ABSTRACT
A glutathione peroxidase (GPx) gene, designated as PsGPx, was cloned from Antarctic psychrotrophic bacterium Pseudoalteromonas sp. ANT506 and expressed in Escherichia coli. The full-length PsGPx contained a 585-bp encoding 194 amino acids with predicted molecular masses of approx. 21.7 kDa. Multiple sequence alignments revealed that PsGPx belonged to the thioredoxin-like superfamily. PsGPx was heterologously overexpressed in E. coli, purified and characterized. The maximum catalytic temperature and pH value for recombinant PsGPx (rPsGPx) were 30°C and pH 9.0, respectively. rPsGPx retained 45% of the maximum activity at 0°C and exhibited high thermolability with a half-life of approx. 40 min at 40°C. In addition, the enzymatic activity of rPsGPx was still manifested under 3 M NaCl. The Km and Vmax values of the recombinant enzyme using GSH and H2O2 as substrates were 1.73 mM and 16.28 nmol/mL/min versus 2.46 mM and 21.50 nmol/mL/min, respectively.
KEYWORDS: glutathione peroxidase, expression, psychrotrophic, Antarctic sea ice, gene
Introduction
Glutathione peroxidase (GPx, EC 1.11.1.9) is an enzyme family with peroxidase activity, and its biological function is the control of the organism defenses against oxidative damage. GPx can reduce lipid hydroperoxides to the corresponding alcohols and free hydrogen peroxide (H2O2) to H2O. GPxs were reported to be the first seleno enzymes discovered in mammals.1,2 The “classical” GPx, now referred to as GPx-1, is the most abundant isoform, found in the cytoplasm of almost all mammalian tissues, whose preferred substrate is also H2O2. It was first described as an erythrocyte enzyme that metabolized H2O2, long-chain fatty acid peroxides, and cholesterol, and controlled the protection of human red blood cells from hemolysis by the oxidation.3 In addition, GPxs have been characterized in many organisms, such as mammals, trematodes, nematodes, yeasts, bacteria, and even plants. Several different isozymes vary in substrate specificity and cellular location. To date, eight different isoforms GPx1-8 of humans have been separated and identified. Most of the human GPxs, such as GPx 1, 2, 3 and 4, contain a selenocysteine (Sec)-Cys residue at its active site and catalyze the substrate glutathione (GSH).4 Some members of the GPx family have been identified and classified into different isoforms according to their sequence similarity, conserved sequence regions, biochemical characterization, and substrate specifics.
Noteworthy are the protection mechanisms against oxidative damage used by organisms in the Antarctic sea ice, wherein the environment is characterized by the increased oxygen concentration correlated to low temperature and high salt concentration. The respective environmentally induced oxidative stress is assumed to be adjusted by the production of free radicals and reactive oxygen species (ROS).5,6 To eliminate the influence of ROS, Antarctic microorganism has developed a thorough defense system containing both high molecular mass antioxidants and low-molecular mass scavengers, also including antioxidant enzymes.5 In recent works, Antarctic fungi Penicillium sp. were found to resist the high concentration oxygen by means of increasing antioxidant enzymes' activity, for instance catalase (CAT) and superoxide dismutase (SOD).7 Other antioxidant enzymes from the Antarctic bacterium, such as thioredoxin, thioredoxin reductase, glutathione reductase and SOD were also studied through the recombinant protein characterization.8,9,10 Our earlier studies have revealed that glutathione S-transferase (GST) and SOD play the main roles in the protection mechanism against high salt concentration and low-temperature in the Antarctic bacterium Pseudoalteromonas sp.11,12 Among other antioxidant enzymes, GPxs are the ubiquitous cytosolic ones, which can catalyze the reduction of H2O2 and other organic peroxides. However, there are few reports about biochemical characterizations of the GPx protein. In this study, the cloning, expression, and biochemical properties of the GPx from Antarctic bacteria are discussed in detail.
Materials and methods
Bacterial strains, media, and growth conditions
Pseudoalteromonas sp. ANT506, isolated from sea ice in Antarctica (68°30′E, 65°00′S), was cultivated in the 2216E sea water medium with agitation of 150 rpm at 8°C. After incubation for 96 h, cells were collected by centrifugation at 12,000 × g for 15 min at 4°C, then were stored at −20°C.
