposes.
Total sequence numbers generated were similar among the treatment groups with the cells treatment group resulting in 23.9 million reads, tissue treatment group with 25.9 million, and size selected treatment group with 21.5 million reads, as shown in Table 2. After cleaning, 18.7 million reads remained in the cells treatment group, 20.3 million in the tissue treatment group, and 12 million in the size selected group (Table 2). Mapping was performed as described in the materials and methods for both the IgH and Igκ loci and VH- and Vκ-gene segments. Locus mapping returned higher levels of probable Ig sequences than V-gene segment specific mapping. V-gene segment IgH- and Igκ-mapped sequences were lowest in the cells treatment group (1.56% IgH and 1.38% Igκ) and highest in the size selected treatment group (3.1% IgH and 1.99% IGκ).
Table 2. Sequencing and mapping results from the cells, tissue, and size selected treatment groups.
| Cells | Tissue | Size Selected | |
|---|---|---|---|
| Total Reads | 23.9 M | 25.9 M | 21.5 M |
| Post Cleaning | 18.7 M | 20.3 M | 12M |
| IgH Mapped | 278318 | 313194 | 327015 |
| VH Mapped | 12851 | 26559 | 42375 |
| Igκ Mapped | 261037 | 273562 | 264938 |
| Vκ Mapped | 20776 | 35719 | 57493 |
| Heavy Chain Productive | 2036 | 4991 | 8939 |
| Heavy Chain Unknown | 6139 | 11374 | 14047 |
| Light Chain Productive | 3439 | 6799 | 24812 |
| Light Chain Unknown | 6894 | 10462 | 45530 |
After submission to IMGT and cleaning, antibody sequences were identified as potentially functional or of unknown functionality by IMGT. Unknown functionality sequences were comprised of partial sequences lacking CDR3 information to determine functionality. The total number of antibody sequences obtained for the IgH and Igκ was lowest in the cells treatment group and highest in the size selected treatment group, (Table 2) mirroring the V-gene segment mapping results. Both productive and unknown IgH and Igκ antibody sequence counts were also lowest in the cells treatment group and highest in the size selected treatment group. This trend also described Igκ sequences, with more unknown than productive sequences being identified. The size selected treatment group produced both the highest number of productive antibody sequences and the highest total number of identified Ig sequences among treatment groups.
Comparison of Immunoglobulin Gene Segment Usage Among Treatment Groups
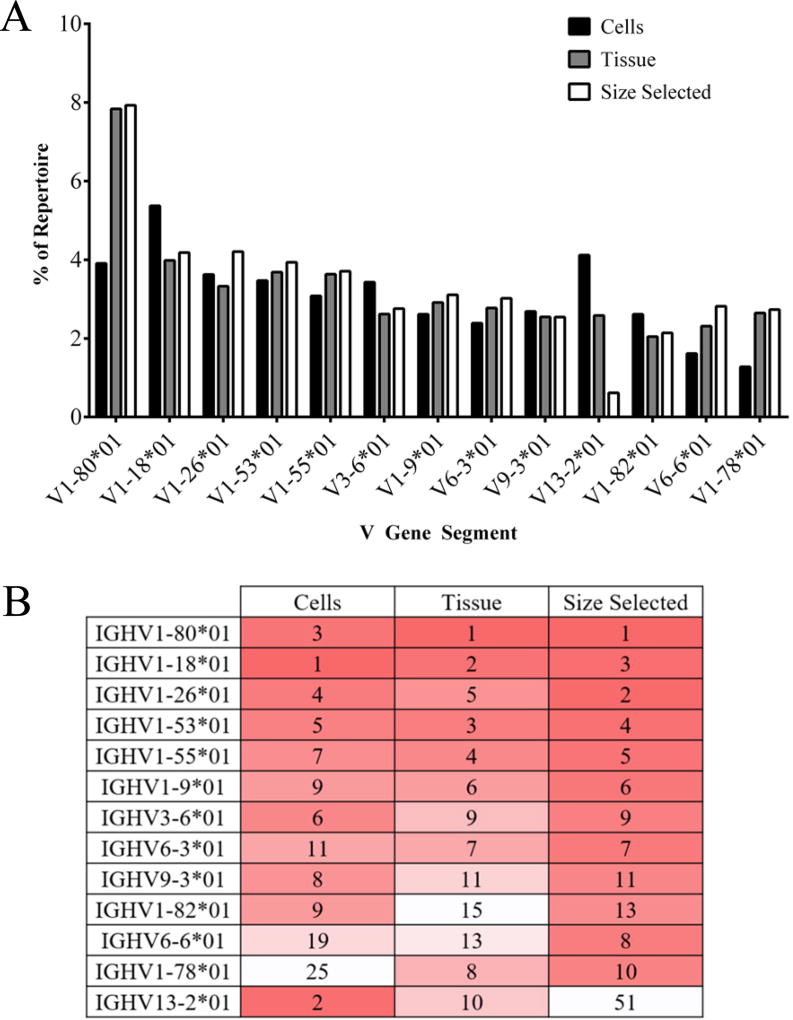
We compared IgH and Igκ gene segment frequency using multiple metrics across all three treatment groups; the first of which is V gene segment usage. To assess the frequency of each VH- and Vκ- gene segment in normal mouse spleen, the total frequency of each V-gene segment was tabulated in Figures 3 and 4 as a percentage of the total repertoire for our cells, tissue, and size selected treatment groups. VH-gene segment usage was similar among all three treatment groups (Figure 3A). V-gene segment V1–80 was detected most frequently, followed by V1–18, and V1–26 gene segments. The gene segment V1–80 was ranked either first or third as a percentage of the total repertoire in all three treatment groups (Figure 3B). The gene segment V1–18 ranged between the first and third most frequently used gene segment. V1–26 was the second to fifth most frequently used VH-gene segment. While gene frequency detection rankings among cells, tissue and size selected treatment groups were not identical, there was high similarity in overall VH-gene segment detection and in repertoire usage. Correlations between treatment groups produced an R2 of at least 0.7562 (p<0.0001, data not shown) between the cells and size selected treatment groups. The R2 between cells and tissue treatment group was higher (R2=0.8149, p<0.0001, data not shown). Tissue and size selected treatment groups had the highest correlation (R2= 0.9645, p<0.0001 data not shown).
Figure 3. Top ten VH gene segments used among treatment groups.
(A) The top ten VH gene segments for each treatment group are presented as a percent of repertoire with corresponding percent of repertoire in other treatment groups listed. (B) Top ten VH gene segments are listed by rank order (most frequent to least frequent). Dark red indicates higher rank moving to white, of lower rank. VH-gene segments with identical ranks are displayed as ties.
