Abstract
Kava is a plant traditionally used for making beverages in Pacific Basin countries and has been used for the treatment of nervous disorders in the United States. The pharmacological activity of kava is achieved through kavalactones in kava extract, which include kawain, 7,8-dihydrokawain, yangonin, 5,6-dehydrokawain, methysticin, and 7,8-dihydromethysticin. Recent studies have shown that kava extract induces hepatic CYP1A1 enzyme; however, the mechanisms of CYP1A1 induction have not been elucidated, and the kavalactones responsible for CYP1A1 induction have not yet been identified. Using a combination of biochemical assays and molecular docking tools, we determined the functions of kava extract and kavalactones and delineated the underlying mechanisms involved in CYP1A1 induction. The results showed that kava extract displayed a concentration-dependent effect on CYP1A1 induction. Among the six major kavalactones, methysticin triggered the most profound inducing effect on CYP1A1 followed by 7,8-dihydromethysticin. The other four kavalactones (yangonin, 5,6-dehydrokawain, kawain, and 7,8-dihydrokawain) did not show significant effects on CYP1A1. Consistent with the experimental results, in silico molecular docking studies based on the aryl hydrocarbon receptor (AhR)-ligand binding domain homology model also revealed favorable binding to AhR for methysticin and 7,8-dihydromethysticin compared with the remaining kavalactones. Additionally, results from a luciferase gene reporter assay suggested that kava extract, methysticin, and 7,8-dihydromethysticin were able to activate the AhR signaling pathway. Moreover, kava extract-, methysticin-, and 7,8-dihydromethysticin-mediated CYP1A1 induction was blocked by an AhR antagonist and abolished in AhR-deficient cells. These findings suggest that kava extract induces the expression of CYP1A1 via an AhR-dependent mechanism and that methysticin and 7,8-dihydromethysticin contribute to CYP1A1 induction. The induction of CYP1A1 indicates a potential interaction between kava or kavalactones and CYP1A1-mediated chemical carcinogenesis.
Keywords: aryl hydrocarbon receptor, CYP1A1, kava extract, kavalactone, molecular docking
In recent years, there has been a global increase in the use of herbal dietary supplements as complementary and alternative medicines (CAMs) for chronic disease treatment and general health promotion. In the 1990’s, approximately 10–40% of the general North American population used herbal products (Nelson and Perrone, 2000); this number has grown sharply during the 20th Centuries (Su and Li, 2011). There are potential problems and concerns with their use due to potential toxicity, adulteration, contamination, undesirable herb-drug interactions, and lack of standardization (De Smet, 1995, 2004; Koh and Woo, 2000).
Kava, historically used as a beverage in the South Pacific Islands, is currently one of the best selling and available in health food stores in the United States, in spite of the fact that it has been banned in several Western countries because of liver injury (Ernst, 2001; Fu et al., 2008). Studies have indicated that kava as well as other popularly used herbal dietary supplements, such as St John’s wort (Hypericum perforatum), ginseng (Panax ginseng), garlic (Allium sativum), ginkgo (Ginkgo biloba), echinacea (Echinacea spp.), and saw palmetto (Serenoa repens), may alter plasma concentrations of coadministered drugs and cause undesirable responses via the modulation of drug metabolizing enzymes, such as cytochrome P450 (CYP) and/or P-glycoprotein (Fugh-Berman, 2000; Hu et al., 2005; Yang et al., 2006).
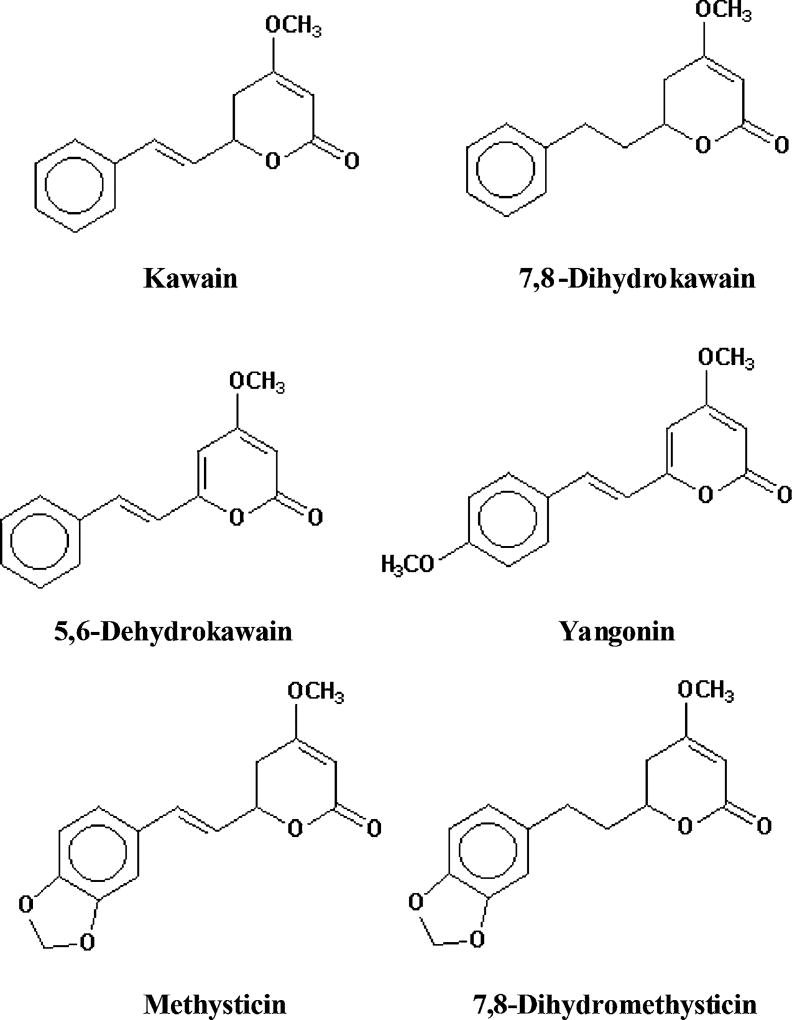
Kavalactones are the major constituents in kava extract. Kawain, 7,8-dihydrokawain, yangonin, 5,6-dehydrokawain, methysticin, and dihydromethysticin (Fig. 1) are responsible for 95% of the total therapeutic activity of kava (Gautz et al., 2006; Jiang et al., 2007; Singh and Singh, 2002). Clinical trials indicated that both kava extract and kavalactones can be used as a remedy for anxiety and insomnia (Ernst, 2004; Singh and Singh, 2002; Volz and Kieser, 1997). Besides having therapeutic effects, toxicities (including hepatotoxicity and carcinogenicity) have been reported for kava and some of its constituents (Nerurkar et al., 2004; NTP, 2011; Olsen et al., 2011; Tang et al., 2011; Zhou et al., 2010); however, the mechanisms of kava-associated toxicities are not well understood. One of the proposed mechanisms is that kava induces hepatotoxicity through the modulation of drug metabolizing enzymes, thus affecting drug metabolism. When taken concomitantly with therapeutic drugs, kava can lead to herb-drug interactions (Bressler, 2005; Clouatre, 2004). Accordingly, the effects of kava extract and kavalactones on drug metabolizing enzymes have been examined. Studies have shown that preincubation of kava extract with human liver microsomes inhibited CYP1A2, 2C9, 2C19, 2D6, 3A4, and 4A9/11 (Mathews et al., 2002, 2005). In a study to investigate the actions of kavalactones on CYP450 (Mathews et al., 2002), Mathews et al. found that significant inhibitions of CYP2C9 by 5,6-dehydrokawain (42%), methysticin (58%), and 7,8-dihydromethysticin (69%); of 2C19 by 7,8-dihydromethysticin (76%); of 2D6 by methysticin (44%); and of 3A4 by 5,6-dehydrokawain (40%), methysticin (27%), and 7,8-dihydromethysticin (54%). Similarly, Zou et al. (Zou et al., 2002) reported that 5,6-dehydrokawain, 7,8-dihydromethysticin, and methysticin were potent inhibitors of CYP2C9 and CYP2C19. 5,6-dehydrokawain and 7,8-dihydromethysticin exerted inhibitory effects on CYP2A23 (Ma et al., 2004). In vivo studies revealed that the expression of hepatic CYP2D1 decreased in rats fed kava extract and that the expression of CYP1A2, 2B1, and 3A1 increased (Clayton et al., 2007). Clinical studies have shown that kava had an inhibitory effect on CYP2E1 (Gurley et al., 2005, 2008). Collectively, these results indicate that kava and its constituents act as CYP modulators. When acting as an enzyme inhibitor, kava can reduce the metabolism of coadministered drugs, thus leading to higher plasma concentrations and toxicities, especially for the coadministered drugs having a narrow therapeutic window (Guo et al., 2010a).
FIG. 1.
Structures of six major kavalactones.
