ABSTRACT
The production of poly-3-hydroxybutyrate (PHB) by photosynthetic cyanobacteria is a potentially sustainable production method for the biodegradable plastics industry. β-Ketoacyl-ACP reductase (FabG), from the photosynthetic cyanobacterium Synechocystis sp. PCC 6803 (SpFabG), is the first NADPH-dependent reductase in the fatty acid biosynthesis pathway. Its structure is similar to that of acetoacetyl-CoA reductase (SpPhaB), which is critical for PHB synthesis and can replace SpPhaB for acetoacetyl-CoA reduction in vitro. However, the specific function of SpFabG in fatty acid synthesis and whether SpFabG could participate in PHB synthesis in vivo were not yet clear. In this study, the role of SpFabG in fatty acid synthesis was first verified in vivo by knocking down and overexpressing of fabG. It was shown that SpFabG was essential yet not rate-limiting for fatty acid biosynthesis. The biochemical characterization of SpFabG using acetoacetyl-CoA as the substrate showed that the optimum temperature, optimum pH, Km and kcat were 30°C, 7, 2.30 mM, and 19.85 s−1, respectively, which exemplified the ability of SpFabG to reduce acetoacetyl-CoA with a relatively low affinity and weak catalytic efficiency. Functional analysis of SpFabG in vivo indicated that SpFabG was able to partially complement SpPhaB under nitrogen-deprived conditions, and overexpression of fabG led to the diversion of partial carbon flux from fatty acid toward PHB synthesis.
KEYWORDS: β-Ketoacyl-ACP reductase, Acetoacetyl-CoA reductase, Poly-3-hydroxybutyrate, Fatty acid, Synechocystis
Introduction
Poly-3-hydroxybutyrate (PHB), the most common type of polyhydroxyalkanoate (PHA), is a biodegradable polymer of biologic origin that has increasingly wide applications in the materials field.1 PHB production in photosynthetic cyanobacteria has been regarded as a sustainable way for developing the biodegradable plastics industry.2 PHB has been found to act mainly as an energy and carbon reserve inside microorganisms and algal cells in response to stressful conditions. In the cyanobacterium Synechocystis sp. PCC 6803, PHB biosynthesis can be triggered under N- or P-deprived conditions.3,4 PHB synthesis in Synechocystis sp. PCC 6803 involves 3 steps. First, 2 molcules of acetyl-CoA are converted by acetoacetyl-CoA thiolase (PhaA) to form acetoacetyl-CoA. Next, acetoacetyl-CoA reductase (PhaB) (EC 1.1.1.36) catalyzes NADPH-dependent reduction of acetoacetyl-CoA to yield D-(β)-3-hydroxybutyryl-CoA. Finally, D-(β)-3-hydroxybutyryl-CoA is polymerized to form PHB by PHA synthase (PhaC).5 It was reported that the overexpression of phaAB led to a 2.6-fold increase in PHB content in Synechocystis sp. PCC 6803.6
FabG (EC 1.1.1.100), which is responsible for reducing β-ketoacyl-ACP to β-hydroxyacyl-ACP, is the first NADPH-dependent reductase in the fatty acid (FA) synthesis pathway in bacteria, plants and algae.7,8 It was found that fabG was an essential gene in Escherichia coli,9 and overexpression of fabG led to a 2–3-fold increase in fatty acid production.10 So far, there are no reports on the specific functions of FabG in cyanobacteria in vivo. Acetoacetyl-ACP, the substrate of FabG for fatty acid (FA) synthesis, is structurally similar to acetoacetyl-CoA, the substrate of PhaB in the PHB synthesis pathway. The only difference between them is the thioester bond that connects to ACP and CoA.11 Therefore, the similarities in the substrates of FabG and PhaB could make it possible for FabG to participate in the synthesis of PHB and become the bridge between fatty acid metabolism and the PHB synthesis pathway. In vitro characterization showed that FabGs from some bacteria such as Pseudomonas and Escherichia coli could reversibly convert β-ketoacyl-CoA to D-β-hydroxyacyl-CoA.12,13 An in vivo study also demonstrated that the FabG from Pseudomonas aeruginosa could act as β-ketoacyl-CoA reductase for PHA production in E. coli.12 However, to date, there are no reports of investigations on the potential role of FabG in PHB synthesis in photosynthetic cyanobacteria in vivo.
Our previous study showed that the structures of FabG and PhaB from the photosynthetic cyanobacterium Synechocystis sp. PCC 6803 (SpFabG and SpPhaB, respectively) were very similar, and SpFabG could act as a β-ketoacyl-CoA reductase in vitro.11 These results implied that SpFabG might play a role in PHB synthesis and could participate in carbon flux distribution between FA and PHB in Synechocystis sp. PCC 6803. In this study, the role of SpFabG in fatty acid synthesis was first verified in vivo by knocking down and overexpressing fabG. Next, the kinetic characteristics of SpFabG as a β-ketoacyl-CoA reductase were investigated in vitro. Furthermore, the role of SpFabG in complementing SpPhaB and in regulating carbon flux was revealed in phaB knocked-down and fabG-overexpressed phaB knocked-down mutant strains.