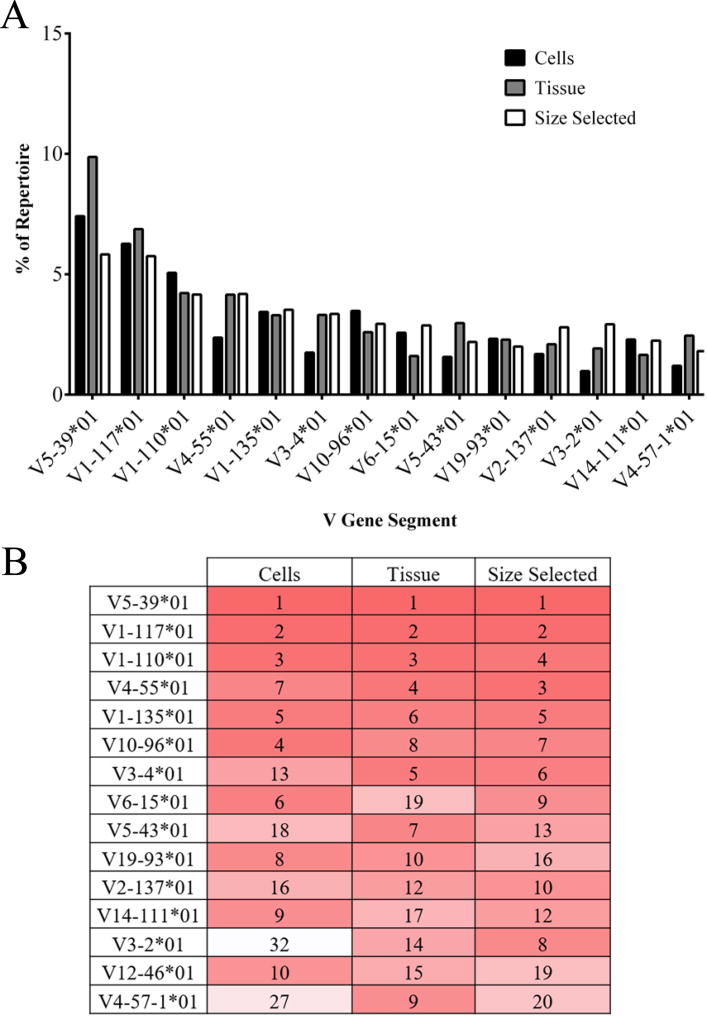
Figure 4. Top ten Vк used among treatment groups.
(A) The top ten Vк gene segments for each treatment group are presented as a percent of repertoire with corresponding percent of repertoire in other treatment groups listed. (B) The top ten Vк gene segments are listed by rank order (most frequent to least frequent). Dark red indicates higher rank moving to white, lower rank. VH-gene segments with identical ranks are displayed as ties.
Kappa chain V-gene segment usage was also compared among the different treatment groups. Figure 4 shows the percent of repertoire for the top ten most abundant Vκ gene-segments of each treatment group. There was significant overlap in the top Vκ of each treatment group when assessed as either percent of repertoire detected (Figure 4A) or when ranked from highest to lowest frequency (Figure 4B). Greater similarities in Vκ existed between tissue and tissue size selected treatments. Correlations between Vκ in treatment groups produced an R2 of at least 0.7813 (p<0.0001, data not shown), with tissue and size selected treatment groups having the highest correlation (R2=0.8335, p<0.0001, data not shown).
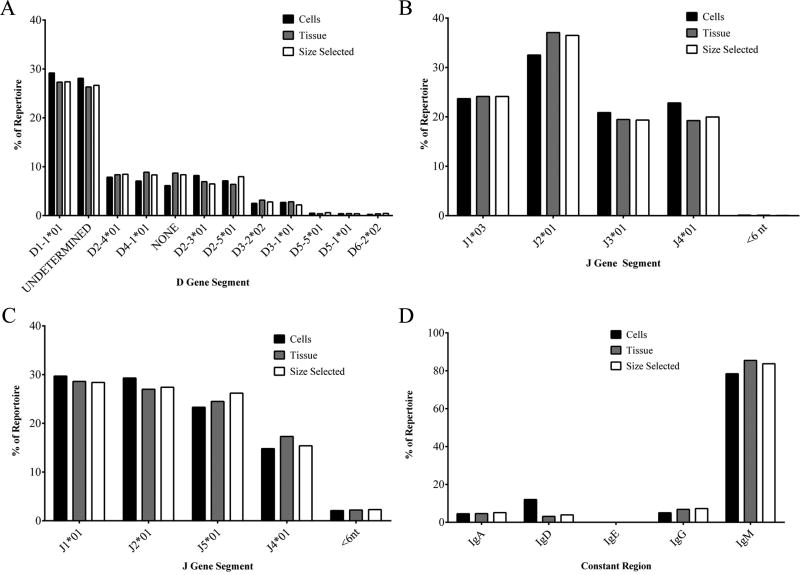
The frequency of D- and JH-gene segment use in normal mice was also assessed. Figure 5 shows that the cells, tissue and size selected D- and JH-gene-segment usage frequency was similar. D1-1 was the most frequently discovered D-gene segment in all three treatment groups, comprising almost 30% of the repertoire (Figure 5A). Due to the short length of the D gene, it was often difficult to properly determine which D gene was used in an antibody. When a D gene was identified, but was attributed to a non-strain-specific D gene, they were labeled “undetermined”. These D-gene segments were also very common, occurring between 26%-28% of the time, in all three treatment groups. Gene segments D2–4, D4-1, D-5, D2–3, and “no” D-gene (labeled NONE) were the next most frequent assignments, ranging from about six percent to eight percent of D-gene frequency.
Figure 5. D, J, and heavy chain constant usage among treatment groups.
(A) DH gene segment usage by percent of repertoire. (B) JH gene segment usage by percent of repertoire. (C) Jк gene segment usage by percent of repertoire. (D) Heavy chain constant region usage by percent of repertoire.
We found that JH-gene segment usage was the same for the cells, tissue and size selected treatment groups (Figure 5B). JH2 was the most frequently used J-gene segment followed by JH1, JH4, and JH3, respectively. Gene segment usage in kappa chains was also similar in all three treatment groups, with Jκ1 as the most frequently used, followed by Jκ5, Jκ2 and Jκ4 respectively (Figure 5C). When less than six nucleotides from the J gene segment were identified, they were marked as <6 nt.
Five heavy chains, IgA, IgD, IgE, IgG (all subfamilies), and IgM are part of the normal mouse Ig repertoire. Almost 80% of the repertoire used the IgM constant region (Figure 5D). IgD, IgA, and IgG were detected at frequencies between three percent and twelve percent of the total repertoire and were evenly distributed among cells, tissue and size selected treatment groups. IgE was not detected in any of the treatment groups.