Cloning of the PsGPx gene
The methodology flowchart is depicted in Fig. 1. The genomic DNA was extracted from strain ANT506 using DNA extraction kit (Tiangen, China). Two degenerated primers (5′-ACAACAACYATAMT AAGCVATGTAC-3′ and 5′-AKAAHTYTGATTAGTGATTGGCA-3′) were designed based on the upstream and downstream of the coding sequences of GPx from the Pseudoalteromonas genus. The PCR reaction parameters were as follows: one cycle of 94°C for 5 min, followed by 30 cycles of 94°C for 1 min, 50°C for 1 min, 72°C for 1.5 min, and 72°C for 10 min.
Figure 1.
The methodology flowchart for the cloning and heterologous expression of the PsGPx gene.
Sequence analysis
The sequence identity and similarity were analyzed via the DNAman software package, version 6.1 developed by Lynnon BioSoft (www.lynnon.com) and the basic local alignment search tool (BLAST) from the NCBI databases, respectively. The molecular mass and theoretical pI values were predicted by the ExPASy ProtParam tools, which were available at http://us.expasy.org/tools/protparam.html.
Overexpression and purification of PsGPx gene in E. coli
The PsGPx gene encoding the open reading frame (ORF) was amplified from the genomic DNA with the upstream following primers: 5′-GCGGGATCCATGATAAAAATAATATT-3′ (where BamHI site is underlined) and the downstream primer 5′-TTAAAGCTTGTTAATAGCTAATGTCG-3′ (where HindIII site is underlined). The reaction included one cycle of 94°C for 5 min, followed by 30 cycles of 94°C for 1 min, 58°C for 1 min, 72°C for 1.5 min, and 72°C for 10 min. The PCR product was directly cloned into the corresponding sites of the pET-28a (+) expression vector and transformed into E. coli BL21, then the PsGPx gene sequence was verified. The transformants with the PsGPx gene were grown in the Luria–Bertani (LB) medium supplemented with kanamycin (100 mg/L) and cultured in a rotary shaker at 150 rpm at 37°C until the OD600nm reached 0.6–0.8. Then, 0.12 mg/mL of isopropyl-β-D- thiogalactopyranoside (IPTG) was added for induction. Cells were cultured at 37°C for 5 h, and then the culture temperature was changed to 28°C, in order to induce the protein expression for additional 6 h.
The induced cells were centrifuged with 7,500 × g for 15 min at 4°C and disrupted by ultrasonication. The insoluble debris were removed by centrifuging with 12,000 × g for 15 min at 4°C, and the supernatant was collected as a crude protein. The Histidine-tagged protein purification of rPsGPx was conducted using Ni–NTA resins affinity chromatography, being previously equilibrated with the buffer. rPsGPx was eluted with the buffer (250 mM NaCl, 50 mM Tris-HCl, 100 mM imidazole, pH 8.0) at a flow rate of 1.0–1.2 mL/min. The rPsGPx purity and molecular mass were determined by SDS-PAGE using 12.5% polyacrylamide gels. SDS-PAGE under nonreducing condition (without 2-mercapto ethanol) was conducted as conducted previously.13
The rPsGPx activity assay and total protein concentration
GPx activity was determined by measuring the oxidation of NADPH in the presence of GSH reductase, which catalyzed the oxidation of GSH formed by GPx.14 One unit of GPx activity was defined as 1 µM NADPH oxidized per minute at 25°C and 340 nm. The protein content was measured according to the Bradford method using the BSA as a standard.15
Properties of purified recombinant PsGPx
To evaluate the temperature effect on the enzymatic activity, purified rPsGPx was assayed at different temperatures ranging from 0 to 60°C, and Tris-HCl (pH 7.0) was used as a standard reaction mixture. After incubating the enzyme for the designated time period (10–60 min), the thermal stability of purified rPsGPx was examined using the standard assay at 30°C, 40°C and 50°C. To investigate the pH effect on the activity of rPsGPx, the latter was measured at 25°C in the pH range from 5.0 to 11.0. To meaure the pH stability of rPsGPx, the latter was incubated for various pH values at 20°C for 1 h, and the residual activity was examined via the standard method. The following buffers (50 mM) were used: sodium acetate/acetic acid (pH 5.0); KH2PO4/Na2HPO4 (pH 6.0, 7.0); Tris-HCl (pH 8.0, 9.0) and Na2HPO4/NaOH (pH 10.0, 11.0). The effect of NaCl on the enzyme activity was assayed by adding NaCl (0–3.0 M) into the reaction mixture and incubating at 25°C for 1 h. The effect of different metal ions and various other reagents on the rPsGPx activity was investigated, with the concentration of the reagent shown in Table 1. The rPsGPx-containing compounds were incubated at pH 7.0 for 1 h at 25°C. The residual activity was examined via the standard method, whereas the activity assayed with no reagents was defined as the control (100% of the relative activity).
