Abstract
A key goal of HIV-1 vaccine development is the induction of broadly neutralizing antibodies (bnAbs) targeted to the vulnerable regions of the HIV envelope. BnAbs develop over time in ∼50% of HIV-1-infected individuals. However, to date, no vaccines have induced bnAbs and few or none of these vaccine-elicited HIV-1 antibodies carry the high frequencies of V(D)J mutations characteristic of bnAbs. Do the high frequencies of mutations characteristic of naturally induced bnAbs represent a fundamental barrier to the induction of bnAbs by vaccines? Recent studies suggest that high frequencies of V(D)J mutations can be achieved by serial vaccination strategies. Rather, it appears that, in the absence of HIV-1 infection, physiologic immune tolerance controls, including a germinal center process termed affinity reversion, may limit vaccine-driven bnAb development by clonal elimination or selecting for mutations incompatible with bnAb activity.
Great Debates
What are the most interesting topics likely to come up over dinner or drinks with your colleagues? Or, more importantly, what are the topics that don't come up because they are a little too controversial? In Immune Memory and Vaccines: Great Debates, Editors Rafi Ahmed and Shane Crotty have put together a collection of articles on such questions, written by thought leaders in these fields, with the freedom to talk about the issues as they see fit. This short, innovative format aims to bring a fresh perspective by encouraging authors to be opinionated, focus on what is most interesting and current, and avoid restating introductory material covered in many other reviews.
The Editors posed 13 interesting questions critical for our understanding of vaccines and immune memory to a broad group of experts in the field. In each case, several different perspectives are provided. Note that while each author knew that there were additional scientists addressing the same question, they did not know who these authors were, which ensured the independence of the opinions and perspectives expressed in each article. Our hope is that readers enjoy these articles and that they trigger many more conversations on these important topics.
How can HIV-1 broadly neutralizing antibodies (bnAbs) acquire so many V(D)J mutations? How is it possible that that B cells carrying V(D)J gene rearrangements mutated at most codons survive? Corollary questions are as follows: (1) Whether the mutational burden of HIV-1 bnAbs is strictly necessary for bnAb activity? (2) Whether extensive V(D)J mutation is a limiting factor for bnAb generation by vaccines?
It is generally presumed that the “arms race” between rapidly mutating HIV-1 populations and germinal center (GC) B cells result in waves of somatic evolution as strain-specific neutralizing antibody selects for resistant virus mutant that, in turn, drives a new round of Darwinian selection in GC B cells to produce a more avid neutralizing antibody. These cycles of humoral response followed by virus escape eventually lead to the generation of bnAbs from which the virus can no longer escape, that is, the bnAbs react with virus epitopes that cannot be altered without drastic losses of viral fitness (Liao et al. 2013; Haynes et al. 2016).
All bnAbs possess one or more of the unusual antibody traits: a long heavy-chain third complementarity-determining region (HCDR3), significant poly- and/or autoreactivity for host proteins and lipids, and exceptional frequencies of V(D)J mutations—all characteristics associated with control by immunological tolerance. Indeed, a number of knockin (KI) mouse lines that express the bnAb B-cell antigen receptors (BCRs) show that bnAb B cells and/or their unmutated precursors are subject to control by central and/or peripheral tolerance as defined by clonal deletion, receptor editing, and anergy (Haynes and Verkoczy 2014). In this brief review, we discuss bnAb induction in the setting of HIV-1 infection, and evaluate the prospects for eliciting bnAb bearing the characteristic high frequencies of somatic mutation by HIV-1 envelope (Env) vaccines.
ANTIBODY AFFINITY MATURATION
Antibody affinity to T-dependent antigens can increase 1000-fold over the 2 months following immunization (Eisen and Siskind 1964), a process of affinity maturation that was first believed to reflect clonal competition, but later shown to be the consequence of somatic mutations in less avid germline V(D)J sequences (Bothwell et al. 1981). Affinity maturation is the product somatic hypermutation (SHM) of transcribed V(D)J rearrangements and Darwinian selection for higher-affinity B-cell mutants in GCs (Berek et al. 1991; Jacob et al. 1991). Populations of GC B cells proliferate rapidly, with cell-cycle times between 6 and 12 h (Victora and Nussenzweig 2012). Bcl-6, a transcriptional repressor, is a master regulator of GC B cells and is up-regulated by the time nascent GCs are present. Absent Bcl-6 expression, the GC reaction and antibody affinity maturation are lost. Although the role of Bcl-6 in GC B cells is not yet fully understood, this repressor establishes at least two critical properties, a proapoptotic state that may be helpful in driving affinity-dependent selection and suppression of Blimp-1, a regulatory transcriptional factor that promotes plasmacytic differentiation and terminates the GC reaction (Victora and Nussenzweig 2012).
