Abstract
The subtelomere, a telomere-adjacent chromosomal domain, contains species-specific homologous DNA sequences, in addition to various genes. However, the functions of subtelomeres, particularly subtelomeric homologous (SH) sequences, remain elusive. Here, we report the first comprehensive analyses of the cellular functions of SH sequences in the fission yeast, Schizosaccharomyces pombe. Complete removal of SH sequences from the genome revealed that they are dispensable for mitosis, meiosis and telomere length control. However, when telomeres are lost, SH sequences prevent deleterious inter-chromosomal end fusion by facilitating intra-chromosomal circularization. Surprisingly, SH-deleted cells sometimes survive telomere loss through inter-chromosomal end fusions via homologous loci such as LTRs, accompanied by centromere inactivation of either chromosome. Moreover, SH sequences function as a buffer region against the spreading of subtelomeric heterochromatin into the neighboring gene-rich regions. Furthermore, we found a nucleosome-free region at the subtelomeric border, which may be a second barrier that blocks heterochromatin spreading into the subtelomere-adjacent euchromatin. Thus, our results demonstrate multiple defense functions of subtelomeres in chromosome homeostasis and gene expression.
INTRODUCTION
Telomeres, which are specialized chromatin structures at the ends of eukaryotic linear chromosomes, are crucial for genome stability. Telomere repeat DNA-binding proteins participate in activities crucial for cell viability, including protection of chromosome ends, regulation of telomere length, formation of heterochromatin adjacent to telomeres and regulation of chromosome movements in mitosis and meiosis (1–4). Many eukaryotic chromosomes possess subtelomere regions adjacent to telomeres and close to chromosome ends. Within a specific organism, subtelomeres generally contain multiple sequence segments that share high similarity with each other, and that are distinct from the telomere repeat sequence. In the budding yeast, Saccharomyces cerevisiae, the subtelomeres contain X and Y’ elements, which are highly variable in length; the Y’ element includes the open reading frame (ORF) of a helicase gene (5). In humans, the subtelomeres are composed of a mosaic of multiple common segments (more than 40 types in total) containing various ORFs (6,7).
Recent studies have shown that microdeletions in human subtelomeres cause intellectual disability and/or multiple congenital anomalies (subtelomere deletion syndrome, STDS) (8). Importantly, ∼5% of patients with intellectual disability exhibit subtelomeric rearrangement. The major cause of STDS is thought to be haploinsufficiency of subtelomeric genes. Furthermore, expression of subtelomeric genes is influenced by telomere position effects (TPEs). Facioscapulohumeral muscular dystrophy (FSHD) is an age-related disease caused by a striking increase in expression of the DUX4 gene, which is located at the subtelomere of chromosome 4q and is affected by TPE (9). Thus, maintenance of expression of subtelomeric genes is critical for human health.
The fission yeast Schizosaccharomyces pombe possesses three chromosomes (Figure 1A). Although some parts of the subtelomeres have not yet been sequenced (http://www.pombase.org/status/sequencing-status), they are mainly composed of two distinct parts (Figure 1B). One is the telomere-adjacent SH region, spanning ∼50 kb, which contains a mosaic of multiple segments of homologous sequences (10). The other is the telomere-distal subtelomere, containing unique sequences and spanning ∼50 kb. The telomere-proximal regions, although not completely overlapping the homologous regions, form heterochromatin, in which histone H3 methylated at Lys9 (H3K9me) is enriched (11,12). The telomere-distal region forms a highly condensed chromatin structure, designated a ‘knob,’ which is distinct from heterochromatin (13). In the knob region, which overlaps so-called subtelomeric (ST) chromatin, the levels of histone modifications are very low compared with the neighboring euchromatin and subtelomeric heterochromatin regions (14). Interestingly, a shugoshin family protein, Sgo2, which is recruited to the centromeres and contributes to precise chromosome segregation during mitosis (15), is relocated to the subtelomeres during interphase and plays a crucial role in knob formation (16).
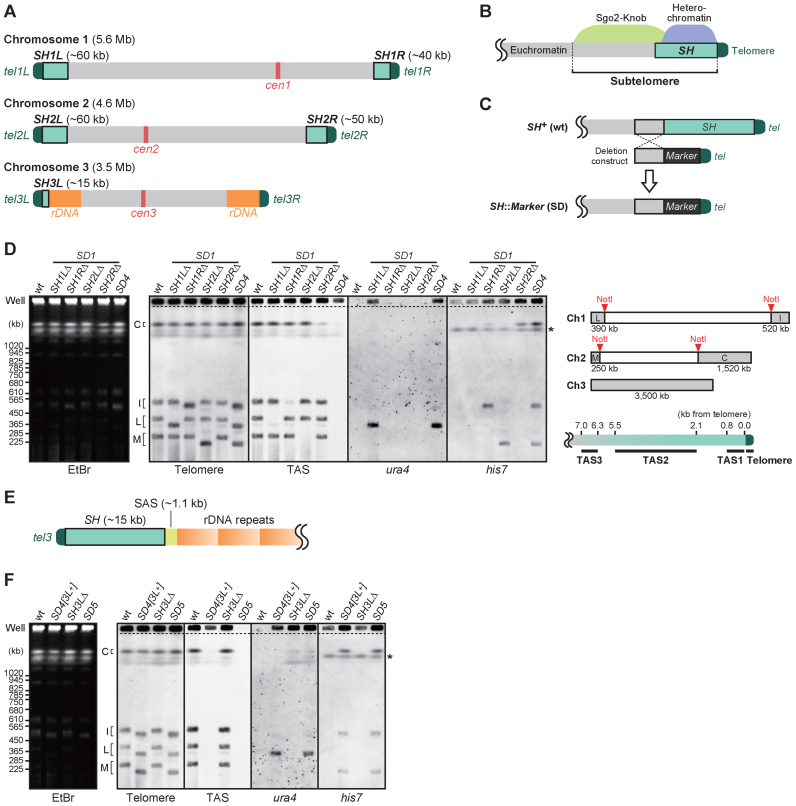
Figure 1.
Deletion of all of the SH regions from the Schizosaccharomyces pombe genome. (A) Schematic illustration of S. pombe chromosomes. Boxes in light green adjacent to telomeres (dark green) represent subtelomeric homologous (SH) regions, which share high sequence identity (>98%) with more than one other subtelomere. (B) Schematic illustration of the subtelomeres of chromosomes 1 and 2 in S. pombe. Heterochromatin is formed around SH regions (∼50 kb), which are adjacent to telomeres. A highly condensed knob structure (∼50 kb) is generated by Sgo2, adjacent to the heterochromatin. The Sgo2-knob region contains unique sequences, where various histone modifications are maintained at low levels. In this paper, the term ‘subtelomere’ refers to the region from the telomere-proximal end of an SH region to the telomere-distal end of the Sgo2-knob region. A euchromatin region, where gene transcription is active, is adjacent to the subtelomere. (C) Deletion strategy for SH sequences. Target SH regions were replaced with marker genes by homologous recombination to obtain SH deletion (SD) mutants. (D) Pulsed-field gel electrophoresis (PFGE) of NotI restriction fragments of the S. pombe genome followed by Southern blotting using telomere, TAS (telomere-associated sequence), ura4 or his7 as probes. Left panel, ethidium bromide staining of the gel after PFGE. The asterisk indicates the endogenous his7 gene with a point mutation (his7–366), which was also detected by the his7 probe. SD1, single SH deletion mutant series. SD4, SH1LΔ1RΔ2LΔ2RΔ. The upper right panel illustrates the telomere-containing NotI restriction fragments. Note that fragments L, I, M and C contain SH1L, SH1R, SH2L and SH2R, respectively, and that chromosome 3, which contains no Not1 site, was mostly retained in the wells of gels due to its long length. The lower right panel illustrates the positions of the DNA fragments detected by probes for telomere and TAS (TAS1–3) sequences. (E) Schematic illustration of the DNA composition of at least one of the two ends of chromosome 3. SAS, subtelomere-associated sequence (24). (F) Analyses of the chromosome end structures in the SD5 mutant in which all five SH regions were deleted. PFGE-Southern analyses performed as described in (D). Asterisk indicates the position of the endogenous his7 gene with a point mutation (his7–366).
Although some of the functions of subtelomeres have been clarified, their overall functions in influencing gene expression and maintaining stability at the whole chromosome scale have yet to be investigated. In particular, the physiological significance of the multiple copies of SH sequences remains obscure. Complete removal of SH sequences from the genome is the most appropriate strategy to clarify its roles in vivo; however, it is difficult to delete all subtelomere regions because most eukaryotes possess a large number of subtelomeres. Schizosaccharomyces pombe is an ideal model organism for studying subtelomeres because it normally proliferates as a haploid organism and possesses only three chromosomes and a maximum of six subtelomeres. Deleting all SH sequences from the S. pombe genome by a conventional method, homologous recombination, was practicable because no essential ORFs are present in its subtelomere regions (17).
In this study, we successfully deleted all of the SH sequences from the S. pombe genome, revealing novel chromosome defense systems important for cell survival during various chromosomal crises. Unexpectedly, the complete deletion of SH sequences did not affect telomere length, mitosis or meiosis under normal conditions. However, when telomeres were eliminated by telomerase deletion in SH-deleted cells, survivors primarily harbored inter- or intra-chromosomal circularized chromosomes, where fusion had occurred within homologous genomic regions around the subtelomeres. The inter-chromosomal fused chromosomes carried two centromeres, one of which remained functional, whereas the other was inactivated. Furthermore, SH sequence deletion allowed heterochromatin invasion of the SH-adjacent subtelomeric regions, thereby repressing the expression of genes in these regions. Thus, the SH region serves as a buffer zone to maintain the active expression of subtelomeric genes. In addition, the telomere-distal border of the subtelomere exhibited a nucleosome-free configuration, which may prevent further spread of heterochromatin into the euchromatin region. These results reveal for the first time the in vivo functions of eukaryotic subtelomeres in chromosome homeostasis, cell survival and gene regulation.
MATERIALS AND METHODS
Strains, media and general techniques for S. pombe
The S. pombe strains used in this study are listed in Supplementary Table S2. Growth media and basic genetic and biochemical techniques were as described previously (18–20). The primers listed in Supplementary Table S3 were used to generate DNA fragments for strain construction. The details of the generation of SH deletions are described below.
Calculating the distance from a telomere
The distance from telomeric repeat DNA was calculated as described previously (16). If a genomic site of interest was located within the gap between the contigs c1348 and pB10D8 at subtel2L, the distance was calculated using the following equation:
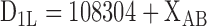 |
where, XAB is the nucleotide position number in sequence data determined previously (21) (accession number AB325691).
