Abstract
Several strictly anaerobic bacteria that are Gram-stain-positive have the ability to use uric acid as the sole source of carbon and energy. The phylogeny of three such species, Clostridium acidurici, Clostridium purinilyticum, and Eubacterium angustum, members of the Clostridium cluster XII that ferment purines, but not most amino acids or carbohydrates, has been re-examined, taking advantage of their recently sequenced genomes. Phylogenetic analyses, based on 16S rRNA gene sequences, protein sequences of RpoB and GyrB, and on a concatenated alignment of 50 ribosomal proteins, revealed tight clustering of C. acidurici and C. purinilyticum. Eubacterium angustum showed consistent association with C. acidurici and C. purinilyticum , but differed from these two in terms of the genome size, G+C content of its chromosomal DNA and its inability to form spores. We propose reassigning C. acidurici and C. purinilyticum to the novel genus Gottschalkia as Gottschalkia acidurici gen. nov. comb. nov. (the type species of the genus) and Gottschalkia purinilytica comb. nov., respectively. Eubacterium angustum is proposed to be reclassified as Andreesenia angusta gen. nov. comb. nov. Furthermore, based on the phylogenetic data and similar metabolic properties, we propose assigning genera Gottschalkia and Andreesenia to the novel family Gottschalkiaceae. Metagenomic sequencing data indicate the widespread distibution of organisms falling within the radiation of the proposed family Gottschalkiaceae in terrestrial and aquatic habitats from upstate New York to Antarctica, most likely due to their ability to metabolize avian-produced uric acid.
Keywords: Clostridium, taxonomy, sporulation, purine degradation, 16S rRNA, uric acid
For historical reasons, the genus Clostridium includes a large number of diverse bacteria whose only common features are obligately anaerobic growth, a Gram-positive type cell wall, the absence of sulfate reduction and the ability to form endospores [1–3]. In 1994, based on the studies of clostridial 16S rRNA gene sequences, Collins and colleagues divided it into 19 clusters that roughly represented family-level taxa; each cluster included several proposed genera [4]. Over the past 20 years, many former Clostridium spp. have been reassigned to new genera, some have been moved to novel families, orders and even to the novel classes, Erysipelotrichia and Negativicutes [1, 5]. An important step towards streamlining clostridial classification has been made in the latest edition of Bergey’s Manual of Systematic Bacteriology [1, 6], which reclassified a large number of Clostridium spp. based on phylogenetic criteria, along the lines of the work of Collins et al. [4].
In 2016, Lawson and Rainey [7] proposed limiting the genus Clostridium to the members of Clostridium sensu stricto (Clostridium cluster I [4]), which includes approximately 70 species that are sufficiently close to the type species Clostridium butyricum. Adoption of this proposal means that species of the genus Clostridium that do not belong to cluster I need to be reclassified. Here, we propose such a reclassification for three species of bacteria with validly published names, Clostridium acidurici, Clostridium purinilyticum and Eubacterium angustum, members of the Clostridium cluster XII [4, 8]. Based on the phylogenetic analyses presented here and in a previous work [9], we propose re-assigning these three organisms to two novel genera, Gottschalkia and Andreesenia, within the novel family Gottschalkiaceae.
In their original description of Clostridium cluster XII, Collins et al. [4] identified two loosely connected branches. One of them included a tight cluster of C. acidurici and C. purinilyticum, which shared approximately 94 % similarity with respect to 16S rRNA gene sequences and were put into the same genus. A subsequent paper from the same authors added E. angustum to the same genus [8]. The other branch included Clostridium hastiforme, Clostridium sp. strain BN11, and ‘Clostridium filamentosum’. The first two were later reclassified as Tissierella praeacuta and Tissierella creatinini, respectively [8, 10]. ‘Clostridium filamentosum’ has not been validly named but is available under this name in some culture collections (e.g., ATCC 25785 = JCM 6585). Based on its 16S rRNA gene sequence, it probably belongs to the genus Anaerosalibacter and is listed as Anaerosalibacter sp. in the DSMZ catalog (https://www.dsmz.de/catalogues/details/culture/DSM-6645.html). Because of the ambiguous phylogeny of Tissierella-related organisms, in the 2009 edition of Bergey’s Manual of Systematic Bacteriology these organisms, along with the members of Clostridium cluster XIII, were assigned to Clostridiales Family XI Incertae Sedis [6]. More recently, members of cluster XIII have been assigned to the family Peptoniphilaceae [11], whereas the genera Tissierella and Soehngenia (and potentially also Sporanaerobacter and Tepidimicrobium) have been proposed to form the novel family Tissierellaceae in the order Tissierellales [12]. These changes still left three members of the original Clostridium cluster XII without a correct assignment: C. acidurici, C. purinilyticum and E. angustum [4, 8], and these are the subjects of the present study.
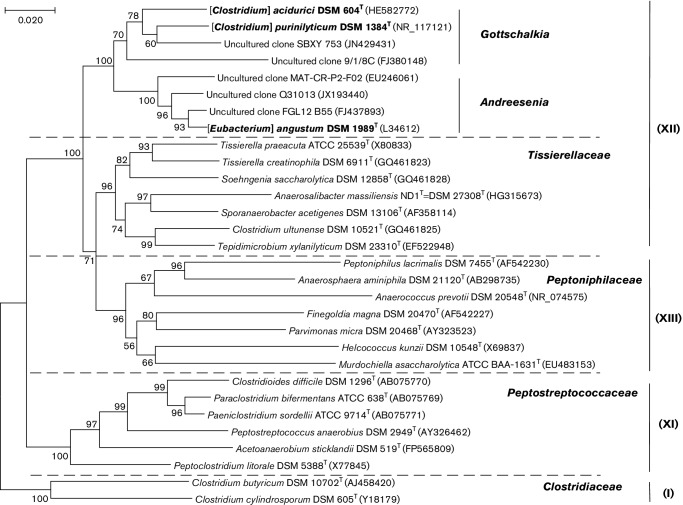
Phylogenetic analyses, based upon the 16S rRNA gene sequences of C. acidurici, C. purinilyticum and E. angustum and their neighbours from clusters I, XI, XII, and XIII were performed using the neighbour-joining (Fig. 1) and maximum likelihood methods (Fig. S1, available in the online Supplementary Material). The 16S rRNA gene sequences of the type strains were obtained either from GenBank or from the NCBI RefSeq Targeted Loci project [13] (see the online Supplementary Material for details). Sequences were aligned with ClustalW [14], as implemented in the mega7 software suite [15], and the neighbour-joining and maximum likelihood trees were reconstructed using mega7.
