Abstract
Sertraline is used for the treatment of depression, and is also used for the treatment of panic, obsessive-compulsive, and post-traumatic stress disorders. Previously, we have demonstrated that sertraline caused hepatic cytotoxicity, with mitochondrial dysfunction and apoptosis being underlying mechanisms. In this study, we used microarray and other biochemical and molecular analyses to identify endoplasmic reticulum (ER) stress as a novel molecular mechanism. HepG2 cells were exposed to sertraline and subjected to whole genome gene expression microarray analysis. Pathway analysis revealed that ER stress is among the significantly affected biological changes. We confirmed the increased expression of ER stress makers by real-time PCR and Western blots. The expression of typical ER stress markers such as PERK, IRE1α, and CHOP was significantly increased. To study better ER stress-mediated drug-induced liver toxicity; we established in vitro systems for monitoring ER stress quantitatively and efficiently, using Gaussia luciferase (Gluc) and secreted alkaline phosphatase (SEAP) as ER stress reporters. These in vitro systems were validated using well-known ER stress inducers. In these two reporter assays, sertraline inhibited the secretion of Gluc and SEAP. Moreover, we demonstrated that sertraline-induced apoptosis was coupled to ER stress and that the apoptotic effect was attenuated by 4-phenylbutyrate, a potent ER stress inhibitor. In addition, we showed that the MAP4K4-JNK signaling pathway contributed to the process of sertraline-induced ER stress. In summary, we demonstrated that ER stress is a mechanism of sertraline-induced liver toxicity.
Keywords: Apoptosis, Endoplasmic reticulum stress, Drug-induced liver toxicity, Reporter gene assay, Sertraline, MAPK pathway
1. Introduction
Sertraline, a selective serotonin reuptake inhibitor (SSRI) class antidepressant, is the most widely prescribed psychiatric medication in the United States (Kaplan and Zhang, 2012). Although generally considered safe, acute liver failure has been associated with the use of sertraline (Carvajal Garcia-Pando et al., 2002; Collados et al., 2010; Fartoux-Heymann et al., 2001; Galan Navarro, 2001; Hautekeete et al., 1998; Kim et al., 1999; Persky and Reinus, 2003; Tabak et al., 2009; Verrico et al., 2000).
Previously, we reported that sertraline disrupted liver mitochondrial function in rat hepatocytes (Li et al., 2012) and caused apoptosis in HepG2 cells (Chen et al., 2014b). We also demonstrated that the MAPK signaling pathway, in particular, the TNF-initiated-MAP4K4-JNK cascade was activated and involved in sertraline-induced apoptosis (Chen et al., 2014b).
In this study, using microarray analysis, we identified new molecular mechanisms of sertraline’s toxicity. HepG2 cells were exposed to sertraline at various concentrations for 6 h and then subjected to whole genome gene expression microarray analysis. Pathway analysis revealed that endoplasmic reticulum (ER) stress and the MAPK signaling pathway are among the significantly affected biological changes.
Endoplasmic reticulum, a membranous organelle, performs numerous functions including protein folding, trafficking, and post-modulation, and regulation of the intracellular calcium homeostasis. Disturbances of ER function by stimuli such as altered cellular redox, oxidative stress, unbalanced calcium homeostasis, and energy deprivation lead to ER stress response and activate a specific signaling pathway, namely, unfolded protein response (UPR) as adaptive mechanisms. Excessive and prolonged ER stress causes apoptosis and eventually necrotic cell death. Three major UPR branches, PERK (PKR-like endoplasmic reticulum kinase), IRE1α (inositol-requiring enzyme 1α), and ATF6 (activation of transcription factor 6) have been described to promote cell survival by preventing and/or removing misfolded proteins. These three UPR transducers are activated by phosphorylating PERK and IRE1α or by translocating ATF6 to the Golgi when the ER is stressed. Subsequently, a cascade of reactions follows, activating numerous ER stress molecules that serve as measurable mechanism-based ER stress markers. For example, phosphorylation of elF2α (eukaryotic initiation factor 2) and activation of CHOP (C/EBP CCAAT/enhancer binding protein homologous protein) by increasing the translation of ATF4 (activation of transcription factor 4) lead to attenuation of protein synthesis. A transcriptional factor, XBP (X-box binding protein 1), is activated by IRE1α through splicing of XBP mRNA, resulting in increased expression of genes involved in restoring protein folding or degrading unfolded proteins (Dara et al., 2011; Xu et al., 2005).
Based on the microarray results in this study, further experiments were focused on ER stress using biochemical and molecular approaches. The commonly studied markers (CHOP, p-eIF2a, p-PERK, ATF-4, and PDI) of ER stress were examined by Western blot, the expression of ER stress related genes was measured by real-time PCR, and the splicing of XBP mRNA was determined by PCR amplification. In order to efficiently monitor ER stress-mediated liver toxicity, we established two in vitro reporter assays in HepG2 cells. The establishment and utility of the assays are described and discussed.
2. Materials and methods
2.1. Chemicals and reagents
Sertraline, Williams’ medium E, penicillin, streptomycin, dimethysulfoxide (DMSO), brefeldin A, and thapsigargin were from Sigma–Aldrich (St. Louis, MO). Fetal bovine serum (FBS) was obtained from Atlanta Biologicals (Lawrenceville, GA). 4-Phenylbutyrate (4-PBA) was from BioVision (Milpitas, CA). Blasticidin S HCl and puromycin dihydrochloride were from Life Technologies (Grand Island, NY). Luciferase cell lysis buffer was from New England Biolabs (Ipswich, MA).
2.2. Cell culture
HepG2 cells were grown in Williams’ medium E as described previously (Chen et al., 2013). Unless otherwise specified, HepG2 cells were seeded at a concentration of 2–5 × 105 cells/ml in volumes of 100 µl in the wells of 96-well tissue culture plates, in volumes of 5 ml in 60 mm tissue culture plates, or in volumes of 10 ml in 100 mm tissue culture plates. Cells were cultured for approximately 24 h prior to treatment with the indicated concentrations of sertraline or the vehicle control, DMSO (final concentration 0.1%).
The 293T cell line used for lentivirus packaging was purchased from Biosettia (San Diego, CA) and maintained in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% FBS in the presence of 1 mM sodium pyruvate and non-essential amino acids.
2.3. Vector construction and stable cell line establishment
The gene expression lentiviral vector pLv-EF1α–MCS-IRES-Puro and the lentiviral labeling plasmid plv-EF1α-Firefly luciferase (Fluc)-IRES-Bsd were purchased from Biosettia (San Diego, CA). CMV-SEAP plasmid carrying secreted embryonic alkaline phosphatase (SEAP) (as template to amplify SEAP) was purchased from Addgene (Cambridge, MA) and pSV40-Gluc plasmid carrying secreted Gaussia luciferase (Gluc) (as template to amplify Gluc) was purchased from New England Biolabs. The cDNAs of SEAP and Gluc were amplified by PCR and subcloned into BamHI and NheI restriction sites of pLv-EF1α-MCS-IRES-Puro expression vector following the manufacturer’s instructions. The generated lentiviral vectors and viral packaging plasmids (pMDL-G, pRSV-REV, and pVSV-G) were co-transfected into 293T cells to produce lentiviral stocks. The titrations of lentivirus stocks were measured with a functional lentivirus titering kit from Biosettia. HepG2 cells were infected with lentivirus carrying Fluc at a multiplicity of infection (MOI) of 10. Infected cells were selected with 6 µg/ml blasticidin (Bsd) to generate a polyclone of HepG2 cells with stable expression of firefly luciferase (Fluc). Subsequently, the cells with Fluc gene integrated were infected with lentivirus carrying either SEAP or Gluc at a MOI of 10. Infected cells were purified with 2 µg/ml puromycin. Thus, two stable cell lines (HepG2-Fluc-Gluc and HepG2-Fluc-SEAP) were generated.
2.4. Luciferase activity
Gluc activity was measured by adding 50 µl Gaussia luciferase assay reagent (Nanolight Technology, Pinetop, AZ) to 5 µl of the conditioned cell-free medium and immediately determined the bioluminescence using a Synergy 2 Multi-Mode Microplate Reader (BioTek, Winooski, VT). For the measurement of Fluc activity, the enzyme was first extracted from treated cells by adding 20 µl luciferase cell lysis buffer (New England Biolabs) to each well and then incubated at room temperature for 15 min on an orbital shaker. Then, 50 µl of the firefly luciferase assay reagent (Nanolight Technology) was added directly to the cell lysate in a 96 well plate, mixed well, and read immediately in the Synergy 2 Microplate Reader (BioTek).
2.5. Secreted alkaline phosphatase (SEAP) assay
The SEAP detection assay was performed by using the phospha-Light-EXPSEAP reporter gene assay system (Life Technologies) according to the manufacturer’s instructions. In brief, an aliquot of 5 µl cell free conditioned medium was mixed with 50 µl of assay buffer in a white 96-well assay plate and incubated at 65 °C for 5 min. Then, 50 µl of reaction buffer was added and the mixture was incubated at room temperature for 20 min. The chemiluminescence was determined by the Synergy 2 Microplate Reader (BioTek).
2.6. Caspase-3/7 activity measurement
The enzymatic activity of caspase-3/7 was measured using a luminescent assay kit (Caspase-Glo® 3/7 Assay Systems, Promega) using a Synergy 2 Microplate Reader (BioTek). The induction of activity was calculated by comparing the luminescent of the treated cells to that of the DMSO controls.
