Abstract
Fluorescence microscopy is an essential tool for the exploration of cell growth, division, transcription and translation in eukaryotes and prokaryotes alike. Despite the rapid development of techniques to study bacteria, the size of these organisms (1–10 μm) and their robust and largely impenetrable cell envelope present major challenges in imaging experiments. Fusion-based strategies, such as attachment of the protein of interest to a fluorescent protein or epitope tag, are by far the most common means for examining protein localization and expression in prokaryotes. While valuable, the use of genetically encoded tags can result in mislocalization or altered activity of the desired protein, does not provide a readout of the catalytic state of enzymes and cannot enable visualization of many other important cellular components, such as peptidoglycan, lipids, nucleic acids or glycans. Here, we highlight the use of biomolecule-specific small-molecule probes for imaging in bacteria.
While bacteria do not have the distinct internal compartments seen in eukaryotes, these unicellular organisms possess highly organized cellular structures that are essential to orchestrating cell growth, division, motility, DNA transcription, protein translation and secretion in a well-defined synchrony1. Visualization of interactions between the constituents of this organization is critical to our understanding of bacterial growth and pathogenesis, and although great progress has been made in these areas in the past decade, many questions cannot be addressed using existing techniques. Imaging-based methods are and will continue to be essential for the study of the biomolecules involved in cellular and physiological processes in their native environment, with fluorescence microscopy being particularly valuable for examination of the subcellular structures of bacteria.
Until the application of fluorescent tagging techniques in bacteria, these cells were considered amorphous bags in which chromosomes and proteins were randomly distributed. However, work over the last several decades has revealed that bacteria are well-organized multicomponent systems and that individual proteins, such as the cytoskeletal proteins MreB and FtsZ and the chemoreceptors, localize to specific sites of the cell in a dynamic fashion1,2. Fluorescent protein fusion (for example, with green fluorescent protein, GFP; Fig. 1a)3 and epitope tagging (for example, with FLAG or c-Myc; Fig. 1b)4,5 are the most widely used strategies for monitoring protein expression and localization in bacteria3,5,6. Small protein and peptide tags have also been used to facilitate labeling, whereby a fluorescent tag is conjugated to the protein of interest by association with a tetracysteine motif (CCXXCC; FlAsH or ReAsH)7,8 or through the action of enzymes such as biotin ligase or sortase9. Alternatively, the protein target is fused to an enzyme that can be specifically labeled with a small dye-containing substrate (such as HaloTag or SNAP-tag)9.
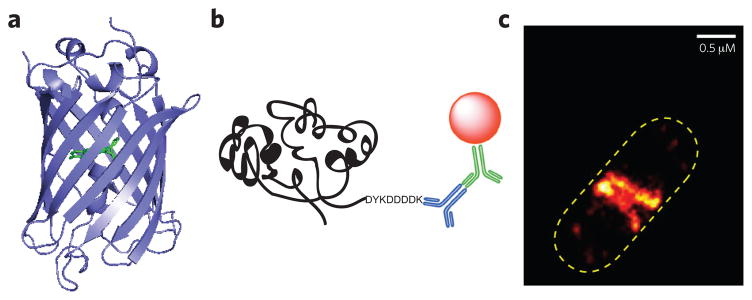
Figure 1. Genetically encoded tags for bacterial imaging.
(a) Crystal structure of green fluorescent protein (GFP) (PDB 1GFL). Ser65-Tyr66-Gly67 residues that form the chromophore (p-hydroxybenzylideneimidazolinone) are highlighted in green. (b) Schematic representation of FLAG epitope tag. The FLAG tag is an octapeptide with a sequence of DYKDDDDK. Cartoon representation shows detection of the FLAG sequence by an anti-FLAG primary antibody (blue), followed by the introduction of secondary antibody (green) carrying a fluorophore (red sphere). (c) E. coli expressing FtsZ-mEos2 imaged with PALM. The number of molecules used to construct the PALM image was 2,843. Image in c reproduced from ref. 18.
Ideally, the addition of this appendage, which can be rather large (for example, 27 kDa for GFP), does not affect the functionality or localization of the targeted protein. However, recent work has demonstrated that the fusion component can dramatically affect protein localization10,11. For instance, the actin-like protein MreB in Bacillus subtilis and Escherichia coli was thought to be organized in a helical structure, on the basis of microscopy results obtained with fluorescent-protein-tagged constructs (GFP and YFP, respectively). However, it has subsequently been shown that MreB does not form a helical pattern during exponential growth and that this pattern was an artifact of the protein tag12–14. Although fluorescent protein tags have been undeniably valuable for protein localization studies, it is now clear that target proteins need to be stringently tested for functionality (such as native-like catalytic activity and growth phenotype) with various fluorescent tags.
Another critical barrier that must be overcome in the study of bacteria arises from the fact that they are so small (1–10 μm). The spatial resolution of conventional fluorescence microscopes is insufficient for this, being limited by the diffraction of light to 200–300 nm and 500–700 nm in the lateral and axial dimensions, respectively. Thus, to adequately visualize the subcellular structures of these organisms, the use of super-resolution (SR) microscopy has become essential (Box 1)15,16. SR imaging has already enabled elucidation of several bacterial processes that was not possible with conventional optical microscopy. For example, the actions of FtsZ, a tubulin-like GTPase involved in cell division that recruits other proteins to the division site for cytokinesis, have been mapped in multiple organisms. In vitro polymerization of FtsZ showed that this protein could assemble into short, single-stranded protofilaments17. These protofilaments were thought to form bundles to produce a ring, called the Z-ring, during division in vivo, but the optical resolution of conventional microscopes was insufficient to validate this hypothesis in live cells. When, more recently, PALM was used to image the Z-ring of E. coli, it showed that protofilaments make randomly overlapping bundles and form a Z-ring with a width of ~110 nm (FtsZ-GFP or FtsZ-mEos2 fusions; Fig. 1c)18,19. Similarly, this technique was used to examine the organization of FtsZ in live Caulobacter crescentus. FtsZ in this organism localizes mainly to the midcell as a patchy band because of the random distribution of protofilaments (FtsZ-Dendra2 fusion)20. Finally, 3D-SIM was used to examine FtsZ localization and Z-ring structure in B. subtilis and Staphylococcus aureus, showing that there is heterogeneous FtsZ distribution in the Z-ring and the ring has a bead-like discontinuous pattern (FtsZ-GFP)21.
Box 1. Super-resolution microscopy.
Several platforms for super-resolution (SR) microscopy have been devised. Importantly, SR microscopy techniques often require a limited set of fluorophores, with each technique necessitating fluorescent molecules with different optical properties.