Fig. 1.
16S rRNA gene-based phylogenetic tree of Clostridium acidurici and related organisms and metagenomic samples. The names of the characterized members of the proposed genera Gottschalkia and Andreesenia are shown in bold in square brackets. The sequences from type strains (indicated with T) were used and listed under their DSM accession numbers; where available. GenBank accession numbers are listed in parentheses. Roman numerals on the right indicate the clostridial cluster assignments of Collins et al. [4]. Clostridioides difficile, Acetoanaerobium sticklandii and Peptoclostridium litorale are the recently assigned names of formerly misclassified Clostridium spp. [45, 46]. The tree was inferred using the neighborhood-joining method, based on the Tamura-Nei model [47] as implemented in mega7 [15]. The evolutionary distances were computed using the Jukes-Cantor method and are in the units of the number of base substitutions per site. The tree was rooted using sequences from C. butyricum and C. cylindrosporum, which are members of Clostridium sensu stricto (cluster I).
The 16S rRNA gene-based phylogenetic trees (Figs 1 and S1) showed that C. acidurici, C. purinilyticum and E. angustum form a distinct cluster, separate from other species of cluster XII (members of Tissierellaceae), as well as from representatives of clusters I, XI, and XIII (members of Clostridiaceae, Peptostreptococcaceae and Peptoniphilaceae, respectively). As noted previously, C. acidurici and C. purinilyticum are particularly closely related [4, 16–18]. E. angustum forms a separate branch in the same cluster, as it did in the trees presented in several earlier reports [2, 8, 19–21].
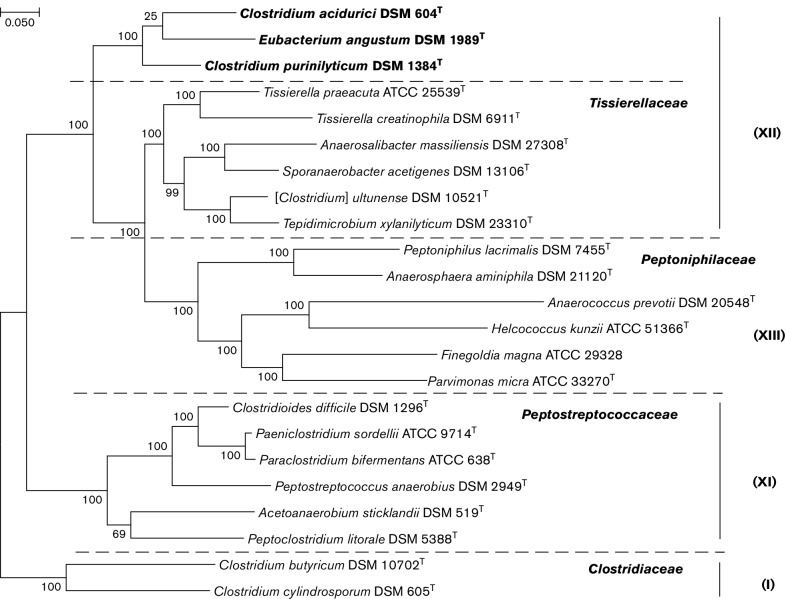
To further evaluate the phylogenetic relationships of C. acidurici, C. purinilyticum and E. angustum, we have analyzed protein trees reconstructed from ribosomal proteins (Fig. 2) and from sequences of the DNA-directed RNA polymerase beta subunit (RpoB) and DNA gyrase subunit B (GyrB) of various members of clostridial clusters I, XI, XII, and XII, using sequences from selected organisms with completely or partially sequenced genomes, where available (Table S1). The ribosomal proteins-based phylogenetic tree was reconstructed from a concatenated alignment of 50 widespread ribosomal proteins, as described earlier [9, 22], (see online Supplementary Material for details). On this tree, C. acidurici, C. purinilyticum and E. angustum again formed a tight cluster with well-supported branches (Fig. 2). Clustering of these organisms was also seen in the phylogenetic trees for RpoB and GyrB subunits (Fig. S2a, b). The assignment of C. acidurici and C. purinilyticum to a single genus satisfies both rRNA similarity-based [23] and protein overlap-based [24] criteria. Based on these data, we formally propose reassigning C. acidurici and C. purinilyticum to the novel genus Gottschalkia.
Fig. 2.
Ribosomal protein-based phylogenetic tree of Clostridium acidurici and related species. Members of the proposed genera Gottschalkia and Andreesenia are shown in bold. Roman numerals on the right indicate the clostridial cluster assignments of Collins et al. [4]. The tree was reconstructed essentially as described previously [9, 22]. Fifty sets of ribosomal proteins (L1–L7, L9–L11, L13–L24, L27–L29, L31–L36 and S2–S20) were extracted from the respective genomic entries (listed in Table S1) and aligned using muscle [48]; gapped columns (with more than 30 % of gaps) and columns with low information content were removed from the alignments. Individual ribosomal protein alignments were concatenated, giving a total of 6238 positions, and a maximum-likelihood tree was reconstructed using the PhyML program [49], the latest version of which (http://www.atgc-montpellier.fr/phyml-sms/) includes automatic selection of the best-fit substitution model for a given alignment and calculation of branch support values using aBayes algorithm [50]. The tree was rooted using the sequences from C. butyricum and C. cylindrosporum.
