SUMMARY
Mechanistic target of rapamycin (MTOR) cooperates with Hedgehog (HH) signaling, but the underlying mechanisms are incompletely understood. Here, we provide genetic, biochemical, and pharmacologic evidence that MTOR complex 1 (mTORC1)-dependent translation is a prerequisite for HH signaling. The genetic loss of mTORC1 function inhibited HH signaling–driven growth of the cerebellum and medulloblastoma. Inhibiting translation or mTORC1 blocked HH signaling. Depleting 4EBP1, an mTORC1 target that inhibits translation, alleviated the dependence of HH signaling on mTORC1. Consistent with this, phosphorylated 4EBP1 levels were elevated in HH signaling–driven medulloblastomas in mice and humans. In mice, an mTORC1 inhibitor suppressed medulloblastoma driven by a mutant SMO that is inherently resistant to existing SMO inhibitors, prolonging the survival of the mice. Our study reveals mTORC1-mediated translation is a key component of HH signaling and an important target for treating medulloblastoma and other cancers driven by HH signaling.
eTOC Blurb
Wu et al. show that Hedgehog (HH) signaling promotes protein synthesis via a non-canonical, mTORC1/4EBP1-dependent pathway, and that mTORC1/4EBP1-dependent translation is a prerequisite for canonical HH signaling. They show further that inhibition of mTORC1 signaling suppresses HH-dependent cerebellar growth and the development of HH-driven medulloblastoma.
INTRODUCTION
Hedgehog (HH) signaling regulates many aspects of animal development from early embryonic patterning to tissue homeostasis in adults (Briscoe and Therond, 2013). Defective HH signaling causes various developmental malformations, whereas aberrantly active HH signaling can lead to tumors, including medulloblastomas, basal cell carcinomas, rhabdomyosarcomas, meningiomas, and odontogenic tumors (Amakye et al., 2013). Elucidating the molecular mechanism of HH signal transduction is critical for understanding normal development and diseases, including congenital defects and tumors arising from abnormal HH signaling.
In vertebrates, canonical HH signaling is triggered by one of three HH proteins (Sonic Hedgehog [SHH], Indian Hedgehog, or Desert Hedgehog) and culminates in changes in the transcriptional program. In the absence of HH ligands, Patched1 (PTCH1), a 12-transmembrane receptor, inhibits ciliary accumulation and the activation of a G protein–coupled receptor (GPCR)-like protein Smoothened (SMO) (Rohatgi et al., 2007). HH binds to and inhibits PTCH1, leading to ciliary accumulation and activation of SMO (Corbit et al., 2005; Rohatgi et al., 2007). Activated SMO promotes the activation of GLI transcription factors (GLI2 and GLI3) by inhibiting their binding partner Suppressor of Fused (SUFU) and protein kinase A (PKA) (Hui and Angers, 2011). Activated GLI2 and GLI3 induce the expression of HH target genes, including Ptch1 and Gli1.
During perinatal development, HH signaling derives massive proliferation of cerebellar granule neuron precursor cells (GNPs) to produce the cerebellar granule neurons that constitute more than half of the neurons in the brain (Dahmane and Ruiz i Altaba, 1999; Wallace, 1999; Wechsler-Reya and Scott, 1999). Mutations that lead to constitutive activation of HH signaling in GNPs result in unconstrained GNP proliferation and the development of SHH-group medulloblastoma (Taylor et al., 2012). Medulloblastoma is the most common pediatric brain cancer, and the SHH group constitutes a third of these tumors (Taylor et al., 2012). Aberrant activation of HH signaling in GNPs also causes medulloblastoma in mice (Oliver et al., 2005; Schuller et al., 2008; Yang et al., 2008). Accordingly, SMO inhibitors have demonstrated efficacy in treating SHH-group medulloblastoma in both clinical trials and mouse models (Berman et al., 2002; Buonamici et al., 2010; Kieran et al., 2017; Robinson et al., 2015; Rodon et al., 2014; Romer et al., 2004; Rudin et al., 2009). However, medulloblastoma cells inevitably acquire resistance to SMO inhibitors in humans and mice (Buonamici et al., 2010; Dijkgraaf et al., 2011; Robinson et al., 2015; Rudin et al., 2009; Yauch et al., 2009). Furthermore, tumors driven by alterations downstream from SMO do not respond to SMO inhibitors (Buonamici et al., 2010; Dijkgraaf et al., 2011; Robinson et al., 2015). Therefore, new therapies are needed to overcome these resistances and target tumors with mutations downstream of SMO.
Mechanistic target of rapamycin (MTOR) is a kinase that regulates the homeostasis and growth of cells and organisms in response to diverse extracellular and intracellular cues (Laplante and Sabatini, 2012). MTOR forms two multi-protein complexes, MTOR complex 1 (mTORC1) and mTORC2 (Figure S1A). mTORC1 is a key regulator of protein translation (Ma and Blenis, 2009). The two major downstream effectors of mTORC1 in translational control are ribosomal protein S6 (RPS6) kinase 1 (RPS6KB1, also known as S6K1) and eukaryotic translation initiation factor 4E (EIF4E) binding protein 1 (EIF4EBP1, also known as 4EBP1). 4EBP1 binds to EIF4E bound at the 5′ cap of mRNAs and prevents the formation of the translation initiation complex (Sonenberg and Hinnebusch, 2009). Phosphorylation of 4EBP1 by mTORC1 dissociates 4EBP1 from EIF4E, enabling the formation of the translation initiation complex. Deregulated protein translation is critical for the development of many cancers. Oncogenic signaling pathways often aberrantly increase EIF4E activity by enhancing its transcription and/or by inhibiting 4EBP1 through mTORC1-dependent phosphorylation (Truitt and Ruggero, 2016). HH signaling increases EIF4E transcription in GNPs (Mainwaring and Kenney, 2011); however, it is unclear whether HH signaling controls 5′ cap–dependent translation and whether oncogenic HH signaling depends on the mTORC1/4EBP1/EIF4E axis.
Here, we report that HH signaling promotes mTORC1/4EBP1-dependent translation and mTORC1/4EBP1-dependent translation is essential for HH signaling. Disrupting mTORC1 function inhibited GNP expansion and medulloblastoma growth driven by a mutant SMO that is resistant to an SMO inhibitor currently in clinical use. mTORC1 may be an important target for treating HH signaling–driven medulloblastoma and other cancers and for overcoming resistance to SMO inhibitors.
RESULTS
HH signaling requires mTORC1 to drive cerebellar and medulloblastoma growth
Earlier studies concluded that HH signaling does not require MTOR because rapamycin (an allosteric inhibitor of MTOR) does not block HH signaling from inducing target-gene expression (Mainwaring and Kenney, 2011; Riobo et al., 2006). However, rapamycin partially inhibits MTOR activities (Choo et al., 2008; Thoreen et al., 2009). Moreover, MTOR cooperates with HH signaling in several cancer types and contributes to the development of resistance to SMO inhibitors (Buonamici et al., 2010; Filbin et al., 2013; Sharma et al., 2015; Syu et al., 2016; Wang et al., 2012). To scrutinize the function of MTOR in HH signaling in vivo, we removed Raptor and Rictor, essential components of mTORC1 and mTORC2, respectively (Figure S1A), in GNPs by using GFAP::Cre (Hara et al., 2002; Jacinto et al., 2004; Kim et al., 2002; Sarbassov et al., 2004). GFAP::Cre results in recombination in GNPs but does not induce recombination in Purkinje neurons and meningeal cells that non-cell-autonomously affect GNP proliferation (Spassky et al., 2008; Zhuo et al., 2001). The cerebella of GFAP::Cre; RaptorloxP/loxP mice were much smaller than those of wild-type (WT) mice and contained far fewer granule neurons (Figure 1A). Although we cannot rule out the non-cell-autonomous effects of the loss of Raptor in GFAP::Cre; RaptorloxP/loxP mice, the small cerebellum containing fewer granule neurons is a stereotypical phenotype that results from defective HH signaling in GNPs due to the loss of Gli2, Smo, or ciliogenic genes (Kif3a and Ift88) (Chizhikov et al., 2007; Corrales et al., 2006; Spassky et al., 2008). Consistent with defective HH signaling, GNPs isolated from GFAP::Cre; RaptorloxP/loxP mice had reduced levels of GLI1, a direct target of HH signaling, and SMO (Figures 1B and S1B). In contrast to the loss of Raptor, the loss of Rictor did not affect cerebellar growth in GFAP::Cre; RictorloxP/loxP mice (Figure 1A). Thus, mTORC1, but not mTORC2, was required for HH signaling to expand GNPs.
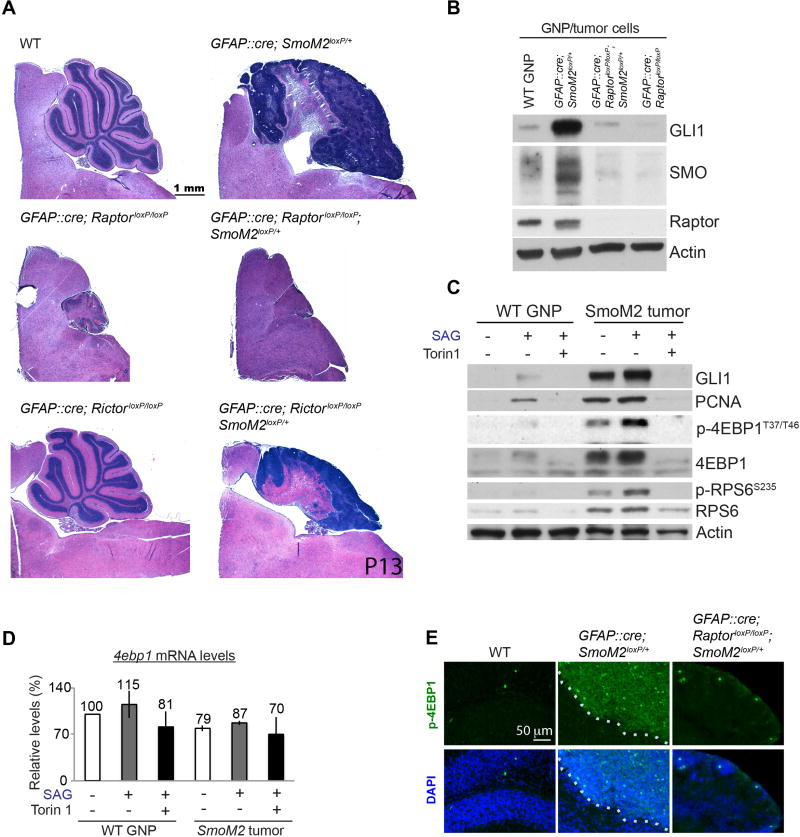
Figure 1. HH-driven cerebellar and medulloblastoma development require mTORC1.
(A) Hematoxylin and eosin (H&E)–stained cerebellar sections. The loss of Raptor in GNPs inhibited cerebellar and medulloblastoma development. (B) The loss of Raptor decreased GLI1 and SMO protein levels in GNPs and in tumor cells purified from the cerebella of P7 WT and mutant mice. (C) Analysis of the indicated protein levels in WT GNPs and tumor cells isolated from P7 WT and GFAP::Cre; SmoM2loxP/+ mice, respectively. Isolated cells were grown in culture for 36 h with or without SAG and Torin1. Actin was used as a loading control. (D) SAG and Torin1 did not affect the 4ebp1 mRNA levels in GNPs or SmoM2 tumor cells in culture. The bars show the fold changes in mRNA levels relative to those in untreated WT cells 24 h after treatments (mean ± standard deviation [SD] of three independent experiments [n=3]). (E) Immunofluorescence labeling showing the high p-4EBP1 levels in GFAP::Cre; SmoM2loxP/+ mice, which were absent in WT or GFAP::Cre; RaptorloxP/loxP; SmoM2loxP/+ mice. Dotted lines mark the boundary of tumors. Scattered mitotic cells were strongly positive for p-4EBP1.
Next, we investigated whether mTORC1 is required for oncogenic HH signaling. GFAP::Cre; SmoM2loxP/+ mice, which express in their GNPs an oncogenic mutant SMO (SMOM2) found in patients with HH-driven tumors (Brastianos et al., 2013; Lam et al., 1999; Xie et al., 1998), develop medulloblastoma by postnatal day (P) 7 to 10 (Han et al., 2009). Remarkably, disrupting mTORC1 in GFAP::Cre; SmoM2loxP/+; RaptorloxP/loxP mice prevented medulloblastoma development (Figure 1A). SMO and GLI1 levels were greatly reduced in GNPs isolated from GFAP::Cre; SmoM2loxP/+; RaptorloxP/loxP mice (Figures 1B and S1B). In contrast, the loss of Rictor in GFAP::Cre; SmoM2loxP/+; RictorloxP/loxP mice did not prevent tumor development, although the tumors were smaller than those in GFAP::Cre; SmoM2loxP/+ mice (Figure 1A). Thus, mTORC1, but not mTORC2, was required for oncogenic HH signaling to induce medulloblastoma.
To directly examine the role of MTOR in HH signaling in GNPs and medulloblastoma cells, we treated primary WT GNPs and medulloblastoma cells with SAG (a SMO agonist) and Torin1 (an active-site mTOR inhibitor) in culture. Consistent with the in vivo phenotype, Torin1 dramatically decreased the SAG- or SMOM2-induced levels of GLI1 and proliferating cell nuclear antigen (PCNA) in WT GNPs and in GFAP::Cre; SmoM2loxP/+ medulloblastoma cells in culture (Figures 1C and S1C). Thus, MTOR was required for HH signaling in GNPs and in SMOM2-driven medulloblastoma cells to increase GLI1 and PCNA levels.