Structured-illumination microscopy (SIM) and stimulated emission depletion (STED) achieve improved resolution by reducing the size of the effective point spread function (PSF), the diffraction pattern produced by an infinitely small point source94–96. SIM stands out among the SR microscopy methods in that it is not dependent upon specialized fluorescent molecules. It can be used with the same variety of fluorescent protein tags and commercially available fluorophores as wide-field fluorescence microscopes, making this method perhaps the most amenable to the utilization of small-molecule probes15,97. SIM enables 100- to 130-nm lateral and 250- to 340-nm axial resolution. Because STED microscopy requires fluorophores with high-intensity brightness and photostability, ATTO dyes have been widely used for this technique, and resolutions of ~50 nm lateral and ~100 nm axial are possible with this combination.
Methods that are based on the sequential detection of discrete fluorophores, such as photoactivated localization microscopy (PALM) and stochastic optical reconstruction microscopy (STORM), enable single-molecule imaging as they can achieve resolutions of ~20 nm lateral and ~50 nm axial98–100. These techniques necessitate the use of specialized fluorophores, with PALM requiring photoactivatable fluorescent protein tags, such as Dendra, Dronpa, Eos and mCherry, and STORM using photoswitchable organic fluorophores including Cy3, Cy5 and Alexa Fluor 647.
The vast majority of studies to examine bacterial substructures have used fluorescent protein fusions and epitope tags to shed light on protein expression and localization. While important, genetically encoded tags are not applicable to the visualization of cellular components such as peptidoglycan (PG), lipids, nucleic acids, metabolites or glycans22. Nucleic-acid-based probes have been used with much success and have been described elsewhere23–27. Here, we focus on small-molecule probes that target the cell envelope components and/or provide a readout of enzymatic activity. These compounds are much less likely to perturb the native system than encoded tags28. However, a major obstacle in their application is the largely impenetrable cell envelope of these organisms. Because of that, the majority of studies using these compounds to date have focused on visualization of the extracellular components (Table 1).
Table 1.
Highlighted probes and their cellular targets
| Probe | Target of probe | Cell type and labeling conditions |
|---|---|---|
| BOCILLIN-FL | PBP activity | Live Gram positive and Gram negative |
| Ceph C-TAMRA | PBP activity | Live Gram positive |
| Van-FL | PG stem peptide | Live Gram positive |
| Ramoplanin-FL | PG stem peptide | Live Gram positive |
| UDP-MurNAc pentapeptide-FL | Nascent PG | Live Gram positive, permeabilized Gram negative |
| L-Alanyl-γ-D-glutamyl-L-lysine analog | PG stem peptide | Live Gram negative |
| FDAA | PG stem peptide | Live and permeabilized Gram positive and Gram negative |
| Trehalose analogs | Mycomembrane | Live mycobacterium |
| KDO | LPS | Live Gram negative (as no LPS in Gram positive) |
| Rhodamine spirolactam, Alexa Fluor 488 succinimidyl ester, Cy3-Cy5-succinimidyl ester | Cell surface amines | Live Gram positive and Gram negative |
| Noncanonical amino acids | Newly synthesized and secreted proteins | Live or permeabilized Gram negative |
| Nitro-aryl fluorogen | Nitroreductase activity | Live Gram positive and Gram negative |
Antibiotic-inspired chemical probes
PG, also called murein, is a major component of the bacterial cell wall, which maintains the organism’s shape and protects it from turgor pressure. PG is a polymeric structure composed of alternating β-(1,4)-linked N-acetylglucosamine (GlcNAc) and N-acetylmuramic (MurNAc) acid glycan chains that are crosslinked by pentapeptides (Fig. 2)29. Although most bacteria have PG, the length of the glycan chains and the amino acid composition of the stem peptides vary between organisms30,31. This structure is unique to bacteria, and inhibition of its biosynthesis by antibacterial agents leads to cell lysis32. A particularly fruitful strategy for probe design has been to harness the power of known antibiotic scaffolds that interfere with cell wall construction.
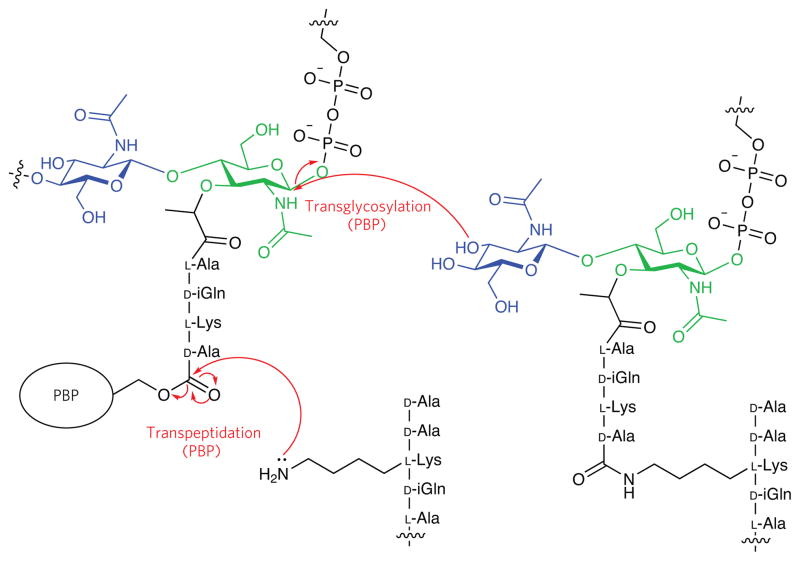
Figure 2. Schematic representation of bacterial peptidoglycan (PG).
PG is composed of alternating β-(1,4)-linked N-acetylglusosamine (blue) and N-acetylmuramic acid (green) glycan chains that are crosslinked by peptides. Polymerization (transglycosylation) and crosslinking (transpeptidation) between glycan chains are catalyzed by the PBPs. The peptides shown are those from S. pneumoniae.
The PG precursors (glycan chains) are polymerized and crosslinked by membrane-anchored proteins called the penicillin-binding proteins (PBPs)33,34. Although their catalytic functions are conserved, most organisms possess a suite of PBPs that are postulated to be critical in different stages of cellular growth and division. The peptide crosslink formed by the PBPs is required for to cellular integrity, and inhibition of PBP transpeptidation activity is the basis for the antibacterial activity of penicillin and other β-lactams. The β-lactam ring is a structural mimetic of the terminal portion of the PG stem peptide substrate, D-alanyl-D-alanine (D-Ala-D-Ala), and covalently labels the PBPs in an activity-dependent fashion35,36. Accordingly, the β-lactam scaffold has served as inspiration for a number of probes devised to examine bacterial protein function.
Fluorescent β-lactams such as BODIPY-conjugated penicillin V analogs, BOCILLIN FL (1; Fig. 3a) and BOCILLIN 650/665 (derived from BODIPY FL and BODIPY 650/665, respectively)37,38 have been used in gel-based studies to examine global PBP activity in both membrane preparations and live cells, a strategy also known as activity-based protein profiling39,40. Additionally, a set of β-lactam probes based on both natural and synthetic compounds appended with an alkyne tag have been reported. Following labeling of target proteins with these molecules, the bacteria are lysed and a fluorophore or biotin conjugated to an azide handle is attached using the Hüisgen 1,3-dipolar cycload-dition reaction (‘click chemistry’) for in-gel fluorescence detection or mass spectrometry (MS) analysis, respectively. Natural β-lactam analogs label PBPs, whereas synthetic monocyclic β-lactam probes have affinity for virulence-associated enzymes41. Similarly, a library of β-lactone probes has been synthesized and applied to several Gram-positive and -negative bacteria. Identification of the labeled proteins by MS revealed that most of the probe-labeled proteins are involved in primary and secondary metabolism, virulence, antibiotic resistance and detoxification42–44.
