Eosinophilic esophagitis (EoE) is associated with eosinophil and mast cell accumulation, which promotes dysphagia and esophageal dysmotility.1, 2 Cytokines and chemokines implicated in eosinophil-, mast cell–, and basophil-mediated EoE pathogenesis include interleukin (IL)-5, IL-13, thymic stromal lymphopoietin, and eotaxin-3.3, 4, 5, 6 A correlation between IL-5 and eotaxin-3 expression and eosinophil infiltration has been observed.7 Nevertheless, it is not clear if eotaxin-3 is the only chemokine, or one of several, that directs eosinophil accumulation in EoE. Herein, we present evidence that a neuroimmune pathway is involved in eosinophil and mast cell accumulation and degranulation in human EoE.
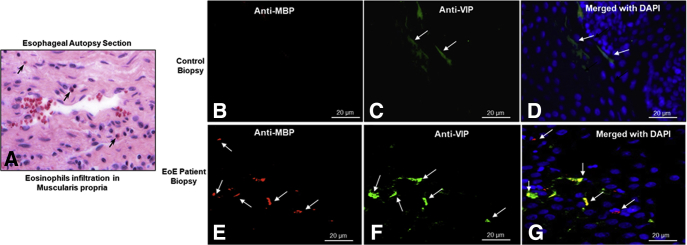
Morphologically, eosinophils accumulate near nerves within the muscular mucosa of the esophagus (Figure 1A). This spatial association suggests that nerve cells may release chemoattractants in EoE. We hypothesize that one such chemoattractant might be vasoactive intestinal peptide (VIP), which has been implicated in eosinophil recruitment during allergic disease (eg, asthma). VIP expression was low in control esophageal biopsy specimens (ie, without eosinophils) (Figure 1B–D). In contrast, eosinophils accumulated adjacent to VIP-expressing nerve cells in EoE biopsy specimens (Figure 1E–G).
Figure 1.
Eosinophils accumulate adjacent to nerve cells in the muscular mucosa in patients with a gastrointestinal disorder. (A) Eosinophil accumulation in the esophageal muscularis propria of an undiagnosed human autopsy sample. (B–D) Anti-major basic protein–positive eosinophils (red) were not detected in control esophageal biopsy specimens, and VIP expression (green) in nerves was low. (E–G) Increased eosinophil numbers and VIP expression were observed in EoE. Arrows indicate anti-major basic protein– and anti-VIP– immunostained eosinophils and nerve cells, respectively. DAPI, 4′,6-diamidino-2-phenylindole. n = 1 for esophageal autopsy, n = 4 for control, and n = 6 for EoE patient biopsies.
VIP performs its immunologic functions via binding to 3 receptors: vasoactive intestinal peptide receptor 1 (VPAC-1), VPAC-2, and chemoattractant receptor homologous molecule expressed on Th2 (CRTH2) lymphocytes.8, 9 Our in vitro analyses indicated that eosinophils mainly express CRTH2 receptor, not VAPC1, VAPC2, and VIP (Supplementary Figure 1A–D). Furthermore, eosinophil motility in response to VIP is comparable with that induced by eotaxin (Supplementary Figure 1E), and that anti-CRTH2 pretreatment restricted human eosinophil motility (Supplementary Figure 1F). In EoE biopsy specimens, we show that CRTH2-expressing eosinophils accumulated in the epithelial mucosa (Supplementary Figure 2A–F). Furthermore, mast cell contribution to the pathogenesis of EoE also has been reported, but the mechanism of mast cell recruitment to the esophagus is undefined. Immunofluorescence analyses confirmed CRTH2 receptor on tryptase-positive mast cells in the esophageal mucosa of EoE patients (Supplementary Figure 2G–L). These findings suggest that, similar to eosinophils, mast cells accumulate via interaction of the CRTH2 receptor with neurally derived VIP. The details of control and EoE patient clinical characteristics are provided in Supplementary Table 1.
