Abstract
Low water availability is the major environmental factor limiting growth and productivity of plants and crops and is therefore considered of high importance for agriculture affected by climate change. Identifying regulatory components controlling the response and tolerance to drought stress is thus of major importance. The NAC transcription factor (TF) JUNGBRUNNEN1 (JUB1) from Arabidopsis thaliana extends leaf longevity under non-stress growth conditions, lowers cellular hydrogen peroxide (H2O2) level, and enhances tolerance against heat stress and salinity. Here, we additionally find that JUB1 strongly increases tolerance to drought stress in Arabidopsis when expressed from both, a constitutive (CaMV 35S) and an abiotic stress-induced (RD29A) promoter. Employing a yeast one-hybrid screen we identified HD-Zip class I TF AtHB13 as an upstream regulator of JUB1. AtHB13 has previously been reported to act as a positive regulator of drought tolerance. AtHB13 and JUB1 thereby establish a joint drought stress control module.
Keywords: Arabidopsis, transcription factor, drought, JUB1, HB13
Introduction
Low water availability is the major environmental factor limiting growth and productivity in plants and crops. Global changes of climate will probably reduce the availability of water even more in a larger part of the world (Hamdy et al., 2003) increasing the need for drought tolerant crops. About 80–95% of the plant’s biomass is water and, thus, water is vital for plant growth and development. Soil water is taken up by the roots and transported through the xylem to leaves for various physiological processes including photosynthesis (Fang and Xiong, 2015). Eventually, water returns back to the atmosphere via transpiration.
Homeodomain-leucine zipper (HD-Zip) proteins are plant-specific transcription factors (TFs) encoded by 47 genes in Arabidopsis thaliana; they fall into four distinct groups defined by their primary sequences (HD-Zip I–IV). HD-Zip proteins have two functional domains: a homeodomain (HD) for DNA binding and a leucine zipper (Zip) domain located C-terminal to the HD and involved in protein–protein interactions (for homo- or heterodimerization). HD-Zip proteins participate in a variety of developmental processes and are involved in the plant’s response to environmental factors (Ariel et al., 2007; Harris et al., 2011).
In Arabidopsis, the HD-Zip class I family includes seventeen members with proteins harboring a well conserved HD domain and a less conserved Zip motif. HD-Zip I TFs typically bind to the dyad-symmetric sequence CAAT(A/T)ATTG (Palena et al., 1999; Johannesson et al., 2001; Capella et al., 2015), and they are mainly involved in the response to abiotic stresses, abscisic acid (ABA) and blue light treatment, and they affect seedling de-etiolation (Perotti et al., 2017).
Members of the HD-Zip I fall into six different clades, α to φ (Henriksson et al., 2005). γ-Clade HD-Zip I TFs are typically induced by ABA treatment and/or water deficit, and include Arabidopsis AtHB7 and AtHB12, sunflower (Helianthus annuus) HaHB4, Medicago truncatula MtHB1, Nicotiana attenuata NaHD20, and rice OsHOX6 (Oryza sativa Homeobox 6), OsHOX22, and OsHOX24 (Harris et al., 2011). Experimental evidence shows that AtHB12 reduces growth during water deficit by inhibiting GA biosynthesis and thereby cell elongation (Son et al., 2010). Similarly, growth is reduced in AtHB7 overexpressors, although no evidence for an involvement of GA was reported (Hjellström et al., 2003). Expression of the two paralogous genes is regulated in a coordinated manner, depending on the developmental stage of the plant and the environmental conditions (Ré et al., 2014).
Expression of the β-clade members AtHB5 and AtHB6 is also affected by water deficit and both genes appear to regulate growth in response to ABA treatment and/or water limitation (Söderman et al., 1999; Himmelbach et al., 2002; Henriksson et al., 2005). Furthermore, δ-clade genes AtHB21, AtHB40, and AtHB53 are induced by ABA treatment and salinity stress; the three TFs are involved in controlling axillary bud development (González-Grandío et al., 2017).
Arabidopsis AtHB13, an α-clade HD-Zip I TF, is upregulated by low temperature, drought, and salinity, similar to its sunflower homologue HaHB1. Overexpression of both genes confers tolerance to these stresses which involves the stabilization of the cell membrane (Cabello et al., 2012; Cabello and Chan, 2012). Plants overexpressing AtHB13 or HaHB1 achieve an improved yield under normal and mild stress conditions suggesting both TFs may be employed as tools for establishing crops with enhanced tolerance to multiple stresses and increased yield (Cabello and Chan, 2012; Silva et al., 2016). Recently, AtHB13 and its paralog AtHB23 were shown to negatively affect stem elongation (Ribone et al., 2015).
TFs of the plant-specific NAC (NAM/ATAF/CUC) family play diverse roles in development and stress responses (Shao et al., 2015; Kim et al., 2016) and have been suggested as tools to improve stress tolerance in crops (Tran et al., 2010; Wang et al., 2016). NAC TFs harbor a well conserved, 60-amino acid-long N-terminal DNA-binding domain (NAM domain) and a variable downstream segment through which they interact with other proteins, including other NACs (to form heterodimers) or other regulatory proteins (Olsen et al., 2005).
JUNGBRUNNEN1 (JUB1; ANAC042) is a multifunctional member of the NAC TF family in A. thaliana acting as a negative regulator of senescence and a positive regulator of the tolerance to heat and salinity stress. While JUB1 overexpressor (JUB1Ox) plants are tolerant to both stresses, the jub1-1 knockdown mutant exhibits hypersensitivity (Shahnejat-Bushehri et al., 2012; Wu et al., 2012). JUB1 directly regulates the expression of stress-responsive TFs such as DREB2A and it reduces the cellular levels of reactive oxygen species (ROS), which contributes to the enhanced stress tolerance (Shahnejat-Bushehri et al., 2012; Wu et al., 2012). We and others recently reported that overexpression of JUB1 enhances drought tolerance in both, tomato (Thirumalaikumar et al., 2017) and banana (Tak et al., 2017).
Besides its direct impact on stress regulatory genes, JUB1 also affects growth by negatively and directly regulating genes encoding key enzymes of gibberellin (GA) and brassinosteroid (BR) biosynthesis, namely GA3ox1 and DWF4, respectively, as demonstrated for Arabidopsis (Shahnejat-Bushehri et al., 2016). Furthermore, overexpression of JUB1 leads to the accumulation of DELLA proteins which are master repressors of growth, but enhance stress tolerance (Davière and Achard, 2016; Shahnejat-Bushehri et al., 2016). We furthermore demonstrated that JUB1 exerts a conserved control over GA and BR metabolism and signaling genes in tomato (Shahnejat-Bushehri et al., 2017).
Here, we report JUB1 as a drought-induced gene whose overexpression enhances drought tolerance in Arabidopsis. To unravel the regulatory integration of JUB1 during drought stress we performed a yeast one-hybrid screen using a promoter fragment conferring H2O2- and drought-responsiveness to JUB1, and identified AtHB13 as its upstream transcription regulator. We demonstrate that AtHB13 confers its role in protecting plants against drought stress in part by regulating the expression of JUB1.
Materials and Methods
General
Oligonucleotides (Supplementary Table S1) were obtained from Eurofins MWG Operon (Ebersberg, Germany). Tools provided by the National Center for Biotechnology Information1, the Arabidopsis Information Resource2, the Plant Transcription Factor Database3, and PLAZA 3.04 were used for computational analyses.
Growth Conditions
Arabidopsis thaliana (L.) Heynh. (Col-0) was used as the wild type; transgenic lines are based on this accession. For experiments at seedling stage, seeds were surface sterilized and sown on half-strength Murashige and Skoog (MS) agar medium containing 1% (w/v) sucrose. Plants were grown in soil (Einheitserde GS90; Gebrüder Patzer) under a 12-h day (120 mmol m-2 s-1; 22°C) : 12-h night regime (22°C). For experiments shown in Figures 4, 5 and Supplementary Figures S1, S2 plants were grown at 21°C under a 16-h day and 8-h dark regime. 35S:JUB1 (JUB1Ox), RD29A:JUB1, and jub1-1 (SALK_ID 036474) plants were described previously (Wu et al., 2012). AtHB13Ox plants and athb13 mutants were reported in (Cabello and Chan, 2012; Cabello et al., 2012).
FIGURE 4.