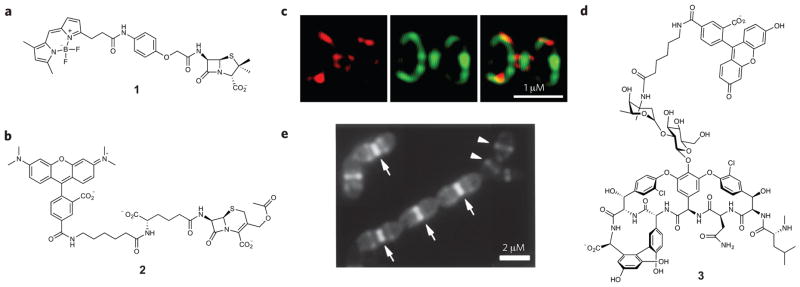
Figure 3. Antibiotic-inspired probes and images obtained with these compounds.
(a,b) Structures of PBP imaging probes. Probe 1 (a) labels all PBPs that are present in bacteria, whereas molecule 2 (b) selectively targets only a subset of PBPs in B. subtilis and S. pneumoniae. (c) 3D-SIM images of PBPs in S. pneumoniae after dual labeling with 2 (red) and 1 (green). (d) Vancomycin-fluorescein (3) enables visualization of nascent PG synthesis. (e) Nascent PG in S. pneumoniae cells is labeled with molecule 3. Arrows indicate heavily stained division septum. Arrowheads indicate lightly stained equators of the daughter cells that will become division sites. Figures reproduced with permission from: c, ref. 45; e, ref. 49.
To enable visualization of the actions of discrete PBPs, we reported the design and synthesis of cephalosporin-based probes that target a PBP subset, the high-molecular-weight homologs. We synthesized cephalosporin C–based probes (Ceph C–carboxyte-tramethylrhodamine (Ceph C-T), 2; Fig. 3b) and demonstrated that they selectively label a small subset of PBPs in B. subtilis and Streptococcus pneumoniae. Fluorescence imaging with 3D-SIM indicated that tagged PBPs localized at the division site in both organisms45,46. Dual labeling experiments were performed by incubating whole cells with Ceph C-T to label Ceph C–sensitive PBPs, and then BOCILLIN FL was applied to saturate the remainder of the PBPs, enabling simultaneous analysis of two PBP subsets. Tagged PBPs were localized at the division site but were not mixed; instead, they displayed distinct localization patterns (Fig. 3c). Such details were not observed with wide-field fluorescence microscopes before our study. Recently, we reported PBP selectivity profiling of 20 β-lactam antibiotics in E. coli and S. pneumoniae that will serve as a foundation for future probe generation47,48.
Vancomycin is a clinically important antibiotic that targets PG biosynthesis by binding to the D-Ala-D-Ala motif of the stem peptide. Vancomycin binds to nascent PG, and it was postulated that a fluorescent version of this antibiotic (Van-FL, 3) could be used as a tool to visualize PG biosynthesis in live cells. Exponentially growing B. subtilis cells treated with 3 show the brightest staining at the division site where nascent PG synthesis was predicted to occur, along with less apparent sidewall staining in a helical pattern (Fig. 3d,e)49. Van-FL has been applied in other Gram-positive bacteria such as S. pneumoniae, Streptomyces coelicolor and Corynebacterium glutamicum and now serves as an important marker for active PG biosynthesis. Another antibiotic, ramoplanin, targets cell wall biosynthesis by sequestering lipid II. Fluorescein-conjugated ramoplanin was used as a chemical probe on B. subtilis cells and displayed dose-dependent labeling. At low probe concentration, staining at the divisional septa and the cell poles, as well as helix-like sidewall patterns, were observed50. The fluorescent variants of both vancomycin and ramoplanin have been valuable tools for understanding PG biosynthesis, but their use is limited to Gram-positive bacteria because the outer membrane of Gram-negative bacteria acts as a barrier to these large molecules.
Peptidoglycan-inspired chemical probes
Imaging of the bacterial cell wall has also been accomplished through the incorporation of unnatural biosynthetic precursors or substrate mimics51. For example, fluorophore-tagged UDP-MurNAc pentapeptide was synthesized and tested for incorporation into bacterial cells52. Fluorescence microscopy images show that this UDP-MurNAc analog is readily incorporated by Gram-positive bacteria. Pretreatment of Gram-negative bacteria with EDTA weakened the thick liposaccharide layer and enabled incorporation of the probe in these cells53. In another study, a derivative of the PG tripeptide, L-alanyl-γ-D-glutamyl-L-lysine, was generated with N-7-nitro-2,1,3-benzoxadiazol-4-yl in the lysine position for in situ detection of nascent PG synthesis in E. coli54.
The use of fluorophore-conjugated D-amino acids that can be incorporated into the PG stem peptide has been reported by several research groups (fluorescent D-amino acid (FDAA) labeling; Fig. 4a). D-Alanine and D-lysine analogs containing small fluorophores that enable detection of newly synthesized PG in live cells by fluorescence microscopy have been developed (hydroxycoumarin-carbonyl-amino-D-alanine, HADA; 4)55. These probes were initially tested on various Gram-positive and -negative bacteria such as B. subtilis, E. coli, S. aureus, S. pneumoniae, Agrobacterium tumefaciens and C. crescentus. A clickable alkyne-containing D-alanine analog ((R)-propargylglycine; 5) has also been reported and used to label nascent PG of Listeria monocytogenes during macrophage infection (Fig. 4b)56. More recently, cyclooctyne D-amino acids were combined with near-infrared fluorogenic azides to facilitate visualization of PG without the need for washes to remove unreacted probe (Fig. 4c,d)57. Intriguingly, several studies have demonstrated that the efficiency of incorporation of a number of DAA derivatives is organism specific58,59, information that is useful for probe design.
