Abstract
Purpose
While efforts to improve control of locally-advanced non-small-cell lung cancer (NSCLC) by escalating radiation dose in unselected patients have been unsuccessful, we hypothesized that a subset of patients may derive benefit. Circulating serum microRNAs (c-miRNAs) have shown promise as prognostic and predictive biomarkers. We assessed the utility of c-miRNAs to predict response to high-dose radiotherapy.
Methods and Materials
Data from 80 patients treated from 2004 to 2013 with definitive standard- or high-dose radiotherapy for stages II-III NSCLC as part of four prospective institutional clinical trials were evaluated. Pretreatment serum levels of 62 miRNAs were measured by quantitative reverse-transcription polymerase chain reaction array. We combined miRNA data and clinical factors to generate a Dose Response Score (DRS) for predicting overall survival (OS) following high-dose versus standard-dose radiotherapy. Elastic net Cox regression was used for variable selection and parameter estimation. Model assessment and tuning parameter selection were performed through full cross validation. DRS was also correlated with local progression, distant metastasis, and grade 3 or higher cardiac toxicity using Cox regression; and grade 2 or higher esophageal and pulmonary toxicity using logistic regression.
Results
Eleven predictive miRNAs were combined with clinical factors to generate a DRS for each patient. In patients with low DRS, high-dose radiotherapy was associated with significantly improved OS compared to treatment with standard-dose radiotherapy (HR 0.22). In these patients, high-dose radiation also conferred lower risk of distant metastasis and local progression, although the latter association was not statistically significant. Patients with high DRS exhibited similar rates of OS regardless of dose (HR 0.78). DRS did not correlate with treatment-related toxicity.
Conclusions
Using c-miRNA signature and clinical factors, we developed a DRS that identified a subset of patients with locally-advanced NSCLC who derive an OS benefit from high-dose radiotherapy. This DRS may guide dose-escalation in a patient-specific manner.
INTRODUCTION
Approximately one-third of patients with non-small cell lung cancer (NSCLC) are diagnosed with locally advanced, unresectable disease [1]. These patients are often treated with a combination of radiotherapy and chemotherapy [2]. Despite aggressive multi-modality therapy, outcomes in these patients are poor, with estimated rates of 3-year overall survival (OS) ranging between 5% and 20% [1,2].
Although high-dose radiotherapy, particularly stereotactic body radiotherapy (SBRT), is highly effective in controlling early stage NSCLC [3,4], efforts to dose-escalate fractionated radiotherapy in the setting of locally advanced NSCLC have been less successful. In particular, RTOG 0617 failed to show a benefit from treatment to 74 Gy compared to 60 Gy [5]. However, this study was conducted in an unselected population, and it is possible that a subset of patients do benefit from dose-escalated radiotherapy. With the development of more sophisticated biomarkers, such patients could be identified, allowing for selection of appropriate candidates for high-dose radiation.
MicroRNAs (miRNAs) are a class of noncoding RNAs of approximately 22 nucleotides in length that regulate gene expression post-transcriptionally. MiRNA expression profiles differ between normal and cancer tissues, and several miRNAs have been implicated in regulating tumorigenesis and progression of NSCLC. These include miRNAs that both inhibit [6–9] and promote [10,11] tumor growth. Certain miRNAs have demonstrated prognostic utility in NSCLC [12–14], while others have shown promise in predicting response to various treatments [15]. Additional efforts have focused on using microarray-based techniques to characterize miRNA expression profiles, or “signatures,” to further classify and risk-stratify NSCLC [16–18]. While the studies referenced above were primarily conducted in tissue samples, more recently, stable, extracellular miRNAs have been identified in various body fluids [19,20]. Functional studies have shown that these circulating miRNAs (c-miRNAs) might be protected from degradation by several complementary mechanisms, including inclusion in phospholipid bilayer-encapsulated vesicles [21] or via the formation of RNA-binding protein complexes [22]. Serum-based c-miRNAs have shown promise as prognostic biomarkers in NSCLC [23–26]. Circulating miRNAs have also demonstrated predictive utility, with elevated levels of miR-21 having been found to correlate with increased resistance to platinum-based chemotherapy [27].
A potential role for c-miRNAs in predicting response of NSCLC to radiotherapy has not yet been investigated. We hypothesized that variations in levels of c-miRNAs may predict response to radiotherapy and that characterization of c-miRNA signatures could aid in identification of patients who benefit from dose-escalation.
MATERIALS AND METHODS
Patient population
This work analyzed data from 4 prospective Institutional Review Board-approved lung-cancer studies: (1) a phase 1/2 study of radiation dose escalation with concurrent chemotherapy, (2–3) 2 consecutive studies using functional imaging and biomarkers to assess patient outcome, and (4) a study using midtreatment positron emission tomography (PET) to guide individualized dose escalation. Included in this analysis were patients with stage II-III NSCLC treated with standard fractionation, i.e. not SBRT. Details of these studies are shown in Supplemental Table 1. All clinical data were prospectively collected.
