Abstract
Background and purpose
Radiotherapy guidance based on magnetic resonance imaging (MRI) is currently becoming a clinical reality. Fast 2d cine MRI sequences are expected to increase the precision of radiation delivery by facilitating tumour delineation during treatment. This study compares four auto-contouring algorithms for the task of delineating the primary tumour in six locally advanced (LA) lung cancer patients.
Material and methods
Twenty-two cine MRI sequences were acquired using either a balanced steady-state free precession or a spoiled gradient echo imaging technique. Contours derived by the auto-contouring algorithms were compared against manual reference contours. A selection of eight image data sets was also used to assess the inter-observer delineation uncertainty.
Results
Algorithmically derived contours agreed well with the manual reference contours (median Dice similarity index: ). Multi-template matching and deformable image registration performed significantly better than feature-driven registration and the pulse-coupled neural network (PCNN). Neither MRI sequence nor image orientation was a conclusive predictor for algorithmic performance. Motion significantly degraded the performance of the PCNN. The inter-observer variability was of the same order of magnitude as the algorithmic performance.
Conclusion
Auto-contouring of tumours on cine MRI is feasible in LA lung cancer patients. Despite large variations in implementation complexity, the different algorithms all have relatively similar performance.
Keywords: Lung tumour tracking, Auto-contouring, MRI-guided radiotherapy, Locally advanced lung cancer
Radiotherapy guidance based on magnetic resonance imaging (MRI) is currently becoming a clinical reality. Several hybrid MRI/radiotherapy devices are undergoing advanced testing or are already at the point of clinical service [1], [2], [3], [4]. The arrival of these novel treatment machines could usher in a new era in image-guided radiotherapy. For moving and deforming tumours such as seen in lung cancer patients, real-time adaptive radiotherapy can benefit from repeatedly contouring the tumour and organs-at-risk in an automated and reliable fashion. Hybrid MRI/radiotherapy devices can acquire repeated 2d cine MR images of the internal patient anatomy during treatment [5] thus facilitating a number of suitable motion management strategies for lung cancer patients. These include gating [6], margin assessment and reduction [7], dynamic multi-leaf collimator (MLC) tracking [8], respiratory coaching based on audiovisual feedback [9], and combinations thereof. Through reductions in planning target volumes and better beam-target alignments [8], [10], [11], these techniques are expected to improve treatments for lung cancer by sparing more healthy tissue and allowing for dose escalation when needed.
A number of studies on auto-contouring of lung tumours based on 2d cine MRI have been published (Table 1). Unfortunately, none of these studies explicitly mentions lung tumour staging. Two of the studies resorted to healthy volunteers using normal lung structures as surrogate for tumour motion [12], [13]. This study focusses on tumour auto-contouring in locally advanced non-small cell lung cancer or limited stage inoperable small cell lung cancer (subsequently referred to collectively as locally advanced lung cancer). This patient cohort has a 5-year overall survival of only [14] and needs better treatment options. Tumour auto-contouring is expected to be challenging for these patients as lesions tend to be large and in close proximity to critical structures.
Table 1.
Previous lung auto-contouring studies based on 2d cine MRI.
| Reference | Bo [T] | Sequence | Orientation | Algorithm | Cohort |
|---|---|---|---|---|---|
| Cervino et al. [12] | 3.0 | n.p. | SAG slice | TM, ANN | 5 healthy volunteers |
| Tryggestad et al. [13] | 1.5 | bSSFP | SAG/CORa | TM | 2 healthy volunteersb |
| Shi et al. [15] | 1.5 | bSSFP | SAG stack | TM | 12 NSCLC |
| Yun et al. [16] | 0.5c | bSSFP | SAG slice | PCNN | 4 NSCLCb |
| Paganelli et al. [17] | 3.0 | bSSFP | SAG/CORa | SIFT | 9 lung patients |
| Seregni et al. [18] | 3.0 | bSSFP | SAG/CORa | SIFT | 6 lung patients |
| Mazur et al. [19] | 0.35 | bSSFP | SAG slice | SIFT | 4 lung patientsb |
| Bourque et al. [20] | 1.5 | bSSFP | SAG slice | Part. filt. | 4 NSCLC |
Abbreviations: ANN = artificial neural network. bSSFP = balanced steady-state free precession. COR = coronal. n.p. = not provided. NSCLC = non small cell lung cancer. Part. filt. = particle filter. PCNN = pulse-coupled neural network. SAG = sagittal. SIFT = scale invariant feature transform. TM = template matching.
