Abstract
Alterations in gene dosage due to copy-number variation (CNV) are associated with autism spectrum disorder (ASD), intellectual disability (ID) and other psychiatric disorders. The nervous system is so acutely sensitive to the dose of methyl-CpG-binding protein 2 (MeCP2) that even a two-fold change in MeCP2 protein level—either increased or decreased—results in distinct disorders with overlapping features including ID, autistic behavior and severe motor dysfunction. Rett syndrome is caused by loss-of-function mutations in MECP2, whereas duplications spanning the MECP2 locus result in MECP2 Duplication Syndrome (MDS) which accounts for ~1% of X-linked ID. Despite evidence from mouse models that restoring MeCP2 levels can reverse the course of disease, there are currently no FDA-approved therapies available to clinically modulate MeCP2 abundance. In this study we used a forward genetic screen against all known human kinases and phosphatases to identify druggable regulators of MeCP2 stability. Two putative modulators of MeCP2 levels, HIPK2 and protein phosphatase PP2A, were validated as stabilizers of MeCP2 in vivo. Further, pharmacological inhibition of PP2A in vivo reduced MeCP2 levels within the nervous system and rescued both overexpression and motor abnormalities in a mouse model of MDS. Our findings reveal potential therapeutic targets for treating disorders of altered MECP2 dosage and establish a potent strategy to identify druggable candidates for the broader category of neurologic disease resulting from CNVs.
Introduction
The human brain’s requirement for precise gene dosage is clear from the over-representation of copy-number variants (CNVs) in individuals with neuropsychiatric disorders, such as autism spectrum disorder (ASD), intellectual disability (ID) and schizophrenia (1–3). A prime example of this dosage sensitivity is embodied by Methyl-CpG-binding protein 2 (MECP2) disorders. Devastating neurological disorders characterized by ID, autistic features and motor dysfunction result from both a two-fold increase in MeCP2 protein—as experienced by males with MECP2 Duplication Syndrome (MDS)—and a decrease or loss of the protein in ~50% of cells, occurring in females with Rett syndrome (4). MDS accounts for ~1% of X-linked ID and is further distinguished by epilepsy and premature death (5, 6). Mouse models recapitulate patient symptoms, as male mice expressing twice the normal level of MeCP2, MECP2TG1, exhibit decreased motor activity, alterations in learning and memory, decreased social interactions, increased anxiety, and seizures reminiscent of patients with MDS (7, 8). Phenotypic severity in mice is further increased with threefold overexpression of MeCP2, consistent with the devastating nature of observed triplications spanning the MECP2 locus in humans (7–9). Conversely, classic Rett syndrome is caused in more than 95% of the cases by loss-of-function mutations in MECP2 and occurs in 1/10,000 live female births (10, 11). Male mice with even a 50% reduction of MeCP2 exhibit phenotypes reminiscent of Rett (12). Thus, although it is clinically and experimentally clear that the dose of MeCP2 must be precisely regulated to permit proper neural function, there are currently no FDA-approved avenues to modulate MeCP2 levels (11, 13, 14).
MeCP2 binds preferentially to methylated DNA but localizes broadly across the genome (15, 16). In mature neurons it is present at near histone-octamer levels (15). Loss of MeCP2 results in various chromatin changes including disruption of chromatin architecture, as observed by mislocalization of transcriptional regulator ATRX (17–19) and increased linker histone H1 (15). Expectedly, loss of MeCP2 also results in misregulation of numerous neuronally significant transcripts, such as those encoded by Brain-derived neurotrophic factor (Bdnf) (18, 20, 21) and Corticotropin releasing hormone (Crh) (8). The majority of these molecular alterations are oppositely misregulated in gain-of-function models. At the cellular level, neurons lacking MeCP2 are hypofunctional, exhibiting decreased soma size (22–24) and reduced dendritic branching (25–27). On the other hand, neurons from the MDS mouse model display increased synapse density and dendritic arborization (28, 29). Importantly, neurological phenotypes are largely reversible in both Rett and MDS mouse models by normalization of MeCP2 levels (30, 31), consistent with the absence of neurodegeneration and gross anatomical abnormalities. Previous attempts to correct specific molecular abnormalities identified in Mecp2 mutant mice, such as normalization of BDNF or CRH levels, have resulted in only partial phenotypic rescue (8, 32). We posit that given the broad scope of a chromatin protein’s regulon, it is likely that a constellation of misregulation drives the phenotypes in both the loss- and gain-of-function MECP2 syndromes. Thus, we propose that the most efficacious treatment of these disorders will involve modulating the levels of MeCP2 protein itself.
For a protein whose levels must be tightly regulated, little is known about factors that affect MeCP2 turnover or stability. While regulated post-transcriptionally by various microRNAs (33–35), the impact of MeCP2’s numerous post-translational modifications—including phosphorylation, acetylation, methylation, sumoylation, and ubiquitination—on its stability are largely unknown (36). Given the exquisite sensitivity of brain cells to the amount of MeCP2, we hypothesized that there are multiple endogenous regulators of MeCP2 stability. Thus, the goal of this work was to perform a forward genetic screen to uncover potentially druggable modulators of MeCP2 stability.
Results
Identification of post-translational regulators of MeCP2 stability
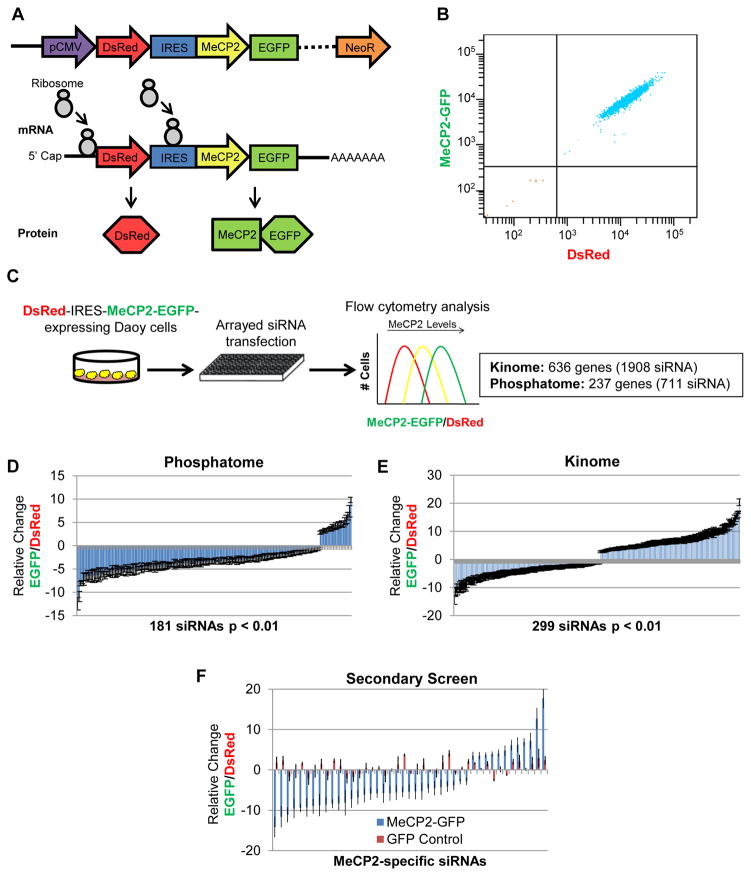
To develop a reporter cell line in which we could monitor MeCP2 levels we selected Daoy human medulloblastoma cells for screening because of their high siRNA transfection efficiency and their endogenous expression of MeCP2, increasing the probability of regulatory circuits being present for perturbation. Daoy cells were transduced with a lentiviral vector that expresses DsRed-IRES-hMECP2-EGFP. This bicistronic transgene allows for unified transcription, but independent translation, of the fluorescent protein DsRed and hMeCP2 with EGFP fused to its C-terminus (Fig. 1A). This system robustly controls for variations in transcription of the transgene by normalization to DsRed fluorescence (37, 38), allowing for precise quantification of MeCP2-specific protein regulation. After infection of Daoy cells with lentivirus containing the transgene, cells were analyzed and sorted by fluorescence-activated cell sorting (FACS) for cells positive for both red and green fluorescence. Multiple stable DsRed-IRES-hMECP2-EGFP-expressing cell lines were assessed for their suitability for the screen based on their expression of both fluorescent proteins in the experimental range and their sensitivity to shRNA and siRNA perturbation of MeCP2-EGFP levels. We chose a clonal cell line with matched dual fluorescence and minimal expression variation for the genetic screen (Fig. 1B).
Fig. 1.
