Abstract
The pTEN/AKT/mTOR signaling pathways play a critical role in balancing cell proliferation, differentiation, and survival. Recent studies researched the associations of core genes in the pTEN/AKT/mTOR pathway polymorphisms with the cancer susceptibility; however, the results are inconclusive. Therefore, a systematically meta-analysis was performed to evaluate the association between the five SNPs (mTOR rs2295080 and rs2536, AKT1 rs2494750 and rs2494752, pTEN rs701848) and cancer risk by systematic review of the literature in 31 eligible studies. The results showed a significant decreased risk between rs2295080 TG, GG genotype, and GG/TG genotypes and overall cancer [TG vs.TT: OR(95% CI) = 0.82(0.76, 0.89), GG/TG vs. TT: OR(95% CI) = 0.82(0.76, 0.88), and GG vs. TG/TT: OR(95% CI) = 0.67(0.51, 0.88)] and the subgroup of urinary system cancer and digestive system cancer. Moreover, the SNP rs701848 CC, TC genotype showed significantly increased the overall cancer risk both in dominant model [CC/TC vs. TT: OR(95% CI) = 1.25(1.15, 1.36)] and recessive model [CC vs. TC/TT: OR(95% CI) = 1.20(1.09, 1.32)], and digestive system cancer and urinary system cancer. In addition, AG genotype and GG/AG genotype of rs2494752 was associated with increased risk of cancer. Therefore, this meta-analysis provided genetic risk factors for carcinogenesis and the most valid cancer prevalence estimate for Asian population.
Introduction
Cancer is a major public health problem around the globe1. It is currently the second leading cause of death, and approximately 1, 658, 370 new cancer cases worldwide, 429000 new cases in China were reported according to the Cancer Statistics 20152. The carcinogenesis is involved in multifactor interaction among environmental exposures, life style and internal factors. In terms of internal factors, the main manifestations are changes in hormone secretion and immune conditions, and genetic variation in the key signaling pathway. In humans, the phosphatase and tensin homolog deleted on chromosome10 (pTEN)/AKT/mammalian target of rapamycin (mTOR) signaling pathway is frequently activated in a variety of cancers, and play a critical role in many cellular processes including proliferation, differentiation, cell cycle progression, cell motility and tumorigenesis, tumor growth, angiogenesis3–5. Therefore, the Single nucleotide polymorphisms (SNPs) of core genes in the pTEN/AKT/mTOR pathway may impact the transcription and expression of the proteins and thus alter the capacity and function of the pathway, which could play a critical role in carcinogenesis6–9.
The mTOR, which is located at the chromosome 1q36.2, plays a significant role in the pTEN/AKT/mTOR pathway. It exerts a prosurvival influence on cells through the activation of factors involved in protein synthesis10–13. pTEN is a tumor suppressor and plasma-membrane lipid phosphatase and which dephosphorylates PIP3 to PIP2, inhibiting the activation of AKT, and negatively regulates the pTEN/AKT/mTOR pathway14. To date, among the pTEN/AKT/mTOR pathway genes, there are more than 1000 coding-region SNPs (cSNPs) (http://www.ncbi.nlm.nih.gov/projects/SNP) reported. Among those cSNPs, a few potential functional SNPs especially located in the 5′-untranslated regions(5′UTR) and 3′UTR of the candidate genes could affect the carcinogenesis by modulating the transcriptional activity of candidate genes or by interacting with the miRNA binding, such as rs2295080 in the mTOR gene promoter region15–23, rs2494750 and rs2494752 in the AKT1 5′UTR region7,15,23–27, rs2536 in the 3′UTR of mTOR 7,15–18,22,23,28–30 and rs701848 the pTEN 3′UTR region7,15,27,31–42. Furthermore, previous studies demonstrated that the mTOR rs2295080 TT genotypes carriers showed a much higher mRNA levels of mTOR transcription by increasing the transcriptional activity of mTOR gene in human gastric cancer cell line SGC-79016. Moreover, carrying the rs2295080 T allele showed increased mTOR mRNA levels compared with the G allele in the patients with renal cell cancer7 and colorectal cancer8. Moreover, another SNP rs2536 located in the mTOR 3′-UTR was predicted to affect miRNA-binding site activity. Li et al.43 found that co-transfection of the rs2536 A allele and G allele with miR-767-3p exhibited different promoter activities. Additionally, the polymorphism of pTEN rs701848 was proposed to involve in affecting the activity of micorRNA binding site36. Therefore, considering the critical role of the genetic variations in the pTEN/AKT/mTOR pathway, understanding the association between these SNPs and cancer susceptibility are urgently required.
To date, numerous studies have investigated the association of genetic polymorphisms of pTEN/AKT/mTOR pathway genes including rs2295080, rs2536 of mTOR gene, rs2494750 and rs2494752 in the AKT1 gene, pTEN rs701848 with cancer susceptibility6–9,15–23,28–42, however, the results were inconclusive. Therefore, this comprehensive meta-analysis was performed in 5 SNPs of pTEN/AKT/mTOR pathway genes included all eligible case-control studies for evaluating the cancer risk and providing more precise estimation of these associations.
Materials and Methods
Literature research and data extraction
A comprehensive literature search was performed independently by three authors (Z.Z., J.Z., and Q.C.C.) in five electronic databases: PubMed database, CNKI, CbmWeb, WanFang Date, BIOSIS Preview, and ClinicalKey. All the searched eligible original studies and review articles were reviewed carefully to identify the relevant articles by using the following search terms “mTOR rs2295080” or “mTOR rs2536” or “pTEN rs701848” or “AKT1 rs2494750” or “AKT1 rs2494752” and “polymorphism or SNP or single nucleotide polymorphism or variation or mutation” and “cancer or carcinoma or tumor or neoplasm”, (the search was updated on Feb 15, 2017). This search was limited to these articles with English or Chinese language, and the results were reviewed and compared by a forth reviewer (Y. L.W.).
In this meta-analysis, selected publications were eligible if they fulfilled the following criteria: (1) a case-control study or cohort study design; (2) evaluated the association of the genetic polymorphisms of mTOR, AKT1, and pTEN gene with the risk of cancer; (3) sufficient genotypic and/or allelic information for estimating the odds ratio (OR) with 95% confidence intervals (CIs) was provided; (4) the samples size of cases or controls were ≥20. Animal studies, case reports, reviews, and unpublished results were excluded. The following data was extracted from each publication in the collection criterion by Z.Z. and Y. L.W. independently: first author, publication year, ethnicity, country, cancer type, control source (population-based controls, or hospital-based controls), genotyping method, the total number of genotyped cases or controls, and the number of each genotype for cases and controls with each SNP for cancer risk assessment.
Statistical analysis
For the genotype frequency of the controls, the Hardy-Weinberg equilibrium (HWE) was assessed by using the Chi-square test or Fisher’s exact test (P > 0.05) in each study. The pooled odds ratios (ORs) and their corresponding 95% confidence intervals (CIs) were estimated to evaluate the strength of association between these selected 5 SNPs and cancer risk. The pooled estimated ORs and CIs were determined by Z-test based on homozygote model, heterozygote model, dominant model, recessive model, and an additive model(significant for P < 0.05). The heterogeneity between-study was assessed across all eligible comparisons by using χ2-based Cochran’s Q-test (significant for P < 0.10). The random-effects model (DerSimonian-Laird method) was chosen If there is statistical heterogeneity, otherwise the fixed-effects model (Mantel–Haenszel method) was used if the studies were homogeneous. The I 2 statistics was also determined from 0% to 100%, which quantified the heterogeneity irrespective of the number of studies. The sensitivity meta-analysis was assessed by leave-one-out each time to reflect the influence of each study to the pooled estimates. The publication bias was evaluated by the Egger’s and Begg-Matzumdar linear regression tests using asymmetry of the funnel plot (significant for P < 0.10). All the statistical analyses of this meta-analysis were performed by using Stata 12.0 software (StataCorp LP, College Station, USA) and Open Meta-Analyst (http://www.cebm.brown.edu/openmeta/).