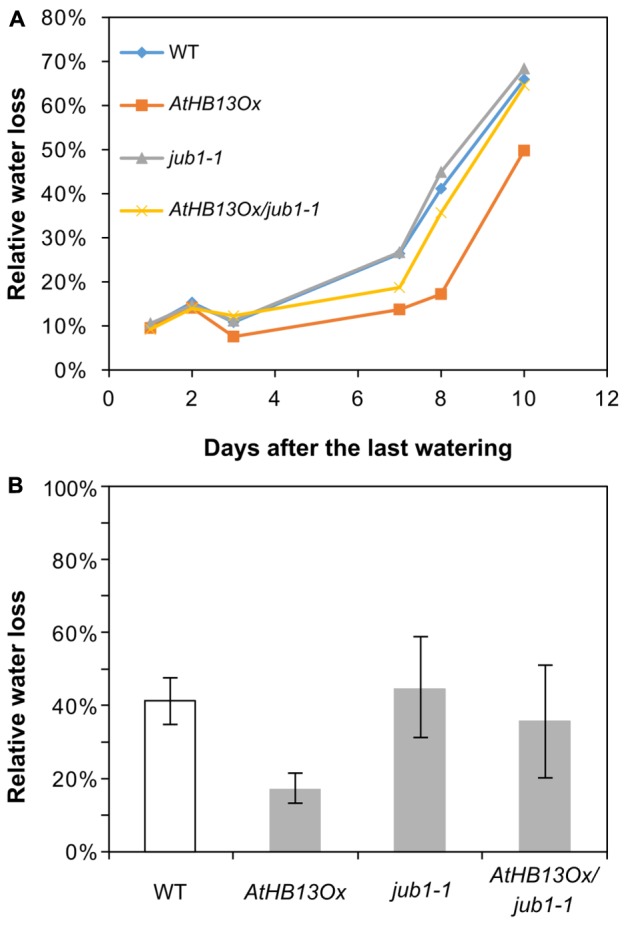
AtHB13Ox/jub1-1 plants behave similar to jub1-1 plants during drought stress. (A) Kinetics of relative water loss of leaves form WT, AtHB13Ox, jub1-1, and AtHB13Ox/jub1-1 plants. Plants were grown under well-watered condition for 20 days; thereafter, irrigation was stopped, gradually leading to severe drought stress. (B) Relative water loss in leaves during 9 days of the treatment. For each genotype, leaves from five plants (one leaf per plant) were analyzed. Bars represent SD. The asterisk indicates significant difference from AtHB13Ox/jub1-1 plants (Student’s t-test, p < 0.05).
FIGURE 5.
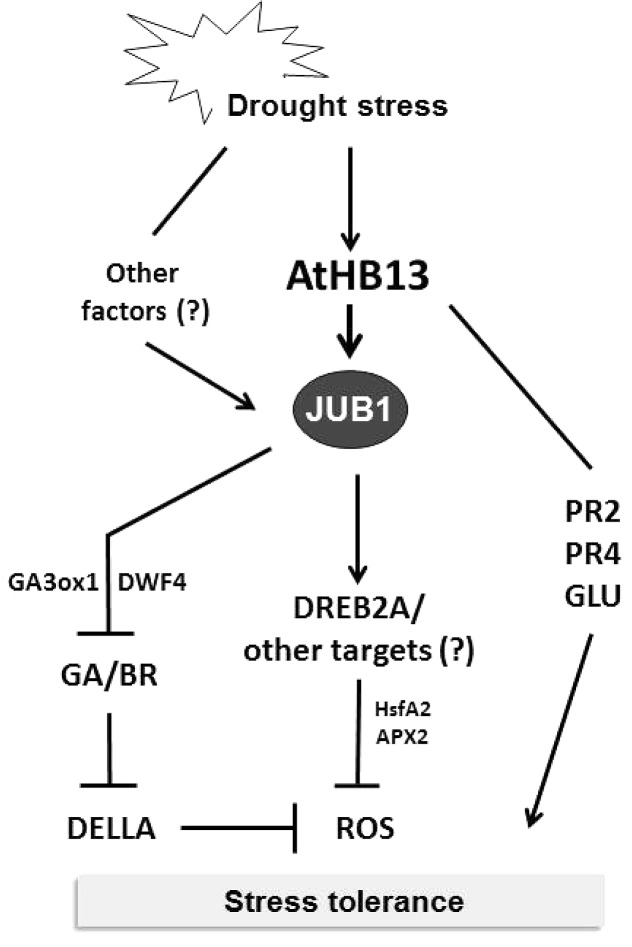
Model for AtHB13-JUB1 regulation of drought stress tolerance. Drought stress induces the expression of both, AtHB13 and JUB1. Increased levels of JUB1 confer enhanced tolerance to drought, in part by lowering cellular reactive oxygen species (hydrogen peroxide) level and by restricting growth via the GA/BR/DELLA pathway. AtHB13 also indirectly induces the expression of glucanase (PR2 and GLU) and chitinase (PR4) genes, each of which enhances drought tolerance when overexpressed. However, control of the PR and GLU genes appears to be independent of JUB1.
Constructs
Gene constructs were generated by polymerase chain reaction (PCR) and directional cloning. All PCR-generated amplicons were checked for correctness by DNA sequence analysis (Seqlab or LGC Genomics). Constructs were transformed into Arabidopsis via Agrobacterium tumefaciens-mediated transformation (floral dip method). To generate the 35S:AtHB13-GFP and AtHB13-GST constructs, the AtHB13 open reading frame (ORF) was PCR-amplified without the stop codon and cloned into pENTR/D-TOPO vector using the pENTR directional TOPO cloning kit (Invitrogen). The AtHB13 ORF was then transferred to the GATEWAY vectors pK7FWG2 (Ghent University) and pDEST24 (Invitrogen), respectively, through LR recombination. To generate the JUB1 promoter deletion constructs (ProJUB1:GUS), 1.0, 0.73, 0.68, 0.31, and 0.21 kb long segments of the JUB1 promoter were PCR-amplified and cloned into pENTR/D-TOPO vector using the pENTR directional TOPO cloning kit (Invitrogen). The promoter fragments were then transferred to the pKGWFS7 GATEWAY vector (Ghent University) by LR recombination. pTUY1H-JUB1-373: The selected 373-bp JUB1 promoter region was first cloned via TA cloning into pCR2.1 entry vector (Invitrogen), and then transferred to the pTUY1H yeast transformation vector (with LEU2 as selection marker) (Castrillo et al., 2011) by restriction enzyme-mediated cloning.
Histochemical GUS Staining
Two-week-old seedlings were carefully transferred from agar media plates to Erlenmeyer flasks containing liquid MS medium (1% [w/v] sucrose) in the absence or presence of 10 mM H2O2, and incubated overnight. For drought treatments, 4-week-old soil-grown plants were not watered for 6 days, and leaves were harvested. Histochemical GUS staining was performed overnight at 37°C in the dark. Chlorophyll was removed by clearing the samples with 70% (v/v) ethanol. Quantification of GUS signal was done as described (Béziat et al., 2017).
Quantitative Real-Time PCR (qRT-PCR)
Total RNA was extracted using the RNeasy Plant Mini kit (Qiagen, Hilden, Germany). Synthesis of cDNA and quantitative real-time PCR (qRT-PCR) using SYBR Green were performed as reported (Balazadeh et al., 2008, 2010) with ACTIN2 (At3g18780) as the reference gene. Primer sequences are given in Supplementary Table S1. Primers were designed using the QuantPrime tool5 (Arvidsson et al., 2008).
Yeast One-Hybrid Screen
The bait construct pTUY1H-JUB1-373 (LEU2 selection marker; JUB1 promoter fragment upstream of HIS3 reporter) was transformed into yeast strain Y187, mating type α. The mating-based Y1H screen was done using a library of approximately 1,200 Arabidopsis TFs, established in vector pDEST22 (TRP1 selection marker) in yeast strain YM4271 (mating type a) (Castrillo et al., 2011). Screening for interaction between TFs and the 373 bp long JUB1 promoter fragment was done on SD medium lacking the essential amino acids Leu, Trp, and His in the absence or presence of different concentrations of 3-amino-1,2,4-triazol (3AT) to prevent false positive interactions.
Electrophoretic Mobility Shift Assay (EMSA)
AtHB13-GST fusion protein was purified from Escherichia coli expression strain BL21 Star (DE3) pRARE, which was generated by transforming the pRARE plasmid isolated from Rosetta (DE3) pRARE cells (Merck) into E. coli BL21 Star (DE3) (Invitrogen). Recombinant GST-fusion protein was purified using GST-agarose beads following the manufacturer’s instructions (Sigma–Aldrich, Taufkirchen, Germany). Electrophoretic mobility shift assays (EMSA) was performed as described (Wu et al., 2012) using the Odyssey Infrared EMSA kit (LI-COR). Sequences of 5′-DY682-labeled fragments are given in Supplementary Table S1.
Chromatin Immunoprecipitation
Chromatin immunoprecipitation (ChIP) was carried out on chromatin extracted from 35S:AtHB13-GFP plants grown (i) well-watered for 4 weeks, (ii) well-watered for 4 weeks and then drought stressed for 6 days (by withholding water), and (iii) well-watered for 50 days. WT plants grown in parallel served as controls in each experiment. Three independent ChIP experiments were performed. The qPCR primers for the JUB1 promoter were designed to flank the AtHB13 binding site (Supplementary Table S1). As negative controls, primers annealing to promoter regions of two Arabidopsis genes lacking an AtHB13 binding site, i.e., AT3G18040 (Neg 1) and AT2G22180 (Neg 2), were used. ChIP-qPCR data were analyzed as described (Kaufmann et al., 2010).
