Abstract
Background
Though normally linked to bone and cartilage development, the Runt-related transcription factor, RUNX2, was reported in the mouse heart during development of the valves. We examined RUNX2 expression and function in the developing avian heart as it related to the epithelial-mesenchymal transition (EMT) in the atrioventricular canal. EMT can be separated into an activation stage involving hypertrophy and cell separation and an invasion stage where cells invade the extracellular matrix. The localization and activity of RUNX2 was explored in relation to these steps in the heart. As RUNX2 was also reported in cancer tissues, we examined its expression in the progression of esophageal cancer in staged tissues.
Results
A specific isoform, RUNX2-I, is present and required for EMT by endothelia of the atrioventricular canal. Knockdown of RUNX2-I inhibits the cell-cell separation that is characteristic of initial activation of EMT. Loss of RUNX2-I altered expression of EMT markers to a greater extent during activation than during subsequent cell invasion. TGFβ2 mediates activation during cardiac endothelial EMT. Consistent with a role in activation, RUNX2-I is regulated by TGFβ2 and its activity is independent of similarly expressed Snai2 in regulation of EMT. Examination of RUNX2 expression in esophageal cancer showed its upregulation concomitant with the development of dysplasia and continued expression in adenocarcinoma.
Conclusions
This data introduces the RUNX2-I isoform as a critical early transcription factor mediating EMT in the developing heart after induction by TGFβ. Its expression in tumor tissue suggests a similar role for RUNX2 in the EMT of metastasis.
Keywords: EMT, RUNX2, heart, AVC
INTRODUCTION
Epithelial-mesenchymal cell transition (EMT) is a regulated cellular process wherein epithelial cells lose cell-cell adhesions, change shape to a stellate or fibroblastic morphology and become invasive (Thiery et al., 2009; Nieto, 2011). In the embryo, EMT provides a mechanism utilized for gastrulation, neural crest cell formation and in other areas of the embryo to produce three-dimensional structures (Hay and Zuk, 1995). EMT reoccurs in the adult as a component of the pathologies of metastasis and fibrosis (Kalluri and Neilson, 2003; Thiery et al., 2009). EMT in the embryonic heart is a prototypical example of this process (Yang and Weinberg, 2008). Cardiac endothelial cells lining the atrioventricular canal (AVC) undergo an EMT, invade the underlying matrix and form valve progenitors (Markwald et al., 1984). This EMT is mediated by TGFβ superfamily members, including TGFβ, TGFβ3 (Potts and Runyan, 1989; Potts et al., 1991; Boyer et al., 1999a), and BMP-2 (Yamagishi et al., 1999), and involves many of the molecules and signal transduction mechanisms common to other EMTs (Person et al., 2005; Nieto, 2011). Studies in the Runyan laboratory utilize an in vitro analysis with collagen gels to explore mechanisms involved in transition from endothelial cells to cardiac mesenchyme (Person et al., 2005; Mercado-Pimentel and Runyan, 2007; Lencinas et al., 2011).
Runt-related transcription factors (RUNX) are a family of regulators that include RUNX1, RUNX2 and RUNX3. All members contain a conserved DNA binding domain, the Runt domain (Ito et al., 2015). RUNX2, also called OSF2 or CBFα1, is expressed in mineralizing tissues, being essential for osteoblast maturation (Komori et al., 1997). This transcription factor is also expressed in other embryonic tissues as well the developing vasculature (Zhao et al., 2002; Bronckers et al., 2005; Jeong et al., 2008). RUNX2 is over-expressed in different types of cancers, including breast cancer and prostate cancer cell lines that metastasize to bone (Pratap et al., 2005; Baniwal et al., 2010; Mendoza-Villanueva et al., 2011). RUNX2 also appears to induce oncogenic transformation of human bone marrow endothelial cells (Vitolo et al., 2007) and of thyroid epithelia (Niu et al., 2012). RUNX2 is also up-regulated in calcified aortic valves (Garg, 2006; Yang et al., 2009). The wide expression of RUNX2 in calcified tissues has led to an emphasis on osteoblastic differentiation and little is known of other roles. There are two RUNX2 isoforms that differ by transcription start site: isoform I (RUNX2-I), expressed in osteoblastic, pre-osteoblastic and non-osteoblastic mesenchymal cells and isoform II (RUNX2-II), a largely bone-specific isoform (Li and Xiao, 2007; Zhang et al., 2009; Okura et al., 2014). Alternative splicing is also responsible for additional and poorly understood isoforms (Terry et al., 2004). Of interest in a TGFβ-mediated cellular process, RUNX2 interacts with SMADs to regulate gene expression (Ito and Miyazono, 2003). We were intrigued by the localization of RUNX2 in the embryonic heart and set out to investigate its function during EMT (Gitler et al., 2003). An initial screen, using siRNA, suggested functional role for RUNX2 during EMT.
In the present study, we explore expression and function of RUNX2 during heart valve development in the chick embryo. RUNX2-I is expressed by the endothelial cells lining the AVC and the mesenchymal cells invading the cardiac jelly during EMT. This transcription factor is TGFβ2-regulated. Inhibition of RUNX2 mRNA expression disrupts EMT at an early stage before loss of cell-cell adhesion. Examination of marker gene expression suggests that RUNX2 regulation can be distinguished from Snai2 regulation of EMT and that both gene transcripts are involved in early stages of EMT. To explore the relevance of the observation to metastatic mechanisms, we examined staged human esophageal tissues in the transition from normal epithelium to metastatic esophageal adenocarcinoma, a carcinoma reported to involve TGFβ signaling (Onwuegbusi et al., 2006; von Rahden et al., 2006). RUNX2-I expression appears coincident with the transition from columnar epithelia (Barrett’s Esophagus) to high-grade dysplasia. This distribution is consistent with a conserved role in an early stage of EMT in both development and pathology. These data suggest that RUNX2 expression in human tumors is related to the process of EMT in metastasis rather than targeting to bone tissue.
RESULTS
RUNX2 is expressed in the AVC region of the developing heart
For a spatiotemporal localization of RUNX2, Hamburger and Hamilton stage 14, 17, 18 and 20 chicken embryo hearts were dissected and processed for immunofluorescent staining. This interval covers the period between initiation of tissue interaction leading to EMT and the population of the cardiac cushions (valvular primordia) (Markwald et al., 1984). A monoclonal antibody against RUNX2 (51.7 kDa) was used in this assay. The antibody detected a band of approximately 50kDa by western blot using total protein of stage 17 chick embryo tissue (data not shown). RUNX2 is required for proper chondrogenesis and osteogenesis (Komori et al., 1997; Stricker et al., 2002) and hair follicle development in mice (Glotzer et al., 2008). As a positive control, tissues of E13 chick embryo wings were immunostained with the antibody. A nuclear protein was marked in osteoblasts in the region of endochondral ossification (arrows in Fig. 1G) and in cells in the feather papilla, homologous to hair follicles in chicken (arrowheads in Fig. 1H), confirming the antibody is recognizing chicken RUNX2.
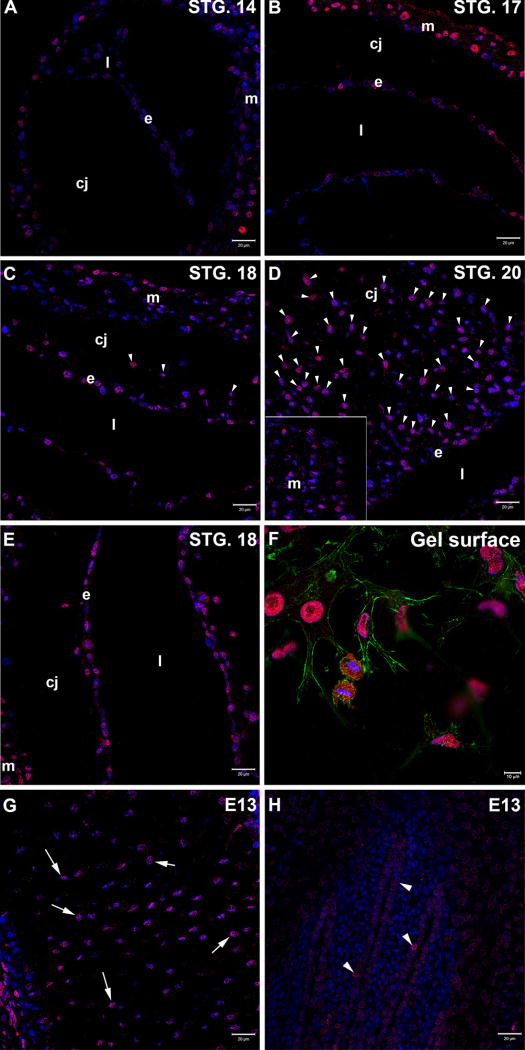
Figure 1. RUNX2 immunolocalization in the chick embryo heart during the timing for EMT.
A-E. Chicken embryo hearts were stained for RUNX2 (red) in the AVC region (A-D) or OFT (E). Nuclei were counterstained with TO-PRO-3 (blue). In the AVC region of stage 14 chicken hearts (A), low expression of RUNX2 is detected in endothelial cells (e) lining the heart lumen (l). This expression is increased by stage 17 (B) and stage 18 (C). RUNX2 is still detected in endothelial cells (e) by stage 20 (D). Mesenchymal cells (arrowheads) invading the heart cardiac jelly (cj) show expression of RUNX2 in stage 18 (C) and stage 20 (D) hearts. A similar expression pattern is observed in stage 18 OFT region (E). RUNX2 is also present in the developing myocardium (m) at all analyzed stages (A-C, inset in D, E). Scale bar: 20µm. F. Activated endothelial cells derived from cultured AVCs on a collagen gel lattice show RUNX2 expression (red) in the nuclei. Low RUNX2 level is also detected in the cytoplasm. Nuclei were counterstained with DAPI (blue) and F-actin with phalloidin (green). Scale bar: 10µm. G-H. E13 chicken wings were stained for RUNX2 (red) as positive controls. The antibody marks osteoblasts in a region of endochondral ossification (arrows in G) and cells in the feather papilla (arrowheads in H). Nuclei were counterstained with TO-PRO-3 (blue). Scale bar: 20µm.
