Abstract
Competition is a key process that determines plant community structure and dynamics, often mediated by nutrients and water availability. However, the role of soil microorganisms on plant competition, and the links between above- and belowground processes, are not well understood. Here we show that the effects of interspecific plant competition on plant performance are mediated by feedbacks between plants and soil bacterial communities. Each plant species selects a singular community of soil microorganisms in its rhizosphere with a specific species composition, abundance and activity. When two plant species interact, the resulting soil bacterial community matches that of the most competitive plant species, suggesting strong competitive interactions between soil bacterial communities as well. We propose a novel mechanism by which changes in belowground bacterial communities promoted by the most competitive plant species influence plant performance and competition outcome. These findings emphasise the strong links between plant and soil communities, paving the way to a better understanding of plant community dynamics and the effects of soil bacterial communities on ecosystem functioning and services.
Introduction
Plant-plant competition plays a key role in defining community structure and dynamics1. The outcomes of this process are often mediated by the availability of nutrients and water in the soil2. However, the role of soil microorganisms on plant-plant competition is less understood3,4 despite evidence for strong links between above- and belowground biota5,6. The interaction between plants and associated soil microbial communities is so close that they can be considered a whole entity7 that jointly responds to environmental conditions8 and which is subject to selection9. Plants shape their rhizosphere microbial communities through changes in soil temperature, moisture, physical structure, litter quality, and root exudates2,10–13. Soil microbial communities, in turn, influence plant community structure by altering plant performance and functional traits14–16, which affect ecosystem functioning through their effects on nutrient cycles and productivity11,17.
Because of this close interaction, assessing the role of soil communities on plant-plant interactions should provide a more realistic view of plant community dynamics and its consequences for ecosystem functioning. In this regard, it has been shown that soil bacteria influence, for instance, facilitation13,14, a key process in community assembly. However, there is less evidence on whether they can mediate the outcome of plant competition3,4,18. We formulated the hypotheses that 1) under intraspecific competition, each plant species will develop a specific bacterial community in its rhizosphere with different composition and activity as a result of different root traits that foster contrasting communities; 2) interspecific competition between two co-existing plant species, similar in size and habitat preferences, will have measurable effects on plant survival, growth and functional traits; and 3) when the two species interact, the final microbial community will resemble the community of the most competitive plant species.
To test these hypotheses, we monitored survival and performance of individuals of two plant species growing with an individual of the same (intraspecific competition) or the other species (interspecific competition) in a greenhouse experiment, using a common soil. We characterized the composition of soil bacterial communities in the different treatments at the end of the experiment by using 16S rDNA sequencing. We also measured enzyme activities to determine both the total microbial activity (through dehydrogenase activity) and the performance of the biogeochemical processes related with carbon (C), nitrogen (N) and phosphorus (P) (β-glucosidase, urease and alkaline phosphatase activities, respectively). Our target species were Maytenus senegalensis subsp. europea Rivas and Lycium intricatum Boiss (Maytenus and Lycium hereafter), two shrub species coexisting in semiarid environments in southern Spain. The two species have differential effects on their associated plant communities2,19, suggesting contrasted effects on microbial communities. Thus, while Maytenus is a facilitator shrub that creates favourable conditions for an understorey community of beneficiary species19, Lycium induces a large decrease in photosynthetically active radiation and soil moisture in its understorey2. Because competition is a common process happening whenever two individuals co-occur1,20,21, the results presented here are applicable to other pairs of co-existing plant species and contribute to better understanding plant community assembly.
Results and Discussion
Plant survival and growth
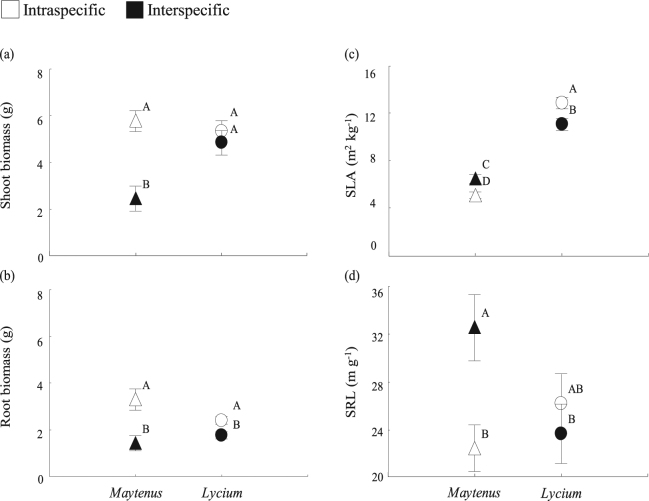
Interspecific competition altered plant survival and growth, supporting our second hypothesis. This was true in particular for Maytenus individuals. Maytenus mortality started 8 months after the beginning of the experiment. By month 12, Maytenus mortality was 60% higher under interspecific competition than under intraspecific competition (Supplementary Fig. S1 and Supplementary Table S1). By contrast, all Lycium individuals survived regardless of treatment. At the end of the experiment, Maytenus individuals also had lower aboveground mass under interspecific than under intraspecific competition (Fig. 1a, Supplementary Table S2). Both species had less root mass in the interspecific treatment than in the intraspecific treatment (Fig. 1b), suggesting strong belowground competition.