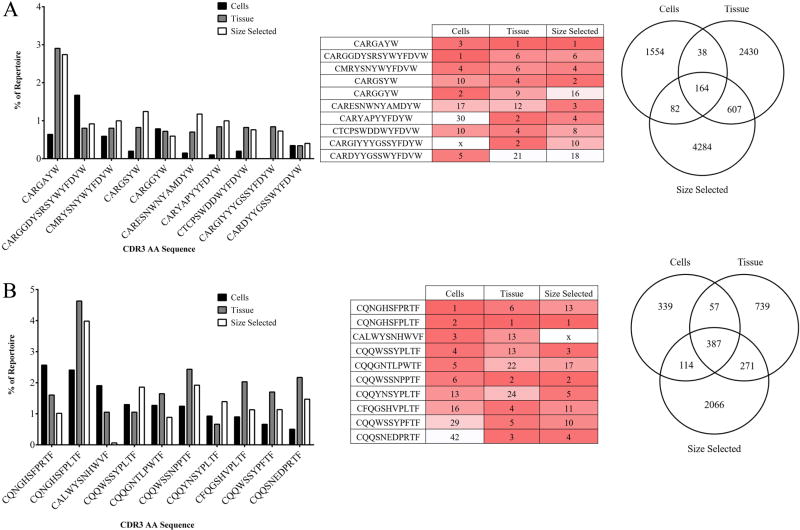
CDR3 AA Sequence Determination
CDR3 is highly variable and it may be critical in determining antigen specificity (Xu and Davis, 2000). Therefore, we assessed individual CDR3 frequency from each treatment group. The top five most common CDR3s from each treatment group for the heavy chain were compiled and shown in Figure 6A, resulting in a total of ten unique CDR3s among treatment groups. The tissue and size selected treatment groups contained all of the most common CDR3s, however, the cells treatment group lacked the CARGIYYGSYFDYW sequence, which ranked as the second most common in the tissue data set (Figure 6B). We detected one hundred sixty-four CDR3 AA sequences in all three data sets at least once (Figure 6C). The tissue and size selected treatment data sets shared 607 CDR3 sequences. Thirty-eight and 82 CDR3 AA sequences were shared between cells and tissue and cells and size selected treatment groups, respectively. Each treatment group data set also contained a large number of unique CDR3 reads.
Figure 6. CDR3 AA sequence usage among treatment groups.
(A) Top five most common heavy chain CDR3 AA sequence usage is presented as percent of repertoire. (B) Top five most common heavy chain CDR3 AA sequence usage is presented by rank. Dark red indicates higher rank moving to white, of lower rank. An x denotes that the AA sequence was not found. (C) Unique heavy chain CDR3 AA sequences identified within and among treatment groups. (D) Top five most common kappa chain CDR3 AA sequence usage is presented as percent of repertoire. (E) Top five most common heavy chain CDR3 AA sequence usage is presented by rank. (F) Unique heavy chain CDR3 AA sequences identified within and among treatment groups.
We found overlap among the top five kappa chain CDR3 of each treatment group (Figure 6D). One CDR3 sequence was not found in all three datasets, CALWYSNHWF, however, all other top CDR3 sequences appeared in at least the top 42 CDR3 sequences of the other treatment groups (Figure 6E). Figure 6F shows the diversity of kappa chain CDR3 sequences. The total number of CDR3 amino acid sequences that were unique to each treatment groups was 957, 1454, and 2838 in cells, tissue, and tissue size selected treatment groups, respectively. Three-hundred and eighty-seven unique kappa chain CDR3 sequences are shared among all three treatment groups.
Application of MiSeq Workflow to RR1 HiSeq Data
The MiSeq workflow was adapted to process the liver Illumina HiSeq data from the RR1 validation flight. Due to short read length (38 nt), only V-gene segment usage was assessed. Due to low IgH read numbers, only Vκ information is presented. The Vκ percent of repertoire from each HiSeq RR1 mouse cohort (CASIS Ground, CASIS Flight, NASA Ground, and NASA Flight) was compared to the Vκ percent abundance of the cell, tissue, and size selected HiSeq datasets. Table 3 shows poor correlation between reference mapped RR1 HiSeq cohorts and the MiSeq datasets described in the previous section. As all mice represented in this comparison are C57BL/6 mice, though ages and experimental conditions varied, we were concerned that the lack of concurrence in Vκ gene-segment usage of the RR1 mice and those used in the workflow discussed above may reflect the bioinformatic treatment of the data. To test this hypothesis, we modified the workflow to map sequencing reads to the entire Mus musculus genome rather than mapping reads to Vκ reference sequences, as genome mapping is commonly employed in transcriptomics analysis. This bioinformatic treatment yielded a higher correlation with the MiSeq datasets. Table 3 shows that the distribution of Vκ percent abundance was dependent on the mapping technique used for the HiSeq datasets.
Table 3. Comparison of mapping techniques in HiSeq datasets.
| Reference2 | Genome3 | Compared4 | ||
|---|---|---|---|---|
|
| ||||
| Cohort | N | R2 (p-value) | R2 (p-value) | R2 (p-value) |
| CASIS G | 3 | 0.026 (.086) | 0.195 (<.0001) | 0.011 (.027) |
| CASIS F | 3 | 0.001 (.776) | 0.282 (<.0001) | 0.042 (.074) |
| NASA G | 7 | 0.021 (.780) | 0.440 (<.0001) | 0.013 (.262) |
| NASA F | 7 | 0.006 (.419) | 0.375 (<.0001) | 0.006 (.476) |
Mapping techniques were compared by assessing the correlation of Vκ usage between multiple HiSeq and Miseq datasets. HiSeq datasets included sequencing data from CASIS and NASA ground (G) or flight (F) RR1 mice. The comparison groups are as follows:
Vκ gene segment usage from reference-mapped HiSeq data versus Vκ gene segment usage of MiSeq sample preparation datasets.
Vκ gene segment usage from genome-mapped HiSeq data versus Vκ gene segment usage of MiSeq sample preparation datasets.
Vκ gene segment usage from reference-mapped HiSeq Data versus Vκ gene segment usage of genome-mapped HiSeq data.
