Abstract
EGFR-TKIs show dramatic treatment benefits for advanced lung adenocarcinoma patients with activating EGFR mutations. Considering the essential role of autophagy in EGFR-TKIs treatments, we hypothesized that genetic variants in autophagy core genes might contribute to outcomes of advanced lung adenocarcinoma treated with gefitinib. We systematically examined 27 potentially functional genetic polymorphisms in 11 autophagy core genes among 108 gefitinib-treated advanced lung adenocarcinoma patients. We found that ATG10 rs10036653, ATG12 rs26538, ATG16L1 rs2241880 and ATG16L2 rs11235604 were significantly associated with survival of lung adenocarcinoma patients (all P < 0.05). Among EGFR-mutant patients, ATG5 rs688810, ATG5 rs510432, ATG7 rs8154, ATG10 rs10036653, ATG12 rs26538, ATG16L1 rs2241880 and ATG16L2 rs11235604 significantly contributed to disease prognosis. We also found that ATG5 rs510432, ATG5 rs688810, ATG10 rs10036653 and ATG10 rs1864182 were associated with primary or acquired resistance to gefitinib. Functional analyses of ATG10 rs10036653 polymorphism suggested that ATG10 A allele might increase transcription factor OCT4 binding affinity compared to the T allele in lung cancer cells. Our results indicate that autophagy core genetic variants show potential clinical implications in gefitinib treatment, especially among advanced lung adenocarcinoma patients, highlighting the possibility of patient-tailored decisions during EGFR-TKIs based on both germline and somatic variation detection.
Introduction
Lung cancer is one of most common and lethal cancers worldwide. Currently, it is classified to two major pathological types1. About 80% of lung cancer patients are characterized as non-small cell lung cancer (NSCLC) and 20% as small cell lung cancer (SCLC). For NSCLC, there are several subtypes, such as adenocarcinoma, squamous cell carcinoma, and large cell carcinoma, and all types can occur in unusual histologic variants2. NSCLC are relatively insensitive to chemotherapy and/or radiotherapy compared to SCLC2. Epidermal growth factor receptor (EGFR) with activating mutations has been proved to be a promising therapeutic target of EGFR tyrosine kinase inhibitors (EGFR-TKIs) for NSCLC3,4. Compared with platinum-based chemotherapy, EGFR-TKIs show great advantages by significantly prolonging progression-free survival (PFS)5. Advanced NSCLC patients, especially ones with adenocarcinoma histology and EGFR active mutations, show great clinical benefits from EGFR-TKIs6. The frequency of EGFR mutations are highest in East Asia populations including Chinese7–9. However, most patients, even cases with EGFR mutations, develop drug resistance after a median PFS of 10–16 months, followed by disease progression after initial EGFR-TKIs treatment10. The detailed mechanisms responsible for EGFR-TKIs resistance are still not fully understood, which greatly limited their application in clinic.
Autophagy is an evolutionarily conserved process which is essential for survival, differentiation, development, and homeostasis. As a lysosomal degradation pathway, autophagy can maintain cell homeostasis through degrading damaged organelles and long-lived proteins11,12. It has been reported that autophagy is involved in multiple diseases, for example cancers, infections, neurodegeneration and aging13–16. During cancer development, autophagy is considered as a non-apoptotic cell death pathway and suppresses tumorigenesis under certain circumstances. However, autophagy facilitates tumorigenesis in most contexts17–19. Autophagosome is a kind of spherical organelle with double layer membranes during autophagy. Establishment of autophagosome is controlled by several autophagy core genes20, which might be involved in cancer initiation and progression21.
Accumulating evidences indicate that germline genetic variants may also play a part in resistance to EGFR-TKIs. For instance, Ng et al. reported that NSCLC patients harboring EGFR mutations showed better clinical response to TKIs if the patients carried a germline deletion polymorphism in BCL2L11 (BIM) at the same time22. Moreover, we also found that EGFR germline polymorphisms (rs2293347 and rs4947492) might be potential predictive markers of overall survival (OS) in advanced lung adenocarcinoma patients treated with gefitinib23. In the current study, we hypothesized that genetic variants of autophagy core genes may contribute to differential prognostic outcomes of advanced lung adenocarcinoma patients treated with gefitinib. To address this, we systematically examined the clinical implications of 23 potentially functional polymorphisms in ten autophagy core genes (ATG2B, ATG3, ATG4C, ATG5, ATG7, ATG9B, ATG10, ATG12, ATG16L2 and BECN) in advanced lung adenocarcinoma who received gefitinib therapy.
Materials and Methods
Study subjects
There is a total of 108 patients with advanced lung adenocarcinoma treated with gefitinib in this study (Supplementary Table 1). Patients were recruited between July 2003 and July 2012 at Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology (Wuhan, Hubei Province, China). As reported previously, eligible patients had at least one measurable lesion with a minimum size in at least one diameter of ≥10 mm for liver, lung, brain or lymph node metastases, WHO performance status of 0-1, and life expectancy of ≥3 months23. Each patient was treated with gefitinib orally at a daily dose of 250 mg as 2nd or 3rd line monotherapy. The exclusion criteria included previous other EGFR-TKIs treatment, pneumonectomy or severe cardio-pulmonary diseases23. This study was approved by the Review Boards of Tongji Hospital, Tongji Medical College and Shandong Cancer Hospital affiliated to Shandong University. Written informed consent from each patient for the use of his/her DNA and clinical information was obtained. The methods were carried out in accordance with the approved guidelines.
Genetic variants selection of autophagy core genes
Single nucleotide polymorphisms (SNPs) of autophagy core genes were selected as previously described24. In briefly, common SNPs (MAF ≥ 0.05 in Chinese Han population) in eleven autophagy core genes (ATG2B, ATG3, ATG4C, ATG5, ATG7, ATG9B, ATG10, ATG12, ATG16L1, ATG16L2 and BECN) were screened in the gene regions including a 10-kb up-stream region of each gene based on the HapMap database. A total of 27 potentially functional SNPs were finally selected according to linkage disequilibrium (LD) analyses with an r 2 threshold of 0.80 as well as prediction with SNPinfo Web Server (http://snpinfo.niehs.nih.gov/).
Genotyping
Genomic DNA was extracted from blood sample which was collected from each patient upon recruitment. The ATG3 rs2705507 polymorphism was excluded from the 27 SNPs since it cannot be analyzed by the MassArray system (Sequenom Inc., San Diego, California, USA). The other 26 SNPs were finally determined to be genotyped as described previously25–28. However, BECN rs9890617, rs9891429 and rs10512488 were excluded because of genotyping failure. As a result, a total of 23 SNPs were successfully genotyped. A 15% blind, random samples were genotyped in duplicates and the reproducibility was 100%.
