Abstract
Purpose
Mutations in the gene encoding 5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase (ATIC), a bifunctional enzyme that catalyzes the final two steps of the purine de novo biosynthetic pathway, were identified in a subject referred for radiation sensitivity testing. Functional studies were carried out to determine if ATIC inhibition was radiosensitizing, and if so, to elucidate the mechanism of this effect and to determine if small molecule inhibitors of ATIC could act as effective radiosensitizing agents.
Methods and Materials
Both siRNA knockdown and small molecule inhibitors were used to inactivate ATIC in cell culture. Clonogenic survival assays, the neutral comet assay, and γH2AX staining were used to assess the effects of ATIC inhibition or depletion on cellular DNA damage responses.
Results
Depletion of ATIC or inhibition of its transformylase activity significantly reduced the surviving fraction of cells in clonogenic survival assays in multiple cancer cell lines. In the absence of ionizing radiation exposure, ATIC knockdown or chemical inhibition activated cell cycle checkpoints, shifting cells to the more radiosensitive G2/M phase of the cell cycle, and depleted cellular ATP, but did not result in detectable DNA damage. Cells in which ATIC was knocked down or inhibited and then treated with ionizing radiation displayed increased numbers of DNA double-strand breaks (DSBs), and a delay in the repair of those breaks relative to irradiated, but otherwise untreated, controls. Supplementation of culture media with exogenous ATP ameliorated the DNA repair phenotypes.
Conclusions
These findings implicate ATIC as an effective, and previously unrecognized, target for chemoradiosensitization and more broadly suggest that purine levels in cells may have an underappreciated role in modulating the efficiency of DNA damage responses that could be exploited in radiosensitizing strategies.
Introduction
In humans, hypersensitivity to ionizing radiation frequently co-occurs with a constellation of clinical and laboratory features that includes increased cancer incidence, immunodeficiency, neurologic abnormalities and DNA breakage and have been termed the XCIND syndrome (X-ray sensitivity, Cancer predisposition, Immunodeficiency, Neurologic involvement, and DNA double-strand break repair deficiency) [9,22]. Several well-defined human autosomal recessive disorders with these clinical features arise from mutations in key DNA damage response molecules [25]. Although these inherited disorders are individually rare, identification of the causative genes has provided broadly applicable insights into the mechanisms and components of cellular DNA damage responses. These include the identification of the Ataxia-telangiectasia Mutated gene (ATM), the primary regulator of cellular responses to DNA double-strand breaks [8,32] that is mutated in Ataxia-Telangiectasia (A-T), Nibrin (NBN), a regulator of the S-phase checkpoint which is mutated in Nijmegen Breakage Syndrome [2,21,42], and the delineation of a novel role for ubiquitination in regulating the cellular response to ionizing radiation through the study of Ring Finger Protein 168 (RNF168) deficiency [5,38]. Not all cases of apparently genetic hypersensitivity to ionizing radiation can be accounted for by these established disorders [12,14,23,27]. Identification of mutations in subjects with unexplained radiation hypersensitivity could point to genes or to cellular pathways not previously implicated in DNA damage responses, providing novel mechanistic insights and potential drug targets for chemoradiosensitization.
We have previously described a panel of subjects referred for diagnostic testing for A-T or NBS [23]. The subjects share clinical features characteristic of these disorders and, where available, the cell lines established from them display radiation hypersensitivity in clonogenic survival assays. However, sequencing of the ATM and NBN genes in these subjects has failed to reveal credible causative mutations. Applying exome sequencing to this panel, we previously identified a subject with a homozygous missense mutation in the gene Mitochondrial Poly(A) Polymerase (MTPAP), which had not been previously implicated in DNA damage responses, and demonstrated that this mutation was responsible for the radiation hypersensitivity by complementation [20]. Here we characterize the product of a second gene mutated in a subject in this panel, ATIC, which encodes the bifunctional purine biosynthetic enzyme 5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase. We demonstrate that depletion of ATIC by siRNA treatment or chemical inhibition of its enzymatic activity impairs the ability of cells to repair DNA double strand breaks, reduces cell survival after irradiation and perturbs intracellular ATP pools.
Methods and Materials
Cell lines and reagents
GM00637, an SV40 transformed human fibroblast line obtained from the NIGMS repository, was maintained in DMEM medium supplemented with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin-glutamine (PSG). HCT116, a human colorectal carcinoma line, was kindly provided by Dr. Robert Hromas. SW48, a colorectal adenocarcinoma line, and U2OS, an osteosarcoma line, were purchased from ATCC (Manassas Virginia). HCT116, SW48, and U2OS were maintained in McCoy’s 5A medium supplemented with 10% FBS and 1% PSG at 37°C with 5% CO2, 95% humidity and a pH of 7.4. Cells were harvested under log growth phase conditions and irradiation was carried out using a GammaCell 40 Exactor Cs-137 sealed source irradiator at a dose rate of ~1 Gy/min.
The chemical inhibitor of ATIC homo-dimerization, Cpd14 [1,37,41], was purchased from EMD Millipore (Billerica, MA) while the ATIC active site inhibitors NSC30171 and NSC326203 were obtained from the National Cancer Institute Developmental Therapeutics repository.
RNAi knockdown and cell viability assay
siRNA sequences (Table E1) directed against three different sites in each of four genes, ATIC, ATM, NBN, and MTPAP, were printed onto a 96-well plate (Qiagen, Valencia CA). The negative control siRNA (siControl), a non-silencing RNA with no homology to any known mammalian gene, was also purchased from Qiagen (Catalogue No. 1027310). Cationic lipid-based transfection reagents were diluted in Opti-MEM (Invitrogen, Carlsbad CA), added to assay plates and 6000 GM00637 cells were added per well. At 48 hours post-transfection, cells were irradiated with 0 or 4 Gy of γ radiation. After an additional 72 hours, cell viability was measured using an MTT assay (Promega, Madison WI) according to the supplier's instructions. Each knockdown was performed in triplicate.
Quantitative PCR
Total cellular RNA was extracted from GM00637 cells 48 hours after siRNA transfection. cDNA was prepared using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Carlsbad CA). All primer pairs used were prefabricated mixtures of forward and reverse primers (Quantitect Primer Assay; QIAGEN, Valencia CA). Relative quantification was performed with SYBR Green (Invitrogen, Carlsbad CA) using GAPDH as an internal control for normalization. Target and reference samples were tested in triplicate for each experiment. PCR efficiency was determined by constructing a standard curve with cDNA containing the target template.