In the AVC region of the chick heart, RUNX2 was detected in the myocardium at all stages (Fig. 1A-D). In endothelial cells, low expression of RUNX2 is detected by stage 14 (Fig 1A). This expression increases by stage 17 (Fig. 1B) and stage 18 (Fig.1C), persisting until stage 20 (Fig.1D). Mesenchymal cells that had delaminated from the endocardium and that were migrating within the underlying extracellular matrix, or cardiac jelly, were also found to express this transcription factor (Fig.1C and D). In the OFT region of the chick heart, RUNX2 expression is also detected in endothelial and mesenchymal cells (Fig.1E).
RUNX2 was also localized in activated endothelial cells (Fig. 1F) and mesenchymal cells (data not shown) derived from AVC explants cultured on a collagen gel lattice. Cells were counterstained with DAPI (nuclei) and phalloidin (F-actin) for cell morphology. RUNX2 co-localizes with DAPI in both cell types (Fig. 1F and data not shown). Although low RUNX2 signal was detected in the cell cytoplasm, the antibody detected RUNX2 mainly in the nuclei of these cells. Taken together, the results argue that RUNX2 is present during the regulation of AVC EMT in endothelial and mesenchymal cells and that changes in endothelial cell expression are concomitant with EMT.
RUNX2-I is the isoform expressed in the chick heart
To further test if RUNX2 is present in the chicken embryo heart at the time of EMT, whole-mount in situ hybridization was performed using an RNA probe specific for chicken RUNX2 mRNA. Figure 2A shows that RUNX2 is expressed in different regions of a stage 18 chicken embryo, including the limb buds (arrowhead) and heart (arrows).
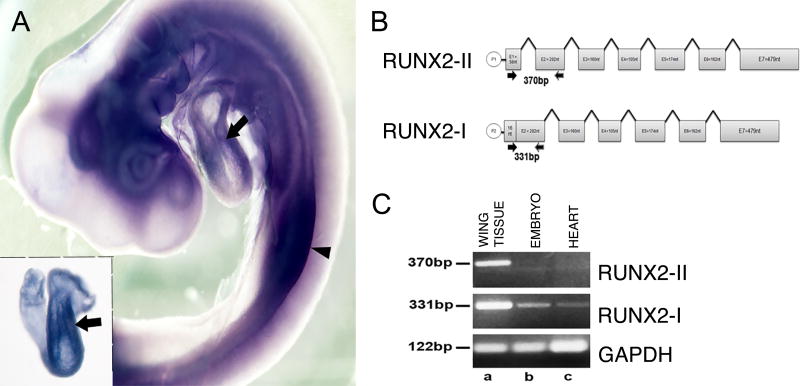
Figure 2. RUNX2-I is the isoform present in the chick heart during EMT.
A. Lateral view of a stage 18 chicken embryo processed for whole-mount in situ hybridization using a probe recognizing chicken RUNX2. There is expression of RUNX2 in the limb buds (arrowhead) and in the heart, including the AVC region (arrows). The inset shows a dissect heart for better visualization. B. Schematic representation of RUNX2 gene. Transcription of RUNX2-II, the bone specific isoform, starts at the distal promoter P1. Transcription of RUNX2-I starts at the proximal promoter P2. RUNX2-II was amplified using primers that produced a 370bp product whereas RUNX2-I was amplified using primers that produced a 331bp product. C. RT-PCR analysis of total RNA from E13 chick wing tissue (a), stage 17 embryos (b) and stage 17 hearts (c) shows RUNX2-II (top lane) is expressed only in E13 chicken wing cells (a) while RUNX2-I (middle lane) is expressed in chicken wing cells (a), stage 17 embryos (b) and stage 17 hearts (c). Bottom lane shows GAPDH expression (122bp) as loading control.
RUNX2 is commonly known to be an osteogenic factor. Type II RUNX2 (RUNX2-II) is the bone-specific isoform while type I RUNX2 (RUNX2-I) is broadly expressed (Li and Xiao, 2007). To verify which isoform is expressed during heart development in the chick embryo, primers specific for each isoform were designed (Fig. 2B). Since only RUNX2-II is sequenced for the chicken (NM_204128), the human and mouse RUNX2-I sequences were used as reference for primer design as well. RT-PCR was performed with tissue from E13 chick wing (Fig. 2C-a), stage 17 chick embryo (Fig. 2C-b) and stage 17 chick hearts (Fig. 2C-c). As expected, at E13, chick wing cells are expressing both isoforms (Fig.2C-a). At stage 17, RUNX2-I is expressed in the chick embryo and heart (Fig.2C-b, c).
RUNX2-I is TGFβ-regulated in vitro
That RUNX2 is expressed by endothelial and mesenchymal cells during the TGFβ-regulated EMT (Potts and Runyan, 1989; Boyer et al., 1999a; Ma et al., 2005) suggests RUNX2 is regulated by TGFβ-family members. To examine whether RUNX2 is involved in TGFβ signaling, cultured AVC explants were co-stained for RUNX2 and pSmad1/5/8 (BMP signaling mediators, Fig. 3A-C) or pSmad2/3 (TGFβ signaling mediators, Fig. 3D-F). Activated endothelial cells on the gel surface showed co-expression of RUNX2 and pSmad1/5/8 (Fig. 3C) and RUNX2 and pSmad2/3 (Fig. 3F) in the cell nuclei showing cells that express RUNX2 are receiving TGFβ and/or BMP signal.
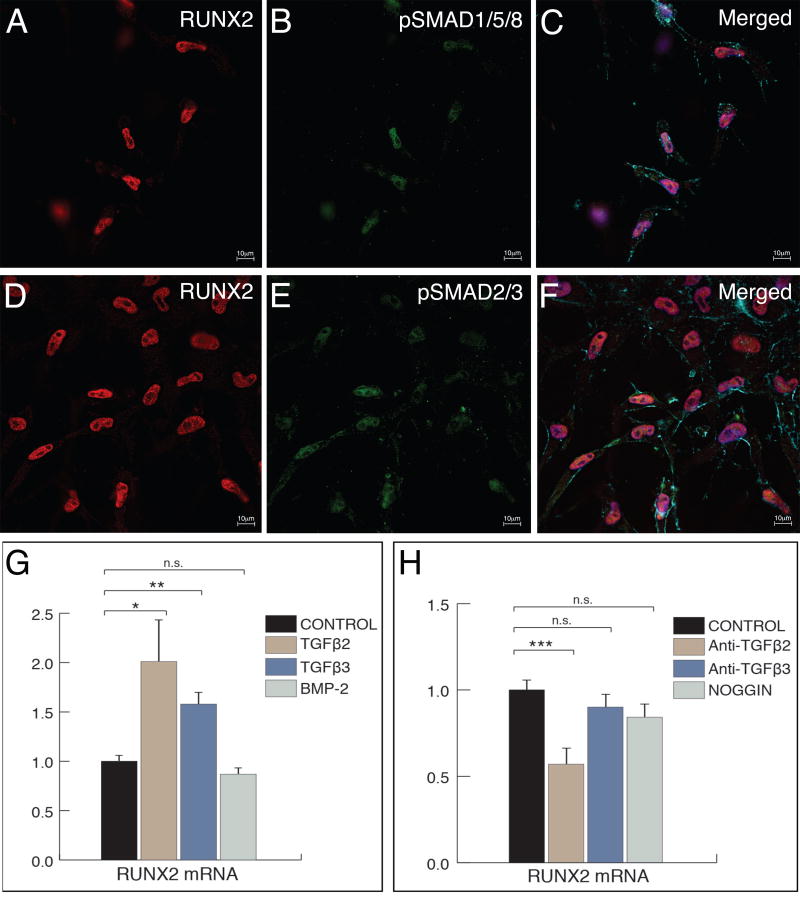
Figure 3. RUNX2-I is induced by TGFβ signaling.
A-F. Activated endothelial cells derived from cultured AVC explants co-express RUNX2 (red in A and D) and pSmad1/5/8 (green in B) or pSmad2/3 (green in E). Merged picture in C and F confirms co-localization of the proteins in the cell nuclei (DAPI, blue). F-actin was stained with phalloidin to show cell morphology (cyan). Scale bar: 10µm. G. qPCR analysis of RUNX2 mRNA in AVC explants cultured in the presence of TGFβ2, TGFβ3 or BMP-2. Addition of TGFβ2 or TGFβ3 to culture media caused a significant increase in RUNX2 message levels whereas addition of BMP-2 caused no significant changes. H. qPCR analysis of RUNX2 mRNA in AVC explants cultured in the presence of neutralizing antibodies against TGFβ2 or TGFβ3 or with the BMP inhibitor, noggin. Blockage of TGFβ2 caused a significant decrease in the level of RUNX2 mRNA whereas blockage of TGFβ3 or BMP signaling did not cause any significant changes. Error bars = S.E.M.; * p < 0.05; ** p < 0.01; *** p < 0.001; n.s. – non-significant.