Notably, the total 4EBP1 and phosphorylated 4EBP1 (p-4EBP1) protein levels, but not the 4ebp1 mRNA levels, were increased in WT GNPs treated with SAG and in medulloblastoma cells (Figures 1C, 1D, S1C). Torin1 abolished these increases in 4EBP1 proteins without decreasing the 4ebp1 mRNA levels (Figures 1C, 1D, S1C). These results suggest that HH signaling posttranscriptionally increases 4EBP1 levels, possibly by mTORC1-dependent translation, and increases their mTORC1-dependent phosphorylation. The level of p-4EBP1 was also greatly increased in medulloblastoma tissues of GFAP::Cre; SmoM2loxP/+ mice (Figure 1E), thereby demonstrating that mTORC1 activities are strong in tumor cells in situ. The role of mTORC1 in medulloblastoma growth in GFAP::Cre; SmoM2loxP/+ mice may be to phosphorylate and antagonize the immensely increased 4EBP1, a negative regulator of 5′ cap–dependent translation.
Canonical HH signaling requires MTOR/4EBP1-dependent translation
To investigate the mechanism by which MTOR functions in HH signaling, we used NIH 3T3 mouse embryonic fibroblast cells, which produce robust and consistent responses to HH signaling. In NIH 3T3 cells, SAG increased the mRNA levels of Gli1, a direct transcriptional target of canonical HH signaling (Figure 2A). Torin1, but not rapamycin, suppressed this Gli1 mRNA induction (Figure 2A). Thus, canonical HH signaling required rapamycin-resistant MTOR activities at the downstream of SMO. SMO must localize to cilia to activate canonical HH signaling (Corbit et al., 2005). SMO localized to cilia within 2 h after SAG stimulation (Figure S2). Torin1 did not inhibit this ciliary accumulation of SMO, thereby ruling out a possible mechanism of mTORC1 function in canonical HH signaling.
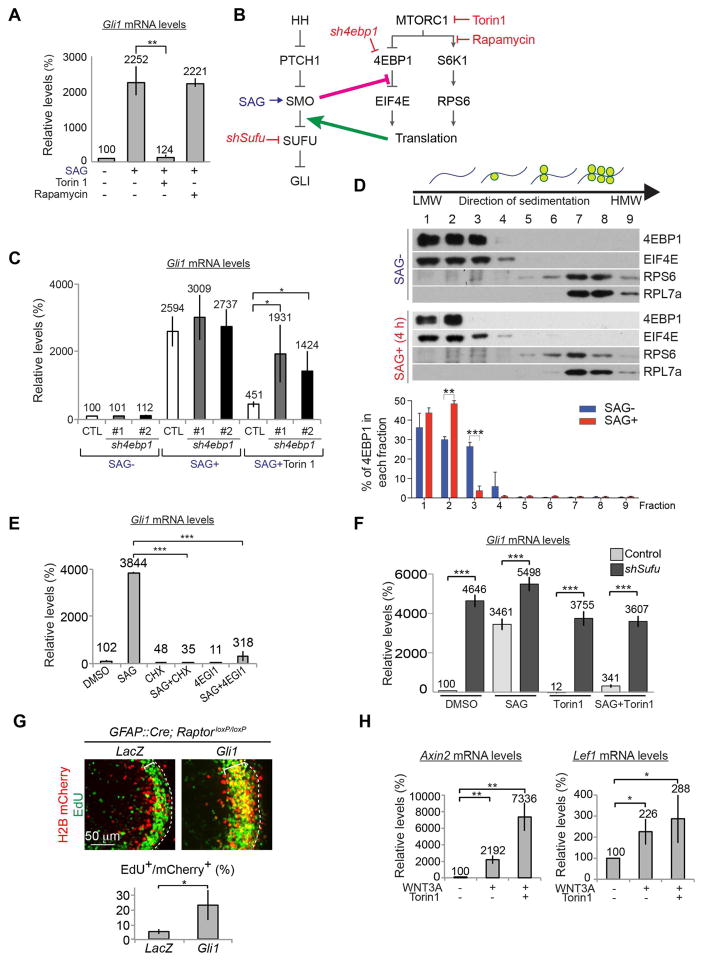
Figure 2. HH signaling requires MTOR/4EBP1-dependent translation.
(A) Torin1, but not rapamycin, suppressed the Gli1 mRNA increase in SAG-stimulated NIH 3T3 cells. (B) Simplified schematic of the HH and mTORC1 signaling cascade with the activators and inhibitors used in the study. We found that activation of SMO inhibits 4EBP1/EIF4E complex formation and that translation is required between SMO and SUFU in canonical HH signaling. (C) Depleting 4EBP1 did not increase Gli1 mRNA levels in unstimulated cells but restored the SAG-induced Gli1 mRNA increase in Torin1-treated NIH 3T3 cells. (D) Western blotting and quantification (graphs) show that 4 h of SAG stimulation shifted 4EBP1 to the lighter cytosolic fractions in cell lysates fractionated on a sucrose gradient. Note the changes in 4EBP1 in fractions 2 and 3 of the SAG-treated NIH 3T3 cell lysates, as compared to the control (SAG−) cell lysates. The graph shows the percentage of the total 4EBP1 in each fraction (mean ± SD, n=3). The levels of ribosomal proteins (RPS6 and RPL7A) were used to determine the distribution of the cytosol, initiation complex, monosome, and polysomes, as shown in the schematic. (E) Translation inhibitors (CHX and 4EGI-1) suppressed the SAG-induced increase in Gli1 mRNA levels in NIH 3T3 cells. (F) Depleting SUFU increased Gli1 mRNA levels in the absence of SAG and in the presence of Torin1. (G) Images of electroporated cells (H2B mCherry+, red) and proliferating cells (EdU+, green) in GFAP::Cre; RaptorloxP/loxP mutant cerebellar slices maintained in culture for 48 h after co-electroporation of H2B mCherry plasmids with either LacZ (control) or Gli1 plasmids at P7. EdU was added to the cultures 24 h before fixation. Dotted lines mark the surface of the cerebellum, and brackets mark the EGL. Bar graphs show the proportion of proliferating cells among the electroporated cells (mean ± the standard error of the mean, n=3). (H) Torin1 did not inhibit WNT3A from inducing the expression of the target genes Axin2 and Lef1. The bar graphs in (A, C, E, F, H) show the fold changes in mRNA levels relative to those in control (CTL) cells 24 h after treatments (mean ± SD, n=3). * P < 0.05; ** P < 0.01; *** P < 0.005.
S6K1 and 4EBP1 are two major substrates of mTORC1 in translational control (Figure 2B). Whereas Torin1 inhibits mTORC1 from phosphorylating both S6K1 and 4EBP1, rapamycin efficiently blocks S6K1 but not 4EBP1 phosphorylation (Choo et al., 2008; Thoreen et al., 2009), suggesting that canonical HH signaling requires inhibition of 4EBP1 by mTORC1-dependent phosphorylation. We tested whether depletion of 4EBP1 can bypass the requirement of MTOR in canonical HH signaling. 4ebp1 knockdown (KD) by two different shRNAs (sh4ebp1 #1 and #2) (Figure S3A) did not increase Gli1 mRNA levels in unstimulated cells (Figure 2C) but did significantly restore the SAG-stimulated Gli1 mRNA increase in the presence of Torin1 (Figure 2C), indicating that HH signaling requires mTORC1-mediated inhibition of 4EBP1.
Inhibitory phosphorylation of 4EBP1 by mTORC1 is a key step in the initiation of 5′ cap–dependent translation. P-4EBP1 dissociates from EIF4E bound at the 5′ cap of the mRNA, enabling the formation of the translation initiation complex. To investigate whether HH signaling affected the complex of 4EBP1, EIF4E, and mRNA, we fractionated cell lysates by using sucrose gradients and examined the distribution of key components of the translational machinery. SAG treatment shifted 4EBP1 to the lighter cytosolic fractions within 4 h (Figures 2D and S3B), suggesting that HH signaling promotes translation by releasing 4EBP1 from the EIF4E-mRNA complex and that Torin1 inhibits canonical HH signaling by suppressing this HH-promoted translation. Indeed, like Torin1, inhibitors of translation initiation (4EGI-1) or elongation (cycloheximide [CHX]) suppressed Gli1 mRNA induction by SAG (Figure 2E). Remarkably, however, mTORC1/4EBP1-dependent translation was dispensable for HH signaling triggered by the loss of SUFU, a key downstream negative regulator of HH signaling (Cooper et al., 2005; Svard et al., 2006) (Figure 2F). Gli1 mRNA and GLI1 protein levels were strongly induced by Sufu KD in the absence of SAG or SHH (Figures 2F and S3C). Sufu KD cells still strongly increased Gli1 mRNA levels in the presence of Torin1 or translation inhibitors (4EGI-1 or CHX) (Figures 2F and S3D). Thus, rapamycin-resistant and mTORC1/4EBP1-dependent translation was required a step between SMO and SUFU in the canonical HH signaling pathway (Figure 2B).
To confirm the requirement of mTORC1 upstream of SUFU, we determined whether activation of HH signaling downstream of SUFU can increase the proliferation of GNPs lacking Raptor and, thus, mTORC1. We chose to express Gli1 instead of removing Sufu in GNPs because complex functions of SUFU in GNPs may complicate interpretation of Sufu loss-of-function experiments (Kim et al., 2011). To closely mimic in vivo GNPs, we electroporated plasmids expressing either LacZ (control) or Gli1 into the cerebella of GFAP::Cre; RaptorloxP/loxP mice ex vivo and cultured slices of the electroporated cerebella. Electroporated cells were labeled by co-electroporation of H2B-mCherry. We added 5-ethynyl-2-deoxyuridine (EdU) to label proliferating cells 24 h after electroporation and analyzed proliferation 24 h after EdU addition. Consistent with the requirement of mTORC1 upstream of SUFU in canonical HH signaling, expression of GLI1 significantly increased GNP proliferation independently of Raptor (Figure 2G).
MTOR activities are dispensable for WNT signaling
Because MTOR may be a general prerequisite for signaling pathways, we determined whether MTOR was specifically required for HH signaling by testing its function in WNT signaling, which shares striking similarities with HH signaling (Kalderon, 2002). In both signaling pathways, common protein kinases (glycogen synthase kinase 3 and casein kinase 1) constitutively promote proteolytic processing of effector transcription factors (β-catenin for WNT and GLI2/3 for HH). The activation of Frizzled-family GPCR-like proteins (Frizzled for WNT and SMO for HH) inhibits this processing, leading to the accumulation and activation of β-catenin in WNT signaling and of GLI2/3 in HH signaling. In NIH 3T3 cells, WNT3A induced the expression of target genes, Axin2 and Lef1; however, blocking MTOR with Torin1 did not inhibit WNT signaling from inducing these WNT target genes but instead augmented their induction (Figure 2H). Thus, MTOR function is specifically required for HH signaling but not for WNT signaling, despite the similarities between the two.
Because MTOR may be a prerequisite for Gli1 transcription, we determined whether MTOR was specifically required for HH signaling to increase Gli1 transcription. We noticed that fibroblast growth factor 2 (FGF2) also increased Gli1 mRNA levels in NIH 3T3 cells (Figure S3E). Although SANT1 (a SMO inhibitor) and Torin1 almost completely blocked SAG from increasing Gli1 mRNA levels, they failed to efficiently block FGF2 from doing so (Figure S3E). Thus, in contrast to HH signaling, MTOR activities were not absolutely required for FGF2 to induce Gli1 expression.
HH signaling increases SMO protein expression via a noncanonical pathway
Because our data suggested that HH signaling promoted 5′ cap–dependent translation and translation was required for canonical HH signaling, we sought a candidate protein that positively regulates HH signaling and whose translation is promoted by HH signaling. We noted that the loss of Raptor dramatically decreased SMO protein levels in the cerebellum (Figures 1B and S1B). Smo is not an established transcriptional target of HH signaling, suggesting that mTORC1 and HH signaling increase or maintain SMO protein levels. Indeed, activation of HH signaling (Figure 3A) increased SMO protein levels in GNPs and NIH3T3 cells (Figures 3B–D). Notably, the increase in SMO occurred much earlier than did the increase in GLI1 (at 8 h vs. 24 h), which is a direct transcriptional target of HH signaling (Figures 3C and 3D). In contrast, fetal bovine serum (FBS), epidermal growth factor (EGF), or FGF2 did not increase SMO protein levels, although FBS and FGF2 increased GLI1 protein levels (Figures 3E and S4A), suggesting that the increase in SMO proteins was more specifically attributable to HH signaling activation than was the increase in GLI1. Consistently, SANT-1 inhibited the SHH-induced increase in SMO and GLI1 proteins but not the FBS-induced increase in GLI1 proteins (Figure 3E).
