ABSTRACT
Many Clostridium perfringens strains produce NanI as their major sialidase. Previous studies showed that NanI could potentiate C. perfringens epsilon toxin cytotoxicity by enhancing the binding of this toxin to host cells. The present study first determined that NanI exerts similar cytotoxicity-enhancing effects on C. perfringens enterotoxin and beta toxin, which are also important toxins for C. perfringens diseases (enteritis and enterotoxemia) originating in the gastrointestinal (GI) tract. Building upon previous work demonstrating that purified trypsin can activate NanI activity, this study next determined that purified chymotrypsin or mouse intestinal fluids can also activate NanI activity. Amino acid sequencing then showed that this effect involves the N-terminal processing of the NanI protein. Recombinant NanI (rNanI) species corresponding to major chymotrypsin- or small intestinal fluid-generated NanI fragments possessed more sialidase activity than did full-length rNanI, further supporting the proteolytic activation of NanI activity. rNanI species corresponding to proteolysis products also promoted the cytotoxic activity and binding of enterotoxin and beta toxin more strongly than did full-length rNanI. Since enterotoxin and beta toxin are produced in the intestines during human and animal disease, these findings suggest that intestinal proteases may enhance NanI activity, which in turn could further potentiate the activity of intestinally active toxins during disease. Coupling these new results with previous findings demonstrating that NanI is important for the adherence of C. perfringens to enterocyte-like cells, NanI sialidase is now emerging as a potential auxiliary virulence factor for C. perfringens enteritis and enterotoxemia.
KEYWORDS: Clostridium perfringens, enterotoxin, beta toxin, sialidase
INTRODUCTION
Clostridium perfringens is an abundant toxin producer, with a current repertoire of ∼20 different toxins. However, individual strains produce various subsets of this imposing toxin arsenal. Consequently, C. perfringens isolates are classified into one of five types (types A to E) based upon their production of four typing toxins, i.e., alpha toxin, beta toxin (C. perfringens beta toxin [CPB]), epsilon toxin (ETX), and iota toxin (1, 2). Some strains also produce C. perfringens enterotoxin (CPE), which is not used for toxin typing classification but is biomedically important, as discussed below.
Besides causing histotoxic infections such as clostridial myonecrosis (1, 3, 4), C. perfringens ranks among the major causes of human and domestic animal diseases that originate in the gastrointestinal (GI) tract (1, 2, 5). C. perfringens illnesses of GI origin involve enteritis or enterotoxemia, with the latter occurring when toxins produced in the intestines are absorbed into the circulation and then damage internal organs such as the brain (1, 6). Each C. perfringens type is associated with particular GI infections; e.g., type A strains producing the NetB toxin cause avian necrotic enteritis (2, 7, 8).
With respect to human GI diseases, C. perfringens type A food poisoning is caused by CPE-producing type A strains (5). This disease currently ranks as the second most common bacterial foodborne illness in the United States, where approximately 1 million cases occur per year (9). CPE-producing type A strains also cause about 5 to 15% of all nonfoodborne human GI diseases, which include sporadic diarrhea and antibiotic-associated diarrhea (10). Molecular Koch's postulate analyses indicated that CPE production is essential for the pathogenesis of both foodborne and nonfoodborne GI infections caused by CPE-positive type A strains (11).
Type C strains of C. perfringens are responsible for both veterinary and human diseases of GI origin (12–15). Human GI infection caused by type C strains is referred to as enteritis necroticans (EN). In the Papua New Guinea highlands during the 1960s and 1970s, EN (known locally as pigbel) was the leading cause of childhood death (16, 17). EN outbreaks still sporadically occur in malnourished individuals in Papua New Guinea and other developing countries. Individual cases of this severe disease have also been reported in developed countries, particularly in diabetics (18, 19). The pathogenesis of EN involves the production of CPB, which is trypsin labile, in the intestines of hosts with reduced trypsin levels due to malnourishment, a diet rich in food (e.g., sweet potato) containing trypsin inhibitors, disease, or coinfection with a parasite producing a trypsin inhibitor (16, 17). For CPE-producing type C strain CN3758, the production of both CPB and CPE was shown to be essential to elicit GI pathology in animal models, indicating that these two toxins can act synergistically when produced together (20).
In addition to potent toxins such as CPE and CPB, C. perfringens strains also produce up to three different sialidases, named NanH, NanI, and NanJ (21, 22). Two of these enzymes, i.e., NanI and NanJ, are exosialidases, while NanH has a cytoplasmic location in early- to mid-log-phase cultures (21). When produced, NanI is responsible for most of the sialidase activity present in C. perfringens culture supernatants (21–24). Studies using C. perfringens single- and multiple-sialidase-null mutants demonstrated that NanI can trim the surface of host cells to significantly enhance C. perfringens adhesion (21, 23). NanI sialidase was also shown to increase the binding and activity of ETX in cultured host cells (21). Finally, those previous studies also demonstrated that an arbitrarily chosen amount of trypsin can increase NanI activity and further promote ETX-induced cytotoxicity (21, 22). This finding led to the proposal that, during GI disease, trypsin may activate NanI to cause, among other effects, a further enhancement of ETX action.
Since C. perfringens uses CPE and CPB, individually or in combination, to cause important human diseases of GI origin (20), the present study had a 2-fold purpose. First, this study aimed to assess whether NanI can increase the binding and cytotoxic activity of these two GI-active toxins. Second, this study directly investigated whether intestinal fluids can activate NanI and, if so, whether this activation potentiates the binding and cytotoxic activity of CPE and CPB.
RESULTS
NanI pretreatment increases cytotoxic and binding activities of CPE.
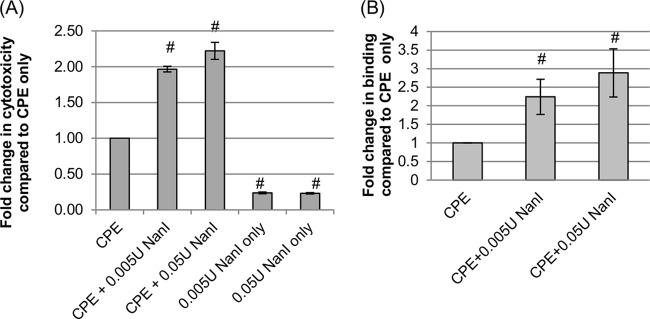
A previous study (21) by our group demonstrated that pretreatment of host cells with purified C. perfringens NanI increases ETX-induced cytotoxic effects in host cells. To determine if NanI can similarly enhance the cytotoxic effects of CPE, Caco-2 cells were pretreated with NanI purified from C. perfringens and dissolved in Hanks' balanced salt solution (HBSS); these cells were then washed and treated with purified CPE. As shown in Fig. 1A, pretreatment of Caco-2 cells with either 0.005 U/ml or 0.05 U/ml of this sialidase significantly increased CPE-mediated cytotoxicity compared to that for cells that were pretreated with HBSS alone (no NanI) prior to CPE challenge. There was a small further increase in CPE-induced cytotoxicity using the 0.05-U/ml dose versus the 0.005-U/ml dose of NanI, but these differences did not reach statistical significance (P > 0.05). The NanI-induced increase in CPE-induced cytotoxic activity shown in Fig. 1A was not due to direct cytotoxic effects from NanI since cells treated with this sialidase in the absence of CPE showed little or no increase in cytotoxicity over background levels of cell death.
FIG 1.
Effects of NanI on cytotoxic activity and binding of CPE. (A) CPE-induced cytotoxicity. Confluent Caco-2 monolayers were pretreated for 1 h at 37°C with buffer or buffer containing purified native NanI at the concentrations indicated. The pretreated monolayers were then washed twice with HBSS and treated with HBSS or HBSS containing 0.5 μg/ml of purified CPE for 1 h at 37°C. After this challenge, the supernatant was collected, and relative cytotoxicity was determined by measuring LDH release. (B) Analysis of CPE binding. To determine if the enhanced cytotoxicity shown in panel A involved an increase in CPE binding, Caco-2 monolayers were pretreated for 1 h at 37°C with buffer or buffer containing purified native NanI at the concentrations indicated, washed twice with HBSS, and then treated for 1 h at 4°C with 5 μg/ml of AF488-CPE. The monolayers were then washed 3 times at 4°C, collected, and lysed, and fluorescence was quantified by using a plate reader. All experiments shown were performed at least in triplicate, and the means are presented. Error bars show standard errors of the means (A) or standard deviations (B). # indicates a significant (P < 0.05) difference compared to cells pretreated with buffer prior to CPE treatment.