With the isolation of the first HIV-1 bnAbs, it became clear that these antibodies carried exceptionally high frequencies of V(D)J mutations. Mutation frequencies in some bnAbs reach as high as 30% (West et al. 2014), well beyond the 4%–6% frequency characteristic of high-affinity human immunoglobulin G (IgG) antibodies for other pathogens (Tiller et al. 2007; Moody et al. 2012). The numbers of mutations in bnAbs include not only the antigen-contacting complementarity-determining regions (CDRs) but also in the structurally conserved framework regions (FWRs) of the immunoglobulin (Ig)-variable regions (Klein et al. 2013). It has been proposed that the high mutation frequencies, including those in FWRs, may be necessary to achieve bnAb activity (Klein et al. 2013).
HYPERMUTATION IN GC B CELLS
The Ig-targeted SHM in GC B cells is initiated by activation-induced cytidine deaminase (AID) (Muramatsu et al. 2000; Revy et al. 2000), an enzyme that deaminates cytidines in the V(D)J and switch regions of Ig genes, leading to SHM and class switch recombination (Di Noia and Neuberger 2007). The rate of mutation in the Ig-variable (V-) regions during SHM in the GC is estimated to be ≥106-fold above background levels (McKean et al. 1984; Berek and Milstein 1987); as AID is not exclusively active at the Ig loci, it can produce off-target mutations and even chromosomal translocations (Liu et al. 2008; Nussenzweig and Nussenzweig 2010).
SHM is active along the full length of the V(D)J template, but replacement mutations that alter the amino acid sequence preferentially accumulate in CDRs as a consequence of affinity-driven selection and the presence of nucleotide motifs that are preferred by AID (Di Noia and Neuberger 2007); in contrast, the prevalence codon degeneracy in the FWRs of V gene segments disfavors replacement mutations (Reynaud et al. 1995; Jolly et al. 1996). Alterations in FWRs are constrained as they conserve the overall structure of antibody, and B cells that cannot express functional Ig quickly die by apoptosis (Rajewsky 1996). Indeed, FWRs appear to have evolved nucleotide-coding sequences resistant to SHM, presumably to reduce structural changes that disrupt antibody structure (Reynaud et al. 1995; Jolly et al. 1996). This pattern of intrinsic mutability is neither absolute nor fully understood, as non-Ig “passenger genes” inserted into active Ig loci acquire mutations at rates and sites comparable but not identical to V(D)J rearrangements (Yelamos et al. 1995; Yeap et al. 2015). Given the evolutionary accommodations to AID-driven SHM, it seems likely that SHM rates and GC B-cell proliferation have also been optimized to balance increasing genetic diversity against the possibility of clonal extinction by genomic damage and/or losses in antigen receptor fitness.
VACCINE DRIVEN V(D)J HYPERMUTATION AND SELECTION
Immunization with protein immunogens elicits GC responses, SHM, affinity maturation of circulating antibody, and high-affinity memory B-cell (Bmem) compartments (Kelsoe 1995). Nonetheless, neither immunization nor the vast majority of infections generate B-cell populations that are as extensively mutated as are bnAbs. Repeated immunization with HIV-1 vaccine glycoproteins also elicit high-affinity antibodies, but these lack both the neutralizing activity and extraordinary mutation frequencies of bnAbs (Haynes et al. 2012). Although SHM in GCs is controlled by molecular processes, affinity-driven selection must be controlled at the level of cell populations, that is, affinity maturation cannot escape the limitations of the cell-to-cell encounters that decide the fate of GC B cells by promoting survival and proliferation or cell death (Victora et al. 2010). In consequence, with regard to affinity, the somatic evolution of GC B cells likely shows both a floor and a ceiling that reflect cellular—not molecular—fitness (Batista and Neuberger 1998).
