Abstract
Proline utilization (Put) systems have been described in a number of bacteria; however, the importance and functionality of the Put system in the intracellular pathogen Brucellaabortus has not been explored. Generally, bacterial Put systems are composed of the bifunctional enzyme proline dehydrogenase PutA and its transcriptional activator PutR. Here, we demonstrate that the genes putA (bab2_0518) and putR (bab2_0517) are critical for the chronic infection of mice by B. abortus, but putA and putR are not required for the survival and replication of the bacteria in naive macrophages. Additionally, in vitro experiments revealed that putR is necessary for the ability of the bacteria to withstand oxidative stress, as a ΔputR deletion strain is hypersensitive to hydrogen peroxide exposure. Quantitative reverse transcription-PCR and putA-lacZ transcriptional reporter studies revealed that PutR acts as a transcriptional activator of putA in Brucella, and electrophoretic mobility shift assays confirmed that PutR binds directly to the putA promoter region. Biochemical analyses demonstrated that a purified recombinant B. abortus PutA protein possesses quintessential proline dehydrogenase activity, as PutA is capable of catalysing the conversion of proline to glutamate. Altogether, these data are the first to reveal that the Put system plays a significant role in the ability of B. abortus to replicate and survive within its host, as well as to describe the genetic regulation and biochemical activity of the Put system in Brucella.
Keywords: Brucella virulence, PutR, PutA, α-proteobacteria
Abbreviations
BCV, Brucella-containing vacuole; CoQ1, coenzyme Q1; EMSA, electrophoretic mobility shift assay; MTT, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide; PMS, 5-methylphenazinium methyl sulfate; RT-PCR, reverse transcription PCR.
Introduction
Brucella spp. are intracellular pathogens primarily residing within host macrophages and dendritic cells, and are able to establish replication cycles in a bacterially modified, host-derived vacuole [1]. This vacuole, termed the Brucella-containing vacuole (BCV), aids Brucella in evading the host immune system and leads to brucellosis, a chronic disease characterized most prominently by a waning and waxing fever in humans. Brucella infections in livestock are a major cause of economic losses from Brucella-induced abortions and sterility, and in humans brucellosis is a leading zoonotic disease globally [2]. While Brucella spp. have evolved numerous strategies to cope with host-generated stresses during infection, many of the genetic mechanisms employed by Brucella to cause disease are still poorly characterized [3]. Given the essential roles that amino acids play in bacterial physiology and pathogenesis [4–8], our group became interested in defining and characterizing the mechanisms of amino acid acquisition and utilization in the brucellae.
In culture, Brucella abortus is capable of surviving by utilizing glutamate as the only amino acid available to serve as a source of energy, carbon and nitrogen [9]. Furthermore, it is presumed that the brucellae possess de novo biosynthetic pathways for the 20 standard proteinogenic amino acids. Disruption of several different amino acid metabolic pathways and transport systems results in the attenuation of B. abortus, Brucella melitensis and Brucella suis in cellular and animal models of infection [10]. In addition, a number of genes linked to nitrogen utilization, including glnA, glnD and nrII, are essential for virulent infection, leading to the suggestion that there is a relative paucity of nitrogen and carbon sources available in the BCV, and that efficient use of resources is necessary for Brucella to colonize the host [11]. Amino acids are a likely source of the essential carbon and nitrogen that the bacteria require while in the host, but to date, little is known about specific amino acids that may be available to and utilized by the brucellae during infection.
In several bacteria, a proline utilization (Put) system is responsible for the enzymatically driven conversion of proline to glutamate, and the Put system is composed of two prominent proteins: PutA and PutR [12]. The putA gene encodes the proline utilization protein PutA, which catalyses the conversion of proline to glutamate through two sequential enzymatic steps [13], while the putR gene encodes an Lrp-family transcriptional regulatory protein that controls the expression of putA [14, 15]. In the α-proteobacteria, putR and putA are often transcribed divergently from one another, and PutR is normally a transcriptional activator of putA expression that binds directly to the putA promoter region to control gene expression [15–18]. While the Put system has been well described in a number of bacterial species, there is currently no information available about a Put system in Brucella spp.
In the present work, we characterize the Put system of B. abortus, and demonstrate that isogenic deletion strains of putA and putR are attenuated in a mouse model of infection, while deletion of putR renders the bacteria more sensitive to oxidative stress compared to the parental strain. Furthermore, in B. abortus, PutR acts as a direct transcriptional activator of putA, and importantly, we demonstrate that PutA from B. abortus is a proline dehydrogenase that is capable of converting proline to glutamate. Overall, these data provide important information about the potential nutrients available to the brucellae during host colonization.
Methods
Bacterial strains and growth conditions
B. abortus 2308 and derivative strains were routinely grown on Schaedler blood agar (SBA), which is Schaedler agar (Acumedia Neogen) containing 5 % defibrinated bovine blood (Quad Five), or in Brucella broth (BD). For cloning and recombinant protein production, Escherichia coli strains (DH5α and BL21) were grown routinely on tryptic soy agar or in Luria–Bertani broth. When appropriate, the growth media were supplemented with kanamycin (45 µg ml−1) or carbenicillin (100 µg ml−1; E. coli strains only).
Construction of ΔputR and Δ putA deletion strains, and reconstruction of ΔputR
The putR locus (bab2_0517; bab_rs28810) in B. abortus 2308 was deleted using a non-polar, unmarked gene excision strategy as described previously [19]. An approximately 1 kb fragment representing the region upstream of the gene extending to the second codon of the coding region was amplified by PCR using the primers ΔputR-Up-For and ΔputR-Up-Rev (Table 1), genomic DNA from B. abortus 2308 as a template, and Pfx polymerase (Invitrogen). Similarly, a fragment containing the last two codons of the coding region extending to approximately 1 kb downstream of putR was amplified with the primers ΔputR-Down-For and ΔputR-Down-Rev (Table 1). The upstream fragment was digested with BamHI, while the downstream fragment was digested with PstI, and both fragments were treated with polynucleotide kinase in the presence of ATP. Both of the DNA fragments were included in a single ligation mix with BamHI/PstI-digested pNPTS138 (M. R. K. Alley, unpublished), which contains a kanamycin resistance marker and sacB gene for counter-selection with sucrose. The resulting plasmid (pC3040) was introduced into B. abortus 2308, and merodiploid transformants were obtained by selection on SBA + kanamycin (Table 2). A single kanamycin-resistant clone was grown for ~6 h in Brucella broth and then plated onto SBA containing 10 % sucrose. Genomic DNA from sucrose-resistant, kanamycin-sensitive colonies was isolated and screened by PCR for loss of the putR gene, and an isogenic putR mutant derived from B. abortus 2308 was named CC129. The putR mutation in this strain was verified by DNA sequence analysis and Southern hybridization.