Treatment regimen
All patients were treated with definitive radiotherapy with or without sequential or concurrent chemotherapy. In cases of sequential treatment, chemotherapy was administered following radiotherapy. Total radiation doses ranged from 66 to 86 Gy in daily fractions, as directed by the respective protocols. Radiation was delivered using three-dimensional conformal radiotherapy (3DCRT) as previously described [28]. Gross tumor volume included the primary tumor and any involved hilar or mediastinal lymph nodes, as determined by tissue diagnosis and/or PET. Uninvolved lymph node regions were not included in the clinical target volume. Tissue inhomogeneity corrections were applied for all plans.
As dose and fractionation varied among patients, we standardized values to biologic effective dose (BED), which normalizes doses of various fractionations by supposing a hypothetical condition of an infinite number of fractions. Tumor BEDs were calculated using the linear-quadratic formula using an alpha-beta ratio of 10 Gy. For toxicity analysis, BED for heart, lung, and esophageal doses were similarly calculated using alpha-beta values of 2.5 Gy, 2.5 Gy, and 10 Gy, respectively.
Sample collection and RNA isolation
Blood samples were collected using red-top tubes with no use of anticoagulant within one week prior to initiation of thoracic radiation. Blood samples were placed on ice immediately after collection and were centrifuged within 4 hours of collection at 3000×g for 30 min, following which the upper one third of the supernatants were collected and stored at −80°C until analysis.
Total RNA was isolated from serum samples using the miRNeasy Mini Kit (Qiagen) following the manufacturer’s protocol with slight modifications. In brief, 250 mL of serum was treated with 950 µL of QIAzol (Qiagen) for denaturation of protein contents and subsequent isolation of RNA. Synthetic cel-miR-39 was added to each sample as a means of normalization. After adding 200 µL chloroform, aqueous and organic phases were separated by centrifugation at 12,000 × g × 15 min at 4 °C. The aqueous phase was treated with 900 µL 100% ethanol and loaded to a miRNeasy Mini column for RNA extraction. After extensive washes, RNA was eluted with 30 µL of water. Post-isolation, RNA concentration and quality were determined using a Nanodrop 2000 instrument (Thermo Scientific, DE).
MiRNA profiling
Serum RNA was reverse transcribed to cDNA using the miScript II RT Kit (Qiagen). After a 1/10-dilution with nuclease-free water, RT products were analyzed for the presence and differential expression of a panel of 62 miRNAs detectable in serum, plasma, and other bodily fluids using Human Serum & Plasma miRNA PCR Arrays (Cat. No MIHS-106Z, Qiagen) as previously described [29,30]. A list of all miRNA species assayed is found in Supplemental Table 2. In order to eliminate potential variation introduced during the isolation and RNA quantification processes, the raw Ct value for each miRNA was normalized to the raw Ct value for spike-in cel-miR-39 obtained from each individual sample using the 2−ΔΔCt method as previously described [31].
Outcome definitions
Our primary endpoint was overall survival (OS) time, which was defined from the first day of treatment to the date of death. Patients alive at their last follow-up were censored at this date. Times to local progression and distant metastasis were similarly defined as the time from the start of treatment to the development of local progression or distant metastasis, respectively. Patients who died prior to documented local progression or distant metastasis were censored at the date of their last clinical visit. Time to grade 3 or greater cardiac toxicity was also defined from start of treatment.
Radiation-induced esophageal toxicity was graded per Common Terminology Criteria for Adverse Events (CTCAE) v3.0 [32]. Radiation-induced lung toxicity, including radiation pneumonitis and clinical fibrosis, was graded as previously described [33]. Cardiac toxicity was initially graded per CTCAE v.3.0, and then for this analysis reviewed, confirmed, and updated to CTCAE v4.03. In addition, cardiac events not previously attributed to radiation were documented, graded, and included in the analysis. All events were confirmed by two independent physicians without knowledge of the treatment plan or cardiac radiation dose.
Statistical analysis
Cox elastic net regression was used to model OS as a function of tumor dose, chemotherapy, and interactions between dose and clinical factors, and dose and miRNA measurements. Tumor dose and chemotherapy were modeled as main effects, while clinical factors and miRNAs were modeled as potential dose-effect modifiers (interactions). Chemotherapy was modeled as a main effect because few of the patients were treated without chemotherapy, and we sought to develop a model applicable to all patients regardless of whether or not they were to receive chemotherapy. Clinical factors and miRNAs were modeled as interactions as our goal was to estimate individual patient sensitivity to RT dose (e.g. HR for dose in OS model) and not simply to estimate patient prognosis. No main effects were included in order to minimize risk of overfitting from including 2 parameters per miRNA term (one for main effect and one for interaction). Elastic net is a hybrid of LASSO and RIDGE regression techniques and retains the good prediction performance of RIDGE while also enforcing sparsity [34].