While unification of C. acidurici and C. purinilyticum has already been proposed by Collins et al. [4] and appears quite straightforward, E. angustum differs from them in having a much higher DNA G+C content, a smaller genome size, and an inability to form spores (Table 1). While the ability to sporulate is not necessarily a reliable taxonomic character [8, 25], as it can be easily lost through a deletion of a single core sporulation gene [12, 26], the smaller genome size of E. angustum compared to its relatives (Table 1) indicates substantial loss of genes in its particular lineage. However, E. angustum still encodes certain sporulation proteins, although far fewer than C. acidurici and C. purinilyticum (Table S2). E. angustum has been reported to be non-motile, but formed flagella [27] and its genome carries more than 30 flagellar genes [28]. Based on the differences listed above, and the lower level of similarity in its 16S rRNA gene sequence (91 %) than that recommended for a single genus [23], E. angustum does not fit into the genus Gottschalkia. Further, the percentages of conserved proteins between E. angustum and C. acidurici and C. purinilyticum, calculated as described by Qin and colleagues [24] (49.2 and 47.7 %, respectively) were lower than the suggested genus boundary of 50 %. Accordingly, we propose placing E. angustum in a separate genus, Andreesenia.
Table 1. Characteristics of Clostridium acidiurici, Clostridium purinilyticum and Eubacterium angustum, members of the proposed novel genera Gottschalkia and Andreesenia.
1, Clostridium acidurici 9aT=DSM 604T [16, 29]; 2, Clostridium purinilyticum WA-1T=DSM 1384T [16, 43]; 3, Eubacterium angustum MK-1T=DSM 1989T [27, 28]; 4, Tissierella praeacuta ATCC 25539T or Tissierellacreatinophila KRE 4T=DSM 69113T [8, 12, 19, 51]; 5, Soehngeniasaccharolytica BOR-YT=DSM 12858T=ATCC BAA-502T [20]; 6, Anaerosalibacterbizertensis C5BELT=DSM 23801T or Anaerosalibacter sp. ND1=DSM 27308 [21, 52]; 7, Clostridium ultunense DSM 10521T [53, 54]; 8, Clostridium cylindrosporum HC1T=DSM 605T [16, 29, 55–57]. ± , weak or variable reaction; nd, no available data.
| Property | Organisms | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | ||
| Genome size (kb) | 3108 | 3397 | 2405 | 3116 | nd | 3198 | 3217 | 2720 | |
| Proteins encoded | 2774 | 3135 | 2397 | 2957 | nd | 3054 | 2863 | 1879 | |
| DNA G+C (mol%) | 29.9 | 28.8 | 43.7 | 30.1 | 43 | 29.7 | 32.8 | 27.9 | |
| Cell width (µm) | 0.5–0.7 | 1.1–1.6 | 1.0–1.5 | 0.6–0.9 | 0.5–0.7 | 0.5–1.0 | 0.5–0.7 | 0.8 | |
| Cell length (µm) | 2.5–4.0 | 2.7–9.6 | 3.0–6.5 | 2–8 | 2–11 | 3–20 | 0.5–7.0 | 3.3 | |
| Gram staining | + | + | + | ± | + | + | – | ± | |
| Flagella | + | + | + | ± | + | + | + | + | |
| Spore formation | + | + | – | ± | + | + | + | + | |
| Optimal temperature for growth (°C) | 31–37 | 36 | 37 | 37 | 30–37 | 40 | 37 | 40–45 | |
| Optimal pH for growth | 7.6–8.1 | 7.3–7.8 | 8.0–8.2 | 7.5 | 7.0 | 7.5 | 7.0 | 7.0–8.0 | |
| Hydrolysis of | |||||||||
| Gelatin | – | – | – | ± | – | + | – | – | |
| Starch | – | – | – | – | + | – | nd | – | |
| Utilization of purines | |||||||||
| Adenine | – | + | – | – | nd | nd | nd | – | |
| Adenosine | – | + | –* | nd | nd | nd | nd | – | |
| Guanine | + | + | + | nd | nd | nd | nd | + | |
| 2-Hydroxypurine | + | + | –* | nd | nd | nd | nd | – | |
| Hypoxanthine | + | + | –† | nd | nd | nd | nd | + | |
| Purine | + | + | –* | nd | nd | nd | nd | – | |
| Uric acid | + | + | + | – | nd | nd | nd | + | |
| Xanthine | + | + | + | – | nd | nd | nd | + | |
| Xanthosine | – | + | –* | nd | nd | nd | nd | – | |
| Utilization of sugars | |||||||||
| l-Arabinose | – | – | –* | – | + | – | – | – | |
| Cellobiose | – | – | –* | – | + | – | – | – | |
| d-Fructose | – | – | –* | – | + | – | – | – | |
| d-Galactose | – | – | –* | – | + | – | – | – | |
| d-Glucose | – | – | –* | – | + | + | + | – | |
| Lactose | – | – | –* | – | + | + | – | – | |
| Maltose | – | – | –* | – | + | – | – | – | |
| d-Mannitol | – | – | –* | – | + | + | – | – | |
| d-Mannose | – | – | –* | – | + | – | – | – | |
| d-Ribose | – | – | –* | – | + | – | – | – | |
| d-Sorbitol | – | – | –* | – | + | nd | – | – | |
| Sucrose | – | – | –* | – | + | – | – | – | |
| d-Xylose | – | – | –* | – | + | nd | – | – | |
| Enzymes | |||||||||
| Catalase | – | – | – | – | – | – | – | – | |
| Lecithinase | – | – | – | – | nd | nd | nd | – | |
| Lipase | – | – | – | – | nd | nd | nd | – | |
| Urease | – | – | – | – | – | – | nd | – | |
| Production of | |||||||||
| Acetate | + | + | + | + | + | + | + | + | |
| Butyrate | – | – | – | + | nd | + | – | – | |
| Formate | + | + | + | – | + | – | + | + | |
| CO2 | + | + | + | + | + | nd | + | + | |
| NH3 | + | + | + | + | ± | nd | nd | + | |
| H2 | – | – | – | nd | + | – | + | – | |
| H2S | nd | – | – | + | + | – | nd | – | |
| Reduction of | |||||||||
| Nitrate | – | – | – | ± | – | ± | – | – | |
| Sulfate | – | – | – | – | – | – | – | – | |
| Sulfite | – | – | nd | nd | ± | – | – | – | |
| Thiosulfate | – | – | nd | – | ± | – | – | – | |
| Major fatty acids‡ | C14 : 0, C16 : 0, C16:1ω7c | C14 : 0, C16 : 0, C16:1ω7c | C14 : 0, C16:1ω7c | iso-C15 : 0, C16 : 0 | nd | iso-C15 : 0, C16 : 0 | nd | nd | |
*Beuscher and Andreesen [27] mention the inability of E. angustum to utilize any carbohydrates or purines from the list of compounds tested by Dürre et al. [16] but do not list their names.
