Abstract
S531 of Escherichia coli RNA polymerase (RNAP) β subunit is a part of RNA binding domain in transcription complex. While highly conserved, S531 is not involved in interactions within the transcription complex as suggested by X-ray analysis. To understand the basis for S531 conservation we performed systematic mutagenesis of this residue. We find that the most of the mutations significantly decreased initiation-to-elongation transition by RNAP. Surprisingly, some changes enhanced the production of full-size transcripts by suppressing abortive loss of short RNAs. S531-R increased transcript retention by establishing a salt bridge with RNA, thereby explaining the R substitution at the equivalent position in extremophilic organisms, in which short RNAs retention is likely to be an issue. Generally, the substitutions had the same effect on bacterial doubling time when measured at 20°. Raising growth temperature to 37° ablated the positive influence of some mutations on the growth rate in contrast to their in vitro action, reflecting secondary effects of cellular environment on transcription and complex involvement of 531 locus in the cell biology. The properties of generated RNAP variants revealed an RNA/protein interaction network that is crucial for transcription, thereby explaining the details of initiation-to-elongation transition on atomic level.
INTRODUCTION
RNA polymerase is a complex molecular machine that transfers genetic information from DNA to messenger RNA. Several events occur in this process (1,2): (i) promoter recognition by RNAP holoenzyme (subunit composition α2ββ′ωσ); (ii) initiation of RNA synthesis; (iii) elongation of RNA transcripts and (iv) termination (release of full-size RNA transcript). A key event is the transition from initiation of synthesis to elongation of the transcript. Examination of X-ray structures and biochemical observations lead to the following scenario. Short RNA oligonucleotides, synthesized de novo in the transcription initiation complex (TIC) at the beginning of the transcription cycle, readily dissociate from the active center and cause cycles of what is called abortive initiation. This process can repeat many times before a synthesized product occasionally acquires new stabilizing contacts with RNAP (Figure 1B), as the transcript enters the hybrid binding site (HBS) of the enzyme (3). Further advancement of growing RNA into the HBS triggers promoter clearance, a process involving displacement of the RNAP σ subunit and a dramatic stabilization of the RNA/DNA/RNAP ensemble as a ternary elongation complex (TEC). Importantly, the efficiency of transcription from each particular promoter depends on how many abortive cycles are performed before RNAP escapes to elongation. Therefore, acquisition of RNA-HBS contacts is a crucial event determining the productivity of RNA synthesis.
Figure 1.
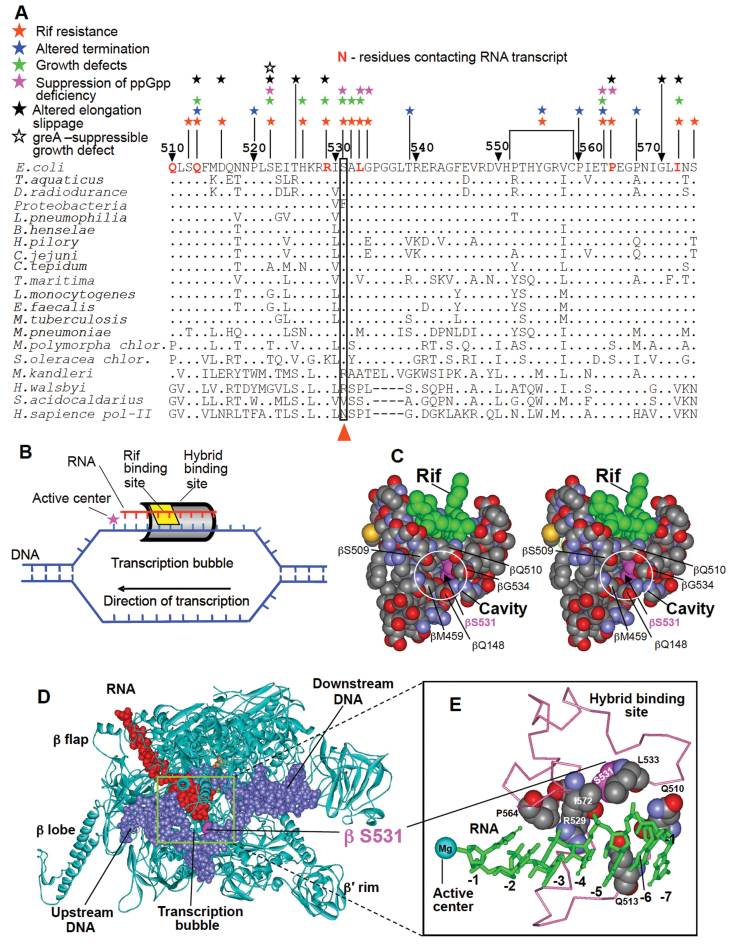
Genetic and structural context of RNA:DNA hybrid-interacting region of RNAP β subunit. (A) Sequence alignment of 510–574 region (Escherichia coli numbering) of RNAP β subunit from various organisms. Amino acid residues whose mutations confer specific phenotypes are indicated. Dots represent identical residues; hyphens mark deletions. S531 or equivalent residues in various sequences are boxed and indicated. (B) The scheme for relative arrangement of nucleic acid scaffold, active center, HBS and rifampicin-binding site in TEC. (C) Stereo view for the surrounding of Rif compound bound to RNAP (PDB code: 1I6V). Bound drug (green) and neighboring amino acid chains are shown in a spacefil rendition. Side chains forming a cavity in the rifampicin binding pocket (element colored) and S531 residue (pink) are indicated. (D) the structure of RNAP TEC. (E) Magnified view of the RNAP region that interacts with RNA transcript in RNA:DNA hybrid (HBS) of the TEC. β subunit residues of HBS contacting RNA are shown in spacefil mode. RNA (green) is in stick representation. Structures are from (42) for (D) and 2O5J for (E).
Part of the HBS includes the central segment of the RNAP β subunit (residues 500–580), a region that is also involved in the binding of rifampicin, an inhibitor of transcription (4–8) (Figure 1A and C). Therefore, many residues constituting rifampicin binding site also contact RNA in the transcription complex (Figure 1E). Rifampicin blocks transcript entrance to HBS, thereby causing the transcription initiation process to stop. The effects of rifampicin-resistance mutations on multiple steps of RNA synthesis (9–12) and the pleiotropic phenotypes of mutants (13) indicate that interactions between the HBS and the RNA transcript (Figure 1D and E) play a central role in the function and regulation of the overall transcription machinery. Consistent with this conclusion HBS also harbors substitutions that (i) suppress effects of a ppGpp deficiency (14); (ii) alter transcription slippage (15), which makes an important contribution to transcription fidelity and transcription regulation and (iii) affect the response of the transcribing complex to major host and phage-encoded termination and anti-termination factors (16–17). A key unanswered question is how the transition from the initiation step to the elongation phase of transcription, a crucial event in RNA synthesis, is mediated by HBS–RNA interactions.
β subunit S531 is located in the central part of HBS and along with neighboring residues constitutes a site for rifampicin binding (Figure 1D and E). This residue is conserved among bacteria and chloroplasts, while natural substitutions at this locus are highly species-specific (see discussion). However, the basis of this conservation is unclear, since S531 is not involved in interactions with RNA transcript or nearby residues in transcription complex as revealed by X-ray analysis 18,19). In previous studies a number of mutants at this position or equivalent loci of other bacteria were selected for rifampicin resistance in laboratory setup or isolated in clinic (9,13,20–22). Remarkably, mutations at this position are among those the most prevalent in clinical isolates of Mycobacterium tuberculosis (20), which justifies their thorough study. However, selection approaches used in the above studies to generate mutations suffer significant limitation, since they do not yield substitutions that require multiple nucleotide changes in the same codon. Indeed, only few substitutions in the 531 locus were recovered.
