ABSTRACT
Ceramides and sphingolipid intermediates are well-established regulators of the cell cycle. In the budding yeast Saccharomyces cerevisae, the complex sphingolipid backbone, ceramide, comprises a long chain sphingoid base, a polar head group, and a very long chain fatty acid (VLCFA). While ceramides and long chain bases have been extensively studied as to their roles in regulating cell cycle arrest under multiple conditions, the roles of VLCFAs are not well understood. Here, we used the yeast elo2 and elo3 mutants, which are unable to elongate fatty acids, as tools to explore if maintaining VLCFA elongation is necessary for cell cycle arrest in response to yeast mating. We found that both elo2 and elo3 cells had severely reduced mating efficiencies and were unable to form polarized shmoo projections that are necessary for cell-cell contact during mating. They also lacked functional MAP kinase signaling activity and were defective in initiating a cell cycle arrest in response to pheromone. Additional data suggests that mislocalization of the Ste5 scaffold in elo2 and elo3 mutants upon mating initiation may be responsible for the inability to initiate a cell cycle arrest. Moreover, the lack of proper Ste5 localization may be caused by the inability of mutant cells to mobilize PIP2. We suggest that VLCFAs are required for Ste5 localization, which is a necessary event for initiating MAP kinase signaling and cell cycle arrest during yeast mating initiation.
Keywords: cell cycle, ceramide, lipid, MAPK, yeast
Introduction
Sphingolipids maintain cell fluidity and act as important signaling intermediates involved in regulating cell cycle progression, apoptosis, and differentiation.1-3 In S. cerevisiae, sphingolipids regulate cell cycle progression during yeast mating,4 under multiple stress conditions,5-9 and during meiotic diploid differentiation.10
S. cerevisiae sphingolipid biosynthesis has been well characterized and the genes have been cloned that are required for biosynthesis and turnover.10 Ceramides are the backbones for all yeast complex sphingolipids and are made up of a long chain base (LCB), a very long chain fatty acid, and a polar head group. There are 2 LCBs in yeast: dihydrosphingosine (DHS) and phytosphingosine (PHS) (Fig. 1). The carbon chain length varies between 16, 18, and 20 carbons for DHS and 18 or 20 carbons for PHS. Dihydro- and phytoceramides are synthesized from these LCBs, respectively (Fig. 1).
Figure 1.
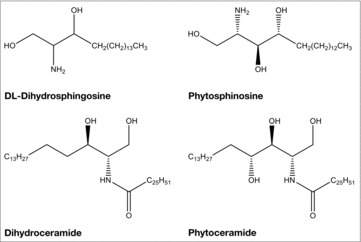
The structures of LCBs and ceramides. Shown are the structures of the LCBs, DL-dihydrosphingosine and phytosphingosine, and the ceramides, dihydroceramide and phytoceramide.
The fatty acids found in yeast complex sphingolipids are very long chain fatty acids (VLCFAs) that are 26 carbons in length, unsaturated, and contain 0–2 hydroxyl groups. Their synthesis is catalyzed by the Fasp complex,11 the Elo1/2/3 fatty acid elongases,12,13 the enoyl reductase Tsc13,14 and the 3-ketoreductase, Ifa38 (YBR159w).11,15 The Fasp complex synthesizes fatty acids that are up to 18 carbons in length. Elo1 synthesizes C16–18 medium chain fatty acids from 12–16 carbons in length. Elo2 substrates include fatty acids up to 24 carbons in length, while Elo3 is required for the synthesis of C20–26 fatty acids from C18-CoAs. Tsc13 (enoyl reductase) catalyzes the last step in each cycle of VLCFA elongation, whereas Ifa38 harbors the 3-keto reductase activity required for the reduction of 3-ketoacyl intermediates during fatty acid elongation. The loss of ELO1 alone does not result in differences in fatty acid composition, suggesting redundancy in function. elo2 elo3 double mutants on the other hand are inviable, and neither can complement the other's loss, indicating separate substrate preference and specificity.12 Tsc13 mutants are inviable, while ifa28 cells grow very poorly.
The inability to elongate fatty acids causes defects in sphingolipid composition.12 elo2 mutants accumulate phytosphingosine, and have an overall reduction in ceramides and complex sphingolipid compositions, while elo3 cells accumulate phytosphingosine and inositolphosphorylceramides (IPCs), but have reductions in the levels of ceramides, mannose inositolphosphorylceramides (MIPCs), and mannose diinositolphosphorylceramides (MIP2Cs). Both tsc13 and Ifa38 mutants accumulate dihydrosphingosine and phytosphingosine, have reduced levels of VLCFAs, and produce medium chain ceramides.11,14,15 Thus, fatty acid chain elongation is essential for producing a normal sphingolipid composition.
S. cerevisiae can exist as either a haploid or diploid. There are 2 haploid mating types, MATa and MATα.16,17 The secretion of a mating-type specific pheromone initiates mating between different haploids and subsequent binding to its cognate receptor on the opposite mating type partner. Once activated, the receptor sends a signal that activates mating-specific MAP kinase signaling, resulting in cell cycle arrest and the induction of several biologic events, all of which are necessary to produce a diploid progeny. Multiple lipid species regulate several mating events, including pheromone/receptor internalization,18,19 mating projection (shmoo) initiation and polarization,20-22 cell-cell fusion,20 sterol distribution,21 and MAP kinase signaling.21 Recently, we showed that ceramide synthesis and subsequent accumulation of ceramides were required for the regulation of cyclin gene expression, cell cycle arrest, MAP kinase signaling, Ste5 localization, proper sterol distribution, and PIP2 (phosphatidylinositol 4,5-bisphosphate) polarization during yeast mating.4 Thus, ceramides act as signaling lipids that regulate mating initiation and diploid generation.
Here, we examined if VLCFA elongation was also required for proficient mating and pheromone-specific cell cycle arrest. Our data supports the idea that maintaining VLCFA elongation is necessary for both events, as loss of Elo2 or Elo3 causes a severe reduction in mating efficiency and the inability to arrest the cell cycle in response to the presence of pheromone. Interestingly, we found that elo2 and elo3 mutants shared many of the mating defects seen in cells deficient in ceramide synthesis (lac1 lag1), including the inability to 1) form shmoo, 2) inhibit mating-specific cyclin gene expression, 3) initiate cell cycle arrest, 4) activate MAP kinase signaling, 4) properly localize Ste5, and 5) polarize PIP2 to sites of shmoo formation. Thus, proper VLCFA chain length must be maintained to initiate a mating-specific cell cycle arrest that is required for diploid progeny generation.
