Key Points
Mutated BRAF accelerates disease and enhances immune abnormalities in murine B-cell leukemia.
This new model will be valuable for understanding and targeting disease-induced immune modulation in MAPK-mutated B-cell malignancies.
Abstract
Mutated mitogen-activated protein kinase (MAPK) pathway components promote tumor survival, proliferation, and immune evasion in solid tumors. MAPK mutations occur in hematologic cancers as well, but their role is less clear and few models are available to study this. We developed an in vivo model of disseminated BRAFV600E B-cell leukemia to determine the effects of this mutation on tumor development and immune evasion. Mice with B-cell–restricted BRAFV600E expression crossed with the Eµ-TCL1 model of chronic lymphocytic leukemia (CLL) developed leukemia significantly earlier (median, 4.9 vs 8.1 months; P < .001) and had significantly shorter lifespan (median, 7.3 vs 12.1 months; P < .001) versus BRAF wild-type counterparts. BRAFV600E expression did not affect B-cell proliferation but reduced spontaneous apoptosis. BRAFV600E-mutant leukemia produced greater T-cell effects, evidenced by exhaustion immunophenotype and CD44+ T-cell percentage, as well as increased expression of PD-L1 on CD11b+ cells. Results were confirmed in syngeneic mice engrafted with BRAFV600E leukemia cells. Furthermore, a BRAFV600E-expressing CLL cell line more strongly inhibited anti-CD3/CD28–induced T-cell proliferation, which was reversed by BRAFV600E inhibition. These results demonstrate the immune-suppressive impact of BRAFV600E in B-cell leukemias and introduce a new model to develop rational combination strategies targeting both tumor cells and tumor-mediated immune evasion.
Visual Abstract

Introduction
The mitogen-activated protein kinase (MAPK) pathway regulates diverse cell functions downstream of many cell-surface receptors by controlling transcription, cell cycle, and apoptosis and is commonly aberrant in tumor cells.1 Mutated MAPK pathway components drive tumor survival and proliferation in the absence of external stimuli, and therapeutic MAPK inhibitors are being pursued to block these effects.2-6 MAPK signaling also induces immune-suppressive behavior of tumor cells via multiple mechanisms, including production and secretion of cytokines, induction of immune checkpoint molecules, and cytoskeletal remodeling.7-9 MAPK pathway activation in tumor cells without MAPK mutations, such as through Toll-like receptor stimulation or transforming growth factor beta signaling, can promote tumor infiltration and immune suppression at least in part via CD200 and programmed death-ligand 1 (PD-L1) induction, and these effects can be reversed by MEK inhibition.10-13 Similar findings are reported in BRAFV600E-mutant melanoma, in which inhibition of BRAF and MEK increases T-cell recognition and dendritic cell function.14,15 This evidence strongly supports that targeting MAPK signaling in cancer patients could reverse tumor-induced immune suppression, leading to longer disease control, improved resistance to opportunistic infections, and enhanced efficacy of immune-based therapies.
The MAPK pathway is mutationally activated in hematologic malignancies, although generally less frequently than in solid tumors. Most notably, nearly 100% of classic hairy cell leukemia (HCL) cases carry the BRAFV600E mutation,16 suggesting a key function for the mutated protein in disease development. Multiple reports demonstrate that HCL can be successfully treated with vemurafenib, further supporting this hypothesis.17 Approximately 10% of chronic lymphocytic leukemia (CLL) patients carry an activating mutation in an MAPK pathway component, including BRAF,18-20 and BRAF mutations were identified as one of the acquired initiating mutations in early hematopoietic cells of CLL patients.21 A recent case report describes the clinical efficacy of MAPK inhibition in a patient with BRAFV600E-mutated multiple myeloma.22 Moreover, a BRAF pseudogene transcript is aberrantly expressed in primary human diffuse large B-cell lymphoma and is positively correlated with BRAF expression, resulting in MAPK signaling activation. Global expression of this pseudogene in mice results in aggressive B-cell lymphoma.23 Together, these findings indicate a role for activated MAPK in a meaningful subset of B-cell malignancies. However, its function with regard to immune suppression in these diseases is unclear, and no suitable mouse model with a B-cell–specific MAPK activating mutation has been available. Here, we describe a new model of BRAF-mutated B-cell leukemia to investigate this critical aspect.
In this study, we generated a murine model of BRAFV600E-mutated B-cell leukemia based on the well-established Eµ-TCL1 strain24-28 and documented the phenotype of the resulting disease. Eµ-TCL1 mice with BRAFV600E-mutated B cells develop an aggressive leukemia significantly earlier than standard Eµ-TCL1 mice and have shortened survival. In addition, BRAFV600E-mutated leukemic B cells exert a greater immunosuppressive effect on T cells, and this can be reversed by pharmacologic inhibition of BRAFV600E. This study provides a new model of MAPK-driven immune defects in B-cell leukemia and supports the introduction of combination strategies using MAPK pathway inhibitors together with Bruton’s tyrosine kinase and/or immune checkpoint inhibitors in patients with BRAFV600E-mutated B-cell malignancies.
Methods
Cell lines and reagents
OSUCLL cells were originally established by Epstein-Barr virus transformation of cells from a CLL patient.29 This cell line displays immunophenotype and cytogenetics similar to the patient’s CLL (trisomy 12 and 19). OSUCLL cells with doxycycline-inducible expression of normal or mutated BRAF were previously described.30 Vemurafenib and dabrafenib were obtained from Selleck Chemicals (Houston, TX) and doxycycline from Clontech (Mountain View, CA).
Transgenic mice
Generation of mice with B-cell–specific BRAFV600E as the result of CD19-driven Cre recombinase is described in the supplemental Data. These or CD19-Cre mice without BRAFV600E were crossed with the established Eµ-TCL1 model.24 Eµ-TCL1 mice with BRAF-mutated B cells are referred to in the text as “BRAFVE×TCL1,” and BRAF wild-type (WT) counterparts are referred to as “BRAFWT×TCL1” (no BRAFV600E allele) or “BRAFCA×TCL1” (conditional allele [CA], no CD19-Cre). Mice were monitored once every 2 weeks by flow cytometry and spleen palpation. Leukemia was defined as enlarged (palpable) spleen and ≥10% CD5+CD19+ cells in the peripheral blood CD45+ population. For adoptive transfer experiments, spleen cells were isolated from transgenic mice with leukemia and assessed by flow cytometry. Live CD5+CD19+ cells (107) were injected intravenously into 8- to 10-week-old WT female C57BL/6 mice. The mice were sacrificed upon reaching predetermined euthanasia criteria (≥20% weight loss, hind limb paralysis, respiratory distress, rough coat, or ≥10% weight loss, with other signs). All mouse experiments were performed under a protocol approved by The Ohio State University Institutional Animal Care and Use Committee.
Cell isolation
Mouse CD19 MicroBeads (Miltenyi Biotec, San Diego, CA), EasySep Mouse Pan-B Cell Isolation Kit, and EasySep Mouse T-Cell Isolation Kit (STEMCELL Technologies, Vancouver, BC, Canada) were used to isolate B and T cells from mouse spleens. Purity was confirmed by flow cytometry and was at least 90% for all experiments.
Immunoblotting
Lysates from B and T cells from spleens were immunoblotted with antibodies for human BRAFV600E (Spring Bioscience, Pleasanton, CA), total BRAF (Santa Cruz Biotechnology, Santa Cruz, CA), phosphorylated and total MEK and ERK (Cell Signaling Technology, Danvers, MA), and actin (Santa Cruz Biotechnology) using standard procedures.
Flow cytometry
Flow cytometry was performed by using either a Gallios (Beckman Coulter, Brea, CA) or a Fortessa (BD Biosciences, San Jose, CA) cytometer, and results were processed using Kaluza software (Beckman Coulter). Antibodies, stains, and sources are listed in supplemental Data.