Western blot analysis
Cells were seeded at 3×105 cells per well in 6-well plates and duplicate wells were treated with siRNA at the indicated concentration for 48 hours. Cell lysates were prepared and total protein concentration determined by the BCA protein assay (ThermoFisher, Waltham MA). 20 µg aliquots of each sample were separated on 5–12% SDS-polyacrylamide gels, transferred to nitrocellulose blotting membrane (Bio-rad, Hercules CA) and immunoblotted with antibodies directed against ATM, ATIC (Abnova, Taiwan), NBN (Cell Signaling, Danvers MA), MTPAP (EMD Millipore, Billeria MA), p53 (Santa Cruz, Dallas TX), p21(Cip1 /WAF1), cyclin B, tubulin, AMPK or Phospho-T172 AMPK (Cell Signaling, Danvers MA), all at a dilution of 1:1000, and incubated at 4 °C overnight. Membranes were washed and incubated with horseradish peroxidase-conjugated secondary antibodies for 1 hour at room temperature, washed and developed using enhanced chemiluminescence protocols (ECL; GE Health Care Life Science, Piscataway Township NJ) and quantified using ImageQuant software (GE Health Care Life Science, Piscataway Township NJ).
Colony survival assay
HCT 116, SW48, and U2OS cells were treated separately with one of three different ATIC siRNAs (siATICa, siATICb, or siATICc, Table E1) or chemical inhibitors at indicated the concentrations for 48 hours. Cells, 250 to 2000 per well in a 6 well plate, were plated in triplicate with McCoy’s 5A medium and sham treated or irradiated. After 3 weeks, colonies were fixed with 10% formaldehyde in PBS, stained with 0.1% crystal violet in PBS and counted. Results were compared to sham treated plates to determine the survival fraction percentage (SF%) and survival curves were generated using SigmaPlot software (Systat Software Inc, Chicago IL). Radiation dose-modifying factors were calculated at 10% clonogenic survival from linear-quadratic curve equations used to plot the curves shown in the figures. The dose-modifying factors reported are the root values for control curves divided by the values for treated curves.
Cell cycle analysis
HCT116, SW48, and U2OS cells were treated with siATICa or Cpd14 at the indicated concentration for 24 or 48 hours, collected and fixed in 70% ethanol at −20°C, stained with propidium iodide/RNAase buffer for DNA content, and analyzed on an LSR II flow cytometer (BD biosciences, San Jose CA). ModFit software (Verity Software House, Topsham ME) was used to assess cell cycle distribution.
Measurement of intracellular ATP
HCT116 colon cancer cells were treated with siATICa or Cpd14 at the indicated concentration. At 12, 24, 36, 48 hours ATP levels were measured using the luciferase/luciferin-based ENLITEN ATP assay system (Promega, Madison, WI). All samples were tested in triplicate.
Irradiation-induced focus assay
HCT116 colon cancer cells were treated with siATICa or Cpd14 for 48 hours followed by radiation exposure at 2 Gy. Cells were collected at 1, 2, 6, 24 and 48 hours, plated on coverslips, fixed with 4% paraformaldehyde, permeabilized with 0.5% Triton-X 100, blocked in 10% FBS, and incubated with anti-γ-H2AX (Santa Cruz, Dallas Texas) (1:300) for 1 hour at room temperature. The coverslips were washed, blocked with 10% FBS and incubated with an AlexaFluor-488 secondary antibody (1:400, Invitrogen, Carlsbad CA) for 45 minutes at room temperature. Coverslips were washed a final time and mounted on slides using Prolong Gold anti-fade reagent containing DAPI (Applied Biosystems, Carlsbad CA). Foci were imaged using a Zeiss fluorescent microscope equipped with an AxioVision camera and software. Cells were scored as positive if they contained 4 or more foci/nuclei. In order to measure the initial induction of DSB following radiation, cells were collected at 1 hour after radiation exposure and foci/nuclei were scored. In order to measure DSB repair, cells were collected at 1, 2, 6, 24, 48 hours after radiation exposure and scored for the reduction of foci relative to the 1 hour time point.
Neutral comet assay
HCT116 cells were treated with siATICa or Cpd14 for 48 hours. Cells were then sham treated or irradiated with 2 Gy, collected at 1 hour post-radiation, re-suspended in 1% low melting point agarose and plated on 2-well comet assay slides. Once the agarose solidified, the slides were immersed in cell lysis solution (Trevigen Inc, Gaithersburg MD) overnight at 4 °C. Slides were electrophoresed and stained using SYBR Green (Invitrogen, Carlsbad CA). Comets were visualized using a Zeiss fluorescent microscope equipped with an AxioVision camera and software. The tail moments (TM) of comets were scored using CometScore software (TriTek, Sumerduck VA). Percent repair (% repair) was determined by monitoring the return to baseline TM levels. Cells were sham treated or irradiated with 2 Gy, collected at 1 and 12 hours postradiation, re-suspended in 1% low melting point agarose and assayed as described above. The initial induction of DSBs was measured 1 hour following 2Gy γ ray exposure, and residual DSBs after repair were measured 12 hours after exposure. TMs were normalized to unirradiated cells. Percent repair (% repair) was calculated as the difference in tail moment between the 1 and 12 hour time points relative to the 1 hour time point, (TM1hr − TM12hrs)/ TM1hr..
Statistical analyses
All data are presented as the mean ± SEM. Statistical significance (P < 0.05) was determined by t test using Graphpad Prism v. 5.0.
Results
ATIC is required for normal cell survival following irradiation
Exome sequencing of a subject referred for testing for A-T or NBS revealed heterozygosity for a complex frameshift mutation, c.130_131insG;131_132insA, and a missense substitution of unknown consequence, c.347C>G, p.Thr116Ser, in the ATIC gene. Although there was no clinical information available on this subject, we noted a previously published report describing a single patient with severe neurologic and developmental features who was heterozygous for the same frameshift mutation in the ATIC gene but had a different and potentially more damaging missense substitution affecting a critical residue on the ATIC transformylase domain [19].