Interleaved acquisition.
Non-lung subjects and phantoms excluded.
Derived from 3.0 T image.
Most previous studies used a balanced steady-state free precession (bSSFP) sequence [13], [15], [16], [17], [18], [19], [20]. The dynamic bSSFP sequence is widely available and captures T2/T1-weighted contrast, highlighting fat and fluids. Slice orientations were limited to the sagittal plane, or in case of interleaved (orthogonal) acquisitions of two slices [13], [17], [18], to the sagittal and coronal planes.
This study compares four auto-contouring algorithms for the task of delineating the primary tumour in six locally advanced lung cancer patients. The auto-contouring algorithms were selected to cover the broad range of previously published algorithms: multi-template matching, a pulse-coupled neural network, intensity-driven deformable image registration, and a feature-driven registration. Besides the ubiquitous bSSFP sequence, this study also uses a spoiled gradient echo sequence more akin in appearance to the familiar CT images [21]. All algorithmically derived contours are compared against manual reference contours to benchmark their validity. We also assessed the inter-observer delineation uncertainty.
Material and methods
Patient cohort
Six locally advanced lung cancer patients were included in this study (Table 2). Although patient #5 suffered from a malignant pleural effusion and was treated with palliative intent, a radical treatment approach might be feasible in the future [22], especially with online MRI guidance.
Table 2.
Patient characteristics. Staging was performed according to the AJCC recommendations [23]. Average 2d GTV size and maximum centroid motion extent (2nd to 98th motion percentile along the principle axis of motion following principle component analysis) were derived from manual contours on the sagittal slice.
| Patient | Sex | Age (a) | GTV (cm2) | Motion (mm) | Pathology | TNM | Stage | Tumour position |
|---|---|---|---|---|---|---|---|---|
| #1 | F | 70 | 16.4 | 4.2 | NSCLC | T2bNIM0 | IIB | Right upper lobe |
| #2 | M | 76 | 14.0 | 6.1 | NSCLC | T4N0M0 | IIIA | Left hilar |
| #3 | M | 50 | 27.9 | 8.4 | SCLC | T4N3M0 | IIIB | Left lower lobe |
| #4 | M | 60 | 22.1 | 22.0 | SCLC | T4N3M0 | IIIB | Right middle lobe |
| #5 | M | 68 | 27.1 | 6.6 | NSCLC | T4N0M1a | IV | Left lower lobe |
| #6 | M | 70 | 86.1 | 1.5 | NSCLC | T4N2M0 | IIIB | Right upper lobe |
Abbreviations: GTV = gross tumour volume. (N)SCLC = (non) small cell lung cancer.
Image acquisition
A total of twenty-two cine MR image series were acquired on a Siemens MAGNETOM Aera (Siemens Healthcare GmbH, Erlangen, Germany) 1.5 T scanner using either a bSSFP (TrueFISP), or a spoiled gradient echo (GRE, FLASH) acquisition technique. Patients were scanned during free breathing in head-first-supine position with arms down. Informed consent was obtained from the patients according to institutional standards. Acquisition parameters are summarised in the supplement. Each image series was acquired for approximately 1 min. A stack of three 2d imaging planes was manually positioned to ensure it intersected with the time-averaged tumour as measured on a 3d golden angle radial VIBE [24] MR image (pixel spacing: 1.5 × 1.5 × 3 mm, field of view: 384 × 384 × 288 mm, TR = 2.51 ms, TE = 1.24 ms, bandwidth = 1085 Hz/Px, acquisition time = 4:58 min, fat-saturation) acquired at the beginning of each imaging session. After scanning, the image plane which intersected best with the tumour (usually the central slice) was selected for further processing, resulting in 1.8 Hz image sampling. Images were positioned in the sagittal and coronal planes, or in case of poor tumour visibility at an oblique angle (perpendicular to the axial plane).
