SUMMARY
Sleep is an essential and evolutionarily conserved behavioral state whose regulation remains poorly understood. To identify genes that regulate vertebrate sleep, we recently performed a genetic screen in zebrafish, and here we report the identification of neuropeptide Y (NPY) as both necessary for normal daytime sleep duration and sufficient to promote sleep. We show that overexpression of NPY increases sleep, whereas mutation of npy or ablation of npy-expressing neurons decreases sleep. By analyzing sleep architecture, we show that NPY regulates sleep primarily by modulating the length of wake bouts. To determine how NPY regulates sleep, we tested for interactions with several systems known to regulate sleep, and provide anatomical, molecular, genetic and pharmacological evidence that NPY promotes sleep by inhibiting noradrenergic signaling. These data establish NPY as an important vertebrate sleep/wake regulator and link NPY signaling to an established arousal-promoting system.
Keywords: Sleep, neuropeptide Y, hypothalamus, locus coeruleus, noradrenaline, locomotor activity, arousal, genetics
eTOC Blurb
Based on a genetic screen, Singh et al identify NPY signaling and npy-expressing neurons as regulators of zebrafish sleep. They show that NPY promotes sleep by inhibiting noradrenergic signaling, thus linking NPY signaling to an established arousal-promoting system.
INTRODUCTION
Sleep is among most basic needs of living organisms, yet mechanisms that regulate sleep remain poorly understood. Several neuropeptides have been implicated in regulating mammalian sleep [1], including hypocretin [2–4], which promotes wakefulness, and galanin [5–8] and melanin concentrating hormone [9–12], which promote sleep, suggesting that examining additional neuropeptides may identify novel mechanisms that regulate sleep. Identifying these mechanisms using mammalian models has been challenging due to their poor amenability for large-scale screens, although such screens are possible [13]. As an alternative approach, several groups have used behavioral criteria to study sleep-like states in simpler model organisms that are amenable to screens, including Drosophila [14–22], C. elegans [23–25], and zebrafish [26–30]. In particular, several groups have demonstrated behavioral, anatomical, genetic and pharmacological conservation of sleep between zebrafish and mammals, establishing zebrafish as a vertebrate sleep model [26–28, 30–33]. We previously described a screen for genes whose overexpression affects zebrafish sleep, and reported that the neuropeptide neuromedin U is necessary and sufficient for normal levels of arousal [29]. Here we demonstrate that another neuropeptide identified in the screen, neuropeptide Y (NPY), is necessary for daytime sleep and sufficient to promote sleep.
NPY is widely expressed in the brain and has been implicated in regulating endocrine, behavioral and circadian processes [34], and is perhaps best known for its role in promoting feeding [35–38]. NPY has also been shown to affect sleep, but its role in this behavioral state remains unclear. Several studies showed that injection of in vitro synthesized NPY into the rodent brain [39–45] or intravenously in young healthy [46] or depressed [47] humans can induce sleep or reduce locomotor activity. However, other rodent studies reported the opposite effect [48–50]. The basis for these disparate reports is unclear, but may be due to different sites and doses of NPY injection, or the use of in vitro synthesized peptide that may vary in different preparations and from endogenous NPY. Understanding the role of NPY in mammalian sleep is also confounded by links between mechanisms that regulate feeding and sleep [48–51]. Indeed, reports of wake-promotion by injected NPY also observed increased feeding [48–50], suggesting that the increased wakefulness may result from increased feeding. npy mutant mice exhibit several phenotypes, including increased anxiety, depression-like behavior, and cognitive deficits [52, 53], and are less susceptible to diet-induced obesity [54]. However, an analysis of sleep in these animals and a role for npy-expressing neurons in sleep has not been described. As a result, the role of NPY in vertebrate sleep remains unclear.
Here we show that NPY is sufficient to promote sleep in zebrafish, whereas loss of npy or npy-expressing neurons results in less daytime sleep. We also show that NPY promotes sleep by inhibiting the wake-promoting noradrenergic system, providing a mechanistic basis for sleep regulation by NPY. Together with the requirement of noradrenergic signaling for the wake-promoting function of hypocretin [55, 56], these results suggest that the noradrenergic system integrates neuropeptidergic signals that regulate sleep/wake states.
RESULTS
Overexpression of human NPY reduces locomotor activity and increases sleep in zebrafish
We previously performed a screen to identify genes that affect larval zebrafish sleep [29]. We injected >1200 unique plasmids in which a heat shock-inducible promoter (hsp) regulates the expression of genes that encode for secreted proteins into wild-type (WT) zebrafish embryos at the one-cell stage. We used human open reading frames (ORFs) encoding secreted proteins from the hORFeome 3.1 library [57] because there was no resource of zebrafish ORFs. Co-injection of each plasmid with tol2 transposase mRNA resulted in incorporation of the hsp-regulated transgene into the genome in many cells of each animal and enabled heat shock-induced overexpression [29]. We then compared sleep/wake behaviors in injected animals before and after heat shock and to negative control animals injected with a hsp:egfp plasmid. One gene whose overexpression increased sleep at night (Z-score=1.8) encoded human NPY (Figure S1A). Even though zebrafish exhibit high levels of sleep at night, NPY-overexpressing animals were 28% less active and slept 34% more than control animals during the night after heat shock (P<0.05 and P<0.01, two-tailed Student’s t test) (Figures S1B–S1G). We observed a similar phenotype during the day before heat shock that did not reach statistical significance, consistent with leaky expression from the hsp promoter that often is observed using this transient injection assay, but is not observed using stable transgenic lines [29].
Overexpression of zebrafish NPY reduces locomotor activity and increases sleep in zebrafish
Using reciprocal BLAST searches, we identified a single zebrafish npy ortholog, which encodes for a preproprotein that generates a predicted 36 amino-acid mature peptide that is 89% identical to the human and mouse orthologs (Figure S1H). npy is widely expressed in the mammalian brain, particularly in the hypothalamus, amygdala, locus coeruleus (LC) and cerebral cortex [58, 59]. Using in situ hybridization (ISH) with an npy-specific probe, immunostaining for total extracellular signal-regulated kinase (t-ERK), and image registration to the Z-brain atlas [60], we found that npy is similarly expressed in several discrete nuclei within the larval zebrafish brain (Figures S1I–S1N and Movie S1). We also observed npy expression in the retina (data not shown) but not in other tissues.
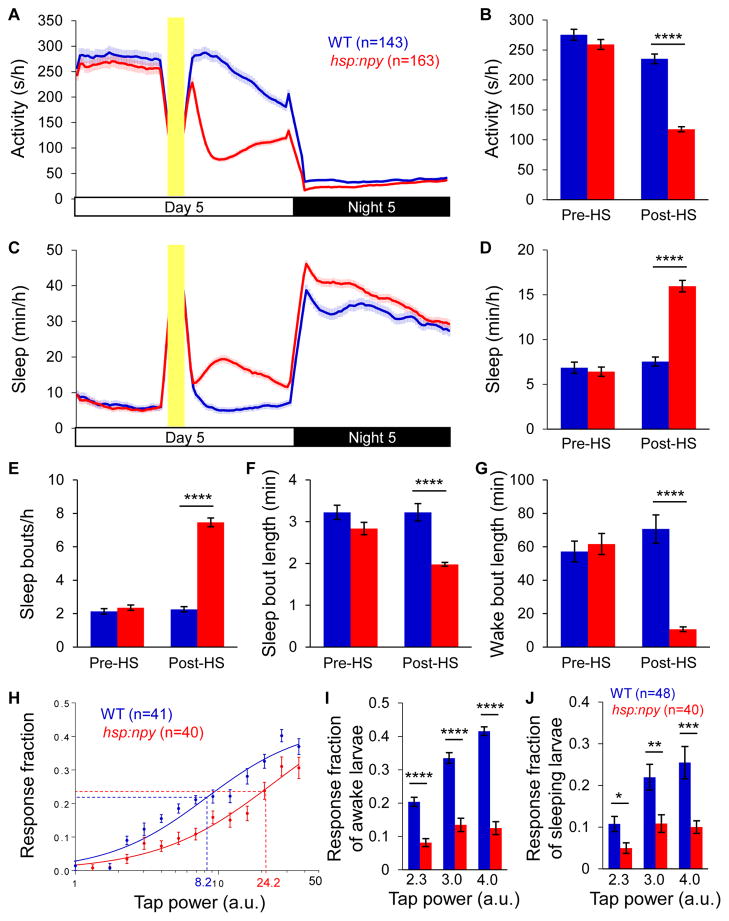
To test whether overexpression of zebrafish NPY affects sleep, we generated Tg(hsp:npy) zebrafish. Tg(hsp:npy) animals and their WT siblings had similar amounts of locomotor activity and sleep before heat-shock (Figures 1A–1D). However, following a heat shock at 3 p.m., Tg(hsp:npy) animals were 50% less active (Figures 1A and 1B) and slept 111% more (Figures 1C and 1D) than their WT siblings for the rest of the day (P<0.0001, two-tailed Student’s t test). The phenotype resulted from a 230% increase in the number of sleep bouts (Figure 1E) and an 85% decrease in the length of wake bouts (Figure 1G) (P<0.0001, two-tailed Student’s t test), with a smaller decrease in the length of sleep bouts (Figure 1F), and thus is primarily due to fragmentation of the wake state.
Figure 1. Overexpression of zebrafish NPY increases sleep and arousal threshold.
(A–G) Overexpression of zebrafish NPY following a heat shock at 3 p.m. resulted in decreased locomotor activity (A,B) and increased sleep (C,D), due to more sleep bouts (E) and shorter sleep (F) and wake (G) bouts. Yellow bars indicate heat shock (HS). Pre-HS and Post-HS quantify data for day 5 before and after heat shock. Mean ± SEM from 4 experiments is shown. (H) Representative stimulus-response curve for Tg(hsp:npy) animals compared to WT siblings following heat shock. Data points represent mean ± SEM. Dashed lines mark ETP50 value for each genotype. Tg(hsp:npy) animals had an ETP50 value of 24.2 vs. 8.2 for WT siblings (293% increase, P< 0.05 by extra sum-of-squares F test). (I,J) Overexpression of NPY reduced the response of Tg(hsp:npy) animals to the stimulus compared to WT siblings during both awake and sleep states. Stimulus intensities of 2.3, 3.0 and 4.0 arbitrary units (a.u.) were tested. A dose-dependent response was observed for WT animals but not their Tg(hsp:npy) siblings. Bar graphs show mean ± SEM. n=number of animals. *P<0.05; **P<0.01; ***P<0.001; ****P<0.0001 by two-tailed Student’s t test. See also Figures S1, S2 and Movie S1.
The increase in sleep after the heat shock-induced pulse of NPY overexpression dampened by nighttime. A previous study showed that the circadian system inhibits sleep in the evening, when homeostatic sleep drive is high [61], suggesting the circadian system might limit NPY overexpression-induced sleep to the day. To test whether NPY overexpression can also increase sleep at night, we heat shocked animals during the last hour of the day. We found that Tg(hsp:npy) animals were 46% less active (Figures S2A–S2C) and slept 54% more (Figures S2D–S2F) than their WT siblings during the night (P<0.0001, two-tailed Student’s t test), similar to the daytime phenotype when NPY overexpression was induced in the afternoon. This phenotype was due to longer sleep bouts (Figure S2H) and shorter wake bouts (Figure S2J), with no change in the number of sleep bouts (Figure S2G). These results suggest that dampening of NPY-induced sleep at night following heat shock in the afternoon is due to declining levels of NPY rather than effects of the circadian clock.
Light affects locomotor activity and sleep in zebrafish [27, 28], as it does in mammals [62]. To determine whether light affects NPY overexpression-induced sleep, we entrained larvae by raising them in 14:10 hour light:dark (LD) conditions for four days, and then transferred them to constant dark before inducing NPY overexpression. NPY-overexpressing animals were 54% less active and slept 80% more than WT siblings during the rest of the subjective day (Figures S2K–S2N) (P<0.0001, two-tailed Student’s t test). This phenotype was due to more sleep bouts and shorter wake bouts, with no change in the length of sleep bouts (Figures S2O–S2Q). Hence, NPY overexpression promotes sleep independent of lighting condition and circadian phase.
