Abstract
Currently, preclinical testing of therapies for hepatoblastoma (HB) is limited to subcutaneous and intrasplenic xenograft models that do not recapitulate the hepatic tumors seen in patients. We hypothesized that injection of HB cell lines into the livers of mice would result in liver tumors that resemble their clinical counterparts. HepG2 and Huh-6 HB cell lines were injected, and tumor growth was monitored with bioluminescence imaging (BLI) and magnetic resonance imaging (MRI). Levels of human α-fetoprotein (AFP) were monitored in the serum of animals. Immunohistochemical and gene expression analyses were also completed on xenograft tumor samples. BLI signal indicative of tumor growth was seen in 55% of HepG2- and Huh-6-injected animals after a period of four to seven weeks. Increased AFP levels correlated with tumor growth. MRI showed large intrahepatic tumors with active neovascularization. HepG2 and Huh-6 xenografts showed expression of β-catenin, AFP, and Glypican-3 (GPC3). HepG2 samples displayed a consistent gene expression profile most similar to human HB tumors. Intrahepatic injection of HB cell lines leads to liver tumors in mice with growth patterns and biologic, histologic, and genetic features similar to human HB tumors. This orthotopic xenograft mouse model will enable clinically relevant testing of novel agents for HB.
Introduction
Hepatoblastoma (HB) is the most common malignant liver tumor seen in children1. The disease is most often diagnosed in patients under five years of age and is usually sporadic but can also be associated with familial adenomatous polyposis, Beckwith-Wiedemann syndrome, or prematurity2. Five-year overall survival (OS) of patients with stage I and II disease is above 95%, but patients with stage IV disease have a five-year OS rate of about 40%3. Standard treatment for HB consists of surgery and high dose, non-targeted chemotherapy, which leads to multiple damaging and long term side effects, including ototoxicity and cardiotoxicity4–6. Thus, new treatment strategies are needed, especially for high-risk patients.
To date, in vivo HB research includes studies with hydrodynamic injection of oncogenes for liver specific expression7, as well as subcutaneous and intrasplenic murine xenograft models8–10. Unfortunately, these models do not recapitulate the disease seen in a majority of patients, which is a large primary tumor encompassing one to four segments of the liver3. Mice with tumors generated with hydrodynamic injection develop multifocal nodules within the liver, and the organ is eventually entirely replaced by tumor. This may be representative of patients that present with tumor in all four segments of the liver, but this is only a small percentage of patients3. With the subcutaneous and intrasplenic xenograft models, tumors can be quickly generated in genetically identical animals from the human HB cell lines Huh-611, HepT18, and HepG212. In the subcutaneous model, injection of all three cell lines led to growth of tumors, depending on the strain of mice and time elapsed since injection of cells8,9. In the intrasplenic model, immunodeficient mice were directly injected with HepG2, Huh-6, or HepT1 cells into the spleen. The Huh-6 and HepT1 tumor cells, but not HepG2 cells, then migrated to the liver, giving rise to intrahepatic tumors9,10. Of note, animals that underwent splenectomy just after injection more readily developed intrahepatic tumors10. These tumors were small, multifocal nodules that again do not represent the disease typically seen in children. Notably, there is one published study of injection of HepG2 cells into the portal vein to generate intrahepatic tumors, but the focus in this work is use of this model for drug testing for hepatocellular carcinoma (HCC)13. Thus, although these models have contributed to the field, none truly recapitulates the disease. For effective preclinical studies to be performed, a true intrahepatic orthotopic xenograft model that accurately replicates the human disease is essential. We have successfully developed an intrahepatic patient-derived xenograft (PDX) model of HB using patient specimens14. Other groups have also examined subcutaneous and intrahepatic growth of patient-derived liver cancer tissues as models of HCC, including an interesting study in which tumors composed of sorted human liver cancer stem cells (hLCSCs) were grown subcutaneously15,16. Since these tissues have limited availability due to the rarity of the disease, we wanted to develop and characterize an intrahepatic, orthotopic xenograft model using commercially available HB cell lines. In addition, cell line derived xenograft models can be better standardized and are not dependent on tissue quality of surgical samples that usually have been exposed to chemotherapy.
In this paper, we describe the development and characterization of such an intrahepatic xenograft HB mouse model. Human HB cells were injected into the livers of immunocompromised mice. Mice were monitored for tumor growth using bioluminescence imaging (BLI), magnetic resonance imaging (MRI), and measurement of serum levels of human α-fetoprotein (AFP). At the conclusion of the study, animals were euthanized and tissues were harvested for protein expression analysis by immunohistochemistry and gene expression profiling by RNA sequencing.
