Abstract
Dietary supplements are consumed by more than 300 million people worldwide, and herbal dietary supplements represent the most rapidly growing portion of this industry. Even though adverse health effects of many herbal dietary supplements have been reported, safety assurances are not being addressed adequately. Toxicological data on the identification of genotoxic and tumorigenic ingredients in many raw herbs are also lacking. Currently, more than 30 herbal dietary supplements and active ingredients have been selected by the National Toxicology Program (NTP) for toxicity and tumorigenicity studies. Due to the complexity of the chemical components present in plant extracts, there are no established methodologies for determining the mechanisms of toxicity (particularly tumorigenicity) induced by herbs, such as Gingko biloba leaf extract (GBE) and other herbal plant extracts. Consequently, the understanding of toxicity of herbal dietary supplements remains limited.
We have proposed that application of DNA microarrays could be a highly practical initial approach for revealing biological pathways and networks associated with toxic-ity induced by herbal dietary supplements and the generation of hypotheses to address likely mechanisms. The changes in expression of subsets of genes of interest, such as the modulation of drug metabolizing genes, can be analyzed after treatment with an herbal dietary supplement. Although levels of gene expression do not represent fully the levels of protein activities, we propose that subsequent biochemical and genomic experiments based on these initial observations will enable elucidation of the mechanisms leading to toxicity, including tumorigenicity. This review summarizes the current practices of microarray analysis of gene expressions in animals treated with herbal dietary supplements and discusses perspectives for the proposed strategy.
Keywords: herbal dietary supplements, DNA microarray, drug metabolizing enzyme, drug metabolizing gene, gene expression, toxicity, tumorigenicity
INTRODUCTION
Since the United States Congress passed the Dietary Supplement Health and Education Act (DSHEA) in 1994, herbal functional products, including herbal dietary supplements, have been the fastest growing segment of the vitamin, mineral supplements, and herbal products industry in the United States. The American Herbal Products Association estimates that there are about 3,000 species of plants, in as many as 50,000 different products, sold as herbal supplements in the United States (1). In the U.S. market, ginseng, ginkgo, saw palmetto, echinacea, soy, bilberry, grape seed, and green tea extracts are the top-selling items (2). Even though a number of herbal dietary supplements has been reported to exert adverse effects in humans and experimental animals (3–6), safety issues concerning potential side effects and toxic contamination of herbal products (i.e., may not contain the correct plant species as claimed or may be contaminated with other herbs, pesticides, or metals) are not being addressed adequately. The evaluation and identification of the toxic ingredients, including genotoxic and tumorigenic ingredients, in many raw herbs are also largely missing (7–13). To ensure consumer health protection, the quality and safety of herbal dietary supplements, as well as the raw herbal plants used for dietary supplement preparations, must be assured (5, 14–17).
To address the issue of human health protection, more than 30 herbal dietary supplements and active ingredients have been nominated by the U.S. Food and Drug Administration (FDA) and the National Institutes of Health (NIH) to the U.S. National Toxicology Program (NTP) for determination of their toxicity and tumorigenicity. These include Ginkgo biloba extract, panax ginseng, kava, Aloe vera, green tea extract, comfrey, symphytine, dong quai, ephedrine alkaloid, L-ephedrine (ma huang), black cohosh, goldenseal root powder, pulegone, usnic acid, and Usnea herb. These herbal dietary supplements are being examined because they are among the most widely used and their consumption continues to increase in the United States. For risk assessment, when a test substance is found to be tumorigenic in the NTP chronic tumorigenicity bioassays, the elucidation of the mechanism of action is useful for establishing the relevance to humans. A tumorigenic compound or herbal product identified in animal bioassays with sufficient documentation of toxicity in humans will be listed as either “known to be a human carcinogen” or “reasonably anticipated to be a human carcinogen” in the “Report on Carcinogens (RoC)”, a U.S. congressionally mandated document prepared by the NTP (18).
In general, approaches for determining the mechanism by which a pure chemical induces toxicity or tumorigenicity have been relatively well established. Unfortunately, these approaches could not be readily extended to the studies of a mixture of different chemical components, such as herbal plants and herbal dietary supplements (19, 20). The chemical complexity of herbal compositions makes it difficulty to evaluate not only the efficacy but also the mechanism of toxicity and tumorigenicity. There are no simple and clear-cut approaches to determine the mechanism of action leading to toxicity and tumorigenicity of herbs. For instance, if an herbal supplement is hepatotoxic, it is difficult to determine if a single constituent is solely responsible for the hepatotoxicity of the whole herb. In an herb with multiple chemical constituents, the relative abundance of each compound may have an impact on the contribution of a specific chemical to hepatotoxicity, producing additive, synergetic, or antagonistic effects. This means that the toxic activities of a pure chemical constituent of an herb plant may not contribute significantly to toxic activity of the whole herb plant. For risk assessment, it is pertinent that toxicity data from the dietary supplement investigation can be used to extrapolate toxicity and tumorigenicity to humans. Thus, there is a need for elucidating mechanisms by which herbal dietary supplements exert toxicity and tumorigenicity.
Analysis of gene expression changes induced by exposure to a chemical both in vitro and in vivo can provide information about its mechanism of action (21–23). Toxicogenomics combines toxicology with genomics or other high throughput molecular profiling technologies such as transcriptomics, proteomics, and metabolomics (24) has been applied broadly in toxicant or toxin characterization, toxicity predication, biomarker discovery and identification, toxicant classification, environmental chemical/compound prioritization, and underlying mechanism elucidation (22, 23, 25–27). Although toxicogenomics has been applied to study toxicology since the application of DNA chips to toxicology from the late 1990s (28), use of a toxicogenomic approach for mechanistic study of toxicity induced by herbal dietary supplements has only recently been initiated.
We have long been interested in studying the toxicity and tumorigenicity induced by a variety of herbal dietary supplements. Our primary studies included the identification of toxic contaminants and tumorigenic components, determination of the mechanisms leading to hepatotoxicity and tumorigenicity in experimental animals, and DNA microarray study of gene expression profiles (12, 29–36). For example, we used DNA microarray technology to identify gene expression alterations induced by exposures of kava extract and GBE to rats and mice and elucidated their potential roles in liver toxicity (31–33). We have proposed that analysis of alterations in gene expression profiles can be an initial approach for determining the mechanisms by which herbal plants and herbal dietary supplements induced toxicity and/or tumorigenicity. In this review, we highlight this strategy with examples illustrating different types of mechanism-associated information, including modulation of drug metabolizing enzymes and changes of pathways/networks. From this information, the likely mechanism(s) of toxicity can be hypothesized and validated by subsequent chemical, biological, and genomic experiments.
MODULATION OF DRUG METABOLIZING ENZYMES
1. Cytochrome P450 (CYP) Enzymes
The drug metabolizing enzymes, located in the smooth endoplasmic reticulum, play a major role in the metabolism, elimination, and detoxification of xenobiotics, including drugs introduced into our bodies. Drug metabolizing enzymes include phase I enzymes, which are involved in oxidation, reduction, and hydrolysis reactions; phase II enzymes, which usually involve conjugation reactions; and phase III transporters. Although hepatotoxicity associated with herbal dietary supplement intake has been frequently reported (3–6, 9), the mechanisms of hepatotoxicity are not yet been elucidated. In addition to an herb’s intrinsic toxicity, herb-drug interactions may also contribute to safety concerns. It has been shown that herbal dietary supplements can induce or inhibit the activities of drug metabolizing enzymes, thus affecting both the pharmacokinetics and pharmacodynamics of co-administrated drugs (37, 38). This may result in decreased pharmacological effects or increased toxic effects of the co-administrated drugs (39) .
Study of herb-drug interactions has primarily focused on CYP450-based interactions, although recent studies also investigated the roles of transporters (37). Conventional approaches for assessment of the modulation of drug metabolizing enzymes include in vitro and in vivo studies, mRNA and protein expression, and enzymatic activity. For instance, the inhibition of CYP450 activity is frequently examined with liver microsomal preparations or recombinant CYP enzymes. Enzyme induction potential is evaluated using freshly isolated, cryopreserved hepatocytes, or precision-cut slices (40, 41). Nevertheless, these approaches are targeted for one or only a few individual CYPs. DNA microarrays has the advantage which allows the simultaneous monitoring of expression of thousands of genes, including many drug metabolizing genes. For example, there are more than 50 probes coding for CYP family genes on human whole genome expression arrays (e.g., HumanRef-6, Illumina) and 300 probes corresponding to phase I, II, and III drug metabolizing genes on mouse whole genome expression arrays (e.g., Mouse OneArray, Phalanx Biotech Group). It is reasonable to anticipate that many drug metabolizing enzymes can be modulated by treatment with herbal dietary supplements. Moreover, changes in gene expression levels often reflect alterations of enzyme levels and this is normally observed for the CYPs except for CYP2E1 where protein stabilization plays a critical role in the inductive response (40, 42–44). For the above reasons, applying microarrays to investigate the induction or inhibition of drug metabolizing enzymes is important and feasible.
1.1 Kava
Kava, the rhizome of the kava tropical shrub plant Piper methysticum Forst. F., has been used in Europe for treatment of anxiety and nervous disorders, such as stress and restlessness, and in the United States as a natural alternative to anti-anxiety drugs and sleeping pills. However, the potential hepatotoxicity of kava in humans has been reported recently (45–51). The association of kava product use with liver-related risks prompted regulatory agencies in many countries to take actions, from warning consumers to removing kava-containing products from the marketplace. The U.S. FDA issued a Consumer Advisory entitled “Kava-containing dietary supplements may be associated with severe liver injury” in 2002 (16).
