Abstract
Background and Aims Certain micro-organisms can improve plant protection against pathogens. The protective effect may be direct, e.g. due to antibiotic compounds, or indirect, by priming of plant defence as induced systemic resistance (ISR). The plant growth-promoting rhizobacterium Bacillus amyloliquefaciens UCMB5113 shows potential for disease management of oilseed rape. To investigate the mode of action of this protection, especially in relation to jasmonic acid-dependent ISR, Bacillus UCMB5113 was tested with Arabidopsis thaliana mutants and several important fungal pathogens of Brassica species.
Methods Secreted lipopeptide fractions from Bacillus UCMB5113, together with synthetic peptide mimics, were evaluated for their effects on fungal phytopathogens and A. thaliana. The structures of secreted lipopeptides were analysed using mass spectrometry. Plant mutants and reporter lines were used to identify signalling steps involved in disease suppression by lipopeptides.
Key Results In plate tests Bacillus UCMB5113 and lipopeptide extracts suppressed growth of several fungal pathogens infecting Brassica plants. Separation of secreted lipopeptides using reversed-phase high-performance liquid chromatography revealed several fractions that inhibited fungal growth. Analysis by mass spectrometry identified the most potent compounds as novel linear forms of antifungal fengycins, with synthetic peptide mimics confirming the biological activity. Application of the lipopeptide extracts on Arabidopsis roots provided systemic protection against Alternaria brassicicola on leaves. Arabidopsis signalling mutants and PDF1.2 and VSP2 promoter-driven GUS lines indicated that the lipopeptide fraction involved jasmonic-acid-dependent host responses for suppression of fungal growth indicative of ISR.
Conclusions The ability of Bacillus UCMB5113 to counteract pathogens using both antagonistic lipopeptides and through ISR provides a promising tool for sustainable crop production.
Keywords: Antagonism, Arabidopsis thaliana, Bacillus amyloliquefaciens, beneficial bacteria, biocontrol, lipopeptide, rhizosphere
INTRODUCTION
Plant exposure to biotic stresses such as microbial pathogens and insect pests hamper crop production. Although many chemical pesticides have been developed to control different biotic stress factors, insect pests and pathogens may develop resistance over time, following repeated pesticide use. Plant stress management can also be improved by breeding. As development of plant resistance based on genetic engineering approaches are currently unfavourable in the European Union, crop development is reliant upon conventional breeding, which can be slow, costly and assumes the presence of the desired trait in the available germplasm (Seifi et al., 2013).
An alternative strategy to manage stress factors is based on the use of biological control agents (BCAs). BCAs are naturally occurring organisms that can counteract pathogens and pests attacking plants. BCAs are considered environmentally friendly tools that can produce healthier crops and improve yield (Bhattacharyya and Jha, 2012). Plant growth-promoting rhizobacteria (PGPR) were first described due to their ability to support plant growth but many PGPR isolates were also found to serve as BCAs. Micro-organisms are prime candidates as BCAs because of their potential wide spectrum of antagonistic activity to different pathogens (Beneduzi et al., 2012). Furthermore, many BCAs can also enhance disease protection by priming induced systemic resistance (ISR) in the host plant (Pieterse et al., 2014).
Priming of enhanced defence refers to the sensitization of plants by certain non-pathogenic micro-organisms or chemicals that causes stronger and more rapid defence responses upon stress challenge (Conrath et al., 2015). A number of bacteria and fungi as well as natural or synthetic chemicals have been shown to prime long-lasting plant defence to a variety of pathogens (Pozo and Azcón-Aguilar, 2007; Pieterse et al., 2014). Several commercial products based on priming micro-organisms are available for control of diseases on various crops (e.g. Bhattacharyya and Jha, 2012). Root colonization of the micro-organisms is a key step to initiate the beneficial interaction and in general the colonizers remain as epiphytes on the rhizoplane followed by embedding in a biofilm. ISR is a jasmonic acid (JA)-dependent process in which enhanced sensitivity to JA rather than increased JA levels seems to be the case (Pieterse et al., 2002). In contrast, plant defence as systemic acquired resistance (SAR) mounted in response to successful recognition of biotrophic pathogens, is salicylic acid (SA)-dependent (Durrant and Dong, 2004). JA signalling is also involved in plant defence to necrotrophic pathogens and insect herbivores (Pieterse et al., 2012). JA metabolism is complex and several different metabolites occur in plants depending on the stimulus that may fine tune the response to be more appropriate (Wasternack and Hause, 2013). In reality, however, numerous hormones and extensive cross-talk regulate defence responses, in which, for example, JA and SA can antagonize each other and JA responses are also modulated by other host factors such as ethylene and abscisic acid (ABA) (e.g. Shigenaga and Argueso, 2016). A number of Arabidopsis mutants and reporter lines have been developed to facilitate analysis of hormonal involvement in different processes, with, for example, activation of JA-dependent VSP2 and PDF1.2 indicating the involvement of MYC/ABA and ERF/ethylene, respectively (Pieterse et al., 2012). While several steps leading to SAR are well characterized, the molecular mechanisms behind the priming of ISR are less well known. The SAR and ISR responses can generally be clearly distinguished with respect to involvement of key regulators such as NPR3 and NPR4 (SAR) or COI1, MYB72 and MYC2 (ISR) as well as activation of defence genes, such as pathogenesis-related (PR) genes (SAR) or PDF1.2 and VSP2 (ISR) (Pieterse et al., 2014). However, both SAR and ISR depend on the redox-sensitive transcription cofactor NPR1 (Pieterse et al., 2014; Conrath et al., 2015). NPR1 is activated by SA and then translocated to the nucleus, functioning as coactivator of PR genes providing SAR, while during development of ISR, NPR1 seems to act in the cytosol although its exact role remains to be established.
Many Bacillus species have great potential as PGPR and BCAs because of their ability to enhance plant growth and antagonize phytopathogens (Kloepper et al., 2004; Cawoy et al., 2011). Several Bacillus products are available on the market for crop protection against various pathogens (Cawoy et al., 2011). Bacillus species produce numerous secondary metabolites with antibiosis properties, especially cyclic lipopeptides (LPs) that are classified based on their amino acid composition, the structure of their peptide ring and the attached fatty acid (Ongena and Jacques, 2008). LPs are synthesized non-ribosomally by large multi-enzyme complexes, non-ribosomal peptide synthetases (NRPSs) (Ongena and Jacques, 2008). The NRPSs are complex structures in which each module incorporates a monomer and the enzymatic machinery functions as a templated pathway (Strieker et al., 2010). The three main groups of LPs formed by members of the Bacillus subtilis and Bacillus amyloliquefaciens species are surfactins, iturins and fengycins. These LPs have been studied intensively because of their antagonistic activity toward phytopathogens (Ongena and Jacques, 2008). Surfactins are heptapeptides linked to a β-hydroxy fatty acid, acting as efficient biosurfactants with haemolytic activity, with antagonism to viruses and bacteria but with limited antifungal activity. Iturins are also heptapeptides but linked to a β-amino fatty acid. These, by contrast, display pronounced antifungal activity but rather limited antiviral and bactericidal activities. Fengycins are decapeptides linked to a β-hydroxy fatty acid that show marked activity against filamentous fungi but with less haemolytic activity when compared to surfactins and iturins. The ability of Bacillus species to produce many LPs with complementing and synergistic activities make these bacteria promising tools for biotechnological application (Cochrane and Vederas, 2016).
The widely cultivated oil crop oilseed rape (Brassica napus) is exposed to many phytopathogens, many of which are controlled through agrochemical approaches, in the absence of appropriate resistant germplasm. A shift towards BCAs to control these common diseases would cause less environmental impact and allow promotion of more organic cultivation. In previous studies, several strains of B. amyloliquefaciens were shown to protect oilseed rape against several phytopathogens (Danielsson et al., 2007; Sarosh et al., 2009). In this context, it is important to increase information on the structural basis of the components involved in conferring such protection and their requirements, to further evaluate their potential practical application. One of the issues we wished to address was if the secreted compounds supported plant protection in a direct (microbial antagonism) or indirect (ISR) manner, or in both ways. In this study, therefore, an LP-enriched fraction secreted from the strain Bacillus UCMB5113 was evaluated for its antagonistic effects towards common Brassica fungal pathogens. The most interesting LPs were structurally characterized via mass spectrometry analysis and synthetic peptide mimics tested for determination of their effect. Furthermore, we studied the potential indirect protective effect of the secreted LP compounds on Brassicaceae plant defence using the model plant Arabidopsis thaliana and various mutants and reporter lines, enabling the involvement of common defence-related hormones in this response to be evaluated.
