Abstract
pH-responsive diblock copolymers provide tailorable nanoparticle (NP) architecture and chemistry critical for siRNA delivery. Here, diblock polymers varying in first (corona) and second (core) block molecular weight (Mn), corona/core ratio, and core hydrophobicity (%BMA) were synthesized to determine their effect on siRNA delivery in murine tenocytes (mTenocyte) and murine and human mesenchymal stem cells (mMSC and hMSCs, respectively). NP-mediated siRNA uptake, gene silencing, and cytocompatibility were quantified. Uptake is positively correlated with first block Mn in mTenocytes and hMSCs (p ≤ 0.0005). All NP resulted in significant gene silencing that was positively correlated with % BMA (p < 0.05) in all cell types. Cytocompatibility was reduced in mTenocytes compared to MSCs (p < 0.0001). %BMA was positively correlated with cytocompatibility in MSCs (p < 0.05), suggesting stable NP are more cytocompatible. Overall, this study shows that NP-siRNA cytocompatibility is cell type dependent, and hydrophobicity (%BMA) is the critical diblock copolymer property for efficient gene silencing in musculoskeletal cell types.
Graphical Abstract
INTRODUCTION
The delivery of nucleic acids, specifically small interfering RNA (siRNA), has benefitted immensely from nanoparticle-based delivery approaches.1,2 siRNA mediates degradation of complementary cytosolic mRNA via the RNA interference (RNAi) pathway. The ability to silence any gene by simply knowing its nucleic acid sequence confers immense therapeutic potential to siRNA. However, there are many barriers to siRNA delivery,3,4 necessitating development of delivery systems to achieve therapeutic efficacy. siRNA is susceptible to degradation by nucleases, resulting in serum instability,5 and due to its anionic nature and large molecular weight, intact siRNA cannot passively diffuse through cell membrane, leading to poor internalization. Most commonly, nanoparticles (NP) contain a cationic component that serves to complex and protect anionic siRNA molecules from nuclease degradation and allow for interaction with negatively charged cell membranes to facilitate cellular uptake.6 However, as endocytosis is the typical mechanism of cellular uptake of such nanocomplexes, escape from endolysosomal trafficking is another delivery hurdle often addressed via NP delivery systems.7
Many materials, including lipids,8 as well as natural9 and synthetic polymers,10 have been utilized to form NP-siRNA complexes. In particular, polymers provide an attractive platform for the development of siRNA delivery systems due to flexibility with respect to architecture and chemical functionality. Both natural and synthetic polymers can incorporate a variety of cationic components that allow for complexation with siRNA and interaction with the cell membrane. Chitosan, a polysaccharide, is the most widely utilized natural polymer for siRNA delivery owing to its cationic charge, biocompatibility, and biodegradability.11 However, chitosan has inherent limitations, such as poor water solubility at physiological pH leading to NP instability and low buffering capacity, which result in inefficient siRNA delivery. Substantial improvements have recently been made to chitosan by employing synthetic polymer modifications.9,12 Poly-(ethylenimine) (PEI) is also commonly employed for NP-mediated delivery of siRNA. PEI is cationic, and bestows endosomal escape through a combination of the proton sponge effect and polymer swelling upon protonation during endolysosomal traficking.2,13 However, cytotoxicity and immunostimulation of PEI has been well documented in a multitude of cell types in vitro and in vivo,14–17 which has led to the exploration of a variety of PEI modifications and alternative polymers to overcome these significant limitations.18,19
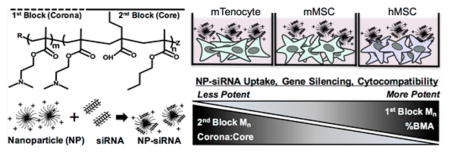
We pioneered the development of diblock copolymers composed of a cationic block and a pH-responsive endosomolytic block to achieve successful siRNA delivery to a variety of cell types20–23 (Scheme 1). The cationic block is composed of poly(dimethylaminoethyl methacrylate) (pDMAEMA), which contains tertiary amines that are 50% protonated at physiological pH to allow electrostatic complexation with anionic siRNA and facilitates cell membrane trafficking. The second block is a tercopolymer composed of DMAEMA, 2-propylacrylic acid (PAA), and butyl methacrylate (BMA), which confers pH-dependent endosomal escape properties. Further, the second block is hydrophobic, which mediates self-assembly of NP in aqueous neutral pH conditions.20 Increasing the amount of hydrophobic BMA, up to 50% of the core block, with equimolar ratios of DMAEMA and PAA, leads to more efficient siRNA delivery in HeLa cells.20 NP-siRNA delivery was drastically improved by increasing the molecular weight of the tercopolymer block with the same composition, which caused polymers to self-assemble into stable micellar NP due to increased hydrophobic content.24 Furthermore, delivery of radioprotective siRNA to mouse salivary glands showed robust in vivo efficacy.22,23 Additionally, PEGylated polymers allow for systemic NP-siRNA delivery in vivo.25 Recent efforts have further adapted this NP-siRNA delivery system to control gene expression for regenerative medicine applications. For example, the NP-delivery system is capable of modulating gene expression in human mesenchymal stem cells (MSCs) in vitro without affecting multipotential differentiation capacity.21 Moreover, the optimal hMSC:NP-siRNA ratios resulting in maximal siRNA delivery while maintaining hMSC function were identified.26
Scheme 1. Diblock Copolymers Form Cationic Nanoparticles (NP) via Self-Assembly That Can Complex with Negatively Charged siRNAa.
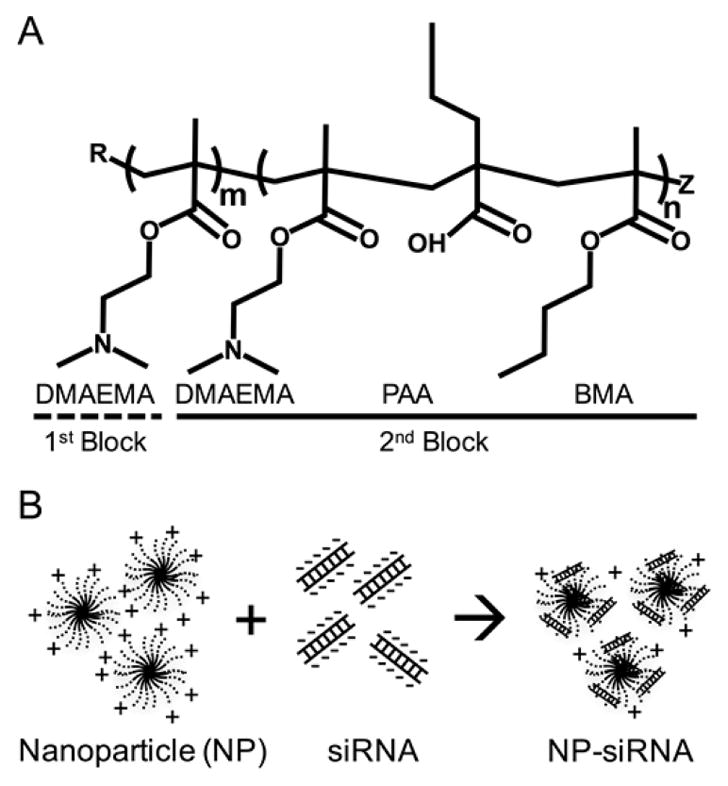
a(A) Diblock copolymer structure showing cationic first block composed of dimethylaminoethyl methacrylate (DMAEMA), and a pH-responsive hydrophobic tercopolymer second block composed of DMAEMA, propylacrylic acid (PAA), and butyl methacrylate (BMA) that drives self-assembly and allows endosomal escape. R and Z are functional end groups. (B) Self-assembled cationic NP electrostatically complex with negatively charged siRNA.
Despite these efforts, it remains unknown how the diblock copolymer properties can be manipulated to control NP-siRNA delivery efficiency and cytotoxicity, as previous studies have not varied the overall and block molecular weights. We hypothesized that changing the overall and block molecular weights would alter siRNA delivery outcomes, such as uptake, cytotoxicity, and gene silencing, and that these effects would be dependent on cell type, species, and differentiation state. Therefore, a library of diblock copolymers varying in first block number-average molecular weight (Mn), second block Mn, the ratio of the two block Mn (corona/core ratio), and the %BMA content were synthesized. These NP were then tested for siRNA delivery efficacy in murine tenocytes (mTenocytes), murine MSCs (mMSC), and human MSCs (hMSC). After measuring uptake, cytotoxicity, and gene silencing of NP-siRNA treatments, we utilized a multiparameter regression analysis to statistically evaluate how polymer properties alter siRNA delivery capabilities in each cell type and whether these structure–function relationships are consistent across cell types and species.
EXPERIMENTAL SECTION
Polymer Synthesis and Characterization
Synthesis of Chain Transfer Agent (CTA)
The CTA used for reversible addition–fragmentation chain transfer (RAFT) polymerization, 4-cyano-4-[(ethylsulfanylthiocarbonyl)sulfanyl]pentanoic acid (ECT), was synthesized as previously described.27,28
Synthesis of Cationic siRNA Complexation Block (First Block, Corona)
ECT and the radical initiator 2,2'-azobis(2-methylpropioni-trile) (AIBN) were used to polymerize poly(dimethylaminoethyl methacrylate) via RAFT polymerization. The degree of polymerization (DP), or [monomer]/[CTA], was varied according to Table 1 to control block Mn, and [ECT]/[AIBN] = 10 for all polymerizations except the first blocks of NP C where [CTA]/[initiator] = 1 and NP G, where [CTA]/[initiator] = 5. Distilled DMAEMA, ECT, and AIBN were mixed in dimethylformamide (DMF) at 25 wt %. The reaction vessel was purged with N2 for 40 min and polymerized for 6 h at 60 °C in an oil bath. Exposing to atmospheric oxygen terminated the reaction. The product was precipitated and washed four times in 80:20 pentane/diethyl ether with centrifugation and dried under vacuum overnight.
Table 1.