A powerful strategy to address the role of a particular residue in an enzyme’s function is saturating mutagenesis. However, it has been applied to RNAP only occasionally (23). Following this strategy in the present study we have generated 19 S531 substitutions and tested their effect on RNAP in vitro transcription and on bacterial growth. We have shown that 531 substitutions affected transcription and displayed mutation-specific effect on bacterial doubling time, reflecting significant and complex involvement of S531 in transcription and cell functioning. Thus most of the substitutions had negative effect on RNA synthesis as measured in in vitro transcription assays with purified RNAP variant enzymes. Surprisingly, 531-G, H, R and K substitutions enhanced productive transcription through reducing abortive loss of short RNA products in TEC. In particular 531-R substitution suppressed the loss by presumably establishing a salt bridge with RNA. Strikingly, the R substitution is present at 531-equivalent position of extremophilic organisms, which function at high temperature or in the media of high salinity. These factors are expected to strongly accelerate short RNA dissociation from inherently unstable TIC. Therefore, the substitution might alleviate the destabilizing effect of the extreme conditions through increasing RNA retention in TIC by the same mechanism.
Biochemical testing of generated RNAP variants also revealed a RNA/HBS interaction crucial for promoter clearance, which includes βR529 salt-bridging with growing transcript.
MATERIALS AND METHODS
All chemicals were from Sigma-Aldrich (St Louis, MO, USA). Ultrapure ribonucleoside-5′-triphosphates were from Pharmacia. [α-32P]CTP (cytidine-5′-triphosphate) and [α-32P]UTP (uridine-5′-triphosphate) were from MP Biomed. T7A2 promoter DNA template was obtained as in (24), pTZ plasmid containing T7A1 promoter was as in (25). Radioactive products were resolved by electrophoresis and quantified by phosphoimagery using a Molecular Dynamics device (GE Healthcare) and Image Quant software. Molecular modeling was performed using WebLab ViewerLight 4.0 (Molecular Simulations Inc.).
RNAP mutations
Mutations at positions 531 or 529 of the RNAP β subunit were introduced with site-directed mutagenesis using pVS10 plasmid coding for RNAP α, β, β′ and ω subunits (26). The identity of generated mutations was confirmed by nucleotide sequence analysis. Engineered mutations were transferred into the Escherichia coli chromosome using the lambda Red system (27). The mutant strains were selected in the presence of rifampicin. Our attempts to select 531-T as well as some other mutant strains using this protocol were not successful, likely to sensitivity of the mutants to Rifampicin. Additional substitutions at position 531 were recovered during selection of slow growing mutants. Thus in the case of the 531-D and -E variants, larger grown colonies contained 531-H, N and 531-A and G substitutions respectively. This result may have been due to the presence of trace amounts of nucleotide changes in the original DNA used for transduction, which were evidently introduced by polymerizing enzyme used to generate polymerase chain reaction fragments for subsequent transduction.
Purification of RNAP
BL21(DE3):pVS10 cells were grown in 3 l LB, 37°C to OD600 = 0.6, induced with 0.5 mM isopropyl-β-D-thiogalactoside (IPTG) for 3 h, harvested at 4°C. All the further procedures were performed either on ice or in a cold room (10°C). Cells were resuspended in 70 ml of buffer A (100 mM Tris pH = 8.0, 500 mM NaCl, 2 mM ethylenediaminetetraacetic acid (EDTA), 1 mM phenylmethylsulfonyl fluoride (PMSF), 5 mM Imidazole) with dounce homogenizer and disrupted with a French Press. The lysate was clarified by centrifugation at 43 000 g for 2 h. One milliliter of Fast Flow Nitrilo-triacetic acid (NTA) Sepharose (GE Healthcare) was added to the supernatant fluid and gently agitated by rotation on a horizontal platform at 50 rpm in 50 ml Falcon tube for 1 h. NTA sepharose was concentrated by centrifugation at 2000 g and washed twice with 10 ml of buffer B (buffer A + 10% Glycerol–PMSF) and loaded onto a gravity column. The column was washed with 15 ml buffer C (Buffer B + 20 mM Imidazole). Protein was eluted with buffer D (Buffer B + 200 mM Imidazole) in 2 ml (about 10 mg of protein). Two milliliter of eluate from the NTA agarose column were diluted with buffer E (50 mM HEPES pH 7.4, 1 mM EDTA, 10% Glycerol) to 10 ml and loaded onto a 1-ml HiTrap Heparin column, washed with 5 ml buffer F (Buffer E + 300 mM NaCl) and eluted with buffer G (Buffer E with NaCl at 700 mM). 2.5 ml of eluate were concentrated using an Amicon filtration units Ultra 50 000 MWCO (20 min, 4000 g) to about 200 μl and loaded onto a Superose-6 Gel Filtration column prewashed with Buffer H and then eluted with the same buffer (Phosphate buffered saline (PBS) + 1 mM EDTA + 10% Glycerol). Fractions containing RNAP were collected and concentrated in Amicon Ultra 50 000 MWCO to about 200 μl (20 min, 4000 g). Protein was distributed to microtubes in 10-μl portions, flash-frozen in liquid nitrogen and stored at −80°C. Purified proteins were analyzed using gradient SDS PAGE (4–30%). The presence and identity of the mutations was confirmed by MS analysis.
Effect of rifampicin on RNA synthesis by engineered mutant enzymes
The reaction mixture (8 μl) containing 2 pmol RNAP, 0.05 pmol pTZ plasmid with cloned T7A1 promoter and 2 pmol purified σ subunit in transcription buffer (TB) (20 mM Tris–HCl pH 8.0, 0.1 M NaCl and 10 mM MgCl2) was incubated for 5 min at 37°C and supplemented with 1 μl of rifampicin to a final concentration 25 μg/ml, 1 μl of 0.25 mM adenosine-5′-triphosphate (ATP), UTP, guanosine-5′-triphosphate (GTP) and [α-32P]CTP (specific activity 30 Ci/mmol). After incubation for 30 min at 37°C the reaction was quenched by addition of 10 M urea and the aliquots (5 μl) were applied to Whatman 3 MM paper discs that were washed by 5% trichloroacetic acid (3 × 100 ml, 20 min each wash). The discs were rinsed with ethanol, dried and the remaining radioactive material quantified by phosphoimagery.
Determination of the bacterial growth rate
Suspensions of bacterial strains in phosphate buffered saline, PBS were adjusted to the same turbidity measured at 600 nm. Aliquots (0.1 ml) of the cells were transferred to the wells of a microtiter plate filled with 0.1 ml of LB medium. The plates were kept at either 20°, 37° or 42°C during shaking. The time course for bacterial growth was monitored by measuring absorption at 600 nm using a plate reader. This testing as well as the assays with purified RNAP mutants was repeated in triplicate.
Abortive initiation assay
In the multi-round version of the assay the reaction mixture (9 μl) containing 0.5 pmol T7A2 promoter, 2 pmol RNAP, 2 pmol purified σ subunit in TB (20 mM Tris–HCl pH 8.0, 0.1 M NaCl and 10 mM MgCl2) was incubated 5 min at 37°C. The reaction was initiated by addition of 1 μl of a mixture of 2 mM GTP, 20 μM [α-32P]CTP (specific activity 30 Ci/mmol) and after 10 min of incubation at 37°C it was stopped by addition of 10 μl of 10 M urea with Bromphenol Blue dye. Aliquots of the reaction mixture (1 μl) were applied to a PEI cellulose sheet (Sigma-Aldrich) and the reaction products were separated by thin-layer chromatography (TLC) using 0.5 M KH2PO4 as the developing solvent. The products were quantified by phosphoimagery. The reaction mixture for the single-round assay was essentially the same, but the final concentration of [α-32P]CTP was 3 nm (specific activity 3000 Ci/mmol). The reaction time was 1 min.