Absent competition, the floor for receptor affinity in GC B cells is very low. Transgenic B cells expressing BCRs with affinities too low to measure accurately (1 × 105 m−1 and 3 × 104 m−1) are fully competent to mount robust humoral responses similar to control mice (Dal Porto et al. 2002). Moreover, recent single-cell cloning of GC B cells showed that even when receptor affinity is not artificially restrained, interclonal receptor avidities of GC B cells vary by 100-fold and intraclonal avidities 40-fold, reaching levels for which antigen binding could not be shown (Kuraoka et al. 2016). In contrast, the upper limit to affinity maturation in GC B cells is remarkably high. Ex vivo studies that correlated antigen presentation and B-cell receptor affinity (Batista and Neuberger 1998) suggest that affinities greater than 1 × 10−10 m−1 may represent an affinity ceiling for GC B cells. Given the time frames over which GC T and B cells interact (Victora et al. 2010), it is unlikely that affinity-dependent selection could discern dissociation half-lives ≥1 h even in the presence of rare, higher-affinity competitors. Indeed, it is widely accepted that affinity-driven selection of GC B cells rests on their capacity to present antigen to GC helper T cells (TFH) (Crotty 2011). For many non-HIV protein antigens, this affinity ceiling can be achieved with mutation frequencies of a few percent; in the context of HIV-1 vaccines, the far more relevant concern is not affinity per se, but the capacity to neutralize multiple HIV-1 clades.
WHY ARE bnAbs SO HEAVILY MUTATED?
Many factors have been proposed to explain the difficulty in eliciting bnAbs; these include, epitope masking (glycan shields), conformational flexibility of the HIV-1 Env glycoprotein, the genetic plasticity of HIV-1, and molecular mimicry of host antigens (Haynes et al. 2012; Haynes and McElrath 2013). If ceiling affinities can be regularly achieved by GC B cells to non-HIV protein antigens with relatively few V(D)J mutations, why is the mutational burden of bnAbs so high? Klein et al. (2013) have suggested that somatic mutations in the Ig FWRs in addition to those in CDRs are required for bnAb potency. Similarly, high frequencies of V(D)J mutation in bnAbs could become necessary when improbable mutations at CDR sites that are “cold spots” for AID activity dictate neutralizing activity (Yeap et al. 2015; Bonsignori et al. 2017).
Although the molecular preferences of AID-driven SHM remain constant, clonal selection in GCs depends on BCR fitness (affinity and specificity) and changes in intensity and focus over the course of clonal maturation. Although possible, we believe it unlikely that a bnAb clone arises in a single GC during HIV-1 infection but rather that mutated Bmem cells periodically return to GCs and undergo subsequent rounds of mutation and selection. This return is itself regulated as most Bmem cells differentiate into plasmacytes following reactivation rather than enter GCs (Kurosaki et al. 2015; Shinnakasu et al. 2016). These divergent fates are controlled by counterpoised transcriptional regulators, Blimp-1 and Bach2, but how each fate is established remains unclear (Shinnakasu et al. 2016). HIV-1 Env vaccine antigens induce specific and high-affinity antibody and, especially when serial vaccine strategies are used (Haynes et al. 2012), are capable of driving high frequencies of V(D)J mutation (Zhang et al. 2016). Why then do HIV-1 Env vaccines not routinely induce bnAb responses?
IMMUNE PERTURBATIONS THAT OCCUR IN THE SETTING OF HIV INFECTION
Clues to answering this question have come recently from two studies. The first identified CD4 binding site (CD4bs) bnAb was in an HIV-1-infected individual with systemic lupus erythematosus (SLE) (Bonsignori et al. 2014). Both bnAb activity and double-stranded (ds)DNA antibody were present in this individual, and an isolated CD4bs bnAb, CH98, recovered from Bmem cells, was heavily mutated and possessed both bnAb activity and dsDNA reactivity. This finding showed that bnAbs and autoantibodies can be derived from the same B-cell pools and even from the same B cell (Bonsignori et al. 2014).
Second, studies of infected individuals making bnAbs show that HIV-1 infection induces a perturbed immune state characterized by increased frequencies and levels of autoreactive antibody, increased TFH cell numbers, and decreased numbers of T regulatory cells (Treg)—a phenotype similar to the immune dysfunction observed in SLE (Gray et al. 2009; Moody 2016). Combined with the autoreactive phenotypes commonly observed in bnAb KI mice (Verkoczy et al. 2010; Chen et al. 2013; Doyle-Cooper et al. 2013; Haynes and Verkoczy 2014) and the identification of host molecules mimicked by HIV-1 Env determinants (Yang et al. 2013; Liu et al. 2015), these observations suggest that the reason bnAbs are not elicited by vaccines is not the inability to drive sufficient levels of V(D)J mutation but rather limitations of on-target mutation and directed affinity maturation imposed by central and peripheral immune tolerance mechanisms.