Reference/IgH Locus Mapping is Comparable to Whole Genome RNA-Seq Methods
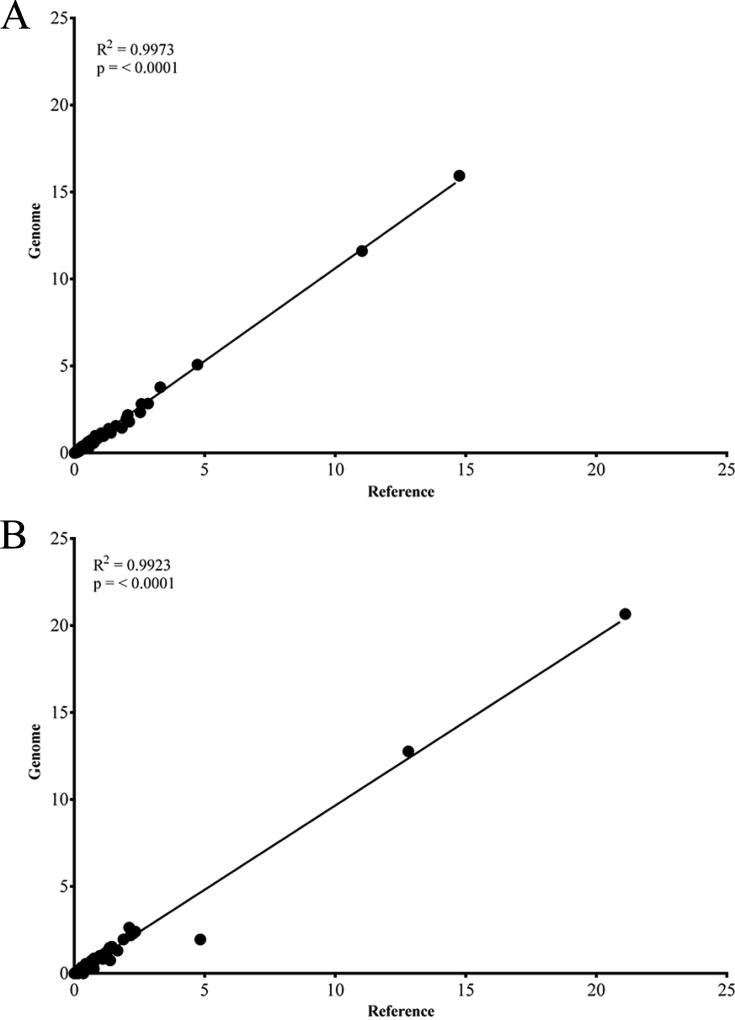
Because the data obtained from the HiSeq data set using reference mapping techniques were less correlative to Vκ gene use to our previously obtained MiSeq data for normal mice we were concerned that our initial bioinformatics techniques for MiSeq data may not be appropriate. To validate the accuracy of our bioinformatic treatments of the sequencing data that were submitted to IMGT, two different mapping methods were compared. The reference mapping approach, used previously, mapped sequences to both the IgH V-gene segments (251 segments) and the entire IgH locus (2.8Mb) obtained from NCBI (NC_000078.6, 113258768 to 116009954) or Igκ V-gene segments (164 segments) and the entire Igκ locus (3.2Mb) obtained from NCBI (NC_000072.6, 67555636 to 70726754). Therefore, we used the whole genome mapping outlined above to compare to our reference mapping. Results were obtained by using the RNA-Seq analysis tool in CLC to map reads to the entire genome with the IgH and Igκ loci selected for submission to IMGT. The IMGT output was processed similarly for both (genome vs. reference) mapping strategies. The median frequency of all VH- and Vκ-gene segments was compared if it was detected in at least one of the three treatment groups (cells, tissue or size-selected), and the data were compiled for both reference- and genome-mapping options. V-gene segments not detected in a treatment group were assigned a “zero” frequency. Assessment of the median frequencies of the two methods by linear regression in Figure 7 revealed that the frequency data for VH-gene segment usage was very similar regardless of the mapping technique (R2 = 0.9973, p = <0.0001) (Figure 7A). There was also a strong correlation of Vκ usage between the two methods (R2=0.9923, p<0.0001) (Figure 7B). Comparisons of D-gene segment, J-gene segment, constant region frequency and CDR3 lengths were also highly correlated using both techniques (data not shown). Therefore, we are confident that our reference mapping bioinformatics strategy is providing an accurate picture of V-gene usage.
Figure 7. Correlation of V gene segments between genome and reference mapping.
(A) Linear regression of median VH gene segment usage from genome and reference mappings. R2=0.9973, p<0.0001. (B) Linear Regression of median Vκ gene segment usage from genome and reference mapping. R2=0.9923, p<0.0001.
DISCUSSION
Spaceflight presents unique difficulties in the collection, preparation and preservation of cells and tissues. Normal preparation methods such as the creation of single-cell suspensions are difficult and normal tissue preservations methods such as the use of liquid nitrogen are unavailable. In an effort to determine the acceptability of whole tissue preparations compared to more traditional single cell suspensions, we examined the differences in Ig sequences obtained from both treatment groups. We were concerned that tissue isolation methods may introduce artifacts into the data since many studies specifically focus on single cell suspensions; often sorted, to isolate B cells specifically (Greiff et al., 2014; Kaplinsky et al., 2014; Kramer et al., 2016; Yang et al., 2015). Our data indicate that comparable results were obtained from both the tissue and the cells treatment groups. There were strong correlations in V-gene usage and the CDR3 sequences identities were very similar.
In an effort to reduce Illumina bias towards short reads seen in the cells and tissue treatment groups, ten months later, we sequenced the same tissue total RNA using size selection. This extended storage time after initial sequencing and additional freeze/thaw cycles are likely the cause of reduced numbers of post-cleaning reads due to RNA degradation. Nevertheless, the size-selected data set still provided the highest number of productive and unknown antibodies. Subsequent preparations have verified that size selection is helpful in providing the highest number of antibody sequences (data not shown). Therefore, we have chosen to include size selection in our protocol for NGS assessment of Ig V-gene usage.
The antibody repertoires from numerous species have been analyzed using a variety of amplification, sequencing, and analysis techniques (Benichou et al., 2012; Greiff et al., 2014). We chose to assess Ig gene usage without using amplification. Although many studies use amplification in order to obtain a higher number of reads, this may lead to bias into the repertoire (Benichou et al., 2012; Georgiou et al., 2014). Bias may be introduced due to primers or to errors created during the PCR reaction (Benichou et al., 2012; Lu et al., 2014). The large number of primers needed to amplify all the V genes in mice also presents some obstacles. Some have used 5’ RACE with primers based on the constant region (Benichou et al., 2012; Wang et al., 2006). However, in order to amplify the entire repertoire, multiple 5’ RACE primers are required, which still increases the chances for primer bias and increases costs significantly. Our goal was to examine the breadth of the antibody repertoire by gathering information about V, D, J, constant region usage and CDR3 composition. The detected V-gene and CDR3 sequences appear to parallel the repertoires reported using more focused amplification methods. Therefore, we have a methodology for future studies that will examine the immune response to vaccination.