Cell proliferation assay
Mice were intraperitoneally injected with 50 μg of 5-ethynyl-2′-deoxyuridine (EdU; Santa Cruz Biotechnology) per gram of body weight. After 24 hours, spleen cells were collected and stained with anti-CD5 fluorescein isothiocyanate, anti-CD19 phycoerythrin, and anti-CD45 Alexa700, followed by Click-iT EdU labeling (ThermoFisher). Cells were analyzed immediately by flow cytometry.
Histology
Details of histologic examinations are provided in the supplemental Data.
Functional assays
T cells from peripheral blood of healthy adults were negatively selected to enrich for CD3, CD4, or CD8 T cells stained with carboxyfluorescein succinimidyl ester (Thermo Fisher Scientific) and stimulated with plate-bound anti-CD3 and soluble anti-CD28 antibodies (BD Biosciences). Human T cells were cocultured with OSUCLL cells expressing normal BRAF or BRAFV600E. All conditions, including T cells alone, also received 1 µg/mL doxycycline. For cytokine analyses, supernatants from cocultures of stimulated healthy T cells with or without leukemic B cells were analyzed by cytometric bead array (BD Biosciences) per the manufacturer’s protocol. Cytokines in mouse plasma were also assessed using MILLIPLEX Multiplex Assays (EMD Millipore, Billerica, MA) according to the manufacturer’s instructions.
Statistics
Kaplan-Meier estimates and the log-rank test were used to compare time from birth to development of leukemia and overall survival from birth among BRAFWT×TCL1, BRAFVE×TCL1, and BRAFCA×TCL1 groups. Organ weight comparisons were made by using two-sample Student t tests, assuming unequal variances; for the spleen data, the nonparametric Wilcoxon rank sum test was used instead. Differences in EdU incorporation and annexin-V binding were assessed by using two-sample Student t tests or Wilcoxon rank sum tests. For the messenger RNA (mRNA) data, mixed effects models were applied to the ΔCT values (difference in cycle numbers at which detection thresholds are reached), and estimated differences were transformed back into fold changes. Changes in F4/80+ macrophages, myeloid-derived suppressor cells (MDSCs), patrolling monocytes, cytokine levels, and T-cell proliferation data (division index [DI]31) were estimated by using mixed effects models to allow for correlations among observations from the same donors. All analyses were performed using SAS/STAT software Version 9.4 (SAS Institute Inc., Cary NC).
Results
B-cell selective expression of BRAFV600E in Eμ-TCL1 mice accelerates disease and shortens survival
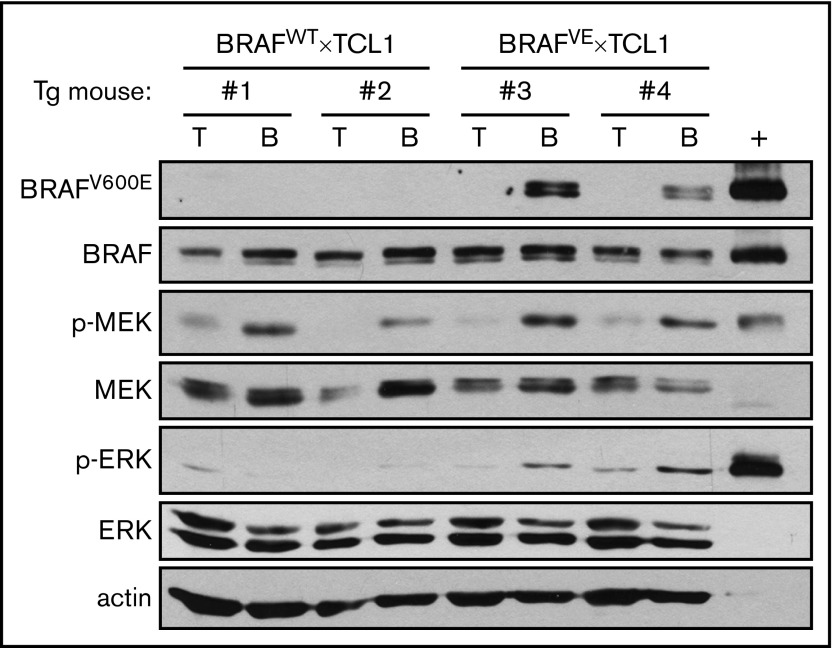
Mouse strains carrying CD19-driven Cre recombinase and immunoglobulin (Ig) Eµ-driven human TCL1, without or with a conditional Cre-driven BRAFV600E allele (BRAFWT×TCL1 and BRAFVE×TCL1, respectively) were generated and genotypically confirmed. Immunoblot analysis showed that BRAFV600E protein is expressed in B cells but not T cells from the spleens of BRAFVE×TCL1 mice, and this expression is associated with increased phosphorylation of downstream kinases MEK and ERK (Figure 1). The onset of leukemia, as defined by at least 10% CD19+CD5+ double-positive cells among the CD45+ population in the peripheral blood, was evaluated once every 2 weeks by flow cytometry starting at 6 weeks. BRAFVE×TCL1 mice (n = 34) developed leukemia at a median of 4.9 months (95% confidence interval [CI], 4.5-5.6 months), significantly earlier than BRAFWT×TCL1 mice (median, 8.1 months; 95% CI, 7.6-9.2 months; n = 46) or BRAFCA×TCL1 mice (median, 8.1 months; n = 22) (P < .001 for both comparisons; Figure 2A). Leukemia onset in the control groups was comparable to that in standard Eµ-TCL1 mice.24,32 There was no evidence of leukemia during this period in BRAFVE×CD19 Cre mice as expected, because BRAFV600E by itself is not sufficient to induce malignancy.33,34 This group was therefore not included in the remaining experiments. BRAFVE×TCL1 mice exhibited significantly shorter survival (median, 7.3 months; 95% CI, 6.9-8.0 months; n = 34) compared with BRAFWT×TCL1 mice (median, 12.1 months; 95% CI, 11.5-12.7 months; n = 46) or BRAFCA×TCL1 mice (median, 11.3 months; 95% CI, 9.9-11.9 months; n = 22) (Figure 2B; P < .001 for both comparisons).
Figure 1.
BRAFV600Eprotein is expressed in the B cells of BRAFVE×TCL1 mice. Lysates from purified B or T cells from spleens of BRAFWT×TCL1 or BRAFVE×TCL1 transgenic (Tg) mice were analyzed by immunoblot for mutant and normal BRAF as well as total and phosphorylated MEK and ERK. Lysate from OSUCLL-BRAFV600E+doxycycline was included as a positive control (+).
Figure 2.
BRAFV600Eproduces more aggressive disease in the Eμ-TCL1 mouse model. B-cell–specific expression of BRAFV600E significantly shortens (A) leukemia onset and (B) overall survival (P < .001 comparing BRAFVE vs BRAFWT and BRAFCA for both onset and overall survival).
Phenotype of disease in BRAFVE×TCL1 mice
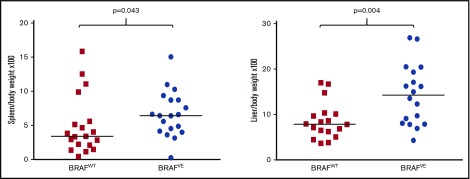
Detailed necropsies were performed on mice meeting euthanasia criteria as a result of advanced disease. Spleens and livers were enlarged in BRAFVE vs BRAFWT TCL1 mice (n = 19 per group for spleen [P = .043]; n = 18 per group for liver [P = .004]; Figure 3). These are shown as organ:body weight ratios to control for body size because the BRAFWT mice were older by the time they reached euthanasia criteria; absolute weights are shown in supplemental Figure 1A. Gross appearances of the spleen and liver are shown in supplemental Figure 1B-C. BRAFVE animals showed thymic enlargement and reduced hematocrit relative to BRAFWT mice, although differences did not reach statistical significance (n = 15 per group for thymus [P = .058]; n = 16 and 13, respectively, for hematocrit [P = .079]), as well as larger submandibular, mediastinal, and intra-abdominal lymph nodes.