MATERIALS AND METHODS
Bacterial and fungal strains and growth conditions
The root colonizing B. amyloliquefaciens subsp. plantarum strain UCMB5113 (Reva et al., 2004) was maintained on Luria–Bertani (LB) medium. The fungal strains Alternaria brassicicola 20297, Alternaria brassicae 980:3 and Botrytis cinerea 30158 were grown on potato-dextrose agar (PDA; Difco) at 22 °C, 16/8-h light and dark photoperiod. Verticillium longisporum D11 and Sclerotinia sclerotiorum 13MM were also kept on PDA but grown in darkness. Spores were isolated from A. brassicicola cultures grown for 3–4 weeks by scraping off the surface layer into water followed by filtration through miracloth. Spore counts were determined using a haemocytometer, with a fresh spore suspension (5 × 105 mL−1) used to inoculate plant leaves.
Growth inhibition test of phytopathogenic fungi by Bacillus UCMB5113
The phytopathogens were inoculated at the centre of a PDA plate and a single colony of Bacillus UCMB5113 was streaked in the periphery of the Petri dish. The samples were incubated at 22 °C, 16/8 h light and dark photoperiod while V. longisporum and S. sclerotiorum were grown in darkness. Growth inhibition of the fungal pathogen strains was recorded 3–14 d later and enlarged images of the samples were measured (mm). The experiment was repeated at least three times with similar results obtained in each. The results of a representative experiment are shown.
Isolation of LPs from Bacillus UCMB5113 exudate
An LP fraction was prepared from Bacillus UCMB5113 exudate by a protocol described by Kim et al. (2004). Bacillus UCMB5113 was grown in LB medium for 5 d at 28 °C and 150 r.p.m. reaching a stationary phase. The bacterial culture was centrifuged at room temperature for 25 min at 1000 g. The supernatant was filtered through a 0·45-µm filter membrane and precipitated by adding 3 m HCl to pH 2 followed by incubation at 4 °C for 30 min and centrifugation at 10 000 g for 20 min at 4 °C. The precipitates were collected and dissolved in chloroform/methanol (2 : 1, v/v) and the solvents were then removed using a rotary vacuum evaporator. The pellet was dissolved in methanol and filtered through a 0·2-µm filter membrane creating the LP extract. Approximately 0·5 mg of crude lipopeptides per litre of original medium was obtained. Further, the LP extract was fractionated using a reversed-phase high-performance liquid chromatography (RP-HPLC) preparative column (nucleosil C18, 16 × 125 mm, 7-μm beads, 120-Å pore size, Knauer Gmbh, Berlin, Germany) with a flow rate of 5 mL min−1. The eluants contained 0·1 % trifluoroacetic acid (TFA) in (A) water or (B) HPLC-grade acetonitrile. The starting concentration of B was 5 % followed by a linear gradient to 20 % B at 15 min with hold for 5 min, linear gradients to 50 % B at 70 min, 75 % B at 90 min and 85 % B at 100 min, hold to 110 min, a linear gradient to 95 % at 120 min and hold to 130 min before going back to 5 % B. The eluate was monitored at 215 nm and the major peak fractions were collected.
Fungal growth inhibition tests using the LP extract
The phytopathogens A. brassicicola, A. brassicae, B. cinerea, S. sclerotiorum and V. longisporum were grown to cover one-quarter of a Petri dish with PDA and then inoculated with 20 µL (50 ng) Bacillus LP extract at one spot on the periphery of the Petri dish and methanol as negative control on the opposite side. The plates were incubated at 22 °C, 16/8 h light and dark photoperiod except for V. longisporum and S. sclerotiorum that were grown in darkness. The peak fractions obtained from RP-HPLC were also examined for antagonistic activity against these phytopathogens in a similar way. The experiment was repeated at least three times with similar results and the results of a representative experiment are shown.
Thermal stability of LP extract compounds
The LP extract was incubated in a heating block at 55, 75 or 95 °C for 0, 12, 24, 48 and 72 h. Aliquots (50 µL, 100 ng) of the heat-exposed compounds were tested for antagonistic activity towards A. brassicicola and effects recorded 7 d later. The experiment was repeated at least twice with similar results and the results of a representative experiment are shown.
Antifungal bioassay using leaf treatment
Leaves from 4-week-old Arabidopsis thaliana Col-0 plants were detached and dipped in 20 mL (2·5 ng mL−1) of Bacillus LP extract (dissolved in 5 % methanol) and/or 5 % methanol and water as control. Leaves were then kept on top of a wet filter paper in a Petri dish. Three microlitres of A. brassicicola fungal spore suspensions was dropped on the leaves and the plates were incubated at 22 °C, 16/8 h light and dark photoperiod. Disease symptoms on the leaves were recorded 4 d after pathogen inoculation. The experiment was repeated at least twice with similar results. The results of a representative experiment are shown.
Mass spectrometry (MS) analysis of LPs
LP profiling was performed by reversed-phase ultrahigh-performance liquid chromatography-MS (UPLC; Waters, Acquity class H) coupled with a single quadrupole mass spectrometer (SQDetector, Waters, Acquity) on an Acquity UPLC BEH C18 (Waters) 2·1 × 50 mm, 1·7 µm column, which allowed detection of all three major LP families. Elution was initiated with 30 % acetonitrile (0·60 mL min−1). After 2·43 min, the percentage of acetonitrile was raised to 95 % and held until 5·2 min. The column was then stabilized at an acetonitrile percentage of 30 % for 1·7 min. Compounds were identified on the basis of their retention times compared with authentic standards (98 % purity, Lipofabrik Society, Villeneuve d’Asc, France) and the masses detected in the SQDetector. Ionization and source conditions were set as follows: source temperature, 130 °C; desolvation temperature, 400 °C; nitrogen flow, 1000 L h−1; cone voltage, 120 V.
Exact mass measurements and tandem MS experiments were performed with a Fourier transform ion cyclotron resonance (FT-ICR) mass spectrometer (9·4T Solarix; Bruker Daltonics, Bremen, Germany) equipped with an electro-spray ionization matrix-assisted laser desorption ionization (ESI/MALDI) dual ion source including SmartbeamII laser (355 nm). For MS/MS experiments, a MALDI time-of flight (TOF)/TOF mass spectrometer was used (UltraFlex II, Bruker Daltonics). The selection window in the LIFT cell was set to optimize the isolation (depending on the presence of adducts or other peaks close to the peak of interest). Several spectra were accumulated to improve the signal-to-noise ratio of the MS/MS spectrum and facilitate interpretation.
Fungal growth inhibition test using synthetic peptides
Synthetic peptides with the l-form of GluOrnTyrThrGluValProGlnTyrLeu with acetylated (‘AcePEP’) or myristoylated (‘MyrPEP’) amino terminus were supplied by ProteoGenix SAS (Schiltigheim, France). The fungus was grown as previously described and 200 ng of synthetic LP with fatty acid or acetyl group, LP extract or methanol as control was inoculated in the periphery of the growth medium. Plates were incubated at 22 °C, 16/8 h light and dark photoperiod. Fungal growth inhibition was recorded after 5 d. The experiment was repeated at least twice and the result of a representative experiment is shown.
Effects of LP extract on fungal disease suppression in plants
Arabidopsis thaliana wild type Col-0 and mutants were employed in this study. These comprised coi1-16 with JA receptor deficiency (Ellis and Turner, 2002), sid2 with aberrant SA synthesis (Wildermuth et al., 2001), jar1 with inability to form the active jasmonyl–isoleucine conjugate (Staswick et al., 1992), myc2 defective in ISR signalling (Lorenzo et al., 2004), myb72 defective in ISR signalling (Van der Ent et al., 2008) and npr1 deficient in SAR response (Cao et al., 1994). β-Glucuronidase (GUS) marker lines driven by VSP2 and PDF1.2 promoters were also used. Seeds were surface sterilized (1 min in ethanol, 5 min in 10 % bleach containing chlorine and tween 20, and 3 × 5 min in sterile water). The seeds were then germinated in Petri dishes containing half-strength MS medium (M0222, Duchefa) with 0·6 % agar, with incubation at 22 °C, 16/8 h light and dark photoperiod. One-week-old seedlings were transplanted to 10 × 10-cm square plates containing half-strength MS medium with 0·8 % agar. Each plate contained four seedlings with a distance of 2·5 cm between the plants. The roots were treated with 5 µL (2·5 ng mL−1) LP extract and/or 5 % methanol and water. The plates were incubated at 22 °C, 16/8 h light and dark photoperiod and positioned vertically.