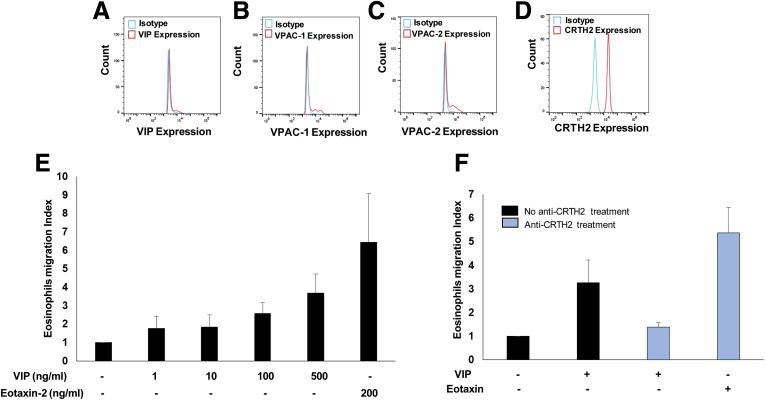
Supplementary Figure 1.
Eosinophils highly express the VIP-receptor CRTH2 compared with VPAC-1 or VPAC-2. An earlier report indicated that intestinal eosinophils produce VIP10; therefore, we examined whether blood eosinophils of EoE patients also produce VIP and express VIP-specific receptors. Accordingly, human eosinophils were examined for the expression of VIP and VIP-associated receptors using anti-VIP, anti-VAPC1, anti-VACP2, and anti-CRTH2 antibodies. (A–C) The flow cytometer analysis indicated none to very low expression of VIP and other VIP receptors VPAC-1 and VPAC-2 on eosinophils. (D) However, our analysis detected highly expressed VIP-receptor CRTH2 on eosinophils. Furthermore, we tested the hypothesis of whether the CRTH2 receptor has a critical role in VIP-induced eosinophil motility. Notably, eotaxins and the interaction of its receptor CCR3 are reported to be major motility factor for eosinophils.11, 12 Accordingly, we kinetically examined human eosinophil motility in response to different concentrations of VIP recombinant protein (0, 1, 10, 100, and 500 ng/mL) using Transwell chemotactic chambers for 4 hours. Established eosinophil chemokine eotaxin-2 (200 ng/mL) recombinant protein was used as a positive control. (E) The analysis indicated that VIP indeed has eosinophil chemoattraction activity similar to eotaxin-2. The critical role of CRTH2 in eosinophil motility was established by examining anti-CRTH2 pretreated eosinophil motility in response to VIP. The eosinophils were treated with anti-CRTH2 for 1 hour, and both treated and nontreated eosinophil motility was again examined using Transwell chambers in response to VIP protein (500 ng/mL) and eotaxin-2 protein (200 ng/mL). (F) Analysis indicated that anti-CRTH2 antibody blocks in vitro migration of eosinophils in response to VIP; whereas, anti-CRTH2 treatment did not restrict eosinophil motility in response to eotaxin-2. The analysis establishes the critical role of CRTH2 and VIP interaction in eosinophil motility. Data are expressed as means ± SD (n = 3 experiments).
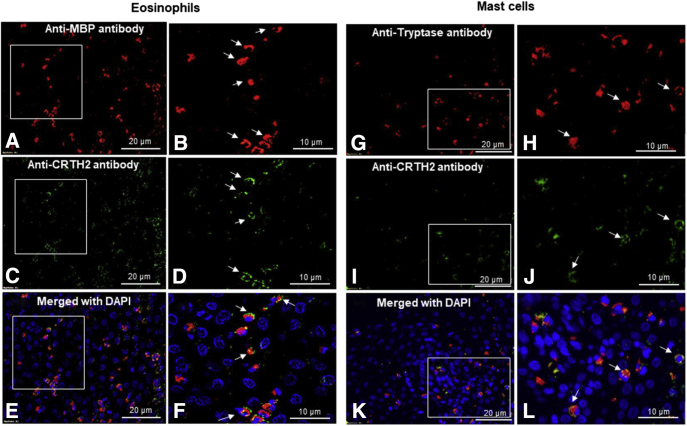
Supplementary Figure 2.