†Hypoxanthine was utilized by E. angustum only in the presence of uric acid [27].
‡Fatty acid analyses of C. acidurici, C. purinilyticum, and E. angustum were carried out by the Identification Service of the DSMZ, Braunschweig, Germany, using Sherlock Microbial Identification System [58] of MIDI Inc. (Newark, DE, USA). Myristic acid C14 : 0 clearly predominated, making up at least 32 %, 25 %, and 36 %, respectively, of the total fatty acid content in these organisms.
Despite certain differences, the high degree of 16S rRNA gene sequence similarity, consistent clustering on 16S rRNA gene-based and protein-based trees (Figs 1, 2 and S2), and the similar metabolic properties justify unification of C. acidurici, C. purinilyticum and E. angustum into a higher-level taxon. It is important to note that Tissierella- and Peptoniphilus-containing clusters on both 16S rRNA gene-based and protein-based trees (Figs 1 and 2) correspond to family-level groupings, Tissierellaceae and Peptoniphilaceae, respectively [11, 12]. Thus, based on the available phenotypic, chemotaxonomic, and phylogenetic information, we propose the designation of Gottschalkiaceae fam. nov., to accommodate the genera Gottschalkia and Andreesenia. The novel family is easily distinguished by the ability of its members to use uric acid as the sole carbon and energy source and the predominance of myristic acid among the fatty acids. As a sister group of Tissierellaceae and Peptoniphilaceae, the proposed family Gottschalkiaceae could be tentatively assigned to the order Tissierellales within the class Tissierellia [12], although the high-order taxonomy of these organisms probably merits further study.
While the proposed family Gottschalkiaceae includes just three species with validly published names, representatives of this family appear to be widespread in nature. In their original description of C. acidurici, Barker and Beck [29] mentioned isolating very similar uric acid-degrading anaerobic bacteria from ten different soil samples from various places in California. They also isolated similar organisms from San Francisco bay mud and from sandy soil collected near Provo, Utah, and stated ‘No soil tested has ever failed to harbour the organisms’ [29]. Further, they found anaerobic uric acid-degrading bacteria in fecal material of the yellow-shafted flicker (Colaptes auratus auratus), an observation in line with uric acid being ‘the main nitrogenous end product of avian metabolism, which may be decomposed mainly by bacteria of this type’ [29].
Accordingly, a search of metagenomic sequence data identified C. acidurici-related 16S rRNA gene sequences in samples taken from a variety of habitats all over the world. These include, among others, the uncultured clones SBXY_753 and MAT-CR-P2-F02, collected from hypersaline microbial mats in the Guerrero Negro lagoon in Mexico [30] and in the Candeleria lagoon in Cabo Rojo, Puerto Rico [31], respectively; clone FGL12_B55 from an anoxygenic phototrophic community in Fayetteville Green Lake in upstate New York [32], and clone Q31013 from an intertidal sediment along the coast of Qinhuangdao in PR China [33] (Fig. 1). Metagenomic sequencing also revealed the presence of the uncultured clones, closely related to C. acidurici, C. purinilyticum and/or E. angustum, in ornithogenic soils of the Ross Sea region and King George Island in Antarctica, which form on land under the rookeries of Adélie penguins (e.g. clone 9/1/8C on Fig. 1) and Chinstrap and Gentoo penguins [34, 35]. This correlates with the finding of a closely related clone 1219A (GenBank accession no. FJ393497) in the fecal flora of Adélie penguins [36]. Finally, although not shown on Fig 1, 16S rRNA gene sequences falling within the radiation of the proposed family Gottschalkiaceae have been amplified from the samples taken from Artemia-associated microbiota in the solar salterns of Eilat, Israel [37], bovine mastitis milk [38], and anaerobic digesters treating poultry litter [39, 40], and other sources (see the https://www.arb-silva.de/browser/ssu-128/HE582772/ entry in the silva database [41] for more examples). These findings indicate the widespread distribution of Gottschalkiaceae-related organisms in both terrestrial and aquatic habitats, most likely due to their ability to metabolize avian-produced uric acid, as originally proposed by Barker and Beck [29].
Description of Gottschalkia gen. nov.
Gottschalkia (Gott.schal′ki.a. N.L. fem. dim. n. Gottschalkia named after Professor Dr Gerhard Gottschalk in recognition of his important contributions to the studies of Clostridia).
Gram-stain-positive, obligately anaerobic, straight or slightly curved rods, 0.5–1.5×2.5–10 µm. Motile by means of lateral flagella. Growth occurs from 18–19 °C and up to 37–42 °C. The optimum temperature for growth is 30–37 °C. The pH range for growth is from 6.5 to 7.0 and up to 9.0; the optimum pH for growth is between 7.5 and 8.1. Form spores that are round to oval and terminal or subterminal. Chemoorganotrophs that require purines for growth, but do not utilize carbohydrates and most amino acids. In the presence of 0.1 % (w/v) yeast extract, can grow using uric acid as the sole carbon and energy source. Can also utilize guanine, purine, 2-hydroxypurine, xanthine and hypoxanthine. Major products of metabolism are acetate, formate, CO2 and NH3. Oxidase-, catalase-, lipase- and urease-negative. Nitrate and sulfate are not reduced. Cell walls contain meso-diaminopimelate. Isolated from soil, marine and freshwater sources and avian droppings.