Diblock Copolymer and Nanoparticle (NP) Characterizationa
| polymer/NP | 1st block
|
2nd block
|
diblock
|
nanoparticle
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DP | Mn1 (kg/mol) | PDI | DP | Mn1 (kg/mol) | %DMAEMA2 | %PAA2 | %BMA2 | Mn1 (kg/mol) | PDI | corona/core | diameter3 (nm) | zeta potential4 (mV) | |
| A | 100 | 11.1 | 1.03 | 300 | 15.2 | 24 | 22 | 54 | 26.3 | 1.17 | 0.7 | 24 ± 5 | 13 ± 7 |
| B | 400 | 23.3 | 1.10 | 250 | 14.7 | 10 | 20 | 70 | 38.0 | 1.17 | 1.6 | 31 ± 7 | 14 ± 5 |
| C | 600 | 46.9 | 1.08 | 250 | 17.7 | 24 | 29 | 47 | 64.6 | 1.09 | 2.6 | 34 ± 10 | 10 ± 5 |
| D | 100 | 11.2 | 1.00 | 300 | 8.3 | 33 | 29 | 38 | 19.5 | 1.01 | 1.3 | 18 ± 4 | 18 ± 4 |
| A | 100 | 11.1 | 1.03 | 300 | 15.2 | 24 | 22 | 54 | 26.3 | 1.17 | 0.7 | 24 ± 5 | 13 ± 7 |
| E | 100 | 11.1 | 1.03 | 500 | 23.5 | 29 | 19 | 52 | 34.6 | 1.21 | 0.5 | 23 ± 6 | 15 ± 4 |
| A | 100 | 11.1 | 1.03 | 300 | 15.2 | 24 | 22 | 54 | 26.3 | 1.17 | 0.7 | 24 ± 5 | 13 ± 7 |
| F | 150 | 15.7 | 1.03 | 200 | 9.8 | 26 | 30 | 44 | 25.5 | 1.21 | 1.6 | 16 ± 4 | 13 ± 4 |
| G | 100 | 18.3 | 1.03 | 175 | 6.3 | 21 | 34 | 45 | 24.6 | 1.02 | 2.9 | 20 ± 4 | 8 ± 14 |
| H | 275 | 22.8 | 1.04 | 325 | 9.8 | 34 | 28 | 27 | 32.6 | 1.04 | 2.3 | 9 ± 2 | 21 ± 4 |
| I | 275 | 22.8 | 1.04 | 325 | 10.9 | 21 | 24 | 55 | 33.7 | 1.06 | 2.1 | 22 ± 6 | 24 ± 4 |
| B | 400 | 23.3 | 1.10 | 250 | 14.7 | 10 | 20 | 70 | 38.0 | 1.17 | 1.6 | 31 ± 7 | 14 ± 5 |
| X | 250 | 22.7 | 1.01 | 300 | 12.8 | 0 | 0 | 100 | 35.5 | 1.01 | 1.8 | 29 ± 8 | 17 ± 10 |
Verified by 1GPC, 2NMR, 3dynamic light scattering, or 4electrophoretic light scattering. Abbreviations: DP, degree of polymerization; PDI, polydispersity index (Mw/Mn); corona/core -1st block Mn/2nd block Mn.
Synthesis of pH-Responsive Endosomal Escape Block (Second Block, Core)
DP was varied using the pDMAEMA macroCTA according to Table 1 to control second block Mn, while maintaining [CTA]/[initiator] = 10 (except for NP G, where [CTA]/[initiator] = 5). Monomer feeds of DMAEMA/propylacrylic acid (PAA)/butyl methacrylate (BMA) were used at 25%:25%:50% for all polymers except NP H (35%:35%:30%) and NP B (15%:15%:70%), where % BMA was intentionally varied. macroCTA, monomers, and initiator were dissolved in DMF at 25 wt %. The reaction vessel was purged with N2 for 40 min and polymerized at 60 °C for 24 h. Exposing to atmospheric oxygen terminated the reaction. The product was precipitated in 80:20 pentane/diethyl ether twice, resolubilized in a minimal amount of acetone, and precipitated/washed three more times in 80:20 pentane/diethyl ether. The resulting polymer was dried overnight under vacuum.
Polymer Characterization
Gel permeation chromatography (GPC; Shimadzu Technologies) was used to obtain molecular weight and polydispersity (PDI, Mw/Mn) of first block and diblock copolymers using a TSKgel Guard SuperH-H guard column (Tosoh Biosciences) and a TSKgel Super HM-N for separation using a column oven at 60 °C. The mobile phase consisted of HPLC grade DMF containing 0.05 M LiCl (0.2 μm filtered) and used at a flow rate of 0.35 mL/min. The GPC system was equipped with a miniDAWN TREOS multiangle light scattering detector (Wyatt Technologies) and an Optilab T-rEX differential refractometer (Wyatt Technologies) to determine absolute molecular weights using previously reported dn/dc value for p(DMAEMA) in DMF (0.06 mL/g).29,30 1H NMR spectroscopy (Bruker Avance400) was used to verify diblock copolymer composition, as previously described.24,28
NP Self-Assembly and Characterization
Raw polymer was dissolved in 100% EtOH and added to an equal volume of 1× Dulbecco’s Phosphate Buffered Saline (DPBS, Invitrogen). This mixture was dialyzed against ultrapure water (dH2O, Barnstead NanoPure Diamond) using 3500 Da molecular weight cutoff dialysis tubing (Spectrum Laboratories) for at least 24 h with multiple dH2O changes. The dialyzed polymer was 0.2 μm filtered, frozen at –80 °C, and lyophilized for long-term storage. When needed, lyophilized polymer was dissolved in 100% EtOH at 20 mg/mL and diluted 10× in dH2O to 2 mg/mL. Polymer solutions were further purified using PD-10 desalting columns (GE Healthcare), lyophilized, dissolved in 100% EtOH at 20 mg/mL, and diluted 10× in dH2O to 2 mg/mL. Zetasizer Nano ZS (Malvern) was used to measure diameter and zeta potential, as previously described.26 Briefly, NP solutions were diluted 10× in 1× DPBS and NP diameter was measured in disposable cuvettes for diameter measurements. NP were diluted 10× in dH2O and zeta potential was measured in folded capillary cells.
Cell Culture
Murine Tenocyte (mTenocyte) Isolation and Culture
Flexor digitorum longus tendons were obtained from the hind limbs of freshly sacrificed 6-week-old C57Bl/6J male mice and processed for mTenocyte harvest and cell culture as previously described.31 Briefly, specimens were isolated and cleared of surrounding soft tissue. In sterile conditions, tendons were washed in 1× DPBS with 1% Pen Strep (Gibco), minced into 1 mm pieces, and placed in 0.25% Trypsin-EDTA (Gibco) for 1 h at room temperature. The tendon fragments were then cultured in MEM-α (no nucleosides, Gibco), supplemented with 20% FBS (Sigma-Aldrich), 1% Pen Strep, and 6.5 μL/L of 2-mercaptoethanol (Sigma-Aldrich). mTenocytes emerge from the tissue over 10–14 days. Isolated mTenocytes were passaged five times and then cryopreserved at –80 °C in 50% MEM-α, 40% FBS, and 10% DMSO (Sigma-Aldrich). The tenocytes were later thawed, plated, and expanded for experimental use at passage 7.
Murine Mesenchymal Stem Cell (mMSC) Culture
Bone marrow-derived OriCell Strain C57BL/6 Mouse Mesenchymal Stem Cells (mMSC) were purchased from Cyagen. mMSCs arrive cryopreserved at passage 6. Vials were thawed and expanded in low-glucose Dulbecco’s Modified Eagle’s Medium (DMEM) containing 10% FBS (Atlanta Biologicals) and 1% Antibiotic-Antimycotic (Gibco). mMSCs were cultured to subconfluent levels and used for experiments or trypsinized using 0.25% trypsin-EDTA for further passaging. mMSCs were used at passages 7–10 for experiments.
Human Mesenchymal Stem Cell (hMSC) Isolation and Culture
Human bone marrow isolates were purchased from Lonza, and hMSCs were isolated as previously described.32 Briefly, bone marrow aspirate was plated at 10 μL/cm2 in hMSC growth media (hGM) composed of low-glucose DMEM containing 1 ng/mL human recombinant basic fibroblast growth factor-2 (bFBF-2, Corning), 10% FBS, and 1% antibiotic-antimycotic (hMSC GM). Plated aspirates were left untouched for 7 days in a cell culture incubator kept at 37 °C and 5% CO2, after which marrow aspirate and media were removed. Nonadherent cells were removed by a single 1× DPBS wash and fresh hGM was added. Adherent MSCs were grown until colonies were subconfluent. hMSC colonies were trypsinized with 0.25% trypsin-EDTA, seeded at 5000 cells/cm2 in cell culture flasks, and grown to subconfluence. hMSCs were cryopreserved at 1 × 106 cells/mL in freezing media composed of low-glucose DMEM, 20% FBS, and 10% DMSO at passage 1. hMSC were thawed and subcultured for experiments at passages 2–4.
Preparation of NP-siRNA Treatments
The critical charge ratio, or the ratio of positive charges of first block Mn assuming 50% protonation of DMAEMA at pH 7.4 to the negative charges on siRNA at which there is no free siRNA, was determined using gel retardation assays for each NP as previously described.21,26,28 All NPs had a critical charge ratio ranging from 1–2, indicating that all NPs complex siRNA in a similar capacity, as the variability is likely due to, and accounts for, error associated with measured molecular weights, NP concentration, and the assumptions that 50% of DMAEMA residues are protonated at pH 7.4 and siRNA length (21 basepairs). Determination of this critical charge ratio confirms that all NP treatments complex 100% of the siRNA dose. NP-siRNA treatments were prepared by diluting siRNA in 1× DPBS, adding NP, and incubating the solution for at least 25 min at room temperature to allow for complexation. Treatments were made at 10×, the desired final concentration of 30 nM siRNA complexed to NP at a charge ratio of 4:1 beyond the critical charge ratio to maintain an overall cationic charge, which was shown to be ideal for delivery to HeLa cells28 and hMSCs21 and added directly to cells in Opti-Mem I Reduced Serum Media (Gibco) and incubated for 24 h. Untreated cells received an equal treatment volume of 1× DPBS. Lipofectamine2000 was used as a positive control according to the manufacturer’s protocol using 30 nM siRNA.