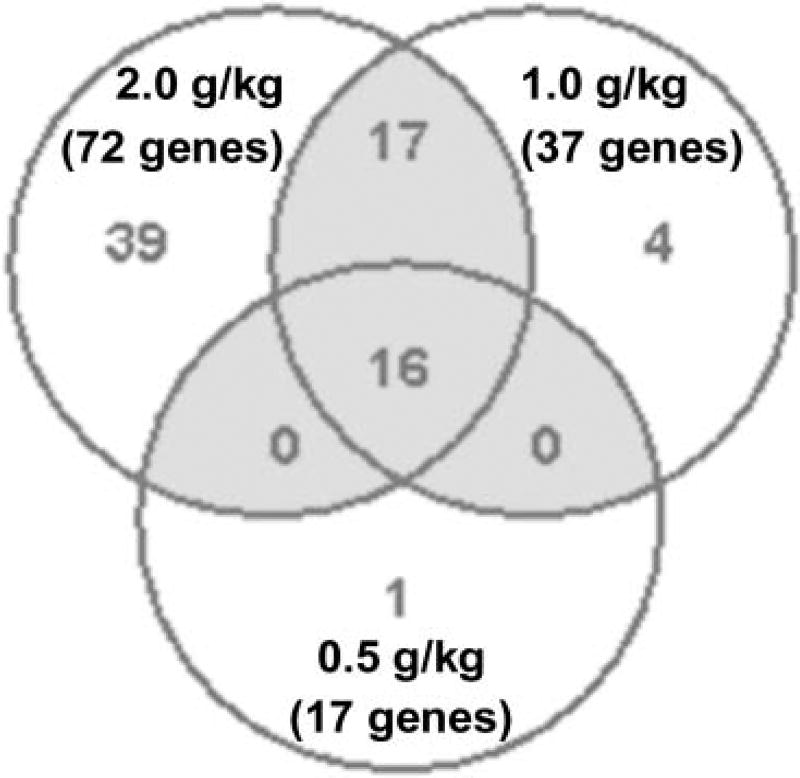
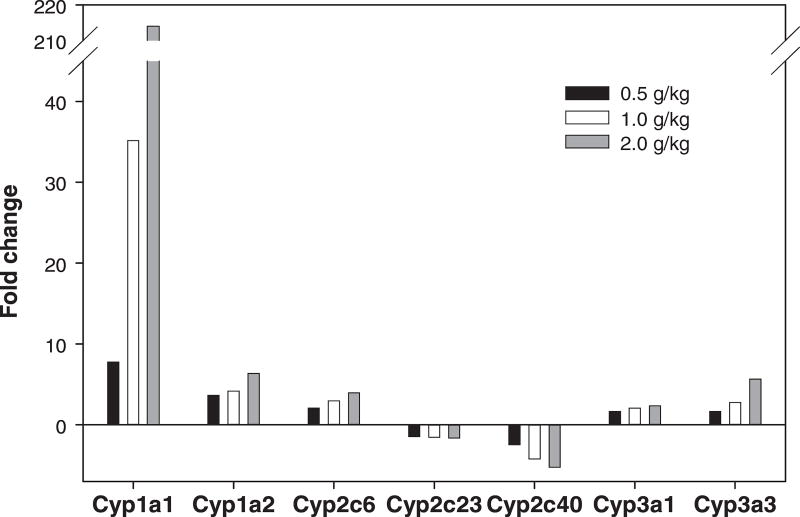
Kava extract was nominated to the NTP for a chronic tumorigenicity bioassay conducted. As part of a range finding study, the NTP conducted 14-week studies to characterize the toxicology of kava exposure in Fischer 344 rats and B6C3F1 mice. Groups of 10 male and 10 female rats and mice were administered kava extract by gavage at 0, 0.125, 0.25, 0.5, 1.0, or 2.0 g/kg/day. Using the tissues from these animals, Guo et al. (31) studied the induced changes in expression of genes, with emphasis on drug metabolizing genes, in the livers of Fischer 344 male rats administered kava. Analysis of 22,226 genes revealed that there were 17, 37, and 72 drug metabolizing genes significantly altered in the 0.5, 1.0, and 2.0 g/kg treatment groups, respectively (Figure 1). Among 72 drug metabolizing genes whose expression were significantly changed in the 2.0 g/kg treatment group, 19 genes were associated with Phase I metabolizing enzymes; 21 genes with Phase II metabolizing enzymes; and 32 genes with transporters (Phase III) (Table 1). Sixteen of the 19 significantly altered Phase I metabolism-associated genes were CYPs. While gene expression of CYP1A1, 1A2, 2C6, 3A1, and 3A3 increased, CYP 2C23 and 2C40 decreased, all in a dose-dependent manner (Figure 2).
Figure 1.
Numbers of drug metabolizing genes altered by 0.5, 1.0 and 2.0 g/kg kava treatments in male rat liver. A gene was identified as differentially expressed if the fold-change was greater than 1.5 (up or down) and the P -value was less than 0.05 in comparison to the control group. The color in gray refers to the number of genes overlapped in three treatments. The data were from Guo et al. (31).
Table 1.
Drug Metabolizing Genes Altered by 2.0 g/kg Kava Treatment in Male Rat Liver ¶
| Gene Symbol | Gene Description | Fold Change* |
|---|---|---|
| Phase I metabolism | ||
| Aldh1a1 | aldehyde dehydrogenase family 1, member A1 | 2.8 |
| Cyp17a1 | cytochrome P450, family 17, subfamily a, polypeptide 1 | −2.2 |
| Cyp1a1 | cytochrome P450, family 1, subfamily a, polypeptide 1 | 214.0 |
| Cyp1a2 | cytochrome P450, family 1, subfamily a, polypeptide 2 | 6.4 |
| Cyp2c23 | cytochrome P450, family 2, subfamily c, polypeptide 23 | −1.7 |
| Cyp2c37 | cytochrome P450, 2c37 | 2.1 |
| Cyp2c40 | cytochrome P450, family 2, subfamily c, polypeptide 40 | −5.3 |
| Cyp2c55 | cytochrome P450, family 2, subfamily c, polypeptide 55 | 4.0 |
| Cyp2c6 | Cytochrome P450, subfamily IIC6 | 4.0 |
| Cyp2t1 | cytochrome P450 monooxygenase CYP2T1 | −1.7 |
| Cyp3a1 | cytochrome P450, family 3, subfamily a, polypeptide 1 | 2.4 |
| Cyp3a13 | cytochrome P450, family 3, subfamily a, polypeptide 13 | −3.7 |
| Cyp3a3 | cytochrome P450, subfamily 3A, polypeptide 3 | 5.7 |
| Cyp4a12 | cytochrome P450, 4a12 | 1.5 |
| Cyp4f6 | cytochrome P450 4F6 | −1.7 |
| Cyp7b1 | cytochrome P450, family 7, subfamily b, polypeptide 1 | −2.2 |
| Cyp8b1 | cytochrome P450, family 8, subfamily b, polypeptide 1 | −1.6 |
| Fmo1 | Flavin containing monooxygenase 1 | −3.8 |
| Gzma | granzyme A | −1.6 |
| Phase II metabolism | ||
| Acsl4 | acyl-CoA synthetase long-chain family member 4 | 1.7 |
| Ces2 | carboxylesterase 2 (intestine, liver) | 8.5 |
| Ces6 | carboxylesterase 6 | 2.6 |
| Ephx1 | epoxide hydrolase 1, microsomal | 3.6 |
| Gnmt | glycine N-methyltransferase | −2.1 |
| Gsta2 | glutathione-S-transferase, alpha type2 | 2.9 |
| Gsta4 | glutathione S-transferase, alpha 4 | 2.0 |
| Gstm1 | glutathione S-transferase, mu 1 | 1.8 |
| Gstm2 | glutathione S-transferase, mu 2 | 1.6 |
| Gstm3 | glutathione S-transferase, mu type 3 | −1.6 |
| Gstp1 | glutathione-S-transferase, pi 1 | 1.6 |
| Nqo1 | NAD(P)H dehydrogenase, quinone 1 | 11.4 |
| Sulf2 | sulfatase 2 | −1.6 |
| Sult1b1 | sulfotransferase family 1B, member 1 | 1.8 |
| Sult1c1 | sulfotransferase family, cytosolic, 1C, member 1 | −2.0 |
| Ugt1a1 | UDP glycosyltransferase 1 family, polypeptide A1 | 1.7 |
| Ugt1a6 | UDP glycosyltransferase 1 family, polypeptide A6 | 10.0 |
| Ugt1a7 | UDP glycosyltransferase 1 family, polypeptide A7 | 3.5 |
| Ugt2b10 | UDP glycosyltransferase 2 family, polypeptide B10 | 2.1 |
| Ugt2b4 | UDP glycosyltransferase 2 family, polypeptide B4 | 1.5 |
| Ugt2b4 | UDP glycosyltransferase 2 family, polypeptide B4 | 1.8 |
| Phase III metabolism | ||
| Abca8 | ATP-binding cassette, sub-family A (ABC1), member 8a | −1.8 |
| Abcb6 | ATP-binding cassette, sub-family B (MDR/TAP), member 6 | 1.9 |
| Abcb9 | ATP-binding cassette, sub-family B (MDR/TAP), member 9 | −2.0 |
| Abcc2 | ATP-binding cassette, sub-family C (CFTR/MRP), member 2 | 1.7 |
| Abcc3 | ATP-binding cassette, sub-family C (CFTR/MRP), member 3 | 15.9 |
| Abcc6 | ATP-binding cassette, sub-family C (CFTR/MRP), member 6 | −1.8 |
| Abcc8 | ATP-binding cassette, sub-family C (CFTR/MRP), member 8 | −1.7 |
| Abcc9 | ATP-binding cassette, sub-family C (CFTR/MRP), member 9 | −1.8 |
| Slc11a1 | solute carrier family 11 (proton-coupled divalent metal ion transporters), member 1 | −1.7 |
| Slc13a3 | solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 3 | −1.5 |
| Slc16a1 | solute carrier family 16 (monocarboxylic acid transporters), member 1 | 1.5 |
| Slc16a10 | solute carrier family 16 (monocarboxylic acid transporters), member 10 | −1.9 |
| Slc16a11 | solute carrier family 16 (monocarboxylic acid transporters), member 11 | −1.6 |
| Slc16a6 | solute carrier family 16 (monocarboxylic acid transporters), member 6 | 1.5 |
| Slc17a1 | Solute carrier family 17 (sodium phosphate), member 1 | 1.8 |
| Slc17a3 | Solute carrier family 17 (sodium phosphate), member 3 | 1.9 |
| Slc22a7 | solute carrier family 22 (organic anion transporter), member 7 | −1.6 |
| Slc22a8 | solute carrier family 22 (organic anion transporter), member 8 | −1.6 |
| Slc25a17 | solute carrier family 25 (mitochondrial carrier, peroxisomal membrane protein), member 17 | 1.5 |
| Slc27a5 | Solute carrier family 27 (fatty acid transporter), member 5 | −1.9 |
| Slc28a2 | solute carrier family 28 (sodium-coupled nucleoside transporter), member 2 | −1.5 |
| Slc2a2 | solute carrier family 2 (facilitated glucose transporter), member 2 | −1.6 |
| Slc34a2 | Solute carrier family 34 (sodium phosphate), member 2 | −4.8 |
| Slc38a3 | Solute carrier family 38, member 3 | −1.8 |
| Slc38a4 | Solute carrier family 38, member 4 | −1.7 |
| Slc39a4 | Solute carrier family 39 (zinc transporter), member 4 | 1.8 |
| Slc3a2 | solute carrier family 3 (activators of dibasic and neutral amino acid transport), member 2 | 2.9 |
| Slc4a2 | Solute carrier family 4, member 2 | 1.6 |
| Slc6a9 | solute carrier family 6 (neurotransmitter transporter, glycine), member 9 | 1.5 |
| Slc7a5 | solute carrier family 7 (cationic amino acid transporter, y+ system), member 5 | 1.7 |
| Slco2b1 | Solute carrier organic anion transporter family, member 2b1 | −1.6 |
| Tap1 | transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) | −1.7 |
The data were from Guo et al. (31).
All changes are greater than 1.5 (up and down) with a P -value less than 0.05 compared to the controls and the symbol of minus (−) indicates down-regulation.
Figure 2.
Gene expression of the common cytochrome P450 drug metabolizing genes altered by three kava treatments (0.5, 1.0, and 2.0 g/kg) in male rat livers.
Using the liver tissues from same animals, Clayton et al. (52) determined the expression of a number of hepatic CYP enzymes by immunohistochemical methods. They found that there was decreased expression of CYP 2D1 (human CYP2D6 homolog) in 2.0 g/kg females and increased expression of CYP1A2, 2B1, and 3A1 in the 1.0 and 2.0 g/kg groups of both sexes. These authors proposed that kava-induced hepatic functional changes in the rat might be relevant to human clinical cases of hepatotoxicity following exposure. Because of the limitation of using the immunohistochemical method, i.e., only limited numbers of antibodies were used for the studies (52), it was not clear how many drug metabolizing enzymes were altered in response to the administration of kava.