The type species is Gottschalkia acidurici [basonym Clostridiumacidurici (Barker 1938) Approved List 1980]. The G+C content of the chromosomal DNA ranges from 28 to 30 mol%.
Description of Gottschalkia acidurici comb. nov.
A.ci.du′ri.ci. N.L. gen. n. adj. acidurici of uric acid, referring to the preferred carbon source.
Basonym: Clostridiumacidurici (Liebert 1909) [29] (Approved List 1980).
The description of Gottschalkia acidurici is identical to that proposed for Clostridiumacidurici [2, 29, 42]. In addition to those described for the genus, has the following properties. Capable of growing in a salt medium containing 0.3 % (w/v) uric acid as the sole source of carbon, energy and nitrogen [42]. On an enrichment medium containing uric acid, forms whitish colonies 1–2 mm in diameter with irregular edges. Forms terminally located oval spores (0.9×1.1 µm in size) that cause a swelling of the cell.
The type strain G. acidurici 9aT(=ATCC 7906T=DSM 604T) was isolated from garden soil in California [29]. Its complete genome sequence [18] is available in GenBank under the accession no. CP003326. The G+C content of the genome is 29.9 mol% (27.8 % by the thermal denaturation method).
Description of Gottschalkia purinilytica comb. nov.
Pu.ri.ni.ly′ti.ca. N.L. fem. adj. purinilytica lysing the purine ring.
Basonym: Clostridiumpurinilyticum Dürre, Andersch and Andreesen 1981.
The description of Gottschalkia purinilytica is identical to that for Clostridiumpurinilyticum [2, 16]. In addition to those described for the genus, has the following properties. Forms spherical terminally located endospores (0.8 to 1.2 µm in size) that result in swollen cells. Requires selenium compounds and thiamine for growth. Can use adenine, adenosine, inosine, or xanthosine as the sole source of carbon and energy. In the presence of purines, is able to utilize glycine, formiminoglycine, benzoylglycine, glycyl-glycine, glycyl-glycyl-glycine and glycyl-leucine.
The type strain WA-1T(=ATCC 33906T=DSM 1384T) was isolated from farm soil containing chicken manure in Bovenden-Eddigehausen, Germany [16]. 43The G+C content of the genome is 28.8 % [43].
Description of Andreesenia gen. nov.
Andreesenia (An.dree.se′ni.a. N.L. fem. n. Andreesenia named after Professor Dr Jan Andreesen in recognition of his contributions to the studies of Clostridia).
Strictly anaerobic obligately purinolytic, Gram-stain-positive, non-spore-forming straight rods, 1.0–1.5×3–7 µm. Growth occurs from 18 to 45 °C (optimum temperature is 30–37 °C). The pH range for growth is from 6.5 to 10.0 (the optimum pH is between 7.5 and 8.5). In the presence of 0.1 % (w/v) yeast extract, can grow using uric acid as the sole carbon and energy source. Do not utilize carbohydrates, alcohols, amino acids, or organic acids. Do not grow on milk or chopped meat medium. The major products of metabolism are acetate, formate, CO2 and NH3. Oxidase-, catalase-, lipase- and urease-negative. Nitrate and sulfate are not reduced. Cell walls contain meso-diaminopimelate. Can be isolated from sewage, hypersaline microbial mats and avian droppings.
The type species is Andreesenia angusta (basonym Eubacterium angustum Beuscher and Andreesen 1985).
Description of Andreesenia angusta comb. nov.
An.gus′ta. L. fem. adj. angusta, restricted, referring to the narrow substrate range.
Basonym: Eubacterium angustum Beuscher and Andreesen 1985.
The description of Andreesenia angusta is identical to that for Eubacterium angustum [27, 44]. In addition to those described for the genus, has the following properties. Non-motile but produces lateral flagella. Requires thiamine for growth, but does not require selenium, tungstate or molybdate. Nutritionally restricted to grow only on uric acid, guanine, or xanthine; in the presence of uric acid, can utilize hypoxanthine. Cells can grow in the presence of 2 % (w/v) bile extract. Colonies are nonpigmented, flat, circular, and 0.5–1.5 mm in diameter. Myristic (tetradecanoic) acid C14 : 0 makes up more than 36 mol% of all fatty acids. A draft genome sequence of the type strain has been deposited in the GenBank with the accession no. MKIE00000000 [28].
The type strain MK-1T(=ATCC 43737T=DSM 1989T) was isolated from sewage plant sludge in Göttingen, Germany [27]. The G+C content of the genome is 43.6 % (40.3 mol% by the thermal denaturation method).
Description of Gottschalkiaceae fam. nov.
Gottschalkiaceae (Gott.schal.ki.a.ce′ae. N.L. fem. dim. n. Gottschalkia type genus of the family; L. suff. –aceae ending to denote a family; N.L. fem. pl. n. Gottschalkiaceae the family of the genus Gottschalkia).
Strictly anaerobic bacteria that can only grow by metabolizing purines. Gram-stain-positive, straight or slightly curved rods, 0.5–1.5×2–10 µm. Produces lateral and subterminal flagella. Growth occurs from 18–19 °C to 37–42 °C, the optimum growth temperature is 30–37 °C. The pH range is from 6.5 to 7.0 to 9.0, with the optimum pH between 7.5 and 8.2. May be spore-forming or asporogenous. In the presence of 0.1 % (w/v) yeast extract, can grow using uric acid, guanine, or xanthine as the sole carbon and energy sources; some representatives may also utilize other purines. Do not utilize carbohydrates and most amino acids; in the presence of purines, may use glycine, serine, or glycine-containing peptides. Major products of metabolism are acetate, formate, CO2 and NH3. Oxidase-, catalase-, lipase- and urease-negative. Nitrate and sulfate are not reduced. Cell walls contain meso-diaminopimelate. Predominant fatty acids are C14 : 0 and C16 : 1. Often associated with avian droppings and can be isolated from soil, and aquatic marine and freshwater sources.