NP-siRNA Uptake
Cells were seeded at 8000 cells/cm2 and allowed to adhere overnight. The next day, NP-siRNA treatments were prepared using nontargeting Silencer FAM-labeled Negative Control siRNA 1 (Ambion) and cells were washed 2× with 1× DPBS. NP-siRNA treatments were added directly to wells containing fresh Opti-Mem media and incubated for 24 h at 37 °C and 5% CO2. After treatment, cells were washed 2× with 1× DPBS, trypsinized, quenched with media, and transferred to 1.5 mL tubes. Tubes were centrifuged for 5 min at 1000 rpm to pellet cells and the supernatant was removed. The cell pellet was resuspended in 1× DPBS, centrifuged to pellet washed cells, and the supernatant was aspirated. Cells were resuspended in 100 μL flow buffer (0.5 w/v% bovine serum albumin, and 0.01 v/v% trypan blue to quench extracellular fluorescence33). Cells were analyzed on an Accuri C6 flow cytometer (BD). Propidium Iodide (PI, Molecular Probes) was added to each sample (1:500) immediately prior to analysis for discrimination of dead cells. 5000 cells were gated for analysis using single stained samples for compensation. Analysis was performed using FlowJo software. Median fluorescence intensities (MFI) of treated cells were normalized to untreated cells (–, negative control) to determine relative siRNA uptake levels.
NP-Mediated GAPDH Gene Knockdown
Cells were seeded at 8000 cells/cm2 and allowed to adhere overnight. The next day, NP-siRNA treatments were prepared using Silencer GAPDH siRNA (human, mouse, rat; Ambion) and cells were washed 2× with 1× DPBS. NP-siRNA treatments were added directly to wells containing fresh Opti-Mem media and incubated for 24 h at 37 °C and 5% CO2. The next day, NP-siRNA containing media was replaced with fresh growth media and cells were incubated for an additional 24 h. Total RNA from hMSCs was purified according to manufacturer’s protocols using E.Z.N.A. Total RNA Kit I (Omega Biotek) and from mMSCs and mTenocytes using NucleoSpin RNA kit (Macherey-Nagel). Both kits include on-membrane DNase digestion. RNA quality (260/280 > 1.8) and quantity was determined using a NanoDrop 1000 (Thermo Scientific) or NanoVue Plus (GE Healthcare Life Sciences) spectrophotometer. A maximum of 1 μg of RNA was reverse transcribed to cDNA using iScript cDNA synthesis kit (Bio-Rad) and all cDNA concentrations were normalized such that each PCR reaction contained equal amounts of cDNA. For mTenocyte gene expression, RT-PCR was performed using 10 μL SYBR Green FastMix (QuantaBio), 5 μL of forward and reverse primers at 300 nM final concentrations, and 5 μL of cDNA as previously described.31 Target gene expression was detected using Rotor-Gene Q RT-PCR system (Qiagen). hMSC and mMSC target gene expression was quantifioed via RT-PCR using 10 μL Sso-Fast EvaGreen Supermix (Bio-Rad), 5 μL of primers at final concentration of 500 nM (Bio-Rad), and 5 μL of cDNA on a CFX96 Real-time PCR detection System (Bio-Rad) as previously described.26 Master mixes of SYBR Green or EvaGreen and primers were used to decrease pipetting errors. In all cases, each biological sample was run in triplicate. RT-PCR was run for 40 cycles and amplification of a single product was confirmed using melt curve analysis. Primer efficiencies were calculated from each well as previously described34 using 3% and 6% of the maximum amplification to set two thresholds. Relative GAPDH expression was calculated using the Pfafll equation35 relative to untreated samples and untreated samples and normalized to β-Actin (murine) or peptidylprolyl isomerase B (PPIB, human) expression. Primer sequences used in this study are listed in Table S1.
Hemolysis Assay for Evaluation of pH-Responsive NP Membrane Lysis Behavior
A hemolysis assay was performed to assess pH-responsive membrane disruption due to NP, as previously described.36 Briefly, 25 mL of blood was collected directly in K2-EDTA-coated vacutainer tubes from a consenting anonymous human donor by a trained phlebotomist in accordance with the University of Rochester’s Institutional Review Board. Blood was subsequently centrifuged at 500g for 5 min and the plasma layer aspirated and replaced with 150 mM NaCl. The tube was inverted to mix, and this washing step was repeated. The supernatant was aspirated and replaced with 1× DPBS. Erythrocytes were split into four tubes for each pH that was tested (7.4, 6.8, 6.2, 5.6) and centrifuged at 500g for 5 min. The supernatant was aspirated and replaced with phosphate buffer at the appropriate pH. For each pH, 1 mL of erythrocytes in was added to 49 mL of phosphate buffer at the appropriate pH. NP-siRNA complexes were prepared according to “Preparation of NP-siRNA Treatments” in the main text at 20× concentrations tested. A total of 10 μL of each NP-siRNA sample in quadruplicate was pipetted into wells of a clear U-bottom 96 well plate. 1% Triton X-100 was used as a positive control at each pH to define 100% hemolysis, and 1× DPBS was used as a negative control to subtract background hemolysis at each pH. One plate was loaded per pH tested. A total of 190 μL of erythrocytes diluted at each pH were gently mixed by inversion and loaded to plates containing NP-siRNA treatments using a multi-channel pipet. Plates were incubated for 1 h at 37 °C, centrifuged for 5 min at 500g to collect intact erythrocytes, 100 μL of supernatant from each well was transferred into a clear flat bottom 96-well plate, and absorbance of released hemoglobin, which is indicative of hemolysis, was measured at 541 nm using a plate reader.
Cytotoxicity via Assessment of Metabolic Activity Using AlamarBlue
Cells were seeded at 8000 cells/cm2 and allowed to adhere overnight. The next day, NP-siRNA treatments were prepared using nontargeting Silencer Negative Control siRNA 1 (Ambion) and cells were washed 2× with 1× DPBS. NP-siRNA treatments were added directly to wells containing fresh Opti-Mem media and incubated for 24 h at 37 °C and 5% CO2. Treated cells were washed 2× using 1× DPBS. A mixture of 10% AlamarBlue in the appropriate cell growth media was added to each well and incubated for 2 h at 37°C and 5% CO2. Blank samples were included by adding the AlamarBlue containing media to wells containing no cells. Fluorescence of the cell culture media resulting from the metabolism of the AlamarBlue compound was measured for each sample in triplicate in a black 96-well plate using a plate reader with excitation = 570 nm, emission = 600 nm, and optimal gain. Blank measurements were subtracted from all samples and relative metabolic activity was calculated by normalizing treated to untreated samples.
Statistical and Multiparameter Regression Analysis
Multi-parameter regression was performed in JMP with first block Mn, second block Mn, corona/core ratio, and %BMA as model inputs to describe the behavior of uptake, gene inhibition, and viability results within each cell type as a function of the individual polymer parameters. This approach limits the influence of confounding factors and allows analysis across all NPs. Model fit significance was set to p < 0.05. If overall model fit was significant for a given outcome measure within a cell type, the individual parameter effect (E) of the NP property was assessed to identify parameters that best described the behavior.
Each experiment was performed in triplicate in two independent experiments (n = 6), except for NP-siRNA uptake in mTenocytes, in which a third independent experiment of triplicate samples was performed (n = 9) to increase statistical power due to variability. One-way ANOVA was used with the appropriate post hoc test as indicated in figure legends to assess significant differences ((α = 0.05) in NP-siRNA means, compared to controls. Two-way ANOVA was used to compare siRNA delivery outcomes across cell types ((α = 0.05) using Tukey’s post hoc test to correct for increased type I error due to multiple comparisons. Pearson’s correlation ((α = 0.05, two-tailed) was used to determine if gene silencing and cytotoxicity are correlated with NP-siRNA uptake. Statistical analyses were performed using GraphPad Prism 6.0 unless otherwise indicated. For all plots, the mean is represented with standard deviation shown as error bars.
RESULTS
Polymer NP Synthesis and Characterization
Diblock copolymers were synthesized using reversible addition–fragmentation chain transfer polymerization (RAFT) as previously described.20,21,26 The degree of polymerization or the ratio of monomer to chain transfer agent was varied to synthesize a library of polymers varying in first block Mn, second block Mn, and the ratio of the 1st/2nd block Mn (corona/core), in addition to the BMA content in the second block (see Table 1). NP X was synthesized as a control with 100% BMA in the core and therefore lacked pH-responsive behavior. All polymers had narrow polydispersities (PDI ≤ 1.21), regardless of reaction conditions. The resulting polymers self-assembled into micellar nanoparticles with diameters ranging from 9 ± 2 to 34 ± 10 nm. All nanoparticles maintained cationic surface charge of 8 ± 14 to 24 ± 4 mV in phosphate buffered saline (PBS).
NP-Mediated siRNA Uptake
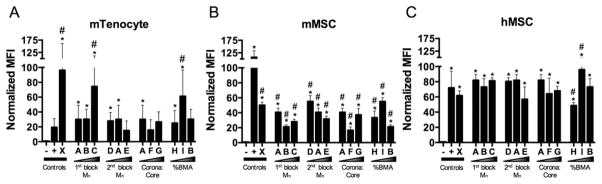
To quantify NP-mediated siRNA uptake, cells were treated for 24 h with NP complexed with 30 nM fluorescent siRNA at a charge ratio (±) of 4:1 to form NP-siRNA complexes, and NP-siRNA uptake was quantified using flow cytometry. In general, uptake was significantly greater in hMSCs than both murine cell types, and multiparameter regression (MPR) analysis shows that first block Mn positively correlated with uptake in mTenocytes and hMSCs. mTenocytes and mMSCs showed highly variable siRNA uptake, as illustrated in Figure 1A, B. In mTenocytes, NP A, B, C, D, and I resulted in significantly increased MFI, while NP E, F, G, and H were not significantly increased compared to untreated. Furthermore, NP C and I resulted in the highest MFI compared to Lipofectamine2000, a commercial transfection reagent (Figure 1A). All NP resulted in significantly increased MFI in mMSCs, compared to untreated cells; however, uptake was lower than that achieved using Lipofectamine2000 (Figure 1B). In hMSCs, all NP resulted in increased MFI, and all NP except H and I performed similarly to the positive control, Lipofecamine2000. Control NP X was taken up by all cell types. A two-way ANOVA comparing effects of cell type and species on MFI means with Tukey’s post hoc test to correct for multiple comparisons shows overall uptake was significantly greater in hMSCs compared to both mMSCs (p < 0.0001) and mTenocytes (p < 0.0001), while there was no difference between murine cell types (p > 0.05).
Figure 1.
