Abstract
To acquire CRISPR–Cas immunity against invasive mobile genetic elements, prokaryotes must first integrate fragments of foreign DNA into their genomic CRISPR arrays for use in future invader silencing. Here, we found that the hyperthermophilic archaeaon, Pyrococcus furiosus, actively incorporates DNA fragments (spacers) from both plasmid (foreign) and host genome (self) sequences into its seven CRISPR loci. The majority of new spacers were derived from DNA immediately downstream from a 5′-CCN-3′ protospacer adjacent motif (PAM) that is critical for invader targeting. Interestingly, spacers were preferentially acquired from genome or plasmid regions corresponding to active transposons, CRISPR loci, ribosomal RNA genes, rolling circle origins of replication, and areas where plasmids recombined with the host chromosome. A common feature of the highly sampled spacers is that they arise from DNA regions expected to undergo DNA nicking and/or double-strand breaks. Taken together with recent results from bacterial systems, our findings indicate that free DNA termini and PAMs are conserved features important for CRISPR spacer uptake in diverse prokaryotes and CRISPR–Cas systems. Moreover, lethal self-targeting by CRISPR systems may contribute to host genome stability by eliminating cells undergoing active transposon mobility or chromosomal uptake of autonomously replicating foreign mobile genetic elements.
INTRODUCTION
In bacteria and archaea, CRISPR–Cas (clustered regularly interspaced short palindromic repeats and CRISPR-associated genes) systems provide RNA-guided adaptive immunity against invaders such as viruses and plasmids (1–5). CRISPR–Cas systems are diverse, and are grouped into two classes, six major types (I–VI) and at least 16 subtypes (6,7). In all these CRISPR–Cas systems, adaptive immunity consists of three stages: adaptation to novel invaders, CRISPR RNA (crRNA) biogenesis, and invader silencing. During adaptation, short fragments of DNA called ‘protospacers’ are recognized and excised from invader DNA and then processed and integrated into CRISPR arrays (where the same sequences are called ‘spacers’) for use in future invader silencing. CRISPR arrays are composed of a leader sequence followed by short direct repeats that are separated by the spacers, which are of common length but variable sequence (8). During crRNA biogenesis, CRISPR arrays are transcribed and processed into crRNAs, which then form a complex with the Cas effector proteins (9). During invader silencing, the effector complex binds to target complementary DNA or RNA and degrades it (1,3,10).
While the steps of crRNA biogenesis and invader silencing are relatively well studied, the molecular mechanisms underlying adaptation are only beginning to be characterized. Adaption mechanisms are being investigated in several diverse systems in a variety of model organisms (11–13), with, the majority of adaptation studies to date focusing on Type I-E and Type I-F systems from bacteria. From some of these studies, evidence indicated that naïve spacer acquisition primarily occurs at sites of double-stranded breaks, which form at stalled DNA replication forks (14). This process is mostly dependent on the DNA repair nuclease–helicase complex, RecBCD (14,15). The proposed model suggests that DNA fragments of RecBCD breakdown products are incorporated into the Cas1–Cas2 adaptation complex and are processed to become new spacers. Sequence features called Chi sites interrupt the activity of the RecBCD complex and thus limit breakdown products. Since these Chi sites are more abundant in the prokaryotic genome than in the invader DNA, they provide a mechanism by which the system can be more preferentially targeted to exogenous DNA (14). In contrast, analysis of the Type II-A system in Streptococcus thermophilus revealed that the majority of new spacers are acquired from the host chromosome versus invader DNA and that lethal self-targeting normally eliminates cells from the population that have acquired chromosomal (self) spacers (16). CRISPR acquisition of spacers also requires the presence of a 2–5 nt flanking sequence called the protospacer adjacent motif (PAM) in the selected protospacer sequences (17–20). In Escherichia coli, the Cas1–Cas2 complex recognizes the PAM, processes protospacers into the proper size (19,21,22), and integrates them at the leader end of the CRISPR array (20,23,24). In the Type I-F system from Pectobacterium atrosepticum, Cas2 is fused with the interference protein Cas3; this novel form of Cas2 complexes with Cas1 and together they are sufficient to recognize protospacer PAMs, process spacers, and integrate them into the array (25–27).
Here we build on what is known about adaptation using the hyperthermophile, Pyrococcus furiosus. This Achaean has three CRISPR–Cas immune effector crRNA-Cas protein complexes, including two type I systems, a type I-A Csa and a type I-G Cst, and a type III-B Cmr system (Figure 1A) (26–30). The Cas1, Cas2 and Cas4 adaptation proteins are encoded in one locus together with the Cst and Cmr effector complex proteins and the Cas6 CRISPR RNA processing endonuclease, while the Csa effector complex genes are located at a separate locus and are not associated with adaptation genes (Figure 1A, 1B). There is also a second Cas4 gene in P. furiosus, Cas4–2, which is not located near any other CRISPR related genes (28). Pyrococcus furiosus contains seven CRISPR arrays (Figure 1A) with 200 spacers among them (31). All seven loci have a highly conserved leader and repeat sequence. Spacers from all seven loci are processed by Cas6 and the mature crRNAs become associated with all three immune effector crRNP complexes (27,28,32,33). Molecular mechanisms of crRNA biogenesis and invader silencing steps are well-characterized (26,27,29,30,32,34). However, new spacer acquisition has not yet been reported in P. furiosus.
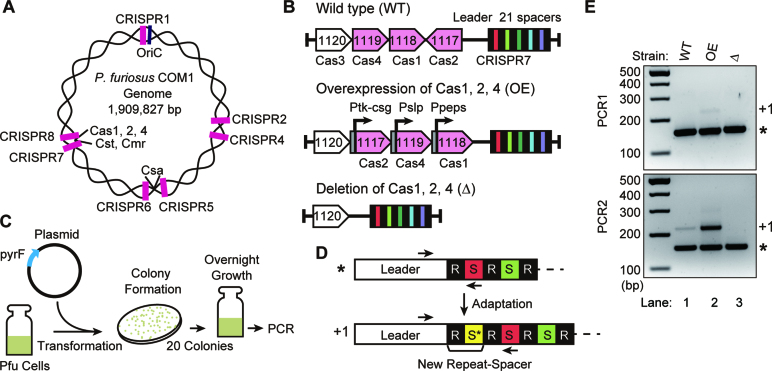
Figure 1.
P. furiosus actively takes up DNA into CRISPR loci. (A) Positions of the seven CRISPR loci (pink), the Cas gene clusters and the DNA replication origin (oriC) in the P. furiosus genome. (B) Overview of Cas gene loci of P. furiosus strains: WT (wild type strain), OE (Cas1, Cas2, Cas4 protein overexpression strain) and Δ (cas1, cas2, cas4 deletion strain). Predicted adaptation genes (pink) are indicated. (C) Graphic representation of the adaptation assay. (D) Illustration of CRISPR locus and adaptation. The CRISPR leader sequence is followed by alternating repeat (R, black) and spacer (S, colored) units. Arrows indicate the locations of primers used in PCR for detection of new spacer (S*)/repeat units (+1). (E) Analysis of adaptation in wild type, OE and Δ strains. The leader-first spacer region of CRISPR7 was amplified with primers indicated in D. The PCR product corresponding to parental arrays and addition of one repeat-spacer unit are indicated with an asterisk and +1, respectively.
In this study, we observed new spacer acquisition in P. furiosus both in strains with endogenous levels of Cas1, Cas2 and Cas4 and in strains overexpressing these proteins. We found that overexpression of Cas1, Cas2 and Cas4 elevates adaptation. We also found that all seven CRISPR arrays are active for adaptation, and that the size, PAM, and distribution of protospacers are consistent across all CRISPR arrays in both strains. The majority of new spacers were acquired from P. furiosus chromosomal DNA (i.e. are self-targeting), but spacers also came from introduced plasmid DNA, preferentially from rolling circle replication plasmids rather than theta replication plasmids. Within the chromosome, hotspots of spacer acquisition were observed at active transposase genes, CRISPR arrays, highly transcribed rDNA gene loci, OriC DNA replication origin, and in regions where there is homology between the chromosome and the rolling circle replication plasmid. Our results show that this in vivo system can actively adapt and that the resulting new spacers bear the size and PAM signatures of functional crRNAs. Furthermore, we consistently observed that spacers are preferentially acquired from DNA regions where nicking or double-strand break events are common, suggesting that free DNA ends are targeted by the adaptation machinery for conversion into new spacers.
MATERIALS AND METHODS
P. furiosus strains and growth conditions
Pyrococcus furiosus strain JFW02 (29) was used as wild type in this study. P. furiosus was grown anaerobically in a defined medium with cellobiose as the carbon source (30) at 90°C for 16–20 h in anaerobic culture bottles or on medium solidified with 1% (wt/vol) Gelrite (Research Products International) for 65 h. For growth of uracil auxotrophic strains, the defined medium contained 20 μM uracil. Plasmid transformation was performed as described previously (30). Pyrococcus furiosus strains were cultured anaerobically at 90°C to mid-to-late log phase in defined liquid medium. Pyrococcus furiosus culture was mixed with DNA at a concentration of 2.5 ng/μl culture and spread in 35 μl aliquots onto defined solid medium.