To test whether TGFβ signaling regulates RUNX2 expression, we examined the effect of exogenous TGFβ2, TGFβ3 or BMP-2 on RUNX2 mRNA expression. Culture media with TGFβ2, TGFβ3 or BMP-2 were added to AVC explant cultures and the level of RUNX2 mRNA was assessed by quantitative real-time PCR (qPCR) (Fig. 3G). TGFβ2 addition caused a significant ~2-fold increase in RUNX2 mRNA (p < 0.05). Addition of TGFβ3 also caused a significant ~1.5-fold increase in RUNX2 message (p < 0.01). BMP-2 did not cause significant changes in RUNX2 mRNA (p=0.116). Next, we used neutralizing antibodies against TGFβ2 and TGFβ3 to block these ligands. To inhibit BMP signaling, cultures were treated with Noggin. The level of RUNX2 mRNA present was compared with control cultures by qPCR (Fig. 3H). Only anti-TGFβ2 caused a significant effect on RUNX2 message (~40% decrease of RUNX2 mRNA, p < 0.001; anti-TGFβ1, p=0.273; Noggin, p=0.837). Taken together, the data shows that RUNX2 is induced by TGFβ2 and suggests that this factor is part of the chick AVC EMT activation step in vitro.
RUNX2-I inhibition disrupts EMT in vitro
That RUNX2 is present in endothelial and mesenchymal cells during the AVC EMT suggests this molecule is required for proper EMT initiation. Therefore, we decided to assay the function of this transcription factor using specific siRNAs. Stage 14 chicken AVC explants were transfected with 3 different siRNAs against RUNX2 and placed on a collagen gel lattice. Control explants were transfected with a scrambled siRNA. The level of RUNX2 mRNA inhibition varied from 40% to 80% between experiments (~60% decrease in RUNX2 message, p < 0.001, Fig. 4H).
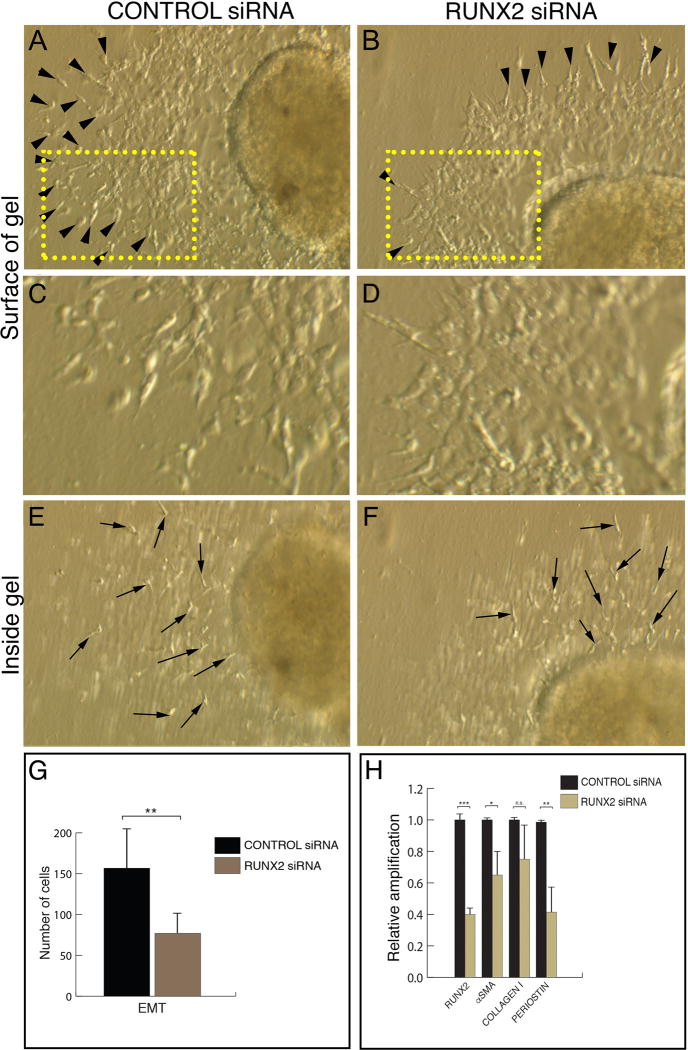
Figure 4. RUNX2-I is required for the activation step of chick heart AVC EMT.
A-F. Stage 14 chicken AVC explants cultured for 24h in the presence of a scrambled siRNA (A, C, E) or a siRNA against RUNX2 (B, D, F). In control explants, multiple activated endothelial cells (arrowheads in A) are present on the gel surface. C represents a higher magnification of the boxed area in A showing the endothelial cell outgrowth. Inside the gel, multiple invasive mesenchymal cells (arrows in E) are observed. RUNX2 siRNA-transfected explants show fewer activated endothelial cells (arrowheads in B) on the gel surface. D represents a higher magnification of the boxed area in B showing a compact endothelial cell outgrowth. Mesenchymal cells (arrows in F) are present in treated explants. G. Quantitation of the number of EMT-derived cells in control and RUNX2 siRNA-transfected explants. There is a significant decrease in the number of activated endothelial cells and mesenchymal cells in treated explants compared to control explants (Average cell number in control explants = 156.71 ± 18.83; average cell number in RUNX2 siRNA-treated explants = 77.00 ± 9.07; ~51% decrease, p < 0.01). n=14 (7, control; 7, experimental). H. qPCR analysis of RUNX2 and mesenchymal cell markers in control and RUNX2 siRNA-transfected explants. There is a significant decrease (~60%) in RUNX2 mRNA after siRNA treatment. RUNX2 downregulation is followed by a decrease in the levels of αSMA (~35% decrease) and PERIOSTIN (POSTN) (~60% decrease). COLLAGEN I is unchanged in treated explants. Error bars = S.E.M.; * p < 0.05; ** p < 0.01; *** p < 0.001; n.s. – non-significant.
RUNX2 siRNA-transfected explants showed inhibition of EMT when analyzed at 24h (Fig. 4A-F) and 48h after explants were placed on gels (data not shown). In control explants an average of 156.71 activated endothelial cells and mesenchymal cells were counted on the gel surface (S.E.M.=18.83; arrowheads in Fig. 4A, C, G) and inside the gel (arrows in Fig. 4E, G). In RUNX2-siRNA treated explants, on the gel surface, fewer activated endothelial cells (arrowheads in Fig. 4B, D, G) and mesenchymal cells (arrows in Fig. 4F, G) were observed (average=77.00 cells; S.E.M.=9.07; ~51% decrease; p < 0.01). RUNX2-siRNA treatment had a greater effect in the number of activated endothelial cells (~58% decrease; p < 0.01, data not shown) than in the number of mesenchymal cells (~47% decrease; p < 0.05, data not shown). These results suggest that RUNX2 is part of the signaling pathway that controls EMT initiation (activation of endothelial cells).
RUNX2-I inhibition alters markers of EMT
To confirm chicken AVC EMT is being disrupted after RUNX2 knock down, expression of smooth muscle alpha actin (αSMA) (Boyer et al., 1999b), Collagen 1 (COL1) (de Vlaming et al., 2012) and Periostin (POSTN) (Inai et al., 2008) messages were assessed by qPCR (Fig. 4H). There was a significant downregulation of αSMA (~35% decrease, p < 0.001) and POSTN (~60% decrease, p < 0.01). COL1 was not significantly affected (p=0.275). These results show that RUNX2 is required for proper EMT in vitro.
Snai2 and RUNX2-I regulate different aspects of EMT activation
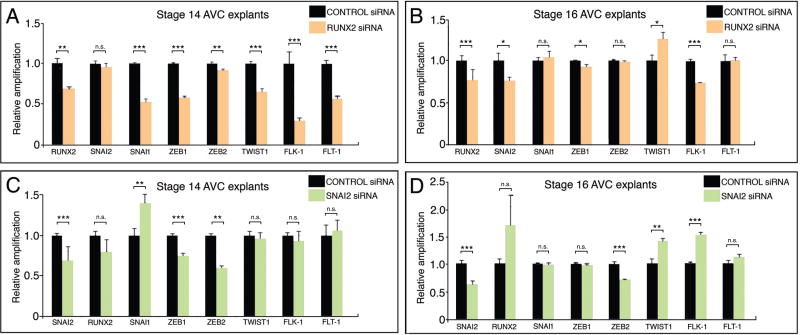
To further our analysis of RUNX2 function in early (activation) and later (invasion) steps of the chick AVC EMT, the mRNA expression of different EMT markers in control and RUNX2 siRNA-treated stage 14 and 16 explants were checked by qPCR (Fig. 5A-B). In stage 14 explants cultured for 16 hours (Fig. 5A), RUNX2 downregulation (~30%, p < 0.01) caused significant decrease in the zinc-finger transcription factors SNAI1 (~47% decrease, p < 0.001), ZEB1 (~42% decrease, p < 0.001) and ZEB2 (~10% decrease, p < 0.01) while SNAI2 showed no significant change (p=0.307). The bHLH transcription factor TWIST1 was decreased (~34% decrease, p < 0.001) in transfected explants as well. Additionally, a significant decrease in endothelial VEGF receptors FLK-1(VEGFR2) (~70% decrease, p < 0.001) and FLT-1(VEGFR1) (~40% decrease, p < 0.001) was observed in transfected explants. In stage 16 explants cultured for 16 hours (Fig. 5B), RUNX2 downregulation (~25%, p < 0.001) caused a significant decrease in SNAI2 (~25% decrease, p < 0.05) and ZEB1 (~10%, p < 0.05) while SNAI1 (p=0.434) and ZEB2 (p=0.316) were not significantly affected. TWIST1 was significantly increased (~1.25-fold, p < 0.05). Finally, FLK-1, but not FLT-1 (p=0.203), was affected (~27% decrease, p < 0.001) after RUNX2 knock down. This molecular analysis confirms RUNX2 is mainly functioning in the activation step of EMT.