Results and discussion
SpFabG was essential but not rate-limiting in fatty acid biosynthesis
To identify the specific functions of fabG, the knockout of fabG was initially attempted. However, the PCR amplification of the fabG::C.K2 mutant genome yielded 2 bands, one corresponding to the wild-type fabG gene, which was 1426 bp long, and the other band that was 2607 bp long which contained 1426 bp of the WT gene and 1181 bp of the kanamycin fragment (Fig. 1A). These results showed that the fabG::C.K2 mutant lacking fabG was never completely segregated even after 6 months of culturing of the transformants in BG11 medium supplemented with kanamycin. The fabG could not be completely knocked out in Synechocystis sp. PCC 6803 suggesting that it was essential for cell growth. Similarly, EcFabG had been proven to be essential for cell growth in E. coli.9
Figure 1.
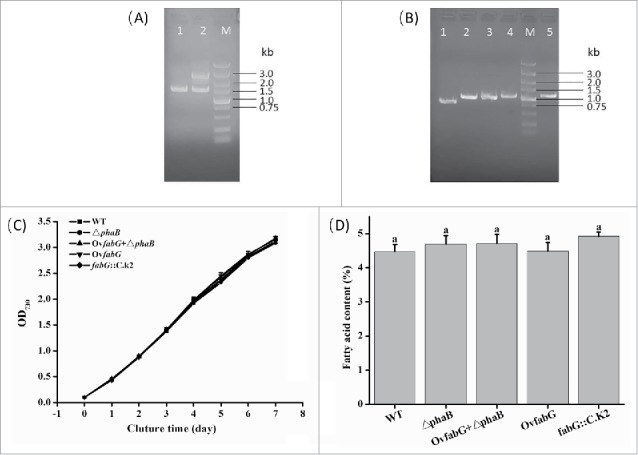
Verification of the mutant genotype (A and B) and characterization of their growth (C) and fatty acid synthesis (D). (A) Verification of the genotype of fabG::C.K2. Lanes 1, 2: The PCR products amplified with the primers KfabG-1 and KfabG-2 using WT and fabG::C.K2 strain DNA. (B) Verification of the genotypes of △phaB, OvfabG and OvfabG + △phaB. Lanes 1, 2, 3: The PCR products amplified with the primers phaB-1 and phaB-2 using WT, △phaB and OvfabG + △phaB DNA, and the correct sizes of the amplicons were 765 bp, 1002 bp and 1002 bp, respectivly. Lanes 4, 5: The PCR products amplified with the primers psbAII-1 and fabG-2 using OvfabG + △phaB and OvfabG strain DNA; the correct size was 1065 bp. (C) Growth curves and (D) fatty acid content of WT, △phaB, OvfabG + △phaB, fabG::C.K2 and OvfabG mutant strains grown under N repletion for 7 d. The letter (a) implies that no significant differences were observed (p >0.05) using Tukey's HSD test. The data indicate the means ± SD (n = 4).
To further characterize the role of fabG in fatty acid biosynthesis in Synechocystis sp. PCC 6803, fabG overexpressed (OvfabG) and fabG knocked-down (fabG::C.K2) strains were constructed (Fig. 1B) and evaluated under N-replete conditions (Fig. 1). The results showed that cell growth was almost identical in WT, OvfabG and fabG::C.K2 strains (Fig. 1C). Furthermore, the fatty acid content in WT, OvfabG and fabG::C.K2 reached an average of 4.7% of the DW and showed no obvious differences (Fig. 1D). It indicated that FabG was not the rate-limiting enzyme in Synechocystis sp. PCC 6803. It was reported that enoyl-ACP reductase (FabI) and β-hydroxyacyl-ACP dehydratase (FabZ) were the rate-limiting enzymes in E. coli fatty acid synthesis, while in another cyanobacterium, Synechococcus sp. PCC 7002, it was β-ketoacyl-ACP synthase III (FabH).14,15 This suggested that although the rate-limiting enzyme in different organisms might be different, the fabG step did not appear to determine the commitment to fatty acid biosynthesis in bacteria and cyanobacteria despite being essential.
Biochemical characterization of SpFabG activity with acetoacetyl-CoA as substrate
To examine the kinetics of the catalysis of acetoacetyl-CoA by SpFabG, the effects of temperature and pH on SpFabG activity were first explored. As shown in Figure 2A, SpFabG showed the highest activity at 30°C, although the enzyme retained 55, 82, 94 and 75% of its peak activity at 20, 40, 50 and 60°C, respectively. The preferred temperature of 30°C for SpFabG was consistent with the optimum temperature for the growth of Synechocystis sp. PCC 6803. The optimum temperature for SpFabG from Synechocystis sp. PCC 6803 was consistent with that of FabG from Streptococcus pneumoniae (Table 1).16 As shown in Figure 2B, SpFabG displayed the highest activity at pH 7, which was also the preferred pH for growth, and the activities at pH 6, 8, 9 and 10 were only 66, 67, 38 and 15% of that at pH 7, respectively. The optimum pH for SpFabG of Synechocystis sp. PCC 6803 was very close to that of FabG from Plasmodium falciparum, which is 6.8 (Table 1).17 The results described above demonstrated that the optimum temperature and pH for SpFabG activity was in line with the optimum culture conditions for Synechocystis sp. PCC 6803.