Our previous study (21) also showed that the NanI enhancement of ETX-induced cytotoxic activity correlated with a NanI-induced increase in the binding of this toxin to host cells. Therefore, to test whether the enhanced cytotoxic effects of CPE shown in Fig. 1A involve effects of NanI on CPE binding, Caco-2 cells were pretreated with purified NanI sialidase or buffer alone (no NanI) prior to washing and challenge with Alexa Fluor 488 (AF488)-labeled CPE. After three washes, the amount of AF488-labeled CPE bound to these pretreated Caco-2 cells was then determined by quantifying bound fluorescence (Fig. 1B). A significant increase in CPE binding was detected when Caco-2 cells were pretreated with HBSS containing NanI compared to Caco-2 cells pretreated with only HBSS (no NanI). CPE binding levels increased significantly by using a 0.05-U/ml dose versus a 0.005-U/ml dose of NanI.
NanI pretreatment increases cytotoxic and binding activity of CPB.
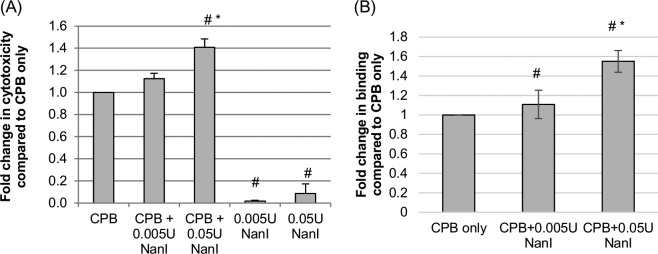
While some C. perfringens type C strains produce CPE, all type C strains must (by definition) produce CPB, as must the type B strains that cause enteritis and enterotoxemia in livestock (20, 25, 26). To evaluate whether NanI enhances the cytotoxic effects of CPB, human umbilical vein endothelial cells (HUVECs) were pretreated with this sialidase, followed by treatment with purified CPB. The results of this analysis, shown in Fig. 2A, demonstrated a significant increase in cytotoxicity when cells were pretreated with 0.05 U/ml of sialidase. While mean cytotoxicity also increased when cells were pretreated with 0.005 U/ml of sialidase, this effect did not reach statistical significance. As with CPE, the enhanced cytotoxicity shown in Fig. 2A was not due to direct cytotoxic effects from NanI, since cells treated with sialidase alone (no CPB) showed only a slight increase in cytotoxicity over background (buffer-alone) levels.
FIG 2.
Effects of NanI on the cytotoxic activity and binding of CPB. (A) CPB-induced cytotoxicity. HUVEC monolayers were grown to confluence and pretreated for 1 h at 37°C with buffer or buffer containing purified native NanI at the concentrations indicated. After these pretreatments, the HUVEC monolayers were rinsed with HBSS twice and then treated with 1.0 μg/ml of purified CPB for 4 h at 37°C. Following this challenge, the supernatant was collected, and relative cytotoxicity was determined by LDH release. (B) Analysis of CPB binding. To determine if the enhanced cytotoxicity observed with NanI pretreatment of HUVECs involved increased toxin binding, HUVEC monolayers were pretreated for 1 h at 37°C with buffer or buffer containing purified native NanI at the concentrations indicated, rinsed twice with HBSS, and then treated for 1 h at 4°C with HBBS or HBSS containing 5 μg/ml of CPB. The monolayers were then washed 3 times at 4°C, and an anti-CPB mouse monoclonal antibody was applied for overnight incubation at 4°C. After two washes with HBSS, an Alexa Fluor 488-labeled anti-mouse IgG antibody was applied for 30 min at room temperature. After 3 washes, fluorescence was quantified by using a plate reader. All experiments were performed at least in triplicate, and the means are presented. Error bars show standard errors of the means (A) or standard deviations (B). # indicates a significant (P < 0.05) difference compared to cells pretreated with buffer prior to treatment with CPB, and * indicates a significant (P < 0.05) difference between cells treated with 0.05 U/ml and those treated with 0.005 U/ml of NanI.
To test if the enhancement in CPB cytotoxicity caused by NanI pretreatment, as shown in Fig. 2A, might involve increased toxin binding, HUVECs were pretreated with purified NanI and then intoxicated with CPB. CPB binding levels were quantified by reacting CPB-treated cells with a CPB mouse monoclonal antibody, followed by the addition of a fluorescently labeled anti-mouse secondary antibody (Fig. 2B). This assay detected a significant increase in CPB binding when cells were pretreated with HBSS containing either 0.005 or 0.05 U/ml of sialidase versus pretreatment with HBSS alone (no NanI). CPB binding also significantly increased when cells were pretreated with 0.05 U/ml of sialidase compared to a 0.005 U/ml sialidase pretreatment.
Exosialidase activity in C. perfringens culture supernatants.
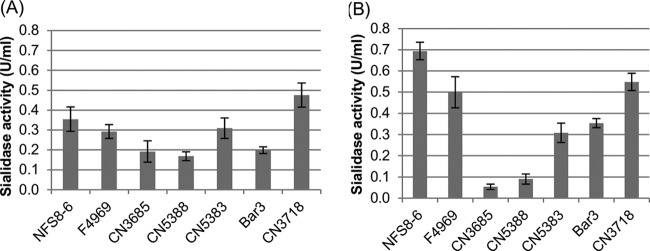
Previous studies (21, 23, 24) indicated that NanI is typically the preeminent exosialidase produced by NanI-positive C. perfringens strains, but the production of exosialidase activity varies among different C. perfringens isolates (21, 23). Therefore, to help assess the physiological relevance of the 0.05-U/ml NanI sialidase dose that significantly enhanced the cytotoxic activities of CPB and CPE in the studies shown in Fig. 1 and 2, exosialidase activity was measured for several NanI-producing C. perfringens strains grown under both vegetative (Fig. 3A) and sporulating (Fig. 3B) growth conditions.
FIG 3.
Exosialidase activities of C. perfringens culture supernatants. Seven C. perfringens strains (Table 1) were grown in either TH broth (A), for vegetative growth, or DS sporulation medium (B) for 14 h at 37°C. Following this incubation, cell-free culture supernatants were prepared by centrifugation, followed by the filtration of the supernatants to remove any bacterial cells. The total exosialidase activity in the culture supernatants was then determined spectrophotometrically, as described previously (23). The experiment was performed in triplicate, and the means are depicted. Error bars indicate the standard errors.
The results of these analyses identified considerable variation in supernatant exosialidase activity between isolates. However, under both growth conditions, all seven tested isolates produced >0.05 U/ml of exosialidase activity, with most isolates producing >0.1 U/ml of exosialidase activity. Since previous studies (21–24) indicated that ∼75% of the exosialidase activity in culture supernatants of NanI-producing C. perfringens strains is typically attributable to NanI, the results shown in Fig. 3 offer support for the physiological relevance of the conditions used for the studies shown in Fig. 1 and 2 by suggesting that the NanI activity present in supernatants from most NanI-producing C. perfringens strains, whether grown under sporulating or vegetative culture conditions, should contain >0.05 U/ml of NanI activity.
Proteolytic processing of NanI by chymotrypsin or mouse small intestinal fluid increases sialidase activity.