Overexpression of NPY increases arousal threshold
Sleep is distinguished from quiet wakefulness by reduced sensory responsiveness [63]. Because NPY overexpression increases sleep, we asked whether it also alters arousal threshold by monitoring responses to mechano-acoustic stimuli. We found that the stimulus intensity at which we observed the half-maximal response (effective tap power 50, ETP50) was 290% higher for Tg(hsp:npy) animals than their WT siblings (Figure 1H) (P<0.05 by extra sum-of-squares F test). Thus, NPY overexpression increases arousal threshold, consistent with increased sleep. We next asked if NPY overexpression affects arousal in awake and/or sleeping animals by allowing 5 minutes between trials. According to the behavioral definition of sleep, we scored animals as awake if they moved during the minute before a stimulus was delivered. We used stimulus intensities of 2.3, 3.0 and 4.0 arbitrary units, which were lower than the ETP50 values of both Tg(hsp:npy) and WT animals. NPY-overexpressing animals were less responsive to these stimuli than WT siblings during both awake (Figure 1I) and sleep (Figure 1J) states. These data suggest that NPY overexpression decreases arousal in awake animals and increases sleep depth in sleeping animals.
npy mutant zebrafish are more active and sleep less during the day
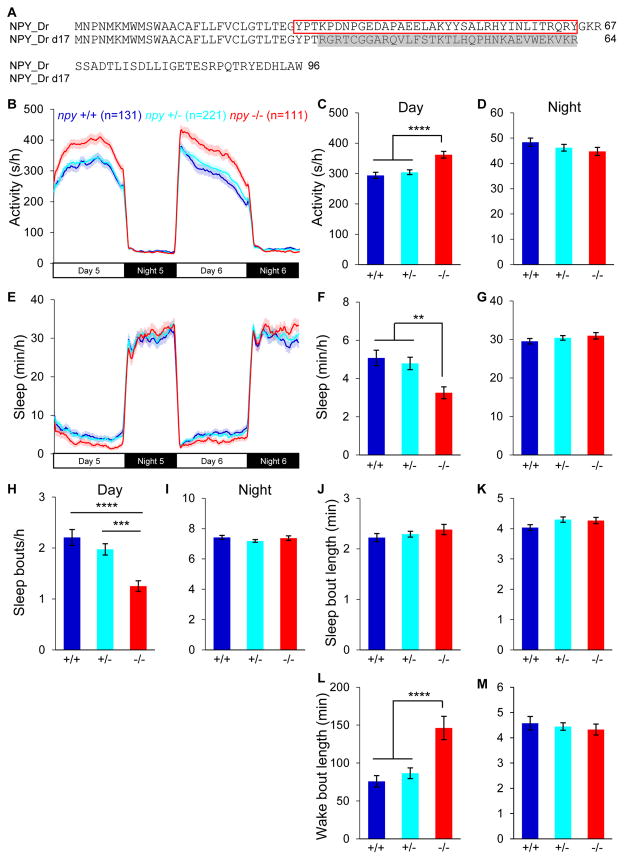
We next asked whether endogenous npy is required for normal sleep/wake behaviors by using the zinc finger nuclease method to generate zebrafish containing a predicted null mutation in the npy open reading frame [64]. We isolated zebrafish containing a 17-nucleotide deletion in the second exon of the npy gene [64], which results in a translational frame shift at the beginning of the mature peptide domain (Figure 2A), generating a protein that lacks the mature peptide domain and thus is likely nonfunctional. Homozygous mutant animals are viable and fertile, and lack obvious developmental defects.
Figure 2. Loss of npy reduces daytime sleep.
(A) Sequences of WT and mutant zebrafish NPY proteins. The mature peptide is indicated with a red box. Altered amino acids in the mutant are shaded grey. (B–M) npy−/− animals were more active (B,C), and slept less (E,F), than their npy+/+ and npy+/− siblings during the day, due to fewer sleep bouts (H) and longer wake bouts (L). Mean ± SEM from 7 experiments is shown. n=number of animals. **P<0.01; ***P<0.001; ****P<0.0001 by one-way ANOVA with Holm-Sidak test.
Consistent with the NPY overexpression phenotype, npy−/− larvae were 23% more active and slept 36% less during the day than their npy+/+ siblings (P<0.0001 and P<0.01, one-way ANOVA, Holm-Sidak test) (Figures 2B, 2C, 2E and 2F). These effects were due to fewer sleep bouts (Figure 2H) and longer wake bouts (Figure 2L), with no effect on the length of sleep bouts (Figures 2J). Thus, reduced daytime sleep in npy−/− animals is due to consolidation of the wake state. We did not observe npy−/− phenotypes at night. These data indicate that endogenous npy is required for normal daytime sleep amounts.
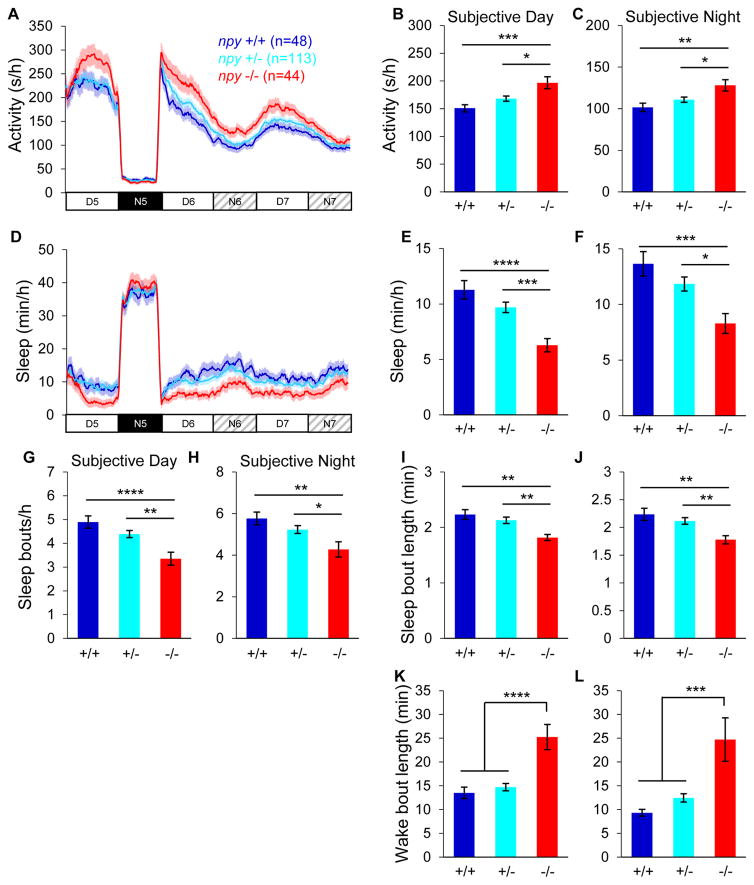
Microinjection of NPY into the hamster suprachiasmatic nucleus phase shifts the locomotor activity circadian rhythm in constant light (LL) [65, 66], suggesting that NPY may regulate entrainment or expression of circadian rhythms. To test whether endogenous npy is required for circadian regulation of locomotor activity and sleep, we tested larvae that were entrained for 4 days in LD, then monitored for 24 hours in LD and then for 48 hours in LL. Absence of npy had no obvious effect on the locomotor activity or sleep circadian period length or phase (Figures 3A and 3D). As expected, in LD npy−/− animals were more active (Figures 3A and S3A) and slept less (Figures 3D and S3C) than their npy+/+ and npy+/− siblings during the day, with no phenotype at night. The daytime phenotype was due to fewer sleep bouts and longer wake bouts (Figures S3E and S3I). Following the shift to LL, npy−/− animals were more active by 30% and 26% during the subjective day and night, respectively, compared to their npy+/+ siblings (P<0.001 and P<0.01, one-way ANOVA, Holm-Sidak test) (Figures 3A–3C). npy−/− larvae also slept ~40% less during the subjective day and night (P<0.0001 and P<0.001, one-way ANOVA, Holm-Sidak test) (Figures 3D–3F). These phenotypes were primarily due to longer wake bouts (Figures 3K and 3L), although there were also fewer (Figures 3G and 3H) and shorter (Figures 3I and 3J) sleep bouts. These results indicate that npy is not required for circadian regulation of locomotor activity or sleep in zebrafish larvae, but rather regulates sleep in a light-dependent manner.
Figure 3. Entrained npy mutants sleep less in constant light.
Larvae were entrained in 14:10 hour LD cycles for the first 4 days and nights of development, then behaviorally monitored for 24 hours in LD and then for 48 hours in LL. npy−/− animals were more active (A–C) and slept less (D–F) than their npy+/− and npy+/+ siblings during subjective day and night, due to fewer (G,H) and shorter (I,J) sleep bouts, and longer wake bouts (K,L). Mean ± SEM from 3 experiments is shown. n=number of animals. *P<0.05; **P<0.01; ***P<0.001; ****P<0.0001 by one-way ANOVA with Holm-Sidak test. See also Figure S3.
Ablation of npy-expressing neurons increases locomotor activity and decreases sleep
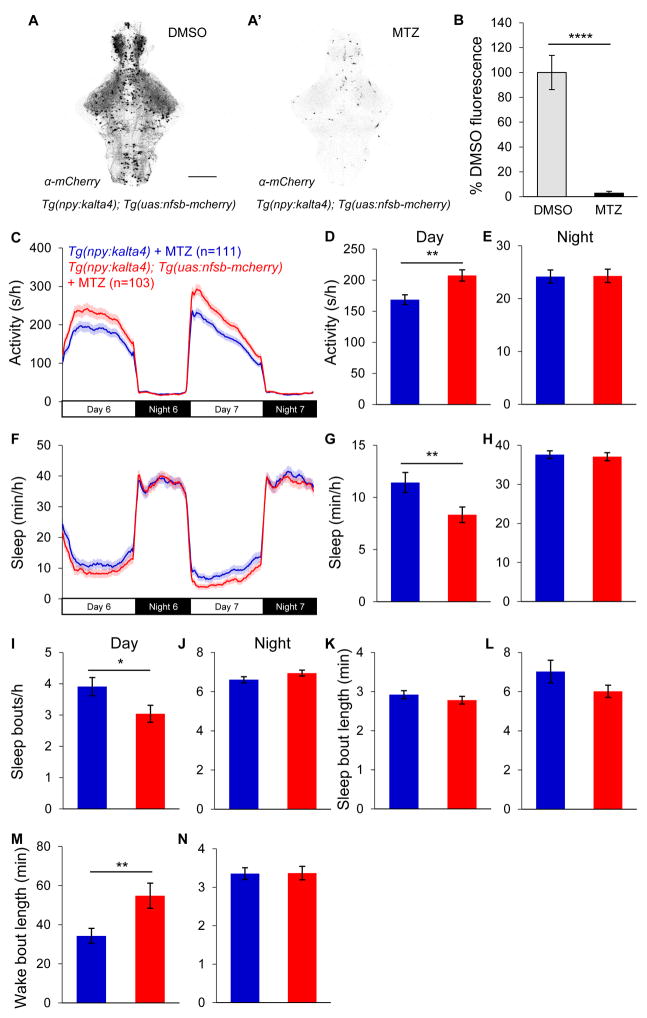
As an alternative approach to test the hypothesis that NPY is necessary for normal sleep duration, we ablated npy-expressing neurons. To this end, we generated Tg(npy:kalta4) zebrafish, in which NPY neurons express an optimized version of the transcriptional activator Gal4 (KalTA4). To verify the specificity of this transgene, we performed double fluorescent ISH (FISH) using probes specific for npy and kalta4. We observed that kalta4 is expressed in >80% of npy-expressing neurons (>95% for some brain regions), and that >92% of kalta4-expressing neurons express npy (Figure S5A and Table S1). We mated these fish to Tg(uas:nfsb-mcherry) animals [67], resulting in the expression of nitroreductase (nfsb) in npy-expressing neurons (Figure 4A). Nitroreductase is a bacterial protein that converts the inert prodrug metronidazole (MTZ) into a cytotoxic DNA crosslinking agent, thus enabling drug-inducible ablation of the targeted cell type [68]. We treated Tg(npy:kalta4);Tg(uas:nfsb-mcherry) and Tg(npy:kalta4) sibling control animals with MTZ or DMSO vehicle control for 48 hours (from 3 days post-fertilization (dpf) to 5 dpf). MTZ treatment almost completely eliminated mCherry-labeled cells in double transgenic animals (Figures 4A–4C), indicating loss of most npy-expressing neurons. Consistent with these observations, we detected TUNEL labeling in npy-expressing neurons in Tg(npy:kalta4);Tg(uas:nfsb-mcherry) animals treated with MTZ, but not in those treated with DMSO (Figures S5B–S5D), indicating that MTZ treatment induces apoptosis of npy-expressing neurons. Consistent with the npy−/− phenotype, npy-ablated animals were 23% more active (Figures 4C and 4D) and slept 28% less (Figures 4F and 4G) (P<0.01 and P<0.05, two-tailed Student’s t test) compared to sibling controls during the day. This phenotype was due to fewer sleep bouts (Figure 4I) and longer wake bouts (Figure 4M), indicating consolidation of the wake state, similar to npy−/− animals. To confirm that the Tg(uas:nfsb-mcherry) transgene alone does not cause a behavioral phenotype, we crossed Tg(npy:kalta4)/+;Tg(uas:nfsb-mcherry)/+ to WT fish, excluded animals that were positive for mCherry, and treated the remaining animals with MTZ. We observed no difference in locomotor activity or sleep among animals of these three genotypes (Figure S4). The cell ablation phenotype was slightly weaker than that of the npy mutant, likely because the npy:kalta4 transgene is not expressed in all npy-expressing neurons. Because a small number of neurons express kalta4 but not npy in some brain regions (8% in the subpallium, <5% in other brain regions; Figure S5A and Table S1), it is possible that ablation of these NPY-negative cells is responsible for the behavioral phenotype. However, this is unlikely to be the case due to the small number of cells involved and because the NPY neuron ablation phenotype is consistent with the npy mutant phenotype, suggesting that both NPY and npy-expressing neurons are necessary for normal daytime sleep amount.
Figure 4. Loss of npy-expressing neurons reduces daytime sleep.