Table 1. Oligonucleotide primers used in this study.
| Designation | Sequence (5′→3′) |
|---|---|
| Δ putR-Up-For | GCGGATCCCTGCCCGCCTGTCGCAA |
| Δ putR-Up-Rev | GTCAGGGTCTATCCGGGCGC |
| Δ putR-Down-For | TCGCATTCTATAAGCCTTCATAGGA |
| Δ putR-Down-Rev | GCCTGCAGCAGAAGTTCACCCATGAC |
| Δ put A-Up-Rev | GGTGCACTGCTCACAAAAAG |
| Δ putA-DN-For | TTTCTTCGCATATGTGAGGCAG |
| Δ putA-DN-Rev | CTGCAGCCTTGATGATGGACATTGAGCC |
| Δ putA-UP-For | GGATCCGGGAGGAGGCACAACGGTAAG |
| Δ putA-Down-Rev 2 | TTCACAGGCGCAGAACCGGACTT |
| rPutR-For | GCGGTCTCAGCGCATGCGAAAGATA |
| rPutR-Rev | GCGGTCTCATATCATCACAACGCGCC |
| rPutA-For | GGATCCATGACCGATAATATTCCTGTT |
| rPutA-Rev | CTGCAGTCAGCCGATCGCCATCAGGCT |
| putA-RT-For | CTTACCCGCCTCATCGCC |
| putA-RT-For 2 | TGGCGCGGCTCATCCAGG |
| putA-RT-Rev | GCGGAAGCCGCGCTCTT |
| putA-RT-Rev 2 | CCGAAGCGATGACATCAAACACC |
| 16S rRNA-RT-For | TCTCACGACACGAGCTGACG |
| 16S rRNA-RT-Rev | CGCAGAACCTTACCAGCCCT |
| putR-A-EMSA-For | TCTATAAGCCTTCATAGGAGG |
| putR-A-EMSA-Rev | GGTGCACTGCTCACAAAA |
| putR-lacZ-For | TAGGATCCTCTATAAGCCTTCATAGGAGGTA |
| putR-lacZ-Rev | TAAAGCTTGGTGCACTGCTCACAAAAAGGAA |
| putA-lacZ-For | TAAAGCTTTCTATAAGCCTTCATAGGAGGTA |
| putA-lacZ-Rev | TAGGATCCGGTGCACTGCTCACAAAAAGGAA |
Underlined sections indicate restriction endonuclease recognition sequences.
Table 2. Plasmids used in this study.
| Plasmid name | Description | Reference |
|---|---|---|
| pNPTS138 | Cloning vector; contains sacB gene; KanR | (M. R. K. Alley, unpublished) |
| pMR15 | Broad-host range vector containing a promoterless lacZ gene; KanR | [20] |
| pC3040 | In-frame deletion of putR plus 1 kb of each flanking region in pNPTS138 | This study |
| pLS010 | In-frame deletion of putA plus 1 kb of each flanking region in pNPTS138 | This study |
| pLS011 | Intact putR gene plus 1 kb of each flanking region in pNPTS138 | This study |
| pLS019 | putR coding region in pASK-IBA7 | This study |
| prPutA | putA coding region in pASK-IBA7 | This study |
| pLS020 | putA-lacZ transcriptional fusion in pMR15 | This study |
| pLS021 | putR-lacZ transcriptional fusion in pMR15 | This study |
The putA locus (bab2_0518; bab_rs28815) was deleted using the strategy described above, but for the generation of the putA deletion construct, the primers ΔputA-Up-For and ΔputA-Up-Rev, and ΔputA-Down-For and ΔputR-Down-Rev were used to amplify the upstream and downstream flanking regions, respectively. The resulting plasmid (pLS010) was introduced into B. abortus 2308, and selection was performed as described above. The isogenic putA mutant derived from B. abortus 2308 was named LS014.
Genetic complementation of the putR was performed by reconstruction of the deleted loci. For this, the putR locus was amplified and isolated using the primers 0517-Dn-Rev and 0517-Up-For, digested using PstI and BamHI, and ligated and transformed in a sacB vector, as described above. The resulting complementation plasmid was named pLS011 and introduced into B. abortus 2308 using the previously described selection process. The resulting complemented putR strain was named LS015.
Construction of transcriptional putA-lacZ and putR-lacZ promoter fusions and β-galactosidase assays
The promoter region of the B. abortus 2308 putA gene was fused to a lacZ reporter as a transcriptional fusion. Approximately 170 bp of the putR–A intergenic region was amplified by PCR using the primers putA-lacZ-For and putA-lacZ-Rev (Table 1) and B. abortus 2308 genomic DNA as a template. For the putR-lacZ promoter fusion, the putR–A intergenic region was amplified using the primers putR-lacZ-For and putR-lacZ-Rev (Table 1). The amplified DNA fragments were sequentially digested with BamHI and HindIII and subsequently ligated into BamHI/HindIII-digested pMR15 [20], which contains a promoterless lacZ gene. The putA-lacZ promoter fusion plasmid (pLS020) or the putR-lacZ promoter fusion plasmid (pLS021) was then electroporated into B. abortus 2308 and its derivative strains. B. abortus strains harbouring the putA-lacZ or the putR-lacZ fusion construct were grown in Brucella broth, and β-galactosidase assays were performed as described previously [21].
Quantitative reverse transcription PCR (RT-PCR)
Quantitative RT-PCR was performed as described previously [19]. Briefly, total Brucella RNA was isolated as above and treated with RNase-free DNase I (Ambion) to remove genomic DNA. cDNA was generated from the final RNA preparation using the SuperScript III cDNA synthesis system (Invitrogen) following the manufacturer’s protocol, and this cDNA was used for real-time PCR, employing a SYBR green PCR supermix (Roche). For these experiments, primers for 16S RNA were used as a control, while gene-specific primers were used for evaluating relative mRNA levels (Table 1). The parameters for PCR included a single denaturing step for 5 min at 95 °C, followed by 40 cycles (denature for 15 s at 95 °C, anneal for 15 s at 50 °C and extend for 15 s at 72 °C) of amplification. Fluorescence from SYBR green incorporation into double-stranded DNA was measured with an iCycler machine (Bio-Rad), and the relative abundance of mRNA was determined using the Pfaffl equation [22].
Purification of recombinant PutR and PutA proteins
The Strep-tag II system (IBA) was used to produce recombinant Brucella PutR and PutA in E. coli strain BL21. The coding region of the putR gene (bab2_0517; bab_rs28810) was amplified using the primers rPutR-For and rPutR-Rev (Table 1), B. abortus 2308 chromosomal DNA as a template, and Taq polymerase (Monserate Biotechnology Group). The amplified DNA fragment was digested with BsaI and ligated into BsaI-digested pASK-IBA7, which encodes an amino-terminal Strep-tag II on the protein of interest, resulting in the plasmid prPutR. Similarly, the coding region of putA (bab2_0518; bab_rs28815) was amplified with the rPutA-For and rPutA-Rev primers (Table 1) from B. abortus 2308 genomic DNA and Phusion High-Fidelity DNA polymerase (New England). The fragment and pASK-IBA7 were digested with EcoRI and SalI and then ligated, creating plasmid prPutA.
The resulting plasmids, prPutR and prPutA, were each transformed into E. coli strain BL21, and the strains harbouring these respective plasmids were grown to an OD600 nm of approximately 0.6 before recombinant gene expression was induced by the addition of anhydrotetracycline (200 µg ml−1 final concentration). Following 3 h of incubation at 37 °C, the cells were collected by centrifugation (4200 g for 10 min at 4 °C) and lysed by treatment with cCelLytic B cell lysis reagent (Sigma) in the presence of the protease inhibitor phenylmethanesulfonylfluoride. The resulting lysed cell suspension was then passed through a French press at 600 p.s.i. and the supernatant from the suspension of lysed cells was collected by centrifugation (14 000 g for 10min at 4 °C). The collected supernatant was passed through an affinity column packed with Strep-Tactin sepharose. The column was washed extensively with buffer W (100 mM Tris-HCl and 150 mM NaCl), and the recombinant protein was eluted with 2.5 mM desthiobiotin in buffer W. The degree of purity of the recombinant PutR and PutA was high as judged by SDS-PAGE.