Results
VLCFA synthesis is required for yeast to mate
Little is known about whether VLCFA elongation is necessary for haploid yeast mating. To address this question, we determined the mating efficiencies of cells defective in VLCFA elongation. Wild type (ELO1 ELO2 ELO3), elo1, elo2, and elo3 mutants were mated to wild-type cells or the opposite mating type partner, and mating efficiencies were determined using quantitative limited mating assays.
The mating efficiency of elo1 cells was similar to that seen for wild-type cells, whether they were mated to the opposite wild type or elo1 mating partner (Table 1). On the other hand, elo2 and elo3 cells had severely reduced mating efficiencies. elo2 mating efficiencies were reduced 9-fold and 8.5-fold when cells were mated to MATa and MATα wild-type cells, respectively. Mating efficiencies were further reduced when MATa elo2 and MATα elo2 cells were mated to the opposite elo2 mating type partner (Table 1). In fact, no mating was observed between elo2 X elo2 crosses. elo3 cells also had severely reduced mating efficiencies when mated to MATa and MATα wild-type cells (∼7-fold and 3.4-fold, respectively). Mating efficiencies were further reduced when cells were mated to MATa elo3 and MATα elo3 partners (∼46-fold and 100-fold, respectively) (Table 1). In all cases, ectopic expression of the corresponding wild-type gene suppressed the observed mating defects (Table 1). Thus, VLCFA elongation is required for maintaining normal mating efficiency.
Table 1.
Quantitative mating efficiencies.
|
MATα tester strain | ||||||
|---|---|---|---|---|---|---|
| MATa | WT | elo1 | elo2 | elo2 pRS-ELO2 | elo3 | elo3 pRS-ELO3 |
| WT | 23.1 ± 2.3 | 18.7 ± 1.6 | 2.6 ± 1.1 | 16.8 ± 2.6 | 3.4 ± 0.9* | 22.6 ± 1.3 |
| mutant | 21.4 ± 2.8 | <0.01 | 17.3 ± 1.5 | 0.5 ± 0.2* | 21.7 ± 1.2 | |
|
MATa tester strain | ||||||
|
MATα |
WT |
elo1 |
elo2 |
elo2 pRS-ELO2 |
elo3 |
elo3 pRS-ELO3 |
| WT | 10.2 ± 0.9 | 9.13 ± 2.1 | 1.2 ± 0.2 | 8.6 ± 0.7 | 2.9 ± 0.5* | 11.9 ± 1.7 |
| mutant | 11.3 ± 3.9 | <0.01 | 6.8 ± 1.8 | 0.1 ± 0.0* | 14.1 ± 2.3 | |
Mating efficiency is represented as the ratio of diploid colonies per haploid colonies counted
(mean ± SEM, n = 5,
p < 0.001). Cultures were grown in YEPD before mating assay.
MATa/α, wild type.
VLCFA synthesis is required for ceramide accumulation during mating
elo2 and elo3 mutants have defects in ceramide and complex sphingolipid compositions.12 They accumulate higher levels of LCBPs and contain less ceramides than wild type cells. To begin to understand the molecular basis for elo2 and elo3 cells reduced mating efficiencies, ceramides and complex sphingolipid compositions were determined in the absence or presence of pheromone. Ceramides and complex sphingolipids were radiolabeled with [3H]DHS and [3H]inositol, respectively. Thin layer chromatography resolved individual lipid species and scintillation counting was used to quantify levels.
In the absence of pheromone, wild type and elo1 cells synthesized similar levels of total [3H]inositol-radiolabeled complex sphingolipids (Fig. 2A), while elo2 and elo3 cells had reduced levels when compared with wild-type cells (Fig. 2A). Comparing the level of individual IPC species between mutant and wild-type cells revealed that elo2 cells did not synthesize IPC-B (Fig. 2B), produced lower levels of MIPC and MIP2C (not shown), and accumulated IPC-D (Fig. 2B), while elo3 cells lacked production of IPC-C (Fig. 2B), had lower levels of MIPC and MIP2C (not shown), and had increased levels of IPC-A and IPC-B species (Fig. 2B).
Figure 2.
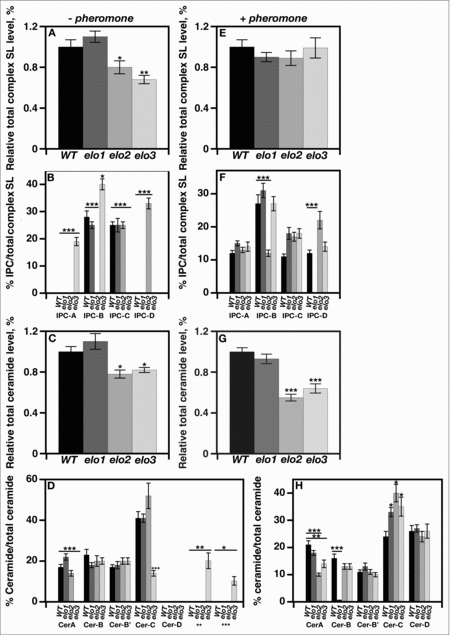
VLCFA elongation is required for maintaining the appropriate sphingolipid composition in response to mating initiation. Radiolabeling of cells and subsequent separation of lipids by TLC determined the levels of various complex sphingolipids, inositolphosphorylceramides, and ceramides. Individual sphingolipid levels were assayed using scintillation counting and were compared with the total level of complex sphingolipid, and represented as a percentage. n = 5, *p < 0.01, **p < 0.001, ***p < 0.0001.
In the absence of pheromone, elo2 and elo3 cells also produced less total [3H]DHS-radiolabeled ceramides than wild-type or elo1 cells (Fig. 2C). elo2 cells had decreased levels of Cer-A (Fig. 2D), while elo3 cells had decreased levels of Cer-A and Cer-C levels, and accumulated 2 unknown [3H]DHS-radiolabeled lipids (Fig. 2D). Cer-D accumulation was not observed in any of the strains tested.
In the presence of pheromone, all cells produced similar levels of total complex sphingolipids after a 4 hr treatment (Fig. 2E). They produced similar levels of IPC-A, which is an IPC species that was not produced by wild type, elo1, and elo2 cells in the absence of pheromone (Fig. 2F). elo1 cells lacked any detectable IPC-D, and elo2 cells had lower levels of IPC-B, when both were compared with wild type cells (Fig. 2F). All IPC species produced by elo3 cells were at levels seen for wild type cells (Fig. 2F).