As no p-values are available from penalized regression techniques such as Elastic Net, we next sought provide empirical evidence that the DRS can distinguish between patients who will have better or worse outcomes following high dose RT. Specifically, Kaplan-Meier estimates of OS in groups of patients stratified by dose (standard vs. high) and DRS group were calculated, along with dose hazard ratios for the high and low DRS groups. For this analysis, DRS group membership was determined using 10-fold cross validation (CV) repeated 20 times (to minimize variability associated with fold choice). All aspects of model fitting (variable selection, parameter estimation and tuning parameter choice) were included in the cross-validation loop. To perform CV, the dataset was randomly divided into 10 folds. For each held-out fold, a separate model was fit and a separate set of predictive miRNAs/clinical factors was obtained using the remaining 9 training folds. The DRS for patients in the held-out testing fold was then calculated using this fitted model.
To gain further insight, DRS, as calculated from the model fitted using all data, was also correlated with local progression, distant metastasis, and grade 3 or higher cardiac toxicity using Cox regression; and grade 2 or higher esophageal and pulmonary toxicity using logistic regression. Cardiac toxicity was evaluated by Cox regression in order to account for censored outcomes, whereas logistic regression was used for esophageal and pulmonary toxicity as these events occurred earlier. All statistical analyses were performed with R version 3.2.1.
RESULTS
Patient characteristics
Of the 173 NSCLC patients treated on the above-described protocols, miRNA measurements were obtained from 99. After excluding patients treated with SBRT and patients with stage I disease, 80 patients remained in our final dataset. Demographic, disease, and treatment characteristics for these 80 patients are shown in Table 1, and were similar to those of the original cohort of 173 (Supplemental Table 3). Sixty-two deaths were recorded, with a median overall survival time of 19.1 months (95% CI 13.8 – 29.0).
Table 1.
Demographic, clinical, and treatment data of analyzed patients.
| Parameter | Value |
|---|---|
|
| |
| Age | |
| Mean (Std. Dev.) | 66.2 (9.6) |
| Median (Range) | 66.0 (43.4 – 84.6) |
|
| |
| KPS | |
| Mean (Std. Dev.) | 84.2 (10.2) |
| Median (Range) | 85.0 (50 – 100) |
| < 70 | 3 |
| >= 70 | 77 |
|
| |
| Target BED (Gy) | |
| Mean (Std. Dev.) | 89.9 (14.0) |
| Median (Range) | 87.1 (40.8 – 110.2) |
|
| |
| Sex (n (%)) | |
| Male | 62 (77.5%) |
| Female | 18 (22.5%) |
|
| |
| Smoking Status (n (%)) | |
| Never-smoker | 2 (2.5%) |
| Former-smoker | 36 (45.0%) |
| Current-smoker | 37 (46.3%) |
| Unknown | 5 (6.3%) |
|
| |
| TNM Simple Stage (n (%)) | |
| II | 7 (8.8%) |
| III | 73 (91.3%) |
|
| |
| Concurrent Chemotherapy (n (%)) | |
| Yes | 71 (88.7%) |
| No | 9 (11.3%) |
KPS = Karnofsky performance status. Std. Dev. = standard deviation. GTV = gross tumor volume. Gy = Gray. TNM = tumor, node, metastasis.
Dose Response Score development
Table 2 lists the variables selected by Cox elastic net and their corresponding estimated hazard ratios (HR) for death. Target dose (BED) and chemotherapy were modeled as main effects, while the selected clinical factors and miRNA measurements were modeled as potential dose modifiers. Covariates were centered and standardized so that the estimated overall dose effect was the estimated dose effect for a patient with all covariates at their average value. For the average patient, the hazard of death decreased by 1% for each 10 Gy increase in tumor dose (HR 0.990). Age and stage negatively interacted with dose, suggesting that high-dose radiation might be less beneficial or even harmful (in terms of predicted OS) for patients with older age or higher stage. Conversely, patients with better than average performance status demonstrated an enhanced dose effect, with their predicted hazard of death decreasing more rapidly as a function of increasing radiation dose in comparison to patients with average or below-average performance status. Eleven miRNAs were identified that predicted increased or decreased effect of dose on OS (Table 2). Based on these results, a predictive “Dose Response Score” (DRS) was defined such that the estimated dose-HR for an individual patient is given by 0.99*exp(DRS), where
Table 2.
Variables selected by Cox elastic net with corresponding hazard ratios for death (HR). For each effect modifier, HR represents the multiplicative change in the dose HR. For each miRNA, HR corresponds to per standard deviation change in normalized miRNA expression
| Variable | HR |
|---|---|
| Main Effect | |
| Dose (per 10 Gy BED) | 0.990 |
| Chemo (Yes vs. No) | 0.410 |
| Dose Effect Modifiers (Per 10 Gy Change in Dose) | |
| Age (per 5 years increase) | 1.002 |
| Stage (III vs II) | 1.005 |
| KPS (per 10 pts increase) | 0.994 |
| miR-10b-5p | 1.293 |
| miR-125b-5p | 1.184 |
| miR-126-3p | 0.992 |
| miR-134 | 0.839 |
| miR-155-5p | 0.943 |
| miR-200b-3p | 1.141 |
| miR-205-5p | 0.964 |
| miR-34a-5p | 1.435 |
| miR-92a-3p | 1.179 |
| miR-145-5p | 0.922 |
| miR-22-3p | 0.993 |
KPS = Karnofsky performance status.