NP facilitate significant and differential siRNA uptake across multiple cell types and species. Murine tenocytes (A), murine MSCs (mMSC) (B), and human MSCs (hMSC) (C) were treated with 30 nM fluorescent siRNA complexed to NP at a charge ratio of 4:1. Median fluorescence intensity (MFI) was detected via flow cytometry and normalized to negative controls within each cell type. *p < 0.05 compared to negative control (–), #p < 0.01 compared to positive (+) control. Negative controls (–) are comprised of untreated cells and positive controls (+) are cells treated with 30 nM siRNA using Lipofectamine2000.
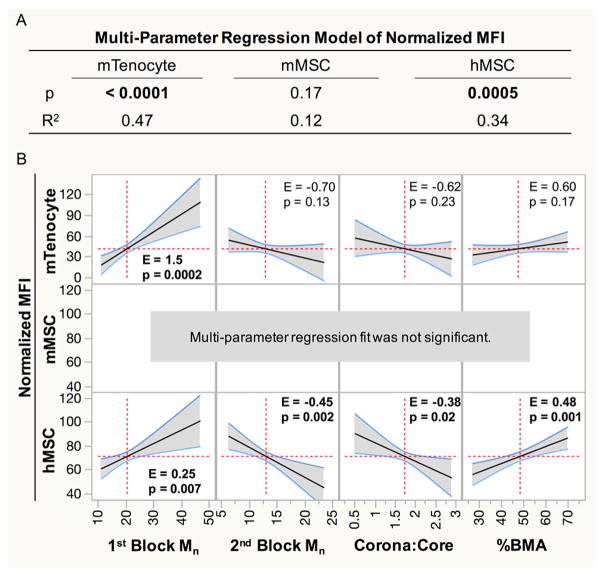
To determine the extent to which the varying NP polymer parameters contributed to siRNA uptake, MPR analysis was applied. This statistical analysis creates a linear regression model of siRNA delivery outcome measures as a function of NP properties. If the overall fit significantly describes the siRNA delivery outcome measure (p < 0.05), indicating dependence on polymer properties, then the analysis can be expanded to determine the effect size (E) of the individual polymer parameters and if the effect is significant (p < 0.05). Figure 2 shows this analysis applied to normalized MFI in each cell type. The model significantly predicts uptake behavior for mTenocytes (p < 0.0001) and hMSCs (p = 0.0005), but not mMSCs (p = 0.1682; Figure 2A). MFI in mTenocytes is primarily correlated positively with first block Mn (effect size (E) = 1.5, p = 0.0002; Figure 2B). All parameters significantly influenced MFI in hMSC, which is positively correlated with % BMA (E = 0.48, p = 0.001) and first block Mn (E = 0.25, p = 0.007). Second block Mn (E = –0.45, p = 0.002) is similar in effect size to %BMA, but negatively correlates with MFI. Additionally, corona/core negatively affects MFI (E = –0.38, p < 0.02).
Figure 2.
Normalized MFI data were fit using a multiparameter linear regression to determine if siRNA uptake is dependent on NP properties. (A) p-values less than 0.05 (bold) indicate that siRNA uptake is dependent on NP parameters, and the R2 values illustrate what percentage of the behavior can be adequately described by the predictive model. (B) Effect sizes (E) from significant regression models show the extent that a single property correlates with MFI. p < 0.05 indicates the polymer property significantly influences MFI. Significant effects are in bold.
NP-siRNA-Mediated Gene Silencing
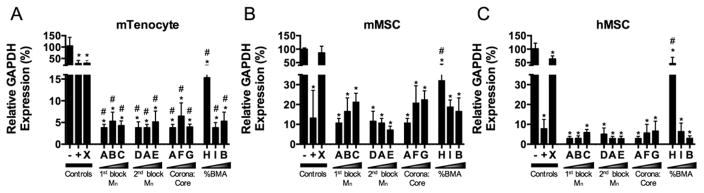
After quantifying uptake of siRNA, cells were treated with NP-siRNA complexes targeting glyceradehyde 3-phosphate dehydrogenase (GAPDH) to quantify the functional gene silencing capabilities of the NP. Cells were treated with NP-siRNA complexes for 24 h and RNA was extracted an additional 24 h later. In general, gene silencing is positively correlated with %BMA content in all cell types. Figure 3A shows that all NP resulted in >90% knockdown in GAPDH expression in mTenocytes, except for NP H, which resulted in ~85% gene knockdown. These effects were greater than the knockdown achieved using Lipofect-amine2000. Control NP X resulted in ~70% gene silencing. All NP except NP X also showed significantly reduced GAPDH expression in mMSCs compared to untreated cells (Figure 3B). However, knockdown using NP C, F, G, H, and I were significantly smaller than that achieved with Lipofecamine2000, indicating differential gene silencing abilities in mMSCs across the NP library. Similarly, all NP achieved significant reductions in GAPDH expression in hMSCs (Figure 3C). All NP except for H and X resulted in >90% reductions compared to untreated controls, and performed no differently than Lip-ofectamine2000. Two-way ANOVA using Tukey’s post hoc test shows that overall gene silencing in mMSCs was significantly reduced compared to mTenocytes (p < 0.0001) and hMSCs (p < 0.0001).
Figure 3.
NP-mediated delivery of siRNA results in robust gene silencing across multiple cell types and species. Murine tenocytes (A), murine MSCs (mMSC) (B), and human MSCs (hMSC) (C) were treated with 30 nM siRNA targeting glyceraldehyde 3-phosphate dehydrogenase (GAPDH) complexed to NP at a charge ratio of 4:1. Gene expression was detected using RT-PCR and normalized to negative controls in each cell type. GAPDH expression was normalized to B-Actin expression for murine cell types and peptidylprolyl isomerase B (PPIB) expression for hMSCs. *p < 0.0001 compared to negative control, #p < 0.001 compared to positive controls (+) control. Negative controls (–) are comprised of untreated cells and positive controls (+) are cells treated with 30 nM anti-GAPDH siRNA using Lipofectamine2000.
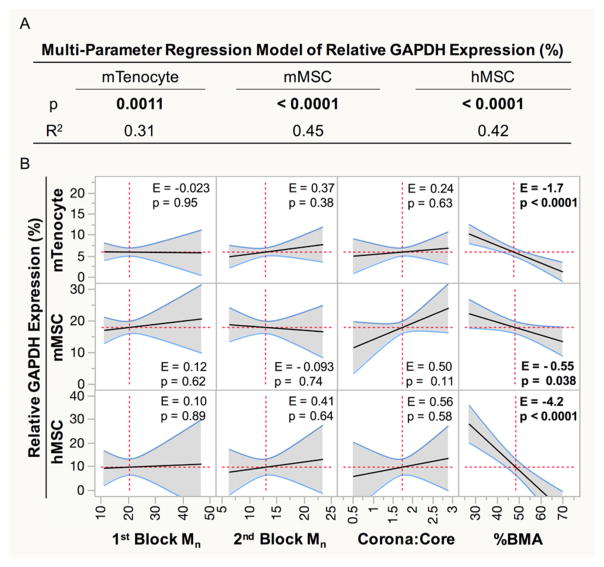
To determine the extent to which polymer parameters control NP-mediated gene silencing, MPR analysis was performed on NP-mediated gene expression. NP gene silencing capability is significantly dependent on NP properties in all three cell types (mTenocyte p = 0.0011, mMSC p < 0.0001, hMSC p < 0.0001; Figure 4A). Furthermore, relative GAPDH expression is significantly dependent on and negatively correlated with %BMA content in all three cell types (mTenocyte E = –1.7, p < 0.0001; mMSC E = –0.55, p = 0.038; hMSC E = –4.2, p < 0.0001; Figure 4B), suggesting % BMA may be an important parameter governing gene silencing.
Figure 4.
Relative GAPDH expression data was fit using a multiparameter linear regression to determine if gene-silencing capability is dependent on NP properties. (A) p values less than 0.05 (bold) indicate that gene silencing is dependent on NP parameters, and the R2 value describes what percentage of the behavior can be adequately described by the predictive model. (B) Effect sizes (E) from significant regression models show the extent that a single property correlates with MFI. p < 0.05 indicates the polymer property significantly influences GAPDH expression. Significant effects are bolded.
pH-Dependent NP Membrane Disruption Behavior
A hemolysis assay was performed to determine the impact of polymer properties on NP pH-dependent membrane disruption ability, as this behavior is required for endosomal escape and subsequent gene knockdown. In this assay, human erythrocytes are isolated and incubated with NP complexed to negative control nontargeting siRNA under normal treatment conditions, and incubated at various pH that mimic the various stages of the endosomal environment. NP-mediated membrane disruption is assayed by measuring absorbance of hemoglobin released by lysed erythrocytes. Triton-X is used as a positive control to set 100% lysis for normalization. Figure 5 shows that all NPs display pH-dependent membrane lysis ability, except the control NP X, which contains no pH responsive core components. A two-way ANOVA using Tukey’s test shows that mean membrane lysis significantly increases as %BMA increases (p < 0.001). No significant differences were observed for the other polymer properties.
Figure 5.
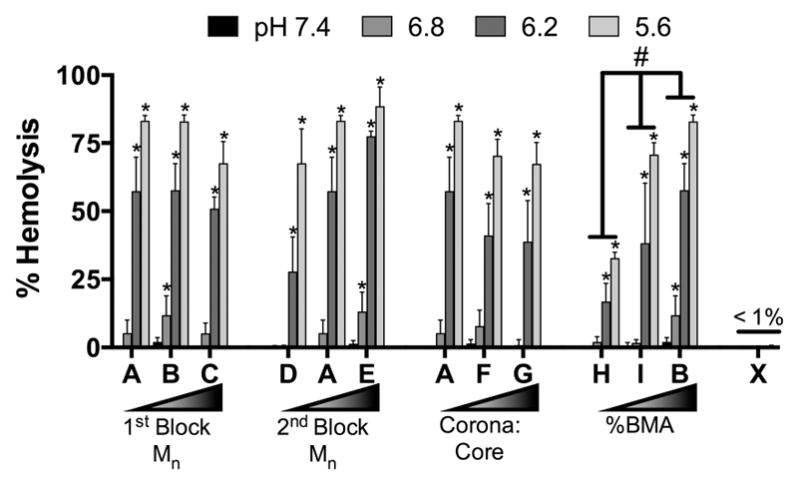
pH-dependent hemolysis ability of NPs. *p < 0.05 compared to pH 7.4 for a given NP using two-way ANOVA with Dunnett’s correction for multiple comparisons. #p < 0.0001 comparing hemolysis across indicated NPs using two-way ANOVA with Tukey’s post hoc test for multiple comparisons. n = 8 from two independent experiments using blood from two separate donors. Error bars represent standard deviation.