Results
Studies extraction and characteristics
Figure 1 summarizes a flowchart presenting the literature review process of study identification, inclusion, exclusion. With the search strategy, a total of 145 published articles were extracted and identified for cancer risk assessment from PubMed, CNKI, CbmWeb, WanFang Date, BIOSIS Preview. After manually screening abstracts and texts of the included 145 studies, 114 were excluded for 2 lack of enough information, 14 abstracts without sufficient data, and 98 duplicated publication or overlapping with other publications for further evaluation. Finally, 31 studies of Asian population were met the inclusion criteria6–9,15–23,28–42, 13 studies of them evaluated the association of mTOR rs2295080 with cancer risks6–9,15–23, 10 studies determined the SNP rs2536 of mTOR gene and cancer susceptibility7,15–18,22,23,28–30, 16 publications were pTEN rs701848 polymorphism7,15,27,31–42, 6 reports studied the AKT1 rs2494750 SNP7,15,23–27 and 3 reports determined AKT1 rs2494752 SNP19,26,27. The distribution of genotypes in the controls for the SNPs of pTEN/AKT/mTOR pathway were in the HWE, except for these 2 publications of Liu, B et al.33 for rs70148 and Fallah et al.24 for rs2494750 (Table 1). In this final meta-analysis 8965 cases and 9868 controls for the mTOR rs2295080, 8411 cases and 8837 controls for the mTOR rs2536, 5882 cases and 6284 controls for the pTEN rs701848, 4332 cases and 4498 controls for the AKT1 rs2494750, and 3187 cases and 3174 controls for the AKT1 rs2494752 were included. The types of cancers mainly include renal cancer, prostate cancer, acute lymphoblastic leukemia (ALL), gastric cancer, hepatocellular cancer, laryngo cancer, colorectal cancer and esophageal squamous cell cancer (ESCC). The ethnicity of the included studies is Asian, and genotyping method includes TaqMan SNP genotyping assay and PCR-RFLP method. The essential characteristics for all studies were shown in Table 1.
Figure 1.
Flow of identification, inclusion, exclusion of the studies.
Table 1.
Characteristics of studies included in the meta-analysis.
| Variant | Author[ref] | Year | Country | Ethnicity | Tumor type | Control Source | Genotyping method | Cases | Controls | HWE (cases) | HWE (controls) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| mTOR rs2295080 | TT | TG | GG | Total | TT | TG | GG | Total | |||||||||
| Cao, Q.7 | 2012 | China | Asian | Renal cancer | HB | TaqMan assay | 454 | 218 | 38 | 710 | 438 | 277 | 45 | 760 | 0.084 | 0.891 | |
| Chen, J. W.15 | 2012 | China | Asian | Prostate cancer | HB | TaqMan assay | 429 | 209 | 28 | 666 | 413 | 259 | 36 | 708 | 0.690 | 0.573 | |
| Huang, L.16 | 2012 | China | Asian | ALL | HB | TaqMan assay | 254 | 140 | 23 | 417 | 353 | 180 | 21 | 554 | 0.523 | 0.742 | |
| Li, Q. X.17 | 2013 | China | Asian | Prostate cancer | PB | PCR-RFLP | 653 | 311 | 40 | 1004 | 617 | 382 | 52 | 1051 | 0.697 | 0.468 | |
| Xu, M.6 | 2013 | China | Asian | Gastric cancer | HB | PCR-RFLP | 482 | 246 | 25 | 753 | 497 | 305 | 52 | 854 | 0.345 | 0.569 | |
| Zhu, M. L.18 | 2015 | China | Asian | ESCC | HB | TaqMan assay | 674 | 390 | 49 | 1113 | 702 | 362 | 49 | 1113 | 0.432 | 0.788 | |
| Xu, M.8 | 2015 | China | Asian | Colorectal cancer | HB | TaqMan assay | 482 | 225 | 30 | 737 | 459 | 273 | 45 | 777 | 0.563 | 0.602 | |
| Wang, M. Y.19 | 2015 | China | Asian | Gastric cancer | HB | TaqMan assay | 568 | 394 | 40 | 1002 | 607 | 355 | 41 | 1003 | 0.005 | 0.221 | |
| Zhao, P.20 | 2015 | China | Asian | ALL | HB | PCR-RFLP | 68 | 50 | 15 | 133 | 173 | 111 | 12 | 296 | 0.221 | 0.263 | |
| Zhao, P.20 | 2015 | China | Asian | AML | HB | PCR-RFLP | 27 | 14 | 6 | 47 | 173 | 111 | 12 | 296 | 0.080 | 0.263 | |
| Zhu, J. H.21 | 2015 | China | Asian | Renal cancer | HB | TaqMan assay | 674 | 390 | 49 | 1113 | 702 | 362 | 49 | 1113 | 0.432 | 0.788 | |
| Zhao, Y.22 | 2016 | China | Asian | Breast cancer | HB | Sequencing | 351 | 197 | 12 | 560 | 345 | 212 | 26 | 583 | 0.009 | 0.358 | |
| Zhang, J.23 | 2016 | China | Asian | Renal cancer | HB | TaqMan assay | 454 | 218 | 38 | 710 | 438 | 277 | 45 | 760 | 0.084 | 0.891 | |
| mTOR rs2536 | TT | TC | CC | Total | TT | TC | CC | Total | |||||||||
| Cao, Q.7 | 2012 | China | Asian | Renal cancer | HB | TaqMan assay | 607 | 99 | 4 | 710 | 628 | 128 | 4 | 760 | 0.001 | 0.353 | |
| Chen, J. W.15 | 2012 | China | Asian | Prostate cancer | HB | TaqMan assay | 565 | 96 | 5 | 666 | 585 | 119 | 4 | 708 | 0.697 | 0.435 | |
| Huang, L.16 | 2012 | China | Asian | ALL | HB | TaqMan assay | 346 | 65 | 6 | 417 | 448 | 103 | 3 | 554 | 0.153 | 0.258 | |
| Li, Q.17 | 2013 | China | Asian | Prostate cancer | PB | PCR-RFLP | 804 | 192 | 8 | 1004 | 894 | 147 | 10 | 1051 | 0.346 | 0.156 | |
| Zhu, M. L.18 | 2013 | China | Asian | ESCC | HB | TaqMan assay | 951 | 165 | 7 | 1123 | 957 | 157 | 7 | 1121 | 0.957 | 0.839 | |
| Mao, L. Q.28 | 2013 | China | Asian | Hepatocellular cancer | HB | TaqMan assay | 849 | 186 | 13 | 1048 | 850 | 188 | 14 | 1052 | 0.439 | 0.330 | |
| He, J.29 | 2013 | China | Asian | Gastric cancer | HB | TaqMan assay | 938 | 179 | 8 | 1125 | 1019 | 170 | 7 | 1196 | 0.865 | 0.975 | |
| Liu, Y. C.30 | 2014 | China | Asian | Hepatocellular cancer | HB | TaqMan assay | 849 | 186 | 13 | 1048 | 850 | 188 | 14 | 1052 | 0.439 | 0.330 | |
| Zhang, J.23 | 2016 | China | Asian | Renal cancer | HB | TaqMan assay | 607 | 99 | 4 | 710 | 628 | 128 | 4 | 760 | 0.987 | 0.353 | |
| Zhao, Y.22 | 2016 | China | Asian | Breast cancer | HB | Sequencing | 453 | 100 | 7 | 560 | 486 | 93 | 4 | 583 | 0.580 | 0.845 | |
| pTEN rs701848 | TT | TC | CC | Total | TT | TC | CC | Total | |||||||||