The family includes the genera Gottschalkia and Andreesenia. The type genus is the genus Gottschalkia. The G+C content of the chromosomal DNA ranges from 28 to 44 mol%.
Funding information
This work was supported by the NIH Intramural Research Program at the U.S. National Library of Medicine (NY, MYG) and by ‘Nds. Ministerium für Wissenschaft und Kultur (MWK)’ of Lower Saxony, Germany (AP, RD).
Acknowledgements
We thank Professor Drs Gerhard Gottschalk and Jan Andreesen for the permission to use their names.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Supplementary Data
Footnotes
GenBank accession numbers : CP003326, LGSS00000000, MKIE00000000.
Two supplementary tables and two supplementary figures are available with the online Supplementary Material.
References
- 1.Ludwig W, Schleifer K-H, Whitman WB. Revised road map to the phylum Firmicutes. In: De Vos P, Garrity GM, Jones D, Krieg NR, Ludwig W, et al., editors. Bergey's Manual of Systematic Bacteriology. 2nd ed. vol. 3. New York: Springer; 2009. pp. 1–24. (editors) The Firmicutes. [Google Scholar]
- 2.Rainey FA, Hollen BJ, Small A. Genus I. Clostridium. In: De Vos P, Garrity GM, Jones D, Krieg NR, Ludwig W, et al., editors. Bergey's Manual of Systematic Bacteriology. New York: Springer; 2009. pp. 738–828. (editors) 2nd ed, vol. 3, The Firmicutes. [Google Scholar]
- 3.Parte AC. LPSN - list of prokaryotic names with standing in nomenclature. Nucleic Acids Res. 2014;42:D613–D616. doi: 10.1093/nar/gkt1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collins MD, Lawson PA, Willems A, Cordoba JJ, Fernandez-Garayzabal J, et al. The phylogeny of the genus Clostridium: proposal of five new genera and eleven new species combinations. Int J Syst Bacteriol. 1994;44:812–826. doi: 10.1099/00207713-44-4-812. [DOI] [PubMed] [Google Scholar]
- 5.Marchandin H, Teyssier C, Campos J, Jean-Pierre H, Roger F, et al. Negativicoccus succinicivorans gen. nov., sp. nov., isolated from human clinical samples, emended description of the family Veillonellaceae and description of Negativicutes classis nov., Selenomonadales ord. nov. and Acidaminococcaceae fam. nov. in the bacterial phylum Firmicutes. Int J Syst Evol Microbiol. 2010;60:1271–1279. doi: 10.1099/ijs.0.013102-0. [DOI] [PubMed] [Google Scholar]
- 6.Ludwig W, Schleifer K-H, Whitman WB. Taxonomic outline of the phylum Firmicutes. In: De Vos P, Garrity GM, Jones D, Krieg NR, Ludwig W, et al., editors. Bergey's Manual of Systematic Bacteriology. 2nd ed. vol. 3. New York: Springer; 2009. pp. 15–17. (editors) The Firmicutes. [Google Scholar]
- 7.Lawson PA, Rainey FA. Proposal to restrict the genus Clostridium (Prazmowski) to Clostridium butyricum and related species. Int J Syst Evol Microbiol. 2015;66:1009–1016. doi: 10.1099/ijsem.0.000824. [DOI] [PubMed] [Google Scholar]
- 8.Farrow JA, Lawson PA, Hippe H, Gauglitz U, Collins MD. Phylogenetic evidence that the gram-negative nonsporulating bacterium Tissierella (Bacteroides) praeacuta is a member of the Clostridium subphylum of the gram-positive bacteria and description of Tissierella creatinini sp. nov. Int J Syst Bacteriol. 1995;45:436–440. doi: 10.1099/00207713-45-3-436. [DOI] [PubMed] [Google Scholar]
- 9.Yutin N, Galperin MY. A genomic update on clostridial phylogeny: Gram-negative spore formers and other misplaced clostridia. Environ Microbiol. 2013;15:2341–2641. doi: 10.1111/1462-2920.12173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bae JW, Park JR, Chang YH, Rhee SK, Kim BC, et al. Clostridium hastiforme is a later synonym of Tissierella praeacuta. Int J Syst Evol Microbiol. 2004;54:947–949. doi: 10.1099/ijs.0.63068-0. [DOI] [PubMed] [Google Scholar]
- 11.Johnson CN, Whitehead TR, Cotta MA, Rhoades RE, Lawson PA, et al. Peptoniphilus stercorisuis sp. nov., isolated from a swine manure storage tank and description of Peptoniphilaceae fam. nov. Int J Syst Evol Microbiol. 2014;64:3538–3545. doi: 10.1099/ijs.0.058941-0. [DOI] [PubMed] [Google Scholar]
- 12.Alauzet C, Marchandin H, Courtin P, Mory F, Lemée L, et al. Multilocus analysis reveals diversity in the genus Tissierella: description of Tissierella carlieri sp. nov. in the new class Tissierellia classis nov. Syst Appl Microbiol. 2014;37:23–34. doi: 10.1016/j.syapm.2013.09.007. [DOI] [PubMed] [Google Scholar]
- 13.Federhen S. Type material in the NCBI taxonomy database. Nucleic Acids Res. 2015;43:D1086–D1098. doi: 10.1093/nar/gku1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Durre P, Andersch W, Andreesen JR. Isolation and characterization of an Adenine-Utilizing, anaerobic sporeformer, Clostridium purinolyticum sp. nov. Int J Syst Bacteriol. 1981;31:184–194. doi: 10.1099/00207713-31-2-184. [DOI] [Google Scholar]
