Abstract
Most small molecules are unable to rapidly traverse the outer membrane of Gram-negative bacteria and accumulate inside these cells, making the discovery of much-needed drugs for these pathogens very challenging. Current understanding of the physicochemical properties that dictate small-molecule accumulation in Gram-negatives is largely based on retrospective analyses of antibacterials that suggest polarity and molecular weight as key factors. Here we assess the ability of over 180 diverse compounds to accumulate in Escherichia coli. Computational analysis of the results reveals major differences from the retrospective studies, namely that the small molecules that are most likely to accumulate contain an amine, are amphiphilic and rigid, and have low globularity. These guidelines were then applied to convert deoxynybomycin, a natural product that is active only against Gram-positive organisms, into an antibiotic with activity against a diverse panel of multi-drug-resistant Gram-negative pathogens. We anticipate these findings will aid in the discovery and development of antibiotics effective against Gram-negative bacteria.
Drug-resistant bacteria are a major public health concern1–3, and Gram-negative bacteria are particularly troubling as they are insensitive to many commonly used antibiotics2,4–8. Exacerbating this problem is the fact that a new class of antibiotics active against Gram-negative bacteria has not been introduced into the clinic since the quinolones in 19684. This void in discovery is not due to a lack of effort; as one example, in 2007 GlaxoSmithKline reported screening around 500,000 synthetic compounds for activity against Escherichia coli, but no tractable hits were identified1.
Gram-negative bacteria have two cellular membranes, and the lipopolysaccharide-coated outer membrane is very challenging for small molecules to cross5,8,9. Compounds that are able to traverse this outer membrane typically do so through narrow β-barrel proteins called porins, channels that are lined with charged amino acids10. Once inside the cell, small molecules are susceptible to efflux pumps5,6,8, thus to accumulate in Gram-negatives to a level that is sufficient for activity, small molecules typically must traverse porins faster than they are pumped out.
Central to the problem of discovering antibiotics effective against Gram-negative bacteria is a limited understanding of the physicochemical properties that enable small-molecule accumulation in Gram-negative bacteria, with current knowledge based largely on retrospective analyses of known antibiotics11,12. In 2008, O’Shea and Moser reported that antibiotics effective against Gram-negative pathogens almost always have a molecular weight less of than 600 Da and tend to be very polar, as measured by ClogD7.4 (the predicted octanol/water distribution coefficient at pH 7.4). These observations are consistent with porin architecture11, and have been reinforced by other retrospective studies12. However, there are antibiotics that meet these polarity and size criteria but are inactive against Gram-negative species, suggesting these properties do not fully encompass the determinants for small-molecule accumulation. In addition, retrospective analyses are highly skewed by the over-representation of certain drug classes. For example, one analysis showed that carboxylic acids were present on up to 40% of Gram-negative-active compounds; however, these carboxylic-acid-containing compounds were almost exclusively β-lactams12. Although some compound accumulation studies in whole cells have been performed13–16, broad conclusions cannot be drawn from these small data sets (10–20 compounds and all within a single structural class). Perhaps most importantly, the canonical view about the importance of ClogD7.4 and molecular weight for Gram-negative activity has not led to general strategies to convert Gram-positive-only compounds into broad-spectrum antibiotics. The seminal observation over 50 years ago that derivatizing penicillin G into ampicillin results in broad-spectrum activity17 has not been generalizable, and important classes of antibiotics have coverage only against Gram-positive organisms despite intensive derivatization efforts. A systematic analysis of the accumulation of an unbiased and structurally diverse set of small molecules in Gram-negative bacteria has not been previously reported. Towards this end, we first assembled a diverse set of 100 compounds and quantified their capacity to accumulate in E. coli. As there are many variables affecting small-molecule accumulation (for example, multiple porins, efflux pumps and varying lipopolysaccharides)5,6,8, compounds were assessed in accumulation assays using whole cells, instead of model systems. From these experiments, subsequent structure–activity relationship (SAR) studies, and computational analyses, predictive guidelines of small-molecule accumulation in Gram-negative bacteria were developed.
Accumulation studies
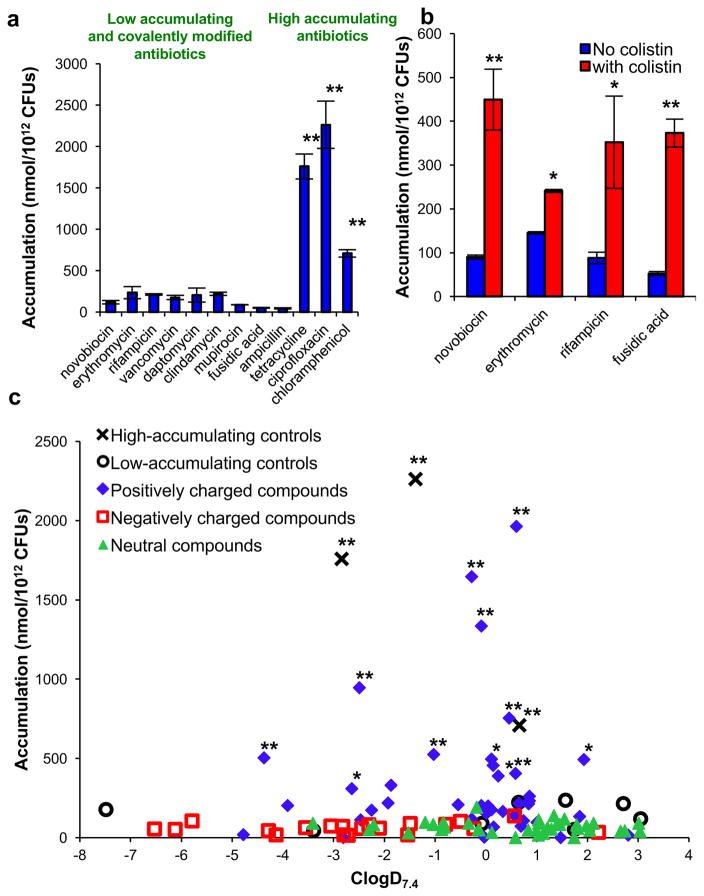
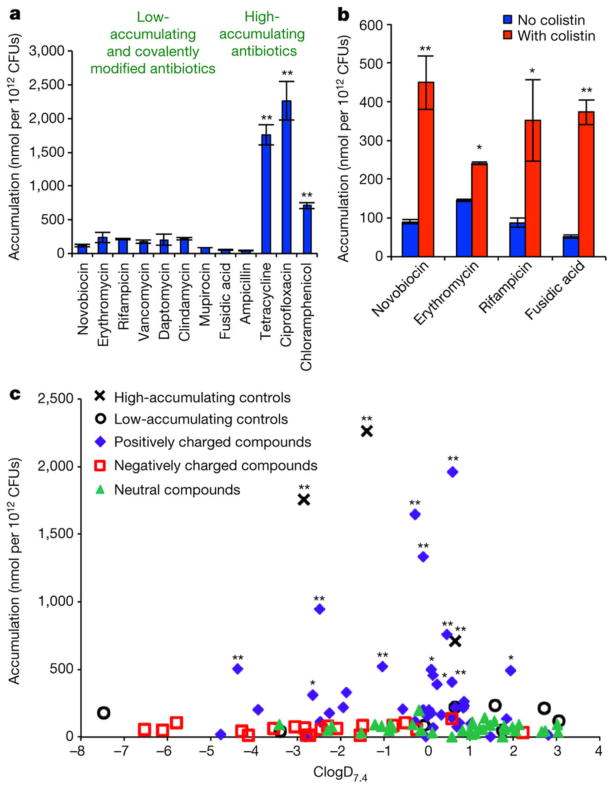
The accumulation method was adapted from known protocols13,14,18 and liquid chromatography with tandem mass spectroscopy (LC–MS/MS) was used to quantify accumulation of each compound. The assay method was evaluated with antibiotics that have known high (tetracycline, ciprofloxacin and chloramphenicol) and low (novobiocin, erythromycin, rifampicin, vancomycin, daptomycin, clindamycin, mupirocin and fusidic acid) levels of accumulation. Ampicillin was also used as a ‘low-accumulation’ control as it is rapidly covalently appended to penicillin-binding proteins, preventing measurement by LC–MS/MS. To account for the possibility of non-specific binding to the outer membrane, Gram-negative-active antibiotics with various charged states at physiological pH were chosen: tetracycline (positively charged), ciprofloxacin (zwitterionic) and chloramphenicol (neutral) (structures shown in Supplementary Table 1). The results show a significantly higher level of accumulation in E. coli for the Gram-negative-active compounds as compared to compounds with low Gram-negative antibacterial activity and ampicillin, consistent with measurement of accumulation as opposed to non-specific binding (Fig. 1a). To further ensure that variations in observed accumulation levels were not due to differences in non-specific affinity for the membrane, penetrance was perturbed by co-treating E. coli with the membrane-permeabilizing agent colistin19; an increase in accumulation for low-accumulating antibiotics (novobiocin, erythromycin, rifampicin and fusidic acid) is observed in this experiment (Fig. 1b).
Figure 1. Small-molecule accumulation in E. coli MG1655.
a, Compounds that are active against E. coli have significantly higher accumulation than low-activity antibiotics and ampicillin; structures in Supplementary Table 1. b, Co-treatment with colistin enhances the accumulation of low-accumulating antibiotics. c, Assessment of 100 compounds for accumulation, labelled by ionic state; structures in Supplementary Table 2. Statistical significance was determined by using a two sample Welch’s t-test (one tailed test, assuming unequal variance) relative to the negative controls. P values relative to the average of the low-accumulating controls; *P < 0.05, **P < 0.01. The s.e.m. for each compound tested is reported in Supplementary Table 2. All experiments were performed in biological triplicate, error bars represent s.e.m.
Towards understanding the physicochemical properties that influence small-molecule accumulation in E. coli, a collection of diverse compounds was required. Given that most antibacterial drugs are natural products or derivatives, critical to this effort would be access to a collection of compounds that possess natural-product-like properties, but also are synthetically accessible so that various properties are tunable, enabling SAR studies. Thus, these compounds were produced using the ‘complexity-to-diversity’ strategy, whereby diverse compounds are constructed from readily available natural products20–23. To begin the analysis, a set of 100 compounds, including positively charged, negatively charged and neutral compounds, were synthesized and tested. The structure, molecular weight, ClogD7.4 and charge of all 100 compounds tested are provided in Supplementary Table 2. As retrospective studies suggest that compound accumulation is related to ClogD7.4 (refs 11, 12), the accumulation data for all 100 compounds were plotted against ClogD7.4 (Fig. 1c). These results differ starkly from the conclusions reached in retrospective studies. Within this set of 100 compounds, charge is the primary factor dictating accumulation in E. coli. The positively charged compounds are the most likely to accumulate, with 12 of 41 positively charged compounds showing a significant level of accumulation compared to the low-accumulating controls. In contrast, 0 of 39 neutral compounds and 0 of 20 negatively charged compounds show significant accumulation. Notably, even carboxylic acids with strongly negative ClogD7.4 values (<−5) do not accumulate.