Sequencing of the subtelomere of chromosome 3
The DNA fragment between the telomere and rDNA of the left arm of chromosome 3 (3L) was amplified by polymerase chain reaction (PCR) using two primers, one (jk386) that hybridizes to the SH region and the other (TB19) to rDNA. The resultant PCR product (10 kb) was subjected to DNA sequencing using the same primers (sequences below):
jk386 5′-TTCCAAGTATGCCAGCTTATCATC-3′
TB19 5′-GGGAACCAGGACTTTTACCTTGA-3′
Deletions of SH regions
Telomeric DNA (0.3 kb) derived from pNSU70 (10) was cloned into the KpnI/ApaI sites of the pBluescript vector, and then the auxotrophic marker genes, ura4+ and his7+, were inserted next to the telomeric DNA to obtain pYN1 and pYN2, respectively. For ura4+, a ura4+-containing HindIII restriction fragment (1.8 kb) was used. For cloning of the his7+ gene, a PCR product (2.3 kb) amplified using the following primer set was treated with BamHI and XhoI.
yn1 5′-TATAAAGGATCCCTCCTATGATGATGTCTTGC-3′
yn2 5′-TATAAACTCGAGACTTGATACCGCAGTATCCTTTG-3′
For SH deletion constructs, a portion of each subtelomeric region with unique DNA sequences adjacent to the SH regions was amplified by PCR using the following primer sets, and then cloned into pYN1 or pYN2.
| - subtel1L (treated with PstI/SmaI and cloned into pYN1 to obtain pYN3) |
| yn31 5′-TATAAACTGCAGAAATAAATCTTTGTACGTTGGATACTC-3′ |
| yn32 5′-TATAAACCCGGGAAAGACGATTTTCATTACTTCTAGCC-3′ |
| - subtel1R (treated with BamHI/SpeI and cloned into pYN2 to obtain pYN4) |
| yn79 5′-TATAAAGGATCCGATATTTTATGGTGCTATCCAGTC-3′ |
| yn80 5′-TATAAAACTAGTCTTGATAGCCATAAAATGTGTTTAG-3′ |
| - subtel2L (treated with BamHI/SpeI and cloned into pYN2 to obtain pYN5) |
| yn35 5′-TATAAAGGATCCGTTCTGCTTCTCGACATTATG-3′ |
| yn36 5′-TATAAAACTAGTGTTCTGCTTCTCGACATTATG-3′ |
| - subtel2R (treated with BamHI/SpeI and cloned into pYN2 to obtain pYN6) |
| yn11 5′-TATAAAGGATCCAGCCGCTTCATCATTACTTACTG-3′ |
| yn12 5′-TATAAAACTAGTGAGCTTATATCTGAAGCTACTCC-3′ |
| - subtel3L (treated with BamHI/XbaI and cloned into pYN1 to obtain pSTk36) |
| st127 5′-TATAAATCTAGAAATGGAATTGTTTGGTTACACAAC-3′ |
| st128 5′-TATAAAGGATCCTGAAGGGATAAATAATGTATCAACAAAT-3′ |
pYN3, pYN4, pYN5, pYN6 and pSTk36 were digested with SacI/SmaI, EcoRI/SpeI, EcoRI/SpeI, EcoRI/SpeI and SacI/XbaI, respectively, to excise the subtelomere–marker–telomere fragments, which were used for transformation. The resultant series of single SH deletion (SD1) mutants was successively crossed to obtain SD5 and other mutant strains with combined SH deletions.
Pulsed-field gel electrophoresis (PFGE)
Pulsed-field gel electrophoresis (PFGE) of NotI-digested chromosomal DNA was performed using a CHEF-DR III Pulsed Field Electrophoresis Systems (Bio-Rad) under the following conditions: 1% SeaKem Gold Agarose (Lonza) or Certified™ Megabase Agarose (BioRad) in 0.5 × TBE; temperature, 10°C; initial switch time, 40 s; final switch time, 80 s; run time, 18 h; voltage gradient, 6.8 V/cm; and angle, 120°. In Supplementary Figure S5A, PFGE of whole chromosomal DNA was performed under the following conditions: 0.8% Certified™ Megabase Agarose in 1 × TAE; temperature, 14°C; first run: initial switch time, 1200 s; final switch time, 1200 s; run time 24 h; voltage gradient, 2 V/cm; angle, 96°; second run: initial switch time, 1500 s; final switch time, 1500 s; run time 24 h; voltage gradient, 2 V/cm; angle, 100°; third run: initial switch time, 1800 s; final switch time, 1800 s; run time 24 h; voltage gradient, 2 V/cm; angle, 106°.
Southern blotting
Restriction-digested genomic DNA was separated by conventional agarose gel electrophoresis or PFGE, and subjected to Southern blotting. For the telomere and telomere-associated sequence (TAS) probes, telomeric DNA and TAS fragments (TAS1, TAS2 and TAS3) were excised from pNSU70. For the 0–10 kb probe, the two ApaI/HindIII fragments of pNSU70 (1.5 and 5.4 kb of DNA covering 0–7 kb from the telomere) and the HindIII/EcoT22I fragment of pNSU56 (10) (2.8 kb of DNA covering 7–10 kb from the telomere) were mixed and then further digested using EcoT22I. For the other subtelomeric probes, PCR products (amplified using JP1225 genomic DNA as a template) digested with the restriction enzymes indicated below were used:
| - subtel 9–20 kb (digested with EcoT22I) |
| jk239 5′-CCGAACAGCTGTTTCCGCTG-3′ |
| jk456 5′-GACCGCTACGCAACCATAAAG-3′ |
| - subtel 20–30 kb (digested with ClaI) |
| jk455 5′-AACGAGTTGTGCAATGTTAGTAAGGT-3′ |
| jk389 5′-TGTTACTCATACTTGAAATACAATTTGAATG-3′ |
| - subtel 30–40 kb (digested with SpeI) |
| jk388 5′-GGCACAATTTCATTTCGTTTAGTTTAC-3′ |
| jk603 5′-CAGTGTGACTGGCACGAACC-3′ |
| - subtel 40–50 kb (digested with EcoT22I) |
| jk602 5′-ACGCCTTTGTTCAATCGAGTAAA-3′ |
| jk607 5′-AGATCAGCCAATGGCAGATGTA-3′ |
In the case of the subtel 50–60-kb probe, two PCR products, jk606-st162 and st161-st152, were fused by PCR with jk606 and st152, and then digested with HindIII:
jk606 5′-TTATCGCGGTGGCTATGGTT-3′
st162 5′-CTGCTTAGCACTTTGGCATTCTT-3′
st161 5′-TTTTCAAGTGTTCTTATTCGCATGA-3′
st152 5′-ATCTCCAACTTGAAGAAAAAGTAGAAC-3′
For the Ch3L and Ch3R probes, the rpc37+ and apl4+ gene loci were amplified by PCR using the following primer sets:
| - rpc37 (Ch3L probe) |
| st171 5′-ATGTCCTTTTCAGAAGATCAAGC-3′ |
| st172 5′-CTAAATAAAGGAGTAATCTTCATCAACAG-3′ |
| - apl4 (Ch3R probe) |
| st173 5′-ATGCAAACAACACATCCAAAGAAC-3′ |
| st174 5′-CTATTGTAAAAGGTCAGATGGCAAC-3′ |
For the ura4 probe, a ura4+-containing HindIII restriction fragment was excised from pYN1. For the his7 probe, pYN2 was treated with ClaI/BamHI to obtain the 5′ half of the his7+ gene. To detect telomere-containing NotI restriction fragments of the S. pombe genome (designated L, I, M and C), the gti1+, mcp3+, fbp1+ and amo1+ gene loci were amplified by PCR using the following primer sets:
| - gti1 (L probe) |
| st163 5′-ATGACCGAACCTGGCAATCTTC-3′ |
| st164 5′-TCATGACAAGGAGCCGCGTTC-3′ |
| - mcp3 (I probe) |
| st241 5′-ATGACTAAAGAAACTCACGAAAATCA-3′ |
| st242 5′-CTATTTGTCTCCAAATGTTTGCTG-3′ |
| - fbp1 (M probe) |
| st245 5′-ATGAAAAAAGATCTCGACGAAATC-3′ |
| st246 5′-CTATTTTATGAAATTAATATATTCCTCGACTTC-3′ |
| - amo1 (C probe) |
| st243 5′-ATGGTCGTTTGTAAGTATTTCCTTC-3′ |
| st244 5′-TTAACAAAATTGTGGTGGAGG-3′ |
For Southern blot analyses, these DNA fragments were labeled with digoxigenin (DIG) using a DIG High Prime DNA Labeling and Detection Starter Kit II (Roche) and signal detection was performed according to the manufacturer’s instructions.
Re-introduction of the trt1+ gene
The trt1+ re-introduction into trt1-deleted cells was conducted by genome integration of a linearized plasmid DNA containing the trt1+ gene and the hygr gene (hygromycin resistant marker). Two regions within the leu1 locus were amplified by PCR using the primer sets, st1-st2 and st3-st4.
st1 5′-GGATTAACAATGCCCTTGCCAGCGATATCG-3′
st2 5′-TATAAAGAGCTCCGTAAGAGTATGGGTGTTTGGGC-3′
st3 5′-TATAAAGAGCTCTTTTAACTCAGGTCGCTTCTTCTC-3′
st4 5′-TGACATTAAAAATTTCGTTTACTAACGTAG-3′
Both PCR products were treated with SacI and Aor51HI, and simultaneously cloned into the SacI site of the pFA6a-hphMX6 vector to obtain pSTk14. Next, a DNA fragment containing the full-length of the trt1+ gene was amplified using the following primer set.
st483 5′-TATAAACCCGGGTTCGCTTACTTTTAATCG-3′
st484 5′-TATAAACCCGGGCTTTTACCAAAATTCG-3′
The PCR product was treated with SmaI and cloned into the SmaI site of pSTk14. The resultant plasmid, pSTk79, was linearized by Aor51HI digestion before integration at the leu1 locus.
Flow cytometry
Cells growing logarithmically in YES liquid medium were subjected to 70% ethanol fixation, followed by 200 μg/ml RNaseA treatment (37°C for 4 h) and 5 μg/ml propidium iodide staining. DNA content was measured by flow cytometry of the propidium iodide-stained cells using FACSCalibur™ (BD), and the data were analyzed by the CellQuest™ (BD) software.
Chromatin immunoprecipitation (ChIP)
Chromatin immunoprecipitation (ChIP) was performed as described previously (16) with anti-H3K9me2 (MAB Institute, MABI 0307), anti-H3 (Abcam, ab1791), anti-H2A (Abcam, ab13923), anti-Flag (Sigma, M2 F3165), and anti-Cnp1 (22) antibodies. Sequences of the primer sets for subsequent quantitative PCR are listed in Supplementary Table S4.
RNA analyses
Total RNA was prepared from exponentially growing cells as described previously (16), followed by the treatment with recombinant DNase I (RNase-free) (TaKaRa Bio). For Reverse transcription (RT)-PCR, complementary DNA was synthesized using a High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems) with random primers and analyzed by quantitative PCR using the StepOne real-time PCR system. Sequences of the primer sets used for quantitative PCR are listed in Supplementary Table S4.
Microarray analysis
Poly(A)-containing RNA was purified from DNase I-treated total RNA using the PolyATtract mRNA Isolation System (Promega). Biotin-labeled cRNAs were synthesized from 0.3 μg of mRNA using GeneChip One-Cycle Target Labeling and Control Reagents (Affymetrix) and were hybridized to a GeneChip Yeast Genome 2.0 Array (Affymetrix), in accordance with the manufacturer's instructions. After staining, GeneChips were scanned using a GeneChip Scanner 3000 G7 and GeneChip Operating Software (GCOS 1.4). Scaling and comparative analyses were performed using GCOS 1.4 software with default settings and S. cerevisiae probe set masking. Data are available under Gene Expression Omnibus (GEO) accession code GSE69714.