Results
Generation of tumors by intrahepatic injection of human HB cell lines
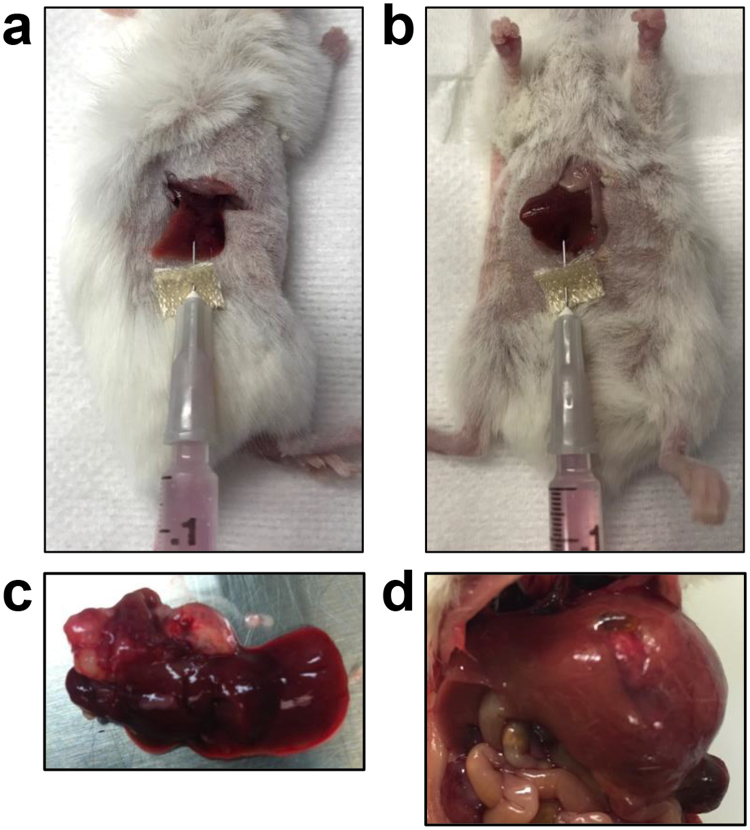
To generate liver tumors, we injected two million HepG2 or Huh-6 human HB cell lines into either the right median lobe with a right flank incision (Fig. 1a) or the left lateral lobe with a midline, abdominal incision (Fig. 1b) of the liver of NOD/Shi-scid/IL-2Rγnull (NOG) mice (see methods). Regardless of injection technique, the tumors grew as large, exophytic masses originating from the injected lobe and invaded multiple adjacent segments of the liver as seen in children with pre-treatment extent of disease (PRETEXT) I, II, and III tumors (Fig. 1c,d) 17.
Figure 1.
Injection of HB cells into the mouse liver to generate xenograft tumors. Cells were injected either into the right median lobe (a) or the left lateral lobe (b). (c,d) Representative gross tumors generated with injection of Huh-6 cells into the right median lobe (c) or HepG2 cells into the left lateral lobe (d).
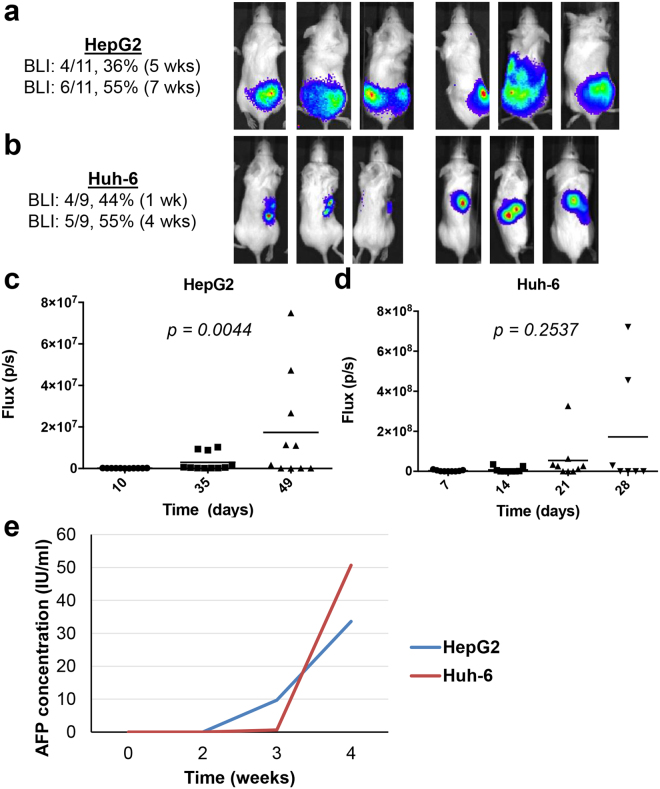
In order to longitudinally monitor the growth of tumors in vivo, we used cells that had been transduced with lentiviral vectors expressing the luciferase gene. The expression of strong luciferase activity (2–3 million relative luminescence (RLU)) was confirmed prior to implantation of tumor cells. Upon intraperitoneal injection of luciferin into the animals, the cells emitted a BLI signal that could be monitored from week to week (Fig. 2a–d). Four of 11 (36%) mice injected with HepG2 cells showed BLI signal at 5 weeks after injection, and 6 of 11 (55%) displayed BLI signal at 7 weeks (Fig. 2a). Four of 9 (44%) mice injected with Huh-6 cells demonstrated BLI signal at 1 week after injection, and 5 of 9 (55%) exhibited BLI signal at 3 weeks after injection (Fig. 2b). Temporal analysis of BLI signals reflects the growth of tumors (Fig. 2c,d) and shows that changes in the HepG2 masses are significant (p = 0.0044). Taken together, the data demonstrates that intrahepatic injection of both HB cell lines successfully leads to the growth of liver tumors in immunocompromised mice.
Figure 2.
BLI signals and serum AFP levels of mice injected with HB cells. (a,b) Three representative mice with HepG2 (a) or Huh-6 (b) xenografted tumors shown at early and late time points with number of mice growing tumors indicated. (c,d) Increase in BLI signals shown at each time point with associated p values determined with the Kruskal-Wallis test to indicate significance. (e) Serum human AFP increases with tumor growth in mice harboring xenograft tumors.
Human AFP is elevated in the serum of mice harboring xenograft tumors
One of the most important indicators used for diagnosis and disease surveillance in patients with HB is levels of AFP in the blood, and 97% of patients show elevated levels18. We hypothesized that levels of human AFP would be elevated in the serum of the mice harboring the xenograft tumors. To test this hypothesis, we measured serum human AFP levels weekly in one mouse representative of each cell line xenograft using an enzyme-linked immunosorbent assay (ELISA). At the time of injection of cells and at one week after injection, levels of human AFP in the blood remained very low (Fig. 2e). By three weeks after injection, animals with both xenograft tumors showed elevated human AFP in their serum (Fig. 2e).