- 17.Dürre P, Andreesen JR. Purine and glycine metabolism by purinolytic clostridia. J Bacteriol. 1983;154:192–199. doi: 10.1128/jb.154.1.192-199.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hartwich K, Poehlein A, Daniel R. The purine-utilizing bacterium Clostridium acidurici 9a: a genome-guided metabolic reconsideration. PLoS One. 2012;7:e51662. doi: 10.1371/journal.pone.0051662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harms C, Schleicher A, Collins MD, Andreesen JR. Tissierella creatinophila sp. nov., a gram-positive, anaerobic, non-spore-forming, creatinine-fermenting organism. Int J Syst Bacteriol. 1998;48:983–993. doi: 10.1099/00207713-48-3-983. [DOI] [PubMed] [Google Scholar]
- 20.Parshina SN, Kleerebezem R, Sanz JL, Lettinga G, Nozhevnikova AN, et al. Soehngenia saccharolytica gen. nov., sp. nov. and Clostridium amygdalinum sp. nov., two novel anaerobic, benzaldehyde-converting bacteria. Int J Syst Evol Microbiol. 2003;53:1791–1799. doi: 10.1099/ijs.0.02668-0. [DOI] [PubMed] [Google Scholar]
- 21.Rezgui R, Maaroufi A, Fardeau ML, Ben Ali Gam Z, Cayol JL, et al. Anaerosalibacter bizertensis gen. nov., sp. nov., a halotolerant bacterium isolated from sludge. Int J Syst Evol Microbiol. 2012;62:2469–2474. doi: 10.1099/ijs.0.036566-0. [DOI] [PubMed] [Google Scholar]
- 22.Yutin N, Puigbò P, Koonin EV, Wolf YI. Phylogenomics of prokaryotic ribosomal proteins. PLoS One. 2012;7:e36972. doi: 10.1371/journal.pone.0036972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yarza P, Yilmaz P, Pruesse E, Glöckner FO, Ludwig W, et al. Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat Rev Microbiol. 2014;12:635–645. doi: 10.1038/nrmicro3330. [DOI] [PubMed] [Google Scholar]
- 24.Qin QL, Xie BB, Zhang XY, Chen XL, Zhou BC, et al. A proposed genus boundary for the prokaryotes based on genomic insights. J Bacteriol. 2014;196:2210–2215. doi: 10.1128/JB.01688-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wiegel J, Tanner R, Rainey FA. An introduction to the family Clostridiaceae. In: Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E, editors. The Prokaryotes. : Bacteria: Firmicutes, Cyanobacteria. vol. 4. 2006. pp. 654–678. (editors) [Google Scholar]
- 26.Galperin MY, Mekhedov SL, Puigbo P, Smirnov S, Wolf YI, et al. Genomic determinants of sporulation in Bacilli and Clostridia: towards the minimal set of sporulation-specific genes. Environ Microbiol. 2012;14:2870–2890. doi: 10.1111/j.1462-2920.2012.02841.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beuscher HU, Andreesen JR. Eubacterium angustum sp. nov., a Gram-positive anaerobic, non-sporeforming, obligate purine fermenting organism. Arch Microbiol. 1984;140:2–8. doi: 10.1007/BF00409763. [DOI] [Google Scholar]
- 28.Poehlein A, Galperin MY, Andreesen JR, Daniel R. Genome sequence of uric acid-fermenting Eubacterium angustum DSM 1989T (MK-1) Genome Announc. 2017;5:e01439-16. doi: 10.1128/genomeA.01439-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barker HA, Beck JV. Clostridium acidi-uridi and Clostridium cylindrosporum, organisms fermenting uric acid and some other purines. J Bacteriol. 1942;43:291–304. doi: 10.1128/jb.43.3.291-304.1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harris JK, Caporaso JG, Walker JJ, Spear JR, Gold NJ, et al. Phylogenetic stratigraphy in the Guerrero Negro hypersaline microbial mat. ISME J. 2013;7:50–60. doi: 10.1038/ismej.2012.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Isenbarger TA, Finney M, Rios-Velazquez C, Handelsman J, Ruvkun G. Miniprimer PCR, a new lens for viewing the microbial world. Appl Environ Microbiol. 2008;74:840–849. doi: 10.1128/AEM.01933-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meyer KM, Macalady JL, Fulton JM, Kump LR, Schaperdoth I, et al. Carotenoid biomarkers as an imperfect reflection of the anoxygenic phototrophic community in meromictic Fayetteville Green Lake. Geobiology. 2011;9:321–329. doi: 10.1111/j.1472-4669.2011.00285.x. [DOI] [PubMed] [Google Scholar]
- 33.Li J, Li F, Yu S, Qin S, Wang G. Impacts of mariculture on the diversity of bacterial communities within intertidal sediments in the Northeast of China. Microb Ecol. 2013;66:861–870. doi: 10.1007/s00248-013-0272-6. [DOI] [PubMed] [Google Scholar]
- 34.Aislabie J, Jordan S, Ayton J, Klassen JL, Barker GM, et al. Bacterial diversity associated with ornithogenic soil of the Ross Sea region, Antarctica. Can J Microbiol. 2009;55:21–36. doi: 10.1139/W08-126. [DOI] [PubMed] [Google Scholar]
- 35.Kim OS, Chae N, Lim HS, Cho A, Kim JH, et al. Bacterial diversity in ornithogenic soils compared to mineral soils on King George Island, Antarctica. J Microbiol. 2012;50:1081–1085. doi: 10.1007/s12275-012-2655-7. [DOI] [PubMed] [Google Scholar]
- 36.Banks JC, Cary SC, Hogg ID. The phylogeography of Adelie penguin faecal flora. Environ Microbiol. 2009;11:577–588. doi: 10.1111/j.1462-2920.2008.01816.x. [DOI] [PubMed] [Google Scholar]
- 37.Tkavc R, Ausec L, Oren A, Gunde-Cimerman N. Bacteria associated with Artemia spp. along the salinity gradient of the solar salterns at Eilat (Israel) FEMS Microbiol Ecol. 2011;77:310–321. doi: 10.1111/j.1574-6941.2011.01112.x. [DOI] [PubMed] [Google Scholar]
