Abstract
Genomes acquire lesions that can block the replication fork and some lesions must be bypassed to allow survival. The nuclear genome of flowering plants encodes two family-A DNA polymerases (DNAPs), the result of a duplication event, that are the sole DNAPs in plant organelles. These DNAPs, dubbed Plant Organellar Polymerases (POPs), resemble the Klenow fragment of bacterial DNAP I and are not related to metazoan and fungal mitochondrial DNAPs. Herein we report that replicative POPs from the plant model Arabidopsis thaliana (AtPolI) efficiently bypass one the most insidious DNA lesions, an apurinic/apyrimidinic (AP) site. AtPolIs accomplish lesion bypass with high catalytic efficiency during nucleotide insertion and extension. Lesion bypass depends on two unique polymerization domain insertions evolutionarily unrelated to the insertions responsible for lesion bypass by DNAP θ, an analogous lesion bypass polymerase. AtPolIs exhibit an insertion fidelity that ranks between the fidelity of replicative and lesion bypass DNAPs, moderate 3′-5′ exonuclease activity and strong strand-displacement. AtPolIs are the first known example of a family-A DNAP evolved to function in both DNA replication and lesion bypass. The lesion bypass capabilities of POPs may be required to prevent replication fork collapse in plant organelles.
INTRODUCTION
The genome of flowering plants encodes at least 10 DNA polymerases (DNAPs), including nuclear replicative DNAPs α, ϵ and δ (1,2), DNAPs involved in translesion DNA synthesis (TLS) and DNA repair (3–7). In contrast to most organisms, plants harbor nuclear, plastidic and mitochondrial genomes. Plastidic and mitochondrial genomes are particularly exposed to DNA damage by reactive oxygen species during respiration and photosynthesis (8,9). Apurinic/apyrimidinic (AP) sites are among of the most common DNA lesions, result from the cleavage of chemically modified bases by DNA glycosylases or by hydrolysis of glycosidic bonds (10,11). AP sites pose strong impediments for replicative DNAPs, and their non-instructional character makes them highly mutagenic (10,12–14).
In fungi and metazoans, a DNAP phylogenetically related to T-odd bacteriophages, DNAP γ, is responsible for both for mitochondrial DNA replication and Base Excision Repair (15,16). AP sites and thymine dimers pose strong blocks to metazoan DNAP γ (17,18). A homologous DNAP γ gene is not present in Arabidopsis thaliana genome or any other plant sequenced to date. In rice and A. thaliana, the function of DNAP γ is provided by DNAPs that resemble the Klenow fragment of Escherichia coli DNAP I (KF-DNAP I) (19,20). These DNAPs have been characterized in tobacco (21), red algae (22) and Tetrahymena termophila (23) and cluster into a group dubbed Plant and Protist Organellar DNA Polymerases (POPs) (22–24). The genomes of flowering plants, like tobacco, rice and Arabidopsis harbor two POP genes, whereas algae and protists only contain one copy. In Arabidopsis and tobacco, both POPs are nuclear encoded and translocate into chloroplast and mitochondria via a dual N-terminal localization targeting sequence (19,21,25). Biochemical studies using recombinant and isolated POPs indicate that they are processive DNAPs with 3′-5′ proofreading activity (19,21,22,26). Arabidopsis thaliana harboring deletions of its two POPs genes (named AtPolIA and AtPolIB) are lethal, indicating an essential role of DNAPs in organellar DNA replication (27). Genetic analyses using single deleted AtPolIB, but not the AtPolIA, increase the frequency of DNA rearrangements in plastid genomes, suggesting a role for AtPolIB in DNA repair (20,27,28).
Although the genes coding AtPolIA and AtPolIB were cloned more than a decade ago (20), potential roles for these enzymes in TLS have yet to be studied. Here, we show that both AtPolIA and AtPolIB are efficient TLS DNAPs, providing the first example of a family-A DNAP with an active exonuclease domain that efficiently bypasses an AP site. Two unique insertions in the thumb and fingers subdomains of AtPolIs account for TLS. These insertions resemble the lesion bypass mechanism of human DNAP θ (POLQ), revealing that amino acid insertions and specialization are part of a convergent evolutionary mechanism for lesion bypass in family-A DNAPs (29–31).
MATERIALS AND METHODS
Sub cloning of full-length and KF-AtPolIs
Synthetic genes harboring nucleotide sequences for AtPolIA and AtPolIB were codon optimized for bacterial expression (Biomatik, Wilmington, DE, USA) and subcloned into a pUC57 vector. Nucleotide sequences lacking the dual targeting sites were digested and subcloned into the Nde I and BamH I restriction sites of a modified pET19b vector. Nucleotide sequences were confirmed by Sanger Sequencing. Primers used for subcloning are listed in Supplementary Table S1.
Site-directed mutagenesis
Mutants were constructed using the Q5 Site-directed mutagenesis protocol (New England Biolabs). The KF-AtPolIs mutants were constructed using the Q5 site-directed mutagenesis protocol and subsequently cloned into the NdeI and BamHI restriction sites of a modified pET19b vector. Primers used for site directed mutagenesis are listed in Supplementary Table S1.
Heterologous expression and purification of recombinant AtPolIs
Protein expression was carried out in an E. coli BL21(DE3) strain supplemented with the pKJE7 plasmid (Takara). Recombinant proteins were induced by adding 0.5 mM of isopropyl-1-thio-β-D-galactopyranoside (IPTG) at an OD600 of 0.6 and 0.5 mg/ml of arabinose. Cell cultures were incubated for 16 h at 16°C. Bacterial cells were harvested by centrifugation at 4°C and resuspended in 40 ml lysis buffer (20 mM HEPES pH 8.0, 500 mM NaCl, 10% glycerol, 10 mM imidazole, 1 mM phenylmethylsulfonyl fluoride (PMSF)). Cell lysates were sonicated on ice for 10 cycles of 30 s. To remove DNA, polyethyleneimine was added and incubated with agitation for 30 min at 4°C. Debris was removed by centrifugation and the supernatant was loaded into a Ni-NTA agarose column. The resin was washed with 15 column volumes of wash buffer A (20 mM HEPES pH 8.0, 500 mM NaCl, 10% glycerol, 30 mM imidazole, 1 mM PMSF). The protein was eluted with buffer A containing 500 mM imidazole. Fractions containing the desired protein were pooled and dialyzed in buffer B (20 mM HEPES pH 8.0, 20 mM NaCl, 10% glycerol, 2 mM ethylenediaminetetraacetic acid (EDTA), 5 mM dithiothreitol (DTT), 1 mM PMSF). The pooled fractions were loaded onto a heparin column and subject to a salt gradient (20–1500 mM NaCl with the same buffer B). Pooled fractions were dialyzed in buffer B and loaded onto a phosphocellulose column and eluted with increasing concentration of NaCl. Finally, pure fractions were pooled and stored in buffer B supplemented with 50% glycerol and stored at −20°C.
Oligonucleotide substrates
Oligonucleotides sequences for subcloning and mutagenesis are given in Supplementary Table S1. Oligonucleotides used to assemble dsDNA substrates were 5′-end-labeled using T4 polynucleotide kinase and [γ32P] adenosine triphosphate (ATP). Primer purification was carried out using Nucleotide Removal kit as recommended by the manufacturer.
DNA polymerase assays
Typical reaction mixtures were incubated at 37°C and contained 5 nM of 32P-end-labeled 24-mer primer annealed to 45-mer template DNA, 1 nM DNAP and 100 μM of each dNTP. Reactions were performed in 10 mM Tris–HCl pH 7.7, 50 mM NaCl, 5% glycerol, 1.5 mM DTT, 0.2 mg/ml bovine serum albumin and 1.8 mM MgCl2. Reactions were stopped at several times by the addition of an equal amount of stop buffer (95% formamide, 10 mM EDTA, 0.1% xylene cyanol, 0.1% bromophenol blue). Reaction mixtures were run on a 15% polyacrylamide sequencing gel and analyzed by phosphorimagery.
For initial DNA primer-template exonucleolysis and nucleotide incorporation reactions, DNAPs were present at 1 nM and double-stranded DNA substrate was present at 5 nM and incubated from 15 s to 4 min. The polymerase activities contained dNTP at 100 μM whereas the exonuclease reaction did not contain dNTPs. Samples were run on a 15% polyacrylamide sequencing gel.
For TLS studies, template DNA was present at 2 nM, DNAPs were present at 0.1 nM for reactions on an undamaged template or 1 nM in reactions containing an AP site.
Strand-displacement activity
Strand-displacement was carried out with a template of 65 oligonucleotides and a 32P-end-labeled 24-mer primer. A blocking oligonucleotide of 35 nts was hybridized to create a gap of 6 nts. Reactions were carried out with 5 nM primer-template, 1 nM of AtPolIs and T7 DNAP from 15 s to 2 min using dNTPs at a concentration of 150 μM.
Steady-state kinetics
Primer extension reactions to quantify steady-state parameters at AP sites and canonical templates contained 500 nM of dsDNA and varying AtPolIs and dNTP concentrations, as indicated in Supplementary Figure S5. Reactions were optimized so less than 10% of the dsDNA substrate was converted to product and assure steady-state conditions. Reactions were carried out at 37°C in a final volume of 10 μl and terminated by the addition of an equal amount of stop buffer (95% formamide, 10 mM EDTA, 0.1% xylene cyanol, 0.1% bromophenol blue). Reactions were performed on 17% polyacrylamide sequencing gels and quantified by phosphorimagery. Kinetic constants, Michaelis-Menten (KM) and kcat, were determined as previously described (32).
Structural modeling
Structural modeling was conducted using crystal structures of the Klenow Fragment and KlenTaq DNAP (PDB codes: 1KLN and 3KTQ). Amino acids corresponding to the Klenow Fragment and KlenTaq were aligned with the amino acid sequence of AtPolIA and AtPolIB. Two structural models were constructed using the Medicinal chemistry and the molecular operating environment (MOE) platform. AtPolIs insertions were constructed de novo using the peptide library of the MOE data base. Ten homology models were constructed using the CHARMM27 force field. The atomic coordinates of the model generated with the Klenow fragment and KlenTaq were superimposed. A composite model was constructed using the atomic coordinates of the exonuclease domain from the model constructed using the Klenow Fragment and the coordinates for the polymerization domain with the model constructed using KlenTaq. Models were adjusted manually and subject to energy minimization.
RESULTS
AtPolIA and AtPolIB present the canonical motifs for 3′-5′ editing and 5′-3′ polymerization domains and three unique insertions at the polymerization domain
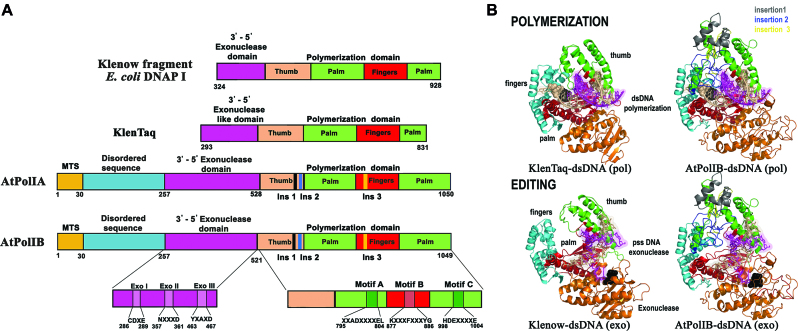
The nuclear genome of A. thaliana encodes two POP genes dubbed AtPolIA (At1g50840) and AtPolIB (At3g20540) (20,33). AtPolIA codes for a protein of 1050 amino acids (AtPolIA) whereas AtPolIB codes for two protein isoforms of 1034 and 1049 amino acids (AtPolIB1 and AtPolIB2), respectively (20,33). As AtPolIB1 was previously cloned, we elected to work with this isoform (20). AtPolIA and AtPolIB share 72% amino acid identity. A sequence alignment of AtPolIA and AtPolIB with the Klenow Fragment of DNAP I from E. coli (KF-DNAP I) (Supplementary Figure S1) illustrates the conservation in three motifs necessary for 3′-5′ exonuclease activity (34). Utilizing AtPolIA as a reference, residues involved in DNA hydrolysis in KF-DNAP I (D355, E357, Y497 and D501) are conserved in AtPolIA (D294, E296, Y470 and D474) (35). Residues L361 and F473 in KF-DNAP I place the single-stranded 3′-end into the exonuclease active site via stacking interactions (35,36). In AtPols, residue L361 is replaced by the synonymous substitution I300, whereas F473 is replaced by M432. Catalytically conserved residues in the polymerization domain corresponding to motifs A, B and C in family A DNAPs (Figure 1A) are also conserved (37,38). Those residues are the invariant motif A aspartic acid (AtPolIA-D799), two carboxylates in motif C (AtPolIA-D1000 and E1001) and a conserved tyrosine (AtPolIA-Y886) that orientates the incoming dNTP (37,38). An amino acid alignment using the polymerization domains of E. coli, Geobacillus stereatermophilus and Thermus aquaticus DNAPs I illustrates three unique amino acid insertions in AtPols (Supplementary Figure S2) with respect to bacterial DNAPs I (22,23). A structural model of AtPolIB, using Klen-Taq in complex with double-stranded DNA and an incoming dNTP, (Figure 1B) depicts POPs as archetypical family-A DNAPs in which the polymerization domain resembles a cupped right hand with thumb, fingers and palm subdomains decorated by three insertions. Insertion 1 corresponds to residues 576–621, insertion 2 is located between residues 648 and 712, and insertion 3 between residues 844 and 869. The first two insertions are in the thumb subdomain and are in an optimal position to encircle DNA, while the third insertion is in the fingers subdomain (Supplementary Figure S2). An amino acid alignment with POPs from algae, protists and protozoans shows that the presence of three insertions in the polymerization domain is a conserved feature in POPs (Supplementary Figure S3). A structural model of AtPolIB, with KF-DNAP I, complexed with double-stranded DNA and a 3′ single-stranded DNA at the exonuclease domain, illustrates that AtPols are poised to perform 3′-5′ exonucleolytic editing of misincorporated bases (39) (Figure 1B).
Figure 1.
AtPolIs are family-A DNA polymerases (DNAPs) with canonical editing and polymerization domains. (A) Domain organization of AtPolIs in comparison to KF-DNAP I and Klen-Taq. AtPolIs contain a conserved 3′-5′ exonuclease and polymerization domains. The mitochondrial targeting sequence (MTS) of the dual targeting element is colored in gold and the predicted N-terminal disorder sequence is colored in cyan. (B) Homology model of AtPolIB in comparison to the crystal structure of Klen-Taq in complex with dsDNA and incoming nucleotide (upper part). Homology model of AtPolIB in comparison to the crystal structure of the KF-DNAP I with dsDNA and extruded 3′ single-stranded DNA in the exonuclease domain (bottom part). In both models the primer strand is colored in magenta and the template strand in gold. The incoming dNTP and the 3′ single-stranded DNA is black colored and in a ball-stick representation. The three unique amino acid insertions in POPs are colored in gray, blue and yellow.
AtPolIs are encoded by a single gene in protists and ancestral plants but are duplicated in flowering plants
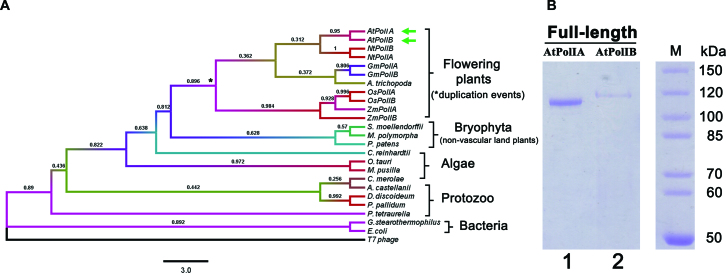
To understand the phylogenetic relationship between POPs and bacterial DNAPs we performed a phylogenetic analysis with representative DNAPs. POPs from flowering plants and POPs from protist and unicellular algae cluster in separate clades (Figure 2A). The amino acid alignment shows that insertion 2 in ancestral plants (Briophyta), algae and protists is ∼20 amino acids shorter than in flowering plants (Supplementary Figure S3). Protozoan, algae and non-vascular plants contain a single POP copy, whereas flowering plants possess a duplicated POP copy (20,21) possibly the result of a whole-genome duplication event in angiosperms. There is more than 70% amino acid identity between the duplicated POPs genes in flowering plants. Bacteriophage T7 DNAP does not cluster with POPs or bacterial DNAPs, underscoring the evolutionary divergence between POPs and DNAPs from T-odd bacteriophages.
Figure 2.
Phylogenetic analysis of POPs and heterologous purification of AtPolIs. (A) Phylogenetic tree constructed from POPs from land plants, algae and protozoan. Representative bacterial and T-odd bacteriophage DNAPs were included for comparison. A multiple sequence alignment was used to construct a phylogenetic tree using the neighbor joining algorithm, bootstrap values were calculated from 1000 trials and the evolutionary distances were constructed using the Poisson correction method. The bar indicates the numbers of substitutions per site. The significance of each branch of the phylogenetic tree is indicated by its bootstrap percentage. (B) 10% Coomassie blue stained SDS-PAGE gel showing the purification of full-length AtPolIA and AtPolIB after three purification steps: IMAC, phosphocellulose and heparin chromatography AtPolIs were purified as a single protein bands of ∼115 kDa.
AtPolIs can be heterologously expressed in bacteria and purified to homogeneity
In silico predictions suggest that the first 30 amino acids of AtPolIA and AtPolIB correspond to a mitochondrial targeting sequence, whereas their predicted chloroplast targeting sequences end at residues 41 and 46 for AtPolIA and AtPolIB respectively (40). We expressed the recombinant AtPolIs starting at residue 31 (Figure 1A). Thus, recombinant full-length AtPolIs reflect processed DNAPs after the cleavage of their predicted mitochondria targeting sequence. After three chromatographic steps, recombinant AtPolIs were purified to homogeneity with the expected molecular weights of 118 and 116 kDa for AtPolIA and AtPolIB respectively. Typical yields were 0.2 and 0.05 mg of pure protein per liter of cell culture for AtPolIA and AtPolIB respectively. AtPolIA was purified with a higher yield than AtPolIB because of increased protein induction (Figure 2B).
AtPolIs are active DNA polymerases, with editing and strand-displacement activities.
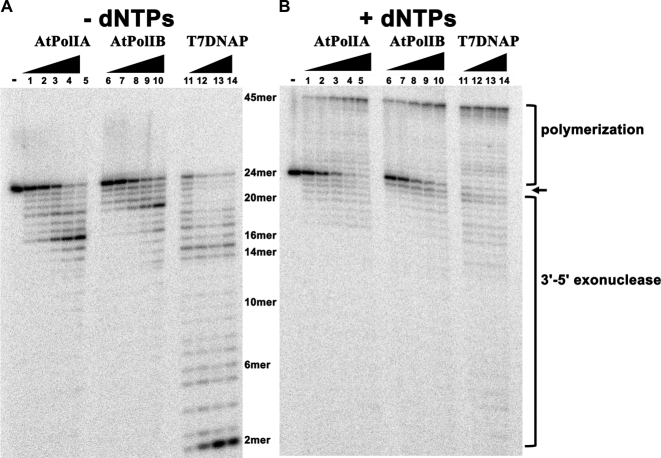
The 3′-5′ exonuclease activity of AtPolIs was measured using double-stranded DNA in the absence of incoming dNTPs and compared to the exonuclease activity of T7 DNAP in 1:1 complex with E. coli thioredoxin. DNAPs degrade duplex DNA even when the 3′ base is perfectly paired (i.e. in the absence of 3′-mismatches) (41). In this experiment, the ratio of dsDNA primer-template to DNAPs was 5:1. At the first time point of 15 s, T7 DNAP degraded the 24-mer primer to a 2-mer, whereas AtPolIA and AtPolIB degraded the primer to an 11-mer and to an 8-mer respectively (Figure 3A). At longer time points, from 1 to 4 min, intermediate degradation products accumulated in AtPolIs, but products smaller than a 6-mer were only observed in AtPolIA. In the presence of dNTPs the fate of the DNA primer was either exonucleolytic degradation or polymerization. This balance was measured by adding dNTPs in a primer elongation assay (Figure 3B). The coupled exonucleolysis/polymerization activities of AtPolIs produced fragments than ranged between 16 and 45 nts. In contrast, fragments from 2 to 45 nts are observed in reactions incubated with T7 DNAP (Figure 3B, lanes 1–14).
Figure 3.
AtPolIs present 3′–5′ exonuclease and 5′–3′ polymerization activities. Exonuclease and polymerization activities measured using a 5′-32P-labeled 24-mer primer annealed to a complementary 45-mer template DNA. (A) Equimolar amounts of each DNAP were incubated with the labeled primer-template and 3′–5′ exonucleolysis was initiated with the addition of MgCl2 in the absence of dNTPs. Reactions were stopped at 15, 30, 60, 120 and 240 s. At first-time point (15 s) exonucleolytic degradation of the 24-mer to a 2-mer is observed in the samples incubated with T7DNAP (lane 11), whereas in the samples incubated with AtPolIA and AtPolIB degradation bands corresponding to a 17-mer and a 20-mer are present. The lane labeled with the minus sign (−) corresponds to the reaction without added MgCl2. (B) Time course reaction from 15 to 240 s showing the polymerase activity of AtPolIA and AtPolIB in comparison to T7DNAP. 5′–3′ polymerization was initiated with the addition of 2 mM MgCl2 and 100 μM of each dNTP. At the first-time point (15 s), full primer extension to a 45-mer is observed in the samples incubated with T7DNAP and dNTPs (lanes 11–14). In contrast the samples incubated with AtPolIA and AtPolIB present a constant accumulation of the 45-mer from 15 s to 4 min (lanes 1–10).
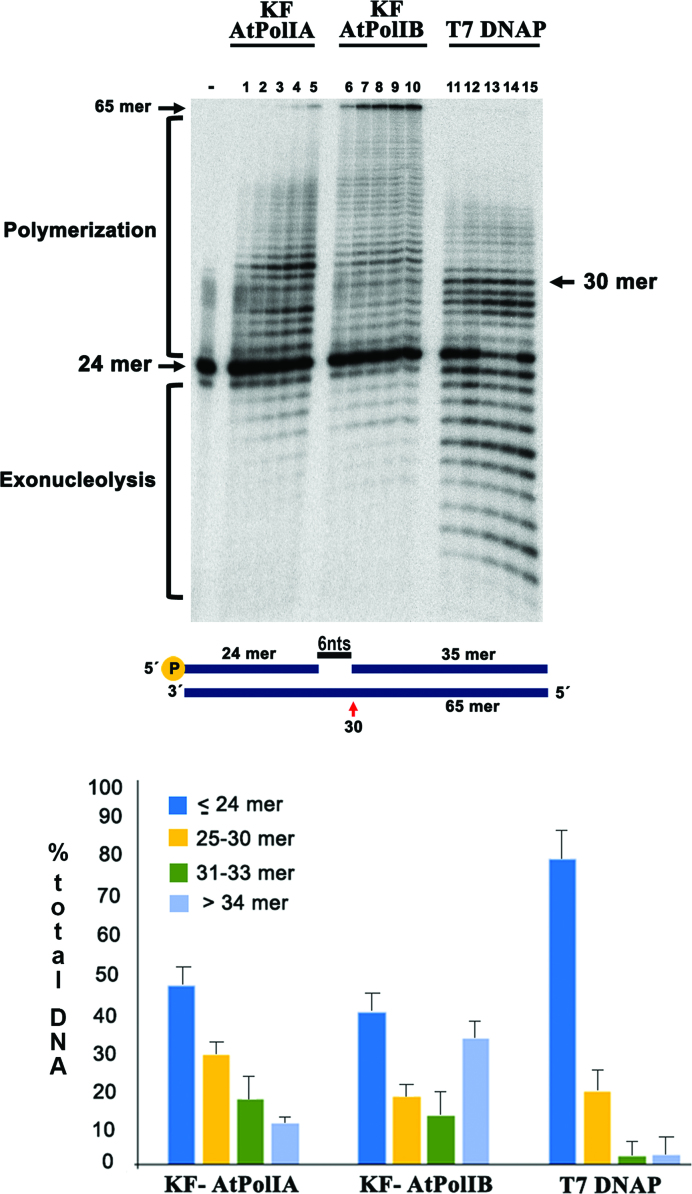
To investigate if AtPolIs execute strand-displacement DNA synthesis, we designed a DNA substrate in which a 65-mer template was annealed to an extending 24-mer primer and a blocking 35-mer. This design created a single-stranded gap of 6 nts between the 3′-OH of the extending primer and the 5′-PO4 of the blocking primer. Thus, primer extension would synthesize a 30-mer before the polymerase encounters the blocking primer (Figure 4). Strand-displacement activity was compared with T7 DNAP that by itself is inefficient in displacing a 5′-blocking oligonucleotide (42). In the control experiment with T7 DNAP, the presence of the blocking template halted the reaction after the synthesis of ∼30 nts (Figure 4). In contrast, both AtPolIs polymerized products of 50 nts. In this substrate, AtPolIA exhibits strand-displacement activity displacing 2 nts and predominantly accumulating a product of 32 nts. After an incubation period of 4 min, the fully extended 65-mer product only represented 9% of the total DNA in the reaction. In contrast, in the reactions synthesized by AtPolB, the full-length product represented 32% of the total DNA in the reaction and this polymerase did not pause during strand-displacement (Figure 4). The strong strand-displacement of AtPolIB correlates with its low exonuclease activity. Clearly, DNPs in which the exonuclease active site is abolished increase strand-displacement by increasing primer partitioning into the polymerase active site (43).
Figure 4.
AtPolIs perform strand-displacement DNA synthesis. (A) Time course nucleotide addition reaction from 15 to 240 s in a substrate assembled by hybridizing a 65-mer template to an extending 24-mer and blocking 35-mer (middle panel). This arrangement creates a gap of 6 nts before the blocking oligonucleotide. In reactions incubated with T7 DNAP a radioactively labeled band of 30 nts is predominantly observed, indicating that this polymerase is deficient in strand-displacement (lanes 11–15). In the reaction with AtPolIA, an accumulation of a product of 32 nts is observed (lanes 1–5) and only 9% of this product is able to reach the end of the template after an incubation of 240 s. In contrast, 32% of the product extended by AtPolIB is able to reach the end of the template after an incubation of 240 s (lanes 6–10). (B) Percentage of products synthetized by DNAPs after 240 s. The bands equal of lower to 24-mer correspond to substrate and exonucleolytic degradation, the bands from 25 to 30 nts correspond to gap filling and bands longer than 30 nts correspond to strand-displacement.
AtPolIs are translesion synthesis DNA polymerases
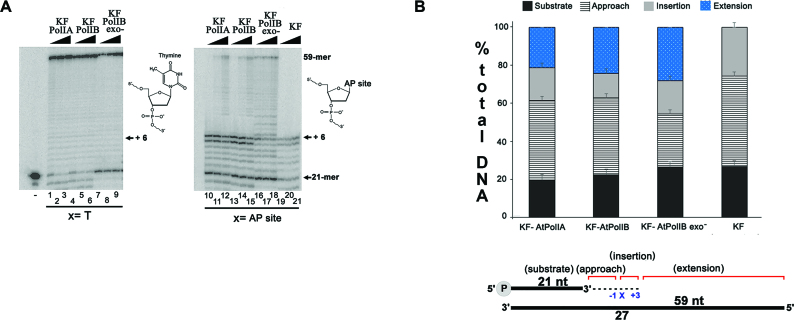
Since AtPolIs are the sole plant DNAPs in organelles, we hypothesized that AtPolIs may perform TLS. To avoid any trace of contaminating bacterial DNAPs in our studies, we constructed AtPolIs that lack the first 257 amino acids corresponding to the N-terminal unstructured region. As these constructs resemble the Klenow Fragment produced by proteolytic cleavage of E. coli DNAP I, we herein dubbed them Klenow Fragment-AtPolIs (KF-AtPolIs) (44,45) (Figure 1A). Eradication of the disordered region increased protein purity without altering polymerization and exonucleolysis (Supplementary Figure S4). We elected to investigate TLS opposite to an AP site because this lesion presents a strong block for replicative DNAPs and is highly mutagenic (13,46). Initial experiments with full-length KF-AtPolIs showed that AtPolIs fully incorporated and extended the base opposite to a template oligonucleotide containing a tetrahydrofuran moiety, that is a stable AP site analog. TLS by AtPolIs thus appears to be independent of the N-terminal disordered region (Supplementary Figure S4). To further corroborate TLS by AtPolIs, we assessed their bypass activity using a template containing an AP site 6 nts after the 3′-OH of the primer. As disruption of the exonuclease active site increases TLS in replicative DNAPs (47,48), we also utilized an exonuclease deficient KF-AtPolIB (KF-AtPolIB exo−) and KF-DNAP I unable to extend past an AP site (Figure 5A). A time course primer-extension reaction from 15 s to 1 min showed that KF-AtPolIA, KF-AtPolIB and KF-AtPolIB exo− incorporated and extended bases opposite an AP site. In contrast, KF-DNAP I was blocked after nucleotide incorporation opposite the AP site (Figure 5A, lanes 10–21 and B). Remarkably AtPolIs perform TLS even in the presence of an active exonuclease domain (Figure 5A, lanes 10–15). The percent of primer extension between KF-AtPolIs and an KF-AtPolIB exo− shows that disruption of the exonuclease activity only presents a moderate increase in TLS (Figure 5A, lanes 16–21 and B). The sum of the products incorporated after the AP site by KF-AtPolIB exo− were <15% more than the observed TLS products for the exonuclease active AtPolIs (Figure 5B). The polymerization product of AtPolIs using a template containing an AP site or an undamaged template migrated at the same position, indicating that TLS occurs by direct base insertion and not by a template-strand misalignment mechanism (43,49).
Figure 5.
AtPolIs bypass an apurinic/apyrimidinic (AP) site. (A) Time-course nucleotide incorporation reaction, from 15 s to 1 min, showing primer extension by AtPolIA, AtPolIB and AtPolIB exo−, in comparison to KF-DNAP I in canonical (lanes 1–9) or damaged templates (lanes 10–21) in the presence of 0.1 nM DNAP and 2 nM DNA. The template strand used for primer extension contained a non-damaged base or an AP site (tetrahydrofuran) 6 nts after the 3′-OH end of the primer. The migration of the 21-mer substrate and full-length extension 59-mer product are indicated in the gel. (B) Graphical representation of the lesion bypass efficiency by AtPolIs in comparison to KF-DNAP I. The values represent the relative amounts of diverse DNAs present in the reactions after 1 min. The products are subdivided into three classes: approach (before the AP site), insertion (incorporation opposite the AP site) and extension (nucleotide incorporation pass the AP site). In this representation the total amount of substrate and products is equal to 100%. Data represent the mean of three independent experiments.
AtPolIs exhibit reduced nucleotide incorporation fidelity
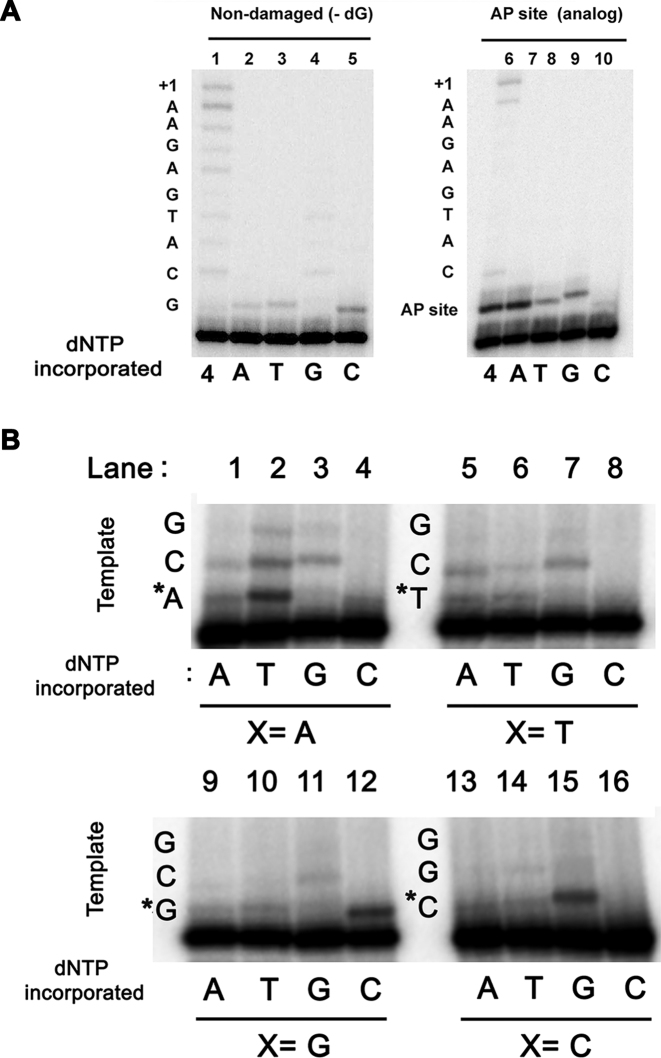
Family-A DNAPs involved in TLS exhibit low fidelity during nucleotide incorporation (29,50). We investigated the selectivity of nucleotide insertion by AtPolIs using a substrate in which the identity of first template base was either a guanosine or an AP site. For these studies, we selected an exonucleolytic deficient AtPolIB (KF-AtPolIB exo−) instead of AtPolIA because the former polymerase is implicated in organellar DNA repair (27). Initial studies indicate that KF-AtPolIB exo− can misincorporate and extend from a mismatch using a canonical guanosine template (Figure 6A, lane 4). Using an AP, KF-AtPolIB exo− preferentially inserted dAMP opposite the lesion, incorporated dGMP and dTMP to a lesser extent, and incorporated dCMP with low efficiency (Figure 6A, lanes 6–10). Low efficiency for dCMP incorporation opposite an APsite and mismatch extension is also exhibited by POLQ (29). To corroborate the low fidelity of AtPolIB exo− we systematically measured its misincorporation on all four template bases using individual deoxynucleotides. AtPolIB exo− misincorporated dAMP, dTMP and dGMP with high frequency, but dCMP misincorporation was less prominent (Figure 6B).
Figure 6.
AtPolIs are low-fidelity DNAPs that bypass an AP site following the A rule. (A) Nucleotide insertion opposite a canonical template and an AP site. (A) Single nucleotide primer extension reactions using a template containing a canonical (dGMP) (lanes 1–5) or an AP site (lanes 6–10) following the 3′-OH end of the primer. In the reactions incubated with AtPolIB in the presence of dATP, dTTP and dGTP opposite a template G a band corresponding to nucleotide addition is synthetized (lanes 1–5). In the presence of dGTP, this band can be further extended (lane 5). In the reaction opposite a non-instructional AP site, dAMP is preferentially incorporated. dGMP and dTTP are incorporated with similar efficiencies, but dCTP is not efficiently used as a substrate (lanes 6–10). (B) AtPolIB is a low fidelity enzyme. Nucleotide incorporation by AtPolIB opposite all template bases. The identity of the template base is indicated by an X and the incorporated dNMP (A, T, G, C) is indicated. In all substrates, erroneous base incorporation and extension is observed. Incoming dNTPs were present at 100 μM.
To quantitatively evaluate AtPolIB exo− misincoporation, we measured its KM and κcat values for dATP incorporation opposite a template thymidine (Supplementary Figure S5) and calculated its misincorporation frequency (finc) using correct and incorrect dNMPs incorporation (Table 1). The misincorporation values for dTMP, dGMP and dCMP opposite thymidine were 4.45 × 10−4, 5.7 × 10−3, 2.4 × 10−4 respectively. These values are 10–100× higher than the misincorporation frequency values reported for replicative DNAP and are 10–100× lower than the values reported by low fidelity TLS polymerases (51–53). Thus, the fidelity during nucleotide incorporation by the polymerization domain of AtPolB exo− is intermediate to high fidelity replicative DNAPs and inaccurate TLS DNAPs. Steady-state analysis also indicated that KF-AtPolIB exo− decreases the incorporation efficiency opposite an AP site by 12.5-fold and decreases extension opposite an AP site by 256-fold (Table 1). These values contrast with dAMP insertion frequency of KF-DNAP I exo− opposite an AP site. KF-DNAP I exo− is 15-fold less efficient than dAMP incorporation opposite an AP site, and 2714-fold less efficient for extension of a dAMP incorporated across from an AP site (54).
Table 1. Kinetic parameters for nucleotide insertion and extension by KF-AtPolIB exo− opposite canonical and AP sites.
| Template | Incoming nucleotide | K M (μM) | k cat (min−1) | k cat/KM | f cat relative thymine-dATP |
|---|---|---|---|---|---|
| Insertion opposite T | |||||
| dATP | 0.7605 ± 0.15 | 1.9722 ± 0.04 | 2.59 ± 0.26 | 1 | |
| dTTP | 261.6 ± 59 | 0.3019 ± 0.02 | 1.154 × 10−3 ± 0.003 | 4.45 × 10−4 (↓ 2247 x) | |
| dGTP | 24.8 ± 5.1 | 0.37 ± 0.01 | 1.48 × 10−2 ± 0.003 | 5.74 × 10−3 (↓ 174 x) | |
| dCTP | 185.3 ± 38.8 | 0.11545 ± 0.01 | 6.23 × 10−4 ± 0.0004 | 2.40 × 10−4 (↓ 4166 x) | |
| Insertion opposite AP site | |||||
| dATP | 7.2 ± 0.57 | 1.464 ± 0.04 | 0.202 ± 0.0633 | 0.08 | |
| dTTP | 40.8 ± 2.99 | 0.8248 ± 0.015 | 2.02 × 10−2 ± 0.004 | 7.7 × 10−3 (↓ 10 x) | |
| dGTP | 51.95 ± 3.52 | 0.9973 ± 0.02 | 1.91x 10−2 ± 0.004 | 7.3 × 10−3 (↓ 11 x) | |
| dCTP | 186.9 ± 30.9 | 0.2071 ± 0.025 | 1.11 × 10−3 ± 0.0004 | 4.2 × 10−4 (↓ 190 x) | |
| Extension with dGTP (X) | |||||
| A-AP site (X)-C | 18.41 ± 2.32 | 0.184 ± 0.006 | 1.0 × 10−2 ± 0.0024 | 3.9 × 10−3 | |
Unique AtPolIs insertions determine lesion bypass
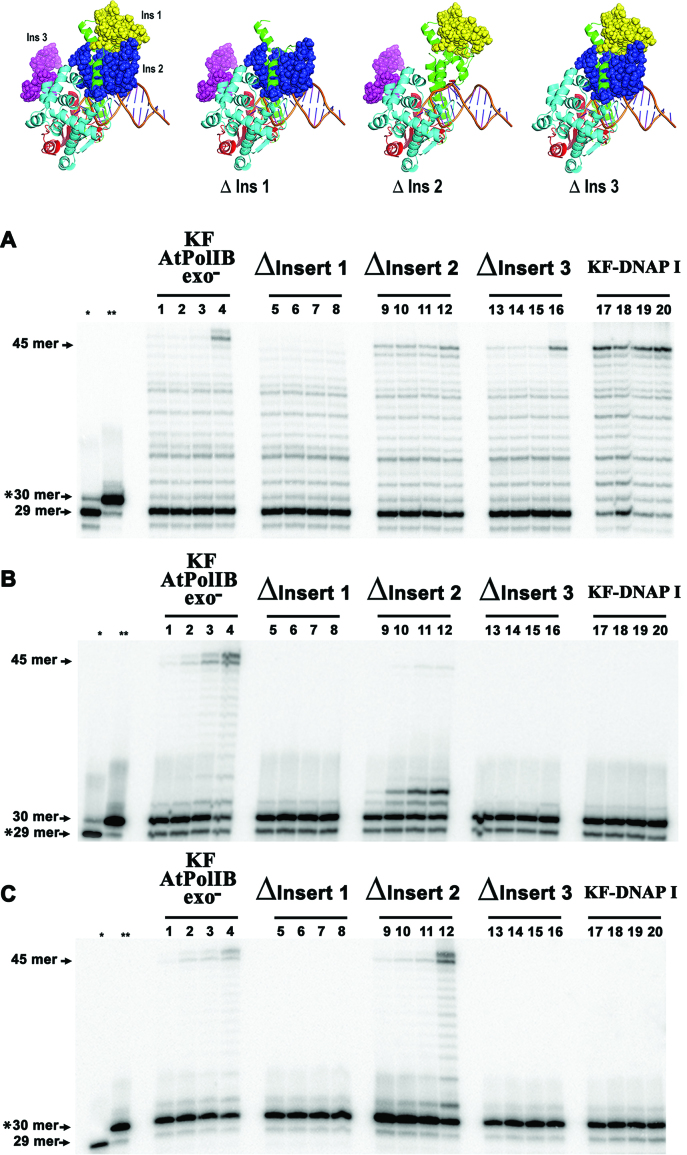
The only family-A DNAP known to perform TLS opposite an AP site, POLQ, exerts this capability due to amino acid insertions in its polymerase domain (30,31). To explore if the amino acid insertions present in AtPolIs are involved in TLS we constructed individual deletion mutants. The amino acids deleted for each construct were 576–621 (Δins1), 648–712 (Δins2) and 844–869 (Δins3). All proteins were expressed and purified with yields similar to KF-AtPolIB exo−, indicating that these insertions are flexible elements that do not contribute to the overall folding process of the protein (Supplementary Figure S6). Primer extension on an undamaged template shows that all deletion mutants are active, suggesting that they fold properly and that the elimination of each insertion does not perturb the polymerase active site (Figure 7A). Δins1 was unable to synthesize the full-extension 45-mer product. This mutant, however, efficiently synthesized products terminating before the end of the template. In contrast to the similar primer extension at a non-damaged base, incorporation opposite an AP site by AtPolIB deletion mutants exhibited clear differences (Figure 7A). As nucleotide incorporation opposite an AP site is less efficient that at an undamaged nucleotide we executed time-course experiment on damaged substrates using 10-fold DNAPs concentrations (Figure 7B and C). AtPolIB exo− deletions performed nucleotide insertion opposite an AP site, as they extended the 29-mer primer to a 30-mer. They exhibit differences; however, in extending from an AP site. Δins1 and Δins3 mutants and the KF-DNAP I control were unable to extend beyond the lesion. In contrast, Δins2 showed full primer extension to a 45-mer. (Figure 7B). A primer extension experiment in which an oligonucleotide containing a 3′-AMP that covers the AP site was not extended by Δins1 and Δins3 mutants (Figure 7C). On the other hand, AtPolIB exo− and the Δins2 mutant showed similar levels of primer extension. TLS thus is dependent on the integrity of insertions 1 and 3. Our data indicate that a replicate POPs evolved to become a TLS DNAP by the acquisition of amino acid insertions that facilitate lesion bypass.
Figure 7.
Specific amino acid insertions in AtPolI allow intrinsic lesion-bypass. Time course (from 15 s to 2 min) primer extension reaction by AtPolIB exo−, individual deletion mutants and KF-DNAP I using an undamaged or an AP site template. (A) primer extension on an undamaged template. (B) Primer extension on a template in which the AP site is the first nucleotide to be replicated. (C) Primer extension on a template in which the AP site is covered by 3′-AMP. Structural models of AtPolIB and each of the deletions mutants are depicted at the upper part of the figure. The amino acids corresponding to each of the three insertions are represented as spheres. The migration of the substrate and products are indicated in the gel. Reactions using a canonical template contained 0.1 nM DNAP and 2 nM DNA, whereas reactions in an AP site contained 1 nM DNAP. The migration of the primers located to hybridized before the lesion (29-mer) or to cover the AP site (30-mer) are indicated by asterisks.
DISCUSSION
To perform efficient DNA synthesis and avoid replication fork collapse, organisms have evolved DNAPs specialized in replication or TLS that switch places at the replication fork. This switching requires an intricate system to exchange DNAPs (55,56). POPs are the sole DNAPs present in plant organelles and therefore are candidates to function as both replicative and TLS polymerases. POPs are highly processive DNAPs able to synthesize ∼1000 nts per binding event (22,57). In the accompanying manuscript (58), we report that organellar DNAPs from A. thaliana efficiently extended RNA primers synthesized by the organellar DNA primase-helicase (AtTwinkle) at a unique single-stranded recognition sequence. The high processivity and specific RNA primer extension of AtPolIs, reveal that these enzymes are biochemically equipped to function as the replicative organellar DNAPs. In this study, we identified three biochemical properties that suggest that AtPolIs may also play a role in DNA repair.
AtPolIs conduct strand displacement
Few DNAPs have an intrinsic strand-displacement activity that allows them to replicate DNA in the absence of a DNA helicase (59,60). Plant organelles contain a gene homologous to the T7 primase-helicase, thus the strand-displacement activity of AtPolIs may not be necessary for DNA unwinding (Figure 4) (61). However, strand displacement is present during DNA repair in at least two processes: (i) the long-patch sub-pathway of Base Excision DNA repair (BER), a sub-pathway in which the strand-displacement activity of a DNAP creates a single-stranded flap cleaved by a flap endonuclease. BER activity is present in plant organelles (62,63). As mammalian mitochondria harbor both short and long-patch BER sub-pathways (64–66) and plant organelles contain flap specific endonucleases, long-patch BER may serve as an active sub-pathway in plant organelles (24). (ii) Microhomology-mediated end-joining (MMEJ), in which strand-displacement stabilizes the pairing of microhomologous regions (67). Plant organellar DNA is subject to DNA rearrangements catalyzed by MMEJ using perfect repeats as short as 6 bp (68–70) suggesting that AtPolIs may be involved in MMEJ. Both AtPolIA and AtPolIB display strand-displacement activity; however AtPolIB exhibit greater strand-displacement activity as this enzyme efficiently displaces the blocking oligonucleotide (Figure 4). This observation correlates with the proposed role of AtPolIB as a DNAP specialized in DNA repair (27).
AtPolIs are TLS polymerases
AtPolIs efficiently bypass an AP site in two independent sequence contexts (Supplementary Figure S4 and Figure 5). AP sites block replicative DNAPs during extension but not at the incorporation step. Steady-state kinetic parameters indicate that dAMP incorporation opposite an AP site decreases the catalytic efficiency by 12.5-fold (Table 1). This value is similar to the differences in catalytic efficiency of 15- and 36-fold by POLH and KF-DNAP I exo− respectively (54,71). In contrast to the permissive incorporation opposite an AP site, extension from an AP site by AtPolIB is 259-fold less efficient. This efficiency is intermediate between the catalytic efficiencies of POLH and KF-DNAP I exo− that are 15- and 2174-fold less efficient (54,71). Thus, steady state kinetic parameters indicate that AtPolB exo− is 14-fold less efficient than POLH in bypassing an AP site.
Family-A DNAPs, like Klen-Taq and POLQ, efficiently incorporate dAMP opposite an AP site. (29,30,54,72). However, to bypass an AP site, POLQ and AtPolIB have evolved mechanisms to elongate from an incorporated dAMP that is not stabilized by base pairing with a template nucleotide. TLS by AtPolIs is surprising because family-A DNAPs capable of lesion bypass are devoid of a proofreading activity (29,50,73). Modulation of the 3′-5′ exonuclease activity in AtPolIs may be a key feature for TLS in plant organelles, as a decrease in exonuclease activity would reduce a futile cycle of nucleotide incorporation-degradation opposite a DNA lesion. It is tempting to suggest that in the organellar replisome, AtPolIs would associate with other protein factors to regulate 3′-5′ proofreading, as is the case for other replicative DNAPs (74,75).
AtPolIs exhibit moderate DNA fidelity during nucleotide incorporation but have an active editing domain
Lesion bypass DNAPs exhibit low fidelity during nucleotide incorporation. For instance, the fidelity of a DNAP can be measured by calculating the frequency of incorporating correct versus incurred nucleotides using steady-state catalytic parameters. POLQ is a low fidelity DNAP characterized by misincorporation frequencies (finc) from 1.7 × 10−3 to 2.0 × 10−2 (29). In contrast, replicative POLG or POLD exhibits misincorporation frequencies from 1.0 × 10−4 to 1 × 10−6 (76,77). The misincorporation frequency of AtPolIB is between approximately between 5 × 10−4 to 5 × 10−3 and therefore is in an intermediate position between TLS and the replicative DNAPs (Table 1). As the misincorporation frequency is reciprocally related to fidelity (78), the fidelity of AtPolIB exo– ranks between one 200 and 2000 errors per incorporated dNTPs. AtPolIs, however, contrast with TLS DNAPs in their intrinsic 3′-5′ exonuclease activity. in POLG, its 3′-5′ exonuclease activity is estimated to increase fidelity by 200-fold (79). Assuming that the 3′-5′ exonuclease of AtPolIB would have a 100-fold increase in fidelity, AtPolIB would synthesize one erroneous nucleotide for every 2 × 104 to 2 × 105 replication event. This value is low in comparison to replicative DNAPs that have an average error frequency of 1 for 106 to 108 replication events (12).
DNAP insertions are a common mechanism to bypass an AP site
To investigate the possible role of AtPolIs insertions in TLS, we constructed independent deletions of the three unique POPs insertions. Constructs expressing individual deletions are active on an undamaged template and all deletions are capable of nucleotide insertion across an AP site. AtPolIB-Δins1 is unable to reach the end of the template, whereas the primer-extensions observed for AtPolIB- Δins2 and AtPolIB- Δins3 are similar to the wild-type polymerase. The decrease in activity by AtPolIB-Δins1 infers that these residues form a surface important for processivity, as in the case of extended loops in T7DNAP and POLG (80,81). Although Δins1 shows a decrease in activity, the data contrast with the absence of TLS by AtPolIB harboring deletions in Δins1 or Δins3 (Figure 7).
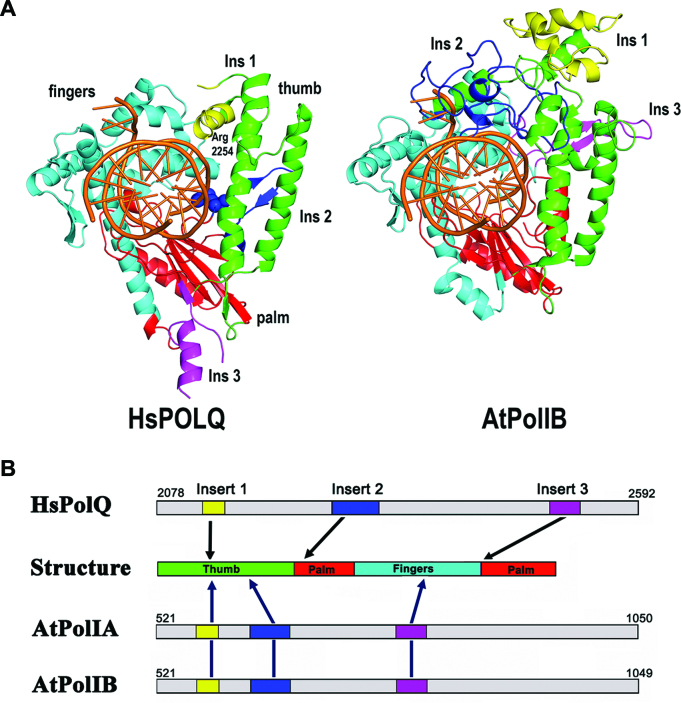
The involvement of specific insertions directing TLS by AtPolIB resembles TLS by bacteriophage Bam35 DNAP, DNAP B2 from Entamoeba histolytica, and POLQ (30,59,82). Bacteriophage Bam35 DNAP uses an extended terminal protein region 2 to drive nucleotide incorporation at a distance in which an AP site is detected (59). Crystal structures of POLQ indicate that Arg2254, located in its second insertion, interacts with the 3′ phosphate of the primer strand and that this interaction drives TLS (31). An amino acid alignment between the AtPolIs and POLQ indicates that residue Arg2254 is not conserved in AtPolIs. Furthermore, this amino acid alignment illustrates that the three amino acid insertions present in POLQ and AtPolIs are not evolutionarily related (Supplementary Figure S7 and Figure 8). In POLQ one insertion is located at the thumb subdomain and the other two are located at the palm subdomain. In AtPolIs two insertions are located at the thumb subdomain and one is located at the finger subdomain. Although AtPolIs and POLQ insertions are in different positions, a structural model suggests that insertion 3 of AtPolIs is in an analogous position than insertion 2 of POLQ. The notion that insertions 1 and 3 are responsible for TLS in AtPolIs is supported by an amino acid alignment between POPs showing that the amino acids sequences at insertions 1 and 3 are conserved (Supplementary Figure S3). This contrasts with the amino acid sequences at insertion 2 that differ in length and sequence. Specifically, in AtPolIB a lysine residue (AtPolIB-K866) in insertion 3 and a cluster of positively charged amino acids (AtPolIB 605–618) in insertion 1 are conserved, suggesting that these amino acids may interact with the primer strand in an analogous fashion to Arg2254 in POLQ. Although further structure-function studies are needed to elucidate the specific amino acid interactions between AtPolIs and the DNA chain, our data strongly support that lesion bypass by AtPolIs is carried out by a mechanism involving high catalytic efficiency during incorporation and extension from an AP site (Table 1).
Figure 8.
AtPolIB and POLQ present structurally equivalent insertions. Structural comparison between POLQ and AtPolIB. (A) Crystal structure of POLQ showing the interaction between R2254 and the 3′ phosphate of the primer strand. No continuous electron density was observed for regions corresponding to insertions 1 and 3. dsDNA is colored in orange and the palm, thumb and fingers subdomains are colored in red, green and cyan respectively. Structural model of AtPolIB showing the structural localization of its three insertions. Insertion 3 of AtPolIB is in a similar region that insertion 2 of POLQ. (B) Graphical representation of the insertions in POLQ and AtPolIB. In Both enzymes, insertion 1 is located at the thumb subdomain. POLQ insertions 2 and 3 are located at the palm subdomain, whereas in AtPolIB they are located at the thumb and fingers subdomain respectively.
TLS by AtPols and its implications for lesion bypass in organelles
The majority of DNAPs have specialized as replicative or TLS polymerases, one exception being, POLD, a replicative family-B DNAP able to execute TLS in vitro and in vivo (83–85). AP sites thus can be bypassed by two mechanisms: specialized TLS enzymes like POLH and by replicative polymerases like POLD (83–85). Herein we show that AtPolIs are replicative family-A DNAP able to efficiently perform TLS. As bacteriophage T7 and metazoan DNA mitochondrial polymerases are inefficient in lesion bypass (17,48), we speculate that avoiding replication fork collapse was a key factor in the selection of POPs over bacteriophage T-odd derived DNAPs in plant organelles.
Furthermore, specialized TLS DNAPs, like POLH, are not located in plant organelles. This observation suggests, that in contrast to nuclear DNA replication, plants only have the TLS mechanisms of AtPolIs to cope with DNA damage. As in other TLS DNAPs, AtPolIs have a decreased nucleotide incorporation fidelity (Table 1). Interestingly, plant organelles harbor unique proteins, like a family of MutS and RecA bacterial homologs, that may eliminate deleterious mutations by DNA mismatch repair and homologous recombination (70,86,87).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Corina Diaz-Quezada for technical support. N.B-.T. thanks CONACYT for Master’s and Doctoral fellowships. We thank Teri Markow, Cei Abreu-Goodger, Gerardo Cisneros, Charles Richardson and Alfredo Hernandez for critical reading.
Author contributions: L.G.B. conceived the project and designed experiments. N.B-.T.: designed, generated new ideas and executed experiments. N.B-.T. and L.G.B. wrote the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
CONACYT-Ciencia Básica [253737]; CONACYT Doctoral Fellowship (N.B-.T.); CONACYT Master’s Fellowship (N.B-.T.). Funding for open access charge: CONACYT-Ciencia Básica [253737].
Conflict of interest statement. None declared.
REFERENCES
- 1. Yokoi M., Ito M., Izumi M., Miyazawa H., Nakai H., Hanaoka F.. Molecular cloning of the cDNA for the catalytic subunit of plant DNA polymerase alpha and its cell-cycle dependent expression. Genes Cells. 1997; 2:695–709. [DOI] [PubMed] [Google Scholar]
- 2. Garcia-Diaz M., Bebenek K.. Multiple functions of DNA polymerases. CRC Crit. Rev. Plant Sci. 2007; 26:105–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Anderson H.J., Vonarx E.J., Pastushok L., Nakagawa M., Katafuchi A., Gruz P., Di Rubbo A., Grice D.M., Osmond M.J., Sakamoto A.N. et al. . Arabidopsis thaliana Y-family DNA polymerase eta catalyses translesion synthesis and interacts functionally with PCNA2. Plant J. 2008; 55:895–908. [DOI] [PubMed] [Google Scholar]
- 4. Furukawa T., Angelis K.J., Britt A.B.. Arabidopsis DNA polymerase lambda mutant is mildly sensitive to DNA double strand breaks but defective in integration of a transgene. Front. Plant Sci. 2015; 6:357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Inagaki S., Suzuki T., Ohto M.A., Urawa H., Horiuchi T., Nakamura K., Morikami A.. Arabidopsis TEBICHI, with helicase and DNA polymerase domains, is required for regulated cell division and differentiation in meristems. Plant Cell. 2006; 18:879–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Takahashi S., Sakamoto A., Sato S., Kato T., Tabata S., Tanaka A.. Roles of Arabidopsis AtREV1 and AtREV7 in translesion synthesis. Plant Physiol. 2005; 138:870–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sakamoto A., Lan V.T., Hase Y., Shikazono N., Matsunaga T., Tanaka A.. Disruption of the AtREV3 gene causes hypersensitivity to ultraviolet B light and gamma-rays in Arabidopsis: implication of the presence of a translesion synthesis mechanism in plants. Plant Cell. 2003; 15:2042–2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Boesch P., Weber-Lotfi F., Ibrahim N., Tarasenko V., Cosset A., Paulus F., Lightowlers R.N., Dietrich A.. DNA repair in organelles: pathways, organization, regulation, relevance in disease and aging. Biochim. Biophys. Acta. 2011; 1813:186–200. [DOI] [PubMed] [Google Scholar]
- 9. Kumar R.A., Oldenburg D.J., Bendich A.J.. Changes in DNA damage, molecular integrity, and copy number for plastid DNA and mitochondrial DNA during maize development. J. Exp. Bot. 2014; 65:6425–6439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lindahl T., Nyberg B.. Rate of depurination of native deoxyribonucleic acid. Biochemistry. 1972; 11:3610–3618. [DOI] [PubMed] [Google Scholar]
- 11. Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993; 362:709–715. [DOI] [PubMed] [Google Scholar]
- 12. Kunkel T.A. DNA replication fidelity. J. Biol. Chem. 2004; 279:16895–16898. [DOI] [PubMed] [Google Scholar]
- 13. Shibutani S., Takeshita M., Grollman A.P.. Translesional synthesis on DNA templates containing a single abasic site. A mechanistic study of the “A rule. J. Biol. Chem. 1997; 272:13916–13922. [DOI] [PubMed] [Google Scholar]
- 14. Berquist B.R., Wilson D.M. 3rd. Pathways for repairing and tolerating the spectrum of oxidative DNA lesions. Cancer Lett. 2012; 327:61–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Oliveira M.T., Haukka J., Kaguni L.S.. Evolution of the metazoan mitochondrial replicase. Genome Biol. Evol. 2015; 7:943–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Shutt T.E., Gray M.W.. Bacteriophage origins of mitochondrial replication and transcription proteins. Trends Genet. 2006; 22:90–95. [DOI] [PubMed] [Google Scholar]
- 17. Pinz K.G., Shibutani S., Bogenhagen D.F.. Action of mitochondrial DNA polymerase gamma at sites of base loss or oxidative damage. J. Biol. Chem. 1995; 270:9202–9206. [DOI] [PubMed] [Google Scholar]
- 18. Kasiviswanathan R., Gustafson M.A., Copeland W.C., Meyer J.N.. Human mitochondrial DNA polymerase gamma exhibits potential for bypass and mutagenesis at UV-induced cyclobutane thymine dimers. J. Biol. Chem. 2012; 287:9222–9229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kimura S., Uchiyama Y., Kasai N., Namekawa S., Saotome A., Ueda T., Ando T., Ishibashi T., Oshige M., Furukawa T. et al. . A novel DNA polymerase homologous to Escherichia coli DNA polymerase I from a higher plant, rice (Oryza sativa L.). Nucleic Acids Res. 2002; 30:1585–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mori Y., Kimura S., Saotome A., Kasai N., Sakaguchi N., Uchiyama Y., Ishibashi T., Yamamoto T., Chiku H., Sakaguchi K.. Plastid DNA polymerases from higher plants, Arabidopsis thaliana. Biochem. Biophys. Res. Commun. 2005; 334:43–50. [DOI] [PubMed] [Google Scholar]
- 21. Ono Y., Sakai A., Takechi K., Takio S., Takusagawa M., Takano H.. NtPolI-like1 and NtPolI-like2, bacterial DNA polymerase I homologs isolated from BY-2 cultured tobacco cells, encode DNA polymerases engaged in DNA replication in both plastids and mitochondria. Plant Cell Physiol. 2007; 48:1679–1692. [DOI] [PubMed] [Google Scholar]
- 22. Moriyama T., Terasawa K., Fujiwara M., Sato N.. Purification and characterization of organellar DNA polymerases in the red alga Cyanidioschyzon merolae. FEBS J. 2008; 275:2899–2918. [DOI] [PubMed] [Google Scholar]
- 23. Moriyama T., Terasawa K., Sato N.. Conservation of POPs, the plant organellar DNA polymerases, in eukaryotes. Protist. 2011; 162:177–187. [DOI] [PubMed] [Google Scholar]
- 24. Moriyama T., Sato N.. Enzymes involved in organellar DNA replication in photosynthetic eukaryotes. Front. Plant. Sci. 2014; 5:480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Christensen A.C., Lyznik A., Mohammed S., Elowsky C.G., Elo A., Yule R., Mackenzie S.A.. Dual-domain, dual-targeting organellar protein presequences in Arabidopsis can use non-AUG start codons. Plant Cell. 2005; 17:2805–2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Udy D.B., Belcher S., Williams-Carrier R., Gualberto J.M., Barkan A.. Effects of reduced chloroplast gene copy number on chloroplast gene expression in maize. Plant Physiol. 2012; 160:1420–1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Parent J.S., Lepage E., Brisson N.. Divergent roles for the two PolI-like organelle DNA polymerases of Arabidopsis. Plant Physiol. 2011; 156:254–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cupp J.D., Nielsen B.L.. Arabidopsis thaliana organellar DNA polymerase IB mutants exhibit reduced mtDNA levels with a decrease in mitochondrial area density. Physiol. Plant. 2013; 149:91–103. [DOI] [PubMed] [Google Scholar]
- 29. Seki M., Masutani C., Yang L.W., Schuffert A., Iwai S., Bahar I., Wood R.D.. High-efficiency bypass of DNA damage by human DNA polymerase Q. EMBO J. 2004; 23:4484–4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hogg M., Seki M., Wood R.D., Doublie S., Wallace S.S.. Lesion bypass activity of DNA polymerase theta (POLQ) is an intrinsic property of the pol domain and depends on unique sequence inserts. J. Mol. Biol. 2011; 405:642–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zahn K.E., Averill A.M., Aller P., Wood R.D., Doublie S.. Human DNA polymerase theta grasps the primer terminus to mediate DNA repair. Nat. Struct. Mol. Biol. 2015; 22:304–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. O’Flaherty D.K., Guengerich F.P.. Steady-state kinetic analysis of DNA polymerase single-nucleotide incorporation products. Curr. Protoc. Nucleic. Acid. Chem. 2014; 59:21–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rhee S.Y., Beavis W., Berardini T.Z., Chen G., Dixon D., Doyle A., Garcia-Hernandez M., Huala E., Lander G., Montoya M. et al. . The Arabidopsis Information Resource (TAIR): a model organism database providing a centralized, curated gateway to Arabidopsis biology, research materials and community. Nucleic Acids Res. 2003; 31:224–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Blanco L., Bernad A., Salas M.. Evidence favouring the hypothesis of a conserved 3′-5′ exonuclease active site in DNA-dependent DNA polymerases. Gene. 1992; 112:139–144. [DOI] [PubMed] [Google Scholar]
- 35. Derbyshire V., Grindley N.D., Joyce C.M.. The 3′-5′ exonuclease of DNA polymerase I of Escherichia coli: contribution of each amino acid at the active site to the reaction. EMBO J. 1991; 10:17–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Beese L.S., Steitz T.A.. Structural basis for the 3′-5′ exonuclease activity of Escherichia coli DNA polymerase I: a two metal ion mechanism. EMBO J. 1991; 10:25–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Steitz T.A. DNA polymerases: structural diversity and common mechanisms. J. Biol. Chem. 1999; 274:17395–17398. [DOI] [PubMed] [Google Scholar]
- 38. Doublie S., Tabor S., Long A.M., Richardson C.C., Ellenberger T.. Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 A resolution. Nature. 1998; 391:251–258. [DOI] [PubMed] [Google Scholar]
- 39. Beese L.S., Derbyshire V., Steitz T.A.. Structure of DNA polymerase I Klenow fragment bound to duplex DNA. Science. 1993; 260:352–355. [DOI] [PubMed] [Google Scholar]
- 40. Sperschneider J., Catanzariti A.M., DeBoer K., Petre B., Gardiner D.M., Singh K.B., Dodds P.N., Taylor J.M.. LOCALIZER: subcellular localization prediction of both plant and effector proteins in the plant cell. Sci. Rep. 2017; 7:44598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Donlin M.J., Patel S.S., Johnson K.A.. Kinetic partitioning between the exonuclease and polymerase sites in DNA error correction. Biochemistry. 1991; 30:538–546. [DOI] [PubMed] [Google Scholar]
- 42. Ghosh S., Marintcheva B., Takahashi M., Richardson C.C.. C-terminal phenylalanine of bacteriophage T7 single-stranded DNA-binding protein is essential for strand displacement synthesis by T7 DNA polymerase at a nick in DNA. J. Biol. Chem. 2009; 284:30339–30349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wang F., Yang W.. Structural insight into translesion synthesis by DNA Pol II. Cell. 2009; 139:1279–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Brutlag D., Atkinson M.R., Setlow P., Kornberg A.. An active fragment of DNA polymerase produced by proteolytic cleavage. Biochem. Biophys. Res. Commun. 1969; 37:982–989. [DOI] [PubMed] [Google Scholar]
- 45. Klenow H., Henningsen I.. Selective elimination of the exonuclease activity of the deoxyribonucleic acid polymerase from Escherichia coli B by limited proteolysis. Proc. Natl. Acad. Sci. U.S.A. 1970; 65:168–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Loeb L.A., Preston B.D.. Mutagenesis by apurinic/apyrimidinic sites. Annu. Rev. Genet. 1986; 20:201–230. [DOI] [PubMed] [Google Scholar]
- 47. Tanguy Le Gac N., Delagoutte E., Germain M., Villani G.. Inactivation of the 3′-5′ exonuclease of the replicative T4 DNA polymerase allows translesion DNA synthesis at an abasic site. J. Mol. Biol. 2004; 336:1023–1034. [DOI] [PubMed] [Google Scholar]
- 48. McCulloch S.D., Kunkel T.A.. Multiple solutions to inefficient lesion bypass by T7 DNA polymerase. DNA Repair (Amst). 2006; 5:1373–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ohashi E., Ogi T., Kusumoto R., Iwai S., Masutani C., Hanaoka F., Ohmori H.. Error-prone bypass of certain DNA lesions by the human DNA polymerase kappa. Genes Dev. 2000; 14:1589–1594. [PMC free article] [PubMed] [Google Scholar]
- 50. Takata K., Shimizu T., Iwai S., Wood R.D.. Human DNA polymerase N (POLN) is a low fidelity enzyme capable of error-free bypass of 5S-thymine glycol. J. Biol. Chem. 2006; 281:23445–23455. [DOI] [PubMed] [Google Scholar]
- 51. Arana M.E., Seki M., Wood R.D., Rogozin I.B., Kunkel T.A.. Low-fidelity DNA synthesis by human DNA polymerase theta. Nucleic Acids Res. 2008; 36:3847–3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Longley M.J., Nguyen D., Kunkel T.A., Copeland W.C.. The fidelity of human DNA polymerase gamma with and without exonucleolytic proofreading and the p55 accessory subunit. J. Biol. Chem. 2001; 276:38555–38562. [DOI] [PubMed] [Google Scholar]
- 53. Bebenek K., Joyce C.M., Fitzgerald M.P., Kunkel T.A.. The fidelity of DNA synthesis catalyzed by derivatives of Escherichia coli DNA polymerase I. J. Biol. Chem. 1990; 265:13878–13887. [PubMed] [Google Scholar]
- 54. Miller H., Grollman A.P.. Kinetics of DNA polymerase I (Klenow fragment exo-) activity on damaged DNA templates: effect of proximal and distal template damage on DNA synthesis. Biochemistry. 1997; 36:15336–15342. [DOI] [PubMed] [Google Scholar]
- 55. O’Donnell M., Langston L., Stillman B.. Principles and concepts of DNA replication in bacteria, archaea, and eukarya. Cold Spring Harb. Perspect. Biol. 2013; 5:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zhao L., Washington M.T.. Translesion synthesis: insights into the selection and switching of DNA polymerases. Genes (Basel). 2017; 8:1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Takeuchi R., Kimura S., Saotome A., Sakaguchi K.. Biochemical properties of a plastidial DNA polymerase of rice. Plant Mol. Biol. 2007; 64:601–611. [DOI] [PubMed] [Google Scholar]
- 58. Peralta-Castro A., Baruch-Torres N., Brieba L.G.. Plant organellar DNA Primase-Helicase synthesizes RNA primers for organellar DNA polymerases using a unique recognition sequence. Nucleic Acids Res. 2017; doi:10.1093/nar/gkx745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Berjon-Otero M., Villar L., de Vega M., Salas M., Redrejo-Rodriguez M.. DNA polymerase from temperate phage Bam35 is endowed with processive polymerization and abasic sites translesion synthesis capacity. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:E3476–E3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Viikov K., Valjamae P., Sedman J.. Yeast mitochondrial DNA polymerase is a highly processive single-subunit enzyme. Mitochondrion. 2011; 11:119–126. [DOI] [PubMed] [Google Scholar]
- 61. Diray-Arce J., Liu B., Cupp J.D., Hunt T., Nielsen B.L.. The Arabidopsis At1g30680 gene encodes a homologue to the phage T7 gp4 protein that has both DNA primase and DNA helicase activities. BMC Plant Biol. 2013; 13:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Boesch P., Ibrahim N., Paulus F., Cosset A., Tarasenko V., Dietrich A.. Plant mitochondria possess a short-patch base excision DNA repair pathway. Nucleic Acids Res. 2009; 37:5690–5700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Gutman B.L., Niyogi K.K.. Evidence for base excision repair of oxidative DNA damage in chloroplasts of Arabidopsis thaliana. J. Biol. Chem. 2009; 284:17006–17012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Liu P., Qian L., Sung J.S., de Souza-Pinto N.C., Zheng L., Bogenhagen D.F., Bohr V.A., Wilson D.M. 3rd, Shen B., Demple B.. Removal of oxidative DNA damage via FEN1-dependent long-patch base excision repair in human cell mitochondria. Mol. Cell. Biol. 2008; 28:4975–4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Szczesny B., Tann A.W., Longley M.J., Copeland W.C., Mitra S.. Long patch base excision repair in mammalian mitochondrial genomes. J. Biol. Chem. 2008; 283:26349–26356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Stierum R.H., Dianov G.L., Bohr V.A.. Single-nucleotide patch base excision repair of uracil in DNA by mitochondrial protein extracts. Nucleic Acids Res. 1999; 27:3712–3719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Bartlett E.J., Brissett N.C., Plocinski P., Carlberg T., Doherty A.J.. Molecular basis for DNA strand displacement by NHEJ repair polymerases. Nucleic Acids Res. 2016; 44:2173–2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Kwon T., Huq E., Herrin D.L.. Microhomology-mediated and nonhomologous repair of a double-strand break in the chloroplast genome of Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:13954–13959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Davila J.I., Arrieta-Montiel M.P., Wamboldt Y., Cao J., Hagmann J., Shedge V., Xu Y.Z., Weigel D., Mackenzie S.A.. Double-strand break repair processes drive evolution of the mitochondrial genome in Arabidopsis. BMC Biol. 2011; 9:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Shedge V., Arrieta-Montiel M., Christensen A.C., Mackenzie S.A.. Plant mitochondrial recombination surveillance requires unusual RecA and MutS homologs. Plant Cell. 2007; 19:1251–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Choi J.Y., Lim S., Kim E.J., Jo A., Guengerich F.P.. Translesion synthesis across abasic lesions by human B-family and Y-family DNA polymerases alpha, delta, eta, iota, kappa, and REV1. J. Mol. Biol. 2010; 404:34–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Obeid S., Welte W., Diederichs K., Marx A.. Amino acid templating mechanisms in selection of nucleotides opposite abasic sites by a family a DNA polymerase. J. Biol. Chem. 2012; 287:14099–14108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Pastor-Palacios G., Azuara-Liceaga E., Brieba L.G.. A nuclear family A DNA polymerase from Entamoeba histolytica bypasses thymine glycol. PLoS Negl. Trop. Dis. 2010; 4:e786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Yoda T., Tanabe M., Tsuji T., Yoda T., Ishino S., Shirai T., Ishino Y., Takeyama H., Nishida H.. Exonuclease processivity of archaeal replicative DNA polymerase in association with PCNA is expedited by mismatches in DNA. Sci. Rep. 2017; 7:44582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Meng X., Zhou Y., Lee E.Y., Lee M.Y., Frick D.N.. The p12 subunit of human polymerase delta modulates the rate and fidelity of DNA synthesis. Biochemistry. 2010; 49:3545–3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Dieckman L.M., Johnson R.E., Prakash S., Washington M.T.. Pre-steady state kinetic studies of the fidelity of nucleotide incorporation by yeast DNA polymerase delta. Biochemistry. 2010; 49:7344–7350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Johnson A.A., Johnson K.A.. Fidelity of nucleotide incorporation by human mitochondrial DNA polymerase. J. Biol. Chem. 2001; 276:38090–38096. [DOI] [PubMed] [Google Scholar]
- 78. Beard W.A., Shock D.D., Vande Berg B.J., Wilson S.H.. Efficiency of correct nucleotide insertion governs DNA polymerase fidelity. J. Biol. Chem. 2002; 277:47393–47398. [DOI] [PubMed] [Google Scholar]
- 79. Johnson A.A., Johnson K.A.. Exonuclease proofreading by human mitochondrial DNA polymerase. J. Biol. Chem. 2001; 276:38097–38107. [DOI] [PubMed] [Google Scholar]
- 80. Hamdan S.M., Marintcheva B., Cook T., Lee S.J., Tabor S., Richardson C.C.. A unique loop in T7 DNA polymerase mediates the binding of helicase-primase, DNA binding protein, and processivity factor. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:5096–5101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Lee Y.S., Kennedy W.D., Yin Y.W.. Structural insight into processive human mitochondrial DNA synthesis and disease-related polymerase mutations. Cell. 2009; 139:312–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Pastor-Palacios G., Lopez-Ramirez V., Cardona-Felix C.S., Brieba L.G.. A transposon-derived DNA polymerase from Entamoeba histolytica displays intrinsic strand displacement, processivity and lesion bypass. PLoS One. 2012; 7:e49964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Hirota K., Tsuda M., Mohiuddin, Tsurimoto T., Cohen I.S., Livneh Z., Kobayashi K., Narita T., Nishihara K., Murai J. et al. . In vivo evidence for translesion synthesis by the replicative DNA polymerase delta. Nucleic Acids Res. 2016; 44:7242–7250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Fazlieva R., Spittle C.S., Morrissey D., Hayashi H., Yan H., Matsumoto Y.. Proofreading exonuclease activity of human DNA polymerase delta and its effects on lesion-bypass DNA synthesis. Nucleic Acids Res. 2009; 37:2854–2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Schmitt M.W., Matsumoto Y., Loeb L.A.. High fidelity and lesion bypass capability of human DNA polymerase delta. Biochimie. 2009; 91:1163–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Culligan K.M., Hays J.B.. Arabidopsis MutS homologs-AtMSH2, AtMSH3, AtMSH6, and a novel AtMSH7-form three distinct protein heterodimers with different specificities for mismatched DNA. Plant Cell. 2000; 12:991–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Arrieta-Montiel M.P., Shedge V., Davila J., Christensen A.C., Mackenzie S.A.. Diversity of the Arabidopsis mitochondrial genome occurs via nuclear-controlled recombination activity. Genetics. 2009; 183:1261–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.