Screening for post-translational regulators of MeCP2. (A) The bicistronic reporter transgene allows for monitoring of MeCP2 stability via C-terminal fusion of EGFP, while controlling for variation in transgene expression by normalization to the independently translated DsRed. (B) Flow cytometry analysis of a clonal Daoy DsRed-IRES-hMECP2-EGFP reporter cell line, indicating dual fluorescence and minimal expression variation. (C) Schematic for arrayed siRNA screen against all known and putative human kinases and phosphatases by flow cytometry analysis of the MeCP2-GFP/DsRed ratio. (D) 181 siRNAs significantly (p <0.01) altered the MeCP2-EGFP/DsRed ratio in the primary screen against human phosphatases (711 siRNAs total). (E) 299 siRNAs significantly (p <0.01) altered the MeCP2-EGFP/DsRed ratio in the primary screen against human kinases (1908 siRNAs total). (F) A secondary screen using independent siRNAs confirmed 120 significant siRNAs affecting the MeCP2-EGFP/DsRed ratio. siRNAs that also significantly altered a GFP control cell line were eliminated, revealing 43 MeCP2-specific siRNAs, representing 33 genes.
Using the developed Daoy DsRed-IRES-hMECP2-EGFP-expressing cell line, we probed the ability of all known human kinases and phosphatases (873 genes/2619 siRNAs) to modulate MeCP2 protein stability. The reporter cell line was transfected in triplicate for each siRNA (3 siRNAs/gene) and the level of MeCP2-EGFP assessed using flow cytometry (Fig. 1C). To control for variance in transgene transcription, the MeCP2- EGFP/DsRed ratio was assessed for each siRNA and compared to the effect of three scrambled control siRNAs. We identified 480 siRNAs that significantly (p<0.01) affected the MeCP2- EGFP/DsRed ratio (Fig. 1, D and E). To enrich for true candidates to follow-up, we required that 2 out of the 3 siRNAs targeting a candidate gene significantly affected the MeCP2-EGFP/DsRed ratio in the same direction and that the candidate gene be expressed in the brain. This resulted in a list of 81 genes for which we wanted to distinguish MeCP2-specific effects from IRES-GFP effects. Thus, for the secondary screen, we compared siRNA effects on the GFP/DsRed ratio in the original DsRed-IRES-hMECP2-EGFP cell line and a second cell line expressing DsRed-IRES-EGFP alone. This secondary screen was also performed with a set of siRNAs independent of those used in the original screen to reduce the likelihood of any off-target effects and to generate greater confidence in the candidates that continued to show a significant effect on MeCP2-EGFP/DsRed ratio. This secondary screen validated 33 candidate modulators of MeCP2 levels (table S1), while eliminating genes whose knockdown altered EGFP levels irrespective of MeCP2 (Fig. 1F).
Validation on endogenous MeCP2 reveals known and unknown regulators
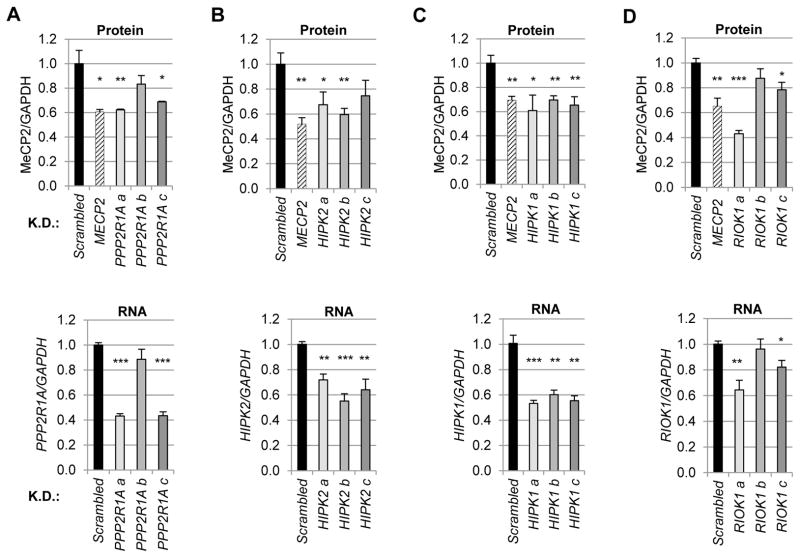
Following the validation of siRNAs that specifically affected MeCP2, we wanted to ensure that these candidate genes could modulate endogenous MeCP2, as opposed to MeCP2 over-expressed from the transgene, and that this modulation could be recapitulated in an independent cell type. To this end we performed a final round of validation in HEK293T cells which exhibit robust endogenous MeCP2 expression, using 3 shRNAs against each candidate gene. Immunoblot analysis was used to determine the effects of candidate knockdown on endogenous MeCP2 levels, whereas RNA analysis enabled evaluation of the efficiency of the knockdown for the candidate modulator. Thus, at this stage we required the extent of knockdown of the candidate to tightly correspond to effects on MeCP2 protein level. Four candidate modulators emerged from this analysis: one phosphatase subunit, PPP2R1A (Fig. 2A), and three kinases (Fig. 2B–D). Interestingly, one of these three kinases is a known kinase of MeCP2, Homeodomain Interacting Protein Kinase 2 (HIPK2) (Fig. 2B) (39) intimating the physiological relevance and power of our screening strategy. However, knockdown or inhibition of HIPK2 has never previously been evaluated for its effect on MeCP2 levels, suggesting our screening approach might be sensitive to direct modifiers of MeCP2 and possibly catch changes in stability hitherto missed.
Fig. 2.
Validation on endogenous MeCP2 reveals known and unknown regulators. The effects of candidate knockdown on endogenous MeCP2 levels were tested in HEK293T cells along with RNA analysis to determine which shRNAs were on-target. For each candidate, three shRNAs (a–c) were transfected along with vectors expressing scrambled shRNA and shRNA targeting MECP2. Two days post-transfection, puromycin was applied to select for cells expressing the silencing plasmid. Selection was maintained for four days then cells were split for parallel immunoblot analysis (top) and RNA analysis (bottom). (A) The effects of Protein Phosphatase 2 Regulatory subunit 1 Alpha (PPP2R1A) knockdown on endogenous MeCP2 levels relative to GAPDH. Coupled RNA analysis (below) indicated that only the shRNAs “a” and “c” which resulted in significant knockdown of PPP2R1A significantly decreased MeCP2 protein levels. (B) As in A, but for Homeodomain Interacting Protein Kinase 2 (HIPK2). (C) As in A and B, but for Homeodomain Interacting Protein Kinase 1 (HIPK1). (D) As in A–C, but for RIO Kinase 1 (RIOK1). K.D., knockdown. n = 3–4 per group. *p< 0.05; **p<0.01; ***p<0.001, two-tailed t-test.
To determine how these four candidates might regulate MeCP2 protein levels, we first determined whether the observed decreases in MeCP2 level occurred post-transcriptionally. For this analysis, RNA from HEK293T cells treated with our candidate shRNAs was assayed for MECP2 transcript levels by quantitative real-time polymerase-chain reaction (qPCR). Despite significant decreases in MeCP2 protein level, no significant decreases in MECP2 transcript were detected, except in cells treated with the shRNA targeting MECP2 (fig. S1). Thus, our screening approach utilizing only the coding sequence of MECP2 faithfully isolated post-translational regulators of MeCP2.
