ABSTRACT
Escherichia coli is a leading cause of bacterial mastitis in dairy cattle. It is most often transient in nature, causing an infection that lasts 2 to 3 days. However, E. coli has been shown to cause a persistent infection in a minority of cases. Mechanisms that allow for a persistent E. coli infection are not fully understood. The goal of this work was to determine differences between E. coli strains originally isolated from dairy cattle with transient and persistent mastitis. Using RNA sequencing, we show gene expression differences in nearly 200 genes when bacteria from the two clinical phenotypes are compared. We sequenced the genomes of the E. coli strains and report genes unique to the two phenotypes. Differences in the wca operon, which encodes colanic acid, were identified by DNA as well as RNA sequencing and differentiated the two phenotypes. Previous work demonstrated that E. coli strains that cause persistent infections were more motile than those that cause transient infections. Deletion of genes in the wca operon from a persistent-infection strain resulted in a reduction of motility as measured in swimming and swarming assays. Furthermore, colanic acid has been shown to protect bacteria from complement-mediated killing. We show that transient-infection E. coli strains were more sensitive to complement-mediated killing. The deletion of genes from the wca operon caused a persistent-infection E. coli strain to become sensitive to complement-mediated killing. This work identifies important differences between E. coli strains that cause persistent and transient mammary infections in dairy cattle.
KEYWORDS: colanic acid, Escherichia coli, mastitis, pathogenesis
INTRODUCTION
Mastitis is the most common health problem in the dairy industry. The approximate economic impact of mastitis in dairy cattle to the U.S. economy is $2 billion of lost revenue per year (1). Numerous microorganisms can cause mastitis, and manifestation of the disease can range from completely subclinical to severe and life threatening.
Escherichia coli is a common cause of mastitis in dairy cows and is a leading cause of acute mastitis (2). Typically, an E. coli mastitis infection is transient in duration, but persistent intramammary infections can occur (3, 4). Research suggests that 5 to 20% of mammary-pathogenic E. coli (MPEC) infections are persistent (4–6). Strains of MPEC that were isolated from persistent intramammary infections have been distinguished from MPEC strains that were isolated from transient infections in several ways. First, MPEC strains that were associated with persistent infections have been shown to invade cultured mammary epithelial cells more effectively than strains that cause transient infections (7). Second, MPEC strains that were associated with persistent infections were found to have greater motility in vitro, as demonstrated by increased rates of swimming and swarming, than MPEC strains that cause transient mastitis (8).
Previously, we have shown that growth of E. coli in milk compared to that in Luria-Bertani (LB) broth caused massive changes in protein expression levels (9). In fact, over 50% of the 1,000 proteins identified had significant expression changes between the E. coli grown in milk and that grown in bacterial growth medium. Some of the proteins downregulated in milk were important for bacterial motility. Because previous swimming and swarming assays were performed in LB broth-based medium, we sought to determine if these phenotypes were affected by milk.
Analysis of protein expression differences between the persistent- and transient-infection strains (8) identified 28 proteins whose change in expression levels correlated with disease phenotype (e.g., persistent versus transient). One group of these proteins consisted of flagellar and chemotaxis proteins. The goal of the transcriptomics experiment in this paper was to identify additional differences between the persistent- and transient-infection strains grown in milk to help explain these pathogenic differences.
The objective of this work is to identify mechanisms that allow a subset of MPEC strains to establish a persistent infection and therefore improve our understanding of bacterial pathogenesis in the mammary gland. Utilizing transcriptomics, genomics, and phenotypic assays, we report that the wca operon plays a critical role in determining whether the E. coli strain will produce a transient or persistent infection.
RESULTS
Motility in milk plates.
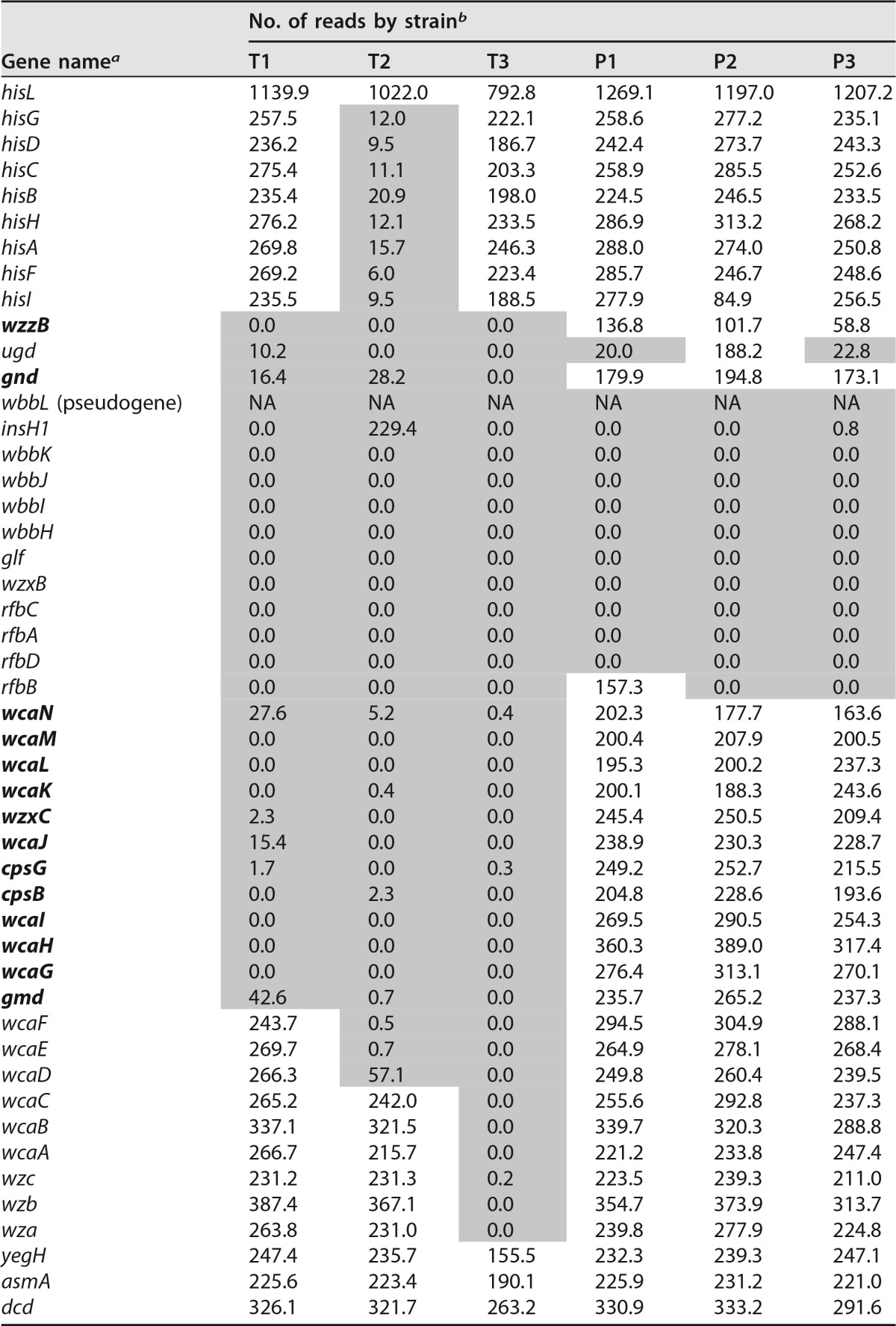
Our previous work showed that three persistent-infection MPEC strains (strains P1 to P3) were more motile in standard LB broth-based swimming and swarming plates than transient-infection MPEC strains (strains T1 to T3) (10). Because milk affects the expression of motility proteins (9), milk-based agar plates were developed to assay swimming and swarming phenotypes among the MPEC isolates. Commercially available milk agar plates are made with tryptone, yeast extract, dextrose, skim milk powder, and agar, providing nutrients not typically found in milk. Therefore, for this swimming assay we developed milk-based agar plates that contained 90% fresh skim milk and a 10× agar solution (final agar concentration, 0.35%). Our plates did not contain typical bacterial growth medium components (e.g., tryptone or yeast extract) in order to generate growth conditions more similar to in vivo conditions. Figure 1A shows a representative photograph of a less motile MPEC strain, and Fig. 1B shows a highly motile MPEC strain. The persistent-infection strains, as a group, were significantly more motile in this assay than the transient-infection strains (P < 0.0001) (Fig. 1C). Additionally, all three persistent-infection strains were significantly more motile than any of the three transient-infection strains (Fig. 1C). These results are consistent with our results previously reported using normal LB swimming plates (10). In the milk and LB swimming assays the time required for the bacteria to swim the same distance was greater for the milk-based plates than for the LB swimming plates (15 versus 5 h).