Figure 3.
Organomegaly in leukemic BRAFWTand BRAFVETCL1 transgenic mice. Weight ratios of spleens (n = 19 for each genotype; P = .043) and livers (n = 18 for each genotype; P = .004) in BRAFWT and BRAFVE TCL1 mice meeting euthanasia criteria. The horizontal bars represent the median.
Unlike Eμ-TCL1 mice with unmutated BRAF, the disease in BRAFVE×TCL1 mice later progressed to a tissue-phase cancer consistent with histiocyte-associated B-cell lymphoma. As shown in supplemental Figure 2A-B, spleens and lymph nodes of BRAFWT×TCL1 mice are infiltrated by a uniform population of small to intermediate size lymphocytes characterized by scant basophilic cytoplasm, dense chromatin, and central round nuclei, as previously reported for this model.24 In contrast, spleens of BRAFVE×TCL1 mice (supplemental Figure 2B) show sheets of a homogeneous population of round cells with abundant eosinophilic cytoplasm and ovoid nuclei with lacy chromatin, consistent with histiocytes or macrophages. Spleens of BRAFVE×TCL1 mice also show histiocytic-origin multinucleated giant cells, and lymph nodes show numerous scattered foci of large, atypical lymphocytes, many of which demonstrate plasmacytoid differentiation, on a background of histiocytic cells. Extended histologic investigation of organs from BRAFVE×TCL1 mice with advanced disease (supplemental Figure 2C-D) showed clear neoplastic cell infiltration, together with increased numbers of cells with B-cell (B220+) and macrophage (F4/80+) lineages. Both genotypes showed small CD3+ lymphocytes throughout the neoplastic population, interpreted as tumor-infiltrating T cells.
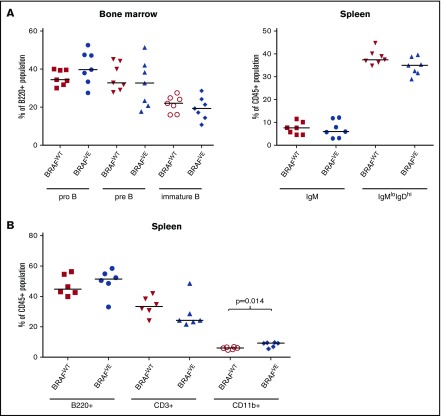
To investigate B-cell development before the onset of leukemia, cells from spleen and marrow from 2-month-old BRAFVE×TCL1 and BRAFWT×TCL1 mice (n = 7 each) were assessed by flow cytometry. Figure 4A shows that pro (B220+CD43+IgM–), pre (B220+CD43–IgM–), immature (B220+CD43–IgM+), and mature (splenic IgMlowIgDhi) B cells35 developed similarly in the 2 groups. BRAFVE×TCL1 mice showed a slightly higher percentage of CD11b+ (myeloid) cells (P = .014) and a trend toward higher B-cell and lower T-cell numbers in spleens compared with BRAFWT×TCL1 mice (Figure 4B). Complete necropsies were also performed on BRAFWT×TCL1 (n = 5) and BRAFVE×TCL1 (n = 6) mice at 2 months of age, before the onset of leukemia. Histologically, spleens from all BRAFVE×TCL1 animals showed increased numbers of atypical lymphocytes throughout the red pulp, which was not observed in the BRAFWT×TCL1 mice (supplemental Figure 3). No gross lesions were observed in either group at this stage. To determine whether this model is recapitulated by adoptive transfer of neoplastic cells to allow larger, more uniform studies, leukemia cells from BRAFVE×TCL1 mice were intravenously injected into 7-week-old syngeneic WT recipients (n = 11). By 2 weeks, CD5+CD19+ leukemia cells were detected in the peripheral blood of all mice. Complete necropsies with histologic evaluation of spleen, bone marrow, lymph nodes, thymus, and liver were performed at 3 weeks. All normal syngeneic mice engrafted with BRAFVE×TCL1 leukemia cells demonstrated histiocyte-associated B-cell leukemia similar to BRAFVE×TCL1 transgenic mice (supplemental Figure 4). This demonstrates the utility of the adoptive transfer strategy of this model to conduct future studies evaluating new treatment combinations and strategies.
Figure 4.
Cell populations in transgenic mice before leukemia onset. (A) Spleen and bone marrow cells from 2-month-old transgenic mice (n = 6 to 7 for each genotype) were analyzed by flow cytometry for pro-B (B220+CD43+IgM–), pre-B (B220+CD43–IgM–), immature B (B220+CD43–IgM+), and mature B (splenic IgMlowIgDhi) cells. Differences between groups were not significant. (B) Spleen cells from 2-month-old BRAFWT or BRAFVE TCL1 transgenic mice (n = 6 per genotype) were analyzed by flow cytometry for B-, T-, and myeloid cell populations. BRAFVE×TCL1 mice showed a minor but significant increase in myeloid (CD11b+) cells before the onset of leukemia and trends toward increased B (B220+) and decreased T (CD3+) cells. The horizontal bars represent the median.
BRAFV600E expression does not increase cell proliferation but reduces cell apoptosis
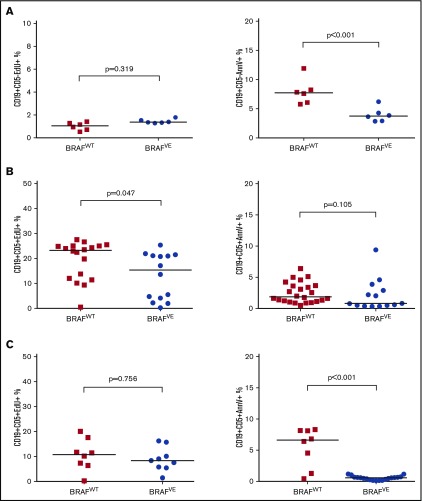
The expression of mutated BRAF in tumor cells alters their proliferative and apoptotic properties, in part via cyclin D1 and BCL2 family proteins, respectively.36 To examine the effect of BRAFV600E expression on these parameters in preleukemic B cells in vivo, EdU was injected into BRAFVE×TCL1 (n = 6) or BRAFWT×TCL1 (n = 5) transgenic mice at approximately 2 months of age, before the onset of leukemia. After 24 hours, spleen cells were collected, and EdU incorporation and annexin-V binding were assessed. Proliferation of CD19+CD5– B cells, as measured by EdU incorporation, was not different between the groups (P = .319). However, B cells from BRAFVE mice showed significantly lower apoptosis as assessed by annexin-V binding (P < .001) (Figure 5A).
Figure 5.
Effect of BRAFV600Eon cell proliferation and apoptosis in vivo. (A) Proliferation (EdU incorporation) and apoptosis (annexin-V binding) were analyzed by flow cytometry in normal B cells (CD5–CD19+) from the spleens of preleukemic transgenic mice (∼2 months old; n = 6 for both BRAFWT×TCL1 and BRAFVE×TCL1). (B) The same experiment was conducted using healthy mice engrafted with leukemia cells pooled from 3 leukemic donor animals after the onset of leukemia (>10% CD19+CD5+ cells in peripheral blood of the CD45+ population) (n = 18 for BRAFWT×TCL1 EdU and n = 24 for annexin-V; n = 14 BRAFVE×TCL1 for both EdU and annexin). (C) The experiment in (B) was repeated using single donors for each genotype (EdU: n = 8 for BRAFWT×TCL1 and n = 9 for BRAFVE×TCL1; annexin-V: n = 8 for BRAFWT×TCL1 and n = 19 for BRAFVE×TCL1). The horizontal bars represent the median.