2.7. RNA isolation
HepG2 cells at a density of 2 × 105 ml−1 were seeded on 60 mm plates as described under Cell culture and treated with various concentrations of sertraline. Total RNA was isolated using the mini RNeasy system (Qiagen, Germantown, MD). The yield of the extracted RNA was determined spectrophotometrically by measuring absorption at 260 nm using a NanoDrop 8000 (Thermo Scientific, Wilmington, DE). The purity and quality of RNA were evaluated using a RNA 6000 LabChip and Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA). High quality RNA with RNA integrity numbers (RINs) greater than 9.0 was used for microarray experiments and PCR assays.
2.8. Quantitative real-time PCR (qPCR) assay
qPCR was used to determine the mRNA expression of the genes involved in ER stress (PERK, IRE1α, HERPUD1, GADD34, and CHOP) with the expression of GAPDH as the endogenous control. Primers used for qPCR are as shown in Table 1.
Table 1.
Primers used for qPCR.
| Gene | Accession number | Forward primer | Reverse primer |
|---|---|---|---|
| PERK | NM_004836 | 5′-ATGAGACAGAGTTGCGACCG-3′ | 5′-TGGATGACACCAAGGAACCG-3′ |
| IRE1α | NM_152461 | 5′-CTCAGAGACAGCGCGAGTAG-3′ | 5′-ATCTCAGCCTAGCTGTCCCA-3′ |
| HERPUD1 | NM_001010990 | 5′-CATGGAGTCCGAGACCGAAC-3′ | 5′-TGCCGTTTTTCCTGCTTTGG-3′ |
| GADD34 | NM_004083 | 5′-TCCTCTGGCAATCCCCCATA-3′ | 5′-TGGTTTTCAGCCCCAGTGTT-3′ |
| CHOP | NM_014330 | 5′-TTGCCTTTCTCCTTCGGGAC-3′ | 5′-CAGTCAGCCAAGCCAGAGAA-3′ |
| GAPDH | NM_002046 | 5′-AGAAGGCTGGGGCTCATTTG-3′ | 5′-AGGGGCCATCCACAGTCTTC-3′ |
qPCR reactions were carried out using SsoAdvanced™ SYBR Green Supermix in a Bio-Rad CFX96™ Real-Time PCR Detection System under universal cycling conditions (10 min at 95°C; 15 s at 95°C, 1 min 60°C, 40 cycles) according to the manufacturer’s instructions (Bio-Rad Laboratories, Hercules, CA). Data normalization and analysis were conducted as described previously (Guo et al., 2009, 2010).
2.9. X-box binding protein1 (XBP1) mRNA splicing assay
The cDNAs were prepared from total RNAs (1 µg) using high capacity cDNA reverse transcription kits (Life Technologies) according to the manufacturer’s protocol. To amplify XBP1 mRNA, the following primers were used: Forward 5’-TTACGAGAGAAAACTCATGGCC-3’ and Reverse 5’-GGGTCCAAGTTGTCCAGAATGC-3’. cDNA obtained with GAPDH primers (Forward 5’AGAAGGCTGGGGCTCATTTG’ and Reverse 5’-AGGGGCCATCCACAGTCTTC-3’) was used as gel loading control. PCR was conducted for 35 cycles (95°C for 30 s; 55°C for 1 min; and 72°C for 1 min) on GeneAmp® PCR system 9700 (Life Technologies). The spliced (263 bp), unspliced (289 bp) XBP1 mRNA, and GAPDH mRNA were analyzed by electrophoresis on 4% (w/v) agarose gels, stained with 0.5 µg/ml ethidium bromide solution and visualized under UV light (ProteinSimple, Santa Clara, CA).
2.10. Microarray analysis
Microarray gene expression analysis was performed at the Microarray Core Facility of University of Texas Southwestern Medical Center (http://microarray.swmed.edu). Illumina Human HT-12 expression BeadChip arrays were used. Microarray data were extracted using BeadStudio v3.1 software, background-subtracted, and normalized using a cubic spline algorithm. Genes differentially expressed between groups were identified using the Illumina custom error model implemented in BeadStudio. A gene was considered significantly differentially expressed when a P value was less than 0.05 and the change was greater than 1.5-fold. Clustering analysis was conducted within ArrayTrack, a software system developed by the FDA’s National Center for Toxicological Research for the management, analysis, visualization and interpretation of microarray data (http://www.fda.gov/nctr/science/centers/toxicoinformatics/ArrayTrack/).
2.11. Western blot analysis
Cells were grown and treated with sertraline in 100 mm tissue culture plates. Standard Western blots were performed using antibodies against PERK, phospho-PERK (Thr 980), phospho-eIF2α (Ser 51), eIF2α, CHOP, PDI, and pro-caspase 4 (Cell Signaling Technology, Danvers, MA), ATF-4, and GAPDH (as internal control, Santa Cruz Biotechnology, Santa Cruz, CA) followed by a secondary antibody conjugated with horseradish peroxidase (HRP) (Santa Cruz Biotechnology). The bands were detected by FluorChem E and M Imagers (ProteinSimple).
2.12. Statistical analysis
Data are presented as mean ± standard deviation (SD) of at least three independent experiments. Analyses were performed using GraphPad Prism 5 (GraphPad Software, San Diego, CA). Statistical significance was determined by one way analysis of variance (ANOVA) followed by the Dunnett’s tests for pairwise-comparisons or two way ANOVA followed by the Bonferroni post-test. The difference was considered statistically significant when P < 0.05.
3. Results
3.1. Analysis of differentially expressed genes
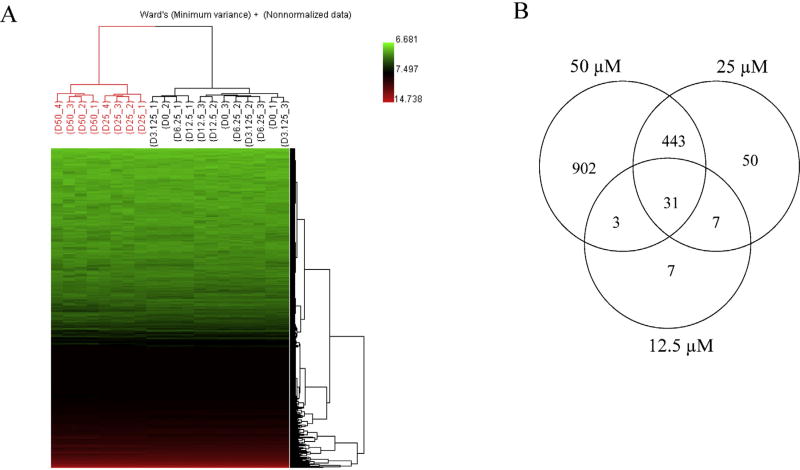
To identify molecular mechanisms of sertraline-induced liver toxicity, HepG2 cells were treated with sertraline at 3.13–50 µM for 6 h; RNA was isolated and gene expression profiles were determined by microarrays. Treatment with sertraline for 6 h at the concentrations of 3.13–50 µM resulted in an IC50 of 25.7 µM as measured by LDH release (Chen et al., 2014b). There were one DMSO control group and five sertraline treatment groups (3.13, 6.25, 12.5, 25, and 50 µM), each group containing 3–4 biological replicates. To explore the treatment effect and to determine the relationship of the samples based on expression profiles, unsupervised Hierarchical Cluster Analysis (HCA) was performed for the entire gene spot set. Results and HCA result displayed in Fig. 1A with the heat map of gene expression depicting the distance between samples. HCA showed that two large clusters (red and black) were formed based on the of treatment concentrations, i.e., higher-dose groups (25 and 50 µM) clustered together (red); control and lower-dose groups (0, 3.13, 6.25, 12.5 µM) clustered together (black), indicating that there are treatment effects at the gene expression level in response to the sertraline exposure.
Fig. 1.
(A) Hierarchical Cluster Analysis (HCA) of gene expression profiles in control and sertraline-treated groups. The values of the log 2 intensities of genes were hierarchically clustered using Ward’s distance metric. The intensity of the entire gene set was used; no specific cut off was applied for the analysis. Each column represents the results from an individual hybridization. Each row represents the log 2 intensity value for one particular gene. Samples are labeled according to the convention of Dose (µM)_Technical replicate number. (B) Numbers of differentially expressed genes in HepG2 cells in response to treatment with 12.5, 25, and 50 µM sertraline. A gene was identified as differentially expressed if the fold-change was greater than 1.5 (up- or down-regulated) and the P-value was less than 0.05 in comparison to the control group. (For interpretation of the references to color in the text, the reader is referred to the web version of the article.)
A differentially expressed gene (DEG) was identified when the criteria of a fold change greater than 1.5 (either up- or down-regulated genes) and a P-value less than 0.05 was used in comparison to the DMSO control group. Based on the defined criteria, 20, 28, 48, 531, and 1379 DEGs were identified in 3.13, 6.25, 12.5, 25, and 50 µM sertraline treatments, respectively. The number of DEGs increased with the increasing concentration, demonstrating there is a dose-response relationship for the number of genes affected. In addition, the commonly altered genes in 12.5, 25, and 50 µM treatments were identified. As shown in the Venn diagrams (Fig. 1B), the majority of DEGs in the groups receiving the two lower concentrations are overlapped with the higher concentration groups. Out of 48 genes identified in 12.5 µM sertraline treatment group, 38 (79.2%) and 34 (70.8%) were found to be overlapped in 25 and 50 µM sertraline treatment groups, and out of 531 genes identified in 25 µM sertraline treatment group, 474 (89.3%) were found in 50 µM sertraline treatment group.