Determination of Ion Leakage
For ion leakage measurements, the first six leaves of the rosette were immersed in 10 ml deionized water and shaken at room temperature for 30 min. Electrical conductivity (σ1) was measured at 25°C, using a conductometer (Schott, Mainz, Germany). Then samples were boiled for 15 min, cooled down to 25°C, and conductivity (σ2) was measured again. Ion leakage was calculated through the expression σ1/σ2 × 100. Three independent experiments were performed.
Determination of Relative Water Content (RWC)
Plant material (five leaves per genotype) was weighed (fresh weight, FW), then put in a Petri dish containing water and kept at room temperature for 3 h. Then, leaves were weighed again (turgid weight, TW). Relative water content (RWC) was calculated using the following formula: RWC [%] = (TW–FW)/(TW) × 100. Three independent experiments were performed.
Establishment of Mild Drought Stress
Plants were grown at a 16-h light : 8-h dark cycle and well watered for 25 days. Thereafter, field capacity was maintained at 50% in all pots by adding the needed quantity of water. Field capacity was determined by weighting the pots. Before the experiment was started, pots were saturated and weighed (100% field capacity). During the experiment, pots were weighed each day and water was added in order to maintain the 50% of the field capacity. The amount of water added to each plant is shown in the figure panels.
Determination of Relative Water Loss during Severe Drought Stress
Plants were grown at a 16-h light : 8-h dark cycle and drought experiments were started by stopping irrigation at day 20. After stopping irrigation, leaves were detached at the indicated times (see figures), weighed (W1), submerged in tap water for 3 h and weighed again (W2). Water loss [%] was calculated using the following formula: (W2–W1)/W2 × 100.
AGI Codes
ACTIN2 (AT3G18780), JUB1 (AT2G43000), ATHB13 (AT1G69780).
Results
Expression of JUB1 Is Induced by Drought
The expression of JUB1 rapidly increases after treatment of Arabidopsis plants with hydrogen peroxide (H2O2) or in the presence of different abiotic stresses such as salinity and heat (Wu et al., 2012; Allu et al., 2014; Shahnejat-Bushehri et al., 2016). To test whether JUB1 is also induced by drought, we analyzed its expression in plants subjected to water shortage. To this end, 4-week-old wild-type Col-0 (WT) plants were subjected to water withholding for 6 days, and whole rosettes were harvested to quantify JUB1 expression by qRT-PCR. As shown in Supplementary Figure S1A, JUB1 expression was considerably higher in drought-stressed plants than in well-watered controls. We also established transgenic Arabidopsis plants expressing the GUS reporter gene from the 1-kb JUB1 promoter (ProJUB1:GUS plants). We grew ProJUB1:GUS plants for 4 weeks under well-watered condition and then subjected them to drought stress (by stopping irrigation) for 6, 9, and 12 days. Histochemical GUS staining revealed drought-induced JUB1 promoter activity (Supplementary Figure S1B).
JUB1 Confers Tolerance to Drought Stress in A. thaliana
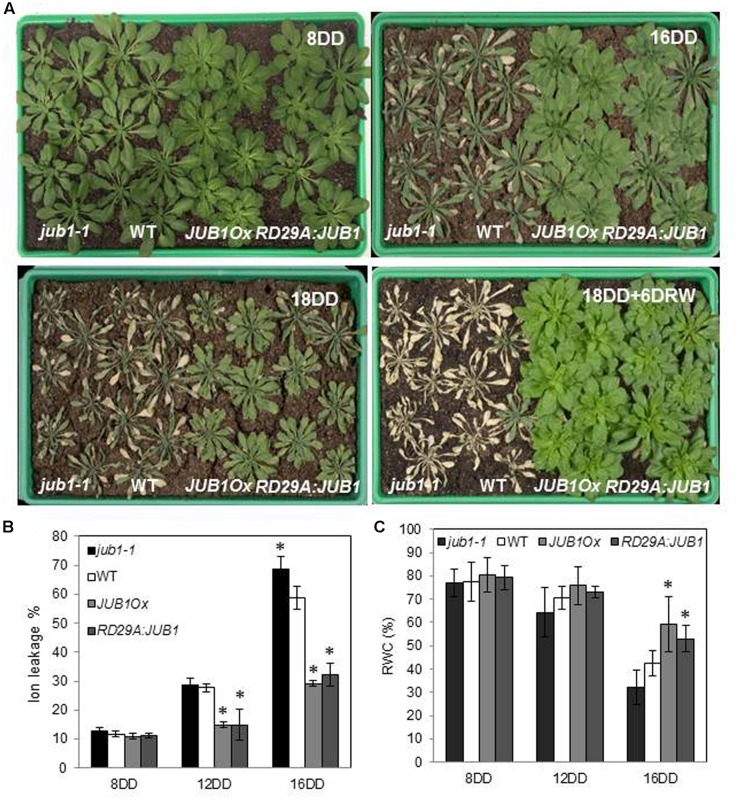
To investigate the function of JUB1 for the response to drought in Arabidopsis, we analyzed the phenotype of JUB1 transgenic lines during drought stress. We previously reported that constitutive overexpression of JUB1 results in reduced growth, delayed flowering, and late leaf senescence (Wu et al., 2012; Shahnejat-Bushehri et al., 2016); we here therefore included transgenic lines expressing JUB1 under the control of the stress-inducible promoter RESPONSIVE TO DESICCATION 29A (RD29A) in our drought assays. The RD29A gene is highly responsive to drought, low temperature, high salt concentration, and desiccation (Yamaguchi-Shinozaki and Shinozaki, 1993, 1994) and only basal expression is observed in non-stressed plants (Yamaguchi-Shinozaki et al., 1999). However, as we reported previously, different RD29A:JUB1 lines displayed different JUB1 expression levels under control conditions (Wu et al., 2012). Here, we selected a RD29A:JUB1 line with lowest expression of JUB1 under non-stress condition for our analyses. The selected transgenic line displayed growth phenotypes similar to WT, in contrast to the plants strongly expressing JUB1 from the constitutive CaMV 35S promoter (JUB1Ox; Shahnejat-Bushehri et al., 2016). Four-week-old RD29A:JUB1, JUB1Ox and WT plants, as well as jub1-1 knockdown mutants were dehydrated for 18 days. As shown in Figure 1A, both, RD29A:JUB1 and JUB1Ox plants exhibited strong drought tolerance, while jub1-1 and WT plants showed severe wilting after 16 days of drought (16DD), and were strongly dehydrated after 18 days (18DD). None of the WT and jub1-1 plants recovered after 18 days of drought followed by re-watering for 6 days (18DD+6DRW) further demonstrating their sensitivity to drought stress. In contrast, JUB1Ox and RD29A:JUB1 plants recovered rapidly after the 18 days of drought stress and showed complete survival after re-watering. Membrane stability under water deficit conditions was assessed by measuring ion leakage in both, control and treated plants. When water was withheld for 12 days, jub1-1 and WT plants showed a higher ion leakage (∼30%) than JUB1Ox and RD29A:JUB1 plants (∼15%). After 16 days of water withholding the differences in electrolyte leakage increased further; while jub1-1 and WT plants showed an ion leakage of ∼70 and ∼58%, respectively, ion leakage was only ∼30% for both, RD29A:JUB1 and JUB1Ox plants upon stress (Figure 1B).
FIGURE 1.
JUB1 confers tolerance to drought. (A) Four-week-old jub1-1, WT, JUB1Ox, and RD29A:JUB1 plants were grown in soil and subjected to drought stress by withholding water for 18 days. Photographs were taken 8 days after start of the drought stress experiment (8DD), 16DD, 18DD, and 18DD + 6 days of re-watering (18DD+6DRW). The experiment was repeated more than three times, and a representative result is shown here. (B) Ion leakage of the first six leaves of WT and transgenic lines after 8, 12, and 16 days of drought stress. (C) Relative water content (RWC) of leaves (%). Means ± SD are shown (n = 3). Asterisks represent statistically significant difference from WT; Student’s t-test (∗p < 0.05).
Furthermore, RWC in leaves was determined after 8, 12, and 16 days of withholding water (Figure 1C). RWC was not significantly different between the genotypes at the early stage of drought (8DD), while at later stages of drought (16DD) a significantly higher RWC was observed in RD29A:JUB1 and JUB1Ox plants than in WT and jub1-1 plants (Figure 1C). Collectively, our results reveal that JUB1, when expressed from the constitutive CaMV 35S promoter or from the abiotic stress-induced RD29A promoter, confers superior tolerance to drought stress in Arabidopsis.
