Abstract
T cell differentiation requires appropriate regulation of DNA methylation. Here we demonstrate that the methylcytosine dioxygenase ten-eleven translocation 2 (TET2) regulates CD8+ T cell differentiation. In a murine model of acute viral infection, TET2 loss promotes early acquisition of a memory CD8+ T cell fate in a cell-intrinsic manner without disrupting antigen-driven cell expansion or effector function. Upon secondary recall, TET2-deficient memory CD8+ T cells demonstrate superior pathogen control. Genome-wide methylation analysis identified a number of differentially methylated regions (DMRs) in TET2-deficient versus wild-type CD8+ T cells. These DMRs did not occur at the loci of differentially expressed memory markers; rather several hypermethylated regions were identified in known transcriptional regulators of CD8+ T cell memory fate. Together, these data demonstrate that TET2 is an important regulator of CD8+ T cell fate decisions.
Keywords: DNA methylation, TET2, CD8 memory
Introduction
In response to infection, naïve CD8+ T cells proliferate and differentiate into a heterogeneous pool of antigen (Ag)-specific cells having divergent cell fates. Following pathogen clearance, most Ag-specific CD8+ T cells die, but a subset persists to become long-lived memory cells, which are able to rapidly respond to rechallenge.
Several cell surface proteins can be used to identify cells with differing memory potential. Ag-specific CD8+ T cells that are CD127hi and KLRG1lo preferentially differentiate into long-lived memory cells; whereas CD127lo and KLRG1hi cells are largely short-lived, terminally differentiated effector cells (1–3). This program of CD8+ T cell differentiation is regulated through the integration of signals from the T cell receptor (TCR), co-stimulatory/-inhibitory receptors and inflammatory cytokines, which direct transcriptional changes that control cell fate. While it is evident that particular transcription factors, such as T-bet, Eomesodermin, Blimp-1, Bcl-6, IRF4 and Runx3, are important in determining the fate of activated cells (4–11), it is also becoming clear that epigenetic programming plays a crucial role in T cell fate determination.
DNA methylation is one epigenetic mechanism by which T cell differentiation is regulated, and recent genome-wide studies have identified coordinated epigenetic changes associated with transcriptional programs during CD4+ or CD8+ T cell differentiation (12–19). It is now appreciated that DNA undergoes regulated demethylation. Recently, the Ten-Eleven Translocation (TET) family of methylcytosine dioxygenases were shown to mediate this process by catalyzing the conversion of 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC), and subsequently to 5-formylcytosine and 5-carboxylcytosine, critical enzymatic steps necessary for generating unmodified cytosines (20–22). The TET family member, TET2, is widely expressed in the hematopoietic system and murine models reveal that TET2 loss leads to expansion of hematopoietic stem cells (HSCs) and myeloid compartments (23–26). In T cells, TET2 contributes to CD4+ T helper cell differentiation (27) and cooperates with TET3 to stabilize FoxP3 expression in regulatory T cells (28).
However, the function of TET2 in CD8+ T cell differentiation is unknown. Here we investigated TET2’s role in directing CD8+ T cell fate following acute LCMV infection. We found that TCR signaling rapidly and dynamically regulates TET2 expression and TET activity. Although mice with selective loss of TET2 in T cells have no overt thymic or peripheral T cell phenotypes at steady-state, following acute viral infection, CD8+ T cells preferentially adopt a memory phenotype in a cell-intrinsic manner and demonstrate superior pathogen control upon re-challenge. Methylation analysis of LCMV-specific CD8+ T cells identified genomic loci that gained 5mC/5hmC in TET2-deficient cells, including several transcriptional regulators known to direct CD8+ T cell effector versus memory differentiation. Together these data demonstrate a novel role for TET2 in directing CD8+ T cell fates.
Materials and Methods
Mice
B6;129S-Tet2tm1.1Iaai/J (TET2fl/fl) mice, C57BL/6J, and CD4Cre+ mice were obtained from Jackson Laboratories and bred at the University of Pennsylvania or the University of Michigan. B6.SJL-Ptprca (CD45.1+) mice were from Taconic. For the LCMV memory phenotyping experiments, the TET2fl/flCD4Cre+ mice were back-crossed twice to C57BL/6J. For all other experiments, TET2fl/flCD4Cre+ mice were backcrossed five to ten times to C57BL/6J. Control mice for experiments included age-matched TET2fl/fl or TET2fl/+CD4Cre− or TET2+/+CD4Cre+, or C57BL/6J. P14 mice (29) were bred to TET2fl/fl CD4Cre+ to generate TET2fl/fl CD4Cre+ P14+ animals. All experiments were performed according to protocols approved by the IACUC of the University of Pennsylvania (#803976 and 803071) and the University of Michigan (PRO00007214).
Infections
Mice were infected with 2×105 plaque-forming units (pfu) of LCMV-Armstrong intraperitoneally, or 0.5–2×105 colony-forming units (cfu) of LM-gp33, as indicated. LM-gp33 was grown and used as previously described (30). LCMV viral titers and Listeria bacterial loads were measured as described (30, 31).
In vitro stimulation
Murine lymphocytes were isolated from spleen and lymph nodes and T cells were purified by negative selection and magnetic separation (Pan T cell Isolation kit, Milltenyi Biotec). T cells were cultured in T cell media (10% FCS, 50 μM 2-mercaptoethanol, 2 mM L-glutamine/penicillin/streptomycin in IMDM) and activated with plate-bound 1μg/ml anti-CD3 (2C11; eBiosciences) and 5μg/ml anti-CD28 (37.51; eBiosciences) for indicated times. Pharmacologic activation of T cells was performed using phorbol-12-myristate-13-acetate (PMA) at 10ng/ml, 25ng/ml or 50ng/ml and ionomycin at 100ng/ml, 250ng/ml or 500ng/ml.
Lymphocyte isolation and adoptive transfer
Lymphoid and non-lymphoid organs were processed and single cell suspensions obtained. Peripheral blood was collected into 4% sodium citrate, purified with a Ficoll gradient (Ficoll-paque Plus; GE Healthcare) and stained for flow cytometric analysis. CD8+ T cells (purified as described above) from “memory” mice were injected into congenic hosts so that 5000 or 7500 CD8+ gp33+ cells were transferred. For P14 adoptive transfer experiments, cells isolated from the peripheral blood were transferred into congenic hosts such that 2000 CD8+ gp33+ Vα2+ cells were transferred.
Flow cytometry and cell sorting
Cells were isolated, washed and stained with indicated antibodies. The following antibodies were used (from BD Biosciences unless otherwise noted): CD8α-Pacific Blue or AlexaFluor (AF)700 (53-6.7, Biolegend); CD4 fluoroscein isothiocyanate (FITC) (GK1.5, eBiosciences), phycoerythrin (PE)-Cy7 (RM4-5, Biolegend), or PE-TexasRed (RM4-5, Invitrogen), TCRβ APC-e780 (H57-597, eBiosciences), CD62L PE-TexasRed (MEL-14, Invitrogen) or Brilliant Violet e605NC (MEL-14, eBiosciences), KLRG1 PE-Cy7, FITC or PerCP-e710 (2F1, eBiosciences), CD127 PE-Cy7 (A7R34, Biolegend) or Pacific Blue (A7R34, eBiosciences), CD27 PE (LG.7F9, eBiosciences), CXCR3 PerCPCy5.5 (CXCR3-173, Biolegend), PD-1 FITC (RMP1-30, eBiosciences) or PE-Cy7 (RMP1-30, Biolegend), 2B4 FITC (eBio244F4, eBiosciences; 2B4, BD), CD160 PE (7H1, Biolegend), CD45.1 PerCP-Cy5.5, PE-Cy5 or PE-Cy7 (A20, eBiosciences) or AF700 (A20, Biolegend), CD45.2 AF700, allophycocyanin (APC)-e780 (104, eBiosciences) or Pacific Blue (104, Biolegend), CD44 AF700 (IM7, Biolegend), IFNγ-PerCPCy5.5 (XMG1.2, Biolegend), TNFα-Pacific Blue (MP6-XT22, eBiosciences), IL-2 APC (JES6-5H4), Granzyme B PE-Cy7 (NGZB, eBiosciences), CD107a FITC or PE (1D4B), human Ki67-FITC (B56), Eomes AF647 (Dan11mag, eBiosciences). Biotinolyted monomers specific for H2-Db restricted gp33-41 of LCMV were obtained from the NIH Tetramer Core Facility and tetramerized using their published protocol. Intracellular staining was performed using either the Cytofix/Cytoperm kit (BD Biosciences) or FoxP3/Transcription Factor Staining Buffer kit (eBiosciences) according to manufacturer’s instructions. Discrimination of live cell populations was performed using Live/Dead Aqua stain (Invitrogen) according to manufacturer’s instructions.
For experiments involving measurement of intracellular 5hmC, T cells were surface stained prior to fixation/permeablization with Cytofix/Cytoperm kit (BD Biosciences), treated with DNaseI (300μg/ml in PBS) at 37°C for one hour and then intracellularly stained with isotype or anti-5hmC (Active Motif #39791, 1μg/ml) antibody for 30 minutes, followed by fluorochrome conjugated goat anti-rabbit secondary antibody (Invitrogen).