Figure 5. RUNX2-I regulates EMT independently of SNAI2.
qPCR analysis of EMT-related transcription factors and receptors in stage 14 (A, C) and 16 (B, D) control and RUNX2 siRNA (A-B) and SNAI2 siRNA (C-D) transfected explants cultured for 16h. A. In cultured stage 14 explants, RUNX2 downregulation (~30%) caused a significant decrease in SNAI1 (~47%), ZEB1 (~42%), ZEB2 (~10%), Twist1 (~34%), FLK-1 (~70%) and FLT-1 (~40%). SNAI2 was not significantly changed in treated explants. B. In cultured stage 16 explants, RUNX2 downregulation (~25%) caused a significant decrease in SNAI2 (~25%), ZEB1 (~10%) and FLK-1 (~27%) and a significant increase in TWIST1 (~1.25-fold increase). All other markers did not show significant changes. C. In cultured stage 14 explants, SNAI2 downregulation (~25%) produced a significant decrease in the levels of ZEB1 (~25%) and ZEB2 (~40%). SNAIL1 was significantly increased after treatment (~1.4 fold). All other genes were unchanged in treated explants. D. In cultured stage 16 explants, SNAI2 downregulation (~40%) caused a significant decrease in ZEB2 (~30%) and a significant increase in TWIST1 (~1.4-fold) and FLK-1 (~1.5-fold). All other markers did not show significant changes. Error bars = S.E.M.; * p < 0.05; ** p < 0.01; *** p < 0.001; n.s. – non-significant.
Next, the changes in RUNX2 siRNA-transfected explants were compared to changes in SNAI2 siRNA-transfected explants (Fig. 5C-D). SNAI2 is a known mediator of TGFβ2 signaling in the activation step of the chick heart EMT (Romano and Runyan, 1999; Romano and Runyan, 2000). SNAI2 siRNA-transfected stage 14 explants (Fig. 5C, ~25% decrease in SNAI2, p < 0.001) presented a significant decrease in ZEB1 (~25% decrease, p < 0.001) and ZEB2 (~40% decrease, p < 0.001) while SNAI1 expression was increased (~1.4-fold increase, p < 0.01). RUNX2 (p=0.053), TWIST1 (p=0.431), FLK-1 (p=0.408) and FLT-1 (p=0.395) were unchanged. SNAI2 knock down (~40% decrease, p < 0.001) in stage 16 transfected explants caused significant changes in ZEB2 (~30% decrease, p < 0.001), TWIST1 (~1.4-fold increase, p < 0.01) and FLK-1 (~1.5-fold increase, p < 0.001). Other markers were unchanged (RUNX2, p=0.061; ZEB1, p=0.172; SNAI, p=0.683; FLT-1, p=0.193). Taken together, these data argue that RUNX2 functions independently of SNAI2 in the chicken AVC EMT activation step.
RUNX2 expression in the progression of esophageal carcinoma is consistent with an early role in pathological EMT
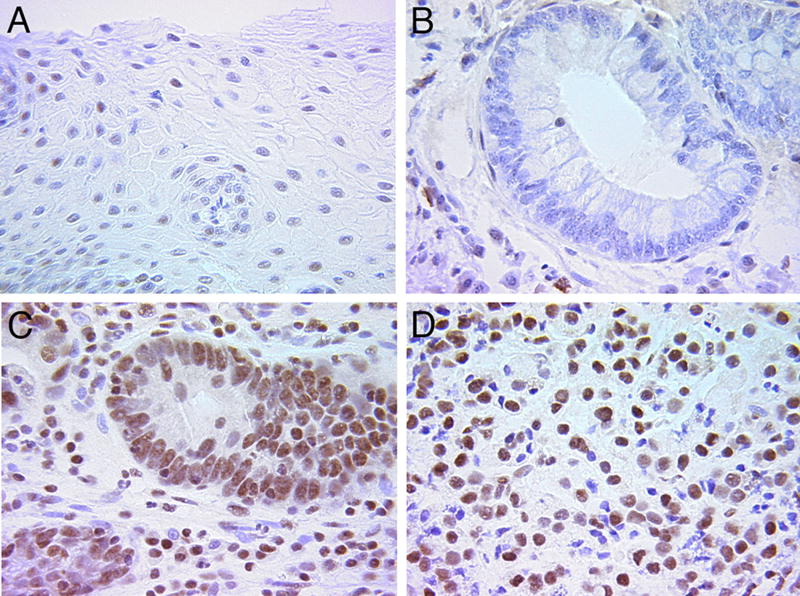
RUNX2 is expressed in breast, prostate and thyroid cancers, functioning to promote EMT and invasion (Baniwal et al., 2010; Chimge et al., 2011; Mendoza-Villanueva et al., 2011; Niu et al., 2012). Because esophageal adenocarcinomas are reported to involve TGFβ signaling (Onwuegbusi et al., 2006; von Rahden et al., 2006), RUNX2 expression was examined in staged human esophageal tissues in the transition from normal squamous epithelium (Fig. 6A) to metastatic esophageal adenocarcinoma (Fig. 6D). RUNX2 expression appears coincident with the transition from columnar epithelia observed in Barrett’s Esophagus (Fig. 6B) to high-grade dysplasia (Fig. 6C). RUNX2 expression persisted in esophageal adenocarcinoma samples (Fig. 6D), suggesting a role for RUNX2 in EMT and invasiveness.
Figure 6. Expression of RUNX2 in staged esophageal adenocarcinoma materials.
A. RUNX2 is not expressed in normal esophageal squamous epithelium from a control patient. B. Columnar esophageal epithelium from a patient with Barrett’s Esophagus does not show RUNX2 expression. C. RUNX2 is highly expressed in esophageal tissue from a patient with high-grade dysplasia. D. Esophageal adenocarcinoma continues to express high levels of RUNX2. RUNX2 positive staining: brown color. Sections were counterstained with H&E.
DISCUSSION
While it is well-known RUNX2 has important roles during osteoblast differentiation in skeletal development, the roles of the individual isoforms are not well understood. Most of the literature on RUNX2 also ignores potential isoform differences and their functions in non-osteogenic tissues of the body. A report showing the expression of RUNX2 in mesenchymal cells of the AV canal in the developing mouse heart was intriguing (Gitler et al., 2003). Based upon communication with the original provider of the published probe, the initial report likely showed localization of POSTN rather than RUNX2. POSTN (periostin) was previously named OSF-2 while RUNX2 was OSF2 and periostin was shown to be expressed in cardiac mesenchymal cells (Kruzynska-Frejtag et al., 2001). However, we found expression of authentic RUNX2 in the chick by rtPCR (Tavares et al., 2006; Mercado-Pimentel and Runyan, 2007). These data prompted us to explore RUNX2’s role in embryonic heart development.
Our current model of EMT by embryonic cardiac endothelia divides the process into at least two separate steps. The first step is an activation process by an extracellular stimulus. This activation stage is characterized by cellular hypertrophy and cell separation (Markwald et al., 1977), a change in cellular polarization (Krug et al., 1985), and an intracellular calcium flux (Runyan et al., 1990). As shown by experiments that block activation, this stage is induced, at least in part, by TGFβ2 and mediated by TBRIII (Brown et al., 1999; Boyer and Runyan, 2001). The SNAI2 transcription factor appears to be involved in EMT at this stage (Romano and Runyan, 2000). Staining data in Figure 3 show that both pSMAD 1/5/8 and pSMAD 2/3 appear to be active at this stage of heart development. The data show that RUNX2-1 expression is unaffected by BMP-2 or Noggin signaling. We note that the ALK-2 receptor is active during cardiac EMT and that ALK-2 binds both TGFβ and BMP ligands and can signal via SMAD1/5 (Mercado-Pimentel and Runyan, 2007; Derynck and Feng, 1998; Daly et al., 2008). The staining pattern may indicate that TGFβ is signaling via both the SMAD 2/3 and SMAD 1/5/8 pathways.
A separate and second step of EMT is characterized by invasion of mesenchymal cells derived from activated endothelial cells into the underlying matrix. Inhibition of TGFβ3 (Potts et al., 1991; Boyer et al., 1999a), TBRII (Brown et al., 1996; Boyer and Runyan, 2001), ADGRL2 (LPHN2, (Doyle et al., 2006), and Olfactomedin-1 (Lencinas et al., 2013) all show normal activation but a loss of cell invasion by explanted tissues in vitro. The invasion step requires elements of cytoskeletal change, alteration of cell adhesion, expression of proteases and secretion of extracellular matrix proteins whose coordination is unclear (Person et al., 2005; Combs and Yutzey, 2009). While all atrioventricular canal endothelial cells appear to be activated, only 7–10% of the cells complete EMT and become invasive. We suggest that completion of EMT may require passage through one or more checkpoints and that incomplete EMT may account for the concept of epithelial plasticity in EMT (Lencinas et al., 2013).
Our present findings indicate that RUNX2-I is the only RUNX2 isoform present in the embryonic heart at the time of EMT and is required for this process to properly initiate after TGFβ2 induction (Fig. 7), as seen in RUNX2 siRNA treated AV canals. The observation that RUNX2-I is significantly down-regulated when treated with anti-TGFβ2, but not by anti-TGFβ3, also supports that RUNX2-I expression is correlated with the activation step of EMT rather than the invasion step (Boyer et al., 1999a). This is also consistent with the loss of normal cell-cell separation observed with the activation step of EMT (Figure 3). When TGFβ3 is added to cultures we see a small increase in RUNX2 mRNA expression, but it is not as strong as when TGFβ2 is added. It is known that differences in potency of exogenous TGFβs can reflect isoform specificity in vivo but that all isoforms can produce a response in vitro (Roberts et al., 1990). These data suggest that, in the heart, TGFβ2 is the principal mediator of RUNX2-I expression.
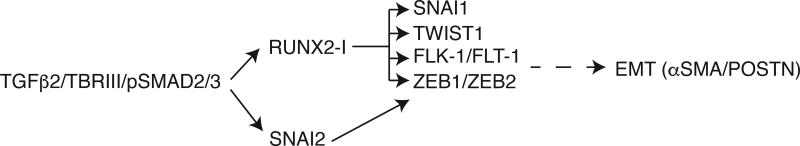
Figure 7. RUNX2-I functions in the chick AVC EMT activation step.