Plasmid construction
Plasmids were constructed using standard cloning techniques. The sequences of oligonucleotides used in this study are shown in Supplemental Table S1. To generate the pYS3-Pslp plasmid, SimR of the pYS3 plasmid (31) was replaced by a P. furiosus slp-promoter (250 bp upstream of PF1399)-pyrF gene expression cassette. The insert for pMS32 plasmid was constructed by overlap PCR using JFW02 genomic DNA and pJE47 plasmid. The genome insertion cassette between PF1120 and CRISPR7, ∼0.7 kb flanking regions immediately upstream and downstream of the Cas2–Cas1–Cas4 gene cluster, with NotI-EcoRV sites was inserted into pHSG298. The Cas overexpression cassette, csg promoter–Cas2 (PF1117)—slp promoter–Cas4 (PF1119)—PRP synthetase promoter–Cas1 (PF1118), was cloned into pJE47. pHSG298 with the genome insertion cassette and the Cas overexpression cassette were digested by NotI and EcoRV and ligated to yield pMS32 plasmid. The plasmids were sequenced to confirm insert sequence and orientation.
Strain construction
To create a Cas1, Cas2, Cas4 protein overexpression strain (OE; TPF43), NruI-linearized pMS32 plasmid was transformed into the JFW02 (wt) strain. Two rounds of colony purification were performed by plating 10−3 dilutions of transformant cultures onto selective medium (without 20 μM uracil) and picking isolated colonies into selective liquid medium. Following marker replacement of the region of interest, 2.75 mM 5-FOA, a toxic PyrF substrate, was used to select for cells that had popped out the pyrF marker by homologous recombination between short regions of homology. The Cas1, Cas2 and Cas4 deletion strain (Δ strain; TPF55) was created using the pop-out marker replacement strategy as described previously (29). The transformed PCR products were generated by overlap PCR. The sequence of oligonucleotide used is showed in Supplemental Table S1.
Spacer acquisition assay and high throughput sequencing
Twenty colonies of P. furiosus strains transformed with plasmid were inoculated in 5 ml defined medium containing 20 μM uracil. Cultures were incubated at 90°C overnight. Genomic DNA was isolated from cells in 1 ml of overnight culture using quick-gDNA miniprep kit (Zymo Research). CRISPR arrays were amplified by PCR using a set of primers in which the forward primer annealed within the leader region of the CRISPR array and the reverse primer annealed to the existing spacer closest to the leader (referred to as the first round of PCR). If a new spacer was integrated into the CRISPR array, the resulting PCR product was longer due to the additional repeat and spacer sequence. These larger, expanded PCR products were separated from unexpanded products by gel electrophoresis followed by DNA recovery (Zymo Research). PCR primers included an overhang corresponding to part of the adapter necessary for Illumina sequencing. After size selection of the first PCR product, a second round of PCR was done to further amplify the expanded product. For very faint products, this second PCR was size selected and then a third round of PCR was done. Lastly, a nested PCR was done to add additional sequences corresponding to Illumina adapters and barcodes. Each experimental condition and replicate received a unique barcode (index) for multiplexing. The sequences of oligonucleotides used in the PCR reactions are listed in Supplemental Table S1.
Final gel-purified amplicon libraries were ranked and pooled by PCR intensity, and the pooled DNA was purified and concentrated by ethanol precipitation. DNA pools were quantitated, normalized according to concentration and number of samples represented in the pool, and then combined to make a final pool for sequencing. Array libraries were sequenced on an Illumina MiSeq set to yield 200 × 100 paired-end reads; the 200 base read 1 sequences were used in this study.
After sequencing, samples were de-multiplexed by index, and the sequence corresponding to a new (expanded) spacer was extracted from each read. To determine the source of these new spacers (i.e. to identify the protospacer sequence) we used Bowtie (32) to align the reads to a reference containing the genome and any plasmids used. For each experiment, the set of aligned protospacer sequences was then characterized with respect to length, GC content, and position on the genome or plasmid. We determined the proportion of new spacers derived from the genome versus a plasmid, and from the plus versus minus strand. Consensus sequences in the DNA upstream and downstream from the protospacer positions were identified by making sequence logos (33) from adjacent genomic sequences which were extracted using bedtools (34).
Next, we created custom genome browser tracks (tools for this are available through the UCSC genome browser) to identify spatial patterns in the distribution of protospacers. Since the presence of a PAM sequence was shown to promote recruitment of a protospacer, we took into account the underlying distribution of PAM sequences along the genome and plasmids. In silico, we created a ‘PAMscape’ track in which a potential 37 bp protospacer is found immediately 3′ to all PAM sequences in the genome. Any clusters or ‘hotspots’ of protospacers that deviated from the background of PAM distribution were assumed to be due to mechanisms other than local sequence motifs. In addition to visually examining the tracks, we used the findPeaks software in the HOMER analysis package (35) to identify statistically significant clusters of protospacers. For this analysis we used a read-count normalized PAMscape as an input control and used the following analysis options to optimize the process for detecting variably-sized spacer peaks: -style histone, -size 500, -strand separate, -fragLength 40, -region.
RESULTS
P. furiosus undergoes naïve adaptation
We tested the ability of P. furiosus to take up new spacers into CRISPR arrays in strains expressing either endogenous levels (WT) or elevated levels (O/E) of Cas1, Cas2 and Cas4 proteins (Figure 1B). As a negative control, we also included a null strain in which these three adaptation cas genes were deleted from the genome (Δ) (see Supplementary Figure S1 for immunoblot analysis of the expression levels of the Cas proteins in each of the three strains and previous work demonstrating that these three cas genes are normally expressed at relatively low levels (36)). Each of the three P. furiosus strains were transformed with an ‘invader’ plasmid (pYS3-Pgdh) and following overnight culturing of the transformants (Figure 1C), CRISPR spacer uptake was assessed by PCR amplification of the region between the leader and first spacer (Figure 1D). If a new spacer has been acquired in the CRISPR array (i.e. the array has been ‘expanded’), the PCR will yield a larger product (+1) because it would include an additional spacer and repeat sequence. After two rounds of PCR amplification PCR products corresponding to the expanded CRISPR array were observed for both WT and OE strains but not for the Cas null (Δ) strain (Figure 1E, lower panel, lanes 1–3). A faint band was observed in the first round of PCR for the OE but not the WT strain (Figure 1E, upper panel, lane 2). Taken together, the results indicate that naïve CRISPR spacer acquisition in P. furiosus occurs under normal growth conditions and with endogenous Cas protein expression levels, but the frequency of spacer uptake can be elevated by increasing the levels of the three Cas proteins (Cas1, Cas2 and Cas4).