Figure 2.
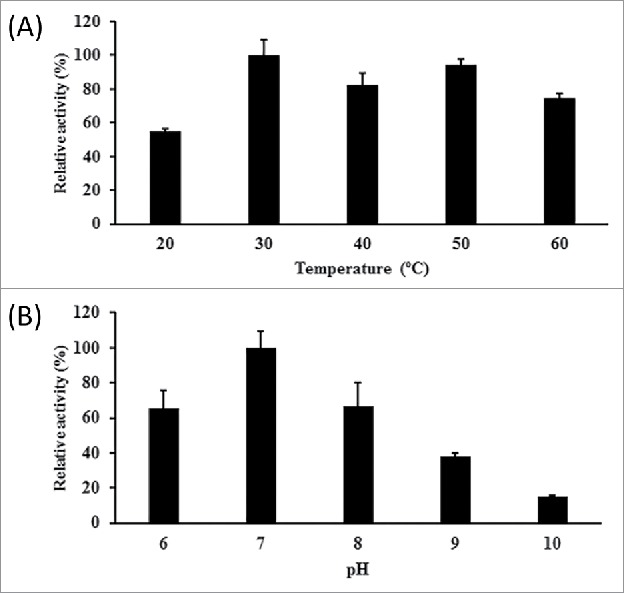
Biochemical characterization of SpFabG with acetoacetyl-CoA as the substrate. (A) Effects of temperature on SpFabG activity. (B) Effects of pH on SpFabG activity.
Table 1.
Kinetic parameters for the catalysis of acetoacetyl-CoA by FabG from Synechocystis sp. PCC 6803 and other microorganisms.
| Organism | Km (mM) | Kcat (s−1) | kcat/Km (mM−1s−1) | Optimum conditions | Reference |
|---|---|---|---|---|---|
| Plasmodium falciparum | 0.08 | 0.01 | 0.19 | pH 7.5 | 18 |
| Streptococcus pneumoniae | 2.20 | 11.00 | 5.00 | pH 7.6, 30°C | 16 |
| Plasmodium falciparum | 0.43 | 259.00 | 602.33 | pH 6.8, 22°C | 17 |
| Synechocystis sp. PCC 6803 | 2.30 | 19.85 | 8.63 | pH 7.0, 30°C | This study |
After confirming the optimum temperature and pH, the kinetics of SpFabG with acetoacetyl-CoA as the substrate was determined. As shown in Table 1, the Km and kcat were 2.3 mM and 19.85 s−1, respectively, which resulted in a kcat/Km value of 8.63 mM−1 s−1. These kinetic parameters were comparable to those obtained with the enzyme from Streptococcus pneumonia under optimal conditions (Table 1).16 However, the FabG from Plasmodium falciparum showed a much lower Km and a higher kcat, as well as a larger kcat/Km (Table 1).17,18 These results indicated that SpFabG was able to utilize acetoacetyl-CoA as its substrate in vitro, although its affinity and catalytic efficiency were relatively weak.
SpFabG was able to partially complement SpPhaB in vivo
To further confirm its role in using acetoacetyl-CoA as a substrate in vivo, the △phaB mutant, in which the phaB gene participating in PHB synthesis was knocked out, was constructed. This was followed by the construction of the OvfabG + △phaB mutant, in which fabG was overexpressed in the △phaB mutant (Fig. 1B). Next, PHB biosynthesis and physiologic features of the △phaB and OvfabG +△phaB mutants as well as the WT strain were tracked under conditions of N deprivation (Fig. 3).
Figure 3.
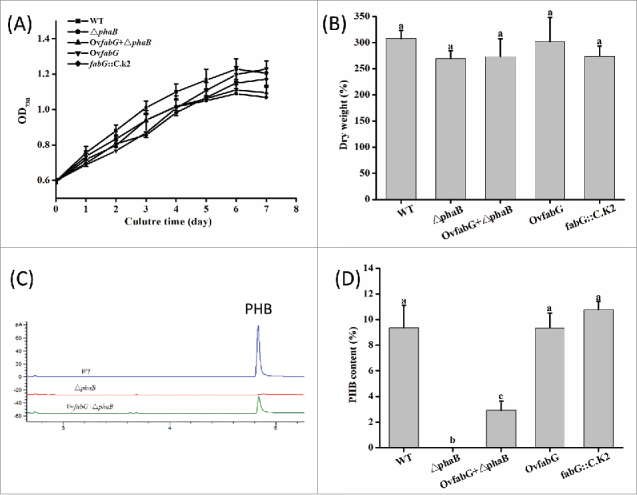
The growth curves (A), dry weights (B), PHB peaks detected on GC (C), and PHB content (D) of WT and mutant strains grown under N deprivation conditions for 7 d. The different letters (a-c) indicate the significance of the differences observed (p < 0.05) using Tukey's HSD test. The data indicate the means ± SD (n = 4).