Electrophoretic mobility shift assays (EMSAs)
ESMAs with recombinant Brucella proteins were performed as described previously [23, 24]. All rPutR EMSA experiments were carried out in a 20 µl total reaction volume containing binding buffer composed of 10 mM Tris-HCl (pH 7.4), 50 mM KCl, 1 mM dithiothreitol, 6 % glycerol, 50 µg ml−1 bovine serum albumin and 50 µg ml−1 salmon sperm DNA. A DNA fragment including the putR–putA intergenic region was amplified by PCR from B. abortus 2308 chromosomal DNA using a specific primer set (Table 1). The amplified DNA fragment was purified by agarose gel electrophoresis, and the fragments were end-labelled with [γ-32P]ATP (PerkinElmer) and polynucleotide kinase (Monserate Biotechnology Group). Increasing amounts of recombinant PutR were mixed with the radiolabelled DNA fragments in binding buffer, and the reactions were incubated at room temperature for 20 min. As controls, ×50 molar concentrations of non-radiolabelled specific DNA (specific competitor) or non-radiolabelled non-specific DNA (i.e. abcR2 promoter) were added to some reactions. The binding reactions were subjected to electrophoresis on 6 % native polyacrylamide gels in 0.5× TBE running buffer for approximately 1 h. Following electrophoresis, gels were dried onto 3 mm Whatman paper using a vacuum gel drier system and visualized by autoradiography.
Assessment of the enzymatic activity of PutA
The l-proline dehydrogenase activity of rPutA was determined as described previously, but with modifications [25, 26]. The reaction was carried out at room temperature (22 °C) in a solution containing 50 mM potassium phosphate buffer (pH 7.5), 25 mM NaCl (pH 7.5), 200 µM NAD+, 240 µM coenzyme Q1 (CoQ1), 20 mM l-proline, 1 mM 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT), 1 mM 5-methylphenazinium methyl sulfate (PMS) and 0.072 µM rPutA. The reaction was initiated by the addition of rPutA and monitored spectrophotometrically at 568 nm. l-Proline dehydrogenase reaction generated NADH [26], which reduced PMS, which in turn reduced MTT, producing formazan and causing a rise in absorbance at 568 nm [25].
To confirm that l-glutamate was the reaction product of the rPutA reaction, the assay mixture was stored at 4 °C overnight and then centrifuged at 14 000 g for 5 min to remove the precipitated formazan. Subsequently, two units of l-glutamic dehydrogenase (Sigma-Aldrich) were added to the supernatant, and the reduction of NAD+ was monitored spectrophotometrically via the subsequent formation of formazan as described above.
Virulence studies
Experiments to test the intracellular survival and replication of Brucella strains in primary, peritoneal murine macrophages were carried out as described previously [27]. Briefly, resident peritoneal macrophages were isolated from BALB/c mice and seeded in 96-well plates in Dulbecco’s modified Eagle’s medium with 5 % foetal bovine serum, and the following day the macrophages were infected with opsonized brucellae at an m.o.i. of 50 : 1. After 2 h of infection, extracellular bacteria were killed by treatment with gentamicin (50 µg ml−1). In some wells, the macrophages were then lysed with 0.1 % deoxycholate in PBS, and serial dilutions were plated on SBA. In other wells, the macrophages were washed with PBS following gentamicin treatment, and fresh cell-culture medium containing gentamicin (20 µg ml−1) was added to the monolayer. After 24 and 48 h of infection with Brucella strains, the macrophages were lysed, and serial dilutions were plated on SBA. Triplicate wells were used for each Brucella strain tested.
The experimental methods for assessing the chronic infection of mice by Brucella strains were described previously [27]. BALB/c mice (five per Brucella strain per time point) were infected intraperitoneally with ~1×105 c.f.u. of each Brucella strain in sterile PBS. The mice were sacrificed at 1, 4 and 8 weeks post-infection, and serial dilutions of spleen homogenates were plated onto SBA.
Results
Brucella abortus Δ putR and ΔputA deletion strains are attenuated in a mouse model of infection
Given that little information is currently available about the role of the proline utilization system in Brucella spp., we sought to begin to define the function of putR and putA by assessing the requirement of these genes for Brucella virulence. Specifically, we tested the capacity of the deletion strains to survive and replicate in murine macrophages (Fig. 1a), and for the ability of the strains to colonize experimentally infected mice (Fig. 1b). Peritoneal-derived macrophages from BALB/c mice were infected with B. abortus 2308 or the ΔputR and ΔputA strains, and the number of viable intracellular brucellae was determined at 2, 24 and 48 h post-infection. All of the strains exhibited similar levels of intracellular bacteria at all of the tested time points, indicating that PutR and PutA are dispensable for the ability of B. abortus to survive and replicate in naive murine macrophages.
Fig. 1.
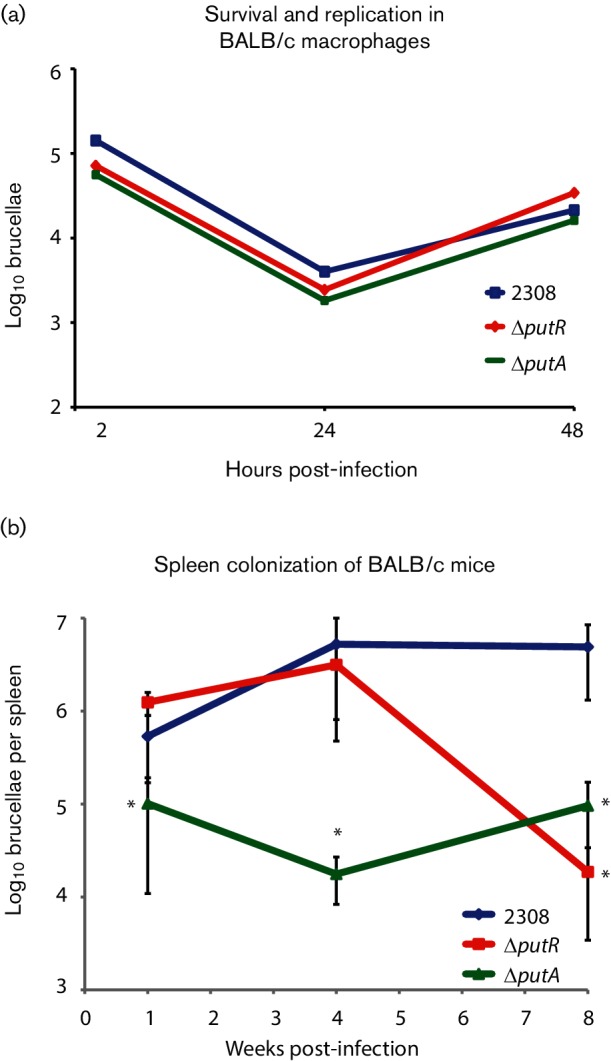
Virulence of the B. abortus Δ putR and ΔputA isogenic strains. (a) B. abortus Δ putR and ΔputA deletion strains are not attenuated in macrophages. Intraperitoneal macrophages were harvested from BALB/c mice and infected with the parental strain B. abortus 2308 and the ΔputR and ΔputA strains. At the times indicated, the macrophages were lysed and serial dilutions were plated to determine the number of intracellular brucellae. (b) B. abortus Δ putR and ΔputA deletion strains were significantly attenuated in a mouse model of chronic infection. BALB/c mice were infected intraperitoneally with 1×105 c.f.u. of B. abortus 2308 or the ΔputR and ΔputA strains. The spleens of the infected mice were collected and homogenized at 1, 4 and 8 weeks post-infection, and the number of colonizing brucellae determined by plating serial dilutions of the homogenates. The data are the average from five mice per strain per time point. A significant difference between a deletion strain and 2308 was determined using a t-test (P<0.05) and is indicated by an asterisk (*).