RESULTS
Complete deletion of subtelomeric homologous (SH) sequences in S. pombe
To investigate the physiological roles of telomere-proximal SH sequences sharing high identity (>98%) with at least one other subtelomere, we set out to delete all of the SH regions from S. pombe chromosomes. The number of SH regions varies from four to six per S. pombe haploid genome, depending on the presence of SH between the telomeres and the rDNA repeats of chromosome 3 (Figure 1A). We first deleted the four SH regions (SH1L, SH1R, SH2L and SH2R) of chromosomes 1 and 2, which span 40–60 kb each (Figure 1A; see ‘Materials and Methods’ section for the calculation of each length). Each SH region was replaced with a marker gene (his7+ or ura4+) by homologous recombination to produce strains in which one SH region was deleted (SD1 strains: SH1LΔ, SH1RΔ, SH2LΔ and SH2RΔ) (Figure 1C). PFGE of the NotI-digested genomic DNA followed by Southern blotting confirmed each replacement and shortening of the subtelomeres (Figure 1D). SD1 strains were then crossed with one another to produce an SD4 strain, in which the four SH regions of chromosomes 1 and 2 were deleted. Southern blot analyses using TASs (TAS1, 0–0.8 kb; TAS2, 2.1–5.5 kb; and TAS3, 6.3–7.0 kb from the telomere of the right arm of chromosome 2, tel2R) (23) within SH regions as probes revealed that the SD4 strain exhibited residual TAS signals at the position of the well in the gel (Figure 1D, well) where chromosome 3, undigested chromosomes 1 and 2, and recombination intermediates remained during electrophoresis because of their large size. These results suggest that additional SH regions exist in chromosome 3.
To identify the remaining SH sequences in SD4, the telomere-proximal regions of chromosome 3 were amplified by PCR (see ‘Materials and Methods’ section). The DNA between the telomere and the rDNA repeats contained a 1.1 kb chromosome 3-specific subtelomere-associated sequence (SAS) (24) and an SH sequence of ∼15 kb with high similarity to the telomere-adjacent part of SH2R (Figure 1E). PFGE-Southern analyses of chromosome 3 indicated that a unit consisting of a short SH and an SAS was located adjacent to the telomere of the left arm of chromosome 3 (3L) of the control strain used in this study (Figure 1A and Supplementary Figure S1A).
To delete all of the SH regions, we replaced the SH3L region with the ura4+ marker gene, and the resultant SH3LΔ strain was crossed with SD4 to produce SD5. PFGE-Southern analyses showed that there was no residual TAS signal in SD5 (Figure 1F, TAS), indicating that all of the TAS sequences were deleted in this strain. Southern analyses for telomere-distal parts of SH other than TAS indicated that there was no SH sequence remaining in the genome of the SD5 strain (Supplementary Figure S1B and C). We confirmed that the marker genes used for the SH deletion, ura4+ and his7+, were stably maintained at the inserted loci with no amplification or translocation (Supplementary Figure S2A). Moreover, the expression of these marker genes was strongly repressed (Supplementary Figure S2B), indicating that replacement of SHs with the marker genes did not interfere with formation of the highly condensed structure of chromosome ends.
SH regions are dispensable for mitotic cell growth, meiotic progression, various stress responses and telomere length control
To investigate the physiological roles of SH sequences, we analyzed the effects of completely deleting them. SD5 cells grew at almost the same rate as wild-type cells and exhibited normal cell morphology in nutrient-rich YES medium at 32°C (Figure 2A and B; Supplementary Figure S3), indicating that the SH regions are dispensable for vegetative cell growth under normal conditions.
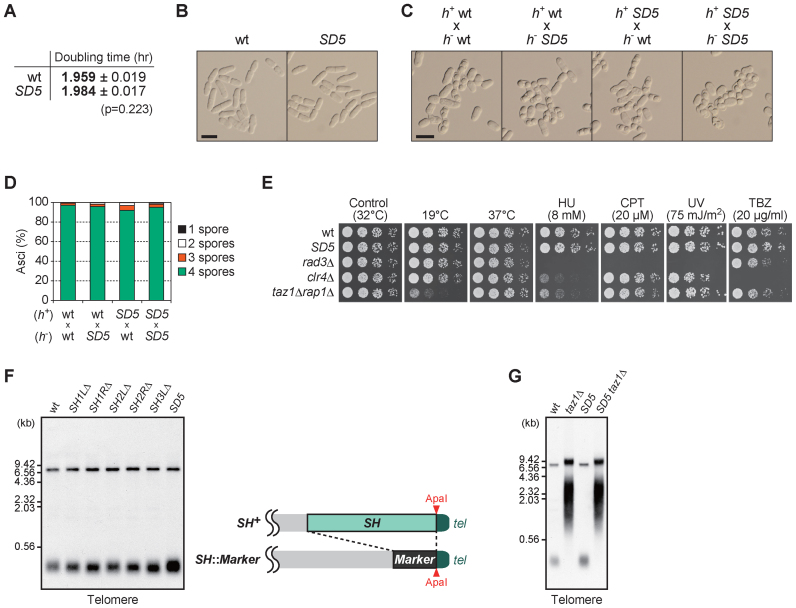
Figure 2.
SH sequences are dispensable for mitosis, meiosis, various stress responses and telomere length control. (A) Growth rate of Schizosaccharomyces pombe strains in YES liquid medium at 32°C. Doubling time is the mean of three independent experiments shown in Supplementary Figure S3. Standard deviation (s.d.) is given as errors. (B) Morphology of wild-type and SD5 cells growing exponentially in YES liquid medium. Shown are DIC images. Bar, 10 μm. (C) Mating and sporulation of the indicated crosses. Normal spore formation was observed in all cases after incubation on MEA plate at 28°C for 2 days. Bar, 10 μm. (D) Spore numbers per ascus in each cross in (C). More than 200 asci were analyzed for each cross. (E) Cell growth assays on YES plates at low (19°C) and high (37°C) temperatures, or on YES plates containing hydroxyurea (HU), camptothecin (CPT) or thiabendazole (TBZ) at the indicated concentrations. For the ultraviolet (UV) sensitivity assay, cells were subjected to UV irradiation immediately after spotting onto YES plates. (F) Analyses of telomere DNA length in SH-deleted cells. Genomic DNA prepared from wild-type, SH1LΔ, SH1RΔ, SH2LΔ, SH2RΔ, SH3LΔ and SD5 strains were digested with ApaI and subjected to Southern blot using telomere DNA as a probe (left panel). Note that ApaI sites at the ends of SHs remain intact in SH-deleted strains as shown in the right panel. The upper bands around at 7 kb correspond to the signals of tel3R and a part of rDNA (Apa1 site is located within the rDNA repeats), and the lower bands around at 0.3 kb correspond to those of tel1L,tel1R, tel2L, tel2R and tel3L. (G) Southern blot analysis (performed as in F) of DNA from wild-type and SD5 cells with or without taz1 deletion.
We next examined whether SH sequences are required for meiosis. Telomere clustering to the spindle pole body to produce the chromosomal bouquet configuration is required for normal meiotic processes, including homologous chromosome pairing, homologous recombination, chromosome segregation and spore formation (25–28). The SD5 strain showed normal mating and spore formation when crossed with wild-type or SD5 strains of the opposite mating type (Figure 2C and D), indicating that the SH regions are dispensable for meiotic progression, and that the difference in the length of subtelomeric DNA (wild-type versus SD5) does not cause severe defects in meiotic chromosome dynamics.
To examine whether the SH regions are required for stress responses, the sensitivity of the SD5 mutant to various stresses was tested. SD5 cells grew at a rate similar to that of wild-type cells at both low (19°C) and high (37°C) temperatures, whereas taz1Δ rap1Δ cells showed slow growth at 19°C, as previously reported (29) (Figure 2E). SD5 cells also grew normally in the presence of various other stresses, including high doses of hydroxyurea, camptothecin, ultraviolet radiation and thiabendazole (Figure 2E). These findings indicate that the SH regions are dispensable for responses to cold or heat treatment, replication block, DNA damage and destabilization of microtubules.
We next determined whether SH sequences are required for the maintenance of telomere DNA length. SD1 and SD5 mutants showed telomere lengths similar to that in the wild type strain (Figure 2F), indicating that the SH regions are dispensable for maintenance of telomere DNA length. Moreover, combined deletion of SH and Taz1 (which causes abnormal telomere elongation by telomerase (23,30)), SD5 taz1Δ, resulted in a telomere length similar to that of a taz1Δ mutant strain (Figure 2G), indicating that lack of SH regions does not prevent telomerase from accessing the telomeres.
Cell survival in the absence of SH sequences through intra- or inter-chromosomal circularization and HAATI after telomere loss
We next investigated whether SH sequences are required for cell survival after the loss of telomere DNA. Telomeres protect the ends of chromosomes and maintain their linear form. In the majority of somatic cells, telomere DNA is shortened by telomerase inactivation after differentiation, or by chance inactivation of telomere-binding proteins. When telomere DNA is lost through lack of telomerase, most S. pombe cells cease growth, presumably due to the activation of the DNA damage checkpoint and inter-chromosomal end fusions; however, some cells survive via a number of mechanisms. The major mechanism is self-circularization of each chromosome, due to the small number of chromosomes (three per S. pombe haploid genome), whereas other mechanisms include amplification of the telomere and/or subtelomere regions by homologous recombination, and amplification and rearrangement of the heterochromatin blocks (comprising parts of SH or rDNA repeats), known as HAATI (23,31). The SH regions of chromosomes 1 and 2 play an important role in chromosome circularization because they contain common DNA sequences that are involved in the single-strand annealing (SSA) reaction that recombines the subtelomeres of the left and right arms of the same chromosome (32). In contrast, the mechanism of self-circularization of chromosome 3 in the absence of telomerase remains obscure; it has been suggested that most frequently, chromosome 3 remains in a linear form via HAATI (31).
To examine whether the SD5 mutant is capable of surviving telomere loss, the trt1+ gene encoding the catalytic reverse transcriptase subunit of telomerase was deleted in the wild-type and SD5 strains, and the colonies were streaked on YES seven times (more than 100 generations) to induce shortening of telomere DNA (Supplementary Figure S4). The SD5 trt1Δ strain exhibited colony formation within the streaks at a frequency comparable to that of the trt1Δ single mutant, indicating that SD5 trt1Δ cells are capable of surviving telomerase loss.
