Abstract
Most bacteria encode a large repertoire of RNA-based regulatory mechanisms. Recent discoveries have revealed that the expression of many genes is controlled by a plethora of base-pairing noncoding small regulatory RNAs (sRNAs), regulatory RNA-binding proteins and RNA-degrading enzymes. Some of these RNA-based regulated processes respond to stress conditions and are involved in the maintenance of cellular homeostasis. They achieve it by either direct posttranscriptional repression of several mRNAs, including blocking access to ribosome and/or directing them to RNA degradation when the synthesis of their cognate proteins is unwanted, or by enhanced translation of some key stress-regulated transcriptional factors. Noncoding RNAs that regulate the gene expression by binding to regulatory proteins/transcriptional factors often act negatively by sequestration, preventing target recognition. Expression of many sRNAs is positively regulated by stress-responsive sigma factors like RpoE and RpoS, and two-component systems like PhoP/Q, Cpx and Rcs. Some of these regulatory RNAs act via a feedback mechanism on their own regulators, which is best reflected by recent discoveries, concerning the regulation of cell membrane composition by sRNAs in Escherichia coli and Salmonella, which are highlighted here.
Keywords: envelope stress, noncoding RNA, RfaH, RpoE sigma factor, stress response, two-component system
Introduction
Small regulatory RNAs (sRNAs) are ubiquitous multipurpose regulators present in all domains of life. They act via complex interactions with DNA/chromatin, proteins, other RNA molecules and metabolites [1]. In bacteria, generally their transcript size varies from 50 to 300 nucleotides. Base-pairing sRNAs can be either cis-acting or trans-acting, depending on their genomic location and with respect to their target mRNAs [2,3]. Many of these sRNAs function by imperfect base pairing with multiple mRNA targets and often require the help of the RNA chaperone Hfq [2]. Base-pairing sRNAs act as major posttranscriptional regulators that can either enhance or repress mRNA decay and/or translation by binding to the 5′ untranslated region (UTR) of mRNA targets [2,3]. However, some sRNAs function by the interaction with regulatory proteins, often acting by sequestration. Initially, sRNAs were identified by the analysis of transcription from intergenic regions [4]. Extensive RNA-seq analysis of transcriptome of various regulators, identification of RNAs by ligating Hfq-bound sRNAs and sequencing sRNA–RNA interactions (RIL-seq) or by UV-cross-linking sRNA–target RNA duplexes to RNase E (CLASH) or immunoprecipitations with Hfq and ProQ have vastly increased the repertoire of sRNAs and their targets [5–8]. These studies have revealed that sRNAs can originate from various genomic regions, including antisense to coding regions [6,8], 3′ UTRs [9–12], 5′ UTRs [13] and even coding regions [6] (Figure 1). Even in bacteria that lack Hfq, like Mycobacterium tuberculosis, the noncoding RNA content can be up to 58% of the total non-rRNA in the stationary phase [14]. Specific sRNAs control the expression of sigma factors like the stationary-phase sigma factor RpoS, the envelope stress-responsive sigma factor RpoE and the activity of house-keeping sigma factor RpoD [2,3,10]. The sRNA-mediated control has also been described for several regulatory factors like FlhDC (motility/flagellar expression), Lrp (amino acid biosynthesis), CRP (catabolite repression) and SoxS (oxidative stress) [15–17]. Thus, sRNAs are deployed by each of the major stress-responsive pathways, controlling the expression of crucial regulatory molecules that can rapidly alter gene expression profiles, allowing fast adjustment to different growth conditions.
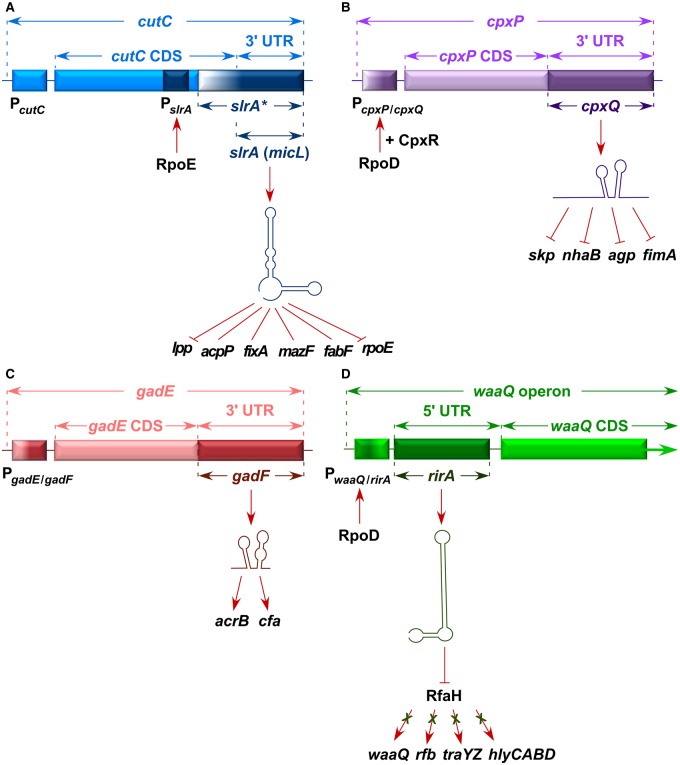
Figure 1. Novel regulatory sRNAs located either in 3′ or 5′ UTR of various mRNAs.
(A) Location of the SlrA (MicL) sRNA within the 3′ UTR of the cutC gene that is transcribed from its own RpoE-regulated promoter. Known and potential targets are indicated [6,10,11]. slrA* depicts its precursor form. (B) The 3′ UTR-located CpxQ sRNA is generated by processing of the cpxP mRNA [12]. Targets that are repressed by CpxQ are shown. (C) A novel 3′ UTR-located gadF sRNA is generated by processing of the gadE mRNA and positively regulates the expression of acrB and cfa genes [6]. (D) The RirA sRNA shares its 5′ end with the waaQ mRNA and interacts with RfaH, preventing target recognition of ops-containing operons [13].