For experiments involving ex vivo stimulation, single cell suspensions were stimulated with 200ng/ml gp33, gp276 or NP396 peptides in the presence of 1mg/ml brefeldin A for 5h and then analyzed for intracellular cytokine staining. Data were acquired using FACS LSR II (BD Biosciences) and analyzed with FlowJo software (TreeStar).
For experiments involving cell sorting, T cells were isolated and sorted on a FACS Aria II (BD Biosciences). For isolation of naïve CD8+ T cells, CD8+ T cells from the spleen and lymph nodes were purified by negative selection and magnetic separation (CD8a+ T cell Isolation Kit II; Miltenyi Biotec) and then sorted for naïve CD8+ T cells (TCRβ+CD8+CD4−CD44−CD62L+). For sorting of gp33+ CD8+ T cells, CD8+ T cells from the spleens of control and TET2fl/flCD4Cre+ mice were negatively isolated as above and sorted for (gp33+CD8+CD4−CD44+) on a FACS Aria II under appropriate biohazardous precautions.
ERRBS
Genomic DNA was bisulfite converted using the EZ DNA Methylation kit (Zymo Research). Base-pair resolution DNA Methylation was performed on control and TET2-deficient gp33+CD8+ T cells (n=4/genotype) as described (32). DMCs were identified using a 25% methylation difference threshold and q-value<0.01 using methylKit (33) and DMRs were determined through eDMR (34).
Pathway analysis
Data were analyzed through the use of Ingenuity Pathways Analysis (Ingenuity Systems, http://www.ingenuity.com, Redwood City, California, USA) specified for ‘Mouse’. The differentially methylated genes containing DMRs that corresponded to at least one pathway annotation in the Ingenuity Knowledge Base were eligible for the analysis. The p-values associated with pathways were calculated using the right-tailed Fisher exact test.
Data accessibility
ERRBS data has been deposited to the NCBI Gene Expression Omnibus under accession number GSE105176 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE105176).
Statistical analysis
Statistical significance was calculated as noted in figure legends. Prism (GraphPad Software) was used for statistical analysis.
Results
TCR signaling dynamically regulates TET2 expression and activity
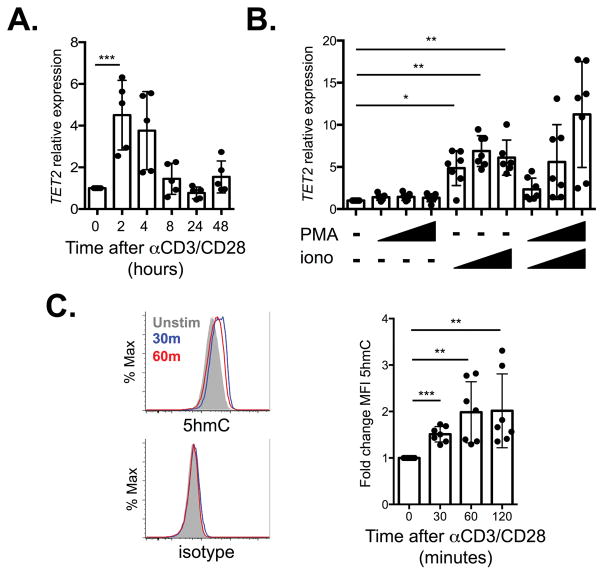
If TET2 plays an important role in T cell differentiation, we reasoned its expression and/or function might be altered in response to TCR ligation. We found that TET2 mRNA expression was rapidly induced in murine T cells after TCR plus CD28 stimulation with maximal expression at two hours (Figure 1A). This upregulation was dependent on Ca2+ signaling as TET2 mRNA was induced by the Ca2+ ionophore ionomycin but not PMA alone, suggesting the protein kinase C pathway is not sufficient for TET2 induction (Figure 1B). In addition to regulation of TET2 mRNA levels, TET2 protein is negatively regulated by the CXXC-type zinc finger domain containing proteins IDAX and CXXC5 (35). We evaluated their expression in primary murine T cells as a possible additional means by which TET2 could be regulated. Whereas IDAX was not detected in T cells, CXXC5 was expressed in resting cells and was down-regulated by TCR stimulation (Supplementary Figure 1A); thus CXXC5 could serve to further regulate TET2 expression. We also assessed if TCR stimulation directs TET activity, specifically the oxidation of 5mC to 5hmC. Using flow cytometry, we found 5hmC levels were rapidly induced in murine T cells after TCR stimulation (Figure 1C). Together these data indicate that TET2 gene expression and TET function are regulated by TCR activation.
Figure 1.
TCR signaling regulates TET2. A) Expression of TET2, relative to β-actin, in cDNA generated from WT T cells stimulated with anti-CD3/CD28 for indicated time periods, normalized to unstimulated cells (n=5, 2 independent experiments). B) TET2 expression, relative to β-actin, in cDNA generated from WT T cells stimulated with increasing concentrations of PMA and/or ionomycin (n=6, 3 independent experiments). C) Left, representative histograms of intracellular 5hmC (top) or isotype control (bottom) in WT T cells stimulated with anti-CD3/CD28 for indicated time periods. Right, fold change in MFI of 5hmC compared to unstimulated cells (n=7, 3 independent experiments). *p<0.05, **p<0.01, ***p<0.001 Dunnett’s multiple comparison test, repeated-measures one-way ANOVA.
TET2fl/flCD4Cre+ mice have intact thymic and peripheral T cell populations
TET2 deficiency in the hematopoietic compartment results in expansion of HSCs with skewing toward the myeloid lineage (23–26, 36). To specifically study TET2 in T cells, we generated TET2fl/flCD4Cre+ mice that lack TET2 in all mature αβ T cells. TET2fl/flCD4Cre+ mice (referred to as TET2cKO) had similar frequencies and numbers of thymic and peripheral T cell populations compared with age-matched control mice, without evidence of substantially altered homeostasis as indicated by similar absolute numbers of naïve (CD44−CD62L+) and CD44+ T cell populations (Supplementary Figure 1B–D). Moreover, we did not see changes in early activation markers (CD25, CD69, Bcl-XL) or proliferation following CD3/CD28 stimulation (Supplementary Figure 1E–F). Together, these data suggest that TET2 does not substantially alter T cell homeostasis or regulate short-term TCR-induced responses in vitro.
TET2cKO CD8+ T cells have intact effector function after acute viral infection
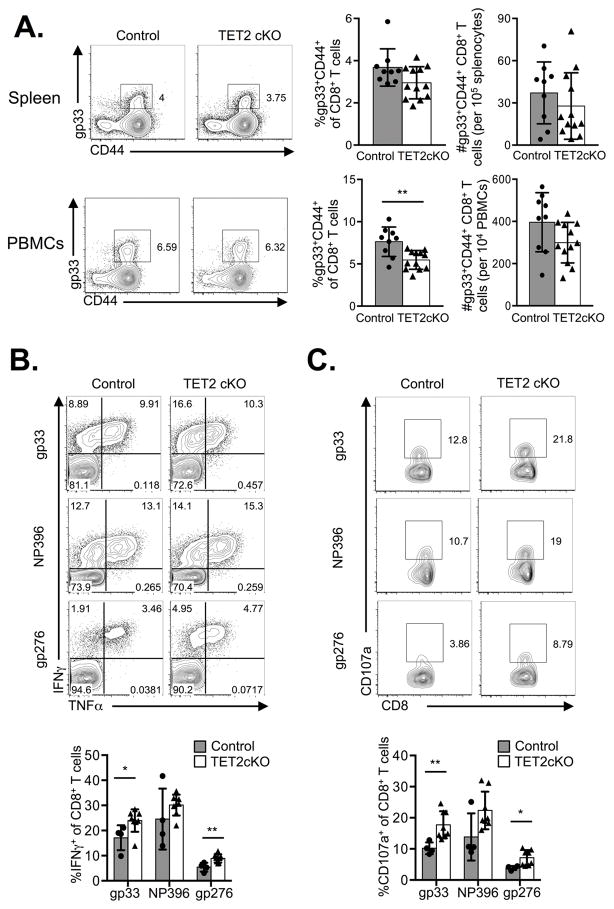
Since DNA methylation regulates the cell fate of T cells and other cell types (37, 38), and TET2 is known to control HSC differentiation, we hypothesized that TET2 might influence CD8+ T cell differentiation. To test this hypothesis, we infected control and TET2cKO mice with LCMV-Armstrong and followed virus-specific effector and memory CD8+ T cell differentiation using tetramers specific for the glycoprotein 33–41 (gp33) epitope of LCMV. At the peak of the CD8+ T cell response (day 8), we found similar frequencies and absolute numbers of gp33+ CD8+ T cells in the spleens, as well as similar absolute numbers (although lower frequencies) of gp33+ CD8+ T cells among peripheral blood lymphocytes of TET2cKO mice compared to control mice (Figure 2A). Additionally, a similar frequency of gp33+ CD8+ T cells expressed the proliferation marker Ki67 on day 8 post-infection (p.i.) in control and TET2cKO mice (data not shown). Together these data suggest TET2 is not required for Ag-specific CD8+ T cell proliferation and expansion following acute viral infection.
Figure 2.