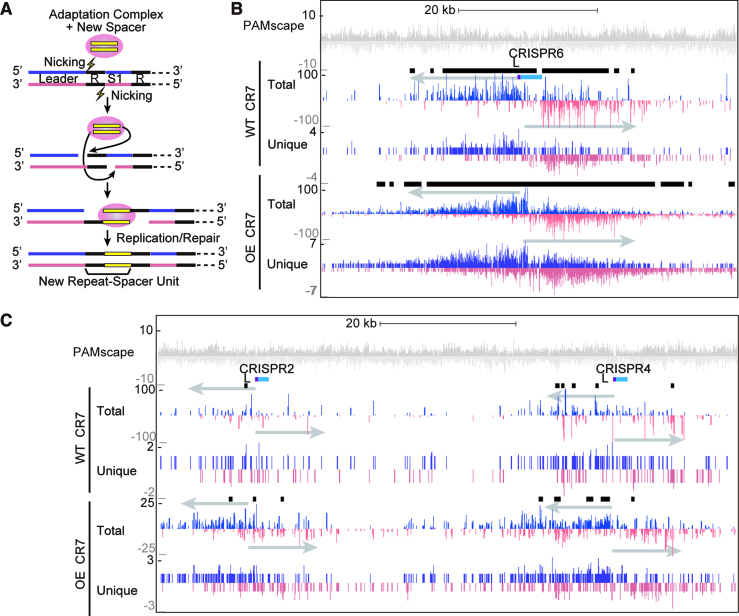
All seven CRISPR loci are active for adaptation
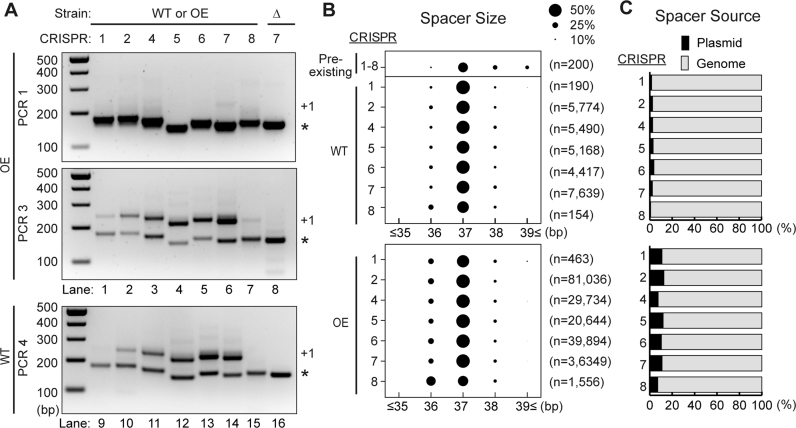
Pyrococcus furiosus has seven CRISPR loci, and all loci are transcribed and active for DNA interference (Figure 1A) (37,38). We tested whether all CRISPR loci are also active for CRISPR spacer uptake. Expanded arrays were observed in all CRISPR loci in both the OE and WT strains indicating that all CRISPR loci are active for adaptation (Figure 2A, lanes 1–7 and 9–15, respectively). We note that the relative efficiencies of spacer uptake at individual CRISPR loci appears to differ for unknown reasons with lower efficiencies of spacer uptake being observed for CRISPR 1 and 8 relative to CRISPR 2–7 loci. Next, we sequenced the products of each of the seven expanded CRISPR arrays from WT and OE strains using an Illumina MiSeq, and characterized the source, distribution, size and flanking sequence of new spacers (Figure 2B and C). In WT, the majority (96–99%) of the unique spacers mapped to the P. furiosus chromosome and the rest (1–4%) mapped to the plasmid (Figure 2C). In the OE strain, acquisition from the plasmid increased to ∼10% (Figure 2C).
Figure 2.
All seven CRISPR loci are active for adaptation. (A) Analysis of adaptation in the WT and OE strains for all seven CRISPR loci. The leader-first spacer region of each CRISPR array was amplified with primers indicated in Figure 1D. Sizes of DNA standards are indicated. (B) Bubble chart showing size distributions for protospacers from all CRISPR loci in WT and OE strains. Length of pre-existing spacers is also shown. The size of the bubbles is directly proportional to the relative proportion in the library. Pooled data from two experiments are presented. (C) Proportion of spacers derived from the plasmid (black) and genome (gray) in WT and OE strains. Pooled data from two experiments are presented.
The reference strain of P. furiosus already harbors 200 spacers across its seven CRISPR loci, and over 70% of the pre-existing spacers are 36–38 bp in length, with 37 bp spacers being the most common (Figure 2B). In WT, over 90% of newly acquired spacers were also 36–38 bp in length with a median of 37 bp in all seven CRISPR loci (Figure 2B). As with WT, most new spacers in the OE strain were 36–38 bp in length and about half of new spacers were 37 bp, though ∼40% of spacers acquired into the CRISPR8 array were 36 bp (Figure 2B). Thus, the size distribution of the newly acquired spacers closely matched that of the naturally acquired spacers both in WT and OE strains, and 37 bp spacers were predominately acquired in all seven CRISPR loci.
Motifs upstream and downstream of protospacers
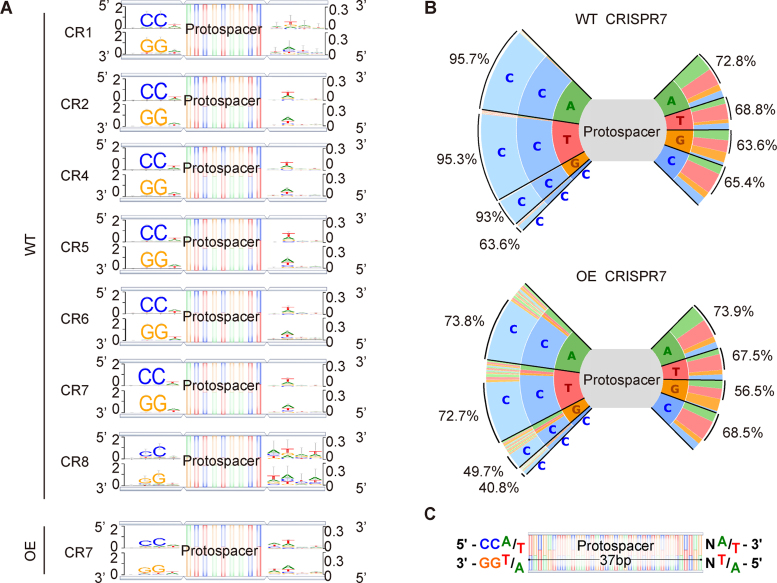
To search for PAM sequences involved in adaptation, we constructed sequence logos (33) of the flanking sequences upstream and downstream of uniquely mapped protospacers. The bioinformatically-predicted PAM for the seven P. furiosus CRISPR loci is 5′-CCN-3′/5′-NGG-3′ (17,39). During DNA interference, wild type P. furiosus can also target a sequence when there is an adjacent 5′-NCN-3′/5′-NGN-3′, 5′-CNN-3′/5′-NNG-3′, 5′-TTN-3′/5′-NAA -3′ or 5′-GTN-3′/5′-NAC-3′ (40,41). Our analysis showed a strong consensus 5′-CCN-3′/5′-NGG-3′ sequence upstream of newly acquired protospacers in WT (Figure 3A). The consensus is slightly weaker for the CRISPR 8 array (Figure 3A, Supplemental Data, Figure S2), perhaps related to its aberrant repeat sequence (37). Unexpectedly, we observed a weak consensus of 5′-NW-3′/5′-WN-3′ (W is A or T) downstream of the protospacer (Figure 3A). Plotting all bases by frequency, we observed that the canonical 5′-CCN-3′ PAM was the most enriched, and we also noted an A/T bias in the third (N) position (Figure 3B). Compared with wild type, the 5′-CCN-3′/5′-NGG-3′ consensus was slightly weaker in OE strain spacers, with ∼70% of spacers having the canonical 5′-CCN-3′/5′-NGG-3′ PAM (Figure 3A and B), perhaps due to unbalanced expression of Cas1, Cas2, Cas4 and other proteins required for protospacer generation. These results indicate that adaptation involves the recognition of a specific sequence both upstream and downstream of protospacers during spacer acquisition (Figure 3C). The results also suggest that the new spacers acquired in our assay could function as guide RNAs in later DNA interference because they were derived from source DNA that had the appropriate PAM for targeting (40,41).
Figure 3.
Analysis of PAM sequences in adaptation. (A) Newly-acquired spacers in each CRISPR array were used to search the genome and plasmids in order to identify the corresponding protospacers, and upstream and downstream sequences were extracted and used to generate consensus motifs on both strands of DNA. Four bp of flanking sequence on each side of the protospacers is shown. (B) Plots show the frequencies of all 3 bp flanking sequences upstream of the protospacer and 2 bp downstream of the protospacer for CRISPR7 in the WT and OE strains. Percentages on left side indicate proportion of spacers with CC at the second and third position and percentages on right side indicate proportion of spacers with W (A/T) at the second position. (C) Based on the observed consensus sequences, a model of the ideal upstream and downstream motifs was made.
Spacers are preferentially acquired from a rolling circle replication plasmid
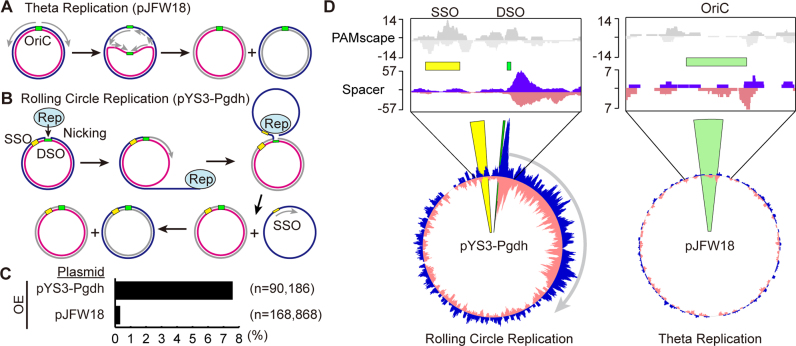
It has been reported that two types of plasmid, pJFW18 and pYS3 can replicate in P. furiosus (31,42). The pJFW18 plasmid has an OriC origin of replication from the P. furiosus genome and replicates by a theta replication mechanism involving bi-directional replication from the origin (Figure 4A). In contrast, the pYS3 plasmid replicates by a rolling circle replication mechanism that first requires nicking of one strand of the DNA (at the double-strand origin of replication) by the plasmid-encoded Rep75 protein followed by unidirectional replication from the free 3′ end of the parental DNA strand (Figure 4B). We used a pYS3-derived plasmid (pYS3-Pgdh) and pJFW18 to examine whether the mode of plasmid replication influences spacer acquisition. As before, we analyzed spacer acquisition by PCR amplification of CRISPR arrays in the plasmid-transformed, OE strain, and observed expanded arrays in both cases (Figure 4C). As described above, in experiments using pYS3-Pgdh, ∼10% of new spacers were acquired from the plasmid. In contrast, when using pJFW18, only 0.3% of spacers were acquired from the plasmid (Figure 4C). Both plasmids are a similar size (pYS3-Pgdh is 6242 bp, pJFW18 is 5400 bp), and it has been reported that the copy number of both pJFW18 and pYS3 is 1–2 copies per chromosome (31,42), so the amount of plasmid DNA is expected to be similar in all experiments. These results indicate that spacers are preferentially acquired from the rolling circle replication plasmid compared to the theta replication plasmid.