3.2. Pathway analysis of the differentially expressed genes
A total of 1379 DEGs identified from 50 µM sertraline treatment was mapped to the Kyoto Encyclopedia of Genes and Genomes (KEGG) database for function classification and pathways analysis. Table 2 lists the significantly altered pathways. Not surprisingly, “Cell cycle” was among the affected pathway, with 23 genes involved and with a Fisher P value of 3.75E-05. We have previously demonstrated that sertraline caused apoptosis and necrosis and altered cell cycle (Chenet al., 2014b; Li et al., 2012). The results of pathway analysis are in good agreement with the data from our previous studies. Interestingly, KEGG pathway analysis identified “protein processing in endoplasmic reticulum” and “MAPK signaling pathway” as affected pathways, with Fisher P values of 0.002 and 0.017, respectively. A total of 23 and 30 genes were characterized as potentially involved in “ER stress” and “MAPK signaling pathway” in 50 µM sertraline treatment (Tables 3 and 4). Changes in these genes started to be seen at 25 µM sertraline treatment and the changes are dose-dependent (Supplementary Table 1). Notably, CHOP, the hallmark of ER stress, displayed a large change with a 16.4-fold increase (Table 3). The changes and the trend of altered genes in ER stress and MAPK signaling pathway for 25 and 50 µM sertraline are detailed in Supplemental Tables 1 and 2 and Supplemental Fig. 2. These results of pathway analysis led us to hypothesize that besides the MAPK signaling pathway, ER stress, is involved in sertraline-induced liver toxicity.
Table 2.
Pathways significantly altered by sertraline treatment. Differentially expressed genes identified from 50 µM sertraline treatment were mapped to the Kyoto Encyclopedia of Genes and Genomes (KEGG) database.
| No. | KEGG pathway | Fisher P value | # of differentially expressed genes |
# of genes in the pathway |
|---|---|---|---|---|
| 1 | Cell cycle | 3.75E–05 | 23 | 124 |
| 2 | Pathogenic Escherichia coli infection | 1.65E–04 | 13 | 55 |
| 3 | TGF-beta signaling pathway | 3.30E–04 | 16 | 80 |
| 4 | p53 signaling pathway | 3.46E–04 | 14 | 68 |
| 5 | Mineral absorption | 8.62E–04 | 11 | 51 |
| 6 | Protein processing in endoplasmic reticulum | 2.00E–03 | 23 | 167 |
| 7 | Ubiquitin mediated proteolysis | 2.29E–03 | 20 | 137 |
| 8 | Oocyte meiosis | 3.27E–03 | 17 | 110 |
| 9 | Osteoclast differentiation | 4.77E–03 | 18 | 132 |
| 10 | Spliceosome | 4.77E–03 | 18 | 131 |
| 11 | Circadian rhythm - mammal | 4.91E–03 | 6 | 30 |
| 12 | Pertussis | 6.46E–03 | 12 | 75 |
| 13 | Toxoplasmosis | 7.14E–03 | 18 | 122 |
| 14 | MAPK signaling pathway | 1.07E–02 | 30 | 259 |
| 15 | Folate biosynthesis | 1.15E–02 | 4 | 14 |
| 16 | Leishmaniasis | 1.53E–02 | 11 | 76 |
| 17 | Measles | 1.63E–02 | 17 | 134 |
| 18 | NOD-like receptor signaling pathway | 2.50E–02 | 9 | 57 |
| 19 | Glutathione metabolism | 2.60E–02 | 8 | 51 |
| 20 | Maturity onset diabetes of the young | 3.11E–02 | 5 | 25 |
| 21 | Pathways in cancer | 3.13E–02 | 33 | 249 |
| 22 | Hepatitis C | 3.23E–02 | 16 | 133 |
| 23 | Shigellosis | 3.34E–02 | 9 | 62 |
| 24 | Small cell lung cancer | 4.22E–02 | 11 | 86 |
| 25 | Arginine and proline metabolism | 4.31E–02 | 8 | 57 |
Table 3.
Genes involved in ER stress. Twenty-three genes were characterized as involved in ER stress in 50 µM sertraline treatment.
| Number of gene | Gene bank ID | Gene name | Fold change | P value |
|---|---|---|---|---|
| 1 | NM_000394 | CRYAA | 2.87 | 0.0005 |
| 2 | NM_003592 | CUL1 | 0.63 | 0.0001 |
| 3 | NM_004083 | CHOP | 16.43 | 0.0001 |
| 4 | NM_004836 | PERK | 1.51 | 0.0016 |
| 5 | NM_152461 | IRE1α | 2.18 | 0.0001 |
| 6 | NM_001010990 | HERPUD1 | 6.68 | 0.0005 |
| 7 | NM_005345 | HSPA1A | 2.78 | 0.004 |
| 8 | NM_005346 | HSPA1B | 1.88 | 0.0001 |
| 9 | NM_002155 | HSPA6 | 3.12 | 0.0001 |
| 10 | NM_006597 | HSPA8 | 0.65 | 0.001 |
| 11 | NM_006164 | NFE2L2 | 2.03 | 0.0002 |
| 12 | NM_014330 | GADD34 | 10.31 | 0.0001 |
| 13 | NM_014248 | RBX1 | 0.54 | 0.0154 |
| 14 | NM_001012456 | SEC61G | 0.67 | 0.0243 |
| 15 | NM_018445 | SELS | 1.74 | 0.0001 |
| 16 | NM_006930 | SKP1A | 0.57 | 0.0014 |
| 17 | XM_945430 | SSR2 | 2.18 | 0.0001 |
| 18 | NM_032431 | SYVN1 | 1.55 | 0.0172 |
| 19 | NM_022085 | TXNDC5 | 0.62 | 0.046 |
| 20 | NM_015983 | UBE2D4 | 0.55 | 0.0001 |
| 21 | NM_003342 | UBE2G1 | 0.66 | 0.0015 |
| 22 | NM_006048 | UBE4B | 0.52 | 0.0001 |
| 23 | NM_053067 | UBQLN1 | 1.52 | 0.0003 |
Table 4.
Genes involved in MAPK signaling pathways. A total of 30 genes were characterized as potentially involved in MAPK signaling pathway in 50 µM sertraline treatment.
| Number of genes | Gene bank ID | Gene name | Fold change | P value |
|---|---|---|---|---|
| 1 | NM_001880 | ATF2 | 1.62 | 0.0012 |
| 2 | NM_001040021 | CD14 | 0.55 | 0.0029 |
| 3 | NM_004083 | CHOP | 16.43 | 0.0001 |
| 4 | NM_144729 | DUSP10 | 2.49 | 0.0012 |
| 5 | NM_007026 | DUSP14 | 1.51 | 0.0011 |
| 6 | NM_030640 | DUSP16 | 2.28 | 0.0001 |
| 7 | NM_004417 | DUSP1 | 14.37 | 0.0001 |
| 8 | NM_004420 | DUSP8 | 2.58 | 0.0002 |
| 9 | NM_005228 | EGFR | 1.60 | 0.0002 |
| 10 | NM_005252 | FOS | 9.25 | 0.0002 |
| 11 | NM_001924 | GADD45A | 4.13 | 0.0001 |
| 12 | NM_015675 | GADD45B | 6.67 | 0.0001 |
| 13 | NM_005345 | HSPA1A | 2.78 | 0.004 |
| 14 | NM_005346 | HSPA1B | 1.88 | 0.0001 |
| 15 | NM_002155 | HSPA6 | 3.12 | 0.0001 |
| 16 | NM_006597 | HSPA8 | 0.65 | 0.001 |
| 17 | NM_002228 | JUN | 27.54 | 0.0005 |
| 18 | NM_005354 | JUND | 2.10 | 0.003 |
| 19 | NM_004721 | MAP3K13 | 1.59 | 0.0335 |
| 20 | NM_005204 | MAP3K8 | 1.67 | 0.0096 |
| 21 | NM_145687 | MAP4K4 | 0.65 | 0.0068 |
| 22 | NM_001040439 | MAPK8IP3 | 1.71 | 0.0263 |
| 23 | NM_002467 | MYC | 3.31 | 0.0015 |
| 24 | NM_003998 | NFKB1 | 1.68 | 0.0001 |
| 25 | NM_001077493 | NFKB2 | 2.73 | 0.0002 |
| 26 | NM_006509 | RELB | 2.60 | 0.0001 |
| 27 | NM_004586 | RPS6KA3 | 0.65 | 0.0063 |
| 28 | NM_203401 | STMN1 | 0.57 | 0.0008 |
| 29 | NM_016151 | TAOK2 | 1.52 | 0.0001 |
| 30 | NM_000594 | TNF | 1.68 | 0.0001 |
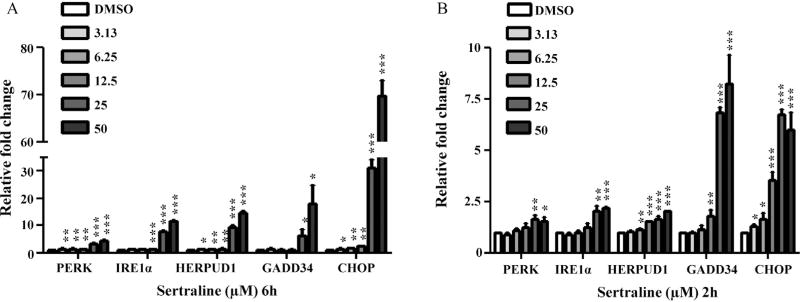
Previously, we examined the role of MAPK signaling pathway and demonstrated that MAPK pathway, particularly the TNF-MAP4K4-JNK cascade signaling pathway, was activated in response to sertraline. The activation of the TNF-MAP4K4-JNK cascade signaling pathway leads to apoptosis and cell death (Chen et al., 2014b). In the current study, we focused on the role of ER stress in sertraline-induced liver toxicity. The changes of ER stress related genes (PERK, IRE1α, HERPUD1, GADD34, and CHOP) were first validated by quantitative real-time PCR (qRT-PCR). As shown in Fig. 2A, upregulation was observed at almost all concentrations at 6 h, especially the two highest concentration groups (25 and 50 µM). It should be noted that the qPCR data correlated well with the microarray results, although real-time PCR is more sensitive than microarray. For example, for the 25 and 50 µM sertraline treatments, CHOP was 8.3-, and 16.4-fold increased as measured with microarray (Table 3 and Supplementary Table 1), but 31.2-, and 69.8-fold increased as measured with qPCR (Fig. 2). Changes of ER stress markers can be detected at lower concentrations by qPCR: CHOP was induced by the lowest concentration of 3.13 µM, a concentration that did not induce cytotoxicity (Fig. 2A) (Chen et al., 2014b). In addition, we also examined the expression of these ER stress related genes at an earlier time point of 2 h by qPCR; the expression of these genes was also found to be increased (Fig. 2B). Taken together, the up-regulation of ER stress related genes at lower concentrations and earlier time point before significant cytotoxicity (Chen et al., 2014b) had occurred indicates that the change in the expression of these genes is more likely represent an initial response to sertraline treatment rather than nonspecific response to cell injury.