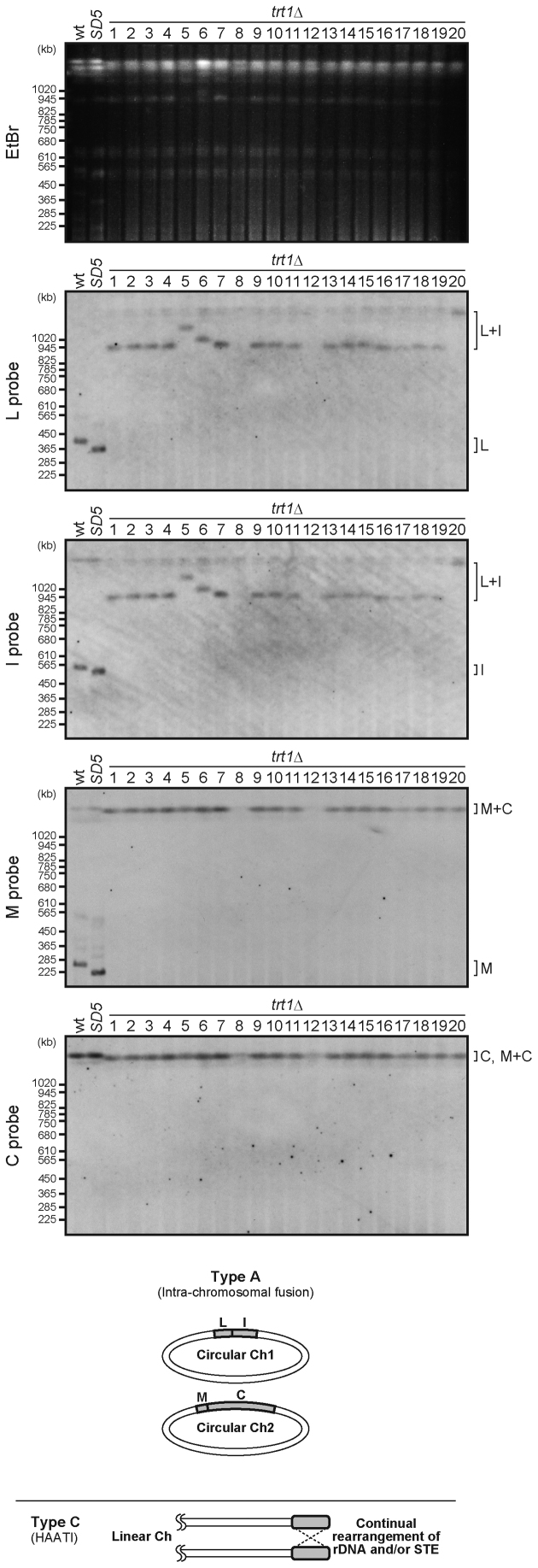
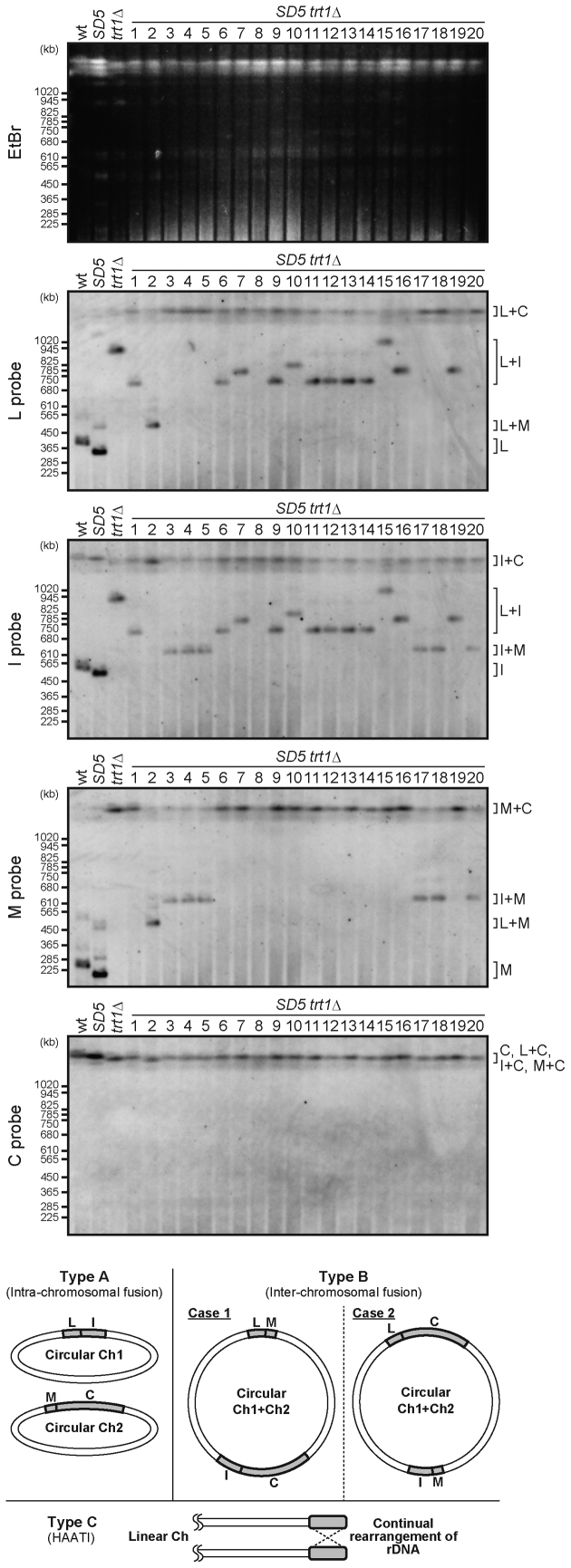
To investigate how SD5 trt1Δ survived telomere loss without SH sequences, the conformations of chromosome ends in trt1Δ and SD5 trt1Δ survivors were analyzed. We performed PFGE followed by Southern blotting to detect NotI fragments (L, I, M and C), corresponding to the ends of chromosomes 1 and 2 (Figures 3 and 4, also see Figure 1D). The results indicated that survivors could be classified into three types: A, B and C. Type A exhibited a L+I fusion band, whereas trt1+ strains (wild-type and SD5) showed two distinct bands for L and I. Type A survivors also exhibited a M+C fusion band, indicating that chromosomes 1 and 2 were self-circularized (Figures 3 and 4, bottom). Type B exhibited bands corresponding to either L+M and I+C (case 1), or I+M and L+C fusions (case 2), indicating that chromosomal fusions occurred between chromosomes 1 and 2, resulting in formation of a circular chromosome (Figure 4, bottom). Type C (trt1Δ #8, #12, and SD5 trt1Δ #8 survivors) exhibited no fast-migrating bands (below the position of 1020 kb, Figures 3 and 4), suggesting the possibility that these strains survived telomere loss by HAATI, which prevents chromosome end regions from entering the gel due to the structures of recombination intermediates. In fact, re-introduction of the trt1+ gene into these type C strains restored migration of the chromosomal DNAs into the gel with amplified SH (TAS) and/or rDNA sequences (Supplementary Figure S5A), confirming that type C strains are HAATI survivors (Figures 3 and 4, bottom).
Figure 3.
Conformation of the chromosomes in trt1Δ survivors. PFGE-Southern analyses of the NotI restriction fragments (I, L, C and M) from trt1Δ survivors (see Figure 1D, right upper panel). Schematic illustrations of the conformations of chromosomes 1 and 2 in survivors are shown beneath the blots. STE, subtelomeric elements (equivalent to SH sequences).
Figure 4.
Conformation of the chromosomes in SD5 trt1Δ survivors. PFGE-Southern analyses wereperformed as described in Figure 3 using DNA from SD5 trt1Δ survivors. Schematic illustrations of the conformations of chromosomes 1 and 2 in survivors are shown beneath the blots.
The majority of trt1Δ survivors, i.e., all except #8 and 12 (type C), exhibited the type A pattern of L+I and M+C fusion bands, indicating that those survived by the self-circularization mechanism (Figure 3). Notably, the trt1Δ strain did not produce type B survivors. In contrast, SD5 trt1Δ survivors included all three types (#1, 6, 7, 9–16 and 19 for type A; #2–5, 17, 18 and 20 for type B; #8 for type C) (Figure 4).
We found that the SfiI-digested fragments of the ends of chromosome 3 from type A, B and C survivors barely entered gels (Supplementary Figure S5B), suggesting that chromosome 3 is maintained via HAATI or other recombination mechanisms in the absence of telomerase, even where chromosomes 1 and 2 are maintained by chromosome circularization, as previously proposed (31). We also found that the survivors (types A, B and C) maintained a haploid state (Supplementary Figure S6), indicating that abnormal chromosome configurations do not affect ploidy.
In trt1Δ mutants, chromosome-end fusions require the SSA repair pathway, which depends on Rad52 (DNA recombination protein), but not Rad51 (RecA family recombinase) (32,33). To determine whether or not the chromosome-end fusions in SD5 trt1Δ were also mediated by the SSA pathway, SD5 diploid cells with trt1+/–rad51+/– or trt1+/–rad52+/– genotypes were generated. After sporulation, the resultant haploid strains, SD5 trt1Δ rad51Δ and SD5 trt1Δ rad52Δ, were subjected to serial streaks on plates to monitor cell survival. The SD5 trt1Δ rad51Δ strain showed high viability (95%), whereas the SD5 trt1Δ rad52Δ strain exhibited a striking decrease in viability (5%), indicating that the SD5 trt1Δ strain requires Rad52, but not Rad51, for survival after telomere loss (Table 1). These findings suggest that the chromosome-end fusions in SD5 trt1Δ are primarily mediated by the SSA repair pathway.
Table 1. Chromosomal end fusions in SD5 trt1Δ cells are primarily mediated by SSA.
| rad51 | rad52 | |
|---|---|---|
| SD5 radΔ trt1Δ clones tested | 20 | 19 |
| Viable over five streaks | 19 | 1 |
| Survival rate | 95% | 5% |
Survival rate of SD5 rad51Δ trt1Δ and SD5 rad52Δ trt1Δ. SD5 diploids with heterozygous rad trt1 deletions (rad+/radΔ::hygr trt1+/trt1Δ::kanr) were sporulated to obtain haploid clones with rad trt1 deletions (SD5 radΔ trt1Δ clones tested). The mutants were successively streaked on YES plates, and the numbers of clones that retained viability over five rounds of streaks were counted (viable over five streaks).
Together, these results demonstrate that S. pombe can survive telomere loss in the absence of the SH regions by chromosome circularization via SSA and by HAATI. Importantly, the SH deletion sometimes induces inter-chromosomal fusions (7 cases out of 20, 35%), which are almost never observed in the presence of the SH regions. Thus, we speculate that the SH regions inhibit inter-chromosomal end fusions, which usually cause deleterious chromosome rearrangements (see below), by facilitating intra-chromosomal end fusions.
Intra- and inter-chromosomal end fusions at non-SH loci
To clarify how the chromosome-end fusions in SD5 trt1Δ occurred in the absence of the SH regions, which are the sites of action of SSA in trt1Δ survivors (32), we analyzed the fusion points of chromosomes 1 and 2 in SD5 trt1Δ survivors. Fusion points in representative SD5 trt1Δ survivors (#1–6) (Figure 4) were amplified by PCR, and their DNA sequences were determined (Supplementary Figure S7). Interestingly, all of the fusion points exhibited high DNA sequence identity between the two chromosome arms in the same 5′ to 3′ direction (Figure 5A). On the other hand, the marker genes (his7+ and ura4+) used for the SH deletion were not utilized for the chromosome fusion sites, probably because the directions of these genes do not match with each other for chromosome end fusions.
Figure 5.
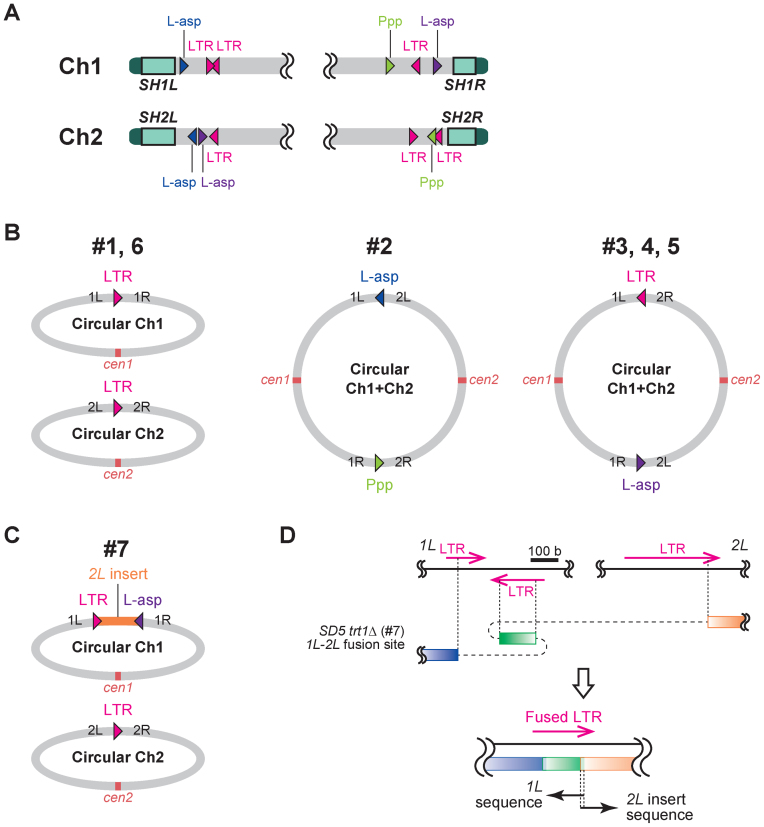
Chromosome-end fusions at homologous genomic loci in SD5 trt1Δ survivors. (A) Map of homologous loci that mediated chromosomal fusions in SD5 trt1Δ survivors #1–7. L-asp, L-asparaginase. Ppp, phosphoprotein phosphatase. Four L-asp genes were classified into two groups colored blue or purple according to their homology (see main text). Triangles indicate the 5′ to 3′ direction of the sequences. Note that positions of the loci are not in scale (see Supplementary Figure S7 for the precise representation). (B) Conformation of chromosomes 1 and 2 in SD5 trt1Δ survivors #1–6. Gene loci at the fusion points are indicated. DNA sequences of the fusion sites are shown in Supplementary Figure S7. (C) Chromosome conformation of SD5 trt1Δ survivor #7. Top panel indicates circularized chromosome 1 using the 2L sequence (2L is inserted). DNA sequences of the fusion sites are shown in Supplementary Figure S7. (D) Schematic illustration of the complex fusion between 1L and 2L in SD5 trt1Δ survivor #7 shown in the top panel in (C). Blue box, IL-specific region; green box, inverted LTR sequence in IL; orange box, 2L-specific region. See Supplementary Figure S7 for details.