Post-translational regulation of MeCP2 by both HIPK1 and HIPK2
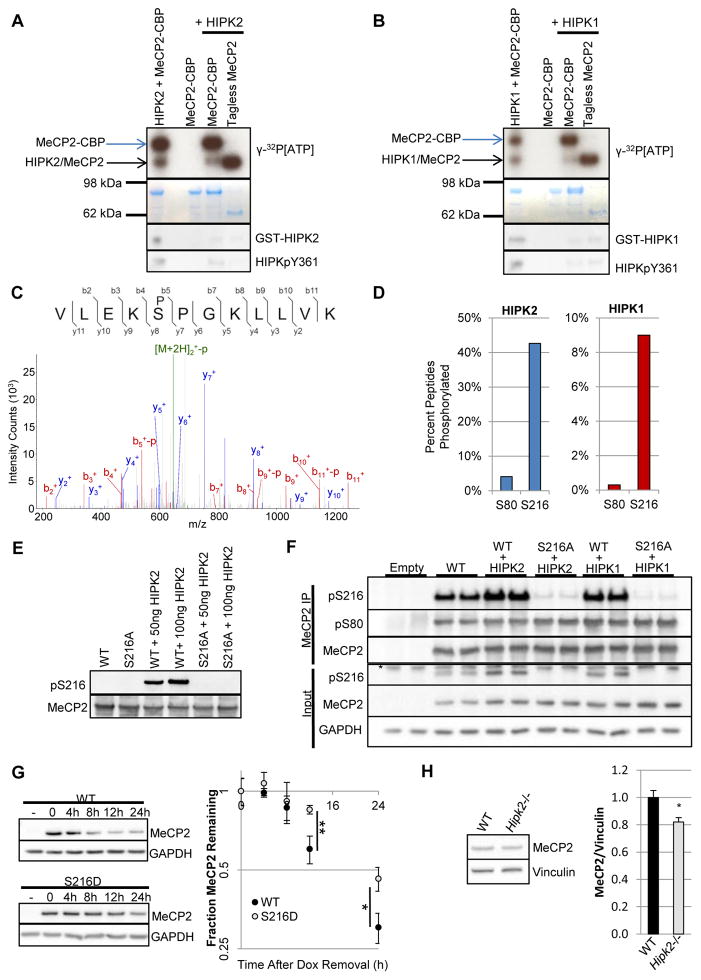
Next we sought to assess which of the kinase candidates might directly modify MeCP2. First we validated our assay by recapitulating previous in vitro results demonstrating the ability of the nuclear kinase HIPK2 to phosphorylate MeCP2 (39). Incubation of recombinant MeCP2 C-terminally tagged with Chitin Binding Protein (CBP) and HIPK2 with γ-32P[ATP] resulted in both HIPK2 autophosphorylation and MeCP2-CBP phosphorylation (Fig. 3A, lane 1). In contrast, incubating MeCP2-CBP only with γ-32P[ATP] resulted in no detectable radioactivity (lane 3). To control for the possibility that the radioactivity associated with MeCP2-CBP was due solely to CBP phosphorylation, chitin-bound MeCP2-CBP from the reaction was washed, such that HIPK2 was depleted (lane 4), and eluted by tag cleavage (lane 5). The radioactivity of tagless MeCP2 clearly demonstrated the specificity of HIPK2’s kinase activity for MeCP2 itself.
Fig. 3.
Both HIPK1 and HIPK2 phosphorylate MeCP2 at S216. (A) Autoradiogram (top) and Coomassie-stained gel (bottom) of kinase reactions with the indicated recombinant proteins in the presence of γ-32P[ATP]. Incubation of HIPK2 and MeCP2-CBP resulted in HIPK2 autophosphorylation and phosphorylation of MeCP2 (lane 1). Whereas MeCP2-CBP without HIPK2 resulted in no detectable radioactivity (lane 3). To control for epitope phosphorylation, chitin-bound MeCP2-CBP incubated with HIPK2 was washed (lane 4) then eluted by tag cleavage (lane 5). n = 3. (B) As in A, but with HIPK1. n = 3. (C) Tandem mass spectrum of pS216-containing peptide resulting from incubation of HIPK2 with purified MeCP2-CBP. Parental ions (green), b-ions (red), y-ions (blue). (D) Percent peptide phosphorylated relative to the sum of modified and unmodified peptides for peptides containing S80 or S216 for HIPK2 (left) and HIPK1 (right). Area under the peptide curves was utilized for relative quantitation. n = 5. (E) Incubation of HIPK2 with purified WT MeCP2-CBP or MeCP2 that could not be phosphorylated at S216, MeCP2S216A-CBP. n = 3. (F) Immunoprecipitation of MeCP2-GFP from HEK293T cells demonstrated a HIPK-dependent increase in pS216 comparable to pS80. Cells were harvested 48h after co-transfection. *, non-specific band. n = 3. (G) Stable Daoy cell lines encoding doxycycline-inducible WT or S216D MECP2-GFP were used to investigate differential protein dynamics. After inducing transgene expression with doxycycline (Dox) for 48h, cells were provided with fresh Dox-lacking media for continued growth (t = 0). -, uninduced. n = 6. (H) MeCP2 levels were decreased relative to vinculin in Hipk2−/− mutant mice whole brain lysates. Mice were 2 – 4 months old. n = 5. *p< 0.05; **p<0.01, two-tailed t-test.
We next tested the ability of HIPK1 to phosphorylate MeCP2 and found that incubation of MeCP2-CBP and HIPK1 with γ-32P[ATP] clearly resulted in both HIPK1 and MeCP2 phosphorylation (Fig. 3B). Given previous work identifying S80 as the primary phosphorylation site on MeCP2 by HIPK2 (39), we sought to determine if this was also true for HIPK1. To our surprise, incorporated radioactivity did not appear decreased when the only substrate provided to either HIPK2 or HIPK1 was MeCP2 that could not be phosphorylated at S80, MeCP2S80A (fig. S2A). This suggested the presence of more HIPK2 and HIPK1 phosphorylation sites on MeCP2. To determine what these additional phosphorylation sites might be, we performed tandem mass spectrometry (MS) on MeCP2 alone and MeCP2 incubated with either HIPK2 or HIPK1. Two primary phosphosites emerged from this analysis for both HIPK2 and HIPK1, pS80 and pS216 (Fig. 3C). Both sites are HIPK kinase consensus sites (S/T-P) (39, 40) and are highly conserved in MeCP2 orthologs from zebrafish to human (fig. S2B). Relative quantification of peptide phosphorylation was achieved by comparing the area under the curve for each phosphorylated peptide to the total area for modified and unmodified forms of the peptide (Fig. 3D). The apparent percent phosphorylation of S216 indicated the robustness of this modification by both HIPK2 and HIPK1.
To investigate the biological relevance of pS216, we performed tandem MS on MeCP2-EGFP immunoprecipitated from brains of transgenic mice expressing the epitope-tagged human gene at approximately endogenous levels (19). Phosphorylation of S216 was detected in immunoprecipitated MeCP2 on ~10% of the corresponding peptides (fig. S2C). Having demonstrated that pS216 occurs in vivo in mouse whole brain, we developed a phospho-specific antibody for pS216 to assess if this phosphorylation was a bona fide target of the HIPK kinases. Repeating in vitro kinase assays with recombinant WT MeCP2 and MeCP2 that could not be phosphorylated at S216 (S216A) demonstrated the ability of both HIPK2 (Fig. 3E) and HIPK1 (fig. S2D) to specifically mediate this phosphorylation. Further, S216 phosphorylation occurred in cells upon expression of either HIPK2 or HIPK1 at similar levels to pS80 (Fig. 3F). Thus, biochemical analyses uncovered both an additional MeCP2 kinase and a new HIPK-mediated MeCP2 phosphorylation site.
To begin to characterize the impact of pS216 on MeCP2’s stability, we generated stable cell lines expressing doxycycline-inducible WT MeCP2 or MeCP2 mutated to mimic constitutive S216 phosphorylation (S216D). After induction of transgenic expression, doxycycline was removed and MeCP2 protein decay was assessed. Phosphomimetic MeCP2S216D exhibited increased stability relative to wild type (Fig. 3G), suggesting pS216 may stabilize MeCP2. Consistent with the model of HIPK2-mediated phosphorylations acting to stabilize MeCP2 levels, MeCP2 levels were decreased in the brains of Hipk2−/− homozygous mice (Fig. 3H).
Unlike the HIPK kinases, RIOK1 is a member of a family of atypical serine-threonine kinases that function in ribosomal 40S biogenesis. RIO kinases lack canonical kinase activation loops and have no known substrates, propelling the model that they act as ATPases in ribosome assembly rather than kinases (41, 42). We did not detect any RIOK1 kinase activity toward MeCP2 (fig. S3). Thus, we believe it unlikely that RIOK1’s stabilization of MeCP2 levels results from phosphorylation by RIOK1.