(A) Ventral views of brains from 5 dpf Tg(npy:kalta4);Tg(uas:nfsb-mcherry) animals stained with anti-DsRed antibody following treatment with DMSO (A) or 10 mM MTZ (A′), showing nearly complete loss of mCherry after MTZ treatment. (B) Mean ± SEM mCherry fluorescence intensity for Tg(npy:kalta4);Tg(uas:nfsb-mcherry) animals treated with DMSO (n=4) or MTZ (n=4). (C–N) Tg(npy:kalta4);Tg(uas:nfsb-mcherry) animals treated with MTZ were more active (C,D) and slept less (F,G) than identically treated Tg(npy:kalta4) siblings during the day, due to fewer sleep bouts (I) and longer wake bouts (M). Mean ± SEM from 3 experiments is shown. n=number of animals. *P<0.05; **P<0.01; ****P<0.0001 by two-tailed Student’s t test. See also Figures S4, S5 and Table S1.
The NPY overexpression phenotype is not blocked by manipulation of several pathways known to regulate sleep
To identify genetic mechanisms through which NPY affects sleep, we tested whether the NPY overexpression phenotype is suppressed in zebrafish containing mutations in other genes implicated in regulating sleep (Table S2). We found that the NPY overexpression phenotype persisted in larvae containing null mutations in histidine decarboxylase (hdc) [69], hypocretin receptor (hcrtr) [27], corticotropin releasing hormone a (crha) (Singh et al., unpublished), crhb (Singh et al., unpublished) or arylalkylamine N-acetyltransferase 2 (aanat2) [70] (data not shown). These data suggest that NPY promotes sleep via other mechanisms.
NPY promotes sleep by inhibiting noradrenergic signaling
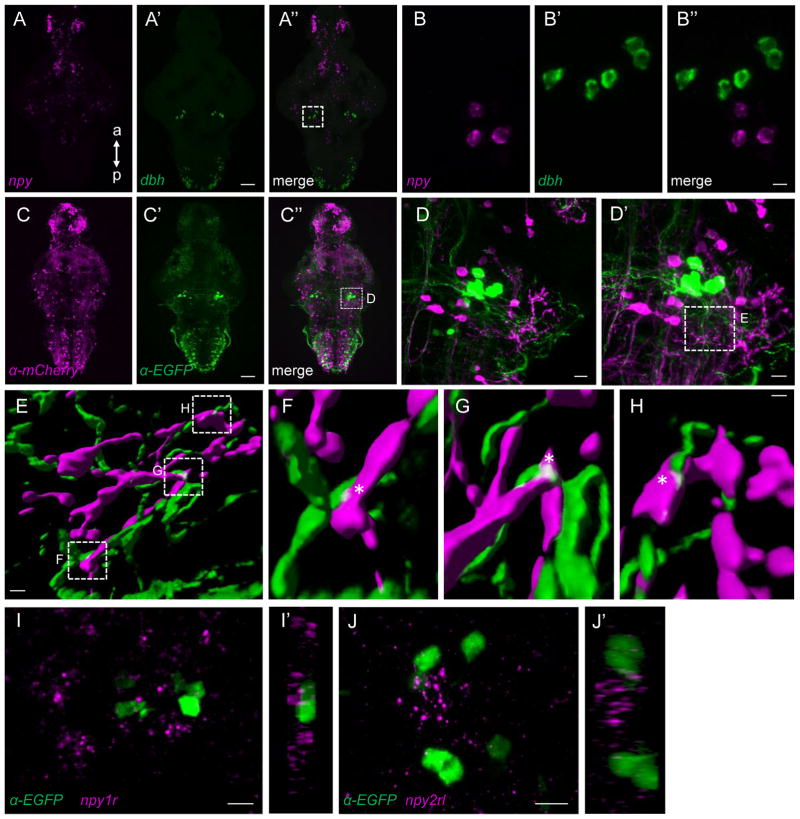
Pharmacological and genetic studies in mammals and zebrafish have shown that norepinephrine (NE) plays an important role in promoting arousal [56, 71], and the LC is the primary source of NE in the brain [72]. We obtained several lines of evidence suggesting that NPY promotes sleep by inhibiting NE signaling. First, a nucleus of 3–5 npy-expressing neurons is located adjacent to, and sends projections that form close contacts with, LC neurons (Figures 5A–5H and Movie S2). While this does not prove a direct interaction between the two neuronal populations, it is consistent with our functional evidence that NPY promotes sleep by inhibiting NE signaling (see below). The zebrafish genome contains seven annotated npy receptor genes [73]. Using FISH, we did not detect npy receptor expression in LC neurons, although we observed expression of npy receptor y1 (npy1r) (Figure 5I) and npy receptor y2 like (npy2rl) (Figure 5J) near the LC. The other npy receptors showed expression in other brain regions (npy8ar and npy8br) or no detectable pattern of expression (npy2r, npy4r and npy7r) (data not shown). These results suggest that NPY indirectly affects NE signaling, although a npy receptor might be expressed in LC neurons at levels too low to be detected using FISH, a common problem for G-protein coupled receptors (GPCRs).
Figure 5. Evidence for anatomical interaction between hindbrain NPY neurons and the LC.
(A) Double FISH using probes specific for npy and dbh show their close proximity in the LC. Boxed region in (A″) is magnified in a 50 μm thick maximum intensity projection in (B). (C) Tg(npy:kalta4);Tg(uas:nfsb-mcherry);Tg(dbh:EGFP) brains labeled using anti-DsRed and anti-EGFP antibodies. Boxed region in (C″) is magnified 25x in (D) and 63x in (D′). Maximum intensity projections 40 μm and 63 μm thick are shown in (D) and (D′). (E) Surface renderings of the boxed region in (D′). Boxed regions are magnified in (F–H). White asterisks show close proximity of NPY and LC neuron projections. (I–J) ISH using npy1r- and npy2rl-specific probes and immunostaining using an anti-EGFP antibody in Tg(dbh:EGFP) brains reveal close proximity of npy1r (I) and npy2rl (J) to dbh-expressing LC neurons. (I′) and (J′) show orthogonal views of the 24 μm and 25 μm thick maximum intensity projections shown in (I) and (J). a, anterior; p, posterior. Samples are 5 dpf brains. Scale bar: (A–C) 50 μm, (B,D) 10 μm, (D′) 7.5 μm, (E) 2.0 μm and (F–H) 0.5 μm. See also Movie S2.
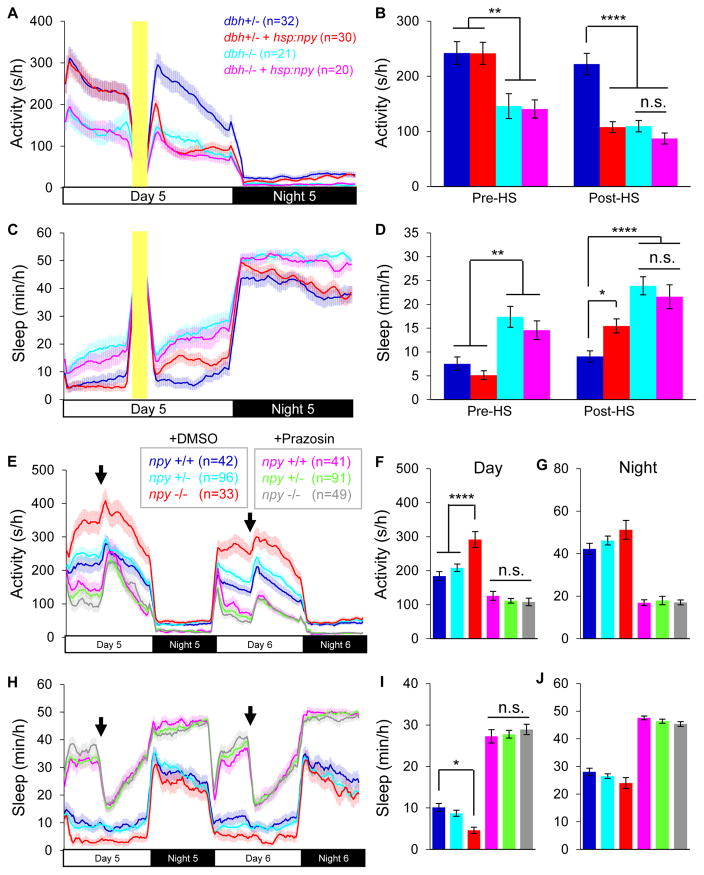
Second, we found that the sedating effects of NPY overexpression and loss of NE signaling are not additive. We made this observation by overexpressing NPY in larvae that lack NE synthesis due to mutation of dopamine beta hydroxylase (dbh) [56], or that lack NE signaling due to treatment with the α-1-adrenergic receptor antagonist prazosin. Both genetic and pharmacological inhibition of NE signaling increase sleep in zebrafish [56]. If NPY promotes sleep by inhibiting NE signaling, then overexpression of NPY should not further increase sleep in dbh−/− larvae or in WT larvae treated with prazosin. Alternatively, if NPY promotes sleep via a NE-independent mechanism, then the combined effects of NPY overexpression and loss of NE signaling on sleep should be additive. Because the behavior of dbh+/− animals is indistinguishable from that of their dbh+/+ siblings [56], we compared dbh+/− and dbh−/− siblings to reduce the number of comparisons in each experiment, and thus increase the number of animals per condition. Prior to heat shock-induced NPY overexpression, dbh−/− larvae were 40% less active and slept >100% more than their dbh+/− siblings for both Tg(hsp:npy) animals and their non-transgenic siblings (Figures 6A–6D) (P<0.01, two-way ANOVA, Holm-Sidak test). NPY overexpression decreased locomotor activity by 54% and increased sleep by 60% in Tg(hsp:npy);dbh+/− animals compared to dbh+/− siblings (Figures 6A–6D) (P<0.0001 and P<0.05, Two-way ANOVA, Holm-Sidak test). However, overexpression of NPY did not further affect the sleep/wake behavior of dbh−/− animals, as activity and sleep amounts were indistinguishable for Tg(hsp:npy);dbh−/− and dbh−/− animals (Figures 6A–6D). We obtained similar results for NPY overexpression in prazosin-treated animals compared to DMSO vehicle-treated controls (Figures S6A–S6D). To confirm that the failure of NPY overexpression to enhance sleep in dbh−/− or prazosin-treated animals is not due to a ceiling effect for sleep, we found that treatment with melatonin, an alternative sedative, enhanced sleep induced by overexpression of NPY (Figures S7A–S7D) or prazosin (Figures S7E–S7H).
Figure 6. Functional evidence that NPY promotes sleep by inhibiting NE signaling.
(A–D) Tg(hsp:npy);dbh−/− and dbh−/− animals were less active (A,B) and slept more (C,D) than dbh+/− siblings during the day before and after heat shock. Tg(hsp:npy);dbh+/− animals were less active and slept more than dbh+/− siblings during the day after heat shock. NPY overexpression in Tg(hsp:npy);dbh−/− animals did not further decrease locomotor activity or increase sleep compared to dbh−/− siblings. Yellow bars indicate heat shock (HS). Pre-HS and Post-HS quantify data on day 5 before and after heat shock. (E–J) npy+/+, npy+/− and npy−/− siblings were treated with either DMSO or prazosin. DMSO-treated npy−/− animals were more active (E,F) and slept less (H,I) than their DMSO-treated npy+/− and npy+/+ siblings during the day. Prazosin decreased activity (E,F) and increased sleep (H,I) to a similar extent for npy−/−, npy+/− and npy+/+ siblings. Arrows indicate behavioral artifacts due to addition of water. Mean ± SEM for 2 (A–D) or 4 (E–J) experiments is shown. n=number of animals. n.s.=not significant, *P<0.05; **P<0.01; ****P<0.0001 by two-way ANOVA, with Holm-Sidak test. See also Figures S6, S7 and Table S2.
Third, we found that the increased locomotor activity and reduced sleep observed in npy−/− animals compared to their npy+/+ siblings was abolished by treatment with prazosin. We made this observation by treating npy+/+, npy+/− and npy−/− larvae with either DMSO or prazosin. If NPY promotes sleep by inhibiting NE signaling, then loss of NPY should not affect prasozin-induced sleep. Alternatively, if NPY promotes sleep via a NE-independent mechanism, then loss of NPY should affect sleep amount in prazosin-treated animals. Consistent with the former possibility, we found that prazosin decreased activity and increased sleep, and this phenotype was indistinguishable for npy+/+, npy+/− and npy−/− siblings (Figures 6E–6J).