The expression of liver genes of kava-treated male mice was studied similarly (33). After exposure to 0.125, 0.25, 0.5, 1.0, and 2.0 g/kg kava, there were 349, 353, 880, 1339, and 1674 genes significantly up or down-regulated. Upon 2.0 g/kg kava treatment, 95 drug metabolizing enzyme-associated genes were significantly altered, among which 28 genes were associated with Phase I metabolizing enzymes, 29 genes with Phase II metabolizing enzymes, and 38 genes with transporters (Phase III).
1.2 Ginkgo Biloba Extract
Ginkgo biloba extract (GBE) has been one of the most widely sold products in health food stores in the United States. Its purported biological effects include scavenging free radicals, lowering oxidative stress, reducing neural damage, reducing platelet aggregation, and anti-inflammation, anti-tumor, and anti-aging activities. Clinically, it has been used to treat CNS disorders, such as Alzheimer disease and cognitive deficits. It also causes allergies and alters bleeding time. While its mutagenicity or carcinogenic activity has not been reported, some of its constituents, quercetin, kaempferol, and rutin have been shown to be genotoxic. However, whether GBE exhibits hepatotoxicity or carcinogenicity remains unclear.
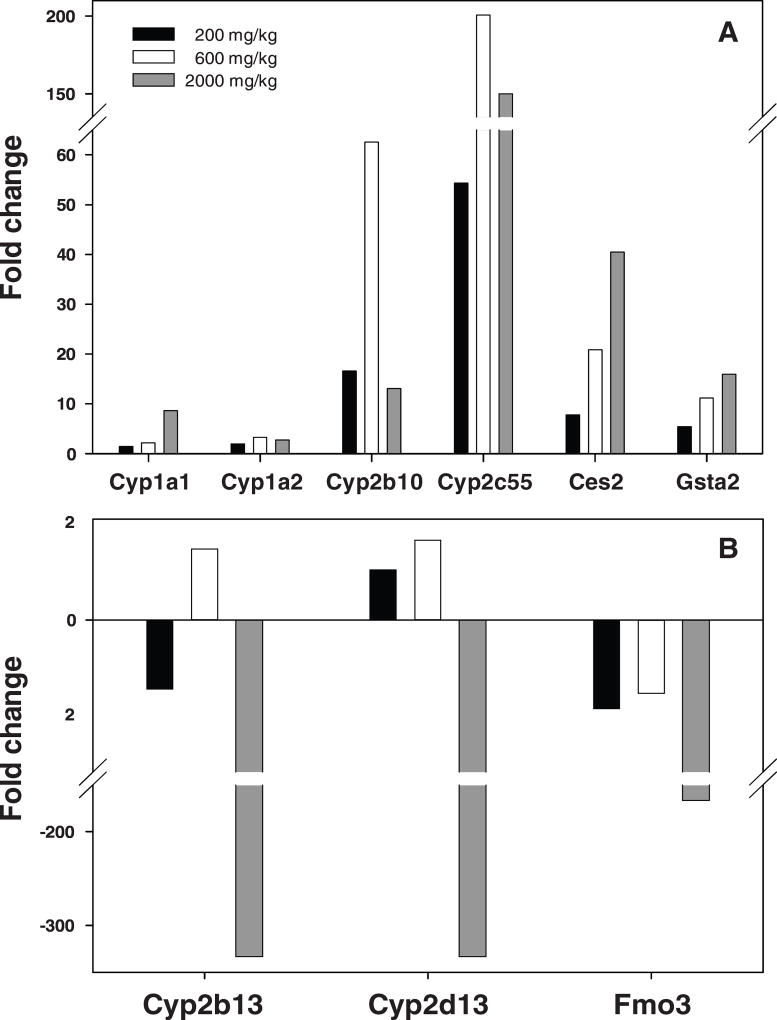
Guo et al. (32) determined gene expression changes in the livers of B6C3F1 mice administered GBE chronically. Among the 31,802 genes analyzed, there were 129, 289, and 2011 genes significantly altered in the 200, 600, and 2,000 mg/kg treatment groups, respectively. Significant numbers of drug metabolizing genes were altered in response to GBE treatments. At 2,000 mg/kg GBE treatment, there were 68 drug metabolizing genes whose expression was significantly altered, of which 33 genes were Phase I metabolizing enzymes, 18 genes were Phase II metabolizing enzymes, and 17 genes were transporters (Phase III). It is worth noting that 23 of the 33 expressed Phase I metabolism associated genes were CYPs, in which 2 genes belong to the CYP1 superfamily (CYP1A1 and 1A2), 15 genes belonged to the CYP2 superfamily, and 2 genes belonged to the CYP3 superfamily (Table 2) (32). TaqMan assays were used to verify some of the results of the gene expression changes measured using microarrays. Nine drug metabolizing genes were selected for the TaqMan analysis to validate the microarray data. Six genes, CYP1A1, CYP1A2, CYP2B10, CYP2C55, CES2, and GSTA2, were found to be overexpressed, and the changes in CYP1A1, CES2, and GSTA2 were dose dependent (Figure 3).
Table 2.
Drug Metabolizing Genes Altered by 2,000 mg/kg Ginkgo biloba Extract Treatment in Male Mouse Liver ¶
| Gene Symbol | Gene Description | Fold Change* |
|---|---|---|
| Phase I metabolism | ||
| Adh7 | alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide | 2.5 |
| Aldh5a1 | aldehyde dehydrogenase family 5, subfamily A1 | −2.9 |
| Aldh8a1 | aldehyde dehydrogenase 8 family, member A1 | −2.8 |
| Cyp1a1 | Cytochrome P450, family 1, subfamily a, polypeptide 1 | 6.8 |
| Cyp1a2 | Cytochrome P450, family 1, subfamily a, polypeptide 2 | 2.3 |
| Cyp2a12 | Cytochrome P450, family 2, subfamily a, polypeptide 12 | −2.2 |
| Cyp2a5 | Cytochrome P450, family 2, subfamily a, polypeptide 5 | 2.9 |
| Cyp2b10 | Cytochrome P450, family 2, subfamily b, polypeptide 10 | 17.3 |
| Cyp2b13 | Cytochrome P450, family 2, subfamily b, polypeptide 13 | −38.5 |
| Cyp2b20 | Cytochrome P450, family 2, subfamily b, polypeptide 10 | 14.6 |
| Cyp2b9 | Cytochrome P450, family 2, subfamily b, polypeptide 9 | −18.9 |
| Cyp2c29 | Cytochrome P450, family 2, subfamily c, polypeptide 29 | 3.6 |
| Cyp2c55 | Cytochrome P450, family 2, subfamily c, polypeptide 55 | 50.2 |
| Cyp2c66 | Cytochrome P450, family 2, subfamily c, polypeptide 66 | 4.4 |
| Cyp2d13 | Cytochrome P450, family 2, subfamily d, polypeptide 13 | −29.4 |
| Cyp2f2 | Cytochrome P450, family 2, subfamily f, polypeptide 2 | −13.2 |
| Cyp2g1 | Cytochrome P450, family 2, subfamily g, polypeptide 1 | −2.1 |
| Cyp2j5 | Cytochrome P450, family 2, subfamily j, polypeptide 5 | −7.1 |
| Cyp2j9 | Cytochrome P450, family 2, subfamily j, polypeptide 9 | −14.9 |
| Cyp2u1 | Cytochrome P450, family 2, subfamily u, polypeptide 1 | −17.9 |
| Cyp3a16 | Cytochrome P450, family 3, subfamily a, polypeptide 16 | −8.4 |
| Cyp3a41 | Cytochrome P450, family 3, subfamily a, polypeptide 41 | −2.3 |
| Cyp46a1 | Cytochrome P450, family 46, subfamily a, polypeptide 1 | −8.9 |
| Cyp4f14 | Cytochrome P450, family 4, subfamily f, polypeptide 14 | −4.8 |
| Cyp4f16 | Cytochrome P450, family 4, subfamily f, polypeptide 16 | 2.9 |
| Cyp51 | Cytochrome P450, family 51 | 2.0 |
| Dhrs3 | dehydrogenase/reductase (SDR family) member 3 | −2.0 |
| Dhrs9 | dehydrogenase/reductase (SDR family) member 9 | 3.6 |
| Dpyd | dihydropyrimidine dehydrogenase | −3.8 |
| Fmo3 | flavin containing monooxygenase 3 | −76.9 |
| Maoa | Monoamine oxidase A | 6.3 |
| Maob | Monoamine oxidase B | −2.4 |
| Ptgs1 | prostaglandin-endoperoxide synthase 1 | −2.5 |
| Phase II metabolism | ||
| Acsl1 | acyl-CoA synthetase long-chain family member 1 | −3.1 |
| Acsl4 | acyl-CoA synthetase long-chain family member 4 | 5.2 |
| Ces2 | carboxylesterase 2 | 24.7 |
| Ephx1 | epoxide hydrolase 1, microsomal | 2.5 |
| Gsta2 | glutathione S-transferase, alpha 2 (Yc2) | 34.6 |
| Gsta4 | glutathione S-transferase, alpha 4 | 2.6 |
| Gstk1 | glutathione S-transferase kappa 1 | −2.2 |
| Gstm1 | glutathione S-transferase, mu 1 | 2.8 |
| Gstm2 | glutathione S-transferase, mu 2 | 4.4 |
| Gstm3 | glutathione S-transferase, mu 3 | 11.5 |
| Gstm4 | glutathione S-transferase, mu 4 | 4.4 |
| Gstm6 | glutathione S-transferase, mu 6 | 5.2 |
| Gsto1 | glutathione S-transferase omega 1 | −4.7 |
| Gstp1 | glutathione S-transferase, pi 1 | 4.4 |
| Nnmt | nicotinamide N-methyltransferase | −10.4 |
| Nqo3a2 | Cytochrome b5 reductase 1 | 3.2 |
| Sult1d1 | sulfotransferase family 1D, member 1 | −2.7 |
| Sult3a1 | sulfotransferase family 3A, member 1 | −35.7 |
| Phase III metabolism | ||
| Abcb1a | ATP-binding cassette, sub-family B (MDR/TAP), member 1A | 2.3 |
| Abcc12 | ATP-binding cassette, sub-family C (CFTR/MRP), member 12 | 5.5 |
| Abcc2 | ATP-binding cassette, sub-family C (CFTR/MRP), member 2 | 3.1 |
| Abcc4 | ATP-binding cassette, sub-family C (CFTR/MRP), member 4 | 18.9 |
| Abcc5 | ATP-binding cassette, sub-family C (CFTR/MRP), member 5 | 7.5 |
| Abcc6 | ATP-binding cassette, sub-family C (CFTR/MRP), member 6 | −3.0 |
| Abcd3 | ATP-binding cassette, sub-family D (ALD), member 3 | −2.3 |
| Aqp6 | aquaporin 6 | 2.2 |
| Aqp7 | aquaporin 7 | 17.8 |
| Mvp | major vault protein | 2.7 |
| Slc10a1 | solute carrier family 10 (sodium/bile acid cotransporter family), member 1 | −4.4 |
| Slc22a3 | solute carrier family 22 (organic cation transporter), member 3 | 10.2 |
| Slc22a7 | solute carrier family 22 (organic anion transporter), member 7 | −4.4 |
| Slc29a1 | solute carrier family 29 (nucleoside transporters), member 1 | −2.4 |
| Slc2a1 | solute carrier family 2 (facilitated glucose transporter), member 1 | 2.0 |
| Slc3a1 | solute carrier family 3, member 1 | −4.2 |
| Slc7a5 | solute carrier family 7 (cationic amino acid transporter, y+ system), member 5 | 3.3 |
The data were from Guo et al. (32).