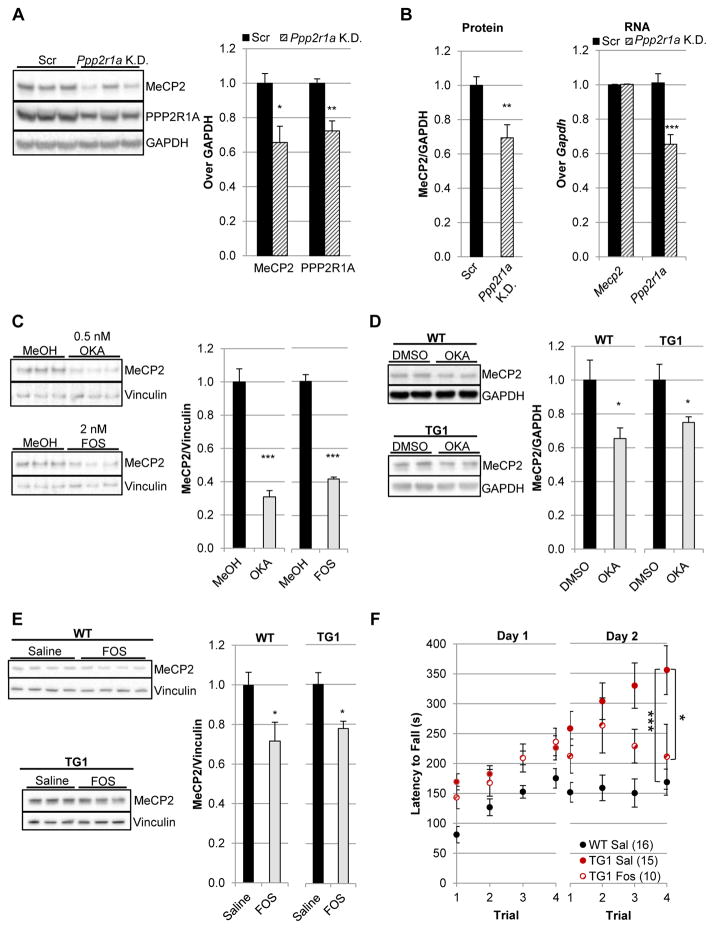
Inhibition of PPP2CA decreases MeCP2 in vivo
Protein phosphatase 2 (PP2A) functions as an obligate dimer of the catalytic subunit and the scaffold, the most abundant isoform of which is encoded by Protein Phosphatase 2 Regulatory subunit 1 Alpha (PPP2R1A) (43). The scaffold further binds regulatory proteins which modulate the enzymatic activity and specificity of the catalytic subunit (44). To determine the effect of Ppp2r1a knockdown in the mouse brain, we optimized a viral delivery system in which adeno-associated virus (AAV8) encoding shRNA is injected into the lateral ventricles of neonates, resulting in widespread cortical and hippocampal infection (45). To perform this experiment, two different AAV vectors were utilized: one encoding shRNA against the candidate, while the other encodes scrambled shRNA. Knockdown of Ppp2r1a potently decreased MeCP2 levels in the cortex, with the decrease in MeCP2 tightly corresponding to the decrease in PPP2R1A (Fig. 4A). This regulation was not specific to cortical tissue, as MeCP2 levels were also decreased in hippocampal lysates (fig. S4). Importantly, we performed a subsequent series of injections to test whether the observed decrease in MeCP2 levels occurred due to transcriptional or post-transcriptional regulation. For this analysis, cortical lysates were fractionated for protein and RNA analyses on the exact same tissue. Consistent with the findings of the cell-based screen, knockdown of Ppp2r1a decreased MeCP2 protein levels without affecting Mecp2 transcript levels (Fig. 4B). Thus, PPP2R1A appeared to regulate MeCP2 post-transcriptionally.
Fig. 4.
Inhibition of PP2A decreases MeCP2 in vivo. (A) P0 intraventricular injection of AAV8 encoding shRNA targeting Ppp2r1a resulted in decreased PPP2R1A and MeCP2 protein compared to animals injected with virus expressing a scrambled shRNA (Scr). Cortex tissue was harvested at two weeks post-injection. n = 6. (B) As in A, but an independent cohort in which cortical lysates were generated in Trizol for protein and RNA analysis of the same tissue. n = 8. (C) Decreased MeCP2 levels occurred upon treatment of cerebellar granule neuron precursors with PP2A inhibitors okadaic acid (OKA, IC50: 0.1–0.5 nM) and fostriecin (FOS, IC50: 1.5 nM). Neurons were harvested after nine days of treatment. n = 3. (D) Decreased MeCP2 levels in cortical tissue upon intraventricular injection of 40ng of okadaic acid (OKA) compared to injection with solvent (2% DMSO) in both WT (n = 8) and MDS model mice, MECP2TG1 (n = 6). Cortices were harvested two weeks post-injection. (E) Decreased MeCP2 levels in cortical tissue upon intraventricular injection of 240 uM fostriecin (FOS) compared to injection with solvent (Saline) in both WT (n = 4) and MECP2TG1mice (n = 6). Cortices were harvested two weeks post-injection. (F) Fostriecin treatment of MECP2TG1mice partially rescued abnormal motor persistence on the accelerating rotating rod as evidenced by decreased latency to fall compared to MECP2TG1mice treated with saline alone (Sal). Mice were assessed two weeks post-injection. n = 10–16. *p< 0.05; **p<0.01; ***p<0.001, A–E, two-tailed t-test. F, repeated-measures two-way analysis of variance (ANOVA) by day followed with post hoc t-tests.
To test whether this regulation by PPP2R1A was occurring through the PP2A holoenzyme (catalytic subunit and scaffold), we determined the effect of two distinct PP2A catalytic inhibitors, okadaic acid and fostriecin (44), on endogenous MeCP2 levels in cultured cerebellar granule neurons. Treatment with either inhibitor significantly decreased MeCP2 levels (Fig. 4C), suggesting that the observed decrease in MeCP2 levels upon PPP2R1A/Ppp2r1a knockdown was the result of decreased PP2A catalytic activity. Okadaic acid’s activity was not restricted to murine MeCP2, as it also decreased human MeCP2 in medulloblastoma cells (fig. S5A). Importantly, this decrease in human MeCP2 occurred at an okadaic acid dose sufficient to result in the hyperphosphorylation of PP2A’s neuronal substrate Tau (fig. S5B) (46). Given the resultant feasibility of pharmacological intervention, we next sought to determine if these findings from cultured neurons would hold true in the mature mouse brain.
Cognizant of the potential limitations of any single drug, we moved in vivo with both okadaic acid and fostriecin. Each PP2A inhibitor was intracerebroventricularly injected into seven-week-old mice and the effects on MeCP2 levels relative to its solvent-matched control detected by immunoblotting. In vivo treatment with either okadaic acid or fostriecin decreased MeCP2 levels (Fig. 4, D and E, and fig. S6), strongly suggesting that the decrease in MeCP2 occurred due to their shared inhibition of PP2A rather than any divergent off-target activities. Next we sought to determine the effect of PP2A inhibition in a mouse model of MDS, MECP2TG1. Importantly, the extra copy of MECP2 in MECP2TG1 mice is the human gene, therefore these mice are ideal models for assessing regulation of the human MeCP2 in vivo (7). Inhibition of PP2A via okadaic acid or fostriecin significantly decreased MeCP2 levels in MDS mice (Fig. 4, D and E), partially rescuing MeCP2 overexpression. Although mice treated with okadaic acid, which is a more potent PP2A inhibitor than fostriecin, maintained healthy brain weights (fig. S7A), we chose to move forward with fostriecin, the more selective PP2A inhibitor (47), to determine if partial rescue of MeCP2 overexpression might result in behavioral benefit. Given the short time scale of treatment because of the limitations using fostriecin long term (invasive), we assayed treated mice for the behavior which is the most sensitive to MeCP2 levels and the first to be rescued upon genetic normalization of MeCP2 levels in MECP2TG1 mice, the accelerating rotating rod (rotarod) (30). MECP2TG1 mice exhibit abnormal perseverance on the rotarod with the greatest deviation from WT occurring on the second day of trials (7, 30). A 20–25% decrease in MeCP2 levels in MECP2TG1 mice treated with fostriecin was sufficient to limit their rotarod persistence on day 2 relative to MECP2TG1 mice treated with sterile saline (vehicle). Indeed by the final trial of day 2, fostriecin-treated MECP2TG1 mice were not distinguishable from WT, but were significantly different from MECP2TG1 mice treated with sterile saline (Fig. 4F). Unlike MECP2TG1 mice, WT mice treated with fostriecin exhibited no decrease in rotarod performance (fig. S7B). Thus, in vivo genetic and pharmacological inhibition of PP2A destabilized MeCP2, partially rescuing both MeCP2 overexpression and motor abnormalities in a mouse model of MDS.
Discussion
The role of altered gene dosage is increasingly being recognized in neuropsychiatric disorders. Examples include a variety of aberrations from single duplications (PMP22) and deletions (SHANK3) to classic aneuploidies, such as Down syndrome (1). The exponential rise in diagnosed cases of autism and ID, coupled with our increasing awareness of de novo CNVs, begs for innovation in our approaches to genetic disorders. To evaluate the feasibility of incisively isolating modifiers of dose-sensitive disease drivers, we began with MECP2 due to the wealth and validity of available mouse models of disease (48). Additionally, even without a direct genetic basis, abnormalities in MeCP2 levels have been observed in ASD, Prader-Willi (49, 50) and ID patients with NUDT21-spanning CNVs (51), suggesting modulation of MeCP2 levels may serve a broader therapeutic purpose. Here we have identified four regulators of MeCP2 stability, validating regulation of MeCP2 by PP2A and HIPK2 in vivo and demonstrating that pharmacological inhibition of PP2A was sufficient to partially rescue MeCP2 overexpression and motor abnormalities in a mouse model of MDS.