To date, most of studies have reported the inhibitory effects of kava. Fewer studies have demonstrated that kava exerts an inductive effect on drug metabolizing enzymes. A study using human hepatocytes demonstrated that kava increased CYP3A4 mRNA level and activated pregnane X receptor (PXR) to mediate CYP3A4 induction (Raucy, 2003). Most recently, studies, including ours, have reported that kava induced the expression of CYP1A1 (Guo et al., 2009, 2010b; Yamazaki et al., 2008; Yueh et al., 2005). For example, kava markedly enhanced CYP1A1 mRNA expression (75–220 fold) as well as CYP1A1 protein expression with a high dose treatment (equivalent to approximately 380 mg/kg/day of kavalactones) in rats for 8 days (Yamazaki et al., 2008) and moderately induced CYP1A1 in HepG2 cells (Yueh et al., 2005). Previously, we have analyzed the gene expression profiles in the livers of mice and rats exposed to kava extract for 14 weeks and found that CYP1A1 gene expression was significantly upregulated (Guo et al., 2009, 2010b). Activation of CYP1A1 is generally mediated by the aryl hydrocarbon receptor (AhR) signaling pathway. Upon ligand binding, AhR translocates to the nucleus and binds to AhR nuclear translocator (Arnt) to form a heterodimer. This heterodimer binds to the xenobiotic responsive element (XRE), the core DNA-binding motif located in promoter regions of AhR-regulated downstream genes. The binding of AhR/Arnt to the promoters allows the transactivation of downstream genes, such as CYP1A1. CYP1A1 is known to be involved in the metabolism of polycyclic aromatic hydrocarbons (PAHs) to their reactive metabolites responsible for their carcinogenicity (Whitlock, 1999). Elevated CYP1A1 activity can increase the production of toxic metabolites of such chemicals; thus any agent that induces CYP1A1 may exaggerate the carcinogenicity and toxicity of PAHs. For such reasons, induction of CYP1A1 by drugs, dietary supplements, and environmental pollutants raises significant concerns regarding toxicity.
We have demonstrated that kava as a whole extract induces the expression of CYP1A1 in vivo. As a continuation of our studies, we examined the underlying molecular mechanisms of CYP1A1 induction by kava and determined the active kavalactones contributing to CYP1A1 induction. Toward these goals, we first analyzed the effects of kava extract and six major kavalactones on the induction of CYP1A1 in mouse hepatic Hepa1c1c7 cells. We then applied an in silico molecular docking approach to gain an initial insight into the interaction between individual kavalactones and AhR. Lastly, we used a luciferase reporter assay, an AhR antagonist (3′,4′-dimethoxyflavone), and an AhR-deficient cell line (CRL-2710) to validate our molecular docking results. Our findings clearly indicate that (1) kava activates CYP1A1 via the AhR signaling pathway and (2) among the six kavalactones, methysticin, and 7′ 8′-dihydromethysticin are the most potent CYP1A1 inducers.
MATERIALS AND METHODS
Chemicals and reagents
Powdered kava extract was a gift from Dr Po-Chuen Chan at the National Institute of Environmental Health Sciences (Research Triangle Park, NC). The preparation of kava extract has been described previously (Guo et al., 2010b). The extract contains about 30% kavalactones, including kawain (8.11%), 7,8-dihydrokawain (8.04%), yangonin (4.18%), 7,8-dihydromethysticin (3.90%), methysticin (3.44%), and 5,6-dehydrokawain (2.39%). Six pure kavalactones, namely kawain, 7,8-dihydrokawain, 5,6-dehydrokawain, yangonin, methysticin, and 7,8-dihydromethysticin, were purchased from LKT Laboratories (St Paul, MN). Fetal bovine serum (FBS) was obtained from Atlanta Biologicals (Lawrenceville, GA). α-Minimum Essential Medium (α-MEM), penicillin, streptomycin, dimethysulfoxide (DMSO), 3′,4′-dimethoxyflavone (3′,4′-DMF), 7-ethoxyresorufin, and dicumarol were purchased from Sigma-Aldrich (St Louis, MO).
Cell culture
The mouse hepatoma cell line Hepa1c1c7 and AhR-deficient cell line CRL-2710 were obtained from American Type Culture Collection (Manassas, VA). Cells were grown in α-MEM supplemented with 10% FBS and antibiotics (50 U/ml penicillin and 50 µg/ml streptomycin) and maintained at 37°C in a humidified atmosphere with 5% CO2. Unless otherwise specified, Hepa1c1c7 and CRL-2710 cells were seeded at a density of 1.5 × 10 cells/ml in 96-well plates with each well containing 100 µl culture medium. Cells were cultured for about 24 h prior to treatment with indicated concentrations of kava extract, individual kavalactones, or DMSO vehicle (final concentration 0.1%) control.
Cytotoxicity determination
The cytotoxicity of kava extract and kavalactones was assessed using the tetrazolium reduction cell viability assay (MTS, CellTiter 96 AQueous Non-Radioactive Cell Proliferation Assay) from Promega Corporation (Madison, WI). After treatment for 24 h, the MTS assay was performed. The viability of the cells was calculated by comparing the absorbance of the treated cells with that of the DMSO controls.
7-Ethoxyresorufin O-dealkylation assay
CYP1A1-mediated 7-ethoxyresorufin O-dealkylation (EROD) was measured according to a method described previously (Donato et al., 1993). Fluorescence was recorded with a Synergy 2 Multi-Mode Microplate Reader (BioTek, Winooski, VT) with excitation/emission wavelengths of 530/590 nm. Protein content from each well was determined using the BCA method (Bio-Rad Laboratories, Hercules, CA) to normalize the enzymatic activity.
RNA isolation
Cells were grown in 6-cm tissue culture plates and treated with kava extract and individual kavalactones. Total RNA from cells was isolated using the RNeasy system (Qiagen, Valencia, CA). The yield of the extracted RNA was determined spectrophotometrically by measuring the optical density at 260 nm using a NanoDrop 8000 (Thermo Scientific, Wilmington, DE). The purity and quality of the RNA were evaluated using an RNA 6000 LabChip on an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA).
Real-time TaqMan PCR assay
Real-time TaqMan PCR assays were conducted using a Bio-Rad CFX96 Real-Time PCR Detection System (Bio-Rad Laboratories). The manufacturer’s instructions for TaqMan Gene Expression Assays were followed to prepare reactions. The TaqMan assay probes were CYP1A1 (Mm00487218_m1) and endogenous control β-actin (ACTB, Mm00607939_s1). Assays were run with Universal Master Mix (2X) without AmpErase UNG, using universal cycling conditions (10 min at 95°C; 15 s at 95°C, 1 min 60°C, 40 cycles). Data normalization and analysis were as described previously (Guo et al., 2009).
Western blot analysis
Cells were grown in 10-cm tissue culture plates and treated with kava extract and individual kavalactones. Standard Western blots were performed using a CYP1A1 antibody (Abcam Inc., Cambridge, MA) followed by a secondary antibody conjugated to horseradish peroxidase. The intensity of each band was quantified with ImageJ software (NIH, Bethesda, MD).
Transient transfection and promoter activity assay
pGL3-Basic and pGL4.74 [hRluc/TK] vectors were purchased from Promega. pGudluc 6.1, encoding a CYP1A1 promoter-driven luciferase reporter gene, was a generous gift from Dr Michael R. Stallcup (University of Southern California, Los Angeles, CA). Hepa1c1c7 cells were seeded in 6-well plates at a density of 1.0 × 10 cells/ml in a volume of 2.5 ml culture medium. After 24 h of incubation, 2 µg of pGudluc6.1 or pGL3-Basic (empty control vector, firefly luciferase) along with 0.2 µg of pGL4.74 [hRluc/TK] (Renilla luciferase as internal control) were mixed with 5 µl of FuGENE HD transfection reagent (Roche Applied Science, Mannheim, Germany) and transfected into cells according to the manufacturer’s instructions. The cells were then treated with kava extract and kavalactones at indicated concentrations (or DMSO) for 24 h. After treatment, firefly and Renilla luciferase activities were determined with the Dual-Luciferase Report Assay System (Promega) according to the manufacturer’s manual. The signals were recorded using a Synergy 2 Multi-Mode Microplate Reader (BioTek).
Statistical analysis
Data are presented as mean ± SD of at least three independent experiments. Analyses were performed using GraphPad Prism 5 (La Jolla, CA). Statistical significance was determined by one- way ANOVA followed by the Dunnett’s tests for pairwise comparisons or two-way ANOVA followed by the Bonferroni post-test. The difference was considered statistically significant when the p < 0.05.
