Abstract
Understanding the control of antigen restimulation-induced T cell death (RICD), especially in cancer immunotherapy where highly proliferating T cells will encounter potentially large amounts of tumor antigens, is important now more than ever. It has been known that growth cytokines make T cells susceptible to RICD, but the precise molecular mediators that govern this in T cell subsets is unknown until now. Signal transducer and activator transcription (STAT) proteins are a family of transcription factors that regulate gene expression programs underlying key immunological processes. In particular, STAT5 is known to favor the generation and survival of memory T cells. Here we report an unexpected role for STAT5 signaling in the death of effector memory T (TEM) cells in mice and humans. TEM cell death was prevented with neutralizing anti-IL-2 antibody or STAT5/JAK3 inhibitors, indicating that STAT5 signaling drives RICD in TEM cells. Moreover, we identified a unique patient with a heterozygous missense mutation in the coiled-coil (CC) domain of STAT5B that presented with autoimmune lymphoproliferative syndrome (ALPS)-like features. Similar to Stat5b−/− mice, this patient exhibited increased CD4+ TEM cells in the peripheral blood. The mutant STAT5B protein dominantly interfered with STAT5-driven transcriptional activity, leading to global downregulation of STAT5-regulated genes in patient T cells upon IL-2 stimulation. Notably, CD4+ TEM cells from the patient were strikingly resistant to cell death by in vitro TCR restimulation, a finding that was recapitulated in Stat5b−/− mice. Hence, STAT5B is a crucial regulator of RICD in memory T cells in mice and humans.
Introduction
Antigen restimulation-induced T cell death (RICD) has been known to play an important role in peripheral T cell homeostasis and tolerance and may potently influence anti-tumor responses and the success of immunotherapy where highly activated and cycling T cells will be restimulated by large amounts of specific tumor antigens. It is essential to understand how RICD mechanisms are balanced so as to prevent T cells from dying in battle against tumor antigens or pathogenic agents but, on the other hand, promote T cell turnover to prevent collateral damage to self-tissue (1,2). Cytokines, especially the common gamma chain cytokines such as IL-2, are known to program T cells for RICD, but the precise molecular mediators that govern this in distinct T cell subsets are unknown until now. STAT proteins are crucial mediators of cytokine signals involved in cell-cell communication in the immune system. Once a cognate ligand binds to its receptor, it initiates a cascade of signaling events from the phosphorylation of a tyrosine residue on a STAT molecule by a Janus Kinase (JAK) to dimerization with another STAT protein, culminating in the translocation of activated STAT dimers to genes in the nucleus (3). STATs regulate gene transcription by recognizing consensus DNA-binding sequences to influence a diverse array of cellular functions such as proliferation, apoptosis, differentiation, reproduction and lipid metabolism (4–7).
In mammals, there are seven STATs: STAT1, STAT2, STAT3, STAT4, STAT5A, STAT5B and STAT6 (8). The term STAT5 actually refers to two paralogs, STAT5A and STAT5B, derived from a recent gene duplication event in evolution (7). Despite a high degree of homology (96% protein homology in humans), studies in mice have revealed distinct biological functions. Most notably, Stat5a-deficient (Stat5a−/−) mice have a defect in mammary gland lactation, whereas Stat5b-deficient (Stat5b−/−) mice exhibit dwarfism due to growth hormone insensitivity (7). Compound Stat5 deficiency (Stat5a−/−Stat5b−/−) in mice is perinatal lethal, indicative of essential roles during early development and the few mice that survive to adulthood exhibit severe combined immunodeficiency (7,9–11). Similar to Stat5-deficient mice, loss-of-function mutations in the STAT5B gene in humans also causes severe growth hormone insensitivity and immunodeficiency resulting in hypergammaglobulinemia, T and NK cell lymphopenia, and impaired regulatory T cell frequency and function (12–15). Notably, many of the immunological phenotypes resulting from Stat5 deficiency have been ascribed to its role in the activation, proliferation and survival of immune cell lineages that require signaling from common gamma-chain cytokines, including IL-2, IL-7 and IL-15.
Upon TCR stimulation, IL-2/STAT5 signaling promotes T cell differentiation, which is the first key step to generating effector T cells that can target pathogens (16,17). T cell activation results in exponential expansion of antigen-specific clones that must be properly controlled in order to abate the ever-present threat of autoimmunity and malignancy. Programmed cell death, and specifically a mechanism of RICD, is critical for achieving this balance, as evidenced by the severe inflammatory phenotypes seen in patients with genetic defects in pro- or anti-apoptotic molecules (17–19). Although highly efficient, the death process allows for the survival of long-lived, self-renewing memory T cells that are capable of mounting a rapid response upon re-challenge (20). Memory T cell homeostasis is dependent on STAT5-activating cytokines, chiefly IL-7 and IL-15, both of which are known to promote survival and proliferation in conjunction with low-level ‘tonic’ TCR-signaling (21). However, in the context of more robust TCR stimulation, TEM cells, a short-lived migratory subset, are highly susceptible to apoptosis while central memory T (TCM) cells, a long-lived lymphoid-tissue resident subset, are resistant (22). The current study further extends these findings by identifying STAT5B as a key mediator of TEM apoptosis in mice and humans. We show that STAT5B signaling is necessary for RICD of mouse and human TEMs in vitro, and that STAT5B deficiency in mice leads to accumulation of TEM cells in the spleen. Importantly, we discovered a patient with combined immunodeficiency and lymphoproliferative disease that has a novel heterozygous missense mutation in the CC domain of STAT5B. Unexpectedly, this mutant allele dominantly interfered with STAT5B transcriptional activity resulting in a loss of STAT5B-induced gene expression in patient T cells stimulated with IL-2. Similar to mice deficient in Stat5b, the patient exhibited increased TEM cells in the peripheral blood which were resistant to RICD in vitro. Overall, these findings describe an unexpected role for STAT5B in humans and mice to control both proliferation and death of memory T cells
Materials and Methods
Human Subjects
The patient, his mother, and healthy controls provided informed consent in accordance with the Declaration of Helsinki under institutional review board-approved protocol (clinical trial identifier NTC00001355) of the NIAID, NIH. Blood was obtained under approved protocols, which also allow for the collection and use of the patient’s family history and pedigrees for publication. Mutations will be archived by Online Mendelian Inheritance in Man and genomic data will be deposited in NIH databases in accordance with policy. (dbGaP accession number: phs001479.v1.p1, https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs001479.v1.p1).
Mice
All protocols were approved by the NIAID Animal Care and Use Committee and followed NIH guidelines for using animals in intramural research. Stat5b−/− mice were a kind gift from Dr. Lothar Hennighausen (NIDDK, NIH, Bethesda, MD) (Udy et al., 1997). Littermate C57BL/6 mice were used as wild-type (WT) controls. Eight- to twelve-week-old gender-matched mice were used for all experiments described in this study.
DNA sequencing
For WES, genomic DNA was extracted from peripheral blood and submitted for whole exome capture (SureSelect Human All Exon 50Mb Kit, Agilent Technologies) followed by HiSeq next-generation sequencing (Illumina). For WGS, genomic DNA was isolated from peripheral blood and submitted to the Sidra Medical and Research Center for DNA sequencing using the Illumina HiSeq X sequencer. DNA sequencing by the Sanger method was performed to confirm selected variants detected by whole exome sequencing (WES), using purified PCR products amplified by exon-specific primers and GoTaq Hot Start Polymerase (Promega). PCR products were directly sequenced using BigDye Terminators (version 1.1) and analyzed on a 3130xL Genetic Analyzer (Applied Biosystems).