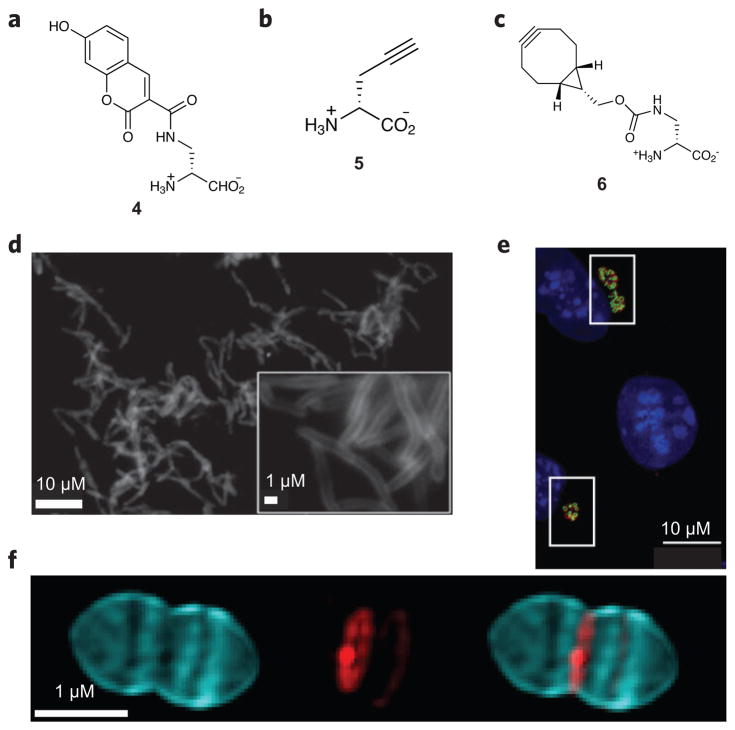
Figure 4. Peptidoglycan-inspired probes and images obtained with these compounds.
(a,b) Fluorescent D-amino acids hydroxycoumarin-carbonyl-amino-D-alanine (4; a) and (R)-propargylglycine (5; b) for tracking PG production. (c) The cyclooctyne D-alanine analog endobcnDala (6) used for no-wash PG labeling. (d) Fluorescence image of M. smegmatis treated with 6 and labeled with bis-sulfated glycol-substituted azido silicon rhodamine. (e) Fluorescence image of mouse fibroblast cells (L2 cells) infected with C. trachomatis and labeled with the dipeptide ethynyl-D-alanyl-D-alanine followed by an azide-modified Alexa Fluor 488 (green). An antibody for chlamydial major outer membrane protein (red) was used to label chlamydial elementary bodies and reticulate bodies, and 4′,6-diamidino-2-phenylindole (DAPI; blue) was used for nuclear staining. (f) 3D-SIM images of S. pneumoniae PG pulse-chase labeled with 4 (blue) and tetramethylrhodamine 3-amino-D-alanine (red). Using this strategy, older PG (blue) can be differentiated from areas of new PG synthesis (red). Figures reproduced with permission from: d, ref. 57; e, ref. 65; f, ref. 67.
Since their introduction, FDAAs have been used in a number of investigations60,61, including elegant studies to determine if Chlamydiae produce PG, a question that has been investigated for decades using indirect techniques62,63. Probes based on D-alanine and D-alanyl-D-alanine were used in Protochlamydia amoebophila and Chlamydia trachomatis, respectively (Fig. 4e). Fluorescence microscopy imaging demonstrates conclusively that these Chlamydia species do in fact synthesize PG64,65. PG assembly in ovoid-shaped bacteria (ovococci) has also been examined with FDAAs in combination with other tools, including Van-FL. These experiments revealed that ovococcus cells have a unique PG structure that is dissimilar to those of other Gram-positive bacteria (Fig. 4f)66,67. None of these studies would have been possible without the generation of applicable small-molecule probes as they required direct visualization of the PG biopolymer.
Carbohydrate-derived chemical probes
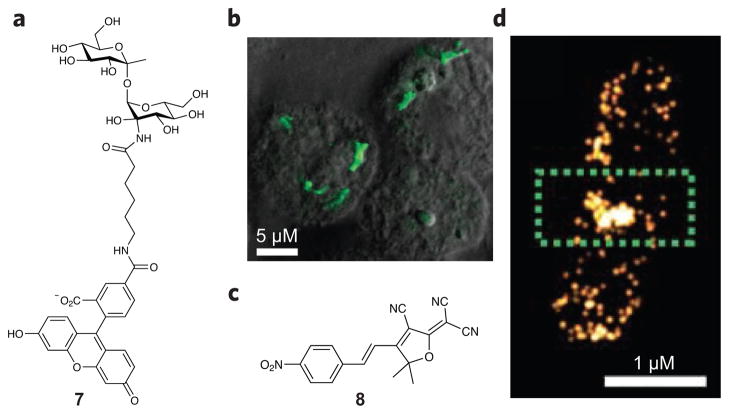
Bacterial cells are surrounded by glycan structures that have been shown to have important functions in pathogenesis. For example, Gram-positive bacteria have teichoic acids on their cell surface, and organisms lacking these sugar moieties exhibit an attenuated ability to cause infection68. Mycobacterial species have a thick PG layer, similar to that of Gram-positive bacteria, and an unique outer membrane called the mycomembrane, which is composed of a layer of arabinogalactan and mycolic acids that is coated with the non-mammalian disaccharide trehalose69. Absence of trehalose causes proliferation defects70. As a specific and sensitive strategy to detect mycobacteria such as Mycobacterium tuberculosis, a fluorescein- conjugated trehalose analog (7) has been developed and used for visualization of M. tuberculosis within infected macrophages (Fig. 5a,b)71. Azide-functionalized trehalose analogs have been generated as bioorthogonal chemical reporters for imaging72,73. This strategy is suitable for studying mycobacterial glycolipid function and dynamics in living cells.
Figure 5. Carbohydrate-inspired and turnover-dependent probe designs.
(a) Fluorescein-conjugated trehalose (7) for selective detection of M. tuberculosis. (b) Fluorescence image of M. tuberculosis–infected macrophages labeled with fluorescein-trehalose. (c) Nitroreductases converts a nitro-aryl fluorogen (8) into a bright fluorophore. (d) Enzymatic-turnover-activated localization microscopy (ETALM) images of nitroreductase localization in B. subtilis. Figures reproduced with permission from: b, ref. 71; d, ref. 93.
Gram-negative bacteria have an outer membrane that is associated with a dense lipopolysaccharide (LPS), which distinguishes them from Gram-positive bacteria. Modification of the LPS layer enables the host immune system to more readily combat these organisms. A clickable version of 3-deoxy-D-mannooctulosonic acid (KDO), thought to be an essential component of LPS, has been reported. Following incorporation, azido-KDO was conjugated to a fluorophore, and the resulting labeling pattern demonstrated that KDO localized at the inner core of LPS in Gram-negative bacteria (E. coli, Salmonella typhimurium, Legionella pneumophila). No labeling was observed in Gram-positive organisms (Shawanella oneidensis, B. subtilis, S. aureus), demonstrating that the LPS can be selectively targeted and used for bacterial imaging74.
Several additional strategies have been reported for the visualization of extracellular components in bacteria, including global conjugation of cell surface amines to a fluorophore followed by SR microscopy75–77 and utilization of the bacterial enzyme sortase to incorporate non-native small molecules into the cell wall for fluorescence microscopy, flow cytometry, electron microscopy and MS applications78.