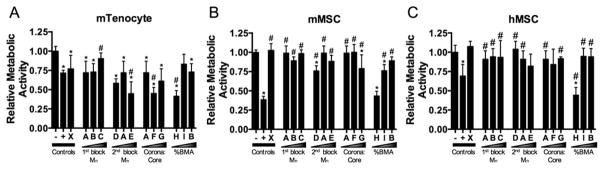
NP-siRNA Cytocompatibility
After determining NP-mediated uptake of siRNA and subsequent gene silencing, metabolic activity normalized to untreated controls (Figure 6) was quantified to determine how NP properties affect cell viability as a measure of cytocompatibility. A nontargeting negative control siRNA was complexed to NP and incubated with cells for 24 h. In general, viability was significantly higher in both MSC cell types and positively correlated with %BMA content. mTenocyte viability was reduced by all NP, except C and I, and Lipofectamine2000 treatments (Figure 6A). Further, NP E, F, and H resulted in significantly reduced viability compared to Lipofectamine2000 (Figure 6A). mMSC viability was only reduced by NP D, NP H, and Lipofectamine2000 (Figure 6B), indicating the NP are well tolerated by mMSCs compared to mTenocytes. hMSCs also showed strong cytocompatibility, as only NP H and Lipofectamine2000 significantly reduced hMSC viability compared to untreated cells (Figure 6C). Two-way ANOVA using Tukey’s post hoc test shows that mTenocyte viability is significantly lower than both mMSC and hMSC (p < 0.0001).
Figure 6.
NP-siRNA cytocompatibility via assessment of metabolic activity. Murine tenocytes (mTenocyte) (A), murine MSCs (mMSC) (B), and human MSCs (hMSC) (C) were treated with 30 nM nontargeting negative control siRNA complexed to NP at a charge ratio of 4:1. Metabolic activity was quantified as a measure of cytocompatibility and normalized to negative controls. *p < 0.05 compared to negative (–) control, #p < 0.05 compared to positive (+) control. Negative controls (–) are comprised of untreated cells and positive controls (+) are cells treated with 30 nM nontargeting negative control siRNA using Lipofectamine2000.
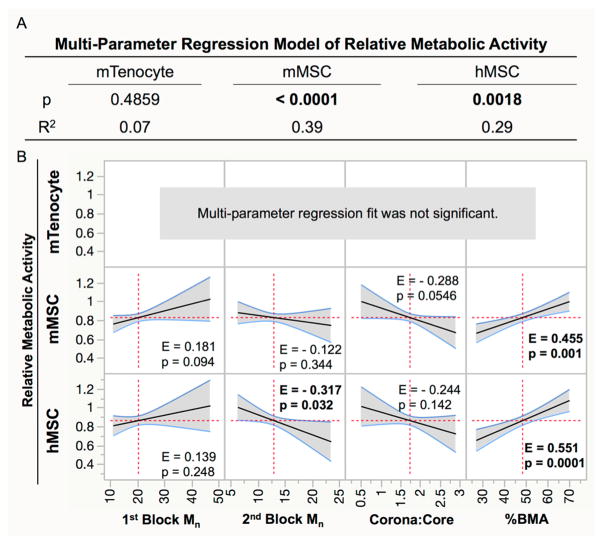
MPR analysis was used to determine which NP polymer properties affected cell viability as measured by the relative metabolic activity (Figure 7) and shows that decreased viability in mTenocytes is independent of NP properties (p = 0.10; Figure 7A). Viability was dependent on NP properties in mMSCs (p < 0.0001) and hMSCs (p = 0.0018). In both cell types, %BMA content was positively correlated with viability (mMSC: E = 0.455, p = 0.001; hMSC: E = 0.551, p = 0.0001). Furthermore, second block Mn was negatively correlated with viability in hMSCs (E = –0.317, p = 0.032). DNA content was quantified as a secondary measure of cytocompatibility, in each cell type (Figure S1) and the MPR analysis was applied (Figure S2). Overall, these data also show that viability was lowest in mTenocytes and was dependent on NP properties in hMSCs, which agrees with metabolic activity analyses. High assay variability in mMSC DNA content measurements led to inconclusive results in this cell type (Figure S1).
Figure 7.
Relative metabolic activity in treated cells was fit using a multiparameter linear regression to determine if cell viability is dependent on NP properties. (A) p values less than 0.05 (bold) indicate that cell viability is dependent on NP parameters, and the R2 value describes what percentage of the behavior can be adequately described by the predictive model. (B) Effect sizes (E) from significant regression models show the extent that a single property correlates with cell viability. p < 0.05 indicates the polymer property significantly influences cell viability. Significant effects are bolded.
DISCUSSION
Despite the successful implementation of polymeric NP-mediated siRNA delivery in a number of applications,21,23,28,30 it remains largely unknown how various polymer properties contribute to siRNA delivery outcomes. This information is necessary for the rational design of safe and effective treatments. Therefore, a library of polymers was synthesized via RAFT polymerization, wherein a number of parameters were varied systematically, including first block Mn, second block Mn, corona/core ratio (first block Mn to second block Mn), and %BMA content, while maintaining narrow poly-dispersity (PDI < 1.21). Results show that NP-mediated siRNA uptake is significantly more robust in hMSCs compared to both murine cell types, which suggests that uptake is species dependent. The MPR model adequately predicts uptake behavior in hMSCs and mTenocytes, and it is positively correlated with first block Mn. All NP treatments mediated robust gene silencing, which is positively correlated with % BMA content in all cell types, regardless of species. Furthermore, NP were significantly more cytocompatible with MSCs compared to mTenocytes, suggesting the influence of differentiation state on NP cytotoxicity. Finally, %BMA content is positively correlated with cytocompatibility in hMSCs, indicating NP stability is critical for effective treatment.
NP were used to assess siRNA delivery in mTenocytes, mMSCs, and hMSCs as RNAi has gained traction to direct cell fate toward multiple musculoskeletal lineages for tissue regeneration.37,38 MSCs are an excellent cell source for musculoskeletal tissue engineering due to ease of isolation, ability to differentiate to multiple musculoskeletal lineages, and immunomodulatory trophic factor secretion that modulates tissue repair.39–42 Furthermore, NP have recently been used for drug delivery to tendon,43 including delivery of TGF-β miRNA plasmids to augment tendon healing by preventing the formation of adhesions.44 However, to our knowledge, this report is the first to describe NP-mediated delivery of siRNA to tenocytes in vitro.
NP enter cells through endocytosis45,46 or direct membrane translocation.47,48 Multiple reviews show the majority of spherical cationic NP with diameters similar to those used herein undergo clathrin-mediated endocytosis, and blocking this pathway results in uptake using compensatory endocytic pathways, such as macropinocytosis.49,50 Regardless of endocytic mechanism, uptake ultimately leads to endosomal trafficking in acidic vesicles (pH 6.8–4.9).51–53 Here, results show that the design parameters of the NP differentially affect siRNA uptake in murine and human cells. Differences in uptake mechanisms are unknown, but could be partly attributed to species and/or cell type differences in lipid bilayer composition.54,55 Cationic drug delivery systems, such as PEI and Lipofectamine, are endocytosed after interaction with anionic surface receptors.45,56–58 However, differences in endocytosis receptors have not been characterized in these cell types or across different species. Uptake is positively correlated with first block Mn in mTenocytes and hMSCs, indicating the influence of the cationic nature of the NP, which is reported to increase NP uptake.59 Furthermore, uptake was positively correlated with %BMA. Generally, increased micelle core hydrophobicity (here, %BMA) increases stability,60 which could explain the improved uptake with increased %BMA. To help support this claim, critical micelle concentration (CMC) was measured for NPs B, I, and H, which have varied %BMA core content (Figure S3). Results show that NP B, which has the highest BMA content (70%) displayed the lowest CMC (0.37 μM) compared to NP I (55% BMA, CMC = 1.0 μM) and NP H (27% BMA, CMC = 1.1 μM). On the other hand, uptake negatively correlated with Corona:Core ratio, and it is known that increases in this ratio decrease NP stability.61 Interestingly, corona/core positively correlated with hydrophobic drug release rate within biofilm microenvironments,30,61 yet here, the resulting instability had adverse effects on siRNA uptake. Additionally, second block Mn is negatively correlated with uptake in hMSCs. A recent study using dissipative particle dynamics simulation shows “soft” NP, composed of self-assembled amphiphilic molecules similar to the NP used herein, deform upon interacting with the cell membrane. This deformation allows hydrophobic NP core components to incorporate into the hydrophobic interior of the bilayer, thereby slowing or inhibiting NP internalization.62 Our results suggest that high Mn second blocks result in greater overall NP hydrophobicity, possibly limiting NP uptake through this mechanism. This deformation could also explain the positive correlation of first block Mn with uptake, as deformation of the NP might allow more positive charges in the polymer chain to interact with the cell membrane, rather than just those at the surface of the NP.
Differences in NP-mediated gene silencing reveal further insights into the NP physicochemical properties that affect siRNA delivery. All NP resulted in significant GAPDH knockdown across all cell types tested, yet gene silencing is not correlated with uptake (Table S2). This finding is interesting because not all NP resulted in significant uptake in mTenocytes. To interpret this outcome, a control NP (X) was synthesized with a core composed of 100% BMA (Table 1), thereby providing significant siRNA uptake in all cell types (MFI = 96 ± 65 in mTenocytes, 50 ± 4 in mMSCs, and 62 ± 6 in hMSCs; Figure 1), but eliminating its pH-dependent membrane disruption properties as verified using a hemolysis assay (Figure 5).36 NP X-mediated GAPDH gene knockdown was robust in mTenocytes (71% ± 13%) compared to hMSCs (<30%) and mMSCs (~0%) (Figure 3). This result indicates gene silencing in mTenocytes is independent of pH-responsive endosomal escape, suggesting NP-mediated uptake and endosomal escape via passive mechanisms, such as direct membrane translocation.47,63 The MPR analysis shows that %BMA content positively influences NP-mediated gene knockdown in all cell types, similar to previous results in HeLa cells.28 Furthermore, pH-dependent NP destabilization, which is required for membrane destabilization,28,36 is not inhibited in NP with up to 70% BMA in the core block, and that greater hydrophobic characteristic provides increased membrane lysis at endosomal pH (Figure 5).