Manual contouring and inter-observer variability
In each cine MR sequence primary tumour contours were manually generated by one of four expert observers, all following the same delineation procedural protocol. For a selection of image data sets two additional secondary tumour contour sets were generated independently by expert observers (giving a total of three manual contour sets) to assess the inter-observer delineation variability. The selection consisted of eight image data sets representing all combinations of imaging sequences, slice orientation, and small or large tumour motion amplitude (threshold: 5 mm). The observers generating secondary contours used the same window/level setting as was used to generate the primary contours. Only the first 30 images in each series were contoured by the additional observers to lighten the work load. A STAPLE delineation [25] approximating the hidden, unobservable ground truth delineation was generated from the three manual contours. The inter-observer variability was assessed by measuring the distance from each individual contour to the STAPLE contour.
Auto-contouring
Each image data set was divided into a training data set (the first ten images) and a test data set of 90–120 images (all subsequent images). Manual contours from the test data set were only used for evaluation purposes and not for algorithm training. Ten training images were chosen to capture at least one complete respiratory cycle.
The following auto-contouring algorithms were implemented.
Multi-template matching (MTM)
This algorithm was adapted from previous work [26], [27]. A rectangular template is cropped from each training image using the smallest tumour-enclosing (axis-aligned) bounding box extended by three pixels in all directions. The test image pixel with the highest normalised cross-correlation score from any of the ten templates is then selected as best match. The contour from the respective best training image is subsequently copied to the test image.
Pulse-coupled neural network (PCNN)
This algorithm was previously presented by Yun et al. [16]. It attempts to improve grey-value contrast between tumour and lung tissue by means of a pulse-coupled neural network (PCNN) approach. Within the PCNN algorithm each image pixel is treated as a neuron. The internal activity of each neuron is influenced by the image intensity and other adjacent neurons. Neurons with similar levels of activity pulse together yielding tumour contours if PCNN parameters are selected appropriately. PCNN parameters are optimised based on the training images and associated manual contours.
Intensity-driven deformable image registration (DIR)
This algorithm uses a two-stage approach to propagate a contour from the training image set to a test image. First, the image similarity is calculated between all training images and the current test image by the means of local normalised cross-correlation [28]. Second, the most similar training image is then transformed by a B-spline deformable image registration (DIR [29], [30]) to match the test image. The resulting displacement vector field is subsequently applied to deform the contour from the corresponding training image to the current test image.
Feature-driven registration (SIFT)
Similar to earlier approaches (e.g. [17]) this algorithm uses image features to select, deform and propagate a training contour onto each test image. Feature extraction [31] and scale invariant feature transform (SIFT) description [32] is performed for all training images and the test image within a region of interest. The best feature set combination is selected and used to generate a transformation from the corresponding training image to the test image. The training contour is then propagated according to the established transformation.
Contour comparison and statistical tests
Automatically generated contours were compared with manual ground-truth contours for all test images using the Dice similarity index, Hausdorff distance, centroid distance, and mean contour distance (c.f. supplement). The centroid distance is especially important for applications such as dynamic tumour tracking [8]. The Hausdorff and mean contour distances are relevant when treatment is adapted to tumour deformations [33].
Differences between contouring methods were tested for significance in terms of contour similarity using a linear mixed effects model. The model intends to separate the random variation due to the finite number of image sets, from the potentially systematic variation introduced by the auto-contouring algorithm. It was constructed in R (R Core Team, v3.4.0) using the lme4 package [34] with the contouring algorithm as predictor. A second model was applied to the data from each contouring algorithm to separate out the impact of different predictors: imaging sequence, slice orientation, maximum motion, and observer. A third model was applied to the inter-observer data to investigate the impact of imaging sequence, slice orientation, and maximum motion on contouring consistency.
Results
Fig. 1 shows examples of the manually and automatically generated contours for the eight cine MR sequences that were selected for inter-observer analysis.
Fig. 1.
Manual (red) and automatically generated contours (green). The selected cine MR sequences correspond to the criteria specified in Section “Manual contouring and inter-observer variability”. The upper two rows show images acquired with the bSSFP sequence and the lower two rows those acquired with the GRE sequence. The left column shows sagittal and the right column coronal or oblique image plane orientations. Images in row one and three were classified as small motion and those in rows two and four as large motion.
Inter-observer variability
The eight inter-observer cases were evaluated by comparing the STAPLE delineation with each observer using all similarity metrics (Fig. 2 and supplement). For the Hausdorff distance and the mean contour distance, the bSSFP sequence resulted in significantly lower values . No other significant differences were found.