Homology modeling
A fragment (amino acids 230–421) containing the full ligand binding activity and specificity of the mouse AhR (GI:7304873) (Coumailleau et al., 1995) was used for homology modeling. Homologous proteins were identified by scanning the Protein Data Bank database using PSI-BLAST, and three proteins (1P97, 1WA9, and 2B02) with the Per-AhR/Arnt-Sim (PAS) domain were used as template molecules to model the ligand binding domain of mouse AhR. Among the three, the highest sequence identity with mouse AhR is about 30%. The homology modeling was carried out using MODELLER 8v1 program (Sali and Blundell, 1993). In brief, homology modeling began with an alignment of the sequence to be modeled (target) with related known 3D structures (templates). Then, distance and dihedral angle restraints on the target sequence were calculated from its alignment with template 3D structures. The restraints were obtained empirically from a database of protein structure alignments (Sali and Overington, 1994). Finally, energy minimization was done by satisfying restraints on bond distances and dihedral angles extracted from the templates. In this study, the homology model obtained was further optimized by 200 ps molecular dynamics simulation using TIP3P explicit water.
Molecular docking
Surflex docking, which has been successful in eliminating false positive results and especially useful in virtual screen research (Jain, 2007), was used to dock six kavalactones to the ligand-binding domain of AhR using Sybyl 8.1 (Tripos Inc, St Louis, MO). Surflex-dock employs an empirical scoring function based on the Hammerhead docking system and a so-called “protomol” to guide the docking process. The protomol is a computational representation of the intended binding site, used to direct the initial placement of the ligand during the docking process. First, three molecular probes: CH4, C=O, and N–H were tessellated in the protein active site and optimized based on the Surflex scoring function. After that, high-scoring fragments were retained and redundant fragments were eliminated. This protomol obtained intend to mimic the ideal interactions made by an ideal ligand to the protein active site. The protomol was further used to direct the initial placement of the ligand during the docking process by a morphological similarity function (Jain, 2003). Surflex-Dock score is expressed in —log10(Kd) units to represent binding affinity. A higher score represents stronger binding affinity. Before docking, six kavalactones were charged with the Gasteiger-Huckel method and optimized with the Tripos force field.
RESULTS
Effects of Kava Extract and Kavalactones on Cell Viability
The MTS cell viability assay was used to determine the effects of kava extract and kavalactones on cell viability in mouse hepatic cells. Hepa1c1c7 cells were treated with various concentrations of kava extract (0–50 µg/ml) and six kavalactones (0–100µM) for 24 h. The results indicated that kava extract at concentrations up to 50 µg/ml and kavalactones up to 100lM did not induce cell death (Supplementary table 1). For the following studies, kava extract at 0.78–6.25 µg/ml and kavalactones at 0.78–25µM, concentrations that caused no damage to cells, were used.
Effects of Kava Extract and Kavalactones on Induction of CYP1A1 at Gene Expression, Protein Expression, and Enzymatic Activity
In our previous study, kava extract was found to induce CYP1A1 (Guo et al., 2009). In the present study, we first examined the gene expression of CYP1A1 in mouse hepatoma cell line Hepa1c1c7 in response to kava challenge and then determined the individual kavalactones responsible for the induction. Hepa1c1c7 cells were cultured with kava extract and purified kavalactones for 24 h, and gene expression, protein expression, and enzymatic activity of CYP1A1 were measured.
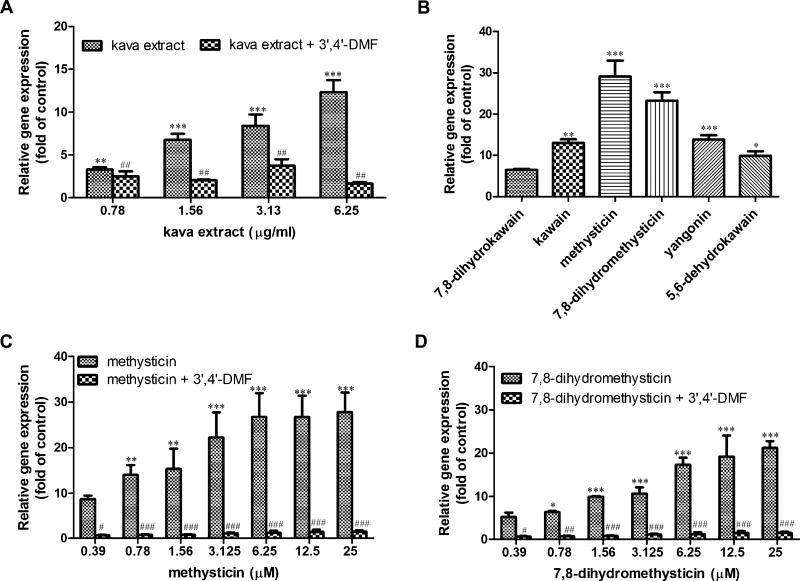
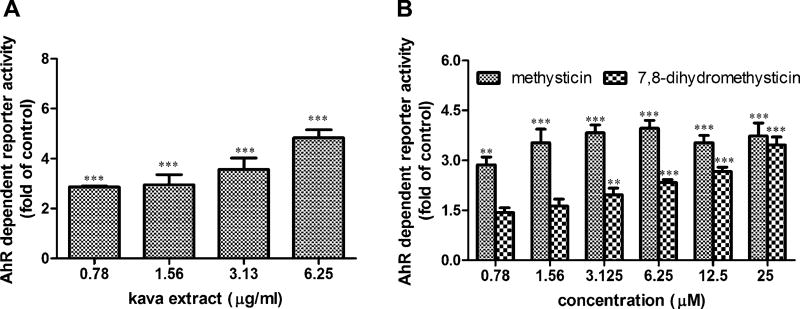
Induction of CYP1A1 gene expression was examined using TaqMan assays. As shown in Figure 2A, kava extract caused a concentration-dependent gene expression induction of CYP1A1. At the concentration of 25µM tested for kavalactones, methysticin and 7,8-dihydromethysticin caused prominent inductions (29- and 23-fold, respectively), followed by yangonin (13.8-fold), kawain (13-fold), 5,6-dehydrokawain (9.9-fold), and 7,8-dihydrokawain (6.5-fold) (Fig. 2B). Further investigation demonstrated that the induction of CYP1A1 gene expression was in a concentration-dependent manner for the treatments of methysticin and 7,8-dihydromethysticin (Figs. 2C and 2D) (Data with addition of 3′,4′-DMF will be described later).
FIG. 2.
Gene expression induction of CYP1A1 mRNA by kava extract and kavalactones in Hepa1c1c7 cells. (A) Induction of CYP1A1 by kava extract (0– 6.25 µg/ml) in the absence or presence of 1µM 3′,4′-DMF. (B) Induction of CYP1A1 by six kavalactones (25µM). (C) Induction of CYP1A1 by methysticin in the absence or presence of 1 µM 3′,4′-DMF. (D) Induction of CYP1A1 by 7,8-dihydromethysticin in the absence or presence of 1µM 3′,4′-DMF. Hepa1c1c7 cells were exposed to kava extract (0–6.25 µg/ml), methysticin (0–25µM), or 7,8-dihydromethysticin (0–25µM) in the absence or presence of 1µM 3′,4′-DMF for 24 h. RNA was isolated and TaqMan assays were performed. Fold change was calculated by normalizing to control (fold change of control is 1). All the data are represented as at least three independent experiments. The values shown are mean ± SD. *p < 0.05; **p < 0.01; ***p < 0.001 versus control. #p < 0.05; ##p < 0.01; ###p < 0.001, significantly different from the treatment without 3′,4′-DMF.
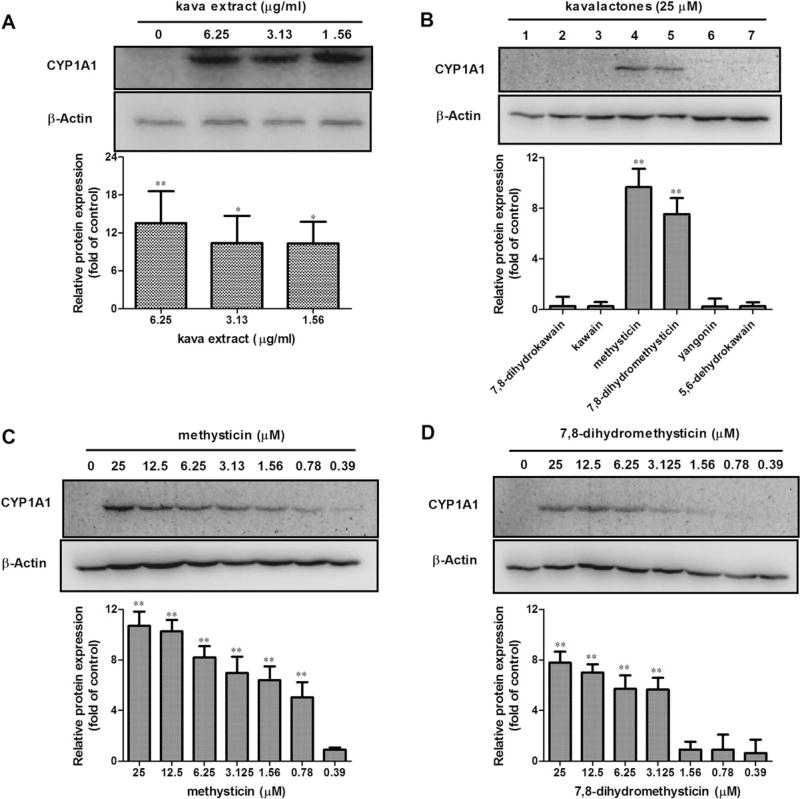
The magnitude of the CYP1A1 protein induction was studied by Western blot analysis. CYP1A1 protein content was increased when Hepa1c1c7 cells were exposed to kava extract (1.56, 3.13, and 6.25 µg/ml) in comparison with their untreated counterpart, although there was no obvious concentration-dependent increase. A representative Western blot result is shown in Figure 3A. While most kavalactones tested showed inducing effects on CYP1A1 at the gene expression level (Fig. 2B), only methysticin and 7,8-dihydromethysticin induced at the protein level (Fig. 3B). To study further the inductive ability of methysticin and 7,8-dihydromethysticin, different concentrations of methysticin and 7,8-dihydromethysticin were tested. As shown in Figures 3C and 3D, both methysticin and 7,8-dihydromethysticin induced CYP1A1 protein expression in a concentration-dependent manner, and the maximum inductions were 10.8-fold by methysticin and 7.9-fold by 7,8-dihydromethysticin compared with the control (Figs. 3C and 3D). The minimum concentration of methysticin needed to trigger CYP1A1 induction was 0.78µM, whereas the same concentration 7,8-dihydromethysticin did not induce CYP1A1 protein expression (Figs. 3C and 3D).