Figure 1.
Maytenus plants grew less and showed increased SLA and SRL when growing with Lycium. Shoot (a) and root (b) mass, specific leaf area (SLA; c) and specific root length (SRL; d) in Maytenus and Lycium plants growing under intra- or interspecific interaction. Different letters in a graph indicate significant differences (p < 0.05) among treatments. Data are mean ± 1 SE; n = 6.
Plant traits
As expected, we found changes in key plant functional traits as a response of one species interacting with another (Fig. 1, Supplementary Table S2). This supports the hypothesis that plant interactions are major drivers of trait plasticity22. The specific leaf area (SLA) has been shown to respond to the presence of a competitor23, and in our study, both Maytenus and Lycium individuals changed SLA when in interspecific competition in contrast to intraspecific competition (Fig. 1c). Interestingly, these changes in SLA followed opposite directions for the two plant species. On one hand, Lycium responded with a decrease in SLA under interspecific competition compared to intraspecific competition, which is often related to increased leaf longevity and C investment in secondary compounds24. On the other hand, Maytenus responded with an increase in SLA under interspecific competition, which is often linked to an increase in leaf N concentration and photosynthetic rate24. Maytenus individuals also increased specific root length (SRL), i.e. produced thinner roots, when growing with Lycium (Fig. 1d). The increase in both SLA and SRL in Maytenus suggests a strategy to maximize N uptake24, a nutrient that was most limited in the interspecific interaction treatment (available NH4 +; Fig. 2). Thinner root production is often triggered by the presence in the soil of root exudates of other species25. Our results support the tenet that plants display different functional responses to optimize performance during competition26. Because trait changes imply changes in ecosystem properties11, the consequences of a competitive environment might translate into altered ecosystem functioning through species turnover.
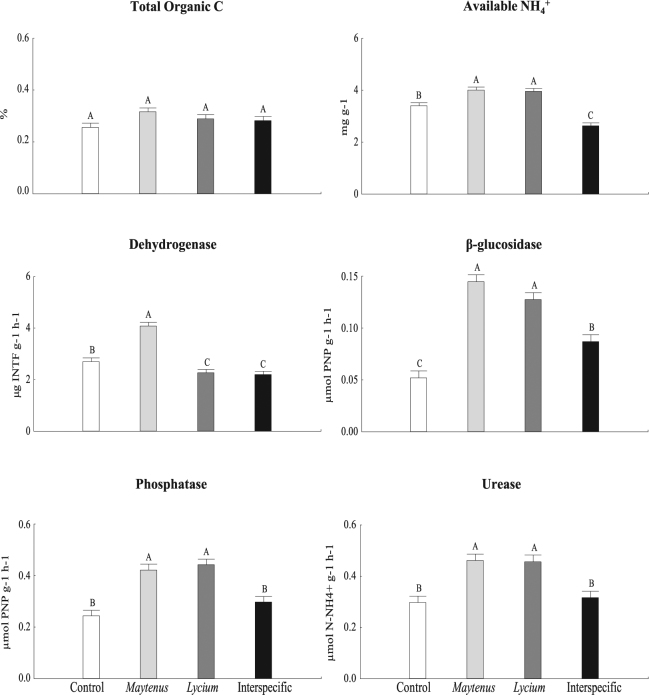
Figure 2.
Soil enzyme activity was lower in interspecific soils than in intraspecific ones. Soil TOC, available NH4 + and enzyme activities in soils of the different plant interaction treatments, i.e. without plants (control), soils with two Maytenus individuals (Maytenus-intraspecific), with two Lycium individuals (Lycium-intraspecific) or with one individual of each plant species (interspecific). Different letters in a graph indicate significant differences (p < 0.05) among treatments. Data are mean ± 1 SE; n = 6. Graph columns are coloured according to the four different plant interaction treatments (control, Maytenus, Lycium, interspecific).
Soil enzyme activities
There was a decrease in most soil enzyme activity in the interspecific treatment compared to soils in the intraspecific treatment, either with only Lycium or only Maytenus (Fig. 2, Supplementary Tables S2 and S3). This lower activity should be an indirect consequence of lower plant growth and less production of root exudates that stimulate bacterial activity. Indeed, responses to plant competitors are likely to alter the quantity, quality and availability of resources supplied by a host plant to its microbiome27. The reduced soil N in pots of the interspecific interaction compared to control pots with no plants (while having similar levels of urease activity, Fig. 2) suggests that plants under interspecific competition actively took up most of the available N. Consequently, it is very likely that soil microbes were also competing amongst themselves and also with the plants for N28. The decrease in microbial activity, and therefore the slow nutrient cycling in the interspecific treatment, may have contributed to the observed poor Maytenus performance in this treatment. Our results support the suggestion that root exudates, nutrient availability and soil microbial community form a tripartite relationship that simultaneously affects plant growth and competitive ability26.