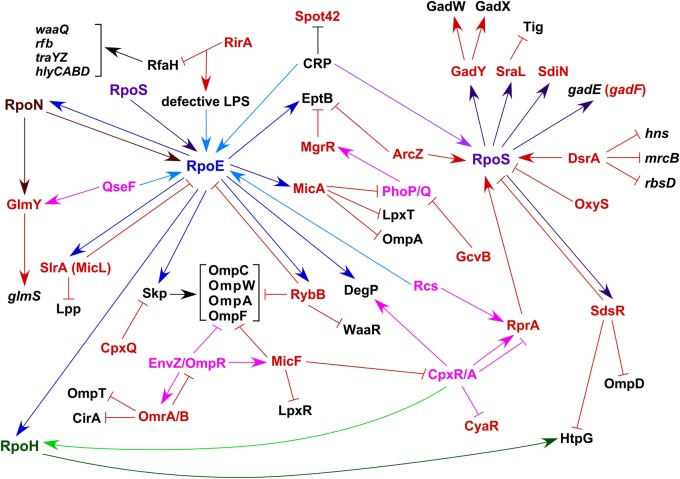
Transcription of most trans-acting sRNAs is controlled by transcriptional factors that include global regulators, two-component systems (TCSs) and specific stress-responsive sigma factors [17,18]. One of the well-studied cases is the regulation of envelope stress response by the RpoE sigma factor, the Cpx TCS and regulatory sRNAs that are members of their regulons [12,18]. RpoE is specifically induced in response to outer membrane protein (OMP) misfolding, defects in the lipopolysaccharide (LPS) composition and an imbalance between phospholipids and LPSs [10]. Interestingly, RpoS, RpoN, RpoD, RpoE sigma factors and TCSs, like Rcs and QseE/F, positively regulate transcription of the rpoE gene in response to specific stress conditions (Figure 2) [13]. However, the RpoE availability/activity is also subjected to a negative regulation by the RseA anti-sigma factor and a feedback control by RpoE-regulated sRNAs [10,19,20]. As several sRNAs, belonging to stress-responsive regulons, also act in a negative feedback mechanism, this suggests functional overlaps and co-integration of signals to achieve homeostasis. Consistent with integration of diverse signals, the expression of many stress-responsive regulators and sigma factors are regulated by sRNAs (Figure 2) [17,18]. In this review, we address how major stress-responsive regulatory systems engage sRNAs to regulate the envelope stress response and co-ordinate the gene expression upon other specific stresses.
Figure 2. Stress-responsive sigma factors regulated sRNAs.
The network of the repressing arm of RpoE/Cpx systems exerted via its sRNA regulon members and the hub of some of sRNAs that either regulate the RpoS expression or are regulated by RpoS. sRNAs are shown in red color and pink color refers to two-component systems. The repressive arm of RpoE by RybB, MicA and SlrA (MicL) that repress OMP synthesis is presented. Suppression of Skp synthesis by the CpxQ sRNA is also shown. RpoS-regulated sRNAs are indicated and their major targets are pointed.
Regulation of the RpoE-dependent extracytoplasmic stress response by sRNAs
The cell envelope of Gram-negative bacteria, such as Escherichia coli or Salmonella, is composed of an asymmetric outer membrane (OM) that is separated from the inner membrane (IM) by the periplasmic space containing the peptidoglycan. The content and assembly of OM components, such as OMPs, lipoproteins and LPSs, are tightly regulated. This involves control mechanisms exerted by sRNAs and cell envelope stress-responsive pathways that detect and respond to any dysfunction in the assembly or an imbalance in different components of the cell envelope. Hence, defects in either OMP assembly or LPS synthesis or exposure to stress conditions induce cellular responses that are transcriptionally regulated by the alternative sigma factor RpoE and TCSs like CpxR/A, Rcs, PhoP/Q, EnvZ/OmpR and BasS/R [21,22]. Some of these regulons often encode sRNAs that serve to provide a rapid response and can either amplify the signal or act by a negative feedback. Furthermore, some response regulators are subjected to a posttranscriptional regulation by sRNAs, as is the case with the RpoE-regulated MicA-mediated repression of phoP mRNA synthesis linking RpoE and the PhoP/Q TCS to control OM constituents: OMPs and LPSs [23]. MicA is a major sRNA initially identified as the posttranscriptional repressor of ompA mRNA synthesis [24]. Similarly, two OmpR-regulated sRNAs, OmrA and OmrB, regulate the expression of certain OMPs like OmpT and also provide a negative feedback to the OmpR regulator [25].