MRI of xenograft tumors
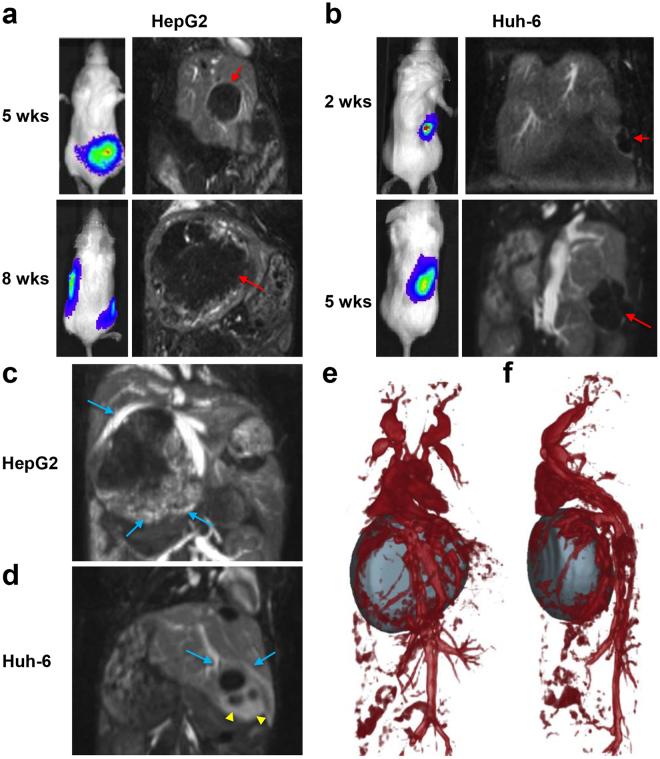
MRI was performed at early and late time points to confirm the presence of tumor and for 3D assessment of tumor burden. Intrahepatic tumors were visible as hypo-intense regions in the lobes of the liver (Fig. 3a,b). Contrast-enhanced T1-weighted imaging was performed using a long circulating blood pool liposomal contrast agent for assessing intra-tumoral vasculature and spatial relationships between tumor and major hepatic blood vessels. The blood pool agent showed the presence of intra-tumoral vasculature with a high degree of vascularity specificity at the tumor periphery (Fig. 3c–f). As the tumors grew, major hepatic vessels, including the inferior vena cava (IVC), were pushed from their regular anatomical orientation.
Figure 3.
MRI of xenograft tumors and tumor-associated vasculature. (a,b) Representative contrast-enhanced T1-weighted coronal images showing the presence of tumor masses (red arrows) at early and late time points. Representative BLI images acquired on the same day are shown for comparison. (c,d) Representative contrast-enhanced T1-weighted coronal thick slab maximum intensity projection (MIP) abdominal images demonstrating the presence of increased vascularity (blue arrows) and highly permeable vessels (yellow arrowheads) in the peripheral regions of the tumors. (e,f) Coronal (e) and sagittal (f) 3D volume rendering MRI of HepG2 tumor and major hepatic blood vessels.
Histological examination of xenograft tumors
At the conclusions of these studies, animals were euthanized and samples were harvested for histological and immunohistochemical analyses. Histological review of the xenografted tumors indicated that those generated with both HB cell lines resemble primary human HB samples to varying degrees (Fig. 4a,b). HepG2-derived tumors most closely resemble human HBs of homogeneous, embryonal phenotype (Fig. 4a). In contrast, Huh-6 tumors had morphologic characteristics that differ from the general histology of primary tumors (Fig. 4b). Huh-6 xenotransplants demonstrated an usual pattern with tumor cells organized in papillary structures generally uncharacteristic of HB (Fig. 4b). Of note, histology was similar to Huh-6 tumors obtained previously in the subcutaneous model19.
Figure 4.
Histological examination of xenograft tumors. Samples representative of each xenograft tumor were stained with H&E (a,b) and antibodies recognizing β-catenin (c,d), AFP (e,f), and GPC3 (g,h), three proteins known to be elevated in HB. (a,b) HepG2 (a) shows classic HB embryonal morphology while Huh-6 (b) demonstrates areas of embryonal HB histology and a papillary pattern. (c,d) Cytoplasmic and nuclear β-catenin expression in HepG2 (c) and Huh-6 (d) samples. (e,f) AFP expression is elevated in HepG2 samples (e) but not in Huh-6 samples (f). (g,h) High GPC3 expression in HepG2 (g) and Huh-6 (h) tumors. Scale bars represent 100 μm in 10X images and 20 μm in 40X images (a–d,g,h) and 25 μm in 10X and 40X images (e,f).
An immunohistochemistry panel, including AFP, β-catenin, and Glypican-3 (GPC3), commonly used in HB diagnosis and classification20 was employed to evaluate protein expression in the mouse xenograft tumors. AFP is variable in HB specimens since this protein is secreted and the key measurement is the levels of AFP in the serum20,21. Of the two cell line xenografts, only HepG2 tumors showed clear positive AFP staining throughout; Huh-6 tumors were predominantly negative with scattered positive patches of cells (Fig. 4e,f, Supplementary Fig. S1a–c). GPC3 is a reliable marker that is expressed in epithelial, fetal, and embryonal components and is negative in normal liver and benign tumor tissues22. Both HepG2 and Huh-6 xenograft tumors showed strong cytoplasmic and membrane staining for this marker (Fig. 4g,h). As a second assessment of expression of AFP and GPC3, we performed quantitative reverse transcription polymerase chain reaction (RT-PCR) (qPCR) experiments to measure levels of mRNA expression of these two HB markers in HepG2 and Huh-6 cells, in comparison to expression in the terminally differentiated hepatic cell line HepRG and in human fibroblasts (HFs). AFP and GPC3 expression are both significantly elevated in HepG2 and Huh-6 cells (Supplementary Fig. S1d,e).
Finally, β-catenin is arguably the most important marker of HB as nuclear staining is used as a surrogate for the presence of mutations in the β-catenin gene, CTNNB1, which are commonly found in cases of HB23. Nuclear β-catenin expression is only seen in malignant hepatocytes24. In both the HepG2 and Huh-6 xenograft tumors, β-catenin staining was strong throughout the tumor nuclei (Fig. 4c,d). Taken together, histological analyses show that the xenograft tumors resemble primary HB tumors.