FIG 1.
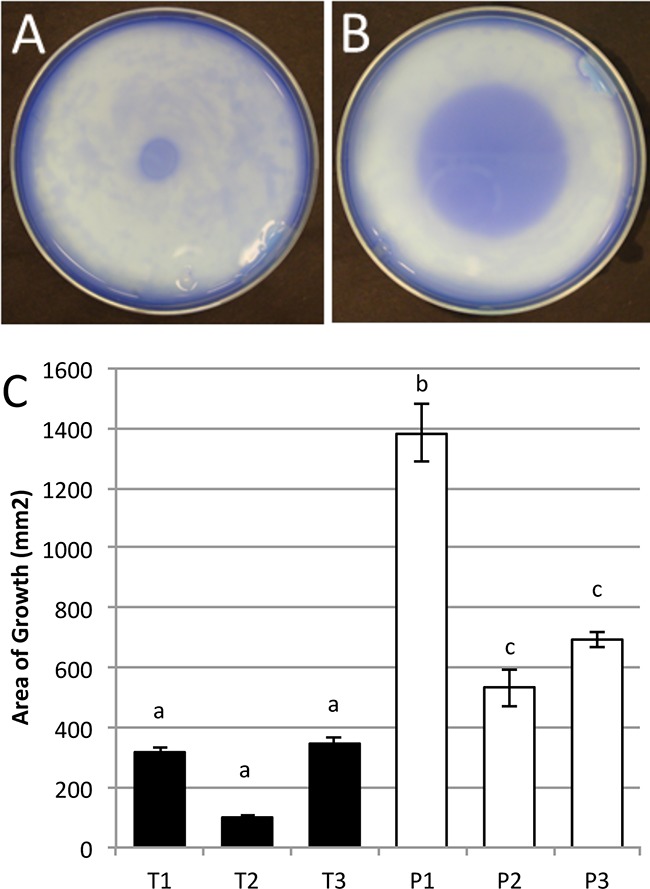
Milk swimming plates. Shown are examples of two E. coli strains on milk swimming plates. The first is strain T2 (A), which is a slower swimmer, and the second is P1 (B), a faster swimmer. The relative swimming abilities of these two strains on the milk swimming plates are consistent with data on the traditional swimming plate with the exception that growth is much slower. (C) Combined results from three swimming assays on skim milk plates. There is a significant difference between the persistent- versus transient-infection groups (P < 0.001). In addition, all three P strains are significantly different from all three T strains. Different lowercase letters indicate significantly different means. Error bars represent the standard errors of the means.
Gene expression differences between transient- and persistent-infection strains.
Each of the six MPEC strains was grown in fresh milk to mid-log phase as previously described (8). This experiment was repeated three times, and RNA from each strain was independently isolated and sequenced. Transcriptome sequencing (RNA-seq) data between replicate experiments for individual strains were consistent, as shown by a principal-component analysis (PCA) plot (Fig. 2). The variation between replicates for each strain was highly consistent (r = 0.98 or greater for all 6 strains).
FIG 2.
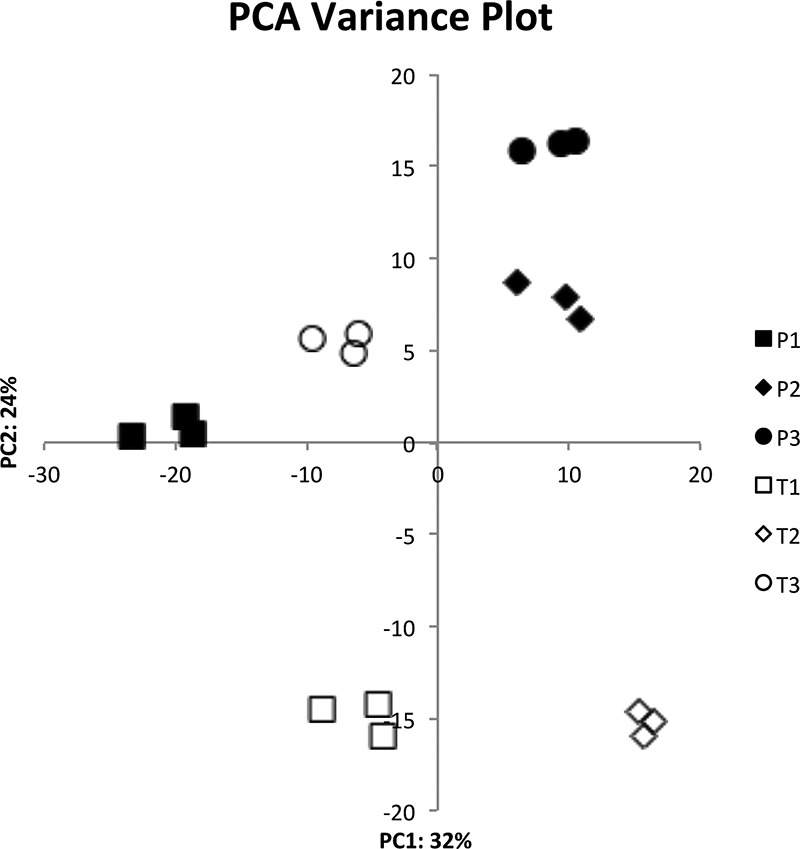
Principal-component analysis (PCA) plot of RNA-seq data. The PCA plot shows the variance of the three experimental replicates of each of the three transient-infection strains (T1, T2, and T3) and the three persistent-infection strains (P1, P2, and P3). The percentages on each axis represent the percentages of variation explained by the principal components.