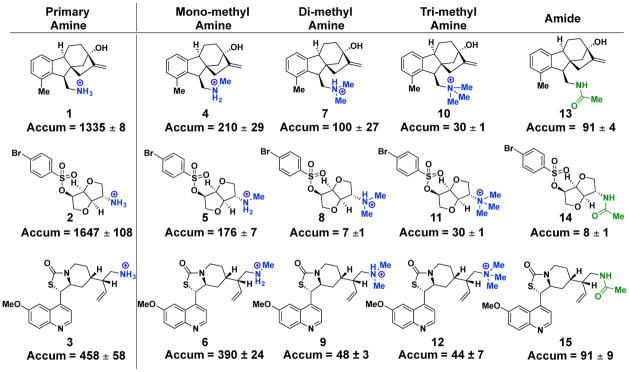
All 12 accumulating compounds contain amines, and most of these compounds (8 out of 12) are primary amines. To further examine the importance of the primary amine, a SAR analysis was performed for multiple different classes of accumulating compounds. Replacement of the amine with a carboxylic acid, amide, ester, nitrile, azide or alcohol on multiple different scaffolds markedly reduces accumulation (Supplementary Table 3). Even conversion of the primary amine to an amine with more substitutions has a deleterious effect on accumulation. Shown in Fig. 2 are three primary amines (1–3) alongside their methylated (4–6), di-methylated (7–9), tri-methylated (10–12) and acetylated derivatives (13–15). In all cases, the primary amine shows the highest level of accumulation. Additional primary-amine-containing compounds were then obtained and their accumulation in E. coli was assessed; these compounds are both ‘complexity-to-diversity’ compounds and commercially available primary amines that more closely mimic the types of compounds found in commercial screening libraries. For the total set of 68 primary amines (all structures in Supplementary Table 4), accumulation does not increase with lower ClogD7.4 (Extended Data Fig. 1a), and there is also no correlation between accumulation and molecular weight (Extended Data Fig. 1b). SAR analysis on several of these compounds also showed the importance of the amine to accumulation (Supplementary Table 3).
Figure 2. Primary amines aid small-molecule accumulation in E. coli.
Modification of a high-accumulating primary amine to a mono-methyl, di-methyl, tri-methyl or acylated derivative has a deleterious effect on compound accumulation in E. coli MG1655. Accumulation reported in nmol per 1012 colony-forming units (CFUs). All experiments were performed in biological triplicate. The s.e.m. is reported for accumulation values.
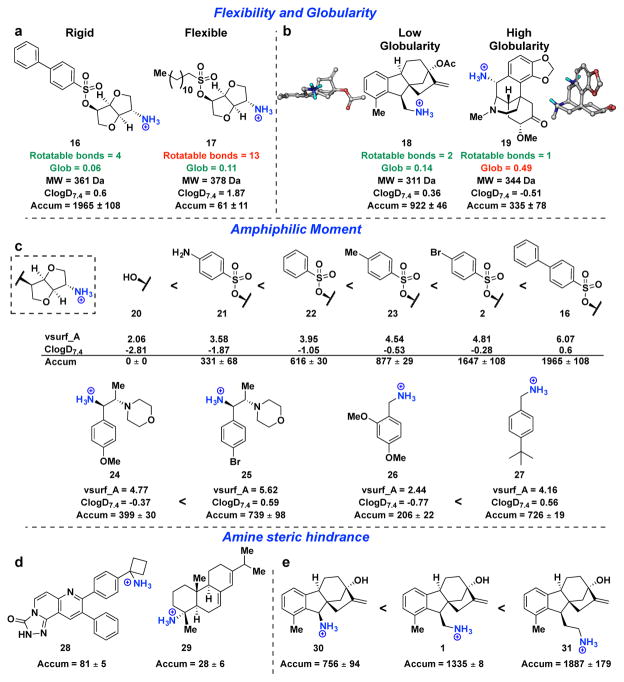
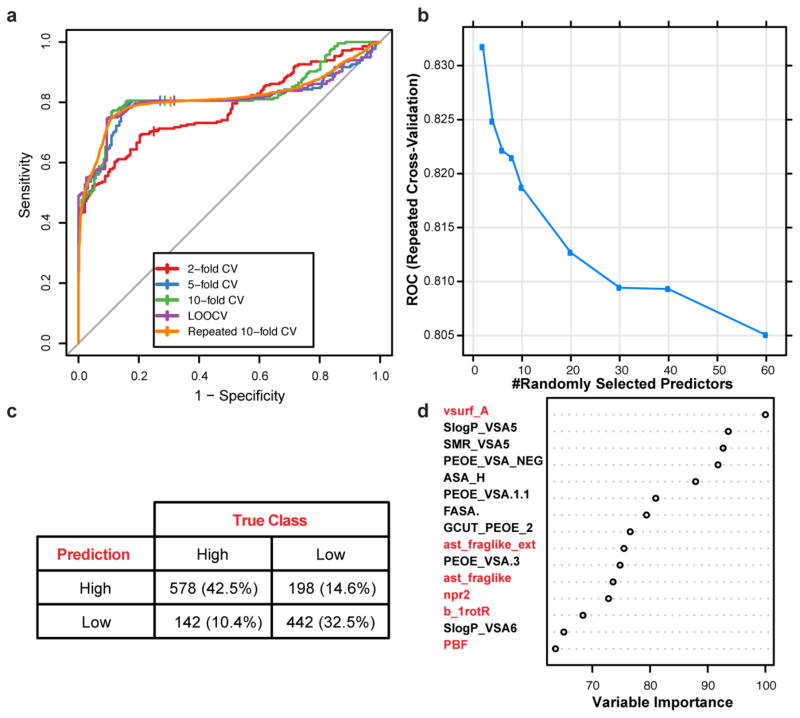
Although the presence of a primary amine is clearly important for accumulation in E. coli, it is not sufficient: although 36 of the primary amines within the test set do accumulate, 32 of them do not. Therefore, a chemoinformatic approach was implemented to understand which factors contribute to amine accumulation. For this set of 68 primary amines, 297 molecular descriptors were calculated for each compound (see Supplementary Table 5 for descriptors). These descriptors were used to train a random forest classification model that predicts amine accumulation (Extended Data Fig. 2). Through this analysis, it was revealed that the flexibility (measured by the number of rotatable bonds) and shape of a compound are important factors that govern accumulation. Shape was best described by globularity24, a term used to provide information on the three-dimensionality of compounds25,26, where a completely flat compound (for example, benzene) has a globularity of 0 and a spherical compound (for example, adamantane) has a globularity of 1. Case studies demonstrate the importance of flexibility and globularity for accumulation of the primary amines: as shown in Fig. 3a, amine 16 with four rotatable bonds accumulates well; however, amine 17, a compound of similar molecular weight and globularity but with 13 rotatable bonds, shows virtually no accumulation. Analogous results are observed in Fig. 3b, when comparing a compound with low globularity (18, globularity = 0.14) to a compound with similar functional groups, molecular weight and rotatable bonds, but with high globularity (19, globularity = 0.49); the globularity of each compound is most easily observed in its three-dimensional model Fig. 3b.
Figure 3. Properties affecting small-molecule accumulation in E. coli.
a, b, Case study comparing the importance of (a) flexibility and (b) globularity. c, Derivative sets demonstrate how accumulation can rise as amphiphilic moment increases within a structural class. d, Two compounds with primary amines attached to tertiary centres, both are low accumulators. e, Systematic separation of the primary amine from a sterically congested ring system results in increased compound accumulation. Accumulation in E. coli MG1655 is reported in nmol per 1012 CFUs. All experiments were performed in biological triplicate. The s.e.m. is reported for accumulation values.
Although, in general, primary amines with five or fewer rotatable bonds and a globularity of 0.25 or less have a markedly higher likelihood of accumulation, two additional factors related to the placement of the primary amine are important. First, the random forest model identified increased amphiphilic moment, which measures the distance between hydrophobic and hydrophilic portions of a compound27, as favouring accumulation. Notably, although mono-amine isomannide (20) does not accumulate, derivatives with increased amphiphilic moment do accumulate (21–23, 2 and 16), and similar trends are observed for other compound classes (24–27) (Fig. 3c). Accordingly, some degree of hydrophobicity appears necessary for accumulation, although in practice most organic compounds possess this feature. Second, two compounds in this set with sterically encumbered primary amines are not high accumulators (28 and 29; Fig. 3d); both have low flexibility and globularity but contain a primary amine on a tetra-substituted carbon. This result is consistent with the increased accumulation of compounds in which the primary amine is systematically extended from a sterically congested ring system (30 < 1 < 31) (Fig. 3e).
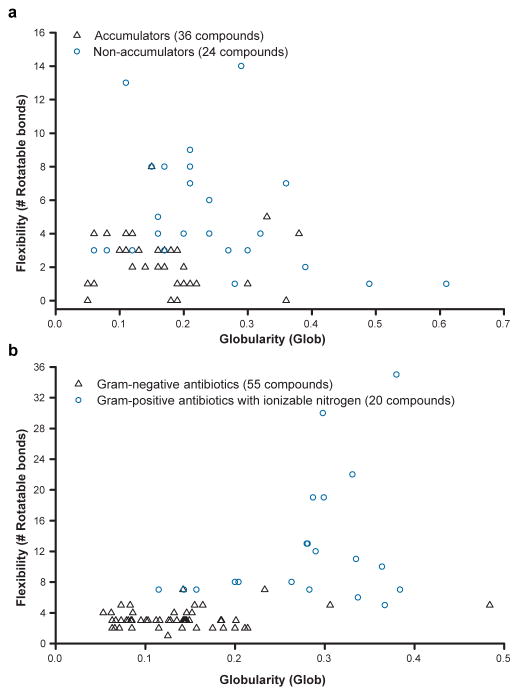
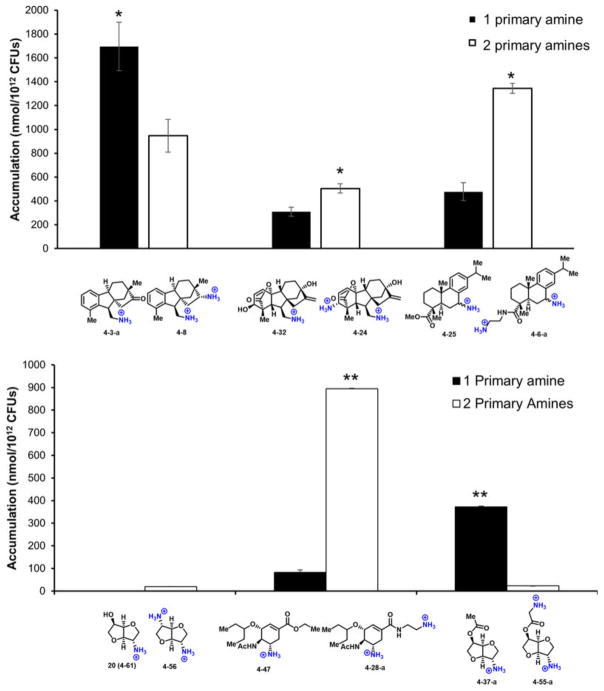
On the basis of these analyses, the following guiding principles for compound accumulation in E. coli were developed: compounds are most likely to accumulate if they contain a non-sterically encumbered amine, some non-polar functionality, they are rigid and have low globularity. As shown in Fig. 4a, the vast majority of compounds that meet these criteria in the test set accumulate in E. coli. Side-by-side comparisons of compounds containing two primary amines to analogues with one amine (six paired sets of compounds) largely follow the same rules for accumulation, with no clear effect of the second primary amine (Extended Data Fig. 3).