Quantitative reverse transcription PCR (qRT-PCR)
After lung cancer A549 cells were transfected with siRNAs of OCT4, MTF1 or SOX5 (Supplementary Table 2), total RNA was isolated from cells with Trizol reagent (Invitrogen) and treated with RNase-Free DNase to remove genomic DNA (Invitrogen). These RNA samples were then reverse transcribed into cDNAs using Revert Ace kit (TOYOBO, Osaka, Japan). OCT4, MTF1, SOX5 ATG5, ATG10 and β-actin mRNAs were measured through the SYBR-Green qRT-PCR. The OCT4, MTF1, SOX5 ATG5 or ATG10 expression was calculated relative to the β-actin expression.
Electrophoretic Mobility-Shift Assays (EMSA)
Synthetic double-stranded and 3′ biotin-labeled oligonucleotides corresponding to the ATG10 rs10036653T or rs10036653A sequences (Supplementary Table 2) and A549 cell nuclear extracts were incubated at 25 °C for 20 min using the Light Shift Chemiluminescent EMSA Kit (Pierce, Rockford, IL). The reaction mixture was separated on 6% PAGE, and the products were detected by Stabilized Streptavidin-Horseradish Peroxidase Conjugate (Pierce).
Statistics
The differences in patients’ characteristics were assessed by Pearson’s χ2 tests or Student’s t test. Univariate and multivariate Cox proportional hazard regression analyses were utilized to calculate hazard ratios (HRs) and 95% confidence intervals (CIs). Sex, age, smoking status, ECGO and stages were used as adjustment factors during multivariate analyses. Survival differences were examined using the log-rank test. P values less than 0.05 were considered significant. All P values represent two-sided statistical tests. All statistical procedures were conducted using the SPSS software (version 19.0) and GraphPad Prism7.
Results
Advanced lung adenocarcinoma patients’ characteristics and clinical outcomes
As shown in Supplementary Table 1, the distribution of demographic and clinical characteristics of patients were summarized. A total of 108 advanced lung adenocarcinoma patients were enrolled in this study. All individuals were ethnic Han Chinese. The mean age of subjects was 56 ranging from 46 to 66. There were 53 males and 55 females. All the patients were treated with gefitinib and 75% patients were detected with EGFR mutations. By the time of the final analysis, the median follow-up time was 29.0 months, and the median PFS and OS was 12.4 and 24.4 months, respectively.
Effects the autophagy core gene SNPs on PFS and OS of gefitinib-treated advanced lung adenocarcinoma patients
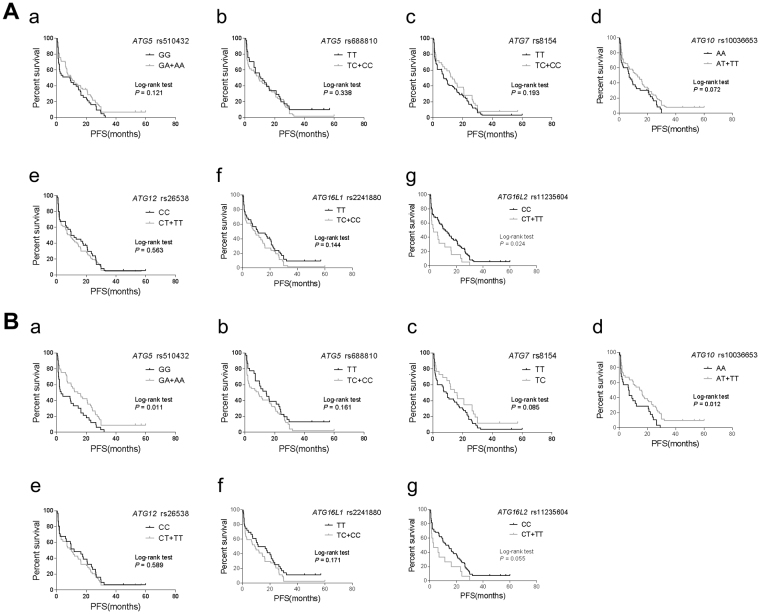
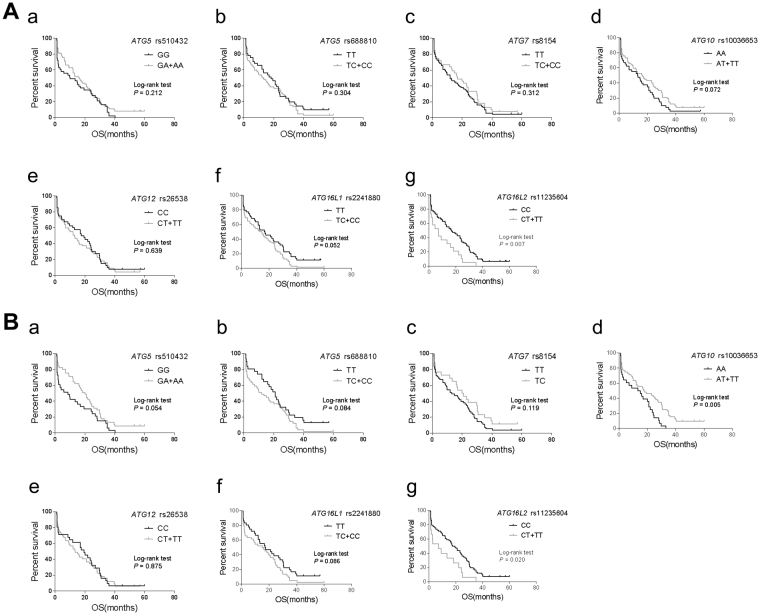
The detailed information of selected 26 autophagy core gene SNPs were summarized in Supplementary Table 3. A total of 23 potentially functional SNPs from 11 autophagy core genes were successfully genotyped. These SNPs were located in introns, exons, 3′ UTR and promoters of autophagy core genes. Associations between the 23 SNPs and PFS or OS were examined using multivariate Cox regression analyses among all patients as well as patients with EGFR mutations. As shown in Table 1, ATG10 rs10036653, ATG12 rs26538, ATG16L1 rs2241880 and ATG16L2 rs11235604 were significantly associated with OS of gefitinib-treated advanced lung adenocarcinoma patients (all P < 0.05). ATG12 rs26538 TT genotype also significantly contributed to increased risk of shorten PFS (HR = 2.47, 95% CI = 1.15-5.31, P = 0.021). In the stratified analyses, ATG5 rs510432 A allele, ATG7 rs8154 T allele and ATG10 rs10036653 C allele were proved to be protective alleles which were significantly associated with good prognosis of patients with EGFR mutations (PFS: HR = 0.55, 95% CI = 0.32-0.92, P = 0.022 for ATG5 rs510432; HR = 0.57, 95% CI = 0.33-0.99, P = 0.045 for ATG7 rs8154; HR = 0.48, 95% CI = 0.29-0.79, P = 0.004 for ATG10 rs10036653; OS: HR = 0.61, 95% CI = 0.37-1.00, P = 0.050; HR = 0.56, 95% CI = 0.32-0.96, P = 0.034; HR = 0.46, 95% CI = 0.27-0.76, P = 0.003). ATG5 rs688810, ATG12 rs26538, ATG16L1 rs2241880 and ATG16L2 rs11235604 were risk SNPs whose minor alleles were significantly associated with bad prognosis of patients with EGFR mutations (PFS: HR = 1.83, 95% CI = 1.08-3.08 for ATG5 rs688810, P = 0.025; HR = 2.51, 95% CI = 1.00-6.30, P = 0.049 for ATG12 rs26538; HR = 1.64, 95% CI = 1.00-2.68, P = 0.050 for ATG16L1 rs2241880; HR = 1.92, 95% CI = 1.04-3.55, P = 0.036 for ATG16L2 rs11235604; OS: HR = 1.76, 95% CI = 1.06-2.91, P = 0.028; HR = 3.17, 95% CI = 1.22-8.23, P = 0.018; HR = 1.72, 95% CI = 1.06-2.79, P = 0.027; HR = 2.28, 95% CI = 1.24-4.22, P = 0.008). However, other genetic variants of autophagy core genes did not significantly affect PFS or OS (all P > 0.05) (Supplementary Table 4 and Supplementary Table 5).
Table 1.
Associations of genetic variants of autophagy core genes with OS and PFS of advanced lung adenocarcinoma patients treated with gefitinib.
| Genes | SNPs | Genotypes | Patients No. (%) | OS | PFS | ||
|---|---|---|---|---|---|---|---|
| HR (95% CI) | P | HR (95% CI) | P | ||||
| ATG5 | rs510432 | 105 | |||||
| GG | 43(40.95) | Reference | Reference | ||||
| GA | 41(39.05) | 0.72 (0.44-1.17) | 0.180 | 0.79 (0.48-1.31) | 0.362 | ||
| AA | 21(20.00) | 0.63 (0.34-1.15) | 0.129 | 0.64 (0.34-1.19) | 0.157 | ||
| GA + AA | 62(59.05) | 0.73 (0.48-1.12) | 0.152 | 0.79 (0.50-1.23) | 0.290 | ||
| ATG5 | rs688810 | 106 | |||||
| TT | 41(38.68) | Reference | Reference | ||||
| TC | 43(40.57) | 1.14 (0.71-1.82) | 0.590 | 1.21 (0.74-1.98) | 0.446 | ||
| CC | 22(20.75) | 1.44 (0.83-2.50) | 0.191 | 1.36 (0.77-2.40) | 0.289 | ||
| TC + CC | 65(61.32) | 1.30 (0.85-1.98) | 0.238 | 1.31 (0.85-2.04) | 0.226 | ||
| ATG7 | rs8154 | 108 | |||||
| TT | 69(63.89) | Reference | Reference | ||||
| TC | 37(34.26) | 0.76 (0.49-1.18) | 0.220 | 0.71 (0.45-1.14) | 0.155 | ||
| CC | 2(1.85) | 1.55 (0.30-8.02) | 0.600 | 1.98 (0.37-10.51) | 0.425 | ||
| TC + CC | 39(36.11) | 0.77 (0.51-1.18) | 0.235 | 0.75 (0.48-1.17) | 0.200 | ||
| ATG10 | rs10036653 | 107 | |||||
| AA | 40(37.38) | Reference | Reference | ||||
| AT | 52(48.60) | 0.70 (0.43-1.11) | 0.130 | 0.66 (0.40-1.07) | 0.090 | ||
| TT | 15(14.02) | 0.43 (0.21-0.89) | 0.022 | 0.56 (0.27-1.14) | 0.108 | ||
| AT + TT | 67(62.62) | 0.67 (0.43-1.03) | 0.068 | 0.66 (0.43-1.04) | 0.074 | ||
| ATG12 | rs26538 | 107 | |||||
| CC | 40(37.38) | Reference | Reference | ||||
| CT | 54(50.47) | 1.08 (0.69-1.70) | 0.738 | 0.99 (0.62-1.57) | 0.966 | ||
| TT | 13(12.15) | 2.83 (1.31-6.15) | 0.008 | 2.47 (1.15-5.31) | 0.021 | ||
| CT + TT | 67(62.62) | 1.19 (0.079-1.81) | 0.404 | 1.12 (0.73-1.71) | 0.608 | ||
| ATG16L1 | rs2241880 | 106 | |||||
| TT | 44(41.51) | Reference | Reference | ||||
| TC | 44(41.51) | 1.63 (1.01-2.61) | 0.044 | 1.42 (0.87-2.32) | 0.158 | ||
| CC | 18(16.98) | 1.62 (0.85-3.08) | 0.142 | 1.69 (0.89-3.22) | 0.109 | ||
| TC + CC | 62(58.49) | 1.62 (1.05-2.49) | 0.029 | 1.42 (0.92-2.20) | 0.118 | ||
| ATG16L2 | rs11235604 | 108 | |||||
| CC | 89(82.41) | Reference | Reference | ||||
| CT | 18(16.67) | 1.78 (1.07-2.96) | 0.028 | 1.54 (0.93-2.56) | 0.094 | ||
| TT | 1(0.93) | N.C. | N.C. | N.C. | N.C. | ||
| CT + TT | 19(17.60) | 1.83 (1.11-3.02) | 0.018 | 1.59 (0.97-2.61) | 0.068 | ||
| Genes | SNPs | Genotypes | Patients with EGFR mutation No. (%) | OS of patients with EGFR mutations | PFS of patients with EGFR mutations | ||
| HR (95% CI) | P | HR (95% CI) | P | ||||
| ATG5 | rs510432 | 78 | |||||
| GG | 33(42.31) | Reference | Reference | ||||
| GA | 28(35.90) | 0.64 (0.36-1.15) | 0.136 | 0.59 (0.32-1.09) | 0.094 | ||
| AA | 17(21.79) | 0.58 (0.30-1.15) | 0.118 | 0.47 (0.23-0.96) | 0.038 | ||
| GA + AA | 45(57.69) | 0.61 (0.37-1.00) | 0.050 | 0.55 (0.32-0.92) | 0.022 | ||
| ATG5 | rs688810 | 80 | |||||
| TT | 31(38.75) | Reference | Reference | ||||
| TC | 31(38.75) | 1.49 (0.84-2.64) | 0.172 | 1.58 (0.87-2.89) | 0.135 | ||