| Zou, J. F.31 | 2006 | China | Asian | Laryngo cancer | HB | PCR-RFLP | 17 | 23 | 12 | 52 | 28 | 52 | 24 | 104 | 0.547 | 0.135 | |
| Liu, B.33 | 2008 | China | Asian | Laryngo cancer | HB | PCR-RFLP | 7 | 20 | 12 | 91 | 13 | 22 | 9 | 104 | 0.578 | 0.008 | |
| Zhai, Y.32 | 2009 | China | Asian | Laryngo cancer | HB | PCR-RFLP | 29 | 45 | 17 | 39 | 26 | 54 | 24 | 44 | 0.144 | 0.074 | |
| Song, Z. X.34 | 2009 | China | Asian | Laryngo cancer | HB | PCR-RFLP | 46 | 74 | 29 | 149 | 26 | 54 | 24 | 104 | 0.791 | 0.073 | |
| Shi, G. L.35 | 2009 | China | Asian | Lung cancer | HB | PCR-RFLP | 21 | 43 | 13 | 77 | 24 | 54 | 26 | 104 | 0.026 | 0.134 | |
| Song, Z. X.42 | 2009 | China | Asian | Gastric cancer | HB | PCR-RFLP | 43 | 67 | 21 | 58 | 65 | 116 | 34 | 104 | 0.311 | 0.253 | |
| Ding, J.36 | 2011 | China | Asian | Hepatocellular cancer | HB | PCR-RFLP | 222 | 338 | 150 | 131 | 277 | 351 | 132 | 215 | 0.797 | 0.788 | |
| Cao, Q.7 | 2012 | China | Asian | Renal cancer | HB | TaqMan assay | 70 | 121 | 35 | 710 | 103 | 90 | 33 | 760 | 0.055 | 0.691 | |
| Chen, J. W.15 | 2012 | China | Asian | Prostate cancer | HB | TaqMan assay | 212 | 329 | 125 | 666 | 235 | 353 | 120 | 708 | 0.789 | 0.956 | |
| Jang, Y.38 | 2013 | China | Asian | ESCC | HB | PCR-RFLP | 91 | 155 | 58 | 304 | 183 | 165 | 65 | 413 | 0.950 | 0.692 | |
| Tang, Q. S.27 | 2014 | China | Asian | Breast cancer | HB | TaqMan assay | 239 | 519 | 212 | 970 | 280 | 486 | 168 | 934 | 0.938 | 0.692 | |
| Zhang, Y. G.41 | 2014 | China | Asian | ESCC | HB | PCR-RFLP | 205 | 182 | 38 | 494 | 243 | 182 | 21 | 494 | 0.894 | 0.519 | |
| Xu, X.39 | 2015 | China | Asian | ESCC | HB | TaqMan assay | 186 | 421 | 173 | 425 | 229 | 397 | 138 | 446 | 0.257 | 0.692 | |
| Lin, L.40 | 2015 | China | Asian | Colorectal cancer | HB | TaqMan assay | 222 | 338 | 150 | 780 | 277 | 351 | 132 | 764 | 0.027 | 0.088 | |
| Liu, N.37 | 2015 | China | Asian | ESCC | HB | PCR-RFLP | 173 | 241 | 80 | 226 | 145 | 248 | 101 | 226 | 0.440 | 0.988 | |
| Zhang, J.23 | 2016 | China | Asian | Renal cancer | HB | TaqMan assay | 17 | 35 | 6 | 710 | 24 | 54 | 26 | 760 | 0.311 | 0.253 | |
| AKT1 rs2494750 | GG | GC | CC | Total | GG | GC | CC | Total | |||||||||
| Cao, Q.7 | 2012 | China | Asian | Renal cancer | HB | TaqMan assay | 300 | 340 | 70 | 710 | 349 | 328 | 83 | 760 | 0.062 | 0.652 | |
| Chen, J. W.15 | 2012 | China | Asian | Prostate cancer | HB | TaqMan assay | 80 | 269 | 317 | 666 | 78 | 299 | 331 | 708 | 0.053 | 0.399 | |
| Fallah, S24 | 2015 | Iran | Asian | Endometrial cancer | HB | PCR-RFLP | 19 | 6 | 5 | 30 | 22 | 5 | 3 | 30 | 0.007 | 0.015 | |
| Zhang, J.23 | 2016 | China | Asian | Renal cancer | HB | TaqMan assay | 300 | 340 | 70 | 710 | 349 | 328 | 83 | 760 | 0.062 | 0.652 | |
| Wang, M. Y.25 | 2016 | China | Asian | Gastric cancer | HB | TaqMan assay | 493 | 480 | 126 | 1099 | 545 | 487 | 112 | 1144 | 0.577 | 0.833 | |
| Zhu, J. H.26 | 2016 | China | Asian | ESCC | HB | TaqMan assay | 555 | 448 | 114 | 1117 | 521 | 460 | 115 | 1096 | 0.098 | 0.371 | |
| AKT1 rs2494752 | AA | AG | GG | Total | AA | AG | GG | Total | |||||||||
| Tang, Q. S.27 | 2014 | China | Asian | Breast cancer | HB | TaqMan assay | 300 | 511 | 159 | 970 | 331 | 464 | 139 | 934 | 0.017 | 0.253 | |
| Wang, M. Y.25 | 2016 | China | Asian | Gastric cancer | HB | TaqMan assay | 547 | 454 | 99 | 1100 | 623 | 430 | 91 | 1144 | 0.730 | 0.167 | |
| Zhu, J. H.26 | 2016 | China | Asian | ESCC | HB | TaqMan assay | 611 | 423 | 83 | 1117 | 597 | 415 | 84 | 1096 | 0.409 | 0.317 | |
HB, Hospital based; PB, Population based; PCR-RFLP, polymorphism chain reaction- restriction fragment length polymorphism; ALL, Acute lymphocytic leukemia; ESCC, Esophageal squamous cell carcinoma; AML, Acute myeloid leukemia.
Meta-analysis results
mTOR rs2295080, rs2536 and cancer risk analysis
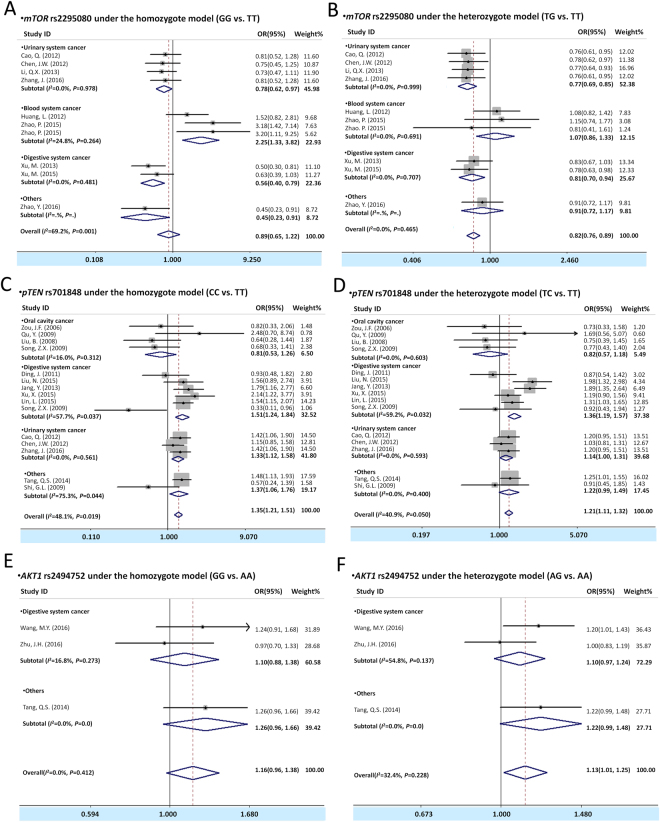
The meta-analysis results for the mTOR rs2295080 and rs2536 polymorphism and cancer susceptibility are illustrated in Tables 2, 3, Fig. 2, and Figure S1. Overall, we observed that carrying mTOR rs2295080 TG or GG genotype and GG/TG genotype showed significant association with decreased cancer risk [TG vs.TT in heterozygote model: OR(95% CI) = 0.82(0.76, 0.89), P < 0.001; GG/TG vs. TT in dominant model: OR(95% CI) = 0.82(0.76, 0.88), P < 0.001; and GG vs.TG/TT in recessive model: OR(95% CI) = 0.67(0.51, 0.88), P = 0.004]. In view of the relative higher heterogeneities, we further analyzed the data by stratification subgroups of urinary system cancer, blood system cancer, and digestive system cancer. Subsequently, we found that rs2295080 GG genotype, TG genotype, and GG/TG genotypes carriers showed a significantly decreased cancer risk in the stratification analysis of urinary system cancer [GG vs.TT: OR(95% CI) = 0.78(0.62, 0.97), P = 0.029; TG vs. TT: OR(95% CI) = 0.77(0.69, 0.85), P < 0.001; GG/TG vs. TT: OR(95% CI) = 0.77(0.69, 0.85), P < 0.001; and GG vs. TG/TT: OR(95% CI) = 0.79(0.63, 0.98), P = 0.035] and digestive system cancer [GG vs.TT: OR(95% CI) = 0.56(0.40, 0.79), P = 0.001; TG vs. TT: OR(95% CI) = 0.81(0.70, 0.94), P = 0.006; GG/TG vs. TT: OR(95% CI) = 0.77(0.67, 0.89), P = 0.001; and GG vs. TG/TT: OR(95% CI) = 0.55(0.40, 0.77), P = 0.001]. However, in the subgroup analysis of blood system cancer, inversely results were found that a significantly increased cancer risk was observed in the carriers of GG genotype [GG vs.TT: OR(95% CI) = 2.25(1.33, 3.82), P = 0.003]. For mTOR rs2536 polymorphism, there was no association was observed both in overall analysis and subgroup analysis (Tables 2 and 3).