The ΔputR and ΔputA strains were also evaluated in a mouse model of chronic Brucella infection, and for this mice were infected intraperitoneally with ~1×105 c.f.u. Following 1, 4 and 8 weeks of infection, the mice were euthanized, and bacterial colonization was determined by measuring the number of brucellae present in the spleens of the mice. The ΔputR deletion strain exhibited significantly reduced spleen colonization at 8 weeks post-infection compared to the parental strain 2308, and the ΔputA mutant was significantly attenuated at 1, 4 and 8 weeks post-infection. Altogether, these data indicate that PutR and PutA are required for the full virulence of B. abortus in an animal model of infection.
B. abortus Δ putR is hypersensitive to oxidative stress in vitro
It has been shown previously that bacterial Put systems are linked to oxidative stress [28–30]. To test the hypothesis that putR and putA in B. abortus are required for the ability of the bacteria to cope with oxidative stress, we cultivated the bacteria in liquid medium, and then the cultures were treated with hydrogen peroxide (H2O2) at a final concentration of 10 mM. It was observed that the ΔputR strain was killed more rapidly than the parental strain 2308, but the ΔputA strain did not deviate significantly from strain 2308 (Fig. 2a). It is important to note that other stresses were also evaluated in these experiments, including exposure to paraquot, polymyxin B and high concentrations of NaCl, but no significant differences were observed between the ΔputR, ΔputA or 2308 strains (data not shown). To ensure the genetic link between putR and the phenotype observed with H2O2 stress, the putR locus was reconstructed in the ΔputR strain, and the ΔputR-RC strain was tested in the H2O2 exposure assay (Fig. 2b). As expected, reconstruction of putR rescued the deletion strain from the increased sensitivity to H2O2-mediated killing. These in vitro experiments demonstrate that the B. abortus Δ putR deletion strain, but not the ΔputA deletion strain, is hypersensitive to H2O2 stress.
Fig. 2.
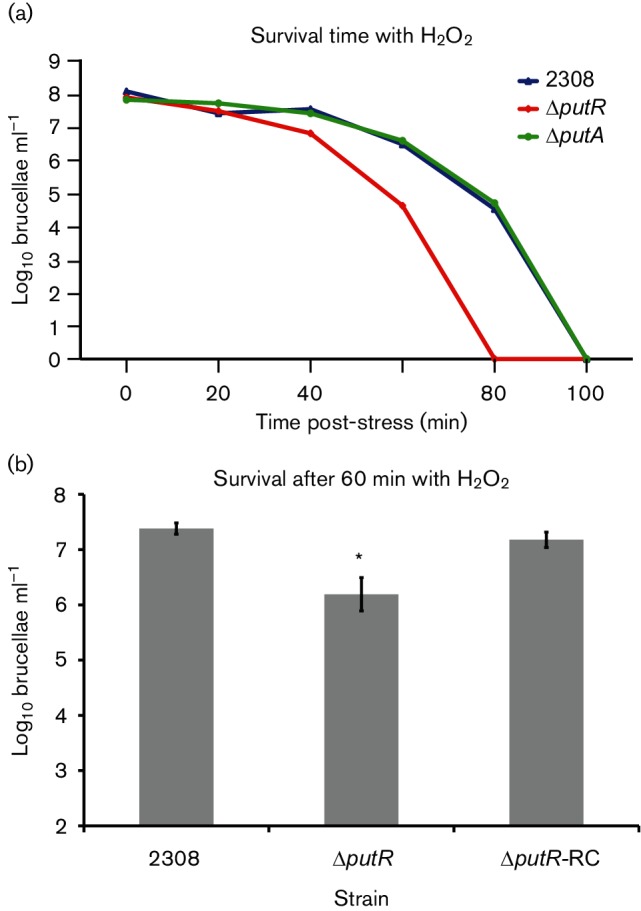
Sensitivity of B. abortus Δ putR and ΔputA strains to hydrogen peroxide (H2O2) exposure. (a) The B. abortus Δ putR deletion strain exhibits increased sensitivity to H2O2 stress compared to the parental strain 2308. The Brucella strains were grown in rich medium (i.e. brucella broth) for 72 h, and subsequently the cultures were diluted into fresh brucella broth at a concentration of ~1×108 c.f.u. ml−1. H2O2 was added to the cultures at a final concentration of 10 mM, and the cultures were incubated at 37 °C with constant shaking. At the indicated time points, aliquots were isolated, serially diluted and plated to enumerate the viable bacteria in each culture. (b) The increased sensitivity of the ΔputR deletion strain to H2O2 can be rescued by genetic complementation. The full-length putR locus was reconstructed on the chromosome of the ΔputR strain, and this strain (ΔputR-RC) was tested for its capacity to withstand H2O2 exposure. For this, the Brucella strains were cultured and exposed to H2O2 as described above, and the number of viable bacteria was determined following 60 min of H2O2 exposure. The asterisk (*) denotes a statistically significant difference (t-test, P<0.05) between the ΔputR strain and the parental strain 2308.
PutR is a transcriptional activator of putA and putR gene expression
The putR and putA genes of B. abortus 2308 are located on chromosome II and are designated bab2_0517 (also known as bab_rs28810) and bab2_0518 (also known as bab_rs28815) in the genome sequence, respectively (Fig. 3a). Since the regulatory aspects of the Put system vary across bacterial species, the ability of PutR to regulate putA in B. abortus was examined using β-galactosidase assays and quantitative RT-PCR (qRT-PCR). The putR–putA intergenic region was cloned into pMR15, a plasmid harbouring a promoterless lacZ cassette, to generate a putA-lacZ transcriptional fusion. The putA-lacZ plasmid was transformed into B. abortus 2308, ΔputR and ΔputR-RC, and the β-galactosidase activity from each stain was measured (Fig. 3b). The ΔputR deletion strain showed significantly lower levels of β-galactosidase activity compared to the parental strain 2308, and the ΔputR-RC strain exhibited similar levels of β-galactosidase activity to those observed in 2308, indicating that PutR is an activator of putA gene expression. To further examine the regulation of putA by PutR, qRT-PCR was conducted with RNA isolated from B. abortus 2308, ΔputR and Δ putR-RC grown to the late exponential phase in Brucella broth (Fig. 3c). This experiment confirmed the results of the putA-lacZ studies, as putA mRNA levels were significantly reduced in the ΔputR deletion strain, and genetic complementation of putR expression restored the levels of putA mRNA to wild-type quantities.
Fig. 3.