| CC | 18(22.50) | 2.03 (1.09-3.79) | 0.026 | 1.98 (1.04-3.77) | 0.039 | ||
| TC + CC | 49(61.25) | 1.76 (1.06-2.91) | 0.028 | 1.83 (1.08-3.08) | 0.025 | ||
| ATG7 | rs8154 | 81 | |||||
| TT | 55(67.90) | Reference | Reference | ||||
| TC | 26(32.50) | 0.56 (0.32-0.96) | 0.034 | 0.57 (0.33-0.99) | 0.045 | ||
| CC | 0(0.00) | N.C. | N.C. | N.C. | N.C. | ||
| TC + CC | 26(32.50) | 0.56 (0.32-0.96) | 0.034 | 0.57 (0.33-0.99) | 0.045 | ||
| ATG10 | rs10036653 | 80 | |||||
| AA | 28(35.00) | Reference | Reference | ||||
| AT | 43(53.75) | 0.49 (0.29-0.84) | 0.009 | 0.51 (0.30-0.86) | 0.012 | ||
| TT | 9(11.25) | 0.23 (0.08-0.87) | 0.007 | 0.39 (0.15-1.03) | 0.057 | ||
| AT + TT | 52(65.00) | 0.46 (0.27-0.76) | 0.003 | 0.48 (0.29-0.79) | 0.004 | ||
| ATG12 | rs26538 | 80 | |||||
| CC | 31(38.75) | Reference | Reference | ||||
| CT | 40(50.00) | 0.95 (0.56-1.82) | 0.858 | 1.00 (0.58-1.71) | 0.991 | ||
| TT | 9(11.25) | 3.17 (1.22-8.23) | 0.018 | 2.51 (1.00-6.30) | 0.049 | ||
| CT + TT | 49(61.25) | 1.12 (0.69-1.82) | 0.657 | 1.14 (0.70-1.86) | 0.608 | ||
| ATG16L1 | rs2241880 | 80 | |||||
| TT | 36(45.00) | Reference | Reference | ||||
| TC | 31(38.75) | 1.83 (1.07-3.13) | 0.027 | 1.76 (1.02-3.03) | 0.044 | ||
| CC | 13(16.25) | 1.65 (0.81-3.35) | 0.168 | 1.68 (0.82-3.42) | 0.156 | ||
| TC + CC | 44(55.00) | 1.72 (1.06-2.79) | 0.027 | 1.64 (1.00-2.68) | 0.050 | ||
| ATG16L2 | rs11235604 | 81 | |||||
| CC | 66(81.48) | Reference | Reference | ||||
| CT | 14(17.28) | 2.24 (1.20-4.19) | 0.012 | 1.88 (1.01-3.52) | 0.047 | ||
| TT | 1(1.23) | N.C. | N.C. | N.C. | N.C. | ||
| CT + TT | 15(18.51) | 2.28 (1.24-4.22) | 0.008 | 1.92 (1.04-3.55) | 0.036 | ||
Note: PFS, progression-free survival; OS, overall survival; HR, hazard ratio; CI, confidence interval; N.C., not calculated.
Hazard ratios (HRs) and 95% confidence intervals (CIs) for the association between SNPs and PFS as well as OS were estimated by Cox regression adjusted by sex, age, smoking status, ECGO and stages.
We also compared the PFS or OS of patients with different genotypes of the aforementioned seven autophagy SNPs (Fig. 1 and Fig. 2). Log-rank tests demonstrated that patients harboring ATG16L2 rs11235604 T allele had significantly shorten PFS compared to the C allele (3.0 months vs 11.8 months, P = 0.024) (Fig. 1A). Among EGFR mutant patients, ATG5 rs510432 A allele or ATG10 rs10036653 T allele showed significantly prolonged PFS compared to ATG5 rs510432 G allele or ATG10 rs10036653 A allele (4.37 months vs 3.07 months, P = 0.011 for ATG5 rs510432; 15.37 months vs 6.82 months, P = 0.012 for ATG10 rs10036653) (Fig. 1B). Similarly, carriers of ATG16L2 rs11235604 T allele had significantly shorten OS after gefitinib treatment compared to the C allele (7.1 months vs 17.0 months, P = 0.007 for all patients; 7.1 months vs 18.4 months, P = 0.020 for EGFR mutant patients) (Fig. 2A,B). Also, EGFR mutant patients with ATG10 rs10036653 T allele showed significantly prolonged OS compared to the A allele (17.85 months vs 14.18 months, P = 0.005) (Fig. 2B). These results elucidated that ATG5 rs510432, ATG10 rs10036653 and ATG16L2 rs11235604 germline polymorphisms might be independent prognostic marker of gefitinib treatment besides somatic EGFR mutations.
Figure 1.
Kaplan-Meier curves of PFS for advanced lung adenocarcinoma patients treated with gefitinib. (A) Kaplan-Meier curves of PFS for all NSCLC patients harboring ATG5 rs510432 (a), ATG5 rs6888810 (b), ATG7 rs8154 (c), ATG10 rs10036653 (d), ATG12 rs26538 (e), ATG16L1 rs2241880 (f) and ATG16L2 rs11235604 (g). (B) Kaplan-Meier curves of PFS for EGFR mutant NSCLC patients harboring ATG5 rs510432 (a), ATG5 rs6888810 (b), ATG7 rs8154 (c), ATG10 rs10036653 (d), ATG12 rs26538 (e), ATG16L1 rs2241880 (f) and ATG16L2 rs11235604 (g). Long-rank analysis was performed, and P values less than 0.05 were considered significant.
Figure 2.