Table 2.
Meta-analysis of the association between genetic polymorphisms of PTEN/AKT/mTOR pathway and cancer risk.
| Variables | No. of cases/controls | P z* | Homozygous OR(95% CI) | P het# | I 2# (%) | P z* | Heterozygous OR(95% CI) | P het# | I 2# (%) |
|---|---|---|---|---|---|---|---|---|---|
| mTOR rs2295080 | GG vs. TT | TG vs. TT | |||||||
| Urinary system cancer† | 4203/4392 | 0.029 | 0.78(0.62, 0.97) | 0.978 | 0.0 | 0.000 | 0.77(0.69, 0.85) | 0.999 | 0.0 |
| Blood system cancer§ | 5971/1146 | 0.003 | 2.25(1.33, 3.82) | 0.264 | 24.8 | 0.574 | 1.07(0.86, 1.33) | 0.691 | 0.0 |
| Digestive system cancer¶ | 3605/3747 | 0.001 | 0.56(0.40, 0.79) | 0.481 | 0.0 | 0.006 | 0.81(0.70, 0.94) | 0.707 | 0.0 |
| Overall | 8965/9868 | 0.456 | 0.89(0.65, 1.22) | 0.001 | 69.2 | 0.000 | 0.82(0.76, 0.89) | 0.465 | 0.0 |
| mTOR rs2536 | CC vs. TT | TC vs. TT | |||||||
| Urinary system cancer | 3090/3279 | 0.954 | 1.02(0.56, 1.86) | 0.976 | 0.0 | 0.729 | 0.95(0.69, 1.29) | 0.001 | 81.0 |
| Digestive system cancer | 417/554 | 0.975 | 0.99(0.64, 1.53) | 0.97 | 0.0 | 0.473 | 1.04(0.93, 1.17) | 0.789 | 0.0 |
| Blood system cancer | 4344/4421 | 0.181 | 2.59(0.64, 10.43) | — | — | 0.245 | 0.82(0.58, 1.15) | — | — |
| Overall | 8411/8837 | 0.555 | 1.10(0.80, 1.53) | 0.968 | 0.0 | 0.998 | 1.00(0.89, 1.13) | 0.022 | 53.7 |
| pTEN rs701848 | CC vs. TT | TC vs. TT | |||||||
| Oral cavity cancer¥ | 331/356 | 0.359 | 0.81(0.53, 1.26) | 0.312 | 16.0 | 0.292 | 0.82(0.57, 1.18) | 0.603 | 0.0 |
| Digestive system cancer | 2418/2662 | 0.000 | 1.51(1.24, 1.84) | 0.037 | 57.7 | 0.000 | 1.36(1.19, 1.57) | 0.032 | 59.2 |
| Urinary system cancer | 2086/2228 | 0.001 | 1.33(1.12, 1.58) | 0.561 | 0.0 | 0.049 | 1.14(1.00, 1.31) | 0.539 | 0.0 |
| Overall | 5882/6284 | 0.000 | 1.35(1.21, 1.51) | 0.019 | 48.1 | 0.000 | 1.21(1.11, 1.32) | 0.050 | 40.9 |
| AKT1 rs2494750 | CC vs. GG | GC vs. GG | |||||||
| Urinary system cancer | 2086/2228 | 0.830 | 0.97(0.76, 1.24) | 0.671 | 0.0 | 0.050 | 3.60(1.00, 12.97) | 0.000 | 96.0 |
| Reproductive system cancer$ | 30/30 | 0.621 | 0.93(0.70, 1.24) | — | — | 0 | 0.01(0.01, 0.03) | — | — |
| Digestive system cancer | 2216/2240 | 0.264 | 1.13(0.91, 1.41) | 0.304 | 5.3 | 0.292 | 1.31(0.79, 2.17) | 0.000 | 97.5 |
| Overall | 4332/4498 | 0.727 | 1.03(0.89, 1.18) | 0.658 | 0.0 | 0.943 | 1.03(0.49, 2.17) | 0.000 | 97.5 |
| AKT1 rs2494752 | GG vs. AA | AG vs. AA | |||||||
| Digestive system cancer | 2217/2240 | 0.397 | 1.10(0.88, 1.38) | 0.273 | 16.8 | 0.151 | 1.10(0.97, 1.24) | 0.137 | 54.8 |
| Other cancer | 970/934 | 0.098 | 1.26(0.96, 1.66) | — | — | 0.057 | 1.22 (0.99, 1.48) | — | — |
| Overall | 3187/3174 | 0.090 | 1.16(0.96, 1.38) | 0.412 | 0.0 | 0.026 | 1.13(1.01, 1.25) | 0.228 | 32.4 |
* P z: the significance of the pooled OR was determined by Z-test, and P < 0.05 was considered as statistically significant.
# P het and I 2 were calculated by Chi square-based Q-test.
†Urinary system cancer: renal cancer, prostate cancer; §Blood system cancer: acute lymphocytic leukemia, acute myeloid leukemia; ¶Digestive system cancer: gastric cancer, ESCC, hepatocellular cancer, colorectal cancer; ¥Oral cavity cancer: laryngo cancer; $Reproductive system cancer: endometrial cancer.
Table 3.
Meta-analysis of the association between genetic polymorphisms of PTEN/AKT/mTOR pathway and cancer risk by recessive and dominant models.