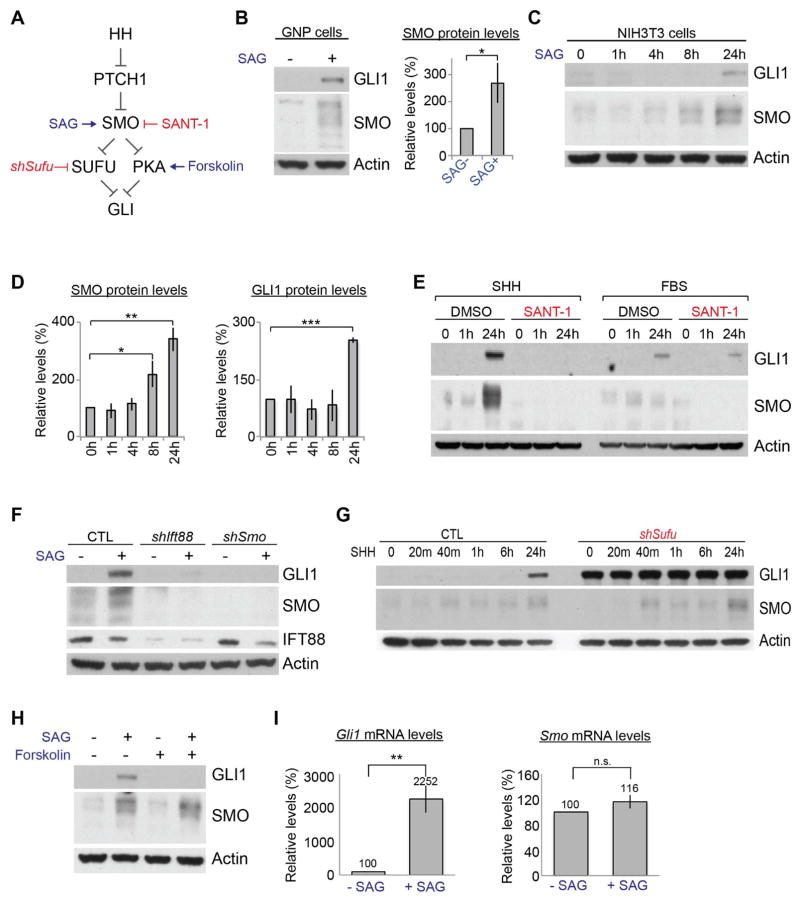
Figure 3. HH signaling increases SMO protein levels via a noncanonical pathway.
(A) Schematic showing a simplified HH signaling pathway with reagents used to modulate the signaling. (B) SAG increased GLI1 and SMO protein levels in GNPs in culture within 24 h. The bars show the fold changes in the SMO levels relative to those in untreated cells (SAG−) (mean ± SD, n=3). (C, D) Western blots and quantifications of GLI1 and SMO protein levels in NIH 3T3 cells at the indicated times after SHH treatment. The bars show the fold changes in the protein levels relative to those in untreated cells (mean ± SD, n=3). (E) SMO and GLI1 protein levels in NIH 3T3 cells stimulated with SHH or FBS in the presence or absence of SANT-1. (F) SAG-induced GLI1 and SMO protein levels in NIH 3T3 cells were suppressed in cells expressing shRNA against Ift88 (shIft88) or Smo (shSmo). (G) GLI1 and SMO protein levels in NIH 3T3 cells expressing shSufu at the indicated times after SHH treatment. Note that SMO, but not GLI1, was induced by SHH. (H) Forskolin blocked SAG from increasing GLI1 but not SMO protein levels. (I) SAG increased Gli1 but not Smo mRNA levels in NIH 3T3 cells. The bars show the fold changes in the mRNA levels relative to those in untreated cells (mean ± SD, n=3). *P < 0.05; ** P < 0.01; *** P < 0.005; n.s., P > 0.05.
We detected multiple SMO bands on immunoblots, which may represent different stages of SMO maturation and posttranslational modifications. Consistently, those multiple bands were reduced in Smo KD cells, also confirming the specificity of the antibody (Figure 3F). SMO undergoes N-linked glycosylation during maturation (Marada et al., 2015). PNGase F (N-glycosidase that removes N-glycans from proteins) downshifted the SMO bands (Figure S4B). Thus, the SMO antibody detected multiple forms of SMO.
HH-induced SMO protein increase preceded the increase of GLI1 (Figures 3C and 3D). Therefore, we examined whether central components of canonical HH signaling are required to increase SMO protein levels (Figure 3A). Primary cilia are essential for canonical HH signaling in mammals (Goetz and Anderson, 2010). The KD of Intraflagellar transport 88 (Ift88), an essential ciliogenic gene (Pazour et al., 2000), blocked SAG from inducing the GLI1 and SMO proteins (Figure 3F). Thus, HH signaling required primary cilia to increase SMO protein levels. Depleting SUFU, a downstream negative regulator, increased GLI1 levels in the absence of SHH or SAG (Figures 3G and S3C). However, SMO protein levels were increased only after SHH treatment in the Sufu KD cells (Figure 3G). Forskolin that activates PKA, another negative regulator of canonical HH signaling (Wang et al., 2000; Wang et al., 1999), completely blocked the increase in GLI1 protein levels after SAG stimulation; however it failed to inhibit SAG-induced increase in SMO protein levels (Figure 3H). Canonical HH signaling culminates in changes in gene expression as exemplified by strong increase in Gli1 mRNA levels; however, Smo mRNA levels were not increased by SAG (Figure 3I). Furthermore, the loss of cilia in Ift88 KD cells inhibited SAG-induced Gli1 mRNA and GLI1 protein induction; however, it inhibited only SMO protein increase without affecting Smo mRNA levels after SAG stimulation (Figures 3F, S4C). Together, these results demonstrate that HH signaling increased the SMO protein levels posttranscriptionally via a SUFU- and PKA-independent but SMO- and primary cilia–dependent pathway.
HH signaling increases SMO translation in an mTORC1- and 4EBP1-dependent manner
Hh signaling stabilizes Smo proteins in Drosophila (Alcedo et al., 2000; Denef et al., 2000). To test whether HH signaling in mammals increased SMO by inhibiting its degradation, we asked whether interrupting the proteasome and lysosome functions increased SMO protein levels. SMO did not accumulate 8 h after we blocked proteasomes (with MG132), lysosomes (with chloroquine), or both, by which time HH signaling had already increased the levels of SMO proteins (Figure S5A). In contrast, inhibiting translation initiation (with 4EGI-1) or elongation (with CHX) completely abolished SAG-induced SMO upregulation (Figures 4A and S5B), indicating that SMO upregulation is dependent on translation but not on increased stability.
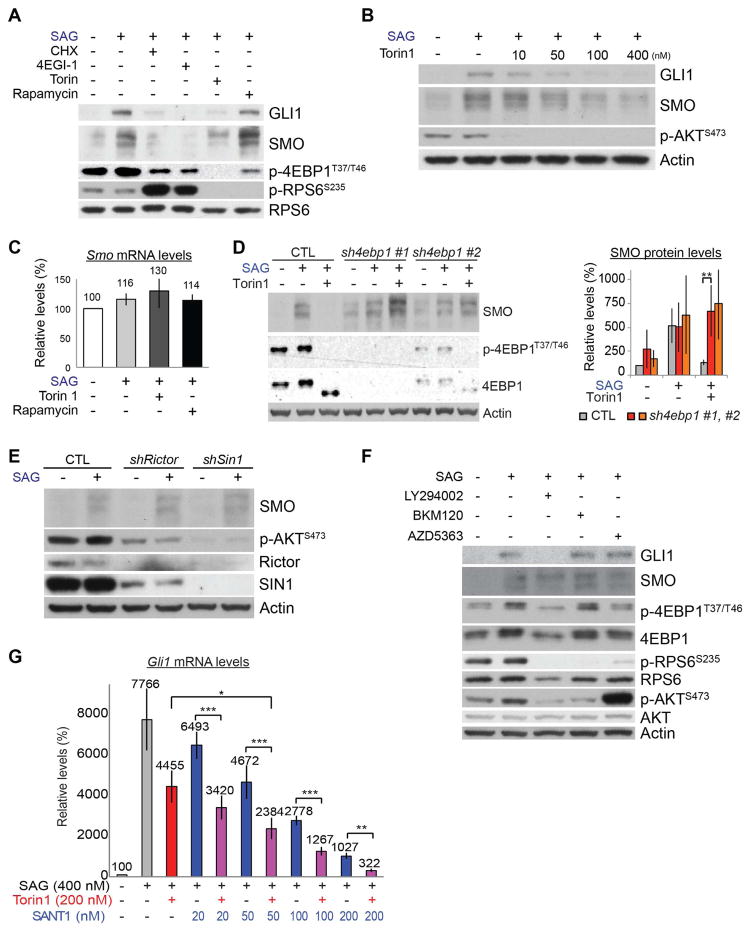
Figure 4. HH signaling increases SMO translation through an mTORC1/4EBP1-dependent mechanism.
(A) Translation inhibitors (CHX, 4EGI-1) and Torin1, but not rapamycin, blocked the SAG-driven increase in SMO protein in NIH 3T3 cells. Torin1, but not rapamycin, completely inhibited the phosphorylation of 4EBP1. (B) Torin1 dose-dependently suppressed SAG-driven increase in GLI1 and SMO. (C) SAG, Torin1, and rapamycin did not affect Smo mRNA levels. The bars show the fold changes in the mRNA levels relative to those in untreated cells (mean ± SD, n=3). (D) Depleting 4EBP1 with two different shRNAs increased the basal SMO protein levels and abolished the inhibitory effect of Torin1 on SMO induction. The downshifted 4EBP1 bands in Torin1-treated lanes reflect un-phosphorylated 4EBP1. The bars show the fold changes (mean ± SD) in the SMO levels relative to those in untreated control cells (n=3 for CTL, 5 for sh4ebp1 #1, and 2 for sh4ebp1 #2). (E) Disruption of mTORC2 by shRictor or shSin1 decreased AKT phosphorylation but not SMO levels in SAG-treated NIH 3T3 cells. (F) PI3K inhibitors (LY294002, BKM120) or an AKT inhibitor (AZD5363) decreased p-RPS6 but not SAG-induced SMO protein levels in NIH 3T3 cells. (G) Torin1 sensitized NIH 3T3 cells to the SMO inhibitor SANT1. The bars show the fold changes in mRNA levels relative to those in untreated cells (mean ± SD, n=3). Cells were treated as indicated in each panel for 24 h. ** P < 0.01; *** P < 0.005.
Notably, Torin1, but not rapamycin, blocked the SAG-induced increase in SMO (Figures 4A and S5B). Torin1 inhibited SAG induction of SMO proteins in a dose-dependent manner but did not affect Smo mRNA levels (Figures 4B and 4C). To test whether Torin1 decreased SMO protein levels by destabilizing the protein, we simultaneously inhibited the proteasome with Torin1 treatment. Proteasome inhibition did not rescue the decrease in SMO proteins caused by Torin1 (Figure S5C). These results show that HH signaling increased SMO protein levels without increasing Smo mRNA levels or SMO protein stability and required translation and rapamycin-resistant MTOR function to do so.
The differential effects of rapamycin and Torin1 on SMO protein levels mirrored their differential effects on canonical HH signaling (Figure 2A), where depletion of 4EBP1 rescued inhibitory effects of Torin1 (Figure 2C), and suggested that inhibitory phosphorylation of 4EBP1 is required for HH signaling to increase SMO translation. Consistent with this, depleting 4EBP1 increased the basal SMO protein levels and restored SMO induction by SAG in the presence of Torin1 (Figure 4D). In contrast to Torin1, 4EGI1, which inhibits cap-dependent translation by directly binding to EIF4E, still inhibited SAG from increasing SMO levels in 4ebp1 KD cells (Figure S5D), indicating that 4ebp1 KD antagonized the effect of Torin1 on SMO levels by increasing cap-dependent translation. Thus, HH signaling required mTORC1-dependent inhibition of 4EBP1 to increase SMO protein synthesis.
Torin1, but not rapamycin, also inhibits mTORC2 (Thoreen et al., 2009). To test whether mTORC2 function is required for HH signaling to increase SMO protein levels, we depleted either Rictor or Mapkap1 (also known as SIN1), two essential components of mTORC2 (Jacinto et al., 2006; Jacinto et al., 2004; Sarbassov et al., 2004; Yang et al., 2006), in NIH 3T3 cells. Depleting Rictor or SIN1 decreased the phosphorylation of AKT serine/threonine kinase (AKT) at Ser473, a direct substrate of mTORC2 (Sarbassov et al., 2005), but failed to inhibit SAG from increasing SMO (Figures 4E and S5E). The function of mTORC2 was dispensable for HH signaling to increase SMO levels, similar to how it is dispensable in HH-driven cerebellar and medulloblastoma development (Figure 1A).
Phosphatidylinositol 3-kinase (PI3K) and AKT are activators of mTORC1 (Laplante and Sabatini, 2012; Ma and Blenis, 2009) (Figure S1A). To determine whether these kinases contributed to mTORC1 effects on SMO, we treated cells with inhibitors of PI3K (LY294002 and BKM120) or AKT (AZD5363). These inhibitors failed to decrease 4EBP1 phosphorylation or the SAG-induced increase in SMO protein (Figures 4F and S5F), although they efficiently inhibited the phosphorylation of RPS6. The high level of p-AKT S473 in cells treated with AZD5363 resulted from feedback hyperphosphorylation after the inhibition of AKT. LY294002 decreased GLI1 but not SMO levels (Figures 4F and S5F). This decrease in GLI1 levels may have resulted from the PI3K-independent action of LY294002 because LY294002 can affect proteins unrelated to PI3K (Gharbi et al., 2007), and BKM120, a more selective PI3K inhibitor, did not affect GLI1 and SMO levels. Another highly specific PI3K inhibitor (GNE-317) does not suppress HH signaling (Metcalfe et al., 2013). These results show that HH signaling required mTORC1 but not mTORC2, PI3K, or AKT to increase SMO protein levels.