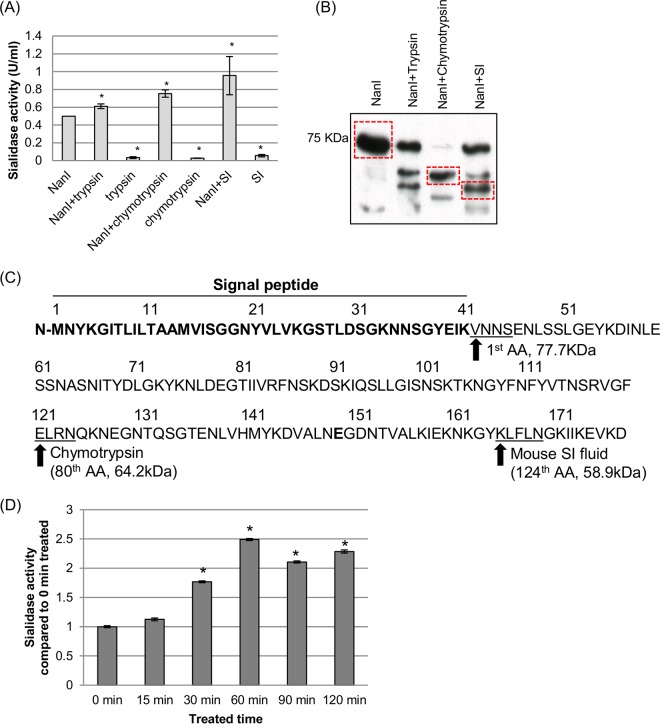
We previously reported that exposure of NanI to purified trypsin increases the activity of this sialidase (21, 23). This effect may have physiological relevance since, when produced in the GI tract, NanI would be exposed to host intestinal proteases such as trypsin. However, the small intestinal (SI) lumen contains a complex mix of many proteases in addition to trypsin, e.g., chymotrypsin and other proteases such as carboxypeptides (27). Therefore, the potential proteolytic activation of the NanI sialidase in the gut was further explored by treating purified NanI with either purified trypsin, purified chymotrypsin, or mouse SI fluid for 30 min and then measuring sialidase activity. As shown in Fig. 4A, this experiment detected a significant increase in sialidase activity following the treatment of NanI with trypsin, chymotrypsin, or mouse SI fluid. This activity was not due to the presence of sialidase activity in trypsin, chymotrypsin, or mouse SI fluid since controls containing these reagents in the absence of the NanI sialidase failed to produce a significant increase in sialidase activity compared to that with buffer alone.
FIG 4.
Proteolytic processing of NanI sialidase. (A and B) Purified NanI (0.5 U/ml) was digested with chymotrypsin (40 ng/ml), trypsin (40 ng/ml), or mouse small intestinal fluid (0.4 μl/μl) at 37°C for 1 h. Following these digestions, samples were assayed for sialidase activity (A) and subjected to SDS-PAGE and NanI Western blotting (B). (C) The dominant digested NanI species present in the chymotrypsin and mouse SI fluid digestions, as noted by boxes in panel B, were selected for N-terminal sequencing to determine the N-terminal cleavage site for each species. AA, amino acid. (D) Kinetics of NanI activity in the presence of mouse SI fluid. The averages of data from three replications are shown in panels A and D, and a representative Western blot is depicted in panel B. In panels A and D, * indicates a significant (P < 0.05) difference compared to wild-type NanI. Error bars show standard deviations.
To better understand the proteolytic processing of NanI, samples of NanI that had been digested with either purified chymotrypsin or mouse SI fluid were analyzed by Western blotting. The results of this assay, shown in Fig. 4B, demonstrated that a primary ∼65-kDa NanI species (indicated by a box) appears after digestion with purified chymotrypsin. A similar digestion of NanI with mouse SI fluid yielded several NanI species. These species included undigested NanI, a 65-kDa species that comigrated with the chymotrypsin-generated NanI species, and a prominent ∼60-kDa NanI species (indicated by boxes) that comigrated with a major NanI species generated by the digestion of NanI with purified trypsin.
To identify the N-terminal cleavage sites for the ∼65-kDa chymotrypsin-generated NanI species and the prominent ∼60-kDa mouse SI fluid-generated NanI species, N-terminal amino acid sequencing was performed on these two NanI fragments after they had been excised from polyvinylidene difluoride (PVDF) membranes containing proteins transferred from acrylamide gels subjected to SDS-PAGE. The results of this sequencing, shown in Fig. 4C, indicated that cleavage induced by purified chymotrypsin occurs 80 amino acids from the N terminus of the mature NanI protein. This cleavage would yield a 64.2-kDa protein, in agreement with the observed 65-kDa molecular mass of this NanI species on Western blots. Sequencing of the predominant mouse SI fluid-generated NanI species indicated that cleavage occurs 124 amino acids from the N terminus of the mature NanI protein. This cleavage would yield a 58.9-kDa protein, in agreement with the observed 60-kDa molecular mass of this NanI species on Western blots. Importantly, analysis of the N-terminal sequence for this ∼60-kDa species indicates that processing had occurred at a trypsin cleavage site, consistent with the Western blot results (Fig. 4B) showing the comigration of this NanI species with a NanI species generated by digestion with trypsin.
An experiment also examined the kinetics of native NanI activation and stability in mouse SI fluid (Fig. 4D). This analysis revealed that significant activation of NanI becomes detectable within 30 min of incubation in mouse SI fluid, and this enhanced activity peaks by 60 min. NanI activity then remained stable throughout a 120-min incubation in SI fluid.
Production of rNanI sialidase species corresponding to proteolytically generated species.
To better understand the sialidase activity of host protease-digested NanI species and rule out concerns about spurious effects in the studies shown in Fig. 1 and 2 that might arise from contaminants in NanI preparations purified from C. perfringens, sequences corresponding to three NanI species (full-length NanI, the 65-kDa chymotrypsin-cleaved NanI fragment, or the prominent 60-kDa mouse SI fluid-cleaved NanI fragment) were cloned into the pTrc-HisB expression system and transformed into Escherichia coli. Nucleotide sequencing confirmed that, compared to wild-type NanI, no point mutations encoding amino acid changes were present in the recombinant NanI (rNanI) species corresponding to full-length NanI, the 65-kDa chymotrypsin-derived NanI fragment, or the rNanI species corresponding to the ∼60-kDa mouse SI fluid-derived NanI fragment.
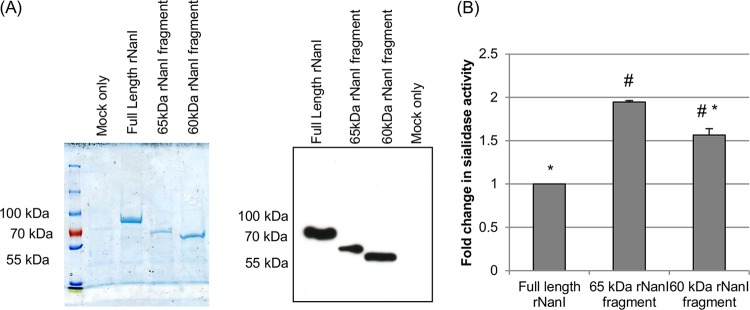
Each recombinant NanI sialidase species was then highly enriched by affinity chromatography (Fig. 5A, left). NanI Western blotting confirmed the identity of the three rNanI species in the enriched preparations (Fig. 5A, right). In contrast, E. coli transformed with pTrc-HisB alone (i.e., empty vector) did not produce any proteins that reacted on the NanI Western blot (Fig. 5A, right).
FIG 5.
Recombinant expression of rNanI species corresponding to protease-cleaved NanI fragments. rNanI species corresponding to full-length NanI or the major 65-kDa chymotrypsin-cleaved or 60-kDa mouse SI fluid-cleaved NanI fragments were recombinantly expressed by using E. coli Top10 cells and the pTrc-HisB expression system. The rNanI species were then affinity enriched from the E. coli culture by using Talon resin and dialyzed overnight against PBS. Panel A shows the dialyzed samples subjected to SDS-PAGE and then either stained with Coomassie blue (left) or Western blotted for NanI (right). The relative amount of each enriched rNanI species was then determined by a protein assay. All experiments were performed in triplicate, and the mean results are shown. Error bars show standard deviations. # indicates a significant (P < 0.05) difference compared to full-length rNanI, and * indicates a significant (P < 0.05) difference compared to the rNanI species corresponding to the ∼65-kDa NanI fragment generated by chymotrypsin treatment.
Equimolar concentrations of each highly enriched rNanI species were then compared for their sialidase activities. The results of this assay (Fig. 5B) demonstrated a significant increase (relative to full-length rNanI) in sialidase activity for both the rNanI species corresponding to the 65-kDa chymotrypsin-derived NanI fragment and the rNanI species corresponding to the ∼60-kDa mouse SI fluid-derived NanI fragment. The sialidase activity of the rNanI species corresponding to the 65-kDa chymotrypsin-generated NanI fragment was also significantly higher than that of the rNanI species corresponding to the ∼60-kDa mouse SI fluid-generated NanI fragment. In comparison, a similarly processed mock-enriched preparation from an equivalent culture volume of E. coli cells carrying the empty vector had nondetectable sialidase activity compared to that of enriched full-length rNanI prepared from an equal volume of culture (not shown).
Finally, full-length rNanI could also be significantly activated (not shown) by 30 min of incubation with purified trypsin or chymotrypsin, or mouse SI fluid, as demonstrated for native NanI in Fig. 4A.
Pretreatment of host cells with rNanI species also enhances CPE and CPB cytotoxicity and binding.