During the course of our studies, we had the opportunity to work with both HiSeq and MiSeq data. While Illumina sequencing (HiSeq and MiSeq) produces a higher volume of sequence reads, they are shorter and more prone to errors than sequencing with 454 or Sanger methods (Benichou et al., 2012). However, Illumina sequencing has improved over time and is arguably now the NGS of choice. Our sequencing with Illumina MiSeq allowed us to obtain reads of up to 560 nt when forward and reverse sequencing ends were paired. This provides enough sequencing length to capture information from the V-gene segment to the constant region of both the heavy and light chains. We also found that as the sequences became longer, there was a drop off in sequencing quality, which has been previously reported (Minoche et al., 2011).
Our workflow for Ig sequence isolation selected for sequences with the most information. This required the merging of overlapping read pairs to provide sequences long enough to identify the V, D, J, and constant regions. In order to collect the highest number of possible Ig sequences, we used multiple mapping techniques to both the V-gene segment and the locus in an effort to collect every possible Ig sequence. Preliminary workflow attempts found that each mapping technique isolated some unique sequences and that locus mapping resulted in a high number of “false positive” sequences. Subsequent sequence removal in Excel selected for antibodies containing the most data retaining productive antibodies with constant regions and high V-gene scores, a measure of the length and accuracy of match to the germline V gene segment. By utilizing multiple mapping methods and subsequent selection for the sequence with the most information, we are able to obtain a relatively large number of antibody sequences without introducing primer bias or PCR-induced sequencing errors.
To the best of our knowledge, this is the first data set of tissue based, non-amplified MiSeq analysis of the antibody repertoire. While our results are not a direct technique match to other published data sets, our normal mouse V-gene usage is consistent with other’s findings. For example, Collins used 5’ RACE from the constant region followed by sequencing using 454 on a splenic cell suspension (Collins et al., 2015). Of the top ten VH-gene segments identified by Collins, we identified five of the same VH genes within our top ten most frequently detected. All except the V1–59 gene segment were among the highest contributors to our repertoire (Collins et al., 2015). In addition, JH-gene segment frequencies were also relatively uniform with the J2 gene segment as the most frequently used (Collins et al., 2015). Yang performed sequencing on cell sorted B cells isolated from the spleen followed by amplification with primers specific for many, but not all, of the V heavy chains of mice and constant region primers to amplify V, D, J and part of the constant region (Yang et al., 2015). Their PCR products were then sequenced on the Illumina platform and aligned to known VH-gene segments (Yang et al., 2015). They identified V1–26 as their most common V-gene segment, which ranked between the second and fifth most common V gene in our data sets. V-gene segments V1–82, V1–-64, and V1–55 were also identified as common V-gene segments in their analysis, all of which were frequently detected in our data (Yang et al., 2015). Kaplinski also examined sorted spleen cells, amplified with PCR and sequenced on MiSeq with 2 × 150 nt reads (Kaplinsky et al., 2014). Sequencing results were analyzed through idAB for identification (Kaplinsky et al., 2014). In contrast to our data analysis, Kaplinski examined only V gene segments found in productive antibodies, where we compiled all V gene segments identified in productive and unknown functionality sequences (Kaplinsky et al., 2014). Of the most common VH-gene segments provided, four, V1–80, V1–26, V1–53, and V1–82 were identified in our top ten grouping (Kaplinsky et al., 2014). Our data sets also isolated D1-1 as the most common D-gene segment. Kramer et al. examined sorted splenic follicular B cells, using IgM restricted PCR and sequenced using the Sanger method (Kramer et al., 2016). As we discovered, Kramer et al.’s most common VH family was V1, followed by V2 and V5 at relatively equal levels (Kramer et al., 2016). In contrast, we found that the V6 gene-segment family was detected at a higher level than found in the Kramer analysis (Kramer et al., 2016). The J4 gene segment was also used more than detected in our data set (Kramer et al., 2016). We both identified D1 and D2 as the most common D gene-segment families.
We also compared our data to Igκ gene family usage. Aoki-Ota assessed skewed Vκ gene segment usage and V-J gene segment usage in unimmunized C57BL/6 mice using primer amplified total RNA of B cells from spleen, bone marrow and lymph node using 454 pyrosequencing (Aoki-Ota et al., 2012). Their sequencing data was analyzed using the NCBI basic local alignment tool with reference sequences for Vκ and Jκ obtained from the IMGT data base. The top seven V gene segments identified in their study were also found to be among the most abundant Vκ gene segments in all of our treatment groups. Additionally, V-J pairing of their top gene segments paralleled our data. Lu examined the effects of primer bias and mouse to mouse variation in Vκ- and Jκ-gene segments and CDR3 regions using primer amplified total RNA isolates of Balb/c splenic B cells on the 454 pyrosequencing platform (Lu et al., 2014). As with our study, sequencing reads were submitted to the IMGT HighV-quest tool, however, only functionally productive immunoglobulins were used in their analysis (Lu et al., 2014). In spite of the strain differences between our studies, of the Vκ-gene segments representing over one-percent of the antibody repertoire reported by Lu et al., at least 80% of those gene segments appearing at a frequency 0.5% or higher in our analyses.
Although there was not 100% agreement among our study and the others, there was a high degree of consistency. Variations in data may result from sequencing and tissue isolation techniques and natural variation among animals, including mouse strain. In addition, since we did not amplify for IgV gene segments, we likely may have missed rarer B cell clones. Primer biases in other studies may have also contributed to some of the differences. Nevertheless, it is clear that our approach provided an unbiased, representative sample of actively transcribing B cells.
Our group utilized liver Rodent Research One sequencing data sequenced on the Illumina HiSeq platform (1 × 50 nt) that was available from the NASA Genelab project. The sequencing length was the largest limitation of these data. Our MiSeq data were sequenced in both directions at a length of 300 nt. Some paired-end reads also contained overlaps, allowing us to merge these sequences and provide reads up to around 560 nt. HiSeq sequencing reads were not of sufficient length to obtain CDR3 composition from the IMGT HighV-Quest tool, limiting the analyses that could be used to assess the antibody repertoire. Therefore, the applicability of publically available datasets to independent research questions is dependent upon the sequencing method used to acquire the data. For Ig gene repertoire studies, we recommend the use of sequencing methods that result in longer reads, though short reads may be useful for other research questions.
Initial comparisons to assess similarity of the different mouse cohorts showed a lack of correlation between the HiSeq RR1 data to the normal mouse MiSeq Vκ usage. We thought that part of the discrepancy may be from problems with the short HiSeq sequences, specifically when forced to align to V-gene segments when multiple matches are excluded. Mapping short HiSeq reads to the entire mouse genome remedied the inconsistencies observed between RR1 and normal mouse MiSeq data, likely due to the limitations of the RNA-Seq analysis employed. This demonstrates that the bioinformatic treatment of the data can impact results. We found that mapping longer MiSeq sequencing reads from RNA isolated from mice within the CASIS ground RR1 cohort to both the whole mouse genome and Vκ reference sequences yielded a strong correlation. This validates the applicability of the MiSeq workflow described in this work on additional MiSeq datasets and reinforces that sequence read length must be taken into account when selecting bioinformatics methods.