These effects were further investigated in vivo using the adoptive transfer model. In the first experiment, pooled leukemic B cells from BRAFVE×TCL1 or BRAFWT×TCL1 mice were engrafted into WT syngeneic mice (BRAFVE×TCL1, n = 14; BRAFWT×TCL1, n = 24). After the onset of leukemia, EdU was injected, and 24 hours later, spleen cells were isolated for flow cytometric analysis as in Figure 5A-B, which shows that BRAFV600E did not increase B-cell proliferation in vivo and, in fact, mildly decreased it (P = .047). Apoptosis was slightly but not significantly reduced (P = .105). A second adoptive transfer experiment using a single leukemic B-cell donor for each genotype confirmed that cell proliferation was not significantly increased in the presence of BRAFV600E, but cell apoptosis was significantly reduced (P = .756 and P < .001, respectively) (Figure 5C; supplemental Figure 5). These experiments support that the introduction of mutated BRAF in B-cell leukemia does not impact proliferation and only moderately reduces spontaneous apoptosis.
BRAFV600E B cells suppress T-cell proliferation and function
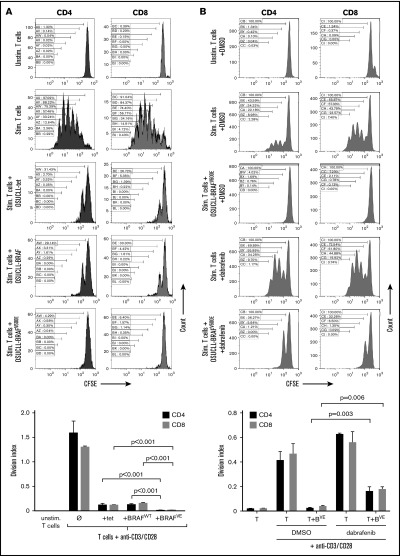
The above results, together with the immunophenotypic characterization, suggest that the BRAFV600E mutation might accelerate disease development in B-cell leukemia by promoting immune evasion or suppression as observed in some solid tumors,10,12-15,37 rather than simply by affecting proliferation or apoptosis. We therefore examined the effects of B cells expressing BRAFV600E on other immune compartments. We first tested this mechanistically in vitro by using OSUCLL cells with doxycycline-inducible expression of normal or mutated BRAF.30 These cells were cocultured with T cells from normal donors that were labeled with carboxyfluorescein succinimidyl ester and stimulated with anti-CD3/anti-CD28 antibodies. OSUCLL cells in general impaired proliferation of stimulated T cells, and this was enhanced in BRAFV600E-expressing cells compared with those transfected with an empty vector (OSUCLL-tet) or WT BRAF. There was a significant decrease in DI, defined as the average number of divisions for all cells in the culture31 between OSUCLL-BRAFVE and OSUCLL-BRAFWT or OSUCLL-tet (P < .001) (Figure 6A). Proliferation was significantly rescued by the BRAFV600E inhibitor dabrafenib (P = .003 and P = .006 for DIs of CD4 and CD8, respectively, in dabrafenib vs vehicle treatment; Figure 6B), further supporting the contribution of mutant BRAF to this effect. Dabrafenib had no direct effect on T-cell proliferation. Interestingly, vemurafenib at the same dose led to a notable impairment of T-cell proliferation in contrast to what has been reported in vitro38 and in support of clinical data39 and was therefore not used.
Figure 6.
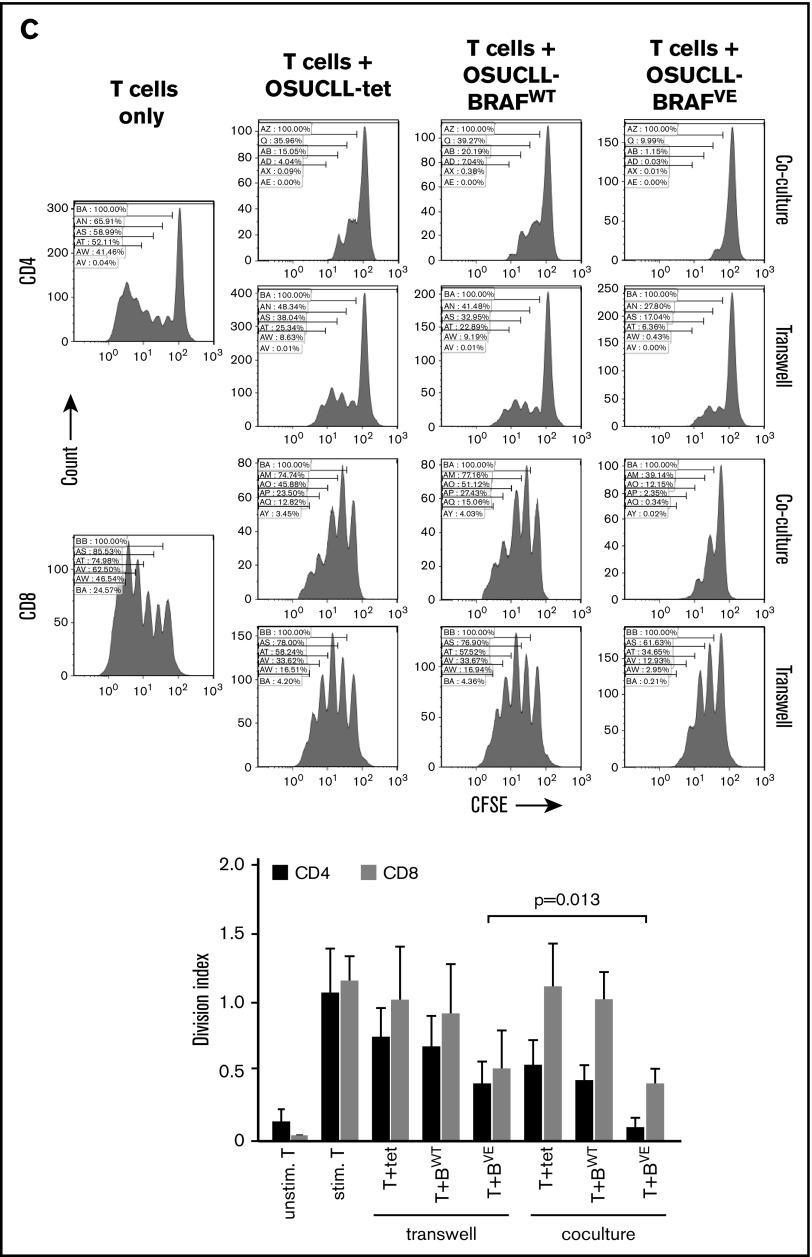
Immunosuppressive effect of BRAFV600Ein B cells in vitro. (A) OSUCLL cells with doxorubicin-induced BRAFV600E expression were incubated 1:1 with normal human CD3 T cells labeled with carboxyfluorescein succinimidyl ester (CFSE) and stimulated with antibodies to CD3 and CD28. After a 4-day incubation, proliferation of CD4+ and CD8+ T cells was determined by CFSE flow cytometry. Proliferation caused dilution of the CFSE and is evidenced by the appearance of lower-intensity peaks. Data are representative of 3 separate experiments using T cells from different donors. Statistical analysis of division indices from the 3 independent experiments are included. (B) The experiment from panel A was repeated with or without dabrafenib (2 µM). Data are representative of 3 similar experiments using T cells from different donors. Statistical analysis of division indices from 3 independent experiments are included. (C) The experiment in panel A was repeated with the different cell types in the same or in opposite chambers of 0.4 µm transwell plates. In this experiment, CD4 and CD8 T cells were seeded separately. Data are representative of 3 separate experiments using T cells from different donors. Statistical analysis of division indices from 3 independent experiments are included. Vertical bars represent mean values, and error bars represent standard error of the mean. Stim., stimulated; Unstim., unstimulated.