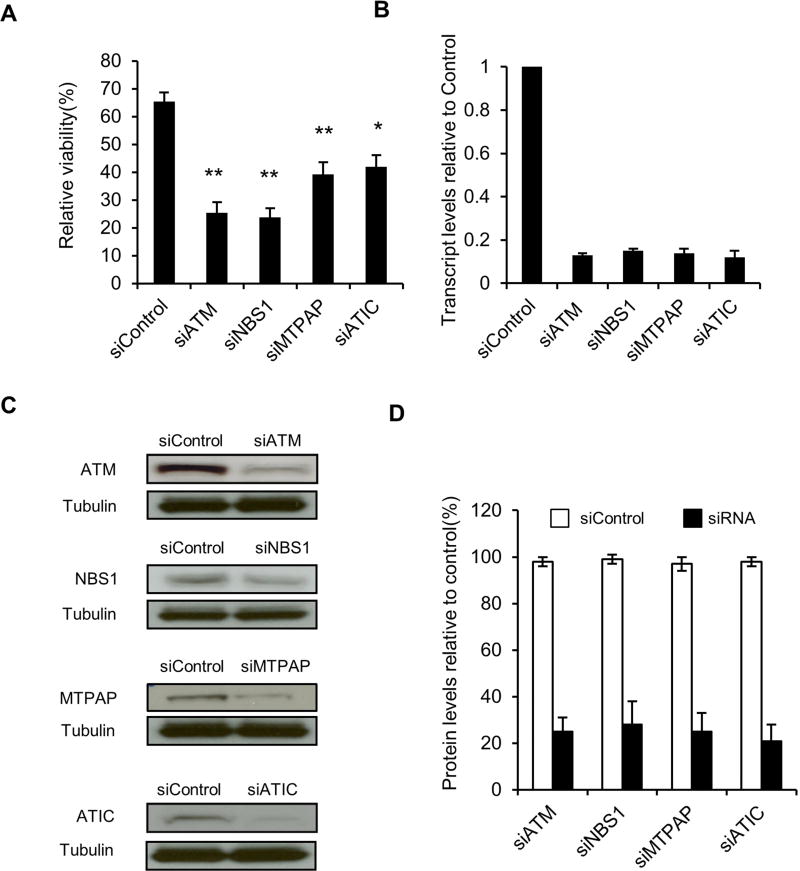
Given the shared genetic features in these two subjects, we tested whether reducing the cellular level of ATIC might affect DNA damage responses. A cell line with normal radiation sensitivity, GM00637, was treated with a pool of 3 siRNAs targeting ATIC, irradiated at 4 Gy and viable cells were counted after 72 hours (Fig. 1A). For comparison purpose, GM00637 cells were treated in parallel with similarly designed pools of siRNAs targeting genes with established roles in cellular responses to ionizing radiation, ATM, NBN, and MTPAP. Effectiveness of the siRNA knockdown of the genes was confirmed for mRNA and protein by quantitative PCR and immunoblotting, respectively (Fig. 1B–D). Knockdown of ATIC significantly reduced cell counts relative to mock treated controls. Despite comparable levels of knockdown (Fig. 1D), siRNA depletion of ATIC had a less significant effect on cell viability than for the well-established DNA damage response genes ATM and NBN (Fig. 1A).
Fig. 1.
siRNA knockdown of ATIC is radiosensitizing. (A) GM00637 cells were transfected with pooled siRNAs targeting ATM (siATM), NBN (siNBS1), MTPAP (siMTPAP) or ATIC (siATIC). The mean percentage ± SEM of viable cells in the irradiated (4Gy) samples relative to mock-irradiated is plotted (*P < 0.05, **P<0.01, compared to siControl). (B) The fraction of transcripts in cells treated with gene-specific siRNAs compared to siControl is plotted. (C) Cells treated with either siControl or the pools of siRNAs directed against the indicated genes were immunoblotted for the products of the targeted genes. (D) Protein levels from (C) quantified and expressed as percentage relative to siControl.
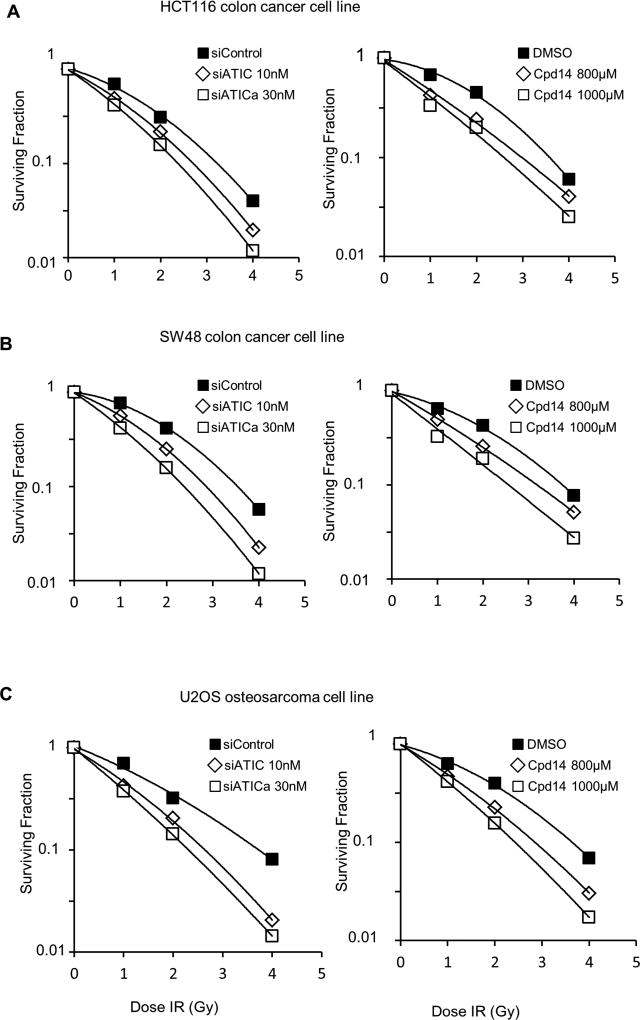
A small molecule inhibitor, Cpd14, that interferes with the homodimerization of ATIC has been identified and characterized. Dimerization is essential to form the active site for ATIC’s transformylase [1,37,41]. Treatment of HCT116, SW48 or U2OS cells with siRNAs directed against ATIC or with Cpd14 48 hours prior to irradiation resulted in a dosage-dependent reduction in cell survival relative to untreated, but irradiated, controls in a clonogenic survival assay (Figs. 2, E1). Efficiency of the knockdowns were confirmed by immunoblotting (Fig. E2). The radiation dose-modifying factors for siATICa treatment of HCT116 cells at 10nM and 30nM were 1.17 and 1.30, and for Cpd14 at 800µM and 1000µM, 1.2 and 1.4, respectively. The radiation dose-modifying factors for siATICa treatment of SW48 cells at 10nM and 30nM were 1.22 and 1.48, respectively, and for Cpd14 at 800µM and 1000µM, 1.17 and 1.47. The radiation dose-modifying factors for siATICa treatment of U2OS cells at 10nM and 30nM were 1.40 and 1.61, respectively, and for Cpd14 at 800µM and 1000µM, 1.28 and 1.52.