The feasibility of pharmacological intervention propelled us to utilize PP2A as our proof-of-principle candidate moving into mouse models of disease. Both knockdown of Ppp2r1a and pharmacological inhibition of PP2A’s catalytic activity decreased MeCP2 levels in vivo. Consistent with the screen’s design, inhibition of PP2A also decreased human MeCP2 levels in the mouse model of MDS, MECP2TG1, in which the extra copy of MECP2 is the human gene. It is of note that two distinct inhibitors of PP2A produced similar effects on MeCP2 levels, supporting the specificity of the observed regulation. Further, having more than one inhibitor to choose from enabled us to then move forward to behavioral studies with the most selective one. Importantly, our results suggest that even partial rescue of MeCP2 overexpression in MECP2TG1 mice can be sufficient to alleviate behavioral abnormalities.
Given that the prime purpose of this study was to demonstrate a potent cell-based screening strategy for druggable regulators of dose-sensitive genes, it is important to note that our in vivo drug studies are preliminary in nature. Okadaic acid and fostriecin are commercially available tool compounds which were used to validate the regulation of MeCP2 by PP2A in vivo. Future work will require in-depth pharmacological studies of optimized drugs, as opposed to tool compounds. Additionally, we believe it may be necessary to delve further into the mechanism of PP2A’s regulation of MeCP2 to enable more precise pharmacological manipulation However, the emergence of PPP2R1A loss-of-function mutations as dominant drivers of ID (52) suggests we have honed in on an important aspect of neuronal physiology.
The recovery of a known MeCP2 kinase was unanticipated, given HIPK2 activity has not been reported to affect the stability of MeCP2. HIPK2 and HIPK1 are nuclear kinases whose phosphorylation of chromatin-associated proteins often modulates their ability to regulate transcription (53–56). In this work, we recapitulated MeCP2 S80 phosphorylation by HIPK2, as previously shown in cultured cells (39). Further, we demonstrated that this phosphorylation can also be mediated by HIPK1. Interestingly, pS80 appears to be a constitutive MeCP2 phosphorylation in rodent brain that when mutated decreases the level of chromatin-associated MeCP2 in vivo (57), consistent with potential regulation of both MeCP2’s chromatin function and possibly its stability. Importantly, we also uncovered a new HIPK-mediated MeCP2 phosphorylation, pS216. This phosphorylation has been detected by numerous groups, both in rodents (57–59) and human (60–64), but never before in brain tissue and thus far has been an orphan MeCP2 phosphorylation site. It is tantalizing to note that this SP motif is conserved from zebrafish to human and falls within MeCP2’s transcriptional repression domain (TRD), a domain critical for mediating MeCP2’s interaction with co-regulators such as the NCoR/SMRT complex (65, 66). Moving forward it will be important to determine if the apparently increased stability of the MeCP2 S216D phosphomimetic is due to a change in MeCP2 interactors.
Consistent with the model of HIPK2-mediated phosphorylations acting to stabilize MeCP2 levels, MeCP2 levels were decreased in the brains of Hipk2−/− homozygous mice. However given the genetic redundancy of Hipk1 and Hipk2 (67), the extent of HIPK-regulation of MeCP2 will not be clear until the function of both genes is restricted simultaneously. Further experimentation will be required to determine if MeCP2 S80 and S216 are HIPK1 and HIPK2 sites in vivo and whether these phosphorylation events mediate the apparent stabilization of MeCP2 by HIPK1 and HIPK2. However, the recovery of both HIPK1 and HIPK2 independently intimates the functional strength of this regulation.
The fact that our screening approach identified multiple regulators of MeCP2 stability may hold promise beyond uncovering individual druggable targets. Recognizing that potently inhibiting any one target to effectively normalize MeCP2 levels might lead to some untoward effects or toxicity, we propose that future studies should focus on partial inhibition of targets that function in two or more independent pathways—a combination therapy approach. In this manner, multiple minor inhibitions would converge on MeCP2 resulting in cumulative MeCP2 normalization without the potential for toxicity from overly inhibiting any one single pathway.
Taken together, we believe we have uncovered four potential biological entry points for therapeutic intervention in patients with MECP2 Duplication Syndrome and other disorders in which MeCP2 levels are increased. Further, greater dissection of these nodes of MeCP2 regulation may enable the identification of genes that when inhibited increase the levels of MeCP2 protein. Such genes could prove very valuable therapeutically as certain missense Rett-causing alleles may be amenable to stabilizing therapy, or treatment aimed at increasing MeCP2 levels. Specifically, the mouse model Mecp2T158A of the single most abundant Rett-causing MECP2 mutation, T158M, exhibits decreased MeCP2 levels (68). However, this mutant allele exhibits only a minor decrease in methyl-CpG binding, suggesting that increasing the nuclear concentration of MeCP2T158A would in theory rescue both association with methyl-CpG and any phenotypes resulting from decreased total protein. Thus the ability to discover both positive and negative regulators of MeCP2, as well as the nuances of the underlying biology, may represent an advantage of genetic screening over gene therapy or antisense strategies.
Materials and Methods
Study design
The primary objective of this study was the identification of genes that modulate MeCP2 protein levels. The number of biological replicates (n) per experiment is noted in each figure legend. All littermates were randomized prior to treatment. All mouse experiments were replicated in an independent cohort. An individual blinded to the genotype and treatment performed the behavioral assay and acquired the data. No outlier removal was performed.
Protein analysis
Cultured cells were washed in cold phosphate-buffered saline (PBS), lysed in 100mM Tris-HCl pH 8.0, 2% SDS supplemented with protease inhibitors (Roche), rotated for 20 min then centrifuged at 13,000 g. After centrifugation, the supernatant was mixed with 2x Laemmli Buffer, heated for 5 min at 95°C, and electrophoresed on a NuPAGE 4–12% Bis-Tris gradient gel. Mouse tissue lysates were similarly prepared but with Dounce homogenization. Following electrophoresis, gels were transferred onto PVDF membranes and probed with rabbit anti-MeCP2 (1:3,000, Zoghbi lab, #0535) (69), rabbit anti-GST (1:2000, Sigma-Aldrich G7781), rabbit anti-HIPK2 pY361 (1:500, Invitrogen), rabbit anti-MeCP2 pS80 (1:500, Active Motif), rabbit anti-PPP2R1A (1:3,000, Abcam, ab154551), mouse anti-Tau (1:2500, Abcam ab80579), rabbit anti-Tau pS356 (1:1000, Abcam ab75603), mouse anti-GAPDH 6C5 (1:20,000, Advanced Immunochemicals, 2-RGM2), and mouse anti-Vinculin (1:10,000, Sigma-Aldrich). Secondary antibodies were mouse anti-rabbit horseradish peroxidase (HRP) (1:3,000, Jackson ImmunoResearch Labs, 211-032-171) and donkey anti-mouse HRP (1:50,000, Jackson ImmunoResearch Labs, 715-035-150). Immunoblot images were acquired with the ImageQuant LAS 4000 (GE Healthcare) and quantified with ImageJ.
Statistical analysis
The number of animals (n), the statistical tests and alphas used are indicated for each experiment in the figure legends. Error bars in all figures indicate SEM. Analysis of variance (ANOVA) was calculated using the aov function in R 3.3.1.
Supplementary Material
Fig. S1. Candidate knockdown did not decrease MECP2 transcript levels.
Fig. S2. MeCP2 pS16 occurred in vivo.
Fig. S3. No RIOK1 kinase activity for MeCP2 was detected.
Fig. S4. Ppp2r1a knockdown also decreased MeCP2 levels in hippocampal tissue.
Fig. S5. Okadaic acid treatment was active against human MeCP2.
Fig. S6. In vivo fostriecin titration.
Fig. S7. In vivo PP2A inhibition resulted in no ill effects in WT at the doses used.
Table S1. MeCP2-specific siRNAs gene list.
Table S2. Primers used.
Table S3. shRNAs used.
Acknowledgments
We are grateful to the following individuals for training: Corinne Spencer on motor behavioral analysis, Yingyao Shao on stereotaxic injections, and Ji-Yeon Kim on P0 intraventricular injections. We also thank Laura Lavery, Trey Westbrook, and Maxime Rousseaux for their scientific critiques and advice. Finally, we thank Johanna Jankowsky and Stacy Grunke for their generous gift of the pAAV-U6-miR-Cag-tdT vector.