Fig. 2.
Results of inter-observer variability study for the complete dataset (all) and selected subsets. The centre-line of the box corresponds to the median, the box signifies the first and third quartiles, the whiskers extend the box by 1.5 times the interquartile range, and crosses are outliers. Significance levels are defined as ∗ (p < 0.05), ∗∗ (p < 0.01), and ∗∗∗ (p < 0.001). The vertical axes are matching with Fig. 3.
Auto-contouring
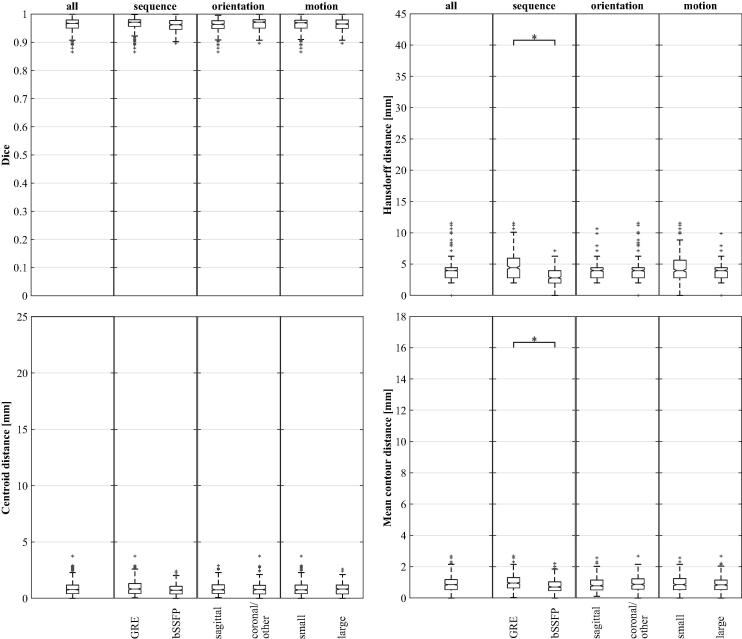
The overall performance of all auto-contouring algorithms is shown in Fig. 3 and the supplement. Contour creation per image took about 1 ms (MTM), 25 ms (PCNN), 150 ms (SIFT), and 500 ms (DIR) on standard workstation computers. Absolute differences in median quantities are generally small when comparing different algorithms. The DIR algorithm performs significantly better than the SIFT or PCNN algorithm for all metrics. The MTM algorithm is significantly better than the PCNN or SIFT algorithm for a subset of metrics. No significant difference could be detected between MTM and DIR.
Fig. 3.
Algorithmic auto-contouring performance is evaluated for the complete dataset (all) and selected subsets. See caption of Fig. 2 for a description of the quantities visualised by the plots.
The complete dataset was then split according to image acquisition sequence, slice orientation, and motion amplitude (Fig. 3). For PCNN, more motion resulted in reduced algorithmic performance for all metrics (all ). The same correlation was observed for the centroid distance using SIFT . Using the bSSFP sequence resulted in an improved mean contour distance for the SIFT algorithm . In addition, the observer was a significant predictor for the centroid distance of DIR and the mean contour distance of PCNN (both ). The magnitude of observer influence was estimated to be 0.87 mm (centroid distance, DIR) and 0.70 mm (mean contour distance, PCNN). No other significant differences were found.
Discussion
Lung tumour patients with locally advanced disease are expected to benefit from online MRI guidance. An essential component for adapted treatment on the basis of continuously acquired cine MR images is the ability to automatically delineate the tumour. To the best of our knowledge, systematic evaluations of auto-contouring algorithms for cine MRI from patients with locally advanced lung cancer were not previously reported in the literature. We thus implemented a broad range of auto-contouring algorithms and compared their performance on image data sets from a cohort of six locally advanced lung cancer patients.
In our study, the MTM and DIR algorithms outperformed the PCNN and SIFT algorithms for all investigated metrics. For the two best performing algorithms median values are 0.94 (Dice similarity index), 4.43 mm (Hausdorff distance), (centroid distance), and (mean contour distance). Centroid and mean contour distances approach a point where pixel resolution is the limiting factor. The MTM algorithm only compensates for translations but not for previously unseen rotations or deformations. The conceptually higher flexibility of the other algorithms might be preferable for longer treatment durations. In this study, scanning for only one minute allowed us to investigate different MR acquisition sequences and slice orientations for the same patient without exceeding the patient-tolerable total scanning time.