Table 1.
Effect of various compounds on the rPsGPx activity.
| Reagent | Concentration | Relative activity (%) | Reagent | Concentration | Relative activity (%) |
|---|---|---|---|---|---|
| None | — | 100.00 | Mn2+ | 5 mM | 101.12 |
| EDTA | 10 mM | 103.75 | Ni2+ | 5 mM | 100.37 |
| Thiourea | 10 mM | 126.59 | Cu2+ | 5 mM | 39.33 |
| SDS | 10 mM | 50.56 | Zn2+ | 5 mM | 88.01 |
| Urea | 10 mM | 120.59 | Fe3+ | 5 mM | 86.36 |
| DTT | 10 mM | 67.72 | K+ | 5 mM | 99.25 |
| Ca2+ | 5 mM | 85.39 | Tween-80 | 0.2% | 98.13 |
| Mg2+ | 5 mM | 98.50 | Triton X-100 | 0.2% | 110.49 |
| Fe2+ | 5 mM | 109.74 | H2O2 | 0.2% | 123.07 |
Kinetic studies
The apparent Km and Vmax values for PsGPx were determined using substrates H2O2 and GSH ranging from 0.5 to 8.0 mM, and the fixed amount of 50 µM NADPH at 25°C. Data were analyzed by the Lineweaver-Burk method.16
Results
Gene cloning and sequence analysis
The GPx gene was amplified from genomic DNA of P. sp. ANT506 and designated as PsGPx. The full-length GPx gene included an ORF of 585 bp, encoding a protein of 194 amino acid residues. The calculated mass of the recombinant PsGPx was approx. 21.7 kDa with a theoretical pI of 8.56. Furthermore, the GPx sequence was deposited to NCBI database as GenBank: KF802238. The multiple sequence alignment of the deduced protein sequence of PsGPx with available GPx sequences is depicted in Fig. 2. Three active site residues (Cys72, Gln103, and Trp154) were predicted in the conserved regions. Additionally, the dimer interface sites (Phe102, Glu105, Asn107, Asp110, Val114 and Val117) were identified in the sequence. The deduced amino acid sequence of rPsGPx has shared 88.8% of the sequence identity with P. haloplanktis GPx (YP_340977), 63.3% with P.agarivorans (WP_004589433), 57.7% with P.luteoviolacea (WP_005493401), 46.9% with S.baltica (YP_001555602), and 44.4% with S. frigidimarina (YP_751531). The protein sequence alignments revealed that rPsGPx belonged to the thioredoxin-like superfamily.
Figure 2.
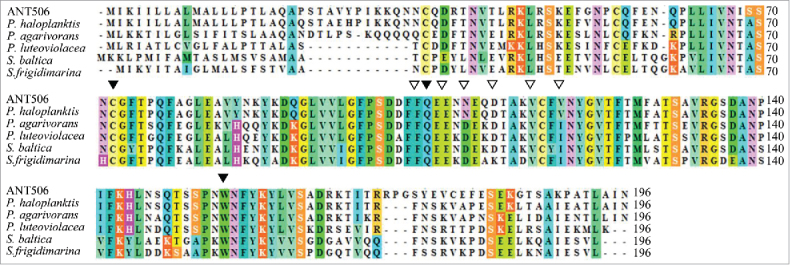
Multiple sequence alignment between the deduced PsGPx and other GPxs. The species names and GenBank accession numbers are as follows: Pseudoalteromonas haloplanktis TAC125 (YP_340977), Pseudoalteromonas agarivorans (WP_004589433), Pseudoalteromonas luteoviolacea (WP_005493401), Shewanella baltica OS155 (YP_001555602), Shewanella frigidimarina (YP_751531). ‘▾’ and ‘▽’ indicate the amino acids defining the active site and the polypeptide binding, respectively.