Figure 4.
Spacers are preferentially acquired from rolling circle replication plasmids. Both theta (A) and rolling circle (B) replication plasmids were used in these studies. Cartoon models shown here illustrate how replication is thought to occur in each plasmid type. (A) Theta plasmid replication initiates synthesis at a replication origin (OriC). Replication extends the leading strand continuously while synthesizing the lagging strand discontinuously. (B) Rolling circle replication is initiated by a Rep protein, which nicks the double-strand origin (43). Replication proceeds using the un-nicked strand as a template displacing the nicked strand. DNA replication initiates at the single strand origin (SSO) to complete the new double-stranded DNA circle. (C) Spacers from expanded arrays were aligned to both the P. furiosus genome and the plasmid used in the assay. The percentage of spacers aligning to the plasmid rather than the genome (black bar) is indicated for CRISPR7 in of the OE strain. Pooled data from two experiments are presented. (D) Alignment of protospacers to the plasmid shows an unequal distribution. Top panels show 5′-CCN-3′/5′-NGG-3′ PAM scape on each plasmid. Green bars show origin of DNA replication. Bottom panels show the distribution of protospacers across plasmids pYS3-Pgdh and pJFW18. Protospacers on the plus and minus strand are indicated in blue and pink, respectively. Protospacers from CRISPR7 in the OE strain are shown here; other CRISPR arrays and WT strains show similar distributions.
We next examined spacer distribution along the plasmids. Since a PAM sequence was consistently observed upstream from a protospacer, we took into account the underlying distribution of PAMs along the genome and plasmids by creating an in silico ‘PAMscape’ in which a 37 bp protospacer is defined immediately 3′ to all 5′-CCN-3′ PAM sequences in the genome and plasmids. Any clusters or ‘hotspots’ of protospacers above the background of PAM distribution were assumed to be due to mechanisms other than the local sequence motif. For pYS3-Pgdh, spacers were acquired throughout the plasmid, consistent with the PAM distribution. In addition, a large peak was observed at the double-strand origin (43) on the plasmid (Figure 4D). On the pJFW18 plasmid, although the spacer distribution was consistent with PAM distribution, the overall spacer number was much lower (Figure 4C and D). The total number of new spacer sequences generated in the pJFW18 experiments was equal to or greater than that in the pYS3-Pgdh experiments but since a larger majority aligned to the genome (Figure 4C) the plasmid was sparsely populated with spacers (Figure 4D). Together these results suggest that certain steps in DNA replication that are unique to the rolling circle mechanism make this type of plasmid a good target for spacer acquisition. Not only was the pYS3-Pgdh more heavily targeted in general (Figure 4D), but pYS3-Pgdh also displayed a large peak at its double-strand origin of replication (Figure 4D). At the origin, DNA nicking initiates replication, and the protospacers map in a pattern wherein they are enriched beginning at the site of nicking by the Rep75 protein and they continue in the direction of replication, suggesting that strand nicking and rolling circle replication positively influence generation of protospacers (Figure 4B and D).
Distribution of self-targeting protospacers across the P. furiosus genome
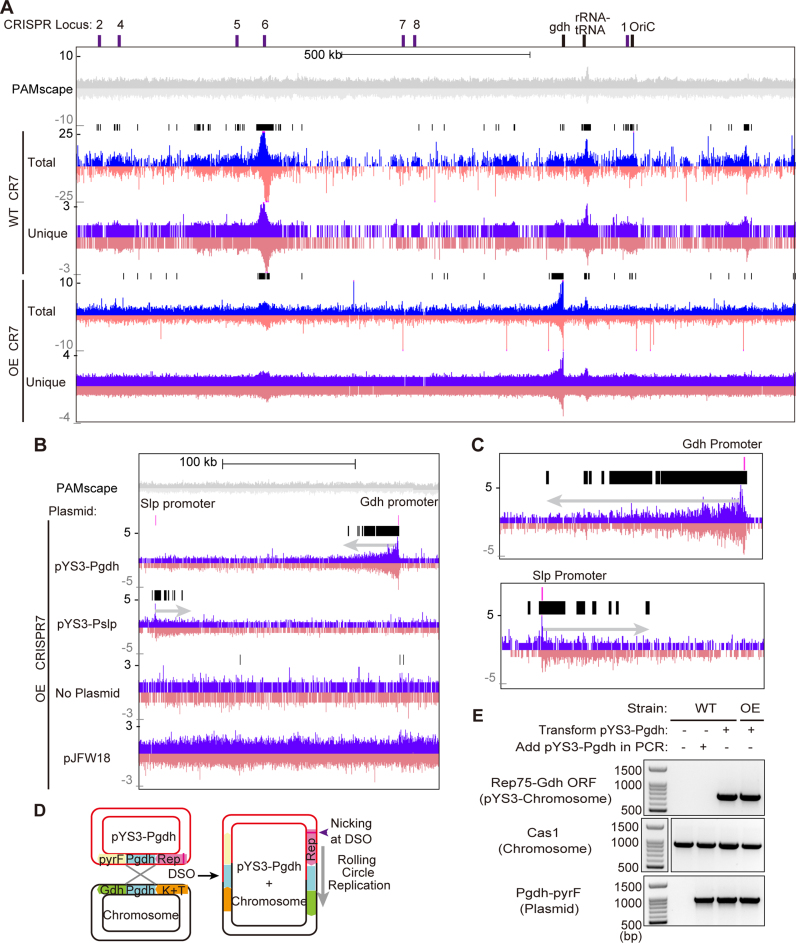
We also looked for protospacer clusters or hotspots along the length of the ∼2 Mb P. furiosus host chromosome. The distribution of acquired protospacers was consistent for all seven CRISPR loci (Supplemental Figure S3), so here we focus the distribution pattern for the CRISPR7 array (Figure 5A). Overall, spacers were equally likely to be derived from the plus and minus strands of both the chromosome and the plasmid (Figures 4D and 5A), but were not evenly distributed, and we noted several clusters that were consistent among replicates. In addition to visually examining the tracks, we used the findPeaks software in the HOMER analysis package (35) to identify statistically significant clusters of protospacers above that expected from PAM density alone (black bars, Figure 5A). As described in more depth below, significant protospacer hotspots in the P. furiosus genome were observed around the replication origin (OriC), CRISPR loci, rRNA and tRNA cluster, transposons, and the Gdh (glutamate dehydrogenase, PF1602) promoter in both WT and OE strains (Figure 5A). DNA next to OriC is replicated first, so a culture contains more copies of origin proximal DNA. Therefore, the average DNA content in the culture can potentially explain a protospacer peak around the OriC origin of DNA replication (Supplementary Figure S4). However, the other peaks could not be explained by DNA content.
Figure 5.
Aligned protospacers are not evenly distributed across the P. furiosus chromosome. (A and B) Distribution of protospacers on the P. furiosus chromosome. Protospacers on the plus and minus strand are indicated in blue and pink respectively. Protospacers were deduced from aligning new spacers acquired into CRISPR7 loci of WT and OE strains. The 5′-CCN-3′/5′-NGG-3′ PAMscape across the chromosome is shown in gray (top). Black bars show regions where protospacers are significantly enriched over the background, as detected by the peak-calling software from the HOMER package. (A) Distribution of protospacers across the full length of the chromosome. (B and C) Protospacers are enriched around areas where homology exists between the chromosome and the plasmid. Prominent clusters of protospacers are observed extending 1000s of bps out from the region of homology. Protospacers from CRISPR7 in the OE strain are shown here; other CRISPR arrays and WT strains show similar distributions. Close-up panels (C) show that the peak of enrichment starts ∼300bp from the location of homology between the plasmid and chromosome, and extends out from that point. (D) Model of homologous recombination occurring at the homologous region (Pgdh) between plasmids and the P. furiosus chromosome. Recombination results in integration of the plasmid into the chromosome. (E) PCR results confirm that homologous recombination between the plasmid and chromosome occurs. Top panel shows amplified products of plasmid-integrated chromosome. Middle panel shows internal control of chromosome and bottom panel shows internal control of the plasmid. Sizes of DNA standards are indicated.