Mutation analysis of cell lines and xenograft tumors
We further characterized the cell lines by performing mutational analyses of CTNNB1 and the Telomerase reverse transcriptase (TERT) promoter gene previously reported to be present in each cell line (Table 1)23. Both cell lines carry mutations in CTNNB1. HepG2 carries a large in-frame deletion, p.W25_I140del (116 codons within exons 3 and 4)25, while Huh-6 carries a CTNNB1 point mutation, p.G34V25. HepG2 is also reported to carry a mutation of the TERT promoter, G228A26.
Table 1.
Mutation analysis of HB cell lines and xenograft samples.
| CTNNB1 | TERT promoter | |
|---|---|---|
| HepG2-C | p.W25_I140del (116 codons within exons 3 and 4) | G228A |
| HepG2-T | p.W25_I140del (116 codons within exons 3 and 4) | G228A |
| Huh-6-C | p.G34V | wild-type |
| Huh-6-T | p.G34V | wild-type |
Two parental cell lines are HepG2-C and Huh-6-C; two cell lines grown in vivo as xenograft tumors are HepG2-T and Huh-6-T.
Gene expression profiling of xenograft tumors
We used RNA sequencing to profile HepG2 and Huh-6 gene expression in vitro, including both parental and luciferase-transduced cell lines, and xenograft tumors generated in vivo. As an initial analysis, we compared transcriptome similarities between the luciferase-transduced cells and the parental cells (Fig. 5d) to show that the luciferase-transduced cells had almost identical gene expression as the parental cells. We then analyzed six normal liver samples, nine primary human untreated HB samples, and four in vitro and in vivo HepG2 and Huh-6 samples by principal component analysis (PCA). PCA of these profiles revealed three main clusters of samples: normal livers, primary tumors, and the in vitro and in vivo HepG2 and Huh-6 samples (Fig. 5a). In vitro and in vivo HepG2 profiles showed greater similarity than the corresponding Huh-6 profiles (Fig. 5a).
Figure 5.
Transcriptome profiling of HB cell lines and tumor samples. (a) PCA of RNA sequencing data of two parental HB cell lines (HepG2-C, Huh-6-C) and two cell lines grown in vivo as xenograft tumors (HepG2-T, Huh-6-T), compared with RNA sequencing data of six normal liver samples and nine primary HB tumor samples. The two HepG2 samples and the Huh-6 sample grown in vitro form a cluster, indicating that their gene expression is similar to each other. The Huh-6 tumor sample is located far from the other clusters, showing that it has more transcriptome differences from the primary tumors and other cell line and xenograft samples. (b) GSEA to reveal transcriptome differences between cells grown in vitro and in vivo as xenograft tumors. (c) Molecular clustering of HB cell lines and tumor samples. Heat map analysis of 16-gene HB signature27 in nine primary HB tumors (yellow) and four HB cell line and xenograft samples (HepG2 (green), Huh-6 (blue)), normalized to six normal liver samples. The cell line and xenograft samples show the C2 signature, which is associated with a worse prognosis. (d) Spearman correlation of 18571 HUGO genes to indicate gene expression similarities of two parental cell lines and two luciferase-transduced cell lines all grown in vitro.
To reveal the major differences in the Huh-6 tumor sample that occur as a result of being grown in vivo, we conducted gene set enrichment analyses (GSEA) of each pair of in vitro cell and in vivo tumor samples with two gene sets from the Broad Molecular Signatures Database (Fig. 5b, Supplementary Table S2). Seventeen pathways in the Hallmark gene set were significantly changed between the two Huh-6 samples while nine pathways were significantly altered between the two HepG2 samples (Fig. 5b). Eleven pathways were significantly enriched in the Huh-6 tumor sample (Fig. 5b), including seven that are involved in the immune system and inflammation (interferon alpha response, interferon gamma response, allograft rejection, inflammatory response, Tumor necrosis factor alpha (TNFα) signaling via Nuclear factor kappa B (NFκB), complement, Interleukin 6 (IL-6)/Janus kinase (JAK)/Signal transducer and activator of transcription 3 (STAT3) signaling). These pathways were not enriched in the HepG2 tumor sample, although four were increased in the HepG2 cell sample (TNFα signaling via NFκB, allograft rejection, IL-6/JAK/STAT3 signaling, inflammatory response) (Fig. 5b). Enrichment for genes that function in the epithelial-mesenchymal transition, KRAS signaling, hypoxia, and apoptosis was also seen in the Huh-6 tumor sample (Fig. 5b), and these changes were not seen in either HepG2 sample. Unique differences seen in the Huh-6 cell sample included higher expression of MYC targets, E2F targets, Wnt/β-catenin signaling, G2M checkpoint, and estrogen response. Higher expression of Mammalian target of rapamycin complex 1 (MTORC1) signaling was seen in both the Huh-6 and HepG2 cell samples. Taken together, these statistical analyses suggest that more gene sets are changed between the Huh-6 in vitro and in vivo samples than the two HepG2 samples, many of which are connected to the immune response.
In a third, specific analysis of the RNA sequencing dataset, we analyzed the expression of a previously published prognostic 16-gene signature that has been reported to differentiate between a low risk, better prognosis HB cluster (C1) versus a high risk, poor prognosis HB cluster (C2)27. We used a heat map to classify expression in the two cell lines and corresponding xenograft tumor samples compared to the nine primary HB patient samples, all normalized to average expression in the normal liver samples (Fig. 5c). In general, all of the human HB tumor samples showed a C2 gene expression profile.