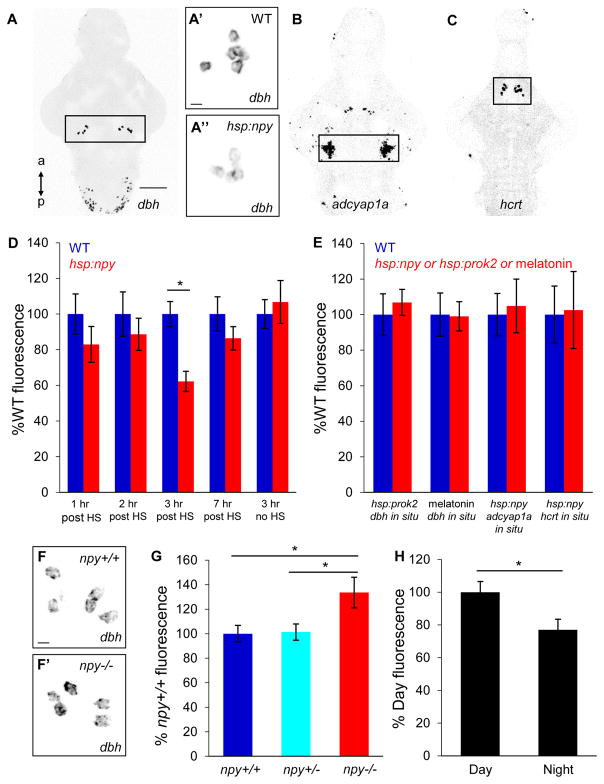
Fourth, we found that NPY regulates dbh expression in the LC. NPY overexpression decreased dbh mRNA in the LC by 38% at 3 hours post-heat shock in Tg(hsp:npy) animals compared to WT siblings (P<0.05, two-tailed Student’s t test) (Figures 7A and 7D). This time point coincides with the maximal effect of NPY overexpression on locomotor activity and sleep (Figures 1A and 1C), suggesting that NPY overexpression-induced sleep may result from reduced dbh expression, and thus reduced NE levels. However, effects of NPY overexpression on behavior begin within the first hour after heat shock, and we only observed a trend of decreased dbh mRNA at 1 and 2 hours post-heat shock that was not statistically significant (Figure 7D). These observations suggest that reduced dbh expression may not be the primary cause of NPY overexpression-induced sleep, but may rather be a secondary effect that maintains NPY-induced sleep, perhaps resulting from decreased LC neuron activity. We also tested whether NPY overexpression affects the level of tyrosine hydroxylase (th), which acts upstream of dbh in the NE synthesis pathway. We found that NPY overexpression did not significantly change th mRNA expression in the LC at 1, 2 or 3 hours post-heat shock (data not shown). Reduced dbh expression was not simply a consequence of increased sleep, as dbh mRNA level was unaffected following overexpression of the sleep-promoting neuropeptide prokineticin 2 (Prok2) [74] (Figure 7E) or treatment with the sedative melatonin (Figure 7E). The interaction between NPY and dbh appears to be specific, as NPY overexpression did not affect expression of other genes involved in promoting arousal, including the neuropeptides hypocretin (hcrt) [28, 56] or adenylate cyclase activating polypeptide 1a (adcyap1a) (Singh and Prober, unpublished) (Figures 7B, 7C and 7E). These results indicate that overexpression of NPY selectively decreases the level of dbh mRNA in the LC, presumably resulting in decreased NE levels and thus increased sleep. In support of this finding, we observed that dbh mRNA level was 33% higher in the LC of npy−/− animals compared to their npy+/− and npy+/+ siblings during the day (Figures 7F and 7G) (P<0.05, one-way ANOVA, Holm-Sidak test). Moreover, dbh mRNA level in the LC of WT animals was 25% lower at night compared to the day (P<0.05, two-tailed Student’s t test) (Figures 7H). This result demonstrates a correlation between the wake circadian phase of this diurnal species and the level of dbh mRNA in the LC, and suggests that changes in NE levels contribute to the regulation of normal sleep/wake states. Taken together, these results are consistent with a model in which NPY promotes sleep by inhibiting NE signaling.
Figure 7. NPY signaling affects dbh mRNA level in the LC.
(A) ISH showing dbh expression in the LC (boxed) and medulla oblongata. dbh mRNA levels were lower in Tg(hsp:npy) animals (A″) compared to WT siblings (A′) after heat shock. ISH using probes specific for adcyap1a (B) and hcrt (C). Boxed regions in (A–C) are quantified in (D,E). (D) dbh mRNA level in the LC is lower in Tg(hsp:npy) animals than WT siblings at 3 hours post-HS, but there is no significant difference at 1, 2, or 7 hours post-HS. (E) Overexpression of Prok2 or treatment with 20 μM melatonin had no effect on dbh expression. NPY overexpression did not affect adcyap1a or hcrt expression. (F–F′) dbh mRNA level in the LC was higher in npy−/− animals (F′) compared to npy+/+ siblings (F). (G) Quantification of dbh mRNA level in the LC of npy−/− animals and sibling controls. (H) dbh mRNA level in the LC of WT animals was lower at night than the day. Mean ± SEM fluorescence intensity from 8–12 brains for each condition is shown. *P<0.05 by two-tailed Student’s t test (D,H) or by one-way ANOVA with Holm-Sidak test (G). a, anterior; p, posterior. Samples are 5 dpf brains. Scale bar: (A,B,C) 100 μm; (A′,A″,F,F′) 10 μm.
DISCUSSION
NPY has been shown to affect sleep in mammals, but its role in sleep has been unclear. Infusion of NPY in rodents has been reported to increase [39–45] or decrease [48–50] sleep. These opposite effects may be due to different sites of injection or dosage, or the use of in vitro synthesized NPY that may lack modifications present on endogenously produced peptide. These studies are also confounded by other functions of NPY. For example, experiments in rats found the wake-promoting effects of NPY to be associated with feeding behaviors [48–50], and NPY can induce hypothermia [75] and increase social interactions [76], which may affect sleep. In agreement with some rodent studies, intravenous NPY injection promoted sleep in both healthy [46] and depressed [47] humans. Reduced NPY was observed in humans with major depression who report sleep disturbances [77] and in humans with primary insomnia [78], consistent with a sleep-promoting role for NPY. Reduced NPY was also found in individuals with post-traumatic stress disorder (PTSD) [79] and could contribute to the insomnia and fragmented sleep experienced by these patients. npy-expressing neurons are also implicated in mammalian sleep. For example, GABAergic cortical interneurons co-expressing neuronal nitric oxide synthase (nnos) and npy express c-fos, a marker of neuronal activity, during sleep in rodents [80]. Furthermore, extracellular single-unit activity in the basal forebrain of anaesthetized rats showed increased firing of npy-expressing neurons during slow wave sleep [81].
To address the role of endogenous NPY in sleep, we performed genetic gain- and loss-of-function studies using zebrafish larvae. These studies are performed before the onset of feeding, when larvae receive nutrients from the yolk sac [82], and before the onset of social interactions [83]. Furthermore, because zebrafish are poikilothermic, thermoregulation is unlikely to be a factor in studies of zebrafish sleep. Thus, zebrafish larvae allow the role of NPY in sleep to be addressed without complications of mammalian models. We found that overexpression of NPY suppresses locomotor activity and increases sleep during the day and night, whereas npy mutant zebrafish are more active and sleep less during the day. Analysis of sleep architecture revealed that NPY overexpression results in shorter wake bouts, whereas npy mutants have longer wake bouts, suggesting that NPY regulates consolidation of the wake state. Consistent with this phenotype, ablation of npy-expressing neurons resulted in decreased sleep during the day, again due to longer wake bouts. The daytime specificity of the loss-of-function phenotype could be explained by the presence of redundant sleep-promoting systems at night, the primary sleep phase of zebrafish. Consistent with our observations, overexpression in Drosophila of neuropeptide F (NPF), a Drosophila homolog of NPY, or its receptor NPFR1, promotes sleep [84], although stimulation of NPF neurons was recently shown to promote wakefulness and feeding [85]. This discrepancy could arise from differences in nutritional status [85]. The Drosophila short neuropeptide F (sNPF) is also thought to promote sleep [86] and has been referred to as an NPY ortholog, but is more likely an ortholog of vertebrate RFamide peptides [87]. In C. elegans, locomotor quiescence during lethargus is abolished in mutants lacking the receptor npr-1 and reduced in mutants lacking the npr-1 ligands flp-18 and flp-21 [88]. npr-1 mutants are also more responsive to oxygen and pheromones, resulting in altered foraging and accelerated locomotion [89–91]. While NPR-1 is related to NPY receptors [92], FLP-18 and FLP-21 are more similar to RFamide peptides [87, 93]. Combined with our results, these studies establish NPY as a conserved sleep promoting neuropeptide, and the human studies described above suggest this function is conserved in humans.
npy is widely expressed in the mammalian brain, particularly in the hypothalamus, amygdala, LC and cerebral cortex [58, 59]. Similar to mammals, NPY is expressed in several discrete brain regions in zebrafish larvae. Because of this broad expression pattern, NPY could act via several known sleep/wake regulators. First, npy-expressing neurons innervate hcrt-expressing neurons, and NPY inhibits hcrt neurons in mouse brain slices [94]. Second, a hypothalamic population of npy-expressing neurons project to the histaminergic tuberomammillary nucleus in rodents [95]. Third, corticotropin releasing hormone (CRH) impairs sleep and enhances vigilance [96], and NPY enhances inhibitory synaptic transmission in crh-expressing neurons in amygdala brain slices [97]. Fourth, melatonin promotes sleep in diurnal vertebrates, including humans [98], and application of NPY to rat pineal explants increases melatonin production [99]. To determine whether any of these pathways underlie the sleep-promoting effects of NPY, we tested whether the NPY overexpression phenotype is blocked in zebrafish mutants in which these pathways are affected, but found this not to be the case. We also found that NPY overexpression increased sleep in WT and melatonin-treated animals to a similar extent. These observations suggest that NPY does not affect sleep by modulating these pathways.
In contrast to these negative results, we made several observations suggesting that NPY promotes sleep by inhibiting NE signaling. Pharmacological and genetic studies in mammals and zebrafish have shown that NE promotes arousal, and that inhibition of NE signaling increases sleep [56, 71, 72]. We found that overexpression of NPY did not enhance the increased sleep observed in dbh−/− animals and prazosin-treated WT animals, suggesting that NPY overexpression promotes sleep by inhibiting NE signaling. Consistent with this possibility, we found that prazosin treatment abolished the decreased sleep observed in npy mutants, suggesting that elevated NE signaling underlies the npy mutant phenotype. In support of these functional interactions, we found that NPY overexpression decreases the level of dbh mRNA in the LC, the primary source of NE in the brain [72], and thus likely reduces NE levels. We observed a trend of reduced dbh mRNA levels at 1 and 2 hours after induction of NPY overexpression, and a significant reduction at 3 hours post-heat shock. These observations suggest that reduced dbh expression may not be the primary cause of NPY overexpression-induced sleep, but rather may be a secondary effect that maintains NPY-induced sleep, perhaps resulting from decreased LC neuron activity. Consistent with this possibility, NPY can inhibit LC neurons in rodent brain slices [100]. However, the maximal effect of NPY overexpression on behavior occurred at ~3 hours post-heat shock, coinciding with a significant reduction in dbh expression in the LC, consistent with NPY directly promoting sleep by decreasing dbh expression, and thus NE production, in the LC. Moreover, we found that npy mutants have elevated dbh expression in the LC, presumably resulting in increased NE levels and increased arousal. It was recently shown that dbh expression undergoes a circadian oscillation in whole zebrafish larvae [101]. Consistent with this observation, we found that the level of dbh mRNA in the LC is lower at night compared to the day, suggesting that NE levels contribute to the diurnal sleep/wake cycle.
Consistent with an interaction between NPY and the LC, we identified a small population of npy-expressing neurons that is adjacent to, and appears to innervate, the LC. This observation contrasts with mammals, where npy and dbh are co-expressed in LC neurons [102, 103]. We were unable to detect expression of NPY receptors in LC neurons, suggesting that NPY may indirectly affect NE signaling. However, expression of GPCRs, the protein class of NPY receptors, is notoriously difficult to detect, and we thus cannot rule out the possibility that a NPY receptor is expressed in LC neurons. We did observe expression of npy1r and npy2rl in cells near the LC, suggesting the possibility of local indirect interactions between NPY neurons and the LC. Thus, while the anatomic interaction between the NPY and NE systems appears to differ in zebrafish and mammals, the functional relationship between the systems may be conserved. Taken together, these observations suggest that NPY could regulate sleep by directly affecting the firing of LC neurons and/or the level of NE. Alternately, the site of interaction between NPY and NE in sleep may lie in a network of neurons near the LC or elsewhere in the brain.
In both mammals and zebrafish, NE is necessary for the wake-promoting functions of Hcrt signaling and hcrt-expressing neurons [55, 56]. Here we provide evidence that NE signaling mediates the sedating effect of NPY, suggesting a central role for the NE system in neuropeptidergic regulation of sleep/wake states. While Hcrt and NPY have opposite effects on sleep via NE signaling, both neuropeptides promote feeding via neurons in the hypothalamus [38, 104], suggesting a segregation of neuronal circuits through which these neuropeptides regulate sleep and feeding. While an interaction between NPY and the LC has been shown to control stress responses in rodents [105], to our knowledge this is the first demonstration of an interaction between NPY and the NE system in the context of sleep.
Finally, we note that cerebrospinal fluid levels of NPY are reduced in individuals suffering from PTSD who have sleep disturbances [79], and treatment with prazosin reduces nightmares and improves sleep in these individuals [106]. Since we found that npy mutant zebrafish have elevated dbh expression, and presumably more NE, the reduced NPY observed in in PTSD might cause increased NE levels, thereby disrupting sleep. These observations suggest that NPY might be therapeutic for at least some aspects of PTSD.
In summary, our results identify NPY as a regulator of sleep/wake behaviors in zebrafish and suggest that NPY promotes sleep by inhibiting NE signaling. These results highlight a central role for NE signaling in regulating sleep, and suggest that modulation of NPY signaling may be a useful therapeutic approach for sleep disorders.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for reagents may be directed to, and will be fulfilled by, the Lead Contact David A. Prober (dprober@caltech.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Zebrafish experiments and husbandry followed standard protocols [107] in accordance with Caltech Institutional Animal Care and Use Committee guidelines. Larval zebrafish were studied before the onset of sexual differentiation and all behavioral experiments were performed using siblings with the same genetic background, differing only in the presence of a transgene, mutation of a specific gene, or treatment with drugs and appropriate vehicle controls. The age of animals used in each experiment is described in the manuscript, in each figure legend, and/or in the STAR Methods.