Bioinformatics analysis of the WES data
For WES analysis, reads with an average coverage of 50- to 100-fold were mapped to the hg19 human genome reference using the Burrows-Wheeler Aligner with default parameters. The alignment was subjected to GATK base quality score recalibration and indel realignment followed by SNP and INDEL discovery and genotyping using the GATK Unified Genotyper and VCFtools with standard hard filtering parameters (qual > 30; depth > 30; RMS mapping quality-Phred score > 20; variant confidence/quality by depth > 5) (23,24). To prioritize genetic variants, we implemented tools in the ANNOVAR package (25), including gene/amino acid annotation, functional prediction scores (SIFT, PolyPhen2, LRT, MutationTaster, FATHMM, RadialSVM, LR, VEST3, CADD), conservation scores (GERP++, phyloP100way_vertebrate, SiPhy_29way_logOdds), and allele frequencies per the Exome Aggregation Consortium [(ExAC), Cambridge, MA (URL: http://exac.broadinstitute.org)(November, 2014)], NCBI dbSNP database, The 1000 Genomes Project (October 2014), and the NHLBI GO Exome Sequencing Project (ESP6500). Only novel non-synonymous, nonsense, indel, and splicing variants were considered further. Variants in segmental duplication regions were initially excluded, which filtered out the STAT5B variant in this report due to high sequence homology between STAT5A and STAT5B (segmental duplication score=0.97295; chr17:40449290). The STAT5B variant was then detected upon a second pass analysis without the segmental duplication filter. Exomic variants were finally prioritized based on clinical correlation. We used Genedistiller 2 (26) and DAVID (27) to highlight genes and related pathways associated with immune response and molecular function. The STAT5B p.Q206R variant is absent from the above-mentioned SNP databases and is predicted to be deleterious by MutationTaster and LRT, and CADD with corroborating high conservation scores (GERP++=4.62, PhyloP=3.315, SiPhy=14.22).
Protein-structure analysis
The structure of the H. sapiens STAT5B monomer (lacking the tetramerization domain) was modeled using I-TASSER (http://zhanglab.ccmb.med.umich.edu/I-TASSER/), with the final model based heavily on a crystal structure of M. musculus STAT5A (PDB: 1Y1U, www.rcsb.org)(28). The relative dimer orientation was modeled by aligning two copies of the monomeric STAT5B homology model onto a crystal structure of the H. sapiens STAT1 dimer (PDB: 1BF5, www.rcsb.org)(29). The STAT5B tetramer was generated by duplication of the STAT5B dimer, with lateral translation along the bound double-stranded DNA helix. The tetramerization domain of M. musculus STAT4 (PDB: 1BGF, www.rcsb.org)(30) was then positioned between the N-terminal ends of the CC domains extending from STAT5B molecules on the same side of the DNA helix (i.e. to link the two adjacent STAT5B dimers into a tetrameric complex).
Plasmids and mutagenesis
Human plasmids pCI-STAT5B, pME18S-IL-2RB, pME18S-IL-2RG and pME18S-JAK3 were a kind gift from Dr. Warren Leonard (NHLBI, NIH, Bethesda, MD). Site-directed mutagenesis was performed to generate pCI-STAT5B Q206R using the forward primer 5′-GGAGACGGCCCTCCAGCGGAAGCAGGTGTCTCTGG-3′ and the reverse primer 5′-CCAGAGACACCTGCTTCCGCTGGAGGGCCGTCTCC-3′. The mutation was confirmed by Sanger sequencing analysis performed by the Genomics Unit of the Rocky Mountain Laboratories Research Technologies Section of the NIAID.
Transfections and reporter assays
The HEK293T cell line (American Type Culture Collection) was maintained in DMEM (Invitrogen) containing 10% FBS, 2 mM glutamine, and penicillin and streptomycin (100 U/mL each; Invitrogen). HEK293T cells were transfected using Turbofect (Thermo Scientific). Luciferase reporter assays used a previously published method (31). Cells were transfected with 1 μg of the reporter plasmid, pGL4.52 [luc2P/STAT5 RE/Hygro] (Promega Corp.), 25 ng of either human WT STAT5B or Q206R STAT5B expression plasmid (pCI-STAT5B), 2 μg of pME18S-IL-2RB, 500 ng of pME18S-IL-2RG, 250 ng of pME18S-JAK3, and 0.1 ng of the transfection control reporter plasmid, pRL-CMV Luciferase (Promega Corp.). For dual luciferase assay, 30 ng of 3xFLAG tagged STAT5B construct (p3xF-STAT5B) and a variable amount of HA-tagged Q206R (pHA-STAT5B Q206R) with a normalizing amount of empty vector (pCI-HA) were mixed with the prepared cocktail for transfection using Turbofect (Thermofisher) following the manufacturer’s instructions. Twenty-four hours after transfection, cell cultures were treated with 100 IU/mL IL-2 for various times and luciferase was assayed with the Dual-Glo luciferase system according to the manufacturer’s protocol (Promega Corp.). All transfection experiments were performed in duplicate or triplicate, and the data are presented as the means +/− standard deviations.
Flow cytometry and cell isolation
For surface immunostaining, PBMCs or mouse splenocytes were washed in PBS and resuspended at 1 × 106 cells/mL in PBS containing 3% FBS. Cells were incubated for 30 min at room temperature in the presence of fluorochrome-labeled monoclonal antibodies or their isotype-matched control antibodies. After two washes with PBS, cells were analyzed using a LSR-II flow cytometer (BD Biosciences). For intracellular staining, PBMCs were fixed and permeabilized after surface staining using FoxP3 staining kit (EBioscience) following the manufacturer’s instructions, and then incubated with anti-FoxP3 for 30 min on ice. For phosphorylated-STAT5 staining, cells were fixed in complete RPMI-1640 medium with BD Lyse-Fix and then permeabilized with Perm Buffer III according to manufacturer’s instructions (BD Biosciences). Intracellular staining of phospho-STAT5 in T cell blasts was performed by incubating cells in PBS containing 5% FBS for 1 hour at room temperature with antibody to STAT5 phosphorylated at Tyr 694. Cells were washed and then analyzed on the LSR-II or Fortessa flow cytometer (BD Biosciences).
The following antibodies were used: anti-human CD4 (OKT4, Biolegend), anti-human CD8α (RPA-T8, Biolegend), anti-human CD3ε (OKT3, Ebioscience), anti-human CD27 (O323, Biolegend), anti-human CD45RA (HI100, Biolegend), anti-human CCR7 (G043H7, Biolegend), anti-human CXCR5 (J252D4, Biolegend), anti-human CD19 (6D5, Biolegend), anti-human CD81 (1D6, Biolegend), anti-mouse CD4 (GK1.5, Ebioscience), anti-mouse CD8a (53–6.7, Ebioscience), anti-mouse CD44 (IM7, Ebioscience), anti-mouse CD62L (MEL-14, Ebioscience) and Alexa Fluor 647 anti-human phospho-STAT5 antibody (TC71E5, Cell Signaling).
RNA sequencing and transcriptome analysis
1–2 × 106 cells were lysed in Trizol reagent and total RNA was isolated by phenol-chloroform extraction with GlycoBlue as co-precipitant (Life Technologies, Grand Island, NY). Single-end libraries were prepared using the TruSeq RNA Sample Preparation Kit V2 and sequenced with a HiSeq 2500 instrument (Illumina, San Diego, CA). >20 million reads per sample were mapped onto human genome build hg19 using TopHat and further processed using Cufflinks (32). All datasets were normalized based on Reads Per Kilobase of transcript per Million mapped reads (RPKM) and purged of micro-RNAs, sno-RNAs and sca-RNAs. To minimize ‘fold change artifacts’ caused by low abundance transcripts, a small offset (equal to the second quartile of all data points) was added to all RPKM values and transcripts with RPKM values of less than 1 in all genotypes/conditions were excluded. Fold change and variance were calculated using the Edge-R algorithm (33). Transcripts were classified as differentially expressed in patient samples if they exhibited a >1.5 fold change and significant pairwise variance (p <0.05) relative to normal controls. The experiment was performed twice (i.e. 2 separate blood draws, 2 sets of normal controls) with similar results.