Discussion
HB is the most common pediatric primary liver tumor. Although generally a rare cancer, it leads to the death of more than half of high-risk patients even with intensive chemotherapy and surgical interventions3. The current clinical trials in the U.S. (AHEP0731) and Europe (SIOPEL-4) all use non-targeted chemotherapy in their regimens including primarily cisplatin and doxorubicin4. These therapies have significant side-effects with about 50% of patients exhibiting ototoxicity and 5% showing cardiotoxicity5,6. Thus, novel therapeutic agents are needed. However, preclinical testing of such therapies has lagged due to the paucity of clinically relevant animal models of the disease. PDX models derived from primary tumors are very promising but have limited availability due to the rarity of the disease14,28. Subcutaneous and intrasplenic models utilizing commercially available cell lines do not accurately recapitulate tumors seen in patients9,10. In addition, a relevant study of intrahepatic xenograft tumors generated with HepG2 cells focused on this as a model of HCC instead of HB13. Hence, there is a compelling need for an intrahepatic xenograft model with human HB cell lines that replicates this complex disease, and, in this paper, we describe such a model that will enhance testing of new therapies.
Importantly, this intrahepatic HB xenograft model recapitulates the key hallmarks of the disease: elevated serum AFP levels, large exophytic tumors with active blood supplies, and embryonal histological phenotype with elevation of AFP, GPC3, and β-catenin. Although elevation of protein levels of AFP were not detected in Huh-6 tumors with immunohistochemistry experiments, significant increases in mRNA expression of both AFP and GPC3 were seen in HepG2 and Huh-6 cells. Because of this discrepancy, we verified our data by staining the Huh-6 tumors with a total of four different AFP antibodies. With all four antibodies, the tissues were predominantly negative with limited scattered patches of positive cells. In the literature, it is still unclear whether detection of AFP with immunohistochemistry assays correlates with serum levels of this protein. In a large immunohistochemistry study of 83 patient samples comprising a mix of diagnostic tumor biopsies and post-chemotherapy, post-surgical specimens, no statistically significant correlation was found between levels of serum AFP at diagnosis and with expression of AFP in resected tumors21. We speculate that such secreted proteins have high rates of protein turnover in cells and thus are not always detectable with immunohistochemistry assays. This study shows that direct intrahepatic orthotopic injection of widely distributed HB cell lines in immunodeficient mice leads to primary hepatic tumors that morphologically mimic human HB tumors, and this is the first report that characterizes such cell line-derived xenograft tumors in relationship to primary HB tumors in order to demonstrate clear clinical relevance.
In our study, we examined the complete gene expression profiles of all cell lines and xenograft tumor samples in comparison to normal liver and HB tumor samples, which has not been described previously. In PCA of the total RNA sequencing dataset, both HepG2 samples and the Huh-6 sample grown in vitro clustered close to each other. However, the Huh-6 sample that was grown in vivo differed from what would be expected given its human HB tumor origin. Interestingly, gene expression changes in the Huh-6 tumor that seem to have occurred as a result of being grown in vivo result in cells with a profile different than the cell line grown in vitro. These results correlate with the histological review, as the Huh-6 xenograft tumor does not resemble primary HB disease as closely as the HepG2 tissue.
We speculate that this inconsistency may be due to the environment of the murine liver affecting the cell line as studies have shown that there are key differences between the human and mouse liver, especially in regards to transcriptional regulation and general gene expression among homologous genes29,30. Previous studies of in vivo models of HB have shown differences in histological appearance of cell line-derived tumors depending on their location of growth in the animal9,10. No previous studies have analyzed gene expression changes in HB cell lines grown in vitro and in vivo, but papers about other types of cancer have shown similar differences depending on whether cells are grown in vitro or in vivo and on location of growth in vivo 31,32.
In support of results from PCA, GSEA suggested more extensive divergence of the Huh-6 in vivo sample, including gene expression changes that indicate immune response and inflammation. It is well accepted that growth of human cells in animals requires the support of animal cells and that human and animal cells interface and interact with each other. Perhaps this contact is leading to unique upregulation of immune pathways within the Huh-6 cells, leading to more widespread alterations in gene expression in Huh-6 cells that are not seen in HepG2 tumors.
Taken together, the results of this study show that the commercially available HB cell lines can be used for in vivo preclinical testing; however, HepG2 more accurately mimics human HB. This study also makes it clear that more cell lines are needed to study HB biology and treatment response; therefore, large centers that treat many patients must come together to develop new patient-derived cell lines and xenografts. With the new international collaborations33,34 that have been formed to study HB, there will be more availability of tissues to develop these valuable resources.
Materials and Methods
All animal procedures used in this study were performed under and in accordance with an animal protocol approved by the Institutional Care and Use Committee of Baylor College of Medicine.
Cells and culture conditions
The HepG2, Huh-6, and HepRG cell lines used in this study were commercially acquired (HepG2: American Type Culture Collection (ATCC), Manassas, VA; Huh-6: Riken Cell Bank, Japan; HepRG: ATCC). Cell lines were grown at 37 °C in 5% CO2 in Eagle’s Minimum Essential Medium (EMEM, Lonza, Allendale, NJ) supplemented with 10% heat-inactivated fetal bovine serum (FBS, SAFC Biosciences, Lenexa, KS), 2 mM glutamine (Invitrogen, Carlsbad, CA), and 100 units/ml streptomycin/penicillin (Invitrogen). Cell lines were validated by DNA mutation analyses for known CTNNB1 and TERT promoter mutations (see below).
Orthotopic mouse model
In vivo studies were performed in female NOD/Shi-scid/IL-2Rγnull (NOG) mice (Taconic Biosciences, Hudson, NY). 2 × 106 HepG2 and Huh-6 cells transduced with luciferase and resuspended in 100 μl phosphate-buffered saline (PBS) were surgically implanted into either the right lobe of the liver through a right flank incision or the left lobe with a midline, abdominal incision. The mice underwent BLI beginning at 10 days after implantation and every week thereafter with the In Vivo Imaging System (IVIS, PerkinElmer, Waltham, MA), and luminescence flux was recorded to assess tumor growth. After seven weeks (HepG2) or four weeks (Huh-6), necropsy was performed, intrahepatic and extrahepatic sites of tumor were noted, and samples were harvested for immunohistochemistry and RNA isolation.