All changes are greater than 2.0 (up and down) with a P -value less than 0.01compared to the controls and the symbol of minus (−) indicates down-regulation.
Figure 3.
Gene expression changes validated by TaqMan assays in liver of mice treated with three Ginkgo biloba extract treatments (200, 600, and 2000 mg/kg). (A), up-regulated genes; (B), the genes with down-regulation. The data were from Guo et al. (32).
2. Other Drug Metabolizing Enzymes
Much attention had been paid to CYP families in the study of drug absorption, distribution, metabolism, and elimination, as well as drug-drug interactions. Recently, the clinical relevance and scientific understanding of other drug metabolism enzymes in these processes, such as membrane transporters, have been recognized (41). In our studies of herbal dietary supplements, in addition to the CYP family, alterations of many other drug metabolizing genes were also observed (31–33). For example, the expression of FMO3 (flavin-containing monooxygenase 3) was dramatically inhibited with 2,000 mg/kg GBE treatment (Table 2). Besides the CYP family, flavin-containing monooxygenase (FMO) is another major family of Phase I drug-metabolizing enzymes catalyzing the oxidative biotransformation of drugs. FMO enzymes break down compounds that contain nitrogen, sulfur, or phosphorus. FMO3, a major FMO isoform, plays an important role in metabolizing drugs such as the anticancer drug tamoxifen, the pain medication codeine, the antifungal drug ketoconazole, and antipsychotic drugs clozapine and olanapine (53–55). In our study, GBE inhibited the expression of FMO3 significantly, with 77-fold reduction detected by microarray and 166-fold reduction validated by TaqMan assay. The data suggest that down-regulated expression of the FMO3 enzyme by GBE may potentially inhibit the metabolism of certain drugs resulting in herb-drug interactions, thus either reducing the therapeutic effects or enhancing the toxicity induced by their metabolites. The expression of many Phase III transporters was altered by kava and GBE treatments (Table 1 and 2). It is, therefore, of great importance to determine the roles of these transporters in herb-drug interactions and what classes of chemicals/drugs are likely affected.
3. Herb-Drug Interactions
One of the most reported adverse effects caused by herbal products is hepatotoxicity. While the mechanisms of hepatotoxicity initiated by herbal products are not clear, modulation of drug-metabolizing enzymes leading to herb-drug interactions has been proposed as one of the major causes (31, 52, 56). When acting as an enzyme inhibitor, the metabolic activity of one or several CYP enzymes can be blocked. In contrast, enzyme inducers enhance CYP activity by increasing the enzyme synthesis. Herbal products can also induce hepatotoxicity through competitive inhibition, e.g., by sharing the same CYP isozyme that is specifically required for metabolism of a drug, and thus, competitively block this drug’s metabolism. This would cause the drug to be present at higher amounts and for a longer time in the liver, potentially leading to hepatotoxicity.
The CYP1, CYP2, and CYP3 superfamilies are the most important enzymes in the metabolism of drugs and the metabolic activation of toxic and carcinogenic xenobiotics (57, 58). The Phase II enzymes can eliminate drugs and carcinogenic metabolites of xenobiotics by forming water soluble conjugated metabolites. The balance between the Phase I and Phase II enzyme components determines the metabolic fate of endogenous and exogenous chemicals. Consequently, co-administration of herbal dietary supplements, such as kava extracts, and therapeutic drugs may result in herb-drug interactions, which can lead to serious clinical as well as toxicological consequences (10, 11, 59– 62). The results obtained from Guo et al. (31) demonstrated that many rat liver drug metabolizing genes were altered in response to kava treatment; this is in good agreement with the report by Lim et al. (63) that kava-induced herb-drug interaction through modulation of metabolizing enzymes is a possible cause of hepatotoxicity.
There are several proposed mechanisms of kava induced hepatotoxicity and the most accepted one is the induction of herb-drug interactions through modulation of metabolizing enzymes by which the drug metabolism can be affected. Guo et al. (31–33) suggested that analysis of the gene expression profiles using microarrays in the livers of rodents treated with herbal dietary supplement is potentially a practical approach for understanding the mechanism of action.
PATHWAY AND NETWORK ANALYSES
Pathway and network analyses can be utilized to investigate the gene relationships, functional clustering, and mechanisms of toxicity. For example, Guo et al. (32) determined the most relevant biological functions, pathways, and networks of the genes altered by 2,000 mg/kg GBE treatment. The top canonical pathways are metabolism of xenobiotics by cytochrome P450; fatty acid metabolism; tryptophan metabolism; LPS/IL-1 mediated inhibition of RXR function; arachidonic acid metabolism; linoleic acid metabolism; aryl hydrocarbon receptor signaling; xenobiotic metabolism signaling; glutathione metabolism; aminosugars metabolism; PXR/RXR activation; NRF2-mediated oxidative stress response; hepatic fibrosis/hepatic stellate cell activation; tyrosine metabolism; acute phase response signaling; FXR/RXR activation; sphingolipid metabolism; G-protein coupled receptor signaling; coagulation system; and glycosaminoglycan degradation. The effects on Nrf2 (nuclear factor erythroid-related factor 2)-mediated oxidative pathway, Myc pathway, and others are described in the following.
1. Nrf2-mediated Oxidative Pathway
Even if there was no clear sign of liver toxicity induced by herbal dietary supplements in experimental animals, distinct gene expression alterations (i.e., gene expression signature) would be a powerful tool for predictive toxicity. Keap1-Nrf2-ARE signaling plays a critical role in protecting cells from endogenous and exogenous stresses, and is involved in antioxidative response, detoxification of xenobiotics, and proteome maintenance. Under normal physiological conditions, the transcription factor Nrf2 localizes in the cytoplasm and interacts with Keap1. Upon oxidative stress, Nrf2 is released from Keap1 and translocates to the nucleus and subsequently activates its various downstream target genes (64). The target genes show a wide spectrum of functions including inactivating oxidants, increasing the levels of glutathione, and enhancing toxin export via transporters to enhance cell survival. Nrf2 knockout mice show higher sensitivity to chemical toxicity. Studies using Nrf2 knock-out mice demonstrated that they were more susceptible to acetaminophen-induced hepatocellular injury (65) and benzo[a]pyrene-induced tumor formation, as indicated by higher levels of DNA adducts (66). This susceptibility is due partly to a reduced level in the expression of detoxification enzymes (64, 67). Activation of detoxification enzymes plays a pivotal role in protecting cells from oxidative insult when cells encounter toxin challenge.
Guo et al. (33) determined that in the liver of male mice treated with kava, the expressions of a group of genes involved in detoxification processes (NQO1, GSTMs, GSTAs, and GSTPs) were elevated. The most prominent changes were observed in the 2.0 g/kg treatment group, with GSTA1, GSTA2, and NQO1 being increased by 50-, 24- and 4-fold, respectively. These altered genes are the well-known target genes controlled by the transcription factor Nrf2, implicating that the enhanced expressions of these genes were caused by Nrf2 activation. Ingenuity Pathway analysis indeed identified Nrf2- mediated oxidative stress response as a significantly changed pathway, although the mechanisms for Nrf2 activation after kava treatment remain elusive. It has been demonstrated that multiple mechanisms are involved in Nrf2 activation, among which, Keap1-dependent pathway and kinase (MAPKs, PKC, and PI3K) signal pathway are the best characterized (68). While the former pathway is usually oxidative stress-driven, the latter could be either oxidative stress-dependent or independent. As to kava effects, a recent study showed that kava ingredients activated Nrf2 by mechanisms of oxidative stress-independent MAPKs, specifically ERK1/2 kinase signal pathway in neural cells (69). Whether this observation can be extended to liver or hepatocytes is worth further investigation.
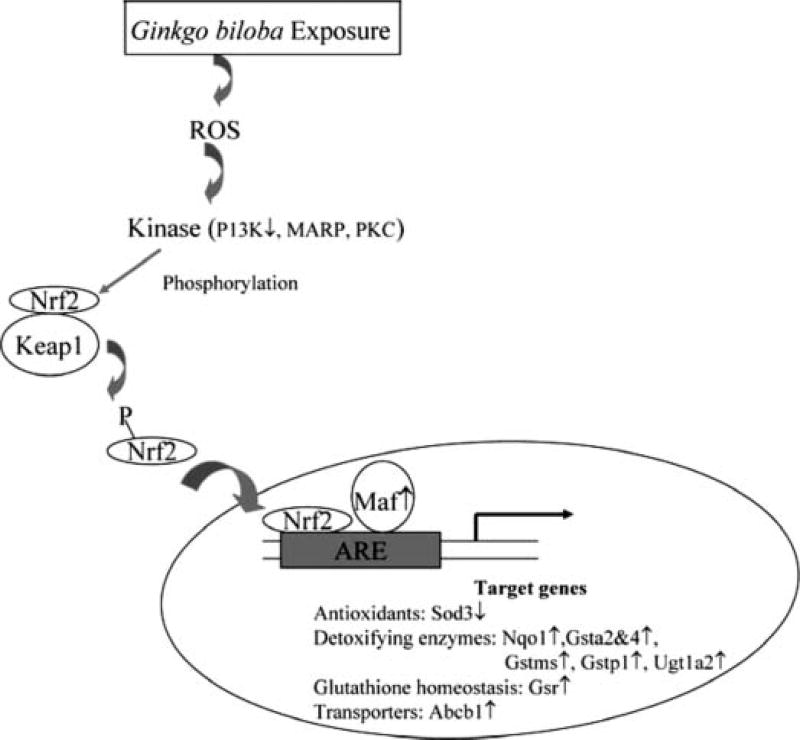
Guo et al. (32) explored pathways and networks in response to Ginkgo biloba extract treatment and found that Ginkgo biloba extract exposure resulted in significant stimulation of the Nrf2-mediated oxidative pathway (Figure 4). Nrf2-mediated oxidative stress response was identified with 23 genes involved. In the pathway regulated by NRF2, most of the genes were up-regulated while two genes were down-regulated. The up-regulated genes included those coding for detoxifying enzymes (NQO1, GSTMs, GSTAs, GSTP1, and UGT1A2), glutathione homeostasis (GSR), and transporter (ABCB1). Maf2, a transcriptional factor that forms heterodimers with Nrf2, was also up-regulated. Two down-regulated genes were P13k, which plays a role in Nrf2 phosphorylation, and Sod3, which is a family member of antioxidants.
Figure 4.
Alteration of NRF2 (nuclear factor erythroid-related factor 2)-mediated oxidative stress response in the liver of male mice treated with Ginkgo biloba extract. The data were from Guo et al. (32).