Recombination between the chromosome and rolling circle replication plasmids generate hotspots of spacer acquisition
A significant protospacer hotspot coincided at one end with the glutamate dehydrogenase (Gdh) promoter (Figure 5A) and extended several thousand bases in one particular direction along the host genome (Figure 5B and C). This hotspot was most pronounced in the OE strain and was also present, though less prominently, in the WT strain, but was completely absent when no plasmid was used (Figure 5B, Supplementary Figure S3), suggesting that some element on the pYS3-Pgdh plasmid induced protospacer acquisition on the chromosome. Because the Gdh promoter (283 bp) exists on both the P. furiosus chromosome and the pYS3-Pgdh plasmid, we hypothesized that this homology drives protospacer acquisition. To test that, we conducted a follow-up experiment using the pYS3-Pslp plasmid, which is identical to pYS3-Pgdh except that the Gdh promoter is replaced by the Slp promoter. When pYS3-Pslp-pyrF was used, the large chromosomal hotspot at the Gdh promoter disappeared, and a new hotspot appeared, coinciding on one end with the Slp promoter and extending several kilobases in a specific direction along the genome (Figure 5B and C), implying that the hotspot is dependent on sequence homology rather than the specific sequence of the Gdh promoter. The directionality of the hotspot changes, likely reflecting that the Slp promoter was cloned into the plasmid in the opposite orientation with respect to the origin of replication. Homology alone does not induce a promoter-associated spacer hotspot: though the pJFW18 plasmid has the same Gdh promoter as pYS3-Pgdh, no hotspot was observed when it was used (Figure 5B). These observations indicate that homology between the chromosome and a rolling circle replication plasmid leads to homologous recombination, and that rolling circle replication initiated by DNA nicking at the double-strand origin (43) of the pYS3 plasmid continues into the chromosome (Figure 5C). We verified that homologous recombination does occur between the plasmid and chromosome at the point of the Gdh promoter, by PCR using a set of primers in which the forward primer anneals within Rep75 of the pYS3 plasmid and the reverse primer within the ORF of the Gdh gene on the chromosome. Amplified products were observed only in strains transformed with the plasmid (Figure 5D and E). This homologous recombination also occurs between the pJFW18 plasmid and the chromosome, but with significantly reduced recombination efficiency (Supplementary Figure S5). Taken together, the results indicate that DNA nicking and rolling circle replication of the plasmid following homologous recombination into the host chromosome induced enhanced protospacer generation of adjacent host genome (self-targeted) sequences.
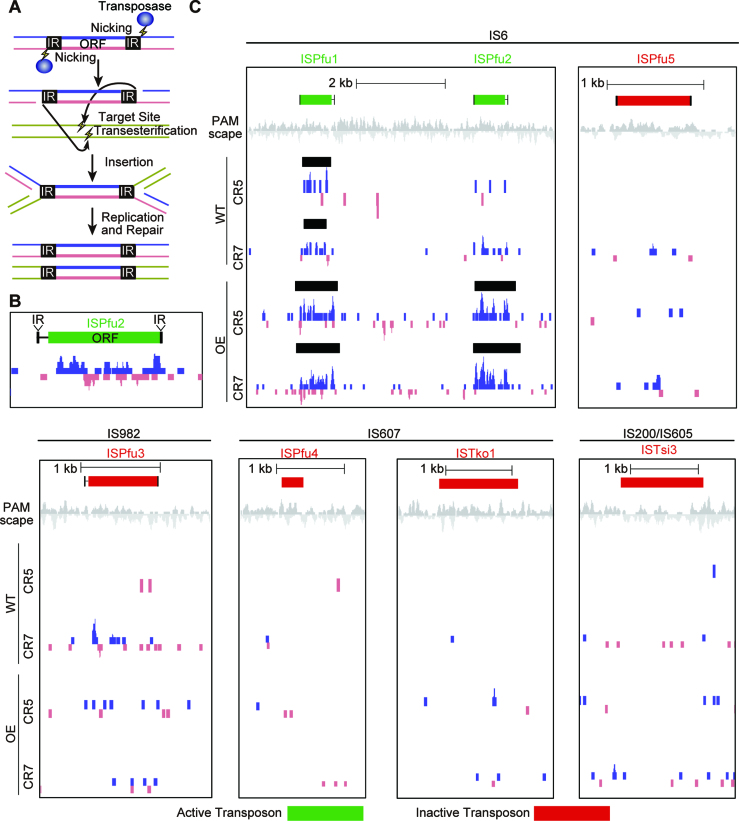
Host genomic protospacer hotspots are located at transposons, CRISPR arrays and the rRNA gene cluster
P. furiosus chromosomal protospacer hotspot peaks were also consistently observed at transposons, CRISPR arrays, and the rRNA gene cluster. The P. furiosus wild type strain contains 39 complete transposons (insertion sequence or IS elements) and 6 partial transposons (44). Most of these transposons are of the IS6 family and the others are IS982, IS607 and IS200/IS605. Interestingly, small protospacer peaks were frequently observed on ISPfu1 and ISPfu2 transposons, and their boundaries almost exactly matched the positions of the transposons (Figure 6B, C). These peaks were observed for all five of the highly active CRISPR arrays and were not influenced by the presence or absence of exogenous (plasmid) DNA (Supplementary Figure S6). The ISPfu1 and ISPfu2 transposons are thought to be active because genome sequencing of P. furiosus lab strains revealed that these gene elements have moved to new places in the genome, while others, namely ISPfu3, ISPfu4 and ISPfu5, are considered inactive (44). Thus, it was particularly interesting that we rarely, if ever, saw the same small protospacer peaks associated with the ISPfu3, ISPfu4 and ISPfu5 transposons. To quantify this discrepancy, we examined all 45 transposons, both active and presumed inactive, for each replicate of the CRISPR5 and CRISPR7 experiments (both WT and OE strains). We used the peaks called by the findPeaks software (35) to identify areas where mapped protospacers were significantly enriched compared to the surrounding region (Figure 6C). In all, 24 of the 29 active transposons had associated protospacer peaks in at least one experiment, while only three of the 16 inactive transposons had such a peak. Taking all replicates together, the probability that an active transposon would have a statistically significant peak was ∼0.27 compared with a probability of 0.027 for inactive transposons (Supplementary Figure S7). This strongly suggests that some aspect of transposon activity promotes protospacer generation. IS6 family transposons transpose by a replicative transposition mechanism wherein each side of the transposon is nicked to yield a 3′-OH, which attacks a phosphate at the DNA target site (45–47) (Figure 6A). A co-integrated product is formed between the donor IS element and the target DNA and is subsequently resolved by recombination to generate a new transposon at a new site while preserving the original transposon copy (47). This mechanism, together with the observation that protospacer clusters overlap with active transposons, again suggests that nicked DNA and/or DNA undergoing recombination provides a good source of protospacers.
Figure 6.
Protospacers are enriched for transposons. (A) Cartoon model of the mechanism for transposon replication. Transposases nick each transposon at the 3′ end. Target DNA is also nicked and the transposon joins to the target. The transposon intermediate is resolved by recombination. The original and target sites are separated, each harboring one copy of the transposon. (B and C) Protospacers are significantly enriched around active transposons. Protospacers on the plus and minus strand are indicated in blue and pink respectively. Protospacers were deduced by aligning new spacers acquired into the CRISPR5 and seven loci of WT and OE strains. Black bars show areas were protospacers are significantly enriched, as detected by peak finding software in the HOMER package. Transposons that were previously described as active (44) were 10 times more likely to have an associated peak than those described as inactive (red).
Another molecular event that requires DNA nicking is CRISPR spacer acquisition, during which both sides of the leader-proximal repeat are nicked to integrate new spacers (21,48) (Figure 7A). Consistent with this, we observed a large protospacer hotspot around the CRISPR6 array and smaller peaks around several other CRISPR loci (Figure 7B and C). Protospacer hotspots were not observed around CRISPR 1 or 8 (Figure 5A), possibly because these loci are less active for spacer acquisition than the others (Figure 2A). In the array hotspots, protospacers were preferentially located on the plus strand before the first spacer and on the minus strand after the first spacer (Figure 7B).
Figure 7.
Protospacers are enriched around CRISPR arrays. (A) Cartoon model showing steps of CRISPR array expansion during acquisition of new spacers. The protospacer 3′-OH group carries out a nucleophilic attack on the leader repeat junction and the repeat-first spacer junction. The protospacer is integrated into the CRISPR array and the array is expanded by a unit of a new spacer and repeat. (B and C) Protospacers on the plus and minus strand are indicated in blue and pink respectively. Protospacers were deduced by aligning new spacers acquired into the CRISPR7 locus of WT and OE strains. New spacers acquired into other CRISPR arrays showed similar trends. Black bars show areas were protospacers are significantly enriched, as detected by peaking finding software in the HOMER package. (B) Protospacers are significantly enriched around the CRISPR6 array. (C) More subtle enrichment was observed around CRISPR2 and CRISPR4 arrays.