FIG. 3.
Protein expression induction of CYP1A1 by kava extract and kavalactones in Hepa1c1c7. (A) Induction of CYP1A1 by kava extract (0–6.25 µg/ml). (B) Induction of CYP1A1 by six kavalactones (25µM). 1–7 represent DMSO control, 7,8-dihydrokawain, kawain, methysticin, 7,8-dihydromethysticin, yangonin, and 5,6-dehydrokawain, respectively. (C) Induction of CYP1A1 protein expression by methysticin. (D) Induction of CYP1A1 by 7,8-dihydromethysticin. Hepa1c1c7 cells were exposed kava or kavalactones for 24 h. Cell lysates (40 µg) were analyzed by western blot for levels of CYP1A1. All the data are generated from at least three independent experiments. The ratios of CYP1A1 protein against β-actin were calculated; values shown are mean ± SD. *p < 0.05; **p < 0.01 versus control.
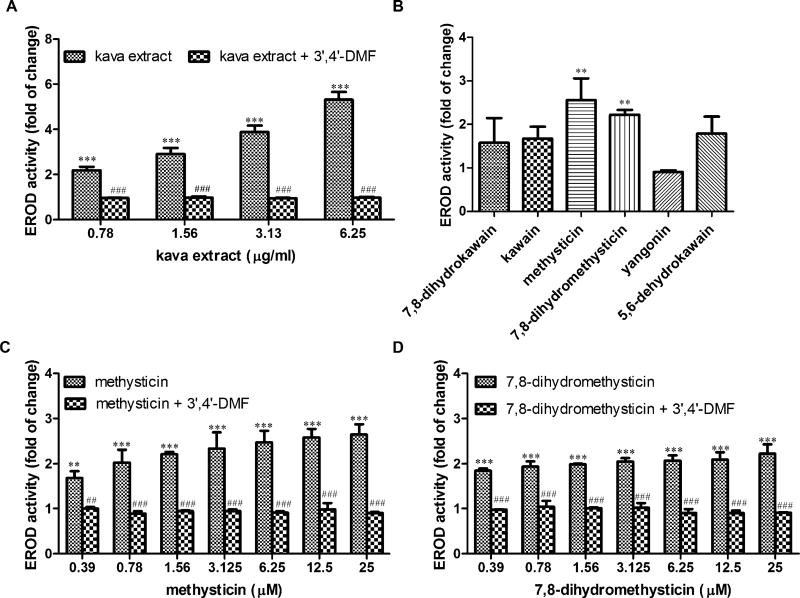
Because kava treatment resulted in elevated CYP1A1 gene expression and protein expression, we investigated whether similar changes were evident at the enzymatic activity level. The effect of kava extract and kavalactones on CYP1A1-mediated enzymatic activity was determined by measuring EROD activity. Both kava extract (0.78–6.25 µg/ml) and kavalactones methysticin and 7,8-dihydromethysticin (25µM) increased CYP1A1 enzymatic activity (Figs. 4A and 4B). Furthermore, kava extract and methysticin had concentration-dependent effects (Figs. 4A and 4C); 7,8-dihydromethysticin showed slight concentration dependency (Fig. 4D). Induction by 7,8-dihydromethysticin was nearly saturated at 0.39µM, with little increase as the concentration was increased to 25µM (Fig. 4D, data with addition of 3′,4′-DMF will be described later).
FIG. 4.
Effects of kava extract and kavalactones on CYP1A1 activity induction. (A) Induction of CYP1A1 activity by kava extract in the absence or presence of 1µM 3′,4′-DMF for 24 h. (B) Induction of CYP1A1 activity by six kavalactones (25µM). (C) Induction of CYP1A1 activity by methysticin. (D) Induction of CYP1A1 activity by 7,8-dihydromethysticin. Hepa1c1c7 cells were exposed to kava extract (0–6.25 µg/ml), methysticin (0 or 25µM), or 7,8-dihydromethysticin (0 or 25µM) in the absence or presence of 3”,4′-DMF for 24 h. Activity were measured by EROD assay. Fold induction was calculated in comparison with DMSO control. The data represent at least three independent experiments. The values shown are mean ± SD. **p < 0.01; ***p < 0.001 versus control. ##p < 0.01; ###p < 0.001, significantly different from the treatment without 3′,4′-DMF.
Effects of Kava Extract and Kavalactones on Interaction with AhR
Induction of CYP1A1 by drugs or chemicals such as β-naphthoflavone, 3-methylcholanthrene, and 2,3,7,8-tetra-chlorodibenzo-p-dioxin (TCDD) has generally been accepted as a ligand-dependent AhR activation (Whitlock, 1999). AhR-independent induction of CYP1A1 has been reported (Delescluse et al., 2000). For example, retinoic acid induced CYP1A1 via the Retinoid Acid Responsive Element (Vecchini et al., 1994, 1995), and protein tyrosine kinase transaction pathway played a role in omeprazole-mediated CYP1A1 activation (Backlund et al., 1997).
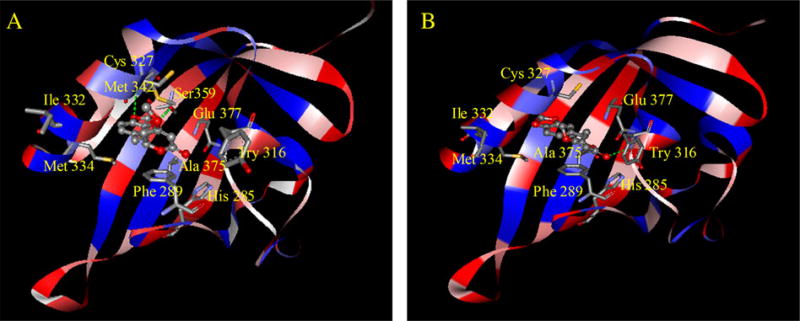
To determine the relationships of kavalactones with AhR, in silico homology modeling and molecular docking were employed. Three proteins, 1P97, 1WA9, and 2B02, were selected as templates for mouse AhR homology modeling based on the results from PSI-BLAST (Supplementary figs. 1 and 2). The protein names, identities (%), E-values of three templates, and sequence alignments are listed in Supplementary table 2. The established homology model is displayed in Figure 5. Previous mutation studies have shown that His 285, Try 316, Cys 327, Ile 332, Met 334, Ala 375, and Glu 377 are the key residues for ligand binding (Bisson et al., 2009; Pandini et al., 2007). As demonstrated in Figure 5, those key residues except Ile 332 were located in the binding pocket with their side chains pointing toward the cavity. Additionally, the binding pocket is characterized by a hydrophilic bottom and a hydrophobic entrance (Fig. 5).
FIG. 5.
The interaction of AhR with methysticin (A) and 7,8-dihydromethysticin (B) with the AhR ligand binding domain, predicted by molecular docking in the homology model. The AhR protein backbone is displayed as a ribbon and colored by hydrophobic properties (blue: hydrophobic; red: hydrophilic); methysticin and 7,8-dihydromethysticin are displayed as ball and stick models, colored by atom type; side chains of amino acids are displayed as sticks and colored by atom type.