Diversity and composition of bacterial communities
Supporting our third hypothesis, the bacterial community in the interspecific treatment resembled the community of the most competitive plant species, i.e. Lycium. As such, the composition of the bacterial community in the interspecific treatment was more similar to soils with only Lycium than to soils with only Maytenus (Fig. 3, Supplementary Fig. S2 and Supplementary Table S4). When in monoculture, each plant species hosted a distinct soil bacterial community (Fig. 3), as expected. The variability in species composition of the community associated to Lycium was higher than that of Maytenus (Fig. 3). Maytenus promoted an increase in species richness compared to control soils with no plants (1037.2 ± 34.7 vs. 894.7 ± 32.1 operational taxonomic units (OTUs) per sample, respectively; Supplementary Table S2) and large modifications in community composition (Fig. 4 and Supplementary Table S5), similar to those reported for other facilitator species such as Retama sphaerocarpa 13,14. The dominant role of Lycium in defining bacterial community structure under interspecific competition could be modulated by the larger decrease in Maytenus root mass and root exudates12 when growing with Lycium. In addition, bacterial groups promoted by Lycium may have competitively displaced29 groups typically associated to Maytenus. Indeed, plant-driven impacts on particular soil microbial taxa may have cascading effects, resulting in altered interaction networks among soil taxa27.
Figure 3.

The soil bacterial community in the interspecific treatment was similar to Lycium but different to Maytenus soils. Ordination of soil bacterial community composition by Non-Metric Multidimensional Scaling (NMDS) in the different plant interaction treatments, i.e. soils without plants (control), with two Maytenus individuals (Maytenus-intraspecific), with two Lycium individuals (Lycium-intraspecific) or with one individual of each plant species (interspecific). Samples are coded by plant interaction treatment, in particular: asterisks = control, triangles = Maytenus-intraspecific, circles = Lycium-intraspecific; black filled squares = interspecific; n = 6. Stress = 0.2.
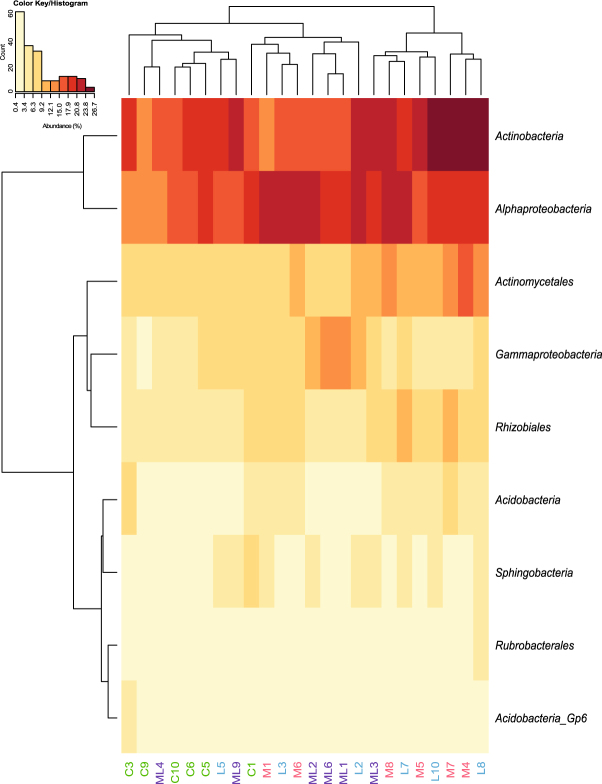
Figure 4.
The abundance of several bacterial groups differed among soils in the different treatments. Heat-map analysis based on hierarchical clustering using the abundance of the main identified bacterial groups that showed significant differences by GLM among plant interaction treatments, i.e. soils without plants (control), with two Maytenus individuals (Maytenus-intraspecific), with two Lycium individuals (Lycium-intraspecific) or with one individual of each plant species (interspecific). Each grid unit in the heat-map is an individual value of abundance of a given bacterial group (rows) per sample (columns); warmer colors represent larger values of abundance and cooler colors represent lower values (see color key/histogram). C, control; M, Maytenus-intraspecific; L, Lycium-intraspecific and ML, interspecific. Numbers following letters denote replicates within each treatment.
Because Maytenus individuals seem to be associated with a specific bacterial community (Fig. 3), changes in the relative abundance of bacterial groups under interspecific competition could have also contributed to its poor performance. There is a need to know which microbial taxa are involved and the mechanisms by which they influence plant competition18. Changes are evident in our experiment (Fig. 4, see Supplementary Table S5 for a complete list) and concern, for instance, Gammaproteobacteria, a group that had higher abundance in the interspecific treatment than in Maytenus soils. This group includes many plant pathogens that could have negatively impacted Maytenus. Higher abundance was also observed for Halomonas and Salinimicrobium, genera that increase under drought conditions in semiarid environments30, and that could be related to greater water shortage in the interspecific treatment. Rhizobiales instead were less abundant in the interspecific treatment than in Maytenus soils, which, given their role on N fixation14, could have contributed to the lower N content and increased N competition in the interspecific treatment.