Promoter clearance assay
The concentration of RNAP and T7A2 promoter, as well as the composition of the TB, was the same as in the abortive initiation assay but the reaction was started by addition of 1 μl of a mixture containing 2 mM ATP, UTP, GTP and 10 μM [α-32P]CTP (specific activity 30 Ci/mmol). Incubation time was 15 min at 37°C. The reaction was stopped as descried above and the reaction products were analyzed by electrophoresis in 20% polyacrylamide gel containing 7 M urea. The products were visualized and quantified by phosphoimagery.
Transcript extension in TEC10
TEC10C, was constructed as previously described (28). Briefly, RNA9 was mixed with equivalent amounts of DNA template strand to a final concentration of 1 μM in TB (20 mM Tris–HCl pH 8.0, 0.1 M NaCl and 10 mM MgCl2) at 40°C and left to cool in a water bath to room temperature for 30 min. This mixture was added to an equivalent amount of His6-tagged RNAP immobilized on NTA agarose and kept at room temperature in a shaker for 10 min followed by addition of the equivalent amount of DNA non-template strand. The mixture was agitated for another 10 min at room temperature and supplemented with TB containing 1 M NaCl. After 10 min, the pellet was separated and washed with regular TB. The resulting TEC9 was elongated to TEC10C by an equivalent amount of [α-32P] CTP (3000 Ci/mmol 10 mCi/ml, MP Biomedicals) for 4 min followed by washing with TB (4 × 1 ml). The reaction mixture containing labeled TEC10 (8 μl) was supplemented with a complete set of NTP substrates (final concentration 20 μM) and MgCl2 (10 mM). The reaction was carried out for 5 min at ambient temperature and stopped as described above. Reaction products were separated by electrophoresis in denaturing PAAG and quantified by phosphoimagery.
Full transcription cycle assay
This was performed at conditions analogous to those for rifampicin sensitivity testing (see above), except that rifampicin was omitted.
RESULTS
Rpoβ-531 substitutions and determination of rifampicin sensitivity of variant enzymes
We introduced into position 531 the amino acid changes specified in Figure 2 and in Table 1, in addition to other substitutions expected to serve as controls, using an expression plasmid encoding all RNAP subunits. A hexahistidine tag sequence was fused to the C-terminus of the RNAP β’ subunit to facilitate purification of the enzyme.
Figure 2.
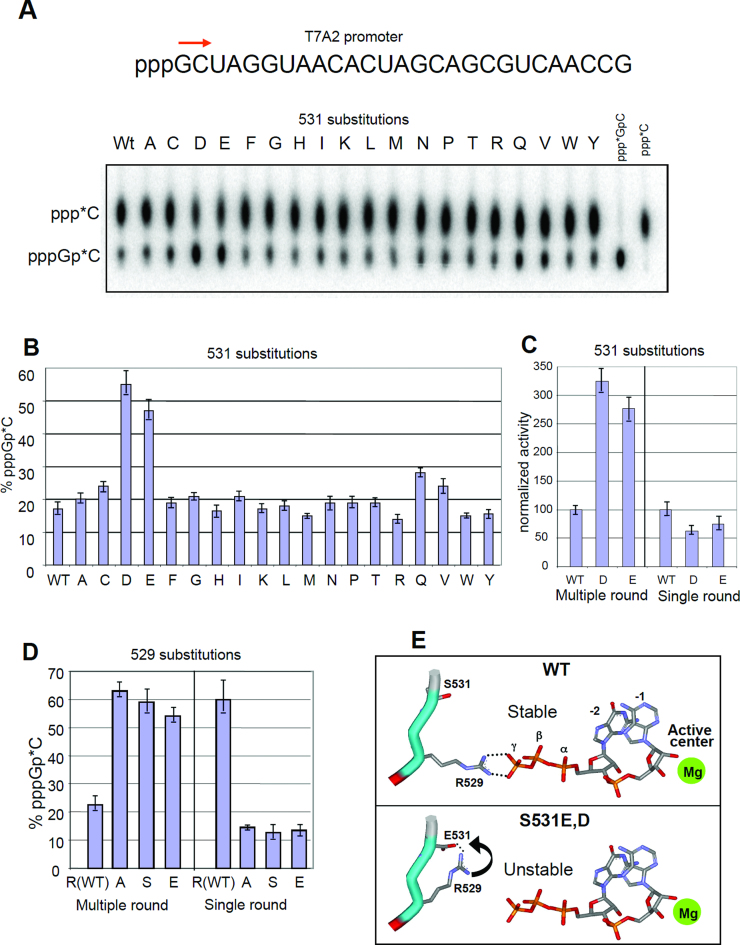
Effect of S531 and R529 substitutions of abortive dinucleotide synthesis by RNAP. (A) Top, initial transcribed sequence of T7A2 promoter. Bottom, phosphoimage of the reaction products (for multi-round pppGpC synthesis) separated by ion-exchange TLC. (B) Quantitation of the results of (A). (C) Performance of the 531-D and -E in multiple- and single-round reactions of dinucleotide synthesis. (D) Effect of 529 substitutions on RNAP abortive dinucleotide synthesis. (E) Proposed mechanism for the influence of 531-D and -E substitutions on dinucleotide synthesis. The structure used for modeling is2O5J. The error bars represent SD from three independent experiments.
Table 1. Sensitivity of β 531 mutant RNAP to Rif (25 μg/ml) in vitro.
| RNAP | Residual activity (%) | Rif resistance* |
|---|---|---|
| 531S (WT) | 1.4 | − |
| 531A | 0.8 | − |
| 531C | 4.0 | + |
| 531D | 76.2 | +++ |
| 531E | 69.0 | +++ |
| 531F | 66.7 | +++ |
| 531G | 2.7 | ± |
| 531I | 7.2 | + |
| 531K | 35.0 | ++ |
| 531L | 45.2 | +++ |
| 531M | 57.0 | +++ |
| 531N | 20.0 | ++ |
| 531P | 9.3 | ++ |
| 531Q | 60.0 | +++ |
| 531R | 15.2 | ++ |
| 531T | 1.2 | − |
| 531V | 4.2 | + |
| 531W | 51.0 | +++ |
| 531Y | 80.0 | +++ |
* ‘−’, <2%; ‘±’, 2–3%; ‘+’, 3–8%; ‘++’, 8–35%; ‘+++’, >35% activity.
In an initial test, we determined the sensitivity of the mutant enzymes to rifampicin. As expected, substitutions with bulky side chains (E, F, K, R, L, M, W and Y) displayed low sensitivity to the drug, while those with smaller groups were more sensitive (Table 1). Resistance to rifampicin is attributed to independent or cumulative action of two factors that cause reduced drug binding: (i) loss of a hydrogen bond between the Ser side chain and rifampicin, and (ii) steric clash between the introduced side chain and the drug. High-level rifampicin sensitivity of RNAP variants having small side chains (e.g. 531-A and -G), which eliminate hydrogen-bonding without creating clashes, suggests that hydrogen bonding has a marginal contribution to rifampicin binding. Therefore, we attribute the strong interfering effect of bulky substitutions to steric clashes.