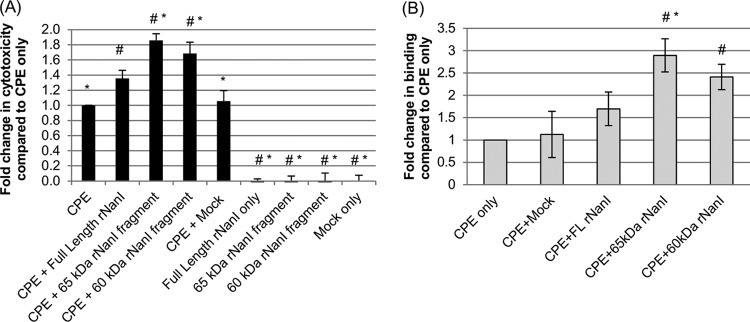
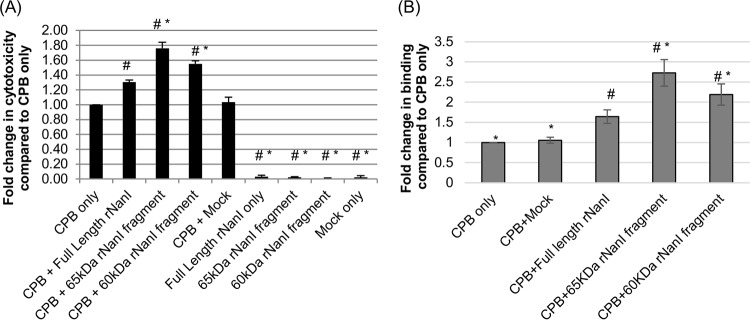
The results shown in Fig. 4 indicated that the proteolytic processing of NanI increases sialidase activity, which should enhance the cytotoxic effects of CPB and CPE on susceptible cells. To definitively evaluate this relationship and compare the relative abilities of different NanI fragments generated by intestinal proteases to promote CPE action, Caco-2 cells were pretreated with equimolar concentrations of full-length rNanI or the rNanI species corresponding to the 65-kDa or 60-kDa NanI fragments generated by the treatment of NanI with chymotrypsin or mouse SI fluid, respectively. When pretreated with equimolar concentrations of any of the three rNanI species, Caco-2 cells exhibited a significant increase in CPE-induced cytotoxicity in comparison to cells treated with CPE only (buffer pretreatment) (Fig. 6A). Moreover, CPE-induced cytotoxicity was further significantly increased by using samples pretreated with the rNanI species corresponding to either the 60- or 65-kDa NanI fragments generated by mouse SI fluid or chymotrypsin, respectively, compared to samples pretreated with full-length rNanI. As controls, CPE cytotoxicity did not significantly change when Caco-2 cells were pretreated with a mock-enriched preparation from an equivalent starting volume of a culture of E. coli cells carrying the empty vector or with any of the enriched rNanI species alone (no CPE).
FIG 6.
Effects of rNanI species on CPE binding and cytotoxicity. (A) CPE-induced cytotoxicity. Confluent Caco-2 monolayers were pretreated with buffer or buffer containing either 0.005 U/ml of full-length rNanI or an equimolar concentration of the other two rNanI species for 1 h at 37°C. Following this pretreatment, the monolayers were washed twice with HBSS and then treated with 0.5 μg/ml of purified CPE for 1 h at 37°C. After this challenge, the supernatant was collected, and relative cytotoxicity was determined by LDH release. (B) Analysis of CPE binding. To determine if increased CPE binding was responsible for the enhanced cytotoxicity observed in panel A, Caco-2 monolayers were pretreated with buffer or buffer containing 0.005 U/ml of full-length rNanI or an equimolar concentration of the other two rNanI species for 1 h at 37°C, washed twice, and challenged for 1 h at 4°C with 5 μg/ml of AF488-CPE. The monolayers were then washed 3 times at 4°C, collected, and lysed, and fluorescence was quantified by using a plate reader. All experiments were performed at least in triplicate, and the means are presented. Error bars depict standard errors of the means (A) or standard deviations (B). # indicates a significant (P < 0.05) difference compared to cells pretreated with buffer prior to CPE treatment. * indicates a significant (P < 0.05) difference compared to cells treated with full-length rNanI before CPE treatment.
The results in Fig. 1 showed that the enhancement of CPE-induced cytotoxicity after pretreatment of Caco-2 cells with purified native NanI involves the increased binding of this toxin to these cells. To determine if the same mechanism contributes to the results shown in Fig. 6A using the three rNanI variants, Caco-2 cells were pretreated with an equimolar concentration of an enriched rNanI species prior to treatment with AF488-labeled CPE. After this toxin treatment, AF488-lableled CPE binding to Caco-2 cells was quantified by using a fluorescence plate reader. A significant increase in AF488-labeled CPE binding was detected for Caco-2 cells pretreated with rNanI species corresponding to the 65-kDa or 60-kDa NanI fragments generated by chymotrypsin or mouse SI fluids, respectively, versus Caco-2 cells pretreated with buffer alone (Fig. 6B). Pretreatment with the rNanI species corresponding to either the 65-kDa or 60-kDa NanI fragments also resulted in higher AF488-labeled CPE binding than with pretreatment with the rNanI species corresponding to full-length NanI, with this difference reaching statistical significance for the NanI species corresponding to the 65-kDa NanI fragment. In contrast, pretreatment of Caco-2 cells with the mock enrichment preparation from E. coli empty vector transformants did not enhance CPE binding over that with the buffer control (Fig. 6B).
Having shown that pretreatment of host cells with any of the three rNanI species increases CPE-induced cytotoxicity and that the rNanI species corresponding to proteolytically generated NanI fragments enhance CPE activity more than do the rNanI species corresponding to full-length NanI, we next sought to determine if similar effects would be observed for CPB. Following pretreatment of HUVECs with an equimolar concentration of one of the three rNanI species, these cells were treated with CPB, and their death was measured by lactate dehydrogenase (LDH) release. The results of this experiment (Fig. 7A) demonstrated a significant increase in cytotoxicity for cells pretreated with equimolar concentrations of any of the three rNanI species compared to that for cells pretreated with buffer alone. This enhanced cytotoxicity in rNanI-pretreated cells was not due to direct cytotoxic effects from the rNanI species since cells pretreated with these species in the absence of CPB did not show significant cytotoxicity. This experiment also revealed a significant increase in CPB-induced cytotoxicity for cells pretreated with the rNanI species corresponding to the 65-kDa or 60-kDa rNanI fragments generated by chymotrypsin or mouse SI fluid, respectively, versus cells pretreated with an equimolar concentration of full-length rNanI, which further supports the proteolytic activation of NanI as an enhancer of CPB activity. In comparison, no significant change in CPB-induced cytotoxicity was observed when HUVECs were pretreated with a mock-enriched preparation from an equal culture volume of E. coli cells carrying the empty vector.
FIG 7.
Effects of rNanI species on CPB binding and cytotoxicity. (A) CPB-induced cytotoxicity. Confluent HUVEC monolayers were pretreated with buffer or buffer containing either 0.05 U/ml of full-length rNanI or an equimolar concentration of the other two rNanI species for 1 h at 37°C. The pretreated monolayers were rinsed twice with HBSS and then challenged with 1 μg/ml of purified CPB for 4 h at 37°C. The culture supernatant was collected, and relative cytotoxicity was determined by LDH release. (B) Analysis of CPB binding. To determine if the enhanced CPB cytotoxicity observed in panel A was due to increased CPB binding, HUVEC monolayers were pretreated with buffer or buffer containing either 0.05 U/ml of full-length rNanI or an equimolar concentration of the other two rNanI species for 1 h at 37°C, washed twice with HBSS, and then challenged for 1 h at 4°C with HBSS or HBSS containing 5 μg/ml of CPB. The monolayers were washed 3 times at 4°C, and an anti-CPB mouse monoclonal antibody was added for overnight incubation at 4°C. After two washes with HBSS, an Alexa Fluor 488-labeled anti-mouse IgG antibody was applied for 30 min at room temperature. After 3 washes, fluorescence was quantified by using a plate reader. All experiments were performed at least in triplicate, and the means are presented. Error bars depict standard errors of the means (A) or standard deviations (B). # indicates a significant (P < 0.05) difference compared to cells pretreated with buffer prior to CPB treatment. * indicates a significant (P < 0.05) difference compared to cells treated with full-length rNanI before CPB treatment.