RpoE-regulated MicA and RybB control of OMP synthesis
The RpoE sigma factor is specialized in its function dedicated to the maintenance of OM homeostasis. The coding arm of the RpoE regulon includes factors that promote OMP synthesis/folding, LPS remodeling and its assembly/transport [18,26]. Defects in OMP or LPS biosynthesis strongly induce the RpoE activity, and here the noncoding arm of the RpoE regulon counteracts deleterious effects of the RpoE expression [13,20–22]. Previously, it was noted that overexpression of RpoE or activation of the RpoE pathway in the absence of the anti-sigma factor RseA leads to repression of OMP synthesis [26,27]. This is now best explained by discoveries of three well-conserved RpoE-transcribed Hfq-binding sRNAs rybB, micA and slrA (micL), which repress the synthesis of abundant OMPs (RybB- and MicA-dependent) and the most abundant lipoprotein Lpp [SlrA (MicL)-dependent] (Figure 2). RybB and MicA together target >30 mRNAs of E. coli [20], providing a repressive arm that alleviates stress and mediating interconnection with other envelope stress response networks. MicA and RybB target all omp mRNAs and some non-omp targets. Thus, MicA represses the synthesis of major OMPs by targeting mRNAs of ompA, ompX and lamB and non-OMP targets like phoP, lpxT and htrG [20,23]. RybB has even more in vivo omp mRNA targets and non-omp targets that include waaR, htrG, fadL and rbsK/B, which are repressed by RybB (Figure 2) [20,22,28]. RybB and MicA bind their target mRNAs using a seed region of 6–8 contiguous bases for base pairing [2,20,29]. This base pairing, involving the seed region of sRNA, can occur close to the ribosome-binding site or within so-called 5-codon region located close to the 5′ end of the coding sequence or located deep within the coding sequence [30]. The interaction within the coding sequence between sRNA and target mRNA can cause mRNA decay without ribosome binding [30]. Overall, the main function of either MicA or RybB sRNA is to protect the cell from the loss of viability when the RpoE activity is inadequate.
The negative feedback control of RpoE by the RpoE-regulated SlrA (MicL) and repression of Lpp synthesis
SlrA (MicL) was identified independently by two groups, during characterization of suppressors of ΔlapB mutants that overproduce LPSs, called SlrA [10], and during examination of RpoE-regulated sRNAs, called MicL [11]. The lapB gene is essential and its absence leads to increased synthesis of LPS at the expense of phospholipids, aggregation of proteins involved in LPS biosynthesis and decoupling of LPS synthesis with LPS transport [10]. Consequently, ΔlapB exhibits hyper-elevated levels of RpoE and Cpx regulons. The main function of SlrA (MicL) was shown to be the Hfq-dependent inhibition of lpp mRNA and repression of the RpoE activity. In support of these results, overproduction of SlrA mimics Δlpp phenotype [10]. Since Lpp contains three acyl chains, reduction in Lpp synthesis by SlrA can liberate pools of phospholipids that can overcome toxicity due to the excess of LPS and depletion of the common precursor for phospholipids. Interestingly, SlrA (MicL) is encoded within the cutC gene and is transcribed from its own RpoE-regulated promoter [10,11]. This sRNA is synthesized as a precursor of 308 nt and is processed to a mature 80-nt RNA located within the 3′ UTR of the cutC gene (Figure 1A). Importantly, overexpression of slrA strongly represses elevated toxic levels of the rpoE regulon in ΔlapB and hence overcomes ΔlapB defects [10]. This negative feedback control of the RpoE expression by increased synthesis of SlrA provides a robust mechanism of maintaining cellular homeostasis by the sRNA arm of RpoE. Indeed, recent global studies have revealed mRNAs of rpoE, acpP, fabF, mazF and fixA as additional base-pairing targets of SlrA (MicL) [6]. If the translation of the rpoE mRNA is blocked by SlrA, this may explain the lack of any significant increase in the RpoE amount under conditions that increase the RpoE transcriptional activity. For instance, in E. coli, severe defects in LPSs known to trigger RpoE induction have unaltered total amount of RpoE [31], since under such conditions transcription of the slrA gene will also be induced counteracting the increase in synthesis of RpoE. However, it is pertinent to point out that burst in the RpoE activity when LPS is defective is due to the liberation of RpoE from RseA [31]. Taken together, three RpoE-regulated sRNAs (RybB, MicA and SlrA) provide a negative feedback arm, as in their excess the induction of RpoE stress response is dampened by reducing unwanted buildup of OMPs under stress conditions and this could allow redirection of RpoE-regulated chaperones to mediate protein folding in the periplasm.
The sRNA-dependent remodeling of LPS and integrated control by RpoE and PhoP/Q
Under RpoE-inducing conditions, E. coli remodels its LPS synthesis that is controlled by RpoE-regulated sRNAs, particularly RybB. Upon the RpoE induction, the RybB sRNA represses the synthesis of WaaR glycosyltransferase, causing truncation in the outer core of LPSs and concomitant incorporation of a third Kdo (WaaZ-dependent) and rhamnose in the inner core [22]. Interestingly, under such conditions, the waaZ mRNA accumulation also increases, contributing to the synthesis of LPS with a third Kdo [22]. This mode of LPS remodeling could be adaptive in function, as the non-essential waaZ gene becomes essential for the bacterial growth when the LPS is tetraacylated and for interaction with the host, since such an LPS cannot ligate O-antigen due to loss of the attachment site [22]. This drastic shift causing the accumulation of a rare form of LPS is accompanied by a nonstoichiometric modification of the second Kdo by phosphoethanolamine (P-EtN). The transfer of P-EtN to the second Kdo is important for resistance to cationic antibiotics like polymyxin B and is mediated by the product of the RpoE-regulated eptB gene [32]. The eptB mRNA synthesis is usually repressed by the PhoP/Q-regulated MgrR and by the ArcZ sRNAs [32]. However, this silencing by MgrR and ArcZ is overcome when RpoE is induced. MicA contributes to remodeling of LPSs by inhibiting phoP and lpxT synthesis at the posttranscriptional level by direct base pairing with these target mRNAs [20,23]. The PhoP/Q TCS regulates LPS nonstoichiometric alterations and is required for transcription of the mgrR sRNA. The MicA-directed repression of LpxT is interesting, since LpxT mediates the phosphorylation of lipid A that causes an increase in the negative charge of LPS. The GcvB sRNA also negatively regulates the phoPQ expression that is independent of MicA [33]. Interestingly, SoxS, a global regulator of the oxidative stress response, is a positive regulator of transcription of the waaY gene, whose product phosphorylates heptose II and is itself subjected to a negative regulation by the MgrR sRNA [16]. Another sRNA that regulates LPS modification is MicF, since base pairing of MicF within the coding sequence of the lpxR mRNA decreases its stability by rendering it susceptible to degradation by RNase E [34]. LpxR is a lipid A deacylase and this modification occurs after LPS translocation. Hence, the regulation of LpxR by MicF contributes to the LPS modification event that occurs in the OM, expanding the role of sRNAs at various steps of the LPS biosynthesis [18]. Such controls exerted by sRNAs on regulators of the stress response that at the same time regulate LPS structural alterations demonstrate rewiring of networks to cope with envelope stress.