Gene expression analysis identified 285 genes that were differentially expressed between the two phenotypic groups (false discovery rate [FDR], P < 0.05) (see Table S2 in the supplemental material). Of the 285 genes, 132 were expressed at higher levels in persistent-infection strains, and 153 were expressed at higher levels in transient-infection strains. To categorize the functions associated with the differentially expressed genes, functional enrichment analysis was performed using DAVID (11, 12). The biological process gene ontology (GO) terms associated with higher expression in persistent-infection strains include flagellum-dependent motility, assembly and organization, lipopolysaccharide and slime layer biosynthetic process, iron ion homeostasis and response to iron, and cell adhesion (Table 1). The biological process terms associated with higher gene expression in transient-infection strains involved the molybdopterin cofactor biosynthetic process (Table 1). In agreement with the phenotypic motility results, 15 flagellar genes were upregulated in the persistent-infection strains compared to their levels in the transient-infection isolates. Additionally, we show that 11 genes involved in iron regulation have increased transcription levels in the persistent-infection isolates. This observation is consistent with previously published data (8, 10). Unique to this data set are the differentially expressed genes within the wca operon. The wcaGHIKLM genes all had greater expression levels in the persistent-infection strains.
TABLE 1.
Enriched gene ontology biological processes in persistent- versus transient-infection MPEC strains based on differentially expressed genes
| Gene group and GO term | No. of genes | P value | Gene names |
|---|---|---|---|
| Genes with higher expression in persistent-infection isolates | |||
| Bacterial-type flagellum-dependent cell motility | 15 | 1.10E−13 | fliF, fliE, flgK, flgL, fliK, fliG, fliM, fliN, fliS, fliJ, flgJ, fliI, fliH, flgI, fliT |
| Lipopolysaccharide biosynthetic process | 15 | 2.10E−10 | arnT, wzzE, wcaH, wecA, wcaK, wcaM, arnA, eptA, wcaI, wcaL, wcaN, wzzB, arnC, arnD, arnB |
| Bacterial-type flagellum assembly | 9 | 7.10E−09 | flgN, fliQ, flhD, flgK, fliK, fliG, fliS, flgJ, fliI |
| Bacterial-type flagellum organization | 7 | 2.50E−07 | flgM, fliO, fliP, flhC, fliJ, fliH, fliT |
| Iron ion homeostasis | 7 | 1.10E−04 | fecI, fecA, fecR, fecE, fecB, fecC, fecD |
| Chemotaxis | 6 | 6.80E−04 | dppA, fliO, fliG, fliN, fliJ, trg |
| Response to iron(III) ion | 4 | 7.00E−04 | arnT, basR, basS, arnD |
| Lipid A biosynthetic process | 6 | 8.40E−04 | arnT, arnA, eptA, arnC, arnD, arnB |
| Slime layer polysaccharide biosynthetic process | 4 | 3.80E−03 | wcaK, wcaM, wcaI, wcaL |
| Ion transport | 6 | 3.90E−03 | fecI, fecR, fecE, fecB, fecC, fecD |
| Gamma-aminobutyric acid catabolic process | 3 | 1.10E−02 | gabT, gabP, gabD |
| Cell adhesion | 5 | 2.40E−02 | fimI, fimD, fimF |
| Genes with higher expression in transient-infection isolates | |||
| Mo-molybdopterin cofactor biosynthetic process | 4 | 8.20E−03 | moaB, moaC, moaE, moaD |
| Molybdopterin cofactor biosynthetic process | 3 | 1.50E−02 | moaB, moaC, moaE |
Genetic differences between transient- and persistent-infection strains.
We reasoned that the difference in gene expression levels between these strains could be due to differential transcription or a difference in gene content at the genome level. To address this second possibility, we sequenced all six strains. Four strains have been previously sequenced (13), and we added two previously unsequenced genomes to the NCBI database for ECA-B and ECC-M. Sequence information was aligned to the E. coli strain MG1655 (GenBank accession number NC_000913). There were 3,782 genes in MG1655 that were found in least one of the three persistent- or three transient-infection strains (Table S3). There were 3,788 genes in all six strains (using a minimal read number of 75). In the MG1655 matched data, there were 14 genes that were present in all three persistent-infection strains but absent in the transient-infection strains. There were three genes that were in the transient-infection strains but not in the persistent-infection strains. De novo-assembled sequences from the six strains that did not match MG1655 were subjected to a BLAST search against the NCBI nonredundant (nr) database. An additional 16 genes from the transient-infection strains were identified that did not exist in the persistent-infection strains (Table S4). Additionally, 90 genes were identified in the persistent-infection strains that did not exist in the transient-infection strains (Table S5). The genes identified by the BLAST search are likely plasmid genes as they included conjugative genes on several contigs. Of all the genetic differences between the strains, the most striking were the 14 genes matched to MG1655 that were present only in the three persistent-infection strains. All 14 of these genes are in the same location on the chromosome, which includes part of the wca operon (Table 2). The wca operon encodes part of the colanic acid metabolic pathway. The absence of the genes within the wca operon in the transient-infection strains is consistent with the transcriptomics data showing differential expression of the colanic acid metabolic process (Table 1).
TABLE 2.
MiSeq read data of six MPEC strains
a Genes in boldface were absent from all three transient strains (T1 to T3) but present in all three persistent strains (P1 to P3).
b Data are reads per kilobase of transcript per million mapped reads. Shading indicates genes with insufficient read numbers to be counted as present. NA, not available.
Effect of gene deletions in the wca operon.
The most motile MPEC strain, P1 (strain ECC-Z), was used to make gene deletion mutants to test the hypothesis that the missing genes are responsible for differences in swimming and swarming between the persistent- and transient-infection MPEC strains. The first deletion removed the genes from gmd through wcaN, making P1 similar to strain T1 in that region of the chromosome (Table 2). The second mutation deleted the motAB genes, which encode proteins that are part of the flagellar motor, thereby allowing the bacteria to make structurally complete but nonfunctional flagella.
Figure 3 shows the data from three independent experiments for the motility assays of swimming (Fig. 3A) and swarming (Fig. 3B). As expected, deletion of the motAB genes caused a loss of both swimming and swarming. The deletion of part of the wca operon that approximates the genes missing from the transient-infection strains caused a 20% reduction in swimming and a 40% reduction in swarming compared to levels of the most motile strain, P1.
FIG 3.