To evaluate a potential mechanism behind the increased CPB cytotoxicity observed after treatment with the three rNanI species, CPB binding assays were performed. In these experiments, HUVECs were pretreated with equimolar concentrations of one rNanI species prior to CPB treatment, and bound CPB was detected by reacting CPB-treated cells with a CPB mouse monoclonal antibody, followed by a fluorescently labeled anti-mouse secondary antibody. These analyses detected significant increases in CPB binding levels in HUVECs pretreated with full-length rNanI or either the 60- or 65-kDa rNanI fragments (Fig. 7B). Significantly more CPB binding was detected for cells pretreated with the rNanI species corresponding to either the 60-kDa or 65-kDa NanI fragments than for cells pretreated with full-length NanI. As a control, pretreatment of HUVECs with the mock enrichment preparation from E. coli empty vector transformants did not affect CPB binding levels compared to those with buffer pretreatment (Fig. 7B).
DISCUSSION
This study contributes several insights into the potential contributions of NanI sialidase to C. perfringens virulence. First, these results showed that NanI can significantly increase the cytotoxic effects of CPB and CPE. Coupling this finding with previous results (21) demonstrating a similar NanI enhancement of ETX activity, it is now clear that this sialidase can potentiate the action of all three toxins proven to have roles in C. perfringens diseases originating in the mammalian GI tract (11, 20, 28–31). While the enhancement of the action of CPB, CPE, and ETX by native NanI is relatively modest (1.5- to 2-fold), this effect may still be important during enteritis or enterotoxemia, since many C. perfringens disease strains produce only small amounts of CPB, CPE, or ETX (20, 32–35). Therefore, augmentation of low toxin activity by NanI, particularly when NanI activity is further increased by proteolytic processing (as discussed below) in the GI tract, could be a critical determinant regarding the development of diseases involving these three toxins. Finally, it should be mentioned that NanI does not only potentiate the action of toxins involved in C. perfringens mammalian enteritis and enterotoxemia. A previous study (24) reported that NanI also enhances the activity of two toxins (alpha toxin and perfringolysin O) involved in gas gangrene caused by this bacterium.
As was previously shown for ETX (21), this study determined that the NanI-induced increase in the action of CPB and CPE involves the enhanced binding of these two toxins to host cells. Whether increased toxin binding is also involved in the NanI-mediated enhancement of perfringolysin O and alpha toxin activities has not yet been determined (24). Since CPE, CPB, and ETX do not share a receptor, the NanI-induced enhancement of cell binding by these three toxins is nonspecific in nature. The nature of the nonspecific enhancement of CPB, CPE, and ETX binding by NanI requires further study, but it could involve several mechanisms, alone or in combination. Sialic acids provide much of the surface charge on mammalian cells, so by removing surface sialic acid residues, NanI sialidase could reduce electrostatic repulsion and increase toxin binding. Alternatively, NanI could remove masking sialic acids from toxin receptors to increase the access of toxins to their receptor. This possibility is difficult to evaluate for CPB and ETX since the receptors for these toxins have not yet been identified; i.e., it is unknown whether these receptors are sialylated. While it is known that CPE uses certain members of the claudin family as receptors (36), there is not yet any evidence that these receptor claudins are sialylated. A third possibility is that NanI might increase toxin binding by trimming glycoproteins or glycolipids adjacent to toxin receptors, thus removing sialic acids that are partially occluding the toxin receptor. Sialidases can increase paracellular permeability (37), so a final possibility is that NanI might facilitate the access of toxins to receptors present on the basolateral surface of host cells, resulting in increased toxin binding.
While the in vitro and ex vivo results reported here further demonstrate that NanI can increase the cellular activity and binding of several C. perfringens toxins with proven involvement in diseases originating in the GI tract, other recent studies (21, 23) suggest that enhancing toxin activity may not be the only potential contribution of NanI to enteritis and enterotoxemia caused by this bacterium. In those previous in vitro studies, it was demonstrated that NanI also trims the surface of enterocyte-like cells to increase C. perfringens adhesion. Interestingly, this effect showed specificity, since NanI was less effective in promoting C. perfringens attachment to kidney cells or fibroblasts than in promoting attachment to enterocyte-like cells (21). Animal model studies are under way to examine directly the contributions of NanI to intestinal diseases.
A second contribution of this work is that it provides new insights into the proteolytic activation of NanI. Previous work (21, 22) showed that purified trypsin can proteolytically activate NanI. The present study found that purified chymotrypsin can also exert NanI activation. Importantly, SI fluid (which is a complex mix of proteases) was also shown to activate NanI ex vivo. These findings suggest direct pathophysiological relevance for NanI proteolytic activation, since during disease, NanI would be produced in the GI tract, where it would be exposed to small intestinal fluid containing a mix of intestinal proteases, including trypsin and chymotrypsin. Thus, in the intestines, the proteolytic activation of NanI should further enhance the action of CPE, ETX, and CPB, possibly contributing to disease.
There is substantial evidence that CPE and ETX, as well as NanI, can be activated by intestinal proteases (27, 38–40), suggesting that these three proteins have evolved to gain greater function via proteolytic processing in the GI tract, where they are naturally produced. In contrast, CPB is very trypsin sensitive and causes disease only in people with low trypsin levels (16, 17). For example, CPB-mediated EN has been very common in the Papua New Guinea highlands, occurring in people with a diet rich in sweet potato, which contains a trypsin inhibitor (16, 17). However, sweet potatoes do not reduce chymotrypsin activity (41), which is notable since the present study found that chymotrypsin can activate NanI. Therefore, protease activation of NanI by chymotrypsin, as shown in the present study, could increase the intestinal activity of CPB and thus contribute to type C (or type B)-mediated diseases even in people with low trypsin levels due to diet, disease, or infection with parasites. It should also be reemphasized that, in vivo, the proteolytic activation of NanI not only may increase toxin binding but also might enhance virulence by promoting the intestinal attachment of cells of enteropathogenic C. perfringens, including type B and C strains.
While a previous study showed that NanI is not necessary for C. perfringens to cause gas gangrene in mice (24), that study also acknowledged that the mouse model for C. perfringens gas gangrene requires a large inoculum that may bypass contributions of factors (perhaps including NanI) involved in early pathogenic steps. Emerging evidence from in vitro and ex vivo studies (including this work) offers support for possible contributions of NanI to C. perfringens diseases originating in the GI tract. Therefore, animal model studies are planned to examine the contributions of NanI to C. perfringens enteritis and enterotoxemia.
MATERIALS AND METHODS
Bacterial strains.
C. perfringens strains used in this work are listed in Table 1. Strains were routinely cultured in fluid thioglycolate broth (FTG) (Becton Dickinson [BD]) at 37°C. Where indicated, strains were cultured at 37°C in Todd-Hewitt (TH) broth (BD) supplemented with 0.1% sodium thioglycolate (Sigma) or Duncan-Strong (DS) broth (0.4% starch [BD], 1.5% proteose peptone [BD], 0.4% yeast extract [BD], 1.0% dibasic sodium phosphate [J. T. Baker], and 0.1% sodium thioglycolate [Sigma]). E. coli Top10 cells (ThermoFisher) were routinely grown on LB medium (Fisher Scientific) supplemented with 100 μg/ml of ampicillin (Fisher Scientific), as indicated.
TABLE 1.
NanI-positivea C. perfringens strains used in this study
| Strain | Description | Type | Reference |
|---|---|---|---|
| NFS8-6 | Normal flora isolate from healthy North American | A | 45 |
| F4969 | Sporadic diarrhea isolate from Europe | A | 46 |
| CN3685 | Ovine isolate from animal known to be experiencing disease | C | 33 |
| CN5388 | Pigbel strain from Papua New Guinea | C | 33 |
| CN5383 | Pigbel strain from Papua New Guinea | C | 33 |
| Bar3 | Pigbel strain from Papua New Guinea | C | 33 |
| CN3718 | Animal disease strain previously shown to express sialidases | D | 21 |
Toxins.
CPE was purified to ∼100% homogeneity from type A strain NCTC8239, as described previously (42). CPB was purified to >95% homogeneity from type C strain CN3685, as described previously (20).
Purified CPE was fluorescently labeled by using an Alexa Fluor 488 protein labeling kit (Molecular Probes, Invitrogen Detection Technologies) according to the manufacturer's instructions. This AF488-labeled CPE and native CPE exhibited similar cytotoxicities for Caco-2 cells using the LDH cytotoxicity detection kit (Roche) (data not shown). Attempts to label purified CPB using the same kit failed, since the labeled CPB toxin aggregated and lost function.