To illustrate interpretation of this model, suppose two hypothetical patients. For patient A, all the terms in Table 2 are at their average values. For patient B, serum miR-10b-5p is elevated by 1 standard deviation above the mean, while all other variables are similar. The estimated hazard ratio associated with 10 Gy increase in dose for patient A is 0.990 whereas for patient B it is 1.280 (=0.990*1.293), reflecting worse predicted survival from high dose radiation.
Kaplan Meier analysis
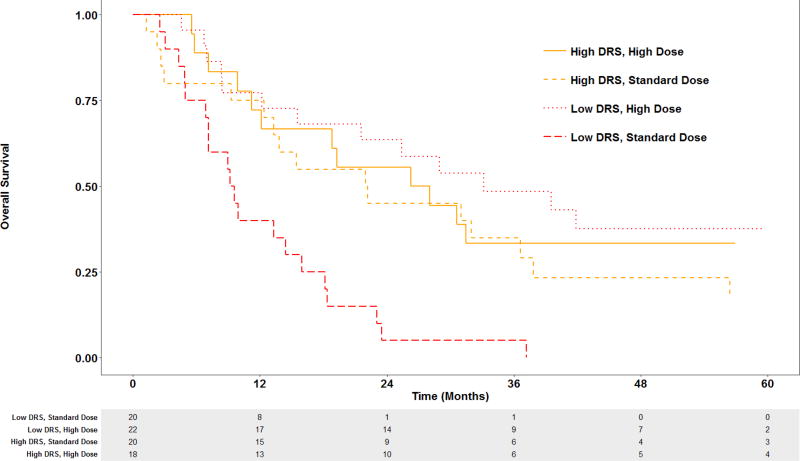
By dichotomizing at median DRS value, we divided patients into two groups: low DRS and high DRS. We further divided each DRS group into two subgroups using median radiation dose (BED 87.1 Gy). Patients receiving radiation dose higher than 87.1 Gy were included in the high-dose group, while those receiving lower dose were included in the standard-dose group. While OS of patients in the high DRS group was similar regardless of radiation dose (HR 0.78, 95% CI 0.37–1.64), patients in the low DRS group showed improved OS after treatment with high-dose compared to standard-dose radiation (HR 0.22, 95% CI 0.10 – 0.48) (Figure 1).
Figure 1.
Kaplan-Meier estimates of OS in low and high DRS groups treated with standard-versus high-dose radiotherapy.
DRS = dose response score. High- and standard-dose defined as greater than and less than 87.1 Gy (BED), respectively.
Association of DRS with local control and distant metastasis
We next investigated the utility of this scoring system in predicting response to standard- versus high-dose radiotherapy in terms of distant metastasis and local progression. The effect of DRS on dose-response was significant for distant metastasis, with higher DRS correlating with higher hazard (Table 3). While higher DRS also correlated with higher hazard for local progression, this effect was not statistically significant.
Table 3.
Cox regression of local progression and distant metastasis.
| Local Progression | Distant Metastasis | |||||
|---|---|---|---|---|---|---|
| HR | 95% CI | P | HR | 95% CI | P | |
| Chemotherapy (Yes vs. No) | 0.657 | 0.206 – 2.094 | 0.48 | 0.877 | 0.180 – 4.272 | 0.87 |
| Target Dose (per 10 Gy BED) | 0.796 | 0.452 – 1.401 | 0.43 | 0.858 | 0.554 – 1.328 | 0.49 |
| DRS:Dose (per 10 Gy increase and per 1 std. dev. increase in DRS) | 1.065 | 0.956 – 1.186 | 0.26 | 1.158 | 1.047 – 1.282 | < 0.01* |
HR = hazard ratio. CI = confidence interval. Gy = Gray. BED = biologic effective dose. DRS = Dose Response Score. Std. Dev. = standard deviation. Statistical significance denoted by *.
Association of DRS with cardiac, esophageal, and pulmonary toxicity
We next investigated the dose-effect modification of DRS on radiation-induced cardiac, esophageal, and pulmonary toxicities. Table 4 summarizes the Cox regression results of time to grade 3 or higher cardiac toxicity by mean heart dose and DRS. Table 5 shows the results from logistic regression of grade 2 or higher esophagitis and pulmonary toxicity by mean esophagus or lung dose and DRS. The results suggest no significant dose-effect modification with cardiac, esophageal, or pulmonary toxicity.
Table 4.