Transgenic and mutant zebrafish
Tg(hsp:npy) ct853Tg
Full-length zebrafish npy cDNA was isolated using 5′ and 3′ RACE (FirstChoice RLM-RACE, AM1700, Thermo Fisher Scientific) and the open reading frame was cloned downstream of the zebrafish hsp70c promoter [28] in a vector containing flanking I-SceI endonuclease recognition sites. The same zebrafish npy gene was cloned in a previous study [108], but the gene isolated in our study contains an arginine residue located C-terminal to the mature peptide domain that was reported as an alanine residue in the previous study [108]. The sequence reported in our study is the same as that reported by the zebrafish genome sequencing project (www.ensembl.org/Danio_rerio). The alanine residue described in the previous report [108] is therefore likely either a sequencing error or a polymorphism in the fish strain used. Stable transgenic lines were generated by injecting plasmids with I-SceI (R0694, New England Biolabs Inc.) into zebrafish embryos at the one-cell stage. Transgenic founders were identified by outcrossing potential founders, heat shocking progeny at 5 dpf, fixing animals 30 minutes after heat shock and performing ISH using an npy-specific probe. Tg(hsp:npy) fish were genotyped using the primers 5′-CCGCCACCATGAATCCA-3′ and 5′-GGTTTGTCCAAACTCATCAATGT-3′, which generate a 370 bp band. We generated two independent Tg(hsp:npy) stable transgenic lines that produced similar phenotypes, but all data shown in the paper are from the line that produced stronger phenotypes.
npy mutant ct811
npy mutant zebrafish were generated using the zinc finger nuclease method [64]. The mutant contains a 17 bp deletion (AGCCCGACAACCCGGGA) after nucleotide 94 of the open reading frame, resulting in a translational frame shift beginning at the fourth amino acid of the mature peptide domain. Mutant animals were genotyped using the primers 5′-ATAAATTGCGCATCAGCACA-3′ and 5′-TGAGGAAGAATTTGAGACTACGC-3′, which produce a 281 or 264 bp band for the WT or mutant allele, respectively. npy heterozygous mutants were outcrossed to the parental TLAB strain for four generations before use in behavioral experiments. Homozygous npy mutants are viable, fertile, lack obvious developmental defects and are morphologically indistinguishable from WT animals.
Tg(npy:kalta4) ct852Tg
We used bacterial artificial chromosome (BAC) recombineering [109] to insert an optimized version of the transcriptional activator Gal4 (KalTA4) [109] at the npy start codon of a BAC (zK50N10SP6; HUKGB735N1050Q, Source BioScience)) containing 288 kb of genomic sequence, including 145 kb upstream and 143 kb downstream of the npy gene. Primers of 70 nucleotides (pIndigoBAC_HA1_iTol2_F and pIndigoBAC_HA1_iTol2_R, Table S3) were used to amplify the long terminal repeats of the medaka Tol2 transposon to enable single-copy integration of the BAC into the zebrafish genome, using the plasmid pIndigoBAC-536 [109] as template. npy-specific primers were designed that contain 50 nucleotide homology arms around the npy start codon (positions −53 to −4 and +4 to +53) with ~20 nucleotide ends (Homology arm F and Homology arm R, Table S3 to amplify a KalTA4_kanamycin cassette from the plasmid pCS2+_kalta4_kanR [109]. These plasmids were a kind gift from Dr. Stefan Schulte-Merker. The modified BAC was purified using the Nucleobond BAC 100 kit (740579, Macherey-Nagel) and injected into zebrafish embryos at the one- or two-cell stage at a concentration of 50 ng/μL, along with tol2 transposase mRNA at a concentration of 50 ng/μL. Transgenic lines were identified by mating potential founders to WT TLAB fish, and progeny were genotyped using the primers 5′-CGCTATCATTTATAGATTTTTGCAC-3′ and 5′-AGTAGCGACACTCCCAGTTG-3′, which produce a 220 bp band in transgenic animals. Transgenic founders were crossed to the Tg(uas:nfsb-mcherry) line [67] and the strongest line was identified by fluorescence microscopy.
Other transgenic and mutant lines
The Tg(dbh:EGFP) transgenic line [110], dbh mutant [56], hcrtr mutant [27], hdc mutant [69], and aanat2 mutant [70] have been previously described. The crha and crhb mutants are unpublished (Singh and Prober unpublished).
METHOD DETAILS
Locomotor activity assay
At 4 dpf, individual larvae were placed into each well of a 96-well plate (7701–1651, GE Healthcare Life Sciences) containing 650 μL of E3 embryo medium (5 mM NaCl, 0.17 mM KCl, 0.33 mM CaCl2, 0.33 mM MgSO4, pH 7.4). Plates were sealed with an optical adhesive film (4311971, Applied Biosystems) to prevent evaporation, except in experiments where drugs were added. The sealing process introduces air bubbles in some wells, which are excluded from analysis. In experiments using transgenic animals, larvae were blindly assigned a position in the plate, and were genotyped after the behavioral experiment was completed. Locomotor activity was monitored using an automated videotracking system (Viewpoint Life Sciences) with a Dinion one-third inch monochrome camera (Dragonfly 2, Point Grey) fitted with a fixed-angle megapixel lens (M5018-MP, Computar) and infrared filter. For heat shock-induced overexpression experiments, larvae were heat shocked at 37°C for 1 hour starting at either 3 p.m. or 10 p.m. at 5 dpf. The movement of each larva was captured at 15 Hz and recorded using the quantization mode in 1-minute time bins. The 96-well plate and camera were housed inside a custom-modified Zebrabox (Viewpoint Life Sciences) that was continuously illuminated with infrared LEDs, and illuminated with white LEDs from 9 a.m. to 11 p.m., except as noted in constant light or constant dark experiments. The 96-well plate was housed in a chamber filled with recirculating water to maintain a constant temperature of 28.5°C. The parameters used for detection were: detection threshold, 15; burst, 29; freeze, 3, which were determined empirically. Data were processed using custom PERL and Matlab (The Mathworks, Inc.) scripts, and statistical tests were performed using Prism 6 (GraphPad).
A movement was defined as a pixel displacement between adjacent video frames preceded and followed by a period of inactivity of at least 67 ms (the limit of temporal resolution). Any one-minute period with no movement was defined as one minute of sleep based on arousal threshold changes [28]. A sleep bout was defined as a continuous string of sleep minutes. Average activity was defined as the average amount of activity in seconds/hour, including sleep bouts.
Arousal threshold assay
The arousal threshold assay was performed as described [56]. Animals were heat shocked at 5 dpf from 12 p.m. to 1 p.m, and taps of 14 different intensities were applied in a random order from 3 p.m. to 10 p.m. Thirty trials were performed at each stimulus intensity, with a 1-minute inter-trial interval. The background probability of movement was calculated by identifying for each genotype the fraction of larvae that moved 5 seconds prior to all stimuli delivered. This value was subtracted from the average response fraction value for each tap event. A response is defined as any movement that occurred within 1 second after a tap was delivered. Data was analyzed using Matlab (Mathworks, Inc.) and dose-response curves were constructed using the Variable Slope log(dose) response curve fitting module of Prism (Graphpad) and fitted using ordinary least squares. The effective tap power 50 (ETP50) was defined as the tapping intensity at which 50% of the maximum number of responding larvae occurs, based on the fitted curve.
Tapping experiments with a 5-minute inter-trial interval were performed using three tap intensities of 2.3, 3.0 and 4.0 arbitrary units to assess the response of awake and sleeping larvae to the stimuli. These stimulus intensities were chosen because they were lower than the ETP50 of animals of both genotypes. Animals were heat shocked at 5 dpf from 12 p.m. to 1 p.m., and thirty-three trials were performed at each stimulus intensity in a random order from 3:00 p.m. to 10:30 p.m. Behavioral responses were analyzed as described above. Three independent experiments for were performed for both 1-minute and 5-minute tapping assays, and one representative experiment for each is shown.
In situ hybridization (ISH)
Animals were fixed in 4% paraformaldehyde (PFA) in phosphate buffered saline (PBS) for 16 hours at room temperature. ISH was performed using digoxygenin (DIG) labeled antisense riboprobes (DIG RNA Labeling Kit, 11175025910, Sigma-Aldrich), followed by incubation with a sheep anti-digoxigenin-POD antibody (1:400; 11207733910, Sigma-Aldrich), and developed using the TSA Plus Fluorescein and Cyanine 3 System (NEL753001KT, PerkinElmer). Double-fluorescent ISH was performed using DIG- and fluorescein-labeled riboprobes (Fluorescein RNA Labeling kit, 11685619910, Sigma-Aldrich), and the TSA Plus Fluorescein and Cyanine 3 System (NEL753001KT, PerkinElmer) using a previously described protocol [28]. Probes specific for npy, dbh, adcyap1a, kalta4, npy1r, npy2r, npy2rl, npy4r, npy7r, npy8ar and npy8br were synthesized using standard protocols [111]. The npy probe was transcribed using a PCR product amplified from a zebrafish cDNA library using the primers Forward: 5′-CCACAGAGCAAGAATTCCAA-3′ and Reverse: 5′-CAGTCATTATTGTTCTCCTTTGC-3′, and then serially amplified with the same Forward primer and the Reverse Primer with a T7 promoter sequence added: 5′-TAATACGACTCACTATAGGGCAGTCATTATTGTTCTCCTTTGC-3′. The kalta4 probe was transcribed using the plasmid pCS2+_kalta4_kanR [109] as a template after linearization with BamH1 and using T7 RNA polymerase (10881767001, Sigma-Aldrich). A probe specific for dbh has been previously described [112]. Probes specific for adcyap1a, npy1r, npy2r, npy2rl, npy4r, npy7r, npy8ar and npy8br were generated as described for the npy-specific probe using the primers listed in Table S3.
Immunohistochemistry (IHC)
Samples were fixed in 4% PFA in PBS overnight at 4°C and then washed with 0.25% Triton X-100/PBS (PBTx). Brains were manually dissected and blocked for at least 1 hour in 2% goat serum/2% dimethyl sulfoxide (DMSO)/PBTx at room temperature or overnight at 4°C. Primary antibody incubations were performed in blocking solution overnight at 4°C using chicken anti-GFP (1:400, GFP-1020, Aves L abs, Inc.) and rabbit anti-DsRed (1:100, 632496, Clontech Laboratories, Inc.). Secondary antibody incubations were performed in blocking solution overnight at 4°C using Alexa Fluo r 488 goat anti-chicken (1:500, A-11039, Thermo Fisher Scientific) and Alexa Fluor 568 goat anti-rabbit (1:500, A-11011, Thermo Fisher Scientific) antibodies. Samples were mounted in 50% glycerol/PBS and imaged using a Zeiss LSM 780 confocal microscope with a 25× 0.8 NA water immersion objective (LD LCI Plan-Apochromat 25x/0.8 1mm Corr DIC M27). Images were processed using Fiji [113].
Z-brain registration
WT larvae were fixed at 6 dpf and ISH was performed using an npy-specific probe on dissected brains as described above, followed by IHC using mouse anti-t-ERK primary antibody (1:500, 4696, Cell Signaling Technology) and Alexa Fluor 488 goat anti-mouse secondary antibody (1:500, A32723, Thermo Fisher Scientific). Imaging was performed using a Zeiss 780 confocal microscope, using a 20× 1.0 NA water dipping objective (W Plan-Apochromat 20x/1.0 DIC CG=0.17 M27 75mm) and imaged at ~0.8/0.8/2 μm voxel size (x/y/z) using the Zeiss tiling function and the pairwise stitching function of Fiji [113]. Non-rigid image registration was performed using the Computational Morphometry Toolkit (CMTK, http://www.nitrc.org/projects/cmtk/) as previously described [60]. t-ERK staining was used to register to the t-ERK reference brain [60], which was then used to align npy ISH labeling. Registered brains were analyzed using the Z-Brain browser (MATLAB) [60] to identify anatomical regions expressing npy. Using Fiji, the registered brain showing npy expression was merged to the database ‘Anti-tERK_6dpf_MeanImageOf193Fish’ from ‘AnatomyLabel DatabaseDownsampled’ from the Z-Brain Downloads [60] to show the expression of npy relative to t-ERK in the reference 6 dpf zebrafish larva. The combined stack was converted into a movie and processed in Windows Movie Maker to add anatomical labels.
Image processing in Imaris and Fiji
Surface rendering to reconstruct projections of npy- and dbh-expressing neurons was performed using Imaris 9 (Bitplane). To perform surface rendering, we used the Volume function followed by the Normal Shading mode to add a depth effect to the 2-dimensional z-stack imaged using a 63× 1.4 NA oil immersion objective (Plan-Apochromat 63x/1.4 oil DIC M27), and then displayed the image in the 3-dimensional isometric view. We then used the Interactive Software Histogram to select a threshold that included as much of the neuronal projections as possible while excluding any background. Areas of overlap between projections from npy- and dbh-expressing neurons were magnified 4-fold and saved as TIFF images.