In the chick heart AVC, TGFβ2 binds to TBRIII expressed by endothelial cells. This signaling pathway through pSMAD2/3 turns on the expression of RUNX2-I and SNAI2 (Romano and Runyan, 2000), two factors required for EMT activation. RUNX2-I induces the expression of EMT-related transcription factors and VEGF receptors independently of SNAI2 with the exception of ZEB1 and ZEB2. Expression of these molecules is followed by delamination of endothelial cells and expression of αSMA and POSTN.
Downregulation of RUNX2-I and reduced activation of AV canal endothelia after treatment with RUNX2 siRNA show that this factor is required for proper EMT. Quantification of activated endothelial cell and mesenchymal cell numbers confirmed this requirement. The observation that EMT was not completely blocked and a few mesenchymal cells could be observed invading the gel in treated explants can be explained by either compensation by other factors or, more likely, by the observation that siRNA treatments reduce mRNA levels with variable efficiency and may cause partial phenotypes (Kurreck, 2006). Though we observed somewhat variable levels of RUNX2 mRNA downregulation between experiments, this downregulation always disrupted EMT and produced changes in marker gene expression. qPCR analysis of selected EMT markers showed downregulation of αSMA and POSTN. Expression of αSMA has been a consistent marker of activation and mesenchymal cell formation. Its expression is detected in endothelial cells after EMT activation (Boyer et al., 1999b) and in endothelial tissues treated with high levels (20 ng/ml) of TGFβ3 (Nakajima et al., 1997). Periostin is an extracellular protein expressed in valvular mesenchymal cells and by metastatic cancer cells (Kruzynska-Frejtag et al., 2001; Inai et al., 2008; Liu et al., 2015). Collagen I is a well-known extracellular matrix component secreted by newly formed mesenchymal cells but is also expressed by embryonic cardiomyocytes during the formation of the cardiac cushions (Sinning et al., 1988; de Vlaming et al., 2012; Hosper et al., 2013). The decrease in αSMA and POSTN levels after RUNX2 downregulation is consistent with the role of these markers in EMT while the lack of significant change in Collagen I may reflect the larger continued synthesis of ECM by myocardial cells.
The finding RUNX2-I is induced by TGFβ2 and therefore functions during EMT initiation led us to investigate changes of EMT markers after RUNX2-I knock down in cultured stage 14 (during EMT initiation) and stage 16 (EMT invasion) explants. Previous in vitro experiments in this lab show that activation elements of EMT predominate in analysis explants collected from stage 14 or 15 embryos and cultured for 24 hr or less. In contrast, invasion components of EMT are predominant in explants derived from stage 16 and 17 chicks and cultured overnight or longer (Potts and Runyan, 1989, Boyer et al., 1999a; Mercado et al., 2006; Lencinas et al., 2013). Zinc-finger and bHLH transcription factors are upregulated in various EMT models and their expression correlates with downregulation of E-cadherin or VE-cadherin prior to cell delamination (Romano and Runyan, 1999; Bolos et al., 2003; Vandewalle et al., 2009; Montserrat et al., 2011). Therefore, EMT inhibition could potentially cause downregulation of these factors. RUNX2 inhibition in stage 14 explants caused downregulation of four EMT-related transcription factors (SNAI1, ZEB1, ZEB2 and TWIST1), whereas inhibition in stage 16 caused downregulation only of SNAI2 and ZEB1. The stronger inhibition at the earlier stage of EMT is consistent with a role for RUNX2-I in EMT initiation or activation (around stage 14) by TGFβ2 (Boyer et al., 1999a).
That SNAI2 is also a transcription factor that is TGFβ2-regulated and required for proper EMT (Romano and Runyan, 1999; Romano and Runyan, 2000), led us to compare the effects of RUNX2-I downregulation to SNAI2 downregulation. We observed that these two transcription factors are regulating different aspects of EMT after TGFβ2 induction. As expected, SNAI2 knockdown caused changes that were more correlated to EMT inhibition (ZEB1 and ZEB2 downregulation) in stage 14 explants (when activation is predominant) rather than in stage 16 explants (when invasion is predominant). This is consistent with both the knockdown of RUNX2-I observed here and SNAI2’s role in the activation step (Romano and Runyan, 1999; Romano and Runyan, 2000). We observed an increase in SNAI1 after SNAI2 downregulation in stage 14 explants while RUNX2-I knockdown caused a decrease in SNAI1 levels. Additionally, TWIST1 was downregulated after RUNX2-I inhibition in stage 14 explants but upregulated after both RUNX2-I and SNAI2 knockdown in stage 16 explants. This likely represents a change in TWIST1 regulation after EMT has been initiated in activated endothelia and mesenchymal cells are invading. Thus, while many of the EMT-associated transcription factors are found in the heart, the interactions between them clearly need more examination.
The most drastic effect was observed in the levels of FLK-1 (VEGR2) and FLT-1 (VEGR1), endothelial markers shown to be up-regulated and down-regulated, respectively, in response to EMT (Stankunas et al., 2010). RUNX2 siRNA treatment of stage 14 explants caused downregulation of both molecules whereas SNAI2 siRNA treatment produced no changes at the same stage. FLK-1 downregulation continued in stage 16 explants after RUNX2 inhibition showing that RUNX2 is required for induction of this receptor during both activation and invasion steps of EMT. Additionally, SNAI2 inhibition in stage 16 explants caused upregulation of FLK-1. Finally, while FLK-1 is behaving in agreement with the pattern previously described, FLT-1 is not (Stankunas et al., 2010). This may mean that the ratio of these markers to one another is important to note, when determining if cells have undergone EMT. Further experiments are underway to explore the utility of FLK-1/FLT-1 ratios as markers of the transition to mesenchyme.
The role of RUNX2 in EMT is not widely explored but may be conserved in both developmental and metastatic EMTs. A developmental role for an ortholog of RUNX2, Zebrafish RUNX2a, is suggested by its localization in neural crest and somites of early embryos, both sites of EMT (Flores et al. 2004). Expression of RUNX2 during the progression from Barrett’s esophagus to dysplasia is consistent with the idea that RUNX2-I may have a common role in the EMT in both metastasis and development at an early or “activation” stage. Progression of esophageal cancer is mediated by TGFβ (von Rahden et al., 2006). Barrett’s esophagus is a metaplasia of the lower esophagus associated with reflux that can progress (0.5% per patient year) to dysplasia and adenocarcinoma. RUNX2 staining is observed in dysplasia and thus appears to be associated with an early stage of cancer progression and may be similarly regulated by TGFβ expression in the tissue. Though not tied to TGFβ signaling, loss of RUNX2 in thyroid carcinoma was associated with loss of a broad spectrum of EMT markers (Niu et al., 2012). RUNX2 expression appears to be linked to a variety of cancers (Voon and Thiery, 2017).
In summary, we find the 2-I isoform of RUNX2 to be specifically expressed in the developing heart and show that it has a role in the activation stage of endothelial EMT as loss of expression inhibits the cell separation characteristic of this stage. It appears to be regulated by the TGFβ2 isoform that is also associated with activation of EMT in the heart. Though it is not clear whether they are direct or indirect targets, loss of RUNX2-I inhibits the expression of the mesenchymal markers, POSTN and α-SMA. Though coincident in expression and regulation by TGFβ2, RUNX2-I is not redundant with SNAI2 in its regulation of the various EMT markers and the activation of EMT. Though Runx2 has been previously observed in cancer, an isoform-specific function of RUNX2 in EMT suggests that this isoform may be linked to metastatic EMT rather than the subsequent progression of cancer to bone tissue.
EXPERIMENTAL PROCEDURES
Embryos
Fertilized White Leghorn eggs were obtained from Texas A & M University, Department of Poultry Sciences and incubated at 37.5°C.
Immunofluorescent Microscopy
Chicken embryos were harvested in Tyrode’s solution at stages 14 to 20 (Hamburger and Hamilton, 1951) and at embryonic day (E)13. The heart region and forelimb of collected embryos were dissected and fixed in solution of −70°C 80% methanol / 20% DMSO and cryosubstituted at −20°C for at least one week. Tissues were then embedded in paraffin and sectioned at 7µm. Sections containing the AV canal region of the heart were processed using an indirect immunofluorescence technique. A mouse monoclonal primary antibody against recombinant RUNX2 was used (Santa Cruz Biotechnology). Specificity of this antibody against chick RUNX2 was tested by western blot (data not shown). Briefly, after deparaffinization, the sections were rinsed in 1X PBS for 10 min, blocked for 1 hour at room temperature with a blocking solution containing 1% bovine serum albumin and 0.1% Tween 20 in PBS and incubated overnight with the primary antibodies at −4°C in a moist chamber. After several rinses in 1X PBS, Alexa fluor 488 or 546-conjugated rabbit anti-mouse secondary antibody (ThermoFisher Scientific) were incubated for 1 hour at room temperature in moist chamber. After rinsing in PBS, the nuclei were stained with TO-PRO-3 (ThermoFisher Scientific) and the sections mounted using Prolong Gold mounting media (ThermoFisher Scientific). The sections were analyzed using a LSM 510 Zeiss Confocal Microscope.
For the in vitro localization of RUNX2 and pSMADs, AVC endocardial cushion explants of stage 17 chicken embryos were collected and incubated for 24 hours (37°C, 5% CO2) with complete medium 199 (1X Medium 199 (Sigma-Aldrich) + Antibiotic/Antimicotic (ThermoFisher Scientific) + ITS Premix (5 µg/ml insulin, 5 µg/ml transferrin, and 5µg/ml selenium – ThermoFisher Scientific) on a type I collagen gel as described previously (Potts and Runyan, 1989). Gels were fixed with 4% paraformaldehyde for 25 min at room temperature, rinsed with 1X PBS for 1 hour at room temperature and processed for RUNX2 and pSmad1/5/8 or pSmad2/3 immunofluorescence microscopy with the antibodies described above and rabbit polyclonal anti-pSmad1/5/8 (Abcam) and rabbit polyclonal anti-pSmad2/3 (Abcam). Nuclei were stained with DAPI (Sigma-Aldrich) and F-actin with Alexa Fluor 488 or 647 phalloidin (ThermoFisher Scientific). Gels were analyzed using a DeltaVision Deconvolution Microscope.