Although the bSSFP sequence has been almost exclusively used in previous cine MRI studies for lung (Table 1), we could not find conclusive evidence showing that bSSFP is preferable over GRE for the task of auto-contouring lung tumours. Previous studies reported that bSSFP images are preferable over GRE [21], [35] for a variety of reasons. It should be noted that our study investigates the consistency between manual and auto contours. Facilitating a medical verdict on whether images from either MRI sequence are preferable is beyond the scope of this study.
In our current implementation the DIR takes substantially longer than the MTM. A GPU-based DIR implementation is expected to reduce the difference substantially [30]. For the PCNN algorithm extensive parameter training is needed. To avoid an exhaustive search of the parameter space, Yun et al. [36] have proposed an adaptive particle swarm optimisation which reduces the training time to 2–4 h. Further improvements are needed to train or re-train the PCNN parameters with manual contours of the day.
All algorithms tested here require manually contoured training images as input for automatically contouring the previously unseen test images. The impact of the choice of expert observer on the measured algorithmic performance was minimal. Statistically significant influences of the observer on the result were identified by the mixed effects model only for two algorithm-metric combinations, and in those cases the estimated inter-observer deviation was well below the image resolution. In our study, training and test images were acquired subsequently without delay. This assumes that the training data can be collected as part of pre-treatment set-up imaging and that reliable manual contours can be created while the patient is being prepared for treatment. Alternatively the training contours could be derived at the time of treatment planning or from a previous treatment fraction. For this approach the risk is that anatomical changes result in deterioration in the performance of the auto-contouring algorithms. A compromise solution might see pre-fraction initial training contours being updated by a human observer with the implicit assumption that updating contours is less time consuming than creating new contours.
Inter-observer variability provides perspective on the achieved auto-contouring performance. The median values for the contouring metrics indicate that inter-observer variability is of similar magnitude as the algorithmic performance: 0.97 (Dice similarity index), 3.96 mm (Hausdorff distance), 0.76 mm (centroid distance), and 0.85 mm (mean contour distance). Given the consistently high performance of all algorithms, future gains in targeting accuracy may only be achieved from improvements in manual contouring or from making the auto-contouring independent from manual contours. In the inter-observer variability study, only the choice of imaging sequence was a significant predictor for observer similarity. Using bSSFP resulted in more similar manual contours with fewer outliers (i.e. reduced mean contour and Hausdorff distances).
A limitation of our study is that we only consider in-plane motion and ignore out-of-plane motion. This limitation is inherent to any 2d cine MRI based target localisation method. To reduce the impact of this issue, slices were always aligned with the cranial-caudal direction. Additionally, the relatively thick 5 or 10 mm slices provide robustness against small out-of-plane motion. Alternative out-of-plane motion can be addressed by matching 2d cine MR images with pre-treatment breath-hold (or navigator-triggered) 3d MR images [37], [38].
Further studies are necessary to investigate whether cine MRI is also useful for automatically contouring the affected mediastinal and hilar lymph nodes in locally advanced lung cancer patients. Studies based on 4d CT have demonstrated differences in phase and amplitude between primary tumours and lymph nodes [39], [40]. Based on the results of this study, automated tracking of the primary disease seems feasible.
Conflict of interest statement
The Institute of Cancer Research is part of the Elekta Atlantic MR-linac Research Consortium and we acknowledge financial and technical support from Elekta AB under a research agreement.
Acknowledgements
Research at The Institute of Cancer Research is supported by Cancer Research UK under Programme C33589/A19727. MFF and BE are supported by Cancer Research UK under Programme C33589/A19908. Research at the Centre for Medical Image Computing, UCL, is supported by the CRUK grant C33589/CRC521. We acknowledge NHS funding to the NIHR Biomedical Research Centre at The Royal Marsden and The Institute of Cancer Research. We would like to thank Hannah Bainbridge, David Collins and Cynthia Eccles (all ICR/RMH) for assisting in the MR image acquisition. Special thanks to Kyle Bromma (University of Victoria, Canada) for assisting in the implementation of MTM and PCNN.