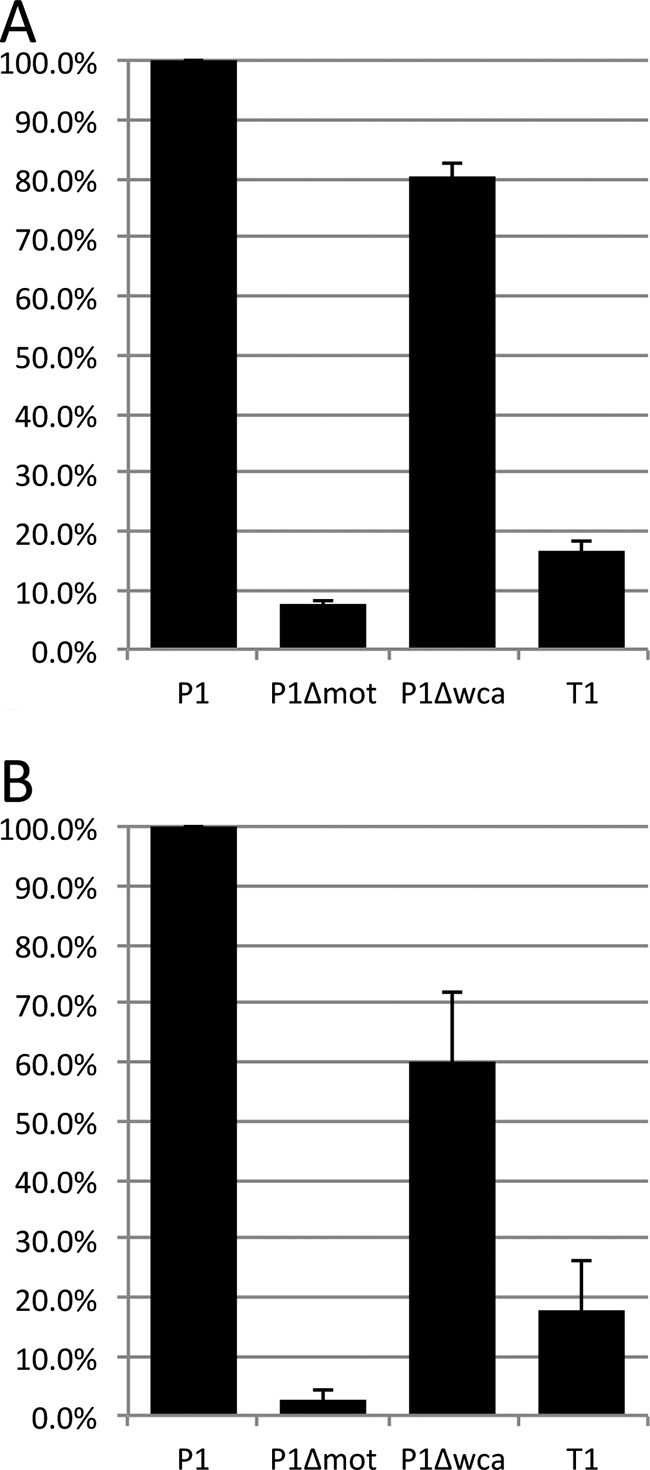
Motility assays with deletion mutants. Results of three independent assays with three replicate plates per assay were averaged. The relative growth for each strain was calculated using the average P1 growth as a reference. (A) Swimming assay. (B) Swarming assay. Error bars represent the standard errors of the means.
A recent paper has shown that colanic acid has a protective role against complement-mediated killing in serum (14). To test whether MPEC bacteria that cause persistent and transient infections have different susceptibilities to complement in serum, the three persistent-infection strains, the three transient-infection strains, and the Δwca mutant were grown in 50% fresh bovine serum and 50% LB broth or in 50% heat-inactivated serum and 50% LB broth (Fig. 4). Growth of strain T2 and the Δwca mutant was completely inhibited in normal serum. Strains T1 and T3 showed growth curves that were repressed in normal serum compared to growth of the persistent-infection strains. Growth of all three persistent-infection strains was unaffected by culture with normal serum. Importantly, heat inactivation of the serum (56°C for 30 min) removed any growth inhibition and allowed all strains to grow at roughly similar rates.
FIG 4.
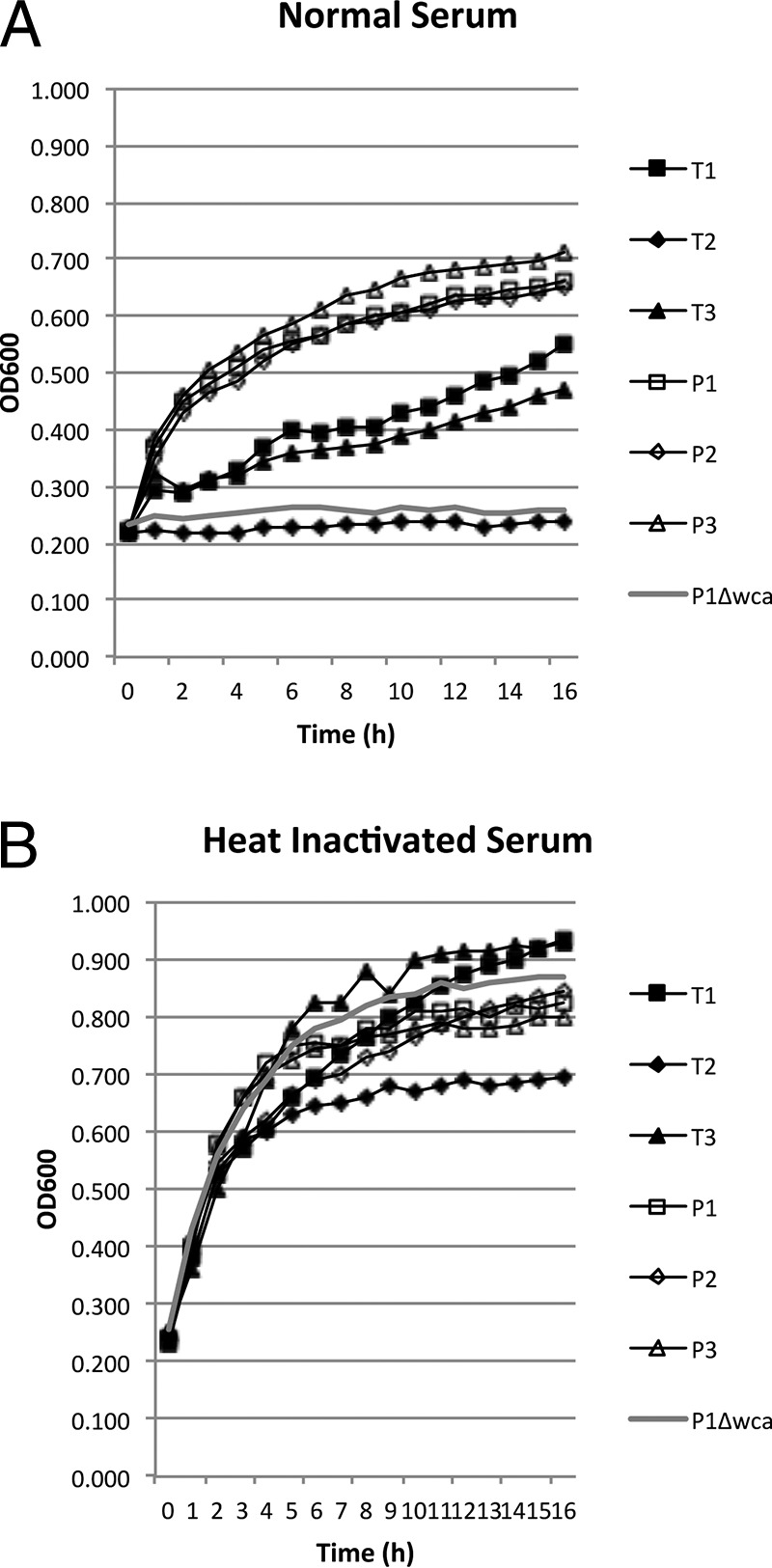
Serum inhibition assay. Persistent- and transient-infection strains and the P1 Δwca mutant were grown in 50% LB broth and either 50% fresh normal bovine serum (A) or heat-inactivated serum (B), as indicated. Optical density (at 600 nm) was monitored in five replicate wells for each bacterial strain, and the experiment was repeated three times. Groups of strains that were statistically different (P < 0.05) are indicated by different lowercase letters.
