Abstract
Most of the genes encoding proteins that function in the mitochondria are located in the nucleus and are called nuclear-encoded mitochondrial genes, or N-mt genes. In Drosophila melanogaster, about 23% of N-mt genes fall into gene families, and all duplicates with tissue-biased expression (76%) are testis biased. These genes are enriched for energy-related functions and tend to be older than other duplicated genes in the genome. These patterns reveal strong selection for the retention of new genes for male germline mitochondrial functions. The two main forces that are likely to drive changes in mitochondrial functions are maternal inheritance of mitochondria and male–male competition for fertilization. Both are common among animals, suggesting similar N-mt gene duplication patterns in different species. To test this, we analyzed N-mt genes in the human genome. We find that about 18% of human N-mt genes fall into gene families, but unlike in Drosophila, only 28% of the N-mt duplicates have tissue-biased expression and only 36% of these have testis-biased expression. In addition, human testis-biased duplicated genes are younger than other duplicated genes in the genome and have diverse functions. These contrasting patterns between species might reflect either differences in selective pressures for germline energy-related or other mitochondrial functions during spermatogenesis and fertilization, or differences in the response to similar pressures.
Keywords: nuclear-encoded mitochondrial genes, gene duplication, male-biased expression, human
Introduction
Mitochondria are organelles that arose from proteobacteria through endosymbiosis (Sagan 1967; Yang et al. 1985) and have undergone drastic reduction in genome size during evolution (e.g., human mtDNA is only ∼16.5 Kb; Berg and Kurland 2000). Currently, most of the genes encoding mitochondrial proteins are located in the nucleus (i.e., nuclear-encoded mitochondrial or N-mt genes). In humans, there are around 1,500 N-mt protein-coding genes that interact with 37 genes encoded by mtDNA (mt genes), 13 of which are protein coding. Together they fulfill all mitochondrial functions, including replication, transcription, translation, transport, mitochondria-nucleus communication and energy production. Only a small fraction of the N-mt genes (10–20%) have a strong support for α-proteobacterial origin whereas the rest originated from prokaryotic (unclear lineage) or eukaryotic organisms’ genomes, and acquired mitochondrial function at some point in time (Gray 2015).
Interestingly, some N-mt genes are duplicates of other N-mt genes and have acquired tissue-biased expression (Emerson et al. 2004; Bai et al. 2007; Porcelli et al. 2007; Wolff et al. 2014). This could be seen as unexpected given that the mitochondrial functions are required in nearly every cell type. An extreme example of this comes from the analysis of the Drosophila melanogaster (D. melanogaster) genome where all duplicates with tissue-biased expression (76%) are testis-biased (Gallach et al. 2010 and this work, see Results). In addition, these genes are 1) older than the average age of duplicated genes from the entire genome, 2) involved in interchromosomal duplication, including an excess of retrogene duplications among N-mt duplicates compared with all duplicates in the genome, 3) overrepresented on autosomes, and 4) enriched for energy-related functions (catabolic and carbohydrate metabolic processes; Gallach et al. 2010). These features reveal strong selection for the retention of these duplicates in the Drosophila genome (Gallach et al. 2010).
We proposed that these duplicates might have evolved in response to intralocus sexual antagonism generated by the appearance of a new male-beneficial and female-harming allele at a single-copy locus. Subsequent duplication of the male-beneficial allele accompanied by the acquisition of male-biased expression could then lead to the resolution of this intralocus sexual antagonism (Gallach et al. 2010; Gallach and Betran 2011). Given that males do not pass mitochondria to their offspring and are under strong male–male competition to fertilize females’ eggs, selection might favor mitochondria that are specialized in high-energy production despite the potential for increased generation of reactive oxygen species and mutations associated with this increase (Waris and Ahsan 2006; Murphy 2009; Gallach et al. 2010; Gallach and Betran 2011).
However, several other nonexclusive hypotheses could explain the patterns observed in Drosophila. New duplicates could have been retained because of their dose effects or have been fixed by chance with subsequent specialization for male germline or sperm functions (See Innan and Kondrashov 2010 for a review of gene duplication models). It has also been suggested that some of these duplicates might compensate for mtDNA male-harming mutations that do not hurt females and cannot be selected against (Rogell et al. 2014). Since mitochondria are inherited only from the mother, mutations in the mitochondrial genome can get fixed even if they are harmful to males (Partridge and Hurst 1998; Rand et al. 2006; Montooth et al. 2010). Mutations that are harmful only to males are likely to affect testis functions (Innocenti et al. 2011; Rogell et al. 2014) and to occur in energy-related genes if selection for energy-related functions is different in males (Gallach et al. 2010; Gallach and Betran 2011).
Under any of the above hypotheses, we argue that selection to improve male germline energy-related mitochondrial function in the presence of male–male competition and/or in the presence of male-deleterious mitochondrial mutations should be common among species. If recruiting N-mt duplicates is the most efficient response to these selective pressures, we would expect to find large number of N-mt duplicates with male germline energy functions in other species. To check this expectation, we analyzed the entire set of N-mt genes in the human genome. Our analyses show that human N-mt duplicates have an entirely different pattern: duplicates show tissue-biased expression in many tissues (not just in testis as in the case of Drosophila), and they fall into various functional categories (not enriched in energy-related functions as in the case of Drosophila). These results suggest that either the selective pressures to improve/maintain energy-related mitochondrial function in human testes are not as strong as in Drosophila or the response to these pressures has been different in the human lineage.