TET2cKO T cells have intact effector function in response to acute LCMV infection. LCMV-specific responses were assessed on D8 p.i. in control and TET2cKO mice. A) Left, representative flow cytometric analysis of gp33 and CD44 on TCRβ+CD8+ splenocytes and peripheral blood mononuclear cells (PBMCs) from control and TET2cKO mice. Right, frequency and absolute number of gp33+CD8+ T cells per spleen and 104 PBMCs. Plots are gated on live, CD8+TCRβ+ lymphocytes. B–C) Representative plots of intracellular TNFα, IFNγ and CD107a expression (top) and frequencies (bottom) of indicated cell populations in CD8+ splenocytes stimulated ex vivo with indicated LCMV-specific peptides. Data representative of n=9/control, 12/TET2cKO, 3 independent experiments (A) or n=4/control, 8/TET2cKO, 2 independent experiments (B,C). *p<0.05, **p<0.01 unpaired t-test.
Next, we examined the functional ability of TET2cKO CD8+ T cells to respond to antigenic stimulation. Based on findings in CD4+ T cells, in which TET2 deficiency led to reduced cytokine production following TH1 and TH17 in vitro skewing (27), we predicted TET2cKO CD8+ T cells would produce less cytokine. However, in contrast to our expectation, TET2cKO LCMV-specific CD8+ T cells had enhanced IFNγ expression and higher expression of CD107a, a surrogate marker for degranulation, in response to gp33 and gp276 peptides with percentages trending higher in response to NP396 peptide stimulation (Figure 2B–C). There was no difference in ‘double-producers’ (ie, IFNγ+TNFα+) or IL-2 producing CD8+ T cells following LCMV-peptide stimulation and both control and TET2cKO mice were able to clear virus by D8 p.i. (data not shown). Together, these data demonstrate that TET2 loss enhances CD8+ T cell effector function at the peak of infection for certain TCR specificities.
TET2 loss promotes memory CD8+ T cell formation
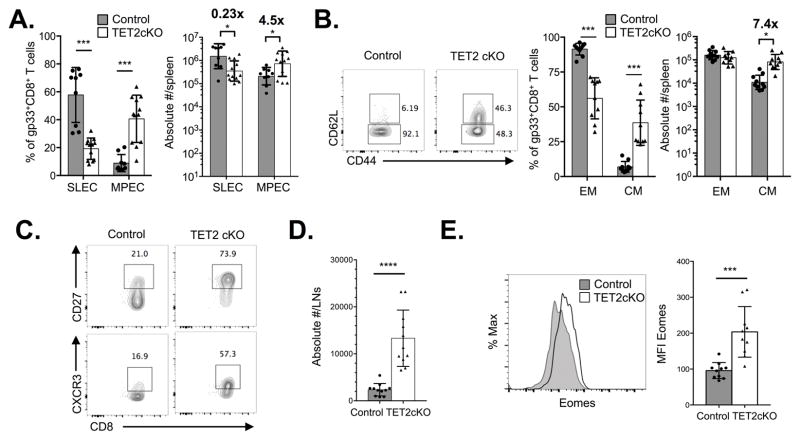
After T cell expansion in response to infection, the majority of Ag-specific CD8+ T cells undergo cell death, while a small subset persists and differentiates into long-lived memory cells. Memory CD8+ T cells have several stem cell-like properties, including long-term survival, self-renewal and multi-potent potential (39), as well as a shared transcriptional program (40). Given these characteristics and the fact that TET2 loss in HSCs leads to an expanded stem/progenitor pool (23–26), we reasoned that TET2 deficiency might skew CD8+ T cells towards a memory phenotype at the expense of short-lived effector cells. We monitored cohorts of mice for persistence of gp33+ CD8+ T cells in the peripheral blood of LCMV-infected animals. The frequency of control and TET2-deficient gp33+ CD8+ T cells in the blood had a similar pattern of expansion, contraction and persistence over time, for >80 days p.i. (Supplementary Figure 2A). However, in the spleens on D8, among TET2cKO gp33+ CD8+ T cells, the memory precursor effector cell (MPEC) population, identified by their KLRG1−CD127+ phenotype, (1–3) was significantly augmented in both frequency and absolute number with a concomitant decrease in the KLRG1+CD127− short-lived effector population (SLEC) (Figure 3A). The phenotype was also evident among gp33+ CD8+ T cells in the peripheral blood (Supplementary Figure 2B). Additionally, during longitudinal monitoring of peripheral blood following LCMV infection, compared to controls, more TET2cKO gp33+ CD8+ T cells expressed the markers CD27, CD62L, and CXCR3 (Supplementary Figure 2C and D), which are highly expressed on central memory CD8+ T cells (TCM) (41–43). Interestingly, the rate of increase over time in the MPEC population among control and TET2-deficient gp33+ CD8+ T cells was similar (Supplementary Figure 2E). Together these data suggest that TET2 loss directs an early decision towards a central memory CD8+ T cell fate.
Figure 3.
TET2 loss enhances memory CD8+ T cell differentiation. Control and TET2cKO mice were infected with LCMV and gp33+ CD8+ T cells isolated from spleen and lymph nodes were evaluated at indicated time-points. A) Frequency (left) and absolute number (right) of SLEC and MPEC populations among gp33+ CD8+ TCRβ+ splenocytes from control and TET2cKO mice isolated D8 p.i. B) Left, representative flow cytometric analysis of CD62L expression on gp33+CD8+ T cells. Right, frequency and absolute number of CD62L− effector memory (EM) and CD62L+ central memory (CM) populations among gp33+ CD8+ TCRβ+ splenocytes from control and TET2cKO mice isolated D45 p.i. C) Representative flow cytometric analysis of CD27 (top) and CXCR3 (bottom) expression on gp33+CD8+ T cells on day 45 p.i.. D) Absolute number of gp33+ CD8+ TCRβ+ lymphocytes isolated from lymph nodes of control and TET2cKO mice on D45 p.i. E) Representative flow cytometric histograms of intracellular Eomes expression in gp33+ CD8+ TCRβ+ splenocytes isolated from control and TET2cKO mice on D45 p.i. Plots are gated on live, gp33+CD8+TCRβ+ lymphocytes. Data representative of n=9/control and 12/TET2cKO, 3 independent experiments (A), n=10/genotype, 2 independent experiments (B–E). *p<0.05, ***p<0.001, unpaired t-test.
To examine if TET2 loss promotes ‘bona fide’ memory formation, we examined control and TET2cKO mice on day 45 p.i.. In the spleens of TET2cKO mice, there was an increase in the frequency and absolute number of TCM gp33+ CD8+ T cells (defined by CD62L and CD44 expression; Figure 3B). Additionally, we saw a significant increase in the expression of other TCM markers, including CD27 and CXCR3 (Figure 3C). Consistent with increased TCM formation, we found an increase in the absolute number of gp33+CD8+ T cells in the lymph nodes of the TET2cKO mice compared to control mice (Figure 3D). To further characterize the TET2-deficient memory CD8+ T cells, we examined expression of transcription factors associated with CD8+ T cell differentiation, including Eomes and T-bet. TET2-deficient gp33+CD8+ T cells had higher expression of Eomes compared to control gp33+CD8+ T cells (Figure 3E), but no significant differences were noted in T-bet expression (data not shown). Together these data show that TET2 loss promotes CD8+ T cell memory formation.
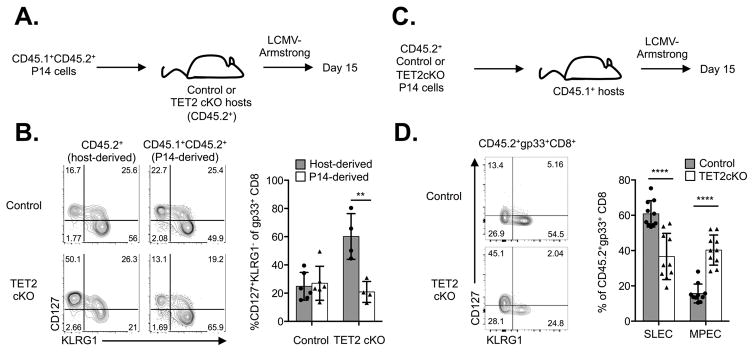
TET2cKO mice lack TET2 in both mature CD4+ and CD8+ T cells, therefore we sought to determine if the preferential CD8+ memory differentiation in TET2cKO mice was intrinsic to TET2 deficiency in the CD8+ T cell population or dependent on an altered environment due to TET2 loss in all T cells. To address this issue, we used two complementary approaches. In one approach, we transferred CD8+ T cells isolated from congenic (CD45.1+CD45.2+) mice expressing the P14 TCR transgene, which is specific for the LCMV gp33 epitope presented by H-2Db, into control and TET2cKO mice, and subsequently infected these mice with LCMV (Figure 4A). As expected, endogenous host (CD45.2+) TET2cKO gp33+ CD8+ T cells rapidly differentiated into MPECs at the expense of SLEC formation (Figure 4B). In contrast, similar frequencies of SLEC and MPEC populations derived from P14 cells were present in the control and TET2cKO mice (Figure 4B; CD45.1+CD45.2+ gate). Additionally, the transferred P14 cells expressed CD62L at similar frequencies in both the control and TET2cKO mice (data not shown). As a second approach, we transferred equal numbers of wild-type or TET2-deficient P14 CD8+ T cells into congenic (CD45.1+) hosts and infected them with LCMV-Armstrong (Figure 4C). Consistent with prior results, a higher frequency of TET2-deficient P14 cells differentiated into MPECs compared to WT P14 cells that predominantly differentiated into SLECs (Figure 4D). Together these data demonstrate that TET2 regulates CD8+ T cell memory differentiation in a cell-intrinsic manner.