These experiments were repeated using transwell plates in which OSUCLL cells and normal T cells were separated by a 0.4 µm filter. As shown in Figure 6C, OSUCLL cells still caused an impairment of T-cell proliferation but to a lesser extent, indicating that soluble factors as well as cell-cell contact contribute to T-cell inhibition. Inhibition of CD4 T-cell proliferation by OSUCLL-BRAFVE cells was greater in coculture than in transwell conditions (P = .013). This suggests a greater role for cell-cell contact in T-cell inhibition by OSUCLL-BRAFVE cells. To further interrogate candidate cell surface factors, OSUCLL cells were analyzed by flow cytometry for differences in expression of known immune suppressive molecules, including CD200 and PD-L1. In contrast to what has been reported in melanoma and acute myeloid leukemia cells,10,11 expression of BRAFV600E did not affect the expression of either of these on OSUCLL cells (data not shown). Next, levels of immunomodulatory cytokines produced by CD3/CD28-stimulated T cells (interleukin-2 [IL-2], IL-4, IL-6, IL-10, tumor necrosis factor [TNF], and interferon-γ [IFN-γ]) were measured. Each of these was strongly induced by anti-CD3/CD28 stimulation, and all except IL-2 were reduced in the presence of OSUCLL cells (supplemental Figure 6). However, these effects were not different in BRAFV600E cells compared with cells transfected with WT BRAF or an empty vector, suggesting these factors are not responsible for the increased inhibitory effect of OSUCLL-BRAFV600E cells on T-cell proliferation. In addition, blocking antibodies against IL-10 or TNF did not rescue T-cell proliferation in cocultures with OSUCLL-BRAFV600E cells (data not shown).
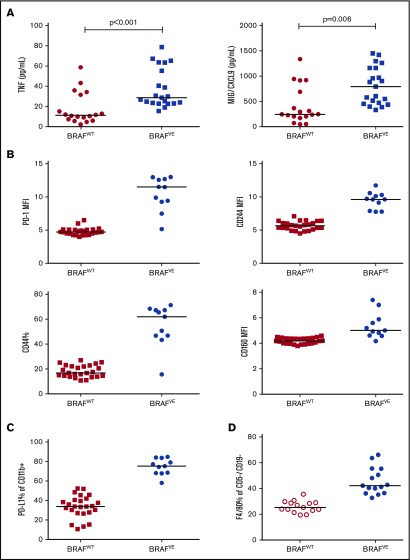
To identify candidate immune suppressive factors in vivo, normal mice were engrafted with spleen cells from leukemic BRAFVE or BRAFWT TCL1 mice. After mice achieved a substantial tumor burden in the periphery, plasma from these animals was analyzed by a multiplex cytokine assay (BRAFWT mice, n = 18; BRAFVE mice, n = 21). Although the level of most analyzed cytokines was similar (IL-1α, IL-1β, IL-2, IL-5, IL-12α, IL-12β, IFN-γ, MCP1/CCL2, MIP2/CXCL2, and VEGF; data not shown), TNF and MIG/CXCL9 were significantly increased in BRAFVE-transplanted mice relative to disease-matched BRAFWT-transplanted mice (Figure 7A). These results were supported by real-time reverse transcription polymerase chain reaction, suggesting that these increases are the result of a transcriptional mechanism rather than just increased secretion (data not shown).
Figure 7.
Immunosuppressive effect of BRAFV600Ein vivo. (A) Plasma from leukemic mice was collected at the point when each mouse exceeded 55% tumor cells in the peripheral blood CD45+ population and was evaluated by using a Milliplex cytokine assay (n = 18 for BRAFWT and n = 21 for BRAFVE). (B) T-cell immune parameters in AT mice were assessed by flow cytometry (n = 25 for BRAFWT; n = 11 for BRAFVE; P < .01 for each). (C) PD-L1 expression on CD11b+ cells (monocytes/macrophages) was assessed in peripheral blood from AT mice by flow cytometry (n = 25 for BRAFWT; n = 11 for BRAFVE; P < .001). (D) The percentage of F4/80+ cells (macrophages) was measured in the nonlymphocyte (CD5–/CD19–) population (n = 15 for each; P = .002). Horizontal lines indicate median. AT, adoptive transfer.
To investigate the effect of BRAFVE leukemic B cells on T cells in vivo, mice were engrafted with leukemia cells from each genotype and monitored until the onset of leukemia. After 3 weeks, peripheral blood T cells were examined by flow cytometry. Relative to BRAFWT leukemia cells, BRAFVE leukemic B cells induced the T-cell activation/exhaustion markers programmed cell death protein 1 (PD-1), CD244, and CD160 (n = 25 and n = 11 for BRAFWT and BRAFVE, respectively; P < .01 for each; Figure 7B). We found no correlation of PD-1, CD44, and CD244 increases with disease load, although there was a minor correlation of CD160 with disease load. In addition, BRAFVE leukemic B cells did not alter the CD4:CD8 ratio but significantly increased the population of CD44-expressing (antigen-experienced) T cells.40 These results demonstrate that BRAF-mutated leukemia cells generally produce a more immune-suppressive T-cell phenotype in vivo compared with WT BRAF leukemia cells.
BRAFVE B-cell leukemia impacts myeloid cells
As previously reported, CLL-associated myeloid cells are aberrant in the Eμ-TCL1 model and show skewing toward a patrolling phenotype, with enhanced PD-L1 expression and secretion of inflammatory and immunosuppressive cytokines.41 The impact of the BRAFVE mutation on the myeloid compartment was therefore evaluated, again using the adoptive transfer model. Mice were engrafted with either BRAFWT or BRAFVE leukemia cells, and upon achieving disease, spleens were evaluated by flow cytometry. Interestingly, the presence of BRAFVE leukemia cells resulted in significant increases in PD-L1 expression on peripheral myeloid (CD11b+) cells (P < .001; Figure 7C). In addition, there was an increase in F4/80+ macrophages (19.8%; P = .002; n = 15 each; Figure 7D). However, in another trial of a smaller cohort (BRAFWT, n = 8; BRAFVE, n = 11), changes were observed only in a subset, and overall differences were not significant (supplemental Figure 7A). In 2 separate experiments, there was a trend toward increased CD11b+Ly6CintLy6Ghi MDSCs and decreased CD11b+Ly6CloLy6Glo patrolling monocytes in spleens of mice with BRAFVE leukemia, although these differences did not reach statistical significance (supplemental Figure 7B). Together, these results suggest that in addition to T cells, myeloid cells may also be affected by BRAFVE, mutated cells to result in additional remodeling of the leukemia microenvironment.
Discussion
BRAF-activating mutations, particularly BRAFV600E, have been most commonly described in solid tumors including melanoma. Such mutations are less frequent across hematologic diseases, with the notable exceptions of HCL,16 Langerhans cell histiocytosis,42 and Erdheim-Chester disease.43 However, the numbers of BRAF-mutant leukemias, lymphomas, and myelomas identified are increasing because of broader and deeper sequencing efforts.18,19,44,45 Besides increased growth and decreased apoptosis, an established effect of aberrantly activated MAPK in tumor cells is enhanced immune suppression, mediated by multiple factors, including immune checkpoint molecules and cytokines.46,47 In melanoma models in vitro, selective BRAFV600E inhibition improves T-cell recognition of tumors.14 In a mouse xenograft melanoma model47 and in patients with metastatic melanoma,48,49 selective BRAF inhibitor therapy leads to improved CD8+ T-cell tumor infiltration; decreases in immunosuppressive cytokines such as IL-6, IL-8, and VEGF; and increased levels of activation/exhaustion markers such as PD-1 and TIM3.48 However, the role of BRAF-activating mutations in hematologic malignancies and how these impact immune function are unclear, and few mouse models are available to evaluate this.