Footnotes
Supplementary data associated with this article can be found, in the online version, athttps://doi.org/10.1016/j.radonc.2017.09.013.
Contributor Information
Martin F. Fast, Email: m.fast@nki.nl.
Björn Eiben, Email: bjoern.eiben.10@ucl.ac.uk.
Supplementary data
References
- 1.Lagendijk J.J., Raaymakers B.W., Raaijmakers A.J., Overweg J., Brown K.J., Kerkhof E.M. MRI/linac integration. Radiother Oncol. 2008;86:25–29. doi: 10.1016/j.radonc.2007.10.034. [DOI] [PubMed] [Google Scholar]
- 2.Fallone B.G., Murray B., Rathee S., Stanescu T., Steciw S., Vidakovic S. First MR images obtained during megavoltage photon irradiation from a prototype integrated linac-MR system. Med Phys. 2009;36:2084–2088. doi: 10.1118/1.3125662. [DOI] [PubMed] [Google Scholar]
- 3.Keall P.J., Barton M., Crozier S. The Australian magnetic resonance imaging-linac program. Semin Radiat Oncol. 2014;24:203–206. doi: 10.1016/j.semradonc.2014.02.015. [DOI] [PubMed] [Google Scholar]
- 4.Mutic S., Dempsey J.F. The ViewRay system: magnetic resonance-guided and controlled radiotherapy. Semin Radiat Oncol. 2014;24:196–199. doi: 10.1016/j.semradonc.2014.02.008. [DOI] [PubMed] [Google Scholar]
- 5.Menten MJ, Wetscherek A, Fast MF. MRI-guided lung SBRT: present and future developments. Phys Medica, in press. https://doi.org/10.1016/j.ejmp.2017.02.003. [DOI] [PubMed]
- 6.Park J.M., Park S.-Y., Kim H.J., Wu H.-G., Carlson J., Kim J.-I. A comparative planning study for lung SABR between tri-Co-60 magnetic resonance image guided radiation therapy system and volumetric modulated arc therapy. Radiother Oncol. 2016;120:279–285. doi: 10.1016/j.radonc.2016.06.013. [DOI] [PubMed] [Google Scholar]
- 7.Liu H.H., Koch N., Starkschall G., Jacobson M., Forster K., Liao Z. Evaluation of internal lung motion for respiratory-gated radiotherapy using MRI: part II-margin reduction of internal target volume. Int J Radiat Oncol Biol Phys. 2004;60:1473–1483. doi: 10.1016/j.ijrobp.2004.05.054. [DOI] [PubMed] [Google Scholar]
- 8.Menten M.J., Fast M.F., Nill S., Kamerling C.P., McDonald F., Oelfke U. Lung stereotactic body radiotherapy with an MR-linac – Quantifying the impact of the magnetic field and real-time tumor tracking. Radiother Oncol. 2016;119:461–466. doi: 10.1016/j.radonc.2016.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee D., Greer P.B., Ludbrook J., Arm J., Hunter P., Pollock S. Audiovisual biofeedback improves cine – magnetic resonance imaging measured lung tumor motion consistency. Int J Radiat Oncol Biol Phys. 2016;94:628–636. doi: 10.1016/j.ijrobp.2015.11.017. [DOI] [PubMed] [Google Scholar]
- 10.Colvill E., Booth J., Nill S., Fast M., Bedford J., Oelfke U. A dosimetric comparison of real-time adaptive and non-adaptive radiotherapy: a multi-institutional study encompassing robotic, gimbaled, multileaf collimator and couch tracking. Radiother Oncol. 2016;119:159–165. doi: 10.1016/j.radonc.2016.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caillet V., Keall P.J., Colvill E., Hardcastle N., O’Brien R., Szymura K. MLC tracking for lung SABR reduces planning target volumes and dose to organs at risk. Radiother Oncol. 2017;124:18–24. doi: 10.1016/j.radonc.2017.06.016. [DOI] [PubMed] [Google Scholar]
- 12.Cervino L.I., Du J., Jiang S.B. MRI-guided tumor tracking in lung cancer radiotherapy. Phys Med Biol. 2011;56:3773–3785. doi: 10.1088/0031-9155/56/13/003. [DOI] [PubMed] [Google Scholar]