Torin1 decreased SMO levels. Thus, we tested whether Torin1 could sensitize cells to SMO inhibitors. Indeed, 200 nM of Torin1, which decreased canonical HH signaling activity to 57% levels (determined by relative Gli1 mRNA levels), significantly enhanced the inhibitory effect of SANT1 on canonical HH signaling (Figure 4G). Notably, the combination of 200 nM Torin1 and SANT1 (50–200nM) was synergistic according to the Bliss Independence model (Bliss, 1939). The observed fractional effect was 0.03–0.04 larger than would be expected if the two drugs acted independently.
p-4EBP1 levels are high in SHH-group medulloblastoma in humans
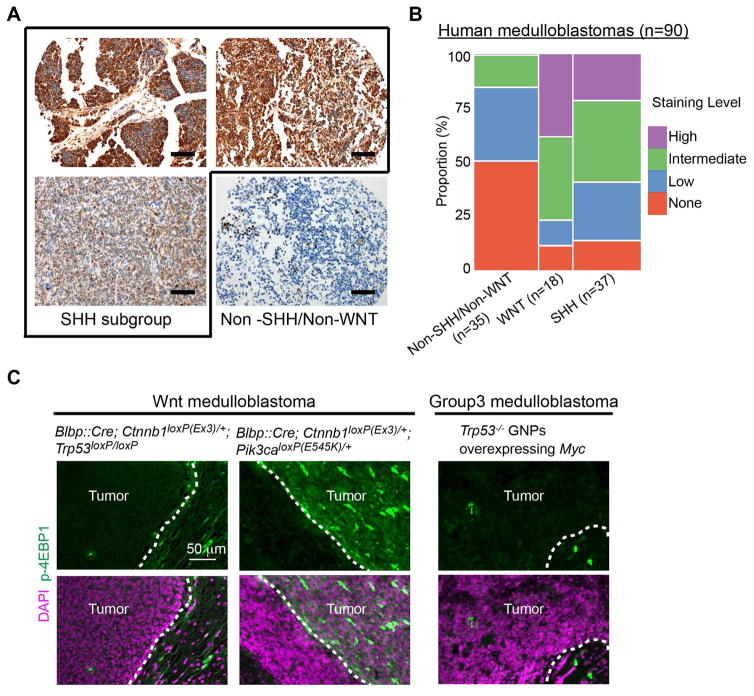
We found that inhibitory phosphorylation of 4EBP1 by mTORC1 is essential for HH signaling. Consistent with this, SMOM2-driven mouse medulloblastoma was dependent on mTORC1 and had high levels of p-4EBP1 (Figures 1A and 1E). Thus, we examined whether p-4EBP1 levels were also high in human medulloblastoma. In humans, medulloblastomas are categorized in four subgroups: SHH, WNT, group 3, and group 4. The subgroups have distinct characteristics, including gene expression profiles, mutations, and signaling pathways that promote cancer (Taylor et al., 2012). Consistent with our mouse model, most SHH-group human medulloblastomas (86%) expressed p-4EBP1 (Figure 5 and Table S1). The staining of p-4EBP1 was significantly enriched in the SHH group, as compared with the non-SHH/non-WNT-group (P = 0.0005 by the chi-square test), and the staining level in the SHH group was greater than in the non-SHH/non-WNT group (P = 0.0005 by the chi-square test) (Figures 5B and S6), suggesting that mTORC1 activities are strong in SHH group. In a subset of SHH-group tumors with identified mutations that affect SHH signaling, p-4EBP1 staining presence or levels were not significantly different between tumors with mutations in PTCH1 or SMO (n=19) and those with mutations in SUFU or GLI2 (n=5) (P= 0.52 and 0.33, respectively) (Table S1). In addition to its role in canonical HH signal transduction, mTORC1 may have other roles in SHH-group medulloblastoma, such as in the insulin-like growth factor (IGF) signaling that is essential for HH signaling–driven tumorigenesis in mice (Hahn et al., 2000). WNT-group tumors also exhibited high levels of p-4EBP1, suggesting strong mTORC1 activity. Some WNT-group medulloblastomas have mutations in PIK3CA that encodes a catalytic subunit of PI3K (Robinson et al., 2012). The lack of mutational information for the WNT tumors that we sampled precluded our ability to correlate p-4EBP1 levels with specific mutations. Notably, p-4EBP1 levels were high in a WNT medulloblastoma mouse model driven by constitutively active PI3K and β-catenin but low in a WNT medulloblastoma model driven by constitutively active β-catenin and loss of Trp53 (Gibson et al., 2010; Robinson et al., 2012) (Figure 5C). Furthermore, p-4EBP1 levels were low in a group 3 medulloblastoma mouse model, which was driven by overexpression of MYC and loss of Trp53 (Kawauchi et al., 2012) (Figure 5C). Together with high p-4EBP1 levels in an SHH group mouse model (Figure 1E), these results show that subgroup-specific patterns of p-4EBP1 levels are conserved in humans and mouse models.
Figure 5. p-4EBP1 levels in human medulloblastoma.
(A) Immunostaining for p-4EBP1 showed high- (top panels) or intermediate-level staining (bottom-left panel) in human SHH-group medulloblastoma, whereas only focal, low-level staining was observed in non-SHH/non-WNT medulloblastoma (bottom-right panel). (B) Mosaic plot for 90 medulloblastomas displaying the proportion of p-4EBP1 staining levels in each subgroup. The p-4EBP1 staining was significantly enriched in the SHH group, as compared with that in the non-SHH/non-WNT group (P = 0.0005 by the chi-square test), and the staining level in the SHH-group was greater than that in the non-SHH/non-WNT group (P = 0.0005 by the chi-square test). (C) Immunostaining for p-4EBP1 in mice. p-4EBP1 levels were high in WNT tumors with a Pik3ca mutation but undetectable in WNT tumors without a Pik3ca mutation and in group 3 tumors. Scale bar = 200 μm (A), 50 μm (B).
INK128, an MTOR inhibitor, inhibits HH signaling in the brain
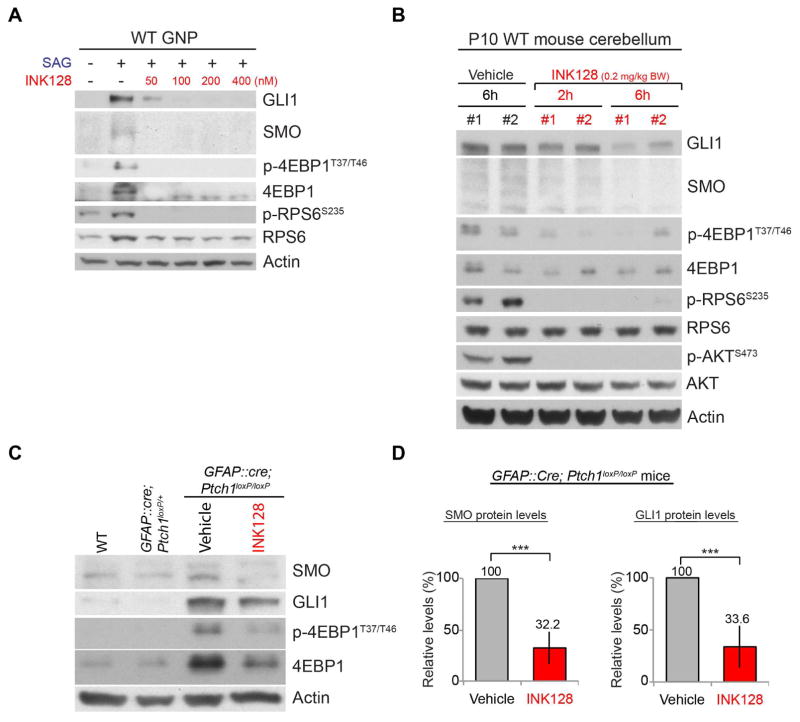
The unexpected critical role of MTOR-dependent translation in HH signaling and the high p-4EBP1 levels in SHH-group medulloblastoma in humans prompted us to investigate the potential of MTOR inhibitors in treating SHH-group medulloblastoma. We chose INK128 (Hsieh et al., 2012) from among the commercially available MTOR inhibitors and tested whether it could inhibit HH signaling in the brain in vivo; INK128 is currently in a clinical trial (NCT02133183) to study its brain penetrance and its effects on glioblastoma. INK128 suppressed the SAG-induced increase in GLI1 and SMO in primary GNPs (Figure 6A). In vivo, INK128 inhibited the phosphorylation of mTORC1 substrates (RSP6 and 4EBP1) and an mTORC2 substrate (AKT) in WT cerebella within 2 h of administration by oral gavage at P10 (Figure 6B). The GLI1 and SMO levels were also decreased within 6 h after treatment (Figures 6B and S7A).
Figure 6. INK128 inhibits MTOR activities and HH signaling in the brain.
(A) INK128 suppressed the SAG-induced increase in GLI1, SMO, 4EBP1, and p-4EBP1 levels in GNPs in culture. Cells were treated for 24 h. (B) Oral gavage of INK128 to WT pups at P10 inhibited the phosphorylation of 4EBP1, RPS6, and AKT within 2 h and decreased the GLI1 and SMO levels within 6 h. (C, D) Administration of INK128 (0.2 mg/kg BW) at P10 decreased the high levels of GLI1 and SMO in the tumors of GFAP::Cre; Ptch1loxP/loxP mice by 24h. The bars show the fold changes in SMO and GLI1 levels in the tumors of GFAP::Cre; Ptch1loxP/loxP mice relative to those in vehicle-treated mice 24 h after treatment (mean ± SD, 4 mice per group). Each lane in panels B and C represents a mouse. *** P < 0.005.
Next, we tested whether INK128 could inhibit HH signaling in medulloblastoma by using GFAP::Cre; Ptch1loxP/loxP mice, in which the loss of PTCH1 results in cerebella filled with tumor cells by P14 (Yang et al., 2008). Remarkably, a single dose of INK128 strongly suppressed the high levels of GLI1 and SMO in the tumors of GFAP::Cre; Ptch1loxP/loxP mice at 24 h after treatment at P10 (Figures 6C and 6D). Thus, INK128 inhibited HH signaling in WT cerebella and SHH-group medulloblastoma in mice.
INK128 inhibits medulloblastoma growth in GFAP::Cre; SmoM2loxP/+ mice and prolongs their survival
Because INK128 inhibited HH signaling in medulloblastoma in GFAP::Cre; Ptch1loxP/loxP mice, we investigated whether INK128 could suppress medulloblastoma growth driven by SMOM2 that is resistant to an SMO inhibitor in the clinic (Dijkgraaf et al., 2011). Currently, no targeted therapy is available for medulloblastoma that is resistant to SMO inhibitors. In vitro, MTOR inhibition suppressed HH signaling in SMOM2-driven medulloblastoma cells (Figure 1C).
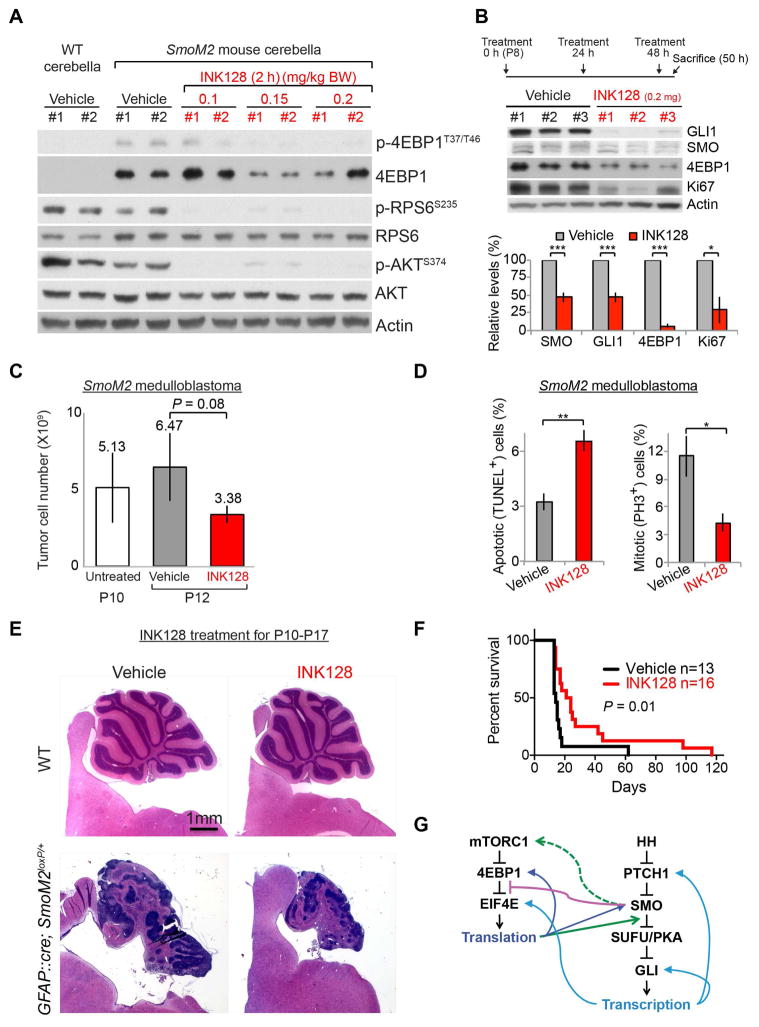
INK128 suppressed MTOR activities in GFAP::Cre; SmoM2loxP/+ medulloblastoma within 2 h of administration at P10 (Figure 7A). Notably, the 4EBP1 and p-4EBP1 levels were dramatically increased in GFAP::Cre; SmoM2loxP/+ tumors but were suppressed by INK128 (Figure 7A), consistent with observations made in vitro (Figures 1C, S1C, and 6A) and in immunostained tumors (Figure 1E, 5A). Daily treatment with INK128 for 3 days decreased the levels of SMO, GLI1, 4EBP1, and Ki67 (a proliferation marker) and the number of tumor cells, as compared with those of mice treated with vehicle (Figures 7B and 7C). Accordingly, INK128 significantly decreased the number of mitotic cells and increased the number of apoptotic cells in the tumors (Figure 7D). Daily treatment of INK128 for 8 days decreased tumor size; however, administering the same treatments to WT mice did not affect the size or histologic appearance of their brains (Figure 7E). Remarkably, daily administration of INK128 to GFAP::Cre; SmoM2loxP/+ mice, starting from P7 (when their entire cerebellum is essentially tumor tissue), significantly prolonged their survival (Figures 7F and S7). All but one GFAP::Cre; SmoM2loxP/+ mouse treated with vehicle died within 18 days of birth (Figure 7F). The GLI1 levels in the tumor of this one vehicle-treated mouse was very low compared to those in tumors of other mice, including those treated with INK128 (Figure S7B), suggesting that this mouse was an unusual outlier. Thus, inhibition of MTOR suppressed HH-driven medulloblastoma growth, and this may be effective against tumors driven by mutant SMO proteins that are resistant to SMO inhibitors.