CRTH2 receptor is expressed on the eosinophils and tissue-accumulated mast cells in the esophageal biopsy specimen of human EoE. The in vitro analysis indicated that VIP-receptor CRTH2 is critical for eosinophil motility. Therefore, we examined whether tissue eosinophils in human EoE similarly express CRTH2 receptors. Accordingly, immunofluorescence analysis was performed using anti-CRTH2 receptor on anti-major basic protein–positive tissue eosinophils in human EoE biopsy specimens. (A and B) The anti-major basic protein– and (C and D) anti-CRTH2–expressed eosinophils are detected in the biopsy specimens of human EoE (A–D, original magnification, 400× and 1000×). The merged photomicrograph of anti-major basic protein– and anti-CRTH2–stained and 4′,6-diamidino-2-phenylindole (DAPI)-mounted tissue sections show that tissue eosinophils expressed VIP-receptor CRTH2 in the esophageal biopsy specimens of human EoE patients (E and F, original magnification, 400× and 1000×). Furthermore, induced mast cell numbers and their role in the pathogenesis of EoE is well established.4, 13 However, it is not clear which chemokine receptors are responsible for mast cell recruitment in human EoE. Therefore, we examined VIP-receptor CRTH2 expression on the tissue-accumulated mast cells in the esophageal biopsy specimens of human EoE by performing immunofluorescence analyses using antitryptase and anti-CRTH2 antibodies. We showed the presence of antitryptase-positive mast cells in the (G and H) esophageal biopsy specimens and (I and J) CRTH2-stained receptors. (K and L) A DAPI-mounted merged tissue section detected the co-localized tryptase-expressed mast cell expressing CRTH2 receptor in esophageal biopsy specimens of EoE patients. The arrows indicate tissue-accumulated mast cells and CRTH2 receptors on mast cells in respective photomicrographs. The photomicrographs presented are (A, C, and E) 400× and (B, D, and F) 1000× of the original magnification photomicrographs presented. Data are expressed means ± SEM (n = 6–7).
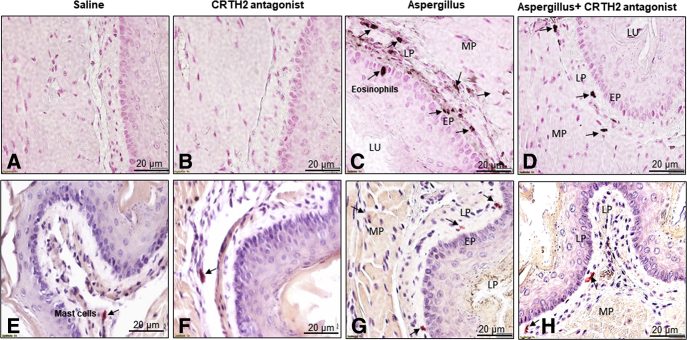
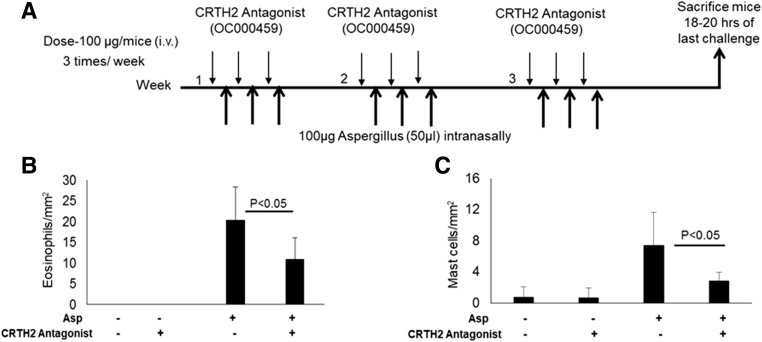
Previous clinical trials have shown a reduction of peak eosinophil levels in adult EoE patients treated with a CRTH2 antagonist.10 However, the effect of the CRTH2 antagonist on mast cells that may be critical to the esophageal functional abnormalities observed in EoE was not examined. Therefore, to better assess the potential therapeutic utility of in vivo CRTH2 blockade, we initially assessed eosinophil and mast cell distributions in mice after induction of experimental EoE (Supplementary Figure 3).11 As in the human study, eosinophil infiltration in each segment of the esophagus was reduced in CRTH2 antagonist-treated mice (Figure 2A–D). This was paralleled by reduced mast cell numbers (Figure 2E–H). Morphometric quantification analysis indicated that CRTH2 antagonist treatment significantly reduced the number of both eosinophils and mast cells (Supplementary Figure 4B and C).