2. Mitochondria Dysfunction
Usnic acid is a prominent secondary lichen metabolite commonly used as a weight-loss dietary supplement in Western countries due to its uncoupling action on mitochondria. However, its use has been associated with severe liver damage in some healthy individuals (9). Most of the studies so far reported focus on determining the effects of usnic acid on mitochondria dysfunctions primarily concerning the rate of oxygen consumption and ATP generation. To determine the effects of usnic acid on mitochondria, Joseph et al. (70) employed a mitochondria-specific microarray (MitoChip) (71) to study the expression levels of a library of 542 genes associated with mitochondrial structure and functions in liver of B6C3F1 female mice. The genes on the MitoChip are involved in oxidative phosphorylation, beta-oxidation of free fatty acids, tri-carboxylic acid cycle, apoptosis, mitochondrial DNA replication, transcription, DNA repair, and others (71). Microarray analysis was performed on livers of mice exposed to usnic acid at 0, 60, 180, and 600 ppm in feed ad libitum for two weeks. The expression of more than 100 genes was altered in the highest dose group. Genes associated with complexes I through IV of the electron transport chain were significantly induced, and a number of genes, which are involved in fatty acid oxidation, the Krebs cycle, apoptosis, and membrane transporters, were over-expressed. Usnic acid is lipophilic and thus can diffuse through mitochondrial membranes and cause a proton leak (uncoupling). Joseph et al. (70) hypothesized that the up-regulation of genes associated with complexes I–IV functions may be a compensatory mechanism to maintain the proton gradient across the mitochondrial inner membrane. Based on this finding, they also suggested that induction of fatty acid oxidation and the Krebs cycle may be an adaptive response to the uncoupling of mitochondria.
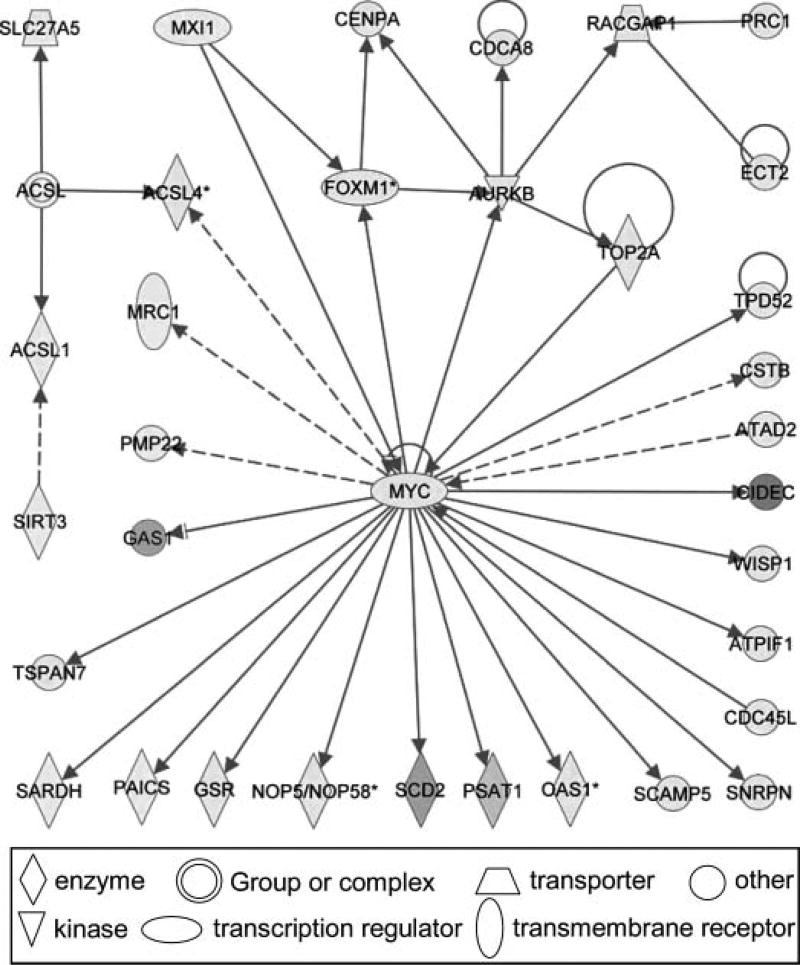
3. Pro-oncogene Myc Pathway
In addition to the effect on the Nrf2- mediated oxidative stress response pathway, Guo et al. (32) found that male mice treated with GBE also showed significant alteration of the MYC regulated pathway (Figure 5). The significantly changed genes after GBE administration were mapped into 87 networks. Each network was associated with specific genes and involved in different functions, of which 23 networks had 20 or more focus molecules. The central gene in this network was the pro-oncogene MYC. Twenty-one genes were found to be directly associated with MYC and these genes function in cell cycle, cell growth/proliferation, and cell death processes. Notably, CIDEC (cell-death-inducing DFFA-like effectors c), which promotes apoptosis, exhibited a 47-fold induction. The expression of genes involved in DNA replication checkpoint (CDCA8 and CDC45l) was also up-regulated. GAS1 (growth arrest-specific 1), the tumor suppressor gene that blocks entry to S phase and prevents cycling of cells, remarkably was down-regulated by 20-fold.
Figure 5.
The first network (cell cycle, cellular movement, and cancer) containing 34 differentially expressed genes in the mice treated with Ginkgo biloba extract. Green indicates down-regulation and red indicates up-regulation. Solid lines indicate direct interactions and dashed lines indicate indirect interactions between two molecules. The data were from Guo et al. (32).
4. Cancer Related Pathway and Comparative Pathway Analysis
Comfrey has been used as a vegetable, a tea, and an herbal medicine for many years, and recently as an herbal dietary supplement. However, comfrey is hepatotoxic in livestock and humans and carcinogenic in experimental animals. Mei et al. (36) studied the comfrey-induced gene expression profile of 26,857 genes in the livers of rats by DNA microarray analysis. Groups of 6 male transgenic Big Blue rats were fed a basal diet (control) or a diet containing 8% comfrey roots, a dose that resulted in liver tumors in a previous carcinogenicity bioassay, for 12 weeks and sacrificed one day after the final treatment. When a two-fold cutoff value and a P-value less than 0.01 were selected, 2,726 genes were identified as differentially expressed in comfreyfed rats compared to control animals. Among these genes, there were 1,617 genes associated by Ingenuity Pathway Analysis with particular functions, and the differentially expressed genes in comfrey-fed rat livers were involved in metabolism, injury of endothelial cells, and liver injury and abnormalities, including liver fibrosis and cancer development. The genes involved in cell death and apoptosis in endothelial cells altered by comfrey treatment in liver are listed in Table 3.
Table 3.
Genes Involved in Cell Death and Apoptosis of Liver Endothelial Cells Altered by 8% Comfrey Treatment in Male Rat ¶
| Gene Symbol | Gene Description | Fold Change* |
|---|---|---|
| Angpt2 | angiopoietin 2 | 3.5 |
| App | amyloid beta (A4) precursor protein | 4.7 |
| Atf3 | activating transcription factor 3 | 6.2 |
| Bcl2 | B-cell cLL/lymphoma 2 | 2.4 |
| Bcl2a1 | Bcl2-related protein A1 | 3.4 |
| Bcl2l | Bcl2-like | 2.5 |
| Casp1 | caspase 1, apoptosis-related cysteine protease (interleukin 1, beta, convertase) | 2.3 |
| Cnp | 2’,3’-cyclic nucleotide 3’ phosphodiesterase | 2.4 |
| Col4a2 | collagen, type IV, alpha 2 | 2.8 |
| Cxcr4 | chemokine receptor (LCR1) | 3.2 |
| Dusp6 | dual specificity phosphatase 6 | 2.3 |
| Edn1 | endothelin 1 | 4.9 |
| Fgf1 | fibroblast growth factor 1 | 2.0 |
| Gpr9 | G protein-coupled receptor 9 (Cxcr3) | 2.0 |
| Hgf | hepatocyte growth factor | 3.2 |
| Hmox1 | heme oxygenase 1 | 3.2 |
| Il2 | interleukin 2 | −3.3 |
| Ins1 | insulin 1 | −5.0 |
| Lrp5 | low density lipoprotein receptor-related protein 5 | −3.3 |
| Mapk9 | mitogen-activated protein kinase 9 | −2.0 |
| Pparg | peroxisome proliferator activated receptor, gamma | −3.3 |
| Serpinf1 | serine (or cysteine) proteinase inhibitor, clade F), member 1 | −2.0 |
| Spp1 | secreted phosphoprotein 1 | 11.6 |
| Tgfb1 | transforming growth factor, beta 1 | 2.1 |
| Tnfrsf6 | tumor necrosis factor receptor superfamily, member 6 | 3.4 |
| Tnfsf10 | tumor necrosis factor (ligand) superfamily, member 10 | 2.2 |
The data were from Mei et al. (36).
All changes are greater than 2.0 (up and down) with a P -value less than 0.01compared to the controls and the symbol of minus (−) indicates down-regulation.
Aristolochic acid is the active component of herbal drugs derived from Aristolochia species that have been used for medicinal purposes since antiquity. Aristolochic acid, however, induces nephropathy and urothelial cancer in people and malignant tumors in the kidney and urinary tract of rodents (12, 72). Although aristolochic acid is bioactivated in both kidney and liver, it only induces tumors in kidney. Chen et al. (29) examined gene expression profiles by DNA microarray analysis in kidney and liver of rats treated with carcinogenic doses of aristolochic acid. The gene expression profiles were significantly altered by aristolochic acid treatment in both kidney and liver. Functional analysis with Ingenuity Pathways Analysis showed that there were many more significantly altered genes involved in cancer-related pathways in kidney than in liver and that these genes were mainly associated with defense response, apoptosis, and immune response in kidney, but not in liver. The differential responses between kidney and liver might be responsible for the tissue-specific toxicity and carcinogenicity of aristolochic acid. These results suggest that analysis of the gene expression profiles can define the differential responses to toxicity and carcinogenicity of aristolochic acid from kidney and liver (29).
USE OF DNA MICROARRAY ANALYSIS FOR CONSEQUENT MECHANISM DETERMINATION
1. Toxicological and Mechanistic Information Obtained from DNA Microarray Analysis
Currently there is no established methodology for determining the mechanisms of toxicity (particularly tumorigenicity) induced by a mixture of many chemical components, such as GBE and other herbal plant extracts. Previously, we proposed that DNA microarray analysis may be a highly practical initial approach for revealing the whole spectrum of gene expression alterations by a chemical mixture (31–33). The experimental results described previously illustrate that useful toxicological effects and mechanistic information can be obtained from analysis of the gene expression changes in the liver of rats and mice treated with herbal dietary supplements (including kava extract, GBE, and comfrey) and herbal components (such as aristolochic acid and usnic acid). The resulting toxicological effects and mechanism information are summarized below.
Kava and GBE modulate CYP metabolizing isozymes, which may lead to herb-drug interactions by affecting the pharmacokinetics of co-administrated drugs, and potentially resulting in hepatotoxicity (31–33). The CYP1, 2, and 3 superfamilies are important in the metabolism of xenobiotics including drugs. CYP1A1, which generally exists in a very low amount in the adult liver, plays a vital role in metabolic activation of xenobiotics, such as tumorigenic polycyclic aromatic hydrocarbons (PAHs) (73). Thus, induction of CYP1A1 by GBE and kava treatments may contribute to their hepatotoxicity, and enhance metabolic activation of tumorigenic xenobiotics, leading to adverse human health effects.