A Promoter Region Central for Drought-Induced Expression of JUB1
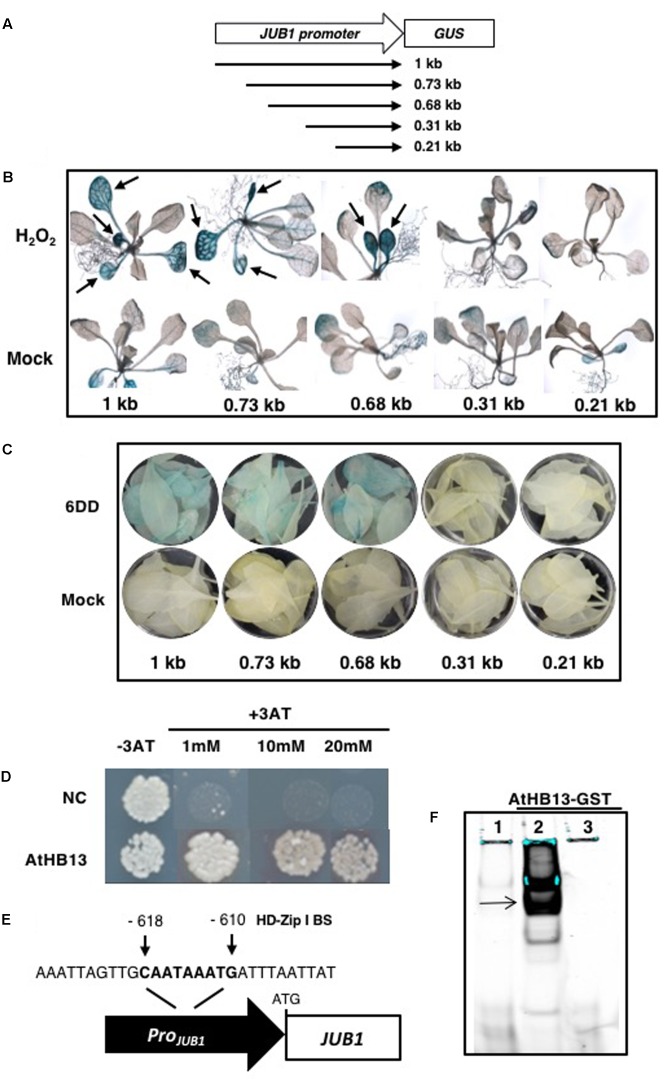
We intended to identify upstream regulatory factors controlling the expression of JUB1 during abiotic stress. To this end, we performed a deletion analysis of the JUB1 promoter and then conducted a yeast one-hybrid (Y1H) screen to identify TFs binding to a functionally relevant promoter segment. Various 5′ deletions of the JUB1 promoter were transcriptionally fused to the Escherichia coli β-GLUCURONIDASE (GUS) reporter gene and the constructs were transformed into Arabidopsis (Figure 2A). The promoter-reporter lines (hereafter, ProJUB1:GUS deletions) were subjected to H2O2 and drought treatments to identify a promoter region relevant for the response of JUB1 to these stresses. To this end, (i) 2-week-old seedlings of ProJUB1:GUS deletion lines were treated with 10 mM H2O2 and (ii) 4-week-old ProJUB1:GUS plants were subjected to desiccation for 6 days (6DD) and leaves were harvested for analysis. Histochemical GUS staining revealed that the 1-kb JUB1 promoter as well as the 0.73-kb and 0.68-kb deletion variants, but not the 0.31-kb and the 0.21-kb promoters, confer stress-inducible activation of the GUS reporter gene, indicating that the region responsive to H2O2 and drought is located between positions -0.68 and -0.31 kb of the JUB1 promoter (Figures 2B,C and Supplementary Figure S1C).
FIGURE 2.
Identification of an upstream regulator of JUB1. (A) A series of 5′ deletions of the JUB1 promoter (including the 5′- untranslated region up to the ATG start codon) were transcriptionally fused to the β-GLUCURONIDASE (GUS) reporter gene and the constructs were transformed into Arabidopsis. (B) Two-week-old seedlings of different ProJUB1:GUS deletion lines (1 kb as well as 0.73, 0.68, 0.31, and 0.21 kb) were subjected to H2O2 (10 mM) treatment overnight and then incubated at 37°C in GUS buffer. Arrows indicate induced GUS staining. Note the lack of induction of GUS activity in the –0.31-kb and –0.21-kb deletion lines. (C) Four-week-old ProJUB1:GUS deletion lines were subjected to drought for 6 days (6DD). Following GUS staining, the lines expressing GUS from the 1, 0.73, and 0.68 kb JUB1 promoter fragments showed higher GUS activity than the corresponding well-watered (control) plants, while no GUS staining was visible in the –0.31-kb and –0.21-kb deletion lines. (D) Yeast-one-hybrid (Y1H) assay demonstrates interaction between the functional 373-bp JUB1 promoter fragment and transcription factor (TF) AtHB13. The JUB1 promoter fragment contains the common binding site of HD-Zip I TFs at positions –618 to –610 bp upstream of the translational start site (ATG). Upon interaction of AtHB13-GAL4AD fusion protein with the binding site, transcription of the yeast HIS3 reporter gene is activated and diploid yeast cells grow on SD medium lacking the three essential amino acids Trp, Leu, and His. The yeast one-hybrid assay was performed three times giving the same result. NC, negative control containing the pTUY1H-JUB1-373 plasmid but no TF as a test for autoactivation. (E) Schematic representation of the HD-Zip I binding site (BS) within the JUB1 promoter. The sequence of the BS as well as the surrounding nucleotides are indicated. (F) EMSA showing binding of purified AtHB13-GST protein to the JUB1 promoter region harboring the HD-Zip I BS. DNA binding reactions were performed with a 40-bp long wild-type fragment derived from the JUB1 promoter containing the HD-Zip I BS. 1, 5′-DY682-labeled, double-stranded oligonucleotide; 2, labeled probe plus AtHB13-GST protein; 3, labeled probe plus AtHB13-GST and 200× competitor (unlabeled oligonucleotide).
AtHB13 Binds to the JUB1 Promoter and Activates Its Expression during Drought Stress
To identify upstream transcriptional regulators of JUB1, we performed a yeast one-hybrid (Y1H) screen using the 0.37-kb promoter fragment (-0.68 and -0.31 kb upstream of the JUB1 start codon) involved in the H2O2- and drought responsiveness of the JUB1 gene as bait. In a screen with nearly 1,200 Arabidopsis TFs we identified HD-Zip I protein AtHB13 (AT1G69780) as a TF binding to the abiotic stress-responsive segment of the JUB1 promoter (Figure 2D). As AtHB13 has previously been reported to affect the response of A. thaliana to various biotic and abiotic stresses including drought (Cabello and Chan, 2012; Cabello et al., 2012; Gao et al., 2014), we investigated whether it is a genuine upstream regulator of JUB1.
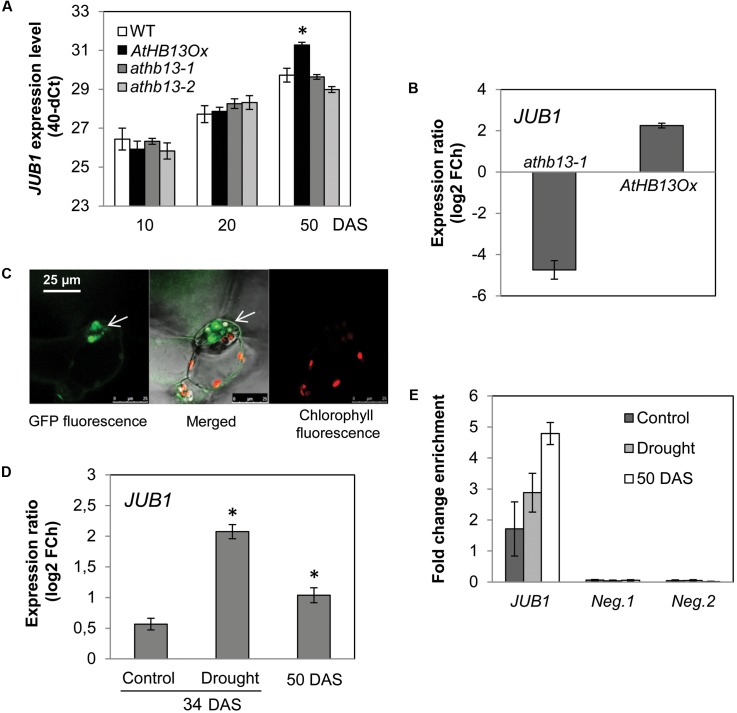
Interestingly, the 373-bp JUB1 fragment employed in the Y1H assay harbors an HD-Zip class I binding site, namely CAATAAATG (Figure 2E). The motif is identical to the one reported by Sessa et al. (1993) for AtHB1 (another HD-Zip I protein from Arabidopsis), with the exception of the underlined A which is a T in the AtHB1 binding site. To test whether AtHB13 binds to the CAATAAATG sequence within the frame of the JUB1 promoter, we performed EMSAs using recombinant AtHB13-GST (glutathione S-transferase) fusion protein and a 5′-DY682-labeled 40-bp JUB1 promoter fragment harboring the HD-Zip class I binding site. As shown in Figure 2F, AtHB13 protein physically interacts in vitro with the respective JUB1 promoter fragment. Next, we checked expression of JUB1 in rosette leaves of transgenic plants overexpressing AtHB13 from the CaMV 35S promoter (AtHB13Ox lines; Cabello et al., 2012) and WT plants at different developmental stages, i.e., at 10, 20, and 50 days after sowing (DAS). Interestingly, expression of JUB1 was not altered in the AtHB13Ox plants at 10 and 20 DAS, but was significantly upregulated in AtHB13Ox plants at 50 DAS (Figure 3A). Thereafter, we analyzed the expression of JUB1 in AtHB13Ox overexpressors and athb13 knockout mutants (Cabello and Chan, 2012; Cabello et al., 2012) during drought stress. To this end, 4-week-old plants were subjected to 6 days of water withholding. As shown in Figure 3B, expression of JUB1 was enhanced in AtHB13Ox plants compared to WT, and strongly repressed in the athb13-1 mutant upon drought stress. Collectively, our data indicate that AtHB13 functions as a positive regulator of JUB1 expression at later stages of development and upon drought stress.