Multivariate Cox regression analysis of effect of DRS on grade 3 or higher cardiac toxicity.
| HR | 95% CI | P | |
|---|---|---|---|
| Baseline Cardiac Disease | 3.558 | 0.929 – 13.632 | 0.06 |
| Concurrent Chemotherapy | 0.303 | 0.0289 – 3.205 | 0.32 |
| Mean Heart Dose (per 10 Gy BED) | 1.941 | 0.898 – 4.195 | 0.09 |
| DRS*Mean Heart Dose (per 10 Gy BED and per 1 std. dev. in DRS) | 1.144 | 0.696 – 1.882 | 0.60 |
HR = hazard ratio. CI = confidence interval. Gy = Gray. BED = biologic effective dose. DRS = Dose Response Score. Std. Dev. = standard deviation.
Table 5.
Multivariate logistic regression analysis of effect of DRS on grade 2 or higher esophageal and pulmonary toxicity.
| Esophagitis, grade 2+ | Lung toxicity, grade 2+ | |||
|---|---|---|---|---|
| Estimate | P | Estimate | P | |
| Concurrent Chemotherapy | 17.470 | 0.99 | −0.556 | 0.50 |
| Mean Dose (per 10 Gy) | 0.104 | 0.73 | 7.291 | 0.04 |
| DRS* Mean Dose (per 10 Gy BED and per 1 std. dev. in DRS) | −0.0494 | 0.60 | −0.483 | 0.41 |
Gy = gray. DRS = Dose Response Score. Std. Dev. = standard deviation. BED = biologic effective dose.
DISCUSSION
We developed a DRS incorporating c-miRNA signature and clinical factors that identified a subset of patients with locally advanced and/or medically inoperable NSCLC who benefit from high-dose radiotherapy. Patients with low DRS treated with high-dose radiotherapy exhibited improved OS compared to those treated with standard-dose. Conversely, patients with high DRS had similar outcomes regardless of radiation dose. Differentiation of those who do and do not benefit from high-dose radiotherapy could allow for patient-specific decision-making regarding dose-escalation. Such prediction ability would benefit not only those allocated to receive high-dose radiotherapy, but also those selected to receive standard dose, as these patients could be spared the increased toxicity from an escalated therapy from which they would not derive benefit.
Improved OS in any study can be associated with multiple factors, including improved local control, improved distant control, and decreased treatment-related toxicity. We found that DRS affected dose-response in terms of distant metastasis in a statistically significant manner, with higher DRS correlating with higher dose-hazard ratios. DRS also predicted dose-effect on local control. Although this relationship did not reach statistical significance, it is possible that a significant association could be identified in a larger study with a greater number of events. We did not find evidence for DRS modifying the interactions between dose and cardiac, esophageal, and pulmonary toxicity.
Of the 11 miRNAs identified in this study as dose-effect modifiers, several have well-described tumor-suppressor activities. These include miR-200, which suppresses the epithelial-to-mesenchymal transition, inhibits tumorigenesis, and reverses chemoresistance [35]; miR-34, which is regulated by TP53 and targets mediators of cell-cycle progression, metastasis, and chemoresistance [36]; miR-126, which inhibits cell proliferation by targeting EGFL7 [37]; miR-205, which suppresses growth in breast and prostate cancer, and is differentially expressed in squamous versus non-squamous NSCLC [38–40]; miR-145, which exhibits anti-tumor activity in breast cancer, colon cancer, and leukemia [41–44]; and miR-22, which targets c-Myc and the estrogen receptor [45,46].
Conversely, three of the miRNA species identified are understood to promote tumorigenesis and progression. These are miR-92a, which promotes cancer-cell survival in leukemia, HCC, and melanoma [47–49]; miR-10b, which promotes proliferation and metastasis in breast and lung cancer, possibly by targeting the transcription factor KLF4 [50,51]; and miR-155, which down-regulates regulators of mismatch repair, including hMSH2, hMSH6, and hMLH1 [52]. One miRNA, miR-125, has been implicated in both tumor suppressor and oncogenic activities [35,53]. The last species, miR-134, does not have well-described cancer-related activity, but is understood to predominantly function in the central nervous system to regulate synaptic development and higher brain functions [54,55].
The mechanisms by which these miRNAs affect response to high-dose radiotherapy are not apparent from these data and should be the focus of future investigation. However, some of the associations identified are hypothesis-generating. For example, elevated serum levels of miRNA-125 and miR-200 have been found to correlate with poor prognosis in NSCLC and other cancers [35,53]. In our model, elevation of these miRNA species was associated with a lack of benefit from high-dose radiotherapy, suggesting that the poor prognosis associated with these miRNAs may not be surmountable by escalation of radiation dose.
The factors incorporated into this scoring system were identified by Elastic net, which has been reported to perform well in such high dimensional scenarios with correlated covariates. A strength of our study is that our findings were fully cross-validated. At each cross validation iteration, the complete model selection process was performed on nine training folds, following which the results were assessed using the held out testing fold. Thus, any overfitting of the data would have led to reduced effects when cross-validated. However, despite the use of rigorous cross-validation, external validation in a larger, prospective cohort is required to confirm predictive accuracy prior to clinical implementation.