Funding: This project was funded by the National Institutes of Health (5R01NS057819 to H.Y.Z.), the Rett Syndrome Research Trust and 401K Project from MDS families, and Howard Hughes Medical Institute (L.M.L. & H.Y.Z.). This work was also made possible by the following Baylor College of Medicine cores: Cell-Based Assay Screening Service (NIH (P30 CA125123)), Cytometry and Cell Sorting Core (NIAID P30AI036211, NCI P30CA125123, and NCRR S10RR024574), Pathway Discovery Proteomics Core, the DNA Sequencing and Gene Vector Core (Diabetes and Endocrinology Research Center (DK079638)), and the mouse behavioral core of the Intellectual and Developmental Disabilities Research Center (NIH U54 HD083092 from the National Institute of Child Health & Human Development).
Footnotes
Author contributions L.M.L. is the leading author and performed all molecular experiments. L.M.L., M.Z., Y.S., S.A.B. and H.Y.Z. contributed to the concept and design of the experiments. M.Z. generated mutant vectors. Y.S. performed the okadaic acid stereotaxic injections. S.A.B. assisted with in vivo immunoblot analysis. T.J.K. generated the doxycycline-inducible stable cell lines. A.A.T. and E.J.H. provided Hipk2−/− mutant brains. L.M.L., M.Z., S.A.B. and H.Y.Z. collected, analyzed and interpreted the data. L.M.L. and H.Y.Z. wrote and edited the paper.
Competing interests The authors declare no competing financial interests.
References and Notes
- 1.Girirajan S, Campbell CD, Eichler EE. Human copy number variation and complex genetic disease. Annual review of genetics. 2011;45:203–226. doi: 10.1146/annurev-genet-102209-163544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gilissen C, Hehir-Kwa JY, Thung DT, van de Vorst M, van Bon BW, Willemsen MH, Kwint M, Janssen IM, Hoischen A, Schenck A, Leach R, Klein R, Tearle R, Bo T, Pfundt R, Yntema HG, de Vries BB, Kleefstra T, Brunner HG, Vissers LE, Veltman JA. Genome sequencing identifies major causes of severe intellectual disability. Nature. 2014;511:344–347. doi: 10.1038/nature13394. [DOI] [PubMed] [Google Scholar]
- 3.Luo R, Sanders SJ, Tian Y, Voineagu I, Huang N, Chu SH, Klei L, Cai C, Ou J, Lowe JK, Hurles ME, Devlin B, State MW, Geschwind DH. Genome-wide transcriptome profiling reveals the functional impact of rare de novo and recurrent CNVs in autism spectrum disorders. American journal of human genetics. 2012;91:38–55. doi: 10.1016/j.ajhg.2012.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chao HT, Zoghbi HY. MeCP2: only 100% will do. Nature neuroscience. 2012;15:176–177. doi: 10.1038/nn.3027. [DOI] [PubMed] [Google Scholar]
- 5.Lugtenberg D, Kleefstra T, Oudakker AR, Nillesen WM, Yntema HG, Tzschach A, Raynaud M, Rating D, Journel H, Chelly J, Goizet C, Lacombe D, Pedespan JM, Echenne B, Tariverdian G, O’Rourke D, King MD, Green A, van Kogelenberg M, Van Esch H, Gecz J, Hamel BC, van Bokhoven H, de Brouwer AP. Structural variation in Xq28: MECP2 duplications in 1% of patients with unexplained XLMR and in 2% of male patients with severe encephalopathy. European journal of human genetics: EJHG. 2009;17:444–453. doi: 10.1038/ejhg.2008.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Van Esch H. MECP2 Duplication Syndrome. Molecular syndromology. 2012;2:128–136. doi: 10.1159/000329580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collins AL, Levenson JM, Vilaythong AP, Richman R, Armstrong DL, Noebels JL, David Sweatt J, Zoghbi HY. Mild overexpression of MeCP2 causes a progressive neurological disorder in mice. Human molecular genetics. 2004;13:2679–2689. doi: 10.1093/hmg/ddh282. [DOI] [PubMed] [Google Scholar]
- 8.Samaco RC, Mandel-Brehm C, McGraw CM, Shaw CA, McGill BE, Zoghbi HY. Crh and Oprm1 mediate anxiety-related behavior and social approach in a mouse model of MECP2 duplication syndrome. Nature genetics. 2012;44:206–211. doi: 10.1038/ng.1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.del Gaudio D, Fang P, Scaglia F, Ward PA, Craigen WJ, Glaze DG, Neul JL, Patel A, Lee JA, Irons M, Berry SA, Pursley AA, Grebe TA, Freedenberg D, Martin RA, Hsich GE, Khera JR, Friedman NR, Zoghbi HY, Eng CM, Lupski JR, Beaudet AL, Cheung SW, Roa BB. Increased MECP2 gene copy number as the result of genomic duplication in neurodevelopmentally delayed males. Genetics in medicine: official journal of the American College of Medical Genetics. 2006;8:784–792. doi: 10.1097/01.gim.0000250502.28516.3c. [DOI] [PubMed] [Google Scholar]
- 10.Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nature genetics. 1999;23:185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- 11.Lombardi LM, Baker SA, Zoghbi HY. MECP2 disorders: from the clinic to mice and back. The Journal of clinical investigation. 2015;125:2914–2923. doi: 10.1172/JCI78167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Samaco RC, Fryer JD, Ren J, Fyffe S, Chao HT, Sun Y, Greer JJ, Zoghbi HY, Neul JL. A partial loss of function allele of methyl-CpG-binding protein 2 predicts a human neurodevelopmental syndrome. Human molecular genetics. 2008;17:1718–1727. doi: 10.1093/hmg/ddn062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gadalla KK, Bailey ME, Cobb SR. MeCP2 and Rett syndrome: reversibility and potential avenues for therapy. The Biochemical journal. 2011;439:1–14. doi: 10.1042/BJ20110648. [DOI] [PubMed] [Google Scholar]
- 14.Chapleau CA, Lane J, Larimore J, Li W, Pozzo-Miller L, Percy AK. Recent Progress in Rett Syndrome and MeCP2 Dysfunction: Assessment of Potential Treatment Options. Future neurology. 2013;8 doi: 10.2217/fnl.12.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Skene PJ, Illingworth RS, Webb S, Kerr AR, James KD, Turner DJ, Andrews R, Bird AP. Neuronal MeCP2 is expressed at near histone-octamer levels and globally alters the chromatin state. Molecular cell. 2010;37:457–468. doi: 10.1016/j.molcel.2010.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen L, Chen K, Lavery LA, Baker SA, Shaw CA, Li W, Zoghbi HY. MeCP2 binds to non-CG methylated DNA as neurons mature, influencing transcription and the timing of onset for Rett syndrome. Proceedings of the National Academy of Sciences of the United States of America; 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nan X, Hou J, Maclean A, Nasir J, Lafuente MJ, Shu X, Kriaucionis S, Bird A. Interaction between chromatin proteins MECP2 and ATRX is disrupted by mutations that cause inherited mental retardation. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:2709–2714. doi: 10.1073/pnas.0608056104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baker SA, Chen L, Wilkins AD, Yu P, Lichtarge O, Zoghbi HY. An AT-Hook Domain in MeCP2 Determines the Clinical Course of Rett Syndrome and Related Disorders. Cell. 2013;152:984–996. doi: 10.1016/j.cell.2013.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heckman LD, Chahrour MH, Zoghbi HY. Rett-causing mutations reveal two domains critical for MeCP2 function and for toxicity in MECP2 duplication syndrome mice. eLife. 2014;3 doi: 10.7554/eLife.02676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ogier M, Wang H, Hong E, Wang Q, Greenberg ME, Katz DM. Brain-derived neurotrophic factor expression and respiratory function improve after ampakine treatment in a mouse model of Rett syndrome. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2007;27:10912–10917. doi: 10.1523/JNEUROSCI.1869-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chahrour M, Jung SY, Shaw C, Zhou X, Wong ST, Qin J, Zoghbi HY. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science. 2008;320:1224–1229. doi: 10.1126/science.1153252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen RZ, Akbarian S, Tudor M, Jaenisch R. Deficiency of methyl-CpG binding protein-2 in CNS neurons results in a Rett-like phenotype in mice. Nature genetics. 2001;27:327–331. doi: 10.1038/85906. [DOI] [PubMed] [Google Scholar]