Fig. 2.
Effect of ATIC inhibition or depletion on radiation survival. HCT116 (A), SW48 (B), and U2OS (C) cells were treated with the indicated concentrations of siATICa or the ATIC inhibitor, Cpd14, for 48 hours prior to irradiation at 0, 1, 2, and 4 Gy. Surviving fraction relative to the 0 Gy treatment is plotted.
In addition to the ATIC dimerization inhibitor Cpd14 [37], a virtual ligand screen has identified a number of active site inhibitors that are specific for the transformylase activity of ATIC [18,43]. We tested two of the compounds (NSC30171 and NSC326203) identified in this screen for their effects on cell survival in irradiated HCT116 cells. Both compounds provided comparable radiosensitization to that obtained with Cpd14 but at tenfold and twofold lower concentrations, respectively (Fig. E3).
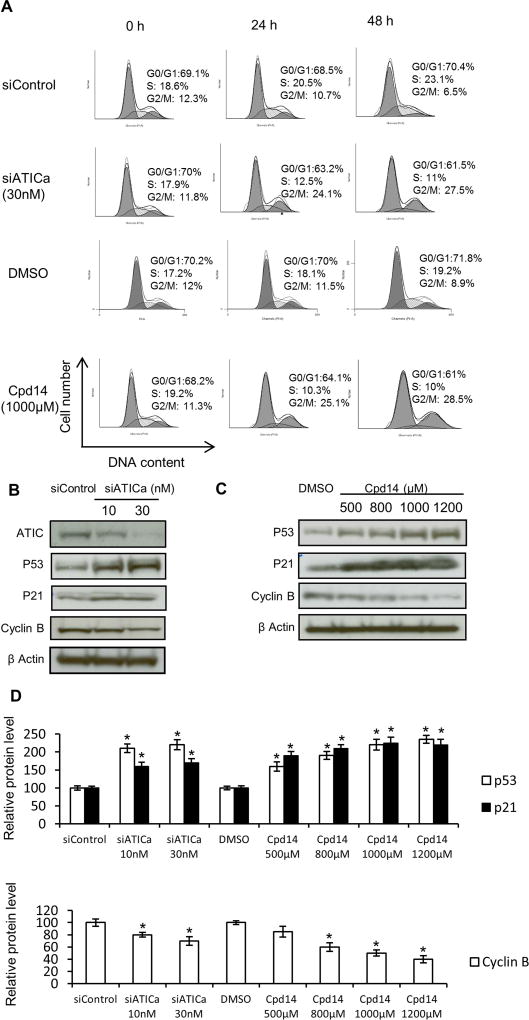
ATIC deficiency or inhibition activates cell cycle checkpoints
Treatment of HCT116 cells with either siATICa or Cpd14 resulted in modest cell cycle redistribution in asynchronous cultures with decreased numbers of cells in S phase and increased number of cells in the more radiosensitive G2/M (Fig. 3A). Relative to mock treated controls, treated cells displayed increased levels of p53 and the cyclin-dependent kinase inhibitor p21, and decreased levels of Cyclin B1, consistent with activation of the G1/S checkpoint and G2/M checkpoints, respectively, and the observed cell cycle distribution (Fig. 3B and 3C). Comparable results were observed in two SW48 and U2OS cells (Fig. E4, E5)
Fig. 3.
ATIC inhibition or depletion activates cell cycle checkpoints. (A) DNA content was assessed by flow cytometry in untreated cells and cells treated with siATICa or Cpd14. Cell cycle distributions are indicated as:
 G1 and G2/M,
G1 and G2/M,
 S phase. HCT116 cells treated with either siATICa (B) or Cpd14 (C) at indicated doses were immunoblotted for checkpoint proteins. β Actin serves as a loading control. (D) Relative protein levels of western blots in (B) and (C) were quantified by ImageQuant. * indicates P<0.05, compared to controls.
S phase. HCT116 cells treated with either siATICa (B) or Cpd14 (C) at indicated doses were immunoblotted for checkpoint proteins. β Actin serves as a loading control. (D) Relative protein levels of western blots in (B) and (C) were quantified by ImageQuant. * indicates P<0.05, compared to controls.
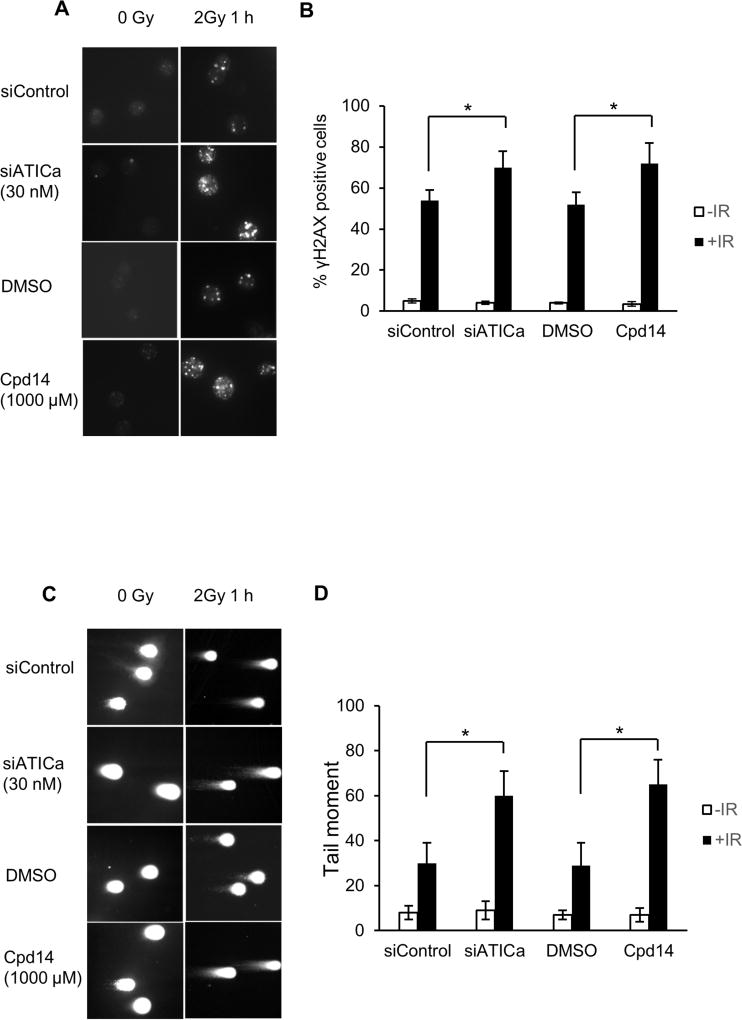
Increased radiation induced DNA double strand breaks in cells deficient in ATIC
The effect of ATIC depletion or inhibition on the repair of DNA double strand breaks was assessed in HCT116 cells by γH2AX focus formation and the neutral comet assay. Perturbation of ATIC, alone, did not result in any significant increase in γH2AX foci (Fig. 4A, 0h time point). However, when combined with ionizing radiation, increased numbers of γH2AX foci per cell were detected (Fig. 4A) and there was a significant increase in the percentage of cells containing γH2AX foci (Fig. 4B). Similar results were obtained with comparably treated cells in the neutral comet assay. Tail moments at one hour post-irradiation were increased relative to mock treated irradiated controls (Fig. 4C and 4D). Taken together, the results from the γH2AX focus assay and the neutral comet assay indicate that depletion of ATIC or inhibition of its transformylase activity results in increased numbers of DNA double strand breaks specifically in irradiated cells.