The dose groups in this study, “standard” and “high”, were defined using median dose of all patients as a dichotomization point. However, it is important to note that many of these patients were treated on dose-escalation protocols. As such, the doses in our standard- and high-dose groups were higher than those in the standard- and high-dose groups in RTOG 0617 (60 Gy and 74 Gy, respectively). This may limit generalizability of our findings, and highlights the need for external validation in a cohort of patients treated with more commonly prescribed radiation doses.
Conclusions
We described a c-miRNA signature that, when combined with clinical factors, identified a subset of patients with locally advanced and/or medically inoperable NSCLC who benefit from high-dose radiotherapy. The findings reported here represent a significant step toward identification of patients for which dose-escalation may be beneficial, which in turn may allow for more optimal individualization of patient care. In addition, characterization of mechanisms by which the predictive miRNA species might influence sensitivity to radiation may suggest targets for the development of novel targeted therapies and radiosensitizers.
Supplementary Material
Summary.
Escalation of radiation dose has not been beneficial in unselected patients with locally-advanced non-small-cell lung cancer. We hypothesized that a subset of patients may derive benefit and sought to identify a dose-response biomarker by analyzing patients treated with varying doses of radiotherapy on four institutional clinical trials. We identified a micro-RNA signature that, when combined with clinical factors, identified a subset of patients who exhibited an overall survival benefit following high-dose radiotherapy.
Acknowledgments
We thank the patients who volunteered to take part in this study; caregivers of the study participants
Funding: This work was supported in part by R0 1CA142840 (Kong) and P01 CA059827 (Ten Haken and Lawrence).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest: The authors declare no potential conflicts of interest.
References
- 1.Torre LA, Siegel RL, Jemal A. Lung cancer statistics. Adv Exp Med Biol. 2016;893:1–19. doi: 10.1007/978-3-319-24223-1_1. [DOI] [PubMed] [Google Scholar]
- 2.Auperin A, et al. Meta-analysis of concomitant versus sequential radiochemotherapy in locally advanced non-small-cell lung cancer. J Clin Oncol. 2010;28:2181–2190. doi: 10.1200/JCO.2009.26.2543. [DOI] [PubMed] [Google Scholar]
- 3.Chang JY, et al. Stereotactic ablative radiotherapy versus lobectomy for operable stage i non-small-cell lung cancer: A pooled analysis of two randomised trials. Lancet Oncol. 2015;16:630–637. doi: 10.1016/S1470-2045(15)70168-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Timmerman R, et al. Stereotactic body radiation therapy for inoperable early stage lung cancer. Jama. 2010;303:1070–1076. doi: 10.1001/jama.2010.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bradley JD, et al. Standard-dose versus high-dose conformal radiotherapy with concurrent and consolidation carboplatin plus paclitaxel with or without cetuximab for patients with stage iiia or iiib non-small-cell lung cancer (rtog 0617): A randomised, two-by-two factorial phase 3 study. Lancet Oncol. 2015;16:187–199. doi: 10.1016/S1470-2045(14)71207-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Webster RJ, et al. Regulation of epidermal growth factor receptor signaling in human cancer cells by microrna-7. J Biol Chem. 2009;284:5731–5741. doi: 10.1074/jbc.M804280200. [DOI] [PubMed] [Google Scholar]
- 7.Li J, et al. Mirna-200c inhibits invasion and metastasis of human non-small cell lung cancer by directly targeting ubiquitin specific peptidase 25. Molecular cancer. 2014;13:166. doi: 10.1186/1476-4598-13-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yamashita R, et al. Growth inhibitory effects of mir-221 and mir-222 in non-small cell lung cancer cells. Cancer medicine. 2015;4:551–564. doi: 10.1002/cam4.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nasser MW, et al. Down-regulation of micro-rna-1 (mir-1) in lung cancer. Suppression of tumorigenic property of lung cancer cells and their sensitization to doxorubicin-induced apoptosis by mir-1. J Biol Chem. 2008;283:33394–33405. doi: 10.1074/jbc.M804788200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 10.Yang Y, et al. Downregulation of microrna-21 expression restrains non-small cell lung cancer cell proliferation and migration through upregulation of programmed cell death 4. Cancer Gene Ther. 2015;22:23–29. doi: 10.1038/cgt.2014.66. [DOI] [PubMed] [Google Scholar]
- 11.Hayashita Y, et al. A polycistronic microrna cluster, mir-17–92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res. 2005;65:9628–9632. doi: 10.1158/0008-5472.CAN-05-2352. [DOI] [PubMed] [Google Scholar]