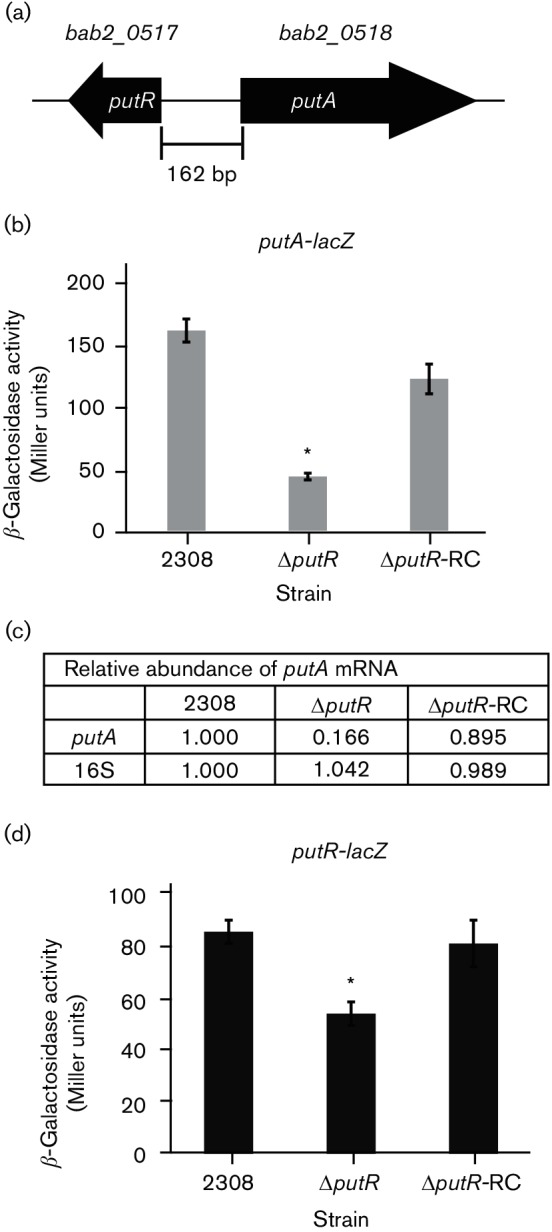
PutR is a transcriptional activator of putA in B. abortus. (a) Genetic organization of putR and putA on chromosome II in B. abortus 2308. putR (bab2_0517) is divergently oriented to putA (bab2_0518). (b) β-Galactosidase activity of a putA-lacZ transcriptional fusion. The indicated Brucella strains carrying a plasmid containing the putA-lacZ transcriptional fusion were grown in Brucella broth to the late exponential phase, and the β-galactosidase activity from each strain was determined. The bar graph shows the average number of Miller units from each strain, and a significant difference between the parental strain 2308 and the ΔputR strain was determined using a t-test (P<0.05) and is indicated by an asterisk (*). (c) Quantitative RT-PCR (qRT-PCR) analysis of putA mRNA levels. The relative levels of putA mRNA in B. abortus 2308, ΔputR and ΔputR-RC were determined by qRT-PCR using RNA isolated from the Brucella strains following cultivation of the bacteria in Brucella broth to the late exponential phase. Oligonucleotide primers specific for either putA or 16S rRNA were employed to amplify the target genes by PCR, and quantification of the amplified DNA fragments was performed using SYBR green incorporation. The values represent the abundance of specific mRNA relative to the level of mRNA from the parental strain 2308 designated as 1.000. (d) β-Galactosidase activity of a putR-lacZ transcriptional fusion. Brucella strains carrying a plasmid containing the putR-lacZ transcriptional fusion were cultured in Brucella broth to the late exponential phase, and the β-galactosidase activity from each strain was measured. The bar graph shows the average number of Miller units from each strain, and a significant difference between the parental strain 2308 and the ΔputR strain was determined using a t-test (P<0.05) and is indicated by an asterisk (*).
The autoregulatory nature of PutR in respect of putR gene expression was also assessed using a putR-lacZ gene fusion in B. abortus 2308, ΔputR and ΔputR-RC (Fig. 3d). It was observed that β-galactosidase activity from the putR-lacZ fusion was significantly reduced in the ΔputR strain compared to the parental strain 2308 and, importantly, reconstruction of the putR locus in the ΔputR strain restored β-galactosidase activity from the putR-lacZ fusion to the levels observed in the parental strain 2308. These results indicate that PutR autoregulates its own expression in a positive manner, and altogether these data demonstrate that PutR is a transcriptional activator of putA and putR gene expression in B. abortus 2308.
PutR binds directly to the putR–putA intergenic region
Given that PutR was determined to be a transcriptional activator of putA expression (Fig. 3), we hypothesized that PutR binds directly to the DNA region encompassing the putA promoter, as PutR proteins in other bacteria have been demonstrated to bind directly to DNA to control gene expression [15, 16]. To test this hypothesis, a recombinant PutR (rPutR) protein was produced and purified from E. coli using affinity chromatography, and subsequently the rPutR protein was used in an EMSA with 32P-labelled DNA corresponding to the B. abortus putR–putA intergenic region (Fig. 4). The EMSAs demonstrated concentration-dependent binding of rPutR to the intergenic region and, importantly, interactions between rPutR and the putR–putA intergenic region were observed at nanomolar concentrations. To ensure the specificity of the rPutR–DNA interactions, competition experiments were performed where excess non-labelled putR–putA intergenic region DNA was added to some reactions, and non-labelled DNA from an unrelated promoter (i.e. abcR2 promoter) was included in other binding reactions. Overall, these experiments determined that PutR from B. abortus binds directly and specifically to the putR–putA intergenic region.
Fig. 4.
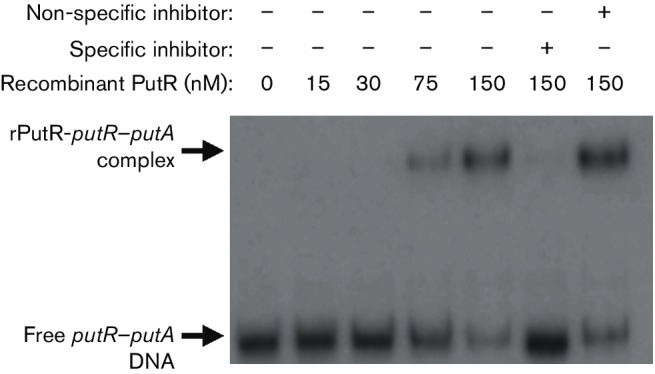
PutR binds directly to the intergenic region between putR and putA. An electrophoretic mobility shift assay (EMSA) was used to assess the binding of a purified recombinant version of PutR (rPutR) to the putR–putA intergenic region. A 336 bp DNA probe corresponding to the putR–putA intergenic region and part of each coding region was amplified by PCR, and the fragment was radiolabelled with [γ-32P]ATP. Increasing nanomolar concentrations of rPutR were incubated with the radiolabelled DNA, and in some binding reactions, unlabelled specific (putR–putA intergenic-region DNA) and non-specific (abcR2 promoter-region DNA) competitor DNA fragments were included as controls. The binding reactions were resolved in 6 % native polyacrylamide gels and visualized by autoradiography.
PutA is an l-proline dehydrogenase that converts l-proline to l-glutamate
Many bacterial PutA proteins are bifunctional enzymes that catalyse the conversion of proline to glutamate with two sequential enzymatic steps [12, 31, 32], and thus we hypothesized that PutA from B. abortus is an authentic proline dehydrogenase. To test this hypothesis, we produced and purified a recombinant version of PutA (rPutA) to a high degree of homogeneity (Fig. 5a) and determined its enzymatic activity. The conversion of proline to glutamate by PutA requires the reduction of NAD+ to NADH [31, 32], and to avoid inhibition of the enzyme by NADH and increase the sensitivity of detection we coupled the reaction to the reduction of MTT with PMS as the intermediate electron carrier [25]. The product of MTT reaction (formazan) was detected at 568 nm. In this assay, we recorded an increase in absorbance at 568 nm, which did not occur if rPutA was not included. This observation demonstrated that NAD+ was reduced by rPutA with l-proline as the substrate (Fig. 5b). To determine if this reaction culminated in the generation of l-glutamate, the respective supernatant was assayed with l-glutamic dehydrogenase, which reduces NAD+ in the presence of l-glutamate. Indeed, the addition of l-glutamate dehydrogenase to the supernatant of the reaction mixture with rPutA caused a rise in the absorbance at 568 nm; the reaction mixture without rPutA did not lead to such an observation (Fig. 5c). Taken together, these data demonstrated that the PutA protein is an l-proline dehydrogenase that catalyses the conversion of l-proline to l-glutamate in B. abortus.