In conclusion, our goals for this project were to examine the breadth of the antibody repertoire gathering information about V, D, J, and constant region usage and CDR3 composition and to lay the foundation for future studies that will examine the immune response to vaccination during space flight. We have determined that whole tissue preparations as will be available from the ISS will yield similar results when examining the antibody repertoire. We also determined that performing a size selection to isolate likely antibody sequences provided the highest number of Ig reads. A novel workflow using multiple mapping methods to characterize NGS data for Ig repertoire data was developed and genome and reference mapping methods were validated through the use of publically available datasets. This novel workflow can be used for future studies on the antibody repertoire regardless of whether they are ISS- or ground-based.
Acknowledgments
This work was supported by NASA grants NNX13AN34G and NNX15AB45G, NIH grant GM103418, the Molecular Biology Core supported by the College of Veterinary Medicine at Kansas State University, and the Kansas State University Johnson Cancer Center. GeneLab data are from the NASA GeneLab Data Repository (http://genelab.nasa.gov/data). We thank Ms. Melissa Gulley for her help in the lab and Mr. Ricky J. Rettig for his help and expertise in Microsoft Excel. We also thank Drs. Ruth Globus and Sungshin Choi at NASA Ames for their help with RR1 data and Dr. Alina Akhunova, Director of the Kansas State University Integrated Genomics Facility, for her help, dedication and expertise.
References
- Alamyar E, Duroux P, Lefranc MP, Giudicelli V. IMGT((R)) tools for the nucleotide analysis of immunoglobulin (IG) and T cell receptor (TR) V-(D)-J repertoires, polymorphisms, and IG mutations: IMGT/V-QUEST and IMGT/HighV-QUEST for NGS. Methods Mol Biol. 2012;882:569–604. doi: 10.1007/978-1-61779-842-9_32. [DOI] [PubMed] [Google Scholar]
- Allebban Z, Gibson LA, Lange RD, Jago TL, Strickland KM, Johnson DL, Ichiki AT. Effects of spaceflight on rat erythroid parameters. J Appl Physiol. 1996;81:117–122. doi: 10.1152/jappl.1996.81.1.117. [DOI] [PubMed] [Google Scholar]
- Allebban Z, Ichiki AT, Gibson LA, Jones JB, Congdon CC, Lange RD. Effects of spaceflight on the number of rat peripheral blood leukocytes and lymphocyte subsets. J Leukoc Biol. 1994;55:209–213. doi: 10.1002/jlb.55.2.209. [DOI] [PubMed] [Google Scholar]
- Aoki-Ota M, Torkamani A, Ota T, Schork N, Nemazee D. Skewed primary Igkappa repertoire and V-J joining in C57BL/6 mice: implications for recombination accessibility and receptor editing. J Immunol. 2012;188:2305–2315. doi: 10.4049/jimmunol.1103484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benichou J, Ben-Hamo R, Louzoun Y, Efroni S. Rep-Seq: uncovering the immunological repertoire through next-generation sequencing. Immunology. 2012;135:183–191. doi: 10.1111/j.1365-2567.2011.03527.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry CA. Summary of medical experience in the Apollo 7 through 11 manned spaceflights. Aerosp Med. 1970;41:500–519. [PubMed] [Google Scholar]
- Bikle DD, Harris J, Halloran BP, Morey-Holton ER. Altered skeletal pattern of gene expression in response to spaceflight and hindlimb elevation. Am J Physiol. 1994;267:E822–E827. doi: 10.1152/ajpendo.1994.267.6.E822. [DOI] [PubMed] [Google Scholar]
- Blanc S, Somody L, Gharib A, Gauquelin G, Gharib C, Sarda N. Counteraction of spaceflight-induced changes in the rat central serotonergic system by adrenalectomy and corticosteroid replacement. Neurochem Int. 1998;33:375–382. doi: 10.1016/s0197-0186(98)00042-4. [DOI] [PubMed] [Google Scholar]
- Chapes SK, Simske SJ, Sonnenfeld G, Miller ES, Zimmerman RJ. Effects of space flight and PEG-IL-2 on rat physiological and immunological responses. J Appl Physiol. 1999;86:2065–2076. doi: 10.1152/jappl.1999.86.6.2065. [DOI] [PubMed] [Google Scholar]
- Cogoli A, Bechler B, Mueller O, Hunzinger E. BioRack on Spacelab D1. Paris: European Space Agency; 1990. Effect of microgravity on lymphocyte activation; pp. 89–100. Vol. ESA SP-1091. [Google Scholar]
- Collins AM, Wang Y, Roskin KM, Marquis CP, Jackson KJ. The mouse antibody heavy chain repertoire is germline-focused and highly variable between inbred strains. Philos Trans R Soc Lond B Biol Sci. 2015;370 doi: 10.1098/rstb.2014.0236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Congdon CC, Allebban Z, Gibson LA, Kaplansky A, Strickland KM, Jago TL, Johnson DL, Lange RD, Ichiki AT. Lymphatic tissue changes in rats flown on Spacelab Life Sciences-2. J Appl Physiol. 1996;81:172–177. doi: 10.1152/jappl.1996.81.1.172. [DOI] [PubMed] [Google Scholar]
- de Bono B, Madera M, Chothia C. VH gene segments in the mouse and human genomes. J Mol Biol. 2004;342:131–143. doi: 10.1016/j.jmb.2004.06.055. [DOI] [PubMed] [Google Scholar]
- Durnova GN, Kaplansky AS, Portugalov VV. Effect of a 22-day space flight on the lymphoid organs of rats. Aviation, Space, and Environmental Med. 1976;47:588–591. [PubMed] [Google Scholar]
- Figliozi GM. NASA's New Rodent Residence Elevates Research To Greater Heights. 2014 http://www.nasa.gov/mission_pages/station/research/news/rodent_research.