FIGURE 3.
AtHB13 directly regulates JUB1. (A) Expression of JUB1 in 10-, 20-, and 50-day-old WT, AtHB13Ox, and athb13-1 and athb13-2 plants in well-watered condition. Transcript levels were determined by qRT-PCR; values are expressed as the difference between an arbitrary value of 40 and dCt, so that high 40-dCt value indicates high gene expression level. Means ± SD calculated from three independent biological experiments (each with nine leaves pooled from three plants). Expression levels were normalized against the expression level of ACTIN2. DAS, days after sowing. Asterisk indicates statistically significant difference (Student’s t-test (∗p < 0.05) from WT. (B) Expression of JUB1 in 35S:AtHB13 (AtHB13Ox) and athb13-1 plants compared to WT upon drought treatment. For drought treatment, 4-week-old plants were subjected to water withholding for 6 days. Whole rosettes of drought-treated and well-watered (control) plants were harvested for gene expression analysis by qRT-PCR. Data represent the means of three biological repetitions ± SD. FCh, fold change. (C) Confocal microscope image showing nuclear localization of the AtHB13-GFP fusion protein in transgenic 35S:AtHB13-GFP Arabidopsis plants. Left, GFP signal; middle, chlorophyll autofluorescence merged with GFP fluorescence; right, chlorophyll autofluorescence. (D) Expression of JUB1 in 35S:AtHB13-GFP plants compared to WT upon drought stress and at a later stage of development (50-day-old plants). For drought treatment, 4-week-old plants were subjected to drought by withholding water for 6 days. Whole rosettes of drought-treated and well-watered (control) plants were harvested for gene expression analysis by qRT-PCR. Data represent the means of three biological repetitions ± SD. FCh, fold change. Asterisks indicate statistically significant difference (p < 0.01; Student’s t-test) from the non-stress control at 34 DAS. (E) ChIP-qPCR showing enrichment of the JUB1 promoter region containing the HD-Zip I binding site, quantified by qPCR. For the ChIP experiment rosettes of 35S:AtHB13-GFP and WT plants were harvested as follows: from 4-week-old control plants (well watered; ‘control’); from plants grown for 4 weeks in well-watered condition and then subjected for 6 d to drought stress by withholding water (‘drought’); and from 50-day-old plants grown under well-watered condition (‘50 DAS’). As negative controls, primers annealing to promoter regions of two Arabidopsis genes lacking an HD-Zip I binding site, i.e., AT3G18040 (Neg. 1) and AT2G22180 (Neg. 2), were used. Data represent the means of three biological repetitions ± SD.
To confirm that AtHB13 interacts with the JUB1 promoter in planta, we performed ChIP assays using Arabidopsis plants stably expressing GFP-tagged AtHB13 protein (hereafter, 35S:AtHB13-GFP). Analysis by confocal microscopy revealed nuclear localization of AtHB13-GFP protein, in accordance with the biological function of AtHB13 as a transcriptional regulator (Figure 3C). Next, we harvested rosette leaves from 35S:AtHB13-GFP and WT plants for gene expression analysis and ChIP-qPCR assays. Plants were grown (i) for 4 weeks in well-watered condition (control); (ii) for 4 weeks in well-watered condition, followed by 6 days without watering (drought); (iii) for 50 days in well-watered condition (50 DAS). Under control condition, JUB1 expression was similar in 35S:AtHB13-GFP and WT plants, while its expression was significantly induced under drought stress in 35S:AtHB13-GFP compared to WT. JUB1 expression was also elevated, although less strongly, in well-watered 50 DAS 35S:AtHB13-GFP plants (Figure 3D). ChIP-qPCR revealed enrichment of the JUB1 promoter fragment harboring the AtHB13 binding site, in particular at 50 DAS control and in drought stress conditions (Figure 3E).
AtHB13 Confers Drought Tolerance in Part via Regulation of JUB1
Expression of AtHB13 and of its sunflower homologue HaHB1 is induced by water deficit and overexpression of the two TFs in transgenic Arabidopsis plants improves drought tolerance (Cabello and Chan, 2012), similar to the overexpression of JUB1 reported here. Considering the fact that AtHB13 binds the JUB1 promoter, and regulates JUB1 in planta, the higher drought tolerance of AtHB13 overexpressors might be conveyed through an upregulation of JUB1 expression by the HD-Zip I TF. To test this hypothesis, we tested the drought tolerance of AtHB13Ox, JUB1Ox, athb13-1, athb13-2, jub1-1, and WT plants grown side by side. Plants were grown under well-watered condition and drought stress was started at day 20, before bolting occurred in all genotypes. Thereafter, irrigation was stopped, which gradually produced severe drought stress. Supplementary Figure S2A shows the phenotype of all plants after eight days of drought stress indicating that the two TF overexpressors (AtHB13Ox, JUB1Ox) are more tolerant than the WT, while the athb13-1, athb13-2, and jub1-1 mutants are less tolerant. We determined water loss in plants of all genotypes at different days after stopping irrigation (Supplementary Figure S2B). Our results show that water loss was significantly lower in JUB1Ox than WT plants, in accordance with the observed increase in drought tolerance of these lines, while jub1-1 and athb13 mutants performed slightly worse than the WT in the experiments performed (Supplementary Figures S2B,C). Similar conclusions can be drawn from water consumption experiments; as shown in Supplementary Figures S2D,E, overexpressors consumed less water than WT and mutant plants during mild drought stress, although the reduction in water consumption was much more prominent in JUB1Ox than AtHB13Ox plants, which may in part be due to the more compact growth phenotype of JUB1 overexpressors compared to AtHB13 overexpressors.
To test whether AtHB13 requires JUB1 for improved drought tolerance we crossed the AtHB13Ox plant with the jub1-1 mutant. As seen in Figure 4A, water loss in the AtHB13Ox/jub1-1 line was similar to that of the jub1-1 mutant and the WT, while water loss was much lower in the AtHB13Ox plant, strongly indicating that AtHB13 confers drought tolerance at least in part through transcriptional control of JUB1 (Figure 4B).
Discussion
The NAC TF JUNGBRUNNEN1 (JUB1) has originally been identified as a positive regulator of leaf longevity in A. thaliana, and to enhance the tolerance toward heat and salinity stress when overexpressed (Wu et al., 2012; Shahnejat-Bushehri et al., 2012). Here, we show that overexpression of JUB1 in transgenic Arabidopsis also enhances tolerance to drought stress. Of importance, this capacity of JUB1 is observed in both, transgenic plants expressing JUB1 from the constitutive CaMV 35S promoter and the stress-induced RD29A promoter. This is an important notion as constitutive overexpression of JUB1 affects plant morphology, due to the fact that JUB1 negatively controls the expression of two key genes of phytohormone biosynthesis, namely GAs (by inhibiting GA3ox1) and BRs (by inhibiting DWF4). However, when JUB1 is expressed from a stress-inducible promoter, no major developmental differences to wild-type plants are observed (Wu et al., 2012), while tolerance to salinity and heat stress is retained, indicating that the developmental effect of JUB1 can be largely separated from the stress tolerance effect. A similar observation we made here with respect to drought tolerance gained through JUB1: while plants expressing JUB1 from the constitutive CaMV 35S promoter are smaller and more compact than wild-type plants (Shahnejat-Bushehri et al., 2016; Figure 1A and Supplementary Figure S2A), development of RD29A:JUB1 plants is virtually indifferent from that of the wild type (Figure 1A). However, strong drought tolerance is observed in both transgenic lines, irrespective of the growth phenotype (Figure 1). This observation indicates that JUB1 can improve tolerance to drought without limiting growth to a large extent.