Figure 4.
TET2 regulates CD8+ T cell differentiation in a CD8+ T cell intrinsic manner. A) Experimental schema: congenically marked P14 CD8+ T cells were adoptively transferred into control or TET2cKO mice that were subsequently infected with LCMV-Armstrong and PBMCs analyzed on D15 post-infection. B) Left, representative flow cytometric analysis of CD127 and KLRG1 expression on host-derived and P14-derived gp33+CD8+TCRb+ PBMCs and right, frequencies of indicated populations. C) Experimental schema: Equal numbers of P14 control and TET2-deficient P14 CD8+ T cells were transferred into congenic mice that were subsequently infected with LCMV-Armstrong and PMBCs analyzed on D15 p.i. D) Left, representative flow cytometric analysis of CD127 and KLRG1 expression on P14-derived gp33+CD8+TCRb+ PBMCs and right, frequencies of SLEC (CD127−KLRG1+) and MPEC (CD127+KLRG1−) populations among adoptively transferred P14 CD8+ T cells. Data representative of n=6/control and 4/TET2 cKO, two independent experiments (A–B), n=10/genotype, 2 independent experiments. **p<0.01, ***p<0.001, unpaired t-test.
Loss of TET2 enhances pathogen control after rechallenge
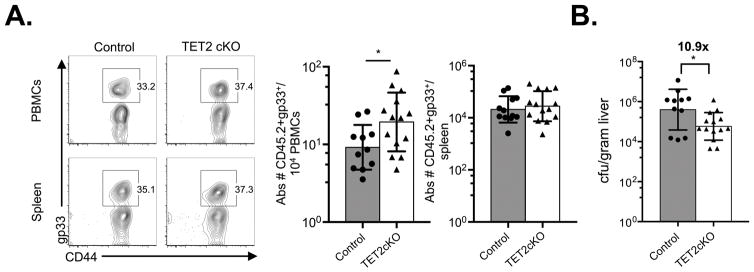
Thus far, we have shown that TET2 loss promotes a central memory phenotype. To determine if TET2cKO cells displayed enhanced functional memory properties, we evaluated the ability of TET2cKO memory CD8+ T cells to mount an effective memory response. Purified CD8+ T cells from control and TET2cKO mice previously infected with LCMV (>100 days p.i.) were transferred into WT congenic hosts such that equal numbers of gp33+ CD8+ T cells were injected. Hosts were subsequently infected with Listeria monocytogenes that expresses the LCMV gp33 epitope (LM-gp33). Five days following LM-gp33 infection, more gp33+ TET2cKO cells than control cells were present in the peripheral blood of infected hosts (Figure 5A). Comparable gp33+ CD8+ T cell numbers were found in the spleen, and frequencies within both organs were similar (Figure 5A, Supplementary Figure 3A). To assess the function of these memory populations, bacterial burden was measured. Mice receiving TET2cKO memory CD8+ T cells had lower Listeria burden in their livers at 5 days p.i. (Figure 5B), indicating that TET2-deficient memory cells exhibit greater pathogen control. This enhanced bacterial control was not accompanied by increased production of IFNγ, TNFα or CD107a expression (Supplementary Figure 3B) but rather was associated with higher pre-transfer expression of CD62L and CXCR3 (Supplementary Figure 3C), two TCM receptors that have been associated with more efficient recall responses (42–44). Together, these data demonstrate that loss of TET2 favors a more effective memory immune response.
Figure 5.
LCMV-specific TET2cKO memory CD8+ T cells reduce LM-gp33 bacterial loads. Equal numbers of gp33+CD8+ T cells from LCMV-immune (>100d p.i.) control and TET2cKO mice were transferred into congenic hosts, which were subsequently infected with LM-gp33. Gp33-specific memory responses were assessed five days later. A) Left, Representative flow plots of gp33 and CD44 expression on CD8+TCRβ+ PBMCs and splenocytes on D5 p.i. LM-gp33. Right, Absolute number of transferred gp33+ of CD8+TCRβ+ PBMCs and splenocytes. B) Bacterial load (colony forming units, cfu) in liver on D5 p.i. LM-gp33 of congenic hosts that received control or TET2cKO gp33+ memory CD8+ T cells. Data representative of n=12/control, 14/TET2cKO, 3 independent experiments. *p<0.05, unpaired t-test.
TET2 loss leads to DNA hypermethylation
We posited that TET2 loss promotes CD8+ T cell memory differentiation through altered epigenetic regulation of gene expression. To examine cytosine methylation differences in control and TET2cKO T cells, we performed enhanced reduced representation bisulfite sequencing (ERRBS), a method that allows for quantitative genome-wide, single base-pair resolution analysis of cytosine methylation, with coverage both within and outside of CpG islands (32, 45). Similar to bisulfite sequencing, ERRBS does not distinguish between 5mC and 5hmC, reading both as a methylated cytosine residue and thus referred to as 5mC/5hmC to indicate detection of either.
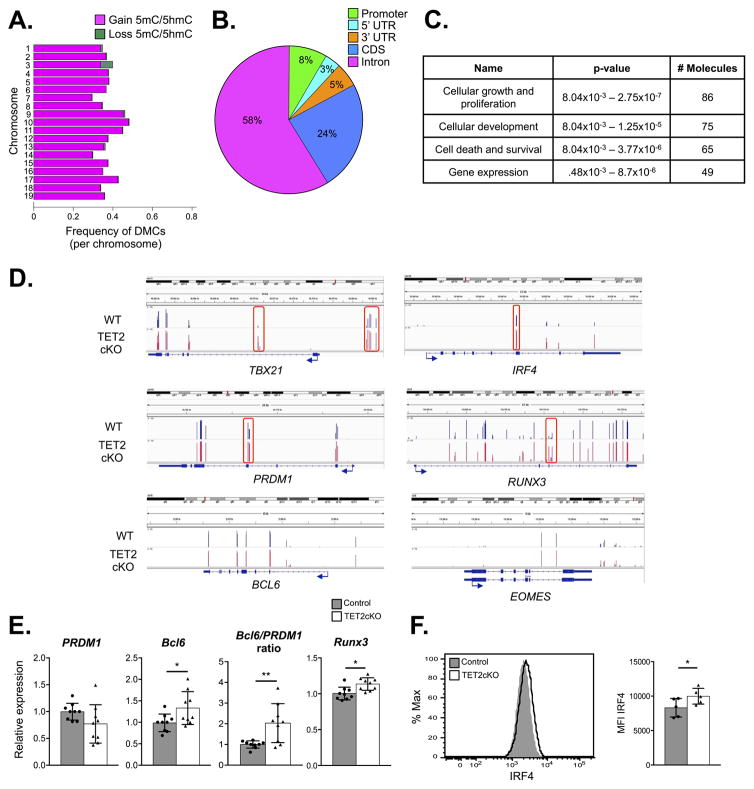
We isolated gp33+ CD8+ T cells from control and TET2cKO mice on D8 p.i. with LCMV. Genomic DNA was digested with a methylation-insensitive restriction enzyme and subjected to standard next-generation sequencing library preparation. Unmethylated cytosines underwent bisulfite conversion prior to final amplification and sequencing. The ERRBS analysis covered an average of 1.54×106 individual CpGs with minimum 10X coverage of the mouse genome. Analysis of differentially methylated cytosines (DMCs; >25% methylation difference) demonstrated that the majority gained 5mC/5hmC in TET2cKO LCMV-specific CD8+ T cells compared to control (Figure 6A). These DMCs occurred primarily in intergenic (35%) and intronic (47%) regions (data not shown). Next, we grouped DMCs into differentially methylated regions (DMRs), in which multiple adjacent CpG sites have differential methylation (≥25% difference in all control versus TET2cKO samples), using the bioinformatic algorithm eDMR (34). Three hundred fifty-five DMRs mapped to 184 unique ReqSeq genes (mm9; Supplementary Table 1). The identified DMRs were mapped to different genomic regions, with the majority located in introns and coding sequences (cds) (Figure 6B), most commonly occurring in genes involved in cellular growth and proliferation, cellular development, cell death and survival, and gene expression according to Ingenuity Pathway Analysis (Figure 6C).
Figure 6.