Regulation of the envelope stress response by the Cpx TCS and mediation by the CpxQ sRNA
The CpxR/A TCS is one of the typical TCSs with the IM-located CpxA acting as a histidine kinase and CpxR being the DNA-binding response regulator. The Cpx system responds to protein folding defects in the periplasm and defects in peptidoglycan, and is induced by variety of stresses that include high pH, hyperosmolarity and alterations in the IM lipid composition, and when LPS is composed of either Kdo2-lipid IVA or only free lipid IVA derivatives or in ΔlapB mutants [10,21,35]. The activity of the Cpx system is further fine-tuned by PrpA (serine/threonine phosphatase) that modulates the phosphorylation status of CpxR [36]. Elucidation of RpoE and Cpx regulon members revealed overlap in the RpoE and Cpx pathways, although certain signals are specific for each system [26,35]. The Cpx system is generally activated upon IM perturbations, while RpoE strongly responds to OM signals. The Cpx activity is regulated by the negative feedback mediated by a periplasmically located CpxP protein [35]. Transcription of the cpxP gene is strongly activated upon the induction of the Cpx pathway. As in the case of RpoE, the sRNA-mediated regulatory control has been discovered, which plays an important role in regulating the Cpx TCS and in interconnections with other envelope stress systems. Importantly, it was shown that the transcript of the cpxP gene includes a novel sRNA cpxQ, located within its 3′ UTR [12]. This sRNA uses the same promoter as the cpxP gene and is generated by the RNase E-mediated processing of the 3′ end of cpxP mRNA (Figure 1B). CpxQ in conjunction with Hfq represses the synthesis of NhaB, Skp, Agp and FimA [12]. NhaB acts as a sodium–proton antiporter and its repression by CpxQ can counteract the loss of proton motive force (PMF) during the IM stress and can provide resistance to dissipation of membrane potential. Consistent with these findings, the loss of PMF is a potent inducer of the Cpx TCS and can thus provide a basis for maintaining homeostasis during envelope stress [12]. Repression of Skp synthesis, a well-known OMP chaperone [37], by CpxQ is another mechanism of counteracting the induction of Skp synthesis upon the RpoE induction that can balance RpoE and Cpx systems. Indeed, in E. coli, it was shown that CpxQ combats toxicity in the IM by down-regulating the synthesis of Skp, preventing insertion of OMPs into the IM by Skp [38]. Thus, the function of cpxQ along with the Cpx system caters to the need of protection against IM damage.
Interestingly, CpxR positively regulates synthesis of OmrA, OmrB, RprA and MicF sRNAs, while repressing the synthesis of CyaR. Targets of OmrA/B sRNAs include mRNAs of several OMP-encoding genes like fecA, ompT, cirA and fepA [25]. In this process, OmrA/B further provides a negative feedback loop to the EnvZ/OmpR system [25]. MicF has been well studied for its negative regulation of OmpF and, in these pathways, the EnvZ/OmpR TCS cross-talks with the Cpx system via RpoE/Cpx-regulated EcfM (MzrA) [26,35]. Since CpxR positively regulates the synthesis of rprA sRNA, it provides a link for Rcs, RpoS and Cpx pathways in the control of stress response [13,35]. The RprA sRNA positively regulates the rpoS expression and is a member of the Rcs regulon. The Rcs system and RpoS further regulate transcription of the rpoE gene in response to LPS defects and entry into the stationary phase by controlling the expression of specific promoters of the rpoE gene [13]. Thus, these noncoding RNAs, sigma factors and TCSs co-regulate the envelope stress response.
The RpoS-regulated stationary-phase response and a role of sRNAs
The RpoS sigma factor is a global regulator of stress response, transcribing nearly 10% of genes, and acts as a major hub for sRNAs, modulating the expression of nearly 23% sRNAs in Salmonella [39]. RpoS regulon members exhibit overlaps with other stress-responsive controlling sigma factors and TCSs (Figure 2). The expression of the rpoS gene is positively regulated by DsrA, RprA and ArcZ and negatively by OxyS sRNAs [40]. Positively regulating base-pairing sRNAs activate the expression of RpoS by binding to the 5′ UTR of rpoS mRNA in an Hfq-dependent manner by releasing the inhibitory secondary structure and enhancing translation [40]. The RprA expression is activated under acid stress conditions, induction of the Rcs system and exposure to chemical stresses like the presence of succinic acid, acetate and itaconic acid [41]. ArcZ overexpression results in differential expression of over 750 genes and loss of motility [42]. The ArcZ sRNA serves as a connector between the general stress response regulated by RpoS and the specific response mediated by the ArcA/B TCS that responds to a redox state of the respiratory chain. RpoS is directly involved in the transcriptional activation of the rpoE gene via the recognition of the rpoEP4 promoter, which is one of its most active promoters [13]. RpoS directly regulates transcription of gadY, omrA, sdsR, sdiN and sraL sRNAs [17,43,44]. RpoS regulates acid resistance by the induction of transcriptional factors GadE/X/W. While the gadY sRNA positively regulates GadX and GadW [2,3], gadE contains a novel sRNA gadF in its 3′ UTR [6]. Overexpression of gadF sRNA up-regulates acrB and cfa expression. Thus, the GadE transcription factor and GadF sRNA are encoded in the same transcript and co-regulate genes in the same pathway (Figure 1C) [6]. The RpoS-regulated SdsR sRNA controls biofilm formation, down-regulation of an abundant porin OmpD in Salmonella and the expression of mismatch repair protein MutS [17]. SdsR could play additional roles in stress response with the identification of 20 new down-regulated targets that include HtpG (conserved heat shock protein), global regulators like CRP (catabolite repression), StpA (nucleoide-associated protein) and TolC (efflux pump protein) [17]. SraL is quite strongly induced in an RpoS-dependent manner, and it targets the tig mRNA in Salmonella [43]. The tig gene encodes the ribosome-associated abundant peptidyl prolyl cis/trans isomerase. Repression of the Tig expression under the stationary phase could be a part of the homeostasis mechanism to reduce protein synthesis. Two small sRNAs, SdsN137 and SdsN178, are transcribed from two RpoS-dependent promoters, but share the same terminator [44]. Overexpression of SdsN137 causes repression of NfsA nitroreductase and hence regulates resistance to nitrofurans [44]. Taken together, RpoS-regulated sRNAs connect various stress regulons like RpoE and RpoH, total protein expression and response to acid resistance under stationary-phase conditions.