To test whether preexposure to an MPEC mammary infection affected the efficacy of serum to inhibit bacterial growth, sera from four cows that were part of an MPEC mastitis study within the last year, as well as sera from four heifers (never lactated), were compared for their ability to inhibit growth of the persistent- and transient-infection MPEC strains (Fig. 5). The growth of the three transient-infection strains was more inhibited in normal serum than that of the persistent-infection strains, consistent with data presented in Fig. 4. There was no difference in the abilities to inhibit growth of the transient-infection strain between the sera from cows with recent known MPEC mastitis and heifers without any known mastitis.
FIG 5.
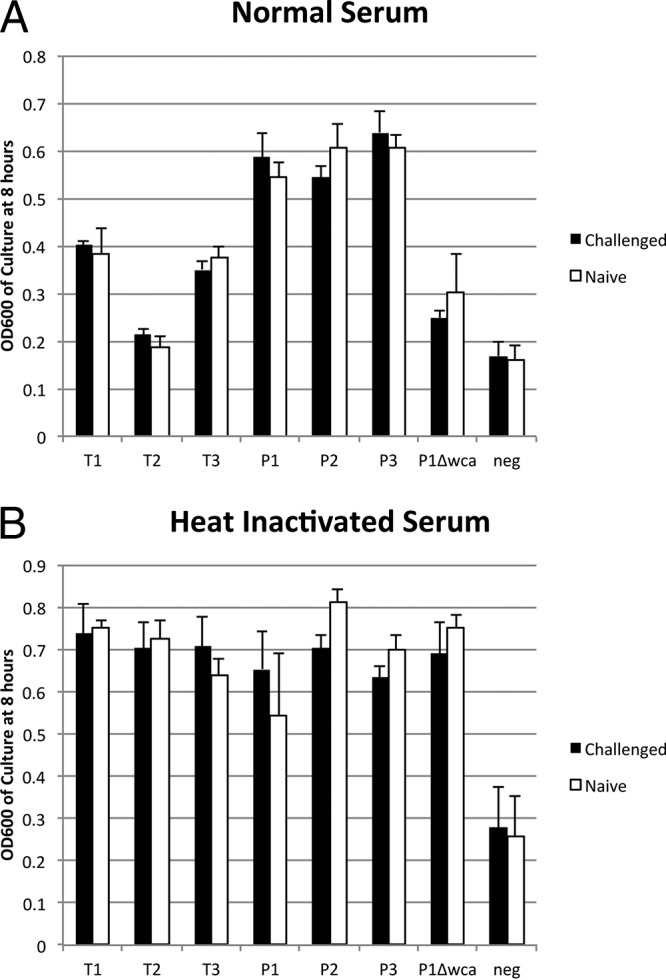
Sera from four previously MPEC-infected cows (Challenged) were compared to sera from four heifers (Naive) for the ability to inhibit growth of various MPEC strains. Normal serum (A) and serum heat inactivated at 56°C for 30 min (B) were used. Error bars represent standard errors of the means.
DISCUSSION
Our research goal is to better understand the pathogenic mechanisms that allow a subset of MPEC strains to persist in the mammary gland of dairy cattle. Our previous work demonstrated that growth of bacteria in whole fresh milk significantly altered the proteome of an MPEC strain compared to that of a strain grown in bacterial culture medium (9). The differentially expressed proteins that were categorized as chemotaxis and motility were all downregulated in the MPEC grown in milk, including the flagellin protein FliC which is the major subunit of flagella. We have shown a correlation between motility and protein expression in persistent-infection MPEC strains, and a greater motility phenotype was determined in swimming and swarming assays of persistent-infection strains than in transient-infection strains (10). We previously used bacterial culture medium-based swimming and swarming plates, and because MPEC growth in milk had a negative effect on motility protein expression, we developed a milk-based motility plate assay. Using semisolid agar motility plates that were 90% fresh skim milk, we have shown that persistent-infection MPEC strains were more motile than the transient-infection strains, consistent with our previous results in standard medium (LB broth). The change in medium did lead to a proportional increase in motility assay time for all the strains (approximately three times as long).
To better understand the differences between the persistent- and transient-infection MPEC strains, we performed RNA sequencing on each strain grown in whole fresh milk. Confirmatory to our previous proteomics data (10), our gene expression data showed higher expression of motility genes in the persistent-infection MPEC strains than in the transient-infection strains. Our RNA sequencing data also demonstrated higher expression levels of genes important for iron acquisition and transport in the persistent-infection strains. It has been shown that iron acquisition and transport genes are important for motility in these MPEC strains (8). However, the largest group of genes with significant expression changes consisted of those involved in colanic acid biosynthesis.
Our subsequent genome sequencing determined that a significant portion of the operon (wca) responsible for colanic acid synthesis is missing in the transient-infection strains. Previous work has shown a connection between motility and colanic acid synthesis (15). In that work, the Rcs signaling system was found to positively regulate colanic acid synthesis and negatively regulate motility. We have deleted part of the wca operon that is absent in all three transient-infection strains from the persistent-infection strain P1 (P1 Δwca). We observed that the deletion of a significant segment of the wca operon (P1 Δwca) inhibited swimming by approximately 20% and swarming by 40% compared to the levels of the wild-type strain (P1).
Colanic acid is induced by culturing bacteria under suboptimal conditions, such as osmotic and oxidative stress (16) and adverse temperature (17). Recently, colanic acid has been shown to have a protective role against complement-mediated killing in serum (14). The three persistent-infection MPEC stains showed little difference in growth rates in either normal bovine serum or heat-inactivated bovine serum. The three transient-infection MPEC strains were inhibited for growth in normal serum. Growth of the transient-infection strain T2 and the deletion mutant P1 Δwca was completely inhibited in normal bovine serum. Growth of the three transient-infection strains and P1 Δwca was comparable to that of the persistent-infection strains when the bovine serum was heat inactivated. Together, these data highlight the dual role of colonic acid synthesis in motility and resistance to serum for the persistent-infection MPEC isolates.
We also have shown that previous exposure to MPEC mastitis does not affect the serum inhibition of the transient-infection strains. Sera from heifers, adult female cattle that have not calved, were equal to sera from cows that were part of a recent MPEC challenge experiment in their ability to inhibit the transient-infection strains. This indicates that an innate immune response that potentially helps to control MPEC infections is inhibited by MPEC that can express colanic acid.
Understanding of the role of complement in mastitis is incomplete. While levels of the major complement proteins are significantly lower in milk than in serum (18), complement proteins can rise dramatically in clinical mastitis in cows (18, 19). MPEC infections have been shown to increase permeability between the vascular system and the mammary gland, as shown by increased bovine serum albumin (BSA) levels in milk in the acute phase of an infection (20, 21). Disruption of the physiological barrier between blood and milk may be how complement can play a role in intramammary infections. The ability of certain MPEC strains to synthesize colanic acid may provide these strains with protection against complement during the acute phase of the infection.