Fig. 2.
qPCR confirmation of microarray data. Genes involved in ER stress response were determined by qPCR. HepG2 cells were treated for 6 (A) and 2 h (B) with increasing concentrations (3.13, 6.25, 12.5, 25, and 50 µM) of sertraline, with DMSO as the vehicle control. Values were means ± SD of three separate experiments. *P <0.05, **P <0.01, and ***P <0.001 as compared with DMSO controls.
3.3. Sertraline triggered ER stress
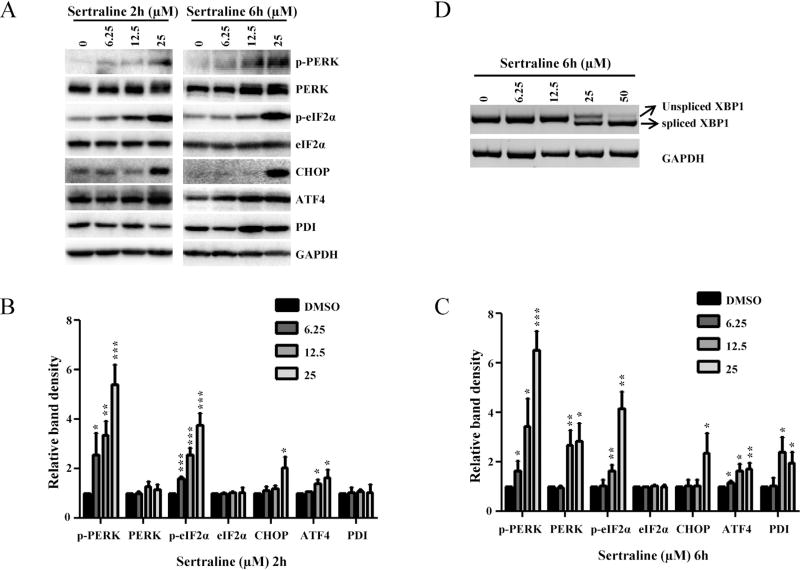
The results of gene expression data indicate that ER stress is one of the mechanisms of sertraline’s toxicity. To confirm further that the treatment with sertraline triggered ER stress response, expression levels of molecules involved in the ER stress were analyzed at the protein level by Western blot analysis. Treatment of HepG2 cells with 12.5 µM sertraline for 2 h, a concentration and time point without a reduction of cell viability as measured by ATP content and LDH release (Chen et al., 2014b), showed increased expression of the main ER stress molecules including PERK, phospho-PERK, and phospho-eIF2α (Fig. 3A). A higher concentration (25 µM) of sertraline treatment for 2 h markedly increased the levels of PERK, phospho-PERK, and phospho-eIF2α, and also elicited enhanced expression of CHOP and ATF-4. A longer treatment, for 6 h, increased the levels of PERK, phospho-PERK, phospho-eIF2α, ATF4, and PDI, and the induction was in a concentration-dependent manner. Another ER stress indicator, the splicing of XBP mRNA, was tested by amplifying spliced (263 bp) XBP as described under Section 2. The XBP splicing observed at 6 h starting from 25 µM and further increased at 50 µM (Fig. 3D).
Fig. 3.
Effects of sertraline in HepG2 cells on the expression of proteins involved in the ER stress response and on splicing of XBP mRNA. (A) Total cellular proteins were extracted at 2 and 6 h after sertraline treatment. The level of ER stress related proteins including phospho-PERK, PERK, phospho-eIF2α, eIF2α, CHOP, ATF-4, and PDI was determined by Western blotting. GAPDH was used as a loading control. Data are typical of three experiments. (B and C) Relative protein level to DMSO control by densitometric analysis. (D) HepG2 cells were treated with the indicated concentration of sertraline for 6 h. The total RNA was isolated and semi-quantitative RT-PCR analysis was performed to detect both spliced (263 bp) and unspliced (289 bp) XBP mRNA. GAPDH was also amplified and used as an internal control. A representative DNA gel from three independent experiments is shown.
3.4. Establishment of Gaussia luciferase or secreted alkaline phosphatase reporter assay for monitoring ER stress in HepG2 cells
The protein secretion is a process to transport and deposit proteins into extracellular space via endoplasmic reticulum and Golgi apparatus. Blocking or decreasing protein secretion is a distinct characteristic of the disruption of ER function; thus, a reduction of protein secretion indicates an ER stress response. Secreted reporters have been used in vitro for monitoring different biological processes in the medium of cultured cells (Badr et al., 2007; Tannous, 2009). The most commonly used reporter is secreted alkaline phosphatase (SEAP) (Berger et al., 1988; Cullen and Malim, 1992; Meng et al., 2005). Recently, the secreted Gaussia luciferase (Gluc) assay has gained popularity due to its high sensitivity (Badr et al., 2007; Tannous, 2009; Wurdinger et al., 2008). Both reporter assays can be used for monitoring ER stress by measuring a change (a decrease) in Gluc or SEAP being excreted into the medium because ER stress blocks or decreases processing in the secretory pathway and trafficking through the secreted Gluc or SEAP activity (Badr et al., 2007). We sought to establish reporter assays in HepG2 cells to monitor ER stress response efficiently and to better study liver toxicity. Two individual assays (Gluc and SEAP) were chosen to quantitatively measure ER stress response. Lentivirus vector carrying the expression cassettes for Gluc or SEAP was used to generate stable cell line; firefly luciferase (Fluc) was used as internal control for normalizing the cell number. HepG2 cells were co-infected with the lentivirus vector carrying Fluc and either Gluc or SEAP. Two stable cell lines, HepG2-Fluc-Gluc and HepG2-Fluc-SEAP were established.
The correlation of cell number and Fluc activity was first assessed. Different numbers of stably infected cells were plated in 96 well plates; 24 h after plating, the luminescent activities of Fluc in the cell lysates were measured. As shown in Supplementary Fig. 1, when cell numbers were plotted against the activity of Fluc, the calculated correlation coefficiency (R2) was 0.993, suggesting a good linearity of Fluc activity with respect to cell number.