Gene set enrichment analysis (GSEA) was performed as described (34). Briefly, unabridged RNA-seq datasets were used to compute a ranked list of genes that were differentially expressed in IL-2-treated patient samples relative to normal controls. Enrichment was then calculated against a user generated gene set that included 98 genes exhibiting 4-fold change in RPKM and significant pairwise variance (p <0.05) when comparing untreated (0 hours) and IL-2-treated (4 hours) normal controls (3 biological replicates per time point; statistics calculated with EdgeR). Enrichment score curves and member ranks were generated by the GSEA software package (Broad Institute, Cambridge, MA). Normalized enrichment score, nominal p value and false discovery rate (FDR) are shown. Comparable enrichment was observed using datasets and gene sets from 1 and 4 hour IL-2 treatments (data not shown). DAVID bioinformatics resources and Ingenuity Systems were used for functional annotation(27,35). First, GSEA was run against a user generated gene set that included 444 genes exhibiting 2-fold change in RPKM and significant pairwise variance (p <0.05) when comparing untreated (0 hours) and IL-2-treated (4 hours) normal controls (3 biological replicates per time point; statistics calculated with EdgeR). This yielded a list of 128 ‘core enrichment’ genes (i.e. both ‘IL-2-regulated’ and differentially expressed in patient samples) that was used as input for querying gene ontology (GO), Swiss-prot and KEGG databases. Volcano plots, bar graphs and MA plots were generated with DataGraph (Visual Data Tools, Inc.). Genome browser files were processed to remove intronic reads using TopHat and tracks displayed with Integrative Genomics Viewer (IGV; Broad Institute, Cambridge, MA).
In vitro TCR RICD assay
Naive T cells (CD4+CD8-CD45RA+CXCR5-), TCM cells (CD4+CD8-CD45RA-CD27+CCR7-CXCR5-) and TEM cells (CD4+CD8-CD45RA-CD27-CCR7-CXCR5-) were isolated from human PBMCs using a BD FACS Aria III cell sorter. Spleens from WT and Stat5b−/− mice were minced, passed through a nylon filter and erythrocytes were lysed with ACK buffer. Mouse naive T cells (CD4+CD8-CD44-CD62L+), TCM cells (CD4+CD8-CD44+CD62L+) and TEM cells (and CD4+CD8-CD44+CD62L-) were isolated by FACS using a BD FACS Aria III cell sorter. T cell purities were greater than 90% after FACS. Isolated T cell subsets were then incubated in round-bottom 96 well plates in triplicates in anti-CD3ε coated wells (anti-mouse 2C11 and anti-human HITα, Ebioscience) in complete RPMI-1640 medium in the presence of either no cytokine, IL-2 (100 IU/mL) or IL-7 (10 ng/mL) as indicated. After 24 hrs, cells were treated with 10 μM of viability dye (TO-PRO-3, Life Technologies) and collected for 35 seconds per sample on a FACS LSR-II flow cytometer (BD Biosciences). Cell loss was quantified as percent cell loss = (1 –[number of viable cells (treated) / number of viable cells (untreated)]) × 100. STAT5 (573108, CAS 285986-31-4, Calbiochem) and JAK3 (Tofacitinib (CP-690550) Citrate, Selleckchem) inhibitors were used prior to RICD assays.
Statistical analysis
P values were calculated with the Students t-test using PRISM software (GraphPad Software), with a two-tailed distribution.
Results
IL-2/STAT5 signaling pathway triggers cell death of CD4+ TEM cells upon TCR restimulation in mice and in humans
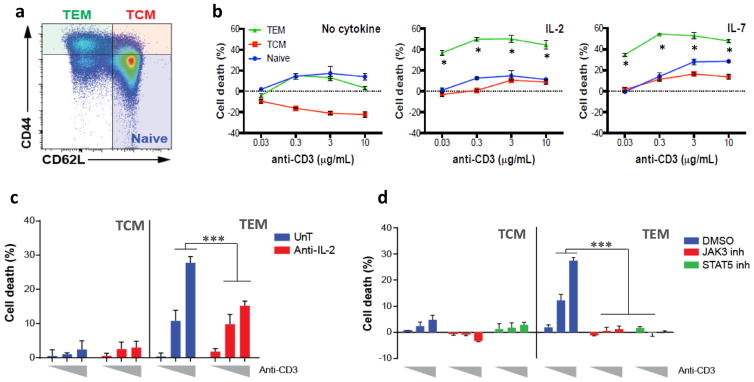
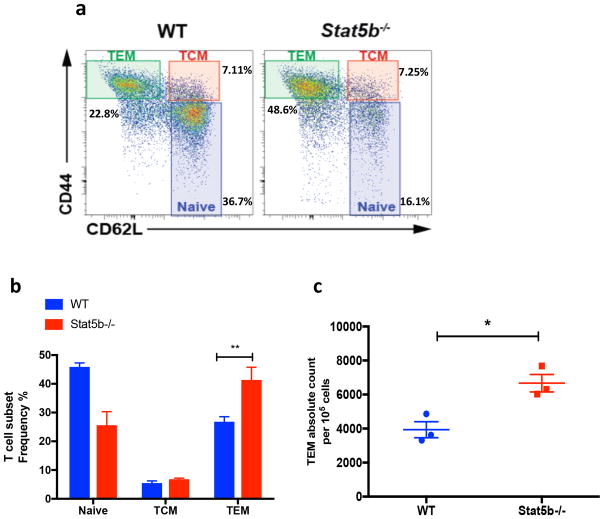
IL-2 and other common gamma-chain cytokines can promote apoptosis of newly activated T cells in the context of TCR reengagement, a process of propriocidal regulation termed RICD (18). More recently, it has been shown that TEM cells are specifically susceptible to RICD as opposed to TCM cells which survive upon TCR stimulation (21,22). Although common gamma-chain cytokines generally promote T cell survival, we first wanted to test their impact on cell death susceptibility of TEM cells during TCR restimulation (17,36). We used CD44 and CD62L staining to identify naïve, TCM, and TEM cell populations (Fig. 1A). Surprisingly, we found that in the absence of common-gamma chain cytokines mouse TEM cells were significantly less susceptible to cell death after TCR reengagement (Fig. 1A and B). In contrast, TEM cells cultured in the presence of IL-2 or IL-7 were strikingly more prone to die by RICD (Fig. 1B). Intrigued by this observation, we asked whether other components of IL-2/STAT5 signaling pathway are required to induce TEM RICD. By first neutralizing the cytokine IL-2, we found that human TEM cells were significantly protected from cell death (Fig. 1C). More strikingly, TEM RICD was completely abrogated when cells were treated with either STAT5 or JAK3 inhibitors (Fig. 1D). Additionally, we investigated how Stat5b could specifically influence the homeostasis of memory cells in vivo (Fig. 2). We found that mice lacking Stat5b exhibited an accumulation of TEM cells in lymphoid organs (Fig. 2A–C). Collectively, these findings indicate that STAT5B signaling pathway plays a crucial and unexpected role in the susceptibility of TEM to TCR restimulation cell death in mice and humans.
Figure 1.
IL-2/STAT5 pathway promotes cell death of effector memory T cells upon RICD in mice and humans.
(a) Expression of CD44 and CD62L (gated on CD4+CD8-) on splenocytes obtained from BALB/c mice, representing naïve T cells (CD44-CD62L+), central memory T cells (TCM; CD44+CD62L+) and effector memory T cells (TEM; CD44+CD62L-). (b) After isolation of the different subsets of naïve and memory T cells from BALB/c spleens by flow sorting, cells were then stimulated in vitro for 24 hours with indicated concentration of plate-bound anti-CD3 in the presence of either no cytokine, IL-2 (100 IU/mL) or IL-7 (10 ng/mL) as indicated. The percent cell loss was calculated at different concentrations of anti-CD3 for each T cell subset using TOPRO-3 staining. * p=<0.05 (c) Human TCM and TEM isolated from healthy control subjects by flow cytometry were stimulated in vitro for 24 hours with plate-bound anti-CD3 at different concentrations (from left to right: 0.1, 1 and 10 μg/mL) and left untreated (blue) or treated either neutralizing anti-IL-2 in combination with blocking anti-CD122 antibodies (10 μg/mL for each antibody) (red) or (d) treated with dimethyl sulfoxide (DMSO) vehicle (blue), JAK3 (red) or STAT5 (green) chemical inhibitors (100 μM). Cell loss percentage was assessed as in (b). Data are representative of at least three independent experiments.