Whole-genome gene expression analysis revealed that kava altered the expression of CYP1A1, which was not studied in prior reports (31). This finding well-illustrates that without obtaining the whole spectrum of gene expression change, some important information, such as CYP1A1 up-regulation, may be missed.
The liver gene expression profile of drug metabolizing enzymes in kava-treated male rats obtained by Guo et al. (31) correlated with the corresponding protein levels determined by immunohistochemical assays (52).
Herbal dietary supplements can modulate Phase I, Phase II, and Phase III metabolizing gene expression. Besides CYP, the flavin-containing monooxygenase (FMO) is another major group of Phase I drug-metabolizing enzyme catalyzing the oxidative biotransformation of drugs and other chemicals. Guo et al. (32) observed that GBE dramatically inhibited the expression of FMO3, which suggests that down-regulated expression of the FMO3 enzyme by GBE potentially inhibits metabolism of certain drugs resulting in drug-drug or drug-food interactions, thus reducing the therapeutic effects or enhancing the toxicity induced by their metabolites. Considerable attention may need to be paid to the likely induction/inhibition of drug metabolizing genes such as CYP1A1 and FMO3 and many others in humans taking kava products.
GBE exposure resulted in the significant stimulation of the Nrf2-mediated oxidative stress pathway and Myc-centered network. These results provide important clues for elucidating the toxic mechanisms of GBE, and may predict the liver lesions caused by GBE exposure.
2. Guidance for Subsequent Mechanistic Studies
The gene expression profile obtained from DNA microarray helps in determining its phenotype, function, and response to the foreign materials entering into the cells. Analysis of the resulting gene expression profile can provide us useful information as described above. Well-planned subsequent mechanistic studies, based on the gene expression profiles, should enable a better understanding of the underlying molecular mechanisms. Several mechanistic approaches are suggested below.
When gene expression is changed by herbal dietary supplement, it is necessary to determine whether protein expression levels and the enzyme activities are also altered. For example, it was determined that hepatic CYP1A1 gene was induced by kava and GBE (31–33). The expression of hepatic CYP enzymes can be determined by immunohistochemical methods or by proteomic analysis. The CYP1A1 enzyme activity, in turn, can be assayed by liver microsomal metabolism of a substrate specifically metabolized only by CYP1A1 enzyme followed by determination of the level of the metabolite formed. For example, benzo[a)pyrene trans-7,8-dihydrodiol is a good substrate of CYP1A1. Benzo[a]pyrene trans-7,8-dihydrodiol is metabolized into the tumorigenic metabolite benzo[a]pyrene trans-7,8-diol anti-9,10-epoxide specifically by CYP1A1. Comparison of the quantities of benzo[a]pyrene trans-7,8-diol anti-9,10-epoxide (by measurement of the corresponding tetrol formation by HPLC analysis) formed from microsomal metabolism of benzo[a]pyrene trans-7,8-dihydrodiol by liver microsomes of untreated mice and mice pretreated with kava will enable us to determine the increase in CYP1A1 enzyme activity caused by kava treatment.
GBE exposure resulted in the significant stimulation of the expression of genes in the Nrf2-mediated oxidative pathway and alteration in the expression of gene in the Myc regulated pathway. These findings suggest that the liver tissues are damaged by reactive oxygen species (ROS) generated by GBE treatment, or by damage of mitochondrial function. This can be validated by examining ROS formation in both in vitro and in vivo systems. ROS can interact with proteins, cellular DNA, and membrane lipid, causing cellular damage of these tissues. This damage can be examined by assaying lipid peroxidation, DNA strand cleavage, DNA adduct formation, and other approaches. Using gene deficient animal models (e.g., Nrf2 knockout mice) or transgenic models (e.g., as Myc overexpressing mice) can help elucidate and determine the roles of the particular genes or pathways in hepatotoxicity and tumorigenesis induced by herbal dietary supplements. These subsequent experiments will provide important clues for elucidating the toxic mechanisms of GBE, which may cause hepatic damage, including severe liver lesions and even liver tumors. Although at present the NTP two-year chronic tumorigenicity bioassay with GBE has been completed, the histopathology results from this study have not been completed. The results of that study will provide the outcomes that may support the predictions.
Integration of gene expression changes with known pathological changes can be used to formulate a mechanistic scheme for induced hepatotoxicity and tumorigenesis. However, due to the complexity of an herbal dietary supplement containing many chemical constituents, this approach cannot provide information concerning which herbal chemical constituents are responsible for toxicity, including tumorigenicity. Consequently, the toxicological effect of its chemical constituents, or closely related analogs, must be studied carefully. An excellent example is the mechanistic study of comfrey-induced hepatotoxicity and tumorigenicity (74). Comfrey contains several tumorigenic pyrrolizidine alkaloids, known to induce liver tumors in rats through a genotoxic mechanism (34, 36, 72). The expression profiles in the liver of Big Blue Fisher 344 rats treated with comfrey or riddelliine, a pyrrolizidine alkaloid, were analyzed and compared. There were 45 and 87 drug metabolizing genes up- or down-regulated by the treatment of riddelliine and comfrey. The expressions of 20 of these genes were altered by both comfrey and riddelliine, and are detailed in Table 4. Very interestingly, the changed expression of each gene was always in the same direction for the two compounds. Among the genes altered by comfrey and riddelliine, there were a number of common genes and functional processes that were related to carcinogenesis (74). There was a strong correlation between the two treatments for fold-change alterations in expression of drug metabolizing and cancer-related genes (74). These results suggest that comfrey induced liver tumors by a genotoxic mechanism and that the pyrrolizidine alkaloid constituents in comfrey were responsible for mutation induction and tumor initiation.
Table 4.
Drug Metabolizing Genes Altered by Both 8% Comfrey and 1.0 mg/kg Riddelliine Treatments in Rat Liver ¶
| Fold Change*
|
|||
|---|---|---|---|
| Gene Symbol | Gene Description | Comfrey | Riddelliine |
| Phase I metabolism | |||
| Ces2 | carboxylesterase 2 (intestine, liver) | 10.4 | 2.4 |
| Cyp2c | cytochrome P450, subfamily IIC | −24.3 | −270.3 |
| Cyp2c40 | cytochrome P450, family 2, subfamily c | 8.7 | 42.3 |
| Cyp2e1 | cytochrome P450, family 2, subfamily e | 2.2 | 2.1 |
| Fmo5 | flavin containing monooxygenase 5 | 4.1 | 4.2 |
| Phase II metabolism | |||
| Gsta3 | glutathione S-transferase, alpha 3 | 22.5 | 13.4 |
| Inmt | indolethylamine N-methyltransferase | −29.3 | −11.7 |
| Nqo1 | NAD(P)H dehydrogenase, quinone 1 | 5.0 | 2.6 |
| Sult1c1 | sulfotransferase family, cytosolic,1C, member1 | −2.7 | −3.2 |
| Phase III metabolism | |||
| Abcb1 | ATP-binding cassette, sub-family B | 97.0 | 11.3 |
| Abcb9 | ATP-binding cassette, sub-family B, member 9 | −5.0 | −2.7 |
| Abcc3 | ATP-binding cassette, sub-family C, member 3 | 25.1 | 10.8 |
| Abcc8 | ATP-binding cassette, sub-family C, member 8 | −9.7 | −2.6 |
| Atp13a5 | ATPase type 13A5 | −3.6 | −4.1 |
| Slc13a5 | solute carrier family 13, member 5 | −2.9 | −2.4 |
| Slc16a4 | solute carrier family 16, member 4 | −2.8 | −4.9 |
| Slc22a6 | solute carrier family 22, member 6 | −3.7 | −2.4 |
| Slc22a8 | solute carrier family 22, member 8 | −12.2 | −833.3 |
| Slc25a21 | solute carrier family 25, member 21 | 4.4 | 2.7 |
| Slc25a30 | solute carrier family 25, member 30 | −4.5 | −2.7 |
The data were from Guo et al. (74).
All changes are greater than 2.0 (up and down) with a P -value less than 0.01compared to the controls and the symbol of minus (−) indicates down-regulation.
PERSPECTIVES
Use of herbal dietary supplements in the United States is rapidly increasing. To ensure consumer health protection, the quality and safety of herbal dietary supplements have to be assured. There are more than 30 herbal dietary supplements that have been, or are being, examined for toxicity and tumorigenicity by the NTP. There is a need of strategic approaches for elucidating mechanisms by which herbal dietary supplements exert toxicity and tumorigenicity. In this review, we provide evidence that analysis of gene expression profiles by DNA microarrays can be an initial approach for determining the mechanisms by which herbal plants and herbal dietary supplements induced toxicity and/or tumorigenicity. We anticipate that, before more chemical-specific and time-consuming methodologies are deployed, this strategy would be employed as a common initial approach for mechanism determination of herbal dietary supplements as well as other types of chemical mixtures.
Herbal dietary supplements consist of multiple chemical constituents and some of these chemical constituents are responsible for induction of toxicity and tumorigenicity. As such, elucidation of the mechanism should determine how the active constituents elicit toxicity at the molecular level. At this time, determination of the principal toxic chemical constituents of an herbal dietary supplement is still a big challenge. Taking the induction of CYP1A1 by kava treatment as an example, it is difficult to determine which constituent(s) are responsible for the CYP1A1 induction. If a single chemical constituent was found to be the most potent inducer, it could be suggested that this constituent would most likely be responsible for the induction effect of the whole kava extract. The relative abundance of each chemical constituent in kava would also have an impact on the contribution of a distinct chemical constituent on the overall biological effect of the herbal dietary supplement. That is, the most potent inducer may be present in the extract at a much lower quantity than a less potent inducer. On the other hand, there is an important clue that can help us determine which constituents are responsible for most, if not all, toxicity of an herbal plant. Based on Traditional Chinese Medicine (TCM), the chemical constituents that exhibit pharmaceutical/functional activities may most likely belong to secondary metabolites in an herbal plant. Those secondary metabolites have been used in humans as medicines. For example, usnic acid is a prominent secondary lichen metabolite that has been used for various purposes including as dietary supplement for weight loss (9).
In contrast to the primary metabolites, secondary metabolites are chemicals produced by plants that are not essential for growth, development, photosynthesis, or reproduction functions. This is because the purpose of forming secondary metabolites in a plant is to against environmental invasion. In order to avoid being attacked by predators such as herbivores and pathogens, etc., it is essential for some of these secondary metabolites to possess toxic properties. As such, selection of the secondary metabolites with claimed pharmaceutical/functional activities for testing of toxicity would become prominently important. For example, there are six kavalactones which account for 30%– 40% composition in the parent kava extract. Study of toxicological and genomic activities of these constituents may enable us to find out the active chemical constituents responsible for kava-induced hepatotoxicity. This approach can search for toxicological constituents responsible for the herb-induced toxicity and enable us to elucidate the underlying molecular mechanism. In addition, this approach can also help us develop safer herbal dietary supplements and carefully monitor those principal toxicological constituents that do not possess the major pharmaceutical/functional activities.
In summary, for human health protection, strategies on determining the molecular mechanisms of toxicity induced by herbal dietary supplements, both on the bioactivation of the chemical constituents and the pathological changes of the cells, have to be established.