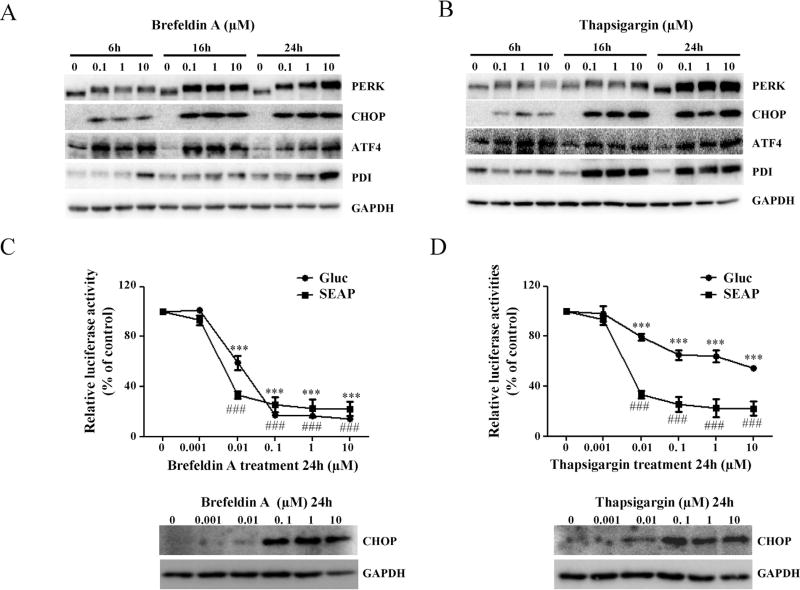
To test the specificity and sensitivity of established Gluc and SEAP systems, two well-known ER stress inducers, brefeldin A (Fishman and Curran, 1992) and thapsigargin (Wong et al., 1993), were used to treat cells. The ER stress was measured by inhibition of Gluc or SEAP secretion and also by Western blot analysis of ER stress marker proteins in parallel. As shown in Fig. 4A and B, treatment with 0.1–10 µM brefeldin A or thapsigargin for 6, 16, and 24 h notably increased the expression levels of PERK, CHOP, ATF4, and PDI in time- and dose-dependent manner, indicating ER stress occurred in response to the brefeldin A or thapsigargin treatment. Accordingly, 0.001–10 µM brefeldin A or thapsigargin treatment for 24 h was used to detect the changes of Gluc or SEAP secretion level. As expected, treatment of these ER stress inducers resulted in significant inhibition of Gluc and SEAP secretion. As shown in Fig. 4C and D, treatment with 10 µM brefeldin A or thapsigargin decreased about 86% or 46% Gluc secretion and decreased about 82% or 78% SEAP secretion, respectively. These data agree well with the results of increased the expression of CHOP, a hallmark of ER stress, indicating that the established Gluc-and SEAP-expressing cell lines can be used for monitoring ER stress.
Fig. 4.
Effects of brefeldin A and thapsigargin on the secretion of Gluc and SEAP proteins in HepG2 cells. As ER stress inducers, brefeldin A and thapsigargin were tested for their inhibitory effects on the molecules involved in ER stress in HepG2 cells and Gluc and SEAP secretion in the Gluc and SEAP reporter cells. (A and B) HepG2 cells were treated with various concentrations of brefeldin A and thapsigargin for 6, 16, and 24 h. The main ER stress response related proteins (PERK, CHOP, ATF4, and PDI) were determined by Western blotting. (C and D) After treatment with increasing concentrations of brefeldin A and thapsigargin for 24 h, the activities of Gluc and SEAP were determined as described in Section 2; the results shown are mean ± SD of four separate experiments. ###P < 0.001 represents relative SEAP activity was significantly different from the control; ***P <0.001 represents relative Gluc activity was significantly different form the control. Relative Gluc or SEAP activity was calculated based on % of each (Gluc or SEAP activity in medium per Fluc activity) over its vehicle control.
3.5. Sertraline induced ER stress as measured by impaired secretion of Gluc and SEAP
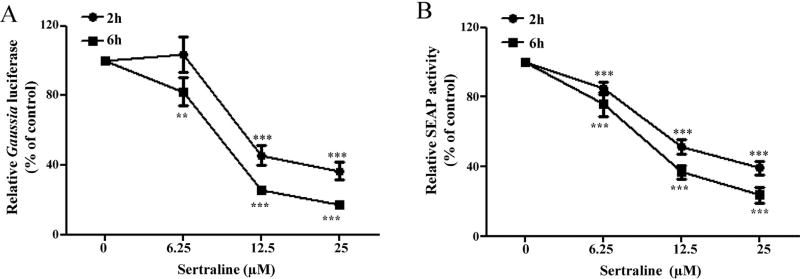
The newly established ER stress monitoring systems were used for examining and quantifying sertraline triggered ER stress. As shown in Fig. 5A, in the cells expressing Gluc (HepG2-Fluc-Gluc), sertraline inhibited the secretion of Gluc in a concentration- and time-dependent manner. Sertraline at 25 µM inhibited Gluc secretion significantly at 2 and 6 h, decreasing Gluc secretion about 63% and 83%, respectively. Similar results were obtained in the cells expressing SEAP (HepG2-Fluc-SEAP), with about 61% and 76% inhibition of SEAP secretion when treated with 25 µM sertraline for 2 and 6 h (Fig. 5B).
Fig. 5.
Effects of sertraline on the secretion of Gluc and SEAP proteins in HepG2 cells. (A) (HepG2-Gluc-Fluc) or (B) (HepG2-SEAP-Fluc) cells were treated with various concentrations of sertraline for 2 and 6 h. The activities of Gluc (A) or SEAP (B) was measured *P <0.05, **P <0.01, and ***P < 0.001 as compared with DMSO controls.
3.6. ER stress response is associated with sertraline-induced apoptosis
In our previous study, we clearly demonstrated that sertraline induced apoptosis in HepG2 cells and that apoptosis is one of the mechanisms of sertraline-associated liver toxicity (Chen et al., 2014b). The role of ER stress in apoptosis has been recognized (Hitomi et al., 2004; Uzi et al., 2013; Zinszner et al., 1998); thus, it was of great interest to examine whether or not sertraline-induced apoptosis is coupled to ER stress.
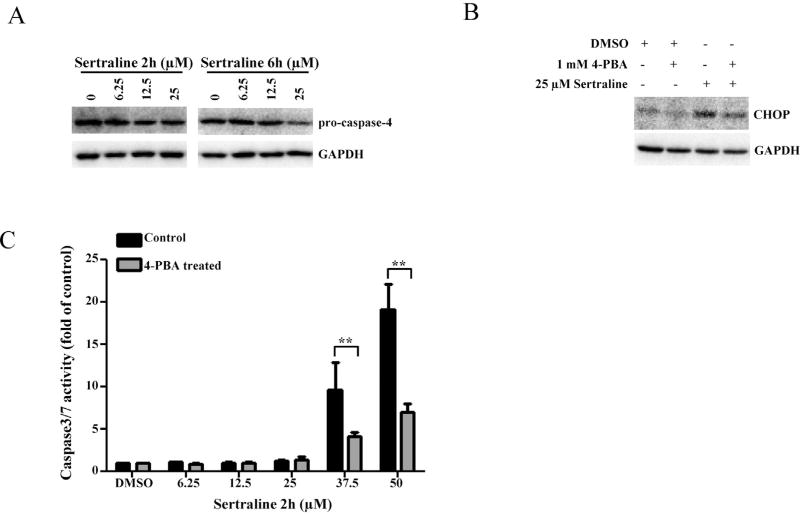
Caspase-4 is known to be specifically involved in ER stress, particularly in the activation of ER stress-induced apoptosis (Hitomi et al., 2004). We first tested if caspase-4 was activated by detecting the decreasing level of pro-caspase-4 as an indicator of activation (cleavage), due to the limited capacity of available antibodies to detect cleaved caspase-4. As demonstrated in Fig. 6A, pro-caspase 4 was decreased (activation of caspase-4) in a time- and dose-dependent manner, indicating that ER stress may be involved in sertraline-induced apoptosis.
Fig. 6.
Effects of ER stress response in the induction of sertraline induced apoptosis. (A) Total cellular proteins were extracted at 2 and 6 h after sertraline treatment. The levels of ER stress related caspase protein pro-caspase 4 was determined by Western blotting. (B) HepG2 cells were pretreated with 1 mM 4-PBA for 2 h prior to 25 µM sertraline treatment for additional 2 h. The expression of CHOP was assessed by Western blotting. GAPDH was used as a loading control. (C) After treatment with 1 mM 4-PBA for 2 h, HepG2 cells were treated with various concentrations of sertraline for additional 2 h. Apoptosis was determined by caspase 3/7 activity. The bar graph shows the mean ± SD of four separate experiments. **P <0.005 versus treatment of sertraline alone.
Accumulation of unfolded proteins in the ER can trigger ER stress. To test whether or not this is a cause for sertraline-induced ER stress, an ER stress modulator, 4-phenylbutyrate (4-PBA) that possesses in vitro chaperone activity to alleviate the unfolded proteins (Yam et al., 2007) was used. As shown in Fig. 6B, pre-treatment with 1 mM 4-PBA for 2 h remarkably suppressed 25 µM sertraline-elicited expression of CHOP. In addition, the effect of 4-PBA on the apoptosis was examined using caspase 3/7 activation as the apoptotic endpoint. Fig. 6C showed that pre-treatment with 4-PBA significantly inhibited caspase3/7 activation induced by 37.5 and 50 µM sertraline with a 2 h treatment, compared to the treatment with sertraline alone. These results suggest that sertraline induced ER stress resulted from the accumulation of unfolded proteins and also ER stress contributed to the process of apoptosis.