To identify the sources of overlapping projections, a 63x z-stack of npy-expressing and dbh-expressing neurons was converted to an 8-bit stack. Projections from a single npy-expressing neuron and a single dbh-expressing neuron were manually traced using the Simple Neurite Tracer plugin in Fiji. Tracings were then filled-in using the same plugin, with an exemplar npy-expressing neuron labeled magenta and an exemplar dbh-expressing neuron labeled green, and saved as individual z-stacks. These z-stacks were then merged with the original z-stack to so that the traced npy-expressing and dbh-expressing neurons were overlaid on the original images. As a result, the traced npy-expressing neuron appears magenta and the traced dbh-expressing neuron appears yellow. This merged image stack is shown in Movie S2.
TUNEL staining
Tg(npy:kalta4);Tg(uas:nfsb-mcherry) larvae were treated with DMSO or 10 mM MTZ for 18 hours starting at 3 dpf, and then were fixed in 4% PFA in PBS for 16 hours at 4°C, and subjected to a TUNEL Assay (In Situ Cell Death Detection Kit, 11684795910, Sigma-Aldrich) according to the manufacturer’s instructions.
Analysis and quantification of dbh expression using ISH
dbh ISH was performed by incubating fixed 5 dpf brains with a DIG-labeled dbh antisense riboprobe, followed by a sheep anti-digoxigenin-POD antibody (1:400; 11207733910, Sigma-Aldrich), and developed using the TSA Plus Cyanine 3 System (NEL753001KT, PerkinElmer). Samples were developed using the cyanine 3 substrate at 1:300 for 5 minutes to avoid saturation. Brains were imaged using a Zeiss LSM 780 confocal microscope using a 561 nm laser and a 25× 0.8 NA water immersion objective (LD LCI Plan-Apochromat 25×/0.8 1mm Corr DIC M27). To quantify dbh expression in Tg(hsp:npy) animals, larvae were heat shocked from 3 p.m. to 4 p.m. and samples were collected at the indicated times after heat shock. To quantify dbh expression in npy mutants, samples were collected at 4 p.m. Both experiments used siblings whose brains were processed for ISH in the same tube, imaged, quantified and then genotyped by PCR. To compare dbh expression levels during the day and night, day samples were collected at 4 p.m. and night samples were collected at 2 a.m. After fixation, a small nick was made in the forebrain of night samples to enable their identification at the end of the experiment. Day and night samples were then placed together in the same tube, processed for ISH, imaged and then quantified. Three independent experiments were performed and images of representative samples are shown. For quantification of dbh mRNA level, confocal z-stacks were obtained as described above. Using Fiji [113], each z-stack was converted into a maximum intensity projection, converted into 8-bit grayscale, and thresholded to select only the fluorescent ISH signal. This function was applied to all images in an experiment to determine a threshold level that was optimal for most images, and this threshold was then used for all images in an experiment. The Analyze-Set Measurements function was used to select Integrated Density as the measurement parameter and Limit to Threshold was selected to measure only the thresholded region. Fluorescent intensity was then measured by the Analyze-Measure function.
QUANTIFICATION AND STATISTICAL ANALYSIS
All line graphs show a 1 hour forward moving average plotted in 10 minute bins, except Figures S1B and S1E, which show data plotted in 10 minute bins. Line and bar graphs show mean ± standard error of the mean (SEM). In all statistical tests, the significance threshold was set to P<0.05. Parametric statistical tests were used because the data followed an approximately normal distribution. For behavioral experiments that compared two genotypes, statistical significance was assessed using a two-tailed Student’s t test. For npy mutant experiments, which compared animals of three different genotypes, one-way ANOVA followed by the Holm-Sidak correction for multiple comparisons was performed to test for significant pair-wise comparisons among all genotypes. The Holm-Sidak test was used to focus on significance but not confidence intervals. For experiments in which NPY was overexpressed in various mutant backgrounds or in which NPY overexpression was combined with drug treatments, statistical significance was assessed using two-way ANOVA followed by the Holm-Sidak correction for multiple comparisons. For experiments in which npy mutants were treated with drugs, statistical significance was assessed using two-way ANOVA followed by Holm-Sidak correction for multiple comparisons. For quantification of ISH data, statistical significance was assessed using a two-tailed Student’s t test for experiments that compared two samples, and one-way ANOVA followed by the Holm-Sidak correction for multiple comparisons for experiments that compared three or more samples. Behavioral data was processed using Matlab (MathWorks), graphs were generated using Excel (Microsoft), and statistical analyses were performed using Prism 6 (Graphpad). The number of animals and statistical test used are stated in each figure or figure legend.
DATA AND SOFTWARE AVAILABILITY
Custom PERL and MATLAB code used for zebrafish behavioral analysis is available upon request.
Supplementary Material
In situ hybridization with an npy-specific probe was performed on 6 dpf larval zebrafish brains, followed by immunostaining for t-ERK. The t-ERK staining was then used to register npy expression to the Z-brain reference brain. Anatomical domains of npy expression were then added using Z-Brain browser annotations. Anterior is to the left and the movie starts from the ventral surface of the brain. The movie does not show the expression of npy in the olfactory bulb or retina.
Projections from hindbrain npy-expressing neurons (blue) and dbh-expressing LC neurons (yellow) form close contacts. An exemplar npy-expressing neuron (highlighted magenta) appears to contact a single dbh-expressing neuron (highlighted green) at least twice (white, indicated by asterisk, magnified 4-fold in insets). Scale bar: 10 μm.
HIGHLIGHTS.
A genetic screen in zebrafish shows that overexpression of NPY promotes sleep
Mutation of npy or ablation of npy-expressing neurons results in decreased sleep
NPY regulates sleep primarily by modulating the length of wake bouts
NPY promotes sleep by inhibiting noradrenergic signaling
Acknowledgments
We thank Daisy Chilin, Alex Mack Cruz, Axel Dominguez and Kenna Molinder for animal husbandry assistance; Viveca Sapin and Uyen Pham for genotyping assistance; Owen Randlett, Ulrich Herget, Caroline Wee, and Marcus Ghosh for assistance with Z-brain registration; and Grigorios Oikonomou for comments on the manuscript. This work was supported by grants from the NIH (DAP: NS070911, NS101665, NS095824, NS101158), the Mallinckrodt, Rita Allen and Brain and Behavior Research Foundations (DAP), and a UCL Excellence Fellowship and European Research Council Starting Grant (JR). We declare no conflicts of interest.
Footnotes
AUTHOR CONTRIBUTIONS
DAP and JR performed the genetic screen. CS and DAP conceptualized and designed the experiments, and generated reagents. CS performed the experiments and analyzed the data. CS and DAP wrote the paper with assistance from JR. DAP supervised the project.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Richter C, Woods IG, Schier AF. Neuropeptidergic control of sleep and wakefulness. Annu Rev Neurosci. 2014;37:503–531. doi: 10.1146/annurev-neuro-062111-150447. [DOI] [PubMed] [Google Scholar]
- 2.Adamantidis AR, Zhang F, Aravanis AM, Deisseroth K, de Lecea L. Neural substrates of awakening probed with optogenetic control of hypocretin neurons. Nature. 2007;450:420–424. doi: 10.1038/nature06310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Lecea L, Kilduff TS, Peyron C, Gao X, Foye PE, Danielson PE, Fukuhara C, Battenberg EL, Gautvik VT, Bartlett FS, 2nd, et al. The hypocretins: hypothalamus-specific peptides with neuroexcitatory activity. Proc Natl Acad Sci U S A. 1998;95:322–327. doi: 10.1073/pnas.95.1.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin L, Faraco J, Li R, Kadotani H, Rogers W, Lin X, Qiu X, de Jong PJ, Nishino S, Mignot E. The sleep disorder canine narcolepsy is caused by a mutation in the hypocretin (orexin) receptor 2 gene. Cell. 1999;98:365–376. doi: 10.1016/s0092-8674(00)81965-0. [DOI] [PubMed] [Google Scholar]
- 5.Saper CB, Fuller PM, Pedersen NP, Lu J, Scammell TE. Sleep state switching. Neuron. 2010;68:1023–1042. doi: 10.1016/j.neuron.2010.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sherin JE, Elmquist JK, Torrealba F, Saper CB. Innervation of histaminergic tuberomammillary neurons by GABAergic and galaninergic neurons in the ventrolateral preoptic nucleus of the rat. J Neurosci. 1998;18:4705–4721. doi: 10.1523/JNEUROSCI.18-12-04705.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sherin JE, Shiromani PJ, McCarley RW, Saper CB. Activation of ventrolateral preoptic neurons during sleep. Science. 1996;271:216–219. doi: 10.1126/science.271.5246.216. [DOI] [PubMed] [Google Scholar]
- 8.Woods IG, Schoppik D, Shi VJ, Zimmerman S, Coleman HA, Greenwood J, Soucy ER, Schier AF. Neuropeptidergic signaling partitions arousal behaviors in zebrafish. J Neurosci. 2014;34:3142–3160. doi: 10.1523/JNEUROSCI.3529-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Konadhode RR, Pelluru D, Blanco-Centurion C, Zayachkivsky A, Liu M, Uhde T, Glen WB, Jr, van den Pol AN, Mulholland PJ, Shiromani PJ. Optogenetic stimulation of MCH neurons increases sleep. J Neurosci. 2013;33:10257–10263. doi: 10.1523/JNEUROSCI.1225-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsunematsu T, Ueno T, Tabuchi S, Inutsuka A, Tanaka KF, Hasuwa H, Kilduff TS, Terao A, Yamanaka A. Optogenetic manipulation of activity and temporally controlled cell-specific ablation reveal a role for MCH neurons in sleep/wake regulation. J Neurosci. 2014;34:6896–6909. doi: 10.1523/JNEUROSCI.5344-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Verret L, Goutagny R, Fort P, Cagnon L, Salvert D, Leger L, Boissard R, Salin P, Peyron C, Luppi PH. A role of melanin-concentrating hormone producing neurons in the central regulation of paradoxical sleep. BMC Neurosci. 2003;4:19. doi: 10.1186/1471-2202-4-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Willie JT, Sinton CM, Maratos-Flier E, Yanagisawa M. Abnormal response of melanin-concentrating hormone deficient mice to fasting: hyperactivity and rapid eye movement sleep suppression. Neuroscience. 2008;156:819–829. doi: 10.1016/j.neuroscience.2008.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Funato H, Miyoshi C, Fujiyama T, Kanda T, Sato M, Wang Z, Ma J, Nakane S, Tomita J, Ikkyu A, et al. Forward-genetics analysis of sleep in randomly mutagenized mice. Nature. 2016;539:378–383. doi: 10.1038/nature20142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shaw PJ, Cirelli C, Greenspan RJ, Tononi G. Correlates of sleep and waking in Drosophila melanogaster. Science. 2000;287:1834–1837. doi: 10.1126/science.287.5459.1834. [DOI] [PubMed] [Google Scholar]
- 15.Hendricks JC, Finn SM, Panckeri KA, Chavkin J, Williams JA, Sehgal A, Pack AI. Rest in Drosophila is a sleep-like state. Neuron. 2000;25:129–138. doi: 10.1016/s0896-6273(00)80877-6. [DOI] [PubMed] [Google Scholar]
- 16.Cavanaugh DJ, Geratowski JD, Wooltorton JR, Spaethling JM, Hector CE, Zheng X, Johnson EC, Eberwine JH, Sehgal A. Identification of a circadian output circuit for rest:activity rhythms in Drosophila. Cell. 2014;157:689–701. doi: 10.1016/j.cell.2014.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shi M, Yue Z, Kuryatov A, Lindstrom JM, Sehgal A. Identification of Redeye, a new sleep-regulating protein whose expression is modulated by sleep amount. Elife. 2014;3:e01473. doi: 10.7554/eLife.01473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nall AH, Sehgal A. Small-molecule screen in adult Drosophila identifies VMAT as a regulator of sleep. J Neurosci. 2013;33:8534–8540. doi: 10.1523/JNEUROSCI.0253-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koh K, Joiner WJ, Wu MN, Yue Z, Smith CJ, Sehgal A. Identification of SLEEPLESS, a sleep-promoting factor. Science. 2008;321:372–376. doi: 10.1126/science.1155942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sathyanarayanan S, Zheng X, Kumar S, Chen CH, Chen D, Hay B, Sehgal A. Identification of novel genes involved in light-dependent CRY degradation through a genome-wide RNAi screen. Genes Dev. 2008;22:1522–1533. doi: 10.1101/gad.1652308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu MN, Koh K, Yue Z, Joiner WJ, Sehgal A. A genetic screen for sleep and circadian mutants reveals mechanisms underlying regulation of sleep in Drosophila. Sleep. 2008;31:465–472. doi: 10.1093/sleep/31.4.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cirelli C, Bushey D, Hill S, Huber R, Kreber R, Ganetzky B, Tononi G. Reduced sleep in Drosophila Shaker mutants. Nature. 2005;434:1087–1092. doi: 10.1038/nature03486. [DOI] [PubMed] [Google Scholar]