In contrast, elo2 and elo3 cells produced lower levels of total ceramides in the presence of pheromone (Fig. 2G). elo2 and elo3 cells had lower levels of Cer-A compared with wild type cells (Fig. 2H), while elo1 cells had severely reduced levels of Cer-B (Fig. 2H). All strains produced similar levels of Cer-B, Cer-B’, and Cer-D (Fig. 2H).
Shmoo formation requires full length VLCFAs
Mating haploids form polarized shmoo projections.23 Shmoo are sites of conjugation and are required for cell-cell contact during mating. We previously found that cells lacking ceramide synthesis harbored defects in shmoo formation.4 Thus, we determined if elo2 and elo3 cells also lacked this ability. Shmoo formation was initiated by treating cells with α-factor, and the percentage formed was determined over time. All strains contained a bar1::URA3 allele, which made them hypersensitive to pheromone.24
There was a time-dependent increase in the percentage of shmoo formed by bar1 (closed circles) and elo1 bar1 cells (open circles) (Fig. 3). 85% and 92% of cells had polarized projections by 4 hr post pheromone induction, respectively (Fig. 3). elo2 bar1 (closed diamonds) and elo3 bar1 cells (open circles) were delayed in forming shmoo, with levels reaching only 52% and 45% of that seen for bar1 cells, respectively. Thus, there is a direct correlation between defects in VLCFA elongation and a delay in shmoo formation.
Figure 3.
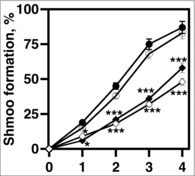
VLCFA elongation is required for shmoo formation in response to mating pheromone. 1 × 107 cells/ml were incubated with 20 μg/ml α factor and cell aliquots were collected at 0, 1, 2, 3, 4, hr. The percentages of shmoo formed per 300 cells were determined using light microscopy. bar1 cells (black circles); elo1 cells (open circles), elo2 cells (closed diamonds); elo3 cells (open diamonds). n = 5, *p < 0.01, ***p < 0.0001.
Mating-specific cell cycle arrest requires VLCFA synthesis
Yeast haploids arrest in G1 in response to pheromone.16 Reducing CLN1/2 G1 cyclin gene expression is necessary for initiating cell cycle arrest. To examine if maintaining VLCFA elongation was required for reducing cyclin gene expression in response to pheromone, CLN1 and CLN2 levels were determined using qRT-PCR. Cells were treated with 20 μg/ml α-factor to initiate cell cycle arrest.
bar1 and elo1 bar1 cells had initial increases in CLN1 (white bars) and CLN2 (black bars) expression levels in response to pheromone (Fig. 4A & B). The increases in expression lasted for 2 hr and decreased thereafter. Levels ended up lower then those seen in the absence of pheromone. On the other hand, elo2 bar1 and elo3 bar1 cells were unable to reduce CLN1/2 expression levels in response to pheromone (Fig. 4C & D). Relative levels were similar to those observed in the absence of pheromone.
Figure 4.
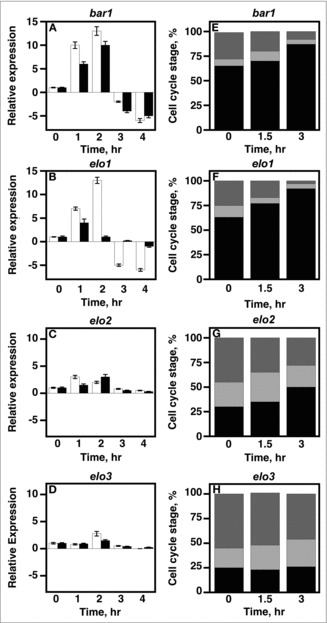
VLCFA elongation is required for cell cycle arrest during yeast mating. A-D, 1 × 107 cells/ml were incubated with 20 μg/ml α factor and cell aliquots were collected at 0, 1, 2, 3, 4, hr. Total RNA was used to determine the mRNA expression levels of CLN1 and CLN2 using qRT-PCR. CLN1 (white bars); CLN2 (black bars). E-H, cells were stimulated with 20 µg/mL α-factor. DNA content was measured using propidium iodide. Data was analyzed using BD CellQuest software. n = 5; G1 phase cells, black bars; S phase cells, light gray; G2M phase cells, dark gray.
The inability to decrease CLN1/2 expression strongly suggested that elo2 and elo3 cells were not inducing a cell cycle arrest in response to pheromone. To see if this was the case, we used FACS analysis to directly look at what stages of the cell cycle cells were arrested at in response to pheromone treatment. ∼85% of bar1 cells were able to arrest at G1 after 3 hr post pheromone treatment (Fig 4E.). The percentage of G1 arrested elo1 bar1 cells was similar (∼90%) (Fig. 4F). elo2 bar1 and elo3 bar1 cells were unable to induce an arrest (Fig. 4G & H). elo2 cells remained asynchronous and were found throughout all phases of the cell cycle, while a high percentage of elo3 cells were found to be in G2 phase.
VLCFA synthesis is required for mating-specific gene expression
Pheromone binding to its receptor increases mating-specific gene expression. It does so by inducing both MAP kinase-dependent protein phosphorylation and Ste12 transcription factor activity.16,17 Ste12 binds to pheromone response elements (PRE) found within the promoters of mating genes. The FUS1 gene promoter has a PRE that is a binding site for Ste12. Determining FUS1 mRNA levels in cells has been routinely used as a read out for mating-specific gene expression. Thus, cells were integrated with a FUS1::FUS1-LacZ::LEU2 allele and β-galactosidase activity was determined in the absence and presence of 20 μg/ml α-factor.
No β-galactosidase activity was seen in cells in the absence of pheromone (Fig. 5A). Upon α-factor treatment, bar1 and elo1 bar1 cells had 35-fold and 30-fold increases in activities, respectively. In contrast, elo2 bar1 and elo3 bar1 cells lacked activity in the presence of α-factor.
Figure 5.