We have previously reported that expression of JUB1 is rapidly induced by H2O2 treatment (Wu et al., 2012). We further observed that overexpression of JUB1 in transgenic Arabidopsis plants lowers the level of cellular H2O2, suggesting that this TF dampens H2O2 accumulation through a gene regulatory network that is currently not known in its details. However, JUB1 acts as a direct upstream transcriptional regulator of DREB2A (DEHYDRATION-RESPONSIVE ELEMENT-BINDING PROTEIN 2A), which encodes an AP2-type TF well known for its involvement in regulating heat and drought responses (Sakuma et al., 2006; Kant et al., 2008). DREB2A itself controls the expression of Heat shock factor A2 (HsfA2) and through this affects several HEAT SHOCK PROTEIN (HSP) genes and genes for H2O2 scavenging enzymes (Schramm et al., 2008; Yoshida et al., 2008). The reduced H2O2 level of JUB1 overexpressor plants may also be causative for their strongly enhanced drought tolerance. The fact that JUB1 is induced by H2O2 in conjunction with the observation that JUB1 dampens cellular H2O2 level suggests the presence of a regulatory loop that helps to protect the plant against overshooting cellular H2O2 levels.
To identify upstream transcriptional regulators controlling the H2O2-dependent activation of JUB1, we performed a yeast one-hybrid screen and identified AtHB13, an HD-Zip I TF, as a positive regulator of JUB1 expression. Notably, the part of the JUB1 promoter that controls responsiveness to H2O2 also controls the responsiveness to drought, suggesting that the drought responsiveness of JUB1 is mediated through H2O2 which accumulates in drought-stressed plants (Kar, 2011; You and Chan, 2015). AtHB13 binds to the CAATAAATG element present within the relevant JUB1 promoter segment. Of importance, AtHB13 has been reported previously to improve drought tolerance when overexpressed in transgenic Arabidopsis plants (Cabello and Chan, 2012). In addition, it was shown that AtHB13 induces the expression of PATHOGENESIS RELATED2 (PR2) and PR4, genes that are able to individually confer drought tolerance (Cabello and Chan, 2012). However, in our experiments transcript levels of PR2 and PR4 were not induced by JUB1 (data not shown), indicating that the drought tolerance conferred by AtHB13 occurs through at least two different mechanisms, one of which involves JUB1 (Figure 5).
Recently, we demonstrated enhanced tolerance to drought stress in transgenic tomato (Solanum lycopersicum) plants overexpressing JUB1 from Arabidopsis. In contrast, inhibition of tomato JUB1 (SlJUB1) by virus-induced gene silencing significantly lowered drought tolerance associated with an increase in the level of H2O2, and a decrease of the expression of various drought-responsive genes including SlDREB1, SlDREB2, and SlDELLA (Thirumalaikumar et al., 2017). Similarly, banana (Musa acuminata) plants overexpressing MusaNAC042 (the closest homologue of JUB1 in this species) revealed increased tolerance to drought stress (Tak et al., 2017). Although levels of H2O2 were not determined in this study, the observation that MusaNAC042 overexpressors contain lower levels of malondialdeyhde (MDA, a marker of lipid peroxidation) than wild-type plants indicates reduced stress-induced oxidative damage (Tak et al., 2017). The molecular control network through which MusaNAC042 lowers oxidative stress damage is unknown at present.
An interesting observation we made is the following: while overexpressing AtHB13 triggers elevated expression of JUB1 in older plants (50 DAS), it does not do so in younger plants (10 DAS; Figure 3A). Furthermore, while JUB1 is not much affected by AtHB13 in well-watered plants, its expression increases in AtHB13Ox plants compared to WT when plants are subjected to drought stress (Figure 3D). Finally, while strong constitutive overexpression of JUB1 reduces growth (Shahnejat-Bushehri et al., 2016), this is not the case for AtHB13 overexpressors (Cabello et al., 2012; Cabello and Chan, 2012), although AtHB13 positively controls JUB1 expression. This indicates that elevated levels of AtHB13 per se may not be sufficient to enhance JUB1 expression under all conditions and suggests that additional mechanisms are needed to trigger transcriptional activation of JUB1 by AtHB13. There are several principle mechanisms that could make AtHB13 competent for activating JUB1, including the following: (i) HD Zip TFs often form heterodimers with other family members in a selective manner (Johannesson et al., 2001; Capella et al., 2015) or with TFs or other families. It may thus be possible that AtHB13 interacts with other TFs at the later stages of leaf development or during drought stress. A possible candidate might be AREB3, a bZIP TF that interacts with AtHB136 and is involved in ABA-dependent signaling and the response to drought stress (Uno et al., 2000). Importantly, JUB1 expression is strongly reduced in the athb13 knockout mutant, clearly indicating that AtHB13 is necessary but not potentially sufficient for full expression of JUB1. (ii) TFs of other families may be required in addition to AtHB13 for enhanced expression of JUB1 under drought stress. (iii) The activity and/or stability of HD Zip TFs may be regulated by posttranslational modification such as phosphorylation, as for example reported for the HD Zip II TF HAT1 in Arabidopsis (Zhang et al., 2014). In a similar way, the stability of the AtHB13 protein may be enhanced in later stages of leaf development or in drought stress conditions. (iv) A TF suppressing JUB1 expression might be active in young leaves, thereby overriding the activation by AtHB13 before the leaves enter a more mature stage. Which of these mechanisms are realized in Arabidopsis has to be revealed in further studies.
Author Contributions
SB and BM-R conceived the idea for the study. SE-M performed the JUB1 promoter deletion study and the Y1H screen. SE-M and VT performed the drought stress experiments. PR made the crosses, confirmed their genotypes, and performed drought stress experiments under the supervision of RC. VT generated the 35S:AtHB13-GFP lines and performed confocal microscopy to check nuclear localization of AtHB13; he also prepared the AtHB13-GST construct. ChIP experiments were done by SE-M. AA and VT performed the AtHB13-GST protein purification and EMSA experiments. SB and BM-R wrote the manuscript, with contributions from SE-M and RC.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Karin Koehl and her team (MPI of Molecular Plant Physiology) for plant care. Financial support by the University of Potsdam and the MPI of Molecular Plant Physiology is gratefully acknowledged.
Funding. SB thanks the Deutsche Forschungsgemeinschaft (DFG) for funding (BA 4769/2-1). The Argentinean group was supported by PICT 2014 3300 (FonCyT) and CONICET. PR is a CONICET fellow and RC a career member of the same institution.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2017.02118/full#supplementary-material
FIGURE S1 | Histochemical GUS staining of ProJUB1:GUS lines upon drought stress. (A) Expression level of JUB1 in leaves of 4-week-old WT plants subjected to 6 days of dehydration (6DD) compared to well-watered (Mock) plants. Transcript levels were determined by qRT-PCR and normalized applying the dCt method. Values are expressed as the difference between an arbitrary value of 40 and dCt, so that high 40 – dCt value indicates high gene expression level. Means ± SD calculated from three independent biological replicates, with nine leaves from three plants in each replicate. ACTIN2 was used as reference gene. Asterisks indicate significant difference in the level of JUB1 between 6DD and mock-treated samples (p < 0.05; Student’s t-test). (B) Induction of JUB1 promoter by drought stress, shown by histochemical GUS staining in leaves of Arabidopsis ProJUB1:GUS plants (1000-bp JUB1 promoter) plants. Plants were grown for 4 weeks under well-watered conditions and then irrigation was stopped for 6, 9, and 12 days. (C) Quantification of GUS signal in leaves of 14-day-old ProJUB1:GUS seedlings drought-stressed on filter paper for 2 h (‘Drought’) or kept in humidified condition (‘Mock’). Promoters of 0.68 and 0.31 kb length were analyzed. GUS signal was quantified as reported (Béziat et al., 2017) by selecting rectangular areas of maximum size of individual leaves. High gray values represent elevated GUS staining. Means ± SD of 20 leaves from 6 to 8 seedlings each.
FIGURE S2 | Behavior of transgenic JUB1 and AtHB13 plants during water deficit. (A) Phenotype of JUB1 and AtHB13 transgenic lines under severe drought stress. WT, JUB1Ox, jub1-1, AtHB13Ox, athb13-1, and athb13-2 plants grown in well-watered condition for 20 days and then subjected to severe drought stress for 8 days by completely stopping watering at day 20. (B) Kinetics of relative water loss of leaves from wild-type (WT) and transgenic Arabidopsis plants. (C) Relative water loss in leaves after day 8 of drought stress. For each genotype, leaves from five plants (one leaf per plant) were analyzed. Error bars represent SD. Plants in (B,C) were grown as in (A). (D) Kinetics of water consumption in WT and transgenic (jub1-1, JUB1Ox, AtHB13Ox) plants during mild drought stress. Plants were grown in soil under well-watered condition until day 25. Then, field capacity was maintained at 50% in all pots by adding the needed quantity of water. The volume of water added to each plant is shown. The graph represents the average of three experiments ± SD calculated from data obtained from five plants per genotype in each experiment. (E) Total amount of water added to each plant line until the end of the experiment. The graph represents the average of three independent experiments ± SD calculated from data obtained from five plants per genotype in each experiment. Asterisks in (C,E) indicate significant difference from WT (Student’s t-test; p < 0.05).