Conclusions
The advent of powerful technologies to identify microbial communities is providing evidence on the widespread effects of microbes on ecological processes, more influential than acknowledged so far. Plant-plant interactions should be assessed not as isolated organisms in the environment but as a complex community of multiple trophic levels within which the soil microbiota is an important component2,3. Altogether, our results suggest that a major driver of the differences in plant performance between the two species was the ability of the better competitor (i.e. Lycium) to promote a preferred soil bacterial community. These data point to a novel mechanism controlling plant-plant competition and support the relevance of feedbacks between soil bacterial communities and plants for community dynamics. Since plant communities determine ecosystem properties, by influencing plant interactions the effect of microbes goes beyond a local process such as competition to impact ecosystem function and services.
Methods
Plant material, soil collection and experimental set-up
Maytenus senegalensis subsp. europaea (Celastraceae) is a thorny evergreen shrub up to 4 m tall with an intricate semispherical canopy. Many other plant species (shrubs, climbers, forbs, grasses) grow within its canopy forming patches with bare soil in between31. It is distributed throughout North Africa and Southeast Spain, where it is found in coastal areas32. Lycium intricatum (Solanaceae) is a thorny shrub with drought-deciduous, succulent leaves and shallow roots33 that can be found in coastal systems either associated to facilitator species such as Maytenus 31 or Ziziphus lotus or isolated2. In the field, Lycium seedlings may establish under the canopy of adult Maytenus shrubs31. Field observations of large dead Maytenus shrubs with large alive Lycium shrubs growing in their understorey (Hortal, personal observation) may suggest that Lycium could be able to outcompete Maytenus in the field. Shifts in the outcome of plant-plant interactions as seedlings become adults have been reported in many other systems34,35.
In March 2011, two-year old Maytenus and Lycium saplings were obtained from a local nursery (Rodalquilar, Almería, Spain). Root systems were carefully washed to remove excess of potting mix and saplings were planted in 6 L pots. Each pot contained two saplings of the same or different species; i.e., two Maytenus plants growing together (Maytenus-intraspecific), two Lycium plants (Lycium-intraspecific) or one Maytenus and one Lycium (interspecific). Control pots without plants were also established resulting in four treatments with 10 replicates each. Pots with only one individual were not included in the experimental design as we were interested on whether inter- and intraspecific competition affected soil bacterial communities in different ways, and not so much on the effect of intraspecific competition. Initial height and basal diameter were recorded for each sapling. Mean height was 29.34 ± 1.17 cm (mean ± standard error) and 33.14 ± 0.85 cm and mean diameter was 0.51 ± 0.03 cm and 0.59 ± 0.02 cm for Maytenus and Lycium, respectively, with no significant differences among treatments. Five additional Maytenus and Lycium individuals were randomly selected to establish initial dry mass after 48 h at 70 °C. Initial mean aboveground dry mass was 2.45 ± 0.28 g and 2.54 ± 0.30 g for Maytenus and Lycium, respectively. Root mass was 1.25 ± 0.18 g and 0.95 ± 0.06 g for Maytenus and Lycium, respectively.
Soil was obtained in March 2011 at Rambla del Toyo (Almería, Spain), a dry riverbed where both species co-exist forming typical coastal shrublands and where Maytenus and Ziziphus lotus are the dominant species. Soil from the first top 10 cm was collected from different bare areas within an approximately 2 ha plot, sieved through 4 mm mesh and mixed. It was sandy, poor in nutrients, with low water holding capacity and pH around 8–8.536. Soil was used to fill the pots containing gravel at the bottom to improve aeration. Three soil samples were taken and stored at 4 °C for a maximum of 30 days37 to determine total N, total C and enzyme activities; additional samples were kept at −80 °C to perform molecular analysis. Total N and C were determined using an Elemental Analyzer (Leco Truspec, St. Joseph, MI, USA). Total N was 0.28 ± 0.03 g kg−1 and total C was 8.80 ± 0.18 g kg−1. Dehydrogenase activity was 5.00 ± 0.20 µg INTF g−1 soil h−1, phosphatase was 0.23 ± 0.01 μmol of p-nitrophenyl phosphate (PNP) g−1 h−1, urease was 0.37 ± 0.06 μmol N-NH4 + g−1 h−1 and β-glucosidase was 0.08 ± 0.01 μmol PNP g−1 h−1.
Pots were randomly distributed in a greenhouse at the Estación Experimental de Zonas Áridas in Almería (Spain) under natural temperature and irradiance regime. Mean annual temperature in the region is 18.8 °C with mean temperatures of 12.5 °C and 26.5 °C on the coldest and warmest months, respectively (www.meteodata.org/koppen). The experiment ran for one year, and for the length of the experiment pots were regularly watered, regularly shifted to avoid gradients, and plant mortality was recorded. To reduce the number of samples for analysis while keeping a reasonable number of replicates, six out of the 10 initial replicates per treatment were randomly selected for plant measurements and soil harvesting in March 2012, during the peak of plant productivity.
Plant traits and growth measurements
Of these 6 replicates (pots with two plants), three replicates per treatment were selected for plant trait measurements following standard protocols24. At harvesting, three leaves per plant were sampled to measure specific leaf area (SLA) and three fine roots (<2 mm diameter, 10 cm in length approximately) per plant were collected to calculate specific root length (SRL). Mean values were obtained per each individual plant and used for statistical analyses. Plant height and stem diameter were also recorded at the end of the experiment. Shoot and root dry mass were weighted after drying at 70 °C for 48 h.