Fig. 4.
ATIC inhibition or depletion increases the incidence of DNA double-strand breaks in HCT116 colon cancer cells after irradiation. (A) γH2AX staining at 0 hour and 1 hour after irradiation (2 Gy). Cells were sham treated, treated with siATICa or with Cpd14 at the indicated concentration for 48 hours prior to radiation exposure (IR). (B) Percentage γH2AX positive cells in treated and control groups. * indicates P<0.05. (C) Neutral comet assay performed at 0 hour and 1 hour after irradiation (2 Gy). Cells were sham treated, treated with siATIC or with Cpd14 at the indicated concentration for 48 hours prior to radiation exposure. (D) Quantitation of tail moments in treated and control groups (*P<0.05).
Effects of ATIC inhibition ATP levels and DNA damage responses
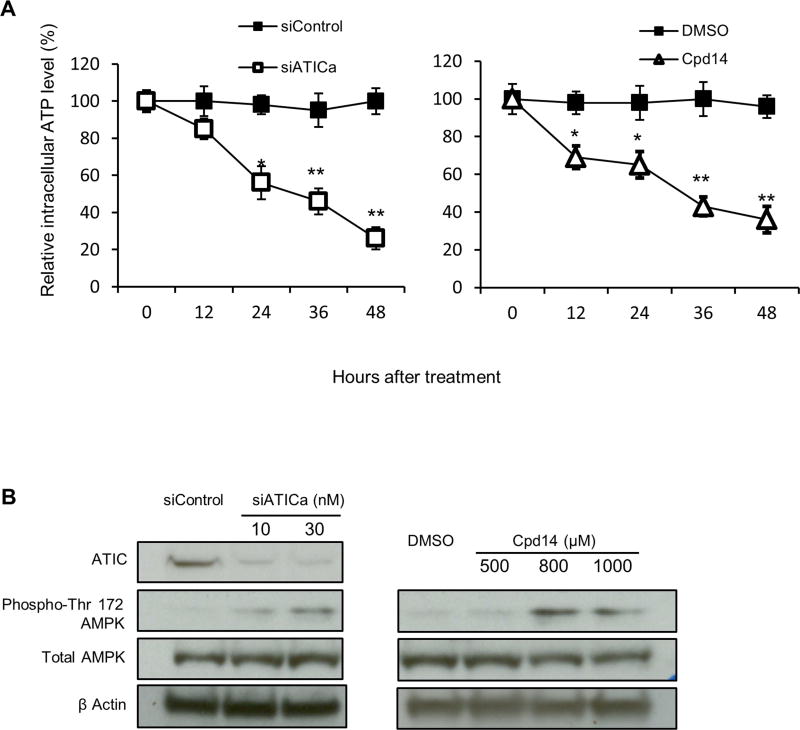
Treatment of cells with Cpd14 has been shown to result in accumulation of ZMP, the substrate for the ATIC transformylase [1], which, because of its structural similarity to AMP, can activate AMP-kinase (AMPK) [11]. Marie et al. reported increased levels of ZMP, ZDP and ZTP in red blood cells from their patient with bi-allelic ATIC mutations [19] but also a significant decline in the ATP/AMP ratio which they attributed to the phosphorylation of ZMP by adenosine kinase, as occurs in Lesch-Nyhan syndrome [34,40]. We observed that treatment of HCT116 cells with either siATICa or Cpd14 also resulted in a significant and sustained reduction in intracellular ATP levels (Fig. 5A) and confirmed that this perturbation of ATP levels was associated with activation of AMPK via phosphorylation on T172 of its catalytic α subunit (Fig. 5B).
Fig. 5.
ATIC inhibition or depletion reduces intracellular ATP levels and activates AMPK (*P < 0.05, **P<0.01, compared to control). (A) HCT116 Cells were treated with siATICa (30nM) or Cpd14 (1000µM) as indicated. At 12, 24, 36, 48 hours ATP levels were measured. (B) HCT116 Cells were treated with siATIC or a chemical ATIC inhibitor at concentration indicated. After 48 hours of treatment total cell lysates were analyzed by western blot for ATIC, total AMPK and phosphorylated AMPK expression level.