Cell culture.
HUVECs (Life Technologies) were grown in medium 200 (Life Technologies) supplemented with a low-serum growth supplement (Life Technologies). To facilitate HUVEC attachment, culture plates were coated for 3 h with 60 μg/ml of bovine collagen (Advanced Biomatrix) prior to seeding. Caco-2 human colonic epithelial cells (ATCC) were routinely cultured in Eagle's minimum essential medium (Lonza) supplemented with 100 μg/ml of penicillin-streptomycin (Fisher Scientific), 10% fetal bovine serum (Fisher Scientific), 1% glutamine, and 1% nonessential amino acids (Sigma). All cell lines were grown at 37°C under atmospheric conditions with supplementation with 5% CO2.
Preparation of cell-free C. perfringens culture supernatants.
Freshly prepared cell-free C. perfringens culture supernatants were used to determine the exosialidase activities of seven NanI-producing C. perfringens strains (Table 1) under vegetative or sporulating growth conditions. Briefly, these C. perfringens strains were inoculated from cooked meat medium (Oxford) freezer stocks into FTG broth and incubated overnight at 37°C. A 0.2-ml aliquot of these cultures grown overnight was inoculated into fresh FTG broth tubes and incubated for 10 h at 37°C. Following this incubation, 0.2-ml and 0.5-ml aliquots of the FTG culture were used to inoculate 10 ml of TH and DS broth tubes, respectively. Cultures were then incubated for 14 h at 37°C. After this incubation, the cultures were centrifuged. Supernatants were collected, passed through a 0.45-μm filter (Millipore), and briefly stored on ice until use.
Sialidase activity assay.
To assay exosialidase activity, a previously described protocol was used (21, 22). Briefly, 20 μl of a sample (cell-free C. perfringens culture supernatant), purified NanI (Roche Applied Science), an enriched rNanI species from E. coli (described below), or a mock enrichment preparation from an equal culture volume of empty vector-carrying E. coli cells was added to 60 μl of 50 mM Tris-HCl buffer (pH 7.2). A 20-μl aliquot of 4 mM 5-bromo-4-chloro-3-indolyl-α-d-N-acetylneuraminic acid (Sigma) was then added to each sample, and the resultant mixtures were incubated at 37°C in the dark for 30 min. Following this incubation, the sample absorbance at 595 nm was measured by using a microplate reader (Bio-Rad). Sialidase activities in the culture supernatants were determined by comparing their activity to that of purified NanI.
Collection and processing of mouse small intestinal contents.
To obtain contents from mouse SI, a healthy anesthetized adult male BALB/c mouse (25 g) was euthanized by cervical dislocation, the abdominal cavity was opened, and the contents were harvested from the jejunum (fresh intestinal fluid) and stored at −80°C (frozen intestinal fluid) until use. This procedure was approved by the University of California—Davis IACUC (permit numbers 16383 and 19186). One gram of fresh or frozen mouse intestinal contents was added to 1 ml of phosphate buffer (pH 7.0). After mixing well, the sample was centrifuged. The supernatant fluid (referred to as mouse SI fluid) was then collected directly for use (fresh intestinal fluid) or aliquoted and stored at −80°C (frozen intestinal fluid) for future experiments. No difference between the abilities of the fresh and frozen intestinal fluids to activate NanI was detected (data not shown).
Effects of NanI or rNanI species on C. perfringens toxin cytotoxicity.
Qualitatively, both CPE and CPB induce cell rounding in sensitive cells, and this effect was not altered by pretreatment with a NanI species. To quantify the effects of purified NanI or an rNanI species (described below) on the ability of CPB or CPE to induce cytotoxicity in host cells, an LDH release-based cytotoxicity assay was utilized, as described previously (21). Briefly, HUVECs or Caco-2 cells (sensitive to CPB and CPE, respectively [26, 43]) were seeded into 24-well plates and grown as described above until confluent monolayers were achieved. Monolayers were rinsed twice with HBSS (Corning) and treated (as described in the figure legends) for 1 h at 37°C with purified NanI, an enriched rNanI species, or a mock enrichment preparation from an equivalent volume of the empty vector-carrying E. coli culture. Monolayers were then rinsed twice with HBSS and treated at 37°C for 4 h with 1 μg/ml of CPB or for 1 h with 0.5 μg/ml of CPE. After this treatment, the cell-free supernatant was collected by centrifugation and assayed for LDH activity by using the LDH cytotoxicity detection kit (Roche) according to the manufacturer's instructions.
Effects of NanI or rNanI species on C. perfringens toxin binding.
To determine whether NanI or rNanI species can affect the ability of CPE to bind to host cells, confluent monolayers of Caco-2 cells were pretreated for 1 h at 37°C with specified amounts of NanI or an enriched rNanI species, buffer alone, or a mock enrichment preparation from an equivalent volume of the empty vector-carrying E. coli culture, prepared as described above. The pretreated monolayers were then rinsed twice with cold HBSS and treated for 1 h at 4°C with buffer or buffer containing 5.0 μg/ml of AF488-CPE. After this treatment, the cells were washed 3 times with cold HBSS. Cells were collected and suspended in 200 μl of radioimmunoprecipitation assay (RIPA) buffer with 2% Benzonase (Novagen). After centrifugation, an aliquot of 100 μl of the supernatant was transferred to a 96-well plate, and the fluorescence in this sample was read at 485/529 nm by using a BioTek Synergy plate reader. Background fluorescence in the buffer-treated samples was subtracted from that of matching samples treated with AF488-CPE.
To quantify CPB binding, confluent monolayers of HUVECs in 6-well plates were rinsed and pretreated with the specified amounts of NanI, an enriched rNanI species, or a mock enrichment preparation from an equivalent volume of the empty vector-carrying E. coli culture, as described above. The pretreated monolayers were then rinsed twice with cold HBSS and treated for 1 h at 4°C with buffer or buffer containing 5.0 μg of CPB/ml. After this treatment, the cells were washed 3 times with cold HBSS. A 1:200 dilution of anti-CPB mouse monoclonal antibody (28) was then applied to these cells. After overnight incubation at 4°C with gentle shaking, the monolayers were washed twice with HBSS, and an AF488-labeled, anti-mouse IgG antibody was applied (1:750 dilution) for 30 min at room temperature. The cells were then washed 3 times with HBSS and suspended in 100 μl of RIPA buffer with 2% Benzonase (Novagen). A 100-μl aliquot of the supernatant was transferred to a 96-well plate, and the fluorescence was read at 428/529 nm by using a BioTek Synergy plate reader. The background fluorescence in the buffer-treated samples was subtracted from that of the matching samples treated with CPB.
Digestion of NanI or rNanI with trypsin, chymotrypsin, or small intestinal fluid.
Purified NanI or full-length rNanI (0.05 U) was digested with trypsin (40 ng/μl; 1.7 μM), chymotrypsin (40 ng/μl; 1.6 μM), or mouse SI fluid (0.4 μl/μl reaction mixture) at 37°C for 30 to 120 min, as specified. Purified trypsin and chymotrypsin were purchased from Sigma.
N-terminal amino acid sequencing of chymotrypsin- or small intestinal fluid-digested NanI.
Four micrograms of purified NanI, which had been digested with chymotrypsin or SI fluid, was electrophoresed on an 8% SDS-PAGE gel and then transferred to a PVDF membrane. After staining with Coomassie blue G250 (Bio-Rad) for 30 s, the membrane was destained until bands became visible. The membrane was washed with double-distilled water (ddH2O) and then air dried. Protein bands of interest were excised from the membrane and submitted to the Molecular Structure Facility, University of California—Davis, for N-terminal amino acid sequencing by Edman degradation.
Expression of rNanI species.
Full-length, chymotrypsin-cleaved, or SI fluid-cleaved rNanI species, as described below, were expressed in recombinant E. coli cells by using the pTrc-HisB expression system (Life technologies), according to the manufacturer's instructions. Briefly, primers were designed to amplify the nanI open reading frame (ORF) starting after the signal sequence (44) for full-length rNanI or at 5′ nucleotides in the nanI ORF that encode the N-terminal amino acids identified by N-terminal amino acid sequencing for chymotrypsin-treated or SI-treated purified NanI. These primers were JRT2014-01 (5′-TTGGATCCGGTAAATAATAGTGAAAAT-3′) (rNanI full-length forward primer), JRT2014-03 (5′-TTGGATCCGAAGCTTTTCCTTAATGGA-3′) (60-kDa rNanI forward primer), JRT2014-05 (5′-TTGGATCCGGAGCTTAGAAATCAAAAAAATGAGGG-3′) (65-kDa rNanI forward primer), and JRT2014-04 (5′-TTGAATTCTTATTTATTAGCTCCACTCTC-3′) (rNanI reverse primer).