Soil sampling and chemical analyses
At harvest, we sampled soil from the first top 10 cm between the two plants (4 cm distance from each plant) in each pot. Soil from control pots without plants was also collected. Each soil sample was homogenised and sieved through 2 mm mesh. Material was cleaned with ethanol 70% between samples. A total of 24 samples were collected, six per each plant interaction treatment (control -no plant-, Maytenus-intraspecific, Lycium-intraspecific, and interspecific). Each soil sample was divided into two subsamples, one was stored at −80 °C for molecular analyses and the other was kept at 4 °C for a maximum of 30 days37 for chemical and enzymatic activity analyses. Available NH4 + and total organic C (TOC) were determined using an Elemental Analyzer (Leco Truspec, St. Joseph, MI, USA). Concentration of available NH4 + in soil was measured using a 1 M KCl solution, in a ratio 1:10 soil:solution, to extract the available fraction of this cation and then determined by a colorimetric method38. Three grams of soil per sample were dried at 105 °C for 24 hours and weighed to analyze soil moisture.
Soil enzyme activities
Soil dehydrogenase activity was determined in 1 g of soil incubated with 0.2 ml of p-iodonitrotetrazolium chloride (INT, 0.4% w/v) at room temperature for 20 hours. The reduction of INT to p-iodonitrotetrazolium formazan (INTF) at soil pH was estimated by a modification of a reported protocol39. The INTF produced was extracted with 10 ml of methanol, and the absorbance of the filtrate was measured in a spectrophotometer (Helios Alpha, Thermo, UK) at 490 nm. The β-glucosidase and alkaline phosphatase activities were determined by following standard methods40,41. Two millilitres of MUB (Modified Universal Buffer), pH 6.5 for the β-glucosidase assay and pH 11 for the alkaline phosphatase assay, and 0.5 ml of p-nitrophenyl substrate (p-nitrophenyl-β-d-glucopyranoside for β-glucosidase and p-nitrophenyl phosphate for alkaline phosphatase) were added to 0.5 g of soil. The mixtures were incubated at 37 °C for 1 h. Then, the p-nitrophenol released was measured by colorimetry in a UV-vis spectrophotometer (Helios Alpha, Thermo, UK) at 400 nm. The urease activity was determined as a previously reported method42. In this procedure, 0.5 ml of a solution of urea (0.48%) and 4 ml of borate buffer (pH 10) were added to 1 g of soil (0.4 g of compost) and then incubated for 2 h at 37 °C. The ammonium concentration of the centrifuged extracts was determined by a modified indophenol-blue reaction and measured at 690 nm by spectrophotometry.
Soil bacterial community composition: 16S rDNA pyrosequencing
DNA was extracted from 0.25 g of homogenised soil of each of the 24 samples using the PowerSoil® DNA Isolation Kit (MO BIO Laboratories, Inc., Carlsbad, USA) following manufacturer’s directions. A 16S rDNA gene fragment corresponding to V1 and V2 regions was amplified19. PCR amplifications were performed in 50 μl reaction volumes containing ultrapure H2O, 2.5 × 5 PRIME MasterMix including 1.5 mM Magnesium, 200 μM dNTPs, 1.25 U Taq polymerase (5 PRIME, Hamburg, Germany), 0.2 μM of primers and 5–10 ng of template DNA. Fragments were amplified under the following conditions: initial denaturation at 94 °C for 3 minutes, followed by 30 cycles with denaturation at 94 °C for 40 seconds, annealing at 52 °C for 40 seconds and extension at 68 °C for 35 seconds, with a final extension at 68 °C for 7 minutes. Each sample was amplified in triplicate, pooled and purified using the QIAquick® PCR Purification Kit (Qiagen, Hilden, Germany). Amplification was checked by electrophoresis in 2% agarose gels stained with SYBR® Safe DNA Gel Stain (Invitrogen™, Carlsbad, USA) and bands were visualized using UV light in a Gel Doc™ EZ Imager (BIO-RAD, Hercules, USA).
DNA concentration of each purified sample was determined as the mean of three lectures in NanoDrop 2000c (Thermo Scientific, Wilmington, USA). Equal amounts of PCR product for each sample were combined in a single tube to obtain an equimolar pool. The pool was pyrosequenced in a Roche Genome Sequencer FLX System (Roche, Basel, Switzerland) using 454 Titanium chemistry at Lifesequencing (Valencia, Spain).