The fusion points in #1 and #6 were not identical; however, loci 1L, 1R, 2L and 2R at ∼111, 101, 119 and 61 kb from the telomeres were fused via retrotransposon LTR (long terminal repeat) sequences (Figure 5A and B; Supplementary Figure S7). LTRs are very common sequences in the S. pombe genome and were also utilized in other fusions, including the 1L-2R fusions in #3–5 (Figure 5B). Another feature of the fusion points was four L-asparaginase gene loci, which were used for the 1L-2L fusion in #2 and the 1R-2L fusions in #3–5 (Figure 5A and B; Supplementary Figure S7). The nucleotide sequences of SPAC977.12 (1L) and SPAP8B6.05 (2L) (indicated in blue in Figure 5A), and SPAC186.03 (1R) and SPBPB21E7.09 (2L) (indicated in purple in Figure 5A) showed 99% identity, whereas the identity between these two groups was ∼70%. Fusions only occurred between L-asp regions of the same groups (Figure 5B). The L-asparaginase genes, SPAC977.12 (1L) and SPAP8B6.05 (2L), were also involved in chromosome-end fusion between chromosomes 1 and 2 in survivors after artificial removal of a centromere (24). The 1R-2R fusion in #2 was mediated by two paralogous phosphoprotein phosphatase genes (SPAC1039.02 and SPBPB2B2.06; indicated as Ppp in Figure 5A and B).
We also analyzed the fusion points of SD5 trt1Δ survivor #7, which exhibited a band in the Southern blot very similar to those of #16 and #19 (Figure 4, L+I). Intriguingly, a part of 2L (106.4–118.5 kb from tel2L) was inserted between 1L and 1R, whereas LTR was used for the 2L-2R fusion, as in the other survivors (Figure 5C). Furthermore, the 2L fragment was fused with 1L in a complex manner involving the three LTRs of 2L and 1L (Figure 5D and Supplementary Figure S7).
These findings demonstrate that S. pombe is capable of surviving telomere DNA loss by fusing chromosome ends for circularization via highly similar DNA sequences around the subtelomeres, other than the SH regions. This system is safe and effective for S. pombe because there are no essential genes around the subtelomeres. As a result, the cells can survive the loss of more than 100 kb of the chromosome ends. Moreover, S. pombe occasionally rearranges two chromosomes during chromosome circularization using these highly similar DNA sequences.
Inter-chromosomal circularization accompanies centromere inactivation and heterochromatin invasion of centromeres
As described above, SD5 trt1Δ survivors #2–5 exhibited fusions between chromosomes 1 and 2. Such inter-chromosomal circularization may lead to chromosome instability, such as chromosome breakage during mitosis, due to the existence of two centromeres in a single chromosome. To clarify how survivors #2–5 survived with dicentric chromosomes, we analyzed the localization of Cnp1, a CENP-A (histone H3 variant) protein required for kinetochore formation (Figure 6A, top). ChIP analyses demonstrated that Cnp1 was completely absent from one of the central core regions of the centromeres of chromosome 1 or 2 (cnt1 or cnt2) in survivors #2–5, whereas the abundance of genomic DNA at those loci in these survivors was comparable to that in the other strains (Figure 6A and Supplementary Figure S8A). In contrast, Cnp1 localization was unaffected in survivors #1 and #6 (Figure 6A). These findings indicate that the dicentric circular chromosomes in survivors #2–5 were stabilized by inactivation of the centromere of either chromosome 1 or 2.
Figure 6.
Inter-chromosomal circularization is accompanied by centromere inactivation. (A) ChIP analyses of Cnp1 (CENP-A) localization at the centromeric central core regions (cnt) of each centromere. Schematic illustration of Schizosaccharomyces pombe centromeres is shown at the top. Note that SD5 trt1Δ survivors #2–5 had inter-chromosomal end fusions between chromosomes 1 and 2. his1, negative control; cen(dg), pericentromeric dg repeat; cnt1, 2 and 3, centromeric central core regions of chromosome 1, 2 and 3, respectively. Error bars indicate the s.d. (n = 3). (B) ChIP analyses of the levels of H3K9me2 at centromeres. Fold enrichment relative to the his1 locus is shown. Error bars indicate the s.d. (n = 3). (C) PFGE-Southern analyses of NotI-digested DNA from SD5 trt1Δ clr4Δ survivors. PFGE-Southern analyses were performed as described in Figure 3. (D) ChIP analyses of the levels of Cnp1 localization at centromeres in SD5 clr4Δ trt1Δ survivors. SD5 clr4Δ trt1Δ survivors #2 and 5 had an inter-chromosomal fusion between chromosomes 1 and 2. Error bars indicate the s.d. (n = 3).
We next investigated how the centromere inactivation is maintained in SD5 trt1Δ survivors #2–5. The central core regions where the kinetochore is formed are flanked by pericentric heterochromatin regions, which are important for the fidelity of chromosome segregation (Figure 6A, top) (34,35). However, heterochromatin inhibits kinetochore formation and targeting of heterochromatin to the kinetochore region causes kinetochore disassembly (36). Moreover, heterochromatin expansion to the centromeric core region of a dicentric linear chromosome after artificial chromosomal fusion prevents reactivation of the inactivated centromere (22). We therefore determined whether the centromere inactivation in survivors #2–5 also involved heterochromatin invasion of the centromeric core region. ChIP analyses demonstrated that H3K9 dimethylation (H3K9me2) was highly enriched at the inactivated centromeric core regions in survivors #2–5, whereas no such heterochromatin invasion was observed in the other strains (Figure 6B). These findings indicate that centromere inactivation in the dicentric circular chromosome was accompanied by heterochromatin invasion of the centromeric core region.
We next examined whether heterochromatin formation at the centromeric core region is a prerequisite for centromere inactivation in a dicentric circular chromosome. The H3K9 methyltransferase, Clr4, was deleted in SD5 trt1Δ to abolish heterochromatin, and the resultant SD5 trt1Δ clr4Δ mutant was serially streaked on plates to monitor cell survival. SD5 trt1Δ clr4Δ cells survived telomerase loss and produced survivors at a frequency comparable to that of the SD5 trt1Δ mutant. Analyses of the DNA structures of SD5 trt1Δ clr4Δ survivors by PFGE-Southern showed that they exhibited both type A and B chromosome-end fusions, namely, chromosomal self-circularization (SD5 trt1Δ clr4Δ survivors #1, 3, 4, 6–8, 12, 14 and 16) and inter-chromosomal circularization (SD5 trt1Δ clr4Δ survivors #2, 5, 9–11, 13 and 15) (Figure 6C). In the type B survivors (#2 and 5), Cnp1 was absent from one of the centromeric core regions, cnt1 or cnt2, whereas Clr4 deletion alone in the SD5 trt1+ strain did not affect Cnp1 localization at centromeres (Figures 6D and Supplementary Figure S8B). These findings indicate that heterochromatin formation at the centromeric core region is not a prerequisite for centromere inactivation of a dicentric circular chromosome.
Collectively, these results demonstrate that S. pombe can survive telomere loss in the absence of SH regions primarily through intra- or inter-chromosomal circularization using highly similar genome sequences around the subtelomeres in a Rad52-dependent manner, and that the fused chromosomes via inter-chromosomal circularization are stabilized by inactivation of either centromere.
Unique distribution of subtelomeric heterochromatin at each chromosome end
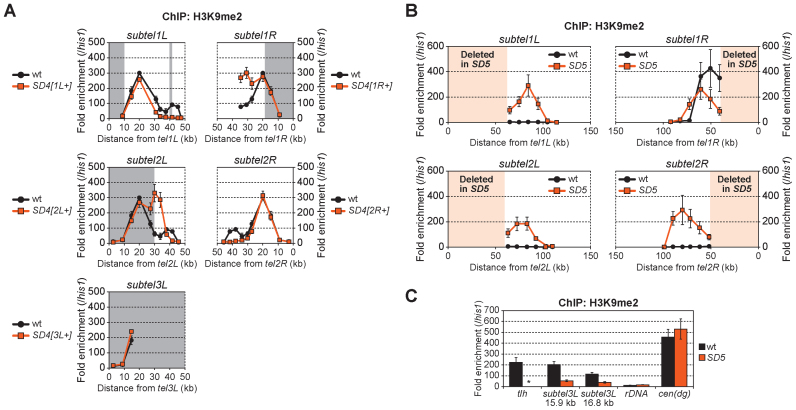
We next investigated the roles of SH sequences in the regulation of heterochromatin structure near telomeres. The H3K9me-mediated subtelomeric heterochromatin adjacent to telomeres in S. pombe (Figure 1B) is established via independent factors, i.e. the telomere-associated shelterin proteins and the RNAi machinery, which acts at the centromere-homologous (cenH) sequence in the SH region (11,12,37). The heterochromatin spreads out around the SH regions, although the subtelomeric heterochromatin on the right arm of chromosome 1 (1R) extends beyond the SH region (11,12,37). However, it was impossible to determine the exact range and level of H3K9me of each chromosome arm by ChIP due to the high similarity of DNA sequences among the SH regions. The series of SD4 mutants, in which only one SH region remained, enabled us to analyze H3K9me2 specifically in each SH region. ChIP analyses revealed that each subtelomeric heterochromatin region exhibited a unique distribution, whereas those at SH1L and SH2R shared a similar pattern of a single high peak, and those at SH1R and SH2L exhibited a double high peak. H3K9me2 was also highly accumulated at the SH3L region (Figure 7A).
Figure 7.
Invasion of heterochromatin of SH-adjacent subtelomeric regions in SD5. (A) ChIP analyses of the distribution of H3K9me2 in the SH regions of wild-type and SD4 series strains. Fold enrichment relative to the his1 locus is shown. Boxes shaded in gray represent chromosomal regions that the Schizosaccharomyces pombe genome database (http://www.pombase.org/status/sequencing-status) does not cover, and that were assumed to have the same sequence as subtel2R in this study. Error bars indicate the s.d. (n = 3). (B) ChIP analyses of the distribution of H3K9me2 in SH-adjacent regions with unique sequences in wild-type and SD5 strains. Fold enrichment relative to the his1 locus is shown. Boxes shaded in light orange represent SH regions deleted in SD5. Error bars indicate the s.d. (n = 3). (C) ChIP analyses of the levels of H3K9me2 in the SAS of chromosome 3 (subtel3L 15.9 or 16.8 kb). Fold enrichment relative to the his1 locus is shown. Asterisk indicates no amplification in quantitative PCR due to the complete absence of SH sequences in the SD5 mutant. Error bars indicate the s.d. (n = 3).