- 13.Tryggestad E., Flammang A., Hales R., Herman J., Lee J., McNutt T. 4D tumor centroid tracking using orthogonal 2D dynamic MRI: implications for radiotherapy planning. Med Phys. 2013;40:091712. doi: 10.1118/1.4818656. [DOI] [PubMed] [Google Scholar]
- 14.Machtay M., Paulus R., Moughan J., Komaki R., Bradley J., Choy H. Defining local-regional control and its importance in locally advanced non-small cell lung carcinoma. J Thorac Oncol. 2012;7:716–722. doi: 10.1097/JTO.0b013e3182429682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shi X., Diwanji T., Mooney K.E., Lin J., Feigenberg S., D’Souza W.D. Evaluation of template matching for tumor motion management with cine-MR images in lung cancer patients. Med Phys. 2014;41:052304. doi: 10.1118/1.4870978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yun J., Yip E., Gabos Z., Wachowicz K., Rathee S., Fallone B.G. Neural-network based autocontouring algorithm for intrafractional lung-tumor tracking using Linac-MR. Med Phys. 2015;42:2296–2310. doi: 10.1118/1.4916657. [DOI] [PubMed] [Google Scholar]
- 17.Paganelli C., Lee D., Greer P.B., Baroni G., Riboldi M., Keall P. Quantification of lung tumor rotation with automated landmark extraction using orthogonal cine MRI images. Phys Med Biol. 2015;60:7165–7178. doi: 10.1088/0031-9155/60/18/7165. [DOI] [PubMed] [Google Scholar]
- 18.Seregni M., Paganelli C., Lee D., Greer P., Baroni G., Keall P. Motion prediction in MRI-guided radiotherapy based on interleaved orthogonal cine-MRI. Phys Med Biol. 2016;61:872–887. doi: 10.1088/0031-9155/61/2/872. [DOI] [PubMed] [Google Scholar]
- 19.Mazur T.R., Fischer-Valuck B.W., Wang Y., Yang D., Mutic S., Li H.H. SIFT-based dense pixel tracking on 0.35 T cine-MR images acquired during image-guided radiation therapy with application to gating optimization. Med Phys. 2016;43:279–293. doi: 10.1118/1.4938096. [DOI] [PubMed] [Google Scholar]
- 20.Bourque A.E., Bedwani S., Filion E., Carrier J.F. A particle filter based autocontouring algorithm for lung tumor tracking using dynamic magnetic resonance imaging. Med Phys. 2016;43:5161–5169. doi: 10.1118/1.4961403. [DOI] [PubMed] [Google Scholar]
- 21.Plathow C., Klopp M., Fink C., Sandner A., Hof H., Puderbach M. Quantitative analysis of lung and tumour mobility: comparison of two time-resolved MRI sequences. Br J Radiol. 2005;78:836–840. doi: 10.1259/bjr/29483804. [DOI] [PubMed] [Google Scholar]
- 22.Lim E., Baldwin D., Beckles M., Duffy J., Entwisle J., Faivre-Finn C., Kerr K. Guidelines on the radical management of patients with lung cancer. Thorax. 2010;65:iii1–iii27. doi: 10.1136/thx.2010.145938. [DOI] [PubMed] [Google Scholar]
- 23.Edge S.B., Compton C.C. The American Joint Committee on Cancer: the 7th edition of the AJCC cancer staging manual and the future of TNM. Ann Surg Oncol. 2010;17:1471–1474. doi: 10.1245/s10434-010-0985-4. [DOI] [PubMed] [Google Scholar]
- 24.Block K.T., Chandarana H., Milla S., Bruno M., Mulholland T., Fatterpekar G. Towards routine clinical use of radial stack-of-stars 3d gradient-echo sequences for reducing motion sensitivity. J Korean Soc Magn Reson Med. 2014;18:87–106. [Google Scholar]
- 25.Warfield S.K., Zou K.H., Wells W.M. Simultaneous truth and performance level estimation (STAPLE): an algorithm for the validation of image segmentation. IEEE Trans Med Imaging. 2004;23:903–921. doi: 10.1109/TMI.2004.828354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wisotzky E., Fast M., Oelfke U., Nill S. Automated marker tracking using noisy X-ray images degraded by the treatment beam. Z Med Phys. 2015;25:123–134. doi: 10.1016/j.zemedi.2014.08.006. [DOI] [PubMed] [Google Scholar]