Additional small-molecule probes
Replacement of an amino acid with an unnatural analog can yield a chemical handle for subsequent detection of a protein of interest9,79,80. For example, co-translational labeling of proteins with noncanonical amino acids that contain an alkyne provides a site for attachment of a fluorogenic dye appended to an azide group. This strategy has been used to visualize newly synthesized proteins in E. coli81 and environmental microbes (L-azidohomoalanine)82. Recently, tagging with azidonorleucine enabled detection of secreted bacterial proteins in host cells83, and the outer-membrane proteins of E. coli have been visualized by STORM following incorporation of an unnatural amino acid (homopropargylglycine) and attachment of a dye by ‘click chemistry’84.
Although probe development for the exploration of bacterial proteins has been an active area of study for many years, relatively few of these compounds have been applied in microscopy-based experiments, a lack that is likely due in part to the difficulty of generating compounds that can penetrate the cell envelope of bacteria. Activitybased probes have been designed to detect a number of bacterial proteins, such as sulfatases85,86, serine proteases87, the cell wall biosynthetic protein MurA88, glycoside hydrolases89, redox-sensitive proteins90,91 and histidine kinases92; however, most of these probes have been used only in gel-based analyses. One additional strategy that has been applied to bacterial imaging is the use of a probe that becomes fluorescent upon turnover by nitroreductase. In this way, the probe (nitro-aryl fluorogen 8, Fig. 5c,d) acts as a readout of enzyme activity in live cells, enabling visualization of the location of nitroreductase in an approach called enzymatic-turnover-activated localization microscopy (ETALM)93.
Summary and future outlook
In this Review, we have provided a survey of small-molecule probes that have been used to explore proteins, carbohydrates and PG in bacterial cells. To date, relatively few of these compounds have been applied in imaging experiments, and only a handful have been utilized with SR imaging technologies. Looking forward, the use of biomolecule-selective small molecules carries both great potential and significant challenges. Perhaps one of the most difficult obstacles will be the development of probes and bright, photostable fluorophores that can readily penetrate the cell envelopes of bacteria, which will be necessary to examine many targets in living cells. To date, most success in the use of smallmolecule probes has been in the examination of extracellular components, making the majority of this work complementary to the use of genetically encoded tags. A true challenge will be to devise strategies to utilize target-selective molecules inside live bacteria. Continued improvements in SR microscopy image acquisition and data analysis processes will also be necessary to increase temporal resolution, which is critical for the visualization of the many events that occur on a biological timescale. These challenges aside, it is clear that much can be learned from biomolecule-specific fluorescent probes and that combination of these molecules with the improved spatial and temporal resolution achievable with SR microscopy will continue to shed light on growth, division, pathogenesis and virulence processes in bacteria. Creative solutions will be required to move the field of probe-promoted bacterial imaging beyond the investigation of ‘readily accessible’ components so as to provide a comprehensive toolkit for visualization of all bacterial biomolecules.
Acknowledgments
This work was supported by NIH DP2OD008592 (E.E.C.), a Pew Biomedical Scholar Award (E.E.C.), Sloan Research Fellow Award (E.E.C.) and Indiana University–Bloomington Department of Chemistry Start-Up Funds and a Marvin Carmack Fellowship (O.K.).
Footnotes
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Shapiro L, McAdams HH, Losick R. Why and how bacteria localize proteins. Science. 2009;326:1225–1228. doi: 10.1126/science.1175685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rudner DZ, Losick R. Protein subcellular localization in bacteria. Cold Spring Harb Perspect Biol. 2010;2:a000307. doi: 10.1101/cshperspect.a000307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Feilmeier BJ, Iseminger G, Schroeder D, Webber H, Phillips GJ. Green fluorescent protein functions as a reporter for protein localization in Escherichia coli. J Bacteriol. 2000;182:4068–4076. doi: 10.1128/jb.182.14.4068-4076.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jarvik JW, Telmer CA. Epitope tagging. Annu Rev Genet. 1998;32:601–618. doi: 10.1146/annurev.genet.32.1.601. [DOI] [PubMed] [Google Scholar]
- 5.Brizzard B. Epitope tagging. Biotechniques. 2008;44:693–695. doi: 10.2144/000112841. [DOI] [PubMed] [Google Scholar]
- 6.Dean KM, Palmer AE. Advances in fluorescence labeling strategies for dynamic cellular imaging. Nat Chem Biol. 2014;10:512–523. doi: 10.1038/nchembio.1556. An overview of the improvements made in fluorescence labeling approaches. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Griffin BA, Adams SR, Tsien RY. Specific covalent labeling of recombinant protein molecules inside live cells. Science. 1998;281:269–272. doi: 10.1126/science.281.5374.269. [DOI] [PubMed] [Google Scholar]
- 8.Zhang J, Campbell RE, Ting AY, Tsien RY. Creating new fluorescent probes for cell biology. Nat Rev Mol Cell Biol. 2002;3:906–918. doi: 10.1038/nrm976. [DOI] [PubMed] [Google Scholar]
- 9.Lang K, Chin JW. Cellular incorporation of unnatural amino acids and bioorthogonal labeling of proteins. Chem Rev. 2014;114:4764–4806. doi: 10.1021/cr400355w. [DOI] [PubMed] [Google Scholar]
- 10.Landgraf D, Okumus B, Chien P, Baker TA, Paulsson J. Segregation of molecules at cell division reveals native protein localization. Nat Methods. 2012;9:480–482. doi: 10.1038/nmeth.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Margolin W. The price of tags in protein localization studies. J Bacteriol. 2012;194:6369–6371. doi: 10.1128/JB.01640-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Domínguez-Escobar J, et al. Processive movement of MreB-associated cell wall biosynthetic complexes in bacteria. Science. 2011;333:225–228. doi: 10.1126/science.1203466. [DOI] [PubMed] [Google Scholar]