Expression and purification of PsGPx
For the overexpression of PsGPx, the recombinant plasmid containing the GPx gene was inserted into the HindIII and BamHI sites of the expression vector, and its expression was induced by IPTG. The PsGPx was found to be stably expressed in E. coli. After a single-step Ni–NTA resins, the rPsGPx was purified from the concentrated culture supernatant with a yield and purification of 18.68% and 3.51-fold, and the specific activity of the final rPsGPx is 21.08 U / mg. The purification procedure yielded an enzyme preparation, which was a clear single band judging from the SDS-PAGE results (Fig. 3). rPsGPx produced the same band under the nonreducing condition, which rPsGPx is a single subunit with no dimer.
Figure 3.
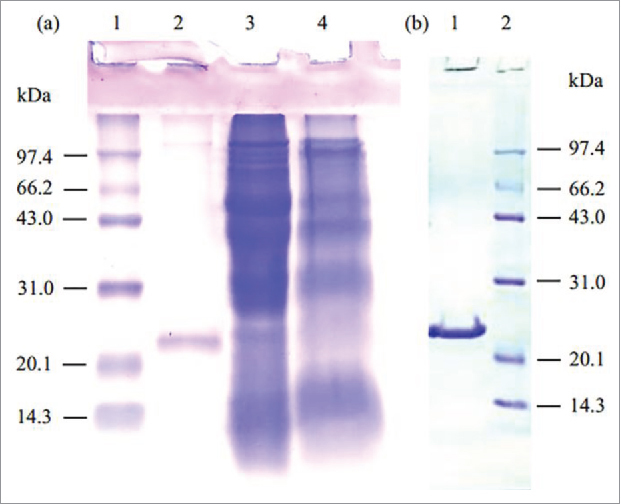
SDS-PAGE (12.5%) analysis of the purified rPsGPx. (a) Lane 1, protein molecular weight marker; Lane 2, the purified rPsGPx under reducing condition (with 2-mercapto ethanol) ; Lane 3, total extracts of the BL21/pETGPx cells with IPTG induction; Lane 4, total extracts of the BL21/pET-28a(+). (b) Lane 1, the purified rPsGPx under nonreducing condition (without 2-mercapto ethanol); Lane 2, protein molecular weight marker.
Biochemical characterization of rPsGPx
The temperature effect on rPsGPx activity and stability is illustrated by Fig. 4. The enzyme activity of rPsGPx was measured between 0°C and 60°C. As shown in Fig. 4a, rPsGPx exhibited the maximum activity at 30°C, and retained about 45% of the maximum activity at 0°C. In the thermal stability study, rPsGPx was also evaluated by measuring the residual activities. A high thermolability with a half-life of approx. 40 min at 40°C was observed (Fig. 4b). The stability of rPsGPx was reduced at 50°C, while the total loss of activity was observed after incubation for 50 min.
Figure 4.
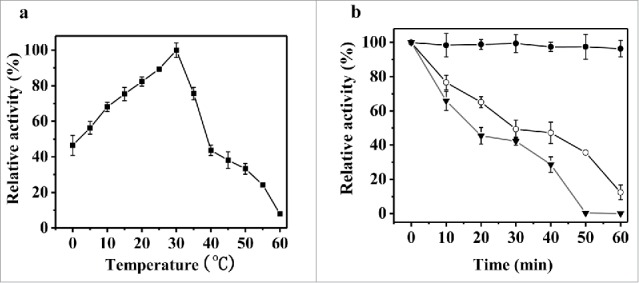
(a) Effect of temperatures on the purified rPsGPx activity. (b) Effect of temperatures on the stability of the purified rPsGPx. •30°C; (○) 40°C; (▾) 50°C. The residual catalytic activity after pre-incubation was assayed at 30°C, 40°C and 50°C for 10, 20, 30, 40, 50, and 60 min.
The pH effect on rPsGPx activity and stability is shown in Fig. 5. The rPsGPx activity was measured at pH 5.0–11.0. As shown in Fig. 5a, rPsGPx exhibited the maximum activity at pH 9.0 and conserved over 45.0% of the maximum activity in the pH range from 7.0 to 10.0. In the stability study, the rPsGPx was stable (over 80% of the maximum activity) in the broad pH range from 6.0 to 9.0 after 1 h at 20°C. Almost 73% of the maximum activity remained stable at pH 10.0 (Fig. 5b)
Figure 5.