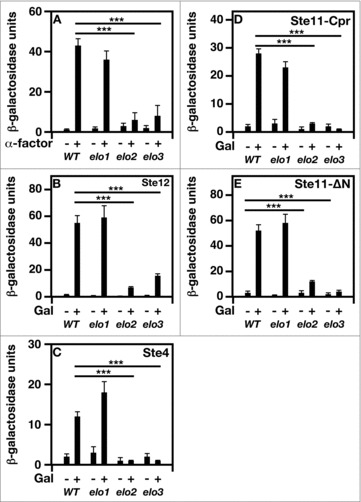
Bypassing the need for MAP kinase signaling does not suppress mating-specific gene expression defect of VLCFA elongation mutants. A FUS1 promoter-driven β-galactosidase reporter assay was used to examine transcriptional activity in the absence and presence of 20 μg/ml α-factor. Plasmid constructs containing the GAL1 promoter were used to overexpress Ste12, Ste4, Ste11ΔN, and Ste11-Cpr. Induction was performed using 2% galactose for 2 hr before addition of pheromone. n = 5; ***p < 0.0001.
Overexpressing MAP kinase signaling components does not restore mating-specific gene expression in elo2 and elo3 cells
We attempted to constitutively activate several steps in the MAP kinase signaling to see if this could suppress the mating defects observed in elo2 and elo3 cells. We first attempted to “bypass” the need for MAP kinase signaling entirely by increasing the level Ste12 in cells. Ste12 was overexpressed using a galactose inducible promoter, and β-galactosidase activity was measured. Overexpressing Ste12 in bar1 and elo1 bar1 cells resulted in the stimulation of β-galactosidase activity (Fig. 5B), whereas elo2 bar1 and elo3 bar1 cells failed to respond (Fig. 5B).
Ste4 is the β subunit of the G protein complex associated with pheromone receptors. It is required for full signaling, and its overexpression has been shown to activate MAP kinase signaling.16,17 Ste4 was overexpressed through a galactose-inducible expression. Overexpressing Ste4 in bar1 or elo1 bar1 cells was able to induce mating specific gene expression (Fig. 5C). On the other hand, both elo2 bar1 and elo3 bar1 cells lacked this response.
Next we overexpressed 2 different forms of the MAPKK kinase, Ste11, which phosphorylates and activates the MAPK kinase, Ste7.16,17 Ste11-Cpr has the plasma membrane targeting sequence of Ras2,25,26 and its expression constitutively activates MAP kinase signaling.25,26 The second, Ste11-ΔN, lacks an auto-inhibitory N-terminal domain.25,26 It is considered a fully activated form of Ste11.
We found that bar1 and elo1 bar1 cells expressing either form of Ste11 activated mating-specific gene expression, whereas elo2 bar1 and elo3 bar1 cells failed to initiate this response (Fig. 5D & E).
VLCFAs are required for mating-specific MAP kinase signaling
We next tested if the results in Figure 5 were truly due to defects in MAP kinase signaling. The MAP kinase Fus3 is phosphorylated in response to pheromone treatment. The time-dependent increase in Fus3 phosphorylation has been used as a read out for the level of MAP kinase signaling activity.27 We therefore assayed MAP kinase activity by determining the level of pheromone-dependent Fus3 phosphorylation. Cells were treated with 20 μg/ml α-factor, and total Fus3-HA and phosphorylated Fus3 were quantitated by immunoblotting using anti-HA monoclonal antibodies and anti-p42 polyclonal antibodies, respectively.
All strains tested lacked detectable levels of total and phosphorylated Fus3 in the absence of pheromone (Fig. 6; 0 time point). Upon pheromone treatment, a time-dependent increase in total Fus3 was observed in bar1 and elo1 bar1 cells (Fig. 6). This was accompanied by a concomitant increase in Fus3 phosphorylation. No increases in total or phosphorylated Fus3 were observed in elo2 bar1 and elo3 bar1 cells in response to pheromone.
Figure 6.
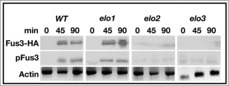
VLCFA elongation mutants lack MAP kinase-dependent activity. The levels of total and phosphorylated Fus3 were analyzed by western analysis using the appropriate antibodies. Actin level was used as a loading control. n = 5; pFus3, phosphorylated Fus3; Fus3-HA, total Fus3 protein.
elo2 and elo3 cells mislocalize Ste5 during mating
Upon pheromone stimulation, the Ste5 scaffold translocates from the nucleus to the plasma membrane where it becomes tethered.28 It acts as a nucleation center that is required for MAP kinase clustering and signal initiation. Thus, we asked if Ste5 was mislocalized in elo2 and elo3 cells. We ectopically expressed a GFP-Ste5 and localized its subcellular location in the presence of pheromone using fluorescence microscopy.
Both bar1 and elo1 bar1 cells properly localized GFP-Ste5 to the plasma membrane in the presence of pheromone (Fig. 7A & B). On the other hand, elo2 bar1 and elo3 bar1 both mislocalized Ste5 to punctate structures within the cytoplasm of the cell (Fig. 7C & D). elo2 bar1 and elo3 bar1 had 4-fold and 8-fold decreases in plasma membrane localized GFP-Ste5, respectively (Fig. 7E).
Figure 7.
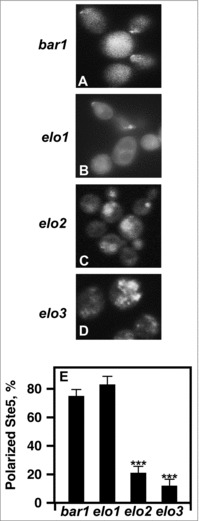
Loss of VLCFA elongation results in Ste5 mislocalization. (A-D), live cells were used to visualize GFP-Ste5. CEN URA3 STE5-GFPx3 was used to overexpress Ste5-GFPX3. Protein localization was visualized using a Leica DRME fluorescence microscope using 100X optics and a GFP filter. E, the percentage values of cellular localized Ste5-GFPX3 were based on examining 500 cells. n = 5; ***p < 0.0001.
Tethering Ste5 to the plasma membrane does not restore pheromone-induced gene expression in elo2 and elo3 cells
The Ste5Q59L mutant is constitutively tethered to the plasma membrane.25 It has been used as a tool to look at the relationship between Ste5 plasma localization and MAP kinase signaling regulation.25,26 To test whether localizing Ste5 to the plasma membrane could restore mating-specific gene expression to elo2 and elo3 cells, we overexpressed Ste5Q59L using a galactose-inducible promoter and assayed β-galactosidase activity in the absence or presence of pheromone.