sRNAs that act by interacting with regulatory proteins
Some sRNAs regulate the gene expression by interacting with regulatory proteins, modifying their activity, and generally act via protein sequestration. Two families of such widely conserved sRNAs, represented by the CsrB/RsmZ family and by 6S RNA, have been well characterized. In addition, a new sRNA RirA interacts with RfaH, and this interaction potentially alters the RfaH conformation, rendering it NusG-like, thereby reducing RfaH's ability to recognize its target operons.
The CsrB family of sRNAs
The CsrB family of sRNAs uses short sequence AUGGA motifs in apical loops of RNA secondary structure to bind multiple copies of carbon storage regulator A (CsrA), leading to its sequestration. CsrA acts as a translational repressor of several mRNAs and regulates a plethora of phenotypes, including carbon storage, secondary metabolism, motility, biofilm formation, peptide uptake, cyclic di-GMP and (p)ppGpp syntheses, quorum sensing and the expression of virulence genes [5,45]. CsrB-like sRNAs contain as many as 18 CsrA-binding motifs. When these sRNAs are in high amounts, CsrA binds to them and hence is not available to bind its target mRNAs [5]. This sequestration causes up-regulation of CsrA targets conferring pleiotropic phenotypes in biofilm, gluconeogenesis, capsule formation and impacting virulence [5]. In E. coli, overexpression of the csrB sRNA mimics a csrA mutation, leading to increased glycogen synthesis [5,45]. The synthesis of CsrB and CsrC is regulated by the BarA/UvrY TCS, and this regulatory cascade is conserved in several bacteria. Furthermore, the McaS sRNA can sequester CsrA, causing repression of PGA exopolysaccharide synthesis [46]. As CsrA and CsrB-like sRNAs are conserved, they serve as a model for study of such sRNA interactions with regulatory proteins.
The regulatory role of 6S RNA
The abundant 6S RNA is conserved in Gram-negative and Gram-positive bacteria and functions by binding at the active site of σ70-RNA polymerase by virtue of its double-stranded RNA hairpin structure that mimics DNA in an open promoter complex [47]. 6S RNA accumulates to high amounts in the stationary phase, sequesters σ70-RNAP and favors transcription by the RpoS-containing RNA polymerase. Recovery from the 6S RNA inhibition, upon transition from the stationary phase into the exponential phase, occurs by the release of 6S RNA from the inhibited RNAP complex involving synthesis of a short transcript of ∼14 nt (pRNA), wherein 6S RNA acts as a template that requires a high NTP concentration. The formation of the pRNA–6S RNA complex induces a structural rearrangement that triggers release of 6S RNA, which is subsequently removed by degradation [48].
The RirA sRNA and its interaction with RfaH
RfaH is a paralog of the NusG family of transcription factors. However, RfaH is unique in its specificity for recognition of only those operons that contain a short 8-nt conserved sequence called the ops pause site in the 5′ UTR. RfaH transcribes genes that play important functions in virulence such as synthesis of the LPS core, O-antigen, hemolysin and capsule, and for conjugation [13,49]. The RirA sRNA (RfaH-interacting RNA) was identified while analyzing signals that induce transcription of the rpoE gene. Overexpression of the 73-nt RirA sRNA, located in the 5′ UTR of the waaQ operon, increases transcription from the rpoEP3 promoter that specifically responds to LPS defects [13]. RirA shares its 5′ end with the waaQ mRNA and contains the conserved ops and JUMPstart sites that are recognized by RfaH (Figure 1D). RirA binds RfaH in the presence of RNAP, and this binding requires the presence of the ops site. RirA overexpression abrogates the synthesis of O-antigen, and causes reduction in LPS amounts and truncation in the LPS core, thus mimicking the ΔrfaH phenotype (Figure 1D) [13]. Thus, RirA could either sequester RfaH or act by inducing conformational change in RfaH that renders it into a NusG-like conformation. Free RfaH without interaction with the ops site exists in an inactive closed conformation with its C-terminal domain (CTD) folded in an α-helical hairpin tightly packed against its N-terminal domain (NTD). In this state, the NTD is not accessible for DNA–RNAP interaction. However, when RfaH binds to the ops site, there is a conformational change causing the opening of CTD and NTD, in which RfaH CTD folds into a β-barrel (like CTD of NusG), allowing recruitment of S10 ribosomal protein to couple transcription with translation and also enhance transcriptional elongation [49].