Figure 4. Flexibility and globularity are important for accumulation in E. coli.
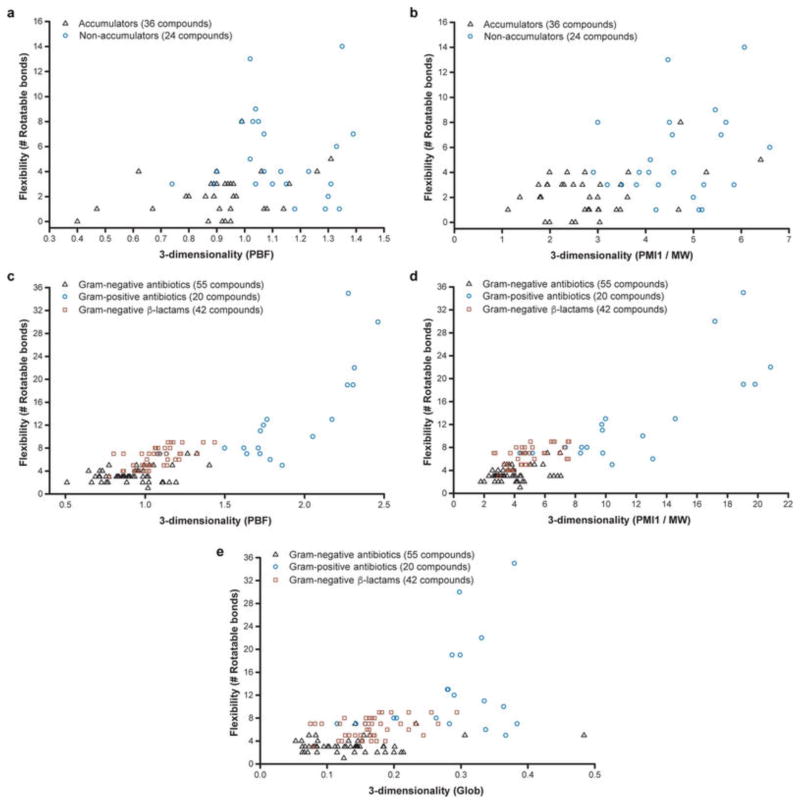
a, Flexibility (as measured by the number of rotatable bonds) plotted against globularity for the primary amines with proper amphiphilic moment and amine steric hindrance; compounds included in this analysis are noted in Supplementary Table 4. b, Flexibility versus globularity for antibiotics. Antibiotics included are Gram-positive-only drugs containing an ionizable nitrogen (circles), and Gram-negative actives that are believed to enter cells through porins (triangles); all compounds included in plot are listed in Supplementary Table 6. For plot including β-lactams, see Extended Data Fig. 4e.
To test the validity of these guidelines, the set of antibacterials assessed by O’Shea and Moser11 was evaluated for charge, flexibility and globularity. For the charge analysis, antibacterial drugs containing an ionizable nitrogen were accepted. For Gram-negative antibacterials, only those predicted to enter through porins were included, and owing to the large number of β-lactams they have been excluded from the graph in Fig. 4b but are included in Extended Data Fig. 4e (the structures of compounds in Fig. 4b and Extended Data Fig. 4e are in Supplementary Table 6 and Supplementary Table 7, respectively). As is shown in Fig. 4b, for drugs with ionizable nitrogens, compounds that are active against Gram-negative bacteria cleanly separate from those with Gram-positive-only activity based on these two physicochemical parameters. No Gram-positive-only antibacterial with an ionizable nitrogen has the correct rigidity and globularity for accumulation in E. coli. Although globularity was found to best predict accumulators and non-accumulators in combination with flexibility, other measures of three-dimensionality also exhibit the same trend (Extended Data Fig. 4a–d).
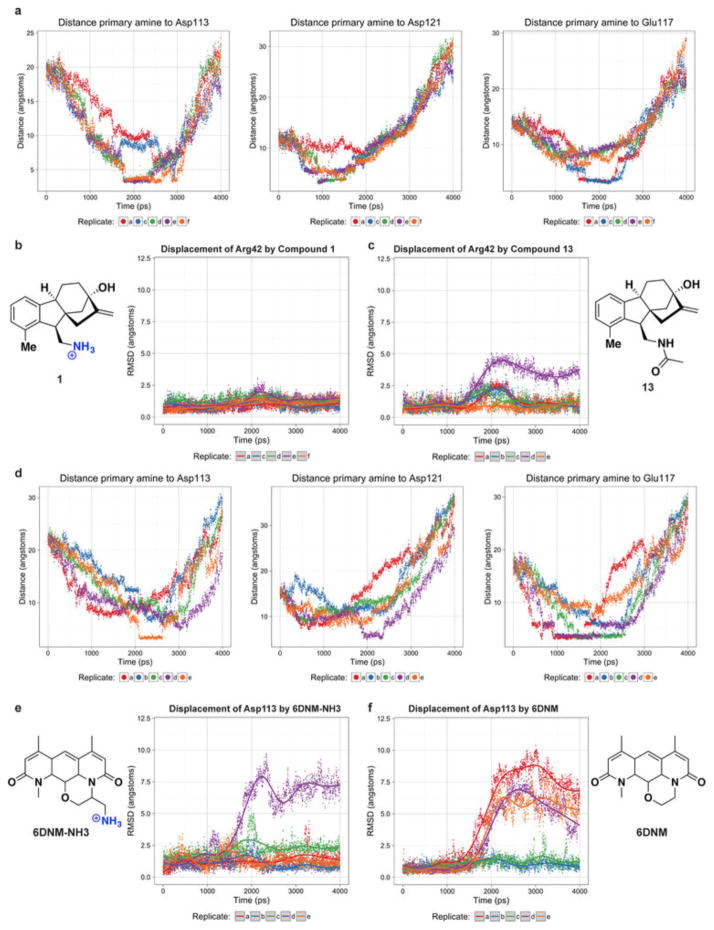
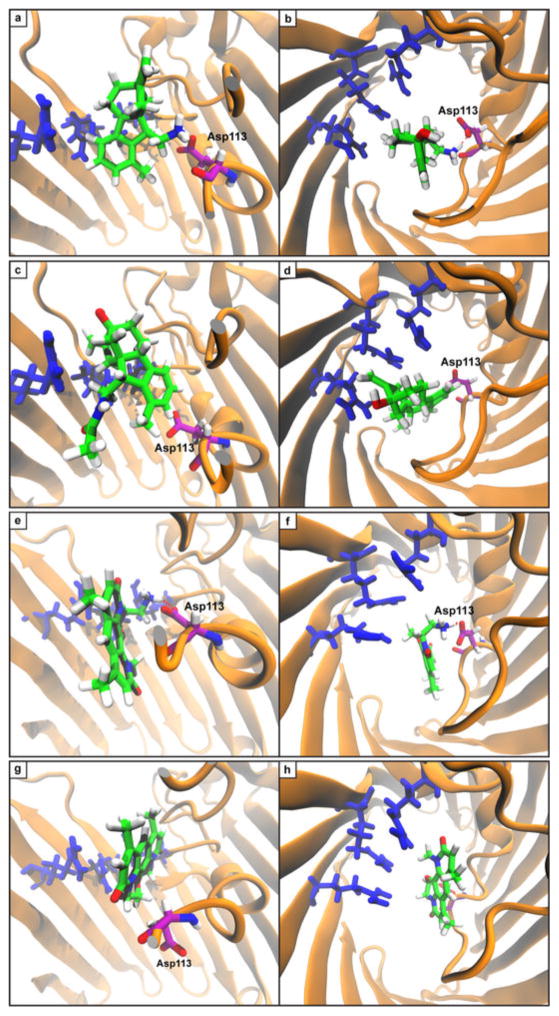
To better understand the observed accumulation trends, the role of porins in accumulation was examined. First, the high-accumulation controls and eight high-accumulating compounds were tested in a strain with a diminished number of porins, and a decrease in accumulation was observed for all compounds tested (Extended Data Fig. 5b; see Methods for a more detailed discussion of this experiment). Second, molecular modelling was performed on a subset of test set compounds and antibacterials as they traverse the major porin of E. coli, OmpF. Steered molecular dynamics (SMD) simulations were performed such that molecules were pulled through the constriction site of OmpF (Extended Data Fig. 6a). The trajectory of high-accumulating compound 1 reveals a key interaction between the pendant amine and acidic residues (most often Asp113) that assisted in traversing the constriction site (Extended Data Fig. 7a, b). This interaction was absent in the trajectory of a low-accumulating analogue of 1 (amide 13) (Extended Data Fig. 7c, d). Further, amide 13 induces greater distortions in constriction-site residues as it is forced through the porin (Extended Data Fig. 6b, c). In addition, minimal variation in permeability was observed for compound accumulation in protoplasts, supporting the hypothesis that traversing the outer membrane is the main barrier to small-molecule accumulation in Gram-negative bacteria28 (see Methods and Extended Data Fig. 5c for more information).
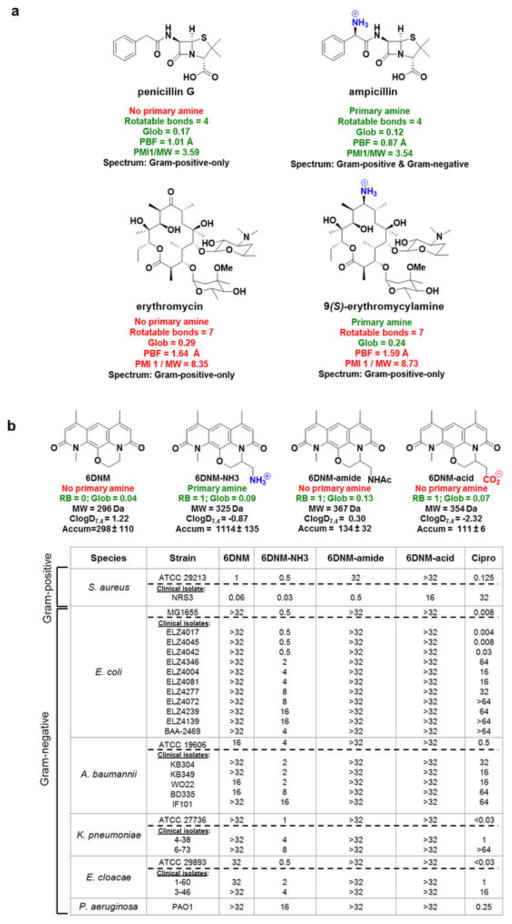
The findings presented here are congruent with what is known about β-lactams and explain why their spectrum could be broadened whereas other classes could not. In previous analyses, positive charge has been found to greatly accelerate penetration of β-lactams through porins, whereas negative charge and bulky substituents impede penetration29. Indeed, early generation β-lactams lacking an amine, such as penicillin G, are inactive against Gram-negative bacteria. Penicillin G has flexibility/shape parameters (rotatable bond = 4, globularity = 0.17) that make it a good candidate for conversion, and addition of an amine results in ampicillin (rotatable bond = 4, globularity = 0.12), which now meets all criteria for accumulation (structures in Extended Data Fig. 8a). Although there are third and fourth generation β-lactams with Gram-negative activity that do not meet the guidelines outlined here, these β-lactams have greatly reduced accumulation in Gram-negative bacteria compared to their positively charged analogues that meet the flexibility/shape parameters but are considerably more stable to β-lactamases than the early generation β-lactams, thus requiring lower levels of accumulation for antibacterial activity29. As delineated herein, to favour Gram-negative accumulation, an amine should be embedded on a compound with proper flexibility and shape parameters; these results explain why the simple addition of an amine has not been a generalizable strategy to increase Gram-negative accumulation and activity for other antibiotic classes. As one example, there are no differences in the spectrum of activity observed for erythromycin versus 9(S)-erythromycylamine30; as shown in Extended Data Fig. 8a, these compounds do not possess the appropriate rotatable bond and/or globularity parameters for accumulation.