The cell growth of △phaB, OvfabG +△phaB and WT as revealed by OD730 was approximately 1.10, and the DW reached approximately 280 mg/L with no significant differences among the 3 groups (Fig. 3A, B). These results indicated that knocking out phaB did not affect biomass accumulation. However, PHB could not be detected in the △phaB strain (Fig. 3C) indicating that △phaB had lost the ability to accumulate PHB. It demonstrated that phaB is an essential gene in the PHB synthesis pathway. Notably, the OvfabG + △phaB strain regained the ability to accumulate PHB, which reached 31% of the WT levels (Fig. 3D). These results demonstrated that SpFabG could complement PhaB when phaB was knocked out, although it could not work as efficiently as PhaB. It was reported that the enzyme activity of SpFabG using acetoacetyl-CoA as the substrate was 16% of that of SpPhaB in vitro,11 which could account for the lower levels of PHB in the OvfabG + △phaB mutant. However, SpFabG did not catalyze acetoacetyl-CoA when phaB was present, since overexpressing (OvfabG) or knocking down (fabG::C.K2) fabG had little impact on PHB content (Fig. 3D). PhaB from various organisms had Km values ranging from 0.002 to 0.037 mM with acetoacetyl-CoA as the substrate (Supplementary Table S2), which was 100–1000 times lower than those of FabGs, indicating that acetoacetyl-CoA was more likely to be used by PhaB due to its much higher affinity than that of FabG. Therefore, the probability of SpFabG functioning as an acetoacetyl-CoA reductase in the presence of SpPhaB was low.
Additionally, the fatty acid content in the OvfabG + △phaB strain decreased by 18% compared with that in the △phaB strain, whereas the carbohydrate content was similar (Fig. 4A, B). These results indicated that the introduction of SpFabG did not disturb carbohydrate synthesis and facilitated PHB instead of fatty acid biosynthesis. This could be due to the down regulation of fatty acid biosynthesis and the contradicting up regulation of the PHB biosynthesis pathway under nitrogen deprived conditions. It had been demonstrated that when Synechocystis sp. PCC 6803 was subjected to nitrogen deprivation, the expression levels of many genes involved in fatty acid biosynthesis, including Acetyl-CoA carboxylase (ACCase) and Malonyl CoA:ACP transacylase (FabD), were decreased, whereas those involved in PHB biosynthesis, such as PhaA and PhaC, increased compared with those under normal conditions.19 As a result, the introduction of SpFabG led to the diversion of partial carbon flux from FA toward PHB synthesis.
Figure 4.
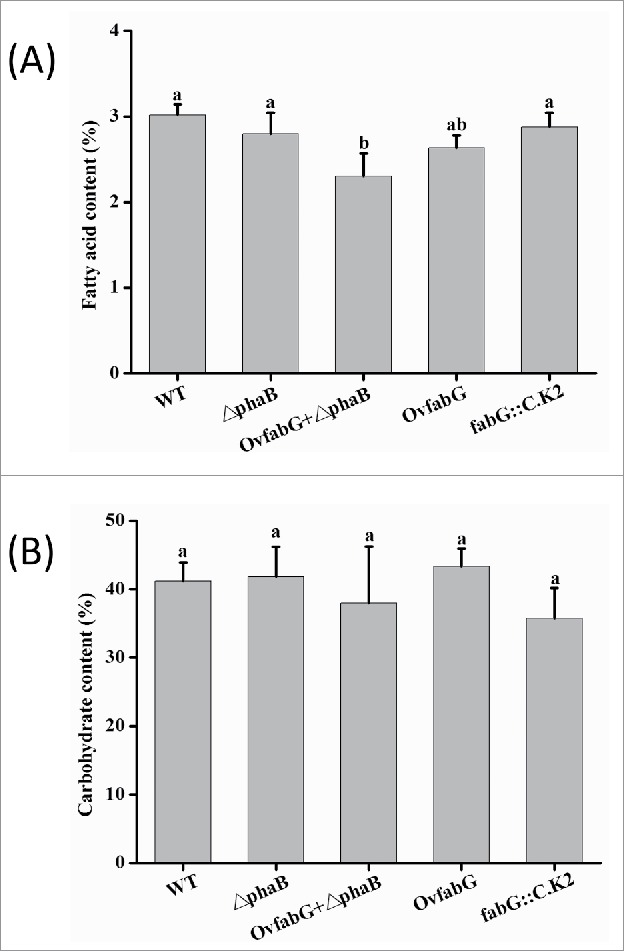
Carbohydrate (A) and fatty acid content (B) of WT and mutant strains under conditions of N deprivation for 7 d. The different letters (a-b) show the significance of the differences observed (p < 0.05) using Tukey's HSD test. The data indicate the means ± SD (n = 4).