Materials and Methods
Data Collection and Statistical Analysis
We used the online data mining tool BIOMART (http://www.biomart.org, last accessed March 10, 2017; Smedley et al. 2009) to mine the ENSEMBL genome database and extract nuclear genes with mitochondrion annotation (N-mt genes) as specified by the Gene Ontology ID GO: 0005739. We retrieved and used information for human (GRCh38.p3) and D. melanogaster (BDGP6) genomes from Ensemble Genes 81 Database including chromosomal position, transcript count, exon count, and protein sequence. Only the longest coding sequence (CDS) of each N-mt gene was used in the following analyses. The final data set included a total of 19,766 genes with 1,640 N-mt genes from human genome and a set of 13,900 genes with 583 N-mt genes from D. melanogaster genome. BlastP searches (Altschul et al. 1997) were performed for every protein against every other protein. Genes were clustered into families using a cutoff levels of 50% identity over 50% of the protein length using Markov Cluster Algorithm (http://micans.org/mcl, last accessed March 12, 2017; Enright et al. 2002; Dongen and Abreu-Goodger 2012). This approach was also used to characterize gene families for complete human and D. melanogaster genomes. Scripts used to perform these steps are provided in Supplementary Material.
The parent and child relationships were assigned using the GenTree database (http://gentree.ioz.ac.cn, last accessed March 17, 2017) which uses the comparative genomic approaches to infer in what branch the particular gene originated. For the human genome, there are 14 different branches (0–13; branch zero is the oldest branch) where genes assigned dating back to the split with Zebrafish (Zhang et al. 2010b). However, there are seven different branches (0–6; branch zero is the oldest branch here too) in Drosophila (Zhang et al. 2010a). When a gene in a gene family is younger than the other closer related copy, this gene is labeled as child and the other as parent. We were not able to assign the child–parent relationship for the genes that originated in the same branch. We consider gene duplication to be tandem if the two genes are on the same chromosome, do not have any type of overlap and the genes are adjacent. We did not assign child–parent relationship for tandem duplications. Retrogenes were identified by looking at the intron number of parental and duplicated genes. Retrogenes were gene copies that had no introns while their parental genes had at least one intron.
The R Stats package (R DevelopmentCoreTeam 2013) and gplots package (Neuwirth 2007; Warnes et al. 2013) were used to perform the statistical analyses and draw heatmap, respectively (http://www.r-project.org, last accessed March 17, 2017). Files needed to draw the heatmaps are provided in the supplementary material.
Expression Data
For D. melanogaster, we used the expression data from FlyAtlas (http://flyatlas.org, last accessed March 18, 2017; Chintapalli et al. 2007) and the approach of Gallach et al. (2010) where a gene was considered tissue biased if the expression level of that gene in a given tissue was higher than the average expression level of that gene in the whole fly (i.e., upregulated in a given tissue) and downregulated in the rest of the adult tissues following the nomenclature from FlyAtlas (Chintapalli et al. 2007).
The expression data for human tissues was downloaded from Human Protein Atlas website (http://proteinatlas.org, last accessed March 18, 2017; Uhlén et al. 2015) and assigned to each gene in the human data set. The assignations of Uhlén and colleagues (Uhlén et al. 2015) were used to assign tissue-biased expression to our gene set. Uhlén and colleagues defined five different tissue expression categories. A gene is “Tissue Enriched”, if the mRNA level of that gene is at least 5-fold higher than in all other tissues. If mRNA level in a particular tissue is at least 5-fold higher than the average levels in all tissues, that gene is called “Tissue Enhanced” and if the mRNA levels is at least 5-fold higher in a group of 2–7 tissues, that gene is “Group Enriched”. In addition, if the gene is expressed in all tissues and the mRNA level of that gene is <1, that gene is placed in “Housekeeping” category. Genes that did not belong to neither of these categories were considered “Mixed”. We considered that a gene has tissue-biased expression if it belongs to Tissue Enriched, Group Enriched or Tissue Enhanced categories.
Gene Ontology Analysis
The gene ontology (GO) analysis was performed using the FatiGO software (Al-Shahrour et al. 2004). All N-mt genes in families were analyzed against the background set of the entire single N-mt genes using the two unranked lists of genes analysis mode. GOslim, GO overview, is reported for this analysis.
Results and Discussion
Duplication of N-Mt Genes
Strong selective pressures appear to be at work in the fly genome to retain new N-mt gene duplicates with testis-biased expression (Gallach et al. 2010). We argue that these pressures should be at work in other genomes and below, we describe features of N-mt duplications in the human genome. To make sure that the results of this study are comparable to those of Drosophila, we analyzed the human genome and reanalyzed the D. melanogaster genome with the same pipeline. This pipeline introduces minor modifications to the one used by Gallach and colleagues (Gallach et al. 2010; See Materials and Methods). The online ENSEMBL genome database was used via the BIOMART data-mining tool. We retrieved N-mt protein-coding genes for the human and the D. melanogaster genomes; 1,640 and 583 genes, respectively (See Materials and Methods for more details and supplementary tables 1–4, Supplementary Material online). Similarities between proteins were assessed using BlastP and proteins were clustered into families using a minimum of 50% amino acid identity and a Markov Cluster Algorithm as described in the Materials and Methods. To see if the patterns of N-mt duplications differ from those observed for gene duplications in the whole genome, we applied the same approach to characterize gene families for the complete human and D. melanogaster genomes (table 1 and supplementary tables 5–8, Supplementary Material online). The reanalysis of Drosophila genes produced results that were consistent with the previous study (Gallach et al. 2010; See table 1).
Table 1.
Characteristics of the N-mt Gene Duplicates and Gene Duplicates in the Whole Genome
| Humana |
Drosophila melanogastera |
|||||
|---|---|---|---|---|---|---|
| Features Compared | N-mt Genes (%) | Whole Genome Gene Set (%) | Inference | N-mt Genes (%) | Whole Genome Gene Set (%) | Inference |
| Gene duplication | ||||||
| Genes in gene families | 298 (18.2) | 8005 (40.5) | N-mt genes have been duplicated less often than nuclear genes (P < 2.2e-16). | 132 (22.6) | 2335 (16.8) | N-mt genes have been duplicated more often than nuclear genes (P = 0.0004). |
| Duplication eventsb | 167 | 5375 | 75 | 1504 | ||
| Retrogenes | 12 (7.2) | 96 (1.8) | RNA-mediated duplications are more prevalent for N-mt genes (P = 9.03e-05). | 26 (34.7) | 92 (6.1) | RNA-mediated duplications are more prevalent for N-mt genes (P =2.613e-12). |
| Duplication agec | Duplications in the whole genome are significantly younger than N-mt duplications (P = 5.201e-07). | Duplications in the whole genome are significantly older than N-mt duplications (P = 0.0006). | ||||
| Older | 86 (87.8) | 1124 (64.2) | 29 (78.4) | 430 (48.9) | ||
| Younger | 12 (12.2) | 626 (35.8) | 8 (21.6) | 449 (51.1) | ||
| Relocation patternd | N-mt duplicates were not significantly more relocated than nuclear gene duplicates (P = 0.4933). | N-mt duplicates have been significantly more relocated than nuclear gene duplicates (P = 4.119e-05). | ||||
| Same chromosome | 40 (33.6) | 686 (37.0) | 17 (45.9) | 972 (77.5) | ||
| Different chromosomes | 79 (66.4) | 1167 (63.0) | 20 (54.1) | 282 (22.5) | ||
Note.—All of the P values are based on Fisher’s exact tests. All of D. melanogaster inferences are consistent with previous observations (Gallach et al. 2010).
The total number of N-mt genes in the human genome is 1,640. The total number of genes in the genome for the genome version used (Ensembl Genes 80, GRCH38.p3) was 19,766. The total number of N-mt genes in the D. melanogaster genome is 583. The total number of genes in the genome for the genome version used (Ensembl Genes 80, BDGP6) was 13,900.
Duplications events were inferred from the number of events needed to explain the number of genes in that gene family. For example, two genes in a gene family requires only one duplication event but three genes requires two duplication events.
An arbitrary 104.7 My cut off (i.e., mammalian duplications vs. older duplication events) was used here for human genome analyses. However, for Drosophila, a 63 My cut off (time of Drosophila genus diversification) was used. Age is from Gentree database (http://gentree.ioz.ac.cn, last accessed March 17, 2017; Zhang et al. 2010).
Only genes with inferred child and parent (See Materials and Methods for more details) were used here. Because we couldn’t assign child–parent relationship to the tandem duplications, we didn’t consider those in this analysis.
We first compared N-mt duplications with duplications in the whole genome. We find that 23% of N-mt genes and 17% of all genes in the genome cluster into gene families in Drosophila. This trend is reversed in humans, where 18% of N-mt genes and 40.5% of all genes cluster into gene families. These comparisons show that N-mt genes have not been duplicated/retained as extensively in the human genome as in Drosophila.
We used information about the lineage of gene origination as described by Zhang and colleagues (Zhang et al. 2010b) to date the duplication events and to assign parent–child relationship to each duplicated pair (See Materials and Methods for details and supplementary tables 1, 3, 5, and 7, Supplementary Material online for assignments). We found that on average, N-mt duplications are older than duplications in the whole genome both in the human and Drosophila genomes (table 1). However, this effect is much more striking in flies where 78% of the N-mt duplicates and 49% of the duplicates in the whole genomes are older than 63 My (time of Drosophila genus diversification). In humans, 88% of N-mt duplicates and 64% of duplicates in the whole genome are older than 104.7 My (cut off that separates mammalian duplicates from older ones; table 1). This result confirms that Drosophila tends to retain its N-mt duplicates for much longer than other duplicates whereas, in the human genome, we observe a trend in the same direction but not as strong.
In Drosophila, N-mt duplicated genes are often found on a different chromosome compared with the parental genes (54%; table 1), but this percentage is much lower for other gene duplications in the genome (22%; table 1). This relocation pattern has been proposed to be a feature that facilitates the acquisition of testis expression and increases N-mt gene retention (Gallach et al. 2010). However, in the human genome N-mt duplicates are relocated as often as nuclear gene duplicates (66% vs. 63%; table 1). These relocation patterns could be explained by the age of duplications since young duplicates tend to be found on their parental chromosomes. On average, N-mt intrachromosomal duplications are younger (have larger branch number; (Zhang et al. 2010b) than interchromosomal duplications. In humans, average branch numbers are 1.5 for old versus 3.0 for young duplicated genes. In flies, average branch numbers are 0.23 for old versus 0.38 for young duplicated genes. In flies, this reveals selection for the retention of interchromosomal duplications because chromosomal rearrangements with time are very rare (i.e., chromosomal arms known as Muller elements have not suffer major rearrangements in the genus; Gallach et al. 2010; Powell 1997). However, interchromosomal rearrangements are frequent between autosomal chromosomes in vertebrates (Murphy et al. 2005; Kemkemer et al. 2009), and that can lead to an apparent excess of relocated genes for the older gene set. So, the interpretation of this result in the human genome is less straightforward as it could be due to chromosomal rearrangements after gene duplication or initial interchromosomal duplication being retained by selection.
We also studied retrogenes (i.e., gene duplications produced through an mRNA intermediate) and found that human N-mt genes are duplicated/retained through RNA-mediated duplication more often than in the whole genome, but not as often as in Drosophila (7% vs. 2% in human and 35% vs. 6% in Drosophila; table 1). In Drosophila, this ∼5-fold excess of retrogenes within N-mt duplicated genes has been interpreted as evidence for relocation as a mechanism that facilitates testis-biased expression and long-term retention (Gallach et al. 2010). In the human genome, we do not see as many N-mt retrogenes and whereas their transcription is testis biased, they are mostly young (See more details and discussion below).
We conclude that N-mt gene duplication in the human genome has not been as extensive as in Drosophila, and these duplicates have not been retained for long periods of time as in flies. In addition, although many N-mt gene duplicates in the human genome relocate to a different chromosome, it is unclear if this feature was selected right after duplication or is a product of chromosomal rearrangements over time.
Expression Analysis of the N-Mt Genes
We studied the expression of the new N-mt genes compared with the parental genes whenever we could assign child–parent relationship. For Drosophila, we used microarray expression data from FlyAtlas (http://flyatlas.org, last accessed March 18, 2017; Chintapalli et al. 2007; See Materials and Methods and supplementary table 3, Supplementary Material online). To study gene expression of N-mt gene duplicates in the human genome, we used expression data (RNAseq, FPKM) from the Human Protein Atlas website (http://proteinatlas.org, last accessed March 18, 2017; Uhlén et al. 2015); See Materials and Methods and supplementary table 1, Supplementary Material online).
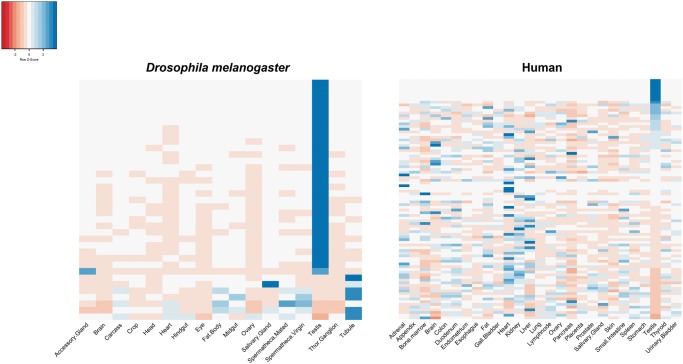
In Drosophila, we were able to assign child–parent relationship (See Materials and Methods and supplementary table 3, Supplementary Material online) for only a fraction (37/75) of the duplication events. Figure 1 shows gene expression profile of new N-mt genes for adult tissues. We find that 76% of N-mt duplicates (28/37) in D. melanogaster can be classified as having tissue-biased expression (See Materials and Methods). All of the genes with tissue-biased expression have testis-biased expression (figs. 1 and 2). The rest of the duplicates are either expressed broadly in different tissues (7/37; fig. 1) or do not have expression data (2/37). The pattern of testis-biased expression holds for gene families even in the instances where we cannot assign the child–parent relationship as we observe that 79% of all the gene families have a testis-biased member (supplementary fig. 1, Supplementary Material online).
Fig. 1.—
Heatmap of gene expression for N-mt duplicated genes from Drosophila (left) and human (right). Only new duplicated N-mt genes (i.e., genes for which the child status was confirmed) are shown on the Y axis ranked by their Z score for testis. Only adult tissues are shown on the X axis (27 out of 32 tissues for human, and 17 out of 27 tissues for D. melanogaster). Raw expression data was transformed into Z scores to show the extent of tissue-biased expression. Expression levels higher/lower than the mean expression across all tissues have positive/negative Z scores as indicated by blue/pink colors.
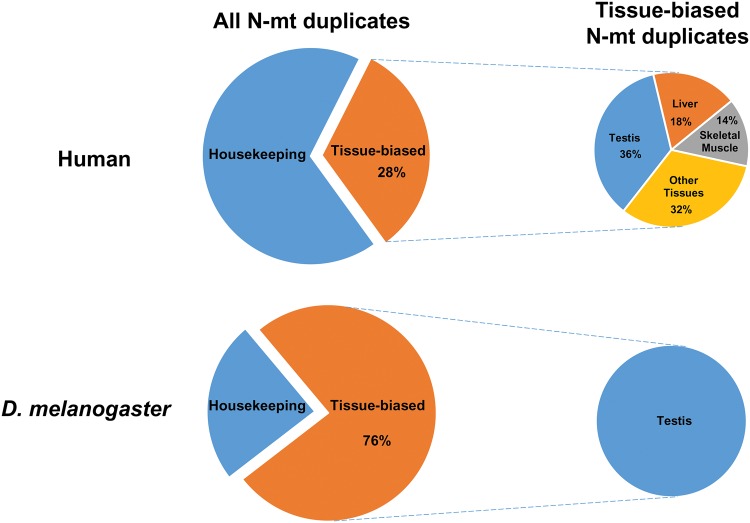
In the human genome, we were able to assign child–parent relationship (See Materials and Methods and supplementary table 1, Supplementary Material online) for only 99 of the 167 duplication events. Figure 1 shows gene expression profile of new N-mt genes. We observe that only 28 (i.e., 28% of these duplications; 28/99) can be classified as tissue-biased while the rest are either housekeeping genes (59.8%) or have mixed/undetected expression in the human genome (11.9%). Among all adult tissues, testis is the tissue with the most tissue-biased N-mt gene duplications (10 out of 28, 35.7%) followed by liver (5 out of 28, 17.8%) and skeletal muscle (4 out of 28, 14.3%; fig. 2). If we consider testis and prostate as male tissues and ovary, fallopian tube and endometrium as female tissues, we observed more genes with male-biased expression than with female-biased expression among N-mt duplicates (11 genes vs. 1 gene, respectively) but not significantly more than single copy N-mt genes male or female biased (23 genes vs. 6 genes, respectively; Fisher’s exact test; P = 0.6515). The testis-biased new genes are often duplicates of housekeeping/mixed parental genes (80%; 8/10; supplementary table 1, Supplementary Material online). These features reveal stronger selection for N-mt testis-biased duplicates in Drosophila than in the human lineage where only 10% of the N-mt duplicates are testis biased compared with 76% in flies. They also reveal stronger selection for N-mt tissue-biased duplicates for male functions than female functions.
Fig. 2.—
Expression pattern of N-mt duplicated genes in human and D. melanogaster. Tissue-biased N-mt duplicated genes in human show expression bias in multiple tissues while all tissue-biased genes in D. melanogaster are testis biased.
Our analysis of gene duplication lineage of origin/age shows that human testis-biased new N-mt genes are younger than other tissue-biased N-mt new genes. On average, they originated in more recent branches (supplementary table 1, Supplementary Material online) and are often retrogenes. We observe 12 retrogenes among all new N-mt genes and, interestingly, 7 of them (54%) have acquired testis-biased expression after duplication, which is higher than testis expression of retroposed copies in the human genome in general (∼40%; Carelli et al. 2016). So, in the human genome, we observe very few testis-biased new N-mt genes, but when relocated by means of retroposition they acquire testis-biased expression as in Drosophila. These testis-biased retogenes tend to be on average younger than any other retrogenes (Human average branch number: 5.6 vs. 3) and also younger than the rest of N-mt duplicates (Human branch number: 5.6 vs. 1.4). In the human genome, some new N-mt genes show biased expression in other tissues such as liver or skeletal muscle revealing tissue specialization of mitochondrial function that does not seem to exist in flies. New N-mt genes with tissue-biased expression in flies are only biased for testis expression out of 17 different adult tissues (fig. 1). This tissue specialization in humans should lead to additional levels of interactions and coevolution between nuclear and mitochondrial genes (Wolff et al. 2014).
Functional Enrichments of N-Mt Duplicate Genes in the Human Genome
Since N-mt duplicates in Drosophila are enriched for energy-related functions (Gallach et al. 2010), we studied the functional enrichments of N-mt duplicate genes set (i.e., genes that are in gene families) in the human genome. We tested the GO enrichment of N-mt duplicated genes against the single copy N-mt genes as was previously done in flies. Based on GOSlim GOA database, transport GO terms such as transmembrane transport (GO:0055085; 28% vs. 8%; P = 4.5e-6) are overrepresented among duplicated genes. Also, carbohydrate metabolic process (GO: 0005975; 15% vs. 4%; P = 0.0014) and kinase activity (GO: 0016301; 14% vs. 4%; P = 0.0040) are overrepresented among the duplicated genes. RNA binding (GO: 0003723; 4% vs. 15%; P = 0.0420) is the only underrepresented term in the GOSlim (See the supplementary table 9, Supplementary Material online for all other GOs enrichments). Since testis-biased genes are a small fraction of all duplicates, we explore the GO enrichment among testis-biased duplicates, but there was not any statistically significant GO term. There is, however, not a lot of power to detect effects in this small gene set (10 genes).
N-mt testis duplicates in human genome have diverse functions, but energy- and catabolism-related functions as well as carbohydrate metabolism are well represented. These functions include an ataxin (ATXN3L; deubiquitinating enzyme), a succinyl-CoA:3-ketoacid coenzyme A transferase 2 (OXCT2; protein involved in energy metabolism of ketone bodies), a glycerol kinase (GK2; protein involved in carbohydrate metabolism), two cytochrome c oxidase subunit duplicates (COX6B2 and COX7B2), a ferritin (FTMT; involved in iron metabolism in the mitochondria), a protein phosphatase (DUSP21), a pyruvate dehydrogenase (PDHA2), ADP/ATP carrier and a cationic and neutral amino acid carriers. Some of these genes are well known to be testis specific (Dahl et al. 1990; Levi et al. 2001; Tanaka et al. 2001; Hood et al. 2002; Dolce et al. 2005; Fornuskova et al. 2010) and some are known to localize in sperm mitochondria (Tanaka et al. 2001). So, some of these duplicates are likely enhancing energy production in testis and/or sperm.
We also studied if human testis-biased N-mt new genes are enriched for N-mt genes that interact with mt genes. We did this as a first step to test the idea that testis-biased N-mt duplicates are selected to compensate for male-deleterious mutations in mtDNA (Rand et al. 2004; Rogell et al. 2014). If this is the case, N-mt genes that duplicate might comprise genes that encode for proteins which interact with mt genes. There are 13 protein-coding mt genes and they are part of four out of five OXPHOS complexes (complexes I, III, IV, and V; Bar-Yaacov et al. 2012). We used the online Mitominer database for human (http://mitominer.mrc-mbu.cam.ac.uk, last accessed May 15, 2017; Tripoli et al. 2005; Smith and Robinson 2016) and MitoDrome for D. melanogaster (Tripoli et al. 2005) and found only 2 genes out of 10 testis-biased new genes in humans and 7 out of 28 testis-biased new genes in flies are duplicates of parental genes in the OXPHOS complexes. The level of interaction of new genes with OXPHOS complexes is not significantly different from testis and nontestis new genes (2/10 vs. 10/89; Fisher’s exact test, P = 0.3481) in the human genome nor in Drosophila (7/28 vs. 5/9; Fisher’s exact test, P = 0.1161). There might be, however, many ways in which testis-biased N-mt duplicates could be selected to compensate for male deleterious mutations in mtDNA (e.g., potentially any gene that increases the flux through complexes), and a negative result might not be enough to reject the hypothesis.
Concluding Remarks
Here, we studied N-mt gene duplications in the human genome to understand if the extent of their contribution to male germline mitochondria-specific functions is similar to that in Drosophila. In Drosophila, 76% of N-mt duplicates have tissue-biased expression, all of which are expressed in testis. In contrast, 28% of human N-mt duplicates have tissue-biased expression, and 36% of these tissue-biased duplicates have testis-biased expression. Expression of other human N-mt duplicates is biased in multiple other tissues, including liver and skeletal muscle (fig. 2). In addition, human tissue-biased N-mt duplicates have diverse functional annotations and are not enriched with genes with energy-related functions as is the case in Drosophila. This result suggests that selective pressures that shape tissue-biased mitochondrial function are different between species. Alternatively, the two species might respond differently to similar selective pressures.
Fertilization and male–male competition for fertilization is expected to drive changes in male germline mitochondria that would improve sperm quality or quantity (Cummins 2009; Pizzari and Parker 2009) These forces are probably stronger in Drosophila, due to larger effective population size and stronger sperm competition (Price et al. 1999; Simmons et al. 2004; Pacey 2009; Pizzari and Parker 2009), and may account for the observed differences in recruitment of testis-biased N-mt duplicates. On the other hand, sperm tail motility is powered differently in flies and humans: mammals do not rely on oxidative phosphorylation as much as on glycolysis and have mitochondria only in the midpiece of the sperm tail (Cummins 2009) and not along the sperm tail like Drosophila (Noguchi et al. 2011). In mammalian sperm, several testis-biased gene duplicates of glycolytic enzymes appear to have unique characteristics to localize along the sperm tail to provide energy for its motility (Krisfalusi et al. 2006). Multiple glycolytic enzymes are tightly bound to the fibrous sheath of mouse spermatozoa (Krisfalusi et al. 2006). These include aldolase A isozymes that are generated by alternative splicing and retrotransposition, and duplicated genes of glyceraldehyde-3-phosphate dehydrogenase, and lactate dehydrogenase A (Krisfalusi et al. 2006; Vemuganti et al. 2007). In addition, mouse Phosphoglycerate kinase 2 (Pgk2) is a duplicate of Pgk1 and is essential for sperm motility and male fertility (Danshina et al. 2010), Thus, mammals appear to improve sperm motility through recruitment of new testis-biased glycolysis genes rather than male-biased N-mt duplicates. In Drosophila, several testis-biased duplicates for glycolytic enzymes (CG5432, CG7069, CG17645, CG32849, CG7024, and CG9961) have been identified, but they have not been found in the sperm proteome (Dorus et al. 2006; Wasbrough et al. 2010) and if there is a role for these genes in sperm motility, it is still unknown. This is not the case for N-mt male-biased duplicates where 17 out of 28 gene products have been detected in the sperm proteome (Dorus et al. 2006; Wasbrough et al. 2010).
Additional species should be studied to understand if energy generation pathways in sperm shape the recruitment of testis-biased N-mt duplicates. For now, the reported pattern indicates that N-mt duplicates contribute to testis-specific MT function to a greater extent in Drosophila than in humans. A recent study in Drosophila demonstrated that the replacement of mitochondria with mitochondria from a separate lineage can have extreme male fertility consequences (Innocenti et al. 2011). This effect is attributed to the accumulation of male-harming mutations in mitochondrial DNA such that introduction of foreign mtDNA disturbs coevolved nuclear-mitochondrial networks responsible for male fertility. If the number of N-mt testis-biased genes reflects the extent to which male germline mitochondria differ from the mitochondria of somatic cells, we would predict a weaker effect of mitochondria replacement on male fertility in humans compared with Drosophila.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
We would like to thank the Betrán lab members for helpful discussions. This work was supported by the National Institutes of Health under award number R01GM071813 to E.B. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Literature Cited
- Al-Shahrour F, Díaz-Uriarte R, Dopazo J.. 2004. FatiGO: a web tool for finding significant associations of Gene Ontology terms with groups of genes. Bioinformatics 20(4):578–580. [DOI] [PubMed] [Google Scholar]
- Altschul SF. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25(17):3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y, Casola C, Feschotte C, Betran E.. 2007. Comparative genomics reveals a constant rate of origination and convergent acquisition of functional retrogenes in Drosophila. Genome Biol. 8(1):R11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Yaacov D, Blumberg A, Mishmar D.. 2012. Mitochondrial-nuclear co-evolution and its effects on OXPHOS activity and regulation. Biochim Biophys Acta 1819(9–10):1107–1111. [DOI] [PubMed] [Google Scholar]
- Berg OG, Kurland CG.. 2000. Why mitochondrial genes are most often found in nuclei. Mol Biol Evol. 17(6):951–961. [DOI] [PubMed] [Google Scholar]
- Carelli FN, et al. 2016. The life history of retrocopies illuminates the evolution of new mammalian genes. Genome Res. 26(3):301–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chintapalli VR, Wang J, Dow JA.. 2007. Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat Genet. 39(6):715–720. [DOI] [PubMed] [Google Scholar]
- Cummins J. 2009. Sperm motility and energetics In: Birkhead TR, Hosken DJ, Pitnick S, editors. Sperm biology: an evolutionary perspective. Oxford, UK: Elsevier; p. 185–206. [Google Scholar]
- Dahl HH, Brown RM, Hutchison WM, Maragos C, Brown GK.. 1990. A testis-specific form of the human pyruvate dehydrogenase E1 alpha subunit is coded for by an intronless gene on chromosome 4. Genomics 8(2):225–232. [DOI] [PubMed] [Google Scholar]
- Danshina PV, et al. 2010. Phosphoglycerate kinase 2 (PGK2) is essential for sperm function and male fertility in mice. Biol Reprod. 82(1):136–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolce V, Scarcia P, Iacopetta D, Palmieri F.. 2005. A fourth ADP/ATP carrier isoform in man: identification, bacterial expression, functional characterization and tissue distribution. FEBS Lett. 579(3):633–637. [DOI] [PubMed] [Google Scholar]
- Dongen S, Abreu-Goodger C.. 2012. Using MCL to extract clusters from networks. Methods Mol Biol. 804:281–295. [DOI] [PubMed] [Google Scholar]
- Dorus S, et al. 2006. Genomic and functional evolution of the Drosophila melanogaster sperm proteome. Nat Genet. 38(12):1440–1445. [DOI] [PubMed] [Google Scholar]
- Emerson JJ, Kaessmann H, Betran E, Long M.. 2004. Extensive gene traffic on the mammalian x chromosome. Science 303(5657):537–540. [DOI] [PubMed] [Google Scholar]
- Enright AJ, Van Dongen S, Ouzounis CA.. 2002. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 30(7):1575–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornuskova D, Stiburek L, Wenchich L, Vinsova K, Hansikova H, Zeman J.. 2010. Novel insights into the assembly and function of human nuclear-encoded cytochrome c oxidase subunits 4, 5a, 6a, 7a and 7b. Biochem J. 428(3):363–374. [DOI] [PubMed] [Google Scholar]
- Gallach M, Betran E.. 2011. Gene duplication might resolve intralocus sexual conflict. Trends Ecol Evol. 26(11):558–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallach M, Chandrasekaran C, Betran E.. 2010. Analyses of nuclearly-encoded mitochondrial genes suggest gene duplication as a mechanism for resolving intralocus sexual antagonistic conflict in Drosophila. Genome Biol Evol. 2:835–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray MW. 2015. Mosaic nature of the mitochondrial proteome: Implications for the origin and evolution of mitochondria. Proc Natl Acad Sci U S A. 112(33):10133–10138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood KL, Tobin JF, Yoon C.. 2002. Identification and characterization of two novel low-molecular-weight dual specificity phosphatases. Biochem Biophys Res Commun. 298(4):545–551. [DOI] [PubMed] [Google Scholar]
- Innan H, Kondrashov F.. 2010. The evolution of gene duplications: classifying and distinguishing between models. Nat Rev Genet. 11(2):97–108. [DOI] [PubMed] [Google Scholar]
- Innocenti P, Morrow EH, Dowling DK.. 2011. Experimental evidence supports a sex-specific selective sieve in mitochondrial genome evolution. Science 332(6031):845–848. [DOI] [PubMed] [Google Scholar]
- Kemkemer C, et al. 2009. Gene synteny comparisons between different vertebrates provide new insights into breakage and fusion events during mammalian karyotype evolution. BMC Evol Biol. 9:84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krisfalusi M, Miki K, Magyar PL, O'Brien DA.. 2006. Multiple glycolytic enzymes are tightly bound to the fibrous sheath of mouse spermatozoa. Biol Reprod. 75(2):270–278. [DOI] [PubMed] [Google Scholar]
- Levi S, et al. 2001. A human mitochondrial ferritin encoded by an intronless gene. J Biol. Chem. 276(27):24437–24440. [DOI] [PubMed] [Google Scholar]
- Montooth KL, Meiklejohn CD, Abt D, Randa DM.. 2010. Mitochondrial-nuclear epistasis affects fitness within species but does not contribute to fixed incompatibilities between species of Drosophila. Evolution 64(12):3364–3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy MP. 2009. How mitochondria produce reactive oxygen species. Biochem J. 417(1):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy WJ, et al. 2005. Dynamics of mammalian chromosome evolution inferred from multispecies comparative maps. Science 309(5734):613–617. [DOI] [PubMed] [Google Scholar]
- Neuwirth E. 2007. RColorBrewer: ColorBrewer palettes. R package version 1.1–2. https://CRAN.R-project.org/package=RColorBrewer; last accessed August, 10, 2017)
- Noguchi T, Koizumi M, Hayashi S.. 2011. Sustained elongation of sperm tail promoted by local remodeling of giant mitochondria in Drosophila. Curr Biol. 21(10):805–814. [DOI] [PubMed] [Google Scholar]
- Pacey AA. 2009. 15 - Sperm, human fertility and society A2 - Birkhead, Tim R In: Hosken DJ, Pitnick S, editors. Sperm biology. London: Academic Press; p. 565–597. [Google Scholar]
- Partridge L, Hurst LD.. 1998. Sex and conflict. Science 281(5385):2003–2008. [DOI] [PubMed] [Google Scholar]
- Pizzari T, Parker GA.. 2009. 6 - Sperm competition and sperm phenotype A2 - Birkhead, Tim R In: Hosken DJ, Pitnick S, editors. Sperm biology. London: Academic Press; p. 207–245. [Google Scholar]
- Porcelli D, Barsanti P, Pesole G, Caggese C.. 2007. The nuclear OXPHOS genes in insecta: a common evolutionary origin, a common cis-regulatory motif, a common destiny for gene duplicates. BMC Evol Biol. 7:215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell JR. 1997. Progress and prospects in evolutionary biology: the Drosophila model. New York: Oxford University Press. [Google Scholar]
- Price CS, Dyer KA, Coyne JA.. 1999. Sperm competition between Drosophila males involves both displacement and incapacitation. Nature 400(6743):449–452. [DOI] [PubMed] [Google Scholar]
- R DevelopmentCoreTeam. 2013. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- Rand DM, Fry A, Sheldahl L.. 2006. Nuclear-mitochondrial epistasis and Drosophila aging: introgression of Drosophila simulans mtDNA modifies longevity in D. melanogaster nuclear backgrounds. Genetics 172:329–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand DM, Haney RA, Fry AJ.. 2004. Cytonuclear coevolution: the genomics of cooperation. Trends Ecol Evol. 19(12):645–653. [DOI] [PubMed] [Google Scholar]
- Rogell B, Dean R, Lemos B, Dowling D.. 2014. Mito-nuclear interactions as drivers of gene movement on and off the X-chromosome. BMC Genomics 15(1):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagan L. 1967. On the origin of mitosing cells. J Theor. Biol. 14(3): 255–274. [DOI] [PubMed] [Google Scholar]
- Simmons LW, Firman RC, Rhodes G, Peters M.. 2004. Human sperm competition: testis size, sperm production and rates of extrapair copulations. Anim Behav. 68(2):297–302. [Google Scholar]
- Smedley D, et al. 2009. BioMart: biological queries made easy. BMC Genomics 10:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AC, Robinson AJ.. 2016. MitoMiner v3.1, an update on the mitochondrial proteomics database. Nucleic Acids Res. 44(D1):D1258–D1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka H, Kohroki J, Iguchi N, Onishi M, Nishimune Y.. 2001. Cloning and characterization of a human orthologue of tetis-specific succinyl CoA: 3-oxo acid CoA tranferase (Scot-t) cDNA. Mol Hum Reprod. 8(1):16–23. [DOI] [PubMed] [Google Scholar]
- Tripoli G, D'Elia D, Barsanti P, Caggese C.. 2005. Comparison of the oxidative phosphorylation (OXPHOS) nuclear genes in the genomes of Drosophila melanogaster, Drosophila pseudoobscura and Anopheles gambiae. Genome Biol. 6(2):R11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlén M, et al. 2015. Tissue-based map of the human proteome. Science 347(6220):1260419. [DOI] [PubMed] [Google Scholar]
- Vemuganti SA, et al. 2007. Three male germline-specific aldolase A isozymes are generated by alternative splicing and retrotransposition. Dev Biol. 309(1):18–31. [DOI] [PubMed] [Google Scholar]
- Waris G, Ahsan H.. 2006. Reactive oxygen species: role in the development of cancer and various chronic conditions. J Carcinogen. 5:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnes GR, et al. 2013. gplots: Various R programming tools for plotting data. R package version 2.12. 1. In: Available online at: http://CRAN. R-project. org/package= gplots, last accessed March 17, 2017.
- Wasbrough ER, et al. 2010. The Drosophila melanogaster sperm proteome-II (DmSP-II). J Proteomics 73(11):2171–2185. [DOI] [PubMed] [Google Scholar]
- Wolff JN, Ladoukakis ED, Enríquez JA, Dowling DK.. 2014. Mitonuclear interactions: evolutionary consequences over multiple biological scales. Philos Trans R Soc Lond B Biol Sci. 369(1646):20130443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D, Oyaizu Y, Oyaizu H, Olsen GJ, Woese CR.. 1985. Mitochondrial origins. Proc Natl Acad Sci U S A. 82(13):4443–4447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YE, Vibranovski MD, Krinsky BH, Long M.. 2010. Age-dependent chromosomal distribution of male-biased genes in Drosophila. Genome Res. 20(11):1526–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YE, Vibranovski MD, Landback P, Marais GAB, Long M, Barton NH.. 2010. Chromosomal redistribution of male-biased genes in mammalian evolution with two bursts of gene gain on the X chromosome. PLoS Biol. 8(10):e1000494. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.