- 23.Raizen DM, Zimmerman JE, Maycock MH, Ta UD, You YJ, Sundaram MV, Pack AI. Lethargus is a Caenorhabditis elegans sleep-like state. Nature. 2008;451:569–572. doi: 10.1038/nature06535. [DOI] [PubMed] [Google Scholar]
- 24.Van Buskirk C, Sternberg PW. Epidermal growth factor signaling induces behavioral quiescence in Caenorhabditis elegans. Nat Neurosci. 2007;10:1300–1307. doi: 10.1038/nn1981. [DOI] [PubMed] [Google Scholar]
- 25.Iannacone MJ, Beets I, Lopes LE, Churgin MA, Fang-Yen C, Nelson MD, Schoofs L, Raizen DM. The RFamide receptor DMSR-1 regulates stress-induced sleep in C. elegans. Elife. 2017;6 doi: 10.7554/eLife.19837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhdanova IV, Wang SY, Leclair OU, Danilova NP. Melatonin promotes sleep-like state in zebrafish. Brain Res. 2001;903:263–268. doi: 10.1016/s0006-8993(01)02444-1. [DOI] [PubMed] [Google Scholar]
- 27.Yokogawa T, Marin W, Faraco J, Pezeron G, Appelbaum L, Zhang J, Rosa F, Mourrain P, Mignot E. Characterization of sleep in zebrafish and insomnia in hypocretin receptor mutants. PLoS Biol. 2007;5:e277. doi: 10.1371/journal.pbio.0050277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prober DA, Rihel J, Onah AA, Sung RJ, Schier AF. Hypocretin/orexin overexpression induces an insomnia-like phenotype in zebrafish. J Neurosci. 2006;26:13400–13410. doi: 10.1523/JNEUROSCI.4332-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chiu CN, Rihel J, Lee DA, Singh C, Mosser EA, Chen S, Sapin V, Pham U, Engle J, Niles BJ, et al. A Zebrafish Genetic Screen Identifies Neuromedin U as a Regulator of Sleep/Wake States. Neuron. 2016;89:842–856. doi: 10.1016/j.neuron.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rihel J, Prober DA, Arvanites A, Lam K, Zimmerman S, Jang S, Haggarty SJ, Kokel D, Rubin LL, Peterson RT, et al. Zebrafish behavioral profiling links drugs to biological targets and rest/wake regulation. Science. 2010;327:348–351. doi: 10.1126/science.1183090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Faraco JH, Appelbaum L, Marin W, Gaus SE, Mourrain P, Mignot E. Regulation of hypocretin (orexin) expression in embryonic zebrafish. J Biol Chem. 2006;281:29753–29761. doi: 10.1074/jbc.M605811200. [DOI] [PubMed] [Google Scholar]
- 32.Renier C, Faraco JH, Bourgin P, Motley T, Bonaventure P, Rosa F, Mignot E. Genomic and functional conservation of sedative-hypnotic targets in the zebrafish. Pharmacogenet Genomics. 2007;17:237–253. doi: 10.1097/FPC.0b013e3280119d62. [DOI] [PubMed] [Google Scholar]
- 33.Kaslin J, Nystedt JM, Ostergard M, Peitsaro N, Panula P. The orexin/hypocretin system in zebrafish is connected to the aminergic and cholinergic systems. J Neurosci. 2004;24:2678–2689. doi: 10.1523/JNEUROSCI.4908-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alfalah M, Michel MC. Neuropeptide Y and related peptides. Berlin; New York: Springer; 2004. [Google Scholar]
- 35.Beck B. Neuropeptide Y in normal eating and in genetic and dietary-induced obesity. Philos Trans R Soc Lond B Biol Sci. 2006;361:1159–1185. doi: 10.1098/rstb.2006.1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sohn JW, Elmquist JK, Williams KW. Neuronal circuits that regulate feeding behavior and metabolism. Trends Neurosci. 2013;36:504–512. doi: 10.1016/j.tins.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yulyaningsih E, Zhang L, Herzog H, Sainsbury A. NPY receptors as potential targets for anti-obesity drug development. Br J Pharmacol. 2011;163:1170–1202. doi: 10.1111/j.1476-5381.2011.01363.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zeltser LM, Seeley RJ, Tschop MH. Synaptic plasticity in neuronal circuits regulating energy balance. Nat Neurosci. 2012;15:1336–1342. doi: 10.1038/nn.3219. [DOI] [PubMed] [Google Scholar]
- 39.Akanmu MA, Ukponmwan OE, Katayama Y, Honda K. Neuropeptide-Y Y2-receptor agonist, PYY3-36 promotes non-rapid eye movement sleep in rat. Neuroscience research. 2006;54:165–170. doi: 10.1016/j.neures.2005.11.006. [DOI] [PubMed] [Google Scholar]
- 40.Fuxe K, Agnati LF, Harfstrand A, Zini I, Tatemoto K, Pich EM, Hokfelt T, Mutt V, Terenius L. Central administration of neuropeptide Y induces hypotension bradypnea and EEG synchronization in the rat. Acta Physiol Scand. 1983;118:189–192. doi: 10.1111/j.1748-1716.1983.tb07261.x. [DOI] [PubMed] [Google Scholar]
- 41.Heilig M, Murison R. Intracerebroventricular neuropeptide Y suppresses open field and home cage activity in the rat. Regul Pept. 1987;19:221–231. doi: 10.1016/0167-0115(87)90278-3. [DOI] [PubMed] [Google Scholar]
- 42.Jolicoeur FB, Michaud JN, Rivest R, Menard D, Gaudin D, Fournier A, St-Pierre S. Neurobehavioral profile of neuropeptide Y. Brain Res Bull. 1991;26:265–268. doi: 10.1016/0361-9230(91)90237-e. [DOI] [PubMed] [Google Scholar]
- 43.Naveilhan P, Canals JM, Valjakka A, Vartiainen J, Arenas E, Ernfors P. Neuropeptide Y alters sedation through a hypothalamic Y1-mediated mechanism. Eur J Neurosci. 2001;13:2241–2246. doi: 10.1046/j.0953-816x.2001.01601.x. [DOI] [PubMed] [Google Scholar]
- 44.Toth A, Hajnik T, Zaborszky L, Detari L. Effect of basal forebrain neuropeptide Y administration on sleep and spontaneous behavior in freely moving rats. Brain Res Bull. 2007;72:293–301. doi: 10.1016/j.brainresbull.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 45.Zini I, Merlo Pich E, Fuxe K, Lenzi PL, Agnati LF, Harfstrand A, Mutt V, Tatemoto K, Moscara M. Actions of centrally administered neuropeptide Y on EEG activity in different rat strains and in different phases of their circadian cycle. Acta Physiol Scand. 1984;122:71–77. doi: 10.1111/j.1748-1716.1984.tb07483.x. [DOI] [PubMed] [Google Scholar]
- 46.Antonijevic IA, Murck H, Bohlhalter S, Frieboes RM, Holsboer F, Steiger A. Neuropeptide Y promotes sleep and inhibits ACTH and cortisol release in young men. Neuropharmacology. 2000;39:1474–1481. doi: 10.1016/s0028-3908(00)00057-5. [DOI] [PubMed] [Google Scholar]
- 47.Held K, Antonijevic I, Murck H, Kuenzel H, Steiger A. Neuropeptide Y (NPY) shortens sleep latency but does not suppress ACTH and cortisol in depressed patients and normal controls. Psychoneuroendocrinology. 2006;31:100–107. doi: 10.1016/j.psyneuen.2005.05.015. [DOI] [PubMed] [Google Scholar]
- 48.Stanley BG, Leibowitz SF. Neuropeptide Y: stimulation of feeding and drinking by injection into the paraventricular nucleus. Life Sci. 1984;35:2635–2642. doi: 10.1016/0024-3205(84)90032-8. [DOI] [PubMed] [Google Scholar]
- 49.Szentirmai E, Krueger JM. Central administration of neuropeptide Y induces wakefulness in rats. Am J Physiol Regul Integr Comp Physiol. 2006;291:R473–480. doi: 10.1152/ajpregu.00919.2005. [DOI] [PubMed] [Google Scholar]
- 50.Ushimura A, Tsuji T, Tanaka S, Kogo M, Yamamoto T. Neuropeptide-Y modulates eating patterns and masticatory muscle activity in rats. Behav Brain Res. 2015;278:520–526. doi: 10.1016/j.bbr.2014.10.031. [DOI] [PubMed] [Google Scholar]
- 51.Brown JA, Woodworth HL, Leinninger GM. To ingest or rest? Specialized roles of lateral hypothalamic area neurons in coordinating energy balance. Frontiers in systems neuroscience. 2015;9:9. doi: 10.3389/fnsys.2015.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bannon AW, Seda J, Carmouche M, Francis JM, Norman MH, Karbon B, McCaleb ML. Behavioral characterization of neuropeptide Y knockout mice. Brain Res. 2000;868:79–87. doi: 10.1016/s0006-8993(00)02285-x. [DOI] [PubMed] [Google Scholar]
- 53.Reichmann F, Wegerer V, Jain P, Mayerhofer R, Hassan AM, Frohlich EE, Bock E, Pritz E, Herzog H, Holzer P, et al. Environmental enrichment induces behavioural disturbances in neuropeptide Y knockout mice. Sci Rep. 2016;6:28182. doi: 10.1038/srep28182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Patel HR, Qi Y, Hawkins EJ, Hileman SM, Elmquist JK, Imai Y, Ahima RS. Neuropeptide Y deficiency attenuates responses to fasting and high-fat diet in obesity-prone mice. Diabetes. 2006;55:3091–3098. doi: 10.2337/db05-0624. [DOI] [PubMed] [Google Scholar]
- 55.Carter ME, Brill J, Bonnavion P, Huguenard JR, Huerta R, de Lecea L. Mechanism for Hypocretin-mediated sleep-to-wake transitions. Proc Natl Acad Sci U S A. 2012;109:E2635–2644. doi: 10.1073/pnas.1202526109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Singh C, Oikonomou G, Prober DA. Norepinephrine is required to promote wakefulness and for hypocretin-induced arousal in zebrafish. Elife. 2015;4:e07000. doi: 10.7554/eLife.07000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lamesch P, Li N, Milstein S, Fan C, Hao T, Szabo G, Hu Z, Venkatesan K, Bethel G, Martin P, et al. hORFeome v3.1: a resource of human open reading frames representing over 10,000 human genes. Genomics. 2007;89:307–315. doi: 10.1016/j.ygeno.2006.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chronwall BM, DiMaggio DA, Massari VJ, Pickel VM, Ruggiero DA, O’Donohue TL. The anatomy of neuropeptide-Y-containing neurons in rat brain. Neuroscience. 1985;15:1159–1181. doi: 10.1016/0306-4522(85)90260-x. [DOI] [PubMed] [Google Scholar]
- 59.Dumont Y, Martel JC, Fournier A, St-Pierre S, Quirion R. Neuropeptide Y and neuropeptide Y receptor subtypes in brain and peripheral tissues. Prog Neurobiol. 1992;38:125–167. doi: 10.1016/0301-0082(92)90038-g. [DOI] [PubMed] [Google Scholar]
- 60.Randlett O, Wee CL, Naumann EA, Nnaemeka O, Schoppik D, Fitzgerald JE, Portugues R, Lacoste AM, Riegler C, Engert F, et al. Whole-brain activity mapping onto a zebrafish brain atlas. Nat Methods. 2015;12:1039–1046. doi: 10.1038/nmeth.3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cavanaugh DJ, Vigderman AS, Dean T, Garbe DS, Sehgal A. The Drosophila Circadian Clock Gates Sleep through Time-of-Day Dependent Modulation of Sleep-Promoting Neurons. Sleep. 2016;39:345–356. doi: 10.5665/sleep.5442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mrosovsky N. Masking: history, definitions, and measurement. Chronobiol Int. 1999;16:415–429. doi: 10.3109/07420529908998717. [DOI] [PubMed] [Google Scholar]
- 63.Siegel JM. Sleep viewed as a state of adaptive inactivity. Nat Rev Neurosci. 2009;10:747–753. doi: 10.1038/nrn2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen S, Oikonomou G, Chiu CN, Niles BJ, Liu J, Lee DA, Antoshechkin I, Prober DA. A large-scale in vivo analysis reveals that TALENs are significantly more mutagenic than ZFNs generated using context-dependent assembly. Nucleic Acids Res. 2013;41:2769–2778. doi: 10.1093/nar/gks1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Albers HE, Ferris CF. Neuropeptide Y: role in light-dark cycle entrainment of hamster circadian rhythms. Neurosci Lett. 1984;50:163–168. doi: 10.1016/0304-3940(84)90480-4. [DOI] [PubMed] [Google Scholar]
- 66.Huhman KL, Gillespie CF, Marvel CL, Albers HE. Neuropeptide Y phase shifts circadian rhythms in vivo via a Y2 receptor. Neuroreport. 1996;7:1249–1252. doi: 10.1097/00001756-199605170-00005. [DOI] [PubMed] [Google Scholar]
- 67.Davison JM, Akitake CM, Goll MG, Rhee JM, Gosse N, Baier H, Halpern ME, Leach SD, Parsons MJ. Transactivation from Gal4-VP16 transgenic insertions for tissue-specific cell labeling and ablation in zebrafish. Dev Biol. 2007;304:811–824. doi: 10.1016/j.ydbio.2007.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Curado S, Anderson RM, Jungblut B, Mumm J, Schroeter E, Stainier DY. Conditional targeted cell ablation in zebrafish: a new tool for regeneration studies. Dev Dyn. 2007;236:1025–1035. doi: 10.1002/dvdy.21100. [DOI] [PubMed] [Google Scholar]