3.7. MAP4K4-JNK pathway regulated sertraline-induced ER stress
We have previously demonstrated that the MAPK signaling pathway regulated sertraline-caused apoptosis (Chen et al., 2014b). Although all three major MAPK signaling pathways (JNK, ERK1/2, and p38) were turned on, using inhibitors of the key molecules in MAPK pathway, we demonstrated that JNK activation was associated with the induction of apoptosis. Silencing MAP4K4, the upstream kinase of JNK, attenuated apoptosis, indicating that sertraline-induced apoptosis via the activation of the TNF-MAP4K4-JNK cascade signaling pathway (Chen et al., 2014b). Using microarray pathway analysis, in this study, with expectation, we found that MAPK signaling pathway altered (Supplemental Fig. 2 and Table 2). It is reasonable to suspect that a cascade of reaction and a cross talk among different signaling pathways can be involved in sertraline’s toxicity. Because we demonstrated in this study that ER stress is involved in sertraline-induced liver toxicity (Figs. 2 and 3) and also showed that ER stress is involved in sertraline-induced apoptosis when an ER stress inhibitor 4-PBA was used (Fig. 6C). To explore whether or not MAP4K4-JNK pathway contributes to sertraline-induced ER stress, a previously generated shRNA-MAP4K4 stable cell line to silence MAP4K4 and a JNK specific inhibitor (SP600125) as described previously were used (Chen et al., 2014b). As shown in Fig. 7A, the increased expression of phospho-PERK, CHOP, and PDI, and the activation of caspase-4 (as prevented the cleavage of pro-caspase-4) induced by 25 µM sertraline was markedly prevented in shRNA-MAP4K4 cells. Furthermore, pre-treatment with 10 µM SP600125 significantly attenuated the reduction of secreted Gluc activities caused by 6 h sertraline treatment, compared to the treatment with sertraline alone (Fig. 7B). These data indicate that the MAP4K4-JNK signaling pathway participated in the regulation of sertraline-induced ER stress and ER stress-induced apoptosis.
Fig. 7.
Effect of silencing MAP4K4 on sertraline-induced ER stress. (A) Two HepG2 cell lines with silenced gene expression of MAP4K4 (sh1-MAP4K4 and sh2-MAP4K4) or a scramble control (SC) were treated with sertraline at 25 µM for 6 h. Total cellular proteins were extracted. The expression levels of MAP4K4 and ER stress related proteins (PERK, p-PERK, CHOP, PDI, and pro-caspase-4) were detected by Western blotting. GAPDH was used as a loading control B) After treatment with 10 µM SP600125 (JNK inhibitor) for 2 h (HepG2-Fluc-Gluc) cells were treated with indicated concentrations of sertraline for 6 h. The relative Gluc activity was calculated; the results shown are mean ± SD of four separate experiments. *P <0.05, **P <0.01 versus treatment with sertraline alone.
4. Discussion
Although sertraline is generally considered safe, liver toxicity related to its use has been reported and the adverse drug reaction is idiosyncratic. Accordingly, we have studied sertraline’s toxicity and the mechanisms of that toxicity. Our previous studies identified mitochondrial dysfunction and apoptosis as underlying mechanisms and that those processes are mediated by MAPK signaling pathway (Chen et al., 2014b; Li et al., 2012). In this study, we showed that ER stress is a novel mechanism of sertraline’s liver toxicity using HepG2 cells. As compared human primary hepatocytes, which are more relevant for studying of pharmaceuticals, HepG2 cells have the disadvantage of lacking some drug metabolizing genes when studying metabolism related toxicity (Guo et al., 2011; Ning et al., 2008; Ramaiahgari et al., 2014); however, the ability to modify genes of interest (silencing or overexpressing) in HepG2 cells is a distinct advantage for the in-depth mechanistic studies at the molecular level (Chen et al., 2014b) and for toxicity screening (in our case, to generate two stable cell lines monitoring ER stress response). Metabolism related toxicity was not a major concern in this study, because in our previous study using rat primary hepatocytes, treatment of a general inhibitor of CYP 450 SKF 525-A enhanced sertraline-associated toxicity, implying that the parent form of sertraline maybe more toxic than its metabolites (Li et al., 2012). Importantly, we used HepG2 cells to study the MAPK signaling pathway and apoptosis induced by sertraline previously (Chen et al., 2014b), and thus felt it was appropriate to use the same cell line in this continuation study to explore the role of the MAPK signaling pathway in ER stress and also the cross talk between different pathways. The concentrations of sertraline used in our study were 3.13–50 µM, which is higher than reported human plasma concentrations (Cmax, 0.3 or 0.74 µM) (Mandrioli et al., 2013; Saletu et al., 1986). It has been suggested that a concentration equivalent to 100-fold of the Cmax should be tested in in vitro studies to identify idiosyncratic hepatotoxic drugs (Laifenfeld et al., 2014; Porceddu et al., 2012; Xu et al., 2008). The maximum concentration of sertraline in our study was 50 µM, which is within the suggested range of 100 times the Cmax; thus, the concentrations used in our study were reasonable.
Protein expression of the major ER stress markers include PERK, p-PERK, p-eIF2α, CHOP, ATF-4, and PDI was induced by exposure of sertraline (Fig. 3A). Besides the upregulation of ER stress markers at the protein level, the induction of PERK, IRE1α, and CHOP was also detected at the gene expression level (Fig. 2 and Table 3). Significant increased splicing of XBP, a key regulator of ER stress, was also observed (Fig. 3D). Collectively, these data clearly demonstrate that sertraline triggers ER stress in hepatic cells.
ER stress is associated with a wide range of diseases, such as neurodegeneration, cancer, inflammatory diseases, and diabetes (Xu et al., 2005). ER stress response is often described as one of the mechanisms in liver diseases, which include nonalcoholic fatty liver disease, alcoholic liver disease, hyperhomocysteinemia, and cholestatic liver disease (Dara et al., 2011). Studying and evaluating ER stress is a growing research area and various experimental approaches have been applied (Badr et al., 2007; Berger et al., 1988; Chen et al., 2014a, 2012; Cullen and Malim, 1992; Tannous, 2009; Weng et al., 2014). The most commonly used methods include ER stress marker measurements at protein and mRNA level, PCR-based assay to detect mRNA splicing of XBP, and reporter assays to measure the activity of ER stress-responding transcriptional factors such as XBP and ATF6 (Badr et al., 2007; Samali et al., 2010). Reporter assays for monitoring protein secretion have also been used to study ER stress because blocked or decreased protein secretion is a characteristic consequence of ER stress; thus, seeing a reduction of protein secretion indicates an ER stress response. Although such experimental approaches have been used in various cells, including 293T and human fibroblast cells HF8 (Badr et al., 2007), fibroblast-like cell line COS (Berger et al., 1988; Cullen and Malim, 1992), clonal mesangial cells SM43 (Meng et al., 2005), human glioma cells Gli36 (Tannous, 2009; Wurdinger et al., 2008), there were no similar systems available for drug-induced liver toxicity study. For this reason, we established two stable cell lines with secreted alkaline phosphatase (SEAP) and secreted Gaussia luciferase (Gluc) as reporters, validated the cell lines by using well-known ER inducers, and compared their sensitivity. As shown in Fig. 4C and D, both cell lines showed significantly decreased activity of SEAP or Gluc upon administration of brefeldin A or thapsigargin, commonly used ER stress inducers, demonstrating the usefulness of both assays. A rapid drop was seen in SEAP activity at a concentration of 0.01 µM and the response saturated at higher concentrations of both brefeldin A and thapsigargin. In contrast, for the Gluc assay, a dose-response effect was observed for a wide range concentrations, from 0.001 to 0.1 µM (covering three orders of magnitudes), suggesting that the sensitivity of the Gluc assay is higher than that of the SEAP assay. In addition, Gluc assay is less time-consuming with respect to experimental procedures (Badr et al., 2007), therefore, the Gluc assay may be superior over SEAP assay.
It should be stated that no single assay will be the most appropriate for ER stress evaluation; however, the Gluc assay can be used as an initial step because of its sensitivity and ease of use. In addition, it can also be used in high-throughput settings to screen drugs for liver toxicity mediated by ER stress. The experiments are ongoing in our laboratory.
In the past, ER stress was underestimated in drug-induced liver toxicity, but a growing body of evidence now indicates that ER stress plays an important role (Apostolova et al., 2013; Auman et al., 2007; Craig et al., 2006; Mohammad et al., 2012; Nagy et al., 2010; Naranmandura et al., 2012; Uzi et al., 2013). In a recent publication, it has been reported that Efavirenz, a non-nucleoside analog reverse transcriptase inhibitor to treat HIV1 infection, induced ER stress as inducing the expression of CHOP and GRP78 expression, phosphorylation of eIF2α, and enhancing the splicing of XBP, and also demonstrated there was an interplay between mitochondrial dysfunction and ER stress (Apostolova et al., 2013). A study on acetaminophen, a well-studied hepatotoxic drug, indicated that the toxicity of acetaminophen resulted in induction of CHOP and phosphorylation of eIF2α, the indicators of ER stress (Nagy et al., 2010). In a most recent study, administration of acetaminophen to mice resulted in activation of all three branches of UPR, with a clear indication of the involvement of PERK-elF2-CHOP signaling (Uzi et al., 2013). Interestingly, unlike previous reports demonstrating that ER stress is an early event of apoptotic cell death and glutathione depletion (Nagy et al., 2007,2010), in this study, it has been demonstrated that the ER stress occurred later in the cascade events after glutathione depletion and oxidative stress (Uzi et al., 2013). The discrepancy is not clear, could be due to different routes of administration, i.p. injection (Nagy et al., 2010) vs. oral administration (Uzi et al., 2013), nevertheless, these studies indicate that ER stress plays roles in acetaminophen-induced liver toxicity and the case is complex regarding the cause and consequence of ER stress and other responses.