Figure 2.
Stat5b−/− mice exhibit an accumulation of effector memory CD4+ T cells.
(a) Flow cytometry analysis of CD44 and CD62L expression (gated on CD4+CD8-) representing naïve T cells (CD44-CD62L+), central memory T cells (TCM; CD44+CD62L+) and effector memory T cells (TEM; CD44+CD62L-) in spleens obtained from WT or Stat5b−/− mice. (b) Frequency of indicated CD4+ T cell subsets and (c) absolute number of CD4+ TEM cells in spleens from WT (blue) or Stat5b−/− (red) mice. * p=<0.05, ** p=<0.01. Data are representative of at least two independent experiments.
Mutation Q206R in STAT5B is associated with accumulation of CD4+ TEM cells in patient’s blood
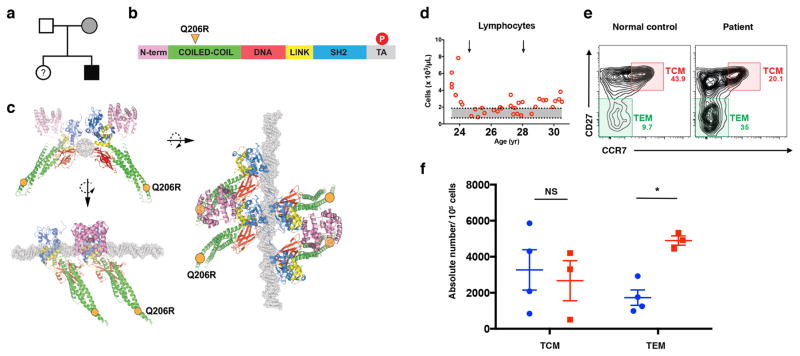
As we were investigating the role of STAT5 in the regulation of memory T cell homeostasis, we identified a 33-year-old African American male who presented with childhood onset autoimmunity in the form of immune thrombocytopenic purpura (ITP) along with chronic lymphadenopathy and splenomegaly (Table S1). Given that clinical manifestations began during early childhood, we predicted a genetic etiology and performed whole exome DNA sequencing on the patient. After filtering out known polymorphisms reported in dbSNP and other databases, we found two novel heterozygous missense variants in CD81 and STAT5B (Fig. S1). CD81 deficiency has been reported in one patient with a homozygous splice site mutation leading to the loss of CD81 and CD19 expression on B cells (37). In that report, the parents were heterozygous for the mutation and their leukocytes expressed intermediate levels of CD81 and CD19, but they showed no clinical signs of immunodeficiency (37). Since the CD81 variant in our patient was also a heterozygous missense mutation, and his B cells expressed normal levels of CD19 and CD81 proteins (data not shown), we excluded this variant as the cause of disease.
STAT5 deficiency in mice is associated with both lymphopenia and lymphoproliferative disease, leading us to focus on the STAT5B variant (10,38). Using Sanger sequencing, we confirmed the presence of the heterozygous missense mutation in STAT5B at position c.617 A>G, p. Q206R in the patient, and also discovered that the mother was a carrier of the STAT5B variant while the father was WT for both alleles (Fig. 3A–C, Fig. S1). The mother of the patient had several immune-related disorders including multiple sclerosis (MS), arthritis, and recurrent infections, and was receiving 17 different medications at the time of blood sampling, including dimethyl fumarate (DMF), which has been shown to selectively decrease memory T cell populations in MS patients (39). For these reasons and the lack of access, we did not further investigate the mother. However, the patient who was first referred to the NIAID, NIH clinical program in 2008 exhibited chronic lymphocytosis (Fig. 3D). Lymphocyte numbers were decreased by treatment with steroids (Fig. 3D, arrows). Interestingly, within the patient’s memory CD4+ T cell population, we observed a specific increase in the frequency and number of TEM cells (Fig. 3E, F). The patient also had a significant increase in the number of circulating follicular helper T cells in the peripheral blood compared to healthy donors (40) (Fig. S2). Although the frequency of CD4+Foxp3+ T regulatory (Treg) cells was normal in the patient, these cells showed diminished expression of the IL-2 receptor alpha chain (CD25) (Fig. S2). Since mice deficient in Stat5b had a similar phenotype for memory T cells, we further investigated how the Q206R mutation could impact STAT5B function as a transcriptional factor and gene regulator.
Figure 3.
Patient with heterozygous Q206R mutation in STAT5B exhibits accumulation of CD4+ TEM cells in blood.
(a) Pedigree of the affected family: black filled symbol, subject with mutation Q206R; grey filled symbol, subject with mutation Q206R and different clinical phenotype; open symbols, unaffected subjects; (?), unscreened subjects. (b) Schematic representation of the STAT5B protein domain structure. N-term, N-terminus domain (pink); Coiled-coil (CC) domain (green); DNA, DNA-binding domain (red); LINK, linker domain (yellow); SH2, src homology 2 domain (blue). Tyrosine phosphorylation (P, red circle) represented in the transcriptional activation (TA) domain (gray). The Q206R residue is located in the CC domain (orange triangle). (c) Three-dimensional provisional models of STAT5B as a tetramer bound to DNA molecule (gray). Three orientations of the STAT5B tetramer are shown, with Q206R positions indicated as orange circles. (d) Absolute numbers of lymphocytes in the patient’s peripheral blood as a function of age. Arrows indicate times of steroid treatment. (e) Flow cytometry plot of peripheral blood from healthy subject (normal control) and patient, stained for CD27 versus CCR7 (gated on CD4+CD45RA- memory T cells), identifying central memory T cells (TCM: CD27+CCR7+) and effector memory T cells (TEM: CD27-CCR7-). (f) Absolute numbers of TCM and TEM cells per 105 cells of PBMCs from healthy donors (blue) and multiple blood draws from the patient (red). ns = not significant; * p= < 0.05
Mutation Q206R in STAT5B leads to a dominant negative effect of STAT5 function and dampens IL-2 STAT5 pathway in patient’s T cells
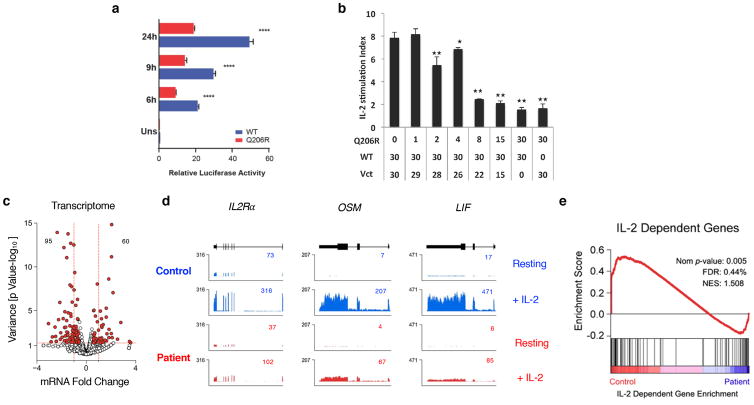
The STAT5B protein is composed of 787 amino acids with six domains: an N-terminal domain, a coiled-coil (CC) domain, a DNA-binding domain, a linker domain, a src homology 2 (SH2) domain and a transcriptional domain (8) (Fig. 3B). Unlike patients with STAT5B deficiency who have homozygous loss of function mutations in residues critical for DNA binding, this mutation results in a glutamine to arginine amino acid substitution within the CC domain (12,14) (Fig. 3B–C). This amino acid substitution mapped to a distal portion of the CC domain that is predicted to be far away from the DNA interaction site, making it unlikely that DNA-binding of mutant STAT5B would be impaired. Although we observed normal expression of STAT5B protein, the activation of STAT5 by phosphorylation upon either IL-2, IL-7 or IL-15 stimulation was reduced in the patient’s T cells, indicating a possible loss-of-function of STAT5B (Fig. S3). Previous studies have shown that heterozygous CC domain mutations tend to generate dominant gain-of-function variants of STAT1 (41). Therefore, we tested whether the Q206R CC mutation resulted in either a gain or loss of function phenotype (8). For these studies, HEK293T cells were co-transfected with either WT or mutant STAT5B and a reporter construct expressing luciferase under the control of a STAT5-regulated IL-2 response element (positive regulatory region III [PRRIII]) derived from the human IL-2Rα promoter (31). Transfected cells were then treated with 100 IU/mL of IL-2 overnight and luciferase activity was measured at the times indicated. Despite the ability of the mutant STAT5B to translocate to nucleus and bind to DNA normally (Fig. S3), its transcriptional function was greatly compromised, suggesting that the mutant allele is hypomorphic (Fig. 4A). We next examined whether Q206R mutation could also lead to a dominant negative effect on total STAT5 function. We co-transfected WT and mutant STAT5B at different ratios and tested STAT5B transcriptional function. As the amount of mutant STAT5B was increased, we observed a reduction in STAT5B-driven transcription upon IL-2 stimulation, demonstrating a dominant-interfering effect of the mutant (Fig. 4B). We also observed that the mutant had a dose-dependent low level of activity in the absence of IL-2 that required a high amount of transfected STAT5B, which may indicate that the mutant has some constitutive unregulated activity (data not shown). These data show that mutant Q206R STAT5B has hypomorphic activity and a dominant-interfering effect that likely compromises STAT5B transcriptional function in our patient.