Figure 7. INK128 inhibits medulloblastoma growth in GFAP::Cre; SmoM2loxP/+ mice and prolongs their survival.
(A) Oral gavage of INK128 at P10 decreased the phosphorylation of 4EBP1, RPS6, and AKT in medulloblastomas of GFAP::Cre; SmoM2loxP/+ mice within 2 h. Note the high levels of 4EBP1 and p-4EBP1 in vehicle-treated tumors. (B, C) The levels of GLI1, SMO, 4EBP1, and Ki67 (B) and the number of tumor cells (C) were decreased after 3 daily treatments with INK128 (0.2 mg/kg BW). The bars show the fold changes in protein levels (B) and tumor cell number (C) after 3 daily treatments (mean ± SD, 3 mice per group). (D) The proportions of TUNEL+ and PH3+ cells relative to the numbers of DAPI+ cells in tumors after 3 daily treatments with either vehicle or INK128 (3 mice per group). (E) H&E–stained sections of cerebella showing the decrease in tumor size after 8 daily treatment with INK128. The same treatments did not affect the size or histologic appearance of WT cerebella. (F) Daily treatments of INK128 from P7 significantly prolonged the survival of GFAP::Cre; SmoM2loxP/+ mice. (G) Schematic summary of crosstalk between HH and mTORC1 pathways. Activation of SMO releases the inhibitory association of EIF4E and 4EBP1 (purple line) in an mTORC1-dependent manner (green dotted arrow), leading to increased translation of proteins, including SMO and 4EBP1 (dark blue arrows). mTORC1/4EBP1-dependent translation is required at a step between SMO and SUFU in HH signaling (green arrow) to induce the transcription of target genes, including Ptch1, Gli1, and Eif4e (light blue arrows). * P < 0.05; ** P < 0.01.
DISCUSSION
Canonical HH signaling activates transcription through a PTCH1-SMO-SUFU/PKA-GLI–dependent pathway. Here, we have shown that mTORC1/4EBP1-dependent translation is required for a step between SMO and SUFU in canonical HH signaling (Figure 7G). Our study reveals mTORC1 to be a potentially important molecular target for treating SHH-group medulloblastomas. Medulloblastoma fully developed in GFAP::Cre; SmoM2loxP/+ pups at a developmental stage similar to that at which medulloblastoma develops in humans. INK128 treatments were tolerated by and effective in these pups whose cerebella consisted mostly of tumor tissue. Until now, SMO has been the only molecule targeted in clinical trials of medulloblastoma. Despite their promising efficacy, SMO inhibitors have shown some limitations in clinical trials; patients with mutations downstream from SMO do not respond to SMO inhibitors, young children treated with SMO inhibitors develop irreversible bony growth plate fusions, and initial responders inevitably acquire resistance (Robinson, 2017; Robinson et al., 2015; Rodon et al., 2014; Rudin et al., 2009). We demonstrated that MTOR inhibitors can overcome some of these limitations and sensitize cells to a SMO inhibitor. INK128 treatment suppressed HH and MTOR signaling in medulloblastomas driven by SMOM2 that is resistant to an SMO inhibitor. Tumor-bearing mice treated with daily INK128 continued to grow. Moreover, the non-overlapping mechanisms of action of an MTOR inhibitor and an SMO inhibitor and their synergistic effects suggest that a combination of these agents represent a promising strategy for SHH-group medulloblastoma and other cancers driven by aberrant HH signaling.
Although mTORC1 activity was required upstream of SUFU in canonical HH signaling, SHH-group tumors that have a SUFU mutation or GLI2 amplification were positive for p-4EBP1 staining, suggesting that mTORC1 may have additional roles in SHH-group tumors. IGF signaling, a well-known activator of mTORC1, synergizes with HH signaling to promote GNP proliferation and is essential for medulloblastoma and rhabdomyosarcoma development in Ptch1 mutant mice (Hahn et al., 2000; Hartmann et al., 2005; Ye et al., 1996). It is unknown whether IGF signaling is required for tumorigenesis driven by mutations downstream of PTCH1. In our cohort, 2 of 4 tumors with GLI2 amplifications also had PTEN loss, suggesting a role for the PTEN/PI3K axis in tumors with GLI2 amplification. Moreover, activation of PI3K signaling is a potential mechanism for the emergence of resistance to SMO inhibitors, and co-inhibition of SMO and PI3K blocks the phosphorylation of RPS6 and delays the appearance of resistance to SMO inhibitors in a mouse medulloblastoma model (Buonamici et al., 2010; Metcalfe et al., 2013). Importantly, a rapamycin derivative also delays the development of resistance in a similar manner to that of PI3K inhibitors (Buonamici et al., 2010), suggesting that a rapamycin-sensitive mTORC1 function is important for resistance development. However, our results suggest that the function of mTORC1 in HH signaling is independent of its role in the development of resistance to SMO inhibitors; PI3K and AKT inhibitors and rapamycin, all of which blocked phosphorylation of RPS6 but not of 4EBP1, failed to inhibit HH signaling. A previous study also showed that a PI3K inhibitor (GNE-317) does not suppress HH signaling (Metcalfe et al., 2013). These results are consistent with the inefficiency of PI3K inhibitors, when administered as a single agent, at suppressing medulloblastoma progression (Buonamici et al., 2010; Metcalfe et al., 2013). Thus, active-site MTOR inhibitors may offer the advantage of directly inhibiting HH signaling, in addition to inhibiting the rapamycin-sensitive MTOR activities that contribute to SMO inhibitor resistance. Although our in vitro studies were performed without IGF or insulin, the loss of Raptor or INK128 treatment in vivo may have also affected IGF signaling and the PTEN/PI3K/MTOR axis. MTOR inhibitors may have additional benefits of blocking IGF signaling and PTEN/PI3K/MTOR activities in HH signaling-driven tumors.
HH signaling promotes transcription by relieving repression and regulates its activities by inducing the expression of positive- and negative-feedback regulators, namely GLI1 and PTCH1. Translational control by HH signaling shows striking similarities to transcriptional control. It promotes translation by relieving the inhibitory function of 4EBP1 and induces the expression of key positive and negative regulators of 5′ cap–dependent translation, namely EIF4E (Mainwaring and Kenney, 2011) and 4EBP1 (this study), respectively. HH signaling also promotes internal ribosome entry site–dependent translation of ornithine decarboxylase, and its inhibition suppresses the growth of SHH-group medulloblastoma allografts (D’Amico et al., 2015). A recent study showed that HH signaling guides axons by inducing local translation at the growth cone (Lepelletier et al., 2017). Together, these results indicate that HH signaling–regulated translation constitutes critical parts of the HH signaling network in development and tumorigenesis. Deregulated translation control is a hallmark of human cancers and is critical for tumorigenesis downstream from multiple oncogenic signaling pathways (Truitt and Ruggero, 2016). Identifying mRNAs whose translation is regulated by HH signaling will deepen our understanding of HH signaling mechanisms and reveal potential therapeutic targets for HH-driven cancers.
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Young-Goo Han (young-goo.han@stjude.org).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell culture and stimulation
NIH 3T3 cells (RRID:CVCL_0594) were grown in culture in DMEM containing 10% calf serum (Sigma) and 1× penicillin-streptomycin-glutamine (PSG; ThermoFisher) at 37°C, 5% CO2. Cells were seeded at 1 million cells per 10-cm dish and typically reached 100% confluency 48 h after seeding. For serum starvation, confluent cells were maintained in DMEM containing 0.5% calf serum and 1× PSG. After 24 h of serum starvation, the cells were treated with the following agents as indicated in the text and figures: SHH peptides (200 ng/mL; R&D Systems), SAG (400 nM; Cayman Chemical, catalog no. 11914), fetal bovine serum (10%; ThermoFisher), FGF2 (100 ng/mL; PeproTech), EGF (100 ng/mL; PeproTech), WNT3A (100 ng/mL; R&D Systems), SANT-1 (400 nM; Santa Cruz Biotechnology), Torin1 (400 nM; Tocris Bioscience), INK128 (400 nM; Selleck Chemicals), rapamycin (100 nM; LC Laboratories), BKM120 (400 nM; Selleck Chemicals), LY294002 (10 μM; LC Laboratories), AZD5363 (100 nM; Selleck Chemicals), 4EGI-1 (15 μM; EMD Millipore), CHX (100 μM; Sigma-Aldrich), forskolin (10 μM; Selleck Chemicals), MG132 (10 μM; Sigma-Aldrich), or chloroquine (10 μM; Sigma-Aldrich). All treatments were done for 24 h with the dose described above except those indicated otherwise in the text and figures. Cells were collected at designated time points for further analysis. 4ebp1, Sufu, Rictor, and Sin1 KD cells were generated by transducing NIH 3T3 cells with lentiviruses expressing shRNAs. We produced replication-incompetent lentiviruses by transfecting 293T cells with helper vectors and shRNA vectors purchased from transOMIC technologies. The shRNA vectors used were TLHSU1400-1978 (4ebp1), TLMSU1400-24069 (Sufu), TLMSU1400-78757 (Rictor), and TLMSU1400-227743 (Sin1).
Purification and culture of GNP and medulloblastoma cells
To purify GNPs and medulloblastoma cells, cerebella of P7 mice without or with tumors were dissociated with Accutase (ThermoFisher, catalog no. 11110501), resuspended in Neurobasal medium (ThermoFisher, catalog no. 21103-049), and separated by Percoll density-gradient (60%–30%) centrifugation (3000 rpm for 15 min). Purified GNPs or medulloblastoma cells were grown in culture in Neurobasal medium containing 1× PSG at 37°C, 5% CO2. GNPs were treated with SAG (200nM; Cayman Chemical, catalog no. 11914), Torin1 (400 nM), or INK128 (400 nM) as indicated in the text or figures. Cells were collected at designated time points for further analysis.
Mice
We used the following mouse strains: SmoM2loxP (Jackson Laboratory, JAX stock # 005130), GFAP::Cre (JAX stock # 004600), RaptorloxP (JAX stock # 013188), RictorloxP (JAX stock # 020649), Ptch1loxP (JAX stock # 012457), Ctnnb1loxP(ex3) (Gibson et al., 2010), and Pik3caE545K (Robinson et al., 2012). A mouse model for group 3 medulloblastoma was generated as described previously (Kawauchi et al., 2012). All mice were maintained on a mixed genetic background. All mice were maintained on a 12 h dark/light cycle and housed with a maximum of five mice of the same sex per cage. We used animals of both sexes for the experiments. All animal procedures were approved by the Institutional Animal Care and Use Committee of St Jude Children’s Research Hospital.
METHOD DETAILS
Polysome purification
NIH 3T3 cells were incubated with CHX (10 μg/mL) for 15 min and lysed in hypotonic buffer (20 mM potassium acetate, 12 mM magnesium acetate, 20 mM Tris-HCl, pH 7.4) by Dounce homogenization (35 strokes). Cell debris and nuclei were removed by centrifugation at 14,000 × g for 10 min. Supernatants were layered on 1 mL of a 17%–47% (wt/vol) sucrose gradient (10 mM sodium chloride, 12 mM magnesium chloride, 20 mM Tris-HCl, pH 7.4) and centrifuged for 1.5 h at 35,000 rpm in a TLS-55 Beckman rotor. Nine fractions were manually collected from each sample by pipetting, and the fractionated samples were concentrated with Vivaspin centrifugal concentrators (Sartorius, Elk Grove, IL) for further analysis.
Oral gavage
INK128 (Selleck Chemicals, catalog no. S2811) was dissolved to 20 mg/mL in vehicle containing 5% n-methylpyrrolidone (Sigma-Aldrich), 15% polyvinylpyrrolidone K 30 (Sigma-Aldrich), and 80% water. INK128 (0.1, 0.15, or 0.2 mg/kg body weight [BW]) or vehicle (2.5 μL/g BW) was administered daily by oral gavage from P10 until the designated time point. For survival analyses, we administered INK128 (0.2 mg/kg BW) or vehicle to mice daily from P7 and euthanized the animals when they became moribund.