- 69.Chen A, Singh C, Oikonomou G, Prober DA. Genetic Analysis of Histamine Signaling in Larval Zebrafish Sleep. eNeuro. 2017;4 doi: 10.1523/ENEURO.0286-16.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gandhi AV, Mosser EA, Oikonomou G, Prober DA. Melatonin is required for the circadian regulation of sleep. Neuron. 2015;85:1193–1199. doi: 10.1016/j.neuron.2015.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Berridge CW, Schmeichel BE, Espana RA. Noradrenergic modulation of wakefulness/arousal. Sleep Med Rev. 2012;16:187–197. doi: 10.1016/j.smrv.2011.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Berridge CW, Waterhouse BD. The locus coeruleus-noradrenergic system: modulation of behavioral state and state-dependent cognitive processes. Brain Res Brain Res Rev. 2003;42:33–84. doi: 10.1016/s0165-0173(03)00143-7. [DOI] [PubMed] [Google Scholar]
- 73.Sundstrom G, Larsson TA, Xu B, Heldin J, Larhammar D. Interactions of zebrafish peptide YYb with the neuropeptide Y-family receptors Y4, Y7, Y8a, and Y8b. Front Neurosci. 2013;7:29. doi: 10.3389/fnins.2013.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chen S, Reichert S, Singh C, Oikonomou G, Rihel J, Prober DA. Light-Dependent Regulation of Sleep and Wake States by Prokineticin 2 in Zebrafish. Neuron. 2017;95:153–168. e156. doi: 10.1016/j.neuron.2017.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Esteban J, Chover AJ, Sanchez PA, Mico JA, Gibert-Rahola J. Central administration of neuropeptide Y induces hypothermia in mice. Possible interaction with central noradrenergic systems. Life Sci. 1989;45:2395–2400. doi: 10.1016/0024-3205(89)90002-7. [DOI] [PubMed] [Google Scholar]
- 76.Sajdyk TJ, Johnson PL, Leitermann RJ, Fitz SD, Dietrich A, Morin M, Gehlert DR, Urban JH, Shekhar A. Neuropeptide Y in the amygdala induces long-term resilience to stress-induced reductions in social responses but not hypothalamic-adrenal-pituitary axis activity or hyperthermia. J Neurosci. 2008;28:893–903. doi: 10.1523/JNEUROSCI.0659-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gjerris A, Widerlov E, Werdelin L, Ekman R. Cerebrospinal fluid concentrations of neuropeptide Y in depressed patients and in controls. J Psychiatry Neurosci. 1992;17:23–27. [PMC free article] [PubMed] [Google Scholar]
- 78.Huang Q, Liao J, Liu Y, Liang H, Ma P, Pan J. Plasma neuropeptide Y levels in Chinese patients with primary insomnia. Sleep Breath. 2015;19:617–622. doi: 10.1007/s11325-014-1059-9. [DOI] [PubMed] [Google Scholar]
- 79.Sah R, Ekhator NN, Strawn JR, Sallee FR, Baker DG, Horn PS, Geracioti TD., Jr Low cerebrospinal fluid neuropeptide Y concentrations in posttraumatic stress disorder. Biol Psychiatry. 2009;66:705–707. doi: 10.1016/j.biopsych.2009.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gerashchenko D, Wisor JP, Burns D, Reh RK, Shiromani PJ, Sakurai T, de la Iglesia HO, Kilduff TS. Identification of a population of sleep-active cerebral cortex neurons. Proc Natl Acad Sci U S A. 2008;105:10227–10232. doi: 10.1073/pnas.0803125105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Duque A, Balatoni B, Detari L, Zaborszky L. EEG correlation of the discharge properties of identified neurons in the basal forebrain. J Neurophysiol. 2000;84:1627–1635. doi: 10.1152/jn.2000.84.3.1627. [DOI] [PubMed] [Google Scholar]
- 82.Clift D, Richendrfer H, Thorn RJ, Colwill RM, Creton R. High-throughput analysis of behavior in zebrafish larvae: effects of feeding. Zebrafish. 2014;11:455–461. doi: 10.1089/zeb.2014.0989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dreosti E, Lopes G, Kampff AR, Wilson SW. Development of social behavior in young zebrafish. Front Neural Circuits. 2015;9:39. doi: 10.3389/fncir.2015.00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.He C, Yang Y, Zhang M, Price JL, Zhao Z. Regulation of sleep by neuropeptide Y-like system in Drosophila melanogaster. PLoS One. 2013;8:e74237. doi: 10.1371/journal.pone.0074237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chung BY, Ro J, Hutter SA, Miller KM, Guduguntla LS, Kondo S, Pletcher SD. Drosophila Neuropeptide F Signaling Independently Regulates Feeding and Sleep-Wake Behavior. Cell Rep. 2017;19:2441–2450. doi: 10.1016/j.celrep.2017.05.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Shang Y, Donelson NC, Vecsey CG, Guo F, Rosbash M, Griffith LC. Short neuropeptide F is a sleep-promoting inhibitory modulator. Neuron. 2013;80:171–183. doi: 10.1016/j.neuron.2013.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nassel DR, Wegener C. A comparative review of short and long neuropeptide F signaling in invertebrates: Any similarities to vertebrate neuropeptide Y signaling? Peptides. 2011;32:1335–1355. doi: 10.1016/j.peptides.2011.03.013. [DOI] [PubMed] [Google Scholar]
- 88.Choi S, Chatzigeorgiou M, Taylor KP, Schafer WR, Kaplan JM. Analysis of NPR-1 reveals a circuit mechanism for behavioral quiescence in C. elegans. Neuron. 2013;78:869–880. doi: 10.1016/j.neuron.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cheung BH, Cohen M, Rogers C, Albayram O, de Bono M. Experience-dependent modulation of C. elegans behavior by ambient oxygen. Curr Biol. 2005;15:905–917. doi: 10.1016/j.cub.2005.04.017. [DOI] [PubMed] [Google Scholar]
- 90.Gray JM, Karow DS, Lu H, Chang AJ, Chang JS, Ellis RE, Marletta MA, Bargmann CI. Oxygen sensation and social feeding mediated by a C. elegans guanylate cyclase homologue. Nature. 2004;430:317–322. doi: 10.1038/nature02714. [DOI] [PubMed] [Google Scholar]
- 91.Macosko EZ, Pokala N, Feinberg EH, Chalasani SH, Butcher RA, Clardy J, Bargmann CI. A hub-and-spoke circuit drives pheromone attraction and social behaviour in C. elegans. Nature. 2009;458:1171–1175. doi: 10.1038/nature07886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.de Bono M, Bargmann CI. Natural variation in a neuropeptide Y receptor homolog modifies social behavior and food response in C. elegans. Cell. 1998;94:679–689. doi: 10.1016/s0092-8674(00)81609-8. [DOI] [PubMed] [Google Scholar]
- 93.Rogers C, Reale V, Kim K, Chatwin H, Li C, Evans P, de Bono M. Inhibition of Caenorhabditis elegans social feeding by FMRFamide-related peptide activation of NPR-1. Nat Neurosci. 2003;6:1178–1185. doi: 10.1038/nn1140. [DOI] [PubMed] [Google Scholar]
- 94.Fu LY, Acuna-Goycolea C, van den Pol AN. Neuropeptide Y inhibits hypocretin/orexin neurons by multiple presynaptic and postsynaptic mechanisms: tonic depression of the hypothalamic arousal system. J Neurosci. 2004;24:8741–8751. doi: 10.1523/JNEUROSCI.2268-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lee EY, Hwang YG, Lee HS. Hypothalamic neuronal origin of neuropeptide Y (NPY) or cocaine- and amphetamine-regulated transcript (CART) fibers projecting to the tuberomammillary nucleus of the rat. Brain Res. 2016 doi: 10.1016/j.brainres.2016.11.025. [DOI] [PubMed] [Google Scholar]
- 96.Steiger A. Sleep and the hypothalamo-pituitary-adrenocortical system. Sleep Med Rev. 2002;6:125–138. doi: 10.1053/smrv.2001.0159. [DOI] [PubMed] [Google Scholar]
- 97.Pleil KE, Rinker JA, Lowery-Gionta EG, Mazzone CM, McCall NM, Kendra AM, Olson DP, Lowell BB, Grant KA, Thiele TE, et al. NPY signaling inhibits extended amygdala CRF neurons to suppress binge alcohol drinking. Nat Neurosci. 2015;18:545–552. doi: 10.1038/nn.3972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Brzezinski A, Vangel MG, Wurtman RJ, Norrie G, Zhdanova I, Ben-Shushan A, Ford I. Effects of exogenous melatonin on sleep: a meta-analysis. Sleep Med Rev. 2005;9:41–50. doi: 10.1016/j.smrv.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 99.Vacas MI, Sarmiento MI, Pereyra EN, Etchegoyen GS, Cardinali DP. In vitro effect of neuropeptide Y on melatonin and norepinephrine release in rat pineal gland. Cell Mol Neurobiol. 1987;7:309–315. doi: 10.1007/BF00711307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Finta EP, Regenold JT, Illes P. Depression by neuropeptide Y of noradrenergic inhibitory postsynaptic potentials of locus coeruleus neurones. Naunyn Schmiedebergs Arch Pharmacol. 1992;346:472–474. doi: 10.1007/BF00171093. [DOI] [PubMed] [Google Scholar]
- 101.Huang J, Zhong Z, Wang M, Chen X, Tan Y, Zhang S, He W, He X, Huang G, Lu H, et al. Circadian modulation of dopamine levels and dopaminergic neuron development contributes to attention deficiency and hyperactive behavior. J Neurosci. 2015;35:2572–2587. doi: 10.1523/JNEUROSCI.2551-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Gray TS, Morley JE. Neuropeptide Y: anatomical distribution and possible function in mammalian nervous system. Life Sci. 1986;38:389–401. doi: 10.1016/0024-3205(86)90061-5. [DOI] [PubMed] [Google Scholar]
- 103.Holets VR, Hokfelt T, Rokaeus A, Terenius L, Goldstein M. Locus coeruleus neurons in the rat containing neuropeptide Y, tyrosine hydroxylase or galanin and their efferent projections to the spinal cord, cerebral cortex and hypothalamus. Neuroscience. 1988;24:893–906. doi: 10.1016/0306-4522(88)90076-0. [DOI] [PubMed] [Google Scholar]
- 104.Inutsuka A, Yamanaka A. The physiological role of orexin/hypocretin neurons in the regulation of sleep/wakefulness and neuroendocrine functions. Front Endocrinol (Lausanne) 2013;4:18. doi: 10.3389/fendo.2013.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zitnik GA. Control of arousal through neuropeptide afferents of the locus coeruleus. Brain Res. 2016;1641:338–350. doi: 10.1016/j.brainres.2015.12.010. [DOI] [PubMed] [Google Scholar]
- 106.Raskind MA, Peskind ER, Hoff DJ, Hart KL, Holmes HA, Warren D, Shofer J, O’Connell J, Taylor F, Gross C, et al. A parallel group placebo controlled study of prazosin for trauma nightmares and sleep disturbance in combat veterans with post-traumatic stress disorder. Biol Psychiatry. 2007;61:928–934. doi: 10.1016/j.biopsych.2006.06.032. [DOI] [PubMed] [Google Scholar]
- 107.Westerfield M. The zebrafish book: a guide for the laboratory use of zebrafish (Brachydanio rerio) Eugene, OR: M. Westerfield; 1993. [Google Scholar]
- 108.Soderberg C, Wraith A, Ringvall M, Yan YL, Postlethwait JH, Brodin L, Larhammar D. Zebrafish genes for neuropeptide Y and peptide YY reveal origin by chromosome duplication from an ancestral gene linked to the homeobox cluster. J Neurochem. 2000;75:908–918. doi: 10.1046/j.1471-4159.2000.0750908.x. [DOI] [PubMed] [Google Scholar]
- 109.Bussmann J, Schulte-Merker S. Rapid BAC selection for tol2-mediated transgenesis in zebrafish. Development. 2011;138:4327–4332. doi: 10.1242/dev.068080. [DOI] [PubMed] [Google Scholar]
- 110.Liu J, Merkle FT, Gandhi AV, Gagnon JA, Woods IG, Chiu CN, Shimogori T, Schier AF, Prober DA. Evolutionarily conserved regulation of hypocretin neuron specification by Lhx9. Development. 2015;142:1113–1124. doi: 10.1242/dev.117424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Thisse C, Thisse B. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat Protoc. 2008;3:59–69. doi: 10.1038/nprot.2007.514. [DOI] [PubMed] [Google Scholar]
- 112.Guo S, Brush J, Teraoka H, Goddard A, Wilson SW, Mullins MC, Rosenthal A. Development of noradrenergic neurons in the zebrafish hindbrain requires BMP, FGF8, and the homeodomain protein soulless/Phox2a. Neuron. 1999;24:555–566. doi: 10.1016/s0896-6273(00)81112-5. [DOI] [PubMed] [Google Scholar]
- 113.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
In situ hybridization with an npy-specific probe was performed on 6 dpf larval zebrafish brains, followed by immunostaining for t-ERK. The t-ERK staining was then used to register npy expression to the Z-brain reference brain. Anatomical domains of npy expression were then added using Z-Brain browser annotations. Anterior is to the left and the movie starts from the ventral surface of the brain. The movie does not show the expression of npy in the olfactory bulb or retina.
Projections from hindbrain npy-expressing neurons (blue) and dbh-expressing LC neurons (yellow) form close contacts. An exemplar npy-expressing neuron (highlighted magenta) appears to contact a single dbh-expressing neuron (highlighted green) at least twice (white, indicated by asterisk, magnified 4-fold in insets). Scale bar: 10 μm.