ANTIBODY AFFINITY REVERSION/ANTIBODY REDEMPTION
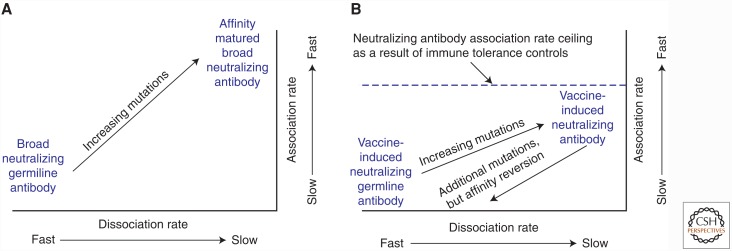
Evidence for immune control of bnAbs in primates comes from a recent study in rhesus macaques whereby immunization with a vaccine immunogen that was avidly bound by the germline, unmutated precursor of the 2F5 bnAb-induced 2F5-like antibodies that were rapidly lost on continued immunization (Zhang et al. 2016). The ability of gp41 membrane proximal bnAbs, including 2F5, to neutralize depends on acquisition of a hydrophobic HCDR3 that mediates a fast association rate for virion binding (Fig. 1A) (Zhang et al. 2016). In this study, immunization elicited alternative forms of gp41-reactive antibodies that bound at/near the 2F5 epitope and accumulated V(D)J mutation frequencies of >10%, but nonetheless exhibited slower association rates than bnAbs (Fig. 1B). Continued immunization resulted in additional mutations (to ∼20% in V(D)J rearrangements) in 2F5-like antibodies but with impaired association rates and lost reactivity for the 2F5 gp41 epitope (Fig. 1B) (Yang et al. 2013; Zhang et al. 2016). This mutation and GC selection to negate autoreactivity has been termed affinity reversion (Verkoczy and Diaz 2014) or clonal redemption (Sabouri et al. 2014; Reed et al. 2016). Reversion/redemption is a newly recognized peripheral tolerance mechanism that acts by the selection for V(D)J mutations that direct the antibody paratope away from autoreactivity; in consequence, this process can “rescue or redeem” anergic B cells that become activated and enter the GC reaction. For the gp41 2F5 epitope, there are at least two autoantigen mimics, the tryptophan enzyme, kynureninase, and the host membrane lipids (Yang et al. 2013; Zhang et al. 2016), that could drive affinity reversion. New studies in KI mice that express other bnAb types suggest that tolerance acts to control bnAb responses to other HIV-1-neutralizing epitopes as well (BF Haynes and L Verkoczy, unpubl.).
Figure 1.
Schema of rate map analysis and ceiling of affinity maturation of gp41 broadly neutralizing antibodies (bnAbs) from HIV-1 infection and from vaccination. Original studies from which these schema are derived used affinity measurements of bnAbs and vaccine-induced antibodies to gp41 peptide liposome virosomes as models of virions. (A) The 2F5 gp41 bnAb in HIV-1 infection with full bnAb development, and (B) the ceiling of affinity maturation limited by immune tolerance when antibodies targeted to the gp41 epitope develop in the setting of vaccination. Studies showed that the ceiling of affinity maturation for gp41 bnAbs was mediated by immune tolerance limiting the development of antibodies with long hydrophobic heavy-chain third complementarity-determining regions (HCDR3s) that are necessary for gp41 bnAbs to neutralize HIV-1. (Panel A is based on data in Zhang et al. 2016, and panel B is based on data in Zhang et al. 2016 and Alam 2009.)