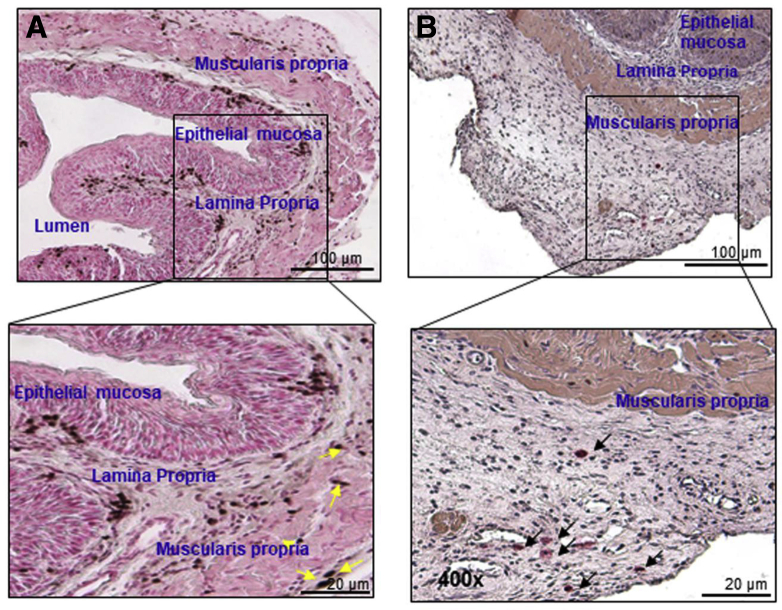
Supplementary Figure 3.
Accumulation of eosinophils and mast cells in muscular mucosa after the induction of experimental EoE. Induced eosinophils and mast cell accumulation is implicated in the induction of esophageal functional abnormalities, including stricture and motility dysfunction in human and experimental EOE.14 However, the accumulation and mechanism of eosinophils and mast cells beyond the epithelial mucosa has not been examined. Because it is difficult to obtain deep mucosal biopsy specimens in human EoE, we examined the accumulation of eosinophils and mast cells in each segment of the mouse esophagus in experimental EoE. The mouse esophageal tissue sections were examined for eosinophils and mast cells after anti-MBP and chloroacetate esterase staining, respectively. Both eosinophils and mast cells were detected in each segment of mouse esophagus after the induction of experimental EoE. (A) A number of eosinophil accumulations in low and high magnification are shown in the epithelial mucosa, lamina propria, and muscular mucosa. Yellow arrows indicates accumulation of eosinophils. (B) Similarly, mast cell accumulation in low and high magnification is shown mostly in the lamina propria and muscular mucosa. Black arrows indicates accumulation of mast cells. Photomicrographs presented are 100× and 400× original magnification, respectively.
Figure 2.
CRTH2 antagonist pretreatment reduces the accumulation of both eosinophils and mast cells in Aspergillus-induced experimental EoE. CRTH2 antagonist pretreatment significantly reduced the number of (D) eosinophils and (H) mast cells relative to (C and H) Aspergillus-challenged mice that were not pretreated. (A and B) No eosinophils and few (E and F) baseline mast cells were detected in sections from mice treated with saline or saline + CRTH2 antagonist. Data are expressed as means ± SD (n = 8–10 mice/group). EP, epithelial mucosa; LP, lamina propria; LU, lumen; MP, muscularis propria.