Acknowledgments
We thank Drs. James C. Fuscoe, Qiang Shi, and Frederick A. Beland from NCTR and Richard D. Irwin and Michael L. Cunningham from NIEHS for their critical review of this manuscript. This research was supported in part by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences.
Footnotes
This article is not subject to US Copyright Laws.
The contents of this article do not necessarily reflect the views and policies of the U.S. Food and Drug Administration or the National Toxicology Program. The mention of trade names or commercial products does not constitute endorsement or recommendation for use.
References
- 1.Zurer P, Hanson D. Chemistry puts herbal supplements to the test. Chemical & Engineering News. 2004;82:16. [Google Scholar]
- 2.Ho CH, Simon JC, Shahidi F, Shao Y. Dietary supplements: an overview. Washington, DC: Dietary Supplements (ACS Symposium Series 987), American Chemical Society; 2008. pp. 2–8. [Google Scholar]
- 3.Arneborn P, Jansson A, Bottiger Y. [Acute hepatitis in a woman after intake of slimming pills bought via Internet] Lakartidningen. 2005;102:2071–2. [PubMed] [Google Scholar]
- 4.Bujanda L, Palacios A, Silvarino R, Sanchez A, Munoz C. [Kava-induced acute icteric hepatitis] Gastroenterol. Hepatol. 2002;25:434–5. doi: 10.1016/s0210-5705(02)70281-1. [DOI] [PubMed] [Google Scholar]
- 5.CDC. Hepatic toxicity possibly associated with kava-containing products—United States, Germany, and Switzerland, 1999–2002. Morbidity and Mortality Weekly Reports (MMWR) 2002:1065–67. [PubMed] [Google Scholar]
- 6.Durazo FA, Lassman C, Han SH, Saab S, Lee NP, Kawano M, Saggi B, Gordon S, Farmer DG, Yersiz H, Goldstein RL, Ghobrial M, Busuttil RW. Fulminant liver failure due to usnic acid for weight loss. Am. J. Gastroenterol. 2004;99:950–2. doi: 10.1111/j.1572-0241.2004.04165.x. [DOI] [PubMed] [Google Scholar]
- 7.Chan PC, Xia Q, Fu PP. Ginkgo biloba leave extract: biological, medicinal, and toxicological effects. J. Environ. Sci. Health C Environ. Carcinog. Ecotoxicol. Rev. 2007;25:211–44. doi: 10.1080/10590500701569414. [DOI] [PubMed] [Google Scholar]
- 8.Fu PP, Xia Q, Guo L, Yu H, Chan PC. Toxicity of kava kava. J. Environ. Sci. Health C Environ. Carcinog. Ecotoxicol. Rev. 2008;26:89–112. doi: 10.1080/10590500801907407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo L, Shi Q, Fang JL, Mei N, Ali AA, Lewis SM, Leakey JE, Frankos VH. Review of usnic acid and Usnea barbata toxicity. J. Environ. Sci. Health C Environ. Carcinog. Ecotoxicol. Rev. 2008;26:317–38. doi: 10.1080/10590500802533392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gurley BJ, Swain A, Barone GW, Williams DK, Breen P, Yates CR, Stuart LB, Hub-bard MA, Tong Y, Cheboyina S. Effect of goldenseal (Hydrastis canadensis) and kava kava (Piper methysticum) supplementation on digoxin pharmacokinetics in humans. Drug Metab. Dispos. 2007;35:240–5. doi: 10.1124/dmd.106.012708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hu Z, Yang X, Ho PC, Chan SY, Heng PW, Chan E, Duan W, Koh HL, Zhou S. Herb-drug interactions: a literature review. Drugs. 2005;65:1239–82. doi: 10.2165/00003495-200565090-00005. [DOI] [PubMed] [Google Scholar]
- 12.Mei N, Guo L, Liu R, Fuscoe JC, Chen T. Gene expression changes induced by the tumorigenic pyrrolizidine alkaloid riddelliine in liver of Big Blue rats. BMC Bioinfor-matics. 2007;(8 Suppl 7):S4. doi: 10.1186/1471-2105-8-S7-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Singh YN. Potential for interaction of kava and st. John’s wort with drugs. J. Ethnopharmacol. 2005;100:108–13. doi: 10.1016/j.jep.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 14.CFSAN. FDA warns consumers not to use the dietary supplement LipoKinetix. 2001 Available at http://wwwcfsanfdagov/~dms/ds-lipohtml.
- 15.CFSAN. Letter to distributor on hazardous dietary supplement LipoKinetix. 2001 Available at http://wwwcfsanfdagov/~dms/ds-ltr26html.
- 16.CFSAN. Center for Food Safety and Applied Nutrition (CFSAN): Kava-containing dietary supplements may be associated with severe liver injury. U.S. Department of Health and Human Services, Food and Drug Administration; Rockville, Maryland: Mar 25, 2002. Available at http://www.cfsan.fda.gov/~dms/ds-warn.html. [Google Scholar]
- 17.FDA. FDA Public Health News, FDA announced major initiatives for dietary supplements. 2004 [Google Scholar]
- 18.Report on carcinogen. Available at http://ntp.niehs.nih.gov/?objectid=72016262-BDB7-CEBA-FA60E922B18C2540.
- 19.Borgert CJ. Predicting interactions from mechanistic information: can omic data validate theories? Toxicol. Appl. Pharmacol. 2007;223:114–20. doi: 10.1016/j.taap.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 20.Borgert CJ, Quill TF, McCarty LS, Mason AM. Can mode of action predict mixture toxicity for risk assessment? Toxicol. Appl. Pharmacol. 2004;201:85–96. doi: 10.1016/j.taap.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 21.Hamadeh HK, Bushel PR, Jayadev S, Martin K, DiSorbo O, Sieber S, Bennett L, Tennant R, Stoll R, Barrett JC, Blanchard K, Paules RS, Afshari CA. Gene expression analysis reveals chemical-specific profiles. Toxicol. Sci. 2002;67:219–31. doi: 10.1093/toxsci/67.2.219. [DOI] [PubMed] [Google Scholar]
- 22.Waring JF, Ciurlionis R, Jolly RA, Heindel M, Ulrich RG. Microarray analysis of hepatotoxins in vitro reveals a correlation between gene expression profiles and mechanisms of toxicity. Toxicol. Lett. 2001;120:359–68. doi: 10.1016/s0378-4274(01)00267-3. [DOI] [PubMed] [Google Scholar]
- 23.Waters MD, Fostel JM. Toxicogenomics and systems toxicology: aims and prospects. Nat. Rev. Genet. 2004;5:936–48. doi: 10.1038/nrg1493. [DOI] [PubMed] [Google Scholar]
- 24.NAP. Communicating toxicogenomics information to nonexperts: a workshop summary. The National Academies Press; 2005. [PubMed] [Google Scholar]
- 25.Boverhof DR, Zacharewski TR. Toxicogenomics in risk assessment: applications and needs. Toxicol. Sci. 2006;89:352–60. doi: 10.1093/toxsci/kfj018. [DOI] [PubMed] [Google Scholar]
- 26.Dix DJ, Houck KA, Martin MT, Richard AM, Setzer RW, Kavlock RJ. The Tox-Cast program for prioritizing toxicity testing of environmental chemicals. Toxicol. Sci. 2007;95:5–12. doi: 10.1093/toxsci/kfl103. [DOI] [PubMed] [Google Scholar]
- 27.Waring JF, Jolly RA, Ciurlionis R, Lum PY, Praestgaard JT, Morfitt DC, Buratto B, Roberts C, Schadt E, Ulrich RG. Clustering of hepatotoxins based on mechanism of toxicity using gene expression profiles. Toxicol. Appl. Pharmacol. 2001;175:28–42. doi: 10.1006/taap.2001.9243. [DOI] [PubMed] [Google Scholar]
- 28.Afshari CA, Nuwaysir EF, Barrett JC. Application of complementary DNA mi-croarray technology to carcinogen identification, toxicology, and drug safety evaluation. Cancer Res. 1999;59:4759–60. [PubMed] [Google Scholar]
- 29.Chen T, Guo L, Zhang L, Shi L, Fang H, Sun Y, Fuscoe JC, Mei N. Gene expression profiles distinguish the carcinogenic effects of aristolochic acid in target (kidney) and non-target (liver) tissues in rats. BMC Bioinformatics. 2006;(7 Suppl 2):S20. doi: 10.1186/1471-2105-7-S2-S20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chou MW, Fu PP. Formation of DHP-derived DNA adducts in vivo from dietary supplements and chinese herbal plant extracts containing carcinogenic pyrrolizidine alkaloids. Toxicol. Ind. Health. 2006;22:321–7. doi: 10.1177/0748233706071765. [DOI] [PubMed] [Google Scholar]
- 31.Guo L, Li Q, Xia Q, Dial S, Chan PC, Fu P. Analysis of gene expression changes of drug metabolizing enzymes in the livers of F344 rats following oral treatment with kava extract. Food Chem. Toxicol. 2009;47:433–42. doi: 10.1016/j.fct.2008.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guo L, Mei N, Liao W, Chan PC, Fu P. Ginkgo biloba extract induces gene expression changes in xenobiotics metabolism and the myc-centered network. OMICS. 2010;14:75–90. doi: 10.1089/omi.2009.0115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guo L, Shi Q, Dial S, Xia Q, Mei N, Li Q, Chan PC, Fu P. Gene expression profiling in male B6C3F1 mouse livers exposed to kava identifies changes in drug metabolizing genes and potential mechanisms linked to kava toxicity. Food Chem. Toxicol. 2010;48:686–696. doi: 10.1016/j.fct.2009.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mei N, Guo L, Fu PP, Heflich RH, Chen T. Mutagenicity of comfrey (Symphytum Officinale) in rat liver. Br. J. Cancer. 2005;92:873–5. doi: 10.1038/sj.bjc.6602420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mei N, Guo L, Tseng J, Dial SL, Liao W, Manjanatha MG. Gene expression changes associated with xenobiotic metabolism pathways in mice exposed to acrylamide. Environ. Mol. Mutagen. 2008;49:741–5. doi: 10.1002/em.20429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mei N, Guo L, Zhang L, Shi L, Sun YA, Fung C, Moland CL, Dial SL, Fuscoe JC, Chen T. Analysis of gene expression changes in relation to toxicity and tumorigenesis in the livers of Big Blue transgenic rats fed comfrey (Symphytum officinale) BMC Bioinformatics. 2006;(7 Suppl 2):S16. doi: 10.1186/1471-2105-7-S2-S16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Driver S, Baxter K, Williamson E. Stockley’s herbal medicines interactios. Pharmaceutical Press. 2009:1–11. [Google Scholar]