The ER stress response is often accompanied by cellular adaptive responses or devastating reactions triggering cell death. Apoptosis, ATP depletion, oxidative stress, and mitochondrial dysfunction are among the effects seen in disease pathogenesis, implying that the stress response involves a complex interplay of different factors and organelles (Dara et al., 2011). Because we previously demonstrated apoptosis occur in response to sertraline’s challenge (Chen et al., 2014b), it was reasonable to explore the relationship between different pathways.
It is a challenge to determine the cause and consequence of different toxic events occurring in drug-induced toxicity. In our case, however, it is likely that ER stress is an earlier response triggering apoptosis and cell death because: (1) ER stress responses include the activation of phospho-PERK and phospho-eIF2α, and the significantly decreased of SEAP and Gluc secretion was present at lower concentrations of 6.25 and 12.5 µM, whereas caspase 3/7 activation, ATP content depletion, and LDH release occurred at higher concentration of 25 or 37.5 µM at the 2 h time point (Figs. 3A, 5B, and 6C) and (Chen et al., 2014b) (2) at 6 h of treatment, CHOP and PERK showed increased expression at as low as 3.13 µM (Fig. 2); whereas neither apoptotic nor necrotic cell death was observed (Chen et al., 2014b); (3) administration of a potent ER stress inhibitor, 4-PBA, significantly attenuated sertraline-caused apoptosis (Fig. 6C) and the increased expression of CHOP (Fig. 6B), whereas caspase inhibitors did not have significant effects on ER induction (data not shown). Additionally, we found that besides the involvement of the MAP4K4-JNK signaling pathway in apoptosis (Chen et al., 2014b), the MAP4K4-JNK signaling pathway also participated in ER stress caused by sertraline; evidenced by (1) silencing of MAP4K4 markedly suppressed increased expression of ER stress markers, phospho-PERK, CHOP, and PDI (Fig. 7A); (2) ER stress associated apoptosis was diminished in MAP4K4 silenced cells (Fig. 7A); and (3) a JNK specific inhibitor efficiently prevented the decreased activity of secreted Gluc (Fig. 7B).
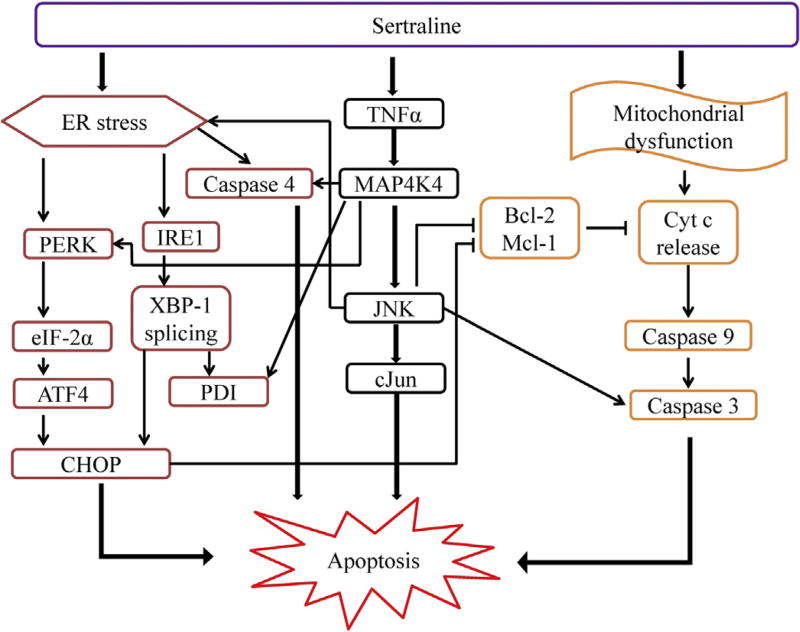
In summary, a concerted cellular response and a complex mechanism regarding sertraline-induced liver toxicity has been demonstrated. Sertraline causes mitochondrial impairment (Li et al., 2012) and apoptosis (Chen et al., 2014b), activates MAPK signaling pathways, particularly the MAP4K4-JNK signaling pathway initiated by the activation of TNFα. Sertraline induces ER stress notably by activating both the PERK and IRE1 branches. Both apoptosis and ER stress are shown to be directly mediated by a MAPK signaling pathway. Sertraline-induced liver toxicity is invoked by stressing or injuring multiple cellular components, such as mitochondria and endoplasmic reticulum, with cross talk between these components. The activation of the MAPK signaling pathway contributes to both apoptosis and ER stress. A scheme depicting the proposed mechanism for sertraline-induced liver toxicity is displayed in Fig. 8.
Fig. 8.
A scheme for proposed mechanism on sertraline-induced liver toxicity.
Supplementary Material
Acknowledgments
SC, JX, and YW were supported by appointments to the Postgraduate Research Program at the National Center for Toxicological Research administered by the Oak Ridge Institute for Science Education through an interagency agreement between the U.S. Department of Energy and the U.S. FDA. AI was supported by NCTR Summer Science Research Program. We thank Drs. William Melchior and Vasily Dobrovolsky for their critical review of this manuscript.
This article is not an official guidance or policy statement of the U.S. FDA. No official support or endorsement by the U.S. FDA is intended or should be inferred.
Footnotes
Conflict of interest
The authors declare that there are no conflicts of interest.
Transparency document
The Transparency document associated with this article can be found in the online version.
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.tox.2014.05.007.
References
- Apostolova N, Gomez-Sucerquia LJ, Alegre F, et al. ER stress in human hepatic cells treated with Efavirenz: mitochondria again. J. Hepatol. 2013;59(4):780–789. doi: 10.1016/j.jhep.2013.06.005. [DOI] [PubMed] [Google Scholar]
- Auman JT, Chou J, Gerrish K, et al. Identification of genes implicated in methapyrilene-induced hepatotoxicity by comparing differential gene expression in target and nontarget tissue. Environ. Health Perspect. 2007;115(4):572–578. doi: 10.1289/ehp.9396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badr CE, Hewett JW, Breakefield XO, Tannous BA. A highly sensitive assay for monitoring the secretory pathway and ER stress. PLoS ONE. 2007;2(6):e571. doi: 10.1371/journal.pone.0000571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger J, Hauber J, Hauber R, Geiger R, Cullen BR. Secreted placental alkaline phosphatase: a powerful new quantitative indicator of gene expression in eukaryotic cells. Gene. 1988;66(1):1–10. doi: 10.1016/0378-1119(88)90219-3. [DOI] [PubMed] [Google Scholar]
- Carvajal Garcia-Pando A, Garcia del Pozo J, Sanchez AS, Velasco MA, Rueda de Castro AM, Lucena MI. Hepatotoxicity associated with the new antidepressants. J. Clin. Psychiatry. 2002;63(2):135–137. doi: 10.4088/jcp.v63n0208. [DOI] [PubMed] [Google Scholar]
- Chen S, Melchior W, Guo L. Endoplasmic reticulum stress in drug- and environmental toxicant-induced liver toxicity. J. Environ. Sci. Health C: Environ. Carcinog. Ecotoxicol. Rev. 2014a;32(1):83–104. doi: 10.1080/10590501.2014.881648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Wan L, Couch L, et al. Mechanism study of goldenseal-associated DNA damage. Toxicol. Lett. 2013;221(1):64–72. doi: 10.1016/j.toxlet.2013.05.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Xuan J, Wan L, et al. Sertraline, an antidepressant, induces apoptosis in hepatic cells through the mitogen-activated protein kinase pathway. Toxicol. Sci. 2014b;137(2):404–415. doi: 10.1093/toxsci/kft254. [DOI] [PubMed] [Google Scholar]
- Chen YW, Yang YT, Hung DZ, Su CC, Chen KL. Paraquat induces lung alveolar epithelial cell apoptosis via Nrf-2-regulated mitochondrial dysfunction and ER stress. Arch. Toxicol. 2012;86(10):1547–1558. doi: 10.1007/s00204-012-0873-8. [DOI] [PubMed] [Google Scholar]
- Collados V, Hallal H, Andrade RJ. Sertraline hepatotoxicity: report of a case and review of the literature. Dig. Dis. Sci. 2010;55(6):1806–1807. doi: 10.1007/s10620-010-1192-7. [DOI] [PubMed] [Google Scholar]
- Craig A, Sidaway J, Holmes E, et al. Systems toxicology: integrated genomic, proteomic and metabonomic analysis of methapyrilene induced hepatotoxicity in the rat. J. Proteome Res. 2006;5(7):1586–1601. doi: 10.1021/pr0503376. [DOI] [PubMed] [Google Scholar]
- Cullen BR, Malim MH. Secreted placental alkaline phosphatase as a eukaryotic reporter gene. Methods Enzymol. 1992;216:362–368. doi: 10.1016/0076-6879(92)16033-g. [DOI] [PubMed] [Google Scholar]
- Dara L, Ji C, Kaplowitz N. The contribution of endoplasmic reticulum stress to liver diseases. Hepatology. 2011;53(5):1752–1763. doi: 10.1002/hep.24279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fartoux-Heymann L, Hezode C, Zafrani ES, Dhumeaux D, Mallat A. Acute fatal hepatitis related to sertraline. J. Hepatol. 2001;35(5):683–684. doi: 10.1016/s0168-8278(01)00159-3. [DOI] [PubMed] [Google Scholar]
- Fishman PH, Curran PK. Brefeldin A inhibits protein synthesis in cultured cells. FEBS Lett. 1992;314(3):371–374. doi: 10.1016/0014-5793(92)81508-j. [DOI] [PubMed] [Google Scholar]
- Galan Navarro JL. Acute cholestatic hepatitis probably caused by sertraline. Rev. Esp. Enferm. Dig. 2001;93(12):822. [PubMed] [Google Scholar]