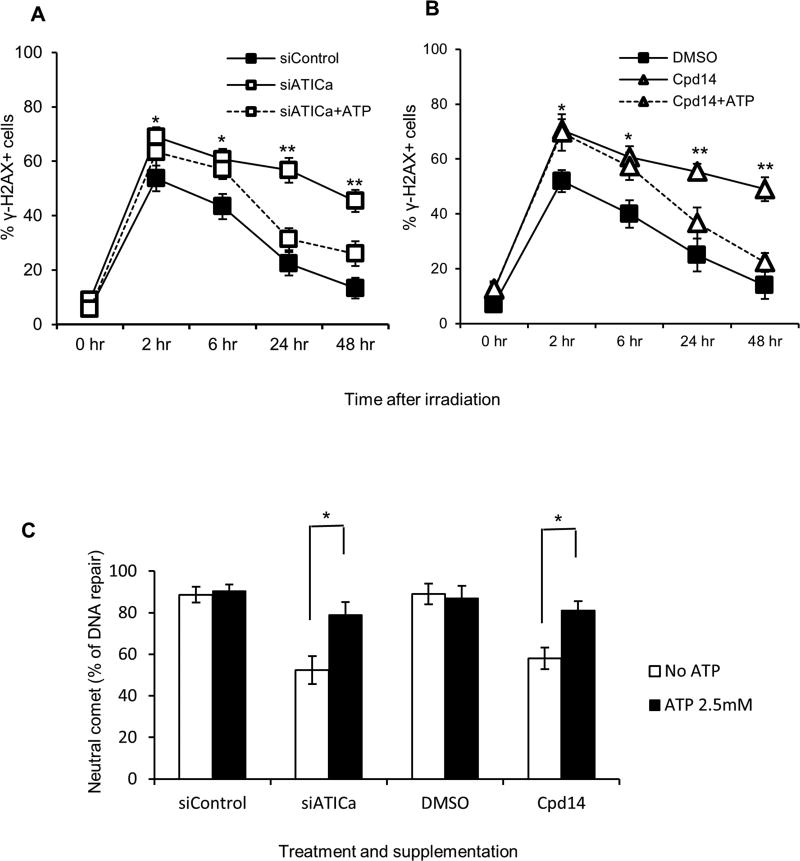
To explore the relationship between radiation sensitivity and alterations in cellular ZMP and ATP levels, the effect of ATP supplementation on the induction and resolution of DNA double strand breaks by ionizing radiation in cells in which ATIC was depleted or inhibited was assessed. The specific phenotype of DNA double-strand break repair was assayed because ATP supplementation might be expected to have general effects on broader phenotypes such as cell viability unrelated to the inhibition or depletion of ATIC. The percentage of γH2AX positive cells in cultures treated with siATICa (Fig. 6A) or Cpd14 (Fig. 6B) was increased, relative to untreated controls, at 2 hours post-irradiation and remained elevated throughout the 48 hours of the assay. The addition of exogenous ATP to the culture media one hour prior to irradiation did not prevent the initial increase in γH2AX foci in cells treated with siATICa or Cpd14 but did reduce the percentage of positive cells at later time points. In the neutral comet assay, ATP supplementation significantly decreased DNA fragmentation in cells treated with siATICa or Cpd14 at later time points relative to that observed at 1 hour after irradiation (Fig. 6C). This effect was observed across a range of supplementary ATP concentrations from 1 mM to 4.5 mM using siATICa (Fig. E6). Supplementation with GTP, CTP or TTP at 2.5 mM did not ameliorate the effect of siATICa on DNA repair in the neutral comet assay (Fig. E7).
Fig. 6.
ATP supplementation reverses the effects of ATIC inhibition or depletion. (*P < 0.05, **P<0.01, compared to control). (A) HCT116 cells were left untreated (siControl) or pretreated for 48 hours with 30nM siATICa and either not supplemented (siATICa) or supplemented with 2.5mM ATP (siATICa+ATP) one hour prior to irradiation (2 Gy). DSB repair were measured via γH2AX staining assay as described in method section. (B) HCT116 cells were left untreated (DMSO) or pretreated for 48 hours with 1000µM Cpd14 and either not supplemented (Cpd14) or supplemented (Cpd14+ATP) with 2.5mM ATP one hour prior to irradiation (2 Gy). DSB repair were measured via γH2AX staining assay as described in method section. (C) Cells were treated as in (A) and (B), DSB repair was measured via the neutral comet assay as described in method section. Percent repair (% DNA repair) was determined by monitoring the return of the tail moments (TM) of comets to baseline levels ((TM1hrs −TM12 hrs)/TM1hrs). * indicates P<0.05.
Discussion
In the current study, we explored the effect of depleting ATIC or inhibiting its transformylase activity on cellular radiation survival and double strand break repair, motivated by the detection of a loss of function mutation in the ATIC gene during exome sequencing of a panel of radiosensitive individuals. A bifunctional enzyme, ATIC possesses separate aminoimidazole carboxamide ribonucleotide transformylase and inosine monophosphate cyclohydrolase activities that catalyze the penultimate and final steps in de novo purine synthesis, respectively [34,40]. Vertebrates rely primarily on the purine salvage pathway, rather than this synthesis pathway, to provide purines for nucleotide and nucleic acid synthesis, and, consistent with this, we observed only a modest effect of ATIC inhibition in human cells on cell cycle progression. When the purine salvage pathway is disrupted, as in the genetic disorder Lesch-Nyhan syndrome [34,40], the transformylase activity of ATIC becomes rate-limiting for purine synthesis, and its substrate, 5-aminoimidazole-4-carboxamide ribotide (ZMP) accumulates. This effect can be mimicked by treatment with the cell-permeable precursor 5-aminoimidazole-4-carboxamide riboside (AICAR), which is converted to ZMP in cells by phosphorylation. Antimetabolites that target folate metabolism, such as methotrexate, can also affect ATIC’s transformylase activity by depleting 10-formyltetrahydrofolate (N10-Formyl-FH4), the one-carbon donor necessary for the formylation of ZMP. Indeed, the ATIC c.347C>G variant detected in the subject studied here has been reported to be associated with adverse responses to methotrexate suggesting a possible deleterious effect on ATIC [17]. Both AICAR and methotrexate have been reported to radiosensitize, although their mechanisms are unresolved [10,13,16]. We demonstrate here that ATIC knockdown, or treatment with any of three different small molecule inhibitors of ATIC’s transformylase activity, acting by two different mechanisms, dimerization inhibition or active site competition, results in radiosensitization. We further demonstrate that this radiosensitization arises, in part, due to deficient DNA double strand break repair in cells in which ATIC’s transformylase activity is inhibited. Considered together, these prior reports and the current findings strongly suggest that the transformylase activity of ATIC plays an important role in radioprotection.
Treatment of cells in culture with the ATIC dimerization inhibitor Cpd14 has been shown to increase levels of ZMP [1] and red blood cells from a previously reported patient with biallelic deleterious mutations in ATIC displayed increased ZMP levels as well as a decline in the ratio of ATP to AMP [19]. AMPK is activated by either of these conditions, but also by DNA damaging agents, including ionizing radiation [29]. We observed that ATIC inhibition activated AMPK, resulting in phosphorylation on the same residue, T172 of the regulatory α subunit, which is also phosphorylated in response to irradiation. This raises the possibility that AMPK could serve as an integrator of independent stress signals arising from combined treatment of cells with ionizing radiation and an ATIC inhibitor, and, indeed, a role for AMPK as an integrator of cellular stress signals has been previously proposed [30]. Upon activation, AMPK phosphorylates multiple substrates that act to balance the requirements of cell growth with the available energy supply through acute effects on metabolism, longer term transcriptional adaptations and regulation of cell cycle progression. It is unclear whether AMPK acts directly on the components of DNA double strand break repair pathways. However, we did observe that ATP supplementation could ameliorate the DNA repair defect caused by ATIC inhibition and there is a long history of reports that highlight the ATP dependence of repair of ultraviolet or ionizing radiation damage to DNA [3,4,6,15,24,33,35]. In addition, prior studies have shown that inhibition of AMPK activity can result in radioresistance [31,39] while AMPK activators can augment cell killing by ionizing radiation [7,26,28,36,44].