- 38.Kaplowitz N. Hepatotoxicity of herbal remedies: insights into the intricacies of plant-animal warfare and cell death. Gastroenterology. 1997;113:1408–12. doi: 10.1053/gast.1997.v113.agast971131408. [DOI] [PubMed] [Google Scholar]
- 39.Zhou S, Gao Y, Jiang W, Huang M, Xu A, Paxton JW. Interactions of herbs with cytochrome P450. Drug. Metab. Rev. 2003;35:35–98. doi: 10.1081/dmr-120018248. [DOI] [PubMed] [Google Scholar]
- 40.Bjornsson TD, Callaghan JT, Einolf HJ, Fischer V, Gan L, Grimm S, Kao J, King SP, Miwa G, Ni L, Kumar G, McLeod J, Obach RS, Roberts S, Roe A, Shah A, Snikeris F, Sullivan JT, Tweedie D, Vega JM, Walsh J, Wrighton SA. The conduct of in vitro and in vivo drug-drug interaction studies: a Pharmaceutical Research and Manufacturers of America (PhRMA) perspective. Drug Metab. Dispos. 2003;31:815–32. doi: 10.1124/dmd.31.7.815. [DOI] [PubMed] [Google Scholar]
- 41.Huang SM, Strong JM, Zhang L, Reynolds KS, Nallani S, Temple R, Abraham S, Habet SA, Baweja RK, Burckart GJ, Chung S, Colangelo P, Frucht D, Green MD, Hepp P, Karnaukhova E, Ko HS, Lee JI, Marroum PJ, Norden JM, Qiu W, Rahman A, Sobel S, Stifano T, Thummel K, Wei XX, Yasuda S, Zheng JH, Zhao H, Lesko LJ. New era in drug interaction evaluation: US Food and Drug Administration update on CYP enzymes, transporters, and the guidance process. J. Clin. Pharmacol. 2008;48:662–70. doi: 10.1177/0091270007312153. [DOI] [PubMed] [Google Scholar]
- 42.Fuhr U. Induction of drug metabolising enzymes: pharmacokinetic and toxicologi-cal consequences in humans. Clin. Pharmacokinet. 2000;38:493–504. doi: 10.2165/00003088-200038060-00003. [DOI] [PubMed] [Google Scholar]
- 43.Iyer KR, Sinz MW. Characterization of Phase I and Phase II hepatic drug metabolism activities in a panel of human liver preparations. Chem. Biol. Interact. 1999;118:151–69. doi: 10.1016/s0009-2797(99)00007-1. [DOI] [PubMed] [Google Scholar]
- 44.Roymans D, Van Looveren C, Leone A, Parker JB, McMillian M, Johnson MD, Koganti A, Gilissen R, Silber P, Mannens G, Meuldermans W. Determination of cytochrome P450 1A2 and cytochrome P450 3A4 induction in cryopreserved human hepatocytes. Biochem. Pharmacol. 2004;67:427–37. doi: 10.1016/j.bcp.2003.09.022. [DOI] [PubMed] [Google Scholar]
- 45.Dentali SJ. Herb safety review: kava, piper methysticum forster f. (Piperaceae) Bethesda, MD: American Herbal Products Association; 1997. [Google Scholar]
- 46.Campo JV, McNabb J, Perel JM, Mazariegos GV, Hasegawa SL, Reyes J. Kava-induced fulminant hepatic failure. J. Am. Acad. Child Adolesc. Psychiatry. 2002;41:631–2. doi: 10.1097/00004583-200206000-00001. [DOI] [PubMed] [Google Scholar]
- 47.Clough AR, Bailie RS, Currie B. Liver function test abnormalities in users of aqueous kava extracts. J. Toxicol. Clin. Toxicol. 2003;41:821–9. doi: 10.1081/clt-120025347. [DOI] [PubMed] [Google Scholar]
- 48.Humberston CL, Akhtar J, Krenzelok EP. Acute hepatitis induced by kava kava. J. Toxicol. Clin. Toxicol. 2003;41:109–13. doi: 10.1081/clt-120019123. [DOI] [PubMed] [Google Scholar]
- 49.Parkman CA. Another FDA warning: kava supplements. Case Manager. 2002;13:26–8. doi: 10.1067/mcm.2002.126437. [DOI] [PubMed] [Google Scholar]
- 50.Russmann S, Barguil Y, Cabalion P, Kritsanida M, Duhet D, Lauterburg BH. Hepatic injury due to traditional aqueous extracts of kava root in New Caledonia. Eur. J. Gastroenterol. Hepatol. 2003;15:1033–6. doi: 10.1097/00042737-200309000-00015. [DOI] [PubMed] [Google Scholar]
- 51.Russmann S, Lauterburg BH, Helbling A. Kava hepatotoxicity. Ann. Intern. Med. 2001;135:68–9. doi: 10.7326/0003-4819-135-1-200107030-00036. [DOI] [PubMed] [Google Scholar]
- 52.Clayton NP, Yoshizawa K, Kissling GE, Burka LT, Chan PC, Nyska A. Immunohis-tochemical analysis of expressions of hepatic cytochrome P450 in F344 rats following oral treatment with kava extract. Exp. Toxicol. Pathol. 2007;58:223–36. doi: 10.1016/j.etp.2006.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Parte P, Kupfer D. Oxidation of tamoxifen by human flavin-containing monooxygenase (FMO) 1 and FMO3 to tamoxifen-N-oxide and its novel reduction back to tamoxifen by human cytochromes P450 and hemoglobin. Drug Metab. Dispos. 2005;33:1446–52. doi: 10.1124/dmd.104.000802. [DOI] [PubMed] [Google Scholar]
- 54.Klick DE, Hines RN. Mechanisms regulating human FMO3 transcription. Drug Metab. Rev. 2007;39:419–42. doi: 10.1080/03602530701498612. [DOI] [PubMed] [Google Scholar]
- 55.Ciraulo DA, Shader RI, Greenblatt DJ, Creelman W. Drug interaction in psychiatry. Baltimore: Lippincott Williams & Wilkins; 2005. [Google Scholar]
- 56.Clouatre DL. Kava kava: examining new reports of toxicity. Toxicol. Lett. 2004;150:85–96. doi: 10.1016/j.toxlet.2003.07.005. [DOI] [PubMed] [Google Scholar]
- 57.Gonzalez FJ, Gelboin HV. Role of human cytochromes P450 in the metabolic activation of chemical carcinogens and toxins. Drug Metab. Rev. 1994;26:165–83. doi: 10.3109/03602539409029789. [DOI] [PubMed] [Google Scholar]
- 58.Gonzalez FJ, Yu AM. Cytochrome P450 and xenobiotic receptor humanized mice. Annu. Rev. Pharmacol. Toxicol. 2006;46:41–64. doi: 10.1146/annurev.pharmtox.45.120403.100007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bressler R. Herb-drug interactions: interactions between kava and prescription medications. Geriatrics. 2005;60:24–5. [PubMed] [Google Scholar]
- 60.Gurley BJ, Gardner SF, Hubbard MA, Williams DK, Gentry WB, Cui Y, Ang CY. Clinical assessment of effects of botanical supplementation on cytochrome P450 phenotypes in the elderly: St John’s wort, garlic oil, Panax ginseng and Ginkgo biloba. Drugs Aging. 2005;22:525–39. doi: 10.2165/00002512-200522060-00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mathews JM, Etheridge AS, Black SR. Inhibition of human cytochrome P450 activities by kava extract and kavalactones. Drug Metab. Dispos. 2002;30:1153–7. doi: 10.1124/dmd.30.11.1153. [DOI] [PubMed] [Google Scholar]
- 62.Singh YN. Kava: an overview. J. Ethnopharmacol. 1992;37:13–45. doi: 10.1016/0378-8741(92)90003-a. [DOI] [PubMed] [Google Scholar]
- 63.Lim ST, Dragull K, Tang CS, Bittenbender HC, Efird JT, Nerurkar PV. Effects of kava alkaloid, pipermethystine, and kavalactones on oxidative stress and cytochrome P450 in F-344 rats. Toxicol. Sci. 2007;97:214–21. doi: 10.1093/toxsci/kfm035. [DOI] [PubMed] [Google Scholar]
- 64.Kensler TW, Wakabayashi N, Biswal S. Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu. Rev. Pharmacol. Toxicol. 2007;47:89–116. doi: 10.1146/annurev.pharmtox.46.120604.141046. [DOI] [PubMed] [Google Scholar]
- 65.Chan K, Han XD, Kan YW. An important function of Nrf2 in combating oxidative stress: detoxification of acetaminophen. Proc. Natl. Acad. Sci. USA. 2001;98:4611–6. doi: 10.1073/pnas.081082098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ramos-Gomez M, Kwak MK, Dolan PM, Itoh K, Yamamoto M, Talalay P, Kensler TW. Sensitivity to carcinogenesis is increased and chemoprotective efficacy of enzyme inducers is lost in nrf2 transcription factor-deficient mice. Proc. Natl. Acad. Sci. USA. 2001;98:3410–5. doi: 10.1073/pnas.051618798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Aleksunes LM, Manautou JE. Emerging role of Nrf2 in protecting against hepatic and gastrointestinal disease. Toxicol. Pathol. 2007;35:459–73. doi: 10.1080/01926230701311344. [DOI] [PubMed] [Google Scholar]
- 68.Shen G, Hebbar V, Nair S, Xu C, Li W, Lin W, Keum YS, Han J, Gallo MA, Kong AN. Regulation of Nrf2 transactivation domain activity. The differential effects of mitogen-activated protein kinase cascades and synergistic stimulatory effect of Raf and CREB-binding protein. J. Biol. Chem. 2004;279:23052–60. doi: 10.1074/jbc.M401368200. [DOI] [PubMed] [Google Scholar]
- 69.Wruck CJ, Gotz ME, Herdegen T, Varoga D, Brandenburg LO, Pufe T. Kavalac-tones protect neural cells against amyloid beta peptide-induced neurotoxicity via extracellular signal-regulated kinase 1/2-dependent nuclear factor erythroid 2-related factor 2 activation. Mol. Pharmacol. 2008;73:1785–95. doi: 10.1124/mol.107.042499. [DOI] [PubMed] [Google Scholar]
- 70.Joseph A, Lee T, Moland CL, Branham WS, Fuscoe JC, Leakey JE, Allaben WT, Lewis SM, Ali AA, Desai VG. Effect of (+)-usnic acid on mitochondrial functions as measured by mitochondria-specific oligonucleotide microarray in liver of B6C3F1 mice. Mitochondrion. 2009;9:149–58. doi: 10.1016/j.mito.2009.02.002. [DOI] [PubMed] [Google Scholar]
- 71.Desai VG, Lee T, Delongchamp RR, Moland CL, Branham WS, Fuscoe JC, Leakey JE. Development of mitochondria-specific mouse oligonucleotide microarray and validation of data by real-time PCR. Mitochondrion. 2007;7:322–9. doi: 10.1016/j.mito.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 72.Fu PP, Xia Q, Lin G, Chou MW. Pyrrolizidine alkaloids—genotoxicity, metabolism enzymes, metabolic activation, and mechanisms. Drug Metab. Rev. 2004;36:1–55. doi: 10.1081/dmr-120028426. [DOI] [PubMed] [Google Scholar]
- 73.Martignoni M, Groothuis GM, de Kanter R. Species differences between mouse, rat, dog, monkey and human CYP-mediated drug metabolism, inhibition and induction. Expert Opin. Drug Metab. Toxicol. 2006;2:875–94. doi: 10.1517/17425255.2.6.875. [DOI] [PubMed] [Google Scholar]
- 74.Guo L, Mei N, Dial S, Fuscoe J, Chen T. Comparison of gene expression profiles altered by comfrey and riddelliine in rat liver. BMC Bioinformatics. 2007;(8 Suppl 7):S22. doi: 10.1186/1471-2105-8-S7-S22. [DOI] [PMC free article] [PubMed] [Google Scholar]