TABLE S1 | Constructs and primer sequences.
References
- Allu A. D., Soja A. M., Wu A., Szymanski J., Balazadeh S. (2014). Salt stress and senescence: identification of cross-talk regulatory components. J. Exp. Bot. 65 3993–4008. 10.1093/jxb/eru173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariel F. D., Manavella P. A., Dezar C. A., Chan R. L. (2007). The true story of the HD-Zip family. Trends Plant Sci. 12 419–426. 10.1016/j.tplants.2007.08.003 [DOI] [PubMed] [Google Scholar]
- Arvidsson S., Kwasniewski M., Riano-Pachon D. M., Mueller-Roeber B. (2008). QuantPrime - a flexible tool for reliable high-throughput primer design for quantitative PCR. BMC Bioinformatics 9:465. 10.1186/1471-2105-9-465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balazadeh S., Riaño-Pachón D. M., Mueller-Roeber B. (2008). Transcription factors regulating leaf senescence in Arabidopsis thaliana. Plant Biol. 10 63–75. 10.1111/j.1438-8677.2008.00088.x [DOI] [PubMed] [Google Scholar]
- Balazadeh S., Siddiqui H., Allu A. D., Matallana-Ramirez L. P., Caldana C., Mehrnia M., et al. (2010). A gene regulatory network controlled by the NAC transcription factor ANAC092/AtNAC2/ORE1 during salt-promoted senescence. Plant J. 62 250–264. 10.1111/j.1365-313X.2010.04151.x [DOI] [PubMed] [Google Scholar]
- Béziat C., Kleine-Vehn J., Feraru E. (2017). Histochemical staining of β-glucuronidase and its spatial quantification. Methods Mol. Biol. 497 73–80. 10.1007/978-1-4939-6469-7_8 [DOI] [PubMed] [Google Scholar]
- Cabello J. V., Arce A. L., Chan R. L. (2012). The homologous HD-Zip I transcription factors HaHB1 and AtHB13 confer cold tolerance via the induction of pathogenesis-related and glucanase proteins. Plant J. 69 141–153. 10.1111/j.1365-313X.2011.04778.x [DOI] [PubMed] [Google Scholar]
- Cabello J. V., Chan R. L. (2012). The homologous homeodomain-leucine zipper transcription factors HaHB1 and AtHB13 confer tolerance to drought and salinity stresses via the induction of proteins that stabilize membranes. Plant Biotechnol. J. 10 815–825. 10.1111/j.1467-7652.2012.00701.x [DOI] [PubMed] [Google Scholar]
- Capella M., Ribone P. A., Arce A. L., Chan R. L. (2015). “Homeodomain–leucine zipper transcription factors: structural features of these proteins, unique to plants,” in Plant Transcription Factors: Evolutionary, Structural and Functional Aspects, ed. González D. (San Diego, CA: Academic Press/Elsevier; ). [Google Scholar]
- Castrillo G., Turck F., Leveugle M., Lecharny A., Carbonero P., Coupland G., et al. (2011). Speeding cis-trans regulation discovery by phylogenomic analyses coupled with screenings of an arrayed library of Arabidopsis transcription factors. PLOS ONE 6:e21524. 10.1371/journal.pone.0021524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davière J.-M., Achard P. (2016). A pivotal role of DELLAs in regulating multiple hormone signals. Mol. Plant 9 10–20. 10.1016/j.molp.2015.09.011 [DOI] [PubMed] [Google Scholar]
- Fang Y., Xiong L. (2015). General mechanisms of drought response and their application in drought resistance improvement in plants. Cell. Mol. Life Sci. 72 673–689. 10.1007/s00018-014-1767-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao D., Appiano M., Huibers R. P., Chen X., Loonen A. E. H. M., Visser R. G. F., et al. (2014). Activation tagging of ATHB13 in Arabidopsis thaliana confers broad-spectrum disease resistance. Plant Mol. Biol. 86 641–653. 10.1007/s11103-014-0253-2 [DOI] [PubMed] [Google Scholar]
- González-Grandío E., Pajoro A., Franco-Zorrilla J. M., Tarancón C., Immink R. G., Cubas P. (2017). Abscisic acid signaling is controlled by a BRANCHED1/HD-ZIP I cascade in Arabidopsis axillary buds. Proc. Natl. Acad. Sci. U.S.A. 114 E245–E254. 10.1073/pnas.1613199114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamdy A., Ragab R., Scarascia-Mugnozza E. (2003). Coping with water scarcity: water saving and increasing water productivity. Irrig. Drain. 52 3–20. 10.1002/ird.73 [DOI] [Google Scholar]
- Harris J. C., Hrmova M., Lopato S., Langridge P. (2011). Modulation of plant growth by HD-Zip class I and II transcription factors in response to environmental stimuli. New Phytol. 190 823–837. 10.1111/j.1469-8137.2011.03733.x [DOI] [PubMed] [Google Scholar]
- Henriksson E., Olsson A. S. B., Johannesson H., Johansson H., Hanson J., Engström P., et al. (2005). Homeodomain leucine zipper class I genes in Arabidopsis. Expression patterns and phylogenetic relationships. Plant Physiol. 139 509–518. 10.1104/pp.105.063461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himmelbach A., Hoffmann T., Leube M., Höhener B., Grill E. (2002). Homeodomain protein ATHB6 is a target of the protein phosphatase ABI1 and regulates hormone responses in Arabidopsis. EMBO J. 21 3029–3038. 10.1093/emboj/cdf316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hjellström M., Olsson A. S. B., Engstrom P., Soderman E. M. (2003). Constitutive expression of the water deficit-inducible homeobox gene ATHB7 in transgenic Arabidopsis causes a suppression of stem elongation growth. Plant Cell Environ. 26 1127–1136. 10.1046/j.1365-3040.2003.01037.x [DOI] [Google Scholar]
- Johannesson H., Wang Y., Engström P. (2001). DNA-binding and dimerisation preferences of Arabidopsis homeodomain- leucine zipper transcription factors in vitro. Plant Mol. Biol. 45 63–73. 10.1023/A:1006423324025 [DOI] [PubMed] [Google Scholar]
- Kant P., Gordon M., Kant S., Zolla G., Davydov O., Heimer Y. M., et al. (2008). Functional-genomics-based identification of genes that regulate Arabidopsis responses to multiple abiotic stresses. Plant. Cell Environ. 31 697–714. 10.1111/j.1365-3040.2008.01779.x [DOI] [PubMed] [Google Scholar]
- Kar R. K. (2011). Plant responses to water stress: role of reactive oxygen species. Plant Signal. Behav. 6 1741–1745. 10.4161/psb.6.11.17729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann K., Muiño J. M., Østerås M., Farinelli L., Krajewski P., Angenent G. C. (2010). Chromatin immunoprecipitation (ChIP) of plant transcription factors followed by sequencing (ChIP-SEQ) or hybridization to whole genome arrays (ChIP-CHIP). Nat. Protoc. 5 457–472. 10.1038/nprot.2009.244 [DOI] [PubMed] [Google Scholar]
- Kim H. J., Nam H. G., Lim P. O. (2016). Regulatory network of NAC transcription factors in leaf senescence. Curr. Opin. Plant Biol. 33 48–56. 10.1016/j.pbi.2016.06.002 [DOI] [PubMed] [Google Scholar]
- Olsen A. N., Ernst H. A., Leggio L., Skriver K. (2005). NAC transcription factors: structurally distinct, functionally diverse. Trends Plant Sci. 10 79–87. 10.1016/j.tplants.2004.12.010 [DOI] [PubMed] [Google Scholar]
- Palena C. M., Gonzalez D. H., Chan R. L. (1999). A monomer-dimer equilibrium modulates the interaction of the sunflower homeodomain leucine-zipper protein Hahb-4 with DNA. Biochem. J. 341 81–87. [PMC free article] [PubMed] [Google Scholar]
- Perotti M. F., Ribone P. A., Chan R. L. (2017). Plant transcription factors from the homeodomain-leucine zipper family I. Role in development and stress responses. IUBMB Life 69 280–289. 10.1002/iub.1619 [DOI] [PubMed] [Google Scholar]
- Ré D. A., Capella M., Bonaventure G., Chan R. L. (2014). Arabidopsis AtHB7 and AtHB12 evolved divergently to fine tune processes associated with growth and responses to water stress. BMC Plant Biol. 14:150. 10.1186/1471-2229-14-150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribone P. A., Capella M., Chan R. L. (2015). Functional characterization of the homeodomain leucine zipper I transcription factor AtHB13 reveals a crucial role in Arabidopsis development. J. Exp. Bot. 66 5929–5943. 10.1093/jxb/erv302 [DOI] [PubMed] [Google Scholar]
- Sakuma Y., Maruyama K., Qin F., Osakabe Y., Shinozaki K., Yamaguchi-Shinozaki K. (2006). Dual function of an Arabidopsis transcription factor DREB2A in water-stress-responsive and heat-stress-responsive gene expression. Proc. Natl. Acad. Sci. U.S.A. 103 18822–18827. 10.1073/pnas.0605639103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schramm F., Larkindale J., Kiehlmann E., Ganguli A., Englich G., Vierling E., et al. (2008). A cascade of transcription factor DREB2A and heat stress transcription factor HsfA3 regulates the heat stress response of Arabidopsis. Plant J. 53 264–274. 10.1111/j.1365-313X.2007.03334.x [DOI] [PubMed] [Google Scholar]