- 12.Takamizawa J, et al. Reduced expression of the let-7 micrornas in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64:3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- 13.Tejero R, et al. Mir-141 and mir-200c as markers of overall survival in early stage non-small cell lung cancer adenocarcinoma. PLoS One. 2014;9:e101899. doi: 10.1371/journal.pone.0101899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma XL, et al. Prognostic role of microrna-21 in non-small cell lung cancer: A meta-analysis. Asian Pacific journal of cancer prevention : APJCP. 2012;13:2329–2334. doi: 10.7314/apjcp.2012.13.5.2329. [DOI] [PubMed] [Google Scholar]
- 15.Garofalo M, et al. Microrna signatures of trail resistance in human non-small cell lung cancer. Oncogene. 2008;27:3845–3855. doi: 10.1038/onc.2008.6. [DOI] [PubMed] [Google Scholar]
- 16.Eder M, Scherr M. Microrna and lung cancer. N Engl J Med. 2005;352:2446–2448. doi: 10.1056/NEJMcibr051201. [DOI] [PubMed] [Google Scholar]
- 17.Bishop JA, et al. Accurate classification of non-small cell lung carcinoma using a novel microrna-based approach. Clin Cancer Res. 2010;16:610–619. doi: 10.1158/1078-0432.CCR-09-2638. [DOI] [PubMed] [Google Scholar]
- 18.Yu SL, et al. Microrna signature predicts survival and relapse in lung cancer. Cancer Cell. 2008;13:48–57. doi: 10.1016/j.ccr.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 19.Mitchell PS, et al. Circulating micrornas as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Turchinovich A, Weiz L, Burwinkel B. Extracellular mirnas: The mystery of their origin and function. Trends Biochem Sci. 2012;37:460–465. doi: 10.1016/j.tibs.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 21.Valadi H, et al. Exosome-mediated transfer of mrnas and micrornas is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 22.Arroyo JD, et al. Argonaute2 complexes carry a population of circulating micrornas independent of vesicles in human plasma. Proc Natl Acad Sci U S A. 2011;108:5003–5008. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen X, et al. Characterization of micrornas in serum: A novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 24.Hu Z, et al. Serum microrna signatures identified in a genome-wide serum microrna expression profiling predict survival of non-small-cell lung cancer. J Clin Oncol. 2010;28:1721–1726. doi: 10.1200/JCO.2009.24.9342. [DOI] [PubMed] [Google Scholar]
- 25.Boeri M, et al. Microrna signatures in tissues and plasma predict development and prognosis of computed tomography detected lung cancer. Proc Natl Acad Sci U S A. 2011;108:3713–3718. doi: 10.1073/pnas.1100048108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keller A, et al. Stable serum mirna profiles as potential tool for non-invasive lung cancer diagnosis. RNA Biol. 2011;8:506–516. doi: 10.4161/rna.8.3.14994. [DOI] [PubMed] [Google Scholar]
- 27.Gao W, et al. Mirna-21: A biomarker predictive for platinum-based adjuvant chemotherapy response in patients with non-small cell lung cancer. Cancer Biol Ther. 2012;13:330–340. doi: 10.4161/cbt.19073. [DOI] [PubMed] [Google Scholar]
- 28.Kong FM, et al. High-dose radiation improved local tumor control and overall survival in patients with inoperable/unresectable non-small-cell lung cancer: Long-term results of a radiation dose escalation study. Int J Radiat Oncol Biol Phys. 2005;63:324–333. doi: 10.1016/j.ijrobp.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 29.Ryu MS, et al. Genomic analysis, cytokine expression, and microrna profiling reveal biomarkers of human dietary zinc depletion and homeostasis. Proc Natl Acad Sci U S A. 2011;108:20970–20975. doi: 10.1073/pnas.1117207108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Philippidou D, et al. Signatures of micrornas and selected microrna target genes in human melanoma. Cancer Res. 2010;70:4163–4173. doi: 10.1158/0008-5472.CAN-09-4512. [DOI] [PubMed] [Google Scholar]
- 31.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative pcr and the 2(-delta delta c(t)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 32.Trotti A, et al. Ctcae v3.0: Development of a comprehensive grading system for the adverse effects of cancer treatment. Seminars in radiation oncology. 2003;13:176–181. doi: 10.1016/S1053-4296(03)00031-6. [DOI] [PubMed] [Google Scholar]
- 33.Kong FM, et al. Final toxicity results of a radiation-dose escalation study in patients with non-small-cell lung cancer (nsclc): Predictors for radiation pneumonitis and fibrosis. International journal of radiation oncology, biology, physics. 2006;65:1075–1086. doi: 10.1016/j.ijrobp.2006.01.051. [DOI] [PubMed] [Google Scholar]