Here, we generated a novel mouse strain in which BRAFV600E is selectively expressed in B cells of the established Eµ-TCL1 B-cell leukemia mouse model and investigated the pathologic role of this mutation in B-cell leukemia development, behavior, and immune evasion/suppression in vivo. The Eµ-TCL1 model was selected because of its extensive prior characterization, including studies that demonstrated its utility in investigating tumor-induced immune defects.50-52 Our results demonstrate that BRAFV600E accelerates Eµ-TCL1 B-cell leukemia development, likely because of a combination of decreased spontaneous apoptosis and enhanced immune suppression rather than increased proliferation. In addition, BRAFVE leukemia cells produced greater T-cell effects than BRAFWT leukemia cells, including an enhanced activated/exhausted phenotype that was previously described in CLL patients.53 Myeloid subsets were affected as well, with increased expression of PD-L1 on circulating monocytes and an elevated percentage of F4/80+ macrophages in spleens. Alterations in monocytic subsets were also observed, with a shift from a patrolling to an MDSC immunophenotype in contrast to what has been reported in Eµ-TCL1 mice.41 Depletion of myeloid cells, including patrolling monocytes in the Eµ-TCL1 adoptive transfer model, controls CLL-like disease development,41 but the patrolling monocytes also have antitumor function for suppressing tumor metastasis to lung by increasing natural killer cell-mediated killing of metastatic tumor cells in multiple metastatic mouse models.54 Although the function of patrolling cells in the cancer context is controversial, the observation of the affected myeloid cells in our mouse model indicates that BRAFV600E leukemia cells can skew these cells to an immune suppressive phenotype. Although functional analyses will be required to fully define these populations, our observations suggest a tumor-mediated shift of innate and adaptive immunity in BRAFVE mice toward a more tumor-tolerant environment.
Selective inhibitors of BRAFV600E have clear clinical benefit in BRAF-mutated cancers, but duration of response is typically short. Thus, efforts are now directed toward combination strategies with additional kinase inhibitors (eg, trametinib) as well as immune checkpoint inhibitors and other immune-based therapies, based on preclinical studies demonstrating activity.55,56 Clinical trials of such combinations are underway, but to evaluate these systematically in hematologic malignancies in which BRAF mutations are less common will be a lengthy process. A relevant model of BRAF-mutated leukemia will be a valuable tool not only to understand the mechanism of immune modulation but to quickly evaluate various combinations to support clinical trial design.
We investigated several factors that might mediate the enhanced immune suppressive effects of BRAFVE-mutated leukemia cells. In agreement with previous reports in solid tumors,57 PD-L1 and CD200 are not further induced in BRAFVE cells compared with BRAFWT cells, despite reports showing that these markers are increased in BRAF inhibitor-resistant cells.13,57 However, we detected increased IL-10 and TNF in a CLL cell line with mutated BRAF, which is in line with the TGF-β–induced IL-10 in the melanoma line A375 with endogenous expression of BRAFV600E 12. Although blocking antibodies to these cytokines failed to reverse T-cell inhibitory effects of BRAFVE-mutant cells in vitro and IL-10 was not changed in vivo, the potential contributions of TNF and other candidate cytokines in vivo remain to be fully evaluated. TNF transcript is increased in mouse BRAFVE leukemia cells, and adoptively transferred BRAFVE leukemia cells increase TNF in recipient mice. TNF exerts both protumor and antitumor activities in different cancers. In CLL, TNF supports proliferation of CLL cells and induces IL-6 expression to protect from spontaneous apoptosis.58,59 Furthermore, we find that MIG/CXCL9 in recipient mice is increased in the presence of BRAFVE leukemia. MIG/CXCL9, a chemoattractant for activated T cells, is secreted by various cell types, including T cells, natural killer cells, dendritic cells, macrophages, and endothelial cells. Similar to TNF, the function of MIG/CXCL9 in tumor regulation is controversial across different cancer types.60 MIG/CXCL9 level is higher in CLL patient sera and is associated with CD38 expression.61 In addition, a high level of the cytokine cluster CL1, including MIG/CXCL9, is correlated with shorter overall survival,61 suggesting that a high level of MIG/CXCL9 might predict a poor prognosis in CLL. The expression of MIG/CXCL9 receptor CXCR3 in CLL and splenic marginal zone lymphoma is higher compared with normal circulating B cells and mediates chemotaxis to support CLL survival and migration.62-64
It has been reported that leukemia B cells induce T-cell immune suppression in Eμ-TCL1 mice by changes in expression of cytokines and immune checkpoint molecules.50 For example, increase in IL-1, IL-4, IL-6, and CTLA4+ regulatory T cells and decrease in IL-2, IFN-γ, IL-12β, and BTLA are seen in the Eμ-TCL1 mice compared with WT control mice.50 IL-10, MIG/CXCL9, IL-1β, CXCL10, TNF, IL-1RN, and CXCL16 transcripts and cytokine levels are significantly increased in CLL-associated splenic monocytes and in the adoptive-transferred TCL1 mice, and elevated TNF, CXCL9, and CXCL16 are reduced upon depletion of myeloid cells.41 Along with this, percentage of CD44+CXCR3+ splenic T cells are increased in the TCL1 mice, indicating that the monocyte-associated cytokines attract these T cells and may contribute to CLL-like disease progression.41 Along with this, however, there is increasing data supporting that various myeloid-derived cells contribute to CLL disease progression indirectly via their effects on T cells.41 Our results are consistent with this hypothesis, in that BRAFV600E-mutant CLL cells might secrete TNF and CXCL9 not only to support survival but also to recruit T cells and macrophages to lymphoid organs and alter their immune function. Whether the immune suppressive effects seen on T cells in the BRAFV600E-mutant leukemia mouse model are produced directly by CLL cells and/or indirectly by CLL-enhanced immunosuppressive myeloid cells remains to be determined. Our data suggest that elevated levels of TNF and MIG/CXCL9 in BRAFVE Eμ-TCL1 mice could contribute to the more aggressive disease and immune evasion. Although additional studies are needed to support this, our experiments indicate that BRAFVE leukemia cells mediate suppression simultaneously through several factors, both soluble and contact-dependent, suggesting therapeutic combinations of agents that block these different pathways are needed for optimal disease control.
The phenotype of the disease at earlier stages is similar in BRAFVE and BRAFWT TCL1 mice, with progressive accumulation of abnormal B lymphocytes in the organs and periphery. However, the leukemia in most of the BRAFVE mice (19 of the 22 examined for histology) transforms to a histiocyte-associated B-cell lymphoma compared with just 2 of the 20 BRAFWT leukemic mice examined for histology. This intriguing transformation was reproduced in healthy syngeneic mice receiving BRAFVE leukemia cells by adoptive transfer, indicating that these effects are the result of an intrinsic property of the tumor cells. This progression to a lymphoma-type disease may in part explain the finding that, despite the relative frequency of MAPK-activating mutations in CLL (∼9%), BRAFV600E mutations are quite rare.65 It is possible that the greater activity of BRAFV600E vs other pathway mutations (even within BRAF) produces a disease phenotype inconsistent with CLL, and such cases are thus excluded from the cohort being studied.
In conclusion, the introduction of BRAFV600E into B-cell leukemia in vivo provides a novel and valuable tool to investigate the impact of this mutation on immune function in disseminated disease, building on the extensive work conducted in melanoma models. Current studies are investigating the impact of BRAF inhibition in these mice on the immune microenvironment. Our goal is to use this model to test novel combination strategies that will simultaneously block tumor growth and restore immune function for long-term disease control.
Supplementary Material
The full-text version of this article contains a data supplement.
Acknowledgments
The authors thank the members of the Byrd and Grever Laboratories for experimental assistance and Carlo Croce for providing Eµ-TCL1 mice.
This work was supported by a Pelotonia fellowship from The Ohio State University (OSU) Comprehensive Cancer Center (Y.-T.T.), by an American Association for Cancer Research–Amgen Fellowship in Clinical/Translational Cancer Research (16-40-11-LAKS) (A. Lakshmanan), by the National Institutes of Health, National Cancer Institute grants P50 CA140158, P30 CA016058, and R35 CA197734, and by the OSU Department of Internal Medicine.