Some anomalies were observed. Moderate rifampicin sensitivity of some variants that are expected to create significant steric clashes with the incoming drug (531-I and -C) can be explained by the ability of the bound drug to force the interfering side chain into a cavity near the drug-binding pocket (Figure 1C) located between β subunit residues Q148, M459, S509, Q510 and G534. This idea also explains the detectable difference in regard to rifampicin-mediated inhibition displayed by isosteric substitutions 531-D and -N; the neutral N side chain can be forced into the cavity as rifampicin enters the binding site. In contrast, the negatively charged D side chain can form a salt bridge with nearby 529-R, which would affect rifampicin binding by placing both side chains into the drug binding pocket. Residual rifampicin-mediated inhibition for variants that are expected to create significant steric clashes with the incoming drug can be explained as structural changes in the backbone of the surrounding polypeptide chains to accommodate drug binding with only reduced affinity (29).
Viability of all generated bacterial strains suggests that introduced mutations do not significantly affect overall structure, which is also confirmed by X-ray analysis of S531L enzyme (29).
Effect of amino acid substitutions at position 531 on abortive dinucleotide synthesis
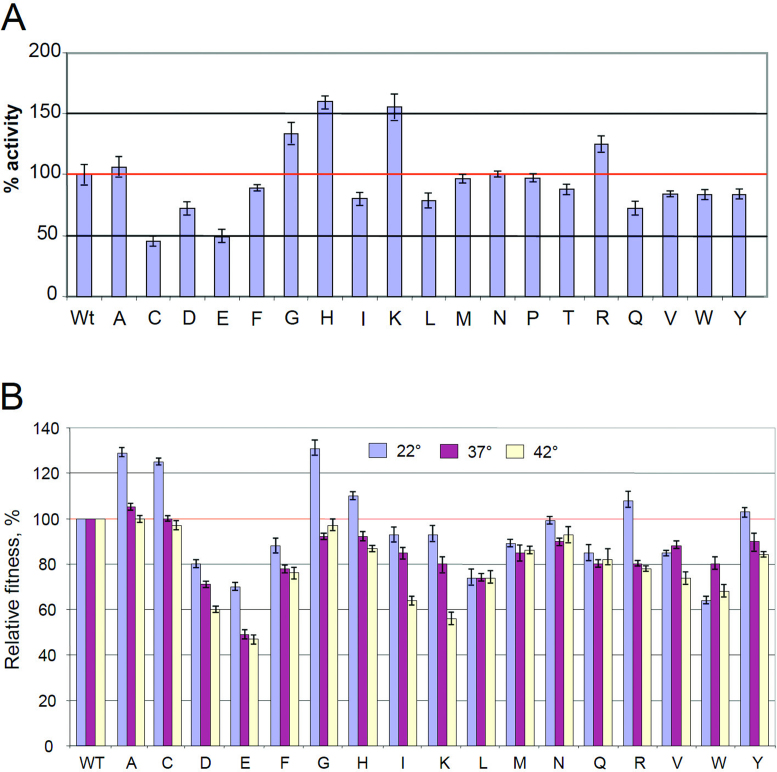
We have also investigated RNA synthesis with purified variant RNAP. First we determined the rate of dinucleotide synthesis, which is the initial step of transcription. The assay was performed in the multi-round initiation mode at the T7A2 promoter using GTP and CTP, substrates that yield dinucleotide pppGpC as the reaction product. As seen from Figure 2A–C, the 531-D and -E enzymes displayed 2.5–3-fold higher reaction rates than other variant enzymes or wild-type (WT) RNAP.
The enhanced reaction rate seen above could be due to either an increased catalytic constant for phosphodiester bond formation or to destabilization of the synthesized dinucleotide at the active center. The latter would promote product dissociation, thereby increasing turnover rate. To distinguish between these two alternatives, we conducted the same reaction, but in single-cycle conditions by using concentrations of radiolabeled CTP elongating substrate that were lower than that of the RNAP transcription complex. As seen from Figure 2C, under these conditions the rate-enhancing effect of the mutations was completely ablated, suggesting that both mutations stimulate abortive synthesis by facilitating dissociation of the synthesized RNA product. Even though, some other effects of the mutations could be involved in the observed stimulation (e.g. increased Kcat) our conclusion is consistent with molecular modeling (Figure 2E), which shows that after formation of the first phosphodiester bond, followed by forward translocation of the enzyme, the 5′ triphosphate group of the synthesized dinucleotide product acquires a stabilizing salt-bridging contact with β-R529. With the 531-D and -E variants this interaction is predicted to be disrupted by the carboxylate side chains that attract the R529 residue (Figure 2E). This attraction should promote dissociation of the RNA product.
Effect of 529 mutations on abortive dinucleotide synthesis
To confirm involvement of the R529 residue in the mechanism proposed above, we substituted a R side chain at this position with a neutral (A and S) or a negatively charged E group and then tested the variants in the two dinucleotide abortive synthesis assays. As expected (Figure 2 D), these substitutions mimicked the effect of the 531-D and -E substitutions, by enhancing the reaction rate by the same magnitude seen in multi-round experiments, but not the rate observed in a single-round experiment. Moreover, in the single-round experiment all variants displayed reduced catalytic activity. Therefore, 529 mutations can have multiple effects on RNA synthesis by promoting transcript dissociation and by affecting kcat. The later effect is consistent with reduced transcript extension rate displayed by the mutants in an elongation RNAP complex where RNA dissociation is not an issue (Figure 3E).
Figure 3.
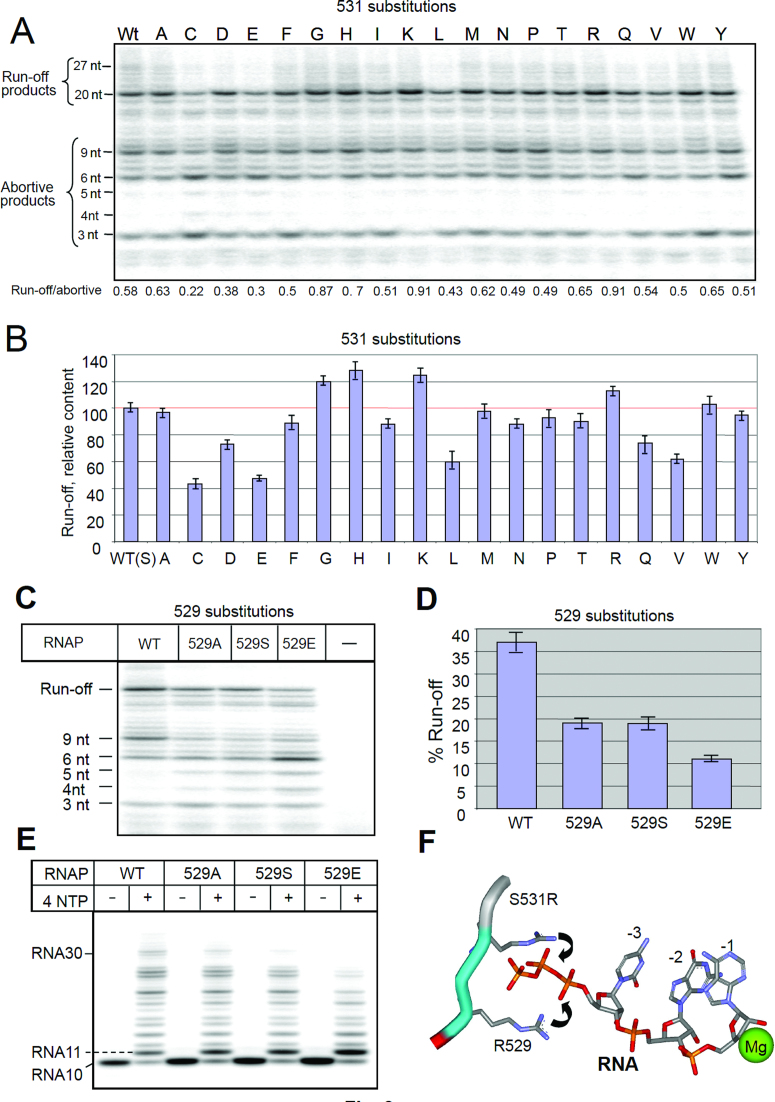
Catalytic properties of 531 and 529 RNAP variants in in vitro transcription assays. (A) The image of radioactive RNA products of initiation after electrophoretic separation. (B) Quantitation of the results of (A). (C) Performance of 529 substitutions in the conditions of (A). (D) Quantitation of (C). (E) Elongation properties of 529 mutants. (F) The mechanism proposed to explain the positive effect of 531-R and -K substitutions on promoter clearance. Error bars represent SD from three independent experiments.