Both bar1 and elo1 bar1 cells responded normally to the presence of pheromone by inducing β-galactosidase activity, while elo2 bar1 and elo3 bar1 cells lacked this response (Fig. 8). The overexpression of Ste5Q59L in bar1 and elo1 bar1 cells also induced activity in the absence of pheromone. Once again, elo2 bar1 and elo3 bar1 cells lacked this response. Interestingly, all strains tested were able to stimulate β-galactosidase activity in the presence of both Ste5Q59L and pheromone (Fig. 8).
Figure 8.
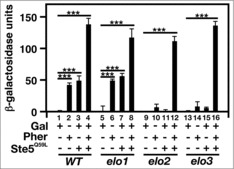
The expression of a plasma membrane-tethered Ste5 cannot re-activate mating-specific gene expression in VLCFA elongation mutants. A FUS1 promoter-driven β-galactosidase reporter assay was used to examine transcriptional activity in the absence and presence of 20 μg/ml α-factor and Ste5Q50L. n = 5; ***p < 0.0001.
elo2 and elo3 cells cannot properly localize plasma membrane-associated PIP2 during mating
Plasma membrane-associated PIP2 clustering is required for tethering Ste5 to the plasma membrane during mating.28,29 Based on the Ste5 localization defects of elo2 bar1 and elo3 bar1 cells, we visualized the localization of PIP2 in in the presence of pheromone.
To visualize PIP2 localization, we expressed a GFP-PHPLCδ1 protein that consists of the PIP2-binding PLCδ1phospholipase C pleckstrin homology domain.30 Both bar1 (63%) and elo1 bar1 (70%) cells mobilized PIP2 to the shmoo tip in response to pheromone treatment (Fig. 9A & B & E), while elo2 bar1 (10%) and elo3 bar1 (5%) cells had reduced efficiencies (Fig. 9C & D & E).
Figure 9.
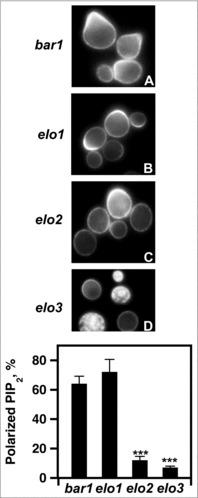
Loss of VLCFA elongation results in PIP2 mislocalization. (A-D), pGAL1-GST-GFP-PHPLCδ was used to express the PHPLCδ protein. For induction, cells were incubated with galactose for 2 hr. 20 μg/ml α factor was added 1 hr before protein visualization. PIP2 polarization was visualized using a Leica DRME fluorescence microscope using 100X optics and a GFP filter. E, the percentage values of properly localized PHPLC were determined using 500 cells. n = 5; ***p < 0.0001.
Discussion
Here, we demonstrated that mating-initiated cell cycle arrest requires that cells produce VLCFAs. The elo2 and elo3 mutants that are defective in C26 fatty acid synthesis had reduced mating efficiencies, lacked MAP kinase activity, and failed to initiate a cell cycle arrest in response to pheromone treatment. They also mislocalized the Ste5 scaffold that acts as a nucleation site for the association and activation of multiple mating-specific MAP kinases, possibly due to an inability to mobilize PIP2.
What is the molecular basis underlying the mating defects seen in mutant cells? Interestingly, we showed that tethering Ste5 to the plasma membrane (Ste5Q59L) alone was not sufficient to induce mating-specific gene expression and MAP kinase signaling in elo2 or elo3 cells. Restoration required that Ste5 be tethered and cells be treated with pheromone, suggesting a possible defect(s) upstream of MAP kinase signaling. Pheromone binding and receptor endocytosis are the first steps in initiating mating.16,17 Loss of these events causes multiple defects in mating signaling. However, elo3 cells have been shown to internalize the Ste2 receptor,31 and its loss restores receptor internalization in cells lacking this ability (rvs161).31 Moreover, it has been shown that loss of function of either Elo2 or Elo3 in cells lacking the v-snare Snc proteins reinitiates endocytosis.32 There could also be a lack of G protein complex association with Ste5 (GPA1/STE4/STE18)25,33. However, we found that overexpressing Ste4 had no effects on gene expression. Neither could we activate MAP kinase signaling by overexpressing activated forms of the Ste11 kinase. We also examined the location of a GFP-Ste18 and found it was properly localized (not shown).
elo2 and elo3 mutants mislocalized Ste5. Moreover, they lacked the ability to mobilize PIP2 in response to pheromone. There are 2 events that aid in activating Ste5 that we examined: 1) Ste5 activation by the βγ subunits in response to pheromone-receptor binding, and 2) plasma membrane tethering due to PIP2 binding. As we alluded to above, overexpressing Ste4 (β) or tethering Ste5 to the plasma membrane alone did not restore gene expression activity. However, we believe that Ste4 is properly localized and able to stimulate Ste5, as the addition of α-factor restored signaling to mutant cells with Ste5 tethered. Garrenton et al.28 have shown that PIP2 binding is absolutely required for Ste5 function and there are mutants in Ste5 that are plasma membrane tethered but have mutations in their PH domain; these mutants do not signal. Thus, PIP2 binding is needed to continuously activate Ste5-dependent signaling. Based on the data presented, we propose that Ste5 binding to PIP2 does not occur in elo2 or elo3 mutants. Why tethering Ste5 to the plasma membrane does not suppress could possibly be due to the fact that STE5Q59L is tethered but not clustered.
Examining the sphingolipid composition data as a whole revealed that there was no single IPC or ceramide species induced or reduced in the presence of pheromone in mating capable strains, when compared with strains unable to mate. However, there was an overall reduction in total ceramides in elo2 and elo3 cells during mating, which directly correlated with reduced mating efficiencies (Table 1). There was also a downward trend in the levels of Cer-A and Cer-D in elo2 and elo3 cells in the presence of pheromone. Previously, we showed that sur2 mutants have no problems mating, suggesting that phytosphingosine derived ceramides (Cer-B, Cer-C, and Cer-D) are not required for mating. On the other hand, Both Cer-A and Cer-B’ are dihydrosphingosine-derived and are produced in sur2 mutants. There were no differences in Cer-B’ levels in any strains analyzed here. This may point to Cer-A being the critical ceramide species regulating yeast mating.