RNA isolation, RT-PCR and Quantitative Real-Time PCR analyses
Total RNA was extracted from isolated E13 chick wings, stage 17 chick embryos, microdissected stage 17 hearts or from incubated explants on gels using the PureLink RNA Mini kit (ThermoFisher Scientific). 1µg of RNA was used from samples to make cDNA using the qScript cDNA Synthesis kit (Quanta Biosciences). Semi-quantitative analysis of RUNX2 isoforms was performed by PCR amplification (30 cycles) using primers designed to specifically identify type I RUNX2 (FW: 5’-TGCGTATTCCCGTAGATCC-3’; RV: 5’-TTAGGAGAAGTGCCCGATGGGA-3’) or type II RUNX2: (FW: 5’-GCATCAAACAGCCTCTTCAGCTC-3’; RV: 5’-TTAGGAGAAGTGCCCGATGGGA-3’). GAPDH was used as loading control. PCR products were analyzed on 1% agarose gels. Quantitative Real-time PCR was performed using the FastStart SYBR Green Master (Sigma-Aldrich) on RotorGene 3000 PCR machine (Corbett Research). Equal amounts of cDNA were added for each reaction, and in each experiment triplicate reactions were performed for each gene to be analyzed. Quantitative data were normalized with HPRT and β-actin. Experiments were repeated three times and results analyzed with Rotor Gene 6 software (Corbett Research) using the comparative quantitation method and Student’s t Test. Primer sequences used in these experiments are shown in Table 1.
Table 1.
qPCR primers used for measurement of EMT markers
| Primer | Forward | Backward |
|---|---|---|
| αSMA | 5’-AGGACAGCACTGCCCTTGTTT-3’ | 5’-CCCATACCAACCATCACACCCT-3’ |
| β-actin | 5’-CCCCGTGCTGTGTTCCCATCTATCG-3’ | 5’-TGATTTTCATTGTGCTAGGTGCCAGG-3’ |
| COL1a1 | 5’-TGTGGATTCTCGGTTACTGCTGTT-3’ | 5’-ATCTGGCACGGTTCGGGTTT-3’ |
| FLK-1 | 5’-CAGCCCTGCGCCTCTTGTGT-3’ | 5’-GGCAAGGAAGGCACATCCCGG-3’ |
| FLT-1 | 5’-CCCAACGAGCGGCCATGGTT-3’ | 5’-CGCAGGGGGAAACCCGCTTT-3’ |
| HPRT | 5’-TGTTGTTGGATACGCCCTCG-3’ | 5’-TCTGCTTCCCCGTCTCACTG-3’ |
| POSTN | 5’-CAGTTGCTCACAATCCTGAAGAAGC-3’ | 5’-TCCCCCTCCCTGACTATTTTGAAC-3’ |
| RUNX2 | 5’-GCGAAATGCTTCTGCTGTGA-3’ | 5’-AGTCGCTACATACCACAGAGC-3’ |
| SNAI1 | 5’-TTGCACTGCGATGCTCAGACCA-3’ | 5’-TCAGCAAAGGCCCGGTTGCAAT-3’ |
| SNAI2 | 5’-GAAAGCCTGCCTTCAAAATGC-3’ | 5’-CCACACAGTAATGGGGTTGTAAGC-3’ |
| TWIST1 | 5’-TGGTTGTTGCTGTCGTTTTGG-3’ | 5’-TGGTTGGGTGCTTTGCTTTC-3’ |
| ZEB1 | 5’-TGACTTGCCAACAGACCACACC-3’ | 5’-CTTTTCCTTCATCTTCCCAGCAG-3’ |
| ZEB2 | 5’-GCCTTGAAAAACCTTTGCTACCTG-3’ | 5’TTGCTCTTCGTTCTTGTTCTCCTC-3’ |
Whole-mount in situ hybridization
Stage 18 chick embryos were fixed in 4% PFA overnight at 4°C. Embryos were processed for hybridization per (Nieto et al. (1996) with minor modifications. Digoxigenin-labelled RNA probe was synthesized with T3 RNA polymerase (Sigma-Aldrich) and DIG RNA labeling mix (Sigma-Aldrich). Probe template (434nt) was amplified by PCR with the following primers: FW: 5’-TTATAAAAGCTTGCGGCCGCAGAATAT-GCTGCGAAATGCTTCTGCTG-3’; RV: 5’-GCTCTAGAAATTAACCCTCACTAAAGG-AAGTCATCTGGCTCAAGTAGGACG-3’
In vitro inhibition and stimulation of TGFβ family members
Neutralizing anti-porcine TGFβ2 (20µg/ml, goat IgG, R&D Systems), anti-TGFβ3 (20µg/ml, mouse IgM, Developmental Studies Hybridoma Bank) antibodies, recombinant human Noggin (50ng/ml), porcine TGFβ2 (0.002ng/µl), recombinant human TGFβ3 (0.002ng/µl), recombinant human BMP-2 (0.002ng/µl) (R&D Systems) were diluted in complete medium. Gels were pre-treated with each solution prior to explantation. Control gels were pre-treated with complete medium alone or complete medium + IgG/IgM. Stage 14 AVC explants were dissected and collected in 1X Medium 199. The explants were placed on the pre-treated gels and incubated overnight for adhesion (37°C, 5% CO2). More treatment was added to gels the following morning and cultures were incubated for a total of 24 hours after the explants were initially placed on the gel. Total RNA was extracted as described above. The samples were then processed for Quantitative Real-Time PCR.
Inhibition of RUNX2 and SNAI2 message by siRNA knockdown
Small interfering RNAs (siRNAs) homologous to the 3’ORF of RUNX2 (NM_204128) were designed and synthesized using the Silencer siRNA Construction Kit (ThermoFisher Scientific) or purchased from ThermoFisher Scientific (Silencer Select Custom Designed siRNA). Silencer Select Negative Control was also purchased from ThermoFisher Scientific. The sequences of RUNX2 siRNA templates were: Silencer Select Custom Designed siRNA: GAGCTTTACTTTGACAATA; Silencer siRNA-1: GGACAGAGTCAGATTACAG; Silencer siRNA-2: TTCTAGTGGCAGGATGGAT; SNAI2 siRNA template was: TCTAGGAAATCGTTCAGCTGCAAGTACTGT. siRNAs were diluted in 1X Medium 199 to a concentration of 50nM and transfected using the siPORT NeoFX Transfection Agent (ThermoFisher Scientific). Briefly, stage 14 or 16 explants were collected in 1X Medium 199 and added to medium containing siRNAs. They were incubated for 1h 30 min at 37°C, 5% CO2 and then placed on gels. After an adhesion period (overnight, 37°C, 5% CO2), new transfection solution was added to cultures and the incubation allowed to continue for up to 16h, 24h or 48 hours after the explants were initially placed on the gel. The gels were either processed for RNA extraction as described above or fixed with 4% paraformaldehyde for 25 min at room temperature and endothelial outgrowth from the explant and mesenchymal cell invasion of the collagen gel were qualitatively and quantitatively analyzed using Hoffman Modulation contrast optics. Activated cells, defined as isolated cells on the surface of the gel, and mesenchymal cells defined as cells wholly within the gel matrix, were counted. A total of 14 explants (7 control explants, 7 experimental explants) were examined and the data analyzed with Student’s t test.
Immunohistochemistry
Samples from control patients, patients with Barret’s esophagus, with high-grade dysplasia or with esophageal adenocarcinoma were obtained from the Biobank core at the Cancer Center at the University of Arizona, Tucson – AZ. They were fixed in 4% PFA, paraffin embedded and processed for RUNX2 immunohistochemistry using a mouse monoclonal primary antibody against recombinant RUNX2 (Santa Cruz Biotechnology). Detection was performed with an HRP-conjugated anti-mouse secondary antibody (ThermoFisher Scientific) and DAB (ThermoFisher Scientific). Sections were counterstained with H&E. Samples from 30 patients for each group were analyzed for RUNX2 expression.