After PCR using DNA from C. perfringens strain CN3718 (Table 1), products were digested with EcoRI and BamHI (New England BioLabs) and ligated into pTrc-HisB, and the ligation mixture was then transformed into E. coli Top10 cells. Successful constructs were confirmed by PCR detection of the presence of nanI gene sequences and by DNA sequencing to confirm that point mutations leading to amino acid changes had not been introduced during PCR cloning. Sequencing was performed at the Genomics Research Core at the University of Pittsburgh.
For the expression of these rNanI species, E. coli strains producing an rNanI variant (or carrying the pTrc-HisB empty vector) were grown overnight at 37°C with aeration in 50 ml of LB broth supplemented with 100 μg/ml of ampicillin. The starter cultures were then used to inoculate 1 liter of LB broth supplemented with 100 μg/ml of ampicillin to an optical density at 600 nm (OD600) of 0.05. These cultures were incubated at 37°C with aeration until a culture OD600 of 0.5 to 0.6 was obtained. Isopropyl-β-d-thiogalactopyranoside (IPTG) was then added to the culture at a final concentration of 1 mM, and incubation continued for 4 h at room temperature. After this final incubation, cells were harvested by centrifugation and frozen at −80°C until further processing.
To enrich the lysates containing an rNanI species, or to enrich lysates from empty vector-carrying E. coli cultures as controls, the cell pellets were thawed on ice, resuspended in phosphate-buffered saline (PBS) containing 1 mg/ml lysozyme (Sigma), and incubated at 25°C for 30 min. Suspensions were then lysed on ice by using a Qsonica sonicator (2-min total run time with 30% maximum output) and centrifuged for 10 min. Lysate supernatants were added to 1 ml of Talon resin (Clontech) and incubated for 30 min at 25°C. Following this incubation, the resin was loaded onto a gravity column and washed with 20 ml of PBS. Bound rNanI was eluted from the column in PBS containing 100 mM imidazole (Sigma). The collected eluent containing rNanI was dialyzed against PBS overnight at 4°C to remove imidazole. The volume of the affinity-enriched preparation containing full-length rNanI was adjusted with PBS to achieve 0.05 U/ml (for CPB experiments) or 0.005 U/ml (for CPE experiments) of sialidase activity, determined as described above. Affinity-enriched preparations of the other two rNanI species were then diluted to achieve a matching (equimolar) concentration as full-length rNanI; these dilutions were based upon a protein bicinchoninic acid (BCA) assay. After similar processing of lysates from an equivalent volume of an empty vector-carrying E. coli control culture, a similar volume of this mock-enriched preparation was used in all studies as a control for background E. coli activities.
Statistical analyses.
All statistical analyses were performed by using GraphPad Prism 6. For more than two groups, ordinary one-way analysis of variance (ANOVA) was applied with post hoc analysis using Dunnett's test or Tukey's multiple-comparison test.
Accession number(s).
The sequence encoding mature NanI was deposited in GenBank (accession number KX290921).
ACKNOWLEDGMENTS
This work was generously supported by grants R21 AI125796-2 (B.A.M., J.L., and F.A.U. [principal investigators {PIs}]), R01 AI019844-34 (B.A.M. [PI]), and R03 AI 105635-2 (J.L.) from the National Institute of Allergy and Infectious Disease. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
We thank Paul Hauer for providing the monoclonal antibody against CPB.
REFERENCES
- 1.McClane BA, Uzal FA, Miyakawa MF, Lyerly D, Wilkins TD. 2006. The enterotoxic Clostridia, p 688–752. In Dworkin M, Falkow S, Rosenburg E, Schleifer H, Stackebrandt E (ed), The prokaryotes, 3rd ed Springer, New York, NY. [Google Scholar]
- 2.Li J, Adams V, Bannam TL, Miyamoto K, Garcia JP, Uzal FA, Rood JI, McClane BA. 2013. Toxin plasmids of Clostridium perfringens. Microbiol Mol Biol Rev 77:208–233. doi: 10.1128/MMBR.00062-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rood JI. 2006. Clostridium perfringens and histotoxic disease, p 753–770. In Dworkin M, Falkow S, Rosenburg E, Schleifer H, Stackebrandt E (ed), The prokaryotes, 3rd ed Springer, New York, NY. [Google Scholar]
- 4.Titball RW, Rood JI. 2002. Clostridium perfringens: wound infections, p 1875–1903. In Sussman M. (ed), Molecular medical microbiology. Academic Press, London, United Kingdom. [Google Scholar]
- 5.McClane BA, Robertson SL, Li J. 2013. Clostridium perfringens, p 465–489. In Doyle MP, Buchanan RL (ed), Food microbiology: fundamentals and frontiers, 4th ed ASM Press, Washington, DC. [Google Scholar]
- 6.Songer JG. 1996. Clostridial enteric diseases of domestic animals. Clin Microbiol Rev 9:216–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Freedman JC, Theoret JR, Wisniewski JA, Uzal FA, Rood JI, McClane BA. 2015. Clostridium perfringens type A-E toxin plasmids. Res Microbiol 166:264–279. doi: 10.1016/j.resmic.2014.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Keyburn AL, Boyce JD, Vaz P, Bannam TL, Ford ME, Parker D, Di Rubbo A, Rood JI, Moore RJ. 2008. NetB, a new toxin that is associated with avian necrotic enteritis caused by Clostridium perfringens. PLoS Pathog 4:e26. doi: 10.1371/journal.ppat.0040026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.CDC. 2011. CDC estimates of foodborne illness in the United States: Clostridium perfringens. CDC, Atlanta, GA: https://www.cdc.gov/foodsafety/diseases/clostridium-perfringens.html. [Google Scholar]
- 10.Carman RJ. 1997. Clostridium perfringens in spontaneous and antibiotic-associated diarrhoea of man and other animals. Rev Med Microbiol 8(Suppl 1):S43–S45. doi: 10.1097/00013542-199712001-00023. [DOI] [Google Scholar]
- 11.Sarker MR, Carman RJ, McClane BA. 1999. Inactivation of the gene (cpe) encoding Clostridium perfringens enterotoxin eliminates the ability of two cpe-positive C. perfringens type A human gastrointestinal disease isolates to affect rabbit ileal loops. Mol Microbiol 33:946–958. doi: 10.1046/j.1365-2958.1999.01534.x. [DOI] [PubMed] [Google Scholar]
- 12.Uzal FA, McClane BA. 2011. Recent progress in understanding the pathogenesis of Clostridium perfringens type C infections. Vet Microbiol 153:37–43. doi: 10.1016/j.vetmic.2011.02.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Uzal FA, McClane BA, Cheung JK, Theoret J, Garcia JP, Moore RJ, Rood JI. 2015. Animal models to study the pathogenesis of human and animal Clostridium perfringens infections. Vet Microbiol 179:23–33. doi: 10.1016/j.vetmic.2015.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lawrence G. 2005. The pathogenesis of pig-bel in Papua New Guinea. 1979. P N G Med J 48:39–49. [PubMed] [Google Scholar]
- 15.Murrell TG, Walker PD. 1991. The pigbel story of Papua New Guinea. Trans R Soc Trop Med Hyg 85:119–122. doi: 10.1016/0035-9203(91)90183-Y. [DOI] [PubMed] [Google Scholar]
- 16.Johnson S, Gerding DN. 1997. Enterotoxemic infections, p 117–140. In Rood JI, McClane BA, Songer JG, Titball RW (ed), The Clostridia: molecular biology and pathogenesis. Academic Press, London, United Kingdom. [Google Scholar]
- 17.Lawrence GW. 1997. The pathogenesis of enteritis necroticans, p 198–207. In Rood JI, McClane BA, Songer JG, Titball RW (ed), The Clostridia: molecular genetics and pathogenesis. Academic Press, London, United Kingdom. [Google Scholar]
- 18.Petrillo TM, Beck-Sague CM, Songer JG, Abramowsky C, Fortenberry JD, Meacham L, Dean AG, Lee H, Bueschel DM, Nesheim SR. 2000. Enteritis necroticans (pigbel) in a diabetic child. N Engl J Med 342:1250–1253. doi: 10.1056/NEJM200004273421704. [DOI] [PubMed] [Google Scholar]