It is well established that the surrounding lipid environment affects membrane protein function.1 elo2 and elo3 mutants produce fatty acids of shorter chain lengths and less ceramides. Defects in the production of either of these lipids could severely affect several cellular events, such as lipid raft-dependent protein transport,34 receptor clustering,35 and endocytosis.1 In the case of yeast mating, Ste5 oligimerizes and this event is required for efficient mating signaling.36 Whether Ste5Q59L is able to aggregate in elo2 or elo3 cells is not known, but the loss of this process could explain why membrane tethering alone is not enough to stimulate gene expression. Moreover, there are studies demonstrating that long chain fatty acids act against PIP2 in regulating ATP-sensitive (K(ATP)) channels.37,38
There are several human fatty acid elongase orthologs, and some of these (ELOVL1/3/7) suppress the loss of Elo3 function in S. cerevisiae.12,39 The role of VLCFAs in cell cycle regulation has not been thoroughly studied. Studies have shown that ELOVL2/4 expression shows a rhythmic oscillation pattern.40 Thus, how changes in the activity of these enzymes may affect the cell cycle needs to be studied. Here, we have demonstrated that VLCFAs play a crucial role in initiating mating-specific cell cycle arrest. We believe that VLCFAs do so by ensuring proper PIP2 mobilization, an event that is necessary for Ste5 clustering in response to pheromone.
Materials and methods
Strains, media, and miscellaneous microbial techniques
The yeast strains used in this study were derived from W303 (MATa leu2–3,112 trp1–1 ura3–1 his3–11,15 can1–100). Yeast strains were grown in YEPD (1% yeast extract, 2% bacto-peptone, 2% glucose), YEP containing the indicated fermentable carbon source, or in synthetic minimal media containing 0.67% Yeast Nitrogen Base (Difco, Sparks, MD) supplemented with the appropriate amino acids, adenine, and uracil. Yeast transformation was performed using the procedure described by Ito et. al.41 E. coli XL1-Blue cells were used for plasmid propagation and were grown in LB medium supplemented with ampicillin (150 μg/ml).
The following yeast strain was a gift from Peter Pryciak: MATa FUS1::FUS1-lacZ::LEU2.42 The following plasmids were gifts from Peter Pryciak: 2µ HIS3 pGAL1-STE12,43 CEN URA3 STE5-GFPx3,42 CEN URA3 pGAL1-STE5Q59L, and CEN LEU2 pGAL1-GST-GFP-STE5Q59. The following plasmid was a gift from Jeremy Thorner: CEN LEU2 pGAL1-GST-GFP-PHPLCδ.
Quantitative limited mating liquid assays
Quantitative limited mating assays were performed as described.44 Mating efficiency is represented as the ratio of diploid colonies per haploid colonies counted. Data are representative of 5 independent experiments.
β-galactosidase assays
To measure effects of α-factor on FUS1-lacZ allele induction, cells were grown in 2% glucose media and induced by the addition of 20 µg/ml α-factor for 2 hr. Induction of the integrated FUS1-lacZ allele by galactose was measured 4 hr after adding 2% galactose to cultures grown in 2% raffinose media. Total cell extracts were obtained and assayed for β-galactosidase activity using Chlorophenol red-β-D-galactopyranoside (CPRG) (Roche, Belvidere, NJ) as the substrate, as described in the CLONTECH Yeast Protocols Handbook. 1 β-gal unit is defined as the amount of β-galactosidase hydrolyzing 1 μmol of CPRG to chlorophenol red and D-galactose per minute per cell. Data are representative of 5 independent experiments.
Microscopy
Visualization of Ste5-GFPx3 andGFP-PHPLCδ1 were performed in live cells (without fixation). Cells expressing Ste5-GFPx3 were treated with 20 µg/ml of α-factor for 1 hr before they were visualized. Cells containing pGAL1-GST-GFP-PHPLCδ1 were grown in media containing 2% raffinose and induced with 2% galactose for 3 hr; 20 µg/ml of α-factor was added during the last hour of galactose induction. Images shown are representative of 5 independent experiments.
Protein extraction and immunoblotting
Cell lysates containing Fus3–3HA protein were isolated using the NaOH/TCA cell lysis/protein extraction procedure. Proteins were resolved by SDS-PAGE and detected by immunoblotting with antibodies. Fus3–3HA was detected using anti-HA (1:000) (clone 16B12) from Covance (Princeton, NJ). HRP-conjugated secondary mouse antibodies (1:2,500) (GE Healthcare, Waukesha, WI) were used for protein detection. Anti-P42/44 polyclonal antibodies were used to visualize total Fus3 (1:1,000) (Abcam, Cambridge, MA) and HRP-conjugated secondary mouse antibodies (1:2,500) were used for protein detection (GE Healthcare, Waukesha, WI).
Radiolabeling of ceramides and sphingolipids
Sphingolipids were radiolabeled, extracted, and analyzed as described previously.45
Statistical analysis
Student's t-tests were used to determine significance of data. All data compared with results from wild-type cells.
Funding Statement
We acknowledge and greatly appreciate the financial support of Genesis Biotechnology Group.
Abbreviations
- LacZ
β-galactosidase
- FUS1
cell FUSion
- MAP
mitogen-activated protein
- VLCFA
very long chain fatty acid
- PH
pleckstrin homology domain
- LCB
long chain sphingoid base
- LCBPs
long chain sphingoid base phosphates
- PIP2
phosphatidylinositol 4,5-bisphosphte
- PRE
pheromone response element
- DHS
dihydrosphingosine
- PHS
phytosphingosine
- GFP
green fluorescent protein
- IPC
inositolphosphorylceramide
- MIPC
mannose inositolphosphoryl ceramide
- MIP2C
mannose diinositolphosphoryl ceramide
- Cer
ceramide
- CLN
cyclin
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We are grateful to Drs. Peter Pryciak and Jeremy Thorner for strains and plasmids. We thank Drs. Martin Adelson, and Eli Mordechai, and those affiliated with the Institute of Metabolic disorders, for helpful discussions.