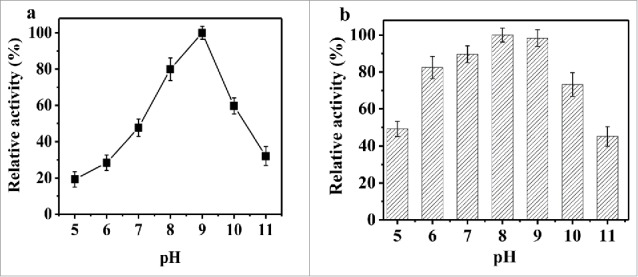
Effect of pH on enzymatic activity (a) and stability (b) of the purified rPsGPx.
The effect of NaCl on rPsGPx activity was measured using 0–3.0 M of NaCl. As shown in Fig. 6, over 90% of the maximum activity is manifested in the range from 0 to 1.0 M NaCl, while 69.10%-activity is observed in 3.0 M NaCl, indicating that rPsGPx has a good salt tolerance.
Figure 6.
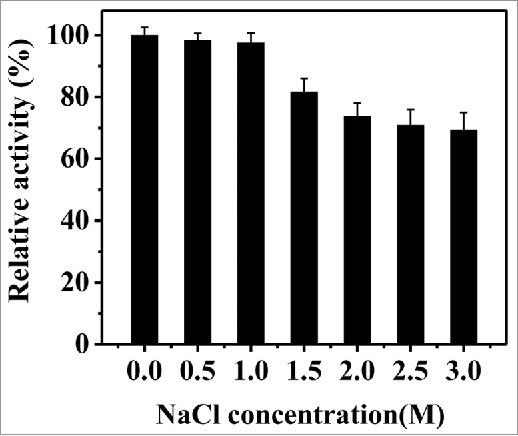
Effect of NaCl on the purified rPsGPx activity.
The effect of various reagents on the rPsGPx activity is presented in Table 1. In contrast to other investigated metal ions, which have no strong effect on the enzyme activity, Cu2+ has a strong inhibitory effect: reduces it by 60.67%. Although Thiourea, Urea, and Triton X-100 have only a slight activating effect on the GPx activity, while SDS and DTT have a strong inhibitory effect on the rPsGPx activity. In this study, H2O2 and Thiourea enhanced the rPsGPx activity by 23.07% and 26.59%, respectively. Moreover, the values of Km and Vmax were assessed as 1.73 mM and 16.28 nmol/mL/min using GSH as a substrate, respectively, while application of H2O2 as a substrate yielded 2.46 mM and 21.50 nmol/mL/min, respectively.
Discussion
The Antarctic extreme environment is the enormous source of novel enzymes. Some reports have focused on the identification and features of novel genes and their recombinant proteins from Antarctic bacterium. In the present study, the GPx was cloned from Antarctic psychrotrophic bacterium P. sp. ANT506, expressed, and characterized. According to the deduced amino acid sequence analysis, PsGPx consists of active site residues, corresponding to the known structures of other GPxs. The catalytic center of GPxs was firstly identified as a triad composed of Cys or Sec, Gln and Trp,17 and later was found to be a tetrad with an additional Asn.18 In this study, rPsGPx contained three active site residues (Cys72, Gln103, and Trp154), and the amino acid residues were conserved in all other members and were vital for active-site structure.17,19,20 Meanwhile, the GPx from adult Haemonchus contortus possessed active site residues Cys38, Gln73, and Trp127,19 whereas the GPx from halotolerant Chlamydomonas sp. W80 contained amino acid residues Cys37, Gln71, and Trp125.20 It illustrates that the catalytic sites of glutathione peroxidase from various sources are different. The Km assay of rPsGPx demonstrated that it could catalyze H2O2 in the presence of GSH, which further proved that rPsGPx was a GPx of the thioredoxin-like family.19
As compared with other reported GPxs from different organisms, rPsGPx exhibits some distinct enzymatic properties. For example, rPsGPx manifested the highest activity at 30°C and retained at least 40% activity even at 0°C (Fig. 4a). This observation implies that rPsGPx is a highly efficient catalyst at low temperatures. However, optimal temperature for the recombinant lvGPx from the white shrimp Litopenaeus vannamei was 55°C,21 while that for GPx from Chlamydomonas sp. W80 was 40°C.20 It was also found that the optimal temperature for the GI-GPx from human was 25°C, but the enzyme was observed to be unstable, with almost total activity loss after 24 h-incubation at 37°C and 72 h-incubation at 4°C.22 In this study, rPsGPx retained 47% of the maximum highest activity at 40°C for 40 min (Fig. 4b). Meanwhile, the reported GPx from