Supplementary Figure 4.
(A) BALB/c mice challenged with Aspergillus were treated with a CRTH2 antagonist (OC000459) as per the protocol of experimental EoE. Morphometric quantification indicated that CRTH2-antagonist treatment reduced the (B) number of eosinophils and (C) mast cells that accumulate in the esophagus of Aspergillus-challenged mice relative to the untreated Aspergillus-challenged control mice. The levels of eosinophils and mast cells in the esophageal sections are expressed as eosinophils/mm2 and mast cells/mm2, respectively. Data are expressed as means ± SD (n = 8–10 mice/group).
Taken together, the current studies identify a novel and important chemoattractant role for VIP in the accumulation of eosinophils and mast cells in the pathogenesis of EoE. Moreover, we suggest that inhibiting the VIP–CRTH2 axis may ameliorate the dysphagia, stricture, and motility dysfunction of chronic EoE.
Footnotes
Author contributions Alok K. Verma and Murli Manohar acquired data and performed the data statistical analysis; Sathisha Upparahalli Venkateshaiah provided technical support, figures, and edited the manuscript; Uwe Blecker provided eosinophilic esophagitis patient material support; Margaret Collins provided human esophageal autopsy specimens; and Anil Mishra was responsible for the study concept and design, interpretation, supervision, and funding support.
Conflicts of interest This author discloses the following: Anil Mishra has served as a consultant for Axcan Pharma, Aptalis, Elite Biosciences, Calypso Biotech SA, and Enumeral Biomedical. The remaining authors disclose no conflicts.
Funding This work was supported in part by National Institutes of Health grant R01 AI080581 (A.M.) and by the Tulane Edward G. Schlieder Educational Foundation.
Supplementary Methods
Clinical Characteristics of EoE and non-EoE patients
Formalin-fixed, paraffin-embedded biopsy samples were obtained from the esophagus of control patients or EoE patients as per an Institutional Review Board–approved protocol. The comparison groups included control patients (non-EoE), EoE, and dysphagia patients, who were selected without regard to age, atopic status, or sex. The diagnosis was established based on the maximum eosinophil count per high-power field (×400). Control patients (non-EoE) were defined as having 0–2 esophageal eosinophils/high-power field and no basal layer expansion. The normal biopsy specimens were obtained from patients who came to the clinic with symptoms typically noticed in EoE, but had completely normal esophageal endoscopic and microscopic analyses. Patients with EoE were defined as having 15 or more esophageal eosinophils/high-power field. The patients with EoE and dysphagia have more esophageal eosinophilia as compared with patients with only EoE along with basal cell hyperplasia. Blood was drawn in citrate-coated tubes from each control patient, EoE patient, and dysphagia patient a the time of scheduled endoscopy (including before and after treatment of EoE patients) at Tulane University School of Medicine. All samples were used according to the patients’ consent and Institutional Review Board–approved protocol at Tulane University School of Medicine (New Orleans, LA; years 2013–2018). Detailed control, EoE, and dysphagia patient characteristics including treatment are listed in Supplementary Table 1.
Eosinophil Detection in the Muscular Mucosa of Autopsy and Biopsy Tissue Sections
Eosinophils were detected in the muscular mucosa of esophageal autopsy (provided by Dr Margaret Collins, Cincinnati Children’s Hospital Medical Center) tissue sections stained with H&E.