Heterochromatin spreads into SH-adjacent regions of subtelomeres after SH deletion
We next examined whether SH deletion caused changes in heterochromatin distribution close to telomeres. In the SD5 mutant, the distribution of H3K9me2 in the SH-adjacent 50 kb regions, (that is, up to ∼100 kb from the original telomeres of chromosomes 1 and 2) was analyzed (Figure 7B). Heterochromatin derived from the telomeres spread to the SH-adjacent regions of the subtelomeres in the absence of SH sequences. In contrast, in the short SH-adjacent region (∼1.1 kb) between the telomere and the rDNA repeats of chromosome 3 (3L), the level of H3K9me2 instead decreased in the SD5 mutant compared with that in the wild-type (Figure 7C), suggesting that SH3L may be important for maintenance of heterochromatin at the end of chromosome 3, or that the ura4+ marker gene used for the replacement of SH3L has an inhibitory effect on heterochromatin formation in its vicinity.
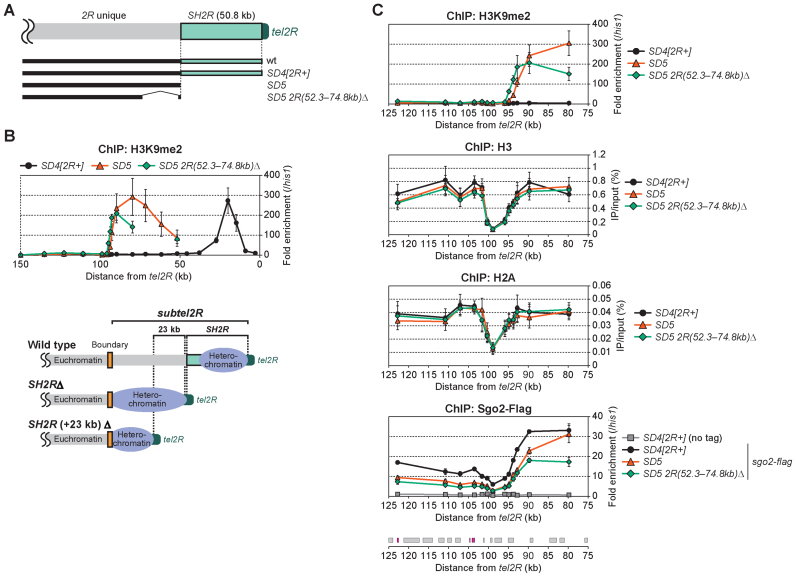
The subtelomeric chromatin boundary blocks further heterochromatin spreading into euchromatin region
A shugoshin family protein, Sgo2, is recruited to subtelomeres during interphase and defines the range of the telomere-distal part of subtelomeres, namely, the distribution of ST-chromatin and highly condensed knob structures (Figure 1B) (16). The localization of Sgo2 exhibited a profile at the subtelomere in the SD4[2R+] and SD5 mutants similar to that of H3K9me2 in SD5 (Figure 8C, top and bottom). The fact that the spreading of heterochromatin in SD5 stopped at ∼100 kb from the original telomeres of chromosomes 1 and 2 (Figure 7B), the positions of which correspond to the subtelomeric borders, suggests that chromatin boundaries to block further heterochromatin spreading into the transcriptionally active euchromatin regions may exist, or that there are limitations on the spreading of heterochromatin during a single cell cycle. To clarify this issue, we deleted the SH-adjacent region (∼23 kb) in addition to SH2R (Figure 8A). We found that heterochromatin spreading in the SD5 2R(52.3–74.8kb)Δ strain stopped at 95 kb from tel2R, as in SD5, suggesting that this region blocks the further spreading of heterochromatin (Figure 8B). Interestingly, the levels of histones H3 and H2A were strikingly reduced in the regions ∼100 kb from tel2R in all strains tested (Figure 8C), suggesting the existence of a mechanism that inhibits the formation of nucleosomes in this region. A similar nucleosome-free configuration was also observed at the boundary of subtel1L (Supplementary Figure S9A), although we were unable to detect this phenomenon at the subtel1R or subtel2L boundaries. These findings suggest the possibility that a lack of nucleosome structure may inhibit the spread of heterochromatin into the inner euchromatin region, at least at the boundaries of subtel1L and subtel2R (Supplementary Figure S9B), which could be important for the protection of expression of genes essential for cell growth.
Figure 8.
Subtelomeric border blocks heterochromatin invasion of the euchromatin region. (A) Schematic illustration of subtel2R in the mutants analyzed in (B). (B) ChIP analyses of the distribution of H3K9me2 at subtel2R in SD4[2R+], SD5 and SD5 2R(52.3–74.8kb)Δ. Fold enrichment relative to the his1 locus is shown (upper). Schematic illustration of the distribution of heterochromatin (H3K9me2) in each strain (lower). Yellow boxes indicate putative subtelomeric chromatin boundaries. Error bars indicate the s.d. (n = 3). (C) ChIP analyses of the distributions of H3K9me2, total H3, total H2A and Sgo2-Flag at 80–123 kb from tel2R. In the graphs for H3K9me2 and Sgo2-Flag, fold enrichment relative to the his1 locus is shown. In the graphs for H3 and H2A, recovery of immunoprecipitated DNA relative to total input DNA is shown. Boxes below the graph indicate the ORFs of genes located in this region. LTRs are shown in magenta. Error bars indicate the s.d. (n = 3).
Heterochromatin spreading into SH-adjacent regions causes gene silencing
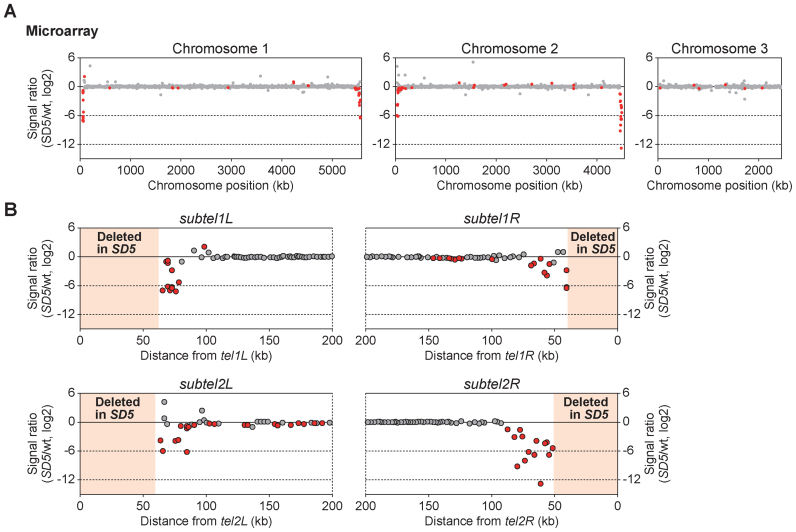
We next investigated the consequences of heterochromatin spreading into SH-adjacent regions in SD5. Limitation of heterochromatin regions in chromosomes is crucial for normal cell activities because heterochromatin structure generally inhibits the expression of genes nearby (38). Moreover, the localization of subtelomeres near the nuclear envelope is important for maintenance of subtelomeric gene expression (39); therefore, the drastic change to the subtelomeric structure in SD5 may alter gene expression. Accordingly, we performed genome-wide microarray analyses to examine the effects of SH deletion on gene expression. We found that the expression of genes located in SH-adjacent regions decreased significantly in the SD5 mutant compared with the wild-type strain, whereas there was little change in the expression levels of genes at other chromosomal loci (Figure 9A, GEO accession number: GSE69714). The range of genes that were affected by the SH deletion correlated with the distribution of the invaded heterochromatin in SD5 (Figures 7B and 9B). These findings indicate that heterochromatin spreading causes silencing of genes in SH-adjacent regions.
Figure 9.
Heterochromatin invasion of SH-adjacent regions inhibits subtelomeric gene expression in SD5. (A) Microarray analysis of genome-wide changes in mRNA expression caused by SH removal. The signal ratio (SD5/wild-type) of each probe on the Schizosaccharomyces pombe gene array is plotted. Red and gray dots indicate significant (P < 0.002 for an increase and P > 0.998 for a decrease) and insignificant differences, respectively. The P-value was calculated as change P-value (two-sided) by Affymetrix GCOS1.4 with the default settings. (B) Magnified views of the subtelomeric regions of chromosomes 1 and 2 in (A).
Of 27 internal genes that showed significant yet relatively small changes upon SH deletion (indicated as red dots located at inner chromosome regions in Figure 9A), six genes are related to metal transport (Supplementary Table S1). How expression of these genes was influenced by the SH deletion is currently unknown; however, those may be indirectly affected by the silencing of genes in the SH-adjacent regions, such as nhe1+ (encoding a plasma membrane Na+ (or Li+)/H+ antiporter (40), also see below), SPAC977.17c (encoding a putative MIP water channel), ght3+ (encoding a hexose transporter) and SPAC750.02c (encoding a putative transmembrane transporter).
Gene silencing in the SH-adjacent region causes high sensitivity to osmotic stress
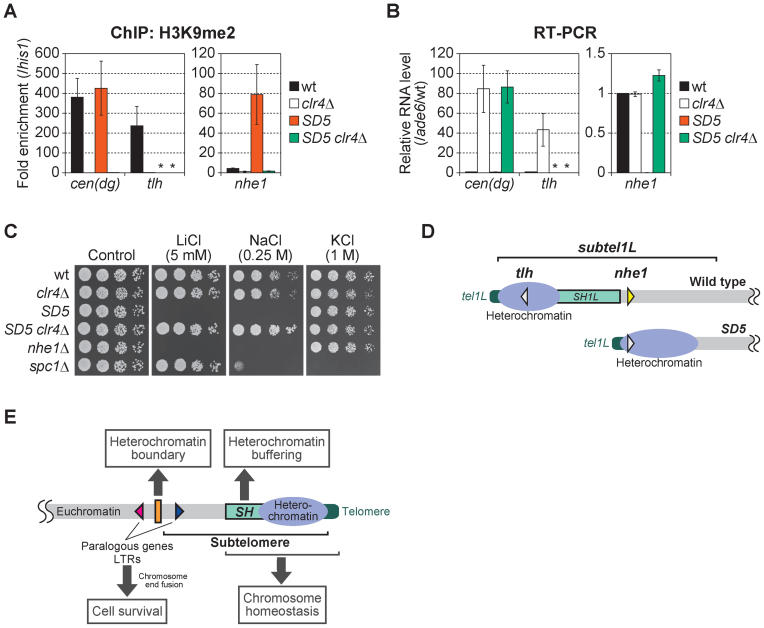
We further investigated whether gene silencing in SH-adjacent regions has consequences for cellular activity. The nhe1+ gene is located in the SH1L-adjacent region (∼65 kb from tel1L). The level of H3K9me2 at the nhe1 locus was strikingly increased and its RNA expression drastically decreased in SD5, whereas no such change was observed at the centromeres (Figure 10A and B). When the H3K9 methyltransferase, Clr4, was deleted in the SD5 mutant to destroy heterochromatin (Figure 10A), nhe1+ RNA expression recovered; however, in contrast to the striking effect of Clr4 deletion on expression of the tlh+ gene located in the SH region, no such effect was observed in the wild-type strain due to the lack of heterochromatin at the nhe1 locus (Figure 10B and D). As a result, the SD5 mutant, but not the SD5 clr4Δ double mutant, exhibited a phenotype similar to that of the nhe1Δ strain, including high sensitivity to LiCl and NaCl (Figure 10C), indicating that the high sensitivity of the SD5 mutant to LiCl and NaCl was caused by heterochromatin spreading into the SH-adjacent region (Figure 10D).