CONCLUSIONS
We propose that the numbers and quality of V(D)J mutations necessary for bnAb activity can be achieved by serial vaccination with vaccine formulations that (1) provide sufficient immune activation to bnAb precursors and induce AID activity sufficient for requisite mutation in AID cold spots, (2) support the reentry of memory B cells into GC and the accumulation of additional rounds of mutation and selection, and (3) promote the conditions of increased TFH numbers (and perhaps decreased Treg activity) associated with HIV-1 infection and natural bnAb induction. These requirements for immunization success can be achieved by the work of antibody–virus coevolution and mapping (Liao et al. 2013; Bonsignori et al. 2016), optimization of sequential immunogens to recreate crucial events in HIV-1 infection and bnAb induction (Haynes et al. 2012), and vaccine formulations that perturb immune responses to facilitate bnAb development (Gray et al. 2009; Moody 2016).
ACKNOWLEDGMENTS
This study is supported by the National Institutes of Allergy and Infectious Diseases (NIAID) and by intramural National Institutes of Health (NIH) support for the NIAID Vaccine Research Center, by a grant from the NIH, NIAID, AI100645 (the Duke Center for HIV/AIDS Vaccine Immunology-Immunogen Discovery [CHAVI-ID]). The authors thank Masayuki Kuraoka, Thomas Kepler, Laurent Verkoczy, Mattia Bonsignori, Munir Alam, Fred Alt, Ming Tian, Peter Kwong, Larry Shapiro, John Mascola, Persephone Borrow, Tony Moody, and Kevin Wiehe for insightful discussions and their participation in the work cited.
REFERENCES
- Batista FD, Neuberger MS. 1998. Affinity dependence of the B cell response to antigen: A threshold, a ceiling, and the importance of off-rate. Immunity 8: 751–759. [DOI] [PubMed] [Google Scholar]
- Berek C, Milstein C. 1987. Mutation drift and repertoire shift in the maturation of the immune response. Immunol Rev 96: 23–41. [DOI] [PubMed] [Google Scholar]
- Berek C, Berger A, Apel M. 1991. Maturation of the immune response in germinal centers. Cell 67: 1121–1129. [DOI] [PubMed] [Google Scholar]
- Bonsignori M, Wiehe K, Grimm SK, Lynch R, Yang G, Kozink DM, Perrin F, Cooper AJ, Hwang KK, Chen X, et al. 2014. An autoreactive antibody from an SLE/HIV-1 individual broadly neutralizes HIV-1. J Clin Invest 124: 1835–1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonsignori M, Zhou T, Sheng Z, Chen L, Gao F, Joyce MG, Ozorowski G, Chuang GY, Schramm CA, Wiehe K, et al. 2016. Maturation pathway from germline to broad HIV-1 neutralizer of a CD4-mimic antibody. Cell 165: 449–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonsignori M, Kreider EF, Fera D, Meyerhoff RR, Bradley T, Wiehe K, Alam SM, Aussedat B, Walkowicz WE, Hwang KK, et al. 2017. Sci Transl Med 9: eaai7514.28298420 [Google Scholar]
- Bothwell AL, Paskind M, Reth M, Imanishi-Kari T, Rajewsky K, Baltimore D. 1981. Heavy chain variable region contribution to the NPb family of antibodies: Somatic mutation evident in a γ2a variable region. Cell 24: 625–637. [DOI] [PubMed] [Google Scholar]
- Chen Y, Zhang J, Hwang KK, Bouton-Verville H, Xia SM, Newman A, Ouyang YB, Haynes BF, Verkoczy L. 2013. Common tolerance mechanisms, but distinct cross-reactivities associated with gp41 and lipids, limit production of HIV-1 broad neutralizing antibodies 2F5 and 4E10. J Immunol 191: 1260–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotty S. 2011. Follicular helper CD4 T cells (TFH). Annu Rev Immunol 29: 621–663. [DOI] [PubMed] [Google Scholar]
- Dal Porto JM, Haberman AM, Kelsoe G, Shlomchik MJ. 2002. Very low affinity B cells form germinal centers, become memory B cells, and participate in secondary immune responses when higher affinity competition is reduced. J Exp Med 195: 1215–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Noia JM, Neuberger MS. 2007. Molecular mechanisms of antibody somatic hypermutation. Annu Rev Biochem 76: 1–22. [DOI] [PubMed] [Google Scholar]