TET2 loss leads to genomic hypermethylation in LCMV-specific CD8+ T cells. Genomic DNA from control and TET2cKO gp33+ CD8+ T cells on D8 p.i. with LCMV was subjected to ERRBS methylation analysis. A) Stacked barplot showing frequency of differentially methylated cytosines (DMCs) of all covered CpGs per autosomal chromosome. Magenta represents gain of 5mC/5hmC and green represents loss of 5mC/5hmC in TET2cKO versus control samples. B) Pie chart representing proportion of annotated DMRs to indicated regions of genomic DNA. C) Top four molecular and cellular functions of 182 genes containing DMRs analyzed by Ingenuity Pathway Analysis (IPA) software with associated p-values and number of differentially methylated genes grouped into functional categories. D) Integrated Genomics Viewer (IGV) browser visualization of loss of 5hmC/5mC methylation measured by ERRBS at indicated genomic loci from representative control (blue) and TET2-deficient (red) samples. DMRs are boxed in red. E) PRDM1 and Bcl6 expression, relative to β-actin, and ratio of Bcl-6 to PRDM1, and Runx3 expression in cDNA generated from control or TET2cKO CD8+ T cells activated for 3 days with plate-bound anti-CD3/CD28 and IL-2. F) Representative flow cytometric analysis of intracellular IRF4 expression (left) and median fluorescence intensity (right) in control and TET2cKO CD8+ T cells activated for 3 days with plate-bound anti-CD3/CD28 and IL-2. Data representative of n=9/genotype, 3 independent experiments (E), n=5/genotype, 2 independent experiments (F).
Several proteins that were differentially expressed on TET2cKO versus control gp33+ CD8+ T cells, including KLRG1 (15), CD127 (46), CD27 (47), CXCR3 (15, 48) and CD62L (15), are known to be regulated by DNA methylation. Thus, mechanistically, we reasoned TET2 could individually regulate the methylation status of the loci of differentially expressed memory markers. Alternatively, TET2 could control the expression of key transcription factors and thus promote a CD8+ T cell memory transcriptional program. Some differentially expressed molecules, including CD127 and CD62L, did not have 10X ERRBS coverage in all eight samples (our cut-off for assessing DMCs) and thus could not be evaluated. However, gene loci encoding KLRG1, CD27, and CXCR3 were appropriately represented. Interestingly, these loci did not contain DMRs, suggesting TET2 does not regulate expression of these genes through direct demethylation. Therefore, we evaluated DMRs associated with transcription factors known to direct CD8+ T cell effector versus memory differentiation, including Tbx21 (encoding T-bet), Eomes, PRDM1 (encoding Blimp-1), Bcl6, Runx3 and IRF4 (4, 6–10, 49). Bcl6 and Eomes did not contain DMRs, whereas Tbx21, PRDM1, IRF4 and Runx3 had associated DMRs that gained 5mC/5hmC in TET2cKO gp33+ CD8+ T cells (Figure 6D). Taken together, these data suggest that acquisition of a memory fate is not due to TET2-mediated loss of 5mC/5hmC at each locus encoding effector/memory markers (ie, KLRG1, CD27, CXCR3) but rather TET2 may regulate transcriptional drivers of effector or memory cell fate.
Since we observed early differences in CD8+ T cell differentiation in TET2cKO CD8+ T cells, we questioned whether methylation differences seen at this time point were a cause or consequence of altered differentiation. Therefore to evaluate early gene expression changes we isolated control and TET2cKO naïve CD8+ T cells, activated them in vitro with αCD3/CD28 and IL-2 for 3 days and examined gene expression using RT-PCR and flow cytometry. Activated TET2cKO CD8+ T cells had diminished Blimp-1 mRNA, though this did not reach statistical significance. Since Blimp-1 and Bcl-6 negatively regulate one another and have opposing effects on CD8+ T cell effector versus memory development (6, 7, 9), we also assessed Bcl-6 mRNA. We found that TET2 loss promotes Bcl-6 expression and increased the Bcl-6/Blimp-1 ratio (Figure 6E). Additionally, we noted a modest increase in Runx3 mRNA and IRF4 protein (Figure 6 E and F), but no significant difference in expression of T-bet or Eomes by flow cytometry at this early time point (data not shown).
Discussion
The TET family of methylcytosine dioxygenases mediates active DNA demethylation. In multiple cell types, including embryonic stem cells (20, 50), HSCs (23–26) and CD4+ T cells (27), TET activity has been shown to regulate cellular differentiation. It is known that DNA methylation dynamically changes in CD8+ T cells following acute viral infection with both gain and loss of DNA methylation at individual loci and across the genome (15), however, the role TET enzymes and active DNA demethylation play in CD8+ T cell fate decisions has not been fully explored. Here, we identify TET2 as a novel regulator of cell fate choice between terminally differentiated effector CD8+ T cells and central memory CD8+ T cell formation.
TET expression and activity is regulated through multiple mechanisms in non-lymphoid cells, including protein stability via IDAX and CXXC5 (35), ascorbic acid (51–53), and acetylation (54). In this study, we find that TET2 gene expression is rapidly and transiently upregulated in primary murine T cells by TCR signaling. This up-regulation occurs in a Ca2+ dependent manner and coincides with a decrease in CXXC5 gene expression, a known inhibitor of TET2 protein stability (35). We also found that TCR signals regulate TET enzymatic activity, although the timing of changes in 5hmC levels appeared to occur faster than changes in TET2 mRNA expression. The discordant timing of 5hmC induction versus TET2 gene expression may be reflective of the sensitivity of the respective assays or indicate that TCR signals regulate TET2 transcription and TET enzymatic activity independently of each other. Together, these data suggest that TET2 expression and TET activity are tightly controlled in T cells, likely through multiple mechanisms, following T cell activation.
Studies of DNA methylation in T cells have focused primarily on regulation of cytokine and effector molecule gene loci. Previous genome-wide analysis of DNA methylation during CD8+ T cell differentiation revealed decreased methylation and increased gene expression at effector gene loci including Gzmb (Granzyme B) and Ifng, (IFNγ) in D8 effector CD8+ T cells following LCMV infection compared to naïve T cells (15). How TET2 might contribute to these changes was not explored. However, based on a study in which diminished cytokine production was noted in TET2-deficient TH1 and TH17 cells (27) and our unpublished results indicating a similar finding in CD4+ cells, we predicted that TET2-deficient CD8+ T cells would have decreased cytokine production and/or effector molecule expression following viral infection. To our surprise, we found increased cytokine production and effector molecule expression in TET2-deficient CD8+ T cells in vivo during acute viral infection. These data indicate that there are alternative mechanisms regulating removal of repressive DNA methylation at these loci, which could include removal by other TET family members or by DNMT1 inhibition. The lack of DMRs associated with cytokine, effector, and individual memory marker gene loci was contrasted with the several DMRs found in known transcriptional regulators of CD8+ cell differentiation. In this study, we noted that rather than occurring at individual loci of differentially expressed memory markers (such as KLRG1, CD27, CXCR3), the DMRs identified occur in known transcriptional regulators of CD8+ T cell fate. To evaluate whether these methylation changes altered gene expression, we evaluated gene and protein expression in control and TET2-deficient CD8+ T cells activated in vitro. We found that there were modest increases in Runx3 mRNA as well as IRF4 protein expression, which may contribute to the increase in IFNγ and cytolytic potential seen early during infection (8, 55). Moreover, we found an alteration of the Bcl-6/PRDM1 ratio in a manner that would be predicted to promote memory CD8+ T cell differentiation. Though some of these expression changes were modest, they could be synergistic in promoting memory CD8+ T cell formation while maintaining strong effector responses. Taken together, these findings suggest that TET2 may direct CD8+ T cell fate via regulation of transcriptional programs.
Following LCMV infection, Ag-specific TET2-deficient CD8+ T cells rapidly acquired surface markers associated with memory potential in a cell intrinsic manner. These differences occurred early and correlated with an increased number of TET2-deficient TCM CD8+ T cells. Interestingly, a recent report identified DNMT3A, another enzyme involved in DNA methylation, as a repressor of CD8+ T cell effector differentiation demonstrating that mice with a T cell specific deletion of DNMT3A have enhanced TCM differentiation following acute infection (56). The similarity of these phenotypes was somewhat surprising at first since DNMT3A catalyzes the de novo methylation of cytosines (57) and TET2 oxidizes 5mC to 5hmC. However, loss of both genes affect HSC differentiation and self-renewal in a similar (although not identical) manner (23–26, 58). Moreover, mutations in both TET2 and DNMT3A occur in an overlapping set of human myeloid and T cell neoplasms (59, 60), as well as age-related clonal hematopoiesis (61–63). Recent work with TET2 and DNMT3A single- and double-deficient HSCs identified methylated regions that were independently and interdependently regulated (64). These observations suggest that loss of either DNMT3A or TET2 may deregulate similar pathways or an overlapping set of targets.
We also found that TET2-deficient memory cells have intact expansion and effector responses, and provide enhanced pathogen clearance upon rechallenge. The increase in the TCM population in the TET2cKO mice could potentially explain the improved pathogen control upon antigenic rechallenge. At day 45 following LCMV-Armstrong infection, there were more central memory gp33+ CD8+ T cells, with high CD62L, CD27 and CXCR3 expression, in the TET2cKO mice compared to WT mice. Prior studies have demonstrated that the TCM subset (defined by CD62L+) have enhanced pathogen clearance compared to the effector memory subset (42). Additionally, TET2-deficient memory CD8+ T cells expressed higher levels of CXCR3, a chemokine receptor that is important for T cell migration within the lymph node and required for rapid control of pathogens by memory CD8+ T cells (44). The role of these receptors in mediating the enhanced pathogen clearance mediated by TET2-deficient memory CD8+ T cells is being explored.