Conclusions and perspectives
Advances in identifying and characterization of bacterial sRNAs have led to our understanding that RNA regulators are ubiquitous. The majority of sRNAs act in trans and can be encoded within almost every possible genomic element. sRNAs, in general, serve as diverse regulators that affect every aspect of bacterial physiology. Many sRNAs have specific functions in relieving OM or IM stress, control of LPS composition and heterogeneity, and regulation of entry into the stationary phase, and in pathogenic bacteria sRNAs could have a specific role during contact with the host. New technologies, identifying sRNA–mRNA interactions like RIL-seq and RNase E-CLASH, have revealed many new potential targets of already known sRNAs and also identified additional potential sRNAs [6,8]. Recent studies continue to discover new sRNAs, as shown by the transcriptome analysis of Salmonella using starvation-stress-responsive conditions that revealed 58 previously unknown sRNAs mostly located in intergenic regions, some of which are conserved in Enterobacteriaceae [50]. Challenges lie ahead in validating these new targets of sRNAs and how these aspects of regulation are integrated into different regulatory circuits. Similarly, future studies are required to understand how a regulatory sRNA like RirA can change the conformation of RfaH that prevents recognition of its target operons that contain the ops site and transform it into a general transcription factor like NusG.
Abbreviations
- CsrA
carbon storage regulator A
- CTD
C-terminal domain
- IM
inner membrane
- LPS
lipopolysaccharide
- NTD
N-terminal domain
- OM
outer membrane
- OMP
outer membrane proteins
- P-EtN
phosphoethanolamine
- PMF
proton motive force
- RpoE
extracytoplasmic function sigma factor
- SlrA(MicL)
suppressor of Lap deficiency and repressor of lpp mRNA
- sRNA
small regulatory RNA
- TCS
two-component system
- UTR
untranslated region.
Funding
This work was supported by National Science Grant [2013/11/B/NZ2/02641 to S.R.].
Competing Interests
The Authors declare that there are no competing interests associated with the manuscript.
References
- 1.Cech T.R. and Steitz J.A. (2014) The noncoding RNA revolution — trashing old rules to forge new ones. Cell 157, 77–94 doi: 10.1016/j.cell.2014.03.008 [DOI] [PubMed] [Google Scholar]
- 2.Storz G., Vogel J. and Wassarman K.M. (2011) Regulation by small RNAs in bacteria: expanding frontiers. Mol. Cell 43, 880–891 doi: 10.1016/j.molcel.2011.08.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wagner E.G.H. and Romby P. (2015) Small RNAs in bacteria and archaea: who they are, what they do, and how they do it. Adv. Genet. 90, 133–208 doi: 10.1016/bs.adgen.2015.05.001 [DOI] [PubMed] [Google Scholar]
- 4.Argaman L., Hershberg R., Vogel J., Bejerano G., Wagner E.G.H., Margalit H. et al. (2001) Novel small RNA-encoding genes in the intergenic regions of Escherichia coli. Curr. Biol. 11, 941–950 doi: 10.1016/S0960-9822(01)00270-6 [DOI] [PubMed] [Google Scholar]
- 5.Holmqvist E., Wright P.R., Li L., Bischler T., Barquist L., Reinhardt R. et al. (2016) Global RNA recognition patterns of post-transcriptional regulators Hfq and CsrA revealed by UV crosslinking in vivo. EMBO J. 35, 991–1011 doi: 10.15252/embj.201593360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Melamed S., Peer A., Faigenbaum-Romm R., Gatt Y.E., Reiss N., Bar A. et al. (2016) Global mapping of small RNA-target interactions in bacteria. Mol. Cell 63, 884–897 doi: 10.1016/j.molcel.2016.07.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smirnov A., Förstner K.U., Holmqvist E., Otto A., Günster R., Becher D. et al. (2016) Grad-seq guides the discovery of ProQ as a major small RNA-binding protein. Proc. Natl Acad. Sci. U.S.A. 113, 11591–11596 doi: 10.1073/pnas.1609981113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Waters S.A., McAteer S.P., Kudla G., Pang I., Deshpande N.P., Amos T.G. et al. (2017) Small RNA interactome of pathogenic E. coli revealed through crosslinking of RNase E. EMBO J. 36, 374–387 doi: 10.15252/embj.201694639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chao Y., Papenfort K., Reinhardt R., Sharma C.M. and Vogel J. (2012) An atlas of Hfq-bound transcripts reveals 3′ UTRs as a genomic reservoir of regulatory small RNAs. EMBO J. 31, 4005–4019 doi: 10.1038/emboj.2012.229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klein G., Kobylak N., Lindner B., Stupak A. and Raina S. (2014) Assembly of lipopolysaccharide in Escherichia coli requires the essential LapB heat shock protein. J. Biol. Chem. 289, 14829–14853 doi: 10.1074/jbc.M113.539494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo M.S., Updegrove T.B., Gogol E.B., Shabalina S.A., Gross C.A. and Storz G. (2014) MicL, a new σE-dependent sRNA, combats envelope stress by repressing synthesis of Lpp, the major outer membrane lipoprotein. Genes Dev. 28, 1620–1634 doi: 10.1101/gad.243485.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chao Y. and Vogel J. (2016) A 3′ UTR-derived small RNA provides the regulatory noncoding arm of the inner membrane stress response. Mol. Cell 61, 352–363 doi: 10.1016/j.molcel.2015.12.023 [DOI] [PubMed] [Google Scholar]