The strong enhancing action of R529 substitutions on multi-round abortive initiation is mechanistically related to an identical effect of rifampicin on this reaction at the same promoter observed in a previous study (30). Indeed, as seen from molecular modeling, the bound drug obstructs the Arg residue, which eliminates the salt bridge formed with the synthesized dinucleotide product.
Effect of RNAP substitutions on promoter clearance
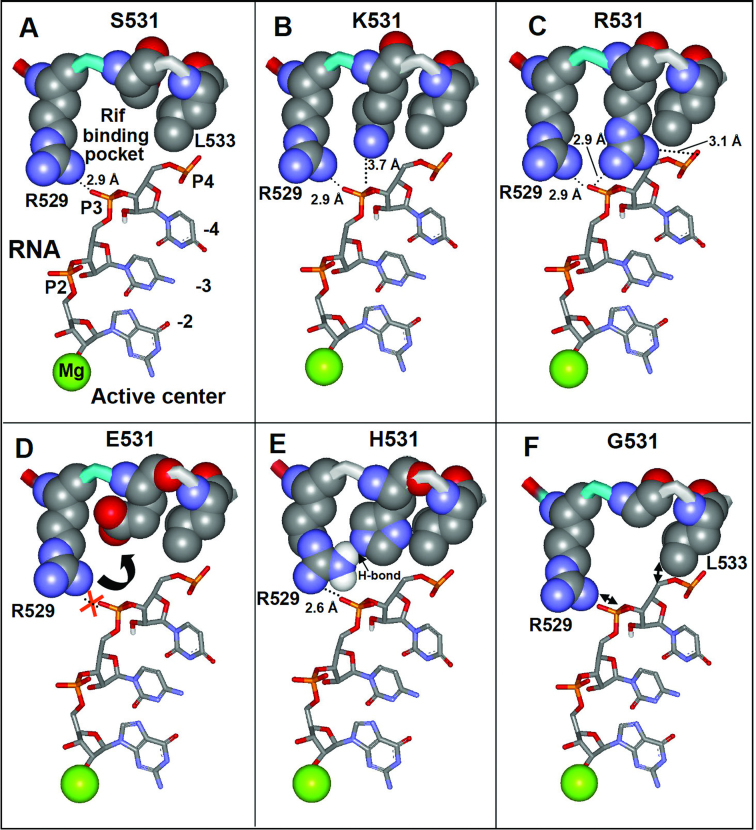
Short RNA products at the initial step of RNA synthesis easily dissociate from the transcription complex, thereby causing cycles of abortive initiation (31). Further growth of the transcripts is accompanied by a dramatic stabilization of the synthesized product in the complex and release of RNAP from the recognition contacts with the promoter, manifesting promoter clearance (32,33). The efficiency of this process, defined as the runoff-to-abortive products ratio, is an important parameter that reflects the ability of the transcription complex to escape the promoter and carry out productive RNA synthesis. Figure 3 shows the performance of variant RNAPs in the promoter escape assay. It is seen that 531-R, K and H substitutions significantly improved promoter escape (115, 125 and 128% respectively), by reducing the amount of abortive products and increasing the yield of run-off transcript. As follows from modeling onto crystallographic images of TIC (4XLN, 18) and TEC (2O5J, 19), the positively charged ε-amino group of the substituted Lys side chain is within salt-bridging distance (3.7 Å) from the RNA phosphodiester group at register −3 (Figure 4B). The guanidinium group of Arg can simultaneously engage in salt-bridging with RNA phosphodiester groups located at the −3 and −4 registers (Figure 4C). Both residues are also within the interaction range with the RNA trinucleotide 5′ triphosphate group (Figure 3F). Therefore the enhanced retention of the short RNA transcript through an additional salt bridging (Figure 4B and C) explains positive effect of these substitutions in promoter clearance. In accord with this assumption, the effect of K and R substitutions was particularly pronounced for the trinucleotide RNA product (Figure 3A). Indeed, the R and K salt-bridging should be particularly effective with highly negatively charged 5′ triphosphate chain of the trinucleotide RNA transcript in TIC (Figure 3F), which would force transition of the trinucleotide to the post-translocated state, followed by its extension to longer products. Stimulating effect of 531-H substitution can be accounted for a hydrogen-bonding interaction with the 529-R side chain, which would reinforce the salt bridge as shown in Figure 4E.
Figure 4.
Molecular modeling of the various side chains in position 531 on the structure of transcription complex:(A) (Ser); (B) (Lys); (C) (Arg); (D) (Glu), (E) (His); (F) (Gly). Active center Mg2+ ion, RNA transcript as well as R529 and L533 residues interacting with RNA transcript are indicated. The modeling was made onto the structure of TEC from Taq (PDB code: 2O5J).
In contrast, substitutions with negatively charged side chains, 531-D and -E, reduced the efficiency of promoter escape (73 and 48% of that seen with WT enzyme, respectively) by promoting abortive initiation (Figure 3A). A direct influence of the substitutions on protein–RNA interactions through electrostatic repulsion with phosphate groups is unlikely, because the carboxylate group of the residues is too far away from RNA (Figure 3D). Apparently the effect of these substitutions involves competitive salt-bridging of D and E side chains (as shown in Figure 3E) with R529, which weakens the interaction of the latter with the RNA transcript. To test involvement of R529 in the proposed mechanism for 531-D and -E substitutions, we performed the promoter clearance assay with R529 mutants. As expected (Figure 3C and D), all 529 substitutions had a strong negative effect on the initiation-to-elongation transition by promoting dissociation of short RNA transcripts. The effect was more severe for 529E substitution, likely due to creating through electric repulsion an additional barrier for RNA exit.
The considerable negative effect of the C, L, V and Q substitutions (43, 60, 62 and 75% activity of WT enzyme, respectively) on promoter clearance, plus the positive effect of Gly substitution (130% of WT enzyme) cannot be easily explained by molecular modeling. The effects of these substitutions may be mediated through a change of backbone conformation. Thus, a Gly substitution could render the backbone more flexible, bringing the neighboring residues (R529 and L534) closer to RNA (Figure 4F), thereby enhancing the interactions. Consistent with this explanation, introduction of a ‘rigid’ Pro substitution at position 531 decreased promoter clearance. Other 531 substitutions moderately reduced or enhanced promoter clearance.
Effect of RNAP substitutions on total RNA synthesis from plasmid pTZ template
We also examined the effect of amino acid substitutions on multiple-round transcription using plasmid pTZ, which contains the bla and T7 A1 promoters. The amount of RNA synthesized in this assay (determined as incorporation of radioactivity from the labeled NTP substrate as acid-insoluble product) reflects the cumulative performance of the enzyme at all steps of the transcription cycle. Remarkably, the performance of the variant enzymes (Figure 5A) was generally close to that observed in the promoter clearance assay (Figure 3B), suggesting that promoter clearance is the bottleneck for the entire transcription cycle.
Figure 5.
Effect of 531 substitutions of multiple-round full-size RNA synthesis from DNA plasmid (A) and on bacterial fitness at various temperatures (B). Error bars represent SD from three independent experiments.