Design of antibiotics for Gram-negative bacteria
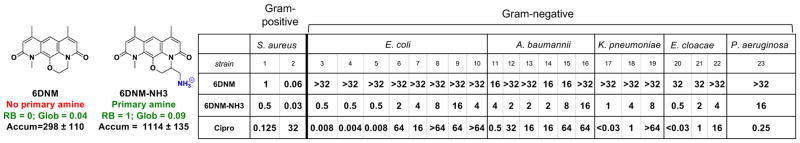
Our findings, in principle, should be able to guide the conversion of certain Gram-positive-only antibacterials to those that also have activity against Gram-negative bacteria. This would be a valuable and directly actionable feature of these results, one that could enable the production of candidate broad-spectrum antibacterials. The most straightforward method to accomplish this conversion would be to identify Gram-positive-only antibacterials that already have the proper flexibility and shape to accumulate in E. coli, and then append a primary amine without disrupting the ability of the compound to interact with its cellular target. Thus, a rotatable bond and globularity analysis of antibacterial agents with Gram-positive-only activity was conducted and used to select the natural product deoxynybomycin (DNM) as a good candidate for conversion to a broad-spectrum agent. DNM has antibacterial activity through inhibition of wild-type and mutant DNA gyrase, and DNM and its derivatives are only active against Gram-positive bacteria31,32. DNM has no rotatable bonds and a globularity of 0.02, suggesting that the addition of an amine to a position that does not alter the DNM antibacterial activity would provide a derivative able to accumulate in and be active against Gram-negative pathogens. To facilitate construction of a derivative with a primary amine, an analogue of DNM was first synthesized where the five-membered ring in DNM is expanded to a six-membered ring (6DNM, Fig. 5). 6DNM is active against Staphylococcus aureus (MIC = 0.06–1 μg ml−1) but shows no activity against E. coli (MIC > 32 μg ml−1, Fig. 5). Assessment by the accumulation assay revealed that 6DNM has low accumulation in E. coli (Fig. 5).
Figure 5. Conversion of a Gram-positive-only antibacterial into a compound that accumulates in E. coli and kills Gram-negative bacteria.
6DNM, 6DNM-NH3 and ciprofloxacin were evaluated against a panel of Gram-positive and Gram-negative organisms. E. coli MG1655 accumulation listed in nmol per 1012 CFUs. MIC values were determined using the micro-dilution broth method as outlined by the Clinical and Laboratory Standards Institute (http://clsi.org/) and are listed in μg ml−1. All experiments were performed in biological triplicate. Strains are listed in the Methods. The s.e.m. is reported for accumulation values.
A derivative of 6DNM was then synthesized that maintains low numbers of rotatable bonds and globularity, but that also contains a primary amine (6DNM-NH3, Fig. 5). 6DNM-NH3 retains activity against S. aureus, but it also accumulates in E. coli in high levels. Consistent with this enhanced accumulation, 6DNM-NH3 shows considerable activity against E. coli MG1655 (MIC = 0.5 μg ml−1) (Fig. 5). Similar to observed patterns of accumulation from the test compounds, when the amine of 6DNM-NH3 is acetylated or replaced with a carboxylic acid, the resulting compounds (6DNM-amide and 6DNM-acid) do not accumulate in E. coli and also show no activity against E. coli (Extended Data Fig. 8b). 6DNM and its derivatives were further evaluated against an expanded panel of Gram-negative pathogens, laboratory strains and clinical isolates of ESKAPE pathogens (E. coli, Acinetobacter baumannii, Klebsiella pneumoniae, Enterobacter cloacae and Pseudomonas aeruginosa) (Fig. 5, Extended Data Fig. 8b). Whereas 6DNM possesses no notable activity against these Gram-negative organisms, 6DNM-NH3 has antibacterial activity against all of these Gram-negative pathogens except P. aeruginosa. Encouragingly, 6DNM-NH3 possesses activity against a multi-drug resistant New Delhi metallo-β-lactamase-1-producing strain of E. coli (ATCC BAA-2469); this clinical isolate is highly resistant to ciprofloxacin (MIC > 64 μg ml−1; Fig. 5) and many other antibiotics, but is killed by 6DNM-NH3 with an MIC of 4 μg ml−1 (Fig. 5). Consistent with other compounds analysed, accumulation of 6DNM-NH3 decreases in the absence of the major porins (Extended Data Fig. 5b). In addition, SMD simulations were performed with 6DNM and 6DNM-NH3 as these molecules moved through the porin OmpF. Similar to SMD simulations for compound 1, the translocation of 6DNM-NH3 was assisted by a key interaction between the primary amine on 6DNM-NH3 and Asp113; 6DNM was incapable of this interaction and instead required distortion in Asp113 and neighbouring residues to allow passage (Extended Data Figs 6d–f, 7e–h).
Discussion
The combined analysis of the test set compounds and antibacterials shows the benefit of an ionizable nitrogen on small-molecule accumulation in E. coli, and suggests that this nitrogen should be embedded in a compound that has strict geometrical constraints (shape and flexibility). Analysis of antibiotics that are active against Gram-negative bacteria suggests that the ionizable nitrogen can take many different forms; for our test set, we observe that accumulation diminishes with increasing substitution on the amine, although some non-primary amines still accumulate.
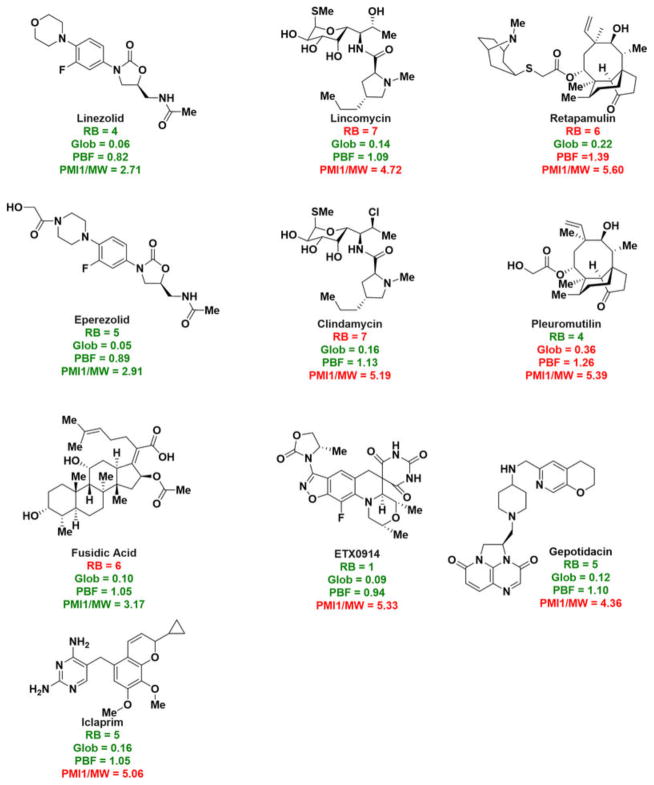
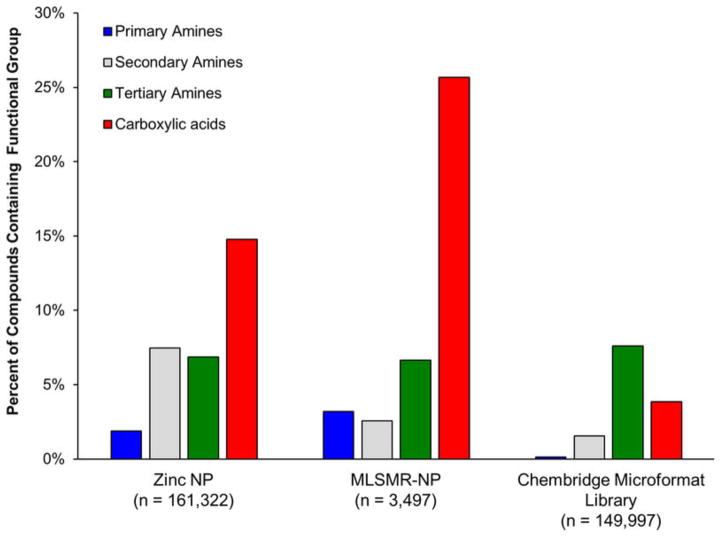
We anticipate the guidelines for predicting small-molecule accumulation in E. coli described herein will be useful in multiple contexts, including: (1) conversion of Gram-positive-only antibacterials into broad-spectrum agents. As shown in Fig. 4b, Gram-positive-only antibiotics with an ionizable nitrogen do not have the proper rigidity/shape parameters for accumulation, and it is also true that those with the proper rigidity/shape do not contain an ionizable nitrogen. As demonstrated with DNM, addition of a primary amine to compounds that meet the rigidity/shape requirements is an effective method to facilitate accumulation in E. coli. Our own chemoinformatic analysis shows that there are multiple Gram-positive-only antibiotics that are outstanding candidates for conversion to broad-spectrum agents (Extended Data Fig. 9). (2) Guiding the construction of collections of compounds that are predisposed for accumulation in Gram-negative bacteria. Primary amines are not common in natural products; only 2% of compounds from the ‘Natural Products’ subset of the ZINC database and 3% of the NIH MLSMR repository natural products contain a primary amine (Extended Data Fig. 10). Furthermore, primary amines are vanishingly rare in standard screening collections with only 0.1% of compounds in the ChemBridge Microformat Set containing a primary amine (Extended Data Fig. 10). These very low percentages are probably a major reason why screening compound collections for novel chemical matter active against Gram-negative pathogens has been unproductive1. These data suggest that assembling and screening compound collections that meet the charge, rigidity and shape requirements may be a fruitful method of discovering novel leads for development of broad-spectrum antibiotics.
P. aeruginosa is known to be even less sensitive to antibiotics than most other Gram-negative bacteria6, consistent with our results with 6DNM-NH3. However, as demonstrated by the FDA-approved antibiotics tigecycline, amoxicillin, and sulfadiazine, drugs without clinical activity against P. aeruginosa but that cover other important Gram-negative pathogens are valuable and highly sought after. A series of experiments similar to those described herein for E. coli will probably be needed to define traits that dictate accumulation of small molecules in P. aeruginosa, and this blueprint can likely also be applied to learn about compound accumulation in other problematic microorganisms; such experiments will help to define how variations in efflux pumps, porins, and lipopolysaccharide structure affect small-molecule penetrance. These analyses will provide much-needed information about compound accumulation in bacteria, aiding in the discovery and development of novel antibacterial agents.