E. coli mastitis is caused by a highly diverse group of MPEC strains (22, 23). No specific virulence factor has been identified that is necessary for infection by E. coli of the mammary gland. The great diversity of clinical outcomes of E. coli mastitis is thought to be due largely to host factors (24). However, prior to our work, there have been no genetic differences identified between E. coli strains that cause persistent infections and those that cause transient infections (22). Our observations of the differences between MPEC strains that cause persistent versus transient infection indicate that persistent-infection strains are more motile, have increased gene expression of the iron acquisition system, and are more resistant to growth inhibition caused by complement. A relationship between iron acquisition and motility has been shown in these strains (8), and a relationship between colanic acid synthesis and motility has been described previously (15). Generation of specific deletion mutants that will test the role of motility, iron acquisition, and colanic acid synthesis in vivo will be necessary to further determine the mechanisms necessary for persistence of E. coli in mammary gland infections.
The use of antibiotics in agriculture is of concern to many consumers, policy makers, and farmers. Current treatment recommendations for dairy cattle with mild to moderate E. coli mastitis include use of anti-inflammatory therapy, frequent milking, and fluid treatment. Antibiotic treatment of E. coli mastitis is not recommended except in severe cases (25). Currently, there are no recommendations for treatment of persistent MPEC infections. Identification of E. coli strains that might progress to persistent infection will lead to better decisions and options for mastitis treatment.
MATERIALS AND METHODS
Bacteria.
E. coli strains used for these experiments were described previously (7). Abbreviations for the bacterial strains used in the manuscript correspond with the published names as follows: T1, ECA-727; T2, ECA-O157; T3, ECA-B; P1, ECC-Z; P2, ECC-M; P3, ECC-1470 (kindly provided by Ynte Schukken).
E. coli deletion mutants were derivatives of the P1 (ECC-Z) strain. Recombineering was performed using a neo cassette encoding kanamycin resistance as previously described (26). Briefly, the oBBI 92/93 neo fragment was amplified using a set of knockout primers containing nucleotide sequences corresponding to the E. coli genomic region targeted for deletion. Following PCR purification, the amplification product was transformed into strain EBB 667 containing pKD46, and knockout mutants were selected on LB medium containing kanamycin. The neo gene was deleted by transforming EBB 671 (motAB::neo) and EBB 683 [(wcaNMLK wzxC wcaJ cpsGB wcaIHG gmd)::neo] with pCP20 and screening the resulting transformants for sensitivity to kanamycin. PCR primers were used confirm gene deletions in EBB 673 (ΔmotAB) and EBB 684 [Δ(wcaNMLK wzxC wcaJ cpsGB wcaIHG gmd); referred to as the Δwca strain in the manuscript for simplicity]. A list of strains and primers is found in Table S1 in the supplemental material.
Animals.
Blood samples from eight Holstein cows were obtained from our herd at the National Animal Disease Center. Milk used in transcriptomic experiments was fresh unprocessed bovine milk obtained from multiple cows and pooled. Milk was plated on bacterial growth plates and incubated overnight at 37°C to ensure clean milk samples prior to use for bacterial assays. The National Animal Disease Center Animal Care and Use Committee approved all animal-related procedures used in this study.
Milk swimming assay.
A modified swimming assay was developed using skim milk as a medium. Fresh unprocessed bovine milk from multiple cows was pooled and then centrifuged at 1,500 × g for 45 min. Fat was scraped off, and milk was transferred to a new bottle that was centrifuged at 10,000 × g for 45 min. The skim milk and the 3.5% (10×) agar solution were equilibrated to 80°C and then mixed for a final agar concentration of 0.35%. Plates were poured and left at room temperature for 1 h and then transferred to 4°C for 1 h. Plates were inoculated by spotting 5 μl of a bacterial overnight culture to the center of the plate, and the bacteria were allowed to grow at 37°C in a humidified incubator for approximately 15 h. In this assay, motile bacteria will swim from the center toward the outside edge of the plates. Bacterial motility zones were visualized by crystal violet staining. Briefly, 10 ml of 100% ethanol was gently overlaid on the surface of the plate for 5 min. After the ethanol was removed, the plates were then stained with 0.002% crystal violet in 10% ethanol for 1 h. Plates were destained with water in three washes. The diameter of the bacterial motility zones was measured from photographs of the plates using ImageJ (NIH). There were four plates per strain used in each experiment, and the diameter for each strain was averaged. The experiment was repeated four times. Statistical analysis of swimming on milk plates comparing all persistent-infection strains to all transient-infection strains was accomplished with an unpaired t test and one-way analysis of variance (ANOVA) with Tukey's multiple-comparison test. The statistical software used was Prism, version 7 (GraphPad, San Diego, CA).
RNA sequencing. (i) Bacterial growth and isolation.
Clean fresh milk was pooled from three cows and inoculated with bacterial stocks for overnight cultures grown at 37°C with shaking at 180 rpm. Overnight cultures were diluted into 100 ml of fresh clean milk and grown to mid-log phase (8). Milk cultures were then poured into 300 ml of ice-cold Hanks balanced salt solution with 10 mM EDTA, and the solutions were cooled in a salted ice bath for 5 min. Diluted milk cultures were centrifuged at 10,000 × g for 30 min at 4°C. The pellets were resuspended in 2 ml of RNA Protect and 1 ml of phosphate-buffered saline (PBS) and held at room temperature for 5 to 30 min. The resuspended pellet was passed through a 5-μm-pore-size syringe filter to remove somatic cells (27). The filtrates were centrifuged at 10,000 × g for 2 min in microcentrifuge tubes, and the supernatants were removed. Cell pellets were stored at −80°C.
(ii) RNA isolation and sequencing.
An RNeasy minikit (Qiagen, Germantown, MD) was used to extract RNA from the bacterial cells, and genomic DNA was subsequently removed using Turbo DNA-free DNase (Ambion, Austin, TX) as per the manufacturer's directions. Quantitation of total RNA was performed using a NanoDrop ND-1000 spectrophotometer (Thermo Scientific, Wilmington, DE). RNA quality was assessed with a 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA). A quantitative PCR (qPCR) assay was performed on all samples using primers for both E. coli gapA and bovine bRPS9 housekeeping genes to identify potential E. coli DNA and bovine RNA contamination. rRNA was depleted using a Ribo-Zero rRNA Removal kit (Bacteria) according to the manufacturer's instructions (Illumina, Inc., San Diego, CA), and rRNA removal was verified on the 2100 Bioanalyzer. The cDNA libraries were constructed using TruSeq Stranded Total RNA Library Prep kits and sequenced on a HiSeq 3000 using a 100-cycle single-end run (Illumina, Inc., San Diego, CA) at the Iowa State University DNA core facility. As per the recommendations of Haas et al. (28), we targeted 10 to 50 million reads per sample; our sequencing resulted in an average of 38.2 ± 5.8 million reads per sample.
RNA-seq analysis.