Immunofluorescence Tissue Staining for Eosinophils, Mast Cells, Nerve Cells, VIP, and CRTH2 Receptors on Esophageal Biopsy Specimens of Human EoE Patients
Immunofluorescence staining of formalin-fixed, paraffin-embedded control patient and EoE esophageal biopsy sections was performed to analyze CRTH2-receptor expression on eosinophils and mast cells. Furthermore, eosinophil accumulation near nerve cells derived VIP after the immunofluorescence tissue staining methods was performed as described earlier.1, 2 Briefly, in formalin-fixed, paraffin-embedded sections from esophageal biopsy specimens, endogenous peroxide was quenched using 0.3% hydrogen peroxide in methanol, followed by antigen retrieval with pepsin, and blocking with 3% goat serum to reduce the nonspecific binding. Esophageal eosinophils were detected using rat anti-MBP (kindly provided by Dr Jamie Lee, Mayo Clinic, Scottsdale, AZ) antibody followed by Tetramethylrhodamine (TRITC)-labeled anti-rat IgG that was used as a secondary antibody. Furthermore, anti-CRTH2 antibody (ProSci) followed by fluorescein isothiocyanate–labeled anti-rabbit IgG (Biolegend, San Diego, CA) were used as a secondary antibody to detect CRTH2-receptor expression on eosinophils on esophageal biopsy specimens. VIP-expressing nerve fibers were detected using goat anti-VIP antibody followed by fluorescein isothiocyanate–anti-goat IgG antibody that were used as secondary antibodies to detect VIP-producing nerve cells. In addition, CRTH2-receptor expression on anti-human tryptase–positive (Bio-Rad) mast cells was detected using antitryptase antibody followed by Phycoerythrin-labeled anti-mouse IgG, which was used as a secondary antibody for mast cell and anti-CRTH2 antibody (ProSci, Poway, CA), followed by fluorescein isothiocyanate–labeled anti-rabbit IgG (Biolegend, San Diego, CA) as a secondary antibody for CRTH2 expression on mast cells in esophageal biopsy specimens. The double-immunostained tissue sections slides were mounted with 4′,6-diamidino-2-phenylindole mounting material (Thermo Fisher Scientific, Waltham, MA), and the images were captured using an Olympus-BX43F (Tokyo, Japan).
CRTH2 Antagonist Treatment in Experimental EoE
Specific pathogen-free BALB/c mice were obtained from The Jackson Laboratory (Bar Harbor, ME). All of the experiments were performed on age- and sex-matched mice 6–8 weeks of age. The Tulane Institutional Animal Care and Use Committee approved the animal protocols. Experimental EoE in mice was induced using methods described previously3, 4, 5 that were used in accordance with National Institutes of Health guidelines. In brief, mice were lightly anesthetized with isoflurane (Iso-Flo; Abbott Laboratories, North Chicago, IL), and 100 μg of Aspergillus fumigatus (Greer Laboratories, Lenoir, NC) in 50 μL normal saline or 50 μL normal saline alone was given intranasally using a micropipette with the mouse held in the supine position 3 times/wk for 3 weeks. In addition, 100 μg/mice CRTH2 antagonist (OC000459) (Cayman Chemical, Ann Arbor, MI) was given intravenously on an alternate day up to the last Aspergillus challenge. The mice were euthanized after 24 hours of their last intranasal allergen or saline challenge. Esophageal tissues sections were analyzed for eosinophils by anti-MBP immunostaining and for mast cells by chloroacetate esterase staining as per the earlier-described protocol.2, 6, 7
Flow Cytometer Analysis for VIP and VIP-Receptor Expression on Blood Eosinophils
VIP-, VPAC-1–, VPAC-2–, and CRTH2-receptor expression on blood eosinophils from EoE patients was tested by flow cytometric analysis as described earlier.8, 9 The blood cells were stained with different florescence-tagged anti-human C-C motif chemokine receptor 3 (hCCR3) (Biolegend, San Diego, CA), anti-human Sialic acid-binding Ig-like lectin 8 (hSiglec-8) (Biolegend), anti-hVIP, anti–hVPAC-1, anti–hVPAC-2, and anti-hCRTH2 antibodies (Santa Cruz Biotechnology, Dallas, TX, and Biolegend). Depending on availability, we used both fluorescence-tagged antibodies or the combination of both primary and secondary antibodies tagged with different florescent-tagged IgG (Santa Cruz Biotechnology, Biolegend, or eBioscience, San Diego, CA). Respective labeled IgG antibodies were used as isotype control. Fluorescence-activated cell sorter analysis was performed using a FACS Calibur (BD Biosciences, San Diego, CA) and analyzed by FlowJo software (BD Biosciences).