METHODS
Accumulation assay
The accumulation assay was performed in triplicate in batches of ten samples, with each batch containing either tetracycline or ciprofloxacin as a positive control. E. coli MG1655 was used for these experiments as this strain has been only minimally altered from its K-12 progenitor33. For each replicate, 2.5 ml of an overnight culture of E. coli was diluted into 250 ml of fresh Luria Bertani (LB) broth (Lennox) and grown at 37 °C with shaking to an optical density (OD600) of 0.55. The bacteria were pelleted at 3,220 r.c.f. for 10 min at 4 °C and the supernatant was discarded. The pellets were re-suspended in 40 ml of phosphate buffered saline (PBS) and pelleted as before, and the supernatant was discarded. The pellets were re-suspended in 8.8 mL of fresh PBS and aliquoted into ten 1.5 ml eppendorf tubes (875 μl each). The number of colony-forming units (CFUs) was determined by a calibration curve. The samples were equilibrated at 37 °C with shaking for 5 min, compound was added (final concentration = 50 μM), and then samples were incubated at 37 °C with shaking for 10 min. A 10 min time point was chosen because it is longer than the predicted amount of time required to reach a steady-state concentration34, but short enough to minimize metabolic and growth changes (no changes in OD600 observed; CFUs were reduced by a factor of five after ciprofloxacin treatment for 10 min, but no other antibiotics had an effect). After incubation, 800 μl of the cultures were carefully layered on 700 μl of silicone oil (9:1 AR20/Sigma High Temperature, cooled to −78 °C). Bacteria were pelleted through the oil by centrifuging at 13,000 r.c.f. for 2 min at room temperature (supernatant remains above the oil); the supernatant and oil were then removed by pipetting. To lyse the samples, each pellet was dissolved in 200 μl of water, and then they were subjected to three freeze-thaw cycle of three minutes in liquid nitrogen followed by three minutes in a water bath at 65 °C. The lysates was pelleted at 13,000 r.c.f. for 2 min at room temperature and the supernatant was collected (180 μl). The debris was re-suspended in 100 μl of methanol and pelleted as before. The supernatants were removed and combined with the previous supernatants collected. Finally, remaining debris was removed by centrifuging at 20,000 r.c.f. for 10 min at room temperature. Supernatants were analysed by LC–MS/MS.
Samples were analysed with the 5500 QTRAP LC/MS/MS system (AB Sciex) with a 1200 series HPLC system (Agilent Technologies) including a degasser, an autosampler, and a binary pump. The liquid chromatography separation was performed on an Agilent SB-Aq column (4.6 × 50 mm, 5 μm) (Agilent Technologies) with mobile phase A (0.1% formic acid in water) and mobile phase B (0.1% formic acid in acetonitrile). The flow rate was 0.3 ml min−1. The linear gradient was as follows: 0–3 min, 100% mobile phase A; 10–15 min, 2% mobile phase A; 15.5–21 min, 100% mole phase AA. The autosampler was set at 5 °C. The injection volume was 15 μl. Mass spectra were acquired with both positive electrospray ionization at the ion spray voltage of 5,500 V and negative electrospray ionization at the ion spray voltage of −4,500 V. The source temperature was 450 °C. The curtain gas, ion source gas 1, and ion source gas 2 were 33, 50 and 65, respectively. Multiple reaction monitoring was used to quantify metabolites.
Power analysis was not used to determine the number of replicates. Error bars represent the standard error of the mean of three biological replicates. All compounds evaluated in biological assays were ≥ 95% pure.
Colistin assay
Assays measuring permeabilization by colistin were performed as above, with the addition of 6.0 μM colistin sulphate immediately before the compound of interest was added.
Bacterial strains
MRSA and P. aeruginosa isolates were from Cubist Pharmaceuticals. E. coli MG1655 was provided by C. Vanderpool (UIUC). E. coli clinical isolated were provided by L. Zechiedrich (Baylor School of Medicine). E. cloacae ATCC 29893 was provided by W. van der Donk (UIUC). A. baumannii isolates were provided by J. Quale. E. cloacae and K. pneumonia clinical isolates were provided by D. Hooper (Massachusetts General Hospital). BAA-2469 (ref. 35) was obtained from ATCC.
Antibiotic susceptibility tests
Susceptibility testing was performed in biological triplicate, using the micro-dilution broth method as outlined by the Clinical and Laboratory Standards Institute. Müller–Hinton broth was used. Strains in Fig. 5: (1) ATCC 29213; (2) NRS3; (3) MG1655, (4) ELZ4017; (5) ELZ4045; (6) ELZ4346; (7) ELZ4081; (8) ELZ4072; (9) ELZ4239; (10) BAA-2469; (11) ATCC 19606; (12) KB304; (13) KB349; (14) WO22; (15) BD335; (16) IF101; (17) ATCC 27736; (18) 4–38; (19) 6–73; (20) ATCC 29893; (21) 1–60; (22) 3–46; (23) PAO1.
Outer membrane protein profiles
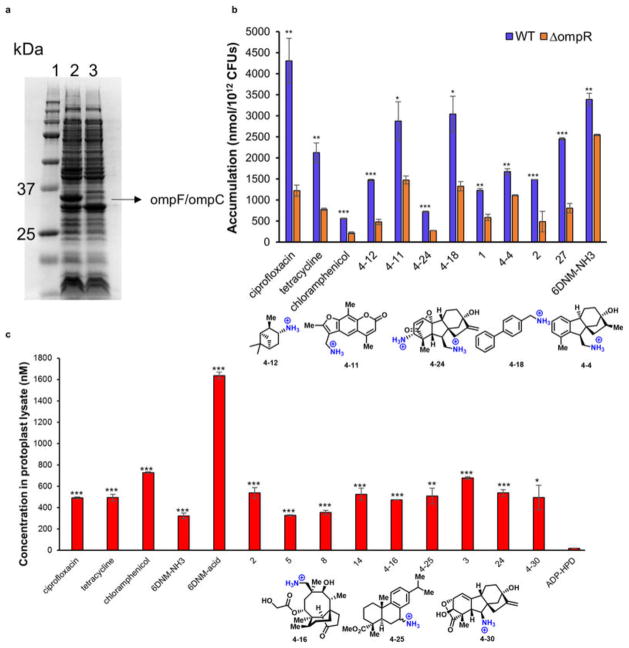
The method used to compare the outer membrane proteins of E. coli BW25113 to outer membrane of the ΔompR E. coli from the KEIO collection was adapted from ref. 36. Briefly, bacteria were grown to OD600 = 1.0 at 37 °C in LB broth. 4 ml were centrifuged for 10 min at 2,350g at 4 °C, washed with 1 ml of 100 mM Tris-HCl, pH 8.0, with 20% sucrose and incubated on ice for 10 min. Cells were pelleted as before and taken up in 1 ml of 100 mM Tris-HCl, pH 8.0, 20% sucrose containing 10 mM sodium ethylenediaminetetraacetate (EDTA). Lysozyme was added to a final concentration of 100 mg ml−1 and incubated on ice for 10 min. MgSO4 was added to 20 mM final concentration and RNaseA and DNaseI were added to a final concentration of 10 mg ml−1. Cells were disrupted with five freeze–thaw cycles in dry ice/ethanol and room temperature/water bath. A sixth freezing sample was left to thaw on ice for 2 h. Membranes were pelleted for 25 min at 16,100g at 4 °C. The supernatants were discarded, and the pellet was washed and pelleted three times in 1 ml of 20 mM NaPO4, pH 7 and 0.5% sarkosyl. The protein extracts were taken up in 60 ml of Laemmli sample buffer (80 mM Tris-HCl, pH 6.8, 3% SDS, 10% glycerol, 5% β-mercaptoethanol, 0.02% bromophenol blue), boiled for 5 min and subjected to SDS–PAGE (12% polyacrylamide). Proteins were visualized by staining with Coomassie blue-G.
Accumulation in a porin-knockout strain
As small molecules that traverse the outer membrane of Gram-negative bacteria predominately cross through porins, knocking out the major porins of E. coli would be expected to decrease compound accumulation. Although there are many porins present in E. coli, the two major non-specific porins are OmpF and OmpC, which are differentially expressed (based on the extracellular osmolarity) by the EnvZ/OmpR two-component regulatory system. A ΔompR strain of E. coli from the KEIO knockout collection was therefore chosen to effectively knockout both OmpF and OmpC (see Extended Data Fig. 5a for validation of knockout). For the control high accumulators ciprofloxacin, tetracycline and chloramphenicol, a significant decrease in accumulation is observed in the ΔompR strain of E. coli compared to the parental strain E. coli BW25113 (Extended Data Fig. 5b), consistent with previous data demonstrating that these antibiotics enter through the OmpF and OmpC porins37–39. Eight high-accumulating compounds from the test set were also evaluated, and accumulation decreased in the ΔompR strain (Extended Data Fig. 5b), suggesting these porins are a major gateway to small-molecule accumulation for the compounds tested.
Inner membrane permeability
A limitation to measuring accumulation in whole cells is that no distinction is made between periplasmic and cytoplasmic accumulation. To reach the cytoplasm of E. coli, compounds must also diffuse through the inner membrane, whose filtering properties may be different from the filtering properties of the outer membrane28. To examine this, high-accumulating compounds and some of their derivatives were tested for accumulation in E. coli protoplasts, cells lacking the outer membrane and peptidoglycan. As shown in Extended Data Fig. 5c, minimal variation in accumulation was observed between compounds in this experiment, supporting the hypothesis that traversing the outer membrane is the main barrier to small-molecule accumulation in Gram-negative bacteria.
Accumulation analysis in protoplasts
The method for preparing protoplasts was adapted from Weiss40. 85 μl of an overnight culture of E. coli MG1655 was diluted into 85 ml of fresh LB broth and grown at 37 °C with shaking to an OD600 = 1.0. The bacteria were pelleted at 3,220 r.c.f. for 10 min at 4 °C and the supernatant was discarded. The pellet was washed three times with 10 ml of 10 mM Tris HCl buffer (pH 8), and the pellet was resuspended in 30 ml of 10 mM Tris HCl (pH 8) containing 0.5 M sucrose. Potassium ethylenediaminetetraacetate (EDTA, 0.5 M, pH 8.0) was added slowly over a period of 20 min to a final concentration of 0.01 M. The bacteria were shaken at 130 r.p.m. for 20 min at 37 °C, and then collected as before. The supernatant was discarded and the pellets were washed two times with SMM buffer (0.5 M sucrose, 20 mM sodium maleate, 20 mM MgCl2, pH 6.5). The bacteria were then resuspended in 30 ml of SMM buffer, 30 mg of lysozyme was added, and the bacteria were shaken at 130 r.p.m. for 1.5 h at 37 °C. The protoplasts were harvested by centrifuging at 2,000 r.c.f. for 20 min at 4 °C.
The protoplast pellet was resuspended in 20 ml of SMM buffer, and protoplast formation was confirmed by diluting an aliquot in water and observing a threefold decrease on OD600. To test accumulation, 500 μl aliquots containing 20 μM compound were shaken at 130 r.p.m. for 5 min at 37 °C. Samples were pelleted at 2,000 r.c.f. for 10 min at room temperature and the supernatants were discarded. The pellets were resuspended in 200 μl of water and incubated at room temperature for 5 min. The lysed protoplasts were pelleted by centrifuging at 21,130 r.c.f. for 10 min, and the supernatant was analysed by LC–MS/MS for compound concentration as before.