Chlamydomonas sp. remained stable up to 40°C with the maximum activity, with a complete stability loss at 70°C, which is higher than the rPsGPx thermal stability.20 It is well known that the general characteristics of the cold active enzymes are low thermal stability and high catalytic activity at low temperatures. Therefore, rPsGPx is a typical cold active enzyme. With respect to pH, rPsGPx exhibited the optimum activity at pH 9.0 (Fig. 5a), which is similar to some other GPxs like Litopenaeus vannamei GPx (pH 9.0),21 Chlamydomonas sp. W80 (pH 8.0),20 Hyalomma dromedarii TEGPx1 and TEGPx2 (pH 7.6 and 8.2, respectively),23 and Oryza sativa GPx (pH 9.3),24 all of which exceed that of the human gastrointestinal GPx (pH 7.4).22 A relatively high activity of PsGPx at pH 9.0 strongly suggests that pH is critical for the regulation of this enzyme.20 PgGPX1 and PgGPX2 exhibited a low activity with 100 mM NaCl treatment.25 However, rPsGPx retained 69.1% of its activity in 3.0 M NaCl (Fig. 6), which indicates that rPsGPx is a low-temperature and salt-tolerant enzyme adapted to the Antarctic sea ice environment with a low-temperature and high salt concentration. In fact, for Antarctic bacterium, the oxidative stress caused by extreme environment can induce the formation of hydroxyl radicals and reactive oxygen species.5 In addition, H2O2 enhanced the enzyme activity by 23.07%. Noteworthy is that rPsGPx exhibited a higher catalytic efficiency of Km and Vmax with H2O2 substrate, as compared to those with the GSH substrate. This can be attributed to the fact that rPsGPx is mainly responsible for reduction of H2O2-detoxifying peroxidases under normal physiological conditions, while rPsGPx can participate in the detoxification of H2O2, when very high concentrations of endogenous H2O2 are accumulated. This strongly suggests that PsGPx is vital for protecting cellular components from oxidative damage caused by the Antarctic sea ice environmental stresses.
Conclusions
In this study, a PsGPx gene was successfully cloned and expressed into E. coli. The expressed target protein was purified by the Ni-NTA affinity chromatography. Properties of rPsGPx were investigated depending on the purity of the enzyme. The maximum catalytic temperature and pH value for rPsGPx were 30°C and pH 9.0, respectively. rPsGPx had 45% of the maximum activity at 0°C and high thermolability with a half-life of approx. 40 min at 40°C. In addition, the enzymatic activity of rPsGPx was still manifested in 3 M NaCl. At present, expression patterns of rPsGPx at low temperature, high salinity and dissolved oxygen are undergoing, in order to further explore the mechanism of Antarctic sea-ice bacteria.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This study was supported by the National Natural Science Foundation of China (31100037), Subject Construction Fund Guided by HIT (WH20150206, WH20160206), Key Research and Development Plan of Shandong Province (2015GSF115016), and the Natural Science Foundation of Shandong Province (ZR2017MC046).
Authors' contributions
Yanhua Hou and Quanfu Wang have conceived this study. Bingqing Cui performed the cloning and heterologous expression of the gene, while Yatong Wang and Yifan Wang performed characterization of the recombinant enzyme. Yanhua Hou, Yatong Wang and Quanfu Wang analyzed the data. Yatong Wang, Han Han and Yanhua Hou wrote the manuscript.