After the establishment of homology model of mAhR (Supplementary fig. 2), six kava constituents were docked into the ligand-binding pocket. As tabulated in Table 1, the surflex docking scores for six kavalactones are greater than 6 with methysticin being the greatest (8.31), followed by 7,8-dihydromethysticin (7.95) (greater score means stronger binding ability). This result suggests that methysticin and 7,8-dihydromethysticin are best able to bind to the AhR pocket. In order to integrate multiple scoring functions for a consensus evaluation, a C-score was also calculated. Multiple scoring functions (C-score, D-score, G-score, ChemScore, PMF score, and Surflex-dock score) can produce a consensus that is more robust and accurate than any single function for evaluating ligand-receptor interactions. The C-score should be 0–5, and the larger value represents the better consensus. In our calculation, C-scores for kava constituents are 4 except for 5,6-dehydrokawain being 2 (Table 1). In addition, both Surflex and C-scores were also calculated for three well-known CYP1A1 inducers, 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD), benzo[a]pyrene (BaP), and 3-methylcholanthene (3-MC).
TABLE 1.
Molecular Docking Scores
| Compound name | Surflex score | C-score |
|---|---|---|
| Methysticin | 8.31 | 4 |
| 7,8-Dihydromethysticin | 7.95 | 4 |
| Yangonin | 6.98 | 4 |
| 7,8-Dihydrokawain | 6.84 | 4 |
| Kawain | 6.42 | 4 |
| 5,6-Dehydrokawain | 6.34 | 2 |
| 2,3,7,8-Tetrachlorodibenzo-p-dioxin (TCDD) | 7.13 | 4 |
| Benzo[a]pyrene (BaP) | 8.28 | 4 |
| 3-Methylcholanthene (3-MC) | 10.45 | 4 |
The optimal docking poses of methysticin and 7,8-dihydromethysticin are shown in Figures 5A and 5B, respectively. Two hydrogen bonds were formed between methysticin (oxygen of the methylenedioxyphenyl [MDP] group) and Met 342 (hydrogen of -NH-) and Ser 359 (hydrogen of -OH), and one hydrogen bond was formed between 7,8-dihydromethysticin (oxygen of C=O) and Try 316 (hydrogen of -NH-), one of the key residues for ligand binding. Both methysticin and 7,8-dihydromethysticin interacted with Phe 289 by Pi-Pi interactions. However, the other four kavalactones did not show such tight binding patterns (Supplementary figs. 3–6).
Our in silico molecular docking results demonstrated that methysticin and 7,8-dihydromethysticin may interact with AhR in the same way as its ligands, suggesting that CYP1A1 induction may be via the activation of the AhR signaling pathway. To experimentally validate the outcomes of in silico calculations, we performed reporter gene assays to investigate the ability of kava and kavalactones to activate the AhR. Briefly, pGudluc 6.1, a DNA plasmid construct that contains a luciferase reporter gene fused to the XRE of the CYP1A1 promoter, was used as the reporter plasmid. Hepa1c1c7 cells were cotransfected with pGudluc 6.1 and an internal control plasmid, pGL4.74 [hRluc/TK] (Renilla luciferase). After transfection, cells were then treated with kava extract and kavalactones at various concentrations for 24 h. A concentration-dependent increase of luciferase activity was observed for kava extract treatments (0.78–6.25 µg/ml) (Fig. 6A). Luciferase activity was significantly increased by methysticin and 7,8-dihydromethysticin treatment (25µM) to 4.2- and 3.5-fold, respectively (Fig. 6B). Although slight increases were observed for treatments with 7,8-dihydrokawain, kawain, 5,6-dehydrokawain, and yangonin in comparison with the control, the differences were not statistically significant (data not shown).
FIG. 6.
Activation of AhR-dependent reporter activity by kava extract and kavalactones. (A) Kava extract. (B) Methysticin and 7,8-dihydromethysticin. Cells were cotransfected with pGudluc 6.1 (a CYP1A1 promoter-driven luciferase reporter plasmid) and an internal control plasmid (pGL4.74 [hRluc/TK]). Twenty-four hours after transfection, kava extract or kavalactones was added at the concentrations as indicated and cells were harvested after 24 h treatment. Firefly luciferase activity level was normalized against Renilla activity level. Fold induction was calculated in comparison with DMSO control. Error bars represent SDs of three replicates. **p < 0.01; ***p < 0.001 versus control.
Effects of an AhR Antagonist on Kava- and Kavalactone-Mediated CYP1A1 Induction
To confirm further whether CYP1A1 induction associated with kava extract, methysticin, and 7,8-dihydromethysticin is mediated by AhR, an AhR antagonist 3′,4′-dimethoxyflavone (3′,4′-DMF) (Lee and Safe, 2000) and an AhR-deficient cell line (CRL-2710) were used.
Hepa1c1c7 cells were cultured with kava extract, methysticin, and 7,8-dihydromethysticin in the presence or absence of 1µM 3′,4′-DMF (1µM 3′,4′-DMF did not alter EROD activity or CYP1A1 gene expression; data not shown). As expected, kava extract-, methysticin-, and 7,8-dihydromethysticin-induced CYP1A1 gene expression (Figs. 2A, 2C, and 2D) as well as EROD activity (Figs. 4A, 4C, and 4D) were significantly blocked by the addition of 3′,4′-DMF.
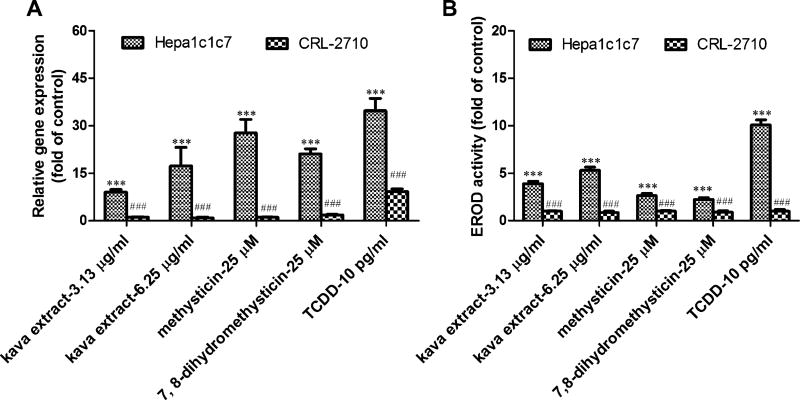
In CRL-2710 cells lacking AhR, kava-, methysticin-, and 7,8-dihydromethysticin-mediated CYP1A1 induction was completely abolished at both gene expression level (Fig. 7A) and enzymatic activity level (Fig. 7B). TCDD, a typical CYP1A1 inducer, was used as a positive control to confirm the lack of response in CRL-2710 cells to AhR.
FIG. 7.
CYP1A1 induction in an AhR-deficient cell line (CRL-2710) is resistant to kava extract and kavalactones. (A) CYP1A1 gene expression induced by kava extract and methysticin, and 7,8-dihydromethysticin was inhibited in CRL-2710 cells. (B) CYP1A1 activity induced by kava extract, methysticin, or 7,8-dihydromethysticin was abolished in CRL-2710 cells. Cells were incubated with kava extract (3.13 and 6.25 µg/ml), methysticin (25µM), or 7,8-dihydromethysticin (25µM) for 24 h. reverse transcription-PCR and EROD assays were used to examine CYP1A1 gene expression and CYP1A1 activity. Ten pg/ml TCDD, a typical AhR-dependent CYP1A1 inducer, was used as a positive control to confirm the behavior of CRL-2710 cells. ***p < 0.001 versus control. ###p < 0.001, significantly different for the treatment in Hepa1c1c7 comparing with that in CRL-2710 cells.
DISCUSSION
We have been interested in studying dietary supplements and have evaluated a number of herbal dietary supplements (Chen et al., 2006; Guo et al., 2007, 2008; Mei et al., 2005, 2006). One of our previous in vivo studies showed that CYP1A1 gene expression was significantly upregulated in response to kava extract treatment (Guo et al., 2009, 2010b). In the present in vitro study, we investigated the effect of kava on CYP1A1 expression and AhR activation. Our data demonstrated that kava extract induced CYP1A1 at the level of gene expression, protein expression, and enzymatic activity (Figs. 2A, 3A, and 4A). Luciferase activity was induced by kava extract in the cells transfected with a reporter DNA plasmid containing the CYP1A1 promoter (Fig. 6A), indicating that kava extract induced CYP1A1 activity at the transcriptional level. We also demonstrated that the induction of CYP1A1 by kava was effectively blocked by an AhR antagonist (Figs. 4A and 4B) and completely abolished in the AhR-deficient cells (Fig. 7). Collectively, our data suggest that kava extract induces CYP1A1 mediated by the AhR signaling pathway.