Cytocompatibility is critical for successful drug delivery. Cationic polymers and NPs have been shown to induce cytotoxicity through membrane disruption64,65 and production of reactive oxygen species45,66 (ROS). Regardless of mechanism, cytotoxicity is dose-dependent and varies with polymer molecular weight and structure (i.e., linear, branched, nano-particle, etc.). This finding has been specifically shown in multiple DMAEMA-containing drug delivery systems.67,68 Here, MSCs, regardless of species, were significantly more resilient to NP treatments than mTenocytes. This trend was consistent when measuring viability via metabolic activity and DNA content, and suggests that differentiation state is an important parameter to consider when designing siRNA delivery systems and optimizing doses. MSCs possess the unique ability to protect themselves from stress-induced cell death by undergoing senescence69,70 or quiescence.71 Additionally, our previous study using RNA-seq and enrichment analysis in hMSCs showed significant upregulation of antiapoptotic genes 24 h after NP-siRNA treatment that is sustained up to 14 days.26 These innate self-protective effects of MSCs could explain the differences in toxicity observed here in mTenocytes compared to MSCs. mTenocyte cytotoxicity is independent of polymer properties (Figure 6, Figure S2) and positively correlated with uptake (Table S2). These data, in concert with uptake and pH-independent gene silencing, lead to the hypothesis that mTenocytes have fewer membrane-associated receptors to coordinate NP uptake leading to nonspecific interaction with membrane lipids that could result in formation of nanoscale pores in lipid-bilayer membranes through which NP can translocate. This premise has been shown in other cationic NP systems, resulting in passive NP uptake and increased cytotoxicity.47,48,72 To our knowledge, the relative amounts of cell surface receptors or diversity of cell receptors have not been characterized in these cell types. Therefore, it is possible that progenitor cells have a wider variety and larger number of surface-associated receptors compared to differentiated cells, which leads to more NP-receptor interactions for uptake and less NP-lipid membrane disrupting interactions. Furthermore, %BMA is positively correlated with mMSC and hMSC viability, indicating that more stable NP are less toxic. % BMA content is also positively correlated with relative DNA content in hMSCs, a secondary assessment of viability (Figure S2), further emphasizing the importance of this parameter with respect to cytocompatibility. These results suggest that unstable NP, which undergo more rapid diblock exchange and have higher CMC (Figure S3) contain more free polymer chains that can disrupt the cell membranes, which has been shown to decrease cytocompatibility through membrane disruption in other pDMAEMA, containing polymer drug delivery systems.68,73–75 These results suggest that mechanism of NP-siRNA-mediated cytotoxicity in mTenocytes should be further investigated, as our previous studies show initiation of, but limited commitment to apoptosis in hMSCs.26 It is also important to note that cytotoxicity does not correlate with polymer mass, as each treatment is based on charge ratio (Table S3).
The study herein identifies polymer parameters that are critical for siRNA delivery. Our results further show the importance of using multifunctional polymers for siRNA delivery in agreement with and expanding upon previous studies.28 Overall, first block molecular weight was shown to increase uptake, while hydrophobic BMA had significant effects on siRNA delivery and gene silencing by increasing polymer stability to increase cytocompatibility while also improving pH-dependent membrane lysis. These results were consistent across multiple species and musculoskeletal cell types indicating the potential universality of these conclusions. These data agree with the current emphasis on efficient endosomal escape as a critical feature for siRNA delivery.7 Recent advancements in siRNA delivery quantification using lipid NP (LNP) reveal that only 1–2% of siRNA taken up by cells could actually escape the endosome,76 while a separate study showed that over 70% of the internalized LNP dose was eventually exocytosed.77 Increasing endosomal escape would allow for significantly lower doses, making siRNA treatments much safer by reducing potential for immunogenic responses and carrier mediated toxicities. There are limitations of this study in that the model only partially describes this siRNA delivery behavior in vitro (R2 ≤ 0.47), indicating the likelihood of other confounding factors, such as additive or synergistic effects of polymer properties, or nonlinear siRNA delivery-polymer parameter correlations. Due to practical synthesis limitations, it was not possible to systematically vary a single property while fixing all others, and therefore conventional multiway ANOVA may be inconclusive. Thus, while the MPR predictions are not causative, they provide an intuitive way to assess the impact of individual polymer properties on NP performance. For example, although NP B has greater BMA content than NP I, the two have similar increases in MFI, even though the MPR model predicts a positive correlation with %BMA content. This observed plateau is likely due to either confounding factors of the other polymer/NP properties, or the assumption of linear behaviors in the MPR model. NP B has greater second block Mn, as a consequence of increasing the %BMA, compared to NP I, and the MPR model predicts that uptake is negatively correlated with second block Mn. This example Illustrates the motivation for the use of MPR analysis to limit the effect of confounding factors and predict the contribution from individual polymer parameters. These relationships show that the most effective NPs will have an optimal combination of these parameters. As mentioned previously, NP zeta potential, and diameter are known to influence siRNA delivery.45,78 Therefore, we also correlated siRNA outcome measures with these NP properties. Zeta potential was not correlated with any siRNA delivery outcomes (Table S4). These results are likely due to identical siRNA-NP formulations with complexation at the same charge ratio (± 4), effectively maintaining similar cationic characteristic across NP treatments. Diameter was positively correlated with viability (DNA content) in mTenocytes (p = 0.01) and hMSC (p = 0.04; Table S5). This trend agrees with other studies that show smaller NPs (diameter ≤ 10 nm) are more likely to create pores in the cell membrane, leading to toxicity.48,79 Furthermore, GAPDH expression was negatively correlated with NP diameter in hMSCs, indicating larger NPs lead to better gene knockdown (Table S5). However, this logic could be confounded with % BMA, which is also correlated with greater gene knockdown, yet increasing core hydrophobicity leads to greater NP diameter.60,61,80
CONCLUSION
Overall, this study determined how polymer properties modulate NP-mediated delivery of siRNA in a previously developed siRNA delivery system using therapeutically relevant musculoskeletal cell types. Our results show larger first block Mn resulted in better NP-siRNA uptake, and %BMA content is a critical parameter that governs gene silencing cytocompatibility in MSCs and tenocytes. These findings lead to the hypothesis that increased %BMA increases NP stability and efficiency of endosomal escape, resulting in effective siRNA delivery and gene silencing across the cell types tested. Additionally, the safety and efficacy siRNA delivery is dependent on the differentiation state of the target cell. We hypothesize that these differences might be attributed to differences in cell membrane composition and alternate uptake mechanisms independent of endolysosomal vesicle trafficking. Our results highlight the importance of undertaking these fundamental characterizations to truly understand NP-mediated siRNA delivery mechanisms. This knowledge could contribute to the development of safer and more effective next-generation siRNA and drug delivery systems via the use of the rationally designed biomaterials.
Supplementary Material
Acknowledgments
The Authors would like to thank the University of Rochester Medical Center’s Flow Cytometry Core for valuable training and guidance, and the University of Rochester’s McGrath Lab for use of facilities. Funding for this work was provided by the National Science Foundation (DMR- 1206219), the New York State Stem Cell Science Program (NYSTEM IDEA-N11G-035), and the National Institutes of Health (R01 AR064200, R01 AR056696, R01 DE018023, and P30 AR069655 (SubProject 5278)). M.T.F. is supported by a NIAMS/NIH Training Grant (T32 AR053459). K.R.S. is supported by NIDCR/NIH Ruth L. Kirschstein National Research Service Award Number F31 DE026944. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Science Foundation, National Institutes of Health, or the other funding sources.
Footnotes
The authors declare no competing financial interest.
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.bio-mac.7b01349
Experimental: Cytotoxicity via assessment of metabolic activity using AlamarBlue; Determination of critical micelle concentration (CMC) by PRODAN assay. Results: Figure S1, NP-siRNA treatments result in varying degrees of cytocompatibility as measured via relative quantification of DNA content; Figure S2, MPR analysis of relative DNA content; Figure S3, Determination of critical micelle concentration (CMC) via PRODAN assay of NP B (A), NP I (B), and NP H (C) containing varying amounts of BMA; Table S1, Primer sequences used in this study; Table S2, Pearson’s correlation of MFI vs GAPDH expression and DNA content; Table S3, Pearson’s correlation of polymer mass used in NP treatments vs DNA content and metabolic activity; Table S4, Pearson’s correlation of zeta potential vs MFI, GAPDH expression, and DNA content; Table S5, Pearson’s correlation of NP diameter vs MFI, GAPDH expression, and DNA content (PDF).