- 23.Marchetto MC, Carromeu C, Acab A, Yu D, Yeo GW, Mu Y, Chen G, Gage FH, Muotri AR. A model for neural development and treatment of Rett syndrome using human induced pluripotent stem cells. Cell. 2010;143:527–539. doi: 10.1016/j.cell.2010.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Y, Wang H, Muffat J, Cheng AW, Orlando DA, Loven J, Kwok SM, Feldman DA, Bateup HS, Gao Q, Hockemeyer D, Mitalipova M, Lewis CA, Vander Heiden MG, Sur M, Young RA, Jaenisch R. Global transcriptional and translational repression in human-embryonic-stem-cell-derived Rett syndrome neurons. Cell stem cell. 2013;13:446–458. doi: 10.1016/j.stem.2013.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kishi N, Macklis JD. MECP2 is progressively expressed in post-migratory neurons and is involved in neuronal maturation rather than cell fate decisions. Molecular and cellular neurosciences. 2004;27:306–321. doi: 10.1016/j.mcn.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 26.Fukuda T, Itoh M, Ichikawa T, Washiyama K, Goto Y. Delayed maturation of neuronal architecture and synaptogenesis in cerebral cortex of Mecp2-deficient mice. Journal of neuropathology and experimental neurology. 2005;64:537–544. doi: 10.1093/jnen/64.6.537. [DOI] [PubMed] [Google Scholar]
- 27.Chapleau CA, Calfa GD, Lane MC, Albertson AJ, Larimore JL, Kudo S, Armstrong DL, Percy AK, Pozzo-Miller L. Dendritic spine pathologies in hippocampal pyramidal neurons from Rett syndrome brain and after expression of Rett-associated MECP2 mutations. Neurobiology of disease. 2009;35:219–233. doi: 10.1016/j.nbd.2009.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chao HT, Zoghbi HY, Rosenmund C. MeCP2 controls excitatory synaptic strength by regulating glutamatergic synapse number. Neuron. 2007;56:58–65. doi: 10.1016/j.neuron.2007.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiang M, Ash RT, Baker SA, Suter B, Ferguson A, Park J, Rudy J, Torsky SP, Chao HT, Zoghbi HY, Smirnakis SM. Dendritic arborization and spine dynamics are abnormal in the mouse model of MECP2 duplication syndrome. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2013;33:19518–19533. doi: 10.1523/JNEUROSCI.1745-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sztainberg Y, Chen HM, Swann JW, Hao S, Tang B, Wu Z, Tang J, Wan YW, Liu Z, Rigo F, Zoghbi HY. Reversal of phenotypes in MECP2 duplication mice using genetic rescue or antisense oligonucleotides. Nature. 2015;528:123–126. doi: 10.1038/nature16159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guy J, Gan J, Selfridge J, Cobb S, Bird A. Reversal of neurological defects in a mouse model of Rett syndrome. Science. 2007;315:1143–1147. doi: 10.1126/science.1138389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chang Q, Khare G, Dani V, Nelson S, Jaenisch R. The disease progression of Mecp2 mutant mice is affected by the level of BDNF expression. Neuron. 2006;49:341–348. doi: 10.1016/j.neuron.2005.12.027. [DOI] [PubMed] [Google Scholar]
- 33.Klein ME, Lioy DT, Ma L, Impey S, Mandel G, Goodman RH. Homeostatic regulation of MeCP2 expression by a CREB-induced microRNA. Nature neuroscience. 2007;10:1513–1514. doi: 10.1038/nn2010. [DOI] [PubMed] [Google Scholar]
- 34.Im HI, Hollander JA, Bali P, Kenny PJ. MeCP2 controls BDNF expression and cocaine intake through homeostatic interactions with microRNA-212. Nature neuroscience. 2010;13:1120–1127. doi: 10.1038/nn.2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Han K, Gennarino VA, Lee Y, Pang K, Hashimoto-Torii K, Choufani S, Raju CS, Oldham MC, Weksberg R, Rakic P, Liu Z, Zoghbi HY. Human-specific regulation of MeCP2 levels in fetal brains by microRNA miR-483-5p. Genes & development. 2013;27:485–490. doi: 10.1101/gad.207456.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bellini E, Pavesi G, Barbiero I, Bergo A, Chandola C, Nawaz MS, Rusconi L, Stefanelli G, Strollo M, Valente MM, Kilstrup-Nielsen C, Landsberger N. MeCP2 post-translational modifications: a mechanism to control its involvement in synaptic plasticity and homeostasis? Frontiers in cellular neuroscience. 2014;8:236. doi: 10.3389/fncel.2014.00236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yen HC, Xu Q, Chou DM, Zhao Z, Elledge SJ. Global protein stability profiling in mammalian cells. Science. 2008;322:918–923. doi: 10.1126/science.1160489. [DOI] [PubMed] [Google Scholar]
- 38.Emanuele MJ, Elia AE, Xu Q, Thoma CR, Izhar L, Leng Y, Guo A, Chen YN, Rush J, Hsu PW, Yen HC, Elledge SJ. Global identification of modular cullin-RING ligase substrates. Cell. 2011;147:459–474. doi: 10.1016/j.cell.2011.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bracaglia G, Conca B, Bergo A, Rusconi L, Zhou Z, Greenberg ME, Landsberger N, Soddu S, Kilstrup-Nielsen C. Methyl-CpG-binding protein 2 is phosphorylated by homeodomain-interacting protein kinase 2 and contributes to apoptosis. EMBO reports. 2009;10:1327–1333. doi: 10.1038/embor.2009.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yamada D, Perez-Torrado R, Filion G, Caly M, Jammart B, Devignot V, Sasai N, Ravassard P, Mallet J, Sastre-Garau X, Schmitz ML, Defossez PA. The human protein kinase HIPK2 phosphorylates and downregulates the methyl-binding transcription factor ZBTB4. Oncogene. 2009;28:2535–2544. doi: 10.1038/onc.2009.109. [DOI] [PubMed] [Google Scholar]
- 41.Ferreira-Cerca S, Kiburu I, Thomson E, LaRonde N, Hurt E. Dominant Rio1 kinase/ATPase catalytic mutant induces trapping of late pre-40S biogenesis factors in 80S-like ribosomes. Nucleic acids research. 2014;42:8635–8647. doi: 10.1093/nar/gku542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Widmann B, Wandrey F, Badertscher L, Wyler E, Pfannstiel J, Zemp I, Kutay U. The kinase activity of human Rio1 is required for final steps of cytoplasmic maturation of 40S subunits. Molecular biology of the cell. 2012;23:22–35. doi: 10.1091/mbc.E11-07-0639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shi Y. Serine/threonine phosphatases: mechanism through structure. Cell. 2009;139:468–484. doi: 10.1016/j.cell.2009.10.006. [DOI] [PubMed] [Google Scholar]
- 44.Lambrecht C, Haesen D, Sents W, Ivanova E, Janssens V. Structure, regulation, and pharmacological modulation of PP2A phosphatases. Methods Mol Biol. 2013;1053:283–305. doi: 10.1007/978-1-62703-562-0_17. [DOI] [PubMed] [Google Scholar]
- 45.Kim JY, Ash RT, Ceballos-Diaz C, Levites Y, Golde TE, Smirnakis SM, Jankowsky JL. Viral transduction of the neonatal brain delivers controllable genetic mosaicism for visualising and manipulating neuronal circuits in vivo. The European journal of neuroscience. 2013;37:1203–1220. doi: 10.1111/ejn.12126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kickstein E, Krauss S, Thornhill P, Rutschow D, Zeller R, Sharkey J, Williamson R, Fuchs M, Kohler A, Glossmann H, Schneider R, Sutherland C, Schweiger S. Biguanide metformin acts on tau phosphorylation via mTOR/protein phosphatase 2A (PP2A) signaling. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:21830–21835. doi: 10.1073/pnas.0912793107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Swingle M, Ni L, Honkanen RE. Small-molecule inhibitors of ser/thr protein phosphatases: specificity, use and common forms of abuse. Methods Mol Biol. 2007;365:23–38. doi: 10.1385/1-59745-267-X:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Katz DM, Berger-Sweeney JE, Eubanks JH, Justice MJ, Neul JL, Pozzo-Miller L, Blue ME, Christian D, Crawley JN, Giustetto M, Guy J, Howell CJ, Kron M, Nelson SB, Samaco RC, Schaevitz LR, St Hillaire-Clarke C, Young JL, Zoghbi HY, Mamounas LA. Preclinical research in Rett syndrome: setting the foundation for translational success. Disease models & mechanisms. 2012;5:733–745. doi: 10.1242/dmm.011007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Samaco RC, Nagarajan RP, Braunschweig D, LaSalle JM. Multiple pathways regulate MeCP2 expression in normal brain development and exhibit defects in autism-spectrum disorders. Human molecular genetics. 2004;13:629–639. doi: 10.1093/hmg/ddh063. [DOI] [PubMed] [Google Scholar]