We and others have seen genome-wide 5mC and/or 5hmC changes in cells with altered TET activity (rev in (65)). However, understanding the functional significance of these changes and linking them mechanistically to cellular outcomes remains a challenge for the field. The majority of the DMRs identified in this study occurred in intragenic regions, with a relatively low frequency (~8%) occurring in promoter regions. These data are consistent with prior studies of DMRs in TET2-deficient embryonic stem cells (66), which suggest that TET2 activity may preferentially occur at intragenic sites. While DNA methylation at promoters is understood to repress gene transcription, the functional significance of DNA methylation/hydroxymethylation at other genomic sites is less well understood. Two recent studies have demonstrated a role for TET2 in maintaining hypomethylation at enhancer regions (67, 68), suggesting that its action at enhancer regions can modulate gene expression. However, in a separate study examining TET2 knockdown cells, a similar number of genes were found to be upregulated and downregulated (66). Taken together, these studies highlight the complexity of TET2-regulated gene expression. Moreover, as is the case in any study of genetic deficiency, there remains a question of whether the DNA methylation changes noted are directly or indirectly mediated by TET2, a question further compounded by technical limitations of determining chromatin occupancy of TET2 with currently available reagents (rev. in (65)). Additionally, there remains the possibility that TET2 may partially (or wholly) regulate CD8+ T cell memory differentiation through a non-catalytic function since other epigenetic modifiers have non-catalytic functions (69, 70). In myeloid cells, TET2 has been shown to regulate cytokine expression and proliferation via mechanisms independent of DNA hydroxymethylation (71, 72); additionally, other TET family members have been shown to function via recruitment of chromatin modifiers (73).
Collectively, our data identify TET2 as a novel epigenetic regulator of CD8+ T cell effector versus memory cell fate decisions following acute viral infection and highlight the importance of epigenetic modifiers in shaping T cell immunity. Further studies of TET2-mediated function, including the significance of 5mC and 5hmC modifications, will advance our understanding of epigenetic regulation of T cell fates and may provide novel methods to modulate immune responses.
Supplementary Material
Acknowledgments
Funding Sources: Work was supported by the NIH grants K08 AI101008 (S.A.C), R37 GM053256 (G.A.K), AI105343, AI112521, AI082630 and AI115712 (E.J.W.) and other support from R01AI082292.
We thank the Cornell Epigenomics Core and Children’s Hospital of Pennsylvania NAPCore facility for data analysis. We thank Edward Behrens for assistance with analysis of publically available microarray data and members of the Jordan, Koretzky, Behrens, Kambayashi and Oliver laboratories for helpful discussions. We also thank Sydney Drury for technical assistance. The authors declare no conflicting financial interests.
References
- 1.Kaech SM, Tan JT, Wherry EJ, Konieczny BT, Surh CD, Ahmed R. Selective expression of the interleukin 7 receptor identifies effector CD8 T cells that give rise to long-lived memory cells. Nat Immunol. 2003;4:1191–1198. doi: 10.1038/ni1009. [DOI] [PubMed] [Google Scholar]
- 2.Sarkar S, Kalia V, Haining WN, Konieczny BT, Subramaniam S, Ahmed R. Functional and genomic profiling of effector CD8 T cell subsets with distinct memory fates. Journal of Experimental Medicine. 2008;205:625–640. doi: 10.1084/jem.20071641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Joshi NS, Cui W, Chandele A, Lee HK, Urso DR, Hagman J, Gapin L, Kaech SM. Inflammation directs memory precursor and short-lived effector CD8(+) T cell fates via the graded expression of T-bet transcription factor. Immunity. 2007;27:281–295. doi: 10.1016/j.immuni.2007.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Intlekofer AM, Takemoto N, Wherry EJ, Longworth SA, Northrup JT, Palanivel VR, Mullen AC, Gasink CR, Kaech SM, Miller JD, Gapin L, Ryan K, Russ AP, Lindsten T, Orange JS, Goldrath AW, Ahmed R, Reiner SL. Effector and memory CD8+ T cell fate coupled by T-bet and eomesodermin. Nat Immunol. 2005;6:1236–1244. doi: 10.1038/ni1268. [DOI] [PubMed] [Google Scholar]
- 5.Intlekofer AM, Banerjee A, Takemoto N, Gordon SM, Dejong CS, Shin H, Hunter CA, Wherry EJ, Lindsten T, Reiner SL. Anomalous type 17 response to viral infection by CD8+ T cells lacking T-bet and eomesodermin. Science. 2008;321:408–411. doi: 10.1126/science.1159806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kallies A, Xin A, Belz GT, Nutt SL. Blimp-1 transcription factor is required for the differentiation of effector CD8(+) T cells and memory responses. Immunity. 2009;31:283–295. doi: 10.1016/j.immuni.2009.06.021. [DOI] [PubMed] [Google Scholar]
- 7.Rutishauser RL, Martins GA, Kalachikov S, Chandele A, Parish IA, Meffre E, Jacob J, Calame K, Kaech SM. Transcriptional repressor Blimp-1 promotes CD8(+) T cell terminal differentiation and represses the acquisition of central memory T cell properties. Immunity. 2009;31:296–308. doi: 10.1016/j.immuni.2009.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cruz-Guilloty F, Pipkin ME, Djuretic IM, Levanon D, Lotem J, Lichtenheld MG, Groner Y, Rao A. Runx3 and T-box proteins cooperate to establish the transcriptional program of effector CTLs. J Exp Med. 2009;206:51–59. doi: 10.1084/jem.20081242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ichii H, Sakamoto A, Hatano M, Okada S, Toyama H, Taki S, Arima M, Kuroda Y, Tokuhisa T. Role for Bcl-6 in the generation and maintenance of memory CD8+ T cells. Nat Immunol. 2002;3:558–563. doi: 10.1038/ni802. [DOI] [PubMed] [Google Scholar]
- 10.Ichii H, Sakamoto A, Kuroda Y, Tokuhisa T. Bcl6 acts as an amplifier for the generation and proliferative capacity of central memory CD8+ T cells. Journal of Immunology (Baltimore, Md : 1950) 2004;173:883–891. doi: 10.4049/jimmunol.173.2.883. [DOI] [PubMed] [Google Scholar]
- 11.Huber M, Lohoff M. IRF4 at the crossroads of effector T-cell fate decision. Eur J Immunol. 2014;44:1886–1895. doi: 10.1002/eji.201344279. [DOI] [PubMed] [Google Scholar]
- 12.Wei G, Wei L, Zhu J, Zang C, Hu-Li J, Yao Z, Cui K, Kanno Y, Roh TY, Watford WT, Schones DE, Peng W, Sun HW, Paul WE, O’Shea JJ, Zhao K. Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity. 2009;30:155–167. doi: 10.1016/j.immuni.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Araki Y, Wang Z, Zang C, Wood WH, Schones D, Cui K, Roh TY, Lhotsky B, Wersto RP, Peng W, Becker KG, Zhao K, Weng NP. Genome-wide analysis of histone methylation reveals chromatin state-based regulation of gene transcription and function of memory CD8+ T cells. Immunity. 2009;30:912–925. doi: 10.1016/j.immuni.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vahedi G, Takahashi H, Nakayamada S, Sun HW, Sartorelli V, Kanno Y, O’Shea JJ. STATs shape the active enhancer landscape of T cell populations. Cell. 2012;151:981–993. doi: 10.1016/j.cell.2012.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scharer CD, Barwick BG, Youngblood BA, Ahmed R, Boss JM. Global DNA methylation remodeling accompanies CD8 T cell effector function. The Journal of Immunology. 2013;191:3419–3429. doi: 10.4049/jimmunol.1301395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Russ BE, Olshanksy M, Smallwood HS, Li J, Denton AE, Prier JE, Stock AT, Croom HA, Cullen JG, Nguyen MLT, Rowe S, Olson MR, Finkelstein DB, Kelso A, Thomas PG, Speed TP, Rao S, Turner SJ. Distinct epigenetic signatures delineate transcriptional programs during virus-specific CD8(+) T cell differentiation. Immunity. 2014;41:853–865. doi: 10.1016/j.immuni.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsagaratou A, Aijo T, Lio CWJ, Yue X, Huang Y, Jacobsen SE, Lahdesmaki H, Rao A. Dissecting the dynamic changes of 5-hydroxymethylcytosine in T-cell development and differentiation. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:E3306–E3315. doi: 10.1073/pnas.1412327111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Komori HK, Hart T, LaMere SA, Chew PV, Salomon DR. Defining CD4 T Cell Memory by the Epigenetic Landscape of CpG DNA Methylation. The Journal of Immunology. 