References
- [1].van Meer G, Voelker DR, Feigenson GW. Membrane lipids: Where they are and how they behave. Nat Rev Mol Cell Biol 2008; 9:112-24; PMID: 18216768; https://doi.org/ 10.1038/nrm2330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Hannun YA, Obeid LM. Many ceramides. J Biol Chem 2011; 286:27855-62; PMID: 21693702; https://doi.org/ 10.1074/jbc.R111.254359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Carroll B, Donaldson JC, Obeid L. Sphingolipids in the DNA damage response. Adv Biol Regul 2015; 58:38-52; PMID: 25434743; https://doi.org/ 10.1016/j.jbior.2014.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Villasmil ML, Francisco J, Gallo-Ebert C, Donigan M, Liu HY, Brower M, Nickels JT. Ceramide signals for initiation of yeast mating-specific cell cycle arrest. Cell Cycle 2016; 15:441-54; PMID: 26726837; https://doi.org/ 10.1080/15384101.2015.1127475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Jenkins GM, Hannun YA. Role for de novo sphingoid base biosynthesis in the heat-induced transient cell cycle arrest of Saccharomyces cerevisiae. J Biol Chem 2001; 276:8574-81; PMID: 11056159; https://doi.org/ 10.1074/jbc.M007425200 [DOI] [PubMed] [Google Scholar]
- [6].Chung N, Jenkins G, Hannun YA, Heitman J, Obeid LM. Sphingolipids signal heat stress-induced ubiquitin-dependent proteolysis. J Biol Chem 2000; 275:17229-32; PMID: 10764732; https://doi.org/ 10.1074/jbc.C000229200 [DOI] [PubMed] [Google Scholar]
- [7].Jenkins GM, Richards A, Wahl T, Mao C, Obeid L, Hannun Y. Involvement of yeast sphingolipids in the heat stress response of Saccharomyces cerevisiae. J Biol Chem 1997; 272:32566-72; PMID: 9405471; https://doi.org/ 10.1074/jbc.272.51.32566 [DOI] [PubMed] [Google Scholar]
- [8].Dickson RC. Roles for sphingolipids in Saccharomyces cerevisiae. Adv Exp Med Biol 2010; 688:217-31; PMID: 20919657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Dickson RC, Nagiec EE, Skrzypek M, Tillman P, Wells GB, Lester RL. Sphingolipids are potential heat stress signals in Saccharomyces. J Biol Chem 1997; 272:30196-200; PMID: 9374502; https://doi.org/ 10.1074/jbc.272.48.30196 [DOI] [PubMed] [Google Scholar]
- [10].Montefusco DJ, Matmati N, Hannun YA. The yeast sphingolipid signaling landscape. Chem Phys Lipids 2014; 177:26-40; PMID: 24220500; https://doi.org/ 10.1016/j.chemphyslip.2013.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Han G, Gable K, Kohlwein SD, Beaudoin F, Napier JA, Dunn TM. The Saccharomyces cerevisiae YBR159w gene encodes the 3-ketoreductase of the microsomal fatty acid elongase. J Biol Chem 2002; 277:35440-9; PMID: 12087109; https://doi.org/ 10.1074/jbc.M205620200 [DOI] [PubMed] [Google Scholar]
- [12].Oh CS, Toke DA, Mandala S, Martin CE. ELO2 and ELO3, homologues of the Saccharomyces cerevisiae ELO1 gene, function in fatty acid elongation and are required for sphingolipid formation. J Biol Chem 1997; 272:17376-84; PMID: 9211877; https://doi.org/ 10.1074/jbc.272.28.17376 [DOI] [PubMed] [Google Scholar]
- [13].Toke DA, Martin CE. Isolation and characterization of a gene affecting fatty acid elongation in Saccharomyces cerevisiae. J Biol Chem 1996; 271:18413-22; PMID: 8702485; https://doi.org/ 10.1074/jbc.271.31.18413 [DOI] [PubMed] [Google Scholar]
- [14].Kohlwein SD, Eder S, Oh CS, Martin CE, Gable K, Bacikova D, Dunn T. Tsc13p is required for fatty acid elongation and localizes to a novel structure at the nuclear-vacuolar interface in Saccharomyces cerevisiae. Mol Cell Biol 2001; 21:109-25; PMID: 11113186; https://doi.org/ 10.1128/MCB.21.1.109-125.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Beaudoin F, Gable K, Sayanova O, Dunn T, Napier JA. A Saccharomyces cerevisiae gene required for heterologous fatty acid elongase activity encodes a microsomal beta-keto-reductase. J Biol Chem 2002; 277:11481-8; PMID: 11792704; https://doi.org/ 10.1074/jbc.M111441200 [DOI] [PubMed] [Google Scholar]
- [16].Chen RE, Thorner J. Function and regulation in MAPK signaling pathways: Lessons learned from the yeast Saccharomyces cerevisiae. Biochim Biophys Acta 2007; 1773:1311-40; PMID: 17604854; https://doi.org/ 10.1016/j.bbamcr.2007.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Wang Y, Dohlman HG. Pheromone signaling mechanisms in yeast: A prototypical sex machine. Science 2004; 306:1508-9; PMID: 15567849; https://doi.org/ 10.1126/science.1104568 [DOI] [PubMed] [Google Scholar]
- [18].D'Hondt K, Heese-Peck A, Riezman H. Protein and lipid requirements for endocytosis. Annu Rev Genet 2000; 34:255-95; PMID: 11092829; https://doi.org/ 10.1146/annurev.genet.34.1.255 [DOI] [PubMed] [Google Scholar]
- [19].Munn AL, Heese-Peck A, Stevenson BJ, Pichler H, Riezman H. Specific sterols required for the internalization step of endocytosis in yeast. Mol Biol Cell 1999; 10:3943-57; PMID: 10564282; https://doi.org/ 10.1091/mbc.10.11.3943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Jin H, McCaffery JM, Grote E. Ergosterol promotes pheromone signaling and plasma membrane fusion in mating yeast. J Cell Biol 2008; 180:813-26; PMID: 18299351; https://doi.org/ 10.1083/jcb.200705076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Villasmil ML, Ansbach A, Nickels JT. The putative lipid transporter, Arv1, is required for activating pheromone-induced MAP kinase signaling in Saccharomyces cerevisiae. Genetics 2011; 187:455-65; PMID: 21098723; https://doi.org/ 10.1534/genetics.110.120725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Bagnat M, Simons K. Lipid rafts in protein sorting and cell polarity in budding yeast Saccharomyces cerevisiae. Biol Chem 2002; 383:1475-80; PMID: 12452424; https://doi.org/ 10.1515/BC.2002.169 [DOI] [PubMed] [Google Scholar]
- [23].Molk JN, Bloom K. Microtubule dynamics in the budding yeast mating pathway. J Cell Sci 2006; 119:3485-90; PMID: 16931596; https://doi.org/ 10.1242/jcs.03193 [DOI] [PubMed] [Google Scholar]