- 27.Menten M.J., Fast M.F., Nill S., Oelfke U. Using dual-energy X-ray imaging to enhance automated lung tumor tracking during real-time adaptive radiotherapy. Med Phys. 2015;42:6987–6998. doi: 10.1118/1.4935431. [DOI] [PubMed] [Google Scholar]
- 28.Cachier P., Bardinet E., Dormont D., Pennec X., Ayache N. Iconic feature based nonrigid registration: the PASHA algorithm. Comput Vision Image Understanding. 2003;89:272–298. [Google Scholar]
- 29.Rueckert D., Sonoda L.I., Hayes C., Hill D.L.G., Leach M.O., Hawkes D.J. Nonrigid registration using free-form deformations: application to breast MR images. IEEE Trans Med Imaging. 1999;18:712–721. doi: 10.1109/42.796284. [DOI] [PubMed] [Google Scholar]
- 30.Modat M., Ridgway G.R., Taylor Z.A., Lehmann M., Barnes J., Hawkes D.J. Fast free-form deformation using graphics processing units. Comput Methods Prog Biomed. 2010;98:278–284. doi: 10.1016/j.cmpb.2009.09.002. [DOI] [PubMed] [Google Scholar]
- 31.Rosten E., Porter R., Drummond T. Faster and better: a machine learning approach to corner detection. IEEE Trans Pattern Anal Mach Intell. 2010;32:105–119. doi: 10.1109/TPAMI.2008.275. [DOI] [PubMed] [Google Scholar]
- 32.Lowe D.G. Distinctive image features from scale-invariant keypoints. Int J Comput Vision. 2004;60:91–110. [Google Scholar]
- 33.Ge Y., O’Brien R.T., Shieh C.-C., Booth J.T., Keall P.J. Toward the development of intrafraction tumor deformation tracking using a dynamic multi-leaf collimator. Med Phys. 2014;41:061703. doi: 10.1118/1.4873682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bates D., Mächler M., Bolker B., Walker S. Fitting linear mixed-effects models using lme4. J Stat Softw. 2015;67(1):1–48. https://www.jstatsoft.org/v067/i01. [Google Scholar]
- 35.Fabel M., Wintersperger B., Dietrich O., Eichinger M., Fink C., Puderbach M. MRI of respiratory dynamics with 2D steady-state free-precession and 2D gradient echo sequences at 1.5 and 3 Tesla: an observer preference study. Eur Radiol. 2009;19:391–399. doi: 10.1007/s00330-008-1148-x. [DOI] [PubMed] [Google Scholar]
- 36.Yun J., Yip E., Gabos Z., Wachowicz K., Rathee S., Fallone B. Improved lung tumor autocontouring algorithm for intrafractional tumor tracking using 0.5 T linac-MR. Biomed Phys Eng Exp. 2016;2:067004. doi: 10.1118/1.4916657. [DOI] [PubMed] [Google Scholar]
- 37.Bjerre T., Crijns S., af Rosenschold P.M., Aznar M., Specht L., Larsen R. Three-dimensional MRI-linac intra-fraction guidance using multiple orthogonal cine-MRI planes. Phys Med Biol. 2013;58:4943–4950. doi: 10.1088/0031-9155/58/14/4943. [DOI] [PubMed] [Google Scholar]
- 38.Brix L., Ringgaard S., Sørensen T.S., Poulsen P.R. Three-dimensional liver motion tracking using real-time two-dimensional MRI. Med Phys. 2014;41 doi: 10.1118/1.4867859. 042302. [DOI] [PubMed] [Google Scholar]
- 39.Donnelly E.D., Parikh P.J., Lu W., Zhao T., Lechleiter K., Nystrom M. Assessment of intrafraction mediastinal and hilar lymph node movement and comparison to lung tumor motion using four-dimensional CT. Int J Radiat Oncol Biol Phys. 2007;69:580–588. doi: 10.1016/j.ijrobp.2007.05.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pantarotto J.R., Piet A.H., Vincent A., de Koste J.R.v.S., Senan S. Motion analysis of 100 mediastinal lymph nodes: potential pitfalls in treatment planning and adaptive strategies. Int J Radiat Oncol Biol Phys. 2009;74:1092–1099. doi: 10.1016/j.ijrobp.2008.09.031. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.