Figure 4.
Mutation Q206R in STAT5B leads to dominant negative effect and dampens IL-2/STAT5 pathway in patient’s T cells.
(a) Dual-luciferase assay of lysates from HEK293T cells transfected with luciferase reporter plasmid containing the IL-2 response element (positive regulatory region III [PRRIII]) from the human IL-2Rα promoter plus pRL-CMV and plasmids encoding WT (blue) or Q206R (red) STAT5B, then treated with 100 IU/mL IL-2 for various times in hours (h) as indicated or unstimulated (uns). **** p=<0.001. (b) Dual-luciferase activity in lysates from HEK293T cells co-transfected with 30 ng of WT, titrations of Q206R STAT5B and normalizing empty vector (Vct) constructs as indicated. IL-2 stimulation index was determined by dividing the normalized luciferase values of IL-2 stimulated cells versus medium controls. Statistical analyses were performed by comparing WT STAT5B only to those with different amounts of Q206R STAT5B, * p<0.05, ** p<0.01. (c) RNA-seq was performed on serum-starved patient T cell blasts treated with 100 IU/mL of IL-2 for 4 hours. Volcano plot shows fold change (log2 transformed) and variance for all transcripts relative to normal controls. Differentially expressed transcripts are highlighted in red. Those with greater abundance in patient (upper left) or controls (upper right) are summed. Dotted red lines indicate 2-fold changes and a p-value > 0.05. (d) Browser tracks show transcripts for selected genes in resting or IL-2-treated cells from the control (blue) or the patient (red). Numbers denote RPKM values for the most detected exon. (e) GSEA plot shows enrichment of IL-2-regulated genes within differentially expressed transcripts. Data are representative of at least three (a–b) and two (c–e) independent experiments.
To assess the impact of mutant STAT5B on global gene expression, we examined the transcriptome of patient T cells cultured in the presence of IL-2. These studies identified a substantial number of transcripts (155) that were affected in the patient, the majority of which were decreased relative to normal controls (Fig. 4C). Among these were transcripts encoded by well-known STAT5 target genes, such as IL2RA, OSM and LIF, suggesting a defect in the activity of STAT5B transcription (Fig. 4D). Next, we used gene set enrichment analysis to confirm that IL-2-regulated genes were selectively impacted. We first identified genes that were highly dependent on IL-2 in normal controls, then ran this gene set against our transcriptome data and found that, indeed, IL-2-dependent genes were selectively reduced in patient cells (Fig. 4E). Taken together, these data confirm that the hypomorphic and dominant-interfering STAT5B Q206R allele mostly impairs genes responsive to the IL-2/STAT5 signaling pathway.
STAT5B deficiency results in CD4+ TEM cell death resistance upon TCR restimulation in mice and humans
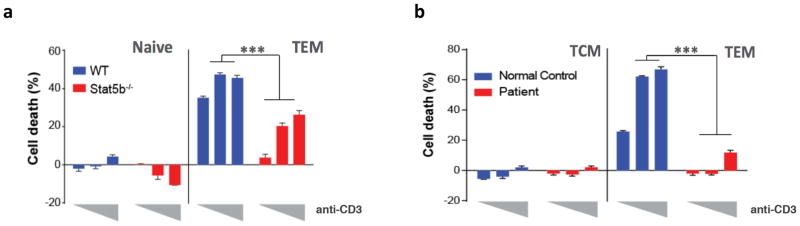
Previous studies have reported an accumulation of effector/memory T cells in STAT5B-deficient mice and humans (10,12). Consistent with these findings, we found that circulating CD4+ TEM cells were increased in the patient (Fig. 3F). This expansion of TEM cells was mirrored in mice deficient in Stat5b, which also exhibited depletion of analogous CD4+CD44lowCD62Lhigh naïve cells (Fig. 2B, C). This striking similarity between Stat5b−/− mice and our patient led us to investigate the mechanism which could explain the specific accumulation of effector memory CD4+ T cells in mice and humans when the STAT5B signaling pathway is dysfunctional. Since chemical inhibition of STAT5 resulted in complete resistance of TEM cells to undergo RICD, we examined how these cells isolated from either Stat5b−/− mice or our patient with the Q206R STAT5B mutation would respond to TCR restimulation in vitro. RICD was compromised in Stat5b−/− TEM cells isolated from mice (Fig. 5A) and even more importantly, TEM cells isolated from the patient were also highly resistant to TCR restimulation (Fig. 5B). Collectively, our data using chemical inhibitors, genetically deficient mice and our patient harboring the Q206R mutation all provide strong evidence that STAT5B is critical to induce RICD of TEM cells in mice and humans.
Figure 5.
STAT5B deficiency results in cell death resistance of CD4+TEM cells upon TCR restimulation in mice and humans.
(a) Naïve and TEM T cells were isolated from WT (blue) or Stat5b−/− (red) mice and then stimulated in vitro for 24 hours with 100U/mL of rhIL-2 and increasing concentration of plate-bound anti-CD3 (from left to right: 0.1, 1 and 10 μg/mL). Cell loss was assessed as in (Fig. 1b). (b) TCM and TEM cells were isolated from peripheral blood from normal control subject (blue) and patient (red), then stimulated in vitro for 24 hours with plate-bound anti-CD3 at different concentrations (from left to right: 0.1, 1 and 10 μg/mL) and cell loss was calculated as in (a). *** p=<0.001. Data are representative of at least two (a) and three (b) independent experiments.
Discussion
T cell homeostasis is tightly regulated via diverse mechanisms that balance cell proliferation and survival or apoptosis (17). Elucidating the molecular dictates of these processes is vital to our understanding of diseases such as autoimmunity and treatments such as immunotherapy for cancer. More than 25 years ago, it was discovered that propriocidal regulation of antigen-driven T cell responses, a process termed RICD, is important for clonal regulation of lymphocyte homeostasis and immunological tolerance as first postulated by Burnet (18,19,42). We now show that STAT5B, a transcription factor downstream of multiple cytokines involved in RICD, is essential for triggering apoptosis of mouse and human TEM cells, and that altered STAT5B transcriptional activity drives immunodeficiency and dysfunction.