Two weeks after LP treatment, plants were inoculated with A. brassicicola spores (5 × 105 mL−1) by adding 3 µL drops onto the surface of four leaves per plant as described (Thomma et al., 1998). Seven days after challenge, disease severity was scored according to Van der Ent et al. (2008) and samples were collected for pathogen quantification. For each pathogen assay, inoculated leaves of three plants were pooled together and taken as one biological replicate, with three biological replicates evaluated. Fungal DNA was isolated from plant leaves using a GenJET genomic DNA purification kit (Thermo Fisher Scientific). SYBR GREEN-based quantitative real-time PCR (qPCR) was conducted as described previously (Fridborg et al., 2013). DNA equivalent to 50 ng was used in a 20-µL PCR in a Bio-Rad iQ cycler with iQ SYBR Green Supermix (Fermentas). A 40-cycle two-step amplification protocol (10 s at 95 °C, 30 s at 60 °C) was used for all measurements. Primer sequences used were: forward primer 5′- GCATGTCCGCTCACCAATATC-3′, reverse primer 5′-GCCTGGGATCTTGGAATGC-3′ (A. brassicicola cutinase, ABU03393). As reference gene, Ubiquitin 5 (At3g62270) from Arabidopsis was used and amplified by the primers 5′-CGATGGATCTGGAAAGGTTC-3′ and 5′-AGCTCCACAGGTTGCGTTAG-3′. The identities of the amplicons and the specificity of the reactions were verified by melting curve analysis. The data were analysed using the comparative CT method with PCR efficiency correction. The average and the standard error of three technical replicates of a pool of three plants and with three different biological samples are shown. The experiment was repeated at least once and the result of a representative experiment is shown.
Histochemical GUS reporter gene expression
To generate JA-responsive GUS reporter plants, promoter regions upstream of the translation start of VSP2 or PDF1.2 were amplified from genomic DNA. PDF1.2 (AT5g44420) was amplified with forward primer TCgagctcAAAAATCTTTGGTGCTTG (SacI restriction site) and reverse primer CGccatggGATGATTATTACTATTTTGTTTTC (NcoI) generating a 1541-bp fragment. VSP2 (AT5g24770) was amplified using forward primer ATggtaccATTTTGTTGCTGCTTGGAC (KpnI) and reverse primer GCaagcttGTTTTTTATGGTATGGTTTAT (HindIII) generating a 1472-bp fragment. The PCR fragments were subsequently digested with the respective restriction enzymes to replace the 2× 35S promoter in the GUS expression vector pJIT166. The pPro-GUS expression cassette were released from PJIT166 with SacI + XhoI or KpnI + XhoI digestion and cloned into the T-DNA vector pBINPLUS. The pBINPLUS plasmids containing the different constructs were transformed into Agrobacterium tumefaciens strain C58, followed by transformation into Arabidopsis using the floral-dip protocol (Davis et al., 2009). A minimum of three homozygous T3 lines were used in each experiment. The resulting GUS transgenic lines (VSP2:GUS and PDF1.2:GUS) were grown on half-strength MS medium with 0·8 % agar for 1 week before being treated with LP extract, methanol or water and challenged with A. brassicicola 2 weeks later. Samples were collected 3 d after inoculation for expression analysis. The GUS staining solution contained 50 mm sodium phosphate buffer (pH 7·2), 10 % Triton X100, 5 mm ferro-cyanide, 5 mm ferri-cyanide and 2 mm X-gluc (5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid cyclohexylammonium salt). The samples were incubated in the GUS staining solution at 37 °C for maximum 24 h, and destained using 70 % ethanol for at least 24 h before photodocumentation.
Statistical analysis
If not stated otherwise, all calculations of mean and standard deviations were based on three or more samples, with t-tests or ANOVA Tukey HSD test conducted using the appropriate application in Minitab. Samples sharing a letter are not significantly different at the 5 % level.
RESULTS
Antifungal activity of B. amyloliquefaciens UCMB5113
The disease suppressive potential of B. amyloliquefaciens UCMB5113 was first assessed as growth inhibitory activity against several Brassica fungal pathogens grown on PDA medium. In all cases (A. brassicicola, A. brassicae, B. cinerea, S. sclerotiorum and V. longisporum), a clear zone of growth inhibition was observed in the configuration with Bacillus cells (Fig. 1A). Whilst the strongest growth inhibition was observed for A. brassicicola, A. brassicae and V. longisporum, the effect was statistically significant in all cases (Supplementary Data, Table S1).
Fig. 1.
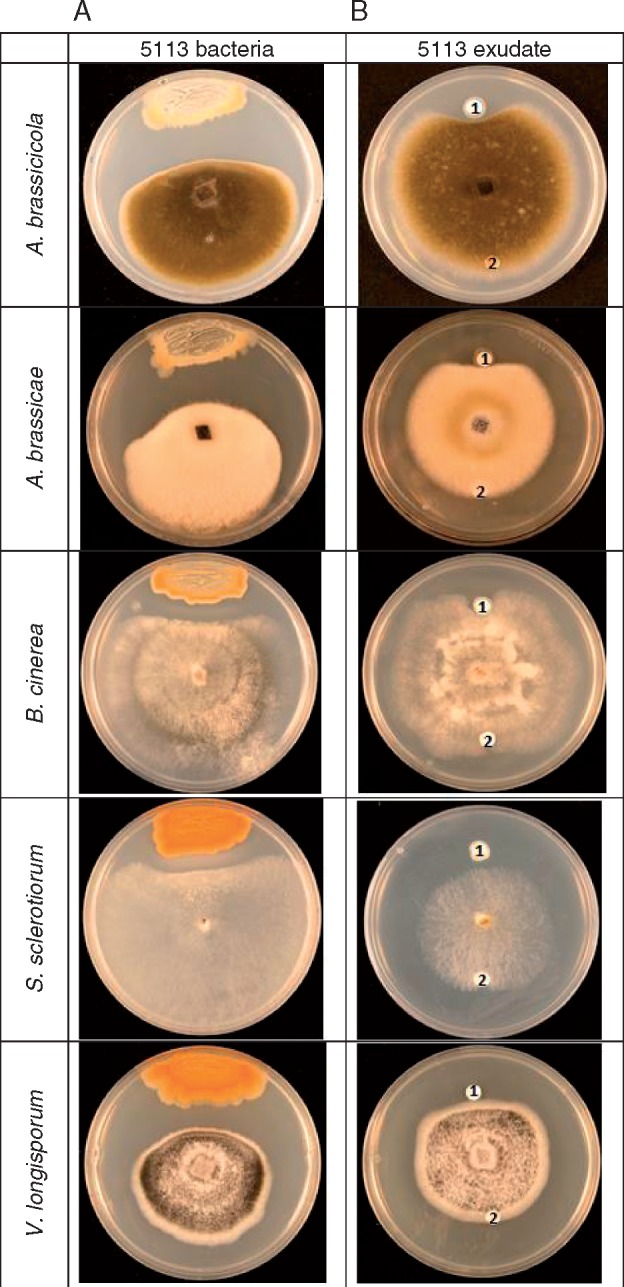
Screening of antagonistic activity of Bacillus amyloliquefaciens UCMB5113 against fungal phytopathogens. (A) A single B. amyloliquefaciens UCMB5113 colony was streaked on one side of the PDA plate and fungal strains were inoculated at the centre. Growth inhibition was studied 3–14 d later. (B) LP extract was tested for antifungal activity to phytopathogens grown on PDA. The pathogen was inoculated at the centre of the plate and aliquots of the LP extract (1) or methanol (solvent control) (2) were added at the periphery of the dish. The plates were incubated at room temperature and growth was recorded after 3–14 d.
Next, the activity of compounds secreted by Bacillus UCMB5113 grown in LB broth was tested after crude fractionation of the cell-free supernatant using acid precipitation. This allowed enrichment of many antibiotic compounds, notably lipopeptides, from the growth medium. When this LP extract was tested for activity against phytopathogens, a significant zone of growth inhibition was observed for A. brassicicola, A. brassicae and B. cinerea (Fig. 1B). The antagonistic effect of the LP extract was weaker than for living bacteria but revealed consistent production and secretion of antifungal compounds occurring in this Bacillus strain.
The antifungal activity of the LP extract was tested on plants by treating leaves of A. thaliana Col-0 prior to inoculation with A. brassicicola. Plants treated with the LP extract showed fewer necrotic symptoms compared to the methanol- and water-treated control leaves, indicating spray application of LP extracts to be useful for plant protection (Supplementary Data, Fig. S1).
Antagonistic activity stability of the Bacillus LP extract
To assess the thermostability of the active compounds, LP extract was heated at 55, 75 and 95 °C for 0, 12, 24, 48 and 72 h. The compounds were shown to maintain activity after exposure to up to 95 °C for 72 hours, as shown by the retainment of antifungal activity (Supplementary Data, Fig. S2, Table S2).