Data analysis and statistics
Statistical significance of accumulation was determined by using a two sample Welch’s t-test (one-tailed test, assuming unequal variance) relative to the negative controls. Variance in accumulation across molecules was found to be unequal by Bartlett’s test. For all other experiments, statistical significance was determined using an unpaired Student’s t-test assuming equal variance as determined by an F-test.
Sources of compound libraries for computational studies
Chemical structure data for the MicroFormat library was obtained from the ChemBridge website. Chemical structure data for the ZINC Natural Products library was obtained from http://zinc.docking.org/browse/catalogs/natural-products. Chemical structure data for the MLSMR-NP library was obtained from PubChem.
Identification of predicted accumulators in commercial libraries
Compounds containing particular functional groups were identified and counted by substructure filtering with Open Babel, using the following SMARTS queries: primary aliphatic amine, [$([N;H2;X3][CX4]),$([N;H3;X4+][CX4])]; secondary aliphatic amine, [$([CX4][N;H1;X3][CX4]),$([CX4][N;H2;X4+][CX4])]; tertiary aliphatic amine, [NX3]([CX4])([CX4])[CX4]; carboxylic acid: [CX3](= O)[OX1H0–, OX2H1].
Calculation of physiochemical properties
Data sets of chemical structures were created and managed using Canvas (Version 2.6, Schrödinger, LLC). Initial structure preparation and 3D minimization was performed with LigPrep (Version 3.6, Schrödinger, LLC) using OPLS_2005 force fields. Tautomeric and protonation states were determined using Epik (Version 3.4, Schrödinger, LLC) at pH 7.4 (refs 41, 42). Generation of ensembles of conformations was performed using Conformational Search in MOE 2015.10 (ref. 43) using the LowModeMD method with default settings.
Physiochemical descriptors (297, both 2D- and 3D-based) were calculated using MOE for each conformation. Descriptors were averaged (unweighted mean) across all conformations for each molecule. Data with descriptors were used to train a random forest classification prediction model using the R package caret. The random forest model offers many advantages for this application including resistance to over-fitting and the ability to measure descriptor importance44. Preprocessing of data removed descriptors with near-zero variance or high co-correlation with other descriptors. Source code for data analysis and model training can be found at https://github.com/HergenrotherLab/GramNegAccum
An additional approximation of three-dimensionality, average distance to the plane of best fit (PBF), was calculated using a custom Python program. The PBF algorithm45 was implemented in Python with SDfile I/O and structure representation being handled by libraries from Schrödinger. For each compound, the PBF algorithm determines the plane that best fits a set of 3D coordinates that represent the positions of all heavy atoms in the molecule using single value decomposition. The distance of each heavy atom to this plane is measured in angstroms and averaged. This Python program was incorporated into the Maestro GUI for convenient use. Source code can be found at https://github.com/HergenrotherLab/GramNegAccum
ClogD7.4 values were calculated using the online compound property calculation software FAFdrugs3 (http://fafdrugs3.mti.univ-paris-diderot.fr/index.html).
Definitions of reported physiochemical properties
Rotatable bonds: count of single bonds, not in a ring, bound to a non-terminal heavy atom. Excluded from the count are C–N amide bonds because of their high rotational energy barrier46. ClogD7.4, the predicted octanol/water distribution coefficient at pH 7.4; globularity, inverse condition number of the covariance matrix of atomic coordinates (a value of 1 indicates a perfect sphere while a value of 0 indicates a two- or one-dimensional object); PBF, average distance in angstroms of each heavy atom in the molecule to the plane that best fits atomic coordinates; PMI 1, first diagonal element of diagonalized moment of inertia tensor47; PMI1/molecular weight, ratio of PMI 1 and molecular weight; vsurf_A, a vector pointing from the centre of the hydrophobic domain to the centre of the hydrophilic domain. The vector length is proportional to the strength of the amphiphilic moment27. Full experimental details and characterization data for all new compounds are included in the Supplementary Information.
All-atom molecular dynamics simulations
Although SMD does not directly provide the free energy landscape, SMD is frequently employed to map the pathway for long timescale processes and has been previously used to study OmpF48. The simulation model was constructed using CHARMM-GUI49–51 and comprised one OmpF monomer (PDB 3POX), 108 (90%) 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE) lipid molecules, 12 (10%) 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1′-rac-glycerol) (POPG)52, and solvated with 8,234 water molecules in 150 mM NaCl (36 Na+ and 15 Cl−) for a total of 45,402 atoms. Hexagonal periodic boundary conditions were applied with a distance of 77.3 Å in the xy-direction and 92.8 Å in the z-direction. Electrostatic interactions were calculated using the particle-mesh Ewald (PME) method. Protein residues E296, D312, and D127 were protonated53–55. The simulations were performed at constant pressure (1 atm) and temperature (303 K) with a time step of 2 fs. Each small-molecule under investigation was manually placed directly above the pore. Restraints were initially applied to protein backbone and small-molecule analyte atoms and then removed to equilibrate the system. For SMD production simulations, each small-molecule analyte was pulled at the molecules centre of mass (5 kcal mol−1 Å−2) at a constant velocity (10 Å ns−1) along the z-axis for 4 ns. The all-atom CHARMM force field was used for protein and lipids56,57. TIP3P was used for water58. All the molecular dynamics simulations were carried out using the NAMD 2.11 scalable molecular dynamics program and run on Stampede at TACC. CHARMM residue topology and parameter files for the small molecules were constructed using CGenFF59. SMD trajectories were analysed and visualized using VMD 1.9.2 and rendered using Pov-Ray 3.6 (ref. 60).
Data availability
Source code for data analysis and model training can be found at https://github.com/HergenrotherLab/GramNegAccum. The authors declare that all other data supporting the findings of this study are available within the paper and the Supplementary Information.
Extended Data
Extended Data Figure 1. ClogD7.4 and molecular weight do not correlate with accumulation of primary amines.
a, b, The accumulation of 68 primary amine containing compounds in E. coli MG1655 compared to (a) ClogD7.4 and (b) molecular weight. High-accumulating controls (black x) = tetracycline, ciprofloxacin and chloramphenicol; low-accumulating controls (open circles) = novobiocin, erythromycin, rifampicin, clindamycin, mupirocin and fusidic acid, clindamycin, and ampicillin. Statistical significance was determined by using a two sample Welch’s t-test (one tailed test, assuming unequal variance) relative to the negative controls. P values are relative to the average of the low-accumulating controls. *P < 0.05, **P < 0.01. Structures of all 68 compounds are shown in Supplementary Table 4. All experiments were performed in biological triplicate and error bars represent s.e.m.
Extended Data Figure 2. Training and performance of random tree classification model.
a, ROC plot comparing cross-validation methods in training classification model. b, Tuning plot optimizing for number of selected predictors. c, Confusion matrix of classification model using repeated tenfold cross-validation (n = 20). d, Relative importance of top 15 predictors. Predictors related to flexibility, shape, and amphiphilicity are highlighted in red.
Extended Data Figure 3.
Accumulation of paired set of compounds containing one or two primary amines. Error bars equal s.e.m. Statistical significance was determined by using a two sample Student’s t-test (two tailed test, assuming equal variance). *P < 0.05, **P < 0.01. All experiments were performed in biological triplicate.
Extended Data Figure 4. Additional scatter plots of small-molecule bacterial accumulators based on molecular shape descriptors.
a, Scatter plot of primary amines (compounds from Fig. 4a) according to rotatable bond count and average distance to the plane-of-best-fit (PBF)45. b, Scatter plot of primary amines (compounds from Fig. 4a) according to rotatable bond count and normalized principal moment of inertia (PMI1/molecular weight)47. c, Scatter plot of antibiotics (same compounds from Fig. 4b, plus β-lactams that are active against Gram-negative bacteria) according to rotatable bond count and PBF45. d, Scatter plot of antibiotics (same compounds from Fig. 4b, plus β-lactams that are active against Gram-negative bacteria) according to rotatable bond count and normalized principal moment of inertia (PMI1/molecular weight). e, Same plot as Fig. 4b, but this time the β-lactams that are active against Gram-negative bacteria are included. Structures of all antibiotics are in Supplementary Tables 6, 7.
Extended Data Figure 5. Mechanistic studies.
a, SDS–PAGE analysis of outer membrane proteins of E. coli BW25113 and E. coli BW25113 ΔompR from the KEIO collection. E. coli outer membrane proteins were separated using SDS-12% PAGE, and visualized with Coomassie stain. Lane 1, protein ladder; lane 2, E. coli BW25113; lane 3, E. coli BW25113 ΔompR. Representative of three replicates. b, Accumulation comparison of 12 compounds for E. coli ΔompR vs parental strain E. coli BW25113. c, Small-molecule accumulation in protoplasts prepared from E. coli MG1655. Test compounds and known antibiotics are able to accumulate in the protoplast. ADP-HPD61,62, a non-cell permeable pyrophosphate, was used as a negative control. Compounds are indicated by the number assigned in the manuscript or the Supplementary Tables. For the compounds in the Supplementary Tables (whose structures are not shown in the manuscript), the structures are shown below the graph. Statistical significance was determined by using a two sample Student’s t-test (two tailed test, assuming equal variance). *P < 0.05, **P < 0.01, ***P < 0.001. All experiments were performed in biological triplicate.
Extended Data Figure 6.
a, Distance of primary amine of compound 1 and constriction site residues in course of simulation. Stabilized distance at ~2 Å is indicative of hydrogen bonding. In repeated simulations molecules adopted similar pathways for traversing the porin while often hydrogen bonding with different hydrophilic residues. b, Displacement of Arg42 by compound 1 relative to initial coordinates was measured during course of simulation. c, Displacement of Arg42 by compound 13. d, Distance of primary amine of 6DNM-NH3 and constriction site residues in course of simulation. e, The displacement of Asp113 by 6DNM-NH3. f, Displacement of Asp113 by 6DNM.
Extended Data Figure 7. Snapshots of SMD simulation of compound translocation through OmpF.
a, Compound 1 is capable of making a key interaction with Asp113 that assists in movement past constriction site. This finding is in accord with previous reports of the importance of Asp113 in producing the cation selectivity of OmpF63,64. b, Same pose as a, viewed from above. c, Compound 13 makes no interactions with Asp113 and thus faces additional barriers to penetrance. d, Same pose as c, viewed from above. e, 6DNM-NH3 makes key H-bonding interactions with Asp113 (purple) that assists in the molecule’s passage through the constriction site. f, Same pose as e, viewed from above. g, 6DNM requires larger movement by peptide backbone in order to pass through constriction site. Asp113 is appreciably displaced by 6DNM. h, Same pose as g, viewed from above. In all simulations, the largest barrier to translocation is a series of stacked arginine residues (Arg 42, 82, and 132) that are depicted in blue.