- 38.Oikonomou G, Machado VS, Santisteban C, Schukken YH, Bicalho RC. Microbial diversity of bovine mastitic milk as described by pyrosequencing of metagenomic 16s rDNA. PLoS One. 2012;7:e47671. doi: 10.1371/journal.pone.0047671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Garcia SL, Jangid K, Whitman WB, das KC. Transition of microbial communities during the adaption to anaerobic digestion of carrot waste. Bioresour Technol. 2011;102:7249–7256. doi: 10.1016/j.biortech.2011.04.098. [DOI] [PubMed] [Google Scholar]
- 40.Smith AM, Sharma D, Lappin-Scott H, Burton S, Huber DH. Microbial community structure of a pilot-scale thermophilic anaerobic digester treating poultry litter. Appl Microbiol Biotechnol. 2014;98:2321–2334. doi: 10.1007/s00253-013-5144-y. [DOI] [PubMed] [Google Scholar]
- 41.Yilmaz P, Parfrey LW, Yarza P, Gerken J, Pruesse E, et al. The SILVA and "All-species Living Tree Project (LTP)" taxonomic frameworks. Nucleic Acids Res. 2014;42:D643–D648. doi: 10.1093/nar/gkt1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barker HA, Peterson WH. The nutritional requirements of Clostridium acidi-urici. J Bacteriol. 1944;47:307–308. doi: 10.1128/jb.47.3.307-308.1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Poehlein A, Bengelsdorf FR, Schiel-Bengelsdorf B, Daniel R, Dürre P. Draft genome sequence of purine-degrading Gottschalkia purinilyticum (formerly Clostridium purinilyticum) WA1 (DSM 1384) Genome Announc. 2015;3:e01088. doi: 10.1128/genomeA.01088-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wade WG. Genus I. Eubacterium. In: De Vos P, Garrity GM, Jones D, Krieg NR, Ludwig W, et al., editors. Bergey's Manual of Systematic Bacteriology. 2nd ed. vol. 3. New York: Springer; 2009. pp. 865–891. (editors) The Firmicutes. [Google Scholar]
- 45.Lawson PA, Citron DM, Tyrrell KL, Finegold SM. Reclassification of Clostridium difficile as Clostridioides difficile (Hall and O'Toole 1935) Prévot 1938. Anaerobe. 2016;40:95–99. doi: 10.1016/j.anaerobe.2016.06.008. [DOI] [PubMed] [Google Scholar]
- 46.Galperin MY, Brover V, Tolstoy I, Yutin N. Phylogenomic analysis of the family Peptostreptococcaceae (Clostridium cluster XI) and proposal for reclassification of Clostridium litorale (Fendrich et al. 1991) and Eubacterium acidaminophilum (Zindel et al. 1989) as Peptoclostridium litorale gen. nov. comb. nov. and Peptoclostridium acidaminophilum comb. nov. Int J Syst Evol Microbiol. 2016;66:5506–5513. doi: 10.1099/ijsem.0.001548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 1993;10:512–526. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]
- 48.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 50.Anisimova M, Gil M, Dufayard JF, Dessimoz C, Gascuel O. Survey of branch support methods demonstrates accuracy, power, and robustness of fast likelihood-based approximation schemes. Syst Biol. 2011;60:685–699. doi: 10.1093/sysbio/syr041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Collins MD, Shah HN. Reclassification of Bacteroides praeacutus Tissier (Holdeman and Moore) in a new genus, Tissierella, as Tissierella praeacuta comb. nov. Int J Syst Bacteriol. 1986;36:461–463. doi: 10.1099/00207713-36-3-461. [DOI] [Google Scholar]
- 52.Dione N, Sankar SA, Lagier JC, Khelaifia S, Michele C, et al. Genome sequence and description of Anaerosalibacter massiliensis sp. nov. New Microbes New Infect. 2016;10:66–76. doi: 10.1016/j.nmni.2016.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schnurer A, Schink B, Svensson BH. Clostridium ultunense sp. nov., a mesophilic bacterium oxidizing acetate in syntrophic association with a hydrogenotrophic methanogenic bacterium. Int J Syst Bacteriol. 1996;46:1145–1152. doi: 10.1099/00207713-46-4-1145. [DOI] [PubMed] [Google Scholar]
- 54.Wei Y, Zhou H, Zhang L, Zhang J, Wang Y, et al. Draft genome sequence of Clostridium ultunense strain BS (DSMZ 10521), recovered from a mixed culture. Genome Announc. 2014;2:e01269-13. doi: 10.1128/genomeA.01269-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schiefer-Ullrich H, Wagner R, Dürre P, Andreesen JR. Comparative studies on physiology and taxonomy of obligately purinolytic clostridia. Arch Microbiol. 1984;138:345–353. doi: 10.1007/BF00410902. [DOI] [PubMed] [Google Scholar]
- 56.Andreesen JR, Zindel U, Durre P. Clostridium cylindrosporum (ex Barker and Beck 1942) nom. rev. Int J Syst Bacteriol. 1985;35:206–208. doi: 10.1099/00207713-35-2-206. [DOI] [Google Scholar]
- 57.Poehlein A, Montoya Solano JD, Bengelsdorf FR, Schiel-Bengelsdorf B, Daniel R, et al. Draft genome sequence of purine-degrading Clostridium cylindrosporum HC-1 (DSM 605) Genome Announc. 2015;3:e00917-15. doi: 10.1128/genomeA.00917-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sasser M. Identification of Bacteria by Gas Chromatography of Cellular Fatty Acids. MIDI Technical Note 101. Newark, DE: MIDI Inc; 2001. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.