Abstract
Wolbachia pipientis from Drosophila melanogaster (wMel) is an endosymbiotic bacterium that restricts transmission of human pathogenic flaviviruses and alphaviruses, including dengue, Zika, and chikungunya viruses, when introduced into the mosquito vector Aedes aegypti. To date, wMel-infected Ae. aegypti have been released in field trials in 5 countries to evaluate the effectiveness of this strategy for disease control. Despite the success in establishing wMel-infected mosquitoes in wild populations, and the well-characterized antiviral capabilities of wMel, transinfecting different or additional Wolbachia strains into Ae. aegypti may improve disease impact, and perhaps more importantly, could provide a strategy to account for the possible evolution of resistant arboviruses. Here, we report the successful transinfection of Ae. aegypti with the Wolbachia strains wMelCS (D. melanogaster), wRi (D. simulans) and wPip (Culex quinquefasciatus) and assess the effects on Ae. aegypti fitness, cytoplasmic incompatibility, tissue tropism and pathogen blocking in a laboratory setting. The results demonstrate that wMelCS provides a similar degree of protection against dengue virus as wMel following an infectious blood meal, and significantly reduces viral RNA levels beyond that of wMel following a direct challenge with infectious virus in mosquitoes, with no additional fitness cost to the host. The protection provided by wRi is markedly weaker than that of wMelCS, consistent with previous characterisations of these lines in Drosophila, while wPip was found to substantially reduce the fitness of Ae. aegypti. Thus, we determine wMelCS as a key candidate for further testing in field-relevant fitness tests and viremic blood feeding challenges in a clinical setting to determine if it may represent an alternative Wolbachia strain with more desirable attributes than wMel for future field testing.
Author summary
Dengue viruses are transmitted by the Aedes aegypti mosquito, with an estimated 390 million human infections occurring per year worldwide. There is no approved antiviral therapeutic, and vaccines described so far have had limited efficacy. Recently, the endosymbiotic bacterium Wolbachia from Drosophila melanogaster (wMel) has been used to infect Ae. aegypti populations as a novel technology for reducing dengue virus transmission. Here we report the generation of three new mosquito lines infected with the Wolbachia strains wMelCS, wRi and wPip. Each line induced cytoplasmic incompatibility and was effectively maternally transmitted, as required for rapid spread through uninfected mosquito populations. Each Wolbachia strain was also found to reside in the salivary glands; a key tissue involved in viral transmission. Perhaps most importantly, wMelCS inhibited dengue virus replication and dissemination in mosquitoes following an infectious blood meal or intrathoracic injection, providing a similar level of protection as that described for wMel. wMelCS therefore warrants further investigation as a potential release strain in future field trials.
Introduction
Arboviruses transmitted by the Aedes aegypti mosquito, including dengue (DENV), Zika (ZIKV) and chikungunya (CHIKV) are emerging threats that impose an increasing health burden on tropical and subtropical regions of the world. While these viruses usually cause self-limiting febrile disease, severe manifestations such as hemorrhagic shock can lead to death. In the absence of specific antiviral therapeutics and suboptimal vaccines [1–4], treatment is supportive only and limiting virus transmission has been largely dependent on vector control. The increasing global incidence of these diseases demonstrates the lack of effectiveness of current control programs and a need for novel efficacious and cost-effective alternatives [5].
In 2011, the first releases of Ae. aegypti mosquitoes carrying the artificially transinfected endosymbiotic bacterium Wolbachia pipientis began in field trials in Northern Australia as part of the Eliminate Dengue Program [6]. Wolbachia is a gram negative, obligate endosymbiont that is maternally transmitted and can impart antiviral properties to arthropod hosts. It is estimated that at least 40% of all terrestrial arthropod species are infected with Wolbachia [7] which can also manipulate host biology to induce feminization, parthenogenesis, cytoplasmic incompatibility (CI) and male-killing [8, 9]. Of these traits, CI is the most common, induced by many (but not all) Wolbachia strains, and enables the spread of Wolbachia into a new host population by providing females with Wolbachia an indirect reproductive advantage. In crosses between uninfected females and Wolbachia-infected males, CI results in the embryonic lethality of offspring. However, Wolbachia-infected females can mate with both uninfected and infected males successfully. CI, coupled with the maternal transmission (MT) of Wolbachia leads to the rapid invasion of the host population [8, 9].
The antiviral activity Wolbachia can provide to its host is a more recently described phenomenon [10]. The mechanism that drives this protection is not well understood but may involve priming of the insect innate immune response pathways in new Wolbachia infections, and competition for resources such as cholesterol [11–13].
Field trials to date have utilized Ae. aegypti transinfected with the Drosophila melanogaster native Wolbachia strain, wMel, and a more pathogenic form, wMelPop-CLA, isolated from a lab stock of D. melanogaster [6, 14, 15]. While these strains have been shown to restrict replication and dissemination of several virus genera including flaviviruses and alphaviruses in Ae. aegypti, due to technical difficulties associated with introducing Wolbachia into a new species, the full potential of using alternative Wolbachia strains has remained largely unexplored.
This study set out to provide an initial examination of Ae. aegypti transinfected with novel Wolbachia strains to identify key candidate strains that could potentially contest, or even improve on the current pathogenic protection afforded to Ae. aegypti by wMel, whilst avoiding fitness costs to the mosquito such as impaired egg viability and larval development as described for wMelPop-CLA infections [10, 16–19]. Given the cost and difficulty in taking alternative strains through a pipeline of more field-relevant laboratory testing and then into comparative field testing the goal of this work was to determine if any of the newly generated transinfected lines reported here could be removed from this pipeline in a first round of comparative testing.
For a Wolbachia-transinfected Ae. aegypti line to be considered for examination in further field-relevant experiments, the Wolbachia strain must provide strong protection against virus replication, demonstrate near-complete MT, and induce CI to enable rapid spread into the wild mosquito population, whilst inducing minimal fitness cost to the mosquito host.
To predict which Wolbachia strains may provide these traits we looked to past studies, most of which have been performed using natively Wolbachia-infected, artificially transinfected, or introgressed Drosophila lines, with the RNA viruses Drosophila C virus (DCV) or Flock House virus (FHV) as models. From these studies, we identified wMelCS (from D. melanogaster) and wRi (from D. simulans) as key candidates [20–24].
Although Ae. aegypti mosquitoes do not naturally carry Wolbachia, a number of other mosquito species have natural infections including the closely related species Aedes albopictus (wAlbA and wAlbB) and Aedes notoscriptus (wNoto) as well as more distantly related species such as Culex quinquefasciatus (wPip). wAlbB has previously been transinfected into Ae. aegypti and shown to provide strong protection against DENV while showing more resilience to heat stress but poorer larval competitive ability compared to wMel [13, 25–29], indicating the difficulty in assessing strains for performance from relatively crude experimental studies and the need for comparative studies to be undertaken under field release conditions. wPip has been reported to provide protection against West Nile virus (WNV) in its natural host, identifying this strain as a possible candidate for transinfection in Ae. aegypti [30].
Here we report the generation of three new Ae. aegypti lines, transinfected with Wolbachia strains wRi, wMelCS and wPip, and describe an initial relative evaluation of these strains, examining the effects on mosquito fitness, MT, CI, tissue tropism and DENV blocking. The findings reveal wMelCS as a potential candidate for future examination under more rigorous field-relevant testing and also indicate that wRi and wPip can be removed as candidates for future testing.
Results
Maternal transmission and cytoplasmic incompatibility
MT of a Wolbachia strain and its ability to induce CI are key features that must be conserved when considering new Wolbachia-infected Ae. aegypti lines for field releases. CI occurs when an uninfected female mated with a Wolbachia-infected male cannot produce viable offspring. This phenomenon ensures all offspring will be Wolbachia-positive, enabling effective spread of Wolbachia throughout a wild population.
MT was assayed by crossing Wolbachia-infected females (at least 5 generations removed from the G0 line; G5) with males from their respective Wolbachia-free tetracycline (Tet)-treated line and assaying for the presence of Wolbachia in adult progeny. The crosses revealed near-complete MT for wMelCS and wPip (99.4 and 98.8%, respectively), similar to reported values for wMel (100% with a 95% confidence interval lower boundary of 89%; [15]), while wRi sustained some breakdown of MT at 87.5% (Table 1).