The rRNA gene cluster (containing 23s rRNA, 16s rRNA, and Ala tRNA) also had an associated protospacer hotspot (Figure 5A, Supplementary Figure S8A). This region has a high density of CCN PAMs, and this may cause a similarly dense localization of protospacers. However, the observed hotspot on the rRNA gene cluster was above the background expected based purely on PAM distribution (Supplementary Figure S8A). 23s rRNA and 16s rRNA are the most highly expressed genes in P. furiosus (36,49), and previous studies in the Pectobacterium atrosepticum type I-F system have implicated high transcription levels in the generation of protospacers (27). During transcription, transcribed RNA can anneal to its DNA to form an R-loop (Supplementary Figure S8B). Moreover, R-loop formation can lead to nicking of the displaced DNA strand as well as double-strand breaks when replication forks stall at R-loop structures (50). Our results reinforce the idea that DNA in an R-loop also provides a potential source of protospacers.
Given that a majority of new spacers in our naïve adaptation assay arose from host rather than invader DNA, we sought to determine whether any of the ∼200 existing spacers in P. furiosus also had a host origin. Using BLAST, we aligned these spacers to the genome and looked for any matches wherein at least 90% of the length of the spacer aligned to the genome with at least 90% nucleotide identity. By this standard, no matches were found.
DISCUSSION
Acquisition of new spacers from invasive DNA into CRISPR loci is what allows CRISPR immune systems to adapt and provide a dynamic, evolving means of protection against phages, plasmids, and other invading mobile genetic elements. It is understood that this acquisition would likely be guided such that the new CRISPR spacers are suitable for later targeting of invaders. However, the steps underlying adaptation are not well studied, and evidence and mechanisms for such guidance is lacking. Here we report the first evidence that P. furiosus actively takes up new spacers into each of seven CRISPR loci under normal growth conditions and endogenous Cas protein expression levels (i.e. this organism undergoes naïve adaptation). Our results indicate that the newly acquired spacers have characteristics that mirror those of the existing, naturally acquired spacers in CRISPR arrays: they are the same average length (37 bp), and they arise from regions of DNA where a PAM (5′-CCN-3′) is located immediately upstream (Figures 2 and 3). The uptake of new spacers was dependent both on the presence and expression level of the presumed set of adaptation proteins: Cas1, Cas2 and Cas4. This work also yields some unexpected results: we find evidence of a degenerate motif (5′-NW-3′) immediately downstream of the new spacers, and we also discovered that most new spacers arise from host DNA (i.e. the P. furiosus genome, rather than invader DNA), so ‘self’ versus ‘non-self’ is not the primary criterion driving selection of new spacers. Instead, observations of spacer distribution along the genome and plasmids suggests that nicked (or broken) DNA may provide the best source of protospacers, irrespective of how the free DNA ends are generated.
Free DNA termini as a potential universal feature for CRISPR DNA uptake
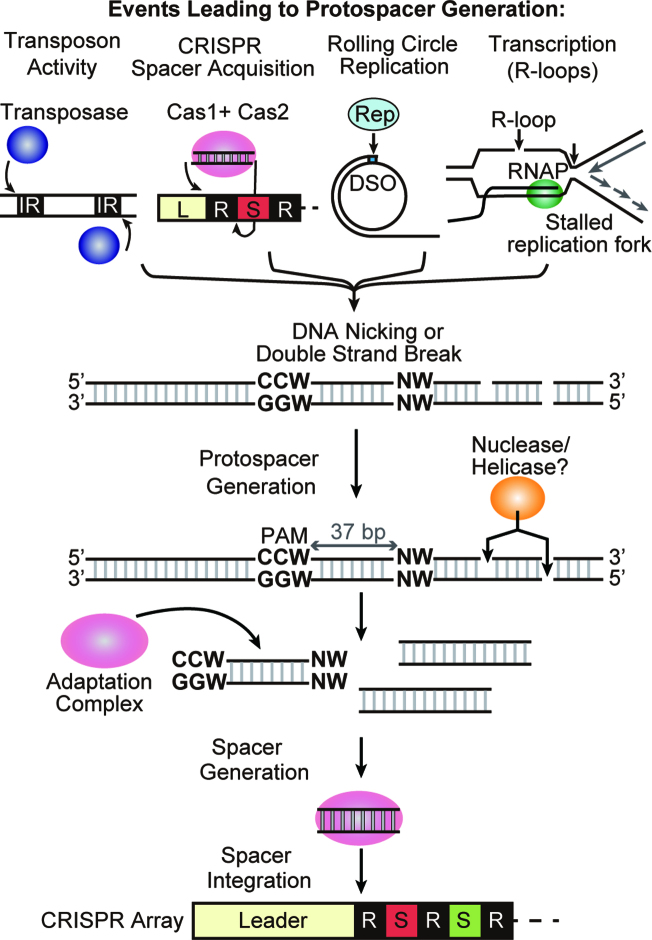
Interestingly, we found that highly sampled protospacers (from either invading plasmid or the host genome) are derived from specific regions of DNA expected or known to experience DNA nicking or double-strand breaks. In particular, site-specific nicking events that apparently contribute to protospacer generation in P. furiosus include transposable element mobilization (Figure 6), CRISPR spacer uptake itself (Figure 7), and rolling circle replication (Figures 4 and 5). Moreover, the observed protospacer hotspot region that overlaps the massively transcribed single rDNA loci of P. furiosus (Supplementary Figure S5 and (49)) is consistent with known R-loop-induced DNA nicking and double-strand breaks at resultant stalled replication forks that typically accompany highly transcribed genomic regions (50).
Our findings that free DNA ends appear to promote protospacer selection are consistent with earlier observations in diverse CRISPR–Cas systems in various bacteria. In the E. coli type I-E system, protospacer hotspots are observed around the chromosomal replication terminus (ter) and around active CRISPR arrays (14). These hotspots are RecBCD-dependent and caused by double-strand breaks at presumed stalled replication forks. In the P. atrosepticum type I-F system, hotspots across the chromosome are observed around highly transcribed regions, suggesting that formation of R-loop structures and resultant DNA damage causes spacer acquisition (27). In the type IIA system, protospacers were preferentially acquired from the DNA termini of injected phage DNA (51). Collectively, the findings to date implicate both PAMs and free DNA termini as highly conserved features important for adaption of highly diverse CRISPR–Cas systems.
Free DNA ends as an imperfect means for detecting mobile genetic elements
Prokaryotes must cope with both intercellular mobile genetic elements (such as viruses and plasmids) as well as intracellular mobile genetic elements (including transposons) that each can harm or kill cells or lead to genome instability. The apparent requirement for free DNA ends for efficient protospacer acquisition may have evolved to more effectively identify both classes of mobile genetic element. Unlike host chromosomes that tend to be circular (i.e. lack free ends), mobile genetic elements often rely on being linear for their transfer into cells by transduction, conjugation, or transformation (52,53). Moreover, specific nicking reactions are often employed to enable transfer of DNA from one cell to the next (e.g. via conjugation), for mobile genetic element replication (e.g. via rolling circle replication mode), or for mobilization from one genomic location to another (e.g. transposon mobilization). This tendency for intracellular mobile genetic element DNA to become linear or nicked within their host cells may make them vulnerable to spacer uptake by CRISPR–Cas adaptation machinery that appears to require free DNA termini for access to protospacers.
Lack of intrinsic discrimination between CRISPR uptake of self and non-self spacers
The apparent requirement for DNA ends for the early stage of the CRISPR spacer uptake process provides an imperfect system of biasing toward acquisition of fragments of foreign DNAs (non-self) vs. host chromosomal DNA (self). Circular host chromosomes also undergo DNA nicks and breaks for a variety of reasons, explaining why self-spacers are also captured into the CRISPR loci of P. furiosus (Figures 2, 5–7 and many other studied CRISPR–Cas systems (14,16,54,55)). The acquisition of self-targeting spacers typically leads to cell death (lethal autoimmunity) in prokaryotes (16,56,57). Our approach of studying spacer acquisition in a cultured population of cells provides us with a steady-state view (i.e. snapshot) of spacer uptake into CRISPR loci and is capable of both detecting rare events (happening in a small number of cells) as well as self-targeting spacer acquisitions that likely eventually lead to cell killing. Using a similar strategy, we previously found that Type II-A (Cas9-based) CRISPR–Cas systems also normally sample both chromosomal and extrachromosomal DNA and that cellular counter-selection through self-targeting and lethal autoimmunity provides the basis for the apparent specificity observed for CRISPR arrays normally lacking significant self-targeting sequences (16). The available information suggests that CRISPR–Cas spacer uptake mechanisms do not have an absolute specificity for taking up only foreign DNA versus self DNA, but different host cells mechanisms and mobile genetic element DNA properties may contribute to biased sampling of invader DNAs to differing degrees in different host cells.