Conclusions
Depletion of ATIC by siRNA knockdown impairs DNA double strand break repair, reducing cellular radiation survival. Small molecule inhibitors specific for the transformylase activity of ATIC have comparable effects suggesting that ATIC may be an effective target for chemoradiosensitization strategies.
Supplementary Material
Summary.
Detection of a frameshift mutation in the gene encoding the purine biosynthetic enzyme ATIC in a panel of radiosensitive individuals prompted studies to elucidate the role of ATIC in DNA damage responses and ascertain whether inhibition of ATIC might be an effective chemoradiosensitization strategy. Knockdown of ATIC or inhibition of its enzymatic activity compromised DNA double strand break repair and reduced cell survival after irradiation.
Acknowledgments
This work was supported by grants R21 ES020521 and R01 ES027121 from the National Institute of Environmental Health Sciences (to PC).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest: none.
References
- 1.Asby DJ, et al. Ampk activation via modulation of de novo purine biosynthesis with an inhibitor of atic homodimerization. Chemistry & biology. 2015;22:838–848. doi: 10.1016/j.chembiol.2015.06.008. [DOI] [PubMed] [Google Scholar]
- 2.Carney JP, et al. The hmre11/hrad50 protein complex and nijmegen breakage syndrome: Linkage of double-strand break repair to the cellular DNA damage response. Cell. 1998;93:477–486. doi: 10.1016/s0092-8674(00)81175-7. [DOI] [PubMed] [Google Scholar]
- 3.Ciarrocchi G, Linn S. A cell-free assay measuring repair DNA synthesis in human fibroblasts. Proceedings of the National Academy of Sciences of the United States of America. 1978;75:1887–1891. doi: 10.1073/pnas.75.4.1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deshpande RA, et al. Atp-driven rad50 conformations regulate DNA tethering, end resection, and atm checkpoint signaling. EMBO J. 2014;33:482–500. doi: 10.1002/embj.201386100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Devgan SS, et al. Homozygous deficiency of ubiquitin-ligase ring-finger protein rnf168 mimics the radiosensitivity syndrome of ataxia-telangiectasia. Cell death and differentiation. 2011;18:1500–1506. doi: 10.1038/cdd.2011.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dresler SL, Lieberman MW. Requirement of atp for specific incision of ultravioletdamaged DNA during excision repair in permeable human fibroblasts. J Biol Chem. 1983;258:12269–12273. [PubMed] [Google Scholar]
- 7.Fritz G, Brachetti C, Kaina B. Lovastatin causes sensitization of hela cells to ionizing radiation-induced apoptosis by the abrogation of g2 blockage. International journal of radiation biology. 2003;79:601–610. doi: 10.1080/09553000310001609233. [DOI] [PubMed] [Google Scholar]
- 8.Gatti RA, et al. Localization of an ataxia-telangiectasia gene to chromosome 11q22-23. Nature. 1988;336:577–580. doi: 10.1038/336577a0. [DOI] [PubMed] [Google Scholar]
- 9.Gatti RA, Boder E, Good RA. Immunodeficiency, radiosensitivity, and the xcind syndrome. Immunologic research. 2007;38:87–101. doi: 10.1007/s12026-007-0018-y. [DOI] [PubMed] [Google Scholar]
- 10.Gupta NK, Pointon RC, Wilkinson PM. A randomised clinical trial to contrast radiotherapy with radiotherapy and methotrexate given synchronously in head and neck cancer. Clinical radiology. 1987;38:575–581. doi: 10.1016/s0009-9260(87)80327-6. [DOI] [PubMed] [Google Scholar]
- 11.Henin N, Vincent MF, Van den Berghe G. Stimulation of rat liver amp-activated protein kinase by amp analogues. Biochimica et biophysica acta. 1996;1290:197–203. doi: 10.1016/0304-4165(96)00021-9. [DOI] [PubMed] [Google Scholar]
- 12.Hiel JA, et al. Nijmegen breakage syndrome in a dutch patient not resulting from a defect in nbs1. J. Med. Genet. 2001;38:E19. doi: 10.1136/jmg.38.6.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Isebaert SF, et al. 5-aminoimidazole-4-carboxamide riboside enhances effect of ionizing radiation in pc3 prostate cancer cells. International journal of radiation oncology, biology, physics. 2011;81:1515–1523. doi: 10.1016/j.ijrobp.2011.06.1964. [DOI] [PubMed] [Google Scholar]
- 14.Izatt L, et al. Autosomal recessive spinocerebellar ataxia and peripheral neuropathy with raised alpha-fetoprotein. Journal of neurology. 2004;251:805–812. doi: 10.1007/s00415-004-0427-y. [DOI] [PubMed] [Google Scholar]
- 15.Kaufmann WK, et al. Requirements for adenosine triphosphate in DNA repair in isolated hepatic nuclei. Biochemical and biophysical research communications. 1982;108:1040–1047. doi: 10.1016/0006-291x(82)92104-0. [DOI] [PubMed] [Google Scholar]
- 16.Knowlton AH, et al. Methotrexate and radiation therapy in the treatment of advanced head and neck tumors. Radiology. 1975;116:709–712. doi: 10.1148/116.3.709. [DOI] [PubMed] [Google Scholar]
- 17.Lee YH, Bae SC. Association of the atic 347 c/g polymorphism with responsiveness to and toxicity of methotrexate in rheumatoid arthritis: A meta-analysis. Rheumatology international. 2016;36:1591–1599. doi: 10.1007/s00296-016-3523-2. [DOI] [PubMed] [Google Scholar]
- 18.Li C, et al. Virtual screening of human 5-aminoimidazole-4-carboxamide ribonucleotide transformylase against the nci diversity set by use of autodock to identify novel nonfolate inhibitors. Journal of medicinal chemistry. 2004;47:6681–6690. doi: 10.1021/jm049504o. [DOI] [PubMed] [Google Scholar]
- 19.Marie S, et al. Aica-ribosiduria: A novel, neurologically devastating inborn error of purine biosynthesis caused by mutation of atic. American journal of human genetics. 2004;74:1276–1281. doi: 10.1086/421475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martin NT, et al. Homozygous mutation of mtpap causes cellular radiosensitivity and persistent DNA double-strand breaks. Cell death & disease. 2014;5:e1130. doi: 10.1038/cddis.2014.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matsuura S, et al. Positional cloning of the gene for nijmegen breakage syndrome. Nat Genet. 1998;19:179–181. doi: 10.1038/549. [DOI] [PubMed] [Google Scholar]