The present study demonstrated the feasibility of SpFabG functioning as an acetoacetyl-CoA reductase for PHB biosynthesis in vivo. This is the first report of a fabG from a photosynthetic microorganism that is not only essential for fatty acid biosynthesis but also plays a potential role in PHB accumulation. In fact, PHB in cyanobacteria, like FA in oleaginous microalgae, had been regarded as a redox-sink to store excess NADPH under imbalanced metabolic conditions such as nutrient starvation.20 The similar physiologic role of PHB and FA might be an indicative of functional multiplicity of FabG. The additional function of SpFabG as a β-ketoacyl-CoA reductase in PHB synthesis might aid cells to acclimate to the stressed conditions more smoothly. This might represent an evolutionary strategy for cyanobacteria to survive complicated situations. Moreover, since FabG is inherently able to reduce β-ketoacyl-ACP with C chain lengths of 4 to 18 in cyanobacteria,21 it is reasonable to extrapolate from the results of this study that it may also work on β-ketoacyl-CoA with medium C chain lengths (mcl) of 6 to 10 that can be obtained from fatty acid β-oxidation, which could provide the substrate for mcl−PHA biosynthesis. This could offer a new method for the photoautotrophic production of PHA in cyanobacteria.
Materials and methods
Construction of plasmids and transformation of Synechocystis sp. PCC 6803
All strains used and constructed in this study are listed in Table 2. All primers used are listed in Supplementary Table S1. The replicative vector pJA2 was kindly provided by Dr. Paul Hudson (KTH Royal Institute of Technology of Sweden).22,23 The fabG gene was amplified by PCR using the Synechocystis sp. PCC 6803 DNA as the template with ovfabG1 and ovfabG2 primers and cloned into the BamHI/XbaI site of pJA2 resulting in the recombinant plasmid pJA-fabG. The △phaB mutant strain of Synechocystis sp. PCC 6803 with phaB deleted was kindly provided by Dr. Weiwen Zhang (Tianjin University).24,25
Table 2.
Synechocystis strains constructed for this study.
| strain | Genome modification | Notes |
|---|---|---|
| OvfabG | fabG was overexpressed by pJA2 in Synechocystis | This study |
| fabG::C.K2 | C.K2 was inserted in ClaI sites of fabG | This study |
| △phaB | phaB was deleted in Synechocystis | Acquired from Dr. Weiwen Zhang24 |
| OvfabG+△phaB | fabG was overexpressed by pJA2 and phaB was deleted | This study |
| WT | Wild type Synechocystis sp. PCC 6803 | Wild type |
The DNA fragment containing the fabG gene (slr0886) was generated by PCR using the primers KfabG1 and KfabG2 with Synechocystis sp. PCC 6803 chromosomal DNA as the template, cloned into pMD18-T, and confirmed by sequencing. The C.K2 fragment, excised from pRL446 (NCBI GenBank accession no. EU346690) by EcoRI, was inserted into the ClaI site of that plasmid, resulting in plasmid dicp1 for the inactivation of fabG in Synechocystis sp. PCC 6803.26 The constructed plasmid dicp1 was introduced into Synechocystis sp. PCC 6803 by natural transformation to generate the Synechocystis strain fabG::C.K2 as described by Jiang et al..27 The constructed plasmid pJA2-fabG was introduced into the strain Synechocystis △phaB to generate the Synechocystis strain OvfabG + △phaB.
Enzyme activity assay and kinetic characterization of SpFabG using the substrate acetoacetyl-CoA
SpFabG was expressed and purified as described previously.11 The fractions containing SpFabG were concentrated using a 10 kDa Amicon concentrator (Millipore) and stored at −80°C until further use. The enzyme activity of SpFabG was determined as described previously.11 Before the reaction, the protein solution was diluted to 0.1 mg/ml using buffer A (50 mM Tris–HCl pH 7.8, 300 mM NaCl, 1 mM EDTA, 5% glycerol (v/v), and 2 mM β-mercaptoethanol), and protein concentrations were determined using the Bradford assay with BSA as the standard. For optimum temperature determination, a mixture containing 200 μM acetoacetyl-CoA and 200 μM NADPH in 0.1 M PBS buffer (pH 7.0) was prepared in a volume of 100 μL at different temperatures ranging from 20°C to 60°C. The mixtures were preincubated for 5 min, and the reaction was initiated by the addition of 2 μL of diluted protein solution containing 0.2 μg SpFabG. The relative activity was determined from the rate of conversion of NADPH to NADP+, which was measured photometrically by the decrease of absorbance at 340 nm (Jasco V630 spectrophotometer, Japan). For determining optimum pH, the same mixture as described above was used except for the use of different pH conditions that were generated with PBS (pH 6–8) and glycine-NaOH (pH 9–10) buffers to obtain a final concentration of 0.1 M. The mixtures were preincubated at 30°C before the reaction was initiated.