- Doyle-Cooper C, Hudson KE, Cooper AB, Ota T, Skog P, Dawson PE, Zwick MB, Schief WR, Burton DR, Nemazee D. 2013. Immune tolerance negatively regulates B cells in knock-in mice expressing broadly neutralizing HIV antibody 4E10. J Immunol 191: 3186–3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen HN, Siskind GW. 1964. Variations in affinities of antibodies during the immune response. Biochemistry 3: 996. [DOI] [PubMed] [Google Scholar]
- Gray ES, Taylor N, Wycuff D, Moore PL, Tomaras GD, Wibmer CK, Puren A, DeCamp A, Gilbert PB, Wood B, et al. 2009. Antibody specificities associated with neutralization breadth in plasma from human immunodeficiency virus type 1 subtype C-infected blood donors. J Virol 83: 8925–8937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes BF, McElrath MJ. 2013. Progress in HIV-1 vaccine development. Curr Opin HIV AIDS 8: 326–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes BF, Verkoczy L. 2014. AIDS/HIV. Host controls of HIV neutralizing antibodies. Science 344: 588–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes BF, Kelsoe G, Harrison SC, Kepler TB. 2012. B-cell-lineage immunogen design in vaccine development with HIV-1 as a case study. Nat Biotechnol 30: 423–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes BF, Shaw GM, Korber B, Kelsoe G, Sodroski J, Hahn BH, Borrow P, McMichael AJ. 2016. HIV-host interactions: Implications for vaccine design. Cell Host Microbe 19: 292–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob J, Kelsoe G, Rajewsky K, Weiss U. 1991. Intraclonal generation of antibody mutants in germinal centres. Nature 354: 389–392. [DOI] [PubMed] [Google Scholar]
- Jolly CJ, Wagner SD, Rada C, Klix N, Milstein C, Neuberger MS. 1996. The targeting of somatic hypermutation. Semin Immunol 8: 159–168. [DOI] [PubMed] [Google Scholar]
- Kelsoe G. 1995. In situ studies of the germinal center reaction. Adv Immunol 60: 267–288. [DOI] [PubMed] [Google Scholar]
- Klein F, Diskin R, Scheid JF, Gaebler C, Mouquet H, Georgiev IS, Pancera M, Zhou T, Incesu RB, Fu BZ, et al. 2013. Somatic mutations of the immunoglobulin framework are generally required for broad and potent HIV-1 neutralization. Cell 153: 126–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuraoka M, Schmidt AG, Nojima T, Feng F, Watanabe A, Kitamura D, Harrison SC, Kepler TB, Kelsoe G. 2016. Complex antigens drive permissive clonal selection in germinal centers. Immunity 44: 542–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurosaki T, Kometani K, Ise W. 2015. Memory B cells. Nat Rev Immunol 15: 149–159. [DOI] [PubMed] [Google Scholar]
- Liao HX, Lynch R, Zhou T, Gao F, Alam SM, Boyd SD, Fire AZ, Roskin KM, Schramm CA, Zhang Z, et al. 2013. Co-evolution of a broadly neutralizing HIV-1 antibody and founder virus. Nature 496: 469–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Duke JL, Richter DJ, Vinuesa CG, Goodnow CC, Kleinstein SH, Schatz DG. 2008. Two levels of protection for the B cell genome during somatic hypermutation. Nature 451: 841–845. [DOI] [PubMed] [Google Scholar]
- Liu M, Yang G, Wiehe K, Nicely NI, Vandergrift NA, Rountree W, Bonsignori M, Alam SM, Gao J, Haynes BF, et al. 2015. Polyreactivity and autoreactivity among HIV-1 antibodies. J Virol 89: 784–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKean D, Huppi K, Bell M, Staudt L, Gerhard W, Weigert M. 1984. Generation of antibody diversity in the immune response of BALB/c mice to influenza virus hemagglutinin. Proc Natl Acad Sci 81: 3180–3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moody MA, Yates NL, Amos JD, Drinker MS, Eudailey JA, Gurley TC, Marshall DJ, Whitesides JF, Chen X, Foulger A, et al. 2012. HIV-1 gp120 vaccine induces affinity maturation in both new and persistent antibody clonal lineages. J Virol 86: 7496–7507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moody MA, PPI, Vandergrift NA, Chui C, Lloyd K, Soderberg KA, Ogbe AT, Cohen MS, Liao HX, Gao F, McMichael AJ, et al. 2016. Immune perturbations in HIV-1-infected individuals who make broadly reactive neutralizing antibodies. Science Immunol 1: aag0851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T. 2000. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell 102: 553–563. [DOI] [PubMed] [Google Scholar]