Effect of amino acid substitutions on E. coli doubling time
For living-cell studies we transferred the mutations described above into the E. coli chromosome, thereby constructing a set of isogenic strains. The desired mutants were selected in the presence of rifampicin and their identity was confirmed by DNA sequence determination. To determine relative bacterial growth, we measured doubling time at various temperatures. Strikingly, at 20° C the doubling time of most of the bacterial strains (Figure 5B) was consistent with the transcription activity of RNAP variants seen with in vitro assays (Figures 3 and 5A). Thus G, H and R substitutions, which had high transcription activity, showed increased growth rate (131, 110 and 108% of WT levels, respectively), while 531-D and -E substitutions, which significantly lowered RNA synthesis with respect to promoter clearance and plasmid transcription assays, displayed reduced growth rate (80 and 70% of the WT respectively). Surprisingly, the 531-C substitution, which strongly suppressed RNA synthesis with in vitro assays, displayed significantly increased bacterial growth rate (125%). Moreover, a 531-K substitution that increased activity with in vitro assays, failed to confer enhanced fitness. Also, a 531-A substitution ‘neutral’ in all in vitro assays produced fast growing bacteria (129% of WT). Unexpectedly, at 37° all mutant strains (except 531-A) grew slower than WT cells. This lack of concordance is currently explained as arising from secondary effects of intracellular environment (e.g. interactions with transcription factors).
DISCUSSION
RNAP represents one of the most complex and highly controlled cellular enzymes. This complexity is justified by the necessity to couple catalytic function of RNA synthesis to regulatory apparatus. Extensive genome sequencing revealed that some RNAP loci are identical among all living organisms, reflecting remarkable evolutional conservation of the enzyme’s features involved in basal RNAP function. However other loci display species-specific conservation, suggesting their involvement in particular function (e.g. regulation of RNA synthesis), specified by an organism's biology and/or life style. The S531 residue of RNAP β subunit is highly conserved among organisms and chloroplasts. Remarkably, mutational substitutions at this or equivalent loci in other organisms are highly species specific. Thus rifampicin-producing bacterial species Mediterranei contain exclusively N substitution at the 531-equivalent position, thereby explaining resistance of the organisms to the produced compound. Notably, the same substitution occurs naturally in many organisms that do not produce rifampicin (e.g. all Spirochetes, Streptomyces and ample Archaea species). Another example is 531-T substitution typical for other organisms (e.g. Thermatogae and green algae). Since the above mutations have not been selected before for rifampicin resistance in E. coli, we set out to determine the effect of N, T as well as all other unknown substitutions at 531 position on this organisms, in order to systematically address the basis for S531 high evolutional conservation. In this way using site-directed mutagenesis we have generated a collection of 19 substitutions and studied the mutations effect on biochemical and bacterial growth indexes.
The results of biochemical testing with purified variant enzymes using transcription assays identified mutations that either decreased, or enhanced productive RNA synthesis by affecting initiation-to-elongation transition. Thus 531-R and -K substitutions, improved enzyme performance by suppressing dissociation of short transcripts likely through establishing a salt bridge with RNA transcript. However, additional experiments are required to confirm this mechanism. In contrast, introduction of negatively charged side chains 531-E and -D increased dissociation of short transcripts, thereby reducing promoter clearance efficiency and the yield of full-size products, apparently by repositioning the neighboring 529R side chain involved in RNA retention. The effect of other substitutions on promoter clearance cannot be easily explained. For example, the 531-C variant displayed a significant negative effect on promoter clearance, while the 531-G substitution enhanced the yield of run-of transcript.
Importantly, the generated substitutions affected both promoter clearance and the activity of RNAP for the entire transcription cycle in the same way, suggesting that the former step is rate-determining for the whole transcription process.
Our biochemical studies of 531 substitutions, combined with molecular modeling, implicated the neighboring R529 residue of HBS in the initiation-to-elongation transition by RNAP through stabilization of the RNA transcript. An exceptional role of this residue in the proposed mechanism was substantiated by site-directed mutagenesis, thereby explaining high evolutional conservation of R529. In accord with our data, previous studies with E. coli (9), Salmonella enterica (22) and M. tuberculosis (34) revealed high cost of the rpoB R529C substitution to culture growth. Notably, homologous Lys substitution in Clostridium difficile at the equivalent position did not affect the fitness, likely by preserving the Lys side chain salt bridge with RNA (35).
Thus far, no detailed comparison of the basal catalytic activity of RNAP variants in vitro and growth rate of corresponding mutant bacterial strains has been reported. The negative effect of the rpoB 529-C substitution on the initiation-to-elongation transition seen with purified RNAP in (12) was consistent with slow bacterial growth. The same trend was observed with many 531 substitutions generated in our study. However, the in vivo effect on bacterial growth was not obvious for substitutions that improve the catalytic activity of RNAP. Our results address this issue by demonstrating that three out of four mutations that increased RNAP activity with the in vitro assays had the same effect on bacterial growth at 20°C. This finding suggests that catalytic performance of RNAP could be a limiting factor for bacterial growth. Few exceptions were observed. Thus the 531-C substitution, which rendered RNAP less active with in vitro tests, displayed the opposite effect on bacterial growth. Another substitution, 531-A did not alter RNAP activity at all steps of RNA synthesis but it significantly increased bacterial growth rate. The later examples could reflect mutation-specific enhancement of the RNAP catalytic activity in the cells.
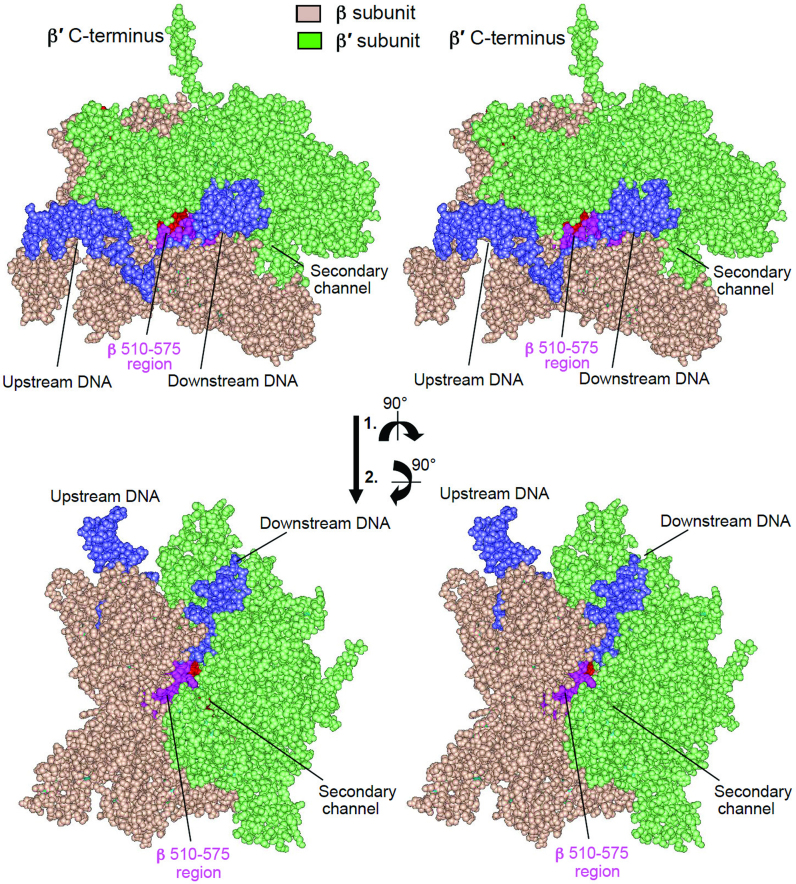
Surprisingly, the positive effect of the 531-A, C, G, H and R substitutions on bacterial growth rate was temperature sensitive: at elevated temperature all mutants except 531-A displayed slower growth than the WT strain. This phenomenon cannot be explained by altered basal transcription, as follows from the experiments shown in Figures 3 and 5A in which RNAP activity was determined at 37°. The enhancing effect of the intracellular environment on growth rates of the 531-C and -A variants, as well as their temperature-dependent behavior, can be explained by involvement of transcription regulatory factors whose binding to RNAP is affected by mutation and/or growth temperature. It is conceivable that protein factors could bind to RNAP and counteract the negative effect of a specific substitution on enzyme performance. The association of such factors with RNAP in turn could be affected by the mutations in temperature-dependent mode, thus rendering the binding temperature sensitive. Possible existence of such factors is consistent with previous analysis of E. coli rifampicin-resistant mutants (16). The results of the later study suggest that some cellular and phage-encoded proteins can bind to the enzyme at the region assembling HBS, thereby modulating termination efficiency. The above conclusion is supported by structural considerations, suggesting that at least part of the HBS, including the S531 residue, would be accessible for such interactions, either through the RNAP main channel (Figure 6, top and Figure 1D) or through the gap between the β′ bridge helix and the downstream DNA duplex (Figure 6, bottom). Intriguingly, the latter pathway is used by transcription inhibitors CBR (36,37) and streptolydigin (38,39), drugs that bind at the interface of the HBS and the β′ bridge helix.