The kinetic parameters Km, Vmax and kcat with acetoacetyl-CoA as the substrate were determined by varying its concentration from 100 μmol/L to 1600 μmol/L under saturating conditions of NADPH at 400 μmol/L in a volume of 100 μL. Each reaction was initiated by the addition of 0.2 μg of the enzyme. The data were evaluated by double reciprocal plots.
Culture conditions and growth measurement
For N-replete cultures, Synechocystis sp. PCC 6803 WT strain was grown in BG11 medium buffered with 10 mM HEPES-NaOH (pH 7.5), and the mutant strains were grown in the same medium containing the corresponding antibiotics as follows: △phaB (chloramphenicol 17 μg/ml), OvfabG+△phaB (chloramphenicol 17 μg/mL, kanamycin 25 μg/mL), OvfabG (kanamycin 25 μg/mL), fabG::C.K2 (kanamycin 25 μg/mL). The cultures with an initial OD730 of 0.2 (determined by a Jasco V530 spectrophotometer, Japan) were incubated aerobically at 30°C under continuous illumination of 40–50 μmol/m2/s on a rotatory shaker operating at 150 r/min. The growth was tracked every day for 7 d.
For N-depleted cultures, the WT and mutant strain cells were first grown in 5 L flasks under N-replete conditions as described above until the OD730 reached 2.4. Next, the cultures were centrifuged at 6800 g for 5 min, and the cell pellets were washed twice with BG11–0 medium (BG11 medium lacking NaNO3) in which ferric ammonium citrate and Co(NO3)2·6H2O were replaced with equimolar concentrations of ferric citrate and CoCl2·6H2O, respectively. The washed cells, with an initial OD730 of 0.6, were inoculated in BG11–0 medium supplemented with sodium acetate at a final concentration of 0.4% (w/v) and cultivated for 7 d under the same conditions as the N-replete culture.
The cells were harvested by centrifugation at 6800 g for 5 min before they were vacuum dried. The dry cell weight (DW, g/L) was measured according to the procedure described by Chi et al.28 Briefly, the cultures (10 mL) were filtered using pre-weighed Whatman GF/C filters (47 mm diameter), washed twice with 2 mL of distilled water, and dried to a constant weight at 60°C. The dry weight of the algal cells was the difference between the final weight and the weight of the clean filter.
Biochemical composition analysis
PHB analysis
Freeze-dried cells (20 mg) were esterified with 1 mL 0.2% H2SO4–methanol (v/v) and 1 mL chloroform at 100°C for 5 h during which the PHB was converted to methyl 3-hydrobutyrate. After cooling to room temperature, 0.5 mL of distilled water was added and mixed for 1 min on a vortex mixer. Next, the mixture was centrifuged at 2000 rpm for 2 min and 200 μL of the chloroform layer was used for analysis by gas chromatography (GC). The PHB analysis was performed by Agilent 6890 GC instrument, as described previously,4 equipped with a DB5-column (Agilent, 30 m × 0.25 mm × 0.25 μm) and a flame-ionization detector (FID) with the temperatures of the injector and detector set to 270°C and 300°C, respectively. The initial oven temperature was maintained at 60°C for 1 min and increased to 120°C at the rate of 10°C /min. Next, the oven temperature was increased to 200°C at the rate of 45°C /min and maintained for 3 min. A PHB standard (Sigma) was used for calibration.
Fatty acid analysis
Freeze-dried cells (5 mg) were transesterified with 5 mL of 0.2% H2SO4–methanol (v/v) in a 10 mL round-bottomed flask at 70°C for 1 h during which the FA was converted to fatty acid methyl esters (FAME). Heptadecanoic acid, the internal standard, was transesterified simultaneously. Next, the FAME content was analyzed by an Agilent 7890 GC instrument equipped with a DB23-column (Aglient, 30 m × 0.32 mm × 0.25 μm) and an FID detector.29
Carbohydrate analysis
Carbohydrate analysis was performed by the sulfuric acid-anthrone method as described by Chi et al..28
Statistical analysis
Statistical calculations were performed using STATISTICA® 7.0 (StatSoft Inc., Tulsa, OK, USA). One-way analysis of variance (ANOVA) was used to determine the effects of treatments, and Tukey's honestly significant difference (HSD) test was conducted to test the statistical significance of the differences between the means of various treatments.
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We appreciate Professor Baosheng Qiu (Central China Normal University) for kindly providing the plasmid PRL446 for the fabG::C.K2 mutant and Professor Weiwen Zhang (Tianjin University) for kindly providing the OvfabG strain, the △phaB strain and the pJA2-fabG plasmid.
Funding
This work was supported by the National Natural Science Foundation of China (NSFC) under Grant (No. 31500294); and NSFC under Grant (No. 31470432).