2015 doi: 10.4049/jimmunol.1401162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Crompton JG, Narayanan M, Cuddapah S, Roychoudhuri R, Ji Y, Yang W, Patel SJ, Sukumar M, Palmer DC, Peng W, Wang E, Marincola FM, Klebanoff CA, Zhao K, Tsang JS, Gattinoni L, Restifo NP. Lineage relationship of CD8(+) T cell subsets is revealed by progressive changes in the epigenetic landscape. Cell Mol Immunol. 2015 doi: 10.1038/cmi.2015.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ito S, D’Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, Rao A. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Z, Cai X, Cai CL, Wang J, Zhang W, Petersen BE, Yang FC, Xu M. Deletion of Tet2 in mice leads to dysregulated hematopoietic stem cells and subsequent development of myeloid malignancies. Blood. 2011;118:4509–4518. doi: 10.1182/blood-2010-12-325241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moran-Crusio K, Reavie L, Shih A, Abdel-Wahab O, Ndiaye-Lobry D, Lobry C, Figueroa ME, Vasanthakumar A, Patel J, Zhao X, Perna F, Pandey S, Madzo J, Song C, Dai Q, He C, Ibrahim S, Beran M, Zavadil J, Nimer SD, Melnick A, Godley LA, Aifantis I, Levine RL. Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell. 2011;20:11–24. doi: 10.1016/j.ccr.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ko M, Bandukwala HS, An J, Lamperti ED, Thompson EC, Hastie R, Tsangaratou A, Rajewsky K, Koralov SB, Rao A. Ten-Eleven-Translocation 2 (TET2) negatively regulates homeostasis and differentiation of hematopoietic stem cells in mice. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:14566–14571. doi: 10.1073/pnas.1112317108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Quivoron C, Couronné L, Della Valle V, Lopez CK, Plo I, Wagner-Ballon O, Do Cruzeiro M, Delhommeau F, Arnulf B, Stern M-H, Godley L, Opolon P, Tilly H, Solary E, Duffourd Y, Dessen P, Merle-Beral H, Nguyen-Khac F, Fontenay M, Vainchenker W, Bastard C, Mercher T, Bernard OA. TET2 Inactivation Results in Pleiotropic Hematopoietic Abnormalities in Mouse and Is a Recurrent Event during Human Lymphomagenesis. Cancer Cell. 2011;20:25–38. doi: 10.1016/j.ccr.2011.06.003. [DOI] [PubMed] [Google Scholar]
- 27.Ichiyama K, Chen T, Wang X, Yan X, Kim BS, Tanaka S, Ndiaye-Lobry D, Deng Y, Zou Y, Zheng P, Tian Q, Aifantis I, Wei L, Dong C. The Methylcytosine Dioxygenase Tet2 Promotes DNA Demethylation and Activation of Cytokine Gene Expression in T Cells. Immunity. 2015;42:613–626. doi: 10.1016/j.immuni.2015.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yue X, Trifari S, Aijö T, Tsagaratou A, Pastor WA, Zepeda-Martínez JA, Lio CWJ, Li X, Huang Y, Vijayanand P, Lähdesmäki H, Rao A. Control of Foxp3 stability through modulation of TET activity. Journal of Experimental Medicine. 2016;213:377–397. doi: 10.1084/jem.20151438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pircher H, Bürki K, Lang R, Hengartner H, Zinkernagel RM. Tolerance induction in double specific T-cell receptor transgenic mice varies with antigen. Nature. 1989;342:559–561. doi: 10.1038/342559a0. [DOI] [PubMed] [Google Scholar]
- 30.Pearce EL, Shen H. Generation of CD8 T cell memory is regulated by IL-12. Journal of Immunology (Baltimore, Md : 1950) 2007;179:2074–2081. doi: 10.4049/jimmunol.179.4.2074. [DOI] [PubMed] [Google Scholar]
- 31.Ahmed R, Salmi A, Butler LD, Chiller JM, Oldstone MB. Selection of genetic variants of lymphocytic choriomeningitis virus in spleens of persistently infected mice. Role in suppression of cytotoxic T lymphocyte response and viral persistence. J Exp Med. 1984;160:521–540. doi: 10.1084/jem.160.2.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Akalin A, Garrett-Bakelman FE, Kormaksson M, Busuttil J, Zhang L, Khrebtukova I, Milne TA, Huang Y, Biswas D, Hess JL, Allis CD, Roeder RG, Valk PJM, Löwenberg B, Delwel R, Fernandez HF, Paietta E, Tallman MS, Schroth GP, Mason CE, Melnick A, Figueroa ME. Base-pair resolution DNA methylation sequencing reveals profoundly divergent epigenetic landscapes in acute myeloid leukemia. PLoS Genet. 2012;8:e1002781. doi: 10.1371/journal.pgen.1002781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Akalin A, Kormaksson M, Li S, Garrett-Bakelman FE, Figueroa ME, Melnick A, Mason CE. methylKit: a comprehensive R package for the analysis of genome-wide DNA methylation profiles. Genome Biol. 2012;13:R87. doi: 10.1186/gb-2012-13-10-r87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li S, Garrett-Bakelman FE, Akalin A, Zumbo P, Levine R, To BL, Lewis ID, Brown AL, D’Andrea RJ, Melnick A, Mason CE. An optimized algorithm for detecting and annotating regional differential methylation. BMC Bioinformatics. 2013;14:S10. doi: 10.1186/1471-2105-14-S5-S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ko M, An J, Bandukwala HS, Chavez L, Aijö T, Pastor WA, Segal MF, Li H, Koh KP, Lähdesmäki H, Hogan PG, Aravind L, Rao A. Modulation of TET2 expression and 5-methylcytosine oxidation by the CXXC domain protein IDAX. Nature. 2013;497:122–126. doi: 10.1038/nature12052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee P, Fitzpatrick DR, Beard C, Jessup H, Lehar S, Makar K, Perez-Melgosa M, Sweetser M, Schilissel M, Nguyen S, Cherry S, Tsai JH, Tucker SM, Weaver WM, Kelso A, Jaenisch R, Wilson CB. A Critical Role for Dnmt1 and DNA methylation in T cell Development, Function and Survival. Immunity. 2001;15:763–774. doi: 10.1016/s1074-7613(01)00227-8. [DOI] [PubMed] [Google Scholar]
- 37.Russ BE, Prier JE, Rao S, Turner SJ. T cell immunity as a tool for studying epigenetic regulation of cellular differentiation. Front Genet. 2013;4:218. doi: 10.3389/fgene.2013.00218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith ZD, Meissner A. DNA methylation: roles in mammalian development. Nature Reviews Genetics. 2013;14:204–220. doi: 10.1038/nrg3354. [DOI] [PubMed] [Google Scholar]
- 39.Cui W, Kaech SM. Generation of effector CD8+ T cells and their conversion to memory T cells. Immunol Rev. 2010;236:151–166. doi: 10.1111/j.1600-065X.2010.00926.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luckey CJ, Bhattacharya D, Goldrath AW, Weissman IL, Benoist C, Mathis D. Memory T and memory B cells share a transcriptional program of self-renewal with long-term hematopoietic stem cells. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:3304–3309. doi: 10.1073/pnas.0511137103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sallusto F, Lenig D, Forster R, Lipp M, Lanzavecchia A. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature. 1999;401:708–712. doi: 10.1038/44385. [DOI] [PubMed] [Google Scholar]
- 42.Wherry EJ, Teichgraber V, Becker TC, Masopust D, Kaech SM, Antia R, von Andrian UH, Ahmed R. Lineage relationship and protective immunity of memory CD8 T cell subsets. Nat Immunol. 2003;4:225–234. doi: 10.1038/ni889. [DOI] [PubMed] [Google Scholar]
- 43.Hikono H, Kohlmeier JE, Takamura S, Wittmer ST, Roberts AD, Woodland DL. Activation phenotype, rather than central- or effector-memory phenotype, predicts the recall efficacy of memory CD8+ T cells. J Exp Med. 2007;204:1625–1636. doi: 10.1084/jem.20070322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sung JH, Zhang H, Moseman EA, Alvarez D, Iannacone M, Henrickson SE, de la Torre JC, Groom JR, Luster AD, von Andrian UH. Chemokine guidance of central memory T cells is critical for antiviral recall responses in lymph nodes. Cell. 2012;150:1249–1263. doi: 10.1016/j.cell.2012.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Garrett-Bakelman FE, Sheridan CK, Kacmarczyk TJ, Ishii J, Betel D, Alonso A, Mason CE, Figueroa ME, Melnick AM. Enhanced reduced representation bisulfite sequencing for assessment of DNA methylation at base pair resolution. J Vis Exp. 2015:e52246. doi: 10.3791/52246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim HR, Hwang KA, Kim KC, Kang I. Down-regulation of IL-7Ralpha expression in human T cells via DNA methylation. Journal of Immunology (Baltimore, Md : 1950) 2007;178:5473–5479. doi: 10.4049/jimmunol.178.9.5473. [DOI] [PubMed] [Google Scholar]