Possible role for CRISPR–Cas self-targeting in maintain host genome stability
Our data provide the first evidence that active transposons trigger generation of self-targeting, chromosomal protospacers (Figure 6 and Supplementary Figure S4). Moreover, analysis of chromosomal protospacer patterns revealed that self-targeting spacers were also induced following recombination of a plasmid that used site-specific nicking and rolling circle replication (Figure 5). In Sulfolobus, conjugative plasmids are known to pass between cells and integrate into the genome of the recipient (58). The plasmids compromise host fitness (58) and to protect against this they are targeted by CRISPR defense - spacers against these plasmids are found in Sulfolobus CRISPR arrays (59). Though these specific events have not been described in Pyrococcus, similar harmful plasmid integration may exist and the recombination inadvertently induced in our assay may mimic such a scenario. Together, these findings raise the interesting possibility that self-targeting by CRISPR–Cas systems, which eliminates specific cells from a population, may contribute to increased genomic stability through copy number control of transposons and prevention of the uptake of autonomously replicating mobile genetic elements into host chromosomes. In this view, self-targeting spacers may serve a positive role at times by eliminating cells that are undergoing detrimental changes to their genomes.
In summary, our results suggest that novel spacer DNA uptake into the CRISPR loci of P. furiosus (and very likely most or all organisms containing CRISPR systems) is stimulated by various mechanisms that generate free DNA ends including transposon hopping, CRISPR spacer uptake, rolling circle replication, transcription-dependent R-loop formation, DNA replication fork stalling, and likely additional events that lead to changes in the covalent structure of DNA (Figure 8). We envision that the nicked or broken DNA molecules serve as entry points for nucleases and helicases that promote formation of an intracellular pool of heterogenous linear DNA fragments. A specific subset of these DNA fragments are then acted upon by the adaptation machinery consisting of Cas1, 2 and 4 proteins that recognize and process the flanking upstream (5′-CCN-3′) and downstream (5′-NW-3′) PAMs that our work identified as being important in protospacer generation. Subsequently, the adaptation machinery catalyzes the polarized addition of processed spacers at the leader end of the CRISPR array. The newly encoded spacers from either foreign or self DNA sources, then provide a means to enable crRNA-mediated targeting of previously encountered mobile genetic elements (immunity) or rapid self-killing (lethal autoimmunity), respectively. Our study has laid the foundation for a detailed mechanistic understanding of how CRISPR spacer capture and integration is executed and regulated.
Figure 8.
Model for spacer acquisition in P. furiosus. DNA nicking is generated by several mechanisms in cells. Transposase catalyzes nicking at the ends of the inverted repeat (IR) in replicative transposition. Adaptation complexes nick leader-proximal repeat junctions of the CRISPR array during spacer integration. Rep protein nicks the double strand origin for DNA replication of a rolling circle replication (RCR) plasmid. R-loop formation induces nicking on the displaced strand and also can block replication fork progression leading to double-strand break. Nuclease / helicase can access DNA through the nicked or double-strand broken site and generate short DNA fragments. Cas1, Cas2 and Cas4 adaptation complex finds DNA fragment with an upstream CCN and downstream NW motif. The complex then processes the protospacer to 37 bp and integrates it as a new spacer into the CRISPR array.
ACCESSION NUMBERS
The DNA sequencing data have been deposited with the Short Read Archive under accession number PRJNA401002.
Supplementary Material
ACKNOWLEDGEMENTS
We thank members of the Terns and Graveley laboratories and Rebecca Terns for helpful discussions, and Joshua Elmore and Michael Ellis for P. furiosus strain construction.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [R35GM118160 to M.P.T., R35GM118140 to B.R.G.]. Funding for open access charge: National Institutes of Health [R35GM118160].
Conflict of interest statement. None declared.
REFERENCES
- 1. Marraffini L.A. CRISPR–Cas immunity in prokaryotes. Nature. 2015; 526:55–61. [DOI] [PubMed] [Google Scholar]
- 2. Terns M.P., Terns R.M.. CRISPR-based adaptive immune systems. Curr. Opin. Microbiol. 2011; 14:321–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. van der Oost J., Westra E.R., Jackson R.N., Wiedenheft B.. Unravelling the structural and mechanistic basis of CRISPR–Cas systems. Nat. Rev. Microbiol. 2014; 12:479–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jiang F., Doudna J.A.. The structural biology of CRISPR–Cas systems. Curr. Opin. Struct. Biol. 2015; 30:100–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jackson R.N., Wiedenheft B.. A conserved structural chassis for mounting versatile CRISPR RNA-guided immune responses. Mol. Cell. 2015; 58:722–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Makarova K.S., Wolf Y.I., Alkhnbashi O.S., Costa F., Shah S.A., Saunders S.J., Barrangou R., Brouns S.J., Charpentier E., Haft D.H. et al. . An updated evolutionary classification of CRISPR–Cas systems. Nat. Rev. Microbiol. 2015; 13:722–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Shmakov S., Abudayyeh O.O., Makarova K.S., Wolf Y.I., Gootenberg J.S., Semenova E., Minakhin L., Joung J., Konermann S., Severinov K. et al. . Discovery and functional characterization of diverse class 2 CRISPR–Cas systems. Mol. Cell. 2015; 60:385–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Grissa I., Vergnaud G., Pourcel C.. The CRISPRdb database and tools to display CRISPRs and to generate dictionaries of spacers and repeats. BMC Bioinformatics. 2007; 8:172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Charpentier E., Richter H., van der Oost J., White M.F.. Biogenesis pathways of RNA guides in archaeal and bacterial CRISPR–Cas adaptive immunity. FEMS Microbiol. Rev. 2015; 39:428–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jackson R.N., van Erp P.B., Sternberg S.H., Wiedenheft B.. Conformational regulation of CRISPR-associated nucleases. Curr. Opin. Microbiol. 2017; 37:110–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fineran P.C., Charpentier E.. Memory of viral infections by CRISPR–Cas adaptive immune systems: acquisition of new information. Virology. 2012; 434:202–209. [DOI] [PubMed] [Google Scholar]
- 12. Sternberg S.H., Richter H., Charpentier E., Qimron U.. Adaptation in CRISPR–Cas systems. Mol. Cell. 2016; 61:797–808. [DOI] [PubMed] [Google Scholar]
- 13. Yosef I., Qimron U.. Microbiology: How bacteria get spacers from invaders. Nature. 2015; 519:166–167. [DOI] [PubMed] [Google Scholar]
- 14. Levy A., Goren M.G., Yosef I., Auster O., Manor M., Amitai G., Edgar R., Qimron U., Sorek R.. CRISPR adaptation biases explain preference for acquisition of foreign DNA. Nature. 2015; 520:505–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Babu M., Beloglazova N., Flick R., Graham C., Skarina T., Nocek B., Gagarinova A., Pogoutse O., Brown G., Binkowski A. et al. . A dual function of the CRISPR–Cas system in bacterial antivirus immunity and DNA repair. Mol. Microbiol. 2011; 79:484–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wei Y., Terns R.M., Terns M.P.. Cas9 function and host genome sampling in Type II-A CRISPR–Cas adaptation. Genes Dev. 2015; 29:356–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mojica F.J., Diez-Villasenor C., Garcia-Martinez J., Almendros C.. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology. 2009; 155:733–740. [DOI] [PubMed] [Google Scholar]
- 18. Shah S.A., Erdmann S., Mojica F.J., Garrett R.A.. Protospacer recognition motifs: mixed identities and functional diversity. RNA Biol. 2013; 10:891–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang J., Li J., Zhao H., Sheng G., Wang M., Yin M., Wang Y.. Structural and mechanistic basis of PAM-dependent spacer acquisition in CRISPR–Cas systems. Cell. 2015; 163:840–853. [DOI] [PubMed] [Google Scholar]
- 20. Yosef I., Goren M.G., Qimron U.. Proteins and DNA elements essential for the CRISPR adaptation process in Escherichia coli. Nucleic Acids Res. 2012; 40:5569–5576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nunez J.K., Harrington L.B., Kranzusch P.J., Engelman A.N., Doudna J.A.. Foreign DNA capture during CRISPR–Cas adaptive immunity. Nature. 2015; 527:535–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nunez J.K., Kranzusch P.J., Noeske J., Wright A.V., Davies C.W., Doudna J.A.. Cas1–Cas2 complex formation mediates spacer acquisition during CRISPR–Cas adaptive immunity. Nat. Struct. Mol. Biol. 2014; 21:528–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Datsenko K.A., Pougach K., Tikhonov A., Wanner B.L., Severinov K., Semenova E.. Molecular memory of prior infections activates the CRISPR/Cas adaptive bacterial immunity system. Nat. Commun. 2012; 3:945. [DOI] [PubMed] [Google Scholar]