- 13.Klein G., Stupak A., Biernacka D., Wojtkiewicz P., Lindner B. and Raina S. (2016) Multiple transcriptional factors regulate transcription of the rpoE gene in Escherichia coli under different growth conditions and when the lipopolysaccharide biosynthesis is defective. J. Biol. Chem. 291, 22999–23019 doi: 10.1074/jbc.M116.748954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arnvig K. and Young D. (2012) Non-coding RNA and its potential role in Mycobacterium tuberculosis pathogenesis. RNA Biol. 9, 427–436 doi: 10.4161/rna.20105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thomason M.K., Fontaine F., De Lay N. and Storz G. (2012) A small RNA that regulates motility and biofilm formation in response to changes in nutrient availability in Escherichia coli. Mol. Microbiol. 84, 17–35 doi: 10.1111/j.1365-2958.2012.07965.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee H.-J. and Gottesman S. (2016) sRNA roles in regulating transcriptional regulators: Lrp and SoxS regulation by sRNAs. Nucleic Acids Res. 44, 6907–6923 doi: 10.1093/nar/gkw358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fröhlich K.S., Haneke K., Papenfort K. and Vogel J. (2016) The target spectrum of SdsR small RNA in Salmonella. Nucleic Acids Res. 44, 10406–10422 doi: 10.1093/nar/gkw632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klein G. and Raina S. (2015) Regulated control of the assembly and diversity of LPS by noncoding sRNAs. BioMed Res. Int. 2015, 1–16 doi: 10.1155/2015/153561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Missiakas D., Mayer M.P., Lemaire M., Georgopoulos C. and Raina S. (1997) Modulation of the Escherichia coli σE (RpoE) heat-shock transcription-factor activity by the RseA, RseB and RseC proteins. Mol. Microbiol. 24, 355–371 doi: 10.1046/j.1365-2958.1997.3601713.x [DOI] [PubMed] [Google Scholar]
- 20.Gogol E.B., Rhodius V.A., Papenfort K., Vogel J. and Gross C.A. (2011) Small RNAs endow a transcriptional activator with essential repressor functions for single-tier control of a global stress regulon. Proc. Natl Acad. Sci. U.S.A. 108, 12875–12880 doi: 10.1073/pnas.1109379108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klein G., Lindner B., Brabetz W., Brade H. and Raina S. (2009) Escherichia coli K-12 suppressor-free mutants lacking early glycosyltransferases and late acyltransferases: minimal lipopolysaccharide structure and induction of envelope stress response. J. Biol. Chem. 284, 15369–15389 doi: 10.1074/jbc.M900490200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klein G., Lindner B., Brade H. and Raina S. (2011) Molecular basis of lipopolysaccharide heterogeneity in Escherichia coli: envelope stress-responsive regulators control the incorporation of glycoforms with a third 3-deoxy-α-d-manno-oct-2-ulosonic acid and rhamnose. J. Biol. Chem. 286, 42787–42807 doi: 10.1074/jbc.M111.291799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Coornaert A., Lu A., Mandin P., Springer M., Gottesman S. and Guillier M. (2010) Mica sRNA links the PhoP regulon to cell envelope stress. Mol. Microbiol. 76, 467–479 doi: 10.1111/j.1365-2958.2010.07115.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Udekwu K.I., Darfeuille F., Vogel J., Reimegård J., Holmqvist E. and Wagner E.G.H. (2005) Hfq-dependent regulation of OmpA synthesis is mediated by an antisense RNA. Genes Dev. 19, 2355–2366 doi: 10.1101/gad.354405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brosse A., Korobeinikova A., Gottesman S. and Guillier M. (2016) Unexpected properties of sRNA promoters allow feedback control via regulation of a two-component system. Nucleic Acids Res. 44, 9650–9666 doi: 10.1093/nar/gkw642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dartigalongue C., Missiakas D. and Raina S. (2001) Characterization of the Escherichia coli σE regulon. J. Biol. Chem. 276, 20866–20875 doi: 10.1074/jbc.M100464200 [DOI] [PubMed] [Google Scholar]
- 27.Kabir M.S., Yamashita D., Koyama S., Oshima T., Kurokawa K., Maeda M. et al. (2005) Cell lysis directed by σE in early stationary phase and effect of induction of the rpoE gene on global gene expression in Escherichia coli. Microbiology 151, 2721–2735 doi: 10.1099/mic.0.28004-0 [DOI] [PubMed] [Google Scholar]
- 28.Papenfort K., Pfeiffer V., Mika F., Lucchini S., Hinton J.C.D. and Vogel J. (2006) σE-dependent small RNAs of Salmonella respond to membrane stress by accelerating global omp mRNA decay. Mol. Microbiol. 62, 1674–1688 doi: 10.1111/j.1365-2958.2006.05524.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Balbontín R., Fiorini F., Figueroa-Bossi N., Casadesús J. and Bossi L. (2010) Recognition of heptameric seed sequence underlies multi-target regulation by RybB small RNA in Salmonella enterica. Mol. Microbiol. 78, 380–394 doi: 10.1111/j.1365-2958.2010.07342.x [DOI] [PubMed] [Google Scholar]
- 30.Pfeiffer V., Papenfort K., Lucchini S., Hinton J.C.D. and Vogel J. (2009) Coding sequence targeting by MicC RNA reveals bacterial mRNA silencing downstream of translational initiation. Nat. Struct. Mol. Biol. 16, 840–846 doi: 10.1038/nsmb.1631 [DOI] [PubMed] [Google Scholar]
- 31.Noor R., Murata M., Nagamitsu H., Klein G., Raina S. and Yamada M. (2009) Dissection of σE-dependent cell lysis in Escherichia coli: roles of RpoE regulators RseA, RseB and periplasmic folding catalyst PpiD. Genes Cells 14, 885–899 doi: 10.1111/j.1365-2443.2009.01318.x [DOI] [PubMed] [Google Scholar]