Kaplan-Meier curves of OS for advanced lung adenocarcinoma patients treated with gefitinib. (A) Kaplan-Meier curves of OS for all NSCLC patients harboring ATG5 rs510432 (a), ATG5 rs6888810 (b), ATG7 rs8154 (c), ATG10 rs10036653 (d), ATG12 rs26538 (e), ATG16L1 rs2241880 (f) and ATG16L2 rs11235604 (g). (B) Kaplan-Meier curves of OS for EGFR mutant NSCLC patients harboring ATG5 rs510432 (a), ATG5 rs6888810 (b), ATG7 rs8154 (c), ATG10 rs10036653 (d), ATG12 rs26538 (e), ATG16L1 rs2241880 (f) and ATG16L2 rs11235604 (g). Long-rank analysis was performed, and P values less than 0.05 were considered significant.
Impacts of autophagy core gene SNPs on gefitinib-resistance
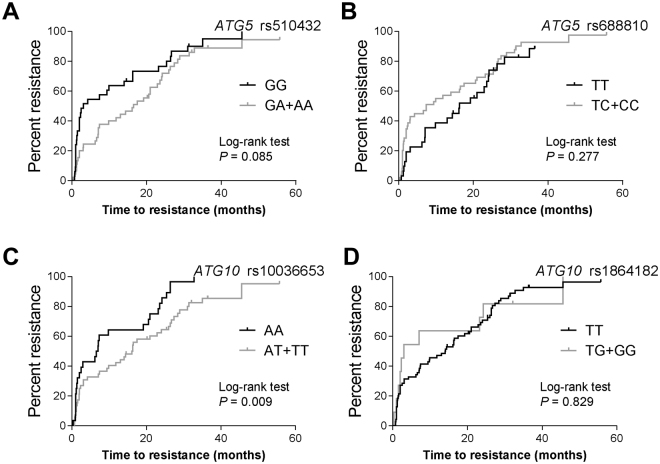
Drug resistance to EGFR-TKIs inevitably develops after a period of effective drug treatment. Here we investigated whether autophagy core gene SNPs could be used as reasonable biomarkers for gefitinib-resistance in advanced lung adenocarcinoma. As shown in Table 2, ATG5 rs510432 acts as a protective SNP significantly associated with 55% decreased risk of primary gefitinib resistance (95% CI = 0.24-0.87, P = 0.017). ATG10 rs1864182 was significantly associated with 2.27-fold elevated risk of primary gefitinib resistance (95% CI = 1.04-4.97, P = 0.040). On the contrary, ATG10 rs1864182 might be a protective SNP for acquired gefitinib resistance (HR = 0.30, 95% CI = 0.09-0.97, P = 0.044). Among patients with EGFR mutations, ATG5 rs510432 and rs688810 genetic variations were significantly associated with primary gefitinib resistance (rs510432 A allele: HR = 0.39, 95% CI = 0.18-0.85, P = 0.018; rs688810 C allele: HR = 3.01, 95% CI = 1.26-7.22, P = 0.014). Additionally, ATG10 rs10036653 was significantly associated with acquired gefitinib resistance of EGFR mutant patients (T allele: HR = 0.37, 95% CI = 0.19-0.72, P = 0.004), which was verified in log-rank test (AT and TT vs. AA: 16 months vs. 6.5 months, P = 0.009) (Fig. 3). However, other autophagy core gene SNPs did not significantly affect primary or acquired gefitinib-resistance (all P > 0.05) (Supplementary Table 6 and Supplementary Table 7).
Table 2.
Association of genetic variants of autophagy core genes with primary resistance or acquired resistance of gefitinib.
| Genes | SNPs | Genotypes | Patients No. (%) | Primary resistance | Acquired resistance | |||
|---|---|---|---|---|---|---|---|---|
| HR (95% CI) | P | HR (95% CI) | P | |||||
| ATG5 | rs510432 | GG | 105 | Reference | Reference | |||
| 43(40.95) | ||||||||
| GA | 41(39.05) | 0.54 (0.25-1.15) | 0.107 | 0.95 (0.55-1.62) | 0.838 | |||
| AA | 21(20.00) | 0.34 (0.13-0.93) | 0.035 | 1.28 (0.67-2.46) | 0.453 | |||
| GA + AA | 62(59.05) | 0.45 (0.24-0.87) | 0.017 | 1.05 (0.65-1.69) | 0.848 | |||
| ATG5 | rs688810 | TT | 106 | Reference | Reference | |||
| 41(38.68) | ||||||||
| TC | 43(40.57) | 1.67 (0.77-3.63) | 0.194 | 0.99 (0.52-1.89) | 0.972 | |||
| CC | 22(20.75) | 1.76 (0.74-4.20) | 0.200 | 0.96 (0.42-2.17) | 0.912 | |||
| TC + CC | 65(61.32) | 1.70 (0.85-3.41) | 0.132 | 1.03 (0.57-1.85) | 0.920 | |||
| ATG10 | rs10036653 | AA | 107 | Reference | Reference | |||
| 40(37.38) | ||||||||
| AT | 52(48.60) | 0.74 (0.35-1.54) | 0.418 | 0.61(0.32-1.18) | 0.142 | |||
| TT | 15(14.02) | 0.66 (0.23-1.86) | 0.432 | 0.39 (0.13-1.20) | 0.101 | |||
| AT + TT | 67(62.62) | 0.77 (0.40-1.50) | 0.446 | 0.59 (0.32-1.10) | 0.099 | |||
| ATG10 | rs1864182 | TT | 113 | Reference | Reference | |||
| 96(84.96) | ||||||||
| TG | 16(14.16) | 2.07 (0.91-4.72) | 0.082 | 0.30 (0.09-0.97) | 0.044 | |||
| GG | 1(0.88) | N.C. | N.C. | N.C. | N.C. | |||
| TG + GG | 17(15.04) | 2.27 (1.04-4.97) | 0.040 | 0.30 (0.09-0.97) | 0.044 | |||
| Genes | SNPs | Genotypes | Patients with EGFR mutation No. (%) | Primary resistance of patients with EGFR mutations | Acquired resistance of patients with EGFR mutations | |||
| HR (95% CI) | P | HR (95% CI) | P | |||||
| ATG5 | rs510432 | GG | 78 | Reference | Reference | |||
| 33(42.31) | ||||||||
| GA | 28(35.90) | 0.56 (0.22-1.41) | 0.217 | 0.65 (0.26-1.62) | 0.353 | |||
| AA | 17(21.79) | 0.24 (0.07-0.85) | 0.026 | 0.77 (0.29-2.05) | 0.605 | |||
| GA + AA | 45(57.69) | 0.39 (0.18-0.85) | 0.018 | 0.74 (0.34-1.60) | 0.448 | |||
| ATG5 | rs688810 | TT | 80 | Reference | Reference | |||
| 31(38.75) | ||||||||
| TC | 31(38.75) | 2.89 (1.05-8.01) | 0.041 | 0.94 (0.45-1.99) | 0.877 | |||
| CC | 18(22.50) | 3.02 (1.11-8.19) | 0.030 | 1.21 (0.46-3.23) | 0.700 | |||
| TC + CC | 49(61.25) | 3.01 (1.26-7.22) | 0.014 | 1.07 (0.54-2.10) | 0.854 | |||
| ATG10 | rs10036653 | AA | 80 | Reference | Reference | |||
| 28(35.00) | ||||||||
| AT | 43(53.75) | 0.66 (0.29-1.53) | 0.332 | 0.41 (0.21-0.83) | 0.013 | |||
| TT | 9(11.25) | 0.60 (0.17-2.12) | 0.423 | 0.29 (0.06-1.38) | 0.120 | |||
| AT + TT | 52(65.00) | 0.66 (0.31-1.40) | 0.278 | 0.37 (0.19-0.72) | 0.004 | |||
| ATG10 | rs1864182 | TT | 81 | Reference | Reference | |||
| 70(86.42) | ||||||||
| TG | 10(12.35) | 1.75 (0.63-4.87) | 0.283 | 0.55 (0.17-1.81) | 0.323 | |||
| GG | 1(1.23) | N.C. | N.C. | N.C. | N.C. | |||
| TG + GG | 11(13.58) | 2.00 (0.78-5.15) | 0.149 | 0.55 (0.17-1.81) | 0.323 | |||
Hazard ratios (HRs) and 95% confidence intervals (CIs) for the association between SNPs and geifitinib-resistance were estimated by Cox regression adjusted by sex, age, smoking status, ECGO and stages. N.C., not calculated.