References
- 1.Zhu L, Mahato RI. Lipid and polymeric carrier-mediated nucleic acid delivery. Expert Opin Drug Delivery. 2010;7(10):1209–1226. doi: 10.1517/17425247.2010.513969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Williford JM, Wu J, Ren Y, Archang MM, Leong KW, Mao HQ. Recent advances in nanoparticle-mediated siRNA delivery. Annu Rev Biomed Eng. 2014;16:347–70. doi: 10.1146/annurev-bioeng-071813-105119. [DOI] [PubMed] [Google Scholar]
- 3.Whitehead KA, Langer R, Anderson DG. Knocking down barriers: advances in siRNA delivery. Nat Rev Drug Discovery. 2009;8(2):129–38. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang J, Lu Z, Wientjes MG, Au JL. Delivery of siRNA therapeutics: barriers and carriers. AAPS J. 2010;12(4):492–503. doi: 10.1208/s12248-010-9210-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hickerson RP, Vlassov AV, Wang Q, Leake D, Ilves H, Gonzalez-Gonzalez E, Contag CH, Johnston BH, Kaspar RL. Stability Study of Unmodified siRNA and Relevance to Clinical Use. Oligonucleotides. 2008;18(4):345–354. doi: 10.1089/oli.2008.0149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nimesh S, Gupta N, Chandra R. Cationic polymer based nanocarriers for delivery of therapeutic nucleic acids. J Biomed Nanotechnol. 2011;7(4):504–20. doi: 10.1166/jbn.2011.1313. [DOI] [PubMed] [Google Scholar]
- 7.Ma D. Enhancing endosomal escape for nanoparticle mediated siRNA delivery. Nanoscale. 2014;6(12):6415–25. doi: 10.1039/c4nr00018h. [DOI] [PubMed] [Google Scholar]
- 8.Sarisozen C, Salzano G, Torchilin VP. Lipid-based siRNA Delivery Systems: Challenges, Promises and Solutions Along the Long Journey. Curr Pharm Biotechnol. 2016;17(8):728–40. doi: 10.2174/1389201017666160401145319. [DOI] [PubMed] [Google Scholar]
- 9.Mokhtarzadeh A, Alibakhshi A, Hashemi M, Hejazi M, Hosseini V, de la Guardia M, Ramezani M. Biodegradable nano-polymers as delivery vehicles for therapeutic small non-coding ribonucleic acids. J Controlled Release. 2017;245:116–126. doi: 10.1016/j.jconrel.2016.11.017. [DOI] [PubMed] [Google Scholar]
- 10.Liu XQ, Sun CY, Yang XZ, Wang J. Polymeric-Micelle-Based Nanomedicine for siRNA Delivery. Part Part Syst Char. 2013;30(3):211–228. [Google Scholar]
- 11.Mao S, Sun W, Kissel T. Chitosan-based formulations for delivery of DNA and siRNA. Adv Drug Delivery Rev. 2010;62(1):12–27. doi: 10.1016/j.addr.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 12.Ragelle H, Vandermeulen G, Preat V. Chitosan-based siRNA delivery systems. J Controlled Release. 2013;172(1):207–18. doi: 10.1016/j.jconrel.2013.08.005. [DOI] [PubMed] [Google Scholar]
- 13.Neuberg P, Kichler A. Recent developments in nucleic acid delivery with polyethylenimines. Adv Genet. 2014;88:263–88. doi: 10.1016/B978-0-12-800148-6.00009-2. [DOI] [PubMed] [Google Scholar]
- 14.Merkel OM, Urbanics R, Bedocs P, Rozsnyay Z, Rosivall L, Toth M, Kissel T, Szebeni J. In vitro and in vivo complement activation and related anaphylactic effects associated with polyethyle-nimine and polyethylenimine-graft-poly(ethylene glycol) block copolymers. Biomaterials. 2011;32(21):4936–42. doi: 10.1016/j.biomaterials.2011.03.035. [DOI] [PubMed] [Google Scholar]
- 15.Kafil V, Omidi Y. Cytotoxic impacts of linear and branched polyethylenimine nanostructures in a431 cells. Bioimpacts. 2011;1(1):23–30. doi: 10.5681/bi.2011.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Godbey WT, Wu KK, Mikos AG. Poly(ethylenimine)-mediated gene delivery affects endothelial cell function and viability. Biomaterials. 2001;22(5):471–80. doi: 10.1016/s0142-9612(00)00203-9. [DOI] [PubMed] [Google Scholar]
- 17.Xue HY, Liu S, Wong HL. Nanotoxicity: a key obstacle to clinical translation of siRNA-based nanomedicine. Nanomedicine (London, U K) 2014;9(2):295–312. doi: 10.2217/nnm.13.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jager M, Schubert S, Ochrimenko S, Fischer D, Schubert US. Branched and linear poly(ethylene imine)-based conjugates: synthetic modification, characterization, and application. Chem Soc Rev. 2012;41(13):4755–67. doi: 10.1039/c2cs35146c. [DOI] [PubMed] [Google Scholar]
- 19.Hobel S, Aigner A. Polyethylenimines for siRNA and miRNA delivery in vivo. Wiley Interdisciplinary Reviews. Nanomedicine and Nanobiotechnology. 2013;5(5):484–501. doi: 10.1002/wnan.1228. [DOI] [PubMed] [Google Scholar]
- 20.Convertine AJ, Benoit DS, Duvall CL, Hoffman AS, Stayton PS. Development of a novel endosomolytic diblock copolymer for siRNA delivery. J Controlled Release. 2009;133(3):221–9. doi: 10.1016/j.jconrel.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Benoit DSW, Boutin ME. Controlling mesenchymal stem cell gene expression using polymer-mediated delivery of siRNA. Biomacromolecules. 2012;13(11):3841–3849. doi: 10.1021/bm301294n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arany S, Xu Q, Hernady E, Benoit DS, Dewhurst S, Ovitt CE. Pro-apoptotic gene knockdown mediated by nano-complexed siRNA reduces radiation damage in primary salivary gland cultures. J Cell Biochem. 2012;113(6):1955–65. doi: 10.1002/jcb.24064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arany S, Benoit DS, Dewhurst S, Ovitt CE. Nanoparticle-mediated gene silencing confers radioprotection to salivary glands in vivo. Mol Ther. 2013;21(6):1182–94. doi: 10.1038/mt.2013.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Convertine AJ, Diab C, Prieve M, Paschal A, Hoffman AS, Johnson PH, Stayton PS. pH-responsive polymeric micelle carriers for siRNA drugs. Biomacromolecules. 2010;11(11):2904–11. doi: 10.1021/bm100652w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nelson CE, Kintzing JR, Hanna A, Shannon JM, Gupta MK, Duvall CL. Balancing cationic and hydrophobic content of PEGylated siRNA polyplexes enhances endosome escape, stability, blood circulation time, and bioactivity in vivo. ACS Nano. 2013;7(10):8870–80. doi: 10.1021/nn403325f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Malcolm DW, Sorrells JE, Van Twisk D, Thakar J, Benoit DS. Evaluating side effects of nanoparticle-mediated siRNA delivery to mesenchymal stem cells using next generation sequencing and enrichment analysis. Bioeng Transl Med. 2016;1(2):193–206. doi: 10.1002/btm2.10035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moad G, Chong YK, Postma A, Rizzardo E, Thang SH. Advances in RAFT polymerization: the synthesis of polymers with defined end-groups. Polymer. 2005;46(19):8458–8468. [Google Scholar]
- 28.Convertine AJ, Benoit DSW, Duvall CL, Hoffman AS, Stayton PS. Development of a novel endosomolytic diblock copolymer for siRNA delivery. J Controlled Release. 2009;133(3):221–229. doi: 10.1016/j.jconrel.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gallow KC, Jhon YK, Genzer J, Loo YL. Influence of gradient strength and composition profile on the onset of the cloud point transition in hydroxyethyl methacrylate/dimethylaminoethyl methacrylate gradient copolymers. Polymer. 2012;53(5):1131–1137. [Google Scholar]
- 30.Horev B, Klein MI, Hwang G, Li Y, Kim D, Koo H, Benoit DSW. pH-Activated Nanoparticles for Controlled Topical Delivery of Farnesol To Disrupt Oral Biofilm Virulence. ACS Nano. 2015;9(3):2390–2404. doi: 10.1021/nn507170s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Farhat YM, Al-Maliki AA, Chen T, Juneja SC, Schwarz EM, O’Keefe RJ, Awad HA. Gene expression analysis of the pleiotropic effects of TGF-beta1 in an in vitro model of flexor tendon healing. PLoS One. 2012;7(12):e51411. doi: 10.1371/journal.pone.0051411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pittenger MF. Mesenchymal stem cells from adult bone marrow. Methods Mol Biol. 2008;449:27–44. doi: 10.1007/978-1-60327-169-1_2. [DOI] [PubMed] [Google Scholar]
- 33.Sahlin S, Hed J, Rundquist I. Differentiation between attached and ingested immune complexes by a fluorescence quenching cytofluorometric assay. J Immunol Methods. 1983;60(1–2):115–24. doi: 10.1016/0022-1759(83)90340-x. [DOI] [PubMed] [Google Scholar]
- 34.Liu W, Saint DA. A new quantitative method of real time reverse transcription polymerase chain reaction assay based on simulation of polymerase chain reaction kinetics. Anal Biochem. 2002;302(1):52–9. doi: 10.1006/abio.2001.5530. [DOI] [PubMed] [Google Scholar]
- 35.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Research. 2001;29(9):e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Evans BC, Nelson CE, Yu SS, Beavers KR, Kim AJ, Li H, Nelson HM, Giorgio TD, Duvall CL. Ex vivo red blood cell hemolysis assay for the evaluation of pH-responsive endosomolytic agents for cytosolic delivery of biomacromolecular drugs. J Visualized Exp. 2013;73:e50166. doi: 10.3791/50166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yau WWY, Rujitanaroj P-o, Lam L, Chew SY. Directing stem cell fate by controlled RNA interference. Biomaterials. 2012;33(9):2608–2628. doi: 10.1016/j.biomaterials.2011.12.021. [DOI] [PubMed] [Google Scholar]
- 38.Ghadakzadeh S, Mekhail M, Aoude A, Tabrizian M, Hamdy RC. Small Players Ruling the Hard Game: siRNA in Bone Regeneration. J Bone Miner Res. 2016;31(7):1481. doi: 10.1002/jbmr.2860. [DOI] [PubMed] [Google Scholar]
- 39.Caplan AI. Adult mesenchymal stem cells for tissue engineering versus regenerative medicine. J Cell Physiol. 2007;213(2):341–7. doi: 10.1002/jcp.21200. [DOI] [PubMed] [Google Scholar]
- 40.Davatchi F, Abdollahi BS, Mohyeddin M, Shahram F, Nikbin B. Mesenchymal stem cell therapy for knee osteoarthritis. Preliminary report of four patients. International journal of rheumatic diseases. 2011;14(2):211–5. doi: 10.1111/j.1756-185X.2011.01599.x. [DOI] [PubMed] [Google Scholar]