- 32.Moon K., Six D.A., Lee H.-J., Raetz C.R.H. and Gottesman S. (2013) Complex transcriptional and post-transcriptional regulation of an enzyme for lipopolysaccharide modification. Mol. Microbiol. 89, 52–64 doi: 10.1111/mmi.12257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Coornaert A., Chiaruttini C., Springer M. and Guillier M. (2013) Post-transcriptional control of the Escherichia coli PhoQ-PhoP two-component system by multiple sRNAs involves a novel pairing region of GcvB. PLoS Genet. 9, e1003156 doi: 10.1371/journal.pgen.1003156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Corcoran C.P., Podkaminski D., Papenfort K., Urban J.H., Hinton J.C.D. and Vogel J. (2012) Superfolder GFP reporters validate diverse new mRNA targets of the classic porin regulator, MicF RNA. Mol. Microbiol. 84, 428–445 doi: 10.1111/j.1365-2958.2012.08031.x [DOI] [PubMed] [Google Scholar]
- 35.Vogt S.L., Evans A.D., Guest R.L. and Raivio T.L. (2014) The Cpx envelope stress response regulates and is regulated by small noncoding RNAs. J. Bacteriol. 196, 4229–4238 doi: 10.1128/JB.02138-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Missiakas D., and Raina S. (1997) Signal transduction pathways in response to protein misfolding in the extracytoplasmic compartments of E. coli: role of two new phosphoprotein phosphatases PrpA and PrpB. EMBO J. 16, 1670–1685 doi: 10.1093/emboj/16.7.1670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Missiakas D., Betton J.-M. and Raina S. (1996) New components of protein folding in extracytoplasmic compartments of Escherichia coli SurA, FkpA and Skp/OmpH. Mol. Microbiol. 21, 871–884 doi: 10.1046/j.1365-2958.1996.561412.x [DOI] [PubMed] [Google Scholar]
- 38.Grabowicz M., Koren D. and Silhavy T.J. (2016) The CpxQ sRNA negatively regulates Skp to prevent mistargeting of β-barrel outer membrane proteins into the cytoplasmic membrane. mBio 7, e00312-16 doi: 10.1128/mBio.00312-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Colgan A.M., Kröger C., Diard M., Hardt W.-D., Puente J.L., Sivasankaran S.K. et al. (2016) The impact of 18 ancestral and horizontally-acquired regulatory proteins upon the transcriptome and sRNA landscape of Salmonella enterica serovar Typhimurium. PLoS Genet. 12, e1006258 doi: 10.1371/journal.pgen.1006258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mandin P. and Gottesman S. (2010) Integrating anaerobic/aerobic sensing and the general stress response through the ArcZ small RNA. EMBO J. 29, 3094–3107 doi: 10.1038/emboj.2010.179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rau M.H., Bojanovič K., Nielsen A.T. and Long K.S. (2015) Differential expression of small RNAs under chemical stress and fed-batch fermentation in E. coli. BMC Genomics 16, 1051 doi: 10.1186/s12864-015-2231-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Papenfort K., Said N., Welsink T., Lucchini S., Hinton J.C.D. and Vogel J. (2009) Specific and pleiotropic patterns of mRNA regulation by ArcZ, a conserved, Hfq-dependent small RNA. Mol. Microbiol. 74, 139–158 doi: 10.1111/j.1365-2958.2009.06857.x [DOI] [PubMed] [Google Scholar]
- 43.Silva I.J., Ortega A.D., Viegas S.C., García-Del Portillo F. and Arraiano C.M. (2013) An RpoS-dependent sRNA regulates the expression of a chaperone involved in protein folding. RNA 19, 1253–1265 doi: 10.1261/rna.039537.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hao Y., Updegrove T.B., Livingston N.N. and Storz G. (2016) Protection against deleterious nitrogen compounds: role of σS-dependent small RNAs encoded adjacent to sdiA. Nucleic Acids Res. 44, 6935–6948 doi: 10.1093/nar/gkw404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vakulskas C.A., Potts A.H., Babitzke P., Ahmer B.M.M. and Romeo T. (2015) Regulation of bacterial virulence by Csr (Rsm) systems. Microbiol. Mol. Biol. Rev. 79, 193–224 doi: 10.1128/MMBR.00052-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jørgensen M.G., Thomason M.K., Havelund J., Valentin-Hansen P. and Storz G. (2013) Dual function of the McaS small RNA in controlling biofilm formation. Genes Dev. 27, 1132–1145 doi: 10.1101/gad.214734.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wassarman K.M. and Storz G. (2000) 6S RNA regulates E. coli RNA polymerase activity. Cell 101, 613–623 doi: 10.1016/S0092-8674(00)80873-9 [DOI] [PubMed] [Google Scholar]
- 48.Panchapakesan S.S.S. and Unrau P.J. (2012) E. coli 6S RNA release from RNA polymerase requires σ70 ejection by scrunching and is orchestrated by a conserved RNA hairpin. RNA 18, 2251–2259 doi: 10.1261/rna.034785.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Burmann B.M., Knauer S.H., Sevostyanova A., Schweimer K., Mooney R.A., Landick R. et al. (2012) An α helix to β barrel domain switch transforms the transcription factor RfaH into a translation factor. Cell 150, 291–303 doi: 10.1016/j.cell.2012.05.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Amin S.V., Roberts J.T., Patterson D.G., Coley A.B., Allred J.A., Denner J.M. et al. (2016) Novel small RNA (sRNA) landscape of the starvation-stress response transcriptome of Salmonella enterica serovar typhimurium. RNA Biol. 13, 331–342 doi: 10.1080/15476286.2016.1144010 [DOI] [PMC free article] [PubMed] [Google Scholar]