FastQC, version 0.11.5, was used to assess the quality of all RNA-seq reads. Bowtie2, version 2.2.9 (29), was then used to align the reads to the E. coli K-12 substrain MG1655 genome (GenBank accession number NC_000913) using the default parameters for the very-sensitive-local setting. The E. coli strain MG1655 was used as a reference genome because of its level of gene functional annotation and coverage. The number of reads that aligned to each gene from the E. coli K-12 substrain MG1655 genome was quantified using HTSeq, version 0.6.1 (30). Gene counts were then normalized, and differentially expressed genes (false discovery rate [FDR] of <0.05) between the persistent- and transient-infection strains were identified using the DESeq2 package, version 1.12.4 (31) in R, version 3.2.5. The regularized logarithm-transformed counts for each E. coli strain were used to generate a principal-component analysis (PCA) plot. To assess the efficiency of the removal of cattle RNA, we aligned the raw RNA-seq reads against the Bos taurus reference genome (GenBank accession number GCA_000003055.3). It was determined that 0.17% ± 0.03% of reads aligned to the Bos taurus genome, indicating that host cell contamination was not a significant issue for downstream analysis.
DNA sequencing and analysis.
DNA was isolated and purified using CsCl gradients as described by Hull et al. (32), with the exceptions that bacteria were harvested from 6 ml of overnight LB cultures and that bacterial lysates were centrifuged at 372,000 × g for 4 h in a Beckman VTi 65 rotor. Isolated DNA was sent to the Iowa State University DNA core facility. DNA was fragmented using a Bioruptor followed by TruSeq (Illumina, Inc.) library preparation with indexing. Insert size was around 700 bp. Samples were run on a MiSeq using a MiSeq reagent kit, version 3 (Illumina, Inc.) (600 cycles). Sequencing resulted in over 3.7 × 106 reads per genome. Sequences were aligned to the reference sequence of E. coli strain MG1655 (GenBank accession number NC_000913) using DNAstar LaserGene NGen, version 14 (Madison, WI). Average read depth was >150×. Genes with few or no reads were considered absent. In addition to the alignment of the MG1655 reference genome, all reads were de novo assembled using Abyss (version 1.9) (33). Sequence reads that were not aligned to the reference strain (MG1655) were de novo assembled using NGen. The resulting contigs were subjected to a BLAST search against the NCBI nr database and were identified as plasmids.
Bacterial swimming and swarming assays.
A 5-μl aliquot of a fresh overnight culture of each E. coli strain was plated on 0.3% swimming agar plates (10 g of Bacto tryptone, 5 g of Bacto yeast extract, 5 g of NaCl, 3 g of Bacto agar in 1 liter of deionized water) or 0.5% swarming agar plates (10 g of Bacto tryptone, 5 g of Bacto yeast extract, 5 g of NaCl, 5 g of Bacto agar, 0.5% [wt/vol] glucose in 1 liter of deionized water) (34, 35). Each E. coli strain was inoculated onto four plates and then incubated for approximately 5 h for swimming and 12 h for swarming in a humidified 37°C incubator. Photographs of all the plates were taken with a fixed distance between the plate and the camera. Swimming and swarming experiments had four replicate plates for each strain, and results were averaged for each experiment. The swimming and swarming experiments were performed three times. ImageJ (NIH) was used to determine the diameter of the swimming plates and area of the outer bacterial growth circle of swarming plates. Differences were assessed among the strains within each motility condition by one-way ANOVA with Tukey's multiple-comparison test using Prism, version 7 (GraphPad).
Serum inhibition test.
Serum was isolated from two cows and was mixed in equal volumes. Half the serum was heat inactivated at 56°C for 30 min. Bacterial strains were grown in Luria-Bertani (LB) broth overnight at 37°C with shaking at 200 rpm. Aliquots of the overnight culture (10 μl) were added to a 1:1 mixture of LB broth and normal cow serum or to a 1:1 mixture of LB broth and heat-inactivated cow serum (190 μl) into a 100-well microtiter plate. Each condition was replicated in five wells. Plates were loaded onto a Bioscreen C growth curve instrument (Growth Curves, Ltd., Raisio, Finland). Plates were incubated at 37°C with continuous shaking, and the optical density at 600 nm (OD600) was read every 15 min. Growth data from five replicates from each plate were averaged, and the complete assay was repeated three times. The area under the curve (AUC) was determined for each growth curve. The replicate AUCs for strains P1 to P3 were compared to the AUCs for strains T1 to T3 using an unpaired t test. Comparisons of results with P1 to P3, T1 to T3, and P1 Δwca were accomplished with one-way ANOVA with Tukey's multiple-comparison test. The AUC and statistics were done using Prism, version 7 (GraphPad).
Accession number(s).
The genome sequences of ECA-B and ECC-M were deposited in the GenBank under accession numbers NHTF00000000 and NHZH00000000, respectively. All RNA-seq files were deposited in the Short Read Archive (SRA) under BioProject accession number PRJNA326931 and BioSample accession numbers SAMN07125434 to SAMN07125451. The genomic sequences were deposited in the SRA under BioSample numbers SAMN07125105 and SAMN07125106.
Supplementary Material
ACKNOWLEDGMENT
We thank Duane Zimmerman, Briony Atkinson, Tera Nyholm, and Kellie Winter for their technical expertise.
Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendations or endorsement by the U.S. Department of Agriculture.
USDA is an equal opportunity provider and employer.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/IAI.00566-17.