Acknowledgments
Funded by NHLBI grant HL082851
References
- Baniwal SK, Khalid O, Gabet Y, Shah RR, Purcell DJ, Mav D, Kohn-Gabet AE, Shi Y, Coetzee GA, Frenkel B. RUNX2 transcriptome of prostate cancer cells: insights into invasiveness and bone metastasis. Mol Cancer. 2010;9:258. doi: 10.1186/1476-4598-9-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolos V, Peinado H, Perez-Moreno MA, Fraga MF, Esteller M, Cano A. The transcription factor Slug represses E-cadherin expression and induces epithelial to mesenchymal transitions: a comparison with Snail and E47 repressors. J Cell Sci. 2003;116:499–511. doi: 10.1242/jcs.00224. [DOI] [PubMed] [Google Scholar]
- Boyer AS, Ayerinskas II, Vincent EB, McKinney LA, Weeks DL, Runyan RB. TGFbeta2 and TGFbeta3 have separate and sequential activities during epithelial-mesenchymal cell transformation in the embryonic heart. Dev Biol. 1999a;208:530–545. doi: 10.1006/dbio.1999.9211. [DOI] [PubMed] [Google Scholar]
- Boyer AS, Erickson CP, Runyan RB. Epithelial-mesenchymal transformation in the embryonic heart is mediated through distinct pertussis toxin-sensitive and TGFbeta signal transduction mechanisms. Dev Dyn. 1999b;214:81–91. doi: 10.1002/(SICI)1097-0177(199901)214:1<81::AID-DVDY8>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Boyer AS, Runyan RB. TGFbeta Type III and TGFbeta Type II receptors have distinct activities during epithelial-mesenchymal cell transformation in the embryonic heart. Dev Dyn. 2001;221:454–459. doi: 10.1002/dvdy.1154. [DOI] [PubMed] [Google Scholar]
- Bronckers AL, Sasaguri K, Cavender AC, D'Souza RN, Engelse MA. Expression of RUNX2/Cbfa1/Pebp2alphaA during angiogenesis in postnatal rodent and fetal human orofacial tissues. J Bone Miner Res. 2005;20:428–437. doi: 10.1359/JBMR.041118. [DOI] [PubMed] [Google Scholar]
- Brown CB, Boyer AS, Runyan RB, Barnett JV. Antibodies to the Type II TGFbeta receptor block cell activation and migration during atrioventricular cushion transformation in the heart. Dev Biol. 1996;174:248–257. doi: 10.1006/dbio.1996.0070. [DOI] [PubMed] [Google Scholar]
- Brown CB, Boyer AS, Runyan RB, Barnett JV. Requirement of type III TGF-beta receptor for endocardial cell transformation in the heart. Science. 1999;283:2080–2082. doi: 10.1126/science.283.5410.2080. [DOI] [PubMed] [Google Scholar]
- Chimge NO, Baniwal SK, Little GH, Chen YB, Kahn M, Tripathy D, Borok Z, Frenkel B. Regulation of breast cancer metastasis by RUNX2 and estrogen signaling: the role of SNAI2. Breast Cancer Res. 2011;13:R127. doi: 10.1186/bcr3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Combs MD, Yutzey KE. Heart valve development: regulatory networks in development and disease. Circ Res. 2009;105:408–421. doi: 10.1161/CIRCRESAHA.109.201566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daly AC, Randall RA, Hill CS. Transforming growth factor beta-induced Smad1/5 phosphorylation in epithelial cells Is mediated by novel receptor complexes and Is essential for anchorage-independent growth. Mol. Cell. Biol. 2008;28:6889–6902. doi: 10.1128/MCB.01192-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vlaming A, Sauls K, Hajdu Z, Visconti RP, Mehesz AN, Levine RA, Slaugenhaupt SA, Hagege A, Chester AH, Markwald RR, Norris RA. Atrioventricular valve development: new perspectives on an old theme. Differentiation. 2012;84:103–116. doi: 10.1016/j.diff.2012.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derynck R, Feng X-H. TGF-b receptor signaling. Biochim Biophys Acta. 1997;1333:F105–F150. doi: 10.1016/s0304-419x(97)00017-6. [DOI] [PubMed] [Google Scholar]
- Doyle SE, Scholz MJ, Greer KA, Hubbard AD, Darnell DK, Antin PB, Klewer SE, Runyan RB. Latrophilin-2 is a novel component of the epithelial-mesenchymal transition within the atrioventricular canal of the embryonic chicken heart. Dev Dyn. 2006;235:3213–3221. doi: 10.1002/dvdy.20973. [DOI] [PubMed] [Google Scholar]
- Flores MV, Tsang VWK, Hu W, Kalev-Zylinska M, Postlewait J, Crosier P, Crosier K, Fisher S. Duplicate zebrafish RUNX2 orthologues are expressed in developing skeletal elements. Gene Express. Patterns. 2004;4:572–581. doi: 10.1016/j.modgep.2004.01.016. [DOI] [PubMed] [Google Scholar]
- Garg V. Molecular genetics of aortic valve disease. Curr Opin Cardiol. 2006;21:180–184. doi: 10.1097/01.hco.0000221578.18254.70. [DOI] [PubMed] [Google Scholar]
- Gitler AD, Lu MM, Jiang YQ, Epstein JA, Gruber PJ. Molecular markers of cardiac endocardial cushion development. Dev Dyn. 2003;228:643–650. doi: 10.1002/dvdy.10418. [DOI] [PubMed] [Google Scholar]
- Glotzer DJ, Zelzer E, Olsen BR. Impaired skin and hair follicle development in RUNX2 deficient mice. Dev Biol. 2008;315:459–473. doi: 10.1016/j.ydbio.2008.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay ED, Zuk A. Transformations between epithelium and mesenchyme: normal, pathological, and experimentally induced. Am J Kidney Dis. 1995;26:678–690. doi: 10.1016/0272-6386(95)90610-x. [DOI] [PubMed] [Google Scholar]
- Hosper NA, van den Berg PP, de Rond S, Popa ER, Wilmer MJ, Masereeuw R, Bank RA. Epithelial-to-mesenchymal transition in fibrosis: collagen type I expression is highly upregulated after EMT, but does not contribute to collagen deposition. Exp Cell Res. 2013;319:3000–3009. doi: 10.1016/j.yexcr.2013.07.014. [DOI] [PubMed] [Google Scholar]
- Inai K, Norris RA, Hoffman S, Markwald RR, Sugi Y. BMP-2 induces cell migration and periostin expression during atrioventricular valvulogenesis. Dev Biol. 2008;315:383–396. doi: 10.1016/j.ydbio.2007.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y, Bae SC, Chuang LS. The RUNX family: developmental regulators in cancer. Nat Rev Cancer. 2015;15:81–95. doi: 10.1038/nrc3877. [DOI] [PubMed] [Google Scholar]
- Ito Y, Miyazono K. RUNX transcription factors as key targets of TGF-beta superfamily signaling. Curr Opin Genet Dev. 2003;13:43–47. doi: 10.1016/s0959-437x(03)00007-8. [DOI] [PubMed] [Google Scholar]
- Jeong JH, Jin JS, Kim HN, Kang SM, Liu JC, Lengner CJ, Otto F, Mundlos S, Stein JL, van Wijnen AJ, Lian JB, Stein GS, Choi JY. Expression of RUNX2 transcription factor in non-skeletal tissues, sperm and brain. J Cell Physiol. 2008;217:511–517. doi: 10.1002/jcp.21524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalluri R, Neilson EG. Epithelial-mesenchymal transition and its implications for fibrosis. J Clin Invest. 2003;112:1776–1784. doi: 10.1172/JCI20530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komori T, Yagi H, Nomura S, Yamaguchi A, Sasaki K, Deguchi K, Shimizu Y, Bronson RT, Gao YH, Inada M, Sato M, Okamoto R, Kitamura Y, Yoshiki S, Kishimoto T. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell. 1997;89:755–764. doi: 10.1016/s0092-8674(00)80258-5. [DOI] [PubMed] [Google Scholar]
- Krug EL, Runyan RB, Markwald RR. Protein extracts from early embryonic hearts initiate cardiac endothelial cytodifferentiation. Dev Biol. 1985;112:414–426. doi: 10.1016/0012-1606(85)90414-2. [DOI] [PubMed] [Google Scholar]
- Kruzynska-Frejtag A, Machnicki M, Rogers R, Markwald RR, Conway SJ. Periostin (an osteoblast-specific factor) is expressed within the embryonic mouse heart during valve formation. Mech Dev. 2001;103:183–188. doi: 10.1016/s0925-4773(01)00356-2. [DOI] [PubMed] [Google Scholar]
- Kurreck J. siRNA efficiency: structure or sequence-that is the question. J Biomed Biotechnol. 2006;2006:83757. doi: 10.1155/JBB/2006/83757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lencinas A, Chhun DC, Dan KP, Ross KD, Hoover EA, Antin PB, Runyan RB. Olfactomedin-1 activity identifies a cell invasion checkpoint during epithelial-mesenchymal transition in the chick embryonic heart. Dis Model Mech. 2013;6:632–642. doi: 10.1242/dmm.010595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lencinas A, Tavares AL, Barnett JV, Runyan RB. Collagen gel analysis of epithelial-mesenchymal transition in the embryo heart: an in vitro model system for the analysis of tissue interaction, signal transduction, and environmental effects. Birth Defects Res C Embryo Today. 2011;93:298–311. doi: 10.1002/bdrc.20222. [DOI] [PubMed] [Google Scholar]
- Li YL, Xiao ZS. Advances in RUNX2 regulation and its isoforms. Med Hypotheses. 2007;68:169–175. doi: 10.1016/j.mehy.2006.06.006. [DOI] [PubMed] [Google Scholar]
- Liu GX, Xi HQ, Sun XY, Wei B. Role of periostin and its antagonist PNDA-3 in gastric cancer metastasis. World J Gastroenterol. 2015;21:2605–2613. doi: 10.3748/wjg.v21.i9.2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Lu MF, Schwartz RJ, Martin JF. Bmp2 is essential for cardiac cushion epithelial-mesenchymal transition and myocardial patterning. Development. 2005;132:5601–5611. doi: 10.1242/dev.02156. [DOI] [PubMed] [Google Scholar]
- Markwald RR, Fitzharris TP, Manasek FJ. Structural development of endocardial cushions. Am J Anat. 1977;148:85–119. doi: 10.1002/aja.1001480108. [DOI] [PubMed] [Google Scholar]