- 47.Tserel L, Kolde R, Limbach M, Tretyakov K, Kasela S, Kisand K, Saare M, Vilo J, Metspalu A, Milani L, Peterson P. Age-related profiling of DNA methylation in CD8+ T cells reveals changes in immune response and transcriptional regulator genes. Sci Rep. 2015;5:13107. doi: 10.1038/srep13107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lleo A, Zhang W, Zhao M, Tan Y, Bernuzzi F, Zhu B, Liu Q, Tan Q, Malinverno F, Valenti L, Jiang T, Tan L, Liao W, Coppel R, Invernizzi P, Lu Q, Adams DH, Gershwin ME PBC Epigenetic Study Group. DNA methylation profiling of the X chromosome reveals an aberrant demethylation on CXCR3 promoter in primary biliary cirrhosis. Clin Epigenetics. 2015;7:61. doi: 10.1186/s13148-015-0098-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Raczkowski F, Ritter J, Heesch K, Schumacher V, Guralnik A, Höcker L, Raifer H, Klein M, Bopp T, Harb H, Kesper DA, Pfefferle PI, Grusdat M, Lang PA, Mittrücker HW, Huber M. The transcription factor Interferon Regulatory Factor 4 is required for the generation of protective effector CD8+ T cells. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:15019–15024. doi: 10.1073/pnas.1309378110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Koh KP, Yabuuchi A, Rao S, Huang Y, Cunniff K, Nardone J, Laiho A, Tahiliani M, Sommer CA, Mostoslavsky G, Lahesmaa R, Orkin SH, Rodig SJ, Daley GQ, Rao A. Tet1 and Tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell Stem Cell. 2011;8:200–213. doi: 10.1016/j.stem.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Minor EA, Court BL, Young JI, Wang G. Ascorbate induces ten-eleven translocation (Tet) methylcytosine dioxygenase-mediated generation of 5-hydroxymethylcytosine. J Biol Chem. 2013;288:13669–13674. doi: 10.1074/jbc.C113.464800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yin R, Mao SQ, Zhao B, Chong Z, Yang Y, Zhao C, Zhang D, Huang H, Gao J, Li Z, Jiao Y, Li C, Liu S, Wu D, Gu W, Yang YG, Xu GL, Wang H. Ascorbic Acid enhances tet-mediated 5-methylcytosine oxidation and promotes DNA demethylation in mammals. J Am Chem Soc. 2013;135:10396–10403. doi: 10.1021/ja4028346. [DOI] [PubMed] [Google Scholar]
- 53.Blaschke K, Ebata KT, Karimi MM, Zepeda-Martínez JA, Goyal P, Mahapatra S, Tam A, Laird DJ, Hirst M, Rao A, Lorincz MC, Ramalho-Santos M. nature12362. Nature. 2013;500:222–226. doi: 10.1038/nature12362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang YW, Wang Z, Xie W, Cai Y, Xia L, Easwaran H, Luo J, Yen RWC, Li Y, Baylin SB. Acetylation Enhances TET2 Function in Protecting against Abnormal DNA Methylation during Oxidative Stress. Mol Cell. 2017;65:323–335. doi: 10.1016/j.molcel.2016.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shan Q, Zeng Z, Xing S, Li F, Hartwig SM, Gullicksrud JA, Kurup SP, Van Braeckel-Budimir N, Su Y, Martin MD, Varga SM, Taniuchi I, Harty JT, Peng W, Badovinac VP, Xue HH. The transcription factor Runx3 guards cytotoxic CD8(+) effector T cells against deviation towards follicular helper T cell lineage. Nat Immunol. 2017;18:931–939. doi: 10.1038/ni.3773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ladle BH, Li KP, Phillips MJ, Pucsek AB, Haile A, Powell JD, Jaffee EM, Hildeman DA, Gamper CJ. De novo DNA methylation by DNA methyltransferase 3a controls early effector CD8+ T-cell fate decisions following activation. Proceedings of the National Academy of Sciences of the United States of America. 2016;113:10631–10636. doi: 10.1073/pnas.1524490113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Okano M, Bell DW, Haber DA, Li E. DNA Methyltransferases Dnmt3a and Dnmt3b Are Essential for De Novo Methylationand Mammalian Development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 58.Challen GA, Sun D, Jeong M, Luo M, Jelinek J, Berg JS, Bock C, Vasanthakumar A, Gu H, Xi Y, Liang S, Lu Y, Darlington GJ, Meissner A, Issa JPJ, Godley LA, Li W, Goodell MA. Dnmt3a is essential for hematopoietic stem cell differentiation. Nat Genet. 2011;44:23–31. doi: 10.1038/ng.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang L, Rau R, Goodell MA. DNMT3A in haematological malignancies. Nat Rev Cancer. 2015;15:152–165. doi: 10.1038/nrc3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ko M, An J, Pastor WA, Koralov SB, Rajewsky K, Rao A. TET proteins and 5-methylcytosine oxidation in hematological cancers. Immunol Rev. 2015;263:6–21. doi: 10.1111/imr.12239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Genovese G, Kähler AK, Handsaker RE, Lindberg J, Rose SA, Bakhoum SF, Chambert K, Mick E, Neale BM, Fromer M, Purcell SM, Svantesson O, Landén M, Höglund M, Lehmann S, Gabriel SB, Moran JL, Lander ES, Sullivan PF, Sklar P, Grönberg H, Hultman CM, McCarroll SA. Clonal Hematopoiesis and Blood-Cancer Risk Inferred from Blood DNA Sequence. N Engl J Med. 2014 doi: 10.1056/NEJMoa1409405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jaiswal S, Fontanillas P, Flannick J, Manning A, Grauman PV, Mar BG, Lindsley RC, Mermel CH, Burtt N, Chavez A, Higgins JM, Moltchanov V, Kuo FC, Kluk MJ, Henderson B, Kinnunen L, Koistinen HA, Ladenvall C, Getz G, Correa A, Banahan BF, Gabriel S, Kathiresan S, Stringham HM, McCarthy MI, Boehnke M, Tuomilehto J, Haiman C, Groop L, Atzmon G, Wilson JG, Neuberg D, Altshuler D, Ebert BL. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med. 2014;371:2488–2498. doi: 10.1056/NEJMoa1408617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xie M, Lu C, Wang J, McLellan MD, Johnson KJ, Wendl MC, McMichael JF, Schmidt HK, Yellapantula V, Miller CA, Ozenberger BA, Welch JS, Link DC, Walter MJ, Mardis ER, Dipersio JF, Chen F, Wilson RK, Ley TJ, Ding L. Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat Med. 2014 doi: 10.1038/nm.3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang X, Su J, Jeong M, Ko M, Huang Y, Park HJ, Guzman A, Lei Y, Huang Y-H, Rao A, Li W, Goodell MA. DNMT3A and TET2 compete and cooperate to repress lineage-specific transcription factors in hematopoietic stem cells. Nat Genet. 2016 doi: 10.1038/ng.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rasmussen KD, Helin K. Role of TET enzymes in DNA methylation, development, and cancer. Genes Dev. 2016;30:733–750. doi: 10.1101/gad.276568.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Huang Y, Chavez L, Chang X, Wang X, Pastor WA, Kang J, Zepeda-Martinez JA, Pape UJ, Jacobsen SE, Peters B, Rao A. Distinct roles of the methylcytosine oxidases Tet1 and Tet2 in mouse embryonic stem cells. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:1361–1366. doi: 10.1073/pnas.1322921111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hon GC, Song CX, Du T, Jin F, Selvaraj S, Lee AY, Yen CA, Ye Z, Mao SQ, Wang BA, Kuan S, Edsall LE, Zhao BS, Xu GL, He C, Ren B. 5mC Oxidation by Tet2 Modulates Enhancer Activity and Timing of Transcriptome Reprogramming during Differentiation. Mol Cell. 2014;56:286–297. doi: 10.1016/j.molcel.2014.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rasmussen KD, Jia G, Johansen JV, Pedersen MT, Rapin N, Bagger FO, Porse BT, Bernard OA, Christensen J, Helin K. Loss of TET2in hematopoietic cells leads to DNA hypermethylation of active enhancers and induction of leukemogenesis. Genes Dev. 2015;29:910–922. doi: 10.1101/gad.260174.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jani A, Wan M, Zhang J, Cui K, Wu J, Preston-Hurlburt P, Khatri R, Zhao K, Chi T. A novel genetic strategy reveals unexpected roles of the Swi-Snf-like chromatin-remodeling BAF complex in thymocyte development. Journal of Experimental Medicine. 2008;205:2813–2825. doi: 10.1084/jem.20080938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chaiyachati BH, Jani A, Wan Y, Huang H, Flavell R, Chi T. BRG1-mediated immune tolerance: facilitation of Treg activation and partial independence of chromatin remodelling. EMBO J. 2013;32:395–408. doi: 10.1038/emboj.2012.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang Q, Zhao K, Shen Q, Han Y, Gu Y, Li X, Zhao D, Liu Y, Wang C, Zhang X, Su X, Liu J, Ge W, Levine RL, Li N, Cao X. Tet2 is required to resolve inflammation by recruiting Hdac2 to specifically repress IL-6. Nature. 2015 doi: 10.1038/nature15252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Montagner S, Leoni C, Emming S, Della Chiara G, Balestrieri C, Barozzi I, Piccolo V, Togher S, Ko M, Rao A, Natoli G, Monticelli S. TET2 Regulates Mast Cell Differentiation and Proliferation through Catalytic and Non-catalytic Activities. Cell Rep. 2016;15:1566–1579. doi: 10.1016/j.celrep.2016.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Williams K, Christensen J, Pedersen MT, Johansen JV, Cloos PAC, Rappsilber J, Helin K. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature. 2011;473:343–348. doi: 10.1038/nature10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
ERRBS data has been deposited to the NCBI Gene Expression Omnibus under accession number GSE105176 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE105176).