- 41.Nooeaid P, Salih V, Beier JP, Boccaccini AR. Osteochondral tissue engineering: scaffolds, stem cells and applications. Journal of cellular and molecular medicine. 2012;16(10):2247–70. doi: 10.1111/j.1582-4934.2012.01571.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Veronesi F, Salamanna F, Tschon M, Maglio M, Nicoli Aldini N, Fini M. Mesenchymal stem cells for tendon healing: what is on the horizon? J Tissue Eng Regener Med. 2016 doi: 10.1002/term.2209. [DOI] [PubMed] [Google Scholar]
- 43.Parchi:PDVittorio O, Andreani L, Battistini P, Piolanti N, Marchetti S, Poggetti A, Lisanti M. Nanoparticles for Tendon Healing and Regeneration: Literature Review. Front Aging Neurosci. 2016;8:202. doi: 10.3389/fnagi.2016.00202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou Y, Zhang L, Zhao W, Wu Y, Zhu C, Yang Y. Nanoparticle-mediated delivery of TGF-beta1 miRNA plasmid for preventing flexor tendon adhesion formation. Biomaterials. 2013;34(33):8269–78. doi: 10.1016/j.biomaterials.2013.07.072. [DOI] [PubMed] [Google Scholar]
- 45.Frohlich E. The role of surface charge in cellular uptake and cytotoxicity of medical nanoparticles. Int J Nanomed. 2012;7:5577–5591. doi: 10.2147/IJN.S36111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Iversen TG, Skotland T, Sandvig K. Endocytosis and intracellular transport of nanoparticles: Present knowledge and need for future studies. Nano Today. 2011;6(2):176–185. [Google Scholar]
- 47.Leroueil PR, Hong S, Mecke A, Baker JR, Jr, Orr BG, Banaszak Holl MM. Nanoparticle interaction with biological membranes: does nanotechnology present a Janus face? Acc Chem Res. 2007;40(5):335–42. doi: 10.1021/ar600012y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Leroueil PR, Berry SA, Duthie K, Han G, Rotello VM, McNerny DQ, Baker JR, Orr BG, Holl MMB. Wide varieties of cationic nanoparticles induce defects in supported lipid bilayers. Nano Lett. 2008;8(2):420–424. doi: 10.1021/nl0722929. [DOI] [PubMed] [Google Scholar]
- 49.Zhao F, Zhao Y, Liu Y, Chang X, Chen C, Zhao Y. Cellular uptake, intracellular trafficking, and cytotoxicity of nanomaterials. Small. 2011;7(10):1322–37. doi: 10.1002/smll.201100001. [DOI] [PubMed] [Google Scholar]
- 50.Bannunah AM, Vllasaliu D, Lord J, Stolnik S. Mechanisms of nanoparticle internalization and transport across an intestinal epithelial cell model: effect of size and surface charge. Mol Pharmaceutics. 2014;11(12):4363–73. doi: 10.1021/mp500439c. [DOI] [PubMed] [Google Scholar]
- 51.Scott CC, Vacca F, Gruenberg J. Endosome maturation, transport and functions. Semin Cell Dev Biol. 2014;31:2–10. doi: 10.1016/j.semcdb.2014.03.034. [DOI] [PubMed] [Google Scholar]
- 52.Tsang AW, Oestergaard K, Myers JT, Swanson JA. Altered membrane trafficking in activated bone marrow-derived macrophages. J Leukoc Biol. 2000;68(4):487–94. [PubMed] [Google Scholar]
- 53.Huotari J, Helenius A. Endosome maturation. EMBO J. 2011;30(17):3481–500. doi: 10.1038/emboj.2011.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mitchell TW, Ekroos K, Blanksby SJ, Hulbert AJ, Else PL. Differences in membrane acyl phospholipid composition between an endothermic mammal and an ectothermic reptile are not limited to any phospholipid class. J Exp Biol. 2007;210(19):3440–50. doi: 10.1242/jeb.007286. [DOI] [PubMed] [Google Scholar]
- 55.Sanders CR, Mittendorf KF. Tolerance to changes in membrane lipid composition as a selected trait of membrane proteins. Biochemistry. 2011;50(37):7858–67. doi: 10.1021/bi2011527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Christianson HC, Belting M. Heparan sulfate proteoglycan as a cell-surface endocytosis receptor. Matrix Biol. 2014;35:51–55. doi: 10.1016/j.matbio.2013.10.004. [DOI] [PubMed] [Google Scholar]
- 57.Rehman ZU, Sjollema KA, Kuipers J, Hoekstra D, Zuhorn IS. Nonviral Gene Delivery Vectors Use Syndecan-Dependent Transport Mechanisms in Filopodia To Reach the Cell Surface. ACS Nano. 2012;6(8):7521–7532. doi: 10.1021/nn3028562. [DOI] [PubMed] [Google Scholar]
- 58.Payne CK, Jones SA, Chen C, Zhuang X. Internalization and trafficking of cell surface proteoglycans and proteoglycan-binding ligands. Traffic. 2007;8(4):389–401. doi: 10.1111/j.1600-0854.2007.00540.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Frohlich E. The role of surface charge in cellular uptake and cytotoxicity of medical nanoparticles. Int J Nanomed. 2012;7:5577–91. doi: 10.2147/IJN.S36111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Owen SC, Chan DPY, Shoichet MS. Polymeric micelle stability. Nano Today. 2012;7(1):53–65. [Google Scholar]
- 61.Zhou JY, Horev B, Hwang G, Klein MI, Koo H, Benoit DSW. Characterization and optimization of pH-responsive polymer nanoparticles for drug delivery to oral biofilms. J Mater Chem B. 2016;4(18):3075–3085. doi: 10.1039/C5TB02054A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li Y, Zhang X, Cao D. Nanoparticle hardness controls the internalization pathway for drug delivery. Nanoscale. 2015;7(6):2758–69. doi: 10.1039/c4nr05575f. [DOI] [PubMed] [Google Scholar]
- 63.Verma A, Uzun O, Hu Y, Hu Y, Han HS, Watson N, Chen S, Irvine DJ, Stellacci F. Surface-structure-regulated cell-membrane penetration by monolayer-protected nanoparticles. Nat Mater. 2008;7(7):588–95. doi: 10.1038/nmat2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fischer D, Li Y, Ahlemeyer B, Krieglstein J, Kissel T. In vitro cytotoxicity testing of polycations: influence of polymer structure on cell viability and hemolysis. Biomaterials. 2003;24(7):1121–31. doi: 10.1016/s0142-9612(02)00445-3. [DOI] [PubMed] [Google Scholar]
- 65.Lv H, Zhang S, Wang B, Cui S, Yan J. Toxicity of cationic lipids and cationic polymers in gene delivery. J Controlled Release. 2006;114(1):100–9. doi: 10.1016/j.jconrel.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 66.Xia T, Kovochich M, Brant J, Hotze M, Sempf J, Oberley T, Sioutas C, Yeh JI, Wiesner MR, Nel AE. Comparison of the abilities of ambient and manufactured nanoparticles to induce cellular toxicity according to an oxidative stress paradigm. Nano Lett. 2006;6(8):1794–1807. doi: 10.1021/nl061025k. [DOI] [PubMed] [Google Scholar]
- 67.Tzankova V, Gorinova C, Kondeva-Burdina M, Simeonova R, Philipov S, Konstantinov S, Petrov P, Galabov D, Yoncheva K. In vitro and in vivo toxicity evaluation of cationic PDMAEMA-PCL-PDMAEMA micelles as a carrier of curcumin. Food Chem Toxicol. 2016;97:1–10. doi: 10.1016/j.fct.2016.08.026. [DOI] [PubMed] [Google Scholar]
- 68.Cai JG, Yue YA, Rui D, Zhang YF, Liu SY, Wu C. Effect of Chain Length on Cytotoxicity and Endocytosis of Cationic Polymers. Macromolecules. 2011;44(7):2050–2057. [Google Scholar]
- 69.Turinetto V, Vitale E, Giachino C. Senescence in Human Mesenchymal Stem Cells: Functional Changes and Implications in Stem Cell-Based Therapy. Int J Mol Sci. 2016;17(7):1164. doi: 10.3390/ijms17071164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Burova E, Borodkina A, Shatrova A, Nikolsky N. Sublethal oxidative stress induces the premature senescence of human mesenchymal stem cells derived from endometrium. Oxid Med Cell Longevity. 2013;2013:474931. doi: 10.1155/2013/474931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cheung TH, Rando TA. Molecular regulation of stem cell quiescence. Nat Rev Mol Cell Biol. 2013;14(6):329–40. doi: 10.1038/nrm3591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ruenraroengsak P, Novak P, Berhanu D, Thorley AJ, Valsami-Jones E, Gorelik J, Korchev YE, Tetley TD. Respiratory epithelial cytotoxicity and membrane damage (holes) caused by amine-modified nanoparticles. Nanotoxicology. 2012;6(1):94–108. doi: 10.3109/17435390.2011.558643. [DOI] [PubMed] [Google Scholar]
- 73.Gary DJ, Min J, Kim Y, Park K, Won YY. The effect of N/P ratio on the in vitro and in vivo interaction properties of PEGylated poly[2-(dimethylamino)ethyl methacrylate]-based siRNA complexes. Macromol Biosci. 2013;13(8):1059–71. doi: 10.1002/mabi.201300046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jones RA, Poniris MH, Wilson MR. pDMAEMA is internalised by endocytosis but does not physically disrupt endosomes. J Controlled Release. 2004;96(3):379–91. doi: 10.1016/j.jconrel.2004.02.011. [DOI] [PubMed] [Google Scholar]
- 75.Schallon A, Jerome V, Walther A, Synatschke CV, Muller AHE, Freitag R. Performance of three PDMAEMA-based polycation architectures as gene delivery agents in comparison to linear and branched PEI. React Funct Polym. 2010;70(1):1–10. [Google Scholar]
- 76.Gilleron J, Querbes W, Zeigerer A, Borodovsky A, Marsico G, Schubert U, Manygoats K, Seifert S, Andree C, Stoter M, Epstein-Barash H, Zhang L, Koteliansky V, Fitzgerald K, Fava E, Bickle M, Kalaidzidis Y, Akinc A, Maier M, Zerial M. Image-based analysis of lipid nanoparticle-mediated siRNA delivery, intra-cellular trafficking and endosomal escape. Nat Biotechnol. 2013;31(7):638–46. doi: 10.1038/nbt.2612. [DOI] [PubMed] [Google Scholar]
- 77.Sahay G, Querbes W, Alabi C, Eltoukhy A, Sarkar S, Zurenko C, Karagiannis E, Love K, Chen D, Zoncu R, Buganim Y, Schroeder A, Langer R, Anderson DG. Efficiency of siRNA delivery by lipid nanoparticles is limited by endocytic recycling. Nat Biotechnol. 2013;31(7):653–8. doi: 10.1038/nbt.2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shang L, Nienhaus K, Nienhaus GU. Engineered nanoparticles interacting with cells: size matters. J Nanobiotechnol. 2014;12:5. doi: 10.1186/1477-3155-12-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ding HM, Tian WD, Ma YQ. Designing nanoparticle translocation through membranes by computer simulations. ACS Nano. 2012;6(2):1230–8. doi: 10.1021/nn2038862. [DOI] [PubMed] [Google Scholar]
- 80.Tyrrell ZL, Shen YQ, Radosz M. Fabrication of micellar nanoparticles for drug delivery through the self-assembly of block copolymers. Prog Polym Sci. 2010;35(9):1128–1143. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.