Figure 10.
High sensitivity to osmotic stress caused by heterochromatin invasion of the SH-adjacent region. (A) ChIP analyses of the levels of H3K9me2 at pericentromeric dg repeats, thetlhloci in the SH regions and the nhe1 locus. Fold enrichment relative to the his1 locus is shown. Asterisks indicate no amplification by quantitative PCR due to the complete absence of SH sequences in the SD5 mutant. Error bars indicate the s.d. (n = 3). (B) Reverse transcription-PCR (RT-PCR) analyses of the transcript levels of cen(dg), tlh+ and nhe1+. Each value was first normalized to that of ade6+ and then to the wild-type value. Asterisks indicate no amplification by quantitative PCR due to the complete absence of SH sequences in the SD5 mutant. Error bars indicate the s.d. (n = 3). (C) Cell growth assay on YES plates supplemented with lithium chloride (LiCl), sodium chloride (NaCl) or potassium chloride (KCl). Cells were incubated at 32°C for 2–3 days. The osmosensitive mutant spc1Δ was used as a control. (D) Schematic illustration of gene silencing at the nhe1 and tlh loci by heterochromatin. The tlh+ genes located in SH regions are silenced by heterochromatin in the wild-type strain. In the SD5 mutant, expression of the nhe1+ gene is silenced by heterochromatin spreading. White triangles indicate that genes are silenced, and a yellow triangle indicates a transcriptionally active phase. (E) Model of the multiple roles of subtelomeres in Schizosaccharomyces pombe. In the wild-type strain, heterochromatin is formed around SH regions. The SH region serves as a buffer zone to protect the expression of subtelomeric genes. The telomere-distal ends of subtelomeres (subtel2R) possess chromatin boundaries that block further heterochromatin spread into euchromatin regions, thereby maintaining the gene expression in the euchromatin. Furthermore, SH regions, together with telomeres, contribute to homeostasis of chromosomes. In the absence of SH sequences, paralogous genes and LTRs located around the subtelomeres mediate chromosome end fusion to ensure cell survival.
Collectively, these results indicate that the SH region serves as a buffer zone against heterochromatin spreading to maintain the expression of genes in SH-adjacent regions. This regulation is particularly important when cells are exposed to osmotic stress. As S. pombe subtelomeres contain only non-essential genes, repression of gene expression in SH-adjacent regions is not critical for cell growth under normal conditions; however, the spread of heterochromatin from the SH-adjacent region (i.e. subtelomere) can lead to cell death by repressing the expression of essential genes. Hence, the subtelomeric border may serve as a crucial barrier to heterochromatin invasion in the absence of SH sequences. Schizosaccharomyces pombe therefore possesses two mechanisms to block heterochromatin spreading: SH regions and the subtelomeric chromatin boundary (Figure 10E).
DISCUSSION
In this study, we successfully deleted all SH sequences from the S. pombe genome. The resulting SD5 mutant is the first eukaryotic organism to completely lack SH sequences. Using this strain, we have uncovered the cryptic roles of subtelomeres in gene expression and chromosome homeostasis. In the SD5 mutant, heterochromatin invaded SH-adjacent regions, thereby severely repressing the expression of local genes. The SH regions thus play an important role as buffer zones to maintain the expression of subtelomeric genes. Our results also suggest the existence of a chromatin boundary at the telomere-distal border of the subtelomere, where no obvious consensus DNA sequence among the subtelomeres exists.
Role of chromatin remodeling at subtelomere borders
The absence of nucleosomes at the subtelomere borders at 1L and 2R suggests that chromatin-remodeling activity is involved in regulating the formation of subtelomeric chromatin boundaries. Recently, it was reported that Fft3, a Fun30 chromatin remodeler protein, binds to LTRs located at the boundaries of subtelomeres, as well as at other chromosomal regions, and maintains the nuclear positioning of subtelomeres (39). Moreover, Fft3 deletion causes changes in histone modifications and upregulation of subtelomeric gene expression (39). Nevertheless, we observed no effect of Fft3 deletion on the distribution of H3K9me and nucleosome vacancy in SD5 2R(52.3–74.8kb)Δ cells (our unpublished data). We also examined whether Sgo2, a factor essential for formation of subtelomeric knob structures (16), regulates the subtelomeric boundaries; however, we detected no effect of Sgo2 deletion on the subtelomeric boundary (our unpublished data). Thus, the subtelomeric boundary in S. pombe is regulated in an Fft3- and Sgo2-independent manner. It is possible that chromatin-remodeling factors other than Fft3, transcription factors or LTRs located near all of the subtelomeric borders of chromosomes 1 and 2 (Figures 5A and 8C; Supplementary Figures S7 and S9A) contribute to nucleosome vacancy and subtelomeric boundary activity.
Mechanisms to ensure survival upon telomere loss
This study has uncovered a novel defense system that ensures survival despite loss of telomeres and SH regions in S. pombe. In addition to intra-chromosomal end fusions and HAATI, SD5 trt1Δ survivors sometimes formed inter-chromosomal end fusions, utilizing highly similar DNA sequences around the subtelomeres. Such inter-chromosomal circularization resulted in loss of substantial regions of genomic DNA, sometimes more than 100 kb. Interestingly, inter-chromosome circularization was never observed in trt1Δ survivors, suggesting that the SH regions somehow inhibit deleterious inter-chromosomal end fusion formation. Thus, the SH regions, together with telomeres, contribute to chromosome homeostasis (Figure 10E).
These observations beg the question of how SH regions can inhibit inter-chromosomal end fusions in the presence of highly similar SH sequences at both ends of chromosomes 1 and 2. Each SH region contains two sets of homologous regions containing blocks (telomere-proximal H1-H5 and telomere-distal H1′-H5′) with opposite sequence directions that are utilized for intra-chromosomal end fusions via SSA (32). Although SH sequences in S. pombe, especially the telomere-proximal regions, have not yet been completely sequenced due to the problem of high sequence identity among the subtelomeres (see Figure 7A), it is possible that the sequence identities between the blocks at the arms of the same chromosome are higher than those of the different chromosomes. The high sequence identity between the SHs of the same chromosome may facilitate intra-chromosomal end fusion rather than inter-chromosomal end fusion, as occurred via the L-asparaginase genes but only of the same groups with 99% identity (Figure 5B). Genomic DNA sequencing of all the SH regions using the SD4 mutants, where a single SH remains, will clarify this issue.
This study demonstrated that S. pombe is an organism highly resistant to telomere shortening; it possesses multiple tools to stabilize its chromosomes including circularization of the chromosomes via many homologous loci, amplification and rearrangement of hererochromatin blocks by HAATI, and expansion of telomere and/or subtelomere sequences by homologous recombination. Intriguingly, circularization of chromosome 3 was never observed in this study. Instead, all the survivors we analyzed maintained chromosome 3 via HAATI after telomerase loss (Supplementary Figure S5B). One obvious reason for this phenomenon is that the SD5 strain does not possess a region with high identity in the same direction with other chromosome ends for SSA; this includes chromosome 3 because the sequence directions of the rDNA repeats at each end do not match each other. On the other hand, the trt1Δ strain containing SH at 3L is theoretically able to fuse chromosome 3 with other chromosomes via SH to form a linear chromosome; however, no such survivor was observed in this study. Thus, although it was reported that chromosome 3 can be self-circularized utilizing micro-homology within the rDNA sequences in the absence of Rad3 (an ATR homolog) and Tel1 (an ATM homolog) (41); however, our data indicate that chromosome 3 has a high preference for HAATI rather than chromosome end fusion. One possible explanation for why chromosome 3 prefers HAATI is that the ends of chromosome 3 containing rDNA repeats are usually isolated in the nucleolus, separate from those of chromosomes 1 and 2. Furthermore, Rad52, which is required for SSA, is mostly excluded from the nucleolus in S. pombe (42). Isolation of the ends of chromosome 3 and deficiency of Rad52 in the nucleolus may considerably decrease the frequency of chromosome 3 end fusions.
In this study, we never obtained trt1Δ or SD5 trt1Δ survivors via expansion of telomere and/or subtelomere by homologous recombination, indicating that this method of cell survival is rarely utilized in S. pombe. To examine the involvement of SH sequences in this mode of cell survival, we employed a Taz1-deleted condition (Supplementary Figure S10), which instead increases the frequencies of cell survival by homologous recombination (23). In the absence of SH sequences (SD5 taz1Δ trt1Δ), we frequently obtained survivors with linear chromosomes that had telomere DNA at each end (4/4 in our experiment), indicating that the SH sequences are not required for cell survival by homologous recombination.
Implications for human chromosome biology
In contrast to S. pombe, some human cancer cells maintain their growth in the absence of telomerase by a process known as alternative lengthening of telomeres (ALT), which is mediated by homologous recombination (43). Saccharomyces cerevisiae also uses a homologous recombination process to extend telomeric or subtelomeric DNA in the absence of telomerase (44,45). These organisms possess large numbers of chromosomes (46 per human diploid, 16 per S. cerevisiae haploid); therefore, it is almost impossible to self-circularize all of the chromosomes while avoiding inter-chromosomal fusions. However, our study suggests that it may be possible for human cells, and cells of other eukaryotes, to survive telomere shortening by inter-chromosomal end fusions through homologous DNA sequences, accompanied by centromere inactivation. In combination with telomere/subtelomere lengthening by homologous recombination, such rearrangement of chromosomes may have contributed to the evolution of primates. In fact, two chromosomes of the ancestor of H. sapiens were fused at their end regions to form the current chromosome 2, whereas great apes, such as chimpanzees, still possess two chromosomes, 2a and 2b (12 and 13) (46). Furthermore, human subtelomeres have acquired segmental duplication via inter-chromosomal recombination, and thus exhibit considerable variation among present-day humans (6).
The highly flexible nature of eukaryotic linear chromosomes to guard against chromosome end crisis brings about two consequences. Dynamic rearrangement of chromosome end regions will sometimes protect cell viability, which is crucial for organismal survival. In addition, it will contribute to evolution of new species, adaptation to various environmental changes and establishment of individuality. On the other hand, alteration of chromosome conformation will sometimes result in cell death, if genes essential for cell proliferation are damaged, or disastrous chromosome end fusions occur. It will also cause various diseases, such as cancer, subtelomere deletion syndrome (8) or facioscapulohumeral muscular dystrophy (9), due to acquisition of unlimited proliferation capacity, or to structural changes in the subtelomeres. Studies on subtelomeres in a wide range of species will further clarify the importance of subtelomeres for responses to telomere loss, with implications for human health.
DATA AVAILABILITY
Data are available under Gene Expression Omnibus (GEO) accession code GSE69714.
Supplementary Material
ACKNOWLEDGEMENTS
We thank H. Maekawa and Y. Murakami for critical reading of the manuscript, Y. Takeshita, H. Asakawa and Y. Hirano for technical assistance, S. Saitoh for the anti-Cnp1 antibody, the NBRP of the MEXT, Japan, for yeast strains, and all former and present lab members for discussion and support.