Extended Data Figure 8.
a, Gram-positive-only antibiotics with proper rotatable bond and globularity scores can be converted to broad-spectrum drugs via strategic placement of a primary amine, as demonstrated by ampicillin. However, for drugs that do not possess the proper flexibility and three-dimensionality parameters, such as erythromycin, addition of the amine does not broaden the antibacterial spectrum. b, Strategic placement of a primary amine (but not an acid or amide) on 6DNM leads to 6DNM-NH3, a broad-spectrum antibacterial. 6DNM and three derivatives were synthesized and evaluated against a panel of Gram-positive and Gram-negative organisms; ciprofloxacin was also evaluated. Flexibility (as measured by the number of rotatable bonds, RB), globularity (Glob), molecular weight, ClogD7.4, and E. coli accumulation (in nmol per 1012 CFUs) is provided. The s.e.m. for accumulation values are reported. MIC values were determined using the micro-dilution broth method as outlined by the Clinical and Laboratory Standards Institute and are listed in μg ml−1. All experiments were performed in biological triplicate.
Extended Data Figure 9.
Some FDA-approved drugs or compounds in phase II or III clinical trials that are good candidates for derivative synthesis and expansion to broad-spectrum agents. Placement of a primary amine at a position that does not alter interaction with the biological target is predicted to provide compounds that accumulate in and are active against Gram-negative bacteria.
Extended Data Figure 10. Primary amines are largely absent from many screening collections.
Comparison of the abundance of primary amines, secondary amines, tertiary amines, and carboxylic acids in the natural products subset of the ZINC database, the Molecular Libraries Small-molecule Repository – Natural Products (MLSMR-NP), and the Chembridge Microformat library.
Supplementary Material
Supplementary Table 1. Data for all control antibiotics. Structures, spectrum of activity, MW, ClogD7.4, number of rotational bonds (RB), globularity (Glob), the plane-of-best-fit score (PBF)45, normalized first principal moment of inertia (PMI1/MW)47, and the accumulation for each antibiotic in E. coli MG1655 is provided, as well as standard error of three replicates.
Supplementary Table 2. Data for initial test set of 100 compounds. Structures, functional groups present, MW, ClogD7.4, number of rotational bonds (RB), globularity (Glob), the plane-of-best-fit score (PBF)45, normalized first principal moment of inertia (PMI1/MW)47, and the accumulation in E. coli MG1655 for each compound is provided, as well as standard error of three replicates.
Supplementary Table 3. Data for compounds involved in structure-activity relationship analysis. Structures, functional groups present, MW, ClogD7.4, number of rotational bonds (RB), globularity (Glob), the plane-of-best-fit score (PBF)45, normalized first principal moment of inertia (PMI1/MW)47, and the accumulation in E. coli MG1655 for each compound is provided, as well as standard error of three replicates.
Supplementary Table 4. Data for all primary amines compounds tested. Structures, functional groups present, MW, ClogD7.4, number of rotational bonds (RB), globularity (Glob), the plane-of-best-fit score (PBF)45, normalized first principal moment of inertia (PMI1/MW)47 and the accumulation in E. coli MG1655 for each compound is provided, as well as standard error of three replicates.
Supplementary Table 5. Definitions and distributions of molecular descriptors used to train classification model, and the complete dataset used to train classification model.
Supplementary Table 6. All antibiotics analyzed in chemoinformatic analysis (derived from the compounds assessed by O’Shea and Moser11). Structures, spectrum of activity, MW, ClogD7.4, number of rotational bonds (RB), globularity (Glob), the plane-of-best-fit score (PBF)45, normalized first principal moment of inertia (PMI1/MW)47, and the spectrum of activity for compound is provided.
Supplementary Table 7. All β-lactams analyzed in chemoinformatic analysis (derived from the compounds assessed by O’Shea and Moser11). Structures, spectrum of activity, MW, ClogD7.4, number of rotational bonds (RB), globularity (Glob), the plane-of-best-fit score (PBF)45, normalized first principal moment of inertia (PMI1/MW)47, spectrum of activity, and generation for compound is provided.
Acknowledgments
We thank L. Li (Metabolomics Center, Roy J. Carver Biotechnology Center, UIUC) for LC–MS/MS analysis, L. Zechiedrich (Baylor College of Medicine) for the E. coli clinical isolates, C. Vanderpool (UIUC) for E. coli MG1655, and W. van der Donk (UIUC) for E. cloacae. We also thank K. Hull, S. Denmark, K. Morrison, R. Hicklin, H. Roth, B. Nakamura, A. Deets and A. Keyes for providing valuable compounds for the test set, and we thank M. Lambrecht for NMR expertise. This work was funded by the UIUC, including funds obtained through the Office of Technology Management Proof-of-Concept award. M.F.R. is a NSF predoctoral fellow. M.F.R., A.G., and R.L.S. are members of the NIH Chemistry-Biology Interface Training Grant (NRSA 1-T32-GM070421). A.P.R. is an NIH postdoctoral fellow. T.S. was supported by the Kao Corporation. Computer time was provided by the Texas Advanced Computing Center through Grant TG-CHE160050 funded by the NSF.
Footnotes
Supplementary Information is available in the online version of the paper.
Author Contributions P.J.H., M.F.R. and B.S.D. conceived the study. M.F.R. performed accumulation analyses. B.S.D. performed the computational analyses. M.F.R., B.S.D, A.P.R., A.G., T.S. and R.L.S. synthesized compounds in the test set. A.P.R. synthesized and tested DNM derivatives. P.J.H. supervised this research and wrote this manuscript with the assistance of M.F.R., B.S.D. and A.P.R.
The authors declare competing financial interests; details are available in the online version of the paper. Readers are welcome to comment on the online version of the paper. Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Reviewer Information Nature thanks P. A. Clemons, S. Khalid, S. L. Schreiber, O. Verho, G. D. Wright and the other anonymous reviewer(s) for their contribution to the peer review of this work.
References
- 1.Payne DJ, Gwynn MN, Holmes DJ, Pompliano DL. Drugs for bad bugs: confronting the challenges of antibacterial discovery. Nat Rev Drug Discov. 2007;6:29–40. doi: 10.1038/nrd2201. [DOI] [PubMed] [Google Scholar]
- 2.Fischbach MA, Walsh CT. Antibiotics for emerging pathogens. Science. 2009;325:1089–1093. doi: 10.1126/science.1176667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boucher HW, et al. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin Infect Dis. 2009;48:1–12. doi: 10.1086/595011. [DOI] [PubMed] [Google Scholar]
- 4.Lewis K. Platforms for antibiotic discovery. Nat Rev Drug Discov. 2013;12:371–387. doi: 10.1038/nrd3975. [DOI] [PubMed] [Google Scholar]
- 5.Nikaido H. Prevention of drug access to bacterial targets: permeability barriers and active efflux. Science. 1994;264:382–388. doi: 10.1126/science.8153625. [DOI] [PubMed] [Google Scholar]
- 6.Silver LL. Challenges of antibacterial discovery. Clin Microbiol Rev. 2011;24:71–109. doi: 10.1128/CMR.00030-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tommasi R, Brown DG, Walkup GK, Manchester JI, Miller AA. ESKAPEing the labyrinth of antibacterial discovery. Nat Rev Drug Discov. 2015;14:529–542. doi: 10.1038/nrd4572. [DOI] [PubMed] [Google Scholar]
- 8.Nikaido H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev. 2003;67:593–656. doi: 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carpenter TS, Parkin J, Khalid S. The free energy of small solute permeation through the Escherichia coli outer membrane has a distinctly asymmetric profile. J Phys Chem Lett. 2016;7:3446–3451. doi: 10.1021/acs.jpclett.6b01399. [DOI] [PubMed] [Google Scholar]
- 10.Cowan SW, et al. Crystal structures explain functional properties of two E. coli porins. Nature. 1992;358:727–733. doi: 10.1038/358727a0. [DOI] [PubMed] [Google Scholar]
- 11.O’Shea R, Moser HE. Physicochemical properties of antibacterial compounds: implications for drug discovery. J Med Chem. 2008;51:2871–2878. doi: 10.1021/jm700967e. [DOI] [PubMed] [Google Scholar]
- 12.Brown DG, May-Dracka TL, Gagnon MM, Tommasi R. Trends and exceptions of physical properties on antibacterial activity for Gram-positive and Gram-negative pathogens. J Med Chem. 2014;57:10144–10161. doi: 10.1021/jm501552x. [DOI] [PubMed] [Google Scholar]
- 13.Bazile S, Moreau N, Bouzard D, Essiz M. Relationships among antibacterial activity, inhibition of DNA gyrase, and intracellular accumulation of 11 fluoroquinolones. Antimicrob Agents Chemother. 1992;36:2622–2627. doi: 10.1128/aac.36.12.2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davis TD, Gerry CJ, Tan DS. General platform for systematic quantitative evaluation of small-molecule permeability in bacteria. ACS Chem Biol. 2014;9:2535–2544. doi: 10.1021/cb5003015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nikaido H, Rosenberg EY, Foulds J. Porin channels in Escherichia coli: studies with β-lactams in intact cells. J Bacteriol. 1983;153:232–240. doi: 10.1128/jb.153.1.232-240.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou Y, et al. Thinking outside the ‘bug’: a unique assay to measure intracellular drug penetration in Gram-negative bacteria. Anal Chem. 2015;87:3579–3584. doi: 10.1021/ac504880r. [DOI] [PubMed] [Google Scholar]
- 17.Brown DM, Acred P. ‘Penbritin’—a new broad-spectrum antibiotic. BMJ. 1961;2:197–198. doi: 10.1136/bmj.2.5246.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cai H, Rose K, Liang LH, Dunham S, Stover C. Development of a liquid chromatography/mass spectrometry-based drug accumulation assay in Pseudomonas aeruginosa. Anal Biochem. 2009;385:321–325. doi: 10.1016/j.ab.2008.10.041. [DOI] [PubMed] [Google Scholar]
- 19.Vaara M. Agents that increase the permeability of the outer membrane. Microbiol Rev. 1992;56:395–411. doi: 10.1128/mr.56.3.395-411.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huigens RW, III , et al. A ring-distortion strategy to construct stereochemically complex and structurally diverse compounds from natural products. Nat Chem. 2013;5:195–202. doi: 10.1038/nchem.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rafferty RJ, Hicklin RW, Maloof KA, Hergenrother PJ. Synthesis of complex and diverse compounds through ring distortion of abietic acid. Angew Chem Int Ed. 2014;53:220–224. doi: 10.1002/anie.201308743. [DOI] [PubMed] [Google Scholar]
- 22.Garcia A, Drown BS, Hergenrother PJ. Access to a structurally complex compound collection via ring distortion of the alkaloid sinomenine. Org Lett. 2016;18:4852–4855. doi: 10.1021/acs.orglett.6b02333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hicklin RW, López Silva TL, Hergenrother PJ. Synthesis of bridged oxafenestranes from pleuromutilin. Angew Chem Int Ed. 2014;53:9880–9883. doi: 10.1002/anie.201404765. [DOI] [PubMed] [Google Scholar]
- 24.Molecular Operating Environment (MOE), 2015, 10. 1010 Sherbooke St. West, Suite #910, Montreal, QC, Canada, H3A 2R7: 2016. [Google Scholar]