| Variables | P z* | Dominant OR(95% CI) | P het# | I 2# (%) | P z* | Recessive OR(95% CI) | P het# | I 2# (%) |
|---|---|---|---|---|---|---|---|---|
| mTOR rs2295080 | GG/TG vs. TT | GG vs. TG/TT | ||||||
| Urinary system cancer† | 0.000 | 0.77(0.69, 0.85) | 1.000 | 0.0 | 0.035 | 0.79(0.63, 0.98) | 0.827 | 0.0 |
| Blood system cancer§ | 0.142 | 1.17(0.95, 1.44) | 0.722 | 0.0 | 0.742 | 0.91(0.52, 1.59) | 0.139 | 49.3 |
| Digestive system cancer¶ | 0.001 | 0.77(0.67, 0.89) | 0.867 | 0.0 | 0.001 | 0.55(0.40, 0.77) | 0.809 | 0.0 |
| Overall | 0.000 | 0.82(0.76, 0.88) | 0.113 | 36.9 | 0.004 | 0.67(0.51, 0.88) | 0.004 | 67.2 |
| mTOR rs2536 | CC/TC vs. TT | CC vs. TC/TT | ||||||
| Urinary system cancer | 0.836 | 0.99(0.86, 1.13) | 0.002 | 79.3 | 0.972 | 1.01(0.55, 1.85) | 0.952 | 0.0 |
| Digestive system cancer | 0.489 | 1.04(0.93, 1.16) | 0.749 | 0.0 | 0.960 | 0.99(0.64, 1.53) | 0.976 | 0.0 |
| Blood system cancer | 0.400 | 0.87(0.62, 1.21) | — | — | 0.165 | 2.68(0.67, 10.78) | — | — |
| Overall | 0.649 | 1.02(0.94, 1.10) | 0.036 | 49.7 | 0.572 | 1.10(0.79, 1.53) | 0.961 | 0.0 |
| pTEN rs701848 | CC/TC vs. TT | CC vs. TC/TT | ||||||
| Oral cavity cancer¥ | 0.674 | 0.82(0.58, 1.15) | 0.568 | 0.0 | 0.250 | 0.92(0.64, 1.34) | 0.409 | 0.0 |
| Digestive system cancer | 0.017 | 1.40(1.22, 1.59) | 0.061 | 52.5 | 0.000 | 1.23(1.04, 1.47) | 0.027 | 60.4 |
| Urinary system cancer | 0.008 | 1.20(1.05, 1.36) | 0.784 | 0.0 | 0.006 | 1.23(1.05, 1.43) | 0.476 | 0.0 |
| Overall | 0.000 | 1.25(1.15, 1.36) | 0.179 | 24.9 | 0.000 | 1.20(1.09, 1.32) | 0.015 | 49.9 |
| AKT1 rs2494750 | CC/GC vs. GG | CC vs. GC/GG | ||||||
| Urinary system cancer | 0.310 | 1.09(0.92, 1.30) | 0.369 | 0.0 | 0.999 | 1.00(0.84, 1.20) | 0.575 | 0.0 |
| Reproductive system cancer$ | 0.312 | 0.92(0.78, 1.08) | — | — | 0.825 | 0.97(0.74, 1.28) | — | — |
| Digestive system cancer | 0.055 | 1.13(1.00, 1.29) | 0.784 | 0.0 | 0.557 | 1.07(0.86, 1.31) | 0.185 | 43 |
| Overall | 0.198 | 1.06(0.97, 1.16) | 0.293 | 18.5 | 0.810 | 1.02(0.90, 1.15) | 0.67 | 0.0 |
| AKT1 rs2494752 | GG/AG vs. AA | GG vs. AG/AA | ||||||
| Digestive system cancer | 0.129 | 1.10(0.97, 1.23) | 0.098 | 63.5 | 0.614 | 1.06(0.85, 1.31) | 0.447 | 0.0 |
| Other cancer | 0.037 | 1.23(1.01, 1.48) | — | — | 0.365 | 1.12(0.88, 1.44) | — | — |
| Overall | 0.017 | 1.13(1.02, 1.25) | 0.157 | 46.0 | 0.329 | 1.08(0.92, 1.28) | 0.704 | 0.0 |
* P z: the significance of the pooled OR was determined by Z-test, and P < 0.05 was considered as statistically significant.
# P het and I 2 were calculated by Chi square-based Q-test.
†Urinary system cancer: renal cancer, prostate cancer; §Blood system cancer: acute lymphocytic leukemia, acute myeloid leukemia; ¶Digestive system cancer: gastric cancer, ESCC, hepatocellular cancer, colorectal cancer; ¥Oral cavity cancer: laryngo cancer; $Reproductive system cancer: endometrial cancer.
Figure 2.
Forest plots of cancer risk with polymorphism of mTOR rs2529080 (A,B), pTEN rs701848 (C,D) and AKT1 rs2494752 (E,F) assessing by subgroup analysis under the homozygoute model (A,C,E), heterozygote model (B,D,F). The estimates of OR(95% CIs) are plotted with a box and a horizontal line for each study. ◇, pooled ORs (95% CIs).
pTEN rs701848 and cancer risk analysis
The effect of pTEN rs701848 polymorphism on cancer risk in overall and subgroup analysis was shown in Tables 2, 3 and Figs 3 and S2.The SNP rs701848 CC or TC genotype and CC/TC genotype were associated with an increased overall cancer risk [CC vs.TT in homozygote model: OR(95% CI) = 1.35(1.21, 1.51), P < 0.001; TC vs. TT in heterozygote model: OR(95% CI) = 1.21(1.11, 1.32), P < 0.001; CC/TC vs. TT in dominant model: OR(95% CI) = 1.25(1.15, 1.36), P < 0.001; and CC vs. TC/TT in recessive model: OR(95% CI) = 1.20(1.09, 1.32), P < 0.001]. In the further stratification analysis, we found that rs701848 CC genotype, TC genotype and CC/TC genotypes were statistically increased association with digestive system cancer [CC vs.TT: OR(95% CI) = 1.51(1.24, 1.84), P < 0.001; TC vs. TT: OR(95% CI) = 1.36(1.19, 1.57), P < 0.001; and CC/TC vs. TT: OR(95% CI) = 1.40(1.22, 1.59), P = 0.017; and CC vs. TC/TT: OR(95% CI) = 1.23(1.04, 1.47), P < 0.001] and urinary system cancer[CC vs. TT: OR(95% CI) = 1.33(1.12, 1.58), P < 0.001; TC vs. TT: OR(95% CI) = 1.14(1.00, 1.31), P = 0.049; and CC/TC vs. TT: OR(95% CI) = 1.20(1.05, 1.36), P = 0.008; and CC vs. TC/TT: OR(95% CI) = 1.23(1.05, 1.43), P = 0.006], however no association was observed between the oral cavity cancer and cancer risk in this concluded studies.
Figure 3.
Forest plots of cancer risk with polymorphism of mTOR rs2529080 (A,B), pTEN rs701848 (C,D) and AKT1 rs2494752 (E,F) assessing by subgroup analysis under the dominant model (A,C,E) and recessive model (B,D,F). The estimates of OR(95% CIs) are plotted with a box and a horizontal line for each study. ◇, pooled ORs (95% CIs).
AKT1 rs2494750, rs2494752 and cancer risk analysis
The association between polymorphisms of AKT1 rs2494750, rs2494752 and cancer risk in overall meta-analysis results was shown in Tables 2, 3 and Fig. 3, and Figure S2. A significant association was observed between AKT1 rs2494752 and overall cancer risk, and the heterozygous genotype AG and GG/AG genotype of AKT1 rs2494752 were associated with increased cancer risk (AG vs. AA in heterozygote model: OR(95% CI) = 1.13(1.01, 1.25), P = 0.026; and GG/AG vs. AA in dominant model: OR(95% CI) = 1.13(1.02, 1.25), P = 0.017). For AKT1 rs2494750 polymorphism, we have not found the correlation with the cancer susceptibility both in overall analysis and subgroup analysis (Tables 2 and 3).
Heterogeneity and sensitivity analysis
No significant heterogeneities were observed for the overall analyses of mTOR rs2536, AKT1 rs2494750 and rs2494752. However, the highest heterogeneity were observed when all the studies were analyzed for all the cases of mTOR rs2295080 under the dominant model (I 2 = 96.1%) and pTEN rs701848 under the dominant model (I 2 = 62.0%) (Tables S1 and S2). The heterogeneity of mTOR rs2295080 was also present in the subgroup of urinary system cancer and digestive system cancer under the dominant model (Table S2). Therefore, the leave-one-out sensitivity analysis and random-effects model was selected for decreasing the heterogeneity. When these 3 publications of Zhu, M. L. (2015), Wang, M.Y. (2015), Zhu, J.H. (2015) were deleted, the value of I 2 decreased from 96.1% to 39.6% under the dominant model. More importantly, after deleting these 2 articles of Zhu, M. L. (2015), Wang, M.Y. (2015) from digestive system cancer, the subgroup of heterogeneity significantly decreased from 97.6% to 0% (Tables 3 and S2). Another article of Zhu, J.H. (2015) from urinary system cancer was deleted, the I 2 decreased from 96.3% to 0% under the dominant model in subgroup analysis (Tables 3 and S2). For pTEN rs701848, we found that the heterogeneity of overall and subgroup was significantly decreased after deleting this article of Zhang Y.G.(2014), except the subgroup under the digestive system cancer (Tables 2, 3, S1 and S2).