- 13.Garner EC, et al. Coupled, circumferential motions of the cell wall synthesis machinery and MreB filaments in B. subtilis. Science. 2011;333:222–225. doi: 10.1126/science.1203285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Swulius MT, Jensen GJ. The helical MreB cytoskeleton in Escherichia coli MC1000/pLE7 is an artifact of the N-terminal yellow fluorescent protein tag. J Bacteriol. 2012;194:6382–6386. doi: 10.1128/JB.00505-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang B, Bates M, Zhuang X. Super-resolution fluorescence microscopy. Annu Rev Biochem. 2009;78:993–1016. doi: 10.1146/annurev.biochem.77.061906.092014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koch AL. What size should a bacterium be? A question of scale. Annu Rev Microbiol. 1996;50:317–348. doi: 10.1146/annurev.micro.50.1.317. [DOI] [PubMed] [Google Scholar]
- 17.Erickson HP, Taylor DW, Taylor KA, Bramhill D. Bacterial cell division protein FtsZ assembles into protofilament sheets and minirings, structural homologs of tubulin polymers. Proc Natl Acad Sci USA. 1996;93:519–523. doi: 10.1073/pnas.93.1.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fu G, et al. In vivo structure of the E. coli FtsZ-ring revealed by photoactivated localization microscopy (PALM) PLoS One. 2010;5:e12682. doi: 10.1371/journal.pone.0012680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Buss J, et al. In vivo organization of the FtsZ-ring by ZapA and ZapB revealed by quantitative super-resolution microscopy. Mol Microbiol. 2013;89:1099–1120. doi: 10.1111/mmi.12331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holden SJ, et al. High throughput 3D super-resolution microscopy reveals Caulobacter crescentus in vivo Z-ring organization. Proc Natl Acad Sci USA. 2014;111:4566–4571. doi: 10.1073/pnas.1313368111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Strauss MP, et al. 3D-SIM super resolution microscopy reveals a bead-like arrangement for FtsZ and the division machinery: implications for triggering cytokinesis. PLoS Biol. 2012;10:e1001389. doi: 10.1371/journal.pbio.1001389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chang PV, Bertozzi CR. Imaging beyond the proteome. Chem Commun (Camb) 2012;48:8864–8879. doi: 10.1039/c2cc31845h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stracy M, Uphoff S, Garza de Leon F, Kapanidis AN. In vivo single-molecule imaging of bacterial DNA replication, transcription, and repair. FEBS Lett. 2014;588:3585–3594. doi: 10.1016/j.febslet.2014.05.026. [DOI] [PubMed] [Google Scholar]
- 24.Song W, Strack RL, Jaffrey SR. Imaging bacterial protein expression using genetically encoded RNA sensors. Nat Methods. 2013;10:873–875. doi: 10.1038/nmeth.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schimak MP, et al. MiL-FISH: Multilabeled oligonucleotides for fluorescence in situ hybridization improve visualization of bacterial cells. Appl Environ Microbiol. 2016;82:62–70. doi: 10.1128/AEM.02776-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Spahn C, Endesfelder U, Heilemann M. Super-resolution imaging of Escherichia coli nucleoids reveals highly structured and asymmetric segregation during fast growth. J Struct Biol. 2014;185:243–249. doi: 10.1016/j.jsb.2014.01.007. [DOI] [PubMed] [Google Scholar]
- 27.Schoen I, Ries J, Klotzsch E, Ewers H, Vogel V. Binding-activated localization microscopy of DNA structures. Nano Lett. 2011;11:4008–4011. doi: 10.1021/nl2025954. [DOI] [PubMed] [Google Scholar]
- 28.Chan J, Dodani SC, Chang CJ. Reaction-based small-molecule fluorescent probes for chemoselective bioimaging. Nat Chem. 2012;4:973–984. doi: 10.1038/nchem.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vollmer W, Blanot D, de Pedro MA. Peptidoglycan structure and architecture. FEMS Microbiol Rev. 2008;32:149–167. doi: 10.1111/j.1574-6976.2007.00094.x. [DOI] [PubMed] [Google Scholar]
- 30.Vollmer W, Seligman SJ. Architecture of peptidoglycan: more data and more models. Trends Microbiol. 2010;18:59–66. doi: 10.1016/j.tim.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 31.Matias VR, Beveridge TJ. Cryo-electron microscopy reveals native polymeric cell wall structure in Bacillus subtilis 168 and the existence of a periplasmic space. Mol Microbiol. 2005;56:240–251. doi: 10.1111/j.1365-2958.2005.04535.x. [DOI] [PubMed] [Google Scholar]
- 32.Bugg TD, Braddick D, Dowson CG, Roper DI. Bacterial cell wall assembly: still an attractive antibacterial target. Trends Biotechnol. 2011;29:167–173. doi: 10.1016/j.tibtech.2010.12.006. [DOI] [PubMed] [Google Scholar]
- 33.Typas A, Banzhaf M, Gross CA, Vollmer W. From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat Rev Microbiol. 2012;10:123–136. doi: 10.1038/nrmicro2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lovering AL, Safadi SS, Strynadka NC. Structural perspective of peptidoglycan biosynthesis and assembly. Annu Rev Biochem. 2012;81:451–478. doi: 10.1146/annurev-biochem-061809-112742. [DOI] [PubMed] [Google Scholar]
- 35.Tipper DJ, Strominger JL. Mechanism of action of penicillins: a proposal based on their structural similarity to acyl-D-alanyl-D-alanine. Proc Natl Acad Sci USA. 1965;54:1133–1141. doi: 10.1073/pnas.54.4.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blumberg PM, Yocum RR, Willoughby E, Strominger JL. Binding of [14C]penicillin G to the membrane-bound and the purified D-alanine carboxypeptidases from Bacillus stearothermophilus and Bacillus subtilis and its release. J Biol Chem. 1974;249:6828–6835. [PubMed] [Google Scholar]
- 37.Zhao G, Meier TI, Kahl SD, Gee KR, Blaszczak LC. BOCILLIN FL, a sensitive and commercially available reagent for detection of penicillin-binding proteins. Antimicrob Agents Chemother. 1999;43:1124–1128. doi: 10.1128/aac.43.5.1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gee KR, Kang HC, Meier TI, Zhao G, Blaszcak LC. Fluorescent bocillins: synthesis and application in the detection of penicillin-binding proteins. Electrophoresis. 2001;22:960–965. doi: 10.1002/1522-2683()22:5<960::AID-ELPS960>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 39.Heal WP, Tate EW. Application of activity-based protein profiling to the study of microbial pathogenesis. Top Curr Chem. 2012;324:115–135. doi: 10.1007/128_2011_299. [DOI] [PubMed] [Google Scholar]
- 40.Puri AW, Bogyo M. Applications of small molecule probes in dissecting mechanisms of bacterial virulence and host responses. Biochemistry. 2013;52:5985–5996. doi: 10.1021/bi400854d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Staub I, Sieber SA. Beta-lactams as selective chemical probes for the in vivo labeling of bacterial enzymes involved in cell wall biosynthesis, antibiotic resistance, and virulence. J Am Chem Soc. 2008;130:13400–13409. doi: 10.1021/ja803349j. [DOI] [PubMed] [Google Scholar]
- 42.Böttcher T, Sieber SA. Beta-lactones as specific inhibitors of ClpP attenuate the production of extracellular virulence factors of Staphylococcus aureus. J Am Chem Soc. 2008;130:14400–14401. doi: 10.1021/ja8051365. [DOI] [PubMed] [Google Scholar]
- 43.Böttcher T, Sieber SA. Structurally refined beta-lactones as potent inhibitors of devastating bacterial virulence factors. ChemBioChem. 2009;10:663–666. doi: 10.1002/cbic.200800743. [DOI] [PubMed] [Google Scholar]