Authorship
Contribution: Y.-T.T. and A. Lakshmanan performed experiments, analyzed data, and wrote the paper; A. Lehman conducted biostatistical analyses; B.K.H., A.D.F., F.J., and K.L.P. performed and interpreted histopathology; F.M.L. and M.L. assisted in designing, running, and interpreting flow cytometry studies; M.T. maintained mouse colonies and assisted with mouse experiments; E.J.S. performed experiments and analyzed data; V.C. and N.M. provided expertise on design and characterization of mouse strains; G.L. interpreted histopathology and conducted flow cytometric examinations; J.C.B., M.R.G., and D.M.L. provided the concept, analyzed data, and oversaw the project; and B.K.H. and D.M.L. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: David M. Lucas, The Ohio State University, 320 West 10th Ave, Room A350B, Columbus, OH 43210; e-mail: david.lucas@osumc.edu.
REFERENCES
- 1.Mandal R, Becker S, Strebhardt K. Stamping out RAF and MEK1/2 to inhibit the ERK1/2 pathway: an emerging threat to anticancer therapy. Oncogene. 2016;35(20):2547-2561. [DOI] [PubMed] [Google Scholar]
- 2.Flaherty KT, Puzanov I, Kim KB, et al. . Inhibition of mutated, activated BRAF in metastatic melanoma. N Engl J Med. 2010;363(9):809-819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chapman PB, Hauschild A, Robert C, et al. ; BRIM-3 Study Group. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364(26):2507-2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Flaherty KT, Robert C, Hersey P, et al. ; METRIC Study Group. Improved survival with MEK inhibition in BRAF-mutated melanoma. N Engl J Med. 2012;367(2):107-114. [DOI] [PubMed] [Google Scholar]
- 5.Long GV, Stroyakovskiy D, Gogas H, et al. . Combined BRAF and MEK inhibition versus BRAF inhibition alone in melanoma. N Engl J Med. 2014;371(20):1877-1888. [DOI] [PubMed] [Google Scholar]
- 6.Robert C, Karaszewska B, Schachter J, et al. . Improved overall survival in melanoma with combined dabrafenib and trametinib. N Engl J Med. 2015;372(1):30-39. [DOI] [PubMed] [Google Scholar]
- 7.Akira S, Takeda K. Toll-like receptor signalling. Nat Rev Immunol. 2004;4(7):499-511. [DOI] [PubMed] [Google Scholar]
- 8.Lee MS, Kim YJ. Signaling pathways downstream of pattern-recognition receptors and their cross talk. Annu Rev Biochem. 2007;76(1):447-480. [DOI] [PubMed] [Google Scholar]
- 9.Zhang E, Lu M. Toll-like receptor (TLR)-mediated innate immune responses in the control of hepatitis B virus (HBV) infection. Med Microbiol Immunol (Berl). 2015;204(1):11-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Petermann KB, Rozenberg GI, Zedek D, et al. . CD200 is induced by ERK and is a potential therapeutic target in melanoma. J Clin Invest. 2007;117(12):3922-3929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berthon C, Driss V, Liu J, et al. . In acute myeloid leukemia, B7-H1 (PD-L1) protection of blasts from cytotoxic T cells is induced by TLR ligands and interferon-gamma and can be reversed using MEK inhibitors. Cancer Immunol Immunother. 2010;59(12):1839-1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Díaz-Valdés N, Basagoiti M, Dotor J, et al. . Induction of monocyte chemoattractant protein-1 and interleukin-10 by TGFbeta1 in melanoma enhances tumor infiltration and immunosuppression. Cancer Res. 2011;71(3):812-821. [DOI] [PubMed] [Google Scholar]
- 13.Jiang X, Zhou J, Giobbie-Hurder A, Wargo J, Hodi FS. The activation of MAPK in melanoma cells resistant to BRAF inhibition promotes PD-L1 expression that is reversible by MEK and PI3K inhibition. Clin Cancer Res. 2013;19(3):598-609. [DOI] [PubMed] [Google Scholar]
- 14.Boni A, Cogdill AP, Dang P, et al. . Selective BRAFV600E inhibition enhances T-cell recognition of melanoma without affecting lymphocyte function. Cancer Res. 2010;70(13):5213-5219. [DOI] [PubMed] [Google Scholar]
- 15.Ott PA, Henry T, Baranda SJ, et al. . Inhibition of both BRAF and MEK in BRAF(V600E) mutant melanoma restores compromised dendritic cell (DC) function while having differential direct effects on DC properties. Cancer Immunol Immunother. 2013;62(4):811-822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tiacci E, Trifonov V, Schiavoni G, et al. . BRAF mutations in hairy-cell leukemia. N Engl J Med. 2011;364(24):2305-2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dietrich S, Glimm H, Andrulis M, von Kalle C, Ho AD, Zenz T. BRAF inhibition in refractory hairy-cell leukemia. N Engl J Med. 2012;366(21):2038-2040. [DOI] [PubMed] [Google Scholar]
- 18.Zhang X, Reis M, Khoriaty R, et al. . Sequence analysis of 515 kinase genes in chronic lymphocytic leukemia. Leukemia. 2011;25(12):1908-1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jebaraj BM, Kienle D, Bühler A, et al. . BRAF mutations in chronic lymphocytic leukemia. Leuk Lymphoma. 2013;54(6):1177-1182. [DOI] [PubMed] [Google Scholar]
- 20.Kanagal-Shamanna R, Luthra R, Luthra M, et al. . RAS/RAF pathway mutations define a distinct subset of chronic lymphocytic leukemia [abstract]. Blood. 2015;126(23). Abstract 1730. [Google Scholar]
- 21.Damm F, Mylonas E, Cosson A, et al. . Acquired initiating mutations in early hematopoietic cells of CLL patients. Cancer Discov. 2014;4(9):1088-1101. [DOI] [PubMed] [Google Scholar]
- 22.Mey UJ, Renner C, von Moos R. Vemurafenib in combination with cobimetinib in relapsed and refractory extramedullary multiple myeloma harboring the BRAF V600E mutation. Hematol Oncol. 2016. [DOI] [PubMed] [Google Scholar]
- 23.Karreth F, Reschke M, Chapuy B, Shipp MA, Chiarle R, Pandolfi PP. The BRAF pseudogene is a proto-oncogenic competitive endogenous RNA [abstract]. Blood. 2014;124(21). Abstract 263. [Google Scholar]
- 24.Bichi R, Shinton SA, Martin ES, et al. . Human chronic lymphocytic leukemia modeled in mouse by targeted TCL1 expression. Proc Natl Acad Sci USA. 2002;99(10):6955-6960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yan XJ, Albesiano E, Zanesi N, et al. . B cell receptors in TCL1 transgenic mice resemble those of aggressive, treatment-resistant human chronic lymphocytic leukemia. Proc Natl Acad Sci USA. 2006;103(31):11713-11718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pekarsky Y, Palamarchuk A, Maximov V, et al. . Tcl1 functions as a transcriptional regulator and is directly involved in the pathogenesis of CLL. Proc Natl Acad Sci USA. 2008;105(50):19643-19648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Herling M, Patel KA, Weit N, et al. . High TCL1 levels are a marker of B-cell receptor pathway responsiveness and adverse outcome in chronic lymphocytic leukemia. Blood. 2009;114(21):4675-4686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hamblin TJ. The TCL1 mouse as a model for chronic lymphocytic leukemia. Leuk Res. 2010;34(2):135-136. [DOI] [PubMed] [Google Scholar]
- 29.Hertlein E, Beckwith KA, Lozanski G, et al. . Characterization of a new chronic lymphocytic leukemia cell line for mechanistic in vitro and in vivo studies relevant to disease. PLoS One. 2013;8(10):e76607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsai YT, Lozanski G, Lehman A, et al. . BRAFV600E induces ABCB1/P-glycoprotein expression and drug resistance in B-cells via AP-1 activation [published online ahead of print 5 September 2015]. Leuk Res. doi:10.1016/j.leukres.2015.08.017. [DOI] [PMC free article] [PubMed]
- 31.Roederer M. Interpretation of cellular proliferation data: avoid the panglossian. Cytometry A. 2011;79(2):95-101. [DOI] [PubMed] [Google Scholar]
- 32.Enzler T, Kater AP, Zhang W, et al. . Chronic lymphocytic leukemia of Emu-TCL1 transgenic mice undergoes rapid cell turnover that can be offset by extrinsic CD257 to accelerate disease progression. Blood. 2009;114(20):4469-4476. [DOI] [PubMed] [Google Scholar]