Identification of the most active antifungal compounds formed by Bacillus UCMB5113
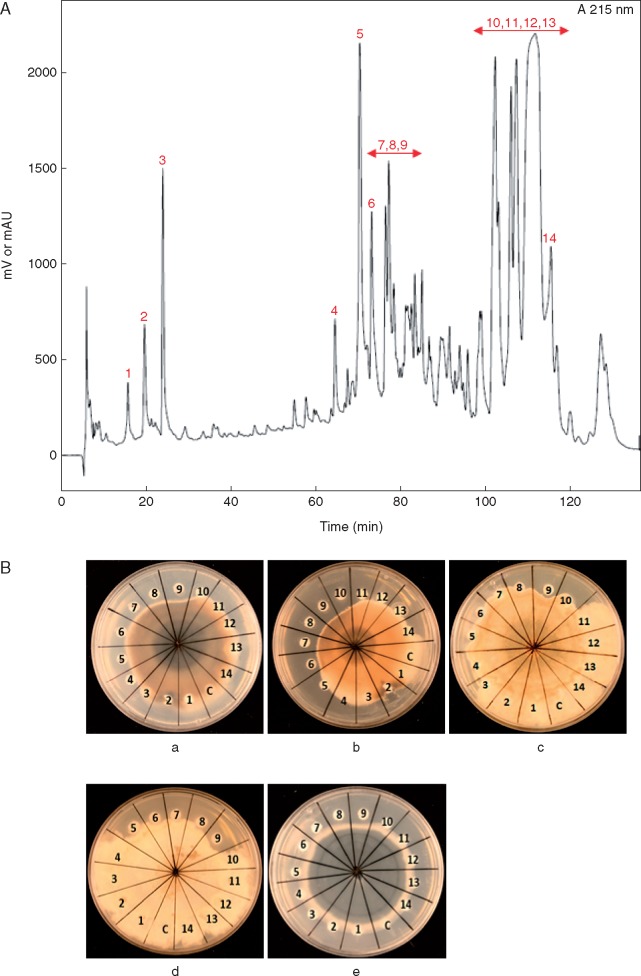
The semi-purified LP extract was further fractioned using preparative RP-HPLC. Fourteen major fractions collected (Fig. 2A) were tested for growth inhibition of Brassica phytopathogens. A number of the fractions showed antagonistic activity, although the relative sensitivity varied among the different pathogens (Fig. 2B) (Supplementary Data, Table S3). Fraction 9 was antagonistic to all pathogens and expressed the highest toxicity.
Fig. 2.
Fractionation of Bacillus amyloliquefaciens UCMB5113 LP extract and antifungal tests. (A) RP-HPLC profile of LP extract obtained after acid precipitation of B. amyloliquefaciens UCMB5113 growth medium was obtained using a C18 reversed phase column using a water/acetonitrile/TFA gradient and the eluent monitored at 215 nm. (B) Growth inhibitory activity of the LP extract fractions isolated by RP-HPLC was tested on the phytopathogens (a) A. brassicicola, (b) A. brassicae, (c) B. cinerea, (d) S. sclerotiorum and (e) V. longisporum. HPLC fractions 1–14 or methanol as control (C) were added to different parts of the Petri dish and growth was studied 3–14 d later.
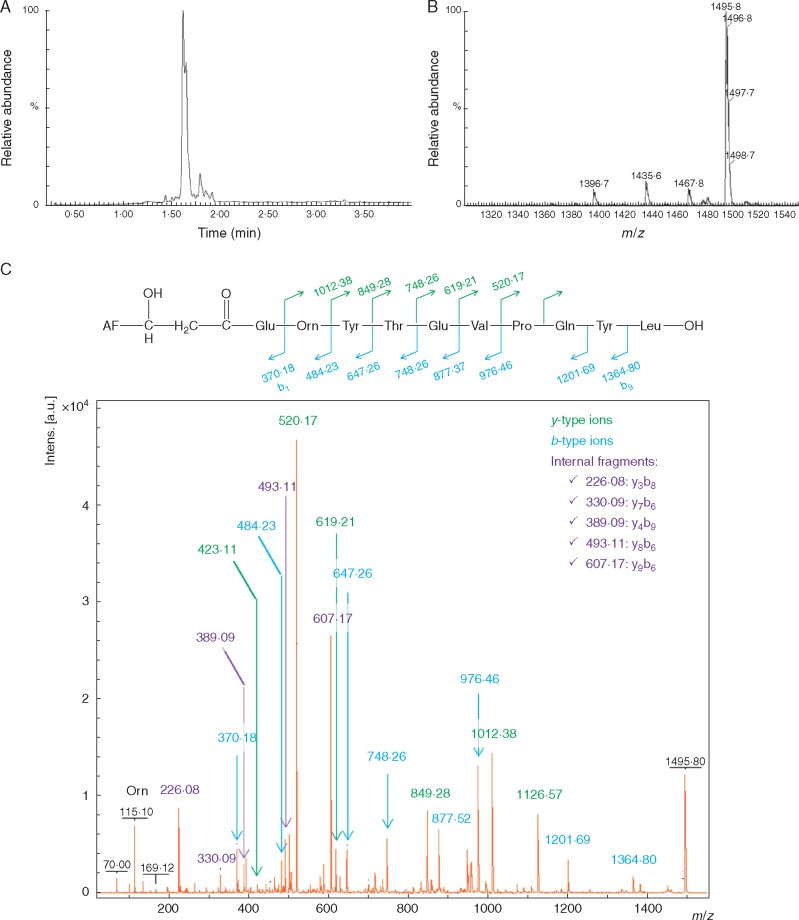
UPLC-ESI-MS analysis of fraction 9 showed one major peak together with several additional peaks of much lower intensities (Fig. 3A). In all, this fraction contained five different compounds with molecular ions detected at m/z 1396·8, 1435·8, 1467·8, 1481·6 and 1495·8. The last ion represented approx. 80 % of the total amount based on the peak area (Fig. 3B) with a molecular mass in the range of those corresponding to antifungal cyclical LPs of the fengycin family. However, it did not match any of the previously described forms. Further structural information on the main compound was obtained by tandem MS analyses. Fragmentation of cyclized fengycins secreted by Bacilli was quite poor, yielding only two main y-type ions (Supplementary Data, Fig. S3). Those fragments correspond to the peptide moiety and may be used as signature ions for discriminating between A, B, C and S variants. By contrast, many ions were generated upon fragmentation of the molecular species m/z 1495·8 (Fig. 3C), strongly suggesting that the peptide chain was linearized. Signature ions were detected at m/z 1126·57 and 1012·38 but did not match any known ions. However, those ions are typical for fengycins from the B1 family taking into account a mass shift of 18 Da due to ring opening (also visible on signature ions). The detection of most of the corresponding y-type, b-type and some internal fragments allowed unambiguous confirmation that the ion at m/z 1495·8 corresponded to the protonated form of the linear C15-fengycin B1. This linear C15-fengycin B1 was also detected in fraction 8, but at much lower relative amounts compared to fraction 9 (Supplementary Data, Fig. S3). This further supports the key role of this fengycin form for antifungal activity, since fraction 8 was also clearly less active at inhibiting the phytopathogens compared to fraction 9. MS/MS analyses revealed that the four other minor ions in fraction 9 also most probably corresponded to fengycin variants based on their mass and fragmentation patterns (Supplementary Data, Fig. S3) but their contribution to the antifungal activity was rather low, if any. As shown in Fig. S3, ions at m/z 1467·8 and 1481·6 were also present in fraction 7, which displayed almost no antifungal activity, while peaks at m/z 1396·8 and 1435·8 were detected in similar relative proportions in fraction 8 as in fraction 9.
Fig. 3.
Analysis of HPLC fraction 9 by UPLC-ESI-MS. (A) LC-MS chromatogram revealing one major peak eluting at 1·95 min. (B) MS spectrum showing that this peak mainly contained a compound with m/z 1495·8 that was further structurally characterized by MS-MS as illustrated in C. (C) Fragmentation pattern obtained and typical y- and b-type ions as well as internal fragments that were unambiguously assigned leading to the linear peptide sequence represented. Note that the analysis cannot distinguish Leu from Ile so Leu was arbitrarily chosen in the molecular structure. Further, the isomery of the fatty acid chain cannot be determined so the chain is written as a linear form for convenience.
Time-course LP profiling over the first days of culture for Bacillus UCMB5113 revealed that this strain also readily produced the iturin and surfactin families of LPs (Supplementary Data, Fig. S4) and showed that the linear C15-fengycin B1 accumulated in the supernatant in a similar kinetic trend as compared to the cyclic fengycins (Supplementary Data, Fig. S5). This linear form was observed as early as 24 h after incubation and increased over time, suggesting that it was actually de novo co-produced together with the other forms. The majority of the fengycins were cyclic in nature and after 72 h the linear B1 form represented only 6 % of the total fengycins.