Ex vivo cerebellar electroporation and organotypic slice culture
P7 cerebella were dissected, soaked in Hank’s buffered salt solution (HBSS), containing a suspension of pCIG2 plasmid DNA at the following concentrations: control (500 ng/μL of H2B-mCherry + LacZ 2 μg/μL) and Gli1 overexpression (500 ng/μL of H2B-mCherry + 2 μg/μL of Gli1). The cerebella were then electroporated in a platinum block petri dish electrode (CUY520-P5, Protech International) with a square wave electroporator (CUY21EDIT, Protech International) with the following program: 5 pulses, 90V, 50 ms pulse, and 500 ms interval. The electroporated cerebella were recovered in ice-cold HBSS for 5 min and embedded in 4% low melting point agarose in HBSS, and 300 μm sagittal sections of the cerebellar vermis were acquired on a vibratome (VT1200, Leica microsystems). The sections were transferred onto Millicell cell culture inserts (Millipore) and maintained for 48 h in Fluorobrite DMEM supplemented with 0.5% Glucose, 2mM L-glutamine, 50 U/mL penicillin, streptomycin, and 1× B27 and 1× N2 supplements (Thermo Fisher). EdU at a final concentration of 10 μM was added to the cerebellar slices during the final 24 h of culture and fixed overnight with 4% paraformaldehyde. The cerebellar slices were permeabilized overnight with 0.2% Triton-X100 at 4°C followed by EdU staining with the Click-it EdU staining kit as recommended by the manufacturer (Thermo Fisher). Stained cerebellar slices were mounted on glass slides with ProLong Gold mounting medium (Thermo Fisher). Cerebellar slices were imaged on a Marianas spinning disk confocal microscope (Intelligent Imaging Innovations) consisting of a Zeiss Axio Observer microscope equipped with 20×/0.8NA Plan-apochromat. Images were captured and analyzed by using Slidebook software (Intelligent Imaging innovations).
Immunoblotting
Cells were disrupted with RIPA lysis buffer (50 mM Tris-HCl pH 7.4, 100 mM NaCl, 5 mM EDTA, 1% Triton X-100) containing protease and phosphatase inhibitors (Roche), incubated on ice for 15 min, and centrifuged for 10 min at 14,000 × g at 4°C. For brain samples, 100 μg of tissue was dissected and homogenized in RIPA buffer with a motor tissue grinder (Fisher Scientific, catalog no. 12-141-361). Tissue debris was pelleted and removed by centrifugation for 10 min at 14,000 × g at 4°C. The protein concentrations in the superna tants were determined by DC Protein Assay (Bio-Rad, catalog no. 5000112). Supernatants containing 3 to 5 μg of protein were subject to SDS-PAGE and analyzed using antibodies to the following: p-4EBP1 T37/T46 (Cell Signaling Technology, catalog no. 2855), 4EBP1 (Cell Signaling Technology, catalog no. 9644), β-actin (Sigma-Aldrich, catalog no. A5441), p-Akt S473 (Cell Signaling Technology, catalog no. 4060), Akt (Cell Signaling Technology, catalog no. 2966), EIF4E (Cell Signaling Technology, catalog no. 2067), GLI1 (Cell Signaling Technology, catalog no. 2534), IFT88 (Proteintech, catalog no. 13967-1-AP), Ki67 (Abcam, catalog no. ab16667), MTOR (Cell Signaling Technology, catalog no. 2983), PCNA (Santa Cruz Biotechnology, catalog no. sc56), Raptor (Cell Signaling Technology, catalog no. 2280), Rictor (Cell Signaling Technology, catalog no. 2114), RPL7A (Cell Signaling Technology, catalog no. 2415), p-RPS6 S235 (Cell Signaling Technology, catalog no. 2211), RPS6 (Cell Signaling Technology, catalog no. 3944), SIN1 (Millipore, catalog no. 05-1044), SMO (Santa Cruz Biotechnology, catalog no. sc166685), and ubiquitin (Santa Cruz Biotechnology, catalog no. sc-8017). We used anti-mouse or rabbit IgG secondary antibodies conjugated with horseradish peroxidase (Jackson ImmunoResearch Laboratories #115-036-006 and #115-036-047) and SuperSignal West Pico chemiluminescent substrate (ThermoFisher #34080) to detect protein bands. The intensity of each protein band was acquired by using ImageJ.
qRT-PCR
Total RNA was extracted using RNeasy Mini Kits (Qiagen). An aliquot of 125–200 ng of total RNA from each sample was reverse transcribed with Superscript III reverse transcriptase (ThermoFisher). Quantitative PCR analysis using SYBR green (ThermoFisher) was performed on an Applied Biosystems 7900 Real-Time PCR System. Transcript levels were normalized to the expression levels of Actb, Gapdh, or Rn18s.
Immunostaining
Mice were perfused with 4% paraformaldehyde (PFA) in PBS. Their brains were dissected out, fixed overnight in 4% PFA at 4°C, washed in PBS ove rnight, cryoprotected in 30% sucrose, embedded in OCT, and sectioned at a slice thickness of 12 μm. Cells grown in culture were fixed with 4% PFA in PBS for 15 min at 4°C then was hed with PBS. Brain sections or fixed cells were incubated with primary antibodies overnight at 4°C then incubated with secondary antibodies at room temperature for 2 h and stained with DAPI (10 μg/mL, Sigma) for 10 min. Coverslips were mounted on slides with Aqua-Poly/Mount (Polysciences). Images were acquired with a Zeiss AxioImager upright microscope or a Zeiss780 microscope and processed using Adobe Photoshop. We used the following antibodies: anti-acetylated tubulin antibody (Sigma, catalog no. T6973, diluted 1:1000), anti-SMO (Santa Cruz Biotechnology, catalog no. sc166685, diluted 1:50), anti-p-4EBP1 (Cell Signaling Technology, catalog no. 2855, diluted 1:200), and Alexa Fluor®–conjugated secondary antibodies (Invitrogen).
Human tissue microarrays and Immunohistochemistry
Molecular subgrouping of medulloblastoma tissue was performed using immunohistochemical staining essentially as previously described but with some slight modification (Ellison et al., 2011). Antibodies to YAP1 (Santa Cruz Biotechnology clone 63.7, diluted 1:50), β-catenin (Ventana Medical Systems clone 14, no dilution required), and GAB1 (Santa Cruz Biotechnology clone H-7, diluted 1:400) were used with appropriate secondary reagents. Detection of phosphorylated-4EBP1 was performed using a monoclonal antibody (Cell Signaling Technology clone 236B4, diluted 1:800). Scoring of phospho-4EBP1 immunolabeling was performed as follows: each tumor was evaluated in a tissue microarray for positive or negative staining, and assigned a qualitative score based on whether there was low, focal staining (staining in less than 50% of the tumor cells), intermediate staining (weak staining in more than 50% of the cells), or high staining (high-intensity staining in more than 50% of the cells). Staining levels were correlated to molecular subgroup and SHH pathway abnormalities (PTCH1/SMO mutations or SUFU/GLI2 mutations).
QUANTIFICATION AND STATISTICAL ANALYSIS
For western blot, the intensity of each protein band was acquired by using ImageJ (NIH). For qPCR analysis, the relative expression of specific mRNAs compared to control groups were calculated according to the measured Ct values. For these quantified results, at least two to three sample sets for each treatment were collected for analysis by the one-tailed, unequal variance Student’s t-test. We used the chi-square test to compare the proportion of tumors stained for p-4EBP1 and the intensity of p-4EBP1 staining in each medulloblastoma subgroup. P-values of less than 0.05 were considered significant.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit monoclonal anti phospho-4EBP1 (T37/T46) (clone 236B4) | Cell Signaling Technology | Cat# 2855S; RRID:AB_560835 |
| Rabbit monoclonal anti 4EBP1 (clone 53H11) | Cell Signaling Technology | Cat# 9644S; RRID:AB_2097841 |
| Mouse monoclonal anti β-actin (AC-15) | Sigma-Aldrich | Cat# A5441; RRID:AB_476744 |
| Rabbit monoclonal anti phospho-AKT S473 (clone D9E) | Cell Signaling Technology | Cat# 4060S; RRID:AB_2315049 |
| Rabbit monoclonal anti AKT (clone 5G3) | Cell Signaling Technology | Cat# 2966S; RRID:AB_10695737 |
| Mouse monoclonal anti β-catenin (clone 14) | Ventana Medical Systems | Cat# 760-4242 |
| Rabbit monoclonal anti EIF4E (clone C46H6) | Cell Signaling Technology | Cat# 2067S; RRID:AB_2097675 |
| Mouse monoclonal anti Gab1 (clone H-7) | Santa Cruz Biotechnology | Cat# sc-133191; RRID:AB_2107855 |
| Rabbit polyclonal anti GLI1 | Cell Signaling Technology | Cat# 2534S; RRID:AB_2294745 |
| Rabbit polyclonal anti IFT88 | Proteintech Group | Cat# 13967-1-AP; RRID:AB_2121979 |
| Rabbit monoclonal anti Ki67 (clone SP6) | Abcam | Cat# ab16667; RRID:AB_302459 |
| Rabbit monoclonal anti mTOR (clone 7C10) | Cell Signaling Technology | Cat# 2983S; RRID:AB_2105622 |
| Mouse monoclonal anti PCNA (clone PC10) | Santa Cruz Biotechnology | Cat# sc-56; RRID:AB_628110 |
| Rabbit monoclonal anti Raptor (clone 24C12) | Cell Signaling Technology | Cat# 2280S; RRID:AB_10694695 |
| Rabbit monoclonal anti Rictor (clone 53A2) | Cell Signaling Technology | Cat# 2114S; RRID:AB_10694641 |
| Rabbit polyclonal anti Ribosomal Protein L7a | Cell Signaling Technology | Cat# 2415S; RRID:AB_2182059 |
| Rabbit polyclonal anti Phospho-S6 Ribosomal Protein (S235/S236) | Cell Signaling Technology | Cat# 2211S; RRID:AB_331679 |
| Mouse monoclonal anti S6 Ribosomal Protein (clone 54D2) | Cell Signaling Technology | Cat# 2317S; RRID:AB_2238583 |
| Mouse monoclonal anti Sin1 (clone 1c7.2) | Millipore | Cat# 05-1044; RRID:AB_1587253 |
| Mouse monoclonal anti Smo (clone E-5) | Santa Cruz Biotechnology | Cat# sc-166685; RRID:AB_2239686 |
| Mouse monoclonal anti Tubulin, Acetylated (clone 6-11B-1) | Sigma-Aldrich | Cat# A6793; RRID:AB_477585 |
| Mouse monoclonal anti Ubiquitin (clone P4D1) | Santa Cruz Biotechnology | Cat# sc-8017; RRID:AB_628423 |
| Mouse monoclonal anti YAP (clone 63.7) | Santa Cruz Biotechnology | Cat# sc-101199; RRID:AB_1131430 |
| Bacterial and Virus Strains | ||
| pZIP-mCMV-ZsGreen-sh4ebp1 | transOMIC technologies | TLHSU1400-1978 |
| pZIP-mCMV-ZsGreen-shSufu | transOMIC technologies | TLMSU1400-24069 |
| pZIP-mCMV-ZsGreen-shRictor | transOMIC technologies | TLMSU1400-78757 |
| pZIP-mCMV-ZsGreen-shSin1 | transOMIC technologies | TLMSU1400-227743 |
| Biological Samples | ||
| Human medulloblastoma tissue samples | This study | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Bovine Calf Serum | Sigma-Aldrich | Cat# 12133C-500ML |
| Recombinant Human Sonic Hedgehog/Shh (C24II) N-Terminus | R&D Systems | Cat# 1845-SH-025/CF |
| SAG | Cayman Chemical | Cat# 11914; CAS: 912545-86-9 |
| Fetal Bovine Serum | ThermoFisher Scientific | Cat# 16000044 |
| Recombinant Human FGF-basic | PeproTech | Cat# 100-18B |
| Animal-Free Recombinant Human EGF | PeproTech | Cat# AF-100-15 |
| Recombinant Human WNT3A Protein | R&D Systems | Cat# 5036-WN-010 |
| SANT-1 | Santa Cruz Biotechnology | Cat# sc-203253; CAS: 304909-07-7 |
| Torin1 | Tocris Bioscience | Cat# 4247; CAS: 1222998-36-8 |
| INK128 | Selleck Chemicals | Cat# S2811; CAS: 1224844-38-5 |
| Rapamycin | LC Laboratories | Cat# R-5000; CAS: 53123-88-9 |
| BKM120 | Selleck Chemicals | Cat# S2247; CAS: 944396-07-0 |
| LY294002 | LC Laboratories | Cat# L-7962; CAS: 154447-36-6 |
| AZD5363 | Selleck Chemicals | Cat# S8019; CAS: 1143532-39-1 |
| 4EGI-1 | EMD Millipore | Cat# 324517-10MG; CAS: 315706-13-9 |
| Cycloheximide | Sigma-Aldrich | Cat# C7698-1G; CAS: 66-81-9 |
| Forskolin | Selleck Chemicals | Cat# S2449; CAS: 66575-29-9 |
| MG-132 | Sigma-Aldrich | Cat# 474787-10MG; CAS: 133407-82-6 |
| Chloroquine diphosphate salt | Sigma-Aldrich | Cat# C6628-25G; CAS: 50-63-5 |
| N-Methylpyrrolidone | Sigma-Aldrich | Cat# PHR1352-2G; CAS: 872-50-4 |
| Polyvinylpyrrolidone K 30 | Sigma-Aldrich | Cat# 234257-5G; CAS: 9003-39-8 |
| DAPI | Sigma-Aldrich | Cat# D9542-1MG; CAS: 28718-90-3 |
| Critical Commercial Assays | ||
| Click-iT EdU Alexa Fluor 488 Imaging Kit | ThermoFisher Scientific | Cat# C10337 |
| DC Protein Assay | Bio-Rad | Cat# 5000112 |
| SuperSignal West Pico Chemiluminescent Substrate | ThermoFisher Scientific | Cat# 34080 |
| RNeasy Mini Kit | Qiagen | Cat# 74106 |
| SuperScript III First-Strand Synthesis System | ThermoFisher Scientific | Cat# 18080051 |
| Power SYBR Green Master Mix | ThermoFisher Scientific | Cat# 4367659 |
| Deposited Data | ||
| Experimental Models: Cell Lines | ||
| NIH 3T3 | ATCC | CRL-1658 |
| Experimental Models: Organisms/Strains | ||
| Mouse: SmoM2loxP | Jackson Laboratory | JAX #: 005130 |
| Mouse: GFAP::Cre | Jackson Laboratory | JAX #: 004600 |
| Mouse: RaptorloxP | Jackson Laboratory | JAX #: 013188 |
| Mouse: RictorloxP | Jackson Laboratory | JAX #: 020649 |
| Mouse: Ptch1loxP | Jackson Laboratory | JAX #: 012457 |
| Mouse: Ctnnb1loxP(ex3) | Gibson et al., 2010 | N/A |
| Mouse: Pik3caE545K | Robinson et al., 2012 | N/A |
| Mouse: Group 3 medulloblastoma | Kawauchi et al., 2012 | N/A |
| Oligonucleotides | ||
| Forward primer for Gapdh, CGTCCCGTAGACAAAATGGT | This paper | N/A |
| Reverse primer for Gapdh, GAATTTGCCGTGAGTGGAGT | This paper | N/A |
| Forward primer for Gli1, GCCACACAAGTGCACGTTTG | This paper | N/A |
| Reverse primer for Gli1, AAGGTGCGTCTTGAGGTTTTCA | This paper | N/A |
| Forward primer for Smo, AATTGGCCTGGTGCTTATTG | This paper | N/A |
| Reverse primer for Smo, TGGTCTCGTTGATCTTGCTG | This paper | N/A |
| Forward primer for Axin2, GCCAATGGCCAAGTGTCTCT | This paper | N/A |
| Reverse primer for Axin2, GCGTCATCTCCTTGGGCA | This paper | N/A |
| Forward primer for 4ebp1 GACCGGAAATTTCTGATGGA | This paper | N/A |
| Reverse primer for 4ebp1 TCATCGCTGGTAGGGCTAGT | This paper | N/A |
| Forward primer for Lef1 AAATGGGTCCCTTTCTCCAC | This paper | N/A |
| Reverse primer for Lef1 TCGTCGCTGTAGGTGATGAG | This paper | N/A |
| Forward primer for Actb TCCTGAGCGCAAGTACTCTGTGT | This paper | N/A |
| Reverse primer for Actb CGGACTCATCGTACTCCTGCTT | This paper | N/A |
| Forward primer for Rn18s AGTAACGGAGCCAGACCTGAGA | This paper | N/A |
| Reverse primer for Rn18s CTTTGAGGTGGAAGACACAGGAG | This paper | N/A |
| Recombinant DNA | ||
| pCIG2-H2B-mCherry | This paper | N/A |
| pCIG2-H2B-LacZ | This paper | N/A |
| pCIG2-H2B-Gli1 | This paper | N/A |
| Software and Algorithms | ||
| Other | ||
Supplementary Material
Supplementary Table 1. Human medulloblastoma samples used for Figure 5.