ELISA to measure circulating AFP in mouse blood
Blood was drawn from the facial veins of mice harboring xenograft tumors at the indicated time points. AFP was measured in the serum of animals with an AFP ELISA kit (EIA-1468, DRG Instruments, Germany).
In vivo MRI
MRI was performed on a 1.0 T permanent MRI scanner (M2 system, Aspect Technologies, Israel). A 35 mm volume coil was used for transmit and receive of radiofrequency (RF) signal. Mice were sedated using 3% isoflurane, setup on the MRI animal bed, and then maintained under anesthesia at 1–1.5% isoflurane delivered using a nose cone setup. Body temperature was maintained by circulating hot water through the MRI animal bed. Respiration rate was monitored using a pneumatically controlled pressure pad placed in the abdominal area underneath the animal. A long circulating liposomal-Gd blood pool contrast agent (SC-Gd liposomes) was systemically administered via the tail vein at a dose of 0.1 mmol Gd/kg and used for contrast-enhanced T1-weighted imaging35. High-resolution contrast-enhanced MRI (CE-MRI) was performed using a T1-weighted 3D gradient echo (GRE) sequence with the following scan parameters: echo time (TE) = 3.5 ms, repetition time (TR) = 20 ms, flip angle = 70, slice thickness = 0.3 mm, field of view = 54 mm, number of slices = 68, matrix = 180 × 180, NEX = 1, spatial resolution = 300 µm2, scan time ~5 min. Images were analyzed and processed in Osirix (version 5.8.5, 64-bit, Pixmeo, Bernex, Switzerland). Tumor volumes were segmented and 3D volume-rendered images were generated in Slicer (version 4.4.0)36.
Immunohistochemistry of tumor tissues
Tissue samples were fixed in 4% paraformaldehyde (PFA, Alfa Aesar, Ward Hill, MA) overnight at 4 °C. Tissues were then dehydrated in 70% ethanol until processing in paraffin. Samples were processed in the Texas Medical Center Digestive Diseases Center (Houston, TX). H&E and immunohistochemical staining of AFP (CP028, Biocare Medical, Concord, CA (Supplementary Fig. S1a); AC-0166, Epitomics, Cell Marque, Rocklin, CA (Supplementary Fig. S1c); ab46799, Abcam, Cambridge, MA (Supplementary Fig. S1b); 180003, Thermo Fisher Scientific, Waltham, MA (Fig. 4e)), β-catenin (ab32573, Abcam), and GPC3 (GPC3, 11395, Santa Cruz Biotechnology, Dallas, TX) were performed. Imaging of tumor sections on slides was done on a DMi8 microscope (Leica, Germany).
Quantitative RT-PCR with cell lines
RNA was extracted from cells with the Direct-zol RNA MiniPrep Kit (Zymo Research, Irvine, CA, USA). RNA purity and quantity were determined using a spectrophotometer measuring absorbance at 260/280 nm. cDNA was generated from total RNA with the SuperScript III First-Strand Synthesis System for RT-PCR (Invitrogen) or the qScript cDNA SuperMix (Quanta Biosciences, Gaithersburg, MD, USA). Taqman qPCR was done with TaqMan Universal Master Mix II (Applied Biosystems, Foster, CA, USA) and with the following primers (Applied Biosystems): AFP (Hs1040598_m1) and GPC3 (Hs01018936_m1). GAPDH (Hs02758991_g1) was used as an internal control in all qPCR experiments. All experiments were run on a StepOnePlus Real-Time PCR System (Applied Biosystems).
Mutation analysis of cell lines and xenograft tumors
DNA from frozen xenograft tissues and cell lines was extracted using the QIAamp DNA Mini Kit (Qiagen, Germany). Samples were treated with RNase A and eluted in Buffer AE. CTNNB1 exon 3 and 4 and TERT promoter mutation status was determined by PCR of genomic DNA with primers listed in Supplementary Table S1 37. Two-directional Sanger sequencing analysis of PCR products was completed with Mutation Surveyor v. 5.0.1 (Softgenetics).
RNA sequencing of hepatic tumors and comparison with parental cell lines and liver cancer samples
RNA from frozen hepatic tumor samples was isolated using the mirVana miRNA isolation kit (Ambion, Austin, TX). Samples were treated with DNase 1 and eluted in nuclease-free water. RNA from xenograft tumors and parental cell lines was isolated using the RNeasy Plus Mini Kit (74134, Qiagen). RNA samples were submitted for RNA sequencing to the Baylor College of Medicine Genomic and RNA Profiling Core (Houston, TX) or to GENEWIZ (South Plainfield, NJ). RNA-seq FastQ files were processed using STAR38 and Cufflinks39, with alignment to Hg19/GRCh37. HB tumor and normal liver samples were processed via Illumuna HiSeq. 250040. HepG2 was sequenced on both platforms, and a multiplicative scaling vector was used to scale the cell line and xenograft values for comparison with the patient tumor and normal liver samples. Fragments per kilobase per million (FPKM) values for 2324 genes were included after filtering for Human Genome Organization (HUGO) Gene Nomenclature Committee (HGNC) protein coding genes with average expression above 25 FPKM and coefficient of variation (CV) above 0.3 over the set of all samples. Spearman correlation of 18571 HUGO genes to indicate gene expression similarities of two parental cell lines and two luciferase-transduced cell lines all grown in vitro. PCA was implemented in R (Team, R. D. C. R: A language and environment for statistical computing, http://www.R-project.org (2014)) using the prcomp function with the scale parameter set to adjust variables to unit variance.