It is important to determine which kava constituents are responsible for the inducing effect on CYP1A1, although it is difficult to single out because kava extract is a mixture composed of many substances. The kava extract used in our study has a high abundance of kavalactones (~30%). Kavalactones are considered to have pharmacological activity (Fu et al., 2008). For example, kawain, dihydrokawain, methysticin, and dihydromethysticin exhibit analgesic effects (Jamieson and Duffield, 1990); and yangonin and methysticin show antioxidant activities (Wu et al., 2002). We, therefore, focused on six major kavalactones to investigate their capacities for CYP1A1 induction. We found that two kavalactones, namely, methysticin and 7,8-dihydromethysticin, were able to induce CYP1A1 (Figs. 2, 3, and 4). Furthermore, the induction by these two kavalactones was significantly inhibited by an AhR antagonist and abolished in AhR-deficient cells. Additionally, our molecular docking results revealed that methysticin and 7,8-dihydromethysticin are the two molecules with the greatest potential to interact with the AhR. Combining biological observations with in silico predictions, our results demonstrate that methysticin and 7,8-dihydromethysticin are the two major kavalactones in kava extract to induce CYP1A1 activity through the AhR signaling pathway.
In this study, the maximum concentration of kava extract tested was 6.25 µg/ml, approximately equivalent to 2.21µM kawain, 2.15µM 7,8-dihydrokawain, 1µM yangonin, 0.66µM 5,6-dehydrokawain, 0.87µM 7,8-dihydromethysticin, and 0.80µM methysticin. We treated cells with kavalactones with numerous concentrations, from 0.39 to 25µM; this broad concentration range enabled us not only to test the concentration dependency but also to compare kavalactones and the kava extract. For example, the induction of CYP1A1 in gene expression, protein induction, and enzymatic activity induction by 0.78µM methysticin or 7,8-dihydromethysticin was similar to that by 6.25 µg/ml kava extract (Figs. 2–4). In contrast, at the same concentration, the other four kavalactones had no effect, indicating that the induction of CYP1A1 by kava may be attributed to methysticin and 7,8-dihydromethysticin and not to the other kavalactones.
Because methysticin and 7,8-dihydromethysticin were identified as the main CYP1A1 inducers of the six major kavalactones, it is worth exploring the features, which distinguish these two constitutes from the others. Our molecular docking results indicate that hydrogen bonds were formed between AhR and the MDP group in methysticin or 7,8-dihydromethysticin. It was reported, in a paper studying chemical structure-activity relationships, that MDP is one of the molecular features important in inducing CYP1A1 (Ryu et al., 1995). Interestingly, among six kavalactones, only methysticin and 7,8-dihydromethysticin (Fig. 1) contain the MDP group. We suspect that this unique characteristic of these two compounds is important in CYP1A1 induction. This is further supported by the findings of our molecular docking study, showing that the bottom of the AhR binding pocket is hydrophilic and the entrance of the pocket is hydrophobic. The MDP group may enhance the binding affinity, possibly by hydrogen bonding or electrostatic interactions between the oxygen of the MDP group and the hydrogen of the amino acids located in the bottom of the pocket (Fig. 5).
CYP1A1 is known to be responsible for the metabolism of procarcinogens such as PAHs to their potent carcinogenic derivatives (Shimada et al., 1996). The basal level of CYP1A1 expression is low in human liver (Edwards, 1998) but can be induced by a variety of compounds. The induction is believed to be regulated via AhR activation (Ma, 2001). Historical studies in metabolic activation of PAHs helped recognize the role of CYP1A1 in chemical-induced toxicity (Conney, 1982; Miller and Miller, 1981; Phillips, 1983). They also led to the idea that prescription drugs, exogenous chemicals, or CAMs involved in CYP1A1 induction may aggravate toxicity by bioactivating procarcinogens (Rajaraman et al., 2009). It was reported that estrogen exposure of MCF-7 human breast cancer cells enhanced the formation of benzo[a]pyrene (BaP)-DNA adducts (Spink et al., 2009), and the authors suggested that enhancement of the AhR-mediated induction of CYP1A1 by estrogen led to a higher capacity for metabolic activation of BaP to an ultimate carcinogen.
Besides the inductive effect of CYP, several studies have showed that kava extract and some kavalactones are potent inhibitors of CYP450 enzymes such as CYP1A2, 2C9, 2C19, 2D6, 3A4, and 4A9/11 (Mathews et al., 2002, 2005; Unger et al., 2002; Zou et al., 2002).
The modulation of these metabolizing enzymes by kava extract may potentially cause liver injury and pharmacokinetic interactions because pharmaceutical agents or chemicals that are metabolized by any of these CYPs might cause elevated and potentially toxic plasma concentrations of their parent compounds when coadministered with kava (Anke and Ramzan, 2004).
Pacific Islanders regularly consume approximately 100–1000 mg kava daily (Kilham, 1996) and the recommended daily intake for kava is equivalent to 120 mg kavalactones (Bundesgesundheitsamt, 1990). Patients taking kava or kavalactones as a remedy for anxiety respond favorably at 60–240 mg kavalactones per day (Pittler and Ernst, 2003). It is postulated that these patients have an average blood/plasma concentration of 80–160µM kavalactones, assuming complete oral absorption and a blood volume of 6 l (Anke and Ramzan, 2004). In humans, a single oral dose of 200 mg synthetic (±)-kawain resulted in a maximum plasma concentration of 18 µg/ml (78µM) (Singh, 2004). The maximum concentration of 25µM (kavalactones) and 6.25 µg/ml (kava extract) used in the present study is within the range of clinical plasma concentration. Thus, the impact of CYP1A1 induction by kava extract and kavalactones is clinically relevant and may raise concern regarding the risks associated with the use of kava or kavalactones. Dietary supplements are used on a traditional basis; long-term consumption is often reported. Therefore, future studies will investigate the secondary effects of long-term consumption of kava or kavalactones on CYP1A1-mediated chemical carcinogenesis in animals and provide further information about the influence of kava on CYP1A1-mediated chemical carcinogenesis.
Supplementary Material
Acknowledgments
We thank Dr William Melchior for his critical review of this manuscript. This article is not an official guidance or policy statement of the U.S. FDA. No official support or endorsement by the U.S. FDA is intended or should be inferred.
FUNDING
Y.L., H.M., and Q.W. were supported by the appointments to the Postgraduate Research Program at the National Center for Toxicological Research administered by Oak Ridge Institute for Science Education through an interagency agreement between the U.S. Department of Energy and the U.S. FDA. S.Z. was supported by the U.S. FDA’s International Scientist Exchange Program.
Footnotes
Supplementary data are available online at http://toxsci.oxfordjournals.org/.