References
- [1].Chen GQ. A microbial polyhydroxyalkanoates (PHA) based bio- and materials industry. Chem Soc Rev 2009; 38(8):2434-46; PMID:19623359; https://doi.org/ 10.1039/b812677c [DOI] [PubMed] [Google Scholar]
- [2].Balaji S, Gopi K, Muthuvelan B. A review on production of poly β hydroxybutyrates from cyanobacteria for the production of bio plastics. Algal Res 2013; 2(3):278-85; https://doi.org/ 10.1016/j.algal.2013.03.002 [DOI] [Google Scholar]
- [3].Panda B, Jain P, Sharma L, Mallick N. Optimization of cultural and nutritional conditions for accumulation of poly-beta-hydroxybutyrate in Synechocystis sp. PCC 6803. Bioresour Technol 2006; 97(11):1296-301; PMID:16046119: https://doi.org/ 10.1016/j.biortech.2005.05.013 [DOI] [PubMed] [Google Scholar]
- [4].Panda B, Mallick N. Enhanced poly-beta-hydroxybutyrate accumulation in a unicellular cyanobacterium, Synechocystis sp. PCC 6803. Lett Appl Microbiol 2007; 44(2):194-8; PMID:17257260; https://doi.org/ 10.1111/j.1472-765X.2006.02048.x [DOI] [PubMed] [Google Scholar]
- [5].Taroncher-Oldenberg G, Nishina K, Stephanopoulos G. Identification and analysis of the polyhydroxyalkanoate-specific beta-ketothiolase and acetoacetyl coenzyme A reductase genes in the cyanobacterium Synechocystis sp. strain PCC6803. Appl Environ Microbiol 2000; 66(10):4440-8; PMID:11010896; https://doi.org/ 10.1128/AEM.66.10.4440-4448.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Khetkorn W, Incharoensakdi A, Lindblad P, Jantaro S. Enhancement of poly-3-hydroxybutyrate production in Synechocystis sp. PCC 6803 by overexpression of its native biosynthetic genes. Bioresour Technol 2016; 214:761-8; PMID:27213577; https://doi.org/ 10.1016/j.biortech.2016.05.014 [DOI] [PubMed] [Google Scholar]
- [7].Beld J, Lee DJ, Burkart MD. Fatty acid biosynthesis revisited: structure elucidation and metabolic engineering. Mol Biosyst 2015; 11(1):38-59; PMID:25360565; https://doi.org/ 10.1039/C4MB00443D [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Price AC, Zhang YM, Rock CO, White SW. Structure of beta-ketoacyl-[acyl carrier protein] reductase from Escherichia coli: Negative cooperativity and its structural basis. Biochem 2001; 40(43):12772-81; https://doi.org/ 10.1021/bi010737g [DOI] [PubMed] [Google Scholar]
- [9].Zhang Y, Cronan JE. Transcriptional analysis of essential genes of the Escherichia coli fatty acid biosynthesis gene cluster by functional replacement with the analogous Salmonella typhimurium gene cluster. J Bacteriol 1998; 180(13):3295-303; PMID:9642179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Jeon E, Lee S, Lee S, Han SO, Yoon YJ, Lee J. Improved production of long-chain fatty acid in Escherichia coli by an engineering elongation cycle during fatty acid synthesis (FAS) through genetic manipulation. J Microbiol and Biotechnol 2012; 22(7):990-9; https://doi.org/ 10.4014/jmb.1112.12057 [DOI] [PubMed] [Google Scholar]
- [11].Liu YH, Feng YB, Cao XP, Li X, Xue S. Structure-directed construction of a high-performance version of the enzyme FabG from the photosynthetic microorganism Synechocystis sp. PCC 6803. FEBS Lett 2015; 589(20):3052-7; PMID:26358291; https://doi.org/ 10.1016/j.febslet.2015.09.001 [DOI] [PubMed] [Google Scholar]
- [12].Nomura CT, Taguchi K, Gan ZH, Kuwabara K, Tanaka T, Takase K, Doi Y. Expression of 3-ketoacyl-acyl carrier protein reductase (fabG) genes enhances production of polyhydroxyalkanoate copolymer from glucose in recombinant Escherichia coli JM109. Appl Environ Microbiol 2005; 71(8):4297-306; PMID:16085817; https://doi.org/ 10.1128/AEM.71.8.4297-4306.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Ren Q, Sierro N, Witholt B, Kessler B. FabG, an NADPH-dependent 3-ketoacyl reductase of Pseudomonas aeruginosa, provides precursors for medium-chain-length poly-3-hydroxyalkanoate biosynthesis in Escherichia coli. J Bacteriol 2000; 182(10):2978-81; PMID:10781572; https://doi.org/ 10.1128/JB.182.10.2978-2981.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Yu XY, Liu TG, Zhu FY, Khosla C. In vitro reconstitution and steady-state analysis of the fatty acid synthase from Escherichia coli. Proc Natl Acad Sci U.S.A. 2011; 108(46):18643-48; PMID:22042840; https://doi.org/ 10.1073/pnas.1110852108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Kuo J, Khosla C. The initiation ketosynthase (FabH) is the sole rate-limiting enzyme of the fatty acid synthase of Synechococcus sp. PCC 7002. Metab Eng 2014; 22:53-9; PMID:24395007; https://doi.org/ 10.1016/j.ymben.2013.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Patel MP, Liu WS, West J, Tew D, Meek TD, Thrall SH. Kinetic and chemical mechanisms of the fabG-encoded Streptococcus pneumoniae beta-ketoacyl-ACP reductase. Biochem 2005; 44(50):16753-65; https://doi.org/ 10.1021/bi050947j [DOI] [PubMed] [Google Scholar]