- Nussenzweig A, Nussenzweig MC. 2010. Origin of chromosomal translocations in lymphoid cancer. Cell 141: 27–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajewsky K. 1996. Clonal selection and learning in the antibody system. Nature 381: 751–758. [DOI] [PubMed] [Google Scholar]
- Reed JH, Jackson J, Christ D, Goodnow CC. 2016. Clonal redemption of autoantibodies by somatic hypermutation away from self-reactivity during human immunization. J Exp Med 213: 1255–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revy P, Muto T, Levy Y, Geissmann F, Plebani A, Sanal O, Catalan N, Forveille M, Dufourcq-Labelouse R, Gennery A, et al. 2000. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the hyper-IgM syndrome (HIGM2). Cell 102: 565–575. [DOI] [PubMed] [Google Scholar]
- Reynaud CA, Garcia C, Hein WR, Weill JC. 1995. Hypermutation generating the sheep immunoglobulin repertoire is an antigen-independent process. Cell 80: 115–125. [DOI] [PubMed] [Google Scholar]
- Sabouri Z, Schofield P, Horikawa K, Spierings E, Kipling D, Randall KL, Langley D, Roome B, Vazquez-Lombardi R, Rouet R, et al. 2014. Redemption of autoantibodies on anergic B cells by variable-region glycosylation and mutation away from self-reactivity. Proc Natl Acad Sci 111: E2567–E2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinnakasu R, Inoue T, Kometani K, Moriyama S, Adachi Y, Nakayama M, Takahashi Y, Fukuyama H, Okada T, Kurosaki T. 2016. Regulated selection of germinal-center cells into the memory B cell compartment. Nat Immunol 17: 861–869. [DOI] [PubMed] [Google Scholar]
- Tiller T, Tsuiji M, Yurasov S, Velinzon K, Nussenzweig MC, Wardemann H. 2007. Autoreactivity in human IgG+ memory B cells. Immunity 26: 205–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkoczy L, Diaz M. 2014. Autoreactivity in HIV-1 broadly neutralizing antibodies: Implications for their function and induction by vaccination. Curr Opin HIV AIDS 9: 224–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkoczy L, Diaz M, Holl TM, Ouyang YB, Bouton-Verville H, Alam SM, Liao HX, Kelsoe G, Haynes BF. 2010. Autoreactivity in an HIV-1 broadly reactive neutralizing antibody variable region heavy chain induces immunologic tolerance. Proc Natl Acad Sci 107: 181–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Victora GD, Nussenzweig MC. 2012. Germinal centers. Annu Rev Immunol 30: 429–457. [DOI] [PubMed] [Google Scholar]
- Victora GD, Schwickert TA, Fooksman DR, Kamphorst AO, Meyer-Hermann M, Dustin ML, Nussenzweig MC. 2010. Germinal center dynamics revealed by multiphoton microscopy using a photoactivatable fluorescent reporter. Cell 143: 592–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West AP Jr, Scharf L, Scheid JF, Klein F, Bjorkman PJ, Nussenzweig MC. 2014. Structural insights on the role of antibodies in HIV-1 vaccine and therapy. Cell 156: 633–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G, Holl TM, Liu Y, Li Y, Lu X, Nicely NI, Kepler TB, Alam SM, Liao HX, Cain DW, et al. 2013. Identification of autoantigens recognized by the 2F5 and 4E10 broadly neutralizing HIV-1 antibodies. J Exp Med 210: 241–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeap LS, Hwang JK, Du Z, Meyers RM, Meng FL, Jakubauskaite A, Liu M, Mani V, Neuberg D, Kepler TB, et al. 2015. Sequence-intrinsic mechanisms that target AID mutational outcomes on antibody genes. Cell 163: 1124–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yelamos J, Klix N, Goyenechea B, Lozano F, Chui YL, Gonzalez Fernandez A, Pannell R, Neuberger MS, Milstein C. 1995. Targeting of non-Ig sequences in place of the V segment by somatic hypermutation. Nature 376: 225–229. [DOI] [PubMed] [Google Scholar]
- Zhang R, Verkoczy L, Wiehe K, Munir Alam S, Nicely NI, Santra S, Bradley T, Pemble CWt, Zhang J, Gao F, et al. 2016. Initiation of immune tolerance-controlled HIV gp41 neutralizing B-cell lineages. Sci Transl Med 8: 336ra362. [DOI] [PMC free article] [PubMed] [Google Scholar]