Fig. 5.
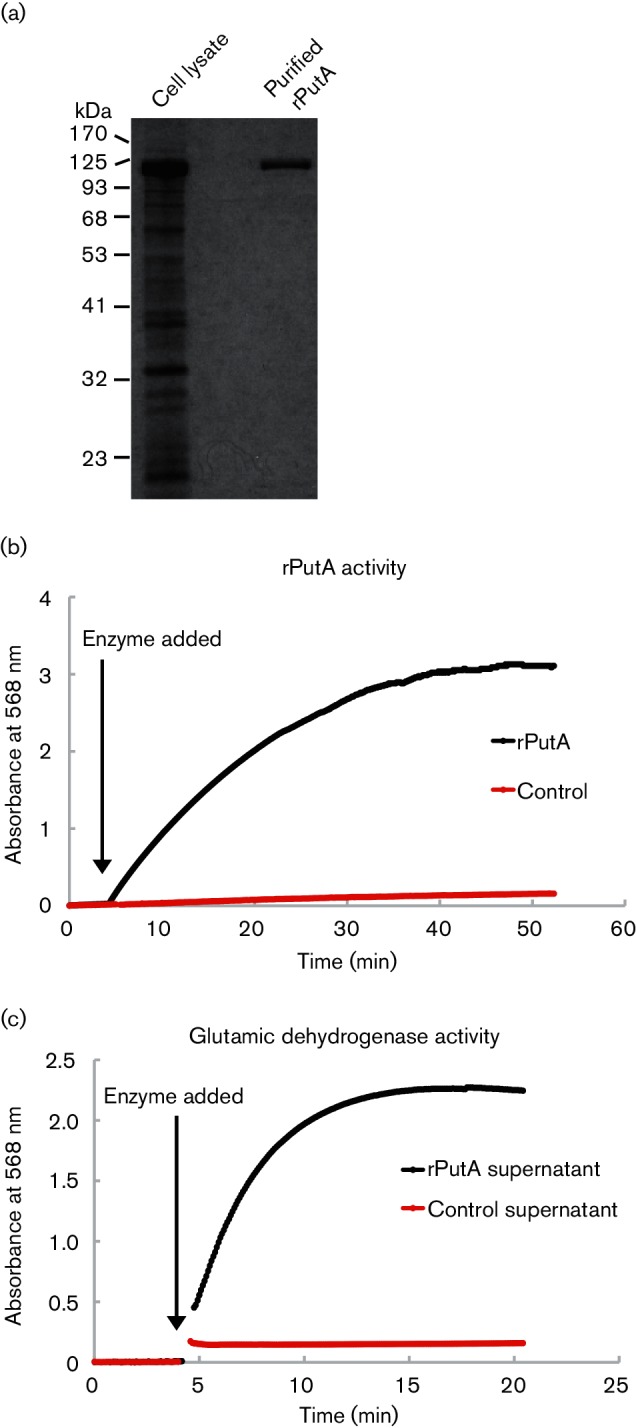
The B. abortus PutA protein is an l-proline dehydrogenase. (a) Purification of a recombinant B. abortus PutA protein (rPutA). The rPutA protein was produced and purified in E. coli using the pASK-IBA7 expression system, and the rPutA protein was purified to a high degree of homogeneity. (b) Enzymatic conversion of l-proline to l-glutamate by rPutA. The reaction was tracked by measuring the increase in absorbance at 568 nm via a spectrophotometer, which monitored the consumption of proline by measuring the reduction of MTT to formazan. The reaction was conducted at room temperature in 50 mM potassium phosphate and 25 mM NaCl (pH 7.5), 20 mM l-proline, 200 µM NAD, 240 µM CoQ1, 1 mM MTT, 1 mM PMS and 0.072 µM rPutA. The baseline of the reaction was recorded for 5 min before the addition of rPutA (indicated by the arrow). The black line depicts the reaction containing rPutA, and the negative control reaction that contains no rPutA is illustrated with the red line. (c) Confirmation of l-glutamate production by rPutA. The reaction mixtures containing rPutA (black line) and the negative control reaction (no rPutA; red line) mixture were subjected to centrifugation at 14 000 g for 5 min to remove previously formed formazan precipitate. The supernatants were then collected and their absorbance was tracked via a spectrophotometer at 568 nm. The baseline of the reaction was recorded for 5 min before the addition of two units of l-glutamic dehydrogenase to the reaction mixture to confirm the presence of l-glutamate produced from the previous reaction. The black arrow indicates the time when l-glutamic dehydrogenase was added.
Interestingly, PutA proteins in some bacteria, such as Salmonella, are trifunctional, as they possess the ability to bind DNA and regulate gene expression in addition to proline dehydrogenase activity [33, 34]. However, the B. abortus PutA protein does not appear to be capable of binding DNA, as it lacks the known DNA-binding motif found in the amino-terminus of trifunctional PutA proteins, and in a direct test PutA did not exhibit DNA-binding activity (data not shown). Taken together, these data demonstrate that the PutA protein is a bifunctional enzyme that catalyses the conversion of proline to glutamate in B. abortus.
Discussion
The data point towards a model in which PutR acts as the transcriptional activator of putA, with PutA acting as a functional proline dehydrogenase in Brucella. Furthermore, it shows that putR and putA are both required for a virulent infection, and may be necessary to help mitigate host-generated stresses in the infection processes, specifically oxidative stress.
Both putA and putR have been described in several α-proteobacteria that are closely related to Brucella. In Rhodobacter capsulatus, the Lrp-type transcriptional protein PutR acts as a transcriptional activator of putA [16]. For Rhodobacter, the Put system is inducible by proline, the addition of which leads to the transcriptional activation of putA via PutR, with PutR subsequently suppressing putR, and PutA being further capable of auto-regulating putA. In contrast, PutR in Agrobacterium tumefaciens is capable of auto-regulation, but PutA is non-essential for its own regulation [15, 17]. In Sinorhizobium meliloti, PutA acts as an autogenous repressor but is non-inducible by proline [18]. This non-conservation of regulatory mechanisms fits with the picture that the Put systems have adapted in response to the individual host–pathogen or environmental conditions, as well as the interactions of specific ecological niches.
The Put system has also been linked to the ability of various pathogens to colonize and infect hosts. In S. meliloti, a ΔputA strain has a weakened ability to form root colonies [35] and, moreover, strains complemented with additional copies of putA are more competitive at establishing a symbiotic relationship with alfalfa [36, 37]. The Put system has also been linked to infectivity in both Helicobacter hepaticas and Helicobacter pylori [28, 29, 38], and the Put system plays a role in regulating the cell cycle within Ehrlichia chaffeensis [39]. Our data demonstrate that PutA and PutR are also required for B. abortus to infect animals (Fig. 1), and thus, it is possible that proline is available to the brucellae during host colonization; however, further work is needed to empirically determine whether proline is an essential host-supplied amino acid for Brucella spp.