Footnotes
Present address: Kazuto Kugou, Department of Frontier Research, Kazusa DNA Research Institute, Kisarazu, Chiba 292–0818, Japan.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Japan Society for the Promotion of Science (JSPS) KAKENHI [16H01310, 26290061, 23114009 to J.K.; 23114001, 23114003 to K.O.]; Asahi Glass Foundation Research Grant (to J.K.); Mitsubishi Foundation Research Grant (to J.K.). Funding for open acccess charge: JSPS KAKENHI [16H01310].
Conflict of interest statement. None declared.
REFERENCES
- 1. Chikashige Y., Ding D.Q., Funabiki H., Haraguchi T., Mashiko S., Yanagida M., Hiraoka Y.. Telomere-led premeiotic chromosome movement in fission yeast. Science. 1994; 264:270–273. [DOI] [PubMed] [Google Scholar]
- 2. Kanoh J., Ishikawa F.. spRap1 and spRif1, recruited to telomeres by Taz1, are essential for telomere function in fission yeast. Curr. Biol. 2001; 11:1624–1630. [DOI] [PubMed] [Google Scholar]
- 3. de Lange T. How telomeres solve the end-protection problem. Science. 2009; 326:948–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fujita I., Nishihara Y., Tanaka M., Tsujii H., Chikashige Y., Watanabe Y., Saito M., Ishikawa F., Hiraoka Y., Kanoh J.. Telomere-nuclear envelope dissociation promoted by Rap1 phosphorylation ensures faithful chromosome segregation. Curr. Biol. 2012; 22:1932–1937. [DOI] [PubMed] [Google Scholar]
- 5. Louis E.J. The chromosome ends of Saccharomyces cerevisiae. Yeast. 1995; 11:1553–1573. [DOI] [PubMed] [Google Scholar]
- 6. Linardopoulou E.V., Williams E.M., Fan Y., Friedman C., Young J.M., Trask B.J.. Human subtelomeres are hot spots of interchromosomal recombination and segmental duplication. Nature. 2005; 437:94–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Riethman H., Ambrosini A., Paul S.. Human subtelomere structure and variation. Chromosome Res. 2005; 13:505–515. [DOI] [PubMed] [Google Scholar]
- 8. de Vries B.B.A., Winter R., Schinzel A., van Ravenswaaij-Arts C.. Telomeres: a diagnosis at the end of the chromosomes. J. Med. Genet. 2003; 40:385–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Stadler G., Rahimov F., King O.D., Chen J.C., Robin J.D., Wagner K.R., Shay J.W., Emerson C.P. Jr, Wright W.E.. Telomere position effect regulates DUX4 in human facioscapulohumeral muscular dystrophy. Nat. Struct. Mol. Biol. 2013; 20:671–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sugawara N.F. DNA sequences at the telomeres of the fission yeast S. pombe. 1988; Harvard University; Ph.D. Thesis. [Google Scholar]
- 11. Cam H.P., Sugiyama T., Chen E.S., Chen X., FitzGerald P.C., Grewal S.I.. Comprehensive analysis of heterochromatin- and RNAi-mediated epigenetic control of the fission yeast genome. Nat. Genet. 2005; 37:809–819. [DOI] [PubMed] [Google Scholar]
- 12. Kanoh J., Sadaie M., Urano T., Ishikawa F.. Telomere binding protein Taz1 establishes Swi6 heterochromatin independently of RNAi at telomeres. Curr. Biol. 2005; 15:1808–1819. [DOI] [PubMed] [Google Scholar]
- 13. Matsuda A., Chikashige Y., Ding D.Q., Ohtsuki C., Mori C., Asakawa H., Kimura H., Haraguchi T., Hiraoka Y.. Highly condensed chromatins are formed adjacent to subtelomeric and decondensed silent chromatin in fission yeast. Nat. Commun. 2015; 6:7753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Buchanan L., Durand-Dubief M., Roguev A., Sakalar C., Wilhelm B., Stralfors A., Shevchenko A., Aasland R., Shevchenko A., Ekwall K. et al. . The Schizosaccharomyces pombe JmjC-protein, Msc1, prevents H2A.Z localization in centromeric and subtelomeric chromatin domains. PLoS Genet. 2009; 5:e1000726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kawashima S.A., Yamagishi Y., Honda T., Ishiguro K., Watanabe Y.. Phosphorylation of H2A by Bub1 prevents chromosomal instability through localizing shugoshin. Science. 2010; 327:172–177. [DOI] [PubMed] [Google Scholar]
- 16. Tashiro S., Handa T., Matsuda A., Ban T., Takigawa T., Miyasato K., Ishii K., Kugou K., Ohta K., Hiraoka Y. et al. . Shugoshin forms a specialized chromatin domain at subtelomeres that regulates transcription and replication timing. Nat. Commun. 2016; 7:10393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sasaki M., Kumagai H., Takegawa K., Tohda H.. Characterization of genome-reduced fission yeast strains. Nucleic Acids Res. 2013; 41:5382–5399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Moreno S., Klar A., Nurse P.. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991; 194:795–823. [DOI] [PubMed] [Google Scholar]
- 19. Bahler J., Wu J.Q., Longtine M.S., Shah N.G., McKenzie A. 3rd, Steever A.B., Wach A., Philippsen P., Pringle J.R.. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998; 14:943–951. [DOI] [PubMed] [Google Scholar]
- 20. Forsburg S.L., Rhind N.. Basic methods for fission yeast. Yeast. 2006; 23:173–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sasaki M., Idiris A., Tada A., Kumagai H., Giga-Hama Y., Tohda H.. The gap-filling sequence on the left arm of chromosome 2 in fission yeast Schizosaccharomyces pombe. Yeast. 2008; 25:673–679. [DOI] [PubMed] [Google Scholar]
- 22. Sato H., Masuda F., Takayama Y., Takahashi K., Saitoh S.. Epigenetic inactivation and subsequent heterochromatinization of a centromere stabilize dicentric chromosomes. Curr. Biol. 2012; 22:658–667. [DOI] [PubMed] [Google Scholar]
- 23. Nakamura T.M., Cooper J.P., Cech T.R.. Two modes of survival of fission yeast without telomerase. Science. 1998; 282:493–496. [DOI] [PubMed] [Google Scholar]
- 24. Ohno Y., Ogiyama Y., Kubota Y., Kubo T., Ishii K.. Acentric chromosome ends are prone to fusion with functional chromosome ends through a homology-directed rearrangement. Nucleic Acids Res. 2016; 44:232–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chikashige Y., Ding D.Q., Imai Y., Yamamoto M., Haraguchi T., Hiraoka Y.. Meiotic nuclear reorganization: switching the position of centromeres and telomeres in the fission yeast Schizosaccharomyces pombe. EMBO J. 1997; 16:193–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nimmo E.R., Pidoux A.L., Perry P.E., Allshire R.C.. Defective meiosis in telomere-silencing mutants of Schizosaccharomyces pombe. Nature. 1998; 392:825–828. [DOI] [PubMed] [Google Scholar]
- 27. Cooper J.P., Watanabe Y., Nurse P.. Fission yeast Taz1 protein is required for meiotic telomere clustering and recombination. Nature. 1998; 392:828–831. [DOI] [PubMed] [Google Scholar]
- 28. Niwa O., Shimanuki M., Miki F.. Telomere-led bouquet formation facillitates homologous chromosome pairing and restricts ectopic interaction in fission yeast meiosis. EMBO J. 2000; 19:3831–3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Miller K.M., Ferreira M.G., Cooper J.P.. Taz1, Rap1 and Rif1 act both interdependently and independently to maintain telomeres. EMBO J. 2005; 24:3128–3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cooper J.P., Nimmo E.R., Allshire R.C., Cech T.R.. Regulation of telomere length and function by a Myb-domain protein in fission yeast. Nature. 1997; 385:744–747. [DOI] [PubMed] [Google Scholar]
- 31. Jain D., Hebden A.K., Nakamura T.M., Miller K.M., Cooper J.P.. HAATI survivors replace canonical telomeres with blocks of generic heterochromatin. Nature. 2010; 467:223–227. [DOI] [PubMed] [Google Scholar]
- 32. Wang X., Baumann P.. Chromosome fusions following telomere loss are mediated by single-strand annealing. Mol. Cell. 2008; 31:463–473. [DOI] [PubMed] [Google Scholar]
- 33. Ivanov E.L., Sugawara N., Fishman-Lobell J., Haber J.E.. Genetic requirements for the single-strand annealing pathway of double-strand break repair in Saccharomyces cerevisiae. Genetics. 1996; 142:693–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Partridge J.F., Borgstrom B., Allshire R.C.. Distinct protein interaction domains and protein spreading in a complex centromere. Genes Dev. 2000; 14:783–791. [PMC free article] [PubMed] [Google Scholar]
- 35. Allshire R.C., Nimmo E.R., Ekwall K., Javerzat J.P., Cranston G.. Mutations derepressing silent centromeric domains in fission yeast disrupt chromosome segregation. Genes Dev. 1995; 9:218–233. [DOI] [PubMed] [Google Scholar]
- 36. Nakano M., Cardinale S., Noskov V.N., Gassmann R., Vagnarelli P., Kandels-Lewis S., Larionov V., Earnshaw W.C., Masumoto H.. Inactivation of a human kinetochore by specific targeting of chromatin modifiers. Dev. Cell. 2008; 14:507–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wang J., Cohen A.L., Letian A., Tadeo X., Moresco J.J., Liu J., Yates J.R. 3rd, Qiao F., Jia S.. The proper connection between shelterin components is required for telomeric heterochromatin assembly. Genes Dev. 2016; 30:827–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ottaviani A., Gilson E., Magdinier F.. Telomeric position effect: from the yeast paradigm to human pathologies?. Biochimie. 2008; 90:93–107. [DOI] [PubMed] [Google Scholar]
- 39. Steglich B., Stralfors A., Khorosjutina O., Persson J., Smialowska A., Javerzat J.P., Ekwall K.. The Fun30 chromatin remodeler Fft3 controls nuclear organization and chromatin structure of insulators and subtelomeres in fission yeast. PLoS Genet. 2015; 11:e1005101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jia Z.P., McCullough N., Martel R., Hemmingsen S., Young P.G.. Gene amplification at a locus encoding a putative Na+/H+ antiporter confers sodium and lithium tolerance in fission yeast. EMBO J. 1992; 11:1631–1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Naito T., Matsuura A., Ishikawa F.. Circular chromosome formation in a fission yeast mutant defective in two ATM homologues. Nat. Genet. 1998; 20:203–206. [DOI] [PubMed] [Google Scholar]
- 42. Irmisch A., Ampatzidou E., Mizuno K., O’Connel M.J., Murray J.M.. Smc5/6 maintains stalled replication forks in a recombination-competent conformation. EMBO J. 2009; 28:144–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Dunham M.A., Neumann A.A., Fasching C.L., Reddel R.R.. Telomere maintenance by recombination in human cells. Nat. Genet. 2000; 26:447–450. [DOI] [PubMed] [Google Scholar]
- 44. Lundblad V., Blackburn E.H.. An alternative pathway for yeast telomere maintenance rescues est1− senescence. Cell. 1993; 73:347–360. [DOI] [PubMed] [Google Scholar]
- 45. Teng S.C., Zakian V.A.. Telomere-telomere recombination is an efficient bypass pathway for telomere maintenance in Saccharomyces cerevisiae. Mol. Cell. Biol. 1999; 19:8083–8093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ventura M., Catacchio C.R., Sajjadian S., Vives L., Sudmant P.H., Marques-Bonet T., Graves T.A., Wilson R.K., Eichler E.E.. The evolution of African great ape subtelomeric heterochrmatin and the fusion of human chromosome 2. Genome Res. 2012; 22:1036–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are available under Gene Expression Omnibus (GEO) accession code GSE69714.