Effects of synthetic peptides on fungal growth
LP mimics of the major component of fraction 9 were synthesized to confirm the biological activity of the natural compound. For this, the synthetic LP EOYTGVPQYL with acetylated (AcePEP) or myristoylated (MyrPEP) N-terminus (Supplementary Data, Fig. S6) were compared to test the effect of a lipophilic fatty acid tail for bioactivity towards A. brassicicola and V. longisporum. MyrPEP but not AcePEP showed a zone of fungal growth inhibition such as that observed for the LP extract (Fig. 4A) (Supplementary Data, Table S4). Such differential activity was also observed by testing these synthetic peptides for systemic disease suppression on Arabidopsis plants grown on MS plates. Both macroscopic disease symptoms and qPCR data for quantification of the pathogen showed a significant suppression of Alternaria growth, but only for plants treated with MyrPEP (Fig. 4B, C).
Fig. 4.
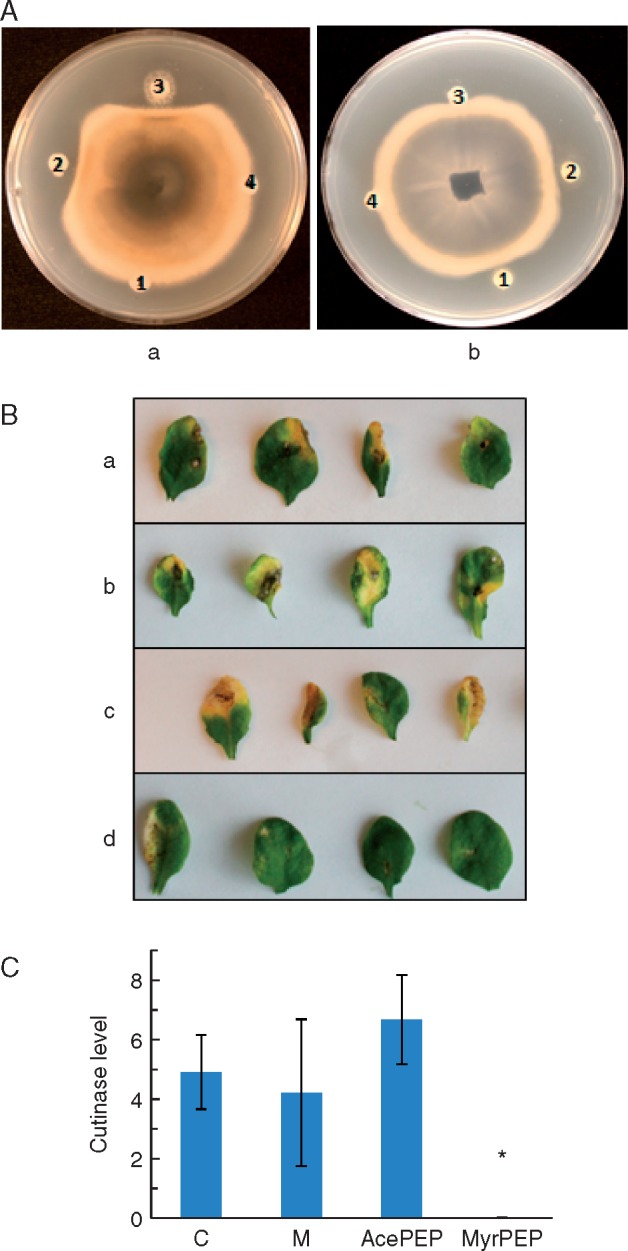
Effects of synthetic peptides on growth of fungal pathogens. (A) Synthetic peptide with acetyl side chain (a1, b3), synthetic peptide with fatty acid side chain (a2, b2), B. amyloliquefaciens UCMB5113 LP extract (a3, b1) or methanol (a4, b4) was dropped on discs in the peripheral part of a PDA plate containing A. brassicicola or V. longisporum in the centre. Effects on fungal growth were recorded 5 d later. (B) Suppressive effect of synthetic peptides against A. brassicicola growth on Arabidopsis thaliana Col-0 leaves. One-week-old seedlings grown on MS media were treated with (a) water, (b) 5 % methanol, (c) synthetic peptide with acetyl side chain and (d) synthetic peptide with fatty acid side chain compounds. Two weeks after treatment, four leaves of each plant were inoculated with A. brassicicola. The presence of necrotic lesions was recorded 1 week after fungal inoculation. (C) Quantification of fungal level in plants treated with synthetic peptides. Five days after fungal inoculation, the level of Alternaria DNA on plants was quantified by qPCR where cutinase levels relative to ubiquitin5 expression are plotted. ANOVA was run to compare DNA levels among different treatments (n = 4). Plants were untreated (‘C’), or treated with methanol as solvent control (‘M’), synthetic peptide with acetyl side chain (‘AcePEP’) and synthetic peptide with fatty acid side chain (‘MyrPEP’).
Effect of Bacillus UCMB5113 LP extract on plant systemic defence
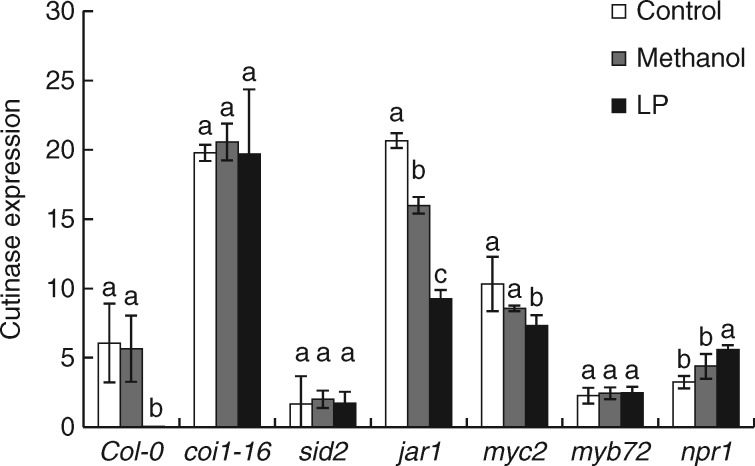
To test the ISR-triggering potential of the LP extract, Arabidopsis roots were treated prior to challenge with A. brassicicola. The coi1-16 and jar1 lines are defective in JA-induced transcriptional regulation, sid2 plants cannot accumulate SA and activate an SAR, myc2 and myb72 lines are defective in transcription factors necessary to transmit the priming of ISR, while npr1 plants display neither SAR nor ISR. The levels of fungal growth in plant leaves were analysed by qPCR specific for an Alternaria cutinase gene. Significantly lower levels of Alternaria were observed in wild type Col-0, jar1 and myc2 plants after LP treatment compared to solvent controls while npr1 plants showed elevated levels (Fig. 5). The level of Alternaria was unaffected by the LP extracts in coi1-16, sid2 and myb72 plants.
Fig. 5.
qPCR analysis of A. brassicicola levels in B. amyloliquefaciens UCMB5113 primed Arabidopsis plants. One-week-old seedlings of Arabidopsis wildtype or the mutants coi1-16, sid2, jar1, myc2, myb72 and npr1 were tested. Roots of one-week-old seedlings grown on MS media were treated with water, 5 % methanol or an LP extract. Two weeks after treatment, four leaves of each plant were inoculated with A. brassicicola. Five days after fungal inoculation, the level of Alternaria DNA was quantified by qPCR. ANOVA was run to compare DNA levels among the treatments for each plant line where different letters indicate significant differences. Plants were untreated (white bar), treated with methanol as solvent control (grey bar) or treated with LP extract (black bar).
Plant defence gene expression after treatment with Bacillus UCMB5113 LP extract
To further investigate the role of JA for the systemic defence responses against A. brassicicola in Arabidopsis, the expression of the JA marker genes VSP2 and PDF1.2 were analysed after pre-treatment of roots with the LP extract. VSP2 expression also depends on ABA and MYC2 while PDF1.2 expression also needs ethylene as well as the ethylene-dependent transcription factors ERF1 and ORA59. VSP2:GUS and PDF1.2:GUS plants pretreated with Bacillus LP extract and subsequently challenged with fungal spores were stained histochemically. Plant roots treated with LP extract showed a significantly elevated systemic expression of VSP2 in leaves and roots after fungal spore inoculation. This was not observed for plants treated with water and/or methanol (Fig. 6A–C). No distinct differences in the expression of PDF1.2:GUS were observed for either treatment (Fig. 6D–E).
Fig. 6.