- [17].Karmodiya K, Surolia N. Analyses of co-operative transitions in Plasmodium falciparum beta-ketoacyl acyl carrier protein reductase upon co-factor and acyl carrier protein binding. FEBS J 2006; 273(17):4093-103; PMID:16934037; https://doi.org/ 10.1111/j.1742-4658.2006.05412.x [DOI] [PubMed] [Google Scholar]
- [18].Pillai S, Rajagopal C, Kapoor M, Kumar G, Gupta A, Surolia N. Functional characterization of beta-ketoacyl-ACP reductase (FabG) from Plasmodium falciparum. Biochem Biophys Res Commun 2003; 303(1):387-92; PMID:12646215; https://doi.org/ 10.1016/S0006-291X(03)00321-8 [DOI] [PubMed] [Google Scholar]
- [19].Huang SQ, Chen L, Te RG, Qiao JJ, Wang JX, Zhang WW. Complementary iTRAQ proteomics and RNA-seq transcriptomics reveal multiple levels of regulation in response to nitrogen starvation in Synechocystis sp. PCC 6803. Mol Biosyst 2013; 9(10):2565-74; PMID:23942477; https://doi.org/ 10.1039/c3mb70188c [DOI] [PubMed] [Google Scholar]
- [20].Hauf W, Schlebusch M, Huge J, Kopka J, Hagemann M, Forchhammer K. Metabolic changes in Synechocystis PCC6803 upon nitrogen-starvation: Excess NADPH sustains polyhydroxybutyrate accumulation. Metabolites 2013; 3(1):101-18; PMID:24957892; https://doi.org/ 10.3390/metabo3010101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Ohlrogge J, Browse J. Lipid Biosynthesis. Plant Cell 1995; 7(7):957-70; https://doi.org/ 10.1105/Tpc.7.7.957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Anfelt J, Hallstrom B, Nielsen J, Uhlen M, Hudson EP. Using transcriptomics to improve butanol tolerance of Synechocystis sp. Strain PCC 6803. Appl Environ Microbiol 2013; 79(23):7419-27; PMID:24056459; https://doi.org/ 10.1128/AEM.02694-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Huang HH, Camsund D, Lindblad P, Heidorn T. Design and characterization of molecular tools for a Synthetic Biology approach towards developing cyanobacterial biotechnology. Nucl Acid Res 2010; 38(8):2577-93; https://doi.org/ 10.1093/nar/gkq164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Wang HL, Postier BL, Burnap RL. Optimization of fusion PCR for in vitro construction of gene knockout fragments. Biotech 2002; 33(1):26-28 [DOI] [PubMed] [Google Scholar]
- [25].Wang YP, Sun T, Gao XY, Shi ML, Wu LN, Chen L, Zhang WW. Biosynthesis of platform chemical 3-hydroxypropionic acid (3-HP) directly from CO2 in cyanobacterium Synechocystis sp. PCC 6803. Metab Eng 2016; 34:60-70; PMID:26546088; https://doi.org/ 10.1016/j.ymben.2015.10.008 [DOI] [PubMed] [Google Scholar]
- [26].Elhai J, Wolk CP. Conjugal transfer of DNA to cyanobacteria. Method Enzymol 1988; 167:747-54 [DOI] [PubMed] [Google Scholar]
- [27].Jiang HB, Lou WJ, Du HY, Price NM, Qiu BS. Sll1263, a unique cation diffusion facilitator protein that promotes iron uptake in the cyanobacterium Synechocystis sp. Strain PCC 6803. Plant Cell Physiol 2012; 53(8):1404-17; PMID:22685083; https://doi.org/ 10.1093/pcp/pcs086 [DOI] [PubMed] [Google Scholar]
- [28].Chi L, Yao CH, Cao XP, Xue S. Coordinated regulation of nitrogen supply mode and initial cell density for energy storage compounds production with economized nitrogen utilization in a marine microalga Isochrysis zhangjiangensis. Bioresour Technol 2016; 200:598-605; PMID:26547809; https://doi.org/ 10.1016/j.biortech.2015.10.059 [DOI] [PubMed] [Google Scholar]
- [29].Liu J, Liu YN, Wang HT, Xue S. Direct transesterification of fresh microalgal cells. Bioresour Technol 2015; 176:284-7; https://doi.org/ 10.1016/j.biortech.2014.10.094 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