STAT5 is commonly thought to promote T cell survival and proliferation, as evidenced by the pronounced lymphoproliferation seen in transgenic mice which overexpress Stat5b (12), and constitutive or exaggerated STAT5 activity is often linked to immunological malignancies, such as leukemias and lymphomas (43,44). Consistent with this pro-proliferative view of STAT5, genetic ablation in mice or loss-of-function mutations in humans are typically associated with lymphopenia and immunosuppression (10). By contrast, we report a heterozygous missense Q206R mutation in STAT5B associated with a hyper-inflammatory phenotype with a unique constellation of clinical symptoms including immune thrombocytopenia, lymphadenopathy, splenomegaly, hypogammaglobulinemia, and necrotizing granulomas. The mutation creates a dominant negative mutant protein that is underphosphorylated in the affected patient and interferes with the transcriptional output of WT STAT5B, particularly downstream of IL-2 and common gamma chain cytokines. These findings provide valuable insights on the genetics, biochemistry and function of STAT5B, particularly in the context of immunological tolerance and lymphocyte homeostasis, and may be applicable to other dimerizing transcription factors (42).
We found that even a single mutant allele encoding the substitution of the amino acid from glutamine to arginine at position 206 in the CC domain of STAT5B could impair transcription of a wide variety of genes in patient T cells, particularly those involved in IL-2/STAT5-mediated cellular functions. Furthermore, the mutation Q206R in STAT5B resulted in a dominant-negative effect on total STAT5 function. How the mutant STAT5B can bind to the WT protein and compromise total STAT5 transcriptional activity remains unclear. Mutant STAT5B exhibits some basal activity in the absence of IL-2 stimulation, and this activity becomes greater as the amount of mutant STAT5B plasmid increases during luciferase reporter assay (data not shown). These data suggest that the mechanism by which mutant STAT5B dominantly interferes with the WT protein may be more complex than a classic dominant interference and further investigation is needed to shed light on how the mutation Q206R impacts STAT5 function. In general, the detailed mechanism by which STAT5 can activate or repress gene expression is not fully understood. STAT5 cannot initiate transcription alone and its interaction with other proteins is necessary to form a gene regulatory complex (45). How a single point mutation in the CC domain can interfere with the transcriptional activity of STAT5B is likely related to the structure of the transcriptional complex on the promoter. Promoting protein-protein interaction due to their extended three-dimensional alpha-helical structure, CC domains could play a prominent role in regulating the transcription complex involving STAT5. However, only a few proteins have been found capable of regulating STAT5 transcriptional activity through interaction with the CC domain. N-myc interactor (Nmi) can enhance STAT5 transcriptional activity by recruiting p300-CREB-binding protein complex (46). Moreover, SMRT can bind to the STAT5 CC domain and repress its activity (47). However, our mutation at position 206 in STAT5B is not part of the domains identified to interact with Nmi (232–275) or SMRT (248–380). Also, we directly examined these factors and found no effect (data not shown). Thus, we conclude that the impairment in mutant Q206R STAT5B transcriptional activity was not due to disruption of the interaction between STAT5B and these two proteins. An unbiased proteome-wide analysis of mutant STAT5B is necessary to identify other ‘missing’ proteins that are a part of the transcriptional holocomplex that may account for its diminished transcriptional capacity. Not only will these studies increase our understanding of the molecular mechanisms underlying STAT5B dysregulation in the patient, but they will also provide insight into the STAT5B interactome necessary for gene activation and repression.
The most striking clinical features of the patient were lymphadenopathy and splenomegaly, which we propose were largely due to cell intrinsic defects in the effector/memory T cell compartment. We found that the number of T lymphocytes in the patient’s blood was greatly elevated with a specific enrichment in CD4+ TEM cells. The frequency of CD8+ TCM and TEM cells was relatively normal, indicating the Q206R mutation preferentially affected the CD4+ T cell compartment. In addition to TEM cells, we observed an elevated number of circulating Tfh cells, within the effector memory T cell compartment (48). Downregulation of STAT5 signaling has been shown to promote Tfh cell differentiation by repressing the function of BLIMP-1, which antagonizes BCL6, the master regulator of Tfh differentiation (49,50). Thus, we hypothesized that the increase in Tfh cells in the patient was likely due to a defect in STAT5 signaling resulting in unfettered expression of BCL6. Despite increased Tfh cells, the patient exhibited reduced IgG+ plasma cells in the spleen in association with hypogammaglobulinemia. Further, our patient had decreased generation of IgG+ plasma cells in vitro, suggesting an intrinsic class-switch defect (data not shown). How a dominant-interfering mutation in STAT5B could lead to reduced class-switched plasma cells remains unclear; previous studies have shown that STAT5 is critical for early mouse B cell development but not for maturation (51). However, these data suggest that STAT5B may also be involved in late-stage differentiation of mature B cells as well as class-switch recombination, and further studies are needed to understand the role of STAT5B in these processes. We also observed reduced CD25 expression on the surface of Treg cells from the patient. CD25 expression on Treg cells is highly dependent on STAT5 and, in turn, Treg cells from humans and mice with loss-of-function STAT5 mutations tend to exhibit reduced CD25 expression (15,52). Further studies should address whether the reduction in CD25 impacts Treg fitness and function, as shown in mouse models, and if this is a causative factor for the autoimmune phenotype of the patient. It is also unclear why Q206R mutation in STAT5B results in granulocytosis. Numerous studies have shown that granulocyte macrophage-colony stimulating factor (GM-CSF), a key cytokine for granulocyte proliferation and homeostasis operates through STAT5 (53). Further investigations would be helpful to understand the impact of Q206R mutation in STAT5B on granulopoiesis. Altogether, these clinical features converge toward the idea that partial STAT5B loss-of-function in our patient results in a spectrum of immunological dysregulatory events. Our data also show that unique heterozygous mutations are gaining prominence in our understanding of the genetic contribution to a wide range of diseases, and these can yield important regulatory insights (54).
Our results in both mice and humans show the different classes of memory T cells have distinct susceptibilities to RICD. TEM cells are phenotypically and functionally distinguishable from TCM cells, by markers such as CCR7 and CD62L, and TCR engagement has varying effects on each subset (21,55,56). Both CD4 and CD8 TEM cells are located in the peripheral tissues, including lung, liver, and gut, and display rapid and potent effector cytokines and cytolytic functions when stimulated. By contrast, TCM remain largely in the secondary lymphoid tissue where they initiate and coordinate immune stimulatory reactions between T cells and antigen presenting cells by producing IL-2 and other trophic cytokines. They are heterogeneous and largely uncommitted but can transition to TEM cells after stimulation. Thus TEM cells driven into cycle by IL-2 may be more potentially harmful when engaging antigens peripheral tissues and thus require more stringent regulation by RICD. Since TCM cells are utilizing IL-2 to coordinate responses in the protective environment of secondary lymphoid tissue, they may have evolved to have a reduced RICD response following IL-2 stimulation. Previous studies have shown that STAT5 is critical for the survival and proliferation of memory T cells (36,44); however, the impact of STAT5 signaling during RICD was unclear. We now show that common gamma chain cytokines, including IL-2 and IL-7, enhanced RICD of TEMs cells in vitro, while TCM and naive cells were unexpectedly resistant to cell death under these conditions. This study further extends these findings to show that RICD of mouse and human TEM cells is dependent upon STAT5B signaling, and that the abnormal accumulation of TEM cells in mice or humans with STAT5B deficiency results from a defect in cell death regulation. It remains unclear whether STAT5A also plays a role in RICD of CD4+ TEM cells, although recent studies have shown that STAT5B is the dominant paralog in this cell type (49). Overall, these findings reveal a vital role of STAT5B in the clonal homeostasis of memory T cells and a healthy immune balance by promoting RICD selectively in different memory subsets.
Supplementary Material
Acknowledgments
We thank the referring physicians, as well as the patient and his family. We thank Warren Leonard, Helen Su, Helen Matthews, Yu Zhang, Qian Zhang and the DNA sequencing center of Sidra Medical and Research Center for experimental advice and assistance. M.B. was an NIH-Oxford-Cambridge Research Scholar and his work was in partial fulfillment of a Ph.D. course at Cambridge University. The content of this publication does not necessarily reflect the views or policies of the U.S. Department of Health and Human Services, nor does mention of trade names, commercial products or organizations imply endorsement by the U.S. Government.1
Footnotes
This work is supported by the Intramural Research Program, NIH Clinical Center and the Division of Intramural Research, NIAID, NIH. J.M.F. was supported in part by a Merck postdoctoral fellowship and a PRAT postdoctoral award from the National Institute of General Medical Sciences, NIH. J.G. was supported by Sigrid Juselius and Emil Aaltonen Foundations and B.A. was supported by the Wellcome Trust.