Highlights.
HH increases protein synthesis via an non-canonical, mTORC1/4EBP1-dependent pathway.
Canonical HH signaling requires mTORC1/4EBP1-dependent translation.
mTORC1 activity is high in HH-driven medulloblastoma in mice and humans.
HH signaling requires mTORC1 to drive cerebellar and medulloblastoma growth.
Acknowledgments
We thank N. Badders, S. Baker, X. Cao, H. Chi, M.-J. Han, H.J. Kim, K.A. Laycock, and S. Ogden for comments on the manuscript. We thank A. Shelat for computationally testing the interaction of inhibitors of SMO and MTOR. Y.-G.H. is supported by NIH/NCI Cancer Center Core Support grant CA021765 (to St. Jude Children’s Research Hospital), the Sontag Foundation Distinguished Scientist Award, a Whitehall Foundation Research Grant, and ALSAC.
Footnotes
AUTHOR CONTRIBUTIONS
C.-C.W. performed most of the experiments. S.H. contributed to the analyses of the Raptor and Rictor mutants. B.A.O. analyzed the p-4EBP1 levels in human medulloblastomas. B.R.K. quantified the number of apoptotic cells and mitotic cells in tumors and contributed to the in vivo analyses. Y.H.Y. quantified cilia containing SMO and contributed to the in vivo analyses. T.O. performed the ex vivo electroporation experiments under the supervision of D.J.S.. F.R. performed the Sin1 and Rictor KD experiments. M.M.T. provided Ctnnb1loxP(ex3) mice. C.G.E. and G.W.R. provided human medulloblastoma samples. R.J.G. and M.R. helped generate mouse models for WNT and group 3 medulloblastomas. C.C.W. and Y.-G.H. analyzed the data and wrote the manuscript with input from their coauthors. All authors reviewed the manuscript. Y.-G.H. supervised the project.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alcedo J, Zou Y, Noll M. Posttranscriptional regulation of smoothened is part of a self-correcting mechanism in the Hedgehog signaling system. Mol Cell. 2000;6:457–465. doi: 10.1016/s1097-2765(00)00044-7. [DOI] [PubMed] [Google Scholar]
- Amakye D, Jagani Z, Dorsch M. Unraveling the therapeutic potential of the Hedgehog pathway in cancer. Nat Med. 2013;19:1410–1422. doi: 10.1038/nm.3389. [DOI] [PubMed] [Google Scholar]
- Berman DM, Karhadkar SS, Hallahan AR, Pritchard JI, Eberhart CG, Watkins DN, Chen JK, Cooper MK, Taipale J, Olson JM, et al. Medulloblastoma growth inhibition by hedgehog pathway blockade. Science. 2002;297:1559–1561. doi: 10.1126/science.1073733. [DOI] [PubMed] [Google Scholar]
- Bliss CI. THE TOXICITY OF POISONS APPLIED JOINTLY. Annals of Applied Biology. 1939;26:585–615. [Google Scholar]
- Brastianos PK, Horowitz PM, Santagata S, Jones RT, McKenna A, Getz G, Ligon KL, Palescandolo E, Van Hummelen P, Ducar MD, et al. Genomic sequencing of meningiomas identifies oncogenic SMO and AKT1 mutations. Nat Genet. 2013;45:285–289. doi: 10.1038/ng.2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briscoe J, Therond PP. The mechanisms of Hedgehog signalling and its roles in development and disease. Nat Rev Mol Cell Biol. 2013;14:416–429. doi: 10.1038/nrm3598. [DOI] [PubMed] [Google Scholar]
- Buonamici S, Williams J, Morrissey M, Wang A, Guo R, Vattay A, Hsiao K, Yuan J, Green J, Ospina B, et al. Interfering with resistance to smoothened antagonists by inhibition of the PI3K pathway in medulloblastoma. Sci Transl Med. 2010;2:51ra70. doi: 10.1126/scitranslmed.3001599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chizhikov VV, Davenport J, Zhang Q, Shih EK, Cabello OA, Fuchs JL, Yoder BK, Millen KJ. Cilia proteins control cerebellar morphogenesis by promoting expansion of the granule progenitor pool. J Neurosci. 2007;27:9780–9789. doi: 10.1523/JNEUROSCI.5586-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choo AY, Yoon SO, Kim SG, Roux PP, Blenis J. Rapamycin differentially inhibits S6Ks and 4E-BP1 to mediate cell-type-specific repression of mRNA translation. Proc Natl Acad Sci U S A. 2008;105:17414–17419. doi: 10.1073/pnas.0809136105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper AF, Yu KP, Brueckner M, Brailey LL, Johnson L, McGrath JM, Bale AE. Cardiac and CNS defects in a mouse with targeted disruption of suppressor of fused. Development. 2005;132:4407–4417. doi: 10.1242/dev.02021. [DOI] [PubMed] [Google Scholar]
- Corbit KC, Aanstad P, Singla V, Norman AR, Stainier DY, Reiter JF. Vertebrate Smoothened functions at the primary cilium. Nature. 2005;437:1018–1021. doi: 10.1038/nature04117. [DOI] [PubMed] [Google Scholar]
- Corrales JD, Blaess S, Mahoney EM, Joyner AL. The level of sonic hedgehog signaling regulates the complexity of cerebellar foliation. Development. 2006;133:1811–1821. doi: 10.1242/dev.02351. [DOI] [PubMed] [Google Scholar]
- D’Amico D, Antonucci L, Di Magno L, Coni S, Sdruscia G, Macone A, Miele E, Infante P, Di Marcotullio L, De Smaele E, et al. Non-canonical Hedgehog/AMPK-Mediated Control of Polyamine Metabolism Supports Neuronal and Medulloblastoma Cell Growth. Dev Cell. 2015;35:21–35. doi: 10.1016/j.devcel.2015.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahmane N, Ruiz i Altaba A. Sonic hedgehog regulates the growth and patterning of the cerebellum. Development. 1999;126:3089–3100. doi: 10.1242/dev.126.14.3089. [DOI] [PubMed] [Google Scholar]
- Denef N, Neubuser D, Perez L, Cohen SM. Hedgehog induces opposite changes in turnover and subcellular localization of patched and smoothened. Cell. 2000;102:521–531. doi: 10.1016/s0092-8674(00)00056-8. [DOI] [PubMed] [Google Scholar]
- Dijkgraaf GJ, Alicke B, Weinmann L, Januario T, West K, Modrusan Z, Burdick D, Goldsmith R, Robarge K, Sutherlin D, et al. Small molecule inhibition of GDC-0449 refractory smoothened mutants and downstream mechanisms of drug resistance. Cancer Res. 2011;71:435–444. doi: 10.1158/0008-5472.CAN-10-2876. [DOI] [PubMed] [Google Scholar]
- Ellison DW, Dalton J, Kocak M, Nicholson SL, Fraga C, Neale G, Kenney AM, Brat DJ, Perry A, Yong WH, et al. Medulloblastoma: clinicopathological correlates of SHH, WNT, and non-SHH/WNT molecular subgroups. Acta Neuropathol. 2011;121:381–396. doi: 10.1007/s00401-011-0800-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filbin MG, Dabral SK, Pazyra-Murphy MF, Ramkissoon S, Kung AL, Pak E, Chung J, Theisen MA, Sun Y, Franchetti Y, et al. Coordinate activation of Shh and PI3K signaling in PTEN-deficient glioblastoma: new therapeutic opportunities. Nat Med. 2013;19:1518–1523. doi: 10.1038/nm.3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gharbi SI, Zvelebil MJ, Shuttleworth SJ, Hancox T, Saghir N, Timms JF, Waterfield MD. Exploring the specificity of the PI3K family inhibitor LY294002. Biochem J. 2007;404:15–21. doi: 10.1042/BJ20061489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson P, Tong Y, Robinson G, Thompson MC, Currle DS, Eden C, Kranenburg TA, Hogg T, Poppleton H, Martin J, et al. Subtypes of medulloblastoma have distinct developmental origins. Nature. 2010;468:1095–1099. doi: 10.1038/nature09587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz SC, Anderson KV. The primary cilium: a signalling centre during vertebrate development. Nat Rev Genet. 2010;11:331–344. doi: 10.1038/nrg2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn H, Wojnowski L, Specht K, Kappler R, Calzada-Wack J, Potter D, Zimmer A, Muller U, Samson E, Quintanilla-Martinez L, et al. Patched target Igf2 is indispensable for the formation of medulloblastoma and rhabdomyosarcoma. J Biol Chem. 2000;275:28341–28344. doi: 10.1074/jbc.C000352200. [DOI] [PubMed] [Google Scholar]
- Han YG, Kim HJ, Dlugosz AA, Ellison DW, Gilbertson RJ, Alvarez-Buylla A. Dual and opposing roles of primary cilia in medulloblastoma development. Nat Med. 2009;15:1062–1065. doi: 10.1038/nm.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara K, Maruki Y, Long X, Yoshino K, Oshiro N, Hidayat S, Tokunaga C, Avruch J, Yonezawa K. Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell. 2002;110:177–189. doi: 10.1016/s0092-8674(02)00833-4. [DOI] [PubMed] [Google Scholar]
- Hartmann W, Koch A, Brune H, Waha A, Schuller U, Dani I, Denkhaus D, Langmann W, Bode U, Wiestler OD, et al. Insulin-like growth factor II is involved in the proliferation control of medulloblastoma and its cerebellar precursor cells. Am J Pathol. 2005;166:1153–1162. doi: 10.1016/S0002-9440(10)62335-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh AC, Liu Y, Edlind MP, Ingolia NT, Janes MR, Sher A, Shi EY, Stumpf CR, Christensen C, Bonham MJ, et al. The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature. 2012;485:55–61. doi: 10.1038/nature10912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui CC, Angers S. Gli proteins in development and disease. Annu Rev Cell Dev Biol. 2011;27:513–537. doi: 10.1146/annurev-cellbio-092910-154048. [DOI] [PubMed] [Google Scholar]
- Jacinto E, Facchinetti V, Liu D, Soto N, Wei S, Jung SY, Huang Q, Qin J, Su B. SIN1/MIP1 maintains rictor-mTOR complex integrity and regulates Akt phosphorylation and substrate specificity. Cell. 2006;127:125–137. doi: 10.1016/j.cell.2006.08.033. [DOI] [PubMed] [Google Scholar]
- Jacinto E, Loewith R, Schmidt A, Lin S, Ruegg MA, Hall A, Hall MN. Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nat Cell Biol. 2004;6:1122–1128. doi: 10.1038/ncb1183. [DOI] [PubMed] [Google Scholar]
- Kalderon D. Similarities between the Hedgehog and Wnt signaling pathways. Trends Cell Biol. 2002;12:523–531. doi: 10.1016/s0962-8924(02)02388-7. [DOI] [PubMed] [Google Scholar]
- Kawauchi D, Robinson G, Uziel T, Gibson P, Rehg J, Gao C, Finkelstein D, Qu C, Pounds S, Ellison DW, et al. A mouse model of the most aggressive subgroup of human medulloblastoma. Cancer Cell. 2012;21:168–180. doi: 10.1016/j.ccr.2011.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieran MW, Chisholm J, Casanova M, Brandes AA, Aerts I, Bouffet E, Bailey S, Leary S, MacDonald TJ, Mechinaud F, et al. Phase I study of oral sonidegib (LDE225) in pediatric brain and solid tumors and a phase II study in children and adults with relapsed medulloblastoma. Neuro Oncol. 2017 doi: 10.1093/neuonc/nox109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, Sarbassov DD, Ali SM, King JE, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell. 2002;110:163–175. doi: 10.1016/s0092-8674(02)00808-5. [DOI] [PubMed] [Google Scholar]