GSEA of xenograft tumors and cell lines
Read FASTQ files were processed with RSEM (version 1.2.17)41 using the read aligner Bowtie242 applied to the combined human and mouse NCBI Refseq (3/21/16) transcriptomes. Next, TPM values from human transcripts were selected and renormalized. Using a gene expression cutoff of 7.5 transcripts per million (TPM) and CV above 0.3 for HGNC protein coding genes, two sample comparison gene set enrichment was implemented using the GSEA (v2.23) program43 in pre-ranked mode with an extended “Signal2Noise” metric score for ranking. The Broad Molecular Signatures Database (MSigDB v5.2) sets h (Hallmark) and c2 (Curated gene sets) were used.
Statistical analysis
The Kruskal-Wallis test was used to determine the statistical significance of in vivo tumor flux differences among the indicated time points.
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Electronic supplementary material
Acknowledgements
We would like to thank Saakshi Bhayana and Dr. Igor Stupin for assistance with MRI. We would like to thank Pamela Parsons in the Texas Medical Center Digestive Diseases Center for completing the immunohistochemistry experiments with our samples. S.A.V. is supported by a Texas Children’s Department of Surgery Seed Award and the Macy Easom Cancer Research Foundation. K.D.B. is supported by the National Heart, Lung, and Blood Institute (NHLBI) grant R01HL134510, the Texas Hepatocellular Carcinoma Consortium (THCC) CPRIT grant RP150587, and the Diane Helis Henry and Adrienne Helis Malvin Medical Research Foundation. The Baylor College of Medicine Dan L. Duncan Cancer Center is supported by NIH grant P30CA125123. The Texas Medical Center Digestive Diseases Center is supported by NIH grant P30DK56338.
Author Contributions
S.A.V. conceived of the study, designed experiments, and procured funding. K.D.B., K.B.G., P.S., D.L.T. and S.A.V. supervised the study. S.E.W., Y.S., R.H.P., J.J., A.M., S.F.S., Z.S., B.Z., S.S.G., Z.C. and A.M.I. performed experiments. S.E.W., Y.S., R.H.P., S.F.S., Z.S., B.Z., K.B.G., P.S., D.L.T. and S.A.V. analyzed and interpreted data. S.E.W. and S.A.V. wrote the manuscript. All authors discussed results, commented on the manuscript, and approved the final manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Sarah E. Woodfield, Yan Shi and Roma H. Patel contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-17665-8.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Darbari A, Sabin KM, Shapiro CN, Schwarz KB. Epidemiology of primary hepatic malignancies in U.S. children. Hepatology. 2003;38:560–566. doi: 10.1053/jhep.2003.50375. [DOI] [PubMed] [Google Scholar]
- 2.Hiyama E. Pediatric hepatoblastoma: diagnosis and treatment. Translational Pediatrics. 2014;3:293–299. doi: 10.3978/j.issn.2224-4336.2014.09.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meyers RL, et al. Predictive power of pretreatment prognostic factors in children with hepatoblastoma: a report from the Children’s Oncology Group. Pediatr. Blood Cancer. 2009;53:1016–1022. doi: 10.1002/pbc.22088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Watanabe K. Current chemotherapeutic approaches for hepatoblastoma. Int J Clin Oncol. 2013;18:955–961. doi: 10.1007/s10147-013-0616-8. [DOI] [PubMed] [Google Scholar]
- 5.Sivaprakasam P, Gupta AA, Greenberg ML, Capra M, Nathan PC. Survival and long-term outcomes in children with hepatoblastoma treated with continuous infusion of cisplatin and doxorubicin. J Pediatr Hematol Oncol. 2011;33:e226–30. doi: 10.1097/MPH.0b013e31821f0eaf. [DOI] [PubMed] [Google Scholar]
- 6.Zsiros J, et al. Dose-dense cisplatin-based chemotherapy and surgery for children with high-risk hepatoblastoma (SIOPEL-4): a prospective, single-arm, feasibility study. Lancet Oncology. 2013;14:834–842. doi: 10.1016/S1470-2045(13)70272-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen X, Calvisi DF. Hydrodynamic transfection for generation of novel mouse models for liver cancer research. The American Journal of Pathology. 2014;184:912–923. doi: 10.1016/j.ajpath.2013.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pietsch T, et al. Characterization of the continuous cell line HepT1 derived from a human hepatoblastoma. Lab Invest. 1996;74:809–818. [PubMed] [Google Scholar]
- 9.Schnater JM, et al. Subcutaneous and intrahepatic growth of human hepatoblastoma in immunodeficient mice. Journal of Hepatology. 2006;45:377–386. doi: 10.1016/j.jhep.2006.03.018. [DOI] [PubMed] [Google Scholar]
- 10.Ellerkamp V, et al. Successful establishment of an orthotopic hepatoblastoma in vivo model in NOD/LtSz-scid IL2Rgammanull mice. PLoS ONE. 2011;6:e23419. doi: 10.1371/journal.pone.0023419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tanaka M, et al. Production of fibronectin by HUH6 C15 cell line established from a human hepatoblastoma. Biochem Biophys Res Commun. 1983;110:837–841. doi: 10.1016/0006-291X(83)91037-9. [DOI] [PubMed] [Google Scholar]
- 12.Doi I. Establishment of a cell line and its clonal sublines from a patient with hepatoblastoma. Gan. 1976;67:1–10. [PubMed] [Google Scholar]
- 13.Ong L-C, et al. Effective inhibition of xenografts of hepatocellular carcinoma (HepG2) by rapamycin and bevacizumab in an intrahepatic model. Mol Imaging Biol. 2009;11:334–342. doi: 10.1007/s11307-009-0213-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bissig-Choisat B, et al. Novel patient-derived xenograft and cell line models for therapeutic testing of pediatric liver cancer. Journal of Hepatology. 2016;65:325–333. doi: 10.1016/j.jhep.2016.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu M, et al. HULC cooperates with MALAT1 to aggravate liver cancer stem cells growth through telomere repeat-binding factor 2. Nature Publishing Group. 2016;6:36045. doi: 10.1038/srep36045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ahmed, S. U. et al. Generation of subcutaneous and intrahepatic human hepatocellular carcinoma xenografts in immunodeficient mice. J Vis Exp e50544, 10.3791/50544 (2013). [DOI] [PMC free article] [PubMed]