References
- 1.Spain L, Diem S, Larkin J. Management of toxicities of immune checkpoint inhibitors. Cancer Treat Rev. 2016;44:51–60. doi: 10.1016/j.ctrv.2016.02.001. [DOI] [PubMed] [Google Scholar]
- 2.O’Connell J. Fas ligand and the fate of antitumour cytotoxic T lymphocytes. Immunology. 2002;105:263–266. doi: 10.1046/j.1365-2567.2002.01379.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.O’Shea JJ, Gadina M, Schreiber RD. Cytokine signaling in 2002: new surprises in the Jak/Stat pathway. Cell. 2002;109(Suppl):S121–131. doi: 10.1016/s0092-8674(02)00701-8. [DOI] [PubMed] [Google Scholar]
- 4.Darnell JE., Jr STATs and gene regulation. Science. 1997;277:1630–1635. doi: 10.1126/science.277.5332.1630. [DOI] [PubMed] [Google Scholar]
- 5.O’Shea JJ, Lahesmaa R, Vahedi G, Laurence A, Kanno Y. Genomic views of STAT function in CD4+ T helper cell differentiation. Nature reviews. Immunology. 2011;11:239–250. doi: 10.1038/nri2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Villarino AV, Kanno Y, Ferdinand JR, O’Shea JJ. Mechanisms of Jak/STAT signaling in immunity and disease. Journal of immunology. 2015;194:21–27. doi: 10.4049/jimmunol.1401867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grimley PM, Dong F, Rui H. Stat5a and Stat5b: fraternal twins of signal transduction and transcriptional activation. Cytokine & growth factor reviews. 1999;10:131–157. doi: 10.1016/s1359-6101(99)00011-8. [DOI] [PubMed] [Google Scholar]
- 8.Levy DE, Darnell JE., Jr Stats: transcriptional control and biological impact. Nature reviews. Molecular cell biology. 2002;3:651–662. doi: 10.1038/nrm909. [DOI] [PubMed] [Google Scholar]
- 9.Liu X, Robinson GW, Gouilleux F, Groner B, Hennighausen L. Cloning and expression of Stat5 and an additional homologue (Stat5b) involved in prolactin signal transduction in mouse mammary tissue. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:8831–8835. doi: 10.1073/pnas.92.19.8831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yao Z, Cui Y, Watford WT, Bream JH, Yamaoka K, Hissong BD, Li D, Durum SK, Jiang Q, Bhandoola A, Hennighausen L, O’Shea JJ. Stat5a/b are essential for normal lymphoid development and differentiation. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:1000–1005. doi: 10.1073/pnas.0507350103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cui Y, Riedlinger G, Miyoshi K, Tang W, Li C, Deng CX, Robinson GW, Hennighausen L. Inactivation of Stat5 in mouse mammary epithelium during pregnancy reveals distinct functions in cell proliferation, survival, and differentiation. Molecular and cellular biology. 2004;24:8037–8047. doi: 10.1128/MCB.24.18.8037-8047.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bernasconi A, Marino R, Ribas A, Rossi J, Ciaccio M, Oleastro M, Ornani A, Paz R, Rivarola MA, Zelazko M, Belgorosky A. Characterization of immunodeficiency in a patient with growth hormone insensitivity secondary to a novel STAT5b gene mutation. Pediatrics. 2006;118:e1584–1592. doi: 10.1542/peds.2005-2882. [DOI] [PubMed] [Google Scholar]
- 13.Hwa V, Nadeau K, Wit JM, Rosenfeld RG. STAT5b deficiency: lessons from STAT5b gene mutations. Best practice & research. Clinical endocrinology & metabolism. 2011;25:61–75. doi: 10.1016/j.beem.2010.09.003. [DOI] [PubMed] [Google Scholar]
- 14.Kofoed EM, Hwa V, Little B, Woods KA, Buckway CK, Tsubaki J, Pratt KL, Bezrodnik L, Jasper H, Tepper A, Heinrich JJ, Rosenfeld RG. Growth hormone insensitivity associated with a STAT5b mutation. The New England journal of medicine. 2003;349:1139–1147. doi: 10.1056/NEJMoa022926. [DOI] [PubMed] [Google Scholar]
- 15.Cohen AC, Nadeau KC, Tu W, Hwa V, Dionis K, Bezrodnik L, Teper A, Gaillard M, Heinrich J, Krensky AM, Rosenfeld RG, Lewis DB. Cutting edge: Decreased accumulation and regulatory function of CD4+ CD25(high) T cells in human STAT5b deficiency. Journal of immunology. 2006;177:2770–2774. doi: 10.4049/jimmunol.177.5.2770. [DOI] [PubMed] [Google Scholar]
- 16.Liao W, Lin JX, Leonard WJ. Interleukin-2 at the crossroads of effector responses, tolerance, and immunotherapy. Immunity. 2013;38:13–25. doi: 10.1016/j.immuni.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ma A, Koka R, Burkett P. Diverse functions of IL-2, IL-15, and IL-7 in lymphoid homeostasis. Annual review of immunology. 2006;24:657–679. doi: 10.1146/annurev.immunol.24.021605.090727. [DOI] [PubMed] [Google Scholar]
- 18.Lenardo MJ. Interleukin-2 programs mouse alpha beta T lymphocytes for apoptosis. Nature. 1991;353:858–861. doi: 10.1038/353858a0. [DOI] [PubMed] [Google Scholar]
- 19.Snow AL, Pandiyan P, Zheng L, Krummey SM, Lenardo MJ. The power and the promise of restimulation-induced cell death in human immune diseases. Immunological reviews. 2010;236:68–82. doi: 10.1111/j.1600-065X.2010.00917.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaech SM, Wherry EJ, Ahmed R. Effector and memory T-cell differentiation: implications for vaccine development. Nature reviews. Immunology. 2002;2:251–262. doi: 10.1038/nri778. [DOI] [PubMed] [Google Scholar]
- 21.Surh CD, Sprent J. Homeostasis of naive and memory T cells. Immunity. 2008;29:848–862. doi: 10.1016/j.immuni.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 22.Ramaswamy M, Cruz AC, Cleland SY, Deng M, Price S, Rao VK, Siegel RM. Specific elimination of effector memory CD4+ T cells due to enhanced Fas signaling complex formation and association with lipid raft microdomains. Cell death and differentiation. 2011;18:712–720. doi: 10.1038/cdd.2010.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, McKenna A, Fennell TJ, Kernytsky AM, Sivachenko AY, Cibulskis K, Gabriel SB, Altshuler D, Daly MJ. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nature genetics. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome research. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic acids research. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seelow D, Schwarz JM, Schuelke M. GeneDistiller--distilling candidate genes from linkage intervals. PloS one. 2008;3:e3874. doi: 10.1371/journal.pone.0003874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 28.Neculai D, Neculai AM, Verrier S, Straub K, Klumpp K, Pfitzner E, Becker S. Structure of the unphosphorylated STAT5a dimer. The Journal of biological chemistry. 2005;280:40782–40787. doi: 10.1074/jbc.M507682200. [DOI] [PubMed] [Google Scholar]
- 29.Chen X, Vinkemeier U, Zhao Y, Jeruzalmi D, Darnell JE, Jr, Kuriyan J. Crystal structure of a tyrosine phosphorylated STAT-1 dimer bound to DNA. Cell. 1998;93:827–839. doi: 10.1016/s0092-8674(00)81443-9. [DOI] [PubMed] [Google Scholar]
- 30.Vinkemeier U, Moarefi I, Darnell JE, Jr, Kuriyan J. Structure of the amino-terminal protein interaction domain of STAT-4. Science. 1998;279:1048–1052. doi: 10.1126/science.279.5353.1048. [DOI] [PubMed] [Google Scholar]