- [24].Jones EW. The synthesis and function of proteases in Saccharomyces: Genetic approaches. Annu Rev Genet 1984; 18:233-70; PMID: 6397123; https://doi.org/ 10.1146/annurev.ge.18.120184.001313 [DOI] [PubMed] [Google Scholar]
- [25].Pryciak PM, Huntress FA. Membrane recruitment of the kinase cascade scaffold protein Ste5 by the Gbetagamma complex underlies activation of the yeast pheromone response pathway. Genes Dev 1998; 12:2684-97; https://doi.org/ 10.1101/gad.12.17.2684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Strickfaden SC, Winters MJ, Ben-Ari G, Lamson RE, Tyers M, Pryciak PM. A mechanism for cell-cycle regulation of MAP kinase signaling in a yeast differentiation pathway. Cell 2007; 128:519-31; PMID: 17289571; https://doi.org/ 10.1016/j.cell.2006.12.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Gartner A, Nasmyth K, Ammerer G. Signal transduction in Saccharomyces cerevisiae requires tyrosine and threonine phosphorylation of FUS3 and KSS1. Genes Dev 1992; 6:1280-92; https://doi.org/ 10.1101/gad.6.7.1280 [DOI] [PubMed] [Google Scholar]
- [28].Garrenton LS, Young SL, Thorner J. Function of the MAPK scaffold protein, Ste5, requires a cryptic PH domain. Genes Dev 2006; 20:1946-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Garrenton LS, Stefan CJ, McMurray MA, Emr SD, Thorner J. Pheromone-induced anisotropy in yeast plasma membrane phosphatidylinositol-4,5-bisphosphate distribution is required for MAPK signaling. Proc Natl Acad Sci U S A 2010; 107:11805-10; PMID: 20547860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Lemmon MA, Ferguson KM, O'Brien R, Sigler PB, Schlessinger J. Specific and high-affinity binding of inositol phosphates to an isolated pleckstrin homology domain. Proc Natl Acad Sci U S A 1995; 92:10472-6; PMID: 7479822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Germann M. Christina Gallo,Tomithy Doanhue, Reza Shirzadi,Joseph Stukey,Silvia Lang,Christoph Ruckenstuhl, Simonetta Oliaro-Bosso,Virginia McDonough,Friederike Turnowsky,Gianni Baliiano, and Joseph T. Nickels, Jr. Characterizing sterol defect suppressors uncovers a novel transcriptional signaling pathway regulating zymosterol biosynthesis. J Biol Chem 2005; 280:35904-13; PMID: 16120615. [DOI] [PubMed] [Google Scholar]
- [32].David D, Sundarababu S, Gerst JE. Involvement of long chain fatty acid elongation in the trafficking of secretory vesicles in yeast. J Cell Biol 1998; 143:1167-82; PMID: 9832547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Whiteway MS, Wu C, Leeuw T, Clark K, Fourest-Lieuvin A, Thomas DY, Leberer E. Association of the yeast pheromone response G protein beta gamma subunits with the MAP kinase scaffold Ste5p. Science 1995; 269:1572-5; PMID: 7667635. [DOI] [PubMed] [Google Scholar]
- [34].Simons K, Ikonen E. Functional rafts in cell membranes. Nature 1997; 387:569-72; PMID: 9177342. [DOI] [PubMed] [Google Scholar]
- [35].Bollinger CR, Teichgraber V, Gulbins E. Ceramide-enriched membrane domains. Biochim Biophys Acta 2005; 1746:284-94; PMID: 16226325. [DOI] [PubMed] [Google Scholar]
- [36].Dohlman HG, Slessareva JE. Pheromone signaling pathways in yeast. Sci STKE 2006; 2006:cm6; PMID: 17148787. [DOI] [PubMed] [Google Scholar]
- [37].Rapedius M, Soom M, Shumilina E, Schulze D, Schonherr R, Kirsch C, Lang F, Tucker SJ, Baukrowitz T. Long chain CoA esters as competitive antagonists of phosphatidylinositol 4,5-bisphosphate activation in Kir channels. J Biol Chem 2005; 280:30760-7; PMID: 15980413; https://doi.org/ 10.1074/jbc.M503503200 [DOI] [PubMed] [Google Scholar]
- [38].Schulze D, Rapedius M, Krauter T, Baukrowitz T. Long-chain acyl-CoA esters and phosphatidylinositol phosphates modulate ATP inhibition of KATP channels by the same mechanism. J Physiol 2003; 552:357-67; PMID: 14561820; https://doi.org/ 10.1113/jphysiol.2003.047035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Ohno Y, Suto S, Yamanaka M, Mizutani Y, Mitsutake S, Igarashi Y, Sassa T, Kihara A. ELOVL1 production of C24 acyl-CoAs is linked to C24 sphingolipid synthesis. Proc Natl Acad Sci U S A 2010; 107:18439-44; PMID: 20937905; https://doi.org/ 10.1073/pnas.1005572107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Wang Q, Tikhonenko M, Bozack SN, Lydic TA, Yan L, Panchy NL, McSorley KM, Faber MS, Yan Y, Boulton ME, et al.. Changes in the daily rhythm of lipid metabolism in the diabetic retina. PLoS One 2014; 9:e95028; PMID: 24736612; https://doi.org/ 10.1371/journal.pone.0095028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol 1983; 153:163-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Strickfaden SC, Winters MJ, Ben-Ari G, Lamson RE, Tyers M, Pryciak PM. A Mechanism for Cell-Cycle Regulation of MAP kinase signaling in a yeast differentiation pathway. Cell 2007; 128:519-31; PMID: 17289571; https://doi.org/ 10.1016/j.cell.2006.12.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Strickfaden SC, Pryciak PM. Distinct Roles for Two G{alpha} G Interfaces in cell polarity control by a yeast heterotrimeric G protein. Mol Biol Cell 2008; 19:181-97; PMID: 17978098; https://doi.org/ 10.1091/mbc.E07-04-0385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Brizzio V, Gammie AE, Nijbroek G, Michaelis S, Rose MD. Cell fusion during yeast mating requires high levels of a-factor mating pheromone. J Cell Biol 1996; 135:1727-39; PMID: 8991086; https://doi.org/ 10.1083/jcb.135.6.1727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Swain E, Stukey J, McDonough V, Germann M, Liu Y, Sturley S, Nickels JT. Yeast cells lacking the ARV1 gene harbor defects in sphingolipid metabolism. Complementation by human ARV1. J Biol Chem 2002; 277:36152-60; PMID: 12145310; https://doi.org/ 10.1074/jbc.M206624200 [DOI] [PubMed] [Google Scholar]