Publication bias analysis
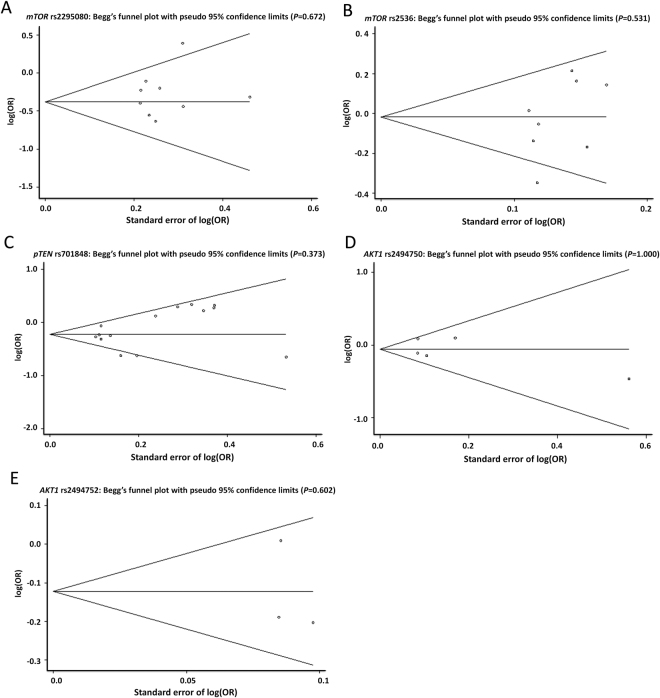
In this meta-analysis, the Begg’s funnel plot and Egger’s test were performed to evaluate the publication bias of the concluded studies. There are no significant publication bias was observed for all the dominant models of the five SNPs (rs2295080: P = 0.200; rs2536: P = 0.176; rs701848: P = 0.218; rs2494750: P = 0.694 and rs2494752: P = 0.696) by the Egger’s test. The funnel plots shapes showed obvious symmetry, which were obtained for the association of the SNPs under the dominant model (rs2295080: P = 0.672; rs2536: P = 0.531; rs701848: P = 0.373; rs2494750: P = 1.000 and rs2494752: P = 602) (Fig. 4). The data indicated that no publication bias might have a significant influence on the observed effect of SNPs located at pTEN/AKT/mTOR pathway on the susceptibility of cancer as assessed.
Figure 4.
Begg’s funnel plots to detect publication bias with pseudo 95% CIs under the dominant model. Each point represents an independent study for the indicated association. (A) mTOR rs2295080; (B) mTOR rs2536; (C) pTEN rs701848; (D and E) AKT1 rs2494750 and rs2494752.
Discussion
Overexpression or mutations of key genes of the pTEN/AKT/mTOR pathways were associated with carcinogenesis, invasion, metastasis, and prognosis of a variety of cancers44,45. Since one group investigated the association of genetic polymorphisms of the pTEN/AKT/mTOR pathway with cancer risk for the first time31, a variety of studies have been performed to explore the possible correlation of the SNPs in this pathway genes on cancer susceptibility6–9,15–23,28–42. The potentially functional SNPs in those key genes, especially in the TFBS or miRNA binding sites may involve in the cancer susceptibility. Therefore, the present meta-analysis analyzed the associations of SNPs in the 5′upstream regulatory or promoter region (mTOR rs2295080, AKT1 rs2494752), and 3′UTR region (mTOR rs2536, pTEN rs701848 and AKT1 rs2494750) in the mTOR signaling pathway (AKT, and PTEN) with cancer risk.
mTOR rs2295080 and rs2536 SNPs and cancer risk
Given the crucial function of mTOR in cellular signals from growth factors and energy status, such as in angiogenesis and cell proliferation, growth, differentiation, and apoptosis3–5,44,45, our findings of an association between the genetic variations in mTOR gene and cancer risk are biologically plausible and wide, including renal cell cancer7,21,23, prostate cancer15,17, breast cancer22, acute lymphocytic leukemia16,20, gastric cancer19,29, esophageal squamous cell cancer18, hepatocellular cancer23,28 and colorectal cancer8. In this meta-analysis of 13 studies including 8965 cases and 9868 controls for rs2295080, we found a significant decreased of rs2295080 TG, GG genotype, G allele and TG/GG genotype on the overall cancer risk, and the stratification subtype of urinary system cancer and digestive system cancer. However, a significant increased association was found on the blood system cancer in the homozygous GG genotype and G allele under the subgroup analysis. Up to now, only two meta-analyses focused on mTOR rs2295080 polymorphism and cancer risk46,47. In one meta-analysis reported that the rs2295080 G allele is associated with decreased risk of cancer46, however, only five studies were included. Recent some opposite findings were reported in gastric cancer6, esophageal cancer18, and especially in acute leukemia16,20. Thereafter, another eight studies meta-analysis found that the rs2295080 G allele was associated with a significantly lower risk of genitourinary cancers in the dominant model, and a higher risk of acute leukemia in the recessive model47, which consistent with our founding’s in overall cancer risk and digestive system cancer. Likewise, we further founded that carrying rs2295080 GG genotype showed increased 2.25-fold association in the blood system cancer including acute lymphocytic leukemia and acute lymphocytic leukemia which was not explicated in the previous meta-analyses. Thus, these data indicated that there was an obvious divergence between the SNP rs2295080 and cancer risk in the digestive system and blood system, which might be partially explained by cancer-specificity of rs2295080 polymorphism.
10 studied of 8411 cases and 8837 controls for rs2536 polymorphism, no significant association was observed between rs2536 and cancer susceptibility after performing stratified analyses by cancer type7,15–18,22,23,28–30 in this pooled meta-analysis. Previously, Shao et al.46 performed a six case-control studies meta-analysis and also reported that there was no association of rs2536 SNP with cancer risk both under dominant and recessive models. Furthermore, this present updated meta-analysis also indicated that rs2536 polymorphism was not an important biomarker for predicting cancer risk, although rs2536 was observed associated with the risk of esophageal cancer18 and prostate cancer15, together with interacting with environmental factors such as body mass index. The previous foundlings were controversial for the SNP rs2536, partially because of insufficient statistical power and further studies of different cancers are needed for providing a more precise estimation of the associations.
pTEN rs701848 and cancer risk
pTEN was originally identified as tumor suppressor gene, considered as a key negative regulator of PI3K/Akt pathway48–50. However, little is known about the association between pTEN rs701848 polymorphism and cancer. Specifically, 15 studies of 5882 cases and 6284 controls for rs701848, CC or TC genotype, C allele and CC/TC genotype were associated with significant increased overall cancer risk and in the subgroup of the digestive system cancer and urinary system cancer, but not in oral cavity cancers. Since Zou et al.31 for the first time reported a significant association of pTEN rs701848 with laryngo cancer risk in 2006, more evidences have accumulated regarding the relationship between SNP rs701848 and the risk of various cancers, such as lung cancer35, esophageal cancer37–39,41, breast cancer27, prostate cancer15, hepatocellular cancer36, renal cancer7,23, gastric cancer42, and colorectal cancer40. It should be noted that SNP rs701848 is located within the 3′ near gene region, which can be targeted by microRNAs to affecting the miRNA binding site activity, thereby altering pTEN expression by influencing the mRNA stability, and then influence cancer susceptibility. However, the hypothesized function about SNP rs701848 still needs to be investigated in future studies and updated meta-analysis.