- 17.Roebuck, D. J. et al. 2005 PRETEXT: a revised staging system for primary malignant liver tumours of childhood developed by the SIOPEL group. Pediatr Radiol37, 123–32– quiz 249–50 (2007). [DOI] [PMC free article] [PubMed]
- 18.Ortega JA, et al. Randomized comparison of cisplatin/vincristine/fluorouracil and cisplatin/continuous infusion doxorubicin for treatment of pediatric hepatoblastoma: A report from the Children’s Cancer Group and the Pediatric Oncology Group. Journal of Clinical Oncology. 2000;18:2665–2675. doi: 10.1200/JCO.2000.18.14.2665. [DOI] [PubMed] [Google Scholar]
- 19.Browne M, et al. Gene transfer of pigment epithelium-derived factor suppresses tumor growth and angiogenesis in a hepatoblastoma xenograft model. Pediatr Res. 2006;60:282–287. doi: 10.1203/01.pdr.0000232789.86632.91. [DOI] [PubMed] [Google Scholar]
- 20.Lopez-Terrada D, et al. Towards an international pediatric liver tumor consensus classification: proceedings of the Los Angeles COG liver tumors symposium. Mod Pathol. 2014;27:472–491. doi: 10.1038/modpathol.2014.27. [DOI] [PubMed] [Google Scholar]
- 21.Purcell R, et al. Potential biomarkers for hepatoblastoma: results from the SIOPEL-3 study. European Journal of Cancer. 2012;48:1853–1859. doi: 10.1016/j.ejca.2011.10.019. [DOI] [PubMed] [Google Scholar]
- 22.Zynger DL, et al. Expression of glypican 3 in hepatoblastoma: an immunohistochemical study of 65 cases. Hum Pathol. 2008;39:224–230. doi: 10.1016/j.humpath.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 23.Eichenmuller M, et al. The genomic landscape of hepatoblastoma and their progenies with HCC-like features. Journal of Hepatology. 2014;61:1312–1320. doi: 10.1016/j.jhep.2014.08.009. [DOI] [PubMed] [Google Scholar]
- 24.Park WS, et al. Nuclear localization of beta-catenin is an important prognostic factor in hepatoblastoma. J Pathol. 2001;193:483–490. doi: 10.1002/1096-9896(2000)9999:9999<::AID-PATH804>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 25.Koch A, et al. Childhood hepatoblastomas frequently carry a mutated degradation targeting box of the beta-catenin gene. Cancer Research. 1999;59:269–273. [PubMed] [Google Scholar]
- 26.Cevik D, Yildiz G, Ozturk M. Common telomerase reverse transcriptase promoter mutations in hepatocellular carcinomas from different geographical locations. World Journal of Gastroenterology. 2015;21:311–317. doi: 10.3748/wjg.v21.i1.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cairo S, et al. Hepatic stem-like phenotype and interplay of Wnt/beta-catenin and Myc signaling in aggressive childhood liver cancer. Cancer Cell. 2008;14:471–484. doi: 10.1016/j.ccr.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 28.Nicolle D, et al. Patient-derived mouse xenografts from pediatric liver cancer predict tumor recurrence and advise clinical management. Hepatology. 2016;64:1121–1135. doi: 10.1002/hep.28621. [DOI] [PubMed] [Google Scholar]
- 29.Yu Y, et al. A comparative analysis of liver transcriptome suggests divergent liver function among human, mouse and rat. Genomics. 2010;96:281–289. doi: 10.1016/j.ygeno.2010.08.003. [DOI] [PubMed] [Google Scholar]
- 30.Odom DT, et al. Tissue-specific transcriptional regulation has diverged significantly between human and mouse. Nature Genetics. 2007;39:730–732. doi: 10.1038/ng2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mitra AK, et al. In vivo tumor growth of high-grade serous ovarian cancer cell lines. Gynecologic Oncology. 2015;138:372–377. doi: 10.1016/j.ygyno.2015.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Camphausen K, et al. Influence of in vivo growth on human glioma cell line gene expression: convergent profiles under orthotopic conditions. Proc. Natl. Acad. Sci. USA. 2005;102:8287–8292. doi: 10.1073/pnas.0502887102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Czauderna P, et al. European Journal of Cancer. 2016. TheChildren’s Hepatic tumors International Collaboration (CHIC): Novel global rare tumor database yields new prognostic factors in hepatoblastoma and becomes a research model; pp. 92–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meyers RL, et al. Risk-stratified staging in paediatric hepatoblastoma: a unified analysis from the Children’s Hepatic tumors International Collaboration. Lancet Oncology. 2017;18:122–131. doi: 10.1016/S1470-2045(16)30598-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ghaghada KB, et al. New dual mode gadolinium nanoparticle contrast agent for magnetic resonance imaging. PLoS ONE. 2009;4:e7628. doi: 10.1371/journal.pone.0007628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fedorov A, et al. 3D Slicer as an image computing platform for the Quantitative Imaging Network. Magn Reson Imaging. 2012;30:1323–1341. doi: 10.1016/j.mri.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sumazin P, et al. Genomic Analysis of Hepatoblastoma Identifies Distinct Molecular and Prognostic Subgroups. Hepatology. 2016 doi: 10.1002/hep.28888. [DOI] [PubMed] [Google Scholar]
- 38.Dobin A, et al. Bioinformatics. 2013. STAR: ultrafast universal RNA-seq aligner; pp. 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trapnell C, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bissig-Choisat B, et al. Novel Patient-Derived Xenograft and Cell Line Models for Therapeutic Testing of Pediatric Liver Cancer. Journal of Hepatology. 2016 doi: 10.1016/j.jhep.2016.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.