- 22.Mizutani S, Takagi M. Xcind as a genetic disease of x-irradiation hypersensitivity and cancer susceptibility. International journal of hematology. 2013;97:37–42. doi: 10.1007/s12185-012-1240-5. [DOI] [PubMed] [Google Scholar]
- 23.Nahas SA, et al. Comprehensive profiling of radiosensitive human cell lines with DNA damage response assays identifies the neutral comet assay as a potential surrogate for clonogenic survival. Radiation research. 2012;177:176–186. doi: 10.1667/rr2580.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pentz M, Vatev R, Goldthwait DA. The effect of preincubation of hela cell nuclei with atp on the degradation of mononucleosomal DNA by micrococcal nuclease. Nucleic acids research. 1986;14:5513–5529. doi: 10.1093/nar/14.13.5513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pollard JM, Gatti RA. Clinical radiation sensitivity with DNA repair disorders: An overview. International journal of radiation oncology, biology, physics. 2009;74:1323–1331. doi: 10.1016/j.ijrobp.2009.02.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rashid A, et al. Resveratrol enhances prostate cancer cell response to ionizing radiation. Modulation of the ampk, akt and mtor pathways. Radiation oncology. 2011;6:144. doi: 10.1186/1748-717X-6-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Resnick IB, et al. Nijmegen breakage syndrome: Clinical characteristics and mutation analysis in eight unrelated russian families. J.Pediatr. 2002;140:355–361. doi: 10.1067/mpd.2002.122724. [DOI] [PubMed] [Google Scholar]
- 28.Sanli T, et al. Lovastatin sensitizes lung cancer cells to ionizing radiation: Modulation of molecular pathways of radioresistance and tumor suppression. Journal of thoracic oncology : official publication of the International Association for the Study of Lung Cancer. 2011;6:439–450. doi: 10.1097/JTO.0b013e3182049d8b. [DOI] [PubMed] [Google Scholar]
- 29.Sanli T, et al. Ionizing radiation activates amp-activated kinase (ampk): A target for radiosensitization of human cancer cells. International journal of radiation oncology, biology, physics. 2010;78:221–229. doi: 10.1016/j.ijrobp.2010.03.005. [DOI] [PubMed] [Google Scholar]
- 30.Sanli T, et al. Amp-activated protein kinase (ampk) beyond metabolism: A novel genomic stress sensor participating in the DNA damage response pathway. Cancer biology & therapy. 2014;15:156–169. doi: 10.4161/cbt.26726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sanli T, et al. Ionizing radiation regulates the expression of amp-activated protein kinase (ampk) in epithelial cancer cells: Modulation of cellular signals regulating cell cycle and survival. Radiother Oncol. 2012;102:459–465. doi: 10.1016/j.radonc.2011.11.014. [DOI] [PubMed] [Google Scholar]
- 32.Savitsky K, et al. A single ataxia telangiectasia gene with a product similar to pi-3 kinase. Science. 1995;268:1749–1753. doi: 10.1126/science.7792600. [DOI] [PubMed] [Google Scholar]
- 33.Seki S, et al. Effects of atp and other nucleotides on DNA repair synthesis in bleomycin-pretreated permeable mouse sarcoma cells. Carcinogenesis. 1987;8:1391–1394. doi: 10.1093/carcin/8.10.1391. [DOI] [PubMed] [Google Scholar]
- 34.Sidi Y, Mitchell BS. Z-nucleotide accumulation in erythrocytes from lesch-nyhan patients. The Journal of clinical investigation. 1985;76:2416–2419. doi: 10.1172/JCI112255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith CA, Hanawalt PC. Phage t4 endonuclease v stimulates DNA repair replication in isolated nuclei from ultraviolet-irradiated human cells, including xeroderma pigmentosum fibroblasts. Proceedings of the National Academy of Sciences of the United States of America. 1978;75:2598–2602. doi: 10.1073/pnas.75.6.2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Song CW, et al. Metformin kills and radiosensitizes cancer cells and preferentially kills cancer stem cells. Scientific reports. 2012;2:362. doi: 10.1038/srep00362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spurr IB, et al. Targeting tumour proliferation with a small-molecule inhibitor of aicar transformylase homodimerization. Chembiochem : a European journal of chemical biology. 2012;13:1628–1634. doi: 10.1002/cbic.201200279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stewart GS, et al. The riddle syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell. 2009;136:420–434. doi: 10.1016/j.cell.2008.12.042. [DOI] [PubMed] [Google Scholar]
- 39.Storozhuk Y, et al. Chronic modulation of amp-kinase, akt and mtor pathways by ionizing radiation in human lung cancer xenografts. Radiation oncology. 2012;7:71. doi: 10.1186/1748-717X-7-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sweetman L, Nyhan WL. Detailed comparison of the urinary excretion of purines in a patient with the lesch-nyhan syndrome and a control subject. Biochemical medicine. 1971;4:121–134. doi: 10.1016/0006-2944(70)90089-x. [DOI] [PubMed] [Google Scholar]
- 41.Tavassoli A, Benkovic SJ. Genetically selected cyclic-peptide inhibitors of aicar transformylase homodimerization. Angewandte Chemie. 2005;44:2760–2763. doi: 10.1002/anie.200500417. [DOI] [PubMed] [Google Scholar]
- 42.Varon R, et al. Nibrin, a novel DNA double-strand break repair protein, is mutated in nijmegen breakage syndrome. Cell. 1998;93:467–476. doi: 10.1016/s0092-8674(00)81174-5. [DOI] [PubMed] [Google Scholar]
- 43.Xu L, et al. Crystal structure of avian aminoimidazole-4-carboxamide ribonucleotide transformylase in complex with a novel non-folate inhibitor identified by virtual ligand screening. J Biol Chem. 2004;279:50555–50565. doi: 10.1074/jbc.M406801200. [DOI] [PubMed] [Google Scholar]
- 44.Zoberi I, et al. Radiosensitizing and anti-proliferative effects of resveratrol in two human cervical tumor cell lines. Cancer letters. 2002;175:165–173. doi: 10.1016/s0304-3835(01)00719-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.