AKT1 rs2494750, rs2494752 and cancer risk
In this pooled meta-analysis, 6 studied of 4332 cases and 4498 controls for rs2494750 polymorphism7,15,23–27 and 3 studied of 3187 cases and 3174 controls for rs2494752 polymorphism19,26,27 were included. A significant association was observed between rs2494752 and overall cancer risk, the heterozygous genotype AG, GG/AG genotype and G allele of rs2494752 SNP was associated with increased cancer risk. However, we have not found the correlation between rs2494750 polymorphism and the cancer susceptibility both in overall analysis and the stratification analysis. Previous study reported15 that the risk effect of rs2494752 AG/GG genotypes was more obvious in the patients of ever-drinkers. Another investigation showed inconsistent results in alcohol consumption patients and risk of gastric cancer25. These results indicated that the environmental factors were interacted with the genetic variants in the aspect of carcinogenesis. Most importantly, the SNP rs2494752 is located at the 5′ UTR of AKT1 gene, a region predicted to be the transcription factor binding region based on the sequence alignments, which may affect the transcription and translation of AKT1. It is plausible that the rs2494752 G allele increased the transcription activity of the promoter in the AKT1 gene, then facilitated the cancer development and progression, which may partially explain the cancer risk associated with this SNP. However, the potential function of this SNP should be necessary to identify in larger sample studies in the future.
There is not previously reported meta-analysis to date that comprehensively elucidated the association between the five SNPs of the pTEN/AKT/mTOR signaling pathway and risk of cancer. However, some limitations need to be addressed in this meta-analysis. First, only Asian population was involved, lack of the samples of other ethnicities such as Caucasian, African in this study, thus a wider spectrum of subjects should be conducted on various ethnicities in the future. Second, only 3 studies were included for the SNP rs2494752, therefore considering the limited small size of rs2494752, we cannot rule out the possibility that the results may be by chance, although the number of study participants met the requirement for analysis. Third, all studies included in the present systematic review were reported in Chinese or English, yet other languages publications may include the relevant studies, which may be the main source of publication bias in our meta-analysis. Finally, only five SNPs located in the 3′UTR or 5′UTR of the candidate genes were chosen, however, the SNPs of exon or intron region was not included. Thus, the limited SNPs were not sufficient to capture most genetic information of pTEN/AKT/mTOR signaling pathway, more SNPs should be included in the study, and interaction with others genes should be investigated in the future updated meta-analysis.
In conclusion, this meta-analysis was performed included the latest publications, and provided a more precise prevalence estimate for associations of five SNPs of pTEN/AK/mTOR pathway with the risk of cancer. We found that mTOR rs2295080 TG, GG genotype and GG/TG genotype carriers showed an decreased in the overall cancer risk, urinary system cancer and digestive system cancer, nevertheless TT genotype of rs2295080 was associated with increased the risk of blood system cancer. Carrying rs701848 CC,TC genotype and CC/TC genotype were associated with an increased overall cancer risk, especially in digestive system cancer and urinary system cancer. Moreover, a significant increased association was observed between rs2494752 AG and GG/AG genotype and overall cancer risk. Therefore, this present study provides the most valid cancer prevalence estimate to date for Asian population, which is a necessary foundational piece for further research on this topic.
Novelty and Impact Statements
In this meta-analysis of Asian population, the carriers of mTOR rs2295080 TG, GG genotype and GG/TG genotypes showed a significantly decreased the risk of overall cancer, the urinary system cancer and digestive system cancer. Furthermore, the SNP rs701848 CC, TC genotype and CC/TC genotype of pTEN were observed increased susceptibility of overall cancer and the subgroup of the urinary and digestive cancer. Moreover, carrying AKT1 rs2494752 AG and GG/AG genotypes showed an increased overall cancer risk.
Electronic supplementary material
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of the People’s Republic of China (Grant No. 31371145, No. 81572613, and No. 81402948), Liaoning Key Laboratory of Molecular Targeted Anti-tumor Drug Development and Evaluation Program, the Educational Commission of Liaoning Province, China (Grant No. L2014315), the Research Fund for the Doctoral Program of Higher Education of Liaoning Province, China (Grant No. 20141032), the Key Laboratory Foundation from Shenyang S&T Projects (F16-094-1-00), Liaoning Province Scientific Research Foundation (2014226033), Program for Liaoning Innovative Research Team in University (No. LT2014016), and the S&T Projects in Shenyang, China (Grant No. F14-232-6-05).
Author Contributions
Huizhe Wu and Minjie Wei conceived the study and edited the paper. Zhen Zhang, Qiuchen Chen, Jing Zhang, Yilin Wang, Haishan Zhao and Weifan Yao searched and collected the data. Xiaoyun Hu, Sainan Yin and Miao He performed the statistical analysis. Zhen Zhang and Huizhe Wu interpreted data and wrote the manuscript. Shu Guan, Wenyan Qin, Qinghuan Xiao and critically revised the manuscript. All authors approved the final version of the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-17250-z.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Huizhe Wu, Email: wuhz@cmu.edu.cn.
Minjie Wei, Email: mjwei@mail.cmu.edu.cn.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 2.Chen W, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–32. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 3.Lim HJ, Crowe P, Yang JL. Current clinical regulation of PI3K/PTEN/Akt/ mTOR signaling in treatment of human cancer. J Cancer Res Clin Oncol. 2015;141:671–89. doi: 10.1007/s00432-014-1803-3. [DOI] [PubMed] [Google Scholar]
- 4.McCubrey JA, et al. Mutations and deregulation of Ras/Raf/MEK/ERK and PI3K/PTEN/Akt/mTOR cascades which alter therapy response. Oncotarget. 2012;3:954–87. doi: 10.18632/oncotarget.652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johnson SM, et al. Novel expression patterns of PI3K/Akt/mTOR signaling pathway components in colorectal cancer. Journal of the American College of Surgeons. 2010;210(767–76):776–8. doi: 10.1016/j.jamcollsurg.2009.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu M, et al. A polymorphism (rs2295080) in mTOR promoter region and its association with gastric cancer in a Chinese population. PLOS One. 2013;8:e60080. doi: 10.1371/journal.pone.0060080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cao Q, et al. A functional variant in the MTOR promoter modulates its expression and is associated with renal cell cancer risk. PloS One. 2012;7:e50302. doi: 10.1371/journal.pone.0050302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu M, et al. Functional promoter rs2295080 T > G variant in mTOR gene is associated with risk of colorectal cancer in a Chinese population. Biomed Pharmacother. 2015;70:28–32. doi: 10.1016/j.biopha.2014.12.045. [DOI] [PubMed] [Google Scholar]