- 34.Zou H, Hastie T. Regularization and variable selection via the elastic net. J Roy Stat Soc B. 2005;67:301–320. [Google Scholar]
- 35.Feng X, et al. Mir-200, a new star mirna in human cancer. Cancer Lett. 2014;344:166–173. doi: 10.1016/j.canlet.2013.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Misso G, et al. Mir-34: A new weapon against cancer? Mol Ther Nucleic Acids. 2014;3:e194. doi: 10.1038/mtna.2014.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sun Y, et al. Mir-126 inhibits non-small cell lung cancer cells proliferation by targeting egfl7. Biochem Biophys Res Commun. 2010;391:1483–1489. doi: 10.1016/j.bbrc.2009.12.098. [DOI] [PubMed] [Google Scholar]
- 38.Wu H, Zhu S, Mo Y. Suppression of cell growth and invasion by mir-205 in breast cancer. Cell Res. 2009;19:439–448. doi: 10.1038/cr.2009.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gandellini P, et al. Mir-205 exerts tumor-suppressive functions in human prostate through down-regulation of protein kinase cepsilon. Cancer Res. 2009;69:2287–2295. doi: 10.1158/0008-5472.CAN-08-2894. [DOI] [PubMed] [Google Scholar]
- 40.Lebanony D, et al. Diagnostic assay based on hsa-mir-205 expression distinguishes squamous from nonsquamous non-small-cell lung carcinoma. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2009;27:2030–2037. doi: 10.1200/JCO.2008.19.4134. [DOI] [PubMed] [Google Scholar]
- 41.Gotte M, et al. Mir-145-dependent targeting of junctional adhesion molecule a and modulation of fascin expression are associated with reduced breast cancer cell motility and invasiveness. Oncogene. 2010;29:6569–6580. doi: 10.1038/onc.2010.386. [DOI] [PubMed] [Google Scholar]
- 42.Larsson E, et al. Discovery of microvascular mirnas using public gene expression data: Mir-145 is expressed in pericytes and is a regulator of fli1. Genome Med. 2009;1:108. doi: 10.1186/gm108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang J, et al. Putative tumor suppressor mir-145 inhibits colon cancer cell growth by targeting oncogene friend leukemia virus integration 1 gene. Cancer. 2011;117:86–95. doi: 10.1002/cncr.25522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sachdeva M, et al. P53 represses c-myc through induction of the tumor suppressor mir-145. Proc Natl Acad Sci U S A. 2009;106:3207–3212. doi: 10.1073/pnas.0808042106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xiong J, Du Q, Liang Z. Tumor-suppressive microrna-22 inhibits the transcription of e-box-containing c-myc target genes by silencing c-myc binding protein. Oncogene. 2010;29:4980–4988. doi: 10.1038/onc.2010.241. [DOI] [PubMed] [Google Scholar]
- 46.Xiong J, et al. An estrogen receptor alpha suppressor, microrna-22, is downregulated in estrogen receptor alpha-positive human breast cancer cell lines and clinical samples. FEBS J. 2010;277:1684–1694. doi: 10.1111/j.1742-4658.2010.07594.x. [DOI] [PubMed] [Google Scholar]
- 47.Venza M, et al. Mir-92a-3p and mycbp2 are involved in ms-275-induced and c-myc-mediated trail-sensitivity in melanoma cells. Int Immunopharmacol. 2016;40:235–243. doi: 10.1016/j.intimp.2016.09.004. [DOI] [PubMed] [Google Scholar]
- 48.Shigoka M, et al. Deregulation of mir-92a expression is implicated in hepatocellular carcinoma development. Pathol Int. 2010;60:351–357. doi: 10.1111/j.1440-1827.2010.02526.x. [DOI] [PubMed] [Google Scholar]
- 49.Sharifi M, Salehi R. Blockage of mir-92a-3p with locked nucleic acid induces apoptosis and prevents cell proliferation in human acute megakaryoblastic leukemia. Cancer Gene Ther. 2016;23:29–35. doi: 10.1038/cgt.2015.63. [DOI] [PubMed] [Google Scholar]
- 50.Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microrna-10b in breast cancer. Nature. 2007;449:682–688. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- 51.Su QL, et al. Effects of microrna-10b on lung cancer cell proliferation and invasive metastasis and the underlying mechanism. Asian Pac J Trop Med. 2014;7:364–367. doi: 10.1016/S1995-7645(14)60056-0. [DOI] [PubMed] [Google Scholar]
- 52.Valeri N, et al. Modulation of mismatch repair and genomic stability by mir-155. Proc Natl Acad Sci U S A. 2010;107:6982–6987. doi: 10.1073/pnas.1002472107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cui EH, et al. Serum microrna 125b as a diagnostic or prognostic biomarker for advanced nsclc patients receiving cisplatin-based chemotherapy. Acta Pharmacol Sin. 2013;34:309–313. doi: 10.1038/aps.2012.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schratt GM, et al. A brain-specific microrna regulates dendritic spine development. Nature. 2006;439:283–289. doi: 10.1038/nature04367. [DOI] [PubMed] [Google Scholar]
- 55.Gao J, et al. A novel pathway regulates memory and plasticity via sirt1 and mir-134. Nature. 2010;466:1105–1109. doi: 10.1038/nature09271. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.