- 31.John S, Vinkemeier U, Soldaini E, Darnell JE, Jr, Leonard WJ. The significance of tetramerization in promoter recruitment by Stat5. Molecular and cellular biology. 1999;19:1910–1918. doi: 10.1128/mcb.19.3.1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nature protocols. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kramer A, Green J, Pollard J, Jr, Tugendreich S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics. 2014;30:523–530. doi: 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hand TW, Cui W, Jung YW, Sefik E, Joshi NS, Chandele A, Liu Y, Kaech SM. Differential effects of STAT5 and PI3K/AKT signaling on effector and memory CD8 T-cell survival. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:16601–16606. doi: 10.1073/pnas.1003457107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Zelm MC, Smet J, Adams B, Mascart F, Schandene L, Janssen F, Ferster A, Kuo CC, Levy S, van Dongen JJ, van der Burg M. CD81 gene defect in humans disrupts CD19 complex formation and leads to antibody deficiency. The Journal of clinical investigation. 2010;120:1265–1274. doi: 10.1172/JCI39748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin JX, Leonard WJ. The role of Stat5a and Stat5b in signaling by IL-2 family cytokines. Oncogene. 2000;19:2566–2576. doi: 10.1038/sj.onc.1203523. [DOI] [PubMed] [Google Scholar]
- 39.Wu Q, Wang Q, Mao G, Dowling CA, Lundy SK, Mao-Draayer Y. Dimethyl Fumarate Selectively Reduces Memory T Cells and Shifts the Balance between Th1/Th17 and Th2 in Multiple Sclerosis Patients. Journal of immunology. 2017;198:3069–3080. doi: 10.4049/jimmunol.1601532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Locci M, Havenar-Daughton C, Landais E, Wu J, Kroenke MA, Arlehamn CL, Su LF, Cubas R, Davis MM, Sette A, Haddad EK, International AVIPCPI, Poignard P, Crotty S. Human circulating PD-1+CXCR3-CXCR5+ memory Tfh cells are highly functional and correlate with broadly neutralizing HIV antibody responses. Immunity. 2013;39:758–769. doi: 10.1016/j.immuni.2013.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu L, Okada S, Kong XF, Kreins AY, Cypowyj S, Abhyankar A, Toubiana J, Itan Y, Audry M, Nitschke P, Masson C, Toth B, Flatot J, Migaud M, Chrabieh M, Kochetkov T, Bolze A, Borghesi A, Toulon A, Hiller J, Eyerich S, Eyerich K, Gulacsy V, Chernyshova L, Chernyshov V, Bondarenko A, Grimaldo RM, Blancas-Galicia L, Beas IM, Roesler J, Magdorf K, Engelhard D, Thumerelle C, Burgel PR, Hoernes M, Drexel B, Seger R, Kusuma T, Jansson AF, Sawalle-Belohradsky J, Belohradsky B, Jouanguy E, Bustamante J, Bue M, Karin N, Wildbaum G, Bodemer C, Lortholary O, Fischer A, Blanche S, Al-Muhsen S, Reichenbach J, Kobayashi M, Rosales FE, Lozano CT, Kilic SS, Oleastro M, Etzioni A, Traidl-Hoffmann C, Renner ED, Abel L, Picard C, Marodi L, Boisson-Dupuis S, Puel A, Casanova JL. Gain-of-function human STAT1 mutations impair IL-17 immunity and underlie chronic mucocutaneous candidiasis. The Journal of experimental medicine. 2011;208:1635–1648. doi: 10.1084/jem.20110958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lenardo M, Chan KM, Hornung F, McFarland H, Siegel R, Wang J, Zheng L. Mature T lymphocyte apoptosis--immune regulation in a dynamic and unpredictable antigenic environment. Annual review of immunology. 1999;17:221–253. doi: 10.1146/annurev.immunol.17.1.221. [DOI] [PubMed] [Google Scholar]
- 43.Bromberg J. Stat proteins and oncogenesis. The Journal of clinical investigation. 2002;109:1139–1142. doi: 10.1172/JCI15617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kelly J, Spolski R, Imada K, Bollenbacher J, Lee S, Leonard WJ. A role for Stat5 in CD8+ T cell homeostasis. Journal of immunology. 2003;170:210–217. doi: 10.4049/jimmunol.170.1.210. [DOI] [PubMed] [Google Scholar]
- 45.Shuai K. Modulation of STAT signaling by STAT-interacting proteins. Oncogene. 2000;19:2638–2644. doi: 10.1038/sj.onc.1203522. [DOI] [PubMed] [Google Scholar]
- 46.Zhu M, John S, Berg M, Leonard WJ. Functional association of Nmi with Stat5 and Stat1 in IL-2- and IFNgamma-mediated signaling. Cell. 1999;96:121–130. doi: 10.1016/s0092-8674(00)80965-4. [DOI] [PubMed] [Google Scholar]
- 47.Nakajima H, Brindle PK, Handa M, Ihle JN. Functional interaction of STAT5 and nuclear receptor co-repressor SMRT: implications in negative regulation of STAT5-dependent transcription. The EMBO journal. 2001;20:6836–6844. doi: 10.1093/emboj/20.23.6836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.He J, Tsai LM, Leong YA, Hu X, Ma CS, Chevalier N, Sun X, Vandenberg K, Rockman S, Ding Y, Zhu L, Wei W, Wang C, Karnowski A, Belz GT, Ghali JR, Cook MC, Riminton DS, Veillette A, Schwartzberg PL, Mackay F, Brink R, Tangye SG, Vinuesa CG, Mackay CR, Li Z, Yu D. Circulating precursor CCR7(lo)PD-1(hi) CXCR5(+) CD4(+) T cells indicate Tfh cell activity and promote antibody responses upon antigen reexposure. Immunity. 2013;39:770–781. doi: 10.1016/j.immuni.2013.09.007. [DOI] [PubMed] [Google Scholar]
- 49.Johnston RJ, Choi YS, Diamond JA, Yang JA, Crotty S. STAT5 is a potent negative regulator of TFH cell differentiation. The Journal of experimental medicine. 2012;209:243–250. doi: 10.1084/jem.20111174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Walker SR, Nelson EA, Frank DA. STAT5 represses BCL6 expression by binding to a regulatory region frequently mutated in lymphomas. Oncogene. 2007;26:224–233. doi: 10.1038/sj.onc.1209775. [DOI] [PubMed] [Google Scholar]
- 51.Dai X, Chen Y, Di L, Podd A, Li G, Bunting KD, Hennighausen L, Wen R, Wang D. Stat5 is essential for early B cell development but not for B cell maturation and function. Journal of immunology. 2007;179:1068–1079. doi: 10.4049/jimmunol.179.2.1068. [DOI] [PubMed] [Google Scholar]
- 52.Villarino A, Laurence A, Robinson GW, Bonelli M, Dema B, Afzali B, Shih HY, Sun HW, Brooks SR, Hennighausen L, Kanno Y, O’Shea JJ. Signal transducer and activator of transcription 5 (STAT5) paralog dose governs T cell effector and regulatory functions. Elife. 2016:5. doi: 10.7554/eLife.08384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kimura A, Rieger MA, Simone JM, Chen W, Wickre MC, Zhu BM, Hoppe PS, O’Shea JJ, Schroeder T, Hennighausen L. The transcription factors STAT5A/B regulate GM-CSF-mediated granulopoiesis. Blood. 2009;114:4721–4728. doi: 10.1182/blood-2009-04-216390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Veltman JA, Brunner HG. De novo mutations in human genetic disease. Nature reviews. Genetics. 2012;13:565–575. doi: 10.1038/nrg3241. [DOI] [PubMed] [Google Scholar]
- 55.MacLeod MK, Kappler JW, Marrack P. Memory CD4 T cells: generation, reactivation and re-assignment. Immunology. 2010;130:10–15. doi: 10.1111/j.1365-2567.2010.03260.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tuma RA, Pamer EG. Homeostasis of naive, effector and memory CD8 T cells. Current opinion in immunology. 2002;14:348–353. doi: 10.1016/s0952-7915(02)00338-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.