- Fitzgerald W, Chen S, Walz C, Zimmerberg J, Margolis L, Grivel JC. Immune suppression of human lymphoid tissues and cells in rotating suspension culture and onboard the International Space Station. In Vitro Cell Dev Biol Anim. 2009 doi: 10.1007/s11626-009-9225-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs BB, Medvedev AE. Countermeasures for ameliorating in-flight immune dysfunction. J Leukoc Biol. 1993;54:245–252. doi: 10.1002/jlb.54.3.245. [DOI] [PubMed] [Google Scholar]
- Gaignier F, Schenten V, De Carvalho Bittencourt M, Gauquelin-Koch G, Frippiat J-P, Legrand-Frossi C. Three Weeks of Murine Hindlimb Unloading Induces Shifts from B to T and from Th to Tc Splenic Lymphocytes in Absence of Stress and Differentially Reduces Cell-Specific Mitogenic Responses. PLoS ONE. 2014;9:e92664. doi: 10.1371/journal.pone.0092664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiou G, Ippolito GC, Beausang J, Busse CE, Wardemann H, Quake SR. The promise and challenge of high-throughput sequencing of the antibody repertoire. Nat Biotechnol. 2014;32:158–168. doi: 10.1038/nbt.2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould CL, Lyte M, Williams J, Mandel AD, Sonnenfeld G. Inhibited interferon-g but normal interleukin-3 production from rats flown on the space shuttle. Aviation, Space, and Environmental Med. 1987;58:983–986. [PubMed] [Google Scholar]
- Greiff V, Menzel U, Haessler U, Cook SC, Friedensohn S, Khan TA, Pogson M, Hellmann I, Reddy ST. Quantitative assessment of the robustness of next-generation sequencing of antibody variable gene repertoires from immunized mice. BMC Immunol. 2014;15:40. doi: 10.1186/s12865-014-0040-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gridley DS, Slater JM, Luo-Owen X, Rizvi A, Chapes SK, Stodieck LS, Ferguson VL, Pecaut MJ. Spaceflight effects on T lymphocyte distribution, function and gene expression. J Appl Physiol. 2009;106:194–202. doi: 10.1152/japplphysiol.91126.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigoriev AI, Bugrov SA, Bogomolov VV, Egorov AD, Polyakov VV, Tarasov IK, Shulzhenko EB. Main medical results of extended flights on Space Station Mir in 1986–1990. Acta Astronautica. 1993;29:581–585. doi: 10.1016/0094-5765(93)90073-6. [DOI] [PubMed] [Google Scholar]
- Grindeland RE, Popova IA, Vasques M, Arnaud SB. COSMOS 1887 mission overview: effects of microgravity on rat body and adrenal weights and plasma constituents. FASEB J. 1990;4:105–109. doi: 10.1096/fasebj.4.1.2295371. [DOI] [PubMed] [Google Scholar]
- Grove DS, Pishak SA, Mastro AM. The effect of a 10-day space flight on the function, phenotype, and adhesion molecule expression of splenocytes and lymph node lymphocytes. Exp Cell Res. 1995;219:102–109. doi: 10.1006/excr.1995.1210. [DOI] [PubMed] [Google Scholar]
- Haidar JN, Zhu W, Lypowy J, Pierce BG, Bari A, Persaud K, Luna X, Snavely M, Ludwig D, Weng Z. Backbone flexibility of CDR3 and immune recognition of antigens. J Mol Biol. 2014;426:1583–1599. doi: 10.1016/j.jmb.2013.12.024. [DOI] [PubMed] [Google Scholar]
- Hughes-Fulford M. Altered cell function in microgravity. Exp Gerontology. 1991;26:247–256. doi: 10.1016/0531-5565(91)90017-g. [DOI] [PubMed] [Google Scholar]
- Ichiki AT, Gibson LA, Jago TL, Strickland KM, Johnson DL, Lange RD, Allebban Z. Effects of spaceflight on rat peripheral blood leukocytes and bone marrow progenitor cells. J Leukoc Biol. 1996;60:37–43. doi: 10.1002/jlb.60.1.37. [DOI] [PubMed] [Google Scholar]
- Jahns G, Meylor J, Fast T, Hawes N, Zarow G. 43rd Congress the Int Astronautical Federation. Washington, DC: 1992. Rodent Growth, Behavior, and Physiology Resulting from Flight on the Space Life Sciences-1 Mission; pp. 1–8. Vol. IAF/IAA-92-0268. [Google Scholar]
- Kaplinsky J, Li A, Sun A, Coffre M, Koralov SB, Arnaout R. Antibody repertoire deep sequencing reveals antigen-independent selection in maturing B cells. Proc Natl Acad Sci U S A. 2014;111:E2622–2629. doi: 10.1073/pnas.1403278111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konstantinova IV, Antropova YN, Legen'kov VI, Zazhirey VD. Study of reactivity of blood lymphoid cells in crew members of the Soyuz-6, Soyuz-7 and Soyuz-8 spaceships before and after flight. Space Biol and Aerospace Med. 1973;7:48–55. [PubMed] [Google Scholar]
- Kramer JM, Holodick NE, Vizconde TC, Raman I, Yan M, Li QZ, Gaile DP, Rothstein TL. Analysis of IgM antibody production and repertoire in a mouse model of Sjogren's syndrome. J Leukoc Biol. 2016;99:321–331. doi: 10.1189/jlb.2A0715-297R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescale C, Schenten V, Djeghloul D, Bennabi M, Gaignier F, Vandamme K, Strazielle C, Kuzniak I, Petite H, Dosquet C, Frippiat JP, Goodhardt M. Hind limb unloading, a model of spaceflight conditions, leads to decreased B lymphopoiesis similar to aging. Faseb j. 2015;29:455–463. doi: 10.1096/fj.14-259770. [DOI] [PubMed] [Google Scholar]
- Lesnyak A, Sonnenfeld G, Avery L, Konstantinova I, Rykova M, Meshkov D, Orlova T. Effect of SLS-2 spaceflight on immunologic parameters of rats. J Appl Physiol. 1996;81:178–182. doi: 10.1152/jappl.1996.81.1.178. [DOI] [PubMed] [Google Scholar]
- Lesnyak AT, Sonnenfeld G, Rykova MP, Meshkov DO, Mastro A, Konstantinova I. Immune changes in test animals during spaceflight. J Leukoc Biol. 1993;54:214–226. doi: 10.1002/jlb.54.3.214. [DOI] [PubMed] [Google Scholar]
- Lu J, Panavas T, Thys K, Aerssens J, Naso M, Fisher J, Rycyzyn M, Sweet RW. IgG variable region and VH CDR3 diversity in unimmunized mice analyzed by massively parallel sequencing. Mol Immunol. 2014;57:274–283. doi: 10.1016/j.molimm.2013.09.008. [DOI] [PubMed] [Google Scholar]