References
- Anke J, Ramzan I. Pharmacokinetic and pharmacodynamic drug interactions with Kava (Piper methysticum Forst. f.) J. Ethnopharmacol. 2004;93:153–160. doi: 10.1016/j.jep.2004.04.009. [DOI] [PubMed] [Google Scholar]
- Backlund M, Johansson I, Mkrtchian S, Ingelman-Sundberg M. Signal transduction-mediated activation of the aryl hydrocarbon receptor in rat hepatoma H4IIE cells. J. Biol. Chem. 1997;272:31755–31763. doi: 10.1074/jbc.272.50.31755. [DOI] [PubMed] [Google Scholar]
- Bisson WH, Koch DC, O’Donnell EF, Khalil SM, Kerkvliet NI, Tanguay RL, Abagyan R, Kolluri SK. Modeling of the aryl hydrocarbon receptor (AhR) ligand binding domain and its utility in virtual ligand screening to predict new AhR ligands. J. Med. Chem. 2009;52:5635–5641. doi: 10.1021/jm900199u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bressler R. Herb-drug interactions: Interactions between kava and prescription medications. Geriatrics. 2005;60:24–25. [PubMed] [Google Scholar]
- Bundesgesundheitsamt. Monographie: Piperis methystici rhizoma (Kava-Kava Wurzelstock). [Federal Office for Medicines and Medicinal Products. Monograph: Kava-Kava] 1990. p. 101. [Google Scholar]
- Chen T, Guo L, Zhang L, Shi L, Fang H, Sun Y, Fuscoe JC, Mei N. Gene expression profiles distinguish the carcinogenic effects of aristolochic acid in target (kidney) and non-target (liver) tissues in rats. BMC Bioinformatics. 2006;7(Suppl. 2):S20. doi: 10.1186/1471-2105-7-S2-S20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton NP, Yoshizawa K, Kissling GE, Burka LT, Chan PC, Nyska A. Immunohistochemical analysis of expressions of hepatic cytochrome P450 in F344 rats following oral treatment with kava extract. Exp. Toxicol. Pathol. 2007;58:223–236. doi: 10.1016/j.etp.2006.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouatre DL. Kava kava: Examining new reports of toxicity. Toxicol. Lett. 2004;150:85–96. doi: 10.1016/j.toxlet.2003.07.005. [DOI] [PubMed] [Google Scholar]
- Conney AH. Induction of microsomal enzymes by foreign chemicals and carcinogenesis by polycyclic aromatic hydrocarbons: G. H. A. Clowes Memorial Lecture. Cancer Res. 1982;42:4875–4917. [PubMed] [Google Scholar]
- Coumailleau P, Poellinger L, Gustafsson JAǺ, Whitelaw ML. Definition of a minimal domain of the dioxin receptor that is associated with Hsp90 and maintains wild type ligand binding affinity and specificity. J. Biol. Chem. 1995;270:25291. doi: 10.1074/jbc.270.42.25291. [DOI] [PubMed] [Google Scholar]
- De Smet PA. Health risks of herbal remedies. Drug Saf. 1995;13:81–93. doi: 10.2165/00002018-199513020-00003. [DOI] [PubMed] [Google Scholar]
- De Smet PA. Health risks of herbal remedies: An update. Clin. Pharmacol. Ther. 2004;76:1–17. doi: 10.1016/j.clpt.2004.03.005. [DOI] [PubMed] [Google Scholar]
- Delescluse C, Lemaire G, De Sousa G, Rahmani R. Is CYP1A1 induction always related to AHR signaling pathway? Toxicology. 2000;153:73–82. doi: 10.1016/s0300-483x(00)00305-x. [DOI] [PubMed] [Google Scholar]
- Donato MT, Gomez-Lechon MJ, Castell JV. A microassay for measuring cytochrome P450IA1 and P450IIB1 activities in intact human and rat hepatocytes cultured on 96-well plates. Anal. Biochem. 1993;213:29–33. doi: 10.1006/abio.1993.1381. [DOI] [PubMed] [Google Scholar]
- Edwards RJ. Targeting antipeptide antibodies towards cytochrome P450 enzymes. Methods Mol. Biol. 1998;107:239–249. doi: 10.1385/0-89603-519-0:239. [DOI] [PubMed] [Google Scholar]
- Ernst E. The Desktop Guide to Complementary and Alternative Medicine: An Evidence-Based Approach. Mosby; St Louis, MO: 2001. [Google Scholar]
- Ernst E. Kava update: A European perspective. N. Z. Med J. 2004;117:1205. [PubMed] [Google Scholar]
- Fu PP, Xia Q, Guo L, Yu H, Chan PC. Toxicity of kava kava. J. Environ. Sci. Health C Environ. Carcinog. Ecotoxicol. Rev. 2008;26:89–112. doi: 10.1080/10590500801907407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fugh-Berman A. Herb-drug interactions. Lancet. 2000;355:134–138. doi: 10.1016/S0140-6736(99)06457-0. [DOI] [PubMed] [Google Scholar]
- Gautz LD, Kaufusi P, Jackson MC, Bittenbender HC, Tang CS. Determination of kavalactones in dried kava (Piper methysticum) powder using near-infrared reflectance spectroscopy and partial least-squares regression. J. Agric. Food Chem. 2006;54:6147–6152. doi: 10.1021/jf060964v. [DOI] [PubMed] [Google Scholar]
- Guo L, Mei N, Dial S, Fuscoe J, Chen T. Comparison of gene expression profiles altered by comfrey and riddelliine in rat liver. BMC Bioinformatics. 2007;8(Suppl. 7):S22. doi: 10.1186/1471-2105-8-S7-S22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L, Shi Q, Fang JL, Mei N, Ali AA, Lewis SM, Leakey JE, Frankos VH. Review of usnic acid and Usnea barbata toxicity. J. Environ. Sci. Health C Environ. Carcinog. Ecotoxicol. Rev. 2008;26:317–338. doi: 10.1080/10590500802533392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L, Li Q, Xia Q, Dial S, Chan PC, Fu PP. Analysis of gene expression changes of drug metabolizing enzymes in the livers of F344 rats following oral treatment with kava extract. Food Chem. Toxicol. 2009;47:433–442. doi: 10.1016/j.fct.2008.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L, Mei N, Xia Q, Chen T, Chan PC, Fu PP. Gene expression profiling as an initial approach for mechanistic studies of toxicity and tumorigenicity of herbal plants and herbal dietary supplements. J. Environ. Sci. Health C Environ. Carcinog. Ecotoxicol. Rev. 2010a;28:60–87. doi: 10.1080/10590500903585416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L, Shi Q, Dial S, Xia Q, Mei N, Li QZ, Chan PC, Fu PP. Gene expression profiling in male B6C3F1 mouse livers exposed to kava identifies-changes in drug metabolizing genes and potential mechanisms linked to kava toxicity. Food Chem. Toxicol. 2010b;48:686–696. doi: 10.1016/j.fct.2009.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurley BJ, Gardner SF, Hubbard MA, Williams DK, Gentry WB, Khan IA, Shah A. In vivo effects of goldenseal, kava kava, black cohosh, and valerian on human cytochrome P450 1A2, 2D6, 2E1, and 3A4/5 phenotypes. Clin. Pharmacol. Ther. 2005;77:415–426. doi: 10.1016/j.clpt.2005.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurley BJ, Swain A, Hubbard MA, Hartsfield F, Thaden J, Williams DK, Gentry WB, Tong Y. Supplementation with goldenseal (Hydrastis canadensis), but not kava kava (Piper methysticum), inhibits human CYP3A activity in vivo. Clin. Pharmacol. Ther. 2008;83:61–69. doi: 10.1038/sj.clpt.6100222. [DOI] [PubMed] [Google Scholar]
- Hu Z, Yang X, Ho PC, Chan SY, Heng PW, Chan E, Duan W, Koh HL, Zhou S. Herb-drug interactions: A literature review. Drugs. 2005;65:1239–1282. doi: 10.2165/00003495-200565090-00005. [DOI] [PubMed] [Google Scholar]
- Jain AN. Surflex: Fully automatic flexible molecular docking using a molecular similarity-based search engine. J. Med. Chem. 2003;46:499–511. doi: 10.1021/jm020406h. [DOI] [PubMed] [Google Scholar]
- Jain AN. Surflex-Dock 2.1: Robust performance from ligand energetic modeling, ring flexibility, and knowledge-based search. J. Comput. Aided Mol. Des. 2007;21:281–306. doi: 10.1007/s10822-007-9114-2. [DOI] [PubMed] [Google Scholar]
- Jamieson DD, Duffield PH. The antinociceptive actions of kava components in mice. Clin. Exp. Pharmacol. Physiol. 1990;17:495–507. doi: 10.1111/j.1440-1681.1990.tb01349.x. [DOI] [PubMed] [Google Scholar]
- Jiang TF, Wang YH, Lv ZH, Yue ME. Determination of kava lactones and flavonoid glycoside in Scorzonera austriaca by capillary zone electrophoresis. J. Pharm. Biomed. Anal. 2007;43:854–858. doi: 10.1016/j.jpba.2006.08.024. [DOI] [PubMed] [Google Scholar]
- Kilham C. Kava: Medicine Hunting in Paradise. Park Street Press; Rochester VT: 1996. [Google Scholar]
- Koh HL, Woo SO. Chinese proprietary medicine in Singapore: Regulatory control of toxic heavy metals and undeclared drugs. Drug Saf. 2000;23:351–362. doi: 10.2165/00002018-200023050-00001. [DOI] [PubMed] [Google Scholar]
- Lee JE, Safe S. 3′,4′-dimethoxyflavone as an aryl hydrocarbon receptor antagonist in human breast cancer cells. Toxicol. Sci. 2000;58:235–242. doi: 10.1093/toxsci/58.2.235. [DOI] [PubMed] [Google Scholar]
- Ma Q. Induction of CYP1A1. The AhR/DRE paradigm: Transcription, receptor regulation, and expanding biological roles. Curr. Drug Metab. 2001;2:149–164. doi: 10.2174/1389200013338603. [DOI] [PubMed] [Google Scholar]