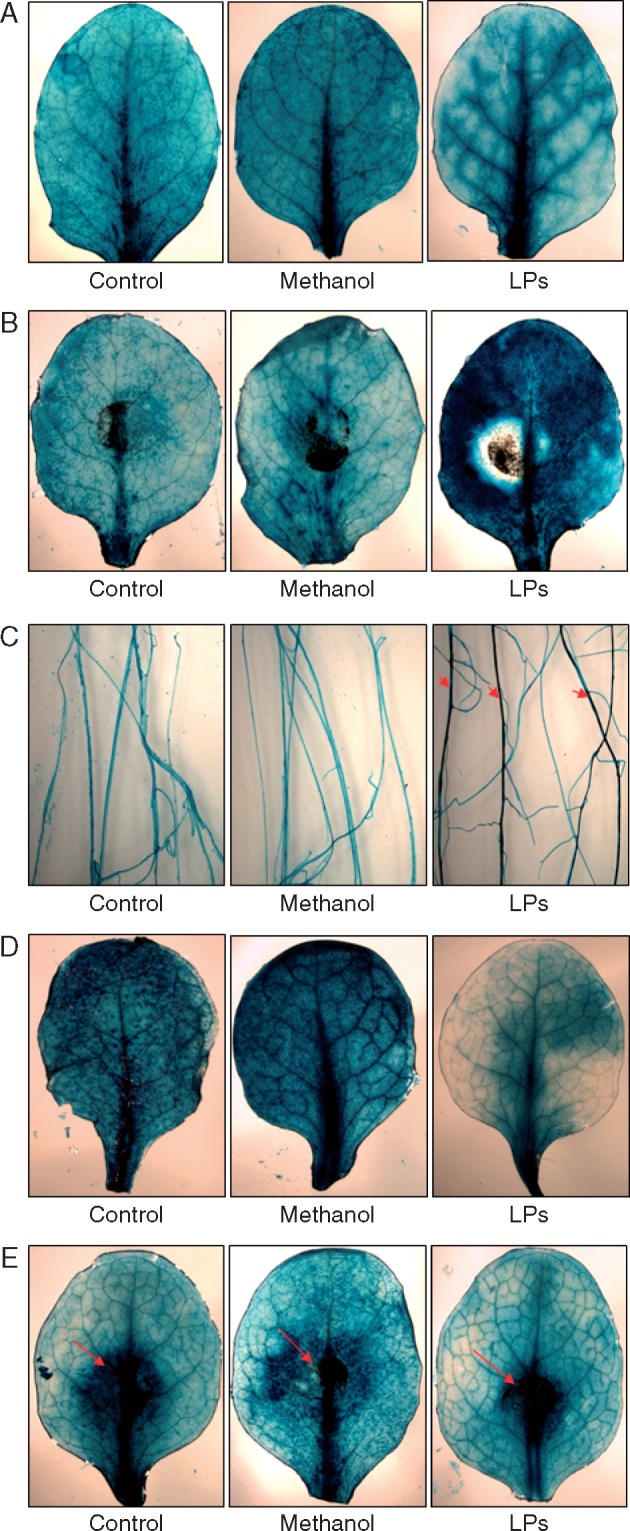
Effects on JA reporter plants by LP extract and A. brassicicola. One-week-old GUS reporter plants with different root treatments by water, methanol or LP extract were challenged with A. brassicicola spores on leaves 2 weeks later. Three days after fungal inoculation, plants were stained for reporter gene GUS expression. (A) VSP2:GUS without fungal treatment; (B) VSP2:GUS with fungal treatment; (C) VSP2:GUS roots after fungal treatment; (D) PDF1.2:GUS without fungal treatments; (E) PDF1.2:GUS with fungal treatment. Arrows indicate site of fungal inoculation on leaves and the enhanced expression in roots.
DISCUSSION
Co-cultivation of Bacillus UCMB5113 with the phytopathogens A. brassicicola, A. brassicae, B. cinerea, S. sclerotiorum and V. longisporum resulted in growth suppression of all pathogens, demonstrating that the bacterium secretes antifungal compounds. The major bioactive molecules produced by this Bacillus species are LPs, which display a wide spectrum of activity. LPs are genetically and biochemically well characterized and important in plant colonization, direct antagonism to phytopathogens and stimulation of plant immunity (Souto et al., 2004; Ongena et al., 2007; Ongena and Jacques, 2008; Cawoy et al., 2014; Farace et al., 2014; Rahman et al., 2015). An LP-enriched extract from Bacillus UCMB5113 culture broth retained antifungal activity as observed also when intact bacteria were used. Bioassays using Arabidopsis plants showed that treatment of leaves with a Bacillus UCMB5113 LP extract provided disease protection. Furthermore, treatment of plant roots with the LP extract resulted in disease suppression when challenged with Alternaria, indicating primed systemic defence as ISR. Other studies have also demonstrated disease suppression by BCAs based on LPs (e.g. Tran et al., 2007; Waewthongrak et al., 2014). Signalling has mostly been studied in Pseudomonas strains (van Loon et al., 1998; Verhagen et al., 2010) for which the ISR concept was coined, although Bacillus-derived ISR has also been demonstrated recently (Falardeau et al., 2013; Farace et al., 2014; Rahman et al., 2015). Analysis with signalling-compromised plants as well as JA-marker GUS reporter lines enabled evaluation of how plant hormones involved in different defence responses responded to Bacillus UCMB5113 LPs. Growth of Alternaria was suppressed by Bacillus UCMB5113 LPs in wildtype, jar1 and myc2 lines, while coi1-16, myb72 and sid2 plants did not respond and the npr1 accession even displayed increased Alternaria levels after LP treatment. Priming of ISR by Pseudomonas acts locally in roots through MYB72 and systemically through MYC2 (Pieterse et al., 2014). JAR1 produces the Ja–Ile conjugate that stimulates COI-dependent degradation of JAZ proteins allowing MYC2 to stimulate G-box containing promoters of JA-responsive genes (Kazan and Manners, 2013). The growth of A. brassicicola in plants is known to be affected by JA (e.g. Thomma et al., 1998). Our data show that the LP-enriched extract suppressed Alternaria without the need for JA–Ile formation but could not rescue plants with compromised JAZ degradation and did not work very well in the absence of MYC2 (where redundancy may assuage the effect). Root treatment of VSP2:GUS plants with the LP extract followed by fungal inoculation resulted in distinct staining systemically in leaves and roots while fungal inoculation alone only raised local VSP2 expression. In contrast, plants expressing PDF1.2:GUS showed low local expression around the fungal inoculation site on leaves both in the control and in the LP extract-treated samples. These results imply JA involvement in the effects observed by the LP extract but with no obvious dependence on ethylene while ABA may contribute to a reaction involving MYC to activate VSP2 transcription. Involvement of ABA in plant defence has been demonstrated for other JA-dependent pathogens with a necrotrophic life style (Adie et al., 2007). These results point to a complex signalling caused by the Bacillus UCMB5113 LPs involving JA.
MS analysis of the most active fractions from the Bacillus UCMB5113 LP extract allowed identification of novel linear forms of antifungal fengycins. Linear forms are unusual, as the standard form is a cyclic structure formed by the multicomplex fengycin biosynthetic apparatus. Many studies have shown the ability of Bacillus species to produce different cyclic fengycins (Vanittanakom et al., 1986; Sun et al., 2006; Deleu et al., 2008; Eeman et al., 2009; Malfanova et al., 2012; Pathak et al., 2012; Farace et al., 2014; O’Connor et al., 2014). However, one study (Pathak et al., 2012) also provided data in support of the presence of linear forms of fengycin in LP extracts of the banyan tree endophyte Bacillus subtilis K1, but no biological activity was reported. That report also found linear fengycins upon direct analysis of cell suspensions, although at low levels. Our time-course analysis suggested strongly that the linear C15 fengycin B1 is co-produced by Bacillus UCMB5113 along with several other structural variants. pH increases slowly during growth of Bacillus UCMB5113 in LB, which could cause non-enzymatic hydrolysis of the bond involved in cyclization. However, in vitro tests where cyclic fengycins were incubated at higher pH did not result in ring opening (our unpublished data). In addition to NRPS templated pathways, non-templated reactions can occur where the arrangement of the peptide is more difficult to predict and various post-synthesis modifications may further change the structure (Giessen and Marahiel, 2012). The linear forms could thus be synthesized as such or result from subsequent modifications (Aleti et al., 2015). The mechanisms forming these linear but highly active forms of fengycins warrant further investigation.