References
- [1].Flohe L, Gunzler W, Schock H. Glutathione peroxidase: a selenoenzyme. Febs Lett. 1973;32:132-134. doi: 10.1016/0014-5793(73)80755-0. PMID:4736708 [DOI] [PubMed] [Google Scholar]
- [2].Sies H, Sharov V, Klotz L, Briviba K. Glutathione peroxidase protects against peroxynitrite-mediated oxidations: A new function for selenoproteins as peroxynitrite reductase. J Biol Chem. 1997;272:27812-27817. doi: 10.1074/jbc.272.44.27812. PMID:9346926 [DOI] [PubMed] [Google Scholar]
- [3].Mills G. Hemoglobin catabolism I. Glutathione peroxidase, an erythrocyte enzyme which protects hemoglobin from oxidative breakdown. J Biol Chem. 1957;229:189-197. PMID:13491573 [PubMed] [Google Scholar]
- [4].Valentina B, Marcus C, Giorgio C, Alessandro N, Slivia Q, Monica R, Antonella R, Stefano T, Fulvio U, Mattia Z, et al.. Protein disulfide isomerase and glutathione are alternative substrates in the one Cys catalytic cycle of glutathione peroxidase 7. Biochim Biophys Acta. 2013;1830:3846-3857. doi: 10.1016/j.bbagen.2013.02.017. PMID:23454490 [DOI] [PubMed] [Google Scholar]
- [5].Chattopadhyay M, Raghu G, Sharma Y, Biju A, Rajasekharan M, Shivaji S. Increase in oxidative stress at low temperature in an Antarctic bacterium. Curr Microbiol. 2011;62:544-546. doi: 10.1007/s00284-010-9742-y. PMID:20730433 [DOI] [PubMed] [Google Scholar]
- [6].Russell N. Psychrophilic bacteria-molecular adaptations of membrane lipids. Comp Biochem Physiol. 1997;118:489-493. doi: 10.1016/S0300-9629(97)87354-9 [DOI] [PubMed] [Google Scholar]
- [7].Gocheva Y, Krumova E, Slokoska L, Miteva J, Vassilev S, Angelova M. Cell response of Antarctic and temperate strains of Penicillium spp. to different growth temperature. Mycol Res. 2006;110:1347-1354. doi: 10.1016/j.mycres.2006.08.007. PMID:17070679 [DOI] [PubMed] [Google Scholar]
- [8].Cotugno R, Ruocco M, Marco S, Falasca P, Evangelista G, Raimo G, Chambery A, Di Maro A, Masullo M, De Vendittis E. Differential cold-adaptation among protein components of the thioredoxin system in the psychrophilic eubacterium Pseudoalteromonas haloplanktis TAC 125. Mol Biosyst. 2009;5:519-528. doi: 10.1039/b818467d. PMID:19381366 [DOI] [PubMed] [Google Scholar]
- [9].Ji M, Barnwell C, Grunden A. Characterization of recombinant glutathione reductase from the psychrophilic Antarctic bacterium Colwellia psychrerythraea. Extremophiles. 2015;19:863-874. doi: 10.1007/s00792-015-0762-1. PMID:26101017 [DOI] [PubMed] [Google Scholar]
- [10].Abrashev R, Feller G, Kostadinova N, Krumova E, Alexieva Z, Gerginova M, Spasova B, Miteva-Staleva J, Vassilev S, Angelova M. Production, purification, and characterization of a novel cold-active superoxide dismutase from the Antarctic strain Aspergillus glaucus 363. Fungal Biol. 2016;120:679-689. doi: 10.1016/j.funbio.2016.03.002. PMID:27109365 [DOI] [PubMed] [Google Scholar]
- [11].Shi Y, Wang Q, Hou Y, Hong Y, Han X, Yi J, Qu J, Lu Y. Molecular cloning, expression and enzymatic characterization of glutathione S-transferase from Antarctic sea-ice bacteria Pseudoalteromonas sp. ANT506. Microbiol Res. 2014;169:179-184. doi: 10.1016/j.micres.2013.06.012 [DOI] [PubMed] [Google Scholar]
- [12].Wang Q, Wang Y, Hou Y, Shi Y, Han H, Miao M, Wu Y, Liu Y, Yue X, Li Y. Cloning, expression and biochemical characterization of recombinant superoxide dismutase from Antarctic psychrophilic bacterium Pseudoalteromonas sp. ANT506. J Basic Microbiol. 2016;56:753-761. doi: 10.1002/jobm.201500444 [DOI] [PubMed] [Google Scholar]