- Ma Y, Sachdeva K, Liu J, Ford M, Yang D, Khan IA, Chichester CO, Yan B. Desmethoxyyangonin and dihydromethysticin are two major pharmacological kavalactones with marked activity on the induction of CYP3A23. Drug Metab. Dispos. 2004;32:1317–1324. doi: 10.1124/dmd.104.000786. [DOI] [PubMed] [Google Scholar]
- Mathews JM, Etheridge AS, Black SR. Inhibition of human cytochrome P450 activities by kava extract and kavalactones. Drug Metab. Dispos. 2002;30:1153–1157. doi: 10.1124/dmd.30.11.1153. [DOI] [PubMed] [Google Scholar]
- Mathews JM, Etheridge AS, Valentine JL, Black SR, Coleman DP, Patel P, So J, Burka LT. Pharmacokinetics and disposition of the kavalactone kawain: Interaction with kava extract and kavalactones in vivo and in vitro. Drug Metab. Dispos. 2005;33:1555–1563. doi: 10.1124/dmd.105.004317. [DOI] [PubMed] [Google Scholar]
- Mei N, Guo L, Fu PP, Heflich RH, Chen T. Mutagenicity of comfrey (Symphytum Officinale) in rat liver. Br. J. Cancer. 2005;92:873–875. doi: 10.1038/sj.bjc.6602420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei N, Guo L, Zhang L, Shi L, Sun YA, Fung C, Moland CL, Dial SL, Fuscoe JC, Chen T. Analysis of gene expression changes in relation to toxicity and tumorigenesis in the livers of Big Blue transgenic rats fed comfrey (Symphytum officinale) BMC Bioinformatics. 2006;7(Suppl. 2):S16. doi: 10.1186/1471-2105-7-S2-S16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller EC, Miller JA. Mechanisms of chemical carcinogenesis. Cancer. 1981;47:1055–1064. doi: 10.1002/1097-0142(19810301)47:5+<1055::aid-cncr2820471302>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- Nelson L, Perrone J. Herbal and alternative medicine. Emerg. Med. Clin. North Am. 2000;18:709–722. doi: 10.1016/s0733-8627(05)70154-1. [DOI] [PubMed] [Google Scholar]
- Nerurkar PV, Dragull K, Tang CS. In vitro toxicity of kava alkaloid, pipermethystine, in HepG2 cells compared to kavalactones. Toxicol. Sci. 2004;79:106–111. doi: 10.1093/toxsci/kfh067. [DOI] [PubMed] [Google Scholar]
- NTP. NTP Technical Report on Toxicology and Carcinogenesis Studies of Kava Kava Extract in F344/N Rats and B6C3F1 Mice. NTP TR 571, NIH Publication No.11-5913, National Toxicology Program; Research Triangle Park NC: 2011. [Google Scholar]
- Olsen LR, Grillo MP, Skonberg C. Constituents in kava extracts potentially involved in hepatotoxicity: A review. Chem. Res. Toxicol. 2011;24:992–1002. doi: 10.1021/tx100412m. [DOI] [PubMed] [Google Scholar]
- Pandini A, Denison MS, Song Y, Soshilov AA, Bonati L. Structural and functional characterization of the aryl hydrocarbon receptor ligand binding domain by homology modeling and mutational analysis. Biochemistry. 2007;46:696–708. doi: 10.1021/bi061460t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips DH. Fifty years of benzo(a)pyrene. Nature. 1983;303:468–472. doi: 10.1038/303468a0. [DOI] [PubMed] [Google Scholar]
- Pittler MH, Ernst E. Kava extract for treating anxiety. Cochrane Database Syst. Rev. 2003;1:CD003383. doi: 10.1002/14651858.CD003383. [DOI] [PubMed] [Google Scholar]
- Rajaraman G, Yang G, Chen J, Chang TK. Modulation of CYP1B1 and CYP1A1 gene expression and activation of aryl hydrocarbon receptor by Ginkgo biloba extract in MCF-10A human mammary epithelial cells. Can. J. Physiol. Pharmacol. 2009;87:674–683. doi: 10.1139/y09-061. [DOI] [PubMed] [Google Scholar]
- Raucy JL. Regulation of CYP3A4 expression in human hepatocytes by pharmaceuticals and natural products. Drug Metab. Dispos. 2003;31:533–539. doi: 10.1124/dmd.31.5.533. [DOI] [PubMed] [Google Scholar]
- Ryu DY, Levi PE, Hodgson E. Regulation of cytochrome P-450 isozymes CYP1A1, CYP1A2 and CYP2B10 by three benzodioxole compounds. Chem. Biol. Interact. 1995;96:235–247. doi: 10.1016/0009-2797(94)03594-x. [DOI] [PubMed] [Google Scholar]
- Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993;234:779–779. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- Sali A, Overington JP. Derivation of rules for comparative protein modeling from a database of protein structure alignments. Protein Sci. 1994;3:1582–1596. doi: 10.1002/pro.5560030923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada T, Yamazaki H, Mimura M, Wakamiya N, Ueng YF, Guengerich FP, Inui Y. Characterization of microsomal cytochrome P450 enzymes involved in the oxidation of xenobiotic chemicals in human fetal liver and adult lungs. Drug Metab. Dispos. 1996;24:515–522. [PubMed] [Google Scholar]
- Singh YN. Pharmacology and toxicology of kava and kavalactones. In: Singh YN, editor. Kava: From Ethnology to Pharmacology. CRC; Boca Raton, FL: 2004. [Google Scholar]
- Singh YN, Singh NN. Therapeutic potential of kava in the treatment of anxiety disorders. CNS Drugs. 2002;16:731–743. doi: 10.2165/00023210-200216110-00002. [DOI] [PubMed] [Google Scholar]
- Spink BC, Bennett JA, Pentecost BT, Lostritto N, Englert NA, Benn GK, Goodenough AK, Turesky RJ, Spink DC. Long-term estrogen exposure promotes carcinogen bioactivation, induces persistent changes in gene expression, and enhances the tumorigenicity of MCF-7 human breast cancer cells. Toxicol. Appl. Pharmacol. 2009;240:355–366. doi: 10.1016/j.taap.2009.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su D, Li L. Trends in the use of complementary and alternative medicine in the United States: 2002–2007. J. Health Care Poor Underserved. 2011;22:296–310. doi: 10.1353/hpu.2011.0002. [DOI] [PubMed] [Google Scholar]
- Tang J, Dunlop RA, Rowe A, Rodgers KJ, Ramzan I. Kavalactones Yangonin and Methysticin induce apoptosis in human hepatocytes (HepG2) in vitro. Phytother. Res. 2011;25:417–423. doi: 10.1002/ptr.3283. [DOI] [PubMed] [Google Scholar]
- Unger M, Holzgrabe U, Jacobsen W, Cummins C, Benet LZ. Inhibition of cytochrome P450 3A4 by extracts and kavalactones of Piper methysticum (Kava-Kava) Planta Med. 2002;68:1055–1058. doi: 10.1055/s-2002-36360. [DOI] [PubMed] [Google Scholar]
- Vecchini F, Lenoir-Viale MC, Cathelineau C, Magdalou J, Bernard BA, Shroot B. Presence of a retinoid responsive element in the promoter region of the human cytochrome P4501A1 gene. Biochem. Biophys. Res. Commun. 1994;201:1205–1212. doi: 10.1006/bbrc.1994.1833. [DOI] [PubMed] [Google Scholar]
- Vecchini F, Mace K, Magdalou J, Mahe Y, Bernard BA, Shroot B. Constitutive and inducible expression of drug metabolizing enzymes in cultured human keratinocytes. Br. J. Dermatol. 1995;132:14–21. doi: 10.1111/j.1365-2133.1995.tb08618.x. [DOI] [PubMed] [Google Scholar]
- Volz HP, Kieser M. Kava-kava extract WS 1490 versus placebo in anxiety disorders-a randomized placebo-controlled 25-week outpatient trial. Pharmacopsychiatry. 1997;30:1–5. doi: 10.1055/s-2007-979474. [DOI] [PubMed] [Google Scholar]
- Whitlock JP., Jr Induction of cytochrome P4501A1. Annu. Rev. Pharmacol. Toxicol. 1999;39:103–125. doi: 10.1146/annurev.pharmtox.39.1.103. [DOI] [PubMed] [Google Scholar]
- Wu D, Yu L, Nair MG, DeWitt DL, Ramsewak RS. Cyclooxygenase enzyme inhibitory compounds with antioxidant activities from Piper methysticum (kava kava) roots. Phytomedicine. 2002;9:41–47. doi: 10.1078/0944-7113-00068. [DOI] [PubMed] [Google Scholar]
- Yamazaki Y, Hashida H, Arita A, Hamaguchi K, Shimura F. High dose of commercial products of kava (Piper methysticum) markedly enhanced hepatic cytochrome P450 1A1 mRNA expression with liver enlargement in rats. Food Chem. Toxicol. 2008;46:3732–3738. doi: 10.1016/j.fct.2008.09.052. [DOI] [PubMed] [Google Scholar]
- Yang XX, Hu ZP, Duan W, Zhu YZ, Zhou SF. Drug-herb interactions: Eliminating toxicity with hard drug design. Curr. Pharm. Des. 2006;12:4649–4664. doi: 10.2174/138161206779010440. [DOI] [PubMed] [Google Scholar]
- Yueh MF, Kawahara M, Raucy J. Cell-based high-throughput bioassays to assess induction and inhibition of CYP1A enzymes. Toxicol. In Vitro. 2005;19:275–287. doi: 10.1016/j.tiv.2004.10.003. [DOI] [PubMed] [Google Scholar]
- Zhou P, Gross S, Liu JH, Yu BY, Feng LL, Nolta J, Sharma V, Piwnica-Worms D, Qiu SX. Flavokawain B, the hepatotoxic constituent from kava root, induces GSH-sensitive oxidative stress through modulation of IKK/NF- B and MAPK signaling pathways. FASEB J. 2010;24:4722. doi: 10.1096/fj.10-163311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L, Harkey MR, Henderson GL. Effects of herbal components on cDNA-expressed cytochrome P450 enzyme catalytic activity. Life Sci. 2002;71:1579–1589. doi: 10.1016/s0024-3205(02)01913-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.