Use of synthetic peptide mimics of the identified Bacillus UCMB5113 linear fengycin showed that the fatty acid side chain is necessary for direct antagonistic activity to phytopathogens and priming of ISR in Arabidopsis plants since replacement with an acetyl group gave no significant inhibition of A. brassicicola and V. longisporum in plate assays or with plants. LPs can serve as membrane-disturbing biosurfactants providing a direct protective effect against phytopathogens (Ongena and Jacques, 2008). The membrane-disturbing properties of LPs imply that many kinds of micro-organisms can be targeted especially when a mixture of LPs is produced. Other surfactin and fengycin LPs have been shown to act as elicitors of ISR in plants (Ongena et al., 2007; Cawoy et al., 2014). Estimates of up to 10 % of the B. amyloliquefaciens genomes may be designated for production of secondary metabolites, providing a plethora of potentially antagonistic molecules (Chen et al., 2009; Niazi et al., 2014). In addition, Bacillus strains can interact with a wide range of plant species, providing potential protection to many different kinds of pathogens (e.g. Alvarez et al., 2012; Garcia-Gutierrez et al., 2013; Adam et al., 2014; Kalai-Grami et al., 2014).
Our results indicate that the LP extract from Bacillus UCMB5113 provides systemic resistance in plants and is dependent on JA, indicative of a primed ISR response. Bacillus UCMB5113 has the ability to produce many secondary metabolites, resulting in a complex cocktail of various antibiotics that can act directly and indirectly, supporting efficient plant protection to phytopathogens. Novel forms of such potentially antifungal linear fengycins may serve as a basis for the development of more powerful plant protective agents. The results obtained suggest that Bacillus UCMB5113 shows potential as a BCA to counteract Brassica phytopathogens. The LP compounds produced by this bacterium are stable under high temperatures, indicating long-term efficacy. Use of ecosystem services such as Bacillus UCMB5113 that could support plant protection through priming of ISR is an important step towards more sustainable crop production, reducing the need for chemical pesticides. Further studies of the mechanisms involved in beneficial plant–microbe interactions will advance our understanding of the complex molecular basis and improve the potential to identify requirements for successful interaction and priming.
SUPPLEMENTARY DATA
Supplementary data are available online at https://academic.oup.com/aob and consist of the following. Figure S1: suppressive effect of B. amyloliquefaciens UCMB5113 LP extract against A. brassicicola growth on A. thaliana Col-0 leaves. Figure S2: thermostability of B. amyloliquefaciens UCMB5113 LP extract in relation to antagonistic activity. Figure S3: MS analyses of fengycins. Figure S4: UPLC-MS profiling of LPs present in the crude culture supernatant of B. amyloliquefaciens UCMB5113 after 48 h of culture. Figure S5: accumulation of a linear form of C15-fengycin. Figure S6: structures of synthetic peptides. Table S1: fungal growth inhibition with bacteria and LP extracts. Table S2: heat stability analysis of LP extracts. Table S3: fungal growth inhibition with LP fractions after HPLC separation. Table S4: fungal growth inhibition by synthetic LP mimics.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by The Swedish Research Council for Environment, Agricultural Sciences and Spatial Planning (FORMAS), Olle-Engkvist Stiftelse, Carl Tryggers Stiftelse, Stiftelsen Tornspiran and the Nilsson-Ehle Stiftelse. Funding for plant growth facilities were provided by KFI-VR. D.D. and M.O. are, respectively, Post-doctoral Researcher and Senior Research Associate at the FRS-FNRS (Fonds National de la Recherche Scientifique, Belgium). This work also received financial support from the programme FRFC no. 2·4567·12 (FRS-FNRS). We are grateful to Dr Laura Grenville-Briggs for helpful comments on the manuscript, to Dr Björn Nicander for assistance with HPLC analysis, Staffan Matzén for growth curve test and to Prof. Christina Dixelius for gifts of the fungal pathogens. The authors declare that they have no conflicts of interest.
LITERATURE CITED
- Adam M, Heuer H, Hallmann J.. 2014. Bacterial antagonists of fungal pathogens also control root-knot nematodes by induced systemic resistance of tomato plants. PLoS One 9: e90402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adie BA, Pérez-Pérez J, Pérez-Pérez MM, et al. 2007. ABA is an essential signal for plant resistance to pathogens affecting JA biosynthesis and the activation of defences in Arabidopsis. The Plant Cell 19: 1665–1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aleti G, Sessitsch A, Brader G.. 2015. Genome mining: prediction of lipopeptides and polyketides from Bacillus and related Firmicutes. Computational and Structural Biotechnology Journal 13: 192–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez F, Castro M, Principe A, et al. 2012. The plant-associated Bacillus amyloliquefaciens strains MEP218 and ARP23 capable of producing the cyclic lipopeptides iturin or surfactin and fengycin are effective in biocontrol of Sclerotinia stem rot disease. Journal of Applied Microbiology 112: 159–174. [DOI] [PubMed] [Google Scholar]
- Beneduzi A, Ambrosini A, Passaglia LM.. 2012. Plant growth-promoting rhizobacteria (PGPR): their potential as antagonists and biocontrol agents. Genetics and Molecular Biology 35(4 Suppl): 1044–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya PN, Jha DK.. 2012. Plant growth-promoting rhizobacteria (PGPR): emergence in agriculture. World Journal of Microbiology and Biotechnology 28: 1327–1350. [DOI] [PubMed] [Google Scholar]
- Cao H, Bowling SA, Gordon AS, Dong X.. 1994. Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell 6: 1583–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cawoy H, Bettiol W, Fickers P, Ongena M.. 2011. Bacillus-based biological control of plant diseases In Stoytcheva M, ed. Pesticides in the modern world - pesticides use and management. InTech. [Google Scholar]
- Cawoy H, Mariutto M, Henry G, et al. 2014. Plant defence stimulation by natural isolates of Bacillus depends on efficient surfactin production. Molecular Plant-Microbe Interactions 27: 87–100. [DOI] [PubMed] [Google Scholar]
- Chen XH, Koumoutsi A, Scholz R, Borriss R.. 2009. More than anticipated - production of antibiotics and other secondary metabolites by Bacillus amyloliquefaciens FZB42. Journal of Molecular Microbiology and Biotechnology 16: 14–24. [DOI] [PubMed] [Google Scholar]
- Cochrane SA, Vederas JC.. 2016. Lipopeptides from Bacillus and Paenibacillus spp.: a gold mine of antibiotic candidates. Medicinal Research Reviews 36: 4–31. [DOI] [PubMed] [Google Scholar]
- Conrath U, Beckers GJ, Langenbach CJ, Jaskiewicz MR.. 2015. Priming for enhanced defence. Annual Review of Phytopathology 53: 97–119. [DOI] [PubMed] [Google Scholar]
- Danielsson J, Reva O, Meijer J. 2007. Protection of oilseed rape (Brassica napus) toward fungal pathogens by strains of plant-associated Bacillus amyloliquefaciens. Microbial Ecology 54: 134–140. [DOI] [PubMed] [Google Scholar]
- Davis AM, Hall A, Millar AJ, Darrah C, Davis SJ.. 2009. Protocol: Streamlined sub-protocols for floral-dip transformation and selection of transformants in Arabidopsis thaliana. Plant Methods 5: 3. doi:10.1186/1746-4811-5-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deleu M, Paquot M, Nylander T.. 2008. Effect of fengycin, a lipopeptide produced by Bacillus subtilis, on model biomembranes. Biophysical Journal 94: 2667–2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrant WE, Dong X.. 2004. Systemic acquired resistance. Annual Review of Phytopathology 42: 185–209. [DOI] [PubMed] [Google Scholar]
- Eeman M, Pegado L, Dufrêne YF, Paquot M, Deleu M. 2009. Influence of environmental conditions on the interfacial organisation of fengycin, a bioactive lipopeptide produced by Bacillus subtilis. Journal of Colloid and Interface Science 329: 253–264. [DOI] [PubMed] [Google Scholar]
- Ellis C, Turner JG.. 2002. A conditionally fertile coi1 allele indicates cross-talk between plant hormone signaling pathways in Arabidopsis thaliana seeds and young seedlings. Planta 215: 549–556. [DOI] [PubMed] [Google Scholar]
- Falardeau J, Wise C, Novitsky L, Avis TJ.. 2013. Ecological and mechanistic insights into the direct and indirect antimicrobial properties of Bacillus subtilis lipopeptides on plant pathogens. Journal of Chemical Ecology 39: 869–878. [DOI] [PubMed] [Google Scholar]