- 19.Gui L, Subramony C, Fratkin J, Hughson MD. 2002. Fatal enteritis necroticans (pigbel) in a diabetic adult. Mod Pathol 15:66–70. doi: 10.1038/modpathol.3880491. [DOI] [PubMed] [Google Scholar]
- 20.Ma M, Gurjar A, Theoret JR, Garcia JP, Beingesser J, Freedman JC, Fisher DJ, McClane BA, Uzal FA. 2014. Synergistic effects of Clostridium perfringens enterotoxin and beta toxin in rabbit small intestinal loops. Infect Immun 82:2958–2970. doi: 10.1128/IAI.01848-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li J, Sayeed S, Robertson S, Chen J, McClane BA. 2011. Sialidases affect the host cell adherence and epsilon toxin-induced cytotoxicity of Clostridium perfringens type D strain CN3718. PLoS Pathog 7:e1002429. doi: 10.1371/journal.ppat.1002429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li J, McClane BA. 2014. The sialidases of Clostridium perfringens type D strain CN3718 differ in their properties and sensitivities to inhibitors. Appl Environ Microbiol 80:1701–1709. doi: 10.1128/AEM.03440-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li J, McClane BA. 2014. Contributions of NanI sialidase to Caco-2 cell adherence by Clostridium perfringens type A and C strains causing human intestinal disease. Infect Immun 82:4620–4630. doi: 10.1128/IAI.02322-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chiarezza M, Lyras D, Pidot SJ, Flore-Diaz M, Awad MM, Kennedy CL, Cordner LM, Phumoonna T, Poon R, Hughes ML, Emmins JJ, Alape-Giron A, Rood JI. 2009. The NanI and NanJ sialidases of Clostridium perfringens are not essential for virulence. Infect Immun 77:4421–4428. doi: 10.1128/IAI.00548-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ma M, Li J, McClane BA. 2012. Genotypic and phenotypic characterization of Clostridium perfringens isolates from Darmbrand cases in post-World War II Germany. Infect Immun 80:4354–4363. doi: 10.1128/IAI.00818-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Theoret JR, Uzal FA, McClane BA. 2015. Identification and characterization of Clostridium perfringens beta toxin variants with differing trypsin sensitivity and in vitro cytotoxicity activity. Infect Immun 83:1477–1486. doi: 10.1128/IAI.02864-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Freedman JC, Li J, Uzal FA, McClane BA. 2014. Proteolytic processing and activation of Clostridium perfringens epsilon toxin by caprine small intestinal contents. mBio 5:e01994-. doi: 10.1128/mBio.01994-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sayeed S, Uzal FA, Fisher DJ, Saputo J, Vidal JE, Chen Y, Gupta P, Rood JI, McClane BA. 2008. Beta toxin is essential for the intestinal virulence of Clostridium perfringens type C disease isolate CN3685 in a rabbit ileal loop model. Mol Microbiol 67:15–30. doi: 10.1111/j.1365-2958.2007.06007.x. [DOI] [PubMed] [Google Scholar]
- 29.Garcia JP, Li J, Shrestha A, Freedman JC, Beingesser J, McClane BA, Uzal FA. 2014. Clostridium perfringens type A enterotoxin damages the rabbit colon. Infect Immun 82:2211–2218. doi: 10.1128/IAI.01659-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garcia JP, Beingesser J, Fisher DJ, Sayeed S, McClane BA, Posthaus H, Uzal FA. 2012. The effect of Clostridium perfringens type C strain CN3685 and its isogenic beta toxin null mutant in goats. Vet Microbiol 157:412–419. doi: 10.1016/j.vetmic.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Garcia JP, Adams V, Beingesser J, Hughes ML, Poon R, Lyras D, Hill A, McClane BA, Rood JI, Uzal FA. 2013. Epsilon toxin is essential for the virulence of Clostridium perfringens type D infection in sheep, goats, and mice. Infect Immun 81:2405–2414. doi: 10.1128/IAI.00238-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Collie RE, Kokai-Kun JF, McClane BA. 1998. Phenotypic characterization of enterotoxigenic Clostridium perfringens isolates from non-foodborne human gastrointestinal diseases. Anaerobe 4:69–79. doi: 10.1006/anae.1998.0152. [DOI] [PubMed] [Google Scholar]
- 33.Fisher DJ, Fernandez-Miyakawa ME, Sayeed S, Poon R, Adams V, Rood JI, Uzal FA, McClane BA. 2006. Dissecting the contributions of Clostridium perfringens type C toxins to lethality in the mouse intravenous injection model. Infect Immun 74:5200–5210. doi: 10.1128/IAI.00534-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sayeed S, Fernandez-Miyakawa ME, Fisher DJ, Adams V, Poon R, Rood JI, Uzal FA, McClane BA. 2005. Epsilon-toxin is required for most Clostridium perfringens type D vegetative culture supernatants to cause lethality in the mouse intravenous injection model. Infect Immun 73:7413–7421. doi: 10.1128/IAI.73.11.7413-7421.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fernandez-Miyakawa ME, Fisher DJ, Poon R, Sayeed S, Adams V, Rood JI, McClane BA, Uzal FA. 2007. Both epsilon-toxin and beta-toxin are important for the lethal properties of Clostridium perfringens type B isolates in the mouse intravenous injection model. Infect Immun 75:1443–1452. doi: 10.1128/IAI.01672-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shrestha A, Uzal FA, McClane BA. 2016. The interaction of Clostridium perfringens enterotoxin with receptor claudins. Anaerobe 41:18–26. doi: 10.1016/j.anaerobe.2016.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cioffi DL, Pandey S, Alvarez DF, Cioffi EA. 2012. Terminal sialic acids are an important determinant of pulmonary endothelial barrier integrity. Am J Physiol Lung Cell Mol Physiol 302:L1067–L1077. doi: 10.1152/ajplung.00190.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Granum PE, Richardson M. 1991. Chymotrypsin treatment increases the activity of Clostridium perfringens enterotoxin. Toxicon 29:445–453. doi: 10.1016/0041-0101(91)90227-I. [DOI] [PubMed] [Google Scholar]
- 39.Kokai-Kun JF, McClane BA. 1997. Deletion analysis of the Clostridium perfringens enterotoxin. Infect Immun 65:1014–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Minami J, Katayama S, Matsushita O, Matsushita C, Okabe A. 1997. Lambda-toxin of Clostridium perfringens activates the precursor of epsilon-toxin by releasing its N- and C-terminal peptides. Microbiol Immunol 41:527–535. doi: 10.1111/j.1348-0421.1997.tb01888.x. [DOI] [PubMed] [Google Scholar]
- 41.Maloney KP, Truong VD, Allen JC. 2014. Susceptibility of sweet potato (Ipomoea batatas) peel proteins to digestive enzymes. Food Sci Nutr 2:351–360. doi: 10.1002/fsn3.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McDonel JL, McClane BA. 1988. Production, purification and assay of Clostridium perfringens enterotoxin. Methods Enzymol 165:94–103. doi: 10.1016/S0076-6879(88)65018-X. [DOI] [PubMed] [Google Scholar]
- 43.Chakrabarti G, Zhou X, McClane BA. 2003. Death pathways activated in CaCo-2 cells by Clostridium perfringens enterotoxin. Infect Immun 71:4260–4270. doi: 10.1128/IAI.71.8.4260-4270.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Traving C, Schauer R, Roggentin P. 1994. Gene structure of the ‘large’ sialidase isoenzyme from Clostridium perfringens A99 and its relationship with other clostridial NanH proteins. Glycoconj J 11:141–151. doi: 10.1007/BF00731154. [DOI] [PubMed] [Google Scholar]
- 45.Carman RJ, Sayeed S, Li J, Genheimer CW, Hiltonsmith MF, Wilkins TD, McClane BA. 2008. Clostridium perfringens toxin genotypes in the feces of healthy North Americans. Anaerobe 14:102–108. doi: 10.1016/j.anaerobe.2008.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Collie RE, McClane BA. 1998. Evidence that the enterotoxin gene can be episomal in Clostridium perfringens isolates associated with non-food-borne human gastrointestinal diseases. J Clin Microbiol 36:30–36. [DOI] [PMC free article] [PubMed] [Google Scholar]