Figure 6.
Surface accessibility of HBS in TEC. Escherichia coli TEC (42) is shown in spacefil rendition (stereo image for crossed-eye view). HBS region is highlighted in pink.
Notably, 531-N mutation common for many bacterial species listed above was essentially ‘neutral’ in all performed assays, suggesting that potential beneficial properties of this substitution could be species specific. Also, our numerous attempts to introduce 531-T substitution (mutation common for many other bacterial species) into E. coli chromosome were not successful. Even though corresponding enzyme purified from the same strain carrying expression plasmid displayed properties nearly identical to WT RNAP. This could be accounted for ‘toxic’ action of the substitution on the used bacterial strain, despite apparent similarity between Ser and Thr residues.
In conclusion, our results suggest that S531 residue does not directly participate in RNA synthesis. This is supported by the absence of the 531-A substitution (which eliminates a hydroxyl group, while preserving the rest of the side chain) effect on transcription in all in vitro assays. The detectable influence of some other 531 mutations on RNA synthesis could be attributed to steric effects of introduced side-chains or/and by interactions of the functional groups in the mutated side-chain with the neighboring aminoacid residues or with RNA product. Remarkable evolutional conservation of S531 could be due to a role in the maintenance of secondary structure of the enzyme’s domain holding RNA transcript. However, recent X-ray analysis (29) argues against this proposal. Indeed, no effect on backbone conformational was observed for 531 RNAP variants. Therefore, S531 can be involved in interactions with external protein factors that regulate the basal transcription machinery.
When the work described above was completed, we were astonished to learn that some organisms, functioning at extreme environmental conditions, contain an R substitution at the position equivalent to 531 (Figure 1A), which, according to our data, provides enhanced RNA retention. One of these organisms, Methanopyrus kandleri, grows at 84–122°C (40). High temperature should exert a strong negative effect on inherently unstable TIC, which can be alleviated by salt-bridging of the R side chain with RNA phosphate groups. The same destabilization is expected for bacteria that grow in media of high salinity. Indeed, intracellular KCl concentration in halophilic bacteria can exceed 1 M, which is required to counteract an external osmotic pressure (41). High salt content is expected to decrease short RNAs retention in TIC, which strongly relies on salt-bridging that is sensitive to ionic strength of the medium. Therefore, additional transcript stabilization through interaction with R residue at 531 equivalent position observed in all halophilic bacteria could be vital.
ACKNOWLEDGEMENTS
We are grateful to Karl Drlica for helpful discussion, suggestions and proofreading the manuscript.
Footnotes
Present address: Vadim Nikiforov, 430 E63-st Street, New York, NY 10065, USA.
FUNDING
NIH Grant [GM115809 to O.Y., V.N., M.O.D.]; PHRI Internal Support Fund (to A.M.). Funding for open access charge: Rutgers University.
Conflict of interest statement. None declared.
REFERENCES
- 1. Chamberlin M. Losick R, Chamberlin M. RNA polymerase: an overview. RNA Polymerase. 1976; NY: Cold Spring Harbor Laboratory Press; 17–67. [Google Scholar]
- 2. Mustaev A., Goldfarb A.. Lennarz W, Lane M. RNA polymerase reaction in bacteria. Encyclopedia of Biological Chemistry. 2004; Academic Press, Elsevier Inc; 775–780. [Google Scholar]
- 3. Korzheva N., Mustaev A., Nudler E., Nikiforov V., Goldfarb A.. Mechanistic model for elongation complex of Escherichia Coli RNA polymerase. Cold. Spring. Harb. Symp. Quant. Biol. 1998; 63:337–345. [DOI] [PubMed] [Google Scholar]
- 4. Hartman G., Honikel K., Nuesch J.. The specific inhibition of the DNA-directed RNA synthesis by rifamycin. Biochem. Biophys. Acta. 1967; 145:843–844. [DOI] [PubMed] [Google Scholar]
- 5. Ovchinnikov Y.A., Monastyrskays G.S., Gubanov V.V., Lipkin V.M., Sverdlov E.D., Kiver I.F., Bass I.A., Mindlin S.Z., Danilevskaya O.N., Khesin R.B.. Primarystructure of Escherichia coli RNA polymerase nucleotide substitution inthe beta subunit gene of the rifampicin resistant rpoB255 mutant. Mol. Gen. Genet. 1981; 184:646–538. [DOI] [PubMed] [Google Scholar]
- 6. Ovchinnikov Y.A., Monastyrskaya G.S., Guriev S.O., Kalinina N.F., Sverdlov E.D., Gragerov A.I., Bass I.A., Kiver I.F., Moiseeva E.P., Igumnov V. et al. RNA polymerase rifampicin resistance mutations in Escherichia coli: sequence changes and dominance. Mol. Gen. Genet. 1983; 190:344–348. [DOI] [PubMed] [Google Scholar]
- 7. Jin D.J., Gross C.. Mapping and sequencing of mutations in the Escherichia coli rpoB gene that lead to rifampicin resistance. J. Mol. Biol. 1988; 202:45–58. [DOI] [PubMed] [Google Scholar]
- 8. Campbell E.A., Korzheva N., Mustaev A., Murakami K., Nair S., Goldfarb A., Darst S.A.. Structural mechanism for rifampicin inhibition of bacterial RNA polymerase. Cell. 2001; 104:901–912. [DOI] [PubMed] [Google Scholar]
- 9. Jin D.J., Walter W., Gross C.. Characterization of the termination phenotypes of Rifampicin resistant mutants. J. Mol. Biol. 1988; 202:245–253. [DOI] [PubMed] [Google Scholar]
- 10. Landick R., Stewart J., Lee D.. Aminoacid changes in conserved regions of the β–subunit of Escherichia coli RNA polymerase alter transcription pausing and termination. Genes Dev. 1990; 4:1623–1636. [DOI] [PubMed] [Google Scholar]
- 11. Jin D.J., Gross C.. RpoB8, a rifampicin-resistant termination-proficient RNA Polymerase Has an increased Km for purine nucleotides during transcription elongation. J. Biol. Chem. 1991; 266:14478–14485. [PubMed] [Google Scholar]
- 12. Jin D.J., Turnbough C.. An Escherichia coli RNA polymerase defective in transcription due to overproduction of abortive initiation products. J. Mol. Biol. 1994; 236:72–80. [DOI] [PubMed] [Google Scholar]
- 13. Jin D.J., Gross C.. Characterization of the pleiotropic phenotypes of rifampicin-resistant rpoB mutants of Escherichia coli. J. Bacteriol. 1989; 171:5229–5231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Murphy H., Cashel M.. Isolation of RNA polymerase Suppressors of a (p)ppGpp Deficiency. Methods Enzymol. 2003; 371:596–601. [DOI] [PubMed] [Google Scholar]