REFERENCES
- 1.USDA Animal and Plant Health Inspection Service. 2007. Highlights of Dairy 2007. Part I: reference of dairy health and management in the United States, 2007. USDA Animal and Plant Health Inspection Service, Fort Collins, CO: https://www.aphis.usda.gov/animal_health/nahms/dairy/downloads/dairy07/Dairy07_is_PartI_Highlights.pdf. [Google Scholar]
- 2.Hogan JS, Smith KL. 2003. Coliform mastitis. Vet Res 34:507–519. doi: 10.1051/vetres:2003022. [DOI] [PubMed] [Google Scholar]
- 3.Roberson JR. 2012. Treatment of clinical mastitis. Vet Clin North Am Food Anim Pract 28:271–288. doi: 10.1016/j.cvfa.2012.03.011. [DOI] [PubMed] [Google Scholar]
- 4.Döpfer D, Barkema HW, Lam TJ, Schukken YH, Gaastra W. 1999. Recurrent clinical mastitis caused by Escherichia coli in dairy cows. J Dairy Sci 82:80–85. doi: 10.3168/jds.S0022-0302(99)75211-2. [DOI] [PubMed] [Google Scholar]
- 5.Bradley AJ, Green MJ. 2001. Adaptation of Escherichia coli to the bovine mammary gland. J Clin Microbiol 39:1845–1849. doi: 10.1128/JCM.39.5.1845-1849.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lam TJ, Lipman LJ, Schukken YH, Gaastra W, Brand A. 1996. Epidemiological characteristics of bovine clinical mastitis caused by Staphylococcus aureus and Escherichia coli studied by DNA fingerprinting. Am J Vet Res 57:39–42. [PubMed] [Google Scholar]
- 7.Dogan B, Klaessig S, Rishniw M, Almeida RA, Oliver SP, Simpson K, Schukken YH. 2006. Adherent and invasive Escherichia coli are associated with persistent bovine mastitis. Vet Microbiol 116:270–282. doi: 10.1016/j.vetmic.2006.04.023. [DOI] [PubMed] [Google Scholar]
- 8.Lippolis JD, Brunelle BW, Reinhardt TA, Sacco RE, Thacker TC, Looft TP, Casey TA. 2016. Differential gene expression of three mastitis-causing Escherichia coli strains grown under planktonic, swimming, and swarming culture conditions. mSystems 1:e00064-16. doi: 10.1128/mSystems.00064-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lippolis JD, Bayles DO, Reinhardt TA. 2009. Proteomic changes in Escherichia coli when grown in fresh milk versus laboratory media. J Proteome Res 8:149–158. doi: 10.1021/pr800458v. [DOI] [PubMed] [Google Scholar]
- 10.Lippolis JD, Brunelle BW, Reinhardt TA, Sacco RE, Nonnecke BJ, Dogan B, Simpson K, Schukken YH. 2014. Proteomic analysis reveals protein expression differences in Escherichia coli strains associated with persistent versus transient mastitis. J Proteomics 108:373–381. doi: 10.1016/j.jprot.2014.06.008. [DOI] [PubMed] [Google Scholar]
- 11.Huang DW, Sherman BT, Lempicki RA. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 12.Seo SW, Kim D, Latif H, O'Brien EJ, Szubin R, Palsson BO. 2014. Deciphering Fur transcriptional regulatory network highlights its complex role beyond iron metabolism in Escherichia coli. Nat Commun 5:4910. doi: 10.1038/ncomms5910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Richards VP, Lefébure T, Bitar PD, Dogan B, Simpson KW, Schukken YH, Stanhope MJ. 2015. Genome-based phylogeny and comparative genomic analysis of intra-mammary pathogenic Escherichia coli. PLoS One 10:e0119799. doi: 10.1371/journal.pone.0119799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miajlovic H, Cooke NM, Moran GP, Rogers TR, Smith SG. 2014. Response of extraintestinal pathogenic Escherichia coli to human serum reveals a protective role for Rcs-regulated exopolysaccharide colanic acid. Infect Immun 82:298–305. doi: 10.1128/IAI.00800-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang S, Shi H, Li Y, Shi Z, Zhang X, Baek CH, Mothershead T, Curtiss R. 2013. A colanic acid operon deletion mutation enhances induction of early antibody responses by live attenuated Salmonella vaccine strains. Infect Immun 81:3148–3162. doi: 10.1128/IAI.00097-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen J, Lee SM, Mao Y. 2004. Protective effect of exopolysaccharide colanic acid of Escherichia coli O157:H7 to osmotic and oxidative stress. Int J Food Microbiol 93:281–286. doi: 10.1016/j.ijfoodmicro.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 17.Navasa N, Rodríguez-Aparicio L, Martínez-Blanco H, Arcos M, Ferrero MÁ. 2009. Temperature has reciprocal effects on colanic acid and polysialic acid biosynthesis in E. coli K92. Appl Microbiol Biotechnol 82:721–729. doi: 10.1007/s00253-008-1840-4. [DOI] [PubMed] [Google Scholar]
- 18.Rainard P. 2003. The complement in milk and defense of the bovine mammary gland against infections. Vet Res 34:647–670. doi: 10.1051/vetres:2003025. [DOI] [PubMed] [Google Scholar]
- 19.Reinhardt TA, Sacco RE, Nonnecke BJ, Lippolis JD. 2013. Bovine milk proteome: quantitative changes in normal milk exosomes, milk fat globule membranes and whey proteomes resulting from Staphylococcus aureus mastitis. J Proteomics 82:141–154. doi: 10.1016/j.jprot.2013.02.013. [DOI] [PubMed] [Google Scholar]
- 20.Harmon RJ, Schanbacher FL, Ferguson LC, Smith KL. 1976. Changes in lactoferrin, immunoglobulin G, bovine serum albumin, and α-lactalbumin during acute experimental and natural coliform mastitis in cows. Infect Immun 13:533–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bannerman DD, Paape MJ, Lee J-W, Zhao X, Hope JC, Rainard P. 2004. Escherichia coli and Staphylococcus aureus elicit differential innate immune responses following intramammary infection. Clin Diagn Lab Immunol 11:463–472. doi: 10.1128/CDLI.11.3.463-472.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zadoks RN, Middleton JR, McDougall S, Katholm J, Schukken YH. 2011. Molecular epidemiology of mastitis pathogens of dairy cattle and comparative relevance to humans. J Mammary Gland Biol Neoplasia 16:357. doi: 10.1007/s10911-011-9236-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leimbach A, Poehlein A, Vollmers J, Görlich D, Daniel R, Dobrindt U. 2017. No evidence for a bovine mastitis Escherichia coli pathotype. BMC Genomics 18:359. doi: 10.1186/s12864-017-3739-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burvenich C, van Merris V, Mehrzad J, Diez-Fraile A, Duchateau L. 2003. Severity of E. coli mastitis is mainly determined by cow factors. Vet Res 34:521–564. doi: 10.1051/vetres:2003023. [DOI] [PubMed] [Google Scholar]
- 25.Suojala L, Kaartinen L, Pyörälä S. 2013. Treatment for bovine Escherichia coli mastitis—an evidence-based approach. J Vet Pharmacol Ther 36:521–531. doi: 10.1111/jvp.12057. [DOI] [PubMed] [Google Scholar]
- 26.Bearson BL, Bearson SM, Uthe JJ, Dowd SE, Houghton JO, Lee I, Toscano MJ, Lay DC Jr. 2008. Iron regulated genes of Salmonella enterica serovar Typhimurium in response to norepinephrine and the requirement of fepDGC for norepinephrine-enhanced growth. Microbes Infect 10:807–816. doi: 10.1016/j.micinf.2008.04.011. [DOI] [PubMed] [Google Scholar]
- 27.Richards VP, Choi SC, Pavinski Bitar PD, Gurjar AA, Stanhope MJ. 2013. Transcriptomic and genomic evidence for Streptococcus agalactiae adaptation to the bovine environment. BMC Genomics 14:920. doi: 10.1186/1471-2164-14-920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haas BJ, Chin M, Nusbaum C, Birren BW, Livny J. 2012. How deep is deep enough for RNA-seq profiling of bacterial transcriptomes? BMC Genomics 13:734. doi: 10.1186/1471-2164-13-734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anders S, Pyl PT, Huber W. 2015. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hull RA, Gill RE, Hsu P, Minshew BH, Falkow S. 1981. Construction and expression of recombinant plasmids encoding type 1 or d-mannose-resistant pili from a urinary tract infection Escherichia coli isolate. Infect Immun 33:933–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Simpson JT, Wong K, Jackman SD, Schein JE, Jones SJM, Birol I. 2009. ABySS: a parallel assembler for short read sequence data. Genome Res 19:1117–1123. doi: 10.1101/gr.089532.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gómez-Gómez J-M, Manfredi C, Alonso J-C, Blázquez J. 2007. A novel role for RecA under non-stress: promotion of swarming motility in Escherichia coli K-12. BMC Biol 5:14. doi: 10.1186/1741-7007-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wozniak CE, Chevance FFV, Hughes KT. 2010. Multiple promoters contribute to swarming and the coordination of transcription with flagellar assembly in Salmonella. J Bacteriol 192:4752–4762. doi: 10.1128/JB.00093-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.