Processing of pyrosequencing data
The 16 S rDNA sequence data were processed using mothur v.1.32.143,44. Sequences were denoised using PyroNoise implemented in the mothur command shhh.flows. Sequences with more than 1 mismatch to the barcode, two mismatches to the primer or homopolymers >8 bp were removed from the dataset. Sequences were trimmed for primers and assigned to samples according to their barcode. Mean length of retained sequences was 269 bp. Sequences were aligned using the SILVA database. To further reduce sequencing errors, sequences that were within 2 bp of a more abundant sequence were merged using the pre.cluster command. Chimeras were identified using UCHIME with the sequences as their own reference and removed from the dataset. Sequences were classified using the mothur version of the RDP Bayesian classifier45. Any sequences classified as Mitochondria, Chloroplast, Archaea, Eukaryota or unknown (i.e. not classified at the Kingdom level) were removed from the dataset. Aligned sequences were clustered into operational taxonomic units (OTUs) defined at 97% similarity cutoff using the average neighbour method. The consensus taxonomy per each OTU was obtained. The number of sequences in each sample was normalized to 2968 (corresponding to the sample with the fewest sequences). We used Past software version 2.1246 to calculate the richness, Shannon diversity index (H’) and evenness for each sample. Relative abundances of the different taxonomic groups in each sample were calculated.
Statistical analyses
Differences among treatments in shoot and root biomass, SLA and SRL were analyzed with general linear models (GLM) full factorial design including species (Maytenus, Lycium), plant-plant interaction (intra- vs. interspecific) as fixed factors and the statistical interaction among these factors. Differences in soil TOC, available NH4 +, enzyme activities, relative abundance of the different taxa, OTU richness and Shannon’s diversity index were analyzed with GLMs that included plant interaction treatment (control, Maytenus-intraspecific, Lycium-intraspecific and interspecific) as fixed factor. Because in three pots of the interspecific treatment the Maytenus individual died before harvest, we repeated the same analyses after excluding those three replicates to ensure results were the same and thus were not explained by the death of Maytenus individuals. Moreover, deaths occurred towards the end of the experiment and were unrelated to transplant; therefore, those Maytenus individuals were alive for most of the experiment. We selected a variance function structure with different coefficients of the variance function for different strata to avoid heteroscedasticity (varIdent R function)47 and a compound symmetry as the spatial autocorrelation structure function (as two plant individuals grew in the same pot). Post-hoc comparisons were performed using the Fisher’s LSD test. Differences in mortality among treatments were assessed with generalized models. Similarity in OTUs composition among treatments was analysed with Non-metric Multidimensional Scaling (NMDS) analysis using Bray-Curtis similarity index and one-way NPMANOVA with 9999 permutations in Past46. The abundance of the main identified bacterial groups showing significant differences among treatments by GLM was used in a heat-map analysis based on hierarchical clustering using the average linkage method with Euclidean distance and bootstrap with 500 iterations48. Statistical analyses were done with R49 using the interface implemented in InfoStat statistical software50. Results are presented as mean values ± 1 SE throughout the text. Differences among treatments were considered significant at p < 0.05.
Data availability
Pyrosequencing data are deposited in the European Nucleotide Archive (ENA) under the accession number PRJEB22755.
Electronic supplementary material
Acknowledgements
This work was funded by the Junta de Andalucía Regional Government (grant P09-RNM-4821). Additional funding was provided by MINECO (grant CGL2014-59010-R). We thank Christian Schöb and Ivan Prieto for field and technical assistance, Jeff R. Powell for fruitful discussions, and David Wardle, Michael J. O’Brien and reviewers for comments on an earlier draft of the manuscript. CA is grateful to the Spanish Government for her “Ramón y Cajal” contract (RYC-2012-12277).
Author Contributions
S.H., F.I.P., C.A. and Y.M.L. designed the experiment with contributions from F.B., J.L.M. and C.G. S.H. carried out the experiment with assistance from Y.M.L. and C.A. F.B. and J.L.M. performed enzyme analyses. S.H. performed molecular analysis. F.I.P. contributed reagents and materials. S.H., Y.M.L. and C.A. analysed the data. S.H. wrote the manuscript with contributions from all co-authors.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-18103-5.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Tilman, D. Plant strategies and the dynamics and structure of plant communities. (Princeton University Press, Princeton, 1988).