- 50.Nagarajan RP, Hogart AR, Gwye Y, Martin MR, LaSalle JM. Reduced MeCP2 expression is frequent in autism frontal cortex and correlates with aberrant MECP2 promoter methylation. Epigenetics: official journal of the DNA Methylation Society. 2006;1:e1–11. doi: 10.4161/epi.1.4.3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gennarino VA, Alcott CE, Chen CA, Chaudhury A, Gillentine MA, Rosenfeld JA, Parikh S, Wheless JW, Roeder ER, Horovitz DD, Roney EK, Smith JL, Cheung SW, Li W, Neilson JR, Schaaf CP, Zoghbi HY. NUDT21-spanning CNVs lead to neuropsychiatric disease and altered MeCP2 abundance via alternative polyadenylation. eLife. 2015;4 doi: 10.7554/eLife.10782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Study TDDD. Large-scale discovery of novel genetic causes of developmental disorders. Nature. 2015;519:223–228. doi: 10.1038/nature14135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sakamoto K, Huang BW, Iwasaki K, Hailemariam K, Ninomiya-Tsuji J, Tsuji Y. Regulation of genotoxic stress response by homeodomain-interacting protein kinase 2 through phosphorylation of cyclic AMP response element-binding protein at serine 271. Molecular biology of the cell. 2010;21:2966–2974. doi: 10.1091/mbc.E10-01-0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang Q, Wang Y. Homeodomain-interacting protein kinase-2 (HIPK2) phosphorylates HMGA1a at Ser-35, Thr-52, and Thr-77 and modulates its DNA binding affinity. Journal of proteome research. 2007;6:4711–4719. doi: 10.1021/pr700571d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ecsedy JA, Michaelson JS, Leder P. Homeodomain-interacting protein kinase 1 modulates Daxx localization, phosphorylation, and transcriptional activity. Molecular and cellular biology. 2003;23:950–960. doi: 10.1128/MCB.23.3.950-960.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mooney SM, Qiu R, Kim JJ, Sacho EJ, Rajagopalan K, Johng D, Shiraishi T, Kulkarni P, Weninger KR. Cancer/testis antigen PAGE4, a regulator of c-Jun transactivation, is phosphorylated by homeodomain-interacting protein kinase 1, a component of the stress-response pathway. Biochemistry. 2014;53:1670–1679. doi: 10.1021/bi500013w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tao J, Hu K, Chang Q, Wu H, Sherman NE, Martinowich K, Klose RJ, Schanen C, Jaenisch R, Wang W, Sun YE. Phosphorylation of MeCP2 at Serine 80 regulates its chromatin association and neurological function. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:4882–4887. doi: 10.1073/pnas.0811648106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu X, Tian L, Li J, Zhang Y, Han V, Li Y, Xu X, Li H, Chen X, Chen J, Jin W, Xie Y, Han J, Zhong CQ. Investigation of receptor interacting protein (RIP3)-dependent protein phosphorylation by quantitative phosphoproteomics. Molecular & cellular proteomics: MCP. 2012;11:1640–1651. doi: 10.1074/mcp.M112.019091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Courcelles M, Fremin C, Voisin L, Lemieux S, Meloche S, Thibault P. Phosphoproteome dynamics reveal novel ERK1/2 MAP kinase substrates with broad spectrum of functions. Molecular systems biology. 2013;9:669. doi: 10.1038/msb.2013.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Olsen JV, Vermeulen M, Santamaria A, Kumar C, Miller ML, Jensen LJ, Gnad F, Cox J, Jensen TS, Nigg EA, Brunak S, Mann M. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Science signaling. 2010;3:ra3. doi: 10.1126/scisignal.2000475. [DOI] [PubMed] [Google Scholar]
- 61.Klammer M, Kaminski M, Zedler A, Oppermann F, Blencke S, Marx S, Muller S, Tebbe A, Godl K, Schaab C. Phosphosignature predicts dasatinib response in non-small cell lung cancer. Molecular & cellular proteomics: MCP. 2012;11:651–668. doi: 10.1074/mcp.M111.016410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kim JY, Welsh EA, Oguz U, Fang B, Bai Y, Kinose F, Bronk C, Remsing Rix LL, Beg AA, Rix U, Eschrich SA, Koomen JM, Haura EB. Dissection of TBK1 signaling via phosphoproteomics in lung cancer cells. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:12414–12419. doi: 10.1073/pnas.1220674110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shiromizu T, Adachi J, Watanabe S, Murakami T, Kuga T, Muraoka S, Tomonaga T. Identification of missing proteins in the neXtProt database and unregistered phosphopeptides in the PhosphoSitePlus database as part of the Chromosome-centric Human Proteome Project. Journal of proteome research. 2013;12:2414–2421. doi: 10.1021/pr300825v. [DOI] [PubMed] [Google Scholar]
- 64.Yi T, Zhai B, Yu Y, Kiyotsugu Y, Raschle T, Etzkorn M, Seo HC, Nagiec M, Luna RE, Reinherz EL, Blenis J, Gygi SP, Wagner G. Quantitative phosphoproteomic analysis reveals system-wide signaling pathways downstream of SDF-1/CXCR4 in breast cancer stem cells. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:E2182–2190. doi: 10.1073/pnas.1404943111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lyst MJ, Ekiert R, Ebert DH, Merusi C, Nowak J, Selfridge J, Guy J, Kastan NR, Robinson ND, de Lima Alves F, Rappsilber J, Greenberg ME, Bird A. Rett syndrome mutations abolish the interaction of MeCP2 with the NCoR/SMRT co-repressor. Nature neuroscience. 2013;16:898–902. doi: 10.1038/nn.3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ebert DH, Gabel HW, Robinson ND, Kastan NR, Hu LS, Cohen S, Navarro AJ, Lyst MJ, Ekiert R, Bird AP, Greenberg ME. Activity-dependent phosphorylation of MeCP2 threonine 308 regulates interaction with NCoR. Nature. 2013;499:341–345. doi: 10.1038/nature12348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Isono K, Nemoto K, Li Y, Takada Y, Suzuki R, Katsuki M, Nakagawara A, Koseki H. Overlapping roles for homeodomain-interacting protein kinases hipk1 and hipk2 in the mediation of cell growth in response to morphogenetic and genotoxic signals. Molecular and cellular biology. 2006;26:2758–2771. doi: 10.1128/MCB.26.7.2758-2771.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Goffin D, Allen M, Zhang L, Amorim M, Wang IT, Reyes AR, Mercado-Berton A, Ong C, Cohen S, Hu L, Blendy JA, Carlson GC, Siegel SJ, Greenberg ME, Zhou Z. Rett syndrome mutation MeCP2 T158A disrupts DNA binding, protein stability and ERP responses. Nature neuroscience. 2012;15:274–283. doi: 10.1038/nn.2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ramocki MB, Peters SU, Tavyev YJ, Zhang F, Carvalho CM, Schaaf CP, Richman R, Fang P, Glaze DG, Lupski JR, Zoghbi HY. Autism and other neuropsychiatric symptoms are prevalent in individuals with MeCP2 duplication syndrome. Annals of neurology. 2009;66:771–782. doi: 10.1002/ana.21715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Meerbrey KL, Hu G, Kessler JD, Roarty K, Li MZ, Fang JE, Herschkowitz JI, Burrows AE, Ciccia A, Sun T, Schmitt EM, Bernardi RJ, Fu X, Bland CS, Cooper TA, Schiff R, Rosen JM, Westbrook TF, Elledge SJ. The pINDUCER lentiviral toolkit for inducible RNA interference in vitro and in vivo. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:3665–3670. doi: 10.1073/pnas.1019736108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Baker SA, Lombardi LM, Zoghbi HY. Karyopherin alpha 3 and Karyopherin alpha 4 Proteins Mediate the Nuclear Import of Methyl-CpG Binding Protein. The Journal of biological chemistry. 2015;2290:22485–22493. doi: 10.1074/jbc.M115.658104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wechsler-Reya RJ, Scott MP. Control of neuronal precursor proliferation in the cerebellum by Sonic Hedgehog. Neuron. 1999;22:103–114. doi: 10.1016/s0896-6273(00)80682-0. [DOI] [PubMed] [Google Scholar]
- 73.Kim JY, Grunke SD, Levites Y, Golde TE, Jankowsky JL. Intracerebroventricular viral injection of the neonatal mouse brain for persistent and widespread neuronal transduction. Journal of visualized experiments: JoVE. 2014:51863. doi: 10.3791/51863. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Candidate knockdown did not decrease MECP2 transcript levels.
Fig. S2. MeCP2 pS16 occurred in vivo.
Fig. S3. No RIOK1 kinase activity for MeCP2 was detected.
Fig. S4. Ppp2r1a knockdown also decreased MeCP2 levels in hippocampal tissue.
Fig. S5. Okadaic acid treatment was active against human MeCP2.
Fig. S6. In vivo fostriecin titration.
Fig. S7. In vivo PP2A inhibition resulted in no ill effects in WT at the doses used.
Table S1. MeCP2-specific siRNAs gene list.
Table S2. Primers used.
Table S3. shRNAs used.