- 44.Zeiler E, Korotkov VS, Lorenz-Baath K, Böttcher T, Sieber SA. Development and characterization of improved β-lactone-based anti-virulence drugs targeting ClpP. Bioorg Med Chem. 2012;20:583–591. doi: 10.1016/j.bmc.2011.07.047. [DOI] [PubMed] [Google Scholar]
- 45.Kocaoglu O, et al. Selective penicillin-binding protein imaging probes reveal substructure in bacterial cell division. ACS Chem Biol. 2012;7:1746–1753. doi: 10.1021/cb300329r. First example of selective PBP imaging using an activity-based probe. This study provides precedent for the use of β-lactam antibiotics to facilitate microscopy-based study of the PBPs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kocaoglu O, Carlson EE. Penicillin-binding protein imaging probes. Curr Protoc Chem Biol. 2013;5:239–250. doi: 10.1002/9780470559277.ch130102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kocaoglu O, Carlson EE. Profiling of β-lactam selectivity for penicillin-binding proteins in Escherichia coli strain DC2. Antimicrob Agents Chemother. 2015;59:2785–2790. doi: 10.1128/AAC.04552-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kocaoglu O, Tsui HCT, Winkler ME, Carlson EE. Profiling of β-lactam selectivity for penicillin-binding proteins in Streptococcus pneumoniae D39. Antimicrob Agents Chemother. 2015;59:3548–3555. doi: 10.1128/AAC.05142-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Daniel RA, Errington J. Control of cell morphogenesis in bacteria: two distinct ways to make a rod-shaped cell. Cell. 2003;113:767–776. doi: 10.1016/s0092-8674(03)00421-5. Beautiful demonstration of the utility of vancomycin as an imaging agent to visualize nascent peptidoglycan. [DOI] [PubMed] [Google Scholar]
- 50.Tiyanont K, et al. Imaging peptidoglycan biosynthesis in Bacillus subtilis with fluorescent antibiotics. Proc Natl Acad Sci USA. 2006;103:11033–11038. doi: 10.1073/pnas.0600829103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gautam S, et al. An activity-based probe for studying crosslinking in live bacteria. Angew Chem Int Edn Engl. 2015;54:10492–10496. doi: 10.1002/anie.201503869. [DOI] [PubMed] [Google Scholar]
- 52.Liu H, Sadamoto R, Sears PS, Wong CH. An efficient chemoenzymatic strategy for the synthesis of wild-type and vancomycin-resistant bacterial cell-wall precursors: UDP-N-acetylmuramyl-peptides. J Am Chem Soc. 2001;123:9916–9917. doi: 10.1021/ja011708w. [DOI] [PubMed] [Google Scholar]
- 53.Sadamoto R, et al. Cell-wall engineering of living bacteria. J Am Chem Soc. 2002;124:9018–9019. doi: 10.1021/ja026133x. [DOI] [PubMed] [Google Scholar]
- 54.Olrichs NK, et al. A novel in vivo cell-wall labeling approach sheds new light on peptidoglycan synthesis in Escherichia coli. ChemBioChem. 2011;12:1124–1133. doi: 10.1002/cbic.201000552. [DOI] [PubMed] [Google Scholar]
- 55.Kuru E, et al. In Situ probing of newly synthesized peptidoglycan in live bacteria with fluorescent D-amino acids. Angew Chem Int Edn Engl. 2012;51:12519–12523. doi: 10.1002/anie.201206749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Siegrist MS, et al. D-Amino acid chemical reporters reveal peptidoglycan dynamics of an intracellular pathogen. ACS Chem Biol. 2013;8:500–505. doi: 10.1021/cb3004995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shieh P, Siegrist MS, Cullen AJ, Bertozzi CR. Imaging bacterial peptidoglycan with near-infrared fluorogenic azide probes. Proc Natl Acad Sci USA. 2014;111:5456–5461. doi: 10.1073/pnas.1322727111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lebar MD, et al. Reconstitution of peptidoglycan cross-linking leads to improved fluorescent probes of cell wall synthesis. J Am Chem Soc. 2014;136:10874–10877. doi: 10.1021/ja505668f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pidgeon SE, et al. Metabolic profiling of bacteria by unnatural C-terminated D-amino acids. Angew Chem Int Edn Engl. 2015;54:6158–6162. doi: 10.1002/anie.201409927. [DOI] [PubMed] [Google Scholar]
- 60.Schirner K, et al. Lipid-linked cell wall precursors regulate membrane association of bacterial actin MreB. Nat Chem Biol. 2015;11:38–45. doi: 10.1038/nchembio.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Monteiro JM, et al. Cell shape dynamics during the staphylococcal cell cycle. Nat Commun. 2015;6:8055. doi: 10.1038/ncomms9055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Garrett AJ, Harrison MJ, Manire GP. A search for the bacterial mucopeptide component, muramic acid, in Chlamydia. J Gen Microbiol. 1974;80:315–318. doi: 10.1099/00221287-80-1-315. [DOI] [PubMed] [Google Scholar]
- 63.Fox A, et al. Muramic acid is not detectable in Chlamydia psittaci or Chlamydia trachomatis by gas chromatography-mass spectrometry. Infect Immun. 1990;58:835–837. doi: 10.1128/iai.58.3.835-837.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pilhofer M, et al. Discovery of chlamydial peptidoglycan reveals bacteria with murein sacculi but without FtsZ. Nat Commun. 2013;4:2856. doi: 10.1038/ncomms3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liechti GW, et al. A new metabolic cell-wall labelling method reveals peptidoglycan in Chlamydia trachomatis. Nature. 2014;506:507–510. doi: 10.1038/nature12892. Demonstration of the presence of peptidoglycan in Chlamydia trachomatis using fluorescent D-amino acids for the first time. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wheeler R, Mesnage S, Boneca IG, Hobbs JK, Foster SJ. Super-resolution microscopy reveals cell wall dynamics and peptidoglycan architecture in ovococcal bacteria. Mol Microbiol. 2011;82:1096–1109. doi: 10.1111/j.1365-2958.2011.07871.x. [DOI] [PubMed] [Google Scholar]
- 67.Tsui HC, et al. Pbp2x localizes separately from Pbp2b and other peptidoglycan synthesis proteins during later stages of cell division of Streptococcus pneumoniae D39. Mol Microbiol. 2014;94:21–40. doi: 10.1111/mmi.12745. Beautiful combination of FDAAs, Van-FL, β-lactams and fusion proteins to explore PBP localization during division. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Weidenmaier C, et al. Lack of wall teichoic acids in Staphylococcus aureus leads to reduced interactions with endothelial cells and to attenuated virulence in a rabbit model of endocarditis. J Infect Dis. 2005;191:1771–1777. doi: 10.1086/429692. [DOI] [PubMed] [Google Scholar]
- 69.Tra VN, Dube DH. Glycans in pathogenic bacteria—potential for targeted covalent therapeutics and imaging agents. Chem Commun (Camb) 2014;50:4659–4673. doi: 10.1039/c4cc00660g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Woodruff PJ, et al. Trehalose is required for growth of Mycobacterium smegmatis. J Biol Chem. 2004;279:28835–28843. doi: 10.1074/jbc.M313103200. [DOI] [PubMed] [Google Scholar]