- 2.Pugnaire FI, Armas C, Valladares F. Soil as a mediator in plant-plant interactions in a semi-arid community. J. Veg. Sci. 2004;15:85–92. doi: 10.1111/j.1654-1103.2004.tb02240.x. [DOI] [Google Scholar]
- 3.Kardol P, et al. Microbe-mediated plant–soil feedback causes historical contingency effects in plant community assembly. Ecol. Monogr. 2007;77:147–162. doi: 10.1890/06-0502. [DOI] [Google Scholar]
- 4.Pendergast TH, Burke DJ, Carson WP. Belowground biotic complexity drives aboveground dynamics: a test of the soil community feedback model. New Phytol. 2013;197:1300–1310. doi: 10.1111/nph.12105. [DOI] [PubMed] [Google Scholar]
- 5.Mangan SA, et al. Negative plant–soil feedback predicts tree-species relative abundance in a tropical forest. Nature. 2010;466:752–756. doi: 10.1038/nature09273. [DOI] [PubMed] [Google Scholar]
- 6.van der Putten WH. Belowground drivers of plant diversity. Science. 2017;355:134–135. doi: 10.1126/science.aal4549. [DOI] [PubMed] [Google Scholar]
- 7.Zilber-Rosenberg I, Rosenberg E. Role of microorganisms in the evolution of animals and plants: the hologenome theory of evolution. FEMS Microbiol. Rev. 2008;32:723–735. doi: 10.1111/j.1574-6976.2008.00123.x. [DOI] [PubMed] [Google Scholar]
- 8.Panke-Buisse K, et al. Selection on soil microbiomes reveals reproducible impacts on plant function. ISME J. 2015;9:980–989. doi: 10.1038/ismej.2014.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guerrero R, Margulis L, Berlanga M. Symbiogenesis: the holobiont as a unit of evolution. Int. Microbiol. 2013;16:133–143. doi: 10.2436/20.1501.01.188. [DOI] [PubMed] [Google Scholar]
- 10.Hartmann A, et al. Plant-driven selection of Microbes. Plant Soil. 2009;321:235–257. doi: 10.1007/s11104-008-9814-y. [DOI] [Google Scholar]
- 11.Bardgett RD, Mommer L, De Vries FT. Going underground: root traits as drivers of ecosystem processes. Trends Ecol. Evol. 2014;29:692–699. doi: 10.1016/j.tree.2014.10.006. [DOI] [PubMed] [Google Scholar]
- 12.Haichar FZ, et al. Root exudates mediated interactions belowground. Soil Biol. Biochem. 2014;77:69–80. doi: 10.1016/j.soilbio.2014.06.017. [DOI] [Google Scholar]
- 13.Rodríguez-Echeverría S, et al. A role for below-ground biota in plant-plant facilitation. J. Ecol. 2013;101:1420–1428. doi: 10.1111/1365-2745.12159. [DOI] [Google Scholar]
- 14.Lozano, Y. et al. Disentangling above- and below-ground facilitation drivers in arid environments: the role of soil microorganisms, soil properties and microhabitat. New Phytol. 216, 1236–1246, 10.1111/nph.14499 (2017). [DOI] [PubMed]
- 15.Cortois R, et al. Plant–soil feedbacks: role of plant functional group and plant traits. J. Ecol. 2016;104:1608–1617. doi: 10.1111/1365-2745.12643. [DOI] [Google Scholar]
- 16.Friesen ML, et al. Microbially mediated plant functional traits. Annu. Rev. Ecol. Evol. Syst. 2011;42:23–46. doi: 10.1146/annurev-ecolsys-102710-145039. [DOI] [Google Scholar]
- 17.Laforest-Lapointe I, et al. Leaf bacterial diversity mediates plant diversity and ecosystem function relationships. Nature. 2017;546:145–149. doi: 10.1038/nature22399. [DOI] [PubMed] [Google Scholar]
- 18.Hodge A, Fitter AH. Microbial mediation of plant competition and community structure. Funct. Ecol. 2013;27:865–875. doi: 10.1111/1365-2435.12002. [DOI] [Google Scholar]
- 19.Tirado R, Bråthen KA, Pugnaire FI. Mutual positive effects between shrubs in an arid ecosystem. Sci. Rep. 2015;5:14710. doi: 10.1038/srep14710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gallien L. Intransitive competition and its effects on community functional diversity. Oikos. 2017;126:615–623. doi: 10.1111/oik.04033. [DOI] [Google Scholar]
- 21.Armas C, Pugnaire FI. Plant neighbour identity matters to belowground contest competition under controlled conditions. PLoS ONE. 2011;6:e27791. doi: 10.1371/journal.pone.0027791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Henneron L, et al. Plant interactions as biotic drivers of plasticity in leaf litter traits and decomposability of Quercus petraea. Ecol. Monogr. 2017;87:321–340. doi: 10.1002/ecm.1252. [DOI] [Google Scholar]
- 23.Burns JH, Strauss SY. Effects of competition on phylogenetic signal and phenotypic plasticity in plant functional traits. Ecology. 2012;93:126–137. doi: 10.1890/11-0401.1. [DOI] [Google Scholar]
- 24.Pérez-Harguindeguy N, et al. Aust. J. Bot. 2013. New handbook for standardised measurement of plant functional traits worldwide; pp. 167–234. [Google Scholar]
- 25.Semchenko M, Saar S, Lepik A. Plant root exudates mediate neighbour recognition and trigger complex behavioural changes. New Phytol. 2014;204:631–637. doi: 10.1111/nph.12930. [DOI] [PubMed] [Google Scholar]
- 26.Pierik R, Mommer L, Voesenek LA. Molecular mechanisms of plant competition: neighbour detection and response strategies. Funct. Ecol. 2013;27:841–853. doi: 10.1111/1365-2435.12010. [DOI] [Google Scholar]