- 9.Yuan T, Cantley L. PI3K pathway alterations in cancer: variations on a theme. Oncogene. 2008;27:5497–510. doi: 10.1038/onc.2008.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zoncu R, Efeyan A, Sabatini DM. mTOR: from growth signal integration to cancer, diabetes and ageing. Nature reviews Molecular cell biology. 2011;12:21–35. doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fingar DC, et al. mTOR controls cell cycle progression through its cell growth effectors S6K1 and 4E-BP1/eukaryotic translation initiation factor 4E. Molecular and cellular biology. 2004;24:200–16. doi: 10.1128/MCB.24.1.200-216.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274–93. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Edinger AL, Thompson CB. Akt maintains cell size and survival by increasing mTOR-dependent nutrient uptake. Molecular biology of the cell. 2002;13:2276–88. doi: 10.1091/mbc.01-12-0584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chung JY, et al. The expression of phospho-AKT, phospho-mTOR, and PTEN in extrahepatic cholangiocarcinoma. Clinical cancer research. 2009;15:660–7. doi: 10.1158/1078-0432.CCR-08-1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen J, et al. Genetic variations in a PTEN/AKT/mTOR axis and prostate cancer risk in a Chinese population. PLoS One. 2012;7:e40817. doi: 10.1371/journal.pone.0040817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang L, et al. Association of genetic variations in mTOR with risk of childhood acute lymphoblastic leukemia in a Chinese population. Leuk Lymphoma. 2012;53:947–51. doi: 10.3109/10428194.2011.628062. [DOI] [PubMed] [Google Scholar]
- 17.Li Q, et al. Polymorphisms in the mTOR gene and risk of sporadic prostate cancer in an Eastern Chinese population. PloS One. 2013;8:e71968. doi: 10.1371/journal.pone.0071968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu ML, et al. Polymorphisms in mTORC1 genes modulate risk of esophageal squamous cell carcinoma in eastern Chinese populations. J Thorac Oncol. 2013;8:788–95. doi: 10.1097/JTO.0b013e31828916c6. [DOI] [PubMed] [Google Scholar]
- 19.Wang MY, et al. Genetic variations in the mTOR gene contribute toward gastric adenocarcinoma susceptibility in an Eastern Chinese population. Pharmacogenet Genomics. 2015;25:521–30. doi: 10.1097/FPC.0000000000000122. [DOI] [PubMed] [Google Scholar]
- 20.Zhao P, et al. Analysis of polymorphism of mTOR gene in children with leukemia. J Clin Pediatr. 2013;33:423–5. [Google Scholar]
- 21.Zhu J, et al. Associations of PI3KR1 and mTOR polymorphisms with esophageal squamous cell carcinoma risk and gene-environment interactions in Eastern Chinese populations. Sci Rep. 2015;5:8250. doi: 10.1038/srep08250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao Y, et al. Impacts of the mTOR gene polymorphisms rs2536 and rs2295080 on breast cancer risk in the Chinese population. Oncotarget. 2016;7:58174–80. doi: 10.18632/oncotarget.11272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang J, et al. Polymorphism in the mTOR promoter is associated with risk of renal cell carcinoma. J Mod Urol. 2015;20:340–6. [Google Scholar]
- 24.Fallah S, Korani M, Hajimirza M, Seifi M. Association between genetic variants of Akt1 and Endometrial Cancer. Biochem Genet. 2015;53:281–90. doi: 10.1007/s10528-015-9690-0. [DOI] [PubMed] [Google Scholar]
- 25.Wang MY, et al. A Functional Polymorphism (rs2494752) in the AKT1 Promoter Region and Gastric Adenocarcinoma Risk in an Eastern Chinese Population. Sci Rep. 2016;6:20008. doi: 10.1038/srep20008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu J, et al. Polymorphisms in the AKT1 and AKT2 genes and oesophageal squamous cell carcinoma risk in an Eastern Chinese population. J Cell Mol Med. 2016;20:666–77. doi: 10.1111/jcmm.12750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tang, Q. S. The genetic polymorphisms of PTEN/PI3K/AKT signaling pathway are associated with the susceptibility and neoadjuvant chemotherapy response and prognosis of breast cancer (Doctoral Thesis). China Medical University, Shenyang, Liaoning, China (2014).
- 28.Mao, L. Q. Association of mTOR polymorphisms and genetic susceptibility of hepatocellular carcinoma (Master’s Thesis). Guangxi Medical University, Xining, Guangxi, China (2013).
- 29.He J, et al. Genetic variations of mTORC1 genes and risk of gastric cancer in an Eastern Chinese population. Mol Carcinog. 2013;52(Suppl 1):E70–9. doi: 10.1002/mc.22013. [DOI] [PubMed] [Google Scholar]
- 30.Liu YC, et al. Association of mTOR polymorphisms with risk of hepatocellular carcinoma. Chin J Public Health. 2014;30:593–7. [Google Scholar]
- 31.Zhou, J. F. Research of association of single nucleotide polymorphisms of PTEN gene with china laryngeal cancer (Master’s Thesis). Jilin University, Jilin, Changchun, China (2006).
- 32.Zhai, Y. Study on the correlationgship of PTEN gene and laryngeal carcinoma (Master’s Thesis). Jilin University, Changchun, Jilin, China (2009).
- 33.Liu B, Liu Y, Zhou JF, Long HZ, Zhu W. Correlation analysis of PTEN gene single nucleotide polymorphism in laryngocarcinoma. Chin J Cancer Prev Treat. 2008;15:1695–7. [Google Scholar]
- 34.Song ZX, Zhu W, Li P, Liu B. Association between anti-oncogene PTEN single nucleotide polymorphism and laryngocarcinoma. Journal of Jilin University (Medicine Edition) 2009;35:918–22. [Google Scholar]
- 35.Shi GL, Qin HY, Chen Q. The association between lung cancer and the single nucleotide polymorphisms of PTEN gene. Modern. Oncology. 2009;17:851–3. [Google Scholar]
- 36.Ding, J. The association of PTEN gene with the development of Hepatocellular Carcinoma (Master’s Thesis). Suzhou University, Suzhou, Jiangsu, China (2011).
- 37.Liu N, Xu C. Association of MDM2 and PTEN gene polymorphisms and susceptibility of esophageal squamous carcinoma in Anyang area. Chin J Digest Med Imageol (Electronic Edition) 2015;5:223–8. [Google Scholar]
- 38.Jang Y, et al. Genetic polymorphisms of CCND1 and PTEN in progression of esophageal squamous carcinoma. Genet Mol Res. 2013;12:6685–91. doi: 10.4238/2013.December.13.2. [DOI] [PubMed] [Google Scholar]
- 39.Xu, X., Chen, G., Wu, L. & Liu, L. Association of genetic polymorphisms in PTEN and additional gene-gene interaction with risk of esophageal squamous cell carcinoma in Chinese Han population. Dis Esophagus (2015). [DOI] [PubMed]
- 40.Lin L, Zhang Z, Zhang W, Wang L, Wang J. Roles of genetic variants in the PI3K/PTEN pathways in susceptibility to colorectal carcinoma and clinical outcomes treated with FOLFOX regimen. Int J Clin Exp Pathol. 2015;8:13314–22. [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang, Y. G. SNPs in micro RNA Binding Site of EGFR Signaling Pathway and Susceptibility to ESCC. (Master’s Thesis). Zhengzhou University, Zhengzhou, Henan, China (2014).
- 42.Song ZX, Liu B, Zhao J, Liu J. Analysis of association between PTEN gene single nucleotide polymorphism and stomach neoplasms. Journal of Jilin University (Medicine Edition) 2009;35:694–7. [Google Scholar]
- 43.Li, Q. Functional genetic variants in the mTORC1 related genes contribute to prostate cancer susceptibility and clinical outcomes (Doctoral Thesis). Fudan University, Shanghai, China (2014).
- 44.Albert V, Hall MN. mTOR signaling in cellular and organismal energetics. Current opinion in cell biology. 2015;33:55–66. doi: 10.1016/j.ceb.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 45.Polivka J, Janku F. Molecular targets for cancer therapy in the PI3K/AKT/ mTOR pathway. Pharmacology & therapeutics. 2014;142:164–75. doi: 10.1016/j.pharmthera.2013.12.004. [DOI] [PubMed] [Google Scholar]
- 46.Shao J, et al. Association of mTOR polymorphisms with cancer risk and clinical outcomes: a meta-analysis. PLoS One. 2014;9:e97085. doi: 10.1371/journal.pone.0097085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zining J, Lu X, Caiyun H, Yuan Y. Genetic polymorphisms of mTOR and cancer risk: a systematic review and updated meta-analysis. Oncotarget. 2016;7:57464–80. doi: 10.18632/oncotarget.10805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Downes CP, et al. Antagonism of PI3-kinase-dependent signaling pathways by the tumour suppressor protein, PTEN. Biochemical Society Transactions. 2001;29:846–90. doi: 10.1042/bst0290846. [DOI] [PubMed] [Google Scholar]
- 49.Hildebrandt MA, et al. Genetic variations in the PI3K/PTEN/AKT/mTOR pathway are associated with clinical outcomes in esophageal cancer patients treated with chemoradiotherapy. J Clin Oncol. 2009;27:857–71. doi: 10.1200/JCO.2008.17.6297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen M, et al. Genetic variations in PI3K-AKT-mTOR pathway and bladder cancer risk. Carcinogenesis. 2009;30(22):2047–52. doi: 10.1093/carcin/bgp258. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.