- 25.Kuenemann MA, Labbé CM, Cerdan AH, Sperandio O. Imbalance in chemical space: How to facilitate the identification of protein-protein interaction inhibitors. Sci Rep. 2016;6:23815. doi: 10.1038/srep23815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuenemann MA, Bourbon LML, Labbé CM, Villoutreix BO, Sperandio O. Which three-dimensional characteristics make efficient inhibitors of protein-protein interactions? J Chem Inf Model. 2014;54:3067–3079. doi: 10.1021/ci500487q. [DOI] [PubMed] [Google Scholar]
- 27.Mannhold R, Kubinyi H, Folkers G, Cruciani G. Molecular Interaction Fields: Applications in Drug Discovery and ADME Prediction. Vol. 27. John Wiley & Sons; 2006. [Google Scholar]
- 28.Silver LL. A Gestalt approach to Gram-negative entry. Bioorg Med Chem. 2016;24:6379–6389. doi: 10.1016/j.bmc.2016.06.044. [DOI] [PubMed] [Google Scholar]
- 29.Yoshimura F, Nikaido H. Diffusion of β-lactam antibiotics through the porin channels of Escherichia coli K-12. Antimicrob Agents Chemother. 1985;27:84–92. doi: 10.1128/aac.27.1.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Massey EH, Kitchell BS, Martin LD, Gerzon K. Antibacterial activity of 9(S)-erythromycylamine-aldehyde condensation products. J Med Chem. 1974;17:105–107. doi: 10.1021/jm00247a018. [DOI] [PubMed] [Google Scholar]
- 31.Hiramatsu K, et al. Curing bacteria of antibiotic resistance: reverse antibiotics, a novel class of antibiotics in nature. Int J Antimicrob Agents. 2012;39:478–485. doi: 10.1016/j.ijantimicag.2012.02.007. [DOI] [PubMed] [Google Scholar]
- 32.Parkinson EI, et al. Deoxynybomycins inhibit mutant DNA gyrase and rescue mice infected with fluoroquinolone-resistant bacteria. Nat Commun. 2015;6:6947. doi: 10.1038/ncomms7947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blattner FR, et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 34.Williams KJ, Piddock LJ. Accumulation of rifampicin by Escherichia coli and Staphylococcus aureus. J Antimicrob Chemother. 1998;42:597–603. doi: 10.1093/jac/42.5.597. [DOI] [PubMed] [Google Scholar]
- 35.Kumarasamy KK, et al. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect Dis. 2010;10:597–602. doi: 10.1016/S1473-3099(10)70143-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Adler M, Anjum M, Andersson DI, Sandegren L. Influence of acquired β-lactamases on the evolution of spontaneous carbapenem resistance in Escherichia coli. J Antimicrob Chemother. 2013;68:51–59. doi: 10.1093/jac/dks368. [DOI] [PubMed] [Google Scholar]
- 37.Thanassi DG, Suh GS, Nikaido H. Role of outer membrane barrier in efflux-mediated tetracycline resistance of Escherichia coli. J Bacteriol. 1995;177:998–1007. doi: 10.1128/jb.177.4.998-1007.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chapman JS, Georgopapadakou NH. Routes of quinolone permeation in Escherichia coli. Antimicrob Agents Chemother. 1988;32:438–442. doi: 10.1128/aac.32.4.438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mortimer PG, Piddock LJ. The accumulation of five antibacterial agents in porin-deficient mutants of Escherichia coli. J Antimicrob Chemother. 1993;32:195–213. doi: 10.1093/jac/32.2.195. [DOI] [PubMed] [Google Scholar]
- 40.Weiss RL. Protoplast formation in Escherichia coli. J Bacteriol. 1976;128:668–670. doi: 10.1128/jb.128.2.668-670.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Greenwood JR, Calkins D, Sullivan AP, Shelley JC. Towards the comprehensive, rapid, and accurate prediction of the favorable tautomeric states of drug-like molecules in aqueous solution. J Comput Aided Mol Des. 2010;24:591–604. doi: 10.1007/s10822-010-9349-1. [DOI] [PubMed] [Google Scholar]
- 42.Shelley JC, et al. Epik: a software program for pKa prediction and protonation state generation for drug-like molecules. J Comput Aided Mol Des. 2007;21:681–691. doi: 10.1007/s10822-007-9133-z. [DOI] [PubMed] [Google Scholar]
- 43.Labute P. LowModeMD—implicit low-mode velocity filtering applied to conformational search of macrocycles and protein loops. J Chem Inf Model. 2010;50:792–800. doi: 10.1021/ci900508k. [DOI] [PubMed] [Google Scholar]
- 44.Breiman L. Random forests. Mach Learn. 2001;45:5–32. [Google Scholar]
- 45.Firth NC, Brown N, Blagg J. Plane of best fit: a novel method to characterize the three-dimensionality of molecules. J Chem Inf Model. 2012;52:2516–2525. doi: 10.1021/ci300293f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sastry M, Lowrie JF, Dixon SL, Sherman W. Large-scale systematic analysis of 2D fingerprint methods and parameters to improve virtual screening enrichments. J Chem Inf Model. 2010;50:771–784. doi: 10.1021/ci100062n. [DOI] [PubMed] [Google Scholar]
- 47.Sauer WH, Schwarz MK. Molecular shape diversity of combinatorial libraries: a prerequisite for broad bioactivity. J Chem Inf Comput Sci. 2003;43:987–1003. doi: 10.1021/ci025599w. [DOI] [PubMed] [Google Scholar]
- 48.Ziervogel BK, Roux B. The binding of antibiotics in OmpF porin. Structure. 2013;21:76–87. doi: 10.1016/j.str.2012.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu EL, et al. CHARMM-GUI Membrane Builder toward realistic biological membrane simulations. J Comput Chem. 2014;35:1997–2004. doi: 10.1002/jcc.23702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee J, et al. CHARMM-GUI input generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM simulations using the CHARMM36 additive force field. J Chem Theory Comput. 2016;12:405–413. doi: 10.1021/acs.jctc.5b00935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jo S, Kim T, Iyer VG, Im W. CHARMM-GUI: a web-based graphical user interface for CHARMM. J Comput Chem. 2008;29:1859–1865. doi: 10.1002/jcc.20945. [DOI] [PubMed] [Google Scholar]
- 52.Lugtenberg EJ, Peters R. Distribution of lipids in cytoplasmic and outer membranes of Escherichia coli K12. Biochim Biophys Acta. 1976;441:38–47. doi: 10.1016/0005-2760(76)90279-4. [DOI] [PubMed] [Google Scholar]
- 53.Im W, Roux B. Ion permeation and selectivity of OmpF porin: a theoretical study based on molecular dynamics, Brownian dynamics, and continuum electrodiffusion theory. J Mol Biol. 2002;322:851–869. doi: 10.1016/s0022-2836(02)00778-7. [DOI] [PubMed] [Google Scholar]
- 54.Im W, Roux B. Ions and counterions in a biological channel: a molecular dynamics simulation of OmpF porin from Escherichia coli in an explicit membrane with 1 M KCl aqueous salt solution. J Mol Biol. 2002;319:1177–1197. doi: 10.1016/S0022-2836(02)00380-7. [DOI] [PubMed] [Google Scholar]
- 55.Varma S, Chiu SW, Jakobsson E. The influence of amino acid protonation states on molecular dynamics simulations of the bacterial porin OmpF. Biophys J. 2006;90:112–123. doi: 10.1529/biophysj.105.059329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Feller SE, Yin D, Pastor RW, MacKerell AD., Jr Molecular dynamics simulation of unsaturated lipid bilayers at low hydration: parameterization and comparison with diffraction studies. Biophys J. 1997;73:2269–2279. doi: 10.1016/S0006-3495(97)78259-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.MacKerell AD, et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 58.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79:926–935. [Google Scholar]
- 59.Vanommeslaeghe K, et al. CHARMM general force field: a force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J Comput Chem. 2010;31:671–690. doi: 10.1002/jcc.21367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 61.Slama JT, Aboul-Ela N, Jacobson MK. Mechanism of inhibition of poly(ADP-ribose) glycohydrolase by adenosine diphosphate (hydroxymethyl)pyrrolidinediol. J Med Chem. 1995;38:4332–4336. doi: 10.1021/jm00021a023. [DOI] [PubMed] [Google Scholar]
- 62.Slama JT, et al. Specific inhibition of poly(ADP-ribose) glycohydrolase by adenosine diphosphate (hydroxymethyl)pyrrolidinediol. J Med Chem. 1995;38:389–393. doi: 10.1021/jm00002a021. [DOI] [PubMed] [Google Scholar]
- 63.Hajjar E, et al. Bridging timescales and length scales: from macroscopic flux to the molecular mechanism of antibiotic diffusion through porins. Biophys J. 2010;98:569–575. doi: 10.1016/j.bpj.2009.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Danelon C, Suenaga A, Winterhalter M, Yamato I. Molecular origin of the cation selectivity in OmpF porin: single channel conductances vs. free energy calculation. Biophys Chem. 2003;104:591–603. doi: 10.1016/s0301-4622(03)00062-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1. Data for all control antibiotics. Structures, spectrum of activity, MW, ClogD7.4, number of rotational bonds (RB), globularity (Glob), the plane-of-best-fit score (PBF)45, normalized first principal moment of inertia (PMI1/MW)47, and the accumulation for each antibiotic in E. coli MG1655 is provided, as well as standard error of three replicates.
Supplementary Table 2. Data for initial test set of 100 compounds. Structures, functional groups present, MW, ClogD7.4, number of rotational bonds (RB), globularity (Glob), the plane-of-best-fit score (PBF)45, normalized first principal moment of inertia (PMI1/MW)47, and the accumulation in E. coli MG1655 for each compound is provided, as well as standard error of three replicates.
Supplementary Table 3. Data for compounds involved in structure-activity relationship analysis. Structures, functional groups present, MW, ClogD7.4, number of rotational bonds (RB), globularity (Glob), the plane-of-best-fit score (PBF)45, normalized first principal moment of inertia (PMI1/MW)47, and the accumulation in E. coli MG1655 for each compound is provided, as well as standard error of three replicates.
Supplementary Table 4. Data for all primary amines compounds tested. Structures, functional groups present, MW, ClogD7.4, number of rotational bonds (RB), globularity (Glob), the plane-of-best-fit score (PBF)45, normalized first principal moment of inertia (PMI1/MW)47 and the accumulation in E. coli MG1655 for each compound is provided, as well as standard error of three replicates.
Supplementary Table 5. Definitions and distributions of molecular descriptors used to train classification model, and the complete dataset used to train classification model.
Supplementary Table 6. All antibiotics analyzed in chemoinformatic analysis (derived from the compounds assessed by O’Shea and Moser11). Structures, spectrum of activity, MW, ClogD7.4, number of rotational bonds (RB), globularity (Glob), the plane-of-best-fit score (PBF)45, normalized first principal moment of inertia (PMI1/MW)47, and the spectrum of activity for compound is provided.
Supplementary Table 7. All β-lactams analyzed in chemoinformatic analysis (derived from the compounds assessed by O’Shea and Moser11). Structures, spectrum of activity, MW, ClogD7.4, number of rotational bonds (RB), globularity (Glob), the plane-of-best-fit score (PBF)45, normalized first principal moment of inertia (PMI1/MW)47, spectrum of activity, and generation for compound is provided.
Data Availability Statement
Source code for data analysis and model training can be found at https://github.com/HergenrotherLab/GramNegAccum. The authors declare that all other data supporting the findings of this study are available within the paper and the Supplementary Information.