- Guo L, Dial S, Shi L, et al. Similarities and differences in the expression of drug-metabolizing enzymes between human hepatic cell lines and primary human hepatocytes. Drug Metab. Dispos. 2011;39(3):528–538. doi: 10.1124/dmd.110.035873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L, Li Q, Xia Q, Dial S, Chan PC, Fu P. Analysis of gene expression changes of drug metabolizing enzymes in the livers of F344 rats following oral treatment with kava extract. Food Chem. Toxicol. 2009;47(2):433–442. doi: 10.1016/j.fct.2008.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L, Mei N, Liao W, Chan PC, Fu PP. Ginkgo biloba extract induces gene expression changes in xenobiotics metabolism and the Myc-centered network. OMICS. 2010;14(1):75–90. doi: 10.1089/omi.2009.0115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hautekeete ML, Colle I, van Vlierberghe H, Elewaut A. Symptomatic liver injury probably related to sertraline. Gastroenterol. Clin. Biol. 1998;22(3):364–365. [PubMed] [Google Scholar]
- Hitomi J, Katayama T, Eguchi Y, et al. Involvement of caspase-4 in endoplasmic reticulum stress-induced apoptosis and Abeta-induced cell death. J. Cell Biol. 2004;165(3):347–356. doi: 10.1083/jcb.200310015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan C, Zhang Y. Assessing the comparative-effectiveness of antidepressants commonly prescribed for depression in the US Medicare population. J. Ment. Health Policy Econ. 2012;15(4):171–178. [PMC free article] [PubMed] [Google Scholar]
- Kim KY, Hwang W, Narendran R. Acute liver damage possibly related to sertraline and venlafaxine ingestion. Ann. Pharmacother. 1999;33(3):381–382. doi: 10.1345/aph.18155. [DOI] [PubMed] [Google Scholar]
- Laifenfeld D, Qiu L, Swiss R, et al. Utilization of causal reasoning of hepatic gene expression in rats to identify molecular pathways of idiosyncratic drug-induced liver injury. Toxicol. Sci. 2014;137(1):234–248. doi: 10.1093/toxsci/kft232. [DOI] [PubMed] [Google Scholar]
- Li Y, Couch L, Higuchi M, Fang JL, Guo L. Mitochondrial dysfunction induced by sertraline, an antidepressant agent. Toxicol. Sci. 2012;127(2):582–591. doi: 10.1093/toxsci/kfs100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandrioli R, Mercolini L, Raggi MA. Evaluation of the pharmacokinetics, safety and clinical efficacy of sertraline used to treat social anxiety. Expert Opin. Drug Metab. Toxicol. 2013;9(11):1495–1505. doi: 10.1517/17425255.2013.816675. [DOI] [PubMed] [Google Scholar]
- Meng Y, Kasai A, Hiramatsu N, et al. Real-time monitoring of mesangial cell-macrophage cross-talk using SEAP in vitro and ex vivo. Kidney Int. 2005;68(2):886–893. doi: 10.1111/j.1523-1755.2005.00471.x. [DOI] [PubMed] [Google Scholar]
- Mohammad MK, Avila D, Zhang J, et al. Acrolein cytotoxicity in hepatocytes involves endoplasmic reticulum stress, mitochondrial dysfunction and oxidative stress. Toxicol. Appl. Pharmacol. 2012;265(1):73–82. doi: 10.1016/j.taap.2012.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy G, Kardon T, Wunderlich L, et al. Acetaminophen induces ER dependent signaling in mouse liver. Arch. Biochem. Biophys. 2007;459(2):273–279. doi: 10.1016/j.abb.2006.11.021. [DOI] [PubMed] [Google Scholar]
- Nagy G, Szarka A, Lotz G, et al. BGP-15 inhibits caspase-independent programmed cell death in acetaminophen-induced liver injury. Toxicol. Appl. Pharmacol. 2010;243(1):96–103. doi: 10.1016/j.taap.2009.11.017. [DOI] [PubMed] [Google Scholar]
- Naranmandura H, Xu S, Koike S, et al. The endoplasmic reticulum is a target organelle for trivalent dimethylarsinic acid (DMAIII)-induced cytotoxicity. Toxicol. Appl. Pharmacol. 2012;260(3):241–249. doi: 10.1016/j.taap.2012.02.017. [DOI] [PubMed] [Google Scholar]
- Ning B, Dial S, Sun Y, Wang J, Yang J, Guo L. Systematic and simultaneous gene profiling of 84 drug-metabolizing genes in primary human hepatocytes. J. Biomol. Screen. 2008;13(3):194–201. doi: 10.1177/1087057108315513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persky S, Reinus JF. Sertraline hepatotoxicity: a case report and review of the literature on selective serotonin reuptake inhibitor hepatotoxicity. Dig. Dis. Sci. 2003;48(5):939–944. doi: 10.1023/a:1023007831047. [DOI] [PubMed] [Google Scholar]
- Porceddu M, Buron N, Roussel C, Labbe G, Fromenty B, Borgne-Sanchez A. Prediction of liver injury induced by chemicals in human with a multi-parametric assay on isolated mouse liver mitochondria. Toxicol. Sci. 2012;129(2):332–345. doi: 10.1093/toxsci/kfs197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaiahgari SC, den Braver MW, Herpers B, et al. A 3D in vitro model of differentiated HepG2 cell spheroids with improved liver-like properties for repeated dose high-throughput toxicity studies. Arch. Toxicol. 2014;88:1083–1095. doi: 10.1007/s00204-014-1215-9. [DOI] [PubMed] [Google Scholar]
- Saletu B, Grunberger J, Linzmayer L. On central effects of serotonin re-uptake inhibitors: quantitative EEG and psychometric studies with sertraline and zimelidine. J. Neural Transm. 1986;67(3–4):241–266. doi: 10.1007/BF01243351. [DOI] [PubMed] [Google Scholar]
- Samali A, Fitzgerald U, Deegan S, Gupta S. Methods for monitoring endoplasmic reticulum stress and the unfolded protein response. Int. J. Cell Biol. 2010;2010:830307. doi: 10.1155/2010/830307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabak F, Gunduz F, Tahan V, Tabak O, Ozaras R. Sertraline hepatotoxicity: report of a case and review of the literature. Dig. Dis. Sci. 2009;54(7):1589–1591. doi: 10.1007/s10620-008-0524-3. [DOI] [PubMed] [Google Scholar]
- Tannous BA. Gaussia luciferase reporter assay for monitoring biological processes in culture and in vivo. Nat. Protoc. 2009;4(4):582–591. doi: 10.1038/nprot.2009.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzi D, Barda L, Scaiewicz V, et al. CHOP is a critical regulator of acetaminophen-induced hepatotoxicity. J. Hepatol. 2013;59(3):495–503. doi: 10.1016/j.jhep.2013.04.024. [DOI] [PubMed] [Google Scholar]
- Verrico MM, Nace DA, Towers AL. Fulminant chemical hepatitis possibly associated with donepezil and sertraline therapy. J. Am. Geriatr. Soc. 2000;48(12):1659–1663. doi: 10.1111/j.1532-5415.2000.tb03879.x. [DOI] [PubMed] [Google Scholar]
- Weng CY, Chiou SY, Wang L, Kou MC, Wang YJ, Wu MJ. Arsenic trioxide induces unfolded protein response in vascular endothelial cells. Arch. Toxicol. 2014;88(2):213–226. doi: 10.1007/s00204-013-1101-x. [DOI] [PubMed] [Google Scholar]
- Wong WL, Brostrom MA, Kuznetsov G, Gmitter-Yellen D, Brostrom CO. Inhibition of protein synthesis and early protein processing by thapsigargin in cultured cells. Biochem. J. 1993;289(Pt 1):71–79. doi: 10.1042/bj2890071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurdinger T, Badr C, Pike L, et al. A secreted luciferase for ex vivo monitoring of in vivo processes. Nat. Methods. 2008;5(2):171–173. doi: 10.1038/nmeth.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Bailly-Maitre B, Reed JC. Endoplasmic reticulum stress: cell life and death decisions. J. Clin. Invest. 2005;115(10):2656–2664. doi: 10.1172/JCI26373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu JJ, Henstock PV, Dunn MC, Smith AR, Chabot JR, de Graaf D. Cellular imaging predictions of clinical drug-induced liver injury. Toxicol. Sci. 2008;105(1):97–105. doi: 10.1093/toxsci/kfn109. [DOI] [PubMed] [Google Scholar]
- Yam GH, Gaplovska-Kysela K, Zuber C, Roth J. Sodium 4-phenylbutyrate acts as a chemical chaperone on misfolded myocilin to rescue cells from endoplasmic reticulum stress and apoptosis. Invest. Ophthalmol. Vis. Sci. 2007;48(4):1683–1690. doi: 10.1167/iovs.06-0943. [DOI] [PubMed] [Google Scholar]
- Zinszner H, Kuroda M, Wang X, et al. CHOP is implicated in programmed cell death in response to impaired function of the endoplasmic reticulum. Genes Dev. 1998;12(7):982–995. doi: 10.1101/gad.12.7.982. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.