- 15. Zhou Y.N., Lubkovska L., Hui M., Court C., Chen S., Court D., Strathern J., Jin D.J., Kashlev M.. Isolation and characterization of RNA polymerase rpoB mutations that alter transcription slippage during elongation in E. coli. J. Biol. Chem. 2013; 288:2700–2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jin D.J., Cashel M., Friedman D., Nakamura Y., Walter W., Gross C.. Effects of rifampicin resistant rpoB mutations on antitermination and interaction with nusA in Escherichia coli. J. Mol. Biol. 1988; 204:247–261. [DOI] [PubMed] [Google Scholar]
- 17. Santangelo T., Mooney R., Landick R., Roberts J.. RNA polymerase mutations that impair conversion to a termination-resistant complex by Q antiterminator proteins. Genes Dev. 2003; 17:1281–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bae B., Feklistov A., Lass-Napiorkowska A., Landick R., Darst S.A.. Structure of a bacterial RNA polymerase holoenzyme open promoter complex. Elife. 2015; 4:e08604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Vassylyev D.G., Vassylyeva M.N., Zhang J., Palangat M., Artsimovitch I., Landick R.. Structural basis for substrate loading in RNA polymerase. Nature. 2007; 448:163–168. [DOI] [PubMed] [Google Scholar]
- 20. Gagneux S., Long C.D., Small PM., Van T., Schoolnik G.K., Bohannan B.J.. The competitive cost of antibiotic resistance in Micobacterium tuberculosis. Science. 2006; 312:1944–1946. [DOI] [PubMed] [Google Scholar]
- 21. O’Neill A.G., Huovinen T., Fishwick C.W.G., Chopra I.. Molecular genetic and structural modeling studies of Staphilicoccus aureus RNA polymerase and the fitness of rifampin resistance genotypes in relation to clinical prevalence. Antimicrob. Agents Chemother. 2006; 60:298–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Brandis G., Wrande M., Lijas L., Hughes D.. Fitness-compensatory mutations in rifampicin-resistant RNA polymerase. Mol. Microbiol. 2012; 85:142–151. [DOI] [PubMed] [Google Scholar]
- 23. Weinzlier R.O.J. The bridge helix of RNA polymerase acts as a central nanomechanical switchboard for coordinating catalysis and substrate movement. Archaea. 2011; 2011:608385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Serpinski O., Karginova E., Mikryukov N., Kravchenko V., Zaychikov E., Maksimova T., Onikienko A., Pletnev A., Mitina Yu.. Cloning of bacteriophage T7 fragment containing T2 promoter. Bioorgan. Chimia (Russ.). 1982; 8:840–846. [Google Scholar]
- 25. Nudler E., Goldfarb A., Kashlev M.. Discontinuous mechanism of transcription elongation. Science. 1994; 265:793–796. [DOI] [PubMed] [Google Scholar]
- 26. Belogurov G., Vassylyeva M., Svetlov V., Klyuyev S., Grishin N., Vassylyev D., Artsimovich I.. Structural basis for converting a general transcription factor into an operon-specific virulence regulator. Mol. Cell. 2007; 26:117–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Court D., Sawitzke J., Thomason L.. Genetic engineering using homologous recombination. Annu. Rev. Genet. 2002; 36:361–388. [DOI] [PubMed] [Google Scholar]
- 28. Vitiello C.L., Kireeva M.L., Lubkowska L., Kashlev M., Gottesman M.. Coliphage HK022 Nun protein inhibits RNA polymerase translocation. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:E2368–E2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Molodtsov V., Scharf N.T., Stefan M.A., Garcia G.A., Murakami K.S.. Structural basis for rifamycin resistence of bacterial RNA polymerase by the three most clinically important RpoB mutations found in Mycobacterium tuberculosis. Mol. Microbiol. 2017; 103:1034–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kumar P., Chatterji D.. Differential inhibition of abortive transcription initiation at different promoters catalysed by E. coli RNA polymerase. FEBS Lett. 1992; 306:46–50. [DOI] [PubMed] [Google Scholar]
- 31. McClure W.R., Cech C.L., Johnston D.E.. A steady state assay for RNA polymerase initiation reaction. J. Biol. Chem. 1978; 253:8941–8948. [PubMed] [Google Scholar]
- 32. McClure W.R. Mechanism and control of transcription initiation in eukaryotes. Ann. Rev. Biochem. 1985; 54:171–204. [DOI] [PubMed] [Google Scholar]
- 33. Carpousis A.J., Gralla J.D.. Cycling of ribonucleic acid polymerase to produce oligoribonucleotides during initiation in vitro at the lac UV5 promoter. Biochemistry. 1980; 19:3245–3253. [DOI] [PubMed] [Google Scholar]
- 34. Gagneux S., Long C.D., Small P.M., Van T., Schoolnik G.K., Bohannan B.J.M.. The competitive cost of antibiotic resistance in Mycobacterium tuberculosis. Science. 2006; 312:1944–1946. [DOI] [PubMed] [Google Scholar]
- 35. Dang U.T., Zamora I., Hevener K., Adhikari S., Wu X., Hurdle J.. Rifamycin resistance in Clostridium difficile is generally associated with a low fitness burden. Antimicrob. Agents Chemother. 2016; 60:5604–5607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Artsimovitch I., Chu C., Lynch A.S., Landick R.. A new class of bacterial RNA polymerase inhibitor affects nucleotide addition. Science. 2003; 302:650–654. [DOI] [PubMed] [Google Scholar]
- 37. Bae B., Nayak D., Mustaev A., Landick R., Darst S.. CBR antimicrobials inhibit RNA polymerase via at least two bridge helix cap-mediated effects on nucleotide addition. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:E4178–E4188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Crum G.F., Devries W.H., Eble T.E., Large C.M., Shell J.W.. Streptolydigin, a new antimicrobial antibiotic. II. Isolation and characterization. Antibiot. Annu. 1955; 3:893–896. [PubMed] [Google Scholar]
- 39. Temiakov D., Zenkin N., Vassylyeva M., Perederina A., Tahirov T., Kashkina E., Savkina M., Zorov S., Nikiforov V., Igarashi N. et al. Structural basis of transcription inhibition by antibiotic streptolydigin. Mol. Cell. 2005; 19:655–666. [DOI] [PubMed] [Google Scholar]
- 40. Takai K., Nakamura K., Toki K., Tsunogai U., Miyazaki M., Miyazaki J., Hirayama H., Nunoura T., Horikoshi K.. Cell proliferation at 122°C and isotopically heavy CH4 production by a hyperthermophilic methanogen under high pressure cultivation. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:10949–10954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Oren A. Life at high salt concentrations, intracellular KCl concentrations, and acidic proteomes. Front. Microbiol. 2013; 4:315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Opalka N., Brown J., Lane W.J., Twist K-A., Landick R., Asturias F.J., Darst S.. Complete structural model of Escherichia coli RNA Polymerase from a hybrid approach. PLoS Biol. 2010; 8:e1000483. [DOI] [PMC free article] [PubMed] [Google Scholar]