There are two likely reasons for the Put system to contribute to virulence: it may provide an efficient respiratory pathway and/or it may aid in inducing anti-oxidative pathways. PutA acts in a two-step process to generate glutamate from proline [12, 31, 32]. In the first step, proline is oxidized to Δ1-pyrroline-5-carboxylate, which is then non-enzymatically hydrolysed to glutamate-γ-semialdehyde. This step leads to the reduction of FAD and the regeneration of ubiquinone in the TCA cycle. In the second step, glutamate-γ-semialdehyde is oxidized to glutamate, generating NADH. From glutamate, α-ketoglutarate can be generated by Brucella to enter into the TCA cycle [40]. Thus, this mechanism represents a potentially efficient system for generating cellular energy products and for using proline as a respiratory metabolite, and given that PutA is an active proline dehydrogenase in Brucella (Fig. 5), this mechanism is highly plausible.
It has also been shown that the PutA acts to induce anti-oxidative stress genes in E. coli through production of reactive oxygen species (ROS) [28, 30]. The exact mechanism for this ROS production was not explored, but it was suggested that ROS were generated through the increased flux through the electron transport chain, leading to the formation of superoxide species and production of H2O2. This rise of endogenous H2O2 would then be sufficient to induce anti-oxidative systems, and the higher basal level of anti-oxidative systems could result in oxidative stress resistance. In Brucella, the two-component system NtrX/NtrY was shown to be a redox sensor, and essential for virulent infection [41], and intriguingly, this system regulates putA in Ehr. chaffeensis [39]. While no connection between the NtrX/NtrY system and the Put system is known of in Brucella, this tentatively shows that the anti-oxidative functions of PutA may be important in Brucella. Interestingly, we did not observe a defect in the ability of the B. abortus Δ putA strain to cope with oxidative stress, but rather, the ΔputR strain exhibited increased sensitivity to H2O2 stress (Fig. 2). This indicates that PutR is involved in the appropriate response of the B. abortus to H2O2 stress, but it is possible that PutR controls the expression of genes other than putA to mediate this oxidative stress response function. This possibility is currently being investigated by our group.
Given that PutR positively regulates the expression of putA in B. abortus (Fig. 3), the observation that the B. abortus Δ putA strain exhibited a more robust attenuation compared to the ΔputR strain was surprising (Fig. 1), because it might be expected that both the putA and putR deletion strains should exhibit similar levels of colonization in the animal model. However, our results show that while putA expression is significantly decreased in the ΔputR strain, the expression of putA is not completely abolished, and thus the ΔputA and ΔputR strains should not display equivalent levels of virulence in the mouse model of infection. Additionally, as postulated in the previous paragraph, it is likely that PutR controls the expression of other genes in B. abortus, and therefore it is not surprising that the ΔputR and ΔputA strains exhibit dissimilar virulence phenotypes.
Ultimately, the findings presented show that the Put system is essential for virulent infection of Brucella, but future work is needed to further characterize the exact roles that proline and the Put system play in establishing infection. Specifically, there is currently no proline-specific uptake system described in the brucellae, and it will be important to identify and characterize such a transport system in order to test the hypothesis that proline is a host-supplied nutrient available to the bacteria during infection. Additionally, a putative proline sensor–regulator system, PrlS/PrlR, has been described previously [42], but it remains to be elucidated if there is a link between PrlS/PrlR, the Put system and Brucella virulence. While these critical areas remain to be investigated, the present work clearly demonstrates the importance of the proline utilization system for B. abortus in vivo.
Funding information
The authors were supported by the Virginia-Maryland College of Veterinary Medicine Summer Veterinary Student Research Program, while the American Veterinary Medical Foundation supported M. C. (AVMA/AVMF 2nd Opportunity Summer Research Scholarship). This work was also supported by the Fralin Life Science Institute at Virginia Tech, the VA-MD College of Veterinary Medicine at Virginia Tech, and a grant from the National Institute of Allergy and Infectious Diseases to C. C. C. (1R15AI117648-01).
Conflicts of interest
The authors declare that there are no conflicts of interest.
Ethical statement
All animal work was reviewed and approved by the Institutional Animal Care and Use Committee (IACUC) at Virginia Tech.
References
- 1.Celli J. The changing nature of the Brucella-containing vacuole. Cell Microbiol. 2015;17:951–958. doi: 10.1111/cmi.12452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Godfroid J, Scholz HC, Barbier T, Nicolas C, Wattiau P, et al. Brucellosis at the animal/ecosystem/human interface at the beginning of the 21st century. Prev Vet Med. 2011;102:118–131. doi: 10.1016/j.prevetmed.2011.04.007. [DOI] [PubMed] [Google Scholar]
- 3.Roop RM, Gaines JM, Anderson ES, Caswell CC, Martin DW. Survival of the fittest: how Brucella strains adapt to their intracellular niche in the host. Med Microbiol Immunol. 2009;198:221–238. doi: 10.1007/s00430-009-0123-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Halvorson H. Utilization of single L-amino acids as sole source of carbon and nitrogen by bacteria. Can J Microbiol. 1972;18:1647–1650. doi: 10.1139/m72-255. [DOI] [PubMed] [Google Scholar]
- 5.Chubukov V, Gerosa L, Kochanowski K, Sauer U. Coordination of microbial metabolism. Nat Rev Microbiol. 2014;12:327–340. doi: 10.1038/nrmicro3238. [DOI] [PubMed] [Google Scholar]
- 6.Price CT, Al-Quadan T, Santic M, Rosenshine I, Abu Kwaik Y. Host proteasomal degradation generates amino acids essential for intracellular bacterial growth. Science. 2011;334:1553–1557. doi: 10.1126/science.1212868. [DOI] [PubMed] [Google Scholar]
- 7.Gouzy A, Poquet Y, Neyrolles O. Amino acid capture and utilization within the Mycobacterium tuberculosis phagosome. Future Microbiol. 2014;9:631–637. doi: 10.2217/fmb.14.28. [DOI] [PubMed] [Google Scholar]
- 8.Barel M, Ramond E, Gesbert G, Charbit A. The complex amino acid diet of Francisella in infected macrophages. Front Cell Infect Microbiol. 2015;5:9. doi: 10.3389/fcimb.2015.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gerhardt P, Tucker LA, Wilson JB. The nutrition of brucellae: utilization of single amino acids for growth. J Bacteriol. 1950;59:777–782. doi: 10.1128/jb.59.6.777-782.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ronneau S, Moussa S, Barbier T, Conde-Álvarez R, Zuniga-Ripa A, et al. Brucella, nitrogen and virulence. Crit Rev Microbiol. 2016;42:507–525. doi: 10.3109/1040841X.2014.962480. [DOI] [PubMed] [Google Scholar]