Eosinophil Migration Assay
The chemoattractant behavior of VIP for eosinophils was analyzed in vitro using Transwell units (24 wells) with 5-μm porosity polycarbonate filters (Corning, Inc, Corning, NY) following the previously described protocol.1 The human blood eosinophils were incubated with the anti-human CCR3 and anti-human Siglec-8 antibodies for 45 minutes, washed, and eosinophils were separated by fluorescence-activated cell sorter. The purified human eosinophils (105 cells/well) in Hank's balance salt solution, pH 7.2 (Life Technologies, Carlsbad, CA) were placed in the upper chamber and different concentrations of recombinant VIP (1, 10, 100, and 500 ng/mL) were added to the lower chamber. Eotaxin-2 (200 ng/mL), a known chemoattractant for eosinophils, was used as a positive control. The Transwell unit was kept at 37°C for 4 hours in a humidified 95% air–5% CO2 atmosphere. After 4 hours, media from the lower chamber was centrifuged at 250 g, and cells were resuspended in phosphate-buffered saline. The number of migrated cells in the lower chamber was counted with a hemocytometer. Each assay was set up in triplicate and repeated 3 times. The data are expressed as an eosinophil migration index, which is defined as the ratio of the migration of eosinophils in the presence of the chemoattractant VIP, and the migration of eosinophils to the medium control.
Statistical Analysis
Parametric data were compared using t tests between 2 groups. Values are reported as means ± SD. P values less than .05 were considered statistically significant.
Supplementary Table 1.
Patient Clinical and Pathologic Characteristics
| Patients | Age, y | Sex | Esophageal disease | Allergic diseases | Other diseases | Eos/HPF | Current treatment | Steroids |
|---|---|---|---|---|---|---|---|---|
| 1 | 9 | M | NL | None | None | 0 | - | |
| 2 | 11 | F | NL | None | None | 0 | - | |
| 3 | 12 | F | NL | None | None | 0 | - | |
| 4 | 9 | F | NL | None | None | 0 | - | |
| 5 | 4 | M | NL | None | None | 0 | - | |
| 6 | 13 | M | NL | None | None | 0 | Elimination diet | Nasocort, Flovent |
| 7 | 11 | M | NL | None | None | 0 | Food trial | - |
| 8 | 2 | M | EoE | None | None | 35 eos/HPF | Elimination | - |
| 9 | 9 | M | EoE | None | None | 40 eos/HPF | Elimination | - |
| 10 | 8 | M | EoE | None | None | 31 eos/HPF | Ad libitum | - |
| 11 | 10 | M | EoE | None | None | 41 eos/HPF | Ad libitum | Flovent (GlaxoSmithKline, Brentford, UK) |
| 12 | 12 | M | EoE | None | None | 63 eos/HPF | Elimination | Nasocort (Chattem, Inc, Chattanooga, TN) |
| 13 | EoE | None | None | 30 eos/HPF | - | Flovent | ||
| 14 | 3 | M | EoE | None | None | 30 Eos/HPF | Ad libitum | Pulmicort (AstraZeneca, Cambridge, UK) |
| 15 | 5 | M | EoE with dysphagia | None | None | 63 eos/HPF | - | Flovent |
| 16 | 10 | F | EoE with dysphagia | None | None | 80 eos/HPF | Ad libitum | - |
| 17 | 6 | M | EoE with dysphagia | None | None | 64 eos/HPF | - | Rhinocort (AstraZeneca), Flovent |
| 18 | 7 | M | EoE with dysphagia | None | Nonspecific colitis with focal cryptitis | 73 eos/HPF | Elimination | Flonase (GlaxoSmithKline) |
| 19 | 15 | M | EoE with dysphagia | None | None | 70 eos/HPF | Elimination | - |
| 20 | 8 | M | EoE with dysphagia | None | None | 50 eos/HPF | Ad libitum | - |
eos, eosinophils; F, female; M, male; NL, normal; HPF, high-power field.