- 71.Backus KM, et al. Uptake of unnatural trehalose analogs as a reporter for Mycobacterium tuberculosis. Nat Chem Biol. 2011;7:228–235. doi: 10.1038/nchembio.539. Describes the use of fluorescent unnatural trehalose for sensitive detection of Mycobacterium tuberculosis in mammalian cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Swarts BM, et al. Probing the mycobacterial trehalome with bioorthogonal chemistry. J Am Chem Soc. 2012;134:16123–16126. doi: 10.1021/ja3062419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Urbanek BL, et al. Chemoenzymatic synthesis of trehalose analogues: rapid access to chemical probes for investigating mycobacteria. ChemBioChem. 2014;15:2066–2070. doi: 10.1002/cbic.201402288. [DOI] [PubMed] [Google Scholar]
- 74.Dumont A, Malleron A, Awwad M, Dukan S, Vauzeilles B. Click-mediated labeling of bacterial membranes through metabolic modification of the lipopolysaccharide inner core. Angew Chem Int Edn Engl. 2012;51:3143–3146. doi: 10.1002/anie.201108127. [DOI] [PubMed] [Google Scholar]
- 75.Lee MK, Rai P, Williams J, Twieg RJ, Moerner WE. Small-molecule labeling of live cell surfaces for three-dimensional super-resolution microscopy. J Am Chem Soc. 2014;136:14003–14006. doi: 10.1021/ja508028h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gunsolus IL, et al. Facile method to stain the bacterial cell surface for super-resolution fluorescence microscopy. Analyst. 2014;139:3174–3178. doi: 10.1039/c4an00574k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Conley NR, Biteen JS, Moerner WE. Cy3-Cy5 covalent heterodimers for single-molecule photoswitching. J Phys Chem B. 2008;112:11878–11880. doi: 10.1021/jp806698p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nelson JW, et al. A biosynthetic strategy for re-engineering the Staphylococcus aureus cell wall with non-native small molecules. ACS Chem Biol. 2010;5:1147–1155. doi: 10.1021/cb100195d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang L, Brock A, Herberich B, Schultz PG. Expanding the genetic code of Escherichia coli. Science. 2001;292:498–500. doi: 10.1126/science.1060077. [DOI] [PubMed] [Google Scholar]
- 80.Charbon G, et al. Subcellular protein localization by using a genetically encoded fluorescent amino acid. ChemBioChem. 2011;12:1818–1821. doi: 10.1002/cbic.201100282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Beatty KE, Xie F, Wang Q, Tirrell DA. Selective dye-labeling of newly synthesized proteins in bacterial cells. J Am Chem Soc. 2005;127:14150–14151. doi: 10.1021/ja054643w. [DOI] [PubMed] [Google Scholar]
- 82.Hatzenpichler R, et al. In situ visualization of newly synthesized proteins in environmental microbes using amino acid tagging and click chemistry. Environ Microbiol. 2014;16:2568–2590. doi: 10.1111/1462-2920.12436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mahdavi A, et al. Identification of secreted bacterial proteins by noncanonical amino acid tagging. Proc Natl Acad Sci USA. 2014;111:433–438. doi: 10.1073/pnas.1301740111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Raulf A, et al. Click chemistry facilitates direct labeling and super-resolution imaging of nucleic acids and proteins. RCS Adv. 2014;4:30462–30466. doi: 10.1039/c4ra01027b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Beatty KE, et al. Sulfatase-activated fluorophores for rapid discrimination of mycobacterial species and strains. Proc Natl Acad Sci USA. 2013;110:12911–12916. doi: 10.1073/pnas.1222041110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Smith EL, Bertozzi CR, Beatty KE. An expanded set of fluorogenic sulfatase activity probes. ChemBioChem. 2014;15:1101–1105. doi: 10.1002/cbic.201400032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gloeckl S, et al. Identification of a serine protease inhibitor which causes inclusion vacuole reduction and is lethal to Chlamydia trachomatis. Mol Microbiol. 2013;89:676–689. doi: 10.1111/mmi.12306. [DOI] [PubMed] [Google Scholar]
- 88.Böttcher T, Sieber SA. Showdomycin as a versatile chemical tool for the detection of pathogenesis-associated enzymes in bacteria. J Am Chem Soc. 2010;132:6964–6972. doi: 10.1021/ja909150y. [DOI] [PubMed] [Google Scholar]
- 89.Chauvigné-Hines LM, et al. Suite of activity-based probes for cellulose-degrading enzymes. J Am Chem Soc. 2012;134:20521–20532. doi: 10.1021/ja309790w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sadler NC, et al. Live cell chemical profiling of temporal redox dynamics in a photoautotrophic cyanobacterium. ACS Chem Biol. 2014;9:291–300. doi: 10.1021/cb400769v. [DOI] [PubMed] [Google Scholar]
- 91.Deng X, et al. Proteome-wide quantification and characterization of oxidation-sensitive cysteines in pathogenic bacteria. Cell Host Microbe. 2013;13:358–370. doi: 10.1016/j.chom.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wilke KE, Francis S, Carlson EE. Activity-based probe for histidine kinase signaling. J Am Chem Soc. 2012;134:9150–9153. doi: 10.1021/ja3041702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lee MK, Williams JC, Twieg RJ, Rao J, Moerner WE. Enzymatic activation of nitro-aryl fluorogens in live bacterial cells for enzymatic turnover-activated localization microscopy. Chem Sci (Camb) 2013;4:220–225. doi: 10.1039/C2SC21074F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gustafsson MG. Surpassing the lateral resolution limit by a factor of two using structured illumination microscopy. J Microsc. 2000;198:82–87. doi: 10.1046/j.1365-2818.2000.00710.x. [DOI] [PubMed] [Google Scholar]
- 95.Hell SW, Wichmann J. Breaking the diffraction resolution limit by stimulated emission: stimulated-emission-depletion fluorescence microscopy. Opt Lett. 1994;19:780–782. doi: 10.1364/ol.19.000780. [DOI] [PubMed] [Google Scholar]
- 96.Schermelleh L, Heintzmann R, Leonhardt H. A guide to super-resolution fluorescence microscopy. J Cell Biol. 2010;190:165–175. doi: 10.1083/jcb.201002018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Coltharp C, Xiao J. Superresolution microscopy for microbiology. Cell Microbiol. 2012;14:1808–1818. doi: 10.1111/cmi.12024. An excellent review comparing super-resolution microscopy techniques for bacterial cell imaging. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Betzig E, et al. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 99.Rust MJ, Bates M, Zhuang X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM) Nat Methods. 2006;3:793–795. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tuson HH, Biteen JS. Unveiling the inner workings of live bacteria using super-resolution microscopy. Anal Chem. 2015;87:42–63. doi: 10.1021/ac5041346. Comprehensive review of cutting-edge strategies to examine bacteria using super-resolution microscopy techniques. [DOI] [PubMed] [Google Scholar]