Table 1. Maternal transmission of Wolbachia assessed from progeny of crosses between infected females and uninfected males.
| No. Parent Females | No. Female progeny | No. Wolbachia positive (Maternal Transmission %) | |
|---|---|---|---|
| wMelCS | 37 | 651 | 647 (99.4%) |
| wRi | 54 | 1253 | 1096 (87.5%) |
| wPip | 49 | 434 | 429 (98.8%) |
CI was determined for each line by measuring the hatch rate from crosses between Tet-treated females and infected males from each respective Wolbachia-infected line (Fig 1B). Control crosses where Tet-treated males and females were mated showed high hatch rates (>80%), while in CI crosses, near-complete sterility was observed in all three lines (0.3, 1.8 and 0% for wMelCS, wRi, and wPip, respectively), comparable to previous reports for wMel (0%) [15, 26]. Note that newly established lines can display some variability between individuals which may explain the small differences in MT and CI shown here for wMelCS and wPip.
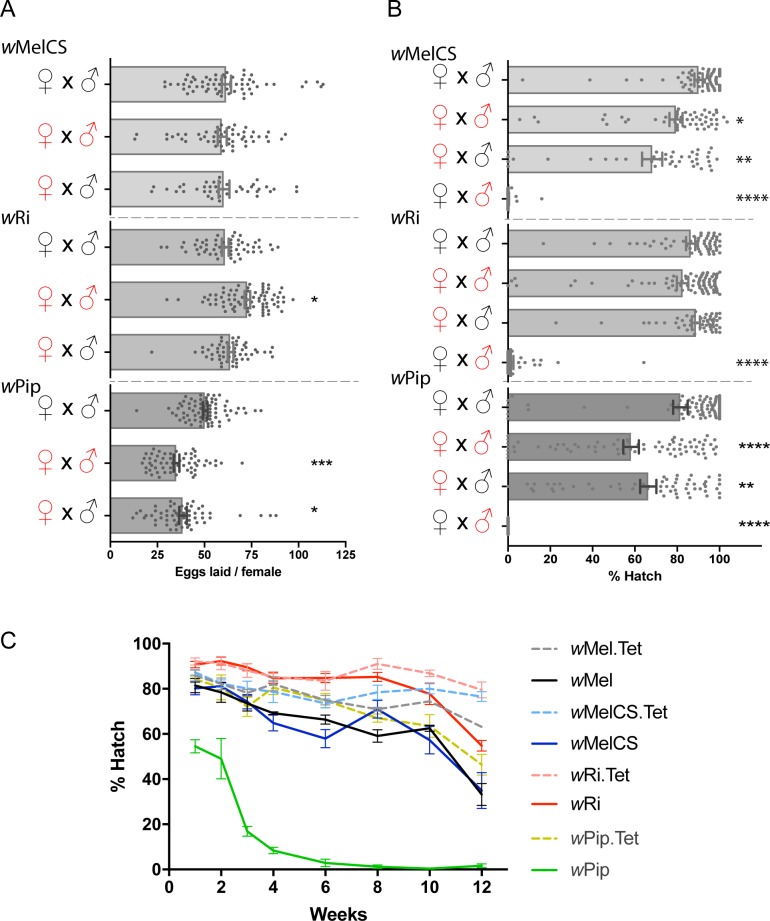
Fig 1. CI, fecundity and egg diapause viability in wMelCS, wRi, and wPip transinfected Ae. aegypti.
(A) Fecundity was determined as a measure of eggs laid per female. Bars are the mean number of eggs laid ± SEM from >40 females (individual data points are superimposed). Symbols for tetracycline-treated mosquitoes (uninfected) are in black, Wolbachia-infected are in red. Asterisks indicate significance compared to Tet x Tet controls (Kruskal-Wallis, Dunn's test, * for <0.05, ** for <0.01, *** for <0.001, **** for <0.0001). (B) Hatch rates for crosses between infected and uninfected mosquitoes show CI and successful hatching. Symbol codes and statistics are as per (A). Data are the mean ± SEM from >40 females (individual data points are superimposed). (C) Eggs from gravid females were collected over 72 h, from 3-days post blood meal. Eggs were dried slowly over 3–5 days then stored in a humid, airtight container. Batches of 100–500 eggs were hatched after 1, 2, 3, 4, 6, 8, 10 and 12 weeks. Hatched larvae were counted at 2nd instar stage until no hatch was observed for a week then the percent hatch calculated. Statistical analysis was performed using 2way ANOVA Sidak's test (n = 4 at each time point). wPip hatch rate was significantly reduced at all weeks compared to wPip.Tet (p<0.0001). wMel hatch rate was significantly reduced at weeks 4, 8, 10, (p<0.05) and 12 (p<0.0001) compared to wMel.Tet. wMelCS had a significantly reduced hatch rate at weeks 10 and 12 compared to wMelCS.Tet (p<0.01 and p<0.0001, respectively). wRi had a significantly reduced hatch rate at week 12 only, compared to wRi.Tet (p<0.0001).
Analysis of mosquito fitness costs induced by each Wolbachia strain
Fecundity, hatch rate and egg diapause viability
Fecundity was measured as the number of eggs laid/female following mating between Wolbachia-infected males and females, and Wolbachia-infected females and Tet-treated males, compared to the control cross of Tet-treated males and females. For wRi and wMelCS, the number of eggs laid/female did not significantly decrease irrespective of these matings, with the wRi-infected male and female cross in fact showing a slight but significant (p<0.05 Kruskal-Wallis, Dunn's test) increase in eggs laid/female, indicative of a positive effect on fecundity (Fig 1A). For wPip, relative to the Tet control cross, the mean number of eggs/female decreased for crosses between infected females and males, and infected females crossed with uninfected males (p<0.001, p<0.05, respectively), suggesting wPip reduces fecundity for this line.
We next examined the egg hatch rates of each cross. No significant reduction in hatch rate was observed for wRi crosses between Wolbachia-infected females and males, when compared to Tet-treated male and female crosses. wMelCS showed a slight although significant reduction in the hatch rate between Wolbachia-infected females and males, compared to the Tet-treated control cross (79% and 90%, respectively; p<0.05, Kruskal-Wallis, Dunn's test) (Fig 1B), similar to that reported for wMel [15, 26]. Hatch rate was more dramatically impaired for wPip, with rates dropping from 82% for the Tet-treated control cross, to 58% for Wolbachia-infected male and female crosses (p<0.0001). For wMelCS and wPip, crosses between infected females and uninfected males also resulted in significant reductions in hatch rate (68 and 66%, compared to respective Tet control cross hatch rates of 90 and 82%, p<0.01), perhaps suggesting there is a small fitness cost for these strains of Wolbachia in eggs, although these reductions were similar to those described previously for wMel [15].
To determine the effect of Wolbachia on egg viability and storage, eggs were collected and stored for 1–12 weeks before determining hatch rates. The benchmark transinfected line, wMel had a slight, but significant reduction in hatch rate following 4, 8, and 10 weeks storage (p<0.05, 2way ANOVA Sidak's test) compared to its Tet control line, with a slightly larger reduction after 12 weeks (p<0.0001) (Fig 1C). wRi and wMelCS lines displayed similar hatch rates (>60%) across all ages until week 10 (no significant difference in hatch rates compared to the respective Tet control lines until week 8). By contrast, wPip eggs appeared to deteriorate rapidly, with < 20% hatch rate observed after 3 weeks of aging (significant reductions at all weeks tested, p<0.0001, compared to wPip.Tet). Together, these data indicate a substantial fitness cost in the wPip line.
Longevity
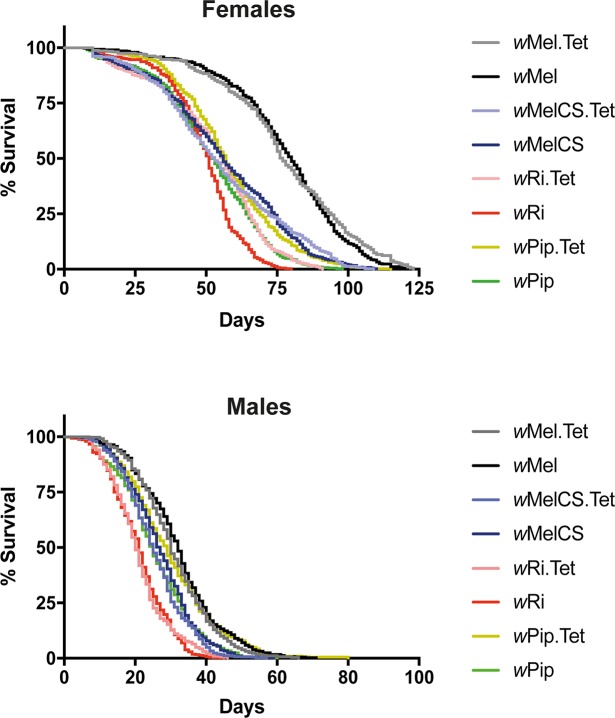
Effects on the survival of mosquitoes from each line were next examined over time (Fig 2). wMel, wMelCS and wRi Wolbachia-infected females tracked closely with their Tet-treated controls suggesting the infection had minimal influence on longevity. Similar relative survival rates were observed for males. Minor, but significant differences were observed for wRi females (p<0.0001, Log-rank (Mantel-Cox) test) and wMel (p<0.05) and wMelCS (p<0.05) males, relative to their respective Tet control lines. wPip was the only line that showed a substantially shorter lifespan for both males and females compared to the Tet-control (p<0.0001). Since <1% of wild female Ae. aegypti are expected to live beyond 3 weeks [31], it’s expected that no infection described here would likely impact on the lifespan of the mosquito in the field.
Fig 2. Longevity in transinfected Ae. aegypti lines.
Triplicate cages of age-controlled (emergence within 24 h) adults (~150 males and ~150 females/cage) were maintained at 26°C, 65% relative humidity and a 12:12 h light:dark cycle in a climate controlled room. The number of dead males and/or females was recorded and carcasses removed daily until all mosquitoes in the cages were dead. Data are the total % survival from the three cages/line. Significant differences were observed for wRi females (p<0.0001), wMel (p<0.05) and wMelCS (p<0.05) males, relative to their respective Tet control line. wPip had a significantly shorter lifespan for both males and females compared its matched Tet-control (p<0.0001). Statistical analysis was performed using a Log-rank (Mantel-Cox) test.
We also examined compatibility between wMel and wMelCS, since wMelCS-infected Ae. aegypti appeared to be the least compromised line from these initial screening experiments. We found these strains to be bidirectionally compatible with crosses between wMelCS females and wMel males, and wMel females and wMelCS males, each resulting in successful hatch rates (71 and 62%, respectively, S1 Fig). These data suggest that wMelCS could not be released over an existing wMel strain and expect to overtake wMel by CI alone. The ability of either of these strains to replace another in a field setting will be dependent upon the which strain is least costly to host fitness.
Wolbachia density and distribution
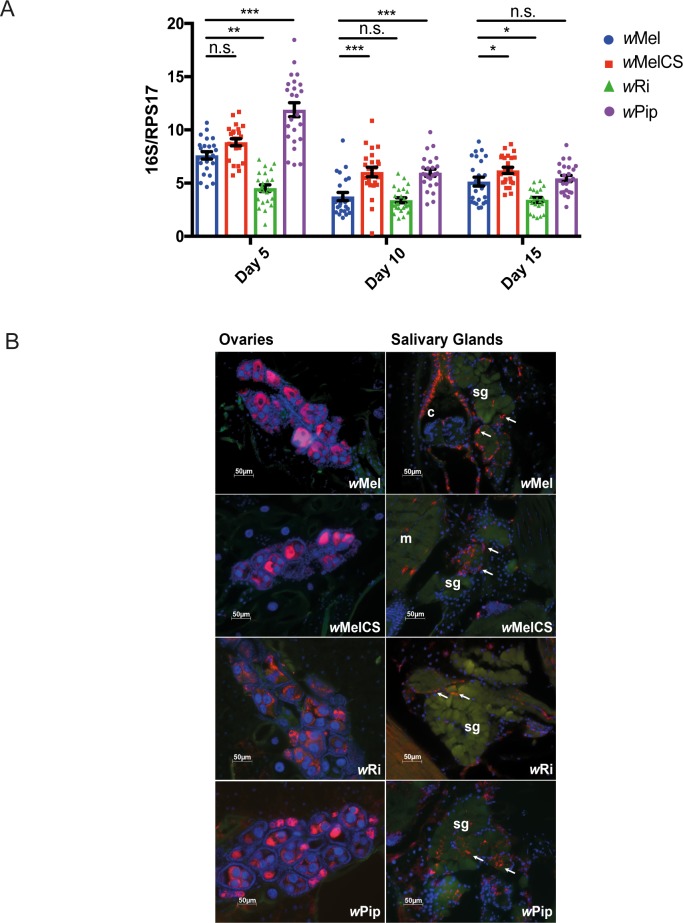
Total Wolbachia density was measured in 5-, 10- and 15-day old adult females of each line (at least G9) and compared to the benchmark line wMel at each time point, using qPCR with primers specific to the highly conserved Wolbachia ribosomal RNA gene, 16S, and Ae. aegypti rps17 gene to normalize for DNA input. Overall, the density of each strain reduced as the mosquitoes aged (Fig 3A). wMelCS density was significantly higher than wMel at days 10 and 15 post emergence (p<0.001, p<0.05, Mann-Whitney test), as was wPip at days 5 and 10 (p<0.001). wRi density was nearly half that of wMel at day 5 and was significantly reduced at day 15 (p<0.01 and p<0.05, respectively), but not significantly different at day 10. Despite these variations, all densities measured were within a 1log10 range of each other, indicating each Wolbachia strain maintains itself at high density.
Fig 3. Wolbachia density and distribution in transinfected Ae. aegypti lines.
(A) Density of Wolbachia within 5-, 10- and 15-day old whole female mosquitoes was determined by qPCR using primers directed to the conserved 16S rRNA gene. Density is expressed as the mean ratio between 16S and the Ae. aegypti host rps17 gene. Data are the mean and SEM of 24 mosquitoes. Asterisks indicate significance compared to wMel at each time point (Kruskal-Wallis, Dunn's test with multiple test corrections; *p<0.05, **p<0.01, ***p<0.001, n.s. not significant). (B) The distribution of wMelCS, wRi and wPip Wolbachia strains in mosquitoes was determined in sections of paraffin-embedded female mosquitoes (5 to 7-day old) using fluorescence in situ hybridisation (FISH). The fluorescently labelled 16S probe detects the 16S rRNA gene from all four Wolbachia strains. Total DNA was stained in blue using DAPI and a green filter was included to increase contrast with surrounding tissues. Sg indicates salivary gland tissue, m indicates muscle, and c indicates cardia. White arrows identify select regions of Wolbachia staining.
The localization of Wolbachia within ovary and salivary gland tissues was next examined in 5–7 day old adult female mosquitoes using fluorescence in situ hybridization (FISH). Mosquitoes were formaldehyde-fixed, and paraffin-embedded tissue sections stained using a probe specific for Wolbachia 16S gene (red, Fig 3B). Total DNA was stained with DAPI (blue). Consistent with the strong MT shown in Table 1 for all lines, each strain was abundant in ovarian tissues, with distribution comparable with wMel (Fig 3B). Similar distribution of Wolbachia was also observed for all lines in the salivary glands, a key tissue involved in viral transmission.
Restriction of DENV-3 replication by wRi and wMelCS in Ae. aegypti
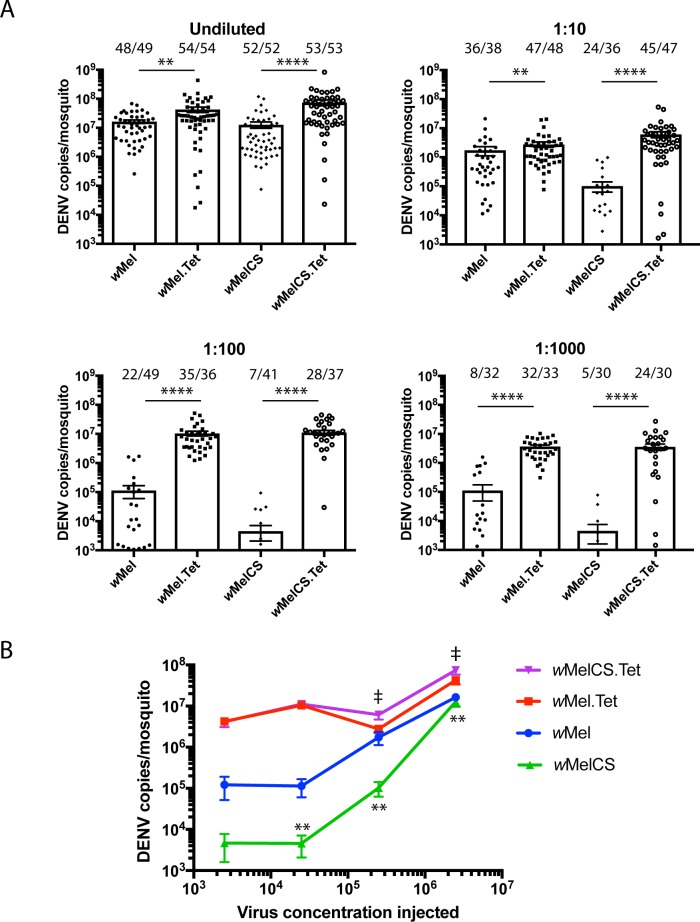
To compare the viral-blocking capacity of these lines, 7 day old mosquitoes were fed an infectious blood meal containing 2.0 x 106 TCID50/ml (DENV-3), then incubated for 14 days at 26°C. Due to the reduced fitness of wPip mosquitoes we elected to exclude this line. Vector competence analyses were performed on wRi and wMelCS lines, and compared to wMel. Mosquitoes were collected, the head separated from each mosquito body, then total RNA extracted to measure rates of infection (as determined by bodies positive for DENV RNA) and dissemination (heads positive for DENV RNA). Consistent with previous findings, wMel reduced the mean DENV RNA levels by 3log10 in mosquito bodies (3 x 104 copies/body compared to 3 x 107 copies/body in the relevant Tet control line, p<0.0001, Mann-Whitney test), as well as reducing the infection rate, with just 9% of wMel mosquito bodies scoring positive for DENV infection compared to 94% in the matched Tet control line (where positive was defined as >1000 copies/body, see Materials & Methods; Table 2 with the same data used to generate Fig 4). By contrast, wRi caused only a slight, although significant (p<0.01) reduction in viral copies (7.6 x 107 copies/body compared to 1.3 x 108 copies/body for the matched Tet control line) with no reduction in infection rate (96% compared to 98%, respectively). wMelCS gave a phenotype intermediate of wMel and wRi, with a wide spread in the number of DENV copies/body observed, with a mean of 1.5 x 107 RNA copies compared to 4.7 x 107 RNA copies for its matched Tet control line (p<0.0001). Interestingly, DENV-3 dissemination, key for viral transmission, was similarly restricted between wMel and wMelCS lines, with ~4log10 reduction in viral RNA copies determined in the head of each line relative to the matched Tet control, and just 2 and 9% infection rates, respectively, compared to 83 and 96% for the Tet controls. While the mean DENV RNA copies/head of wRi-infected mosquitoes was significantly reduced (2 x 106 copies compared to 9.5 x 106 copies for the Tet control line), the dissemination rates were not reduced (94% compared to 95%, respectively). Together, these results indicate that wRi shows weaker DENV-blocking capacity while the ability of wMelCS to restrict viral dissemination appears to be similar to that of wMel.
Table 2. Restriction of DENV-3 infection and dissemination by Wolbachia strains wRi and wMelCS.
| Line | Body | Head | ||
|---|---|---|---|---|
| Number DENV + (total screened) |
% DENV +# | Number DENV + (total screened) | % DENV +# | |
| wMel | 22 (250) | 9 | 5 (250) | 2 |
| wMel.Tet | 78 (83) | 94 | 69 (83) | 83 |
| wMelCS | 135 (240) | 56 | 21 (240) | 9 |
| wMelCS.Tet | 74 (76) | 97 | 73 (76) | 96 |
| wRi | 166 (169) | 98 | 159 (169) | 94 |
| wRi.Tet | 103 (107) | 96 | 102 (107) | 95 |
# Calculated as a percentage of total engorged mosquitoes
Fig 4. wMelCS and wRi Wolbachia strains inhibit DENV-3 replication and dissemination following an infectious blood meal.
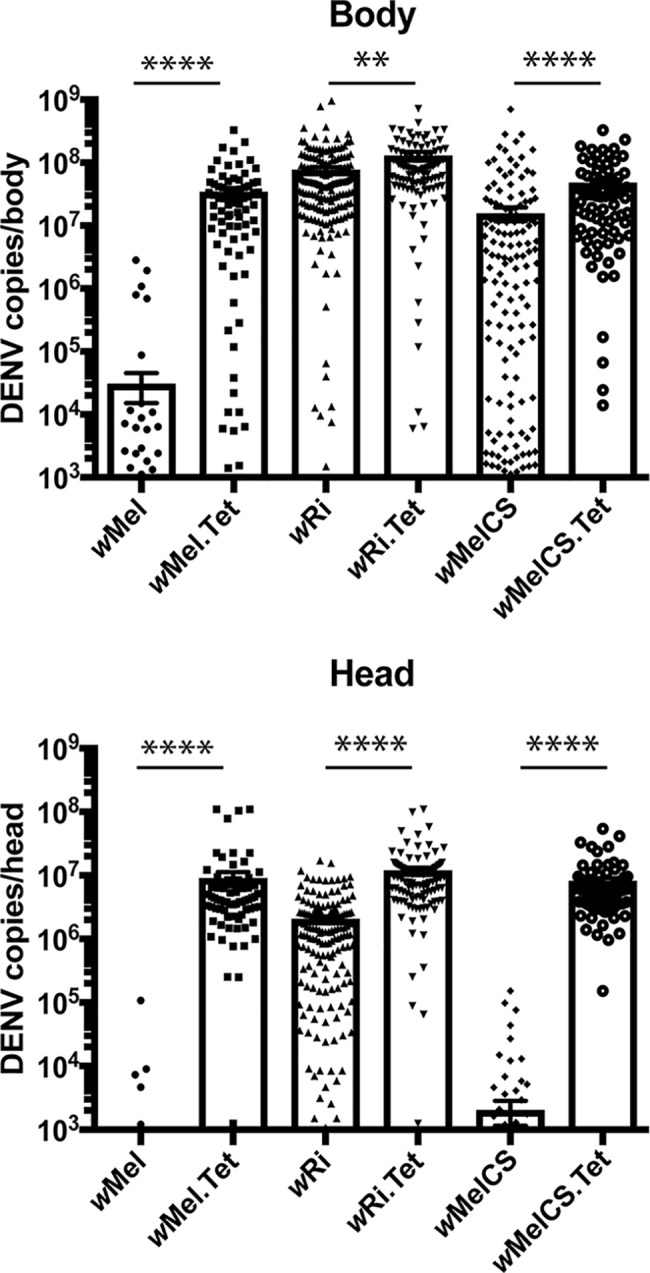
Seven-day old female mosquitoes were fed a blood meal of fresh DENV-3 (2.0 x 106 TCID50/ml) mixed 1:1 with sheep blood (n = 500 per Wolbachia-infected line, n = 200 per respective Tet control line). Mosquitoes were sorted immediately to identify those that took a blood meal, and incubated for 14 days. Mosquito heads were then separated from the thorax and total RNA was extracted from each head and remaining mosquito body. DENV genome copies were determined by qRT-PCR for each body as a measure of infection, and for each head as a measure of viral dissemination. Data are the mean genome copies per mosquito body (top) or head (bottom) ± SEM, and are representative of 3 independent experiments. Statistical analyses were performed using a Mann-Whitney test where ** p < 0.01, ****p<0.0001.
In order to more closely examine the blocking capacity of wMelCS we next used an injection challenge model which bypasses the midgut barrier leading to a higher level of tissue infection, to directly compare viral restriction in wMel and wMelCS lines. Female mosquitoes (7 days old) were injected with 2.5 x 106 TCID50/ml of DENV-3, or 10-fold dilutions thereof. Mosquitoes were incubated for 7 days before total RNA extraction from entire mosquitoes. Note that DENV-injected mosquitoes have virus delivered directly to the thorax, bypassing the midgut barrier to infection, therefore measuring dissemination rates in these mosquitoes is not a relevant test. When mosquitoes were injected with undiluted virus, both Wolbachia lines struggled to restrict viral replication with the number of DENV copies reduced by < 1log10 relative to each matched Tet control line (Fig 5A and 5B). When mosquitoes were injected with 2.5 x 105 TCID50/ml (1:10), wMelCS reduced DENV RNA copies by ~2log10 compared to <1log10 for wMel, indicating that wMelCS may be more effective at restricting replication of DENV-3 than wMel when challenged with a highly infectious setting. This trend continued as the virus was diluted further, with significantly lower viral RNA levels measured in wMelCS mosquitoes when injected with 2.5 x 104 TCID50/ml (1:100) (p<0.01, Mann-Whitney test) and substantially (although not significantly) reduced RNA levels at 2.5 x 103 TCID50/ml (1:1000). Importantly, markedly lower rates of infection were also observed in wMelCS compared to wMel-infected mosquitoes for the three lowest injected virus concentrations (“number of DENV-positive mosquitoes/total injected mosquitoes” are indicated above each bar). Together, the laboratory-based infectious blood feeding and injection challenge systems provide a relative, initial evaluation of the vector competence of these new lines, and identify wMelCS as a potential candidate to progress to more intensive human viremic blood challenge experiments.
Fig 5. wMelCS provides superior blocking of DENV-3 genome replication in a mosquito injection challenge model.
(A) DENV-3 was injected into the thorax of 6 or 7-day old female mosquitoes at 2.5 x106 TCID50/ml (undiluted) or 10, 100 or 1000-fold dilutions thereof. RNA was extracted from whole mosquito bodies 7-days post infection and virus replication was quantified by qRT-PCR. Data are the mean number of genome copies per mosquito ± SEM. Number of DENV-3 positive mosquitoes/total n are indicated above each bar. ** p < 0.01, ****p<0.0001, Mann-Whitney test. (B) The mean DENV genome copies and SEM from (A) are replotted as a function of virus concentration injected. Significant differences in the mean RNA copies of wMel and wMelCS lines are indicated by ** (p<0.01). Significant differences in the mean RNA copies of wMel.Tet and wMelCS.Tet lines are indicated by ‡ (p<0.05), Mann-Whitney test. Injection dilutions were performed as independent experiments, with the data combined to produce the final data series.
Discussion
Field releases of wMel-transinfected Ae. aegypti in order to establish Wolbachia in wild populations have been expanding over the past 6 years in multiple countries and the technology shows significant potential to have a large impact on arbovirus disease. However, there is still a need to further develop and optimize the technology to maximize its impact and to that end testing alternative strains of Wolbachia for greater field effectiveness is a priority. To most effectively measure performance, strains need to be compared in field release experiments. These tests are extremely involved, expensive and time consuming and as a result we have used a panel of laboratory-based tests to determine if a suite of newly developed strains show sufficient promise to be progressed through a development pipeline to final field comparison.
Here we report for the first time the transinfection of Ae. aegypti with Wolbachia strains wMelCS, wRi, and wPip. wMelCS-infection had minimal effect on mosquito fitness, similar to wMel. However, wRi displayed reduced MT, and the introduction of wPip lead to poor egg viability after short storage times, and weaker fecundity and hatch rates; traits that would be likely to compromise effective release strategies when performed in a field setting. Interestingly, Ae. albopictus transinfected with wPip showed similar reductions in female fecundity and egg hatch rate [32], suggesting that wPip may broadly induce this fitness cost when introduced into a non-native host. Although this may be context dependent, as work from Zhang et al. (2015) determined no fitness cost to Ae. albopictus when wPip was introduced into mosquitoes that also contained the native wAlbA and wAlbB strains [33].
When tested for their ability to block DENV, wMelCS and wRi restricted replication and dissemination following an infectious blood meal, although wRi blocking was substantially reduced compared to wMelCS, with wMelCS restricting DENV dissemination to the mosquito head to a similar extent as wMel. In a virus injection challenge model where mosquitoes are essentially overloaded with virus, wMelCS demonstrated superior blocking under all concentrations of virus tested. These findings indicate the blocking provided by wMelCS is robust, and suggests this line is worthy of further consideration in more exhaustive human viremic blood feeding experiments. While these future experiments are both costly and time consuming, they will provide a critical evaluation of vector competence, more likely to closely represent the performance of these mosquitos in the field, and may determine whether wMelCS should be progressed to field release comparisons [26, 34].
It has been inferred that the degree of blocking correlates with density of the Wolbachia strain in key tissues [35–37]. We show here that overall Wolbachia densities are only slightly higher in wMelCS-transinfected mosquitoes compared to wMel, consistent with the similar virus-blocking phenotypes. In wRi-containing mosquitoes, Wolbachia was generally found to be present at a lower density than wMel, and was significantly worse at blocking DENV-3 replication and dissemination. Interestingly, when Osborne et al. (2009) examined blocking of DCV by wRi and transinfected-wMel in D. simulans they observed almost identical densities of wRi and wMel, similar protection against virus-induced mortality, yet wRi did not reduce the viral load [20]. While this supports a role for utilizing Wolbachia densities to predict viral protection in mosquitoes, it highlights the complex nature of this tripartite interaction. Tissue specific quantitation of Wolbachia may provide further insight into the importance of Wolbachia-localization and density in virus blocking by each strain.
Given the substantial time it takes to introduce a new Wolbachia strain into Ae. aegypti, understanding how that strain is likely to behave, and the extent of pathogen protection it will provide once introduced into Ae. aegypti, will be of great benefit for ensuring effective transinfected lines are generated. Here, the increased DENV protection provided to wMelCS-infected mosquitoes compared to wMel under certain infection conditions, is in line with work performed in D. melanogaster and D. simulans using DCV and FHV [22, 23]. Similarly, wRi provided incomplete protection against these viruses in D. simulans [20], indicating that Wolbachia-infected Drosophila lines may be useful predictors for pathogen blocking in Wolbachia-infected mosquitoes. Further analyses of Ae. aegypti lines transinfected with different Drosophila-derived Wolbacha strains is required to determine if this trend holds. It should also be acknowledged that the characterisations described here were not repeated with independently generated lines, and there may be some variation between individual lines, depending on the genome and mitochondrial composition.
With a strong virus blocking phenotype, wMelCS may prove to be an effective candidate for field trials without displaying the fitness costs that prevented field establishment of wMelPop-CLA [18]. To test this prediction these mosquitoes now need to be examined in more realistic challenge settings, such as blood feeding on viremic patients.
Materials and methods
Mosquito rearing
All Ae. aegypti mosquitoes were reared and maintained as described previously [16, 26]. Adult mosquitoes were maintained at 26°C, 65% relative humidity (RH) and a 12 h light:dark cycle in a climate-controlled room. Mosquitoes were blood fed on the arms of human volunteers (Monash University human ethics permit CF11/0766-2011000387). The Wolbachia-infected wMel line included in our experiments has been described previously [15, 38]. We generated uninfected lines for each strain by treatment of the infected lines with the antibiotic tetracycline (Tet) as described earlier [16, 39]. Briefly, wMelCS, wRi and wPip adults were fed on 10% sucrose solution containing 1 mg/ml Tet right after emergence for two generations followed by a recovery period of another two generations, whereby larvae rearing water from the infected lines was added to the respective Tet-treated lines to homogenize their gut flora. Tet-lines were reared for a further minimum of 2 generations before use in maternal transmission, CI, fitness, and vector competence experiments.
To exclude any influence of age and quality of eggs used in experiments, 5-day old females from all lines were fed by the same volunteer in one sitting. The eggs collected from these females were then dried and used as experimental material within 2–3 weeks. For experiments using the wPip line, freshly laid eggs were used due to poor hatch rates following egg storage.
To exclude any influence of mosquito age on our experiments, age-controlled adults emerging within a 24 h window were used [40].
Wolbachia transinfection
The Wolbachia uninfected and inbred PGYP1.Tet line was used as the recipient line for transinfection. Wolbachia strains were not cell line adapted as described previously [16], but were instead directly isolated from donor insects and introduced into Ae. aegypti embryos by cytoplasmic transfer. wMelCS was sourced from Drosophila melanogaster Canton S strain [41], wRi was sourced from Drosophila simulans DSR strain (Riverside) [42], wPip was isolated from Culex quinquefasciatus [43] (a kind gift from Karen Williams, Westmead Hospital, Australia). Embryonic microinjection, isofemale line establishment and selection for stably-infected lines were done as previously described [15, 16] with a few modifications. In brief, the wMelCS, wRi and wPip strains were purified from their respective donor insects, and microinjected into the posterior-pole of pre-blastoderm embryos of the recipient PGYP1.tet using methodology previously described [15, 16]. Surviving G0 adult females from microinjection were mated to PGYP1.tet males, blood fed and set up for oviposition as isofemales. G0 females that laid fertile eggs were screened using quantitative PCR (qPCR) as described below. Once the newly introduced Wolbachia strain was at 100% frequency in the population and maintained itself for at least 2 consecutive generations the line was considered stable. The wMelCS and wRi lines were stable at G3 and wPip at G5.
Cytoplasmic incompatibility and maternal transmission
To investigate the level of Wolbachia-induced cytoplasmic incompatibility (CI) we set up paired crosses between Wolbachia-infected and Tet-treated Ae. aegypti. For each Wolbachia strain four crosses were set up: (1) a CI cross (Tet females x infected males), (2) a maternal transmission (MT) cross (infected females x Tet males), (3) an uninfected control cross (Tet females x Tet males) and (4) an infected control cross (infected females x infected males). Each cross consisted of a group of 70 virgin males and 70 virgin females and each group was allowed to mate for 5 days after emergence. On day five all females were blood fed by one human volunteer. Gravid females were aspirated into individual tubes three-days post blood feeding and allowed to oviposit on wet filter paper [26]. Three-days post oviposition female mosquitoes were removed and tested for presence of Wolbachia by qPCR using strain-specific primers as described in the PCR section.
Any paper containing less than 15 eggs was discarded. The egg papers from the remaining females were photographed and eggs were counted manually. The papers were then submerged in hatching water. Two days later the number of hatched larvae (2nd instar) was counted. Newly hatched larvae were counted daily until no hatching was recorded for three consecutive days. Any unhatched eggs were dried briefly for 48–72 h, hatched and counted again to get the maximum hatch.
To quantify the success of MT of Wolbachia to the next generation, hatched larvae from the MT cross (infected females x Tet males) were reared to adults and screened by qPCR for the presence of Wolbachia.
Fitness determinants
Egg diapause viability
To assess the viability of eggs stored over time, age-controlled adults (emergence within 24 h; 100 males and 100 females) were placed in quadruplet 30x20x20cm cages. One cage of each line was blood-fed by the same human volunteer, to control for the potential variation caused by blood-meal quality and composition. This was repeated 4 times with 4 different volunteers. Female mosquitoes were given a blood meal at 5 days old. Three-days post blood meal, 12 oviposition cups were placed in each cage for three days. Approximately 72 h post oviposition, egg cups were removed, dried slowly over 3–5 days then stored in Whirlpak bags (Sigma) in an airtight container with saturated KCl solution to maintain humidity [16]. Batches of 100–500 eggs were photographed and counted, then hatched after 1, 2, 3, 4, 6, 8, 10 and 12 weeks from each cage. Hatched larvae were counted at 2nd instar stage until no hatch was observed for a week then the percent hatch calculated.
Fecundity and hatch rate
This was performed using the parameters described for CI and MT above. Fecundity was calculated as the average number of eggs laid/female, and hatch rate determined as the number of 2nd instar larvae per total eggs hatched.
Adult longevity
Age-controlled adults (approximately 150 males and 150 females) were placed in triplicate 30x30x30cm cages. Mosquitoes were provided fresh 10% sucrose twice/week and maintained at 26°C, 65% RH and a 12:12 h light:dark cycle in a climate-controlled room. The number of dead males and/or females was recorded and removed daily until all mosquitoes in the cages were dead.
Wolbachia detection by PCR
The presence or absence of Wolbachia in transinfected mosquitoes was confirmed by qPCR using strain-specific primers. wPip was detected by amplifying an IS2 transposon, with 49 identical copies of this sequence in the wPip genome (forward primer: 5’-GCACTTACCCTAACCAAAGGTAAC-3’, reverse primer: 5’ CTAACTTTAGGCCTCTATCGAAGAG-3’), wRi was detected by amplifying a part of the gene WRi_009390, which encodes a hypothetical protein (forward primer: 5’CATGCCAATAACGAAATAGC -3’, reverse primer: 5’-TAGCAACTTTTCTTGCGAAC-3’). A combination of two primer sets was used to differentiate wMelCS from wMel strains. One of the primers sets binds to wMel and wMelCS in the WD0513 region of the genome [34, 44] (forward primer: 5’CAAATTGCTCTTGTCCTGTGG-3’, reverse primer: 5’-GGGTGTTAAGCAGAGTTACGG-3’), Probe Cy5 TGAAATGGAAAAATTGGCGAGGTGTAGG- BHQ3. The second primer set binds to the polymorphic insertion sites of wMelCS at loci IS5-WD1310 (forward primer: 5’- CTCATCTTTACCCCGTACTAAAATTTC-3’, reverse primer: 5’-TCTTCCTCATTAAGAACCTCTATCTTG-3’), Probe HEX TAGCCTTTTACTTGTTTCCGGACAACCT-BHQ1. For each sample, qPCR was performed using a LightCycler 480 II Instrument (Roche) using LightCycler 480 SYBR Green I Master (Roche) for wPip and wRi amplification and Taqman Probes Master for amplifying wMelCS according to the manufacturer’s protocol.
Wolbachia density and distribution
The density of Wolbachia in the three newly introduced strains, wMelCS, wRi and wPip was compared to the density in the long established wMel strain [15, 16]. Total relative Wolbachia densities for the four lines were determined in whole, female mosquitoes at 5-, 10- or 15-days post emergence, using qPCR with primers to amplify a fragment of the gene coding 16S rRNA (16S) (forward primer: 5’-GAGTGAAGAAGGCCTTTGGG-3’, reverse primer: 5’- CACGGAGTTAGCCAGGACTTC-3’, probe 5’ LC640-CTGTGAGTACCGTCATTATCTTCCTCACT-IowaBlackRQ-3’) and the reference Ae. aegypti rps17 gene (forward primer: 5’-TCCGTGGT ATCTCCATCAAGCT-3’, reverse primer: 5’-CACTTCCGGCACGTAGTTGTC-3’, probe 5’FAM- CAGGAGGAGGAACGTGAGCGCAG-BHQ1-3’). Wolbachia densities were quantified relative to rps17 using the delta CT method (2CT(reference)/ 2CT(target)).
The distribution of wMelCS, wRi and wPip Wolbachia in mosquitoes was determined in sections of paraffin-embedded female mosquitoes (5 to 7 days old) using fluorescence in situ hybridisation (FISH), as described in Moreira et al. (2009) [10]. The fluorescently labelled probes used can detect the 16S rRNA from all three Wolbachia strains. Total DNA was stained in blue using DAPI and a green filter was included to increase contrast with surrounding tissues.
Vector competence
DENV-3 Cairns 08/09 strain stocks (Genbank accession number: JN406515.1) were prepared by inoculation of C6/36 cells with a multiplicity of infection (MOI) of 0.1 and collection of culture supernatant 6–7 days later. Virus concentrations were determined by TCID50 as previously described [45] using monoclonal antibody 4G2 [46].
For feeding experiments with DENV-3 (Cairns 08/09) infected blood, 100 seven-day old age-controlled female mosquitoes were placed in 500 mL plastic containers (five containers per Wolbachia line, two containers per Tet line), starved for up to 24 h and allowed to feed on a 50:50 mixture of defibrinated sheep blood and tissue culture supernatant containing freshly harvested 2.0 x 106 TCID50/mL of DENV-3. Feeding was done through a piece of desalted porcine intestine stretched over a water-jacketed membrane feeding apparatus preheated to 37°C. Mosquitoes were left to feed in the dark for approximately 80 min. Fully engorged mosquitoes were placed in 500 mL containers at a density of < 20/container, and incubated for 14 d at 26°C with 65% RH and a 12 h light/dark cycle.
For adult microinjections, 60 six or seven-day old age-controlled female mosquitoes were anesthetized by CO2. The mosquitoes were injected intrathoracically with 69 nL of DENV-3 Cairns 08/09 strain, 2.5 x106 TCID50/ml (or 10, 100 or 1000-fold dilutions thereof) in RPMI media (Life Technologies) using a pulled-glass capillary and a handheld microinjector (Nanoject II, Drummond Scientific). Injected mosquitoes were incubated for 7 days (10 mosquitoes/cup) at 26°C with 65% RH and a 12 h light/dark cycle. To quantify DENV-3 genomic copies, total RNA was isolated from DENV-3 mosquitoes (entire mosquitoes for injection experiments, or head and bodies separately for blood fed mosquitoes) using the RNeasy 96 QIAcube HT kit (Qiagen). DENV-3 RNA was amplified by qRT-PCR (LightCycler Multiplex RNA Virus Master, Roche), using primers to the conserved 3’UTR: Forward 5’-AAGGACTAGAGGTTAGAGGAGACCC; Reverse 5’- CGTTCTGTGCCTGGAATGATG; Probe 5’-HEX- AACAGCATATTGACGCTGGGAGAGACCAGA-BHQ1-3’and absolute copies determined by extrapolation from an internal standard curve generated from plasmid DNA encoding the conserved 3’UTR sequence. Mosquito extracts with ≥1000 copies of DENV per body were scored positive, based on the LOD95 (limit of detection 95%) for DENV-3 with this primer set.
Data availability
All raw data are available within S1 Data.
Ethics statement
Blood feeding by volunteers (Monash University human ethics permit no CF11/0766-2011000387) for this study was approved by the Monash University Human Research Ethics Committee (MUHREC). All adult volunteers provided informed written consent; no child participants were involved in the study.
Supporting information
Bidirectional compatibility between wMel and wMelCS lines was determined by crossing Wolbachia-infected females and males from each line, with control CI crosses performed between uninfected female mosquitoes (WT) and Wolbachia-infected males of each line. Bars are the mean percentage hatch rate ± SEM from 30 females (individual data points are superimposed).
(TIF)
(DOCX)
(XLSX)
Acknowledgments
We wish to thank Breeanna McLean, Johanna Duyvestyn, Etiene C. Pacidônio, Daniela S. Gonçalves, Silk Lin and Rupali Singh for technical assistance, and Prof. Cameron Simmons for his help with manuscript editing.
Data Availability
All raw data has been uploaded as a Supplemental Data Set.
Funding Statement
This work was funded in part by a grant from the Foundation for the National Institutes of Health through the Grand Challenges in Global Health initiative of the Bill & Melinda Gates Foundation and by a Wellcome Trust Award No.102591. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Capeding MR, Tran NH, Hadinegoro SR, Ismail HI, Chotpitayasunondh T, Chua MN, et al. Clinical efficacy and safety of a novel tetravalent dengue vaccine in healthy children in Asia: a phase 3, randomised, observer-masked, placebo-controlled trial. Lancet. 2014;384(9951):1358–65. doi: 10.1016/S0140-6736(14)61060-6 . [DOI] [PubMed] [Google Scholar]
- 2.Hadinegoro SR, Arredondo-Garcia JL, Capeding MR, Deseda C, Chotpitayasunondh T, Dietze R, et al. Efficacy and Long-Term Safety of a Dengue Vaccine in Regions of Endemic Disease. N Engl J Med. 2015;373(13):1195–206. doi: 10.1056/NEJMoa1506223 . [DOI] [PubMed] [Google Scholar]
- 3.Simmons CP. A Candidate Dengue Vaccine Walks a Tightrope. N Engl J Med. 2015;373(13):1263–4. doi: 10.1056/NEJMe1509442 . [DOI] [PubMed] [Google Scholar]
- 4.Pang EL, Loh HS. Towards development of a universal dengue vaccine—How close are we? Asian Pac J Trop Med. 2017;10(3):220–8. doi: 10.1016/j.apjtm.2017.03.003 . [DOI] [PubMed] [Google Scholar]
- 5.Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyes CL, et al. The global distribution and burden of dengue. Nature. 2013;496(7446):504–7. doi: 10.1038/nature12060 ; PubMed Central PMCID: PMCPMC3651993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoffmann AA, Montgomery BL, Popovici J, Iturbe-Ormaetxe I, Johnson PH, Muzzi F, et al. Successful establishment of Wolbachia in Aedes populations to suppress dengue transmission. Nature. 2011;476(7361):454–7. doi: 10.1038/nature10356 . [DOI] [PubMed] [Google Scholar]
- 7.Zug R, Hammerstein P. Still a host of hosts for Wolbachia: analysis of recent data suggests that 40% of terrestrial arthropod species are infected. PLoS One. 2012;7(6):e38544 doi: 10.1371/journal.pone.0038544 ; PubMed Central PMCID: PMC3369835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Serbus LR, Casper-Lindley C, Landmann F, Sullivan W. The genetics and cell biology of Wolbachia-host interactions. Annu Rev Genet. 2008;42:683–707. doi: 10.1146/annurev.genet.41.110306.130354 . [DOI] [PubMed] [Google Scholar]
- 9.Werren JH, Baldo L, Clark ME. Wolbachia: master manipulators of invertebrate biology. Nat Rev Microbiol. 2008;6(10):741–51. doi: 10.1038/nrmicro1969 . [DOI] [PubMed] [Google Scholar]
- 10.Moreira LA, Iturbe-Ormaetxe I, Jeffery JA, Lu G, Pyke AT, Hedges LM, et al. A Wolbachia symbiont in Aedes aegypti limits infection with dengue, Chikungunya, and Plasmodium. Cell. 2009;139(7):1268–78. doi: 10.1016/j.cell.2009.11.042 . [DOI] [PubMed] [Google Scholar]
- 11.Rances E, Johnson TK, Popovici J, Iturbe-Ormaetxe I, Zakir T, Warr CG, et al. The toll and Imd pathways are not required for wolbachia-mediated dengue virus interference. J Virol. 2013;87(21):11945–9. doi: 10.1128/JVI.01522-13 ; PubMed Central PMCID: PMC3807350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Caragata EP, Rances E, Hedges LM, Gofton AW, Johnson KN, O'Neill SL, et al. Dietary cholesterol modulates pathogen blocking by Wolbachia. PLoS Pathog. 2013;9(6):e1003459 doi: 10.1371/journal.ppat.1003459 ; PubMed Central PMCID: PMC3694857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bian G, Xu Y, Lu P, Xie Y, Xi Z. The endosymbiotic bacterium Wolbachia induces resistance to dengue virus in Aedes aegypti. PLoS Pathog. 2010;6(4):e1000833 doi: 10.1371/journal.ppat.1000833 ; PubMed Central PMCID: PMC2848556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Min KT, Benzer S. Wolbachia, normally a symbiont of Drosophila, can be virulent, causing degeneration and early death. Proc Natl Acad Sci U S A. 1997;94(20):10792–6. ; PubMed Central PMCID: PMCPMC23488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walker T, Johnson PH, Moreira LA, Iturbe-Ormaetxe I, Frentiu FD, McMeniman CJ, et al. The wMel Wolbachia strain blocks dengue and invades caged Aedes aegypti populations. Nature. 2011;476(7361):450–3. doi: 10.1038/nature10355 . [DOI] [PubMed] [Google Scholar]
- 16.McMeniman CJ, Lane RV, Cass BN, Fong AW, Sidhu M, Wang YF, et al. Stable introduction of a life-shortening Wolbachia infection into the mosquito Aedes aegypti. Science. 2009;323(5910):141–4. doi: 10.1126/science.1165326 . [DOI] [PubMed] [Google Scholar]
- 17.McMeniman CJ, O'Neill SL. A virulent Wolbachia infection decreases the viability of the dengue vector Aedes aegypti during periods of embryonic quiescence. PLoS Negl Trop Dis. 2010;4(7):e748 doi: 10.1371/journal.pntd.0000748 ; PubMed Central PMCID: PMC2903475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nguyen TH, Nguyen HL, Nguyen TY, Vu SN, Tran ND, Le TN, et al. Field evaluation of the establishment potential of wmelpop Wolbachia in Australia and Vietnam for dengue control. Parasit Vectors. 2015;8:563 doi: 10.1186/s13071-015-1174-x ; PubMed Central PMCID: PMCPMC4625535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yeap HL, Mee P, Walker T, Weeks AR, O'Neill SL, Johnson P, et al. Dynamics of the "popcorn" Wolbachia infection in outbred Aedes aegypti informs prospects for mosquito vector control. Genetics. 2011;187(2):583–95. doi: 10.1534/genetics.110.122390 ; PubMed Central PMCID: PMCPMC3030498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Osborne SE, Leong YS, O'Neill SL, Johnson KN. Variation in antiviral protection mediated by different Wolbachia strains in Drosophila simulans. PLoS Pathog. 2009;5(11):e1000656 doi: 10.1371/journal.ppat.1000656 ; PubMed Central PMCID: PMCPMC2768908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martinez J, Duplouy A, Woolfit M, Vavre F, O'Neill SL, Varaldi J. Influence of the virus LbFV and of Wolbachia in a host-parasitoid interaction. PLoS One. 2012;7(4):e35081 doi: 10.1371/journal.pone.0035081 ; PubMed Central PMCID: PMCPMC3338833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chrostek E, Marialva MS, Esteves SS, Weinert LA, Martinez J, Jiggins FM, et al. Wolbachia variants induce differential protection to viruses in Drosophila melanogaster: a phenotypic and phylogenomic analysis. PLoS Genet. 2013;9(12):e1003896 doi: 10.1371/journal.pgen.1003896 ; PubMed Central PMCID: PMCPMC3861217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martinez J, Longdon B, Bauer S, Chan YS, Miller WJ, Bourtzis K, et al. Symbionts commonly provide broad spectrum resistance to viruses in insects: a comparative analysis of Wolbachia strains. PLoS Pathog. 2014;10(9):e1004369 doi: 10.1371/journal.ppat.1004369 ; PubMed Central PMCID: PMCPMC4169468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martinez J, Ok S, Smith S, Snoeck K, Day JP, Jiggins FM. Should Symbionts Be Nice or Selfish? Antiviral Effects of Wolbachia Are Costly but Reproductive Parasitism Is Not. PLoS Pathog. 2015;11(7):e1005021 doi: 10.1371/journal.ppat.1005021 ; PubMed Central PMCID: PMCPMC4488530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xi Z, Khoo CC, Dobson SL. Wolbachia establishment and invasion in an Aedes aegypti laboratory population. Science. 2005;310(5746):326–8. doi: 10.1126/science.1117607 . [DOI] [PubMed] [Google Scholar]
- 26.Joubert DA, Walker T, Carrington LB, De Bruyne JT, Kien DH, Hoang Nle T, et al. Establishment of a Wolbachia Superinfection in Aedes aegypti Mosquitoes as a Potential Approach for Future Resistance Management. PLoS Pathog. 2016;12(2):e1005434 doi: 10.1371/journal.ppat.1005434 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Axford JK, Ross PA, Yeap HL, Callahan AG, Hoffmann AA. Fitness of wAlbB Wolbachia Infection in Aedes aegypti: Parameter Estimates in an Outcrossed Background and Potential for Population Invasion. Am J Trop Med Hyg. 2016;94(3):507–16. doi: 10.4269/ajtmh.15-0608 ; PubMed Central PMCID: PMCPMC4775882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ross PA, Wiwatanaratanabutr I, Axford JK, White VL, Endersby-Harshman NM, Hoffmann AA. Wolbachia Infections in Aedes aegypti Differ Markedly in Their Response to Cyclical Heat Stress. PLoS Pathog. 2017;13(1):e1006006 doi: 10.1371/journal.ppat.1006006 ; PubMed Central PMCID: PMCPMC5215852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ulrich JN, Beier JC, Devine GJ, Hugo LE. Heat Sensitivity of wMel Wolbachia during Aedes aegypti Development. PLoS Negl Trop Dis. 2016;10(7):e0004873 doi: 10.1371/journal.pntd.0004873 ; PubMed Central PMCID: PMCPMC4961373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Glaser RL, Meola MA. The native Wolbachia endosymbionts of Drosophila melanogaster and Culex quinquefasciatus increase host resistance to West Nile virus infection. PLoS One. 2010;5(8):e11977 doi: 10.1371/journal.pone.0011977 ; PubMed Central PMCID: PMCPMC2916829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brady OJ, Johansson MA, Guerra CA, Bhatt S, Golding N, Pigott DM, et al. Modelling adult Aedes aegypti and Aedes albopictus survival at different temperatures in laboratory and field settings. Parasit Vectors. 2013;6:351 doi: 10.1186/1756-3305-6-351 ; PubMed Central PMCID: PMCPMC3867219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Calvitti M, Moretti R, Lampazzi E, Bellini R, Dobson SL. Characterization of a new Aedes albopictus (Diptera: Culicidae)-Wolbachia pipientis (Rickettsiales: Rickettsiaceae) symbiotic association generated by artificial transfer of the wPip strain from Culex pipiens (Diptera: Culicidae). J Med Entomol. 2010;47(2):179–87. . [DOI] [PubMed] [Google Scholar]
- 33.Zhang D, Zheng X, Xi Z, Bourtzis K, Gilles JR. Combining the sterile insect technique with the incompatible insect technique: I-impact of wolbachia infection on the fitness of triple- and double-infected strains of Aedes albopictus. PLoS One. 2015;10(4):e0121126 doi: 10.1371/journal.pone.0121126 ; PubMed Central PMCID: PMCPMC4388707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ferguson NM, Kien DT, Clapham H, Aguas R, Trung VT, Chau TN, et al. Modeling the impact on virus transmission of Wolbachia-mediated blocking of dengue virus infection of Aedes aegypti. Sci Transl Med. 2015;7(279):279ra37 doi: 10.1126/scitranslmed.3010370 ; PubMed Central PMCID: PMCPMC4390297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lu P, Bian G, Pan X, Xi Z. Wolbachia induces density-dependent inhibition to dengue virus in mosquito cells. PLoS Negl Trop Dis. 2012;6(7):e1754 doi: 10.1371/journal.pntd.0001754 ; PubMed Central PMCID: PMC3404113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Osborne SE, Iturbe-Ormaetxe I, Brownlie JC, O'Neill SL, Johnson KN. Antiviral protection and the importance of Wolbachia density and tissue tropism in Drosophila simulans. Appl Environ Microbiol. 2012;78(19):6922–9. doi: 10.1128/AEM.01727-12 ; PubMed Central PMCID: PMC3457512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Frentiu FD, Robinson J, Young PR, McGraw EA, O'Neill SL. Wolbachia-mediated resistance to dengue virus infection and death at the cellular level. PLoS One. 2010;5(10):e13398 doi: 10.1371/journal.pone.0013398 ; PubMed Central PMCID: PMCPMC2955527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ye YH, Carrasco AM, Frentiu FD, Chenoweth SF, Beebe NW, van den Hurk AF, et al. Wolbachia Reduces the Transmission Potential of Dengue-Infected Aedes aegypti. PLoS Negl Trop Dis. 2015;9(6):e0003894 doi: 10.1371/journal.pntd.0003894 ; PubMed Central PMCID: PMCPMC4482661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dobson SL, Rattanadechakul W. A novel technique for removing Wolbachia infections from Aedes albopictus (Diptera: Culicidae). J Med Entomol. 2001;38(6):844–9. . [DOI] [PubMed] [Google Scholar]
- 40.Yamada R, Floate KD, Riegler M, O'Neill SL. Male development time influences the strength of Wolbachia-induced cytoplasmic incompatibility expression in Drosophila melanogaster. Genetics. 2007;177(2):801–8. doi: 10.1534/genetics.106.068486 ; PubMed Central PMCID: PMCPMC2034644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Riegler M, Sidhu M, Miller WJ, O'Neill SL. Evidence for a global Wolbachia replacement in Drosophila melanogaster. Curr Biol. 2005;15(15):1428–33. doi: 10.1016/j.cub.2005.06.069 . [DOI] [PubMed] [Google Scholar]
- 42.Hoffmann AA, Turelli M, Simmons GM. Unidirectional Incompatibility between Populations of Drosophila Simulans. Evolution. 1986;40(4):692–701. doi: 10.1111/j.1558-5646.1986.tb00531.x . [DOI] [PubMed] [Google Scholar]
- 43.Walker T, Klasson L, Sebaihia M, Sanders MJ, Thomson NR, Parkhill J, et al. Ankyrin repeat domain-encoding genes in the wPip strain of Wolbachia from the Culex pipiens group. BMC Biol. 2007;5:39 doi: 10.1186/1741-7007-5-39 ; PubMed Central PMCID: PMCPMC2045654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Woolfit M, Iturbe-Ormaetxe I, Brownlie JC, Walker T, Riegler M, Seleznev A, et al. Genomic evolution of the pathogenic Wolbachia strain, wMelPop. Genome Biol Evol. 2013;5(11):2189–204. doi: 10.1093/gbe/evt169 ; PubMed Central PMCID: PMCPMC3845649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.O'Brien CA, Hobson-Peters J, Yam AW, Colmant AM, McLean BJ, Prow NA, et al. Viral RNA intermediates as targets for detection and discovery of novel and emerging mosquito-borne viruses. PLoS Negl Trop Dis. 2015;9(3):e0003629 doi: 10.1371/journal.pntd.0003629 ; PubMed Central PMCID: PMCPMC4370754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gentry MK, Henchal EA, McCown JM, Brandt WE, Dalrymple JM. Identification of distinct antigenic determinants on dengue-2 virus using monoclonal antibodies. Am J Trop Med Hyg. 1982;31(3 Pt 1):548–55. . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Bidirectional compatibility between wMel and wMelCS lines was determined by crossing Wolbachia-infected females and males from each line, with control CI crosses performed between uninfected female mosquitoes (WT) and Wolbachia-infected males of each line. Bars are the mean percentage hatch rate ± SEM from 30 females (individual data points are superimposed).
(TIF)
(DOCX)
(XLSX)
Data Availability Statement
All raw data has been uploaded as a Supplemental Data Set.
All raw data are available within S1 Data.