- [13].Hakimi H, Suganuma K, Usui M, Masuda-Suganuma H, Angeles J, Asada M, Kawai S, Lnoue N, Kawazu S. Plasmodium knowlesi thioredoxin peroxidase 1 binds to nucleic acids and has RNA chaperone activity. Parasitol Res. 2014;113:3957-3962. doi: 10.1007/s00436-014-4060-0. PMID:25092384 [DOI] [PubMed] [Google Scholar]
- [14].Flohé L, Günzler W. Assays of glutathione peroxidase. Meth Enzymol. 1984;105:114-121. doi: 10.1016/S0076-6879(84)05015-1. PMID:6727659 [DOI] [PubMed] [Google Scholar]
- [15].Bradford M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248-254. doi: 10.1016/0003-2697(76)90527-3. PMID:942051 [DOI] [PubMed] [Google Scholar]
- [16].Lineweaver H, Burk D. The determination of the enzyme dissociation constants. J Am Chem Soc. 1934;56:658-666. doi: 10.1021/ja01318a036 [DOI] [Google Scholar]
- [17].Epp O, Ladenstein R, Wendel A. The refined structure of the selenoenzyme glutathione peroxidase at 0.2-nm resolution. Eur J Biochem. 1983;133:51-69. doi: 10.1111/j.1432-1033.1983.tb07429.x. PMID:6852035 [DOI] [PubMed] [Google Scholar]
- [18].Tosatto S, Bosello V, Fogolari F, Mauri P, Roveri A, Toppo S, Flohe L, Ursini F, Maiorino M. The catalytic site of glutathione peroxidases. Antioxid Redox Sign. 2008;10:1515-1526. doi: 10.1089/ars.2008.2055 [DOI] [PubMed] [Google Scholar]
- [19].Sun W, Song X, Yan R, Xu L, Li X. Cloning and characterization of a selenium-independent glutathione peroxidase (HC29) from adult Haemonchus contortus. J Vet Sci. 2012;13:49-58. doi: 10.4142/jvs.2012.13.1.49. PMID:22437536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Takeda T, Miyao K, Tamoi M, Kanaboshi H, Miyasaka H, Shigeoka S. Molecular characterization of glutathione peroxidase-like protein in halotolerant Chlamydomonas sp. W80. Physiol Plant. 2003;117:467-475. doi: 10.1034/j.1399-3054.2003.00075.x [DOI] [PubMed] [Google Scholar]
- [21].Wang L, Wu J, Wang W, Cai D, Liu Y, Wang A. Glutathione peroxidase from the white shrimp Litopenaeus vannamei: characterization and its regulation upon pH and Cd exposure. Ecotoxicology. 2012;21:1585-1592. doi: 10.1007/s10646-012-0942-z. PMID:22684731 [DOI] [PubMed] [Google Scholar]
- [22].Guo X, Song J, Guan T, Wang S, Wang Y, Meng Y, Guo J, Li T, Ma C, Wei J. Characterization of recombinant human gastrointestinal glutathione peroxidase mutant produced in Escherichia coli. Free Radical Res. 2015;49:228-235. doi: 10.3109/10715762.2014.995182 [DOI] [PubMed] [Google Scholar]
- [23].Ibrahim M, Mohamed M, Ghazy A, El-Mogy M, Masoud HM. Purification and characterization of two glutathione peroxidases from embryo of the camel tick Hyalomma dromedarii. Russ J Bioorg Chem. 2016;42:272-281. doi: 10.1134/S1068162016030092 [DOI] [Google Scholar]
- [24].Wang Z, Wang F, Duan R, Liu J. Purification and physicochemical characterization of a recombinant phospholipid hydroperoxide glutathione peroxidase from Oryza sativa. J Biochem Mol Biol. 2007;40:412-418. PMID:17562293 [DOI] [PubMed] [Google Scholar]
- [25].Kim Y, Jang M, Noh H, Lee H, Sukweenadhi J, Kim J, Kim S, Kwon W, Yang D. Molecular characterization of two glutathione peroxidase, genes of Panax ginseng, and their expression analysis against environmental stresses. Gene. 2014;535:33-41. doi: 10.1016/j.gene.2013.10.071. PMID:24269671 [DOI] [PubMed] [Google Scholar]