- Sessa G., Morelli G., Ruberti I. (1993). The ATHB-1 and -2 HD-Zip domains homodimerize forming complexes of different DNA binding specificities. EMBO J. 12 3507–3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahnejat-Bushehri S., Allu A. D., Mehterov N., Thirumalaikumar V. P., Alseekh S., Fernie A. R., et al. (2017). Arabidopsis NAC transcription factor JUNGBRUNNEN1 exerts conserved control over gibberellin and brassinosteroid metabolism and signaling genes in tomato. Front. Plant Sci. 8:214. 10.3389/fpls.2017.00214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahnejat-Bushehri S., Mueller-Roeber B., Balazadeh S. (2012). Arabidopsis NAC transcription factor JUNGBRUNNEN1 affects thermomemory-associated genes and enhances heat stress tolerance in primed and unprimed conditions. Plant Signal. Behav. 7 1518–1521. 10.4161/psb.22092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahnejat-Bushehri S., Tarkowska D., Sakuraba Y., Balazadeh S. (2016). Arabidopsis NAC transcription factor JUB1 regulates GA/BR metabolism and signalling. Nat. Plants 2:16013. 10.1038/nplants.2016.13 [DOI] [PubMed] [Google Scholar]
- Shao H., Wang H., Tang X. (2015). NAC transcription factors in plant multiple abiotic stress responses: progress and prospects. Front. Plant Sci. 6:902. 10.3389/fpls.2015.00902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva A. T., Ribone P. A., Chan R. L., Ligterink W., Hilhorst H. W. (2016). A predictive co-expression network identifies novel genes controlling the seed-to-seedling phase transition in Arabidopsis thaliana. Plant Physiol. 170 2218–2231. 10.1104/pp.15.01704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söderman E., Hjellström M., Fahleson J., Engström P. (1999). The HD-Zip gene ATHB6 in Arabidopsis is expressed in developing leaves, roots and carpels and up-regulated by water deficit conditions. Plant Mol. Biol. 40 1073–1083. 10.1023/A:1006267013170 [DOI] [PubMed] [Google Scholar]
- Son O., Hur Y. S., Kim Y. K., Lee H. J., Kim S., Kim M. R., et al. (2010). ATHB12, an ABA-inducible homeodomain-leucine zipper (HD-Zip) protein of Arabidopsis, negatively regulates the growth of the inflorescence stem by decreasing the expression of a gibberellin 20-oxidase gene. Plant Cell Physiol. 51 1537–1547. 10.1093/pcp/pcq108 [DOI] [PubMed] [Google Scholar]
- Tak H., Negi S., Ganapathi T. R. (2017). Banana NAC transcription factor MusaNAC042 is positively associated with drought and salinity tolerance. Protoplasma 254 803–816. 10.1007/s00709-016-0991-x [DOI] [PubMed] [Google Scholar]
- Thirumalaikumar V. P., Devkar V., Mehterov N., Ali S., Ozgur R., Turkan I., et al. (2017). NAC transcription factor JUNGBRUNNEN1 enhances drought tolerance in tomato. Plant Biotechnol. J. 10.1111/pbi.12776 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran L.-S. P., Nishiyama R., Yamaguchi-Shinozaki K., Shinozaki K. (2010). Potential utilization of NAC transcription factors to enhance abiotic stress tolerance in plants by biotechnological approach. GM Crops 1 32–39. 10.4161/gmcr.1.1.10569 [DOI] [PubMed] [Google Scholar]
- Uno Y., Furihata T., Abe H., Yoshida R., Shinozaki K., Yamaguchi-Shinozaki K. (2000). Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc. Natl. Acad. Sci. U.S.A. 97 11632–11637. 10.1073/pnas.190309197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Wang H., Shao H., Tang X. (2016). Recent advances in utilizing transcription factors to improve plant abiotic stress tolerance by transgenic technology. Front. Plant Sci. 7:67. 10.3389/fpls.2016.00067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu A., Allu A. D., Garapati P., Siddiqui H., Dortay H., Zanor M.-I., et al. (2012). JUNGBRUNNEN1, a reactive oxygen species-responsive NAC transcription factor, regulates longevity in Arabidopsis. Plant Cell 24 482–506. 10.1105/tpc.111.090894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K., Kasuga M., Liu Q., Miura S., Shinozaki K. (1999). Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat. Biotechnol. 17 287–291. 10.1038/7036 [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K., Shinozaki K. (1993). Characterization of the expression of a desiccation-responsive rd29 gene of Arabidopsis thaliana and analysis of its promoter in transgenic plants. Mol. Gen. Genet. 236 331–340. 10.1007/BF00277130 [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K., Shinozaki K. (1994). A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell 6 251–264. 10.1105/tpc.6.2.251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T., Sakuma Y., Todaka D., Maruyama K., Qin F., Mizoi J., et al. (2008). Functional analysis of an Arabidopsis heat-shock transcription factor HsfA3 in the transcriptional cascade downstream of the DREB2A stress-regulatory system. Biochem. Biophys. Res. Commun. 11 515–521. 10.1016/j.bbrc.2008.01.134 [DOI] [PubMed] [Google Scholar]
- You J., Chan Z. (2015). ROS regulation during abiotic stress responses in crop plants. Front. Plant Sci. 6:1092. 10.3389/fpls.2015.01092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Ye H., Guo H., Johnson A., Zhang M., Lin H., et al. (2014). Transcription factor HAT1 is phosphorylated by BIN2 kinase and mediates brassinosteroid repressed gene expression in Arabidopsis. Plant J. 77 59–70. 10.1111/tpj.12368 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
FIGURE S1 | Histochemical GUS staining of ProJUB1:GUS lines upon drought stress. (A) Expression level of JUB1 in leaves of 4-week-old WT plants subjected to 6 days of dehydration (6DD) compared to well-watered (Mock) plants. Transcript levels were determined by qRT-PCR and normalized applying the dCt method. Values are expressed as the difference between an arbitrary value of 40 and dCt, so that high 40 – dCt value indicates high gene expression level. Means ± SD calculated from three independent biological replicates, with nine leaves from three plants in each replicate. ACTIN2 was used as reference gene. Asterisks indicate significant difference in the level of JUB1 between 6DD and mock-treated samples (p < 0.05; Student’s t-test). (B) Induction of JUB1 promoter by drought stress, shown by histochemical GUS staining in leaves of Arabidopsis ProJUB1:GUS plants (1000-bp JUB1 promoter) plants. Plants were grown for 4 weeks under well-watered conditions and then irrigation was stopped for 6, 9, and 12 days. (C) Quantification of GUS signal in leaves of 14-day-old ProJUB1:GUS seedlings drought-stressed on filter paper for 2 h (‘Drought’) or kept in humidified condition (‘Mock’). Promoters of 0.68 and 0.31 kb length were analyzed. GUS signal was quantified as reported (Béziat et al., 2017) by selecting rectangular areas of maximum size of individual leaves. High gray values represent elevated GUS staining. Means ± SD of 20 leaves from 6 to 8 seedlings each.
FIGURE S2 | Behavior of transgenic JUB1 and AtHB13 plants during water deficit. (A) Phenotype of JUB1 and AtHB13 transgenic lines under severe drought stress. WT, JUB1Ox, jub1-1, AtHB13Ox, athb13-1, and athb13-2 plants grown in well-watered condition for 20 days and then subjected to severe drought stress for 8 days by completely stopping watering at day 20. (B) Kinetics of relative water loss of leaves from wild-type (WT) and transgenic Arabidopsis plants. (C) Relative water loss in leaves after day 8 of drought stress. For each genotype, leaves from five plants (one leaf per plant) were analyzed. Error bars represent SD. Plants in (B,C) were grown as in (A). (D) Kinetics of water consumption in WT and transgenic (jub1-1, JUB1Ox, AtHB13Ox) plants during mild drought stress. Plants were grown in soil under well-watered condition until day 25. Then, field capacity was maintained at 50% in all pots by adding the needed quantity of water. The volume of water added to each plant is shown. The graph represents the average of three experiments ± SD calculated from data obtained from five plants per genotype in each experiment. (E) Total amount of water added to each plant line until the end of the experiment. The graph represents the average of three independent experiments ± SD calculated from data obtained from five plants per genotype in each experiment. Asterisks in (C,E) indicate significant difference from WT (Student’s t-test; p < 0.05).
TABLE S1 | Constructs and primer sequences.