- Kim JJ, Gill PS, Rotin L, van Eede M, Henkelman RM, Hui CC, Rosenblum ND. Suppressor of fused controls mid-hindbrain patterning and cerebellar morphogenesis via GLI3 repressor. J Neurosci. 2011;31:1825–1836. doi: 10.1523/JNEUROSCI.2166-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam CW, Xie J, To KF, Ng HK, Lee KC, Yuen NW, Lim PL, Chan LY, Tong SF, McCormick F. A frequent activated smoothened mutation in sporadic basal cell carcinomas. Oncogene. 1999;18:833–836. doi: 10.1038/sj.onc.1202360. [DOI] [PubMed] [Google Scholar]
- Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepelletier L, Langlois SD, Kent CB, Welshhans K, Morin S, Bassell GJ, Yam PT, Charron F. Sonic Hedgehog guides axons via Zipcode Binding Protein 1-mediated local translation. J Neurosci. 2017 doi: 10.1523/JNEUROSCI.3016-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma XM, Blenis J. Molecular mechanisms of mTOR-mediated translational control. Nat Rev Mol Cell Biol. 2009;10:307–318. doi: 10.1038/nrm2672. [DOI] [PubMed] [Google Scholar]
- Mainwaring LA, Kenney AM. Divergent functions for eIF4E and S6 kinase by sonic hedgehog mitogenic signaling in the developing cerebellum. Oncogene. 2011;30:1784–1797. doi: 10.1038/onc.2010.564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marada S, Navarro G, Truong A, Stewart DP, Arensdorf AM, Nachtergaele S, Angelats E, Opferman JT, Rohatgi R, McCormick PJ, et al. Functional Divergence in the Role of N-Linked Glycosylation in Smoothened Signaling. PLoS genetics. 2015;11:e1005473. doi: 10.1371/journal.pgen.1005473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metcalfe C, Alicke B, Crow A, Lamoureux M, Dijkgraaf GJ, Peale F, Gould SE, de Sauvage FJ. PTEN loss mitigates the response of medulloblastoma to Hedgehog pathway inhibition. Cancer Res. 2013;73:7034–7042. doi: 10.1158/0008-5472.CAN-13-1222. [DOI] [PubMed] [Google Scholar]
- Oliver TG, Read TA, Kessler JD, Mehmeti A, Wells JF, Huynh TT, Lin SM, Wechsler-Reya RJ. Loss of patched and disruption of granule cell development in a pre-neoplastic stage of medulloblastoma. Development. 2005;132:2425–2439. doi: 10.1242/dev.01793. [DOI] [PubMed] [Google Scholar]
- Pazour GJ, Dickert BL, Vucica Y, Seeley ES, Rosenbaum JL, Witman GB, Cole DG. Chlamydomonas IFT88 and its mouse homologue, polycystic kidney disease gene tg737, are required for assembly of cilia and flagella. J Cell Biol. 2000;151:709–718. doi: 10.1083/jcb.151.3.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riobo NA, Lu K, Ai X, Haines GM, Emerson CP., Jr Phosphoinositide 3-kinase and Akt are essential for Sonic Hedgehog signaling. Proc Natl Acad Sci U S A. 2006;103:4505–4510. doi: 10.1073/pnas.0504337103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson G, Parker M, Kranenburg TA, Lu C, Chen X, Ding L, Phoenix TN, Hedlund E, Wei L, Zhu X, et al. Novel mutations target distinct subgroups of medulloblastoma. Nature. 2012;488:43–48. doi: 10.1038/nature11213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson GW, Kaste SC, Chemaitilly W, Bowers DC, Laughton S, Smith A, Gottardo N, Partap S, Bendel A, Wright KD, Orr BA, Warner WC, Onar-Thomas A, Gajjar A. Irreversible Growth Plate Fusions in Children with Medulloblastoma Treated with Targeted Hedgehog Pathway Inhibitor. Oncotarget. 2017 doi: 10.18632/oncotarget.20619. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson GW, Orr BA, Wu G, Gururangan S, Lin T, Qaddoumi I, Packer RJ, Goldman S, Prados MD, Desjardins A, et al. Vismodegib Exerts Targeted Efficacy Against Recurrent Sonic Hedgehog-Subgroup Medulloblastoma: Results From Phase II Pediatric Brain Tumor Consortium Studies PBTC-025B and PBTC-032. J Clin Oncol. 2015;33:2646–2654. doi: 10.1200/JCO.2014.60.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodon J, Tawbi HA, Thomas AL, Stoller RG, Turtschi CP, Baselga J, Sarantopoulos J, Mahalingam D, Shou Y, Moles MA, et al. A phase I, multicenter, open-label, first-in-human, dose-escalation study of the oral smoothened inhibitor Sonidegib (LDE225) in patients with advanced solid tumors. Clin Cancer Res. 2014;20:1900–1909. doi: 10.1158/1078-0432.CCR-13-1710. [DOI] [PubMed] [Google Scholar]
- Rohatgi R, Milenkovic L, Scott MP. Patched1 regulates hedgehog signaling at the primary cilium. Science. 2007;317:372–376. doi: 10.1126/science.1139740. [DOI] [PubMed] [Google Scholar]
- Romer JT, Kimura H, Magdaleno S, Sasai K, Fuller C, Baines H, Connelly M, Stewart CF, Gould S, Rubin LL, et al. Suppression of the Shh pathway using a small molecule inhibitor eliminates medulloblastoma in Ptc1(+/−)p53(−/−) mice. Cancer Cell. 2004;6:229–240. doi: 10.1016/j.ccr.2004.08.019. [DOI] [PubMed] [Google Scholar]
- Rudin CM, Hann CL, Laterra J, Yauch RL, Callahan CA, Fu L, Holcomb T, Stinson J, Gould SE, Coleman B, et al. Treatment of medulloblastoma with hedgehog pathway inhibitor GDC-0449. N Engl J Med. 2009;361:1173–1178. doi: 10.1056/NEJMoa0902903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarbassov DD, Ali SM, Kim DH, Guertin DA, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr Biol. 2004;14:1296–1302. doi: 10.1016/j.cub.2004.06.054. [DOI] [PubMed] [Google Scholar]
- Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- Schuller U, Heine VM, Mao J, Kho AT, Dillon AK, Han YG, Huillard E, Sun T, Ligon AH, Qian Y, et al. Acquisition of granule neuron precursor identity is a critical determinant of progenitor cell competence to form Shh-induced medulloblastoma. Cancer Cell. 2008;14:123–134. doi: 10.1016/j.ccr.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma N, Nanta R, Sharma J, Gunewardena S, Singh KP, Shankar S, Srivastava RK. PI3K/AKT/mTOR and sonic hedgehog pathways cooperate together to inhibit human pancreatic cancer stem cell characteristics and tumor growth. Oncotarget. 2015;6:32039–32060. doi: 10.18632/oncotarget.5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spassky N, Han YG, Aguilar A, Strehl L, Besse L, Laclef C, Ros MR, Garcia-Verdugo JM, Alvarez-Buylla A. Primary cilia are required for cerebellar development and Shh-dependent expansion of progenitor pool. Dev Biol. 2008;317:246–259. doi: 10.1016/j.ydbio.2008.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svard J, Heby-Henricson K, Persson-Lek M, Rozell B, Lauth M, Bergstrom A, Ericson J, Toftgard R, Teglund S. Genetic elimination of Suppressor of fused reveals an essential repressor function in the mammalian Hedgehog signaling pathway. Dev Cell. 2006;10:187–197. doi: 10.1016/j.devcel.2005.12.013. [DOI] [PubMed] [Google Scholar]
- Syu LJ, Zhao X, Zhang Y, Grachtchouk M, Demitrack E, Ermilov A, Wilbert DM, Zheng X, Kaatz A, Greenson JK, et al. Invasive mouse gastric adenocarcinomas arising from Lgr5+ stem cells are dependent on crosstalk between the Hedgehog/GLI2 and mTOR pathways. Oncotarget. 2016;7:10255–10270. doi: 10.18632/oncotarget.7182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor MD, Northcott PA, Korshunov A, Remke M, Cho YJ, Clifford SC, Eberhart CG, Parsons DW, Rutkowski S, Gajjar A, et al. Molecular subgroups of medulloblastoma: the current consensus. Acta Neuropathol. 2012;123:465–472. doi: 10.1007/s00401-011-0922-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoreen CC, Kang SA, Chang JW, Liu Q, Zhang J, Gao Y, Reichling LJ, Sim T, Sabatini DM, Gray NS. An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J Biol Chem. 2009;284:8023–8032. doi: 10.1074/jbc.M900301200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truitt ML, Ruggero D. New frontiers in translational control of the cancer genome. Nat Rev Cancer. 2016;16:288–304. doi: 10.1038/nrc.2016.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace VA. Purkinje-cell-derived Sonic hedgehog regulates granule neuron precursor cell proliferation in the developing mouse cerebellum. Curr Biol. 1999;9:445–448. doi: 10.1016/s0960-9822(99)80195-x. [DOI] [PubMed] [Google Scholar]
- Wang B, Fallon JF, Beachy PA. Hedgehog-regulated processing of Gli3 produces an anterior/posterior repressor gradient in the developing vertebrate limb. Cell. 2000;100:423–434. doi: 10.1016/s0092-8674(00)80678-9. [DOI] [PubMed] [Google Scholar]
- Wang G, Wang B, Jiang J. Protein kinase A antagonizes Hedgehog signaling by regulating both the activator and repressor forms of Cubitus interruptus. Genes Dev. 1999;13:2828–2837. doi: 10.1101/gad.13.21.2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Ding Q, Yen CJ, Xia W, Izzo JG, Lang JY, Li CW, Hsu JL, Miller SA, Wang X, et al. The crosstalk of mTOR/S6K1 and Hedgehog pathways. Cancer Cell. 2012;21:374–387. doi: 10.1016/j.ccr.2011.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wechsler-Reya RJ, Scott MP. Control of neuronal precursor proliferation in the cerebellum by Sonic Hedgehog. Neuron. 1999;22:103–114. doi: 10.1016/s0896-6273(00)80682-0. [DOI] [PubMed] [Google Scholar]
- Xie J, Murone M, Luoh SM, Ryan A, Gu Q, Zhang C, Bonifas JM, Lam CW, Hynes M, Goddard A, et al. Activating Smoothened mutations in sporadic basal-cell carcinoma. Nature. 1998;391:90–92. doi: 10.1038/34201. [DOI] [PubMed] [Google Scholar]
- Yang Q, Inoki K, Ikenoue T, Guan KL. Identification of Sin1 as an essential TORC2 component required for complex formation and kinase activity. Genes Dev. 2006;20:2820–2832. doi: 10.1101/gad.1461206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang ZJ, Ellis T, Markant SL, Read TA, Kessler JD, Bourboulas M, Schuller U, Machold R, Fishell G, Rowitch DH, et al. Medulloblastoma can be initiated by deletion of Patched in lineage-restricted progenitors or stem cells. Cancer Cell. 2008;14:135–145. doi: 10.1016/j.ccr.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yauch RL, Dijkgraaf GJ, Alicke B, Januario T, Ahn CP, Holcomb T, Pujara K, Stinson J, Callahan CA, Tang T, et al. Smoothened mutation confers resistance to a Hedgehog pathway inhibitor in medulloblastoma. Science. 2009;326:572–574. doi: 10.1126/science.1179386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye P, Xing Y, Dai Z, D’Ercole AJ. In vivo actions of insulin-like growth factor-I (IGF-I) on cerebellum development in transgenic mice: evidence that IGF-I increases proliferation of granule cell progenitors. Brain research. 1996;95:44–54. doi: 10.1016/0165-3806(96)00492-0. [DOI] [PubMed] [Google Scholar]
- Zhuo L, Theis M, Alvarez-Maya I, Brenner M, Willecke K, Messing A. hGFAP-cre transgenic mice for manipulation of glial and neuronal function in vivo. Genesis. 2001;31:85–94. doi: 10.1002/gene.10008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1. Human medulloblastoma samples used for Figure 5.