- 11.Kohler S, Foulongne V, Ouahrani-Bettache S, Bourg G, Teyssier J, et al. The analysis of the intramacrophagic virulome of Brucella suis deciphers the environment encountered by the pathogen inside the macrophage host cell. Proc Natl Acad Sci USA. 2002;99:15711–15716. doi: 10.1073/pnas.232454299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tanner JJ. Structural biology of proline catabolism. Amino Acids. 2008;35:719–730. doi: 10.1007/s00726-008-0062-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ratzkin B, Roth J. Cluster of genes controlling proline degradation in Salmonella typhimurium. J Bacteriol. 1978;133:744–754. doi: 10.1128/jb.133.2.744-754.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Newell SL, Brill WJ. Mutants of Salmonella typhimurium that are insensitive to catabolite repression of proline degradation. J Bacteriol. 1972;111:375–382. doi: 10.1128/jb.111.2.375-382.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cho K, Winans SC. The putA gene of Agrobacterium tumefaciens is transcriptionally activated in response to proline by an Lrp-like protein and is not autoregulated. Mol Microbiol. 1996;22:1025–1033. doi: 10.1046/j.1365-2958.1996.01524.x. [DOI] [PubMed] [Google Scholar]
- 16.Keuntje B, Masepohl B, Klipp W. Expression of the putA gene encoding proline dehydrogenase from Rhodobacter capsulatus is independent of NtrC regulation but requires an Lrp-like activator protein. J Bacteriol. 1995;177:6432–6439. doi: 10.1128/jb.177.22.6432-6439.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jafri S, Evoy S, Cho K, Craighead HG, Winans SC. An Lrp-type transcriptional regulator from Agrobacterium tumefaciens condenses more than 100 nucleotides of DNA into globular nucleoprotein complexes. J Mol Biol. 1999;288:811–824. doi: 10.1006/jmbi.1999.2715. [DOI] [PubMed] [Google Scholar]
- 18.Soto MJ, Jiménez-Zurdo JI, van Dillewijn P, Toro N. Sinorhizobium meliloti putA gene regulation: a new model within the family Rhizobiaceae . J Bacteriol. 2000;182:1935–1941. doi: 10.1128/JB.182.7.1935-1941.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Caswell CC, Gaines JM, Roop RM., 2nd The RNA chaperone Hfq independently coordinates expression of the VirB type IV secretion system and the LuxR-type regulator BabR in Brucella abortus 2308. J Bacteriol. 2012;194:3–14. doi: 10.1128/JB.05623-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bellaire BH, Elzer PH, Hagius S, Walker J, Baldwin CL, et al. Genetic organization and iron-responsive regulation of the Brucella abortus 2,3-dihydroxybenzoic acid biosynthesis operon, a cluster of genes required for wild-type virulence in pregnant cattle. Infect Immun. 2003;71:1794–1803. doi: 10.1128/IAI.71.4.1794-1803.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller JH. Experiments in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1972. pp. 352–355. [Google Scholar]
- 22.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sheehan LM, Budnick JA, Blanchard C, Dunman PM, Caswell CC. A LysR-family transcriptional regulator required for virulence in Brucella abortus is highly conserved among the α-proteobacteria. Mol Microbiol. 2015;98:318–328. doi: 10.1111/mmi.13123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sheehan LM, Budnick JA, Roop RM, 2nd, Caswell CC. Coordinated zinc homeostasis is essential for the wild-type virulence of Brucella abortus. J Bacteriol. 2015;197:1582–1591. doi: 10.1128/JB.02543-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dunigan DD, Waters SB, Owen TC. Aqueous soluble tetrazolium/formazan MTS as an indicator of NADH- and NADPH-dependent dehydrogenase activity. Biotechniques. 1995;19:640–649. [PubMed] [Google Scholar]
- 26.Luo M, Gamage TT, Arentson BW, Schlasner KN, Becker DF, et al. Structures of proline utilization A (PutA) reveal the fold and functions of the aldehyde dehydrogenase superfamily domain of unknown function. J Biol Chem. 2016;291:24065–24075. doi: 10.1074/jbc.M116.756965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gee JM, Valderas MW, Kovach ME, Grippe VK, Robertson GT, et al. The Brucella abortus Cu, Zn superoxide dismutase is required for optimal resistance to oxidative killing by murine macrophages and wild-type virulence in experimentally infected mice. Infect Immun. 2005;73:2873–2880. doi: 10.1128/IAI.73.5.2873-2880.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krishnan N, Becker DF. Oxygen reactivity of PutA from Helicobacter species and proline-linked oxidative stress. J Bacteriol. 2006;188:1227–1235. doi: 10.1128/JB.188.4.1227-1235.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krishnan N, Doster AR, Duhamel GE, Becker DF. Characterization of a Helicobacter hepaticus putA mutant strain in host colonization and oxidative stress. Infect Immun. 2008;76:3037–3044. doi: 10.1128/IAI.01737-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang L, Alfano JR, Becker DF. Proline metabolism increases katG expression and oxidative stress resistance in Escherichia coli. J Bacteriol. 2015;197:431–440. doi: 10.1128/JB.02282-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Menzel R, Roth J. Enzymatic properties of the purified PutA protein from Salmonella typhimurium. J Biol Chem. 1981;256:9762–9766. [PubMed] [Google Scholar]
- 32.Menzel R, Roth J. Purification of the putA gene product. A bifunctional membrane-bound protein from Salmonella typhimurium responsible for the two-step oxidation of proline to glutamate. J Biol Chem. 1981;256:9755–9761. [PubMed] [Google Scholar]
- 33.Ostrovsky de Spicer P, Maloy S. PutA protein, a membrane-associated flavin dehydrogenase, acts as a redox-dependent transcriptional regulator. Proc Natl Acad Sci USA. 1993;90:4295–4298. doi: 10.1073/pnas.90.9.4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Muro-Pastor AM, Ostrovsky P, Maloy S. Regulation of gene expression by repressor localization: biochemical evidence that membrane and DNA binding by the PutA protein are mutually exclusive. J Bacteriol. 1997;179:2788–2791. doi: 10.1128/jb.179.8.2788-2791.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiménez-Zurdo JI, García-Rodríguez FM, Toro N. The Rhizobium meliloti putA gene: its role in the establishment of the symbiotic interaction with alfalfa. Mol Microbiol. 1997;23:85–93. doi: 10.1046/j.1365-2958.1997.1861555.x. [DOI] [PubMed] [Google Scholar]
- 36.van Dillewijn P, Soto MJ, Villadas PJ, Toro N. Construction and environmental release of a Sinorhizobium meliloti strain genetically modified to be more competitive for alfalfa nodulation. Appl Environ Microbiol. 2001;67:3860–3865. doi: 10.1128/AEM.67.9.3860-3865.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Dillewijn P, Villadas PJ, Toro N. Effect of a Sinorhizobium meliloti strain with a modified putA gene on the rhizosphere microbial community of alfalfa. Appl Environ Microbiol. 2002;68:4201–4208. doi: 10.1128/AEM.68.9.4201-4208.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakajima K, Inatsu S, Mizote T, Nagata Y, Aoyama K, et al. Possible involvement of putA gene in Helicobacter pylori colonization in the stomach and motility. Biomed Res. 2008;29:9–18. doi: 10.2220/biomedres.29.9. [DOI] [PubMed] [Google Scholar]
- 39.Cheng Z, Lin M, Rikihisa Y. Ehrlichia chaffeensis proliferation begins with NtrY/NtrX and PutA/GlnA upregulation and CtrA degradation induced by proline and glutamine uptake. MBio. 2014;5:e02141-14. doi: 10.1128/mBio.02141-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marr AG, Olsen CB, Unger HS, Wilson JB. The oxidation of glutamic acid by Brucella abortus. J Bacteriol. 1953;66:606–610. doi: 10.1128/jb.66.5.606-610.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carrica MC, Fernandez I, Martí MA, Paris G, Goldbaum FA. The NtrY/X two-component system of Brucella spp. acts as a redox sensor and regulates the expression of nitrogen respiration enzymes. Mol Microbiol. 2012;85:39–50. doi: 10.1111/j.1365-2958.2012.08095.x. [DOI] [PubMed] [Google Scholar]
- 42.Mirabella A, Yañez Villanueva RM, Delrue RM, Uzureau S, Zygmunt MS, et al. The two-component system PrlS/PrlR of Brucella melitensis is required for persistence in mice and appears to respond to ionic strength. Microbiology. 2012;158:2642–2651. doi: 10.1099/mic.0.060863-0. [DOI] [PubMed] [Google Scholar]