References
- 1.Attwood S.E. Dig Dis Sci. 1993;38:109–116. doi: 10.1007/BF01296781. [DOI] [PubMed] [Google Scholar]
- 2.Martin Martin L. J Gastroenterol Hepatol. 2011;26:1447–1450. doi: 10.1111/j.1440-1746.2011.06770.x. [DOI] [PubMed] [Google Scholar]
- 3.Attwood S.E. Gastroenterol Clin North Am. 2014;43:185–199. doi: 10.1016/j.gtc.2014.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nonevski I. Gastroenterology. 2006;131:2018–2020. doi: 10.1053/j.gastro.2006.10.058. discussion 2020. [DOI] [PubMed] [Google Scholar]
- 5.Mishra A. J Immunol. 2002;168:2464–2469. doi: 10.4049/jimmunol.168.5.2464. [DOI] [PubMed] [Google Scholar]
- 6.Mishra A. J Biol Chem. 2002;277:4406–4412. doi: 10.1074/jbc.M110424200. [DOI] [PubMed] [Google Scholar]
- 7.Blanchard C. J Clin Invest. 2006;116:536–547. doi: 10.1172/JCI26679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dunzendorfer S. J Leukoc Biol. 1998;64:828–834. doi: 10.1002/jlb.64.6.828. [DOI] [PubMed] [Google Scholar]
- 9.El-Shazly A.E. J Biol Chem. 2013;288:1374–1384. doi: 10.1074/jbc.M112.422675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Straumann A. Allergy. 2013;68:375–385. doi: 10.1111/all.12096. [DOI] [PubMed] [Google Scholar]
- 11.Niranjan R. Am J Physiol Gastrointest Liver Physiol. 2013;304:G1087–G1094. doi: 10.1152/ajpgi.00070.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
References
- 1.Mavi P. Am J Physiol Gastrointest Liver Physiol. 2014;307:G499–G507. doi: 10.1152/ajpgi.00141.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rayapudi M. Clin Transl Immunol. 2014;3:e9. doi: 10.1038/cti.2013.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dutt P. Immunol Cell Biol. 2015;93:849–857. doi: 10.1038/icb.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rayapudi M. J Leukoc Biol. 2010;88:337–346. doi: 10.1189/jlb.0110025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rajavelu P. Am J Physiol Gastrointest Liver Physiol. 2012;302:G645–G654. doi: 10.1152/ajpgi.00223.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Niranjan R. Immunol Cell Biol. 2013;91:408–415. doi: 10.1038/icb.2013.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Venkateshaiah S.U. J Allergy Clin Immunol. 2017 Jun 9 pii: S0091-6749(17)30926-0. https://doi.org/10.1016/j.jaci.2017.05.025. [Epub ahead of print] [Google Scholar]
- 8.Zhu X. Am J Physiol Gastrointest Liver Physiol. 2009;297:G550–G558. doi: 10.1152/ajpgi.00148.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu X. Gastroenterology. 2010;139:182–193 e7. doi: 10.1053/j.gastro.2010.03.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Metwali A. J Neuroimmunol. 1994;52:69–78. doi: 10.1016/0165-5728(94)90164-3. [DOI] [PubMed] [Google Scholar]
- 11.Lacy P. Int Arch Allergy Immunol. 2011;156:137–147. doi: 10.1159/000322597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hornung D. J Clin Endocrinol Metab. 2000;85:2604–2608. doi: 10.1210/jcem.85.7.6665. [DOI] [PubMed] [Google Scholar]
- 13.Niranjan R. Am J Physiol Gastrointest Liver Physiol. 2013;304:G1087–G1094. doi: 10.1152/ajpgi.00070.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mavi P. Am J Physiol Gastrointest Liver Physiol. 2012;302:G1347–G1355. doi: 10.1152/ajpgi.00013.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]