Figure 3.
Gefitinib-resistance for EGFR mutant lung adenocarcinoma patients harboring different genotypes of autophagy core gene genes. (A) ATG5 rs510432, (B) ATG5 rs688810, (C) ATG10 rs10036653, (D) ATG10 rs1864182. P values less than 0.05 were considered significant.
ATG5 rs510432 and ATG10 rs10036653 may influence binding of transcription factors
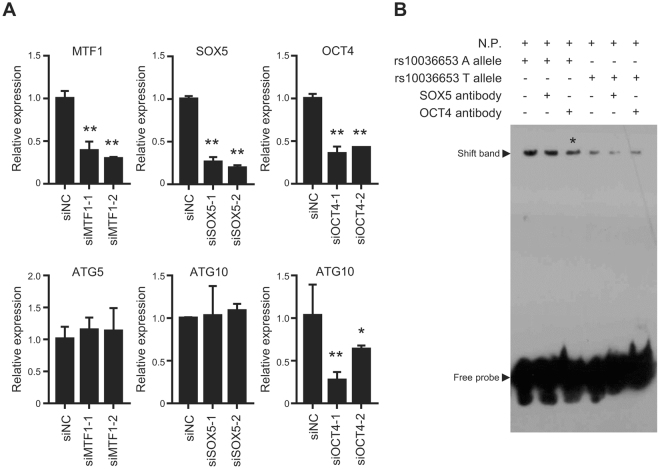
All autophagy core gene SNPs investigated in the current study were selected based on their MAF in Chinese Han population and potential function to their host genes. We found that ATG5 rs510532 and ATG10 rs10036653 contributed to not only survival but also drug resistance among gefitinib-treated advanced lung adenocarcinoma patients. Interestingly, both SNPs are upstream gene variants (Supplementary Table 3), which leads us to examine whether they could change transcription factor (TF) binding affinities to genomic sequences and, thus, affect gene regulation. By using RegulomeDB, an online bioinformatics tool (http://regulome.stanford.edu/)29, we found that ATG5 rs510432 might change the binding ability of Mtf1 to ATG5 promoter, and ATG10 rs10036653 may affect the binding affinities of Sox5 as well as Oct4 to ATG10 promoter (Table 3). We validated the bioinformatics prediction in A549 cells. After silencing endogenous expression of OCT4, MTF1 or SOX5 with siRNAs (siSOX5-1, siSOX5-2, siMTF1-1, siMTF1-2, siOCT4-1 and siOCT4-2), we found that only decreased expression of OCT4 can suppress ATG10 expression (Fig. 4A). These results indicate that OCT4 might acts as important TF impacting ATG10 expression. Because ATG10 rs10036653 SNP is located in a predicted OCT4 binding sequence, we then conducted EMSA to distinguish the differences in binding capacity between the rs10036653T or rs10036653A alleles. As shown in Fig. 4B, we found that OCT4-containing A549 nuclear extracts bound more to the biotin-labeled oligonucleotide probe with the A allele sequence compared to the T allele probe. Interestingly, although we did not find super-shift bands, we did observe attenuated OCT4 binding band with OCT4 antibody used (Fig. 4B). These observations may explain the possible correlations between these SNPs with prognosis of gefitinib treatment.
Table 3.
Transcription factor binding site analyses of ATG5 rs510532 and ATG10 rs10036653.
| SNPs | Score RegulomeDB* | Method | Location | Motif | Reference |
|---|---|---|---|---|---|
| ATG5 rs510432 | 3a | PWM | chr6:106774020..106774034 | Mtf1 | 38 |
| ATG10 rs10036653 | 6 | PWM | chr5:81266375..81266390 | Sox5 | 39 |
| PWM | chr5:81266368..81266383 | Oct-4 (POU5F1) | 38 | ||
| Footprinting | chr5:81266368..81266383 | Oct-4 (POU5F1) | 40 |
Note: PWM, Position-Weight Matrix for TF binding; Footprinting, DNase Footprinting.
All results were from RegulomeDB.
*“3a” means “TF binding + any motif + DNase peak” is supportive for transcription factor binding, while “6” means the results is verified by “other” methods.
Figure 4.
Functional evaluation of ATG5 rs510432 and ATG10 rs10036653 in lung cancer cells. (A) Relative gene expression was examined through qRT-PCR. (B) Electrophoretic mobility-shift assay (EMSA) with biotin-labeled rs10036653T or rs10036653A probes and A549 nuclear extracts (N.P.).