- 27.Bakker MG, et al. Diffuse symbioses: roles of plant-plant, plant-microbe and microbe-microbe interactions in structuring the soil microbiome. Mol. Ecol. 2014;23:1571–1583. doi: 10.1111/mec.12571. [DOI] [PubMed] [Google Scholar]
- 28.Hodge A, Robinson D, Fitter A. Are microorganisms more effective than plants at competing for nitrogen? Trends Plant Sci. 2000;5:304–308. doi: 10.1016/S1360-1385(00)01656-3. [DOI] [PubMed] [Google Scholar]
- 29.Kinkel LL, et al. Sympatric inhibition and niche differentiation suggest alternative coevolutionary trajectories among Streptomycetes. ISME J. 2014;8:249–256. doi: 10.1038/ismej.2013.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bastida F, et al. Combined effects of reduced irrigation and water quality on the soil microbial community of a citrus orchard under semi-arid conditions. Soil Biol. Biochem. 2017;104:226–237. doi: 10.1016/j.soilbio.2016.10.024. [DOI] [Google Scholar]
- 31.Mota JF, et al. Agricultural development vs biodiversity conservation: the Mediterranean semiarid vegetation in El Ejido (Almería, southeastern Spain) Biodivers. Conserv. 1996;5:1597–1617. doi: 10.1007/BF00052118. [DOI] [Google Scholar]
- 32.Mendoza-Fernández AJ, et al. Extreme habitat loss in a Mediterranean habitat: Maytenus senegalensis subsp. europaea. Plant Biosyst. 2015;149:503–511. doi: 10.1080/11263504.2014.995146. [DOI] [Google Scholar]
- 33.Padilla FM, Miranda J, de D, Pugnaire FI. Early root growth plasticity in seedlings of three Mediterranean woody species. Plant Soil. 2007;296:103–113. doi: 10.1007/s11104-007-9294-5. [DOI] [Google Scholar]
- 34.Armas C, Pugnaire FI. Ontogenetic shifts in interactions of two dominant shrub species in a semi-arid coastal sand dune system. J. Veg. Sci. 2009;20:535–546. doi: 10.1111/j.1654-1103.2009.01055.x. [DOI] [Google Scholar]
- 35.Callaway RM, Walker LR. Competition and facilitation: A synthetic approach to interactions in plant communities. Ecology. 1997;78:1958–1965. doi: 10.1890/0012-9658(1997)078[1958:CAFASA]2.0.CO;2. [DOI] [Google Scholar]
- 36.Tirado, R. Interacciones positivas entre plantas: Mecanismos y consecuencias. PhD Thesis. Universidad de Sevilla (2003).
- 37.Cernohlavkova J, et al. Variability of soil microbial properties: effects of sampling, handling and storage. Ecotoxicol Environ Saf. 2009;72:2102e2108. doi: 10.1016/j.ecoenv.2009.04.023. [DOI] [PubMed] [Google Scholar]
- 38.Keeney, D. R. & Nelson, D. W. Nitrogen-inorganic forms. In Agronomy:Methods of soil analysis, number 9part 2 (eds Page, A. L., Miller R.H. & Keeney, D. R.) 643–698 (Madison, Wisconsin, 1982).
- 39.Von Mersi W, Schinner F. An improved and accurate method for determining the dehydrogenase-activity of soils with iodonitrotetrazolium chloride. Boil. Fert. Soils. 1991;11:216–220. doi: 10.1007/BF00335770. [DOI] [Google Scholar]
- 40.Tabatabai MA, Bremmer JM. Use of p-nitrophenyl phosphate for assay of soil phosphatase activity. Soil Biol. Biochem. 1969;1:301–307. doi: 10.1016/0038-0717(69)90012-1. [DOI] [Google Scholar]
- 41.Eivazi F, Tabatabai MA. Phosphatases in soils. Soil Biol. Biochem. 1977;9:167–172. doi: 10.1016/0038-0717(77)90070-0. [DOI] [Google Scholar]
- 42.Kandeler E. Gerber, H. Short-term assay of soil urease activity using colorimetric determination of ammonium. Boil. Fert. Soils. 1988;6:68–72. [Google Scholar]
- 43.Schloss PD. Introducing mothur: Open-Source, Platform-Independent, Community-Supported Software for Describing and Comparing Microbial Communities. Appl. Environ. Microb. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schloss PD, Gevers D, Westcott SL. Reducing the Effects of PCR Amplification and Sequencing Artifacts on 16S rRNA-Based Studies. PLoS ONE. 2011;6:e27310. doi: 10.1371/journal.pone.0027310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Q, et al. Naive Bayesian Classifier for Rapid Assignment of rRNA Sequences into the New Bacterial Taxonomy. Appl. Environ. Microb. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hammer O, et al. PAST: paleontological statistics software package for education and data analysis. Palaeontol. Electronica. 2001;4:9. [Google Scholar]
- 47.Gałecki, A. & Burzykowski, T. Linear Mixed-Effects Models Using R: A Step-by-Step Approach. (Springer, Berlin, 2013).
- 48.Theoretical Biology and Biophysics Group, Los Alamos National Laboratory, NM, LA-UR 13-26007. HIV sequence database, URL http://www.hiv.lanl.gov/ (2016).
- 49.R Development Core Team R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. ISBN 3-900051-07-0, http://www.R-project.org (2016).
- 50.Di Rienzo J. A. et al. InfoStat versión 2016. Grupo InfoStat, FCA, Universidad Nacional de Córdoba, Argentina. http://www.infostat.com.ar (2016).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Pyrosequencing data are deposited in the European Nucleotide Archive (ENA) under the accession number PRJEB22755.