- Farace G, Fernandez O, Jacquens L, et al. 2014. Cyclic lipopeptides from Bacillus subtilis activate distinct patterns of defence responses in grapevine. Molecular Plant Pathology 16: 177–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridborg I, Johansson A, Lagensjö J, et al. 2013. ML3: a novel regulator of herbivory-induced responses in Arabidopsis thaliana. Journal of Experimental Botany 64: 935–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Gutierrez L, Zeriouh H, Romero D, Cubero J, de Vicente A, Perez-Garcia A.. 2013. The antagonistic strain Bacillus subtilis UMAF6639 also confers protection to melon plants against cucurbit powdery mildew by activation of jasmonate and salicylic acid-depentent defence responses. Microbial Biotechnology 6: 264–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giessen TW, Marahiel MA.. 2012. Ribosome-independent biosynthesis of biologically active peptides: Application of synthetic biology to generate structural diversity. FEBS Letters 586: 2065–2075. [DOI] [PubMed] [Google Scholar]
- Kalai-Grami L, Ben Slimane I, Mnari-Hattab M, et al. 2014. Protective effect of Bacillus amyloliquefaciens against infections of Citrus aurantium seedlings by Phoma tracheiphila. World Journal of Microbiology and Biotechnology 30: 529–538. [DOI] [PubMed] [Google Scholar]
- Kazan K, Manners JM.. 2013. MYC2: the master in action. Molecular Plant 6: 686–703. [DOI] [PubMed] [Google Scholar]
- Kim PI, Bai H, Bai D, et al. 2004. Purification and characterization of a lipopeptide produced by Bacillus thuringiensis CMB26. Journal of Applied Microbiology 97: 942–949. [DOI] [PubMed] [Google Scholar]
- Kloepper JW, Ryu C-M, Zhang S.. 2004. Induced systemic resistance and promotion of plant growth by Bacillus spp. Phytopathology 94: 1259–1266. [DOI] [PubMed] [Google Scholar]
- Lorenzo O, Chico JM, Sánchez-Serrano JJ, Solano R.. 2004. JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defence responses in Arabidopsis. Plant Cell 16: 1938–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malfanova N, Franzil L, Lugtenberg B, Chebotar V, Ongena M. 2012. Cyclic lipopeptide profile of the plant-beneficial endophytic bacterium Bacillus subtilis HC8. Archives of Microbiology 194: 893–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niazi A, Manzoor S, Asari S, Bejai S, Meijer J, Bongcam-Rudloff E.. 2014. Genome analysis of Bacillus amyloliquefaciens subsp. plantarum UCMB5113: a rhizobacterium that improves plant growth and stress management. PLoS One 9: e104651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor NK, Hudson AS, Cobb SL, et al. 2014. Noval flourinated lipopeptides from Bacillus sp. CS93 via precursor-directed biosynthesis. Amino Acids 46: 2745–2752. [DOI] [PubMed] [Google Scholar]
- Ongena M, Jacques P.. 2008. Bacillus lipopeptides: versatile weapons for plant disease biocontrol. Trends in Microbiology 16: 115–125. [DOI] [PubMed] [Google Scholar]
- Ongena M, Jourdan E, Adam A, et al. 2007. Surfactin and fengycin lipopeptides of Bacillus subtilis as elicitors of induced systemic resistance in plants. Environmental Microbiology 9: 1084–1090. [DOI] [PubMed] [Google Scholar]
- Pathak KV, Keharia H, Gupta K, Thakur SS, Balaram P. 2012. Lipopeptides from the banyan endophyte, Bacillus subtilis K1: mass spectrometric characterization of a library of fengycins. Journal of the American Society of Mass Spectrometry 23: 1716–1728. [DOI] [PubMed] [Google Scholar]
- Pieterse CMJ, van Wees SCM, Ton J, van Pelt JA, van Loon LC.. 2002. Signalling in Rhizobacteria-induced systemic resistance in Arabidopsis thaliana. Plant Biology 4: 535–544. [Google Scholar]
- Pieterse CM, Van der Does D, Zamioudis C, Leon-Reyes A, Van Wees SC.. 2012. Hormonal modulation of plant immunity. Annual Review of Cell and Developmental Biology 28: 489–521. [DOI] [PubMed] [Google Scholar]
- Pieterse CM, Zamioudis C, Berendsen RL, Weller DM, Van Wees SC, Bakker PA.. 2014. Induced systemic resistance by beneficial microbes. Annual Review in Phytopathology 52: 347–375. [DOI] [PubMed] [Google Scholar]
- Pozo MJ, Azcón-Aguilar C.. 2007. Unraveling mycorrhiza-induced resistance. Current Opinion in Plant Biology 10: 393–398. [DOI] [PubMed] [Google Scholar]
- Rahman A, Uddin W, Wenner NG. 2015. Induced systemic resistance responses in perennial ryegrass against Magnaporthe oryzae elicited by semi-purified surfactin lipopeptides and live cells of Bacillus amyloliquefaciens. Molecular Plant Pathology 16: 546–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reva ON, Dixelius C, Meijer J, Priest FG.. 2004. Taxonomic characterization and plant colonizing abilities of some bacteria related to Bacillus amyloliquefaciens and Bacillus subtilis. FEMS Microbiology Ecology 48: 249–259. [DOI] [PubMed] [Google Scholar]
- Sarosh BR, Danielsson J, Meijer J.. 2009. Transcript profiling of oilseed rape (Brassica napus) primed for biocontrol primed for biocontrol differentiate genes involved in microbial interactions with beneficial Bacillus amyloliquefasciens from pathogenic Botrytis cinerea. Plant Molecular Biology 70: 31–45. [DOI] [PubMed] [Google Scholar]
- Seifi A, Visser RGF, Bai Y.. 2013. How to effectively deploy plant resistances to pests and pathogens in crop breeding. Euphytica 190: 321–334. [Google Scholar]
- Shigenaga AM, Argueso CT.. 2016. No hormone to rule them all: Interactions of plant hormones during the responses of plants to pathogens. Seminars in Cell & Developmental Biology 56: 174–189. [DOI] [PubMed] [Google Scholar]
- Souto GI, Correa OS, Montecchia MS, et al. 2004. Genetic and functional characterization of a Bacillus sp. strain excreting surfactin and antifungal metabolite spartially identified as iturin-like compounds. Journal of Applied Microbiology 97: 1247–1256. [DOI] [PubMed] [Google Scholar]
- Staswick PE, Su W, Howell SH.. 1992. Methyl jasmonate inhibition of root growth and induction of a leaf protein are decreased in an Arabidopsis thaliana mutant. Proceedings of the National Academy of Sciences USA 89: 6837–6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strieker M, Tanović A, Marahiel MA.. 2010. Nonribosomal peptide synthetases: structures and dynamics. Current Opinion in Structural Biology 20: 234–240. [DOI] [PubMed] [Google Scholar]
- Sun L, Zhaoxin L, Bie X, Fengxia L, Yang S.. 2006. Isolation and characterization of a co-producer of fengycins and surfactins, endophytic Bacillus amyloliquefacines ES-2, from Scutellaria baicalensis Gerorgi. World Journal of Microbiology and Biotechnology 22: 1259–1266. [Google Scholar]
- Thomma BPHJ, Eggermont K, Penninckx IAMA, et al. 1998. Separate jasmonate-dependent and salicylate-dependent defence response pathways in Arabidopsis are essential for resistance to distinct microbial pathogens. Proceedings of the National Academy of Sciences USA 95: 15107–15111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran H, Ficke A, Asiimwe T, Höfte M, Raaijmakers JM. 2007. Role of the cyclic lipopeptide massetolide A in biological control of Phytophthora infestans and in colonization of tomato plants by Pseudomonas fluorescens. New Phytologist 175: 731–742. [DOI] [PubMed] [Google Scholar]
- Van der Ent S, Verhagen BW, Van Doorn R, et al. 2008. MYB72 is required in early signalling steps of rhizobacteria-induced systemic resistance in Arabidopsis. Plant Physiology 146: 1293–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanittanakom N, Loeffler W, Koch U, Jung G.. 1986. Fengycin- a novel antifungal lipopeptide antibiotic produced by Bacillus subtilis F-29–3. Journal of Antibiotics (Tokyo) 39: 888–901. [DOI] [PubMed] [Google Scholar]
- van Loon LC, Bakker PA, Pieterse CM.. 1998. Systemic resistance induced by rhizosphere bacteria. Annual Review of Phytopathology 36: 453–483. [DOI] [PubMed] [Google Scholar]
- Verhagen BW, Trotel-Aziz P, Couderchet M, Höfte M, Aziz A.. 2010. Pseudomonas spp.-induced systemic resistance to Botrytis cinerea is associated with induction and priming of defence responses in grapevine. Journal of Experimental Botany 61: 249–260. [DOI] [PubMed] [Google Scholar]
- Waewthongrak W, Leelasuphakul W, McCollum G. 2014. Cyclic lipopeptides from Bacillus subtilis ABS-S14 elicit defence-related gene expression in citrus fruit. PLoS One 9: e109386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasternack C, Hause B.. 2013. Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Annals of Botany 111: 1021–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildermuth MC, Dewdney J, Wu G, Ausubel FM.. 2001. Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414: 562–565. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.