- Markwald RR, Runyan RB, Kitten GT, Funderburg FM, Bernanke DH, Brauer PR. Use of collagen gel cultures to study heart development: proteoglycan and glycoprotein interactions during formation of endocardial cushion tissue. In: Trelstad RL, editor. The role of extracellular matrix in development. New York: Alan R. Liss. Inc.; 1984. pp. 323–350. [Google Scholar]
- Mendoza-Villanueva D, Zeef L, Shore P. Metastatic breast cancer cells inhibit osteoblast differentiation through the RUNX2/CBFbeta-dependent expression of the Wnt antagonist, sclerostin. Breast Cancer Res. 2011;13:R106. doi: 10.1186/bcr3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercado-Pimentel ME, Hubbard AD, Runyan RB. Endoglin and Alk5 regulate epithelial-mesenchymal transformation during cardiac valve formation. Devel. Biol. 2006;304:420–432. doi: 10.1016/j.ydbio.2006.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercado-Pimentel ME, Runyan RB. Multiple transforming growth factor-beta isoforms and receptors function during epithelial-mesenchymal cell transformation in the embryonic heart. Cells Tissues Organs. 2007;185:146–156. doi: 10.1159/000101315. [DOI] [PubMed] [Google Scholar]
- Montserrat N, Gallardo A, Escuin D, Catasus L, Prat J, Gutierrez-Avigno FJ, Peiro G, Barnadas A, Lerma E. Repression of E-cadherin by SNAIL, ZEB1, and TWIST in invasive ductal carcinomas of the breast: a cooperative effort? Hum Pathol. 2011;42:103–110. doi: 10.1016/j.humpath.2010.05.019. [DOI] [PubMed] [Google Scholar]
- Nakajima Y, Mironov V, Yamagishi T, Nakamura H, Markwald RR. Expression of smooth muscle alpha-actin in mesenchymal cells during formation of avian endocardial cushion tissue: a role for transforming growth factor beta3. Dev Dyn. 1997;209:296–309. doi: 10.1002/(SICI)1097-0177(199707)209:3<296::AID-AJA5>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- Nieto MA. The ins and outs of the epithelial to mesenchymal transition in health and disease. Annu Rev Cell Dev Biol. 2011;27:347–376. doi: 10.1146/annurev-cellbio-092910-154036. [DOI] [PubMed] [Google Scholar]
- Nieto MA, Patel K, Wilkinson DG. In situ hybridization analysis of chick embryos in whole mount and tissue sections. Methods Cell Biol. 1996;51:219–235. doi: 10.1016/s0091-679x(08)60630-5. [DOI] [PubMed] [Google Scholar]
- Niu DF, Kondo T, Nakazawa T, Oishi N, Kawasaki T, Mochizuki K, Yamane T, Katoh R. Transcription factor RUNX2 is a regulator of epithelial-mesenchymal transition and invasion in thyroid carcinomas. Lab Invest. 2012;92:1181–1190. doi: 10.1038/labinvest.2012.84. [DOI] [PubMed] [Google Scholar]
- Okura H, Sato S, Kishikawa S, Kaneto S, Nakashima T, Yoshida N, Takayanagi H, Kiyono H. RUNX2-I isoform contributes to fetal bone formation even in the absence of specific N-terminal amino acids. PLoS One. 2014;9:e108294. doi: 10.1371/journal.pone.0108294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onwuegbusi BA, Aitchison A, Chin SF, Kranjac T, Mills I, Huang Y, Lao-Sirieix P, Caldas C, Fitzgerald RC. Impaired transforming growth factor beta signalling in Barrett's carcinogenesis due to frequent SMAD4 inactivation. Gut. 2006;55:764–774. doi: 10.1136/gut.2005.076430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Person AD, Klewer SE, Runyan RB. Cell biology of cardiac cushion development. Int Rev Cytol. 2005;243:287–335. doi: 10.1016/S0074-7696(05)43005-3. [DOI] [PubMed] [Google Scholar]
- Potts JD, Dagle JM, Walder JA, Weeks DL, Runyan RB. Epithelial-mesenchymal transformation of embryonic cardiac endothelial cells is inhibited by a modified antisense oligodeoxynucleotide to transforming growth factor beta 3. Proc Natl Acad Sci U S A. 1991;88:1516–1520. doi: 10.1073/pnas.88.4.1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potts JD, Runyan RB. Epithelial-mesenchymal cell transformation in the embryonic heart can be mediated, in part, by transforming growth factor beta. Dev Biol. 1989;134:392–401. doi: 10.1016/0012-1606(89)90111-5. [DOI] [PubMed] [Google Scholar]
- Pratap J, Javed A, Languino LR, van Wijnen AJ, Stein JL, Stein GS, Lian JB. The RUNX2 osteogenic transcription factor regulates matrix metalloproteinase 9 in bone metastatic cancer cells and controls cell invasion. Mol Cell Biol. 2005;25:8581–8591. doi: 10.1128/MCB.25.19.8581-8591.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts AB, Kondaiah P, Rosa F, Watanabe S, Good P, Danielpour D, Roche NS, Rebbert ML, Dawid IB, Sporn MB. Mesoderm induction in Xenopus laevis distinguishes between the various TGF-beta isoforms. Growth Factors. 1990;3:277–286. doi: 10.3109/08977199009003670. [DOI] [PubMed] [Google Scholar]
- Romano LA, Runyan RB. Slug is a mediator of epithelial-mesenchymal cell transformation in the developing chicken heart. Dev Biol. 1999;212:243–254. doi: 10.1006/dbio.1999.9339. [DOI] [PubMed] [Google Scholar]
- Romano LA, Runyan RB. Slug is an essential target of TGFbeta2 signaling in the developing chicken heart. Dev Biol. 2000;223:91–102. doi: 10.1006/dbio.2000.9750. [DOI] [PubMed] [Google Scholar]
- Runyan RB, Potts JD, Sharma RV, Loeber CP, Chiang JJ, Bhalla RC. Signal transduction of a tissue interaction during embryonic heart development. Cell Regul. 1990;1:301–313. doi: 10.1091/mbc.1.3.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinning AR, Lepera RC, Markwald RR. Initial expression of type I procollagen in chick cardiac mesenchyme is dependent upon myocardial stimulation. Dev Biol. 1988;130:167–174. doi: 10.1016/0012-1606(88)90423-x. [DOI] [PubMed] [Google Scholar]
- Stankunas K, Ma GK, Kuhnert FJ, Kuo CJ, Chang CP. VEGF signaling has distinct spatiotemporal roles during heart valve development. Dev Biol. 2010;347:325–336. doi: 10.1016/j.ydbio.2010.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stricker S, Fundele R, Vortkamp A, Mundlos S. Role of RUNX genes in chondrocyte differentiation. Dev Biol. 2002;245:95–108. doi: 10.1006/dbio.2002.0640. [DOI] [PubMed] [Google Scholar]
- Tavares AL, Mercado-Pimentel ME, Runyan RB, Kitten GT. TGF beta-mediated RhoA expression is necessary for epithelial-mesenchymal transition in the embryonic chick heart. Dev Dyn. 2006;235:1589–1598. doi: 10.1002/dvdy.20771. [DOI] [PubMed] [Google Scholar]
- Terry A, Kilbey A, Vaillant F, Stewart M, Jenkins A, Cameron E, Neil JC. Conservation and expression of an alternative 3' exon of RUNX2 encoding a novel proline-rich C-terminal domain. Gene. 2004;336:115–125. doi: 10.1016/j.gene.2004.04.015. [DOI] [PubMed] [Google Scholar]
- Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- Vandewalle C, Van Roy F, Berx G. The role of the ZEB family of transcription factors in development and disease. Cell Mol Life Sci. 2009;66:773–787. doi: 10.1007/s00018-008-8465-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitolo MI, Anglin IE, Mahoney WM, Jr, Renoud KJ, Gartenhaus RB, Bachman KE, Passaniti A. The RUNX2 transcription factor cooperates with the YES-associated protein, YAP65, to promote cell transformation. Cancer Biol Ther. 2007;6:856–863. doi: 10.4161/cbt.6.6.4241. [DOI] [PubMed] [Google Scholar]
- von Rahden BH, Stein HJ, Feith M, Puhringer F, Theisen J, Siewert JR, Sarbia M. Overexpression of TGF-beta1 in esophageal (Barrett's) adenocarcinoma is associated with advanced stage of disease and poor prognosis. Mol Carcinog. 2006;45:786–794. doi: 10.1002/mc.20259. [DOI] [PubMed] [Google Scholar]
- Voon DC, Thiery JP. The Emerging Roles of RUNX Transcription Factors in Epithelial-Mesenchymal Transition. Adv Exp Med Biol. 2017;962:471–489. doi: 10.1007/978-981-10-3233-2_28. [DOI] [PubMed] [Google Scholar]
- Yamagishi T, Nakajima Y, Miyazono K, Nakamura H. Bone morphogenetic protein-2 acts synergistically with transforming growth factor-beta3 during endothelial-mesenchymal transformation in the developing chick heart. J Cell Physiol. 1999;180:35–45. doi: 10.1002/(SICI)1097-4652(199907)180:1<35::AID-JCP4>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Yang J, Weinberg RA. Epithelial-mesenchymal transition: at the crossroads of development and tumor metastasis. Dev Cell. 2008;14:818–829. doi: 10.1016/j.devcel.2008.05.009. [DOI] [PubMed] [Google Scholar]
- Yang X, Meng X, Su X, Mauchley DC, Ao L, Cleveland JC, Jr, Fullerton DA. Bone morphogenic protein 2 induces RUNX2 and osteopontin expression in human aortic valve interstitial cells: role of Smad1 and extracellular signal-regulated kinase 1/2. J Thorac Cardiovasc Surg. 2009;138:1008–1015. doi: 10.1016/j.jtcvs.2009.06.024. [DOI] [PubMed] [Google Scholar]
- Zhang S, Xiao Z, Luo J, He N, Mahlios J, Quarles LD. Dose-dependent effects of RUNX2 on bone development. J Bone Miner Res. 2009;24:1889–1904. doi: 10.1359/JBMR.090502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao FQ, Sheng ZM, Tsai MM, Hubbs AE, Wang R, O'Leary TJ, Izon DJ, Taubenberger JK. Serial analysis of gene expression in murine fetal thymocyte cell lines. Int Immunol. 2002;14:1383–1395. doi: 10.1093/intimm/dxf103. [DOI] [PubMed] [Google Scholar]