- Mandel AD, Balish E. Effect of space flight on cell-mediated immunity. Aviation, Space, and Environmental Med. 1977;48:1051–1057. [PubMed] [Google Scholar]
- Meehan R, Whitson P, Sams C. The role of psychoneuroendocrine factors on spaceflight-induced immunological alterations. J Leukoc Biol. 1993;54:236–244. doi: 10.1002/jlb.54.3.236. [DOI] [PubMed] [Google Scholar]
- Meehan RT, Neale LS, Kraus ET, Stuart CA, Smith ML, Cintron NM, Sams CF. Alteration in human mononuclear leucocytes following space flight. Immunology. 1992;76:491–497. [PMC free article] [PubMed] [Google Scholar]
- Merrill AH, Wang E, Mullins RE, Grindeland RE, Popova IA. Analyses of plasma for metabolic and hormonal changes in rats flown aboard COSMOS 2044. J Appl Physiol. 1992;73:132S–135S. doi: 10.1152/jappl.1992.73.2.S132. [DOI] [PubMed] [Google Scholar]
- Miller ES, Koebel DA, Sonnenfeld G. Influence of spaceflight on the production of interleukin-3 and interleukin-6 by rat spleen and thymus cells. J Appl Physiol. 1995;78:810–813. doi: 10.1152/jappl.1995.78.3.810. [DOI] [PubMed] [Google Scholar]
- Minoche AE, Dohm JC, Himmelbauer H. Evaluation of genomic high-throughput sequencing data generated on Illumina HiSeq and genome analyzer systems. Genome Biol. 2011;12:R112. doi: 10.1186/gb-2011-12-11-r112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nash PV, Konstantinova IV, Fuchs BB, Rakhmilevich AL, Lesnyak AT, Mastro AM. Effect of spaceflight on lymphocyte proliferation and interleukin-2 production. J Appl Physiol. 1992;73:186S–190S. doi: 10.1152/jappl.1992.73.2.S186. [DOI] [PubMed] [Google Scholar]
- Nash PV, Mastro AM. Variable lymphocyte responses in rats after space flight. Exp Cell Res. 1992;202:125–131. doi: 10.1016/0014-4827(92)90411-z. [DOI] [PubMed] [Google Scholar]
- Pecaut MJ, Simske SJ, Fleshner M. Spaceflight induces changes in splenocyte subpopulations: effectiveness of ground-based models. Am J Physiol Regulatory Integrative Comp Physiol. 2000;279:R2072–R2078. doi: 10.1152/ajpregu.2000.279.6.R2072. [DOI] [PubMed] [Google Scholar]
- Saul FA, Poljak RJ. Crystal structure of human immunoglobulin fragment Fab New refined at 2.0 A resolution. Proteins. 1992;14:363–371. doi: 10.1002/prot.340140305. [DOI] [PubMed] [Google Scholar]
- Serova LV. Weightlessness effects on resistance and reactivity of animals. Physiologist. 1980;23:S22–S26. [PubMed] [Google Scholar]
- Sonnenfeld G, Foster M, Morton D, Bailliard F, Fowler NA, Hakenewerth AM, Bates R, Miller ES., Jr Spaceflight and development of immune responses. J Appl Physiol. 1998;85:1429–1433. doi: 10.1152/jappl.1998.85.4.1429. [DOI] [PubMed] [Google Scholar]
- Sonnenfeld G, Mandel AD, Konstantinova IV, Berry WD, Taylor GR, Lesnyak AT, Fuchs BB, Rakhmilevich AL. Spaceflight alters immune cell function and distribution. J Appl Physiol. 1992;73:191S–195S. doi: 10.1152/jappl.1992.73.2.S191. [DOI] [PubMed] [Google Scholar]
- Sonnenfeld G, Mandel AD, Konstantinova IV, Tylor GR, Berry WD, Wellhausen SR, Lesnyak AT, Fuchs BB. Effects of spaceflight on levels and activity of immune cells. Aviat Space Environ Med. 1990;61:648–653. [PubMed] [Google Scholar]
- Stein TP, Schluter MD. Excretion of IL-6 by astronauts during spaceflight. Am J Physiol. 1994;266:E448–452. doi: 10.1152/ajpendo.1994.266.3.E448. [DOI] [PubMed] [Google Scholar]
- Stowe RP, Mehta SK, Ferrando AA, Feeback DL, Pierson DL. Immune responses and latent herpesvirus reactivation in spaceflight. Aviat Space Environ Med. 2001a;72:884–891. [PubMed] [Google Scholar]
- Stowe RP, Pierson DL, Barrett AD. Elevated stress hormone levels relate to Epstein-Barr virus reactivation in astronauts. Psychosom Med. 2001b;63:891–895. doi: 10.1097/00006842-200111000-00007. [DOI] [PubMed] [Google Scholar]
- Stowe RP, Sams CF, Mehta SK, Kaur I, Jones ML, Feeback DL, Pierson DL. Leukocyte subsets and neutrophil function after short-term spaceflight. J Leukoc Biol. 1999;65:179–186. doi: 10.1002/jlb.65.2.179. [DOI] [PubMed] [Google Scholar]
- Taylor GR, Dardano JR. Human cellular immune responsiveness following space flight. Aviat Space Environ Med. 1983;54:S55–59. [PubMed] [Google Scholar]
- Taylor GR, Neale LS, Dardano JR. Immunological analyses of U.S. Space Shuttle crewmembers. Aviat Space Environ Med. 1986;57:213–217. [PubMed] [Google Scholar]
- Udden MM, Driscoll TB, Gibson LA, Patton CS, Pickett MH, Jones JB, Nachtman R, Allebban Z, Ichiki AT, Lange RD, Alfrey CP. Blood volume and erythropoiesis in the rat during spaceflight. Aviat Space Environ Med. 1995;66:557–561. [PubMed] [Google Scholar]
- Wang Y, Chen W, Li X, Cheng B. Degenerated primer design to amplify the heavy chain variable region from immunoglobulin cDNA. BMC Bioinformatics. 2006;7(Suppl 4):S9. doi: 10.1186/1471-2105-7-S4-S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wronski TJ, Li M, Shen Y, Miller SC, Bowman BM, Kostenuik P, Halloran BP. Lack of effect of spaceflight on bone mass and bone formation in group-housed rats. J Appl Physiol. 1998;85:279–285. doi: 10.1152/jappl.1998.85.1.279. [DOI] [PubMed] [Google Scholar]
- Xu JL, Davis MM. Diversity in the CDR3 region of V(H) is sufficient for most antibody specificities. Immunity. 2000;13:37–45. doi: 10.1016/s1074-7613(00)00006-6. [DOI] [PubMed] [Google Scholar]
- Yang Y, Wang C, Yang Q, Kantor AB, Chu H, Ghosn EE, Qin G, Mazmanian SK, Han J, Herzenberg LA. Distinct mechanisms define murine B cell lineage immunoglobulin heavy chain (IgH) repertoires. Elife. 2015;4:e09083. doi: 10.7554/eLife.09083. [DOI] [PMC free article] [PubMed] [Google Scholar]