- 33.Dankort D, Filenova E, Collado M, Serrano M, Jones K, McMahon M. A new mouse model to explore the initiation, progression, and therapy of BRAFV600E-induced lung tumors. Genes Dev. 2007;21(4):379-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chung SS, Kim E, Park JH, et al. . Hematopoietic stem cell origin of BRAFV600E mutations in hairy cell leukemia. Sci Transl Med. 2014;6(238):238ra71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mauri C, Bosma A. Immune regulatory function of B cells. Annu Rev Immunol. 2012;30(1):221-241. [DOI] [PubMed] [Google Scholar]
- 36.Serasinghe MN, Missert DJ, Asciolla JJ, et al. . Anti-apoptotic BCL-2 proteins govern cellular outcome following B-RAF(V600E) inhibition and can be targeted to reduce resistance. Oncogene. 2015;34(7):857-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ilieva KM, Correa I, Josephs DH, et al. . Effects of BRAF mutations and BRAF inhibition on immune responses to melanoma. Mol Cancer Ther. 2014;13(12):2769-2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Comin-Anduix B, Chodon T, Sazegar H, et al. . The oncogenic BRAF kinase inhibitor PLX4032/RG7204 does not affect the viability or function of human lymphocytes across a wide range of concentrations. Clin Cancer Res. 2010;16(24):6040-6048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schilling B, Sondermann W, Zhao F, et al. ; DeCOG. Differential influence of vemurafenib and dabrafenib on patients’ lymphocytes despite similar clinical efficacy in melanoma. Ann Oncol. 2014;25(3):747-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hofbauer JP, Heyder C, Denk U, et al. . Development of CLL in the TCL1 transgenic mouse model is associated with severe skewing of the T-cell compartment homologous to human CLL. Leukemia. 2011;25(9):1452-1458. [DOI] [PubMed] [Google Scholar]
- 41.Hanna BS, McClanahan F, Yazdanparast H, et al. . Depletion of CLL-associated patrolling monocytes and macrophages controls disease development and repairs immune dysfunction in vivo. Leukemia. 2016;30(3):570-579. [DOI] [PubMed] [Google Scholar]
- 42.Badalian-Very G, Vergilio JA, Degar BA, et al. . Recurrent BRAF mutations in Langerhans cell histiocytosis. Blood. 2010;116(11):1919-1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Haroche J, Charlotte F, Arnaud L, et al. . High prevalence of BRAF V600E mutations in Erdheim-Chester disease but not in other non-Langerhans cell histiocytoses. Blood. 2012;120(13):2700-2703. [DOI] [PubMed] [Google Scholar]
- 44.Chapman MA, Lawrence MS, Keats JJ, et al. . Initial genome sequencing and analysis of multiple myeloma. Nature. 2011;471(7339):467-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Langabeer SE, Quinn F, O’Brien D, et al. . Incidence of the BRAF V600E mutation in chronic lymphocytic leukaemia and prolymphocytic leukaemia. Leuk Res. 2012;36(4):483-484. [DOI] [PubMed] [Google Scholar]
- 46.Khalili JS, Liu S, Rodríguez-Cruz TG, et al. . Oncogenic BRAF(V600E) promotes stromal cell-mediated immunosuppression via induction of interleukin-1 in melanoma. Clin Cancer Res. 2012;18(19):5329-5340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu C, Peng W, Xu C, et al. . BRAF inhibition increases tumor infiltration by T cells and enhances the antitumor activity of adoptive immunotherapy in mice. Clin Cancer Res. 2013;19(2):393-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Frederick DT, Piris A, Cogdill AP, et al. . BRAF inhibition is associated with enhanced melanoma antigen expression and a more favorable tumor microenvironment in patients with metastatic melanoma. Clin Cancer Res. 2013;19(5):1225-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wilmott JS, Long GV, Howle JR, et al. . Selective BRAF inhibitors induce marked T-cell infiltration into human metastatic melanoma. Clin Cancer Res. 2012;18(5):1386-1394. [DOI] [PubMed] [Google Scholar]
- 50.Gorgun G, Ramsay AG, Holderried TA, et al. . E(mu)-TCL1 mice represent a model for immunotherapeutic reversal of chronic lymphocytic leukemia-induced T-cell dysfunction. Proc Natl Acad Sci USA. 2009;106(15):6250-6255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McClanahan F, Hanna B, Miller S, et al. . PD-L1 checkpoint blockade prevents immune dysfunction and leukemia development in a mouse model of chronic lymphocytic leukemia. Blood. 2015;126(2):203-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McClanahan F, Riches JC, Miller S, et al. . Mechanisms of PD-L1/PD-1-mediated CD8 T-cell dysfunction in the context of aging-related immune defects in the Eμ-TCL1 CLL mouse model. Blood. 2015;126(2):212-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Riches JC, Davies JK, McClanahan F, et al. . T cells from CLL patients exhibit features of T-cell exhaustion but retain capacity for cytokine production. Blood. 2013;121(9):1612-1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hanna RN, Cekic C, Sag D, et al. . Patrolling monocytes control tumor metastasis to the lung. Science. 2015;350(6263):985-990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Homet Moreno B, Mok S, Comin-Anduix B, Hu-Lieskovan S, Ribas A. Combined treatment with dabrafenib and trametinib with immune-stimulating antibodies for BRAF mutant melanoma. OncoImmunology. 2015;5(7):e1052212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brauner E, Gunda V, Vanden Borre P, et al. . Combining BRAF inhibitor and anti PD-L1 antibody dramatically improves tumor regression and anti tumor immunity in an immunocompetent murine model of anaplastic thyroid cancer. Oncotarget. 2016;7(13):17194-17211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Atefi M, Avramis E, Lassen A, et al. . Effects of MAPK and PI3K pathways on PD-L1 expression in melanoma. Clin Cancer Res. 2014;20(13):3446-3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Reittie JE, Yong KL, Panayiotidis P, Hoffbrand AV. Interleukin-6 inhibits apoptosis and tumour necrosis factor induced proliferation of B-chronic lymphocytic leukaemia. Leuk Lymphoma. 1996;22(1-2):83-90. [DOI] [PubMed] [Google Scholar]
- 59.Aderka D, Maor Y, Novick D, et al. . Interleukin-6 inhibits the proliferation of B-chronic lymphocytic leukemia cells that is induced by tumor necrosis factor-alpha or -beta. Blood. 1993;81(8):2076-2084. [PubMed] [Google Scholar]
- 60.Ding Q, Lu P, Xia Y, et al. . CXCL9: evidence and contradictions for its role in tumor progression. Cancer Med. 2016;5(11):3246-3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yan XJ, Dozmorov I, Li W, et al. . Identification of outcome-correlated cytokine clusters in chronic lymphocytic leukemia. Blood. 2011;118(19):5201-5210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jones D, Benjamin RJ, Shahsafaei A, Dorfman DM. The chemokine receptor CXCR3 is expressed in a subset of B-cell lymphomas and is a marker of B-cell chronic lymphocytic leukemia. Blood. 2000;95(2):627-632. [PubMed] [Google Scholar]
- 63.Trentin L, Agostini C, Facco M, et al. . The chemokine receptor CXCR3 is expressed on malignant B cells and mediates chemotaxis. J Clin Invest. 1999;104(1):115-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mahadevan D, Choi J, Cooke L, et al. Gene expression and serum cytokine profiling of low stage CLL identify WNT/PCP, Flt-3L/Flt-3 and CXCL9/CXCR3 as regulators of cell proliferation, survival and migration. Hum Genomics Proteomics 2009;2009:453634. [DOI] [PMC free article] [PubMed]
- 65.Landau DA, Tausch E, Taylor-Weiner AN, et al. . Mutations driving CLL and their evolution in progression and relapse. Nature. 2015;526(7574):525-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.