- 24. Swarts D.C., Mosterd C., van Passel M.W., Brouns S.J.. CRISPR interference directs strand specific spacer acquisition. PLoS One. 2012; 7:e35888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fagerlund R.D., Wilkinson M.E., Klykov O., Barendregt A., Pearce F.G., Kieper S.N., Maxwell H.W.R., Capolupo A., Heck A.J.R., Krause K.L. et al. . Spacer capture and integration by a type I-F Cas1–Cas2-3 CRISPR adaptation complex. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:E5122–E5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rollins M.F., Chowdhury S., Carter J., Golden S.M., Wilkinson R.A., Bondy-Denomy J., Lander G.C., Wiedenheft B.. Cas1 and the Csy complex are opposing regulators of Cas2/3 nuclease activity. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:E5113–E5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Staals R.H., Jackson S.A., Biswas A., Brouns S.J., Brown C.M., Fineran P.C.. Interference-driven spacer acquisition is dominant over naive and primed adaptation in a native CRISPR–Cas system. Nat. Commun. 2016; 7:12853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Terns R.M., Terns M.P.. The RNA- and DNA-targeting CRISPR–Cas immune systems of Pyrococcus furiosus. Biochem. Soc. Trans. 2013; 41:1416–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Farkas J., Stirrett K., Lipscomb G.L., Nixon W., Scott R.A., Adams M.W., Westpheling J.. Recombinogenic properties of Pyrococcus furiosus strain COM1 enable rapid selection of targeted mutants. Appl. Environ. Microbiol. 2012; 78:4669–4676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lipscomb G.L., Stirrett K., Schut G.J., Yang F., Jenney F.E. Jr, Scott R.A., Adams M.W., Westpheling J.. Natural competence in the hyperthermophilic archaeon Pyrococcus furiosus facilitates genetic manipulation: construction of markerless deletions of genes encoding the two cytoplasmic hydrogenases. Appl. Environ. Microbiol. 2011; 77:2232–2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Waege I., Schmid G., Thumann S., Thomm M., Hausner W.. Shuttle vector-based transformation system for Pyrococcus furiosus. Appl. Environ. Microbiol. 2010; 76:3308–3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Langmead B., Trapnell C., Pop M., Salzberg S.L.. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009; 10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Crooks G.E., Hon G., Chandonia J.M., Brenner S.E.. WebLogo: a sequence logo generator. Genome Res. 2004; 14:1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Quinlan A.R., Hall I.M.. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010; 26:841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K.. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010; 38:576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yoon S.H., Reiss D.J., Bare J.C., Tenenbaum D., Pan M., Slagel J., Moritz R.L., Lim S., Hackett M., Menon A.L. et al. . Parallel evolution of transcriptome architecture during genome reorganization. Genome Res. 2011; 21:1892–1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hale C.R., Majumdar S., Elmore J., Pfister N., Compton M., Olson S., Resch A.M., Glover C.V. 3rd, Graveley B.R., Terns R.M. et al. . Essential features and rational design of CRISPR RNAs that function with the Cas RAMP module complex to cleave RNAs. Mol. Cell. 2012; 45:292–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Majumdar S., Zhao P., Pfister N.T., Compton M., Olson S., Glover C.V. 3rd, Wells L., Graveley B.R., Terns R.M., Terns M.P.. Three CRISPR–Cas immune effector complexes coexist in Pyrococcus furiosus. RNA. 2015; 21:1147–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Westra E.R., Swarts D.C., Staals R.H., Jore M.M., Brouns S.J., van der Oost J.. The CRISPRs, they are a-changin': how prokaryotes generate adaptive immunity. Annu. Rev. Genet. 2012; 46:311–339. [DOI] [PubMed] [Google Scholar]
- 40. Elmore J., Deighan T., Westpheling J., Terns R.M., Terns M.P.. DNA targeting by the type I-G and type I-A CRISPR–Cas systems of Pyrococcus furiosus. Nucleic Acids Res. 2015; 43:10353–10363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Elmore J.R., Sheppard N.F., Ramia N., Deighan T., Li H., Terns R.M., Terns M.P.. Bipartite recognition of target RNAs activates DNA cleavage by the Type III-B CRISPR–Cas system. Genes Dev. 2016; 30:447–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Farkas J., Chung D., DeBarry M., Adams M.W., Westpheling J.. Defining components of the chromosomal origin of replication of the hyperthermophilic archaeon Pyrococcus furiosus needed for construction of a stable replicating shuttle vector. Appl. Environ. Microbiol. 2011; 77:6343–6349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Vorontsova D., Datsenko K.A., Medvedeva S., Bondy-Denomy J., Savitskaya E.E., Pougach K., Logacheva M., Wiedenheft B., Davidson A.R., Severinov K. et al. . Foreign DNA acquisition by the I-F CRISPR–Cas system requires all components of the interference machinery. Nucleic Acids Res. 2015; 43:10848–10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bridger S.L., Lancaster W.A., Poole F.L. 2nd, Schut G.J., Adams M.W.. Genome sequencing of a genetically tractable Pyrococcus furiosus strain reveals a highly dynamic genome. J. Bacteriol. 2012; 194:4097–4106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Filee J., Siguier P., Chandler M.. Insertion sequence diversity in archaea. Microbiol. Mol. Biol. Rev. 2007; 71:121–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mahillon J., Chandler M.. Insertion sequences. Microbiol. Mol. Biol. Rev. 1998; 62:725–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Shapiro J.A. Molecular model for the transposition and replication of bacteriophage Mu and other transposable elements. Proc. Natl. Acad. Sci. U.S.A. 1979; 76:1933–1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Arslan Z., Hermanns V., Wurm R., Wagner R., Pul U.. Detection and characterization of spacer integration intermediates in type I-E CRISPR–Cas system. Nucleic Acids Res. 2014; 42:7884–7893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. DiRuggiero J., Achenbach L.A., Brown S.H., Kelly R.M., Robb F.T.. Regulation of ribosomal RNA transcription by growth rate of the hyperthermophilic Archaeon, Pyrococcus furiosus. FEMS Microbiol. Lett. 1993; 111:159–164. [DOI] [PubMed] [Google Scholar]
- 50. Aguilera A., Garcia-Muse T.. R loops: from transcription byproducts to threats to genome stability. Mol. Cell. 2012; 46:115–124. [DOI] [PubMed] [Google Scholar]
- 51. Modell J.W., Jiang W., Marraffini L.A.. CRISPR–Cas systems exploit viral DNA injection to establish and maintain adaptive immunity. Nature. 2017; 544:101–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Soucy S.M., Huang J., Gogarten J.P.. Horizontal gene transfer: building the web of life. Nat. Rev. Genet. 2015; 16:472–482. [DOI] [PubMed] [Google Scholar]
- 53. Wagner A., Whitaker R.J., Krause D.J., Heilers J.H., van Wolferen M., van der Does C., Albers S.V.. Mechanisms of gene flow in archaea. Nat. Rev. Microbiol. 2017; 15:492–501. [DOI] [PubMed] [Google Scholar]
- 54. Stern A., Keren L., Wurtzel O., Amitai G., Sorek R.. Self-targeting by CRISPR: gene regulation or autoimmunity?. Trends Genet. 2010; 26:335–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Stachler A.E., Turgeman-Grott I., Shtifman-Segal E., Allers T., Marchfelder A., Gophna U.. High tolerance to self-targeting of the genome by the endogenous CRISPR–Cas system in an archaeon. Nucleic Acids Res. 2017; 45:5208–5216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Vercoe R.B., Chang J.T., Dy R.L., Taylor C., Gristwood T., Clulow J.S., Richter C., Przybilski R., Pitman A.R., Fineran P.C.. Cytotoxic chromosomal targeting by CRISPR/Cas systems can reshape bacterial genomes and expel or remodel pathogenicity islands. PLoS Genet. 2013; 9:e1003454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Yosef I., Goren M.G., Kiro R., Edgar R., Qimron U.. High-temperature protein G is essential for activity of the Escherichia coli clustered regularly interspaced short palindromic repeats (CRISPR)/Cas system. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:20136–20141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Liu G., She Q., Garrett R.A.. Diverse CRISPR–Cas responses and dramatic cellular DNA changes and cell death in pKEF9-conjugated Sulfolobus species. Nucleic Acids Res. 2016; 44:4233–4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Shah S.A., Hansen N.R., Garrett R.A.. Distribution of CRISPR spacer matches in viruses and plasmids of crenarchaeal acidothermophiles and implications for their inhibitory mechanism. Biochem. Soc. Trans. 2009; 37:23–28. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.