Discussions
EGFR-TKIs have been proved to be promising treatment of NSCLC, especially for lung adenocarcinoma patients harboring EGFR mutations. In addition to the EGFR mutations, we and others also found that germline variations might be prognostic markers of gefitinib treatment22,23. In this study, we systematically evaluated 23 SNPs from eleven autophagy core genes and treatment outcomes of advanced lung adenocarcinomas patients. Multiple genetic variations in autophagy core genes, i.e. ATG5 rs510532 and ATG10 rs10036653, were found to be significantly associated with clinical outcomes, especially in those with EGFR mutations. To the best of our knowledge, our study is the first to examine clinical implications of autophagy SNPs in patients with EGFR mutant adenocarcinoma.
Genetic variations of autophagy core genes are investigated in several human cancers. Qin et al. examined 14 potentially functional polymorphisms in six autophagy-related genes (ATG3, ATG5, ATG7, ATG10, ATG12 and LC3) in breast cancer susceptibility and found that ATG10 rs1864182 and rs10514231 were associated with significantly decreased risk of breast cancer24. After genotyping 40 tagging SNPs from 7 core autophagy pathway genes in 458 localized prostate cancer patients, Huang et al. observed the association between ATG16L1 rs78835907 and recurrence of localized disease, which was replicated in more advanced disease30. White et al. examined five SNPs in three ATG genes (ATG5, ATG10 and ATG16L) and found that ATG SNPs might be differentially associated with specific host and melanoma characteristics including age at diagnosis, tumor infiltrating lymphocytes, and stage31. Berger et al. genotyped 12 SNPs in eight autophagy-related genes among patients with mCRC treated with first-line FOLFIRI and bevacizumab in two phase III randomized trials and found that the FIP200 rs1129660 variant showed significant associations with hypertension32. In head and neck squamous cell carcinoma, Fernández-Mateos et al. observed the associations between ATG10 rs1864183 and a higher susceptibility to develop laryngeal cancer, ATG2B rs3759601 and pharyngeal cancer as well as ATG16L1 rs2241880 and oral carcinoma33. However, it is still unclear if genetic variations of autophagy core genes would impact prognosis of advanced lung adenocarcinomas patients.
Accumulated evidences demonstrated that autophagy plays an essential role in escaping from the anti-neoplastic effects of drugs34–36. In NSCLC cells, gefitinib treatment can induce elevated ATG5 expression and increased autophagy34. Cytotoxicity induced by gefitinib was greatly enhanced after autophagy inhibition by ATG5 silencing34, which suggests that ATG5-regulated autophagy inhibition represents a promising approach to improve the efficacy of EGFR-TKIs. Similarly, Sakuma et al. found that depletion of ATG5, an autophagy inhibitor, markedly reduces gefitinib-resistant cell viability of EGFR-mutated lung adenocarcinoma cells under hypoxic conditions36. These results elucidated that ATG5 might be a crucial gene impacting clinical outcomes of gefitinib treatments. As a result, it is biologically plausible that the potential functional ATG5 rs510532 genetic variant may also be a prognostic marker for gefitinib therapy.
ATG10 is an E2-like enzyme involved in E2 ubiquitin-like modifications essential for autophagosome formation. Jo et al. found that ATG10 was increased in colorectal cancer and associated with lymphovascular invasion and lymph node metastasis37. Qin et al. demonstrated that potentially functional polymorphisms in ATG10 were associated with risk of breast cancer in a Chinese population24. These results indicated that ATG10 and its genetic polymorphisms might be an important component during carcinogenesis. In line with this, we observed significant association between the ATG10 rs1864182 SNP with prolonged survival and gefitinib-resistance of EGFR mutant NSCLC patients.
In summary, ATG5 rs510532 and ATG10 rs10036653 genetic variations in autophagy core genes are significantly associated with clinical outcomes of advanced lung adenocarcinoma treated with gefitinib. Genotyping of these genetic variations with detection of EGFR mutations may improve the prediction of the treatment outcomes. Our study also highlights the possibility of patient-tailored decisions especially during EGFR-TKIs based on combination of germline and somatic variation detection.
Electronic supplementary material
Acknowledgements
This study was partially supported by National Natural Science Foundation of China (31671300, 81572934); Taishan Scholars Program of Shandong Province (tsqn20161060); the National High-Tech Research and Development Program of China (2015AA020950); the Science and Technology Planning Project of Huaian (HAS2015013); Foundation of Shandong Academy of Medical Sciences (2015-44); Medicine and Health Science Technology Foundation (2015WSA18054).
Author Contributions
M.Y. and J.Y. conceived and designed the experiments; J.Y., N.Z. and L.Y. performed the experiments; J.Y. and N.Z. analyzed the data; H.Z., L.Zhang. and L.Zhou. contributed materials/analysis tools; M.Y., J.Y. and N.Z. wrote the manuscript. All authors reviewed and approved the manuscript prior to submission.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Jupeng Yuan, Nasha Zhang and Longbin Yin contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-18165-5.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.Chen Z, Fillmore CM, Hammerman PS, Kim CF, Wong KK. Non-small-cell lung cancers: a heterogeneous set of diseases. Nat Rev Cancer. 2014;14:535–546. doi: 10.1038/nrc3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lynch TJ, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 4.Paez JG, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 5.Chan BA, Hughes BG. Targeted therapy for non-small cell lung cancer: current standards and the promise of the future. Transl Lung Cancer Res. 2015;4:36–54. doi: 10.3978/j.issn.2218-6751.2014.05.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhai H, Zhong W, Yang X, Wu YL. Neoadjuvant and adjuvant epidermal growth factor receptor tyrosine kinase inhibitor (EGFR-TKI) therapy for lung cancer. Transl Lung Cancer Res. 2015;4:82–93. doi: 10.3978/j.issn.2218-6751.2014.11.08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shigematsu H, et al. Clinical and biological features associated with epidermal growth factor receptor gene mutations in lung cancers. J Natl Cancer Inst. 2005;97:339–346. doi: 10.1093/jnci/dji055. [DOI] [PubMed] [Google Scholar]
- 8.Tsao AS, et al. Clinicopathologic characteristics of the EGFR gene mutation in non-small cell lung cancer. J Thorac Oncol. 2006;1:231–239. doi: 10.1016/S1556-0864(15)31573-2. [DOI] [PubMed] [Google Scholar]
- 9.Gazdar AF. EGFR mutations in lung cancer: different frequencies for different folks. J Thorac Oncol. 2014;9:139–140. doi: 10.1097/JTO.0000000000000063. [DOI] [PubMed] [Google Scholar]
- 10.Wang J, Wang B, Chu H, Yao Y. Intrinsic resistance to EGFR tyrosine kinase inhibitors in advanced non-small-cell lung cancer with activating EGFR mutations. Onco Targets Ther. 2016;9:3711–3726. doi: 10.2147/OTT.S106399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martini-Stoica H, Xu Y, Ballabio A, Zheng H. The Autophagy-Lysosomal Pathway in Neurodegeneration: A TFEB Perspective. Trends Neurosci. 2016;39:221–234. doi: 10.1016/j.tins.2016.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eskelinen EL, Saftig P. Autophagy: a lysosomal degradation pathway with a central role in health and disease. Biochim Biophys Acta. 1793;664-673:2009. doi: 10.1016/j.bbamcr.2008.07.014. [DOI] [PubMed] [Google Scholar]
- 13.Baehrecke EH. Autophagy: dual roles in life and death? Nat Rev Mol Cell Biol. 2005;6:505–510. doi: 10.1038/nrm1666. [DOI] [PubMed] [Google Scholar]
- 14.Choi AM, Ryter SW, Levine B. Autophagy in human health and disease. N Engl J Med. 2013;368:1845–1846. doi: 10.1056/NEJMra1205406. [DOI] [PubMed] [Google Scholar]
- 15.Komatsu M, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 16.Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature. 2011;469:323–335. doi: 10.1038/nature09782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kondo Y, Kanzawa T, Sawaya R, Kondo S. The role of autophagy in cancer development and response to therapy. Nat Rev Cancer. 2005;5:726–734. doi: 10.1038/nrc1692. [DOI] [PubMed] [Google Scholar]
- 18.Mathew R, Karantza-Wadsworth V, White E. Role of autophagy in cancer. Nat Rev Cancer. 2007;7:961–967. doi: 10.1038/nrc2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.White E. The role for autophagy in cancer. J Clin Invest. 2015;125:42–46. doi: 10.1172/JCI73941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mizushima N, Ohsumi Y, Yoshimori T. Autophagosome formation in mammalian cells. Cell Struct Funct. 2002;27:421–429. doi: 10.1247/csf.27.421. [DOI] [PubMed] [Google Scholar]
- 21.Kruppa AJ, Kendrick-Jones J, Buss F. Myosins, Actin and Autophagy. Traffic. 2016;17:878–890. doi: 10.1111/tra.12410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ng KP, et al. A common BIM deletion polymorphism mediates intrinsic resistance and inferior responses to tyrosine kinase inhibitors in cancer. Nat Med. 2012;18:521–528. doi: 10.1038/nm.2713. [DOI] [PubMed] [Google Scholar]
- 23.Zhang L, et al. Role of EGFR SNPs in survival of advanced lung adenocarcinoma patients treated with Gefitinib. Gene. 2013;517:60–64. doi: 10.1016/j.gene.2012.12.087. [DOI] [PubMed] [Google Scholar]
- 24.Qin Z, et al. Potentially functional polymorphisms in ATG10 are associated with risk of breast cancer in a Chinese population. Gene. 2013;527:491–495. doi: 10.1016/j.gene.2013.06.067. [DOI] [PubMed] [Google Scholar]
- 25.Yang X, et al. Integrative Functional Genomics Implicates EPB41 Dysregulation in Hepatocellular Carcinoma Risk. Am J Hum Genet. 2016;99:275–286. doi: 10.1016/j.ajhg.2016.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu H, et al. Onco-lncRNA HOTAIR and its functional genetic variants in papillary thyroid carcinoma. Sci Rep. 2016;6:31969. doi: 10.1038/srep31969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shi M, et al. The Sp1-mediaded allelic regulation of MMP13 expression by an ESCC susceptibility SNP rs2252070. Sci Rep. 2016;6:27013. doi: 10.1038/srep27013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li Z, et al. Association of a functional RAD52 genetic variant locating in a miRNA binding site with risk of HBV-related hepatocellular carcinoma. Mol Carcinog. 2015;54:853–858. doi: 10.1002/mc.22156. [DOI] [PubMed] [Google Scholar]
- 29.Boyle AP, et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012;22:1790–1797. doi: 10.1101/gr.137323.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang CY, et al. Genetic variants of the autophagy pathway as prognostic indicators for prostate cancer. Sci Rep. 2015;5:14045. doi: 10.1038/srep14045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.White KA, et al. Variants in autophagy-related genes and clinical characteristics in melanoma: a population-based study. Cancer Med. 2016;5:3336–3345. doi: 10.1002/cam4.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Berger MD, et al. Autophagy-related polymorphisms predict hypertension in patients with metastatic colorectal cancer treated with FOLFIRI and bevacizumab: Results from TRIBE and FIRE-3 trials. Eur J Cancer. 2017;77:13–20. doi: 10.1016/j.ejca.2017.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fernandez-Mateos J, et al. Analysis of autophagy gene polymorphisms in Spanish patients with head and neck squamous cell carcinoma. Sci Rep. 2017;7:6887. doi: 10.1038/s41598-017-07270-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Han W, et al. EGFR tyrosine kinase inhibitors activate autophagy as a cytoprotective response in human lung cancer cells. PLoS One. 2011;6:e18691. doi: 10.1371/journal.pone.0018691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nihira K, et al. An activation of LC3A-mediated autophagy contributes to de novo and acquired resistance to EGFR tyrosine kinase inhibitors in lung adenocarcinoma. J Pathol. 2014;234:277–288. doi: 10.1002/path.4354. [DOI] [PubMed] [Google Scholar]
- 36.Sakuma Y, et al. Enhanced autophagy is required for survival in EGFR-independent EGFR-mutant lung adenocarcinoma cells. Lab Invest. 2013;93:1137–1146. doi: 10.1038/labinvest.2013.102. [DOI] [PubMed] [Google Scholar]
- 37.Jo YK, et al. Polypyrimidine tract-binding protein 1-mediated down-regulation of ATG10 facilitates metastasis of colorectal cancer cells. Cancer Lett. 2017;385:21–27. doi: 10.1016/j.canlet.2016.11.002. [DOI] [PubMed] [Google Scholar]
- 38.Badis G, et al. Diversity and complexity in DNA recognition by transcription factors. Science. 2009;324:1720–1723. doi: 10.1126/science.1162327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pique-Regi R, et al. Accurate inference of transcription factor binding from DNA sequence and chromatin accessibility data. Genome Res. 2011;21:447–455. doi: 10.1101/gr.112623.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Matys V, et al. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006;34:D108–110. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.