Abstract
The diversity of Borrelia species discovered in California appears to be particularly high. A divergent group of Borrelia strains collected from Ixodes ticks in California was described by Postic and co-workers and designated ‘genomospecies 2’ (Postic D, Garnier M, Baranton G. Int J Med Microbiol 2007;297:263–271; Postic D, Ras NM, Lane RS, Hendson M, Baranton G. J Clin Microbiol 1998;36:3497–3504). We performed multilocus sequence analysis (MLSA) using eight housekeeping loci (clpA, clpX, nifS, pepX, pyrG, recG, rplB and uvrA) on 12 strains of this Borrelia genospecies to confirm that these strains form a distinct group within the Borreliaburgdorferi s. l. complex (Margos G, Hojgaard A, Lane RS, Cornet M, Fingerle V et al. Ticks Tick Borne Dis 2010;1:151–158). Phylogenetic and genetic distance analyses based on sequences of the MLSA housekeeping genes corroborated the distinctness of this group; genetic distances to all other members of the B. burgdorferi s.l. complex were 96 % or lower. We propose the name Borrelia lanei sp. nov. for this genospecies in honor of Professor Robert S. Lane, University of California Berkeley, for his contributions to Borrelia and tick research. The type strain for Borrelia lanei sp. nov., strain CA28-91T, has been deposited to two culture collections (=DSM 17992T=CIP 109135T).
Keywords: Borrelia burgdorferi sensu lato, tick-borne bacteria, MLSA, species complex
Abbreviations
BSK, Barbour-Stoenner-Kelly; IGS, intergenic spacer; LB, Lyme borreliosis; MLSA, multilocus sequence analysis; MLST, multilocus sequence typing; nov., novel; s.l., sensu lato; sp., species; s.s., sensu stricto; ST, sequence type.
Borrelia burgdorferi sensu lato (s. l.) is a heterogeneous bacterial species complex that contains the Borrelia species causing human Lyme borreliosis (LB). The species complex currently consists of about 20 validated genospecies. In nature, these parasitic bacteria are maintained in transmission cycles among vertebrate reservoir hosts and ticks of the Ixodes persulcatus species complex or other Ixodes species, such as Ixodes spinipalpis [1–3].
The strain designated as the Borrelia lanei sp. nov. type strain, CA28-91, was isolated from a pool of 10 questing Ixodes pacificus ticks collected during a field survey in Southern California by the California Department of Health Services and the Rocky Mountain Laboratories, National Institute of Allergy and Infectious Diseases [4]. Additional strains of this genospecies have been isolated from questing I. spinipalpis and from ticks collected from rabbit vertebrate hosts in Northern California [5, 6]. In 1998, Postic and co-workers undertook molecular analysis of strains from California based on the 16S rRNA gene locus and the 5S–23S intergenic spacer (IGS) region. These investigations suggested that strains CA2 and CA28-91 constituted a new genomic group within the B. burgdorferi s. l. complex. Further molecular investigations using the 16S rRNA gene locus, the 5S–23S IGS, housekeeping genes and an outer surface protein gene confirmed that, indeed, strains CA2 and CA28-91 clustered together in phylogenies based on concatenated sequences of several loci as well as the 16S rRNA gene sequences [5]. This prompted the authors to consider strains CA2 and CA28-91 as representative of a novel Borrelia genospecies, which they designated 'genomospecies 2’. Strain CA28-91 was proposed as the type strain and deposited in two microbial culture collections.
Interestingly, phylogenetic analysis based on sequences of only the 5S–23S IGS showed that the two strains, CA2 and CA28-91 [6], and also in combination with closely related strains (i.e. CA2, CA426, CA388 and CA28, CA400, CA393, CA399) branched in different parts of the tree [5]. These data suggest that this locus alone may not always be reliable as a species marker.
In this study, we used multilocus sequence analysis (MLSA) based on eight chromosomally located housekeeping genes (i.e. clpA, clpX, nifS, pepX, pyrG, recG, rplB and uvrA) to evaluate the status of strains supposed to belong to ‘genomospecies 2’. PCR and sequencing of PCR products was performed as described previously ([7], see also https://pubmlst.org/borrelia/). Sequences were compared to sequences present in the database at https://pubmlst.org/borrelia/ and amongst each other. Sequences that differed by one or more nucleotide from other sequences were considered novel alleles and received a new allelic number. Novel allelic profiles were assigned consecutive new sequence type (ST) numbers. The software mega 5.0 [8] was used for concatenation of sequences, alignment, determination of genetic distances and generation of phylogenetic trees. The Kimura two-parameter model was used to conduct genetic distance analyses [9]. In phylogenetic analysis, the percentage of trees in which the associated taxa clustered together is shown next to the branches. Initial tree(s) for the heuristic search were obtained automatically by applying neighbour-joining (NJ) and BioNJ algorithms to a matrix of pairwise distances estimated using the maximum composite likelihood (MCL) approach, and then selecting the topology with superior log likelihood value. Codon positions included were 1st+2nd+3rd+Noncoding. All positions containing gaps and missing data were eliminated. There were a total of 4768 positions in the final dataset.
Phylogenies were reconstructed using the maximum-likelihood model. The general time-reversible model with uniform rates across sites was chosen as the evolutionary model [10], nearest-neighbour interchange for tree optimization, and 500 bootstrap repetitions to estimate branch support. Sequences of the following LB spirochaete species and strains were included in the genetic distance and phylogenetic analysis: Borrelia afzelii VS461T , B orrelia americana CA-8B-89, ‘Borellia andersonii’ 21123, Borrelia bavariensis PBiT , Borrelia bissettiae DN127T and CA28, Borrelia burgdorferi sensu stricto (s. s.) B31T and Z41293, Borrelia californiensis CA443, Borrelia carolinensis SCW-22T, ‘Borrelia chilensis’ VA1, Borrelia garinii 20047T, Borrelia japonica HO14T, Borrelia kurtenbachii 25015T, Borrelia lusitaniae PoHL1, Borrelia mayonii MN14-1420T, Borrelia sinica CMN3T, Borrelia spielmanii A14S, Borrelia tanukii Hk501T, Borrelia turdi Ya501T, Borrelia valaisiana VS116T, Borrelia yangtzensis QLM4P1, Borrelia sp. CA690 and, as outgroups the relapsing-fever species, Borrelia duttonii Ly , Borrelia hermsii DAH, Borrelia turicatae 91E135, Borrelia miyamotoi USA LB2100 and Borrelia miyamotoi Japan HT31T.
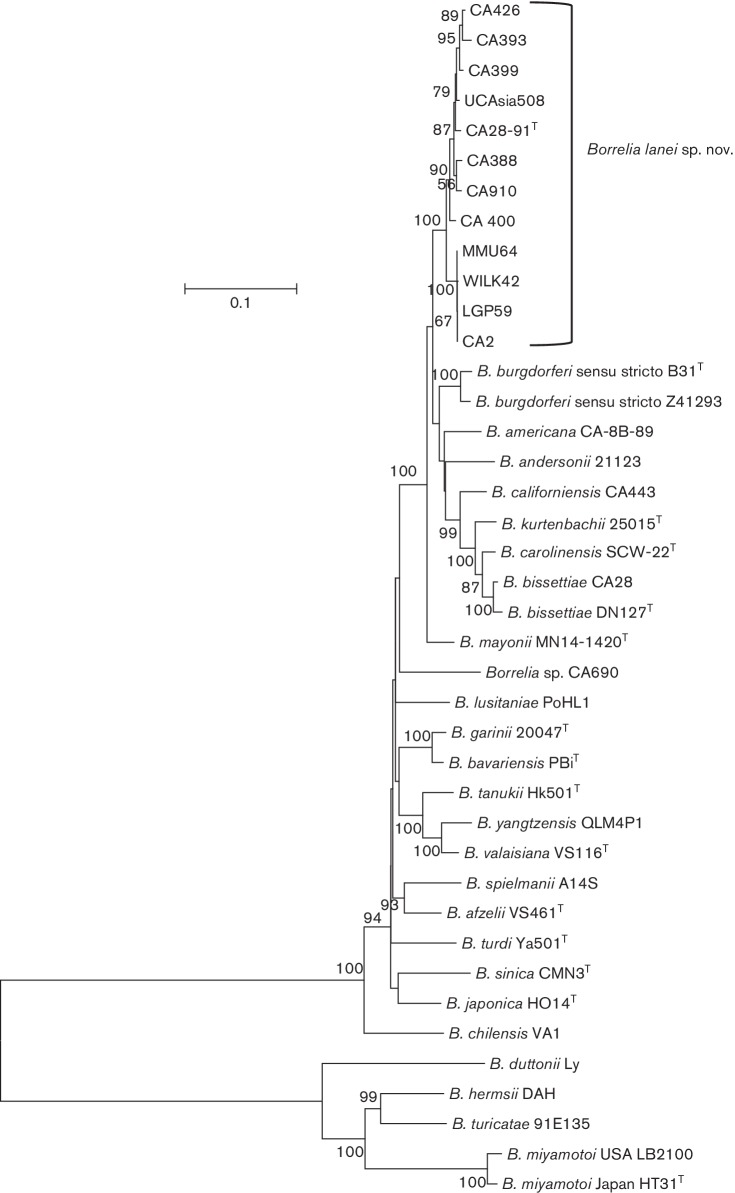
We included some of the strains previously used by Postic and co-workers [5] and added additional strains so that the number of strains included in our study totalled 12 (see Table 1). Phylogenetic analysis of the concatenated sequences of the housekeeping genes confirmed the relationship of strains incorporated in the new Borrelia species and their distinctness from all other previously described Borrelia genospecies. In the phylogeny all strains of the novel genospecies clustered together and apart from all other Borrelia genospecies (Fig. 1). The closest-related genospecies of the B. burgdorferi s.l. species complex were Borrelia species from North America with B. mayonii , a human pathogenic genospecies recently discovered in the Midwest of the USA [11, 12], showing the lowest genetic distance to type strain CA28-91T (0.040, which equals 96 % similarity) followed by B. burgdorferi s. s. strains Z41293 and B31 (0.042 and 0.045, respectively) and B. americana (0.049) (Fig. 1 and Table S1, available in the online Supplementary Material). All other Borrelia genospecies had higher genetic distance values (Table S1).
Table 1. Borrelia lanei sp. nov. strains included into analysis.
| MLST ST | Strain name | Country of origin | Region | Biologic source of isolate | Tick stage | Vertebrate host | Year of collection | Collected by | Isolated by | Reference | GenBank/ MLST ID |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 719 | CA28-91T | USA | Kern Co., California | Ixodes pacificus | Adults | Unknown | 1991 | California Department of Health | California Department of Health | [4, 5] |
DQ393351.1; DQ393371.1; DQ393392.1; DQ393410.1; AJ006375; ID 1949 |
| 720 | LGP59 | USA | Mendocino Co., CA | I. pacificus | Nymph | Unknown | 2004 | R. J. Eisen, L. Eisen | J. Mun | [14] | ID 1950 |
| 721 | MMU64 | USA | Mendocino Co., CA | I. pacificus | Nymph | Unknown | 2004 | R. J. Eisen, L. Eisen | N.Fedorova | [14] | ID 1951 |
| 722 | CA2 | USA | Mendocino Co., CA | Ixodes spinipalpis | Adult female | Jack rabbit | 1986 | R. S. Lane | R. S. Lane | [5] |
DQ393352.1; DQ393391.1; L301231; ID 1952 |
| 723 | WILK42 | USA | Mendocino Co., CA | I. pacificus | Nymph | Unknown | 2004 | R. J. Eisen, L. Eisen | N. Fedorova | [14] | ID 1953 |
| 724 | CA400 | USA | Contra Costa Co., CA | I. spinipalpis | Nymph | Unknown | 1993 | K. A. Padgett | K. A. Padgett | [5] |
AY177638 ID 1954 |
| 725 | CA388 | USA | Contra Costa Co., CA | I. spinipalpis | Nymph | Unknown | 1993 | K. A. Padgett | K. A. Padgett | [5] |
AY176361 ID 1955 |
| 726 | CA426 | USA | Contra Costa Co., CA | Sylvilagus bachmani | Unknown | Brush rabbit | 1995 | C. A. Peavey | C. A. Peavey | [5] |
AY177640 ID 1956 |
| 727 | UCAsia508 | USA | Alameda Co., CA | I. spinipalpis | Nymph | Unknown | 2009 | J. E. Kleinjan | N. Fedorova | Unpublished | ID 1957 |
| 728 | CA399 | USA | Contra Costa Co., CA | I. spinipalpis | Nymph | Unknown | 1993 | K. A. Padgett | K. A. Padgett | [5] |
AY182046 ID 1958 |
| 729 | CA910 | USA | Alameda Co., CA | I. spinipalpis | Nymph | Unknown | 2014 | J. E. Kleinjan | J. E. Kleinjan | Det. N. Fedorova | ID 1959 |
| 730 | CA393 | USA | Contra Costa Co., CA | I. spinipalpis | Nymph | Unknown | 1993 | K. A. Padgett | K. A. Padgett | [5] |
AY177637; ID 1960 |
Fig. 1.
Molecular phylogenetic analysis of B. lanei sp. nov. The evolutionary history was inferred by using the maximum-likelihood method based on the general time reversible model [10]. The tree with the highest log likelihood (−33644, 4319) is shown. A discrete Gamma distribution was used to model evolutionary rate differences among sites [4 categories (+G, parameter=0, 6690)]. The rate variation model allowed for some sites to be evolutionarily invariable [(+I), 32, 8618 % sites]. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site (scale bar). B. lanei sp. nov. strains form a cluster containing two sister clades. Both sister clades form a separate cluster distinct from all other Borrelia genospecies known to date underlining this species is divergent.
Within B. lanei sp. nov. some strains (LGP59, MMU64, CA2, WILK42), all collected in Mendocino County, showed borderline genetic distances (0.017) compared to the type strain CA28-91T and genetic distances just above the species threshold to several other strains of the group (yellow label in Table S1). The same strains formed a distinct branch in the phylogeny (Fig. 1) which, nevertheless, formed a sister clade to B. lanei sp. nov. strains and not to any other Borrelia species. Thus, in spite of slightly high genetic distances for some strains, genetic distances to other strains of the group were below the species threshold. This led us to consider all 12 strains included into our analysis as being members of the new Borrelia species, B. lanei sp. nov.
The strains included here had been isolated from I. pacificus and I. spinipalpis ticks (Table 1) [5, 6, 13, 14]. One tick was collected from a jack rabbit and one strain was isolated from a brush rabbit suggesting a potential transmission cycle between lagomorphs and Ixodes ticks. Interestingly, B. lanei sp. nov. was recently detected in I. spinipalpis ticks also collected from a rabbit, but in Canada [15]. However, solid evidence does not exist for either vector or host adaptations, thus, the transmission cycle(s) maintaining B. lanei sp. nov. need to be explored in future studies.
Description of Borrelia lanei sp. nov.
Borrelia lanei sp. nov. (la.ne′i. N.L. gen. n. lanei in honour of Professor Robert S. Lane for his outstanding contributions to Borrelia and Ixodes research).
The cells are helical, approximately 0.2 µm by 20 µm, and stain well with Giemsa stain. Unstained cells can be visualized by dark-field microscopy. The bacteria are flexible and motile with rotational and forward/backwards movement. Cells can be cultured in vitro under microaerophilic conditions [16] using liquid media such as Barbour–Stoenner–Kelly (BSK) medium. Optimal growth occurs at 33–34 °C. The mean DNA G+C content of the type strain is 27 mol%.
The type strain CA28-91T was isolated from a pool of 10 questing I. pacificus ticks in 1991. It has been deposited at the German Microbial Strain Collection (=DSM 17992T) and at the Institut Pasteur, Paris, France (=CIP 109135T). B. lanei can be distinguished from other Borrelia genospecies via sequences of the rrs locus and by MLSA [5, 6, 17]. This bacterium is maintained in nature in diverse transmission cycles likely involving lagomorph reservoir hosts and certain Ixodes species ticks such as I. spinipalpis and I. pacificus.
Funding information
This work was partially funded by the Robert-Koch-Institute via the German National Reference Centre for Borrelia .
Acknowledgements
The authors gratefully acknowledge Cecilia Hizo-Teufel and Sylvia Stockmeier for technical help in the laboratory and Rebecca J. Eisen, Lars Eisen, Kerry A. Padgett and Chindy A. Peavey for conducting ecological studies in Mendocino and Contra Costa counties.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Supplementary Data
References
- 1.Kurtenbach K, Hanincová K, Tsao JI, Margos G, Fish D, et al. Fundamental processes in the evolutionary ecology of Lyme borreliosis. Nat Rev Microbiol. 2006;4:660–669. doi: 10.1038/nrmicro1475. [DOI] [PubMed] [Google Scholar]
- 2.Brown RN, Lane RS. Lyme disease in California: a novel enzootic transmission cycle of Borrelia burgdorferi. Science. 1992;256:1439–1442. doi: 10.1126/science.1604318. [DOI] [PubMed] [Google Scholar]
- 3.Brown RN, Lane RS. Reservoir competence of four chaparral-dwelling rodents for Borrelia burgdorferi in California. Am J Trop Med Hyg. 1996;54:84–91. doi: 10.4269/ajtmh.1996.54.84. [DOI] [PubMed] [Google Scholar]
- 4.Schwan TG, Schrumpf ME, Karstens RH, Clover JR, Wong J, et al. Distribution and molecular analysis of lyme disease spirochetes, Borrelia burgdorferi, isolated from ticks throughout California. J Clin Microbiol. 1993;31:3096–3108. doi: 10.1128/jcm.31.12.3096-3108.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Postic D, Garnier M, Baranton G. Multilocus sequence analysis of atypical Borrelia burgdorferi sensu lato isolates-description of Borrelia californiensis sp. nov., and genomospecies 1 and 2. Int J Med Microbiol. 2007;297:263–271. doi: 10.1016/j.ijmm.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 6.Postic D, Ras NM, Lane RS, Hendson M, Baranton G. Expanded diversity among Californian Borrelia isolates and description of Borrelia bissettii sp. nov. (formerly Borrelia group DN127) J Clin Microbiol. 1998;36:3497–3504. doi: 10.1128/jcm.36.12.3497-3504.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Margos G, Vollmer SA, Cornet M, Garnier M, Fingerle V, et al. A new Borrelia species defined by multilocus sequence analysis of housekeeping genes. Appl Environ Microbiol. 2009;75:5410–5416. doi: 10.1128/AEM.00116-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 10.Nei M, Kumar S. Molecular Evolution and Phylogenetics. New York: Oxford University Press; 2000. [Google Scholar]
- 11.Kingry LC, Batra D, Replogle A, Rowe LA, Pritt BS, et al. Whole genome sequence and comparative genomics of the novel Lyme Borreliosis causing pathogen, Borrelia mayonii. PLoS One. 2016;11:e0168994. doi: 10.1371/journal.pone.0168994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pritt BS, Respicio-Kingry LB, Sloan LM, Schriefer ME, Replogle AJ, et al. Borrelia mayonii sp. nov., a member of the Borrelia burgdorferi sensu lato complex, detected in patients and ticks in the upper midwestern United States. Int J Syst Evol Microbiol. 2016;66:4878–4880. doi: 10.1099/ijsem.0.001445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fedorova N, Kleinjan JE, James D, Hui LT, Peeters H, et al. Remarkable diversity of tick or mammalian-associated Borreliae in the metropolitan San Francisco Bay area, California. Ticks Tick Borne Dis. 2014;5:951–961. doi: 10.1016/j.ttbdis.2014.07.015. [DOI] [PubMed] [Google Scholar]
- 14.Girard YA, Travinsky B, Schotthoefer A, Fedorova N, Eisen RJ, et al. Population structure of the Lyme Borreliosis spirochete Borrelia burgdorferi in the western black-legged tick (Ixodes pacificus) in Northern California. Appl Environ Microbiol. 2009;75:7243–7252. doi: 10.1128/AEM.01704-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scott JD, Clark KL, Foley JE, Anderson JF, Durden LA, et al. Detection of Borrelia genomospecies 2 in Ixodes spinipalpis ticks collected from a rabbit in Canada. J Parasitol. 2017;103:38–46. doi: 10.1645/16-127. [DOI] [PubMed] [Google Scholar]
- 16.Johnson RC, Schmid GP, Hyde FW, Steigerwalt AG, Brenner DJ. Borrelia burgdorferi sp. nov.: etiologic agent of Lyme disease. Int J Syst Bacteriol. 1984;34:496–497. doi: 10.1099/00207713-34-4-496. [DOI] [Google Scholar]
- 17.Margos G, Hojgaard A, Lane RS, Cornet M, Fingerle V, et al. Multilocus sequence analysis of Borrelia bissettii strains from North America reveals a new Borrelia species, Borrelia kurtenbachii. Ticks Tick Borne Dis. 2010;1:151–158. doi: 10.1016/j.ttbdis.2010.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.