SUMMARY
Understanding how broadly neutralizing antibodies (bnAbs) to HIV envelope (Env) develop during natural infection can help guide the rational design of an HIV vaccine. Here, we described a bnAb lineage targeting the Env V2 apex and the Ab-Env co-evolution that led to development of neutralization breadth. The lineage Abs bore an anionic heavy chain complementarity-determining region 3 (CDRH3) of 25 amino acids, among the shortest known for this class of Abs, and achieved breadth with only 10% nucleotide somatic hypermutation and no insertions or deletions. The data suggested a role for Env glycoform heterogeneity in the activation of the lineage germ-line B cell. Finally, we showed that localized diversity at key V2 epitope residues drove bnAb maturation toward breadth, mirroring the Env evolution pattern described for another donor who developed V2-apex targeting bnAbs. Overall, these findings suggest potential strategies for vaccine approaches based on germline-targeting and serial immunogen design.
In Brief
Understanding the molecular basis of HIV Env-specific broadly neutralizing antibodies (bnAbs) development is key for vaccine design. Landais et al. find that glycan heterogeneity played a role in the activation of V2 apex PCT64 bnAbs precursor and that viral evolution was similar to CAP256, another donor with V2 apex bnAbs.
INTRODUCTION
Elicitation of broadly neutralizing antibodies (bnAbs), i.e., those capable of neutralizing a large fraction of global HIV-1 isolates, is thought to be highly desirable for development of an effective HIV-1 vaccine (reviewed in Burton et al., 2012; Fauci and Marston, 2014). However, this goal has yet to be achieved by a vaccine candidate. During natural HIV infection, a small fraction of individuals (up to 10%) develop broad and potent Ab responses (Doria-Rose et al., 2010; Gray et al., 2007; Hraber et al., 2014; Landais et al., 2016; Rusert et al., 2016; Simek et al., 2009; van Gils et al., 2009). BnAbs isolated from these individuals target at least five conserved epitope regions on Env: the V3-high mannose patch, V2-apex, CD4 binding site (CD4bs), the membrane proximal external region (MPER), and gp120-gp41 interface, including the fusion peptide (Burton and Hangartner, 2016; Kong et al., 2016). In many cases, HIV-1 bnAbs have unusual features such as long heavy chain complementarity-determining regions 3 (CDRH3s), high levels of somatic hypermutation (SHM), and insertions and deletions (indels) (Briney et al., 2012; Klein et al., 2013; Walker et al., 2011). Thus, it is of great importance to understand to what extent these features are required for neutralization breadth.
The developmental pathways of only four bnAb lineages and their interplay with Env have been described to date; these lineages target the CD4bs (Bonsignori et al., 2016; Gao et al., 2014; Liao et al., 2013), V2-apex (Bhiman et al., 2015; Doria-Rose et al., 2015; 2014), and V3-high mannose patch (Bonsignori et al., 2017; MacLeod et al., 2016). These studies demonstrated how key changes in autologous viral Env at specific times during Ab development appeared to be critical for driving the emergence of breadth and potency. Nevertheless, our knowledge of bnAb elicitation during natural infection remains severely limited. In particular, it is necessary to study Ab developmental pathways in a variety of individuals that share the same broad antibody specificity, as similarities among responses in different donors in the co-evolution of Ab and Env, or in the nature of the Env that triggered broad lineages, might reveal promising paths for immunogen design.
BnAbs targeting the V2-apex, such as PG9/16, PGT145/PGDM1400, and CAP256-25, are detected in ~10%–15% of individuals who develop bnAb responses (Doria-Rose et al., 2017; Landais et al., 2016; Rusert et al., 2016) and are among the most potent and broad HIV-1 neutralizing Abs isolated to date (Doria-Rose et al., 2015; Sok et al., 2014; Walker et al., 2011; 2009). Although most V2-apex bnAbs appear to share important structural features (Andrabi et al., 2015; Doria-Rose et al., 2014; Gorman et al., 2016; Pejchal et al., 2011; Sok et al., 2014), it is not clear whether commonalities also arise in their development. Furthermore, a new V2-apex bnAb lineage that does not share such features was recently described (Cale et al., 2017).
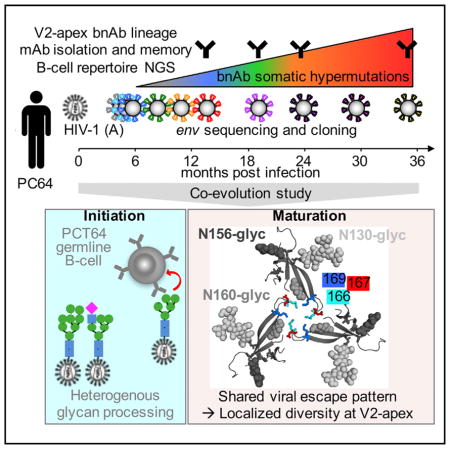
Here we report a longitudinal study of a V2-apex-targeting bnAb lineage, in the context of autologous virus evolution, in an African donor from the International AIDS Vaccine Initiative (IAVI) Protocol C cohort (Landais et al., 2016). Comparison of our findings with CAP256-VRC26 bnAbs targeting the same epitope revealed a common Env escape pathway (Bhiman et al., 2015; Doria-Rose et al., 2015; 2014). In addition, we demonstrated the role of glycoform heterogeneity in early precursor activation and illustrated which Ab structural features were associated with breadth acquisition. Overall, the data provide critical information for vaccine strategies to elicit V2-apex bnAbs based on germline targeting and use of sequential immunogens.
RESULTS
Functional Screening Identifies PCT64, a 25-aa CDRH3 V2-apex bnAb Lineage
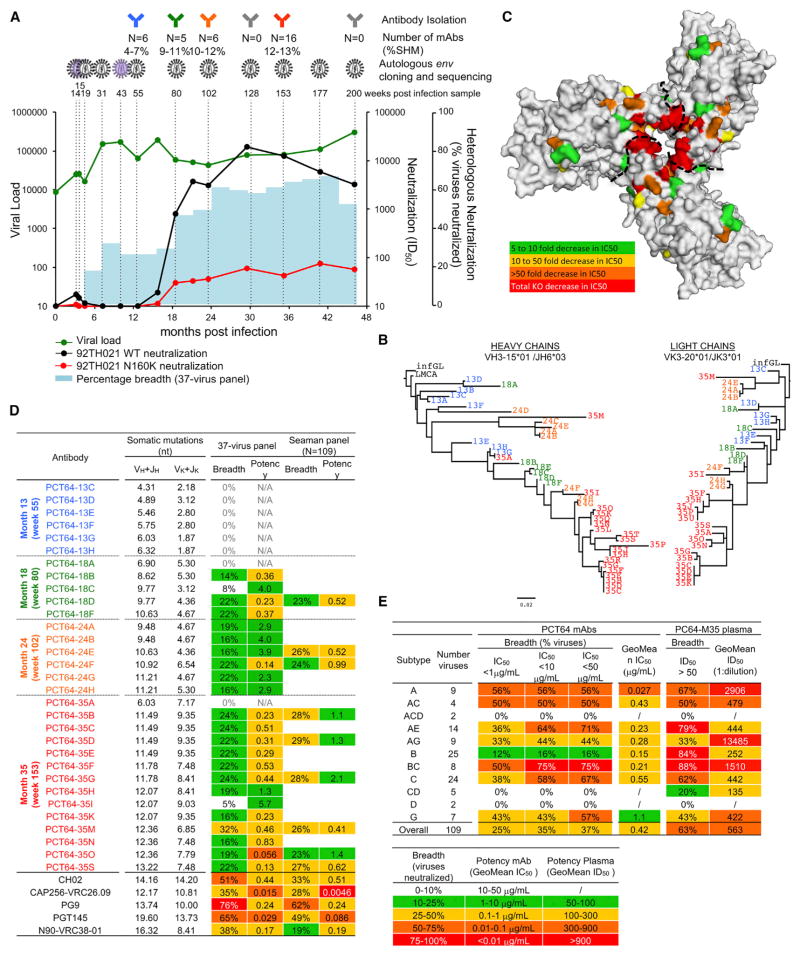
Donor PC64 is an HIV-1 subtype A-infected individual with V2-apex bnAb plasma responses as indicated by sensitivity to N160 glycan knockout (Landais et al., 2016). Peripheral blood mononuclear cells (PBMCs) collected at 13, 18, 24, 29, 35, and 46 months post infection (mpi) (Figure 1A) were used to isolate monoclonal antibodies (mAbs). Following isolation and activation of memory B cells, antibody-containing supernatants were screened for specific neutralizing activity using a high-throughput functional approach (Walker et al., 2011; 2009). Supernatant from month 18 (M18), M24, and M35 samples were screened on wild-type versus N160 glycan knockout viruses to identify V2-targeting bnAbs, while early M13 supernatants were tested against early M3 and M7 autologous viral clones to identify potentially related less-matured mAbs of narrower breadth and lower potency. We identified somatically related clusters of 42 heavy-chain (HC) sequences and 34 light-chain (LC) sequences (Figures 1B, S1A, Table S1) encoded by variable gene alleles VH3-15*01/DH3-3*01/JH6*03 and Vκ3-20*01/Jκ3*01, respectively (Table 1), with a relatively long CRDH3 of 25 amino acids (aa) containing several Tyr residues. The corresponding 33 mAbs isolated between 13 and 35 mpi defined the PCT64 lineage, ranging from 2.6% to 13.5% (VH+JH) and 2.2% to 9.0% (VK+JK) nucleotide mutation.
Figure 1. Functional Screening Identifies a New V2-Apex Specific Broadly Neutralizing Antibody Lineage.
(A) Longitudinal plasma samples from donor PC64 were tested for neutralization against heterologous pseudoviruses. The percent of viruses neutralized (> 50% inhibition of infectivity at the lowest plasma dilution, 1:50) from a cross-clade (A, B, C) 37-virus panel is shown as shaded blue bars. Neutralization Inhibitory Dilution 50 (ID50, plasma dilution giving 50% inhibition of infection) of PC64 longitudinal plasma samples against 92TH021 WT (black circles) and N160K mutant (red circles) pseudoviruses are plotted. The evolution of the viral load (green circles) in the plasma is also plotted. The time points at which PCT64 antibodies were isolated and env sequenced and/or cloned from plasma are indicated by corresponding symbols. The number (N) of PCT64 antibodies isolated and their range of somatic hypermutation frequency (%SHM) is also indicated. See also Table S1.
(B) Evolutionary distance between the PCT64 mAbs is illustrated as a phylogeny for both heavy and light chain nucleotide sequences.
(C) Mapping of the PCT64 epitope. Fold decrease in neutralization IC50 of individual JR-CSF Ala mutants by PCT64-35G as compared to WT is color-coded as indicated and presented on the BG505 SOSIP.664 trimer structure (Julien et al., 2013) based on HxB2 numbering and alignments. Data are representative of at least two independent experiments. See also Table S3.
(D) PCT64 mAbs characteristics. Heavy and light chain V- and J-gene nucleotide (nt) somatic mutation percentages, neutralization breadth (% virus neutralized at IC50<50 μg/mL) and potency (GeoMean IC50 in μg/mL) on a 37- and 109-virus panel are tabulated and color-coded as indicated. See also Tables S1, S5, and S6. Data are representative of at least two independent experiments.
(E) Comparison of PC64 plasma neutralization breadth and potency on the 109-virus panel subdivided by subtype, with that of a theoretical combination of PCT64 mAbs, color-coded as indicated. See also Tables S2, S4, S7, and Figure S1A.
Table 1.
Gene Usage of V2-Apex Specific bnAbs
| mAbs | Putative heavy chain gene alleles | VH+JH (nt) mutation frequency | VH+JH (aa) mutation frequency | CDRH3 aa sequence | CDRH3 length (aa) | ||
|---|---|---|---|---|---|---|---|
| PCT64-35S | VH3-15*01 | DH3-3*01 | JH6*03 | 13.22 | 20.69 | MTGVERGDFWSDDYSQHYNTYLIDV | 25 |
| CAP256-VRC26.09 | VH3-30*18 | DH3-3*01 | JH3*01 | 14.49 | 21.74 | VKDQREDECEEWWSDYYDFGREL PCRKSRGLGLAGIFDM | 39 |
| CH04 | VH3-20*01 | DH3-10*01 | JH2*01 | 12.75 | 22.81 | ARGTDYTIDDQGIRYQGSGTFWYFDV | 26 |
| PG9 | VH3-33*05 | DH3-3*01 | JH6*03 | 13.74 | 17.80 | VREAGGPDYRNGYNYYDFYDGYYN YHYMDV | 30 |
| PGT145 | VH1-8*01 | DH4-17*01 | JH6*02 | 19.60 | 29.57 | LTGSKHRLRDYFLYNEYGPNYEEWGD YLATLDV | 33 |
| N90-VRC38.01 | VH3-13*01 | DH6-19*01 | JH6*02 | 16.32 | 28.57 | GPESGWFYHYYWGLGV | 16 |
| mAbs | Putative light chain gene alleles | VK+JK (nt) mutation frequency | VK+JK (aa) mutation frequency | CDRL3 aa sequence | CDRL3 length (aa) | ||
| PCT64-35S | VK3-20*01 | / | JK3*01 | 7.48 | 14.02 | RQYETSFT | 8 |
| CAP256-VRC26.09 | VL1-51*02 | / | JL1*02 | 10.51 | 15.32 | AVWGVRRGAGAVF | 13 |
| CH04 | VK3-20*01 | / | JK1*01 | 10.80 | 16.67 | QQYGRSPYT | 9 |
| PG9 | VL2-14*01 | / | JL3*02 | 10.00 | 14.55 | KSLTSTRRRV | 10 |
| PGT145 | VK2-28*01 | / | JK1*01 | 13.73 | 21.43 | MQGLHSPWT | 9 |
| N90-VRC38.01 | VK2-28*01 | / | JK4*01 | 8.41 | 11.71 | MEARQTPRLT | 10 |
V2 Apex bnAbs variable genes assignment from IMGT V-Quest analysis.
PCT64 Ab sensitivity to N160-glycan removal was confirmed by testing pseudoviruses in a functional screening assay (Table S2). Neutralization of viruses produced in the presence of kifunensine was completely abrogated, while it was increased with swainsonine or in HEK293 cells lacking N-acetylglucosaminyl-transferase I (293S GnTI−/−), suggesting preferential binding to Man5GlcNAc2 at position N156 and/or N160, and/or interference of neighboring hybrid or complex glycans (Behrens et al., 2016; Cale et al., 2017; Cao et al., 2017; Doores and Burton, 2010; Pancera et al., 2010; Panico et al., 2016). Mapping of the epitope of PCT64 bnAbs using a panel of single Ala mutant JR-CSF pseudoviruses showed that mutation of residues 127, 165, 166, and 167 also knocked out neutralization, while mutation of the N156 PNGS and residue 171 dramatically reduced neutralization by PCT64 mAbs (Table S3). Mutation of residues distal from the epitope, but likely involved in trimer conformation, such as I307, I420, and I423 (located in hydrophobic pockets), also had a dramatic impact on neutralization, consistent with lack of binding of PCT64 mAbs to heterologous or autologous monomeric gp120s (Table S4). Together the data suggested a quaternary V2-apex epitope close to the trimer 3-fold axis (Figure 1C), similar to that of PG9 and PGT145. Overall, the PCT64 mAbs were not polyreactive or autoreactive by ELISA on compiled antigen panel (Wardemann et al., 2003) (Table S4) and by HEp2 (Human Epithelial type 2) cell staining (data not shown). Weak binding to histones was detected for a few PCT64 mAbs likely due to non-specific electrostatic interactions.
Heterologous neutralization by PCT64 mAbs was assessed on medium (N = 37) and large (N = 109) panels of pseudoviruses representing major genetic subtypes and circulating recombinant forms (Landais et al., 2016; Seaman et al., 2010) (Tables S5–S7). PCT64 Abs with < 8% nucleotide SHM (VH+JH) did not neutralize any of the viruses tested, while the more mature PCT64 mAbs showed equivalent breadth and potency (Figure 1D, Table S5), demonstrating: (1) in this lineage, relatively few mutations were necessary to acquire neutralization breadth, and (2) continued maturation over a certain level (~10% nucleotide SHM) did not greatly improve breadth nor potency. Except for subtype B viruses, the PCT64 mAbs recapitulated plasma neutralization breadth across subtypes (Figure 1E, Table S6). PCT64 mAbs neutralized up to 29% (PCT64-35D) of all isolates with moderate potency (Figure 1D, Tables S6 and S7), and up to 56% and 48% of subtype A and C viruses, respectively (up to 56% and 67% of subtype A and C viruses, respectively, as a theoretical combination of all PCT64 somatic variants). Early M13 PCT64 mAbs did not neutralize isolates that were previously identified as potential V2-apex bnAb germline-targeting immunogens (Andrabi et al., 2015; Gorman et al., 2016), such as Q23 (A), WITO (B), ZM233 (C), T250 (AE), except for PCT64-13F and −13H that weakly neutralized Q23 (not shown).
Overall, the PCT64 Ab lineage, with a 25 aa HCDR3 and a moderate level of SHM, recognizes the Env trimer V2-apex with a neutralization pattern that is more subtype-restricted than PG9, PGT145/PGDM1400, CH02, or N90-VRC38 (Bonsignori et al., 2012; Cale et al., 2017) and similar to the CAP256-VRC26 mAbs (Doria-Rose et al., 2015; 2014).
PCT64 Lineage Ontogeny Identifies a Near-Germline Heavy-Chain Sequence and Reveals a Multi-Limb Maturation
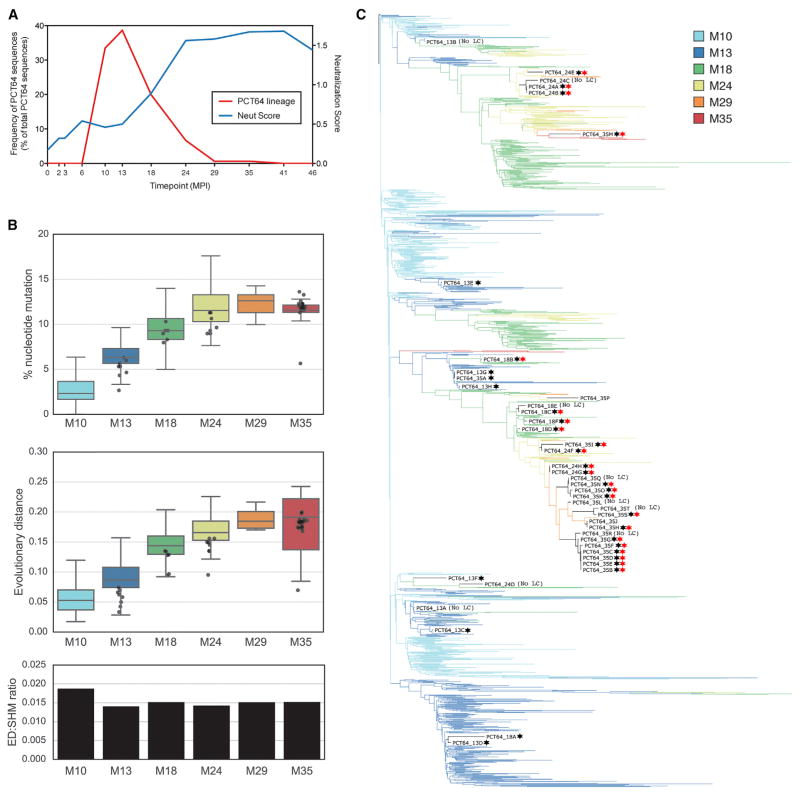
To retrace the genetic development of the PCT64 antibody lineage, we performed next-generation sequencing (NGS) of the circulating immunoglubolin G-positive (IgG+) B cell repertoire at 12 time points between 4 and 46 mpi. The PCT64 lineage was identified by homology with PCT64 mAbs. We first detected the PCT64 lineage at 10mpi, with a striking expansion at 10 and 13 mpi (Figure 2A), similar to that seen in the development of a bnAb lineage targeting the V3-glycan supersite in another Protocol C donor (MacLeod et al., 2016). At 18 mpi, the lineage had declined in frequency, with PCT64 sequences only rarely observed after 24 mpi. The rarity of the PCT64 lineage sequences after 24 mpi corresponded to a slowdown in lineage maturation (Figure 2B), a common feature of developing bnAb lineages (MacLeod et al., 2016; Sheng et al., 2016). In contrast to our previous study showing divergence of nucleotide mutation and evolutionary distance during development of a bnAb lineage targeting the V3-glycan supersite (MacLeod et al., 2016), a slowdown in PCT64 maturation was consistently observed when measuring lineage maturation either by percent nucleotide mutation or evolutionary distance from the VH inferred germline sequence (VH-infGL) (Figure 2C).
Figure 2. Longitudinal Next-Generation Sequencing of the Memory B Cell Repertoire Reveals the Phylogeny of the PCT64 bnAb Lineage.
PC64 IgG libraries prepared from total PBMCs were amplified with IgG-specific primers for all human VH gene families.
(A) Frequency of PCT64 bnAbs at each time point, plotted as a percentage of total PCT64 lineage sequences from all time points. Plasma neutralization score (see Landais et al., 2016) from a heterologous 37-virus panel is plotted as a dashed line.
(B) Somatic hypermutation frequency was calculated for each time point either as divergence (number of amino acid changes compared to LMCA; top panel) or as sum of the evolutionary distance (middle panel). Data are presented as whisker plots showing mean, 95% upper and lower quartiles, SD, and outliers. The evolutionary distance normalized by divergence using mean values is shown in the bottom panel.
(C) Longitudinal phylogeny of PCT64 HC sequences (colored by mpi). The PCT64 mAb sequences are named in black, and star symbols represent autologous (black) and heterologous (red) neutralization. See also Figure S2.
At 10 mpi, we identified a PCT64 HC sequence with very high germline sequence identity (99.4%), encoded by a fully germline V gene (VH3-15*01) and a J gene (JH6*03) containing only two nucleotide mutations (Figure S2A). This sequence was selected as the least mutated common ancestor for the HC (VH_LMCA). Additionally, inferred germline sequences were computed for the heavy chain (VH_infGL) and light chain (VK_infGL) by combining germline V(D)J genes with the N-addition region(s) of the least mutated sequence (Figure S2A).
Phylogenetic analysis of the PCT64 lineage HC sequences (Figure 2C) revealed a highly branched maturation process, with multiple branches maturing independently after initial diversification early in development, consistent with previous reports of HIV bnAb development (Bonsignori et al., 2017; Doria-Rose et al., 2015; MacLeod et al., 2016; Sok et al., 2013). PCT64 mAbs were clustered in two branches, both of which remained detectable by NGS until 35 mpi. The maturing branches (Limbs#1,2,4) differed mostly in CDRH2 and CDRH3 (Figure S2B), where changes in the isolated mAb sequences appear to be concomitant with acquisition of breadth (Figures S1A and S1B). Little similarity was seen in the CDRH2 maturation pathways for each branch. However, in branches that persisted beyond M24 (Limbs#1 and 4), a DD motif in CDRH3 (highlighted in Figure S2B), present in early sequences at positions 100G and 100H (Kabat numbering), changed to positions 100D and 100E usually by M18, effectively inverting the DD around Tyr 100F. Reversion of the DD shift completely abrogated neutralization (data not shown). This shift occurred independently in most maturing branches, suggesting strong selection pressure. In conclusion, PCT64 memory B cells first appeared in the periphery around 10 mpi and the lineage matured through 35 mpi, acquiring breadth via SHM mostly in CDRH2 and CDRH3.
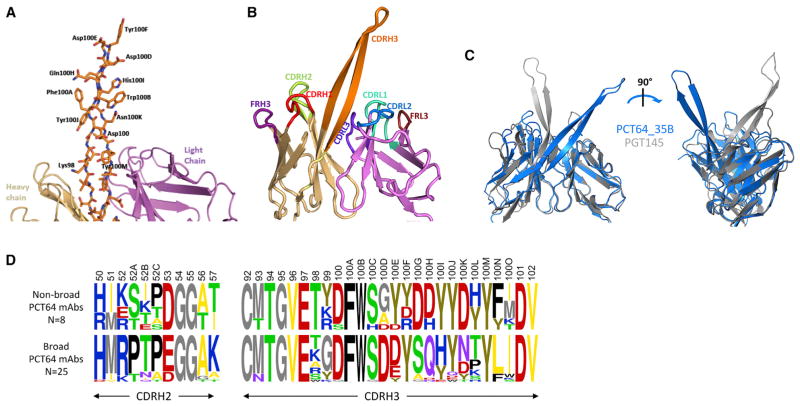
PCT64-35B bnAb CDRH3 Is Sulfated and Adopts a PGT145-like Conformation
The crystal structure for Fab PCT64-35B (5FEH) at 3.1 Å resolution (Figures S3A and S3B), isolated at M35, showed that CDRH3 adopted a β-hairpin conformation (Figure 3A) that protruded above the other CDRs (Figure 3B, S3C). The CDRH3 was rich in aromatic residues that contributed to its stability via a ladder-like stacking (Figure 3A). The two aspartate residues (Asp100D and Asp100E), and a tyrosine at the tip of CDRH3, projected ~28 Å above the other CDRs. Despite the PCT64-35B CDRH3 having 8 fewer residues than PGT145 (McLellan et al., 2011), the vertical distance above the antibody surface to the CDRH3 tip was similar. Additionally, the overall arrangement of PCT64-35B CDRH3 was more similar to those from the PGT145/PDGM1400 lineage than to other V2-apex antibodies, such as PG9 and CAP256.03 (Doria-Rose et al., 2014). However, superimposition of the variable regions of PCT64-35B and PGT145 revealed that the β-hairpins adopted radically different orientations relative to the rest of the Fab variable region (Figure 3C). The electrostatic surface of PCT64-35B (Figure S3D) highlighted the anionic character of the CDRH3, which was similar to CDRH3s in most antibodies targeting this epitope, and is thought to penetrate between the glycans that mask the epitope to contact the positively charged V1/V2 protein surface at the trimer apex. This contrasted with a recently described antibody from the N90-VRC38 lineage, N90-VRC38.01, which instead displays a short, “non-protruding” CDRH3 (16 aa) with near neutral charge (Cale et al., 2017).
Figure 3. Crystal Structure of PCT64-35B bnAb Fab Shows an Extended PGT145-like CDRH3 Conformation.
(A) PCT64-35B Fab crystal structure with CDRH3 side chains as sticks. Heavy chain in beige, CDRH3 in orange, and light chain in purple.
(B) Overall organization of the variable region of PCT64-35B Fab; secondary structure rendering of CDRs and framework (FR) regions.
(C) Superposition of PCT64-35B Fab variable region (blue) and PGT145 variable region (gray), showing differential CDRH3 orientations.
(D) Logograms of CDRH2 and CDRH3 residues (Kabat numbering) for PCT64 mAbs segregated in two groups based on acquisition of heterologous breadth. See also Figures S1B and S3.
Tyrosine sulfation is also important for binding of bnAbs of this class (McLellan et al., 2011; Sok et al., 2014). Despite being unable to confirm the presence of sulfation of Tyr100F in the crystal structure, electrospray mass spectrometry (ESI-MS) of the PCT64-35B Fab suggested at least one sulfotyrosine in the Fab (Figure S3E), which was consistent with computational predictions (Pan et al., 2014) (Figure S3F). There did not appear to be a relationship between increased somatic hypermutation and increased numbers of sulfated tyrosines.
Examination of the LMCA CDRH3 sequence revealed that some tyrosine and aspartic acid residues were present in the LMCA, although their positions differed from those in PCT64-35B CDRH3 (Figure S1A). As previously mentioned, the acquisition of heterologous neutralization in this branch of the lineage was concomitant with significant CDRH2 changes, accompanied by the acidic DD motif switching from one side to the other of the CDRH3 beta-hairpin tip (Figures 3D, S1). In another branch (Limb#5), the DD motif at the tip of CDRH3 did not change position, but PCT64-24D, the only mAb isolated through functional screening in this branch, acquired a 6nt/2aa deletion in CDRH2 (Figures S1A, S2B, Table S1). In contrast, in Limb#2, for which no mAbs was isolated, the CDRH3 DD motif switch was also found in association with a different CDRH2 SHM pattern, further showing that CDRH2 SHMs may have also been key to acquisition of breadth in this lineage. Overall, the position of the DD motif around the tip of CDRH3, together with mutations in CDRH2 and LC residues proximal to CDRH3 in PCT64 bnAbs (Figure S3C), suggested two possible mechanisms for breadth: mutations could alter the angle of the CDRH3 and change the residues that contact the trimer surface, or these mutations could increase the rigidity of the CDRH3, thereby enhancing neutralization.
Localized Diversity at Key V2 Apex Residues Drove PCT64 bnAb Maturation toward Breadth
To understand PCT64 Ab lineage maturation in response to the HIV-1 Env changes over time, we obtained both functional env clones using conventional amplification methods and full-length env NGS sequences (Figures 4, S4) (Laird Smith et al., 2016) from plasma samples collected at ten time points after infection (Figure 1A). During the first three months of infection, env diversity was limited, increasing at around 6 mpi (Figures 4, S4). The Most Recent Common Ancestor (MRCA) virus was thus defined as the consensus of M3 env sequences. We tested representative PC64 mAbs for pseudovirus neutralization of selected autologous envs.
Figure 4. Longitudinal Full Length Env Next Generation Sequencing Allows a Comprehensive Analysis of Env Evolution and Escape from PCT64 Abs.
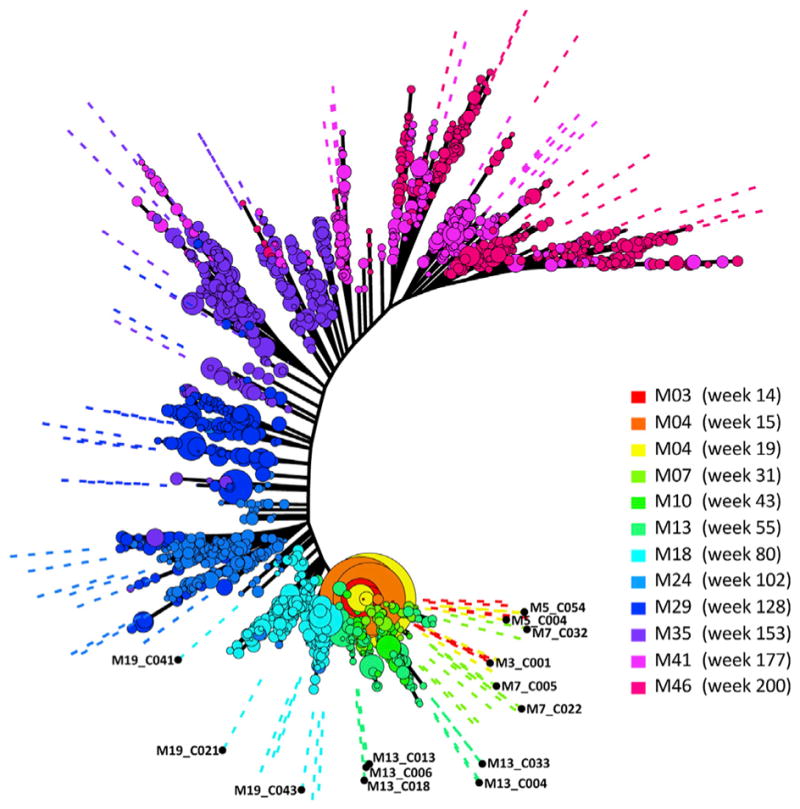
HIV subtype A env phylogeny from donor PC64, colored by sample date, estimated by maximum likelihood from full-length env PacBio high-quality consensus sequences (HQCSs) (with bubbles representing sample proportion) (http://test.datamonkey.org/flea-demo/PC64_kinetics/) and clonal Sanger sequences (Monogram Biosciences Lab Corp) (indicated by dashed lines). Data are representative of one experiment. See also Figure S4.
M3 and M4 viruses were neutralized by the least matured M13 PCT64 mAbs, but most viruses at M7 had escaped M13 to M24 Abs, suggesting that the Ab lineage was elicited within the first 7 months of infection, before corresponding memory B cells in the periphery were detected (M10, Figure 2). In contrast, M35 Abs neutralized viruses up to 18 mpi, when the virus fully escaped the lineage (Figure 5A, S5A). With increasing Ab maturation and breadth, potency against autologous viruses decreased, with the least mutated Abs being at least 10-fold more potent against M3 to M7 viruses than the broadest more mutated Abs against all M3 to M18 viruses (Figures 5A, S5A and S5B). Thus, for PCT64 Abs, maturation caught up with viral escape at the expense of potency. Sequence analysis of env clones with differential neutralization sensitivity to PCT64 mAbs suggested escape mutations arose at residues 166, 167 and 169 (Figure 5A, S5C), which were confirmed by mutational studies of autologous pseudovirus clones (Figures S5D and S5E). As for heterologous viruses, autologous neutralization was strictly dependent on the N160 PNGS.
Figure 5. Analysis of Env Escape from PCT64 Abs Reveals that Localized Diversity at Key V2 Epitope Residues Drove bnAb Maturation toward Breadth.
(A) Left panel: V1/V2 amino acid sequences of PC64 Env cloned at various time points, aligned vertically. The number of clones is indicated and amino-acid identity is color-coded. Middle panel: Logograms of Env C-strand residues 160–171 for each time point. Net charge of the 160–171 peptide for each clone is color-coded in shades of blue (from 0 to 1). Right panel: Autologous neutralization of each pseudotyped Env clone by PCT64 mAbs (grouped and colored by isolation time point). The neutralization IC50 (μg/mL) is tabulated and color-colored as indicated in the legend. The development of the PCT64 bnAb lineage and V1V2-directed heterologous plasma neutralization are indicated on the right.
(B) Evolution of PCT64 mAb neutralization against autologous pseudotyped virus mutants representing various Env immunotypes at residues 160, 166, 167, 169, 181 at several time points. Residues mutated from WT are shaded in pink. The neutralization IC50 (μg/mL) is tabulated and colored as indicated. Gray cells represent the absence of neutralization.
(C) Frequency of PC64 Env immunotypes (combination of residues 160, 166, 167, 169, 181) at several time points, with viral load in plasma (black). Neutralization potency of PCT64 mAbs presented in (B) against these immunotypes is shown as shades of blue, green, orange, and red for PCT64 mAbs isolated at 13, 18, 24, and 35 mpi, respectively. Antibody icons are colored by mpi, with the frequency of PCT64 HC sequences found in the periphery (Figure 2A) shown by icon size. GL: Germline. Development of V2 apex-directed heterologous plasma neutralization (Figure 1A) is indicated in black above. Data are representative of at least two independent experiments. See also Figures S5 and S6.
To more precisely understand the Ab-Env co-evolution that led to emergence of breadth, we selected representative infectious env clones and mutated them to recapitulate the dominant V2 epitope variants observed in the NGS data at each time point (Figures 5B and 5C, S4, S5C). These “immunotypes” represented the diversity of combinations of amino acids for residues 160, 166, 167, 169, and 181, where this latter position also appeared to be under selection pressure from the PCT64 lineage (Figure S5C–S5E). All immunotypes showed consistent neutralization sensitivity when introduced in different autologous clonal backgrounds (Figure 5B), suggesting that mutations outside of the epitope region had minimal impact on PCT64 lineage maturation. Mutations at positions 166, 167, and 169 appeared individually at first, and later in combination, as early as 7 mpi, abrogating neutralization by early PCT64 mAbs (Figures 5B, S5D and S5E), confirming early selective pressure by PCT64 Abs and thus initiation of the lineage prior to M7. Despite resistance to all tested contemporaneous and later PCT64 mAbs, 166S viral variants (immunotypes 3 and 4) disappeared by M18, and were replaced by 166R variants bearing 169I which were sensitive to late M35 PCT64 mAbs (Figure 5B–C). The non-persistence of these early seemingly full escape variants might be explained by pressure from Abs in the lineage that were not tested or not isolated, pressure from a putative helper lineage, or by an eventual strong fitness cost of 166S, which is not found in any subtype A Env sequences in the Los Alamos database. The I181L mutation appears to have only a moderate role in escape. Within the epitope, position 169 sampled the most escape mutations (I, G, K, V, T, E) (Figure S4C), until the autologous virus finally entirely escaped the PCT64 lineage Abs through fixation of K166, leading to full escape, and E169, which did not confer additional resistance, but might be a compensatory mutation restoring viral fitness (Figures 5B and 5C). Little diversity is also found across HIV-1 subtypes at positions 166 and 167, as compared to 169 (Figure S6A). Along with the lack of persistence of 166S escape variants discussed above, this suggests an impact on fitness, possibly from a role of residues 166 and 167 in inter-protomer interactions and trimer stability (Guenaga et al., 2015). Together, the data were similar to that observed in the CAP256 donor (Bhiman et al., 2015) (Figures S6B and S6C) and supported the hypothesis that dynamic localized diversity at residues 166, 167, and especially 169, progressively selected broader PCT64 Abs through waves of viral escape and Ab adaptation (Figure 5C).
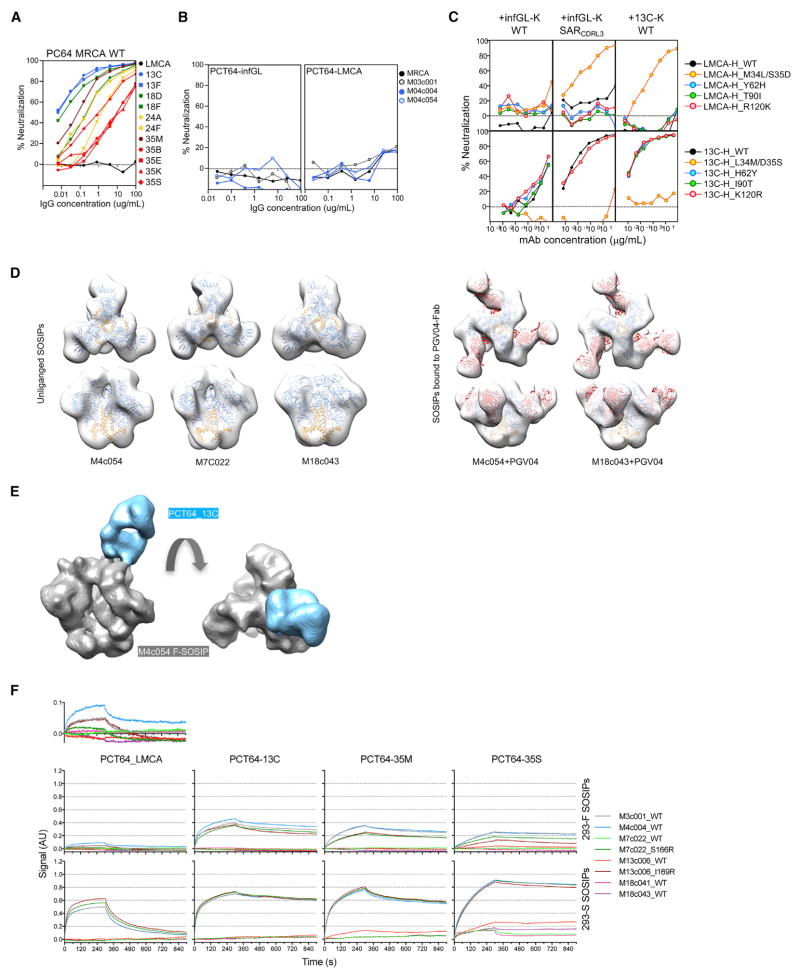
PCT64 LMCA Ab Binds to Early Autologous Env Trimers Only When Devoid of Complex and Hybrid Glycans
To gain better insight into the elicitation of the PCT64 Ab lineage, the PCT64-infGL (VH_infGL+VK_InfGL) and PCT64-LMCA Abs (VH_LMCA+VK_InfGL) were tested against the PC64 pseudoviruses. We found that, in contrast to the CAP256 study (Doria-Rose et al., 2014), the PCT64-LMCA and PCT64-infGL did not neutralize the MRCA (Figure 6A) or early viruses (Figure 6B), while PCT64-13C, the least mutated lineage mAb isolated, showed potent neutralizing activity against these early viruses. In fact, only a subset of the SHM found in PCT64-13C, namely a CRDL3 SAR motif and L34HC+D35HC, was required for the PCT64-LMCA to potently neutralize the MRCA Env (Figure 6C, S1).
Figure 6. Env Glycoform Heterogeneity Played a Role in Elicitation of the PCT64 bnAb Lineage.
(A) Autologous neutralization of the MRCA pseudotyped virus by titrated amounts of PCT64 mAbs.
(B) Autologous neutralization of PC64 Env clones by PCT64-infGL and -LMCA mAbs.
(C) Autologous neutralization of MRCA pseudotyped virus by titrated amounts of WT and mutant PCT64 mAbs. PCT64-LMCA (top) and PCT64-13C (bottom) heavy chain mutants paired with PCT64-infGL WT, PCT64-infGL-CDRL3 mutant(+SAR) or PCT64-13C light chains.
(D) Top and side views of negative-stain EM 3D reconstructions (see Figure S7C) of PC64 unliganded (left) or PGV04-bound (right) SOSIPs produced in 293-F cells. Atomic models of BG505 (gp120 in blue, gp41 in yellow: PBD 5CEZ) in complex with PGV04 (red) (PDB: 3J5M) and with Fabs removed were docked into EM density maps.
(E) Cryo-EM reconstruction of PC64-M4c054 SOSIP with autologous PCT64-13C Fab. Side and top view of the EM density map showing PCT64-13C Fab bound to the V1/V2 apex. The final resolution is ~13Å (Figure S7D).
(F) BLI curves of indicated antibodies immobilized on anti-human IgG Fc sensors and indicated PC64 SOSIP trimers (1 μM) in solution as analytes. PC64 SOSIP trimers were produced in the presence of furin in 293F (top panels; normal glycan processing), or 293S (bottom panels; GnTI−/− with no hybrid/complex glycan processing leading to enrichment in Man5GlcNAc glycans). Data are representative of at least two independent experiments. See also Figure S7.
However, activation of a germline B cell depends on antigen binding and not on neutralization. As the PCT64 mAb epitope is quaternary, we thus aimed to test binding of the PCT64-LMCA to autologous trimers. We first assessed whether PC64 Env sequences could be made into soluble, native-like trimers when modified with SOSIP mutations and purified as described previously (Sanders et al., 2013). Native-like trimers were successfully obtained for the following time points: M3, M4, M6, M13, and M18 (Figure 6D, S7A–S7C). Reference-free 2D class averages revealed that the trimers all adopted native pre-fusion conformations and were, therefore, amenable for structural and antigenic studies. We obtained a 3D-reconstruction of the PC64-M4c054 SOSIP in complex with the autologous PCT64-13C at 13Å resolution (EMDB: 7089) showing clear V2-apex targeting and more diagonal angle of approach than PGT145 but with a similar 1Fab:1Trimer stoichiometry (Figure 6E, S7D).
Binding of PCT64 Abs to autologous SOSIP trimers was overall consistent with neutralization (Figure 6F, S7E). Indeed, PCT64-LMCA binding to all autologous SOSIP trimers was barely detectable, while Abs PCT64-35M and PCT64-35S bound to most Env trimers corresponding to neutralized autologous viruses. However, we noticed a few instances where the neutralization of the Env clone did not match binding to the corresponding SOSIP trimers (e.g.: PCT64-35S on M18c043, PG9 on M13c006, and PGT145 on M18c041) suggesting that binding did not always correlate with neutralization or that the PC64 SOSIPs were not completely reproducing the conformation or more likely the precise glycosylation status or distribution of glycoforms of the native trimers on the virus. The PCT64-LMCA binding on-rate to early autologous SOSIPs was substantially increased when the trimers were produced in 293S cells (GnTI−/−) and therefore lacked hybrid and complex glycans and displayed an increased proportion of N-linked Man5GlcNAc and decreased Man6–9GlcNAc glycans (Doores and Burton, 2010). The data suggested two hypotheses regarding the initial stimulation of the PCT64 lineage: (1) very weak binding to the UCA BCR was sufficient to activate the original naive B cell; (2) due to glycan heterogeneity (Behrens et al., 2016; Cao et al., 2017; Panico et al., 2016), sufficient Env with favorable glycoforms, i.e., with Man5GlcNAc or lack of hybrid or complex glycans at key sites, were present on the early viral spikes that activated the initial naïve B cell.
Higher-resolution structural studies of longitudinal PC64_Env trimers, unliganded and in complex with the PCT64 mAbs, will help elucidate the role of glycoforms and trimer apex flexibility and their impact on the PCT64 lineage maturation.
DISCUSSION
Recent publications offer hope that rational, structure-based HIV vaccine strategies, based on germline targeting followed by sequential immunizations with guide immunogens, might succeed (Briney et al., 2016b; Dosenovic et al., 2015; Escolano et al., 2016; Jardine et al., 2016; Steichen et al., 2016; Tian et al., 2016). This reductionist approach would benefit from a deeper understanding of how Env-Ab coevolution leads to the emergence of bnAbs in HIV-infected individuals. Here we describe how Env evolution drove (and was driven by) the development of bnAbs targeting the V2-apex in a subtype A-infected donor from Africa. The Ab lineage represents the fifth family of bnAbs targeting this region of vulnerability and the second example of V2-apex bnAb ontogeny, enabling comparison with the CAP256-VRC26 lineage (Bhiman et al., 2015; Doria-Rose et al., 2015; 2014). Thus, the relatively early and rapid emergence of both V2-apex bnAb lineages (5 and 7 months respectively) during natural infection is encouraging from a vaccine perspective (Doria-Rose et al., 2015; 2014).
Although neither as broad nor as potent as some other members of the PG9-like family of bnAbs, the PCT64 bnAbs neutralize 50%–70% of subtype A and C viruses, with no indels, and only 10%–12% nucleotide SHM (a level that is achievable through vaccination). Furthermore, their CDRH3 loop is only 25aa long, among the shortest to date in this bnAb class (Cale et al., 2017). The orientation of the β-hairpin component of the CDRH3, either parallel (PG9 and CAP256) or perpendicular (PGT145, PCT64) to the long axis of the Fab, appears to define two structural classes of V2-apex bnAbs, which contrast with the recently described N90-VRC38.01 bnAb lineage displaying a shorter and less protruding CDRH3. These two V2-apex bnAb classes show different dependency on the PCT64 immunotypes, suggesting discrete differences in their recognition of V2-apex strand C (Figure S7B). Defining the fine structural differences in epitope recognition of V2-apex bnAbs will likely be important for immunogen design. Overall, the characteristics of the PCT64 bnAbs, including the absence of significant auto- and poly-reactivity, suggest that PCT64-like bnAbs may be amenable to elicitation through vaccination, making this lineage valuable for design of germline-targeting and sequential immunogens.
Identification of the precise form of Env that activated the naïve B cell at the origin of the bnAb lineage is essential for germ-line-targeting vaccination strategies. In both PC64 and CAP256 lineages, the presence of N160-glycan, 166R and 169R/K appear to be critical features for activation of the relevant germ-line B cell. However, in contrast to the CAP256 lineage, the PCT64 LMCA Ab did not neutralize viruses pseudotyped with autologous Env contemporary with the lineage emergence, and bound very poorly to the corresponding soluble Env trimers. Our data suggest that, in the PC64 donor, glycan heterogeneity might have played an important role in PCT64 germline B cell activation through binding to Env trimer forms bearing, at key positions, Man5GlcNAc glycans (i.e., N160 and/or N156) or a lack of complex or hybrid glycans (N130, N133). Maturation might have allowed adaptation of PCT64 Abs to bind more predominant glycoforms on Env comprising a higher proportion of Man8–9GlcNAc glycans or of hybrid to complex glycans (Behrens et al., 2016; Cao et al., 2017; Panico et al., 2016). Therefore, GnTI−/− produced Env immunogens should be favored for germline-targeting strategies aiming at eliciting PCT64-like bnAbs. Understanding the variation of HIV-1 Env glycoforms during infection and their impact on the PCT64 bnAb lineage initiation and maturation might thus be critical for sequential immunogen design.
Following initial B cell activation, the PCT64 lineage underwent an early dramatic expansion followed by gradual contraction, as observed for other bnAb lineages (Bonsignori et al., 2017; 2016; Doria-Rose et al., 2015; 2014; Liao et al., 2013; MacLeod et al., 2016) and likely resulted from gradual replacement of neutralization-sensitive viral variants with resistant clones, reducing the availability of cognate antigens necessary to sustain maturation. Consistent with other bnAb lineage studies, we also observed a “multi-limb” evolution, with some branches stopping their maturation early (“dead-ends,” Limbs#3,5,6) and others maturing further and eventually leading to bnAbs (Limbs#1,4). As identification of PCT64 mAbs was performed via functional screening, the clustering of the majority of isolated, mature mAbs into one branch of the lineage (limb#4) suggests that other maturing branches might essentially correspond to “off-track” Abs (i.e., with mostly autologous neutralization) (Bhiman et al., 2015). The fact that most Abs isolated using heterologous viruses tended to cluster, while less-matured Abs isolated using autologous viruses were more generally distributed across the PCT64 phylogeny, further supports this hypothesis.
The PCT64 lineage matured toward breadth in about 14 months, which is similar to the CAP256 lineage (20 months for CAP256-VRC26) (Doria-Rose et al., 2015; 2014). However, the two lineages seem to contrast in their evolution pattern with greater initial diversification of the PCT64 lineage. Indeed, from early time points, we observed multiple branches in the PCT64 bnAb lineage, two of which (Limbs#1,4) acquired neutralization breadth, coinciding with concomitant changes in CDRH2 and CDRH3. The mutations selected in response to viral escape might have resulted in substantial structural changes, possibly shifting the Ab angle of approach. Finally, the PCT64 lineage evolution appears to have mostly occurred at the expense of potency and differs from previously described bnAb lineages where SHM led to a concomitant increase of breadth and potency (Doria-Rose et al., 2015; MacLeod et al., 2016; Sok et al., 2013).
Escape pathways in the PC64 and CAP256 viruses were similar. In both, successive mutations at positions 166 and 169 appear to drive the selection of somatic hypermutations that were critical for breadth. Adaptation of Abs to the 166K mutation, which was increased breadth in the CAP256-VRC26 lineage (Bhiman et al., 2015), did not occur in PC64. Instead, 166K corresponded to full neutralization escape for all PC64 mAbs, suggesting that no somatic variant that bound to this Env mutant with sufficient affinity could be selected, terminating the lineage. The continued evolution of the epitope following full escape from the lineage suggests either compensatory mutations to restore fitness, or pressure from another V2-targeting lineage. Overall, the data suggest that minor variations at a few crucial sites drive selection of Abs toward breadth; subtle sequential changes in the epitope might allow the continuous sampling and selection of B cells bearing Abs with sufficient affinity for successive Env mutants, eventually leading to emergence of bnAbs. In contrast, more substantial changes might prevent the Ab lineage from bridging the affinity gap, leading to dead-end branches and eventually terminating further lineage evolution.
In conclusion, our results highlight the importance of Env glycoforms in activating V2-apex germline B cells, and demonstrate common viral escape pathways driving the selection of bnAbs. This is encouraging for the rational design of sequential immunogens aimed at inducing V2 apex bnAbs through vaccination.
STAR★METHODS
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Monoclonal anti-HIV-1 Env PG9 | NIH AIDS Reagent Program; www.hiv.lanl.gov | Cat#12149; RRID: AB_2491030 |
| Monoclonal anti-HIV-1 Env PG16 | NIH AIDS Reagent Program; www.hiv.lanl.gov | Cat#12150; RRID: AB_2491031 |
| Monoclonal anti-HIV-1 Env PGT145 | Dennis R. Burton, The Scripps Research Institute; www.hiv.lanl.gov | RRID: AB_2491054 |
| Monoclonal anti-HIV-1 Env PGDM1400 | Dennis R. Burton, The Scripps Research Institute; www.hiv.lanl.gov | N/A |
| Monoclonal anti-HIV-1 Env CH02 | NIH AIDS Reagent Program; www.hiv.lanl.gov | Cat#12562; RRID: AB_2491056 |
| Monoclonal anti-HIV-1 Env CH04 | NIH AIDS Reagent Program; www.hiv.lanl.gov | Cat#12564; RRID: AB_2491058 |
| Monoclonal anti-HIV-1 Env CAP256.08 (VRC26.08) | John R. Mascola, NIH; www.hiv.lanl.gov | N/A |
| Monoclonal anti-HIV-1 Env CAP256.09 (VRC26.09) | John R. Mascola, NIH; www.hiv.lanl.gov | N/A |
| Monoclonal anti-HIV-1 Env PGT151 | Dennis R. Burton, The Scripps Research Institute; www.hiv.lanl.gov | N/A |
| Monoclonal anti-HIV-1 Env F105 | NIH AIDS Reagent Program; www.hiv.lanl.gov | Cat#857 |
| Monoclonal anti-HIV-1 Env N90-VRC38.01 | John R. Mascola, NIH | |
| Monoclonal anti-HIV-1 Env PCT64-infGL, _LMCA, _13C to _35S | This paper | N/A |
| Alkaline Phosphatase AffiniPure Goat Anti-Human IgG, F(ab’)2 fragment specific | Jackson ImmunoResearch | Cat#109-055-097 |
| Bacterial and Virus Strains | ||
| 37 HIV-1 Env-pseudotyped viruses | Elise Landais IAVI-NAC (elandais@iavi.org) | (Landais et al., 2016) |
| 109 HIV-1 Env-pseudotyped viruses | David C. Montefiori, Duke University (david.montefiori@duke.edu) | (Seaman et al., 2010) |
| Biological Samples | ||
| PBMC from IAVI Protocol C Donor 64 | Protocol C, IAVI | N/A |
| Serum from IAVI Protocol C Donor 64 | Protocol C, IAVI | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| 100mM dNTP set | Thermo Fisher | Cat#10297117 |
| 293Fectin | Thermo Fisher | Cat#12347500 |
| AMPure PB Beads | Pacific Biosciences | Cat#100-265-900 |
| apo-Transferrin human, powder, BioReagent, suitable for cell culture, ≥ 98% | Sigma | Cat#T1147 |
| Capture select LC-kappa(Hu) affinity matrix | Thermo Fisher | Cat#83310 |
| Cardiolipin sodium salt from bovine heart, ≥ 98% (TLC) lyophilized powder | Sigma | Cat#C0563 |
| Complete EDTA free protease inhibitors | Roche | Cat#05056489001 |
| Disialoganglioside GD1a from bovine brain, ≥ 95% (TLC), lyophilized powder | Sigma | Cat#G2392 |
| FUGENE 6 | Promega | Cat#E2692 |
| Galanthus nivalis lectin (snow drop), agarose bound | Vector Labs | Cat#AL-1243 |
| GeneArt® Seamless Cloning and Assembly Enzyme Mix | Thermo Fisher | Cat#A14606 |
| Genomic DNA - Human Adult Normal Tissue: Placenta, from a single donor | BioChain | Cat#D1234200 |
| Hemocyanin from Megathura crenulata (keyhole limpet) | Sigma | Cat#H7017 |
| Histone from calf thymus, Type II-A, lyophilized powder | Sigma | Cat#H9250 |
| HotStarTaq DNA Polymerase | QIAGEN | Cat#203445 |
| Insulin human, recombinant expressed in yeast | Sigma | Cat#I2643 |
| Kifunensine | Cayman Chemical | Cat#10009437 |
| Lipopolysaccharides from Escherichia coli 0111:B4 | Sigma | Cat#L2630 |
| Luciferase Cell Culture Lysis 5X Reagent | Promega | Cat#E1531 |
| Nano-W Stain | Nanoprobes | Cat#2018 |
| Pfu Ultra HF DNA Pol AD | Agilent Technologies | Cat#600389 |
| Phosphatase substrate | Sigma | Cat#S0942 |
| Protein A Sepharose Fast Flow | GE healthcare | Cat#17-1279-03 |
| QC Lightning Mullti Site-Directed Mutagenesis Kit | Agilent Technologies | Cat#210513 |
| Qiaquick PCR purification kit | QIAGEN | Cat#28106 |
| Recombinant HIV-1-gp120 Antigen from 92TH021 strain | Elise Landais, IAVI-NAC (elandais@iavi.org) | Genebank: AY669775 |
| Recombinant HIV-1-gp120 Antigen from PC64_MRCA strain | This paper | N/A |
| Recombinant HIV-1-gp41 Antigen from HxB2 strain | Meridian Life Science, Inc. | Cat#VTI310 |
| RNase OUT | Thermo Fisher | Cat#10777019 |
| RNaseOut | Thermo Fisher | Cat#10777-019 |
| SPRISelect Reagent | Bekman Coulter | Cat#B23317 |
| ssDNA (Deoxyribonucleic acid, single stranded from calf thymus, lyophilized powder) | Sigma | Cat#D8899 |
| SuperScript® III Reverse Transcriptase | Thermo Fisher | Cat#18080-085 |
| Swainsonine | Cayman Chemical | Cat#16860 |
| Transferrin human ≥ 98% | Sigma | Cat#T3309 |
| Critical Commercial Assays | ||
| 4–12% Bis-Tris NuPAGE gel system | Thermo Fisher | Cat#NP0321BOX |
| Advantage® 2 PCR Kit | Clontech | Cat#639206 |
| DNA 1200 Analysis Kit | Agilent Technologies | Cat#5067-1508 |
| HotStarTaq Plus DNA Polymerase Kit | QIAGEN | Cat#203603 |
| Kallestad® HEp-2 Kit – 20 Slides | Bio-Rad | Cat#30472 |
| Luciferase 1000 Assay System | Promega | Cat#E4550 |
| MiSeq Reagent Kit V3 (600-cycle) | Illumina | Cat#MS-102-3003 |
| PacBio RS II C3 Sequencing Kit | Pacific Biosciences | Cat#P/N 100-254-800 |
| QIAamp Viral RNA Mini Kit | QIAGEN | Cat#52906 |
| RNEasy Mini Purification Kit | QIAGEN | Cat#74104 |
| SMRTbell Template Prep Kit 1.0 | Pacific Biosciences | Cat#100-259-100 |
| SuperScript® III First-Strand Synthesis System for RT-PCR | Thermo Fisher | Cat#18080-051 |
| Deposited Data | ||
| PC64 Full-length Env longitudinal PacBio sequences | This Paper | http://test.datamonkey.org/flea-demo/PC64_kinetics/ |
| PC64 Full-length Env longitudinal clonal sequences | This Paper | GenBank: MF565934 - MF566033 |
| PCT64 bnAb lineage HC MiSeq sequences | This Paper | GenBank: SSRP114571 |
| PCT64-infGL, PCT64-LMCA, PCT64-13C to PCT64-35S heavy chain nt sequences | This Paper | GenBank: MF565855 - MF565898 |
| PCT64-infGL, PCT64-13C to PCT64-35S light chain nt sequences | This Paper | GenBank: MF565899 - MF565933 |
| PCT64-35B Fab | This Paper | PDB: 5FEH |
| PC64-M4C054 SOSIP - PCT13C complex CryoEM | This Paper | EMD: 7089 |
| PC64 SOSIPs negative stain EM | This paper | EMD-7104, EMD-7105, EMD-7106, EMD-7107, EMD-7108 |
| Experimental Models: Cell Lines | ||
| Human: HEK293T | ATCC | Cat#CRL-3216, RRID: CVCL_0063 |
| Human: HEK293S GnT1- | ATCC | Cat#CRL-3022, RRID: CVCL_A785 |
| Human: HeLa-derived TZM-bl | NIH AIDS Reagent Program | Cat#8129-442, RRID: CVCL_B478 |
| Human: FreeStyle 293F | Thermo Fisher | Cat#R79007; RRID: CVCL_D603 |
| Oligonucleotides | ||
| IgG NGS Primers | Integrated DNA Technologies | (Briney et al., 2016a) |
| Env-F: GAGCAGAAGACAGTGGCAATGA | Integrated DNA Technologies | N/A |
| Env-R: CCACTTGCCACCCATBTTATAGCA | Integrated DNA Technologies | N/A |
| Recombinant DNA | ||
| Plasmid pcDNA3.1+ | Thermo Fisher | Cat#V790-20 |
| Plasmid pSG3Denv | NIH AIDS Reagent Program | Cat#11051 |
| Software and Algorithms | ||
| AbStar | Bryan Briney (briney@scripps.edu), The Scripps Research Institute | https://github.com/briney/abstar |
| Appion | Bridget Carragher (BCarr@nysbc.org) | http://emg.nysbc.org/redmine/projects/leginon/wiki/Leginon_Homepage |
| Clonify | Bryan Briney (briney@scripps.edu), The Scripps Research Institute | https://github.com/briney/clonify-python |
| Crystallographic Object-Oriented Toolkit (COOT) Software | Paul Emsley (paul.emsley@mrc-lmb.cam. ac.uk), Oxford University | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/; RRID: SCR_014222 |
| CTFfind3 | Nikolaus Grigorieff (niko@grigorieff.org) | http://grigoriefflab.janelia.org/ctf |
| DoG Picker | Bridget Carragher (BCarr@nysbc.org) | http://emg.nysbc.org/redmine/projects/software/wiki/DoGpicker |
| ETE Toolkit | Jaime Huerta-Cepa (jhuerta@crg.es), Centre for Genomic Regulation, Spain | http://ete.cgenomics.org |
| FastTree | Morgan N. Price (MorganNPrice@yahoo.com) Lawrence Berkely University | http://www.microbesonline.org/fasttree; RRID:SCR_015501 |
| FigTree | Andrew Rambaut (andrew.rambaut@zoo.ox. ac.uk) University of Edinburgh | http://tree.bio.ed.ac.uk/software/figtree/; RRID:SCR_008515 |
| Full-Length Env Analysis (FLEA) pipeline | Ben Murrell (murrellb@gmail.com), University of California San Diego | http://test.datamonkey.org/FLEA/ |
| HKL2000 Suite 6 | HKL Research | www.hkl-xray.com |
| IMGT/V-QUEST | International ImMunoGeneTics Information System; Marie-Paule Lefranc (marie-paule. lefranc@igh.cnrs.fr), University of Montpellier, France | www.imgt.org; RRID: SCR_012780 |
| Leginon | Bridget Carragher (BCarr@nysbc.org) | http://emg.nysbc.org/redmine/projects/leginon/wiki/Leginon_Homepage |
| MAFFT | Kazutaka Katoh (kkatoh@kuicr.kyoto-u.ac.jp) Kyoto University, Japan | http://www.biophys.kyoto-u.ac.jp/~katoh/programs/align/mafft; RRID:SCR_011811 |
| Microcal ORIGIN | Microcal Software | http://en.freedownloadmanager.org/Windows-PC/Microcal-Origin.html; RRID: SCR_002815 |
| MolProbity | David C. Richardson (dcr@kinemage.biochem. duke.edu), Duke University | http://molprobity.biochem.duke.edu; RRID: SCR_014226 |
| PacBio SMRTportal Version 2.3 | Pacific Biosciences | http://www.pacb.com/products-and-services/analytical-software/smrt-analysis/; RRID:SCR_002942 |
| PANDAseq | Josh D. Neufeld (ac.oolretawu@dlefuenj) University of Waterloo, Canada | https://github.com/neufeld/pandaseq; RRID:SCR_002705 |
| PHASER | Airlie J.McCoy (ku.ca.mac@102mja), University of Cambridge, UK | https://www.phenix-online.org/documentation/reference/phaser.html; RRID:SCR_014219 |
| Phenix | Paul D. Adams (pdadams@lbl.gov), Lawrence Berkeley Laboratory | www.phenix-online.org; RRID: SCR_014224 |
| PRISM6 | GraphPad | http://www.graphpad.com/; RRID:SCR_002798 |
| PyMol Molecular Graphics Systems Version 1.5.0.4 | Schrodinger LLC | https://www.schrodinger.com/pymol; RRID:SCR_000305 |
| Relion version 1.4 | Sjors Scheres (scheres@mrc-lmb.cam.ac.uk) | http://www2.mrc-lmb.cam.ac.uk/relion/index.php/Main_Page |
| USEARCH | Robert C. Edgar (robert@drive5.com) | http://www.drive5.com/usearch/ |
| Other | ||
| 2100 Bioanalyzer System | Agilent Technologies | Cat#G2939BA |
| 96S Super Magnet Plate | ALPAQUA | Cat#A001322 |
| Agilent 6210 TOF LC/MS System | Agilent Technologies | Cat#G1969A |
| Anti-Human Fc (AHC) Biosensors | ForteBio | Cat#18-5060 |
| Carbon-coated Cu400 mesh grid | Electron Microscopy Sciences | Cat#EMS400-Cu |
| MiSeq Sequencer | Illumina | N/A |
| Mono S 10/100 GL column | GE Healthcare | Cat#17-5169-01 |
| PacBio RS-II Sequencer | Pacific Biosciences | N/A |
| Qubit 3.0 Fluorometer | Thermo Fisher | Cat#Q33216 |
| Superdex 200 HiLoad 16/600 column | GE Healthcare | Cat#28989335 |
CONTACT FOR REAGENT AND RESOURCES SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Elise Landais (elandais@iavi.org).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Ethics Statement, Study Participant and Samples
Donor PC064 was part of the IAVI sponsored Protocol C cohort in sub-Saharan Africa that involved rapid screening of 613 individuals with a recent history of HIV exposure for HIV antibodies in sub-Saharan Africa (Landais et al., 2016). Samples were collected with written, informed consent, and the study was reviewed and approved by the relevant Ethics and Research Committees. We are very grateful to and thank all of the Protocol C participants and clinical investigators, Susan Allen (Emory University, USA and ZERHP, Zambia), William Kilembe (ZEHRP, Zambia), Shabir Lakhi (ZEHRP, Zambia), Mubiana Inambao (ZEHRP, Zambia), Anatoli Kamali (MRC/UVRI Uganda), Eduard J. Sanders (KEMRI, Kenya and Oxford University, UK), Omu Anzala (KAVI, Kenya), Vinodh Edward (The Aurum Institute, South Africa), Linda-Gail Bekker (Cape Town University, South Africa), Jianming Tang (University of Alabama Birmingham, USA), Jill Gilmour (IAVI and London Imperial College, UK) as well as Matt Price (IAVI and University of California San Francisco, USA), Eric Hunter (Emory University, USA) and all of the Protocol C project team members for all of the support provided for this study. Finally, we thank Melissa Simek, Laura Sharpe and Brendan McAtarsney and John Brennan for coordinating the samples transfers and shipments, and Sam Jauregui and Kay Limoli at Monogram Biosciences for their contributions to this study.
Donor PC064 was enrolled in the Protocol C longitudinal primary infection cohort approximately 2 weeks after infection by a subtype A1 HIV-1 virus, and was identified as one of the seven elite neutralizers (Landais et al., 2016) (neutralizing up to 64% of viruses) on a medium virus panel (N = 37, GeoMean ID50 = 390). The neutralizing activity was first detected in the plasma after 1.5 year of infection (18 months) and steadily increased to reach a plateau at 2 years (24 months), neutralizing 62% of viruses of a large (N = 103, GeoMean ID50 = 490) cross clade pseudotyped virus panels (Seaman et al., 2010). The plasma broadly neutralizing activity was mapped to a V2-apex quaternary epitope similar to that of PG9, as it was dependent on the presence of the N160-glycan, was sensitive to virus produced in presence of kifunensine, and was not depleted by absorption with rgp120 monomers (Landais et al., 2016).
Cell Lines
Human embryonic kidney (HEK)-derived 293T, HEK293S N-acetylglucosaminyltransferase I-negative (GnTI-), and HeLa-derived TZM-bl cells were maintained in complete Dulbecco’s Modified Eagle Medium (herein referred to as cDMEM) containing high-glucose Dulbecco’s Modified Eagle Medium (DMEM, Thermo Fisher), 1X Penicillin-Streptomycin (Pen Strep, Thermo Fisher) and 10% fetal bovine serum (FBS, Gemini Bio Products) at 37°C and 5% CO2. FreeStyle 293F cells (Thermo Fisher) were maintained in Freestyle 293 Expression Medium at 37°C and 10% CO2 with shaking at 120 RPM.
METHODS DETAILS
Isolation of PCT64 Monoclonal Antibodies
The method for isolating human monoclonal antibodies from memory B cells in circulation has previously been described (Walker et al., 2011; 2009). Surface IgG1+ B cells seeded at near-clonal density in 384-well microplates were activated in short-term culture. Memory B cell supernatants from week 55, 80, 102, 128, 153, 200 samples were screened at Monogram Biosciences in a micro-neutralization assays using single round of replication pseudoviruses. This assay was based on the 96-well pseudotyped HIV-1 neutralization assay and was modified for screening 15 μL of B cell culture supernatants in a 384-well format. Briefly, pseudoviruses capable of a single round of infection were produced by co-transfection of HEK293 cells with a subgenomic plasmid, pHIV-1lucΔu3, that incorporates a firefly luciferase indicator gene and a second plasmid, PC0XAS that expressed HIV-1 Env clones. Following transfection, pseudoviruses were harvested and used to infect U87 cell lines expressing CD4 plus the CCR5 and CXCR4 coreceptors. Virus infectivity was determined 72h after inoculation by measuring amount of luciferase activity.
Memory B cell supernatants from the week 80, 102, 128, 153, 200 samples were screened for N160K-sensitive neutralizing activity against 92TH021 and JR-CSF pseudoviruses. To identify earlier bnAb precursors, we tested the culture supernatants derived from the week 55 sample for early versus late autologous neutralization (PC64-M03c001 and PC64-M07c032).
Heavy- and light-chain variable regions were isolated from B cell lysates of selected neutralizing hits by reverse transcription from RNA followed by multiplex PCR amplification using family-specific V-gene primer sets. Amplicons from each lysate were uniquely tagged with multiplex identifier (MID) sequences and 454 sequencing regions (Roche). A normalized pooling of gamma, kappa and lambda chains was performed based on agarose gel image quantitation and the pool was analyzed by 454 Titanium sequencing. Consensus sequences of the VH and VL chains were generated using the Amplicon Variant Analyzer (Roche) and assigned to specific B cell culture wells by decoding the MID tags. Selected VH and VL chains were cloned using the Seamless Cloning and Assembly Enzyme mix (Life Technologies) in expression vectors with the appropriate IgG1, Ig kappa or Ig lambda constant domains. Monoclonal antibodies were reconstituted by transient transfection in HEK293F cells followed by purification from serum-free culture supernatants (see below).
Memory B Cell Repertoire Next-Generation Sequencing and Computational Analysis
Library Preparation and Sequencing
RNA was prepared (RNEasy kit, QIAGEN) from total PBMCs and was subjected to reverse transcription using barcoding primers that contain unique Ab identifiers as previously described (Briney et al., 2016a). The cDNA was then amplified using a mix of gene specific primers. Illumina sequencing adapters and sample-specific indexes were added during a second round of PCR using 1μL of purified PCR product in 100μL of total reaction volume and using the following thermal cycling program: 94C for 5 minutes; 10 cycles of 94C for 30 s, 55C for 30 s, 72C for 2 minutes; 72C for 7 minutes. Indexed PCR products were purified using 75μL of SPRIselect beads (Beckman Coulter) and eluted in 50μL of water. Samples were quantified using fluorometry (Qubit; Life Technologies), pooled at approximately equimolar concentrations and the sample pool was requantified. Samples were loaded onto an Illumina MiSeq sequencer with a target loading concentration of 40pM and 10% PhiX and sequenced (MiSeq 600-base v3 reagent kit; Illumina).
Initial Sequence Analysis and UAID Processing
Paired-end MiSeq reads were merged with PANDAseq (Masella et al., 2012). Germline assignment, junction identification, and other basic antibody information was determined using AbStar (www.github.com/briney/abstar), an antibody analysis package that uses BLASTn and Smith-Waterman alignment to calculate the most likely GL genes, identify the junctional region, and annotate SHM-induced substitutions and indels (www.github.com/briney/abstar). As part of the AbStar analysis pipeline, non-functional antibody sequences and other sequencing artifacts were removed and frameshift indels were corrected. Additionally, the AbStar was run with the UAID option set to 20 nucleotides (-u 20), which parses UAIDs from each sequence and populates the appropriate field in the JSON output. Output was stored in a MongoDB database for querying and further analysis. Antibody sequences were then binned by UAID, and all bins containing only a single sequence were discarded. For each bin containing two or more sequences, the appropriate germline variable gene region was added to the bin to serve as a consensus tiebreaker. Consensus sequences were calculated for each UID bin, re-processed with AbStar and stored in a separate MongoDB database.
Clonal Lineage Assignment
Sequences were assigned to clonal lineages using Clonify, a software package specifically developed for antibody lineage assignment (Briney et al., 2016a). Briefly, Clonify uses an antibody-specific distance metric, which incorporates length-normalized CDR3 edit distance, variable and joining gene use, and shared somatic mutations to determine the likelihood of clonal relatedness between each pair of antibody sequences. Following lineage assignment, the PCT64 lineage was identified by calculating the identity of each sequence isolated from PC64 to a panel of PCT64 mAb sequences. The identified PCT64 NGS lineage was the only lineage containing sequences with high amino-acid identity (> 95%) to any of the isolated PCT64 mAb sequences.
Phylogenetic Analysis of the PCT64 Lineage
PCT64 lineage sequences were clustered at 97.5% identity with USEARCH (Edgar, 2010) and the size of each cluster was recorded. Cluster centroids were used to generate a multiple sequence alignment with MAFFT (Katoh et al., 2005) and a tree file was calculated with FastTree using default settings (Price et al., 2010). The phylogenetic tree was drawn in Python using the ETE Toolkit (Huerta-Cepas et al., 2010).
PCT64 Antibody Expression and Purification
Antibodies and Fab fragments were produced with plasmids encoding full-length or truncated HC (after constant region 1, CH1) and full-length LC. For large-scale production, HC constructs and LC were transiently expressed with the FreeStyle 293 Expression System (Invitrogen). Supernatant was collected after 4 days of culture and whole IgGs were purified with Protein A Sepharose (GE Healthcare) while Fabs were purified with CaptureSelect LC-kappa (Hu) Affinity Matrix (Life technologies). Fabs were then further purified using the AKTA pure FPLC system with a Mono S 10/100 GL cation exchange column followed by size exclusion with the HiLoad 16/600 Superdex 200 pg column (GE Healthcare Life Sciences). Purified protein purity and integrity checked by SDS–PAGE.
Fab Crystallization, Data Collection, Structure Determination and Refinement
For crystallization, the Fab of PC64-35B was co-expressed with protein-tyrosine sulfotransferase 1 (TPST1) in FreeStyle HEK293F cells (Invitrogen) to encourage sulfation of sulfation-prone tyrosine residues. The Fab was purified by affinity chromatography (human anti-Kappa) followed by cation exchange chromatography and size exclusion chromatography. Ion-exchange chromatography of PCT64-35B Fab revealed a single peak, which, upon ESI-MS analysis as described previously (Pejchal et al., 2010), reflected a majority population with a molecular weight of 49,377 Da, and a minority population with a mass of 49,296 Da. The difference of 81 Da (the theoretical mass of a sulfate group is 80 Da) supports the presence of at least one sulfotyrosine in the majority of the population (Figure S3E). This same pattern is found when the PCT64-35B Fab was produced in the absence of TPST1, suggesting a high level of endogenous sulfation of the relevant tyrosine. The residual difference between Fab in the minority population and the theoretical mass of the Fab fragment (~245 Da) is likely due to incomplete cleavage of the signal peptide sequence.
Crystallization trials with the protein at 8 mg/ml in 20mM Tris, 150mM NaCl, pH 8.0 (100nL protein with 100nL reservoir solution) were set up on our IAVI/JCSG/TSRI high-throughput robotic CrystalMation system (Rigaku). Crystals were obtained in sitting drops with 0.2M ammonium sulfate pH 6.0, 10% ethylene glycol and 20% PEG 3350, pH 6.0. Data were collected at the APS beamline ID-D23 and processed using HKL-2000 (Otwinowski and Minor, 1997). The structure was solved using PHASER (McCoy et al., 2007) with PDB 1DFB as the initial search model. COOT (Emsley and Cowtan, 2004) and PHENIX (Adams et al., 2010) were used for refinement, with statistics reported in Figure S3A. Pymol (PyMOL Molecular Graphics System, Version 1.5.0.4 Schrödinger, LLC) was used for the rendering of images. The asymmetric unit contained three Fab copies, one of which was well ordered (chains E and F), while the other two were less well resolved.
Envelope Genes Cloning and Mutagenesis
Libraries of full-length envelope genes were isolated by reverse transcription-PCR (RT-PCR) from cryopreserved longitudinal plasma samples from donor PC64. The swarm analysis protocol was described previously (Schweighardt et al., 2007) and is an application of the clonal analysis procedure developed by Monogram Biosciences (South San Francisco, CA). Briefly, the population of viral envelope genes present in the patient plasma was amplified by RT-PCR and then cloned into expression vectors. To test individual clones derived from the envelope population, the DNA was diluted and retransformed in bacteria, and individual clones were selected and screened for infectivity using the Monogram Biosciences co-receptor tropism screening assay. Pseudo-typed viruses containing cloned envelope genes were prepared from each plasma sample in HEK293 cells. Viruses from individual clones were screened for infectivity and chemokine receptor tropism in U87 cells transfected with CD4 and CCR5 or CXCR4 chemokine receptors. Ten to 12 envelopes with high infectivity were selected from each individual and evaluated for autologous neutralization by PCT64 mAbs using the microneutralization method described above (see Isolation of PCT64 monoclonal antibodies).
Full-length autologous env genes were synthesized for representative clones of each time point using GeneArt® gene synthesis services (Life Technologies), then cloned into pcDNA3.1 vector (Life Technologies) for pseudovirus production. Mutagenesis was performed using Quikchange site-directed mutagenesis kit (Agilent Technologies).
Full-length Env Amplification and Sequencing
Library Preparation and Sequencing
HIV-1 envelope genes were amplified and sequenced as described (Laird Smith et al., 2016). Plasma was layered onto 200μl of 20 percent sterile-filtered sucrose in 2 mL screw-cap microcentrifuge tubes. Virions were pelleted through the sucrose cushion at 23,500g for 1 hr at 4°C, and the supernatant discarded. On ice, 140μl sterile phosphate-buffered saline (pH = 7.4) was applied to the remaining viral pellet and allowed to stand for 1 hr. The loosened viral pellets were then resuspended in this volume and used as input into the QIAamp Viral RNA Mini Kit (QIAGEN), which was used to extract total HIV-1 RNA according to the manufacturer’s instructions. Viral RNA was eluted from the QIAmp columns in 55μl AVE buffer. An 8μl sub-aliquot of the eluted viral RNA was added directly to cDNA synthesis reactions to avoid any degradation from freeze/thaw cycles. Remaining viral RNA was aliquoted and stored at −80°C for later use.
cDNA was generated using the SuperScript III First Strand Synthesis System for RT-PCR (Thermo Fisher) using the provided oligo (dT) to prime first-strand synthesis and according to the manufacturer’s protocol. Aliquots of cDNA were stored at 20°C.
HIV-1 env amplification was performed using PCR with HPLC-purified primers: Env-F: GAGCAGAAGACAGTGGCAATGA (corresponding to positions 6,207–6,228 in HXB2); and Env-R: CCACTTGCCACCCATBTTATAGCA (corresponding to positions 8,788–8,811 in HXB2). The primers (IDT) were diluted to 20pmol in 0.1 × TE buffer before use. Each reaction consisted of 2μl HIV-1 cDNA and 48μl of Advantage 2 PCR reaction mixture (Clontech). The reaction mixture comprised 5μl 10 × SA PCR Buffer, containing 2mM magnesium acetate, 1μl of 10 mM dNTP mix, 1μl each of Env-F and Env-R (at 20pmol), 39μl of nuclease free water, and 1μl of Advantage 2 Polymerase Mix. Reactions were heated to 95°C for 1 minute and then subjected to 35 cycles of PCR using the following parameters: 15 s denaturation at 95°C, followed by 30 s annealing at 64°C, followed by 3-min extension at 68°C. After the 35th cycle, the reactions were incubated for 10cmin at 68°C and then held at 4°C.
HIV-1 env amplicons were purified from PCR reactions using the QIAquick PCR Purification Kit (QIAGEN) as described by the manufacturer, and eluted in 30μl EB buffer (10 mM Tris, pH 8). Replicate PCR reactions for each sample were visualized and quantitated using the 2100 Bioanalyzer System with the DNA 12000 kit (Agilent Technologies), and pooled by sample, until a final mass of > 250 ng HIV-1 env amplicon was achieved. To remove any residual PCR reagents and primer dimers, the 250ng of sample was then purified with a 1 × volume of AMPure PB beads (Pacific Biosciences) as described by the manufacturer.
SMRTbell template libraries of ~2.6-kb insert size were prepared according to the manufacturer’s instructions using the SMRTbell Template Prep Kit 1.0 (Pacific Biosciences). A total of 250ng of AMPure PB bead-purified HIV-1 env amplicon was added directly into the DNA damage repair step of the 10-kb Template Preparation and Sequencing (with low-input DNA) protocol. Sequencing primer annealing was performed using the recommended 20:1 primer:template ratio, whereas P5 polymerase binding was performed at a modified polymerase:template ratio of 3:1. HIV-1 env SMRTbell libraries were immobilized onto SMRT cells at a starting concentration of 10pM on chip, loading titrations were performed to achieve optimal sequencing conditions for particular samples as necessary. SMRT sequencing was performed on the PacBio RS II using the C3 sequencing kit with magnetic bead loading and 180-minute movies.
Full-length Env Sequence Analysis
CCS sequences were constructed using the PacBio SMRTportal software (version 2.3), which employs the Quiver algorithm, and .fastq files were used for downstream analysis. The Full-Length Envelope Analysis (FLEA) pipeline (Laird Smith et al., 2016) was used to error correct these CCS reads, and cluster them into near-identical clusters, inferring High Quality Consensus Sequences (HQCSs) for each cluster. Envelope phylogenies, as well as the dynamics of amino acid frequency evolution, were inferred from these HQCSs. MAFFT v7.164b (Katoh and Standley, 2013) with manual curation, was used to create a multiple sequence alignment. Gappy regions were manually removed when reconstructing phylogenies, since their alignment is uncertain. Phylogenies were reconstructed with FastTree v2.1 (Price et al., 2010) and visualized with FigTree (http://tree.bio.ed.ac.uk/software/figtree/). Frequency kinetic plots and similar analyses were created with custom Mathematica scripts.
Neutralization Assay
Plasma and monoclonal antibodies neutralizing activity was assessed using single round of replication in TZM-bl target cells, in absence of DEAE-dextran as described previously (Landais et al., 2016). Briefly, WT and mutant pseudoviruses were generated by co-transfection of HEK293T or 293-S (GnTI−/−) cells with an Env-expressing plasmid and an Env-deficient genomic backbone plasmid (pSG3ΔEnv). Kifunensine-or swainsonine-treated pseudoviruses were produced by treating 293T cells with 25 μM kifunensine or 20 μM swainsonine on the day of transfection (Doores and Burton, 2010). Plasma samples were heat-inactivated at 56 C for 45min prior to use in neutralization assays.
ELISA Assays
Half-area 96-well ELISA plates were coated overnight at 4C with 50μL PBS containing 250 ng of compound per well. The wells were washed four times with PBS containing 0.05% Tween 20 and blocked with 3% BSA at room temperature for 1 h. Serial dilutions of sera were then added to the wells, and the plates were incubated at room temperature for 1 hour. After washing four times, goat anti-human IgG F(ab’)2 conjugated to alkaline phosphatase (Pierce), diluted 1:1000 in PBS containing 1% BSA and 0.025% Tween 20, was added to the wells. The plates were incubated at room temperature for 1 h, washed four times, and developed by adding alkaline phosphatase substrate (Sigma) diluted in alkaline phosphatase staining buffer (pH 9.8), according to the manufacturer’s instructions. The optical density at 405 nm was read on a microplate reader (Molecular Devices). EC50 values were calculated using Prism6 (GraphPad).
HEp-2 Assay
Antibodies were assayed for autoreactivity using a HEp-2 indirect immunofluorescence kit (Bio-Rad) according to manufacturer’s instructions.
Production of Monomeric and Trimeric Recombinant Envelope Glycoproteins
All Env-derived proteins were expressed using the HEK293F cells (ThermoFisher) according to manufacturer’s instructions. In brief, cell culture supernatants of 293-transfected cells were harvested 4–6 days post-transfection, cleared, filtered and two protease inhibitor tablets (Roche) per liter of supernatant were added to limit proteolysis. Secreted Env Proteins were purified using affinity columns. Gp120 proteins were purified from HEK293F supernatants on Galanthus nivalis lectin-bound agarose columns (Vector Laboratories), while PC64 Env trimers were purified over a 2G12 column as previously described (Lee et al., 2015). The eluted glycoproteins were subjected to size exclusion chromatography on HiLoad 16/600 Superdex 200 SEC column (GE Healthcare). Fractions containing the relevant monomers or trimers were concentrated with Amicon Ultra 30,000 MWCO centrifugal filter devices (Millipore, Bedford, MA). Finally, the purified proteins were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis to verify protein purity.
PC64 Env SOSIP constructs were designed according to the published BG505.664 SOSIP to include the A501C, T605C, I559P, R6-cleavage site and 664-deletion modifications (Sanders et al., 2013). The SOSIP constructs were co-transfected with furin protease into 1L of HEK293F or 293S cells cells at 1.2×106 cells/mL. Protein yields per 1L of transfected cells varied for each SOSIP construct; approximately 400 μg of V3C054 SOSIP, 550 μg of V6C022 SOSIP, 600 μg of V12C033 SOSIP, and 1 mg of V18C043 were obtained.
Electron Microscopy
Negative Stain EM
Purified SOSIPs were imaged as unliganded trimers or complexed with PGV04 Fab at 0.025 μg/ml. Complexes were prepared by incubating the Env with Fab overnight at 4°C. 400 mesh Cu grids were glow discharged for 20 s under Ar/O gas flow (Gatan Solarus) prior to sample application, followed by negative staining with NanoW (Nanoprobes). ~160 images were collected for each sample at −1.5μM defocus, electron dose of 25 e−/Å2 and magnification of 52,000X (pixel size 2.05 Å) using an FEI Tecnai Spirit electron microscope operating at 120 keV. Image acquisition was carried out on a Tietz 4k × 4k TemCam-F416 CMOS camera. Image collection was controlled with Leginon automated imaging interface (Potter et al., 1999).
Image processing including particle picking, 2D classification, 3D classification, and 3D refinement were all performed with Relion/2.0 (Kimanius et al., 2016). The contrast transfer function (CTF) was fit to each micrograph using Gctf (Zhang, 2016) and particles were phase-flipped and binned by 2 (pixel size 4.1 Å) during each step of processing. One round reference-free 2D classification followed by subset selection was performed to get a clean set of particles for 3D processing. 2D classification into 25 classes was performed on this particle set to produce the class-averages displayed in the figures. One round of unsupervised 3D classification was performed followed by subset selection to get a final set of clean particles for 3D auto-refinement. 3D auto-refinement of the final set of clean particles was performed under C3 symmetry. Data statistics for each sample are presented in Figure S7C. Resolutions were estimated as the frequency at which the gold-standard Fourier Shell Correlation (FSC) drops below 0.143.
Cryo-EM
PC64M4c054 SOSIP and PCT64-13C Fab were incubated at a 1:3 ratio overnight at RT then plunge frozen onto 1.2×1.3 C-Flat holey carbon grids with the addition of DDM to a final concentration of 0.06 mM. A set of 100 movie micrographs were collected on an FEI Titon Krios operating at 300keV with a Gatan K2 Summit Direct Electron Detector at a nominal magnification of 29K over 8.6 s with 200ms exposures and a per frame dose of ~2e−/Å2. Image collection was controlled with Leginon automated imaging interface (Potter et al., 1999) and movie frames were aligned and dose-weighted with MotionCor2 (Zheng et al., 2017). The CTF was fit to each micrograph using Gctf (Zhang, 2016) and full CTF correction was performed during each round of processing. All Image processing including particle picking, 2D classification, 3D classification, and 3D refinement were all performed with Relion/2.0 (Kimanius et al., 2016). One round of reference-free 2D-classification and subset selection was performed followed by one round of 3D-classification followed by subset selection. A class of 2651 particles was chosen for auto-refinement using a loose mask representing the shape of the complex. Resolutions were estimated as the frequency at which the gold-standard (FSC) drops below 0.143.
Bio-Layer Interferometry (BLI) Binding Analysis
To obtain binding curves by BLI, we used an Octet Red instrument immobilizing IgGs on previously hydrated (PBS pH 7.4) anti-human IgG Fc sensors (Fortebio, Inc.). The SOSIP trimers were analyzed as analytes free in solution (PBS pH 7.4). Briefly, the bio-sensors were immersed in PBS pH 7.4 containing IgGs at 10 μg/mL for 1 minutes and 1000 rpm prior to encounter the analyte (SOSIP trimers at 1 μM). The IgG-immobilized sensor was in contact with the analyte in solution for 5 minutes at 1000 rpm and then removed from the analyte and placed into PBS pH 7.4 for another 10 minutes. These intervals generated association and dissociation binding curves after subtraction of binding signal to a negative control IgG (DEN3, anti-Dengue).
QUANTIFICATION AND STATISTICAL ANALYSIS
For all mAb/serum pseudovirus neutralization and ELISA assays (Figures 1C, 1D, 1E, 5A, 5B, 6A, 6B, S5A, S5B, S5C, S5E, S7A, and S7B, and Tables S2, S4, S5, and S6), the IC50, or concentration of mAb/dilution of serum needed to obtain 50% neutralization against a given pseudovirus, was calculated from the linear regression of the linear part of the neutralization curve. For neutralization assays in which a fold-change in IC50 imparted by a particular virus mutant or virus treatment was reported (Tables S3), the IC50 obtained for one virus/assay condition was divided by the IC50 obtained for the other virus/assay condition, as indicated in the figure legends. All neutralization and ELISA assays were repeated at least twice, and data shown are from representative experiments.
BLI measurements (Figures S6E and S7E) were taken over two independent experiments, and data shown are from representative experiments.
DATA AND SOFTWARE AVAILABILITY
The accession numbers for the PCT64-infGL, PCT64-LMCA, PCT64-13A to PCT64-35T heavy chain sequences reported in this paper are GenBank: MF_565855, MF_565856, ] MF_565857, MF_565858, MF_565859, MF_565860, MF_565861, MF_565862, MF_565863, MF_565864, MF_565864, MF_565865, MF_565866, MF_565867, MF_565868, MF_565869, MF_565870, MF_ 565871, MF_565872, MF_565873, MF_565874, MF_565875, MF_565876, MF_565877, MF_565878, MF_565879, MF_565880, MF_565881, MF_565882, MF_565883, MF_565884, MF_565885, MF_565886, MF_565887, MF_565888, MF_565889, MF_ 565890, MF_565891, MF_565892, MF_565893, MF_565894, MF_565895, MF_565896, MF_565897 and MF_565898.
Accession numbers for the PCT64-infGL, PCT64-13C to PCT64-35U light chain sequences are GenBank: MF_565899, MF_ 565900, MF_565901, MF_565902, MF_565903, MF_565904, MF_565905, MF_565906, MF_565907, MF_565908, MF_565909, MF_ 565910, MF_565911, MF_565912, MF_565913, MF_565914, MF_565915, MF_565916, MF_565917, MF_565918, MF_565919, MF_565920, MF_565921, MF_565922, MF_565923, MF_565924, MF_565925, MF_565926, MF_565927, MF_565928, MF_ 565929, MF_565930, MF_565931, MF_565932, and MF_565933.
The MiSeq PCT64 lineage heavy chain sequences dataset has been deposited to GenBank with ID code Genebank: SRP_114471
Accession number for the Full-length Env clonal sequences PC64M3c001 to PC64M46c043 are GenBank: MF_595934, MF_595935, MF_595936, MF_595937, MF_595938, MF_595939, MF_595940, MF_595941, MF_595942, MF_595943, MF_595944, MF_595945, MF_595946, MF_595947, MF_595948, MF_595949, MF_595950, MF_595951, MF_595952, MF_595953, MF_595954, MF_595955, MF_595956, MF_595957, MF_595958, MF_595959, MF_595960, MF_595961, MF_595962, MF_595963, MF_595964, MF_595965, MF_595966, MF_595967, MF_595968, MF_595969, MF_595970, MF_595971, MF_595972, MF_595973, MF_595974, MF_595975, MF_595976, MF_595977, MF_595978, MF_595979, MF_595980, MF_595981, MF_595982, MF_595983, MF_595984, MF_595985, MF_595986, MF_595987, MF_595988, MF_595989, MF_595990, MF_595991, MF_595992, MF_595993, MF_595994, MF_595995, MF_595996, MF_595997, MF_595998, MF_595999, MF_596000, MF_566001, MF_566002, MF_566003, MF_566004, MF_566005, MF_566006, MF_566007, MF_566008, MF_566009, MF_566010, MF_566011, MF_566012, MF_566013, MF_566014, MF_566015, MF_566016, MF_566017, MF_566018, MF_566019, MF_566020, MF_566021, MF_566022, MF_566023, MF_566024, MF_566025, MF_566026, MF_566027, MF_566028, MF_566029, MF_566030, MF_566031, MF_566032 and MF_566033.
The PacBio PC64 FL Env sequences dataset is publically available online at http://test.datamonkey.org/flea-demo/PC64_kinetics/.
The crystal structure for unliganded PCT64-35B Fab has been deposited in Protein Data Bank with ID code PDB: 5FEH.
The cryo-EM density map of PC64M4c054 SOSIP in complex with the autologous PCT64-13C Fab has been deposited in Electron Microscopy Data Bank with ID code EMD-7089. The negative stain EM density maps have been deposited in the Electron Microscopy Data Bank with ID codes EMD-7104, EMD-7105, EMD-7106, EMD-7107, EMD-7108.
Sequences for antibodies and Env clones were confirmed as being received on July 28th, 2017.
Highlights.
Isolation of PCT64, a PGT145-like, 25-aa CDRH3 HIV Env V2 apex bnAb lineage
Env glycoform heterogeneity plays a role in the lineage precursor B cell activation
Localized diversity at key V2 epitope residues drove bnAb maturation toward breadth
Env evolution pattern is similar to CAP256, another V2 apex broad neutralizer
Acknowledgments
IAVI’s work is made possible by generous support from many donors including: the Bill & Melinda Gates Foundation; the Ministry of Foreign Affairs of Denmark; Irish Aid; the Ministry of Finance of Japan in partnership with The World Bank; the Ministry of Foreign Affairs of the Netherlands; the Norwegian Agency for Development Cooperation (NORAD); the United Kingdom Department for International Development (DFID), and the United States Agency for International Development (USAID). The full list of IAVI donors is available at www.iavi.org. This study is made possible by the generous support of the Bill & Melinda Gates Foundation Collaboration for AIDS Vaccine Discovery and the American people through USAID. The contents are the responsibility of the International AIDS Vaccine Initiative and do not necessarily reflect the views of USAID or the United States Government. This study was supported by Awards Number U19AI090970, Center for HIV/AIDS Vaccine Immunology and Immunogen Discovery Grant UM1AI100663, and in part by R00AI120851 from the National Institute of Allergy and Infectious Diseases (NIH/NIAID). K.E. was supported by T15LM007092 from the National Library of Medicine. Sequence analysis was performed on a cluster supported by U01GM110749 (NIH/NIGMS). This research also used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357. This work was also supported by the International AIDS Vaccine Initiative Neutralizing Antibody Consortium through the Collaboration for AIDS Vaccine Discovery grants OPP1084519 and OPP1115782. The content is solely the responsibility of the authors and does not necessarily represent the official views of NIH, NIAID, NLM, or NIGMS. This is manuscript number 29605 from The Scripps Research Institute.
Footnotes
AUTHOR CONTRIBUTIONS
E.L., B.M., B.B., S.M., K.R., Z.T.B., A.B.W., P.P. conceived and designed the experiments. E.L., B.B., N.d.V., K.R., Z.T.B., S.M., M.L.S., A.R., L.W., A.C., J.U., Y.L., P.A. performed the experiments. E.L., B.M., B.B., N.d.V., K.R., Z.T.B., S.M., K.E., T.W., P.-Y.C.-H., P.P. analyzed the data. E.K., I.P.C.I., and I.A.H.R.N. designed and managed the cohort and collected samples. I.A.W., A.B.W., D.R.B., D.S., S.L.K.P., P.P. provided funding analyzed data, and reviewed the manuscript. E.L., B.M., B.B., S.M., K.R., Z.T.B., P.P. wrote the manuscript.
SUPPORTING CITATIONS
The following references appear in the Supplemental Information: Karplus and Diederichs (2012); Pettersen et al. (2004).
SUPPLEMENTAL INFORMATION
Supplemental Information includes seven figures and seven tables and can be found with this article online at https://doi.org/10.1016/j.immuni.2017.11.002.
CONSORTIA
The members of The IAVI Protocol C Investigators & The IAVI African HIV Research Network are Matt A. Price, Jill Gilmour, Pat Fast, Anatoli Kamali, Eduard J. Sanders, Omu Anzala, Susan Allen, Eric Hunter, Etienne Karita, William Kilembe, Shabir Lakhi, Mubiana Inambao, Vinodh Edward, and Linda-Gail Bekker.
References
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrabi R, Voss JE, Liang CH, Briney B, McCoy LE, Wu CY, Wong CH, Poignard P, Burton DR. Identification of common features in prototype broadly neutralizing antibodies to HIV envelope V2 apex to facilitate vaccine design. Immunity. 2015;43:959–973. doi: 10.1016/j.immuni.2015.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens AJ, Vasiljevic S, Pritchard LK, Harvey DJ, Andev RS, Krumm SA, Struwe WB, Cupo A, Kumar A, Zitzmann N, et al. Composition and antigenic effects of individual glycan sites of a trimeric hiv-1 envelope glycoprotein. Cell Rep. 2016;14:2695–2706. doi: 10.1016/j.celrep.2016.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhiman JN, Anthony C, Doria-Rose NA, Karimanzira O, Schramm CA, Khoza T, Kitchin D, Botha G, Gorman J, Garrett NJ, et al. Viral variants that initiate and drive maturation of V1V2-directed HIV-1 broadly neutralizing antibodies. Nat Med. 2015;21:1332–1336. doi: 10.1038/nm.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonsignori M, Montefiori DC, Wu X, Chen X, Hwang KK, Tsao CY, Kozink DM, Parks RJ, Tomaras GD, Crump JA, et al. Two distinct broadly neutralizing antibody specificities of different clonal lineages in a single HIV-1-infected donor: implications for vaccine design. J Virol. 2012;86:4688–4692. doi: 10.1128/JVI.07163-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonsignori M, Zhou T, Sheng Z, Chen L, Gao F, Joyce MG, Ozorowski G, Chuang GY, Schramm CA, Wiehe K, et al. NISC Comparative Sequencing Program. Maturation pathway from germline to broad hiv-1 neutralizer of a CD4-mimic antibody. Cell. 2016;165:449–463. doi: 10.1016/j.cell.2016.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonsignori M, Kreider EF, Fera D, Meyerhoff RR, Bradley T, Wiehe K, Alam SM, Aussedat B, Walkowicz WE, Hwang K-K, et al. Staged induction of HIV-1 glycan-dependent broadly neutralizing antibodies. Sci Transl Med. 2017;9:eaai7514. doi: 10.1126/scitranslmed.aai7514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briney BS, Willis JR, Crowe JE., Jr Human peripheral blood antibodies with long HCDR3s are established primarily at original recombination using a limited subset of germline genes. PLoS ONE. 2012;7:e36750. doi: 10.1371/journal.pone.0036750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briney B, Le K, Zhu J, Burton DR. Clonify: unseeded antibody lineage assignment from next-generation sequencing data. Sci Rep. 2016a;6:23901. doi: 10.1038/srep23901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briney B, Sok D, Jardine JG, Kulp DW, Skog P, Menis S, Jacak R, Kalyuzhniy O, de Val N, Sesterhenn F, et al. Tailored immunogens direct affinity maturation toward hiv neutralizing antibodies. Cell. 2016b;166:1459–1470. e11. doi: 10.1016/j.cell.2016.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton DR, Hangartner L. Broadly neutralizing antibodies to HIV and their role in vaccine design. Annu Rev Immunol. 2016;34:635–659. doi: 10.1146/annurev-immunol-041015-055515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton DR, Ahmed R, Barouch DH, Butera ST, Crotty S, Godzik A, Kaufmann DE, McElrath MJ, Nussenzweig MC, Pulendran B, et al. A blueprint for HIV vaccine discovery. Cell Host Microbe. 2012;12:396–407. doi: 10.1016/j.chom.2012.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cale EM, Gorman J, Radakovich NA, Crooks ET, Osawa K, Tong T, Li J, Nagarajan R, Ozorowski G, Ambrozak DR, et al. Virus-like particles identify an HIV v1v2 apex-binding neutralizing antibody that lacks a protruding loop. Immunity. 2017;46:777–791. e10. doi: 10.1016/j.immuni.2017.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L, Diedrich JK, Kulp DW, Pauthner M, He L, Park SR, Sok D, Su CY, Delahunty CM, Menis S, et al. Global site-specific N-glycosylation analysis of HIV envelope glycoprotein. Nat Commun. 2017;8:14954. doi: 10.1038/ncomms14954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doores KJ, Burton DR. Variable loop glycan dependency of the broad and potent HIV-1-neutralizing antibodies PG9 and PG16. J Virol. 2010;84:10510–10521. doi: 10.1128/JVI.00552-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doria-Rose NA, Klein RM, Daniels MG, O’Dell S, Nason M, Lapedes A, Bhattacharya T, Migueles SA, Wyatt RT, Korber BT, et al. Breadth of human immunodeficiency virus-specific neutralizing activity in sera: clustering analysis and association with clinical variables. J Virol. 2010;84:1631–1636. doi: 10.1128/JVI.01482-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doria-Rose NA, Schramm CA, Gorman J, Moore PL, Bhiman JN, DeKosky BJ, Ernandes MJ, Georgiev IS, Kim HJ, Pancera M, et al. NISC Comparative Sequencing Program. Developmental pathway for potent V1V2-directed HIV-neutralizing antibodies. Nature. 2014;509:55–62. doi: 10.1038/nature13036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doria-Rose NA, Bhiman JN, Roark RS, Schramm CA, Gorman J, Chuang GY, Pancera M, Cale EM, Ernandes MJ, Louder MK, et al. New member of the V1V2-directed CAP256-VRC26 lineage that shows increased breadth and exceptional potency. J Virol. 2015;90:76–91. doi: 10.1128/JVI.01791-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doria-Rose NA, Altae-Tran HR, Roark RS, Schmidt SD, Sutton MS, Louder MK, Chuang GY, Bailer RT, Cortez V, Kong R, et al. Mapping polyclonal HIV-1 Antibody responses via next-generation neutralization fingerprinting. PLoS Pathog. 2017;13:e1006148. doi: 10.1371/journal.ppat.1006148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dosenovic P, von Boehmer L, Escolano A, Jardine J, Freund NT, Gitlin AD, McGuire AT, Kulp DW, Oliveira T, Scharf L, et al. Immunization for HIV-1 broadly neutralizing antibodies in human Ig knockin mice. Cell. 2015;161:1505–1515. doi: 10.1016/j.cell.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Escolano A, Steichen JM, Dosenovic P, Kulp DW, Golijanin J, Sok D, Freund NT, Gitlin AD, Oliveira T, Araki T, et al. Sequential immunization elicits broadly neutralizing anti-hiv-1 antibodies in Ig knockin mice. Cell. 2016;166:1445–1458. e12. doi: 10.1016/j.cell.2016.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauci AS, Marston HD. Ending AIDS–is an HIV vaccine necessary? N Engl J Med. 2014;370:495–498. doi: 10.1056/NEJMp1313771. [DOI] [PubMed] [Google Scholar]
- Gao F, Bonsignori M, Liao HX, Kumar A, Xia SM, Lu X, Cai F, Hwang KK, Song H, Zhou T, et al. Cooperation of B cell lineages in induction of HIV-1-broadly neutralizing antibodies. Cell. 2014;158:481–491. doi: 10.1016/j.cell.2014.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman J, Soto C, Yang MM, Davenport TM, Guttman M, Bailer RT, Chambers M, Chuang GY, DeKosky BJ, Doria-Rose NA, et al. NISC Comparative Sequencing Program. Structures of HIV-1 Env V1V2 with broadly neutralizing antibodies reveal commonalities that enable vaccine design. Nat Struct Mol Biol. 2016;23:81–90. doi: 10.1038/nsmb.3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray ES, Moore PL, Choge IA, Decker JM, Bibollet-Ruche F, Li H, Leseka N, Treurnicht F, Mlisana K, Shaw GM, et al. CAPRISA 002 Study Team. Neutralizing antibody responses in acute human immunodeficiency virus type 1 subtype C infection. J Virol. 2007;81:6187–6196. doi: 10.1128/JVI.00239-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenaga J, Dubrovskaya V, de Val N, Sharma SK, Carrette B, Ward AB, Wyatt RT. Structure-guided redesign increases the propensity of hiv env to generate highly stable soluble trimers. J Virol. 2015;90:2806–2817. doi: 10.1128/JVI.02652-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hraber P, Seaman MS, Bailer RT, Mascola JR, Montefiori DC, Korber BT. Prevalence of broadly neutralizing antibody responses during chronic HIV-1 infection. AIDS. 2014;28:163–169. doi: 10.1097/QAD.0000000000000106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huerta-Cepas J, Dopazo J, Gabaldón T. ETE: a Python environment for tree exploration. BMC Bioinformatics. 2010;11:24. doi: 10.1186/1471-2105-11-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jardine JG, Kulp DW, Havenar-Daughton C, Sarkar A, Briney B, Sok D, Sesterhenn F, Ereño-Orbea J, Kalyuzhniy O, Deresa I, et al. HIV-1 broadly neutralizing antibody precursor B cells revealed by germline-targeting immunogen. Science. 2016;351:1458–1463. doi: 10.1126/science.aad9195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julien JP, Cupo A, Sok D, Stanfield RL, Lyumkis D, Deller MC, Klasse PJ, Burton DR, Sanders RW, Moore JP, et al. Crystal structure of a soluble cleaved HIV-1 envelope trimer. Science. 2013;342:1477–1483. doi: 10.1126/science.1245625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karplus PA, Diederichs K. Linking crystallographic model and data quality. Science. 2012;336:1030–1033. doi: 10.1126/science.1218231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Kuma K, Toh H, Miyata T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimanius D, Forsberg BO, Scheres SH, Lindahl E. Accelerated cryo-EM structure determination with parallelisation using GPUs in RELION-2. eLife. 2016;5:e18722. doi: 10.7554/eLife.18722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein F, Diskin R, Scheid JF, Gaebler C, Mouquet H, Georgiev IS, Pancera M, Zhou T, Incesu RB, Fu BZ, et al. Somatic mutations of the immunoglobulin framework are generally required for broad and potent HIV-1 neutralization. Cell. 2013;153:126–138. doi: 10.1016/j.cell.2013.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong R, Xu K, Zhou T, Acharya P, Lemmin T, Liu K, Ozorowski G, Soto C, Taft JD, Bailer RT, et al. Fusion peptide of HIV-1 as a site of vulnerability to neutralizing antibody. Science. 2016;352:828–833. doi: 10.1126/science.aae0474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird Smith M, Murrell B, Eren K, Ignacio C, Landais E, Weaver S, Phung P, Ludka C, Hepler L, Caballero G, et al. The IAVI Protocol C Investigators & The IAVI African HIV Research Network. Rapid sequencing of complete env genes from primary HIV-1 samples. Virus Evol. 2016;2:vew018. doi: 10.1093/ve/vew018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landais E, Huang X, Havenar-Daughton C, Murrell B, Price MA, Wickramasinghe L, Ramos A, Bian CB, Simek M, Allen S, et al. Broadly neutralizing antibody responses in a large longitudinal sub-Saharan HIV primary infection cohort. PLoS Pathog. 2016;12:e1005369. doi: 10.1371/journal.ppat.1005369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, de Val N, Lyumkis D, Ward AB. Model building and refinement of a natively glycosylated HIV-1 Env protein by high-resolution cryoelectron microscopy. Structure. 2015;23:1943–1951. doi: 10.1016/j.str.2015.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao HX, Lynch R, Zhou T, Gao F, Alam SM, Boyd SD, Fire AZ, Roskin KM, Schramm CA, Zhang Z, et al. NISC Comparative Sequencing Program. Co-evolution of a broadly neutralizing HIV-1 antibody and founder virus. Nature. 2013;496:469–476. doi: 10.1038/nature12053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLeod DT, Choi NM, Briney B, Garces F, Ver LS, Landais E, Murrell B, Wrin T, Kilembe W, Liang CH, et al. IAVI Protocol C Investigators & The IAVI African HIV Research Network. Early antibody lineage diversification and independent limb maturation lead to broad HIV-1 neutralization targeting the Env high-mannose patch. Immunity. 2016;44:1215–1226. doi: 10.1016/j.immuni.2016.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masella AP, Bartram AK, Truszkowski JM, Brown DG, Neufeld JD. PANDAseq: paired-end assembler for illumina sequences. BMC Bioinformatics. 2012;13:31. doi: 10.1186/1471-2105-13-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLellan JS, Pancera M, Carrico C, Gorman J, Julien JP, Khayat R, Louder R, Pejchal R, Sastry M, Dai K, et al. Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9. Nature. 2011;480:336–343. doi: 10.1038/nature10696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Pan Z, Liu Z, Cheng H, Wang Y, Gao T, Ullah S, Ren J, Xue Y. Systematic analysis of the in situ crosstalk of tyrosine modifications reveals no additional natural selection on multiply modified residues. Sci Rep. 2014;4:7331. doi: 10.1038/srep07331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pancera M, McLellan JS, Wu X, Zhu J, Changela A, Schmidt SD, Yang Y, Zhou T, Phogat S, Mascola JR, Kwong PD. Crystal structure of PG16 and chimeric dissection with somatically related PG9: structure-function analysis of two quaternary-specific antibodies that effectively neutralize HIV-1. J Virol. 2010;84:8098–8110. doi: 10.1128/JVI.00966-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panico M, Bouché L, Binet D, O’Connor MJ, Rahman D, Pang PC, Canis K, North SJ, Desrosiers RC, Chertova E, et al. Mapping the complete glycoproteome of virion-derived HIV-1 gp120 provides insights into broadly neutralizing antibody binding. Sci Rep. 2016;6:32956. doi: 10.1038/srep32956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pejchal R, Walker LM, Stanfield RL, Phogat SK, Koff WC, Poignard P, Burton DR, Wilson IA. Structure and function of broadly reactive antibody PG16 reveal an H3 subdomain that mediates potent neutralization of HIV-1. Proc Natl Acad Sci USA. 2010;107:11483–11488. doi: 10.1073/pnas.1004600107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pejchal R, Doores KJ, Walker LM, Khayat R, Huang PS, Wang SK, Stanfield RL, Julien JP, Ramos A, Crispin M, et al. A potent and broad neutralizing antibody recognizes and penetrates the HIV glycan shield. Science. 2011;334:1097–1103. doi: 10.1126/science.1213256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Potter CS, Chu H, Frey B, Green C, Kisseberth N, Madden TJ, Miller KL, Nahrstedt K, Pulokas J, Reilein A, et al. Leginon: a system for fully automated acquisition of 1000 electron micrographs a day. Ultramicroscopy. 1999;77:153–161. doi: 10.1016/s0304-3991(99)00043-1. [DOI] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS ONE. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusert P, Kouyos RD, Kadelka C, Ebner H, Schanz M, Huber M, Braun DL, Hozé N, Scherrer A, Magnus C, et al. Swiss HIV Cohort Study. Determinants of HIV-1 broadly neutralizing antibody induction. Nat Med. 2016;22:1260–1267. doi: 10.1038/nm.4187. [DOI] [PubMed] [Google Scholar]
- Sanders RW, Derking R, Cupo A, Julien JP, Yasmeen A, de Val N, Kim HJ, Blattner C, de la Peña AT, Korzun J, et al. A next-generation cleaved, soluble HIV-1 Env trimer, BG505 SOSIP. 664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLoS Pathog. 2013;9:e1003618. doi: 10.1371/journal.ppat.1003618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweighardt B, Liu Y, Huang W, Chappey C, Lie YS, Petropoulos CJ, Wrin T. Development of an HIV-1 reference panel of subtype B envelope clones isolated from the plasma of recently infected individuals. J Acquir Immune Defic Syndr. 2007;46:1–11. doi: 10.1097/QAI.0b013e318074eb5a. [DOI] [PubMed] [Google Scholar]
- Seaman MS, Janes H, Hawkins N, Grandpre LE, Devoy C, Giri A, Coffey RT, Harris L, Wood B, Daniels MG, et al. Tiered categorization of a diverse panel of HIV-1 Env pseudoviruses for assessment of neutralizing antibodies. J Virol. 2010;84:1439–1452. doi: 10.1128/JVI.02108-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng Z, Schramm CA, Connors M, Morris L, Mascola JR, Kwong PD, Shapiro L. Effects of darwinian selection and mutability on rate of broadly neutralizing antibody evolution during HIV-1 infection. PLoS Comput Biol. 2016;12:e1004940. doi: 10.1371/journal.pcbi.1004940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simek MD, Rida W, Priddy FH, Pung P, Carrow E, Laufer DS, Lehrman JK, Boaz M, Tarragona-Fiol T, Miiro G, et al. Human immunodeficiency virus type 1 elite neutralizers: individuals with broad and potent neutralizing activity identified by using a high-throughput neutralization assay together with an analytical selection algorithm. J Virol. 2009;83:7337–7348. doi: 10.1128/JVI.00110-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sok D, Laserson U, Laserson J, Liu Y, Vigneault F, Julien JP, Briney B, Ramos A, Saye KF, Le K, et al. The effects of somatic hypermutation on neutralization and binding in the PGT121 family of broadly neutralizing HIV antibodies. PLoS Pathog. 2013;9:e1003754. doi: 10.1371/journal.ppat.1003754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sok D, van Gils MJ, Pauthner M, Julien JP, Saye-Francisco KL, Hsueh J, Briney B, Lee JH, Le KM, Lee PS, et al. Recombinant HIV envelope trimer selects for quaternary-dependent antibodies targeting the trimer apex. Proc Natl Acad Sci USA. 2014;111:17624–17629. doi: 10.1073/pnas.1415789111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steichen JM, Kulp DW, Tokatlian T, Escolano A, Dosenovic P, Stanfield RL, McCoy LE, Ozorowski G, Hu X, Kalyuzhniy O, et al. HIV vaccine design to target germline precursors of glycan-dependent broadly neutralizing antibodies. Immunity. 2016;45:483–496. doi: 10.1016/j.immuni.2016.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian M, Cheng C, Chen X, Duan H, Cheng HL, Dao M, Sheng Z, Kimble M, Wang L, Lin S, et al. Induction of HIV neutralizing antibody lineages in mice with diverse precursor repertoires. Cell. 2016;166:1471–1484. e18. doi: 10.1016/j.cell.2016.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gils MJ, Euler Z, Schweighardt B, Wrin T, Schuitemaker H. Prevalence of cross-reactive HIV-1-neutralizing activity in HIV-1-infected patients with rapid or slow disease progression. AIDS. 2009;23:2405–2414. doi: 10.1097/QAD.0b013e32833243e7. [DOI] [PubMed] [Google Scholar]
- Walker LM, Phogat SK, Chan-Hui PY, Wagner D, Phung P, Goss JL, Wrin T, Simek MD, Fling S, Mitcham JL, et al. Protocol G Principal Investigators. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science. 2009;326:285–289. doi: 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker LM, Huber M, Doores KJ, Falkowska E, Pejchal R, Julien JP, Wang SK, Ramos A, Chan-Hui PY, Moyle M, et al. Protocol G Principal Investigators. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature. 2011;477:466–470. doi: 10.1038/nature10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wardemann H, Yurasov S, Schaefer A, Young JW, Meffre E, Nussenzweig MC. Predominant autoantibody production by early human B cell precursors. Science. 2003;301:1374–1377. doi: 10.1126/science.1086907. [DOI] [PubMed] [Google Scholar]
- Zhang K. Gctf: Real-time CTF determination and correction. J Struct Biol. 2016;193:1–12. doi: 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng SQ